Abstract
Ty3, a retroviruslike element of Saccharomyces cerevisiae, transposes into positions immediately upstream of RNA polymerase III-transcribed genes. The Ty3 integrase (IN) protein is required for integration of the replicated, extrachromosomal Ty3 DNA. In retroviral IN, a conserved core region is sufficient for strand transfer activity. In this study, charged-to-alanine scanning mutagenesis was used to investigate the roles of the nonconserved amino- and carboxyl-terminal regions of Ty3 IN. Each of the 20 IN mutants was defective for transposition, but no mutant was grossly defective for capsid maturation. All mutations affecting steady-state levels of mature IN protein resulted in reduced levels of replicated DNA, even when polymerase activity was not grossly defective as measured by exogenous reverse transcriptase activity assay. Thus, IN could contribute to nonpolymerase functions required for DNA production in vivo or to the stability of the DNA product. Several mutations in the carboxyl-terminal domain resulted in relatively low levels of processed 3′ ends of the replicated DNA, suggesting that this domain may be important for binding of IN to the long terminal repeat. Another class of mutants produced wild-type amounts of DNA with correctly processed 3′ ends. This class could include mutants affected in nuclear entry and target association. Collectively, these mutations demonstrate that in vivo, within the preintegration complex, IN performs a central role in coordinating multiple late stages of the retrotransposition life cycle.
Ty3 is a 5.4-kb retroviruslike element of the yeast Saccharomyces cerevisiae that has a life cycle similar to that of animal retroviruses. Integrated Ty3 DNA is transcribed to produce a 5.2-kb genomic RNA that is translated into Gag3p and Gag3-Pol3p polyproteins (33–35, 48). The polyproteins are processed and assembled together with genomic RNA into viruslike particles (VLPs) analogous to retroviral cores. Ty3 is a member of the gypsylike family of retroelements (used here to collectively refer to retroviruses and retroviruslike elements). The two open reading frames, GAG3 and POL3, encode the major structural proteins capsid (CA) and nucleocapsid (NC) and the enzymes protease (PR), reverse transcriptase (RT), and integrase (IN), respectively. Reverse transcription produces full-length Ty3 DNA. Two extra bases at each 3′ end of this DNA are removed by IN prior to integration (49). This feature is shared with animal retroviruses, but is less common among characterized retrotransposons. Ty3 is also different from most other retroelements in that transposition occurs close to the site of transcription initiation of genes transcribed by RNA polymerase III (11). IN, the subject of this investigation, is required for integration and is thus a candidate for mediating position-specific integration.
Integration into the host genome is a critical step in the life cycle of all retroelements. Integration of retroviruses and some retroviruslike elements is preceded by the removal of two nucleotides 3′ of a conserved CA dinucleotide at the viral DNA 3′ ends (3′-end processing) (6, 26, 42, 69). Integration is a concerted reaction in which the newly exposed 3′ hydroxyls of the viral DNA ends attack the phosphodiester bonds in the host DNA, joining the 3′ ends of the viral DNA to host DNA (strand transfer) (6, 14, 23, 26). The unpaired nucleotides at the 5′ ends of the viral DNA are removed, and the gaps are repaired.
Alignment of retroelement IN proteins shows poorly conserved amino- and carboxyl-terminal regions flanking a conserved central region. Approximately 20 residues within this region are absolutely conserved among retroviral IN proteins, including residues contributing to a zinc finger-like motif (HHCC) and to a D,D(35)E motif required for divalent-metal cation binding in several classes of proteins involved in polynucleotide transfer reactions (3, 10, 15, 20, 25, 39, 46, 51, 70). Mutations in the conserved residues disrupt retrovirus replication in vivo (10, 21, 24, 53, 54, 59, 74, 75, 81, 82). In vitro assays with recombinant IN mutants and with IN subdomains have been useful in dissecting the contributions of subdomains to IN activity. Mutations in the metal finger and in the D,D(35)E motif disrupt both 3′-end processing and strand transfer with oligonucleotide substrates in vitro, showing that both motifs are essential for IN activity and possibly long terminal repeat (LTR) recognition (8, 9, 16–18, 20, 40, 46, 51, 55, 71, 77–79). However, a recombinant protein representing only the core region (without the zinc finger) is competent for disintegration, a reversal of the strand transfer reaction (8, 12). Mutations in the conserved residues of the catalytic triad D,D(35)E further abolish this polynucleotidyl transfer activity in vitro, indicating that this domain represents the catalytic site of IN (8, 9, 19, 20, 51, 55, 77). Not surprisingly, the core region is also implicated in DNA binding and target site selection (22, 30, 38, 43, 44, 65, 73). UV cross-linking studies showed that lysine residues near the active site specifically interact with the deoxyadenosine of the conserved CA dinucleotide at the ends of viral DNA (38). In addition, mutational analyses showed that the core domain recognizes critical features in substrate DNA, such as the CA dinucleotide, the unpaired 5′-end dinucleotide following end processing, and target DNA flanking the site of viral DNA joining (30). In vitro strand transfer experiments with IN chimeras between human immunodeficiency virus type 1 (HIV-1) and feline immunodeficiency virus showed that patterns of insertion site sequence bias correlated with the derivation of the core domain (73). In addition to a role in DNA binding, the core contributes to IN multimer formation. Two-hybrid, mutational, and X-ray crystal analyses have shown that the core and carboxyl-terminal domains mediate IN dimer formation (19, 37, 41).
The carboxyl-terminal domain of several retroviral IN proteins has been shown to mediate nonspecific DNA binding (22, 43, 57, 64, 66, 76, 79, 83, 84). UV cross-linking studies showed that the carboxyl-terminal domain of HIV-1 IN binds DNA nonspecifically in the absence of divalent metal ions whereas the core requires metal ions to bind specific substrate DNA used in the in vitro integration assay (22, 66). The possible functions of nonspecific DNA binding by IN include initial binding of IN to viral DNA prior to 3′-end processing and integration, and interaction of IN with target DNA.
In particular retroelement systems, functions in addition to those directly required for catalysis are performed by IN. In avian sarcoma-leukosis viruses, the major species of RT in the virus is a heterodimer, consisting of an RT subunit (α) and an RT-IN fusion subunit (β) (13, 29, 32, 72). Recent studies have shown that avian sarcoma virus IN (50), HIV-1 IN (28), and Ty1 IN (45, 63) contain functional nuclear localization signals (NLSs) located carboxyl-terminal to the core region. In the case of the retroviral systems, it has been difficult to define the physiological significance of the signal. Avian sarcoma virus is not known to infect nondividing cells, the most obvious instance where function of a signal could be detected. Although HIV infects nondividing cells, mutations in the HIV-1 NLS also disrupt replication in dividing cells, suggesting that the region plays a role in addition to nuclear localization. HIV-1 IN participates in catalysis, reverse transcription, and nuclear localization and is also one of the major nucleus-localized components of the preintegration complex (7, 62). The extent to which it plays a structural role in organizing the preintegration complex is not known.
Ty3 IN is also potentially complex with respect to its functions in the Ty3 life cycle. Immunoblot analysis of Ty3 VLPs showed previously that Ty3 IN is represented in three species, a 115-kDa RT-IN fusion and 61- and 58-kDa IN proteins (35, 48). Previous attempts to mutagenize Ty3 IN by introducing small deletions into the carboxyl-terminal domain resulted in VLPs that lacked DNA (49). This, together with the existence of an RT-IN fusion protein, led to the hypothesis that Ty3 RT might be composed of an α-β heterodimer similar to the avian RTs and therefore that the Ty3 IN domain is essential for RT activity. In addition, because the nuclear membrane of yeast does not break down during the cell cycle, nuclear localization might be mediated by Ty3 IN. The carboxyl-terminal nonconserved region of retroviral IN has been inferred to be involved in target DNA interactions. In comparison to the retroviral carboxyl-terminal domains, the analogous Ty3 domain is larger and does not show significant sequence similarity. If this domain in Ty3 IN is involved in target interactions, it might interact with DNA or RNA polymerase III transcription factors required for target association or both.
The present study was undertaken to investigate the functions of the nonconserved amino- and carboxyl-terminal domains of Ty3 IN by using charged-to-alanine scanning mutagenesis (4, 31). Twenty positions were identified from amino acid positions 1 to 92 and positions 412 to 536 where multiple charged amino acid residues were present within a five-residue region and one to three amino acids in each cluster were converted to alanines. The Ty3 mutants were analyzed for transposition, particle formation, the presence of DNA in the particle, in vitro RT activity, and 3′-end processing of the DNA. The results showed that the nonconserved domains of Ty3 IN contribute essential functions throughout the later stages of the Ty3 life cycle.
MATERIALS AND METHODS
Strains and culture conditions.
Escherichia coli and S. cerevisiae strains were cultured and transformed by standard methods (1). S. cerevisiae yTM443 (61) (previously referred to as TMy18) (MATa trp1-H3 ura3-52 his3-Δ200 ade2-101 lys2-1 leu1-12 can1-100 ΔTy3 bar1::hisG GAL3+), a derivative of yVB110 which contains no endogenous copies of Ty3 (34), was used for transposition assays and whole-cell extract DNA and protein analyses. VLP protein analysis was performed with yTM443 or AGY-9 (MATa ura3-52 his4-539 lys2-801 trp1-Δ63 leu2-Δ1 spt3) (a gift from J. D. Boeke, The Johns Hopkins University). AGY-9 transformed with plasmids expressing Ty3 was used for production of VLPs for in vitro RT activity assays because the mutation in SPT3 reduces Ty1 expression and therefore eliminates the complication of non-Ty3 RT activity. E. coli RZ1032 (lysA[61-62] thi-1 relA1 spoT1 dut-1 ung-1 [Tetr] supE44) was used for production of single-stranded DNA for site-directed mutagenesis by the method of Kunkel (52). Plasmids were amplified in HB101 (F− hsd-20 [rB− mB−] recA13 leuB6 ara-14 proA2 lacY1 galK2 rpsL20 [Smr] xyl-5 mtl-1 supE44 λ−).
Recombinant DNA manipulations.
All recombinant DNA techniques were performed essentially as described in Current Protocols in Molecular Biology (1). Ty3 elements, modified by replacement of the regulatory region with sequences from the GAL1-10 promoter, were used for transposition studies and whole-cell extract and VLP analyses. Plasmid pEGTy3-1 (34), which was used in all experiments, contains a fusion of the GAL1-10 upstream activator sequence upstream of the Ty3 promoter. In addition, pEGTy3-1 contains the 2μm sequence for maintenance at high copy number in S. cerevisiae and the yeast selectable marker URA3, which allows cells containing this plasmid to be selected for by growth on medium lacking uracil or to be selected against on medium containing 5-fluoroorotic acid. The target plasmid, pCH2bo19V (47), which was used in the transposition assays, contains the ARS1 and CEN4 sequences for maintenance at low copy number in S. cerevisiae and the yeast selectable marker HIS3. Plasmid pEGTy3-1 was used for charged-to-alanine scanning mutagenesis of Ty3 IN. Nineteen oligonucleotides (364 to 371 and 373 to 383) (Table 1) were used to insert alanines in place of two or three (oligonucleotide 374) charged amino acids within a window of five residues in the nonconserved amino- and carboxyl-terminal domains of IN spanning amino acid residues 1 to 92 and 412 to 536, respectively. Oligonucleotide 384 was used to change a glutamate to an alanine residue at codon 519 in the polypurine tract of IN. Mutants are referred to by the codon position of the first residue in the mutagenized cluster of charged residues, followed by an A to indicate a change to alanine and a number in parentheses to indicate how many residues were changed in the respective cluster. Mutations in pEGTy3-1 were confirmed by sequence analysis.
TABLE 1.
Oligonucleotides used in charged-to-alanine scanning mutagenesis of Ty3 IN
| Ty3 IN mutanta | Mutation(s) | Oligonucleotide | Sequence |
|---|---|---|---|
| 11A(2) | D11A, E13A | 364 | 5′GATTTCCAGCTTGCTGTGGCGATAGGTCG3′ |
| 20A(2) | K20A, D22A | 365 | 5′CATAATGGGGCTGATGCGTAGTAAG3′ |
| 33A(2) | K33A, E34A | 366 | 5′GTTGTGTCAATGCTGCCATATGAATTAAG3′ |
| 43A(2) | E43A, D44A | 367 | 5′GGCTGACATAGCTGCAGGTGTGAC3′ |
| 53A(2) | K53A, K54A | 368 | 5′GATAGTTCGAGTGCCGCCTGGTAAC3′ |
| 62A(2) | R62A, K63A | 369 | 5′GGGAATAATTCGCTGCGAAGGTCTC3′ |
| 69A(2) | D69A, E70A | 370 | 5′GTAATAGATCATTGCGGCTTCTAGGG3′ |
| 76A(2) | D76A, R77A | 371 | 5′GGTACTACTAGTGCGGCTTGGTAATAG3′ |
| 412A(2) | E412A, D414A | 373 | 5′GAGAGTTTAAAGCTAGTGCGTAGGCGTTATC3′ |
| 419A(3) | K419A, K420A, K421A | 374 | 5′CTCTGTGCGCTGCCGCGTGAGAG3′ |
| 431A(2) | K431A, K432A | 375 | 5′GGTATACAAACGCTGCCAGGAATTG3′ |
| 436A(2) | R436A, D438A | 376 | 5′GGGTACGCGGCTGGAGCGTATACAAAC3′ |
| 442A(2) | K442A, K444A | 377 | 5′GCTGATTGGTGCATTCGCTGGGTACGC3′ |
| 450A(2) | E450A, R451A | 378 | 5′CTCTCTTAATTGCTGCAGTGGAGCTG3′ |
| 453A(2) | K453A, R454A | 379 | 5′CTTCGTGTGCTGCCGCAATTCTTTC3′ |
| 477A(2) | D477A, D479A | 380 | 5′GAAAGTGTTGGGGCTACAGCTTGCATGTG3′ |
| 488A(2) | E488A, E490A | 381 | 5′GGCAAAATGCAGCTGCTGAGTATTC3′ |
| 496A(2) | E496A, R497A | 382 | 5′GATCTTCGTGTTGCTGCGGGAATTTGG3′ |
| 499A(2) | R499A, R500A | 383 | 5′GCTAATATTGATGCTGCTGTTCTTTCG3′ |
| 519A(1) | E519A | 384 | 5′CAACATCTTCCGCTCTCTCAGGG3′ |
Ty3 IN mutants are referred to by the codon position of the first amino acid residue in the mutagenized cluster, followed by an A to indicate a change to alanine and a number in parentheses to indicate how many residues were changed in the respective cluster.
Transposition assays.
Quantitative plasmid-based suppressor target assays were performed essentially as previously described (47). The assay is based on expression of Ty3 under control of the GAL1-10 promoter on a URA3-marked donor plasmid (pEGTy3-1) and subsequent integration of the replicated Ty3 into a HIS3-marked target plasmid (pCH2bo19V). The target plasmid contains two divergent tRNA genes. One of these acts to recruit Ty3 to the target site. The other gene is a transcriptionally inactive ochre suppressor tRNATyr gene (sup2-o), which is activated by Ty3 integration into the target site. Transposition is scored by suppression of the ade2-101 lys2-1 ochre nonsense mutations in yeast strain yTM443. Suppression in cells that have undergone transposition results in papillations on minimal medium supplemented with leucine and tryptophan. YTM443 cells transformed with pEGTy3-1, carrying wild-type Ty3 or 1 of the 20 mutant derivatives, and the target plasmid pCH2bo19V were grown in raffinose-containing liquid medium lacking uracil and histidine to select for cells containing donor and target plasmids, respectively. Raffinose was used as the carbon source to neither induce nor repress Ty3 expression. Cells were grown to mid-log phase and washed, and 106 cells were plated in duplicate or triplicate onto synthetic complete medium containing galactose (SG) or glucose (SD) and lacking uracil and histidine, in order to induce or repress Ty3 expression, respectively. The plates were incubated at 30°C for 29 h on SD medium or 48 h on SG medium, and the cells were replica plated to minimal medium containing leucine and tryptophan. The plates were incubated at 30°C for 6 days, and transposition was scored as papillations. Quantitative transposition assays were performed at least twice with one or two independent transformants for wild-type Ty3 and IN mutants.
In a qualitative patch assay for Ty3 transposition, independent colonies containing yTM443 cells transformed with pEGTy3-1, carrying a wild-type Ty3 or a derivative carrying a mutant Ty3, and pCH2bo19V were patched onto SD medium lacking uracil and histidine. The plate was incubated at 30°C for 24 h, and the cells were replica plated to minimal medium containing leucine and tryptophan (data not shown) and to SG medium lacking uracil and histidine to induce Ty3 transposition. After 48 h (30°C) on SG medium, the patches were replica plated onto minimal medium containing leucine and tryptophan and incubated at 30°C for 6 days. Transposition was scored as papillations on minimal medium containing leucine and tryptophan.
To characterize Ty3 integration sites by nucleotide sequence analysis, colonies were streaked onto medium containing 5-fluoroorotic acid and lacking histidine to select for cells that had lost the URA3-marked donor plasmid but retained the HIS3-marked target plasmid. Target plasmids were isolated from yeast by the method of Hoffman and Winston (36), amplified in E. coli, and prepared for analysis by standard methods (58). Ty3 insertions into the target were characterized by sequence analysis.
VLP preparation.
One-liter cultures of AGY-9 or yTM443 cells transformed with pEGTy3-1, carrying a wild-type Ty3 or a derivative carrying a mutant Ty3, were grown to late log phase in SG medium to induce Ty3 expression. A mock VLP preparation was made with AGY-9 cells that were not transformed with the expression plasmid. VLPs were partially purified from whole-cell extracts as previously described (33). Briefly, the cells were harvested, washed in buffer, digested with Zymolyase, lysed by vortexing with glass beads, and fractionated over a 70%, 30%, and 20% (5, 5, and 15 ml, respectively) sucrose step gradient by centrifugation in an SW28 rotor at 83,000 × g for 3 h at 4°C. A total of 4 ml from the 70%-30% interface of each gradient was collected and divided into two portions. One portion (3 ml) was extracted with phenol-chloroform-isoamyl alcohol (25:24:1), and the nucleic acid was precipitated with ethanol and 0.3 M sodium acetate. The other portion (1 ml) was concentrated by centrifugation in a Ti50 rotor at 100,000 × g for 1 h at 4°C and resuspended in 50 μl of buffer (9 mM HEPES [pH 7.8], 13.5 mM KCl, 4.5 mM MgCl2, 10% glycerol). VLP nucleic acid was used for analysis of Ty3 DNA 3′ termini, and protein was used for RT immunoblot analysis and RT activity assays.
Whole-cell extraction.
Cultures (10 ml) of yTM443 cells transformed with pEGTy3-1 carrying a wild-type Ty3, or a derivative carrying a mutant Ty3, were grown to an absorbance at 600 nm of ∼1.0 in SG medium. The cultures were divided into two equal portions for protein and nucleic acid extraction, and the cells were pelleted. One portion was suspended in 600 μl of whole-cell extract buffer (0.1 mM EDTA, 25 mM HEPES [pH 7.5], 50 mM KCl, 5 mM MgCl2, 10% glycerol) containing 1 μg of leupeptin per ml, 1 μg of pepstatin A per ml, and 1 mM phenylmethylsulfonyl fluoride. The cells were lysed by being vortexed with glass beads at maximum speed for 15 s and plunged into ice for 15 s. This was repeated five times. The extract was centrifuged in an Eppendorf microcentrifuge (Brinkmann). The supernatant was transferred to a new tube. The protein concentration was determined by Bradford assay (5). The other portion of the cell culture was suspended in 200 μl of cell-breaking buffer (1 mM EDTA, 100 mM NaCl, 10 mM Tris [pH 8.0], 1% sodium dodecyl sulfate [SDS], 2% Triton X-100) and extracted with phenol-chloroform-isoamyl alcohol (25:24:1) while being vortexed with glass beads for 5 min at maximum speed. The nucleic acid in the aqueous phase was precipitated with ethanol and 0.3 M sodium acetate.
DNA analysis.
The concentration of nucleic acid isolated from whole-cell extracts was measured by monitoring the absorbance at 260 nm. To detect full-length, replicated DNA, 10 μg of total nucleic acid was treated with 1 μg of RNase A and digested with 10 U of BamHI in a total volume of 20 μl for 1 h at 37°C to linearize the expression plasmid (pEGTy3-1). The samples were separated on a 1% agarose gel by electrophoresis, transferred to nitrocellulose (Duralon UV; Stratagene), immobilized by cross-linking with UV light in a Stratalinker 1800 (Stratagene), and probed with a Ty3 internal-domain-specific probe, produced by BglII-digestion and labeled with [α-32P]dATP. HindIII-digested lambda DNA served as DNA size markers.
Immunoblot analysis.
Proteins from whole-cell extracts or VLPs were fractionated by SDS-polyacrylamide gel electrophoresis, transferred electrophoretically to nitrocellulose membranes (Hybond ECL; Amersham), and probed with antibody to CA or IN (61) for whole-cell extract protein analysis or with antibody to RT (a generous gift from T. M. Menees, University of Missouri, Kansas City, Mo.) for VLP protein analysis. Secondary antibodies to rabbit immunoglobulin G were detected by chemiluminescence, using the ECL system as described by the manufacturer (Amersham).
RT assays.
VLPs were prepared as described above from AGY-9 cells expressing wild-type Ty3 or IN mutant Ty3 or from nontransformed cells. Exogenous RT activity was measured under conditions optimized for Ty3 RT (33), with some modifications to the protocol (60). A 10-μg portion of VLP protein for each sample was mixed with 20 mM Tris (pH 7.8), 20 mM dithiothreitol, 15 mM MgSO4, 4 U of RNasin, 10 μM cold dTTP (Pharmacia Biotech), 1 μg of poly(rA) · p(dT)12–18 template-primer (Pharmacia Biotech), and 1 μCi of [α-32P]dTTP (3000 Ci/mmol; Amersham) in a total reaction volume of 60 μl and incubated at 25°C for 2 h. RT activity was measured as the incorporation of [α-32P]dTTP. The counts per minute (cpm) incorporated was plotted as a function of time, with each datum point representing the average of duplicate samples.
Southern analysis of VLP DNA.
To detect 3′-end processing of Ty3 DNA, the plus-strand sequences in the U5 region of the LTR were analyzed essentially as previously described (49). The concentration of DNA isolated from VLP preparations was measured by fluorometry with a TKO 100 DNA fluorometer (Hoefer Scientific Instruments). We used 0.5 to 4 μg of VLP DNA for the experiment, depending on the relative amount of Ty3 DNA available, as determined by Southern analysis of whole-cell extract DNA. Thus, 0.5 μg of VLP DNA was used for IN mutants with at least wild-type levels of DNA whereas 1 to 4 μg of VLP DNA was used for IN mutants with lower DNA levels to detect 3′-end processing for these mutants. The VLP nucleic acid was treated with RNase A, digested with HinfI, and separated on an 8% polyacrylamide gel containing 6 M urea adjacent to a sequence ladder representing the 3′ terminus of Ty3 DNA (size ladder not shown). The fragments were transferred onto a nylon membrane (GeneScreen; DuPont), immobilized by UV cross-linking (as described above) and baking at 80°C for 2 h, and subjected to Southern analysis with 32P-5′-end-labeled oligonucleotide 202 (5′TACGGGCTCGAGTAATCTCGGAGTGTCTTGACA3′; probe C). Hybridization was performed with 2 × 106 cpm of 5′-end-labeled oligonucleotide at 50°C for 16 h. The blots were washed and exposed to a PhosphorImager screen and analyzed with a PhosphorImager (Molecular Dynamics). The experiment was performed at least twice for IN mutants with near normal DNA levels, as determined by Southern analysis of whole-cell extract DNA, and at least three times for mutants with small amounts of DNA, with the exception of 499A(2), which was tested twice. For these mutants, increasing amounts of VLP DNA were used in each additional experiment.
RESULTS
Mutations in nonconserved regions of Ty3 IN block Ty3 transposition.
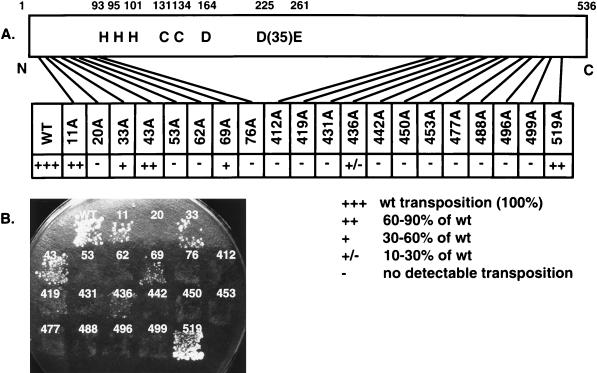
To study the functions of distinct Ty3 IN domains, mutagenesis was used to generate a panel of 20 IN mutants. To produce less disruptive mutations, substitutions of alanine for one to three charged amino acids within a window of five residues were made in the nonconserved amino- and carboxyl-terminal regions (Fig. 1 and Table 1). These regions were defined as the amino-terminal domain upstream of the HHCC motif, spanning amino acid residues 1 to 92, and the carboxyl-terminal domain spanning residues 412 to 536. Eight mutations were made in the amino-terminal domain, and 12 were made in the carboxyl-terminal domain, including one in the polypurine tract. Twenty mutagenic oligonucleotides were used to introduce the mutations by site-directed mutagenesis, and the mutations were confirmed by sequence analysis.
FIG. 1.
Ty3 IN amino acid sequence numbered from the amino-terminal residue (48). Shaded residues indicate the charged amino acid residues converted to alanine. The number above a shaded residue indicates the position of the first amino acid within a cluster of charged amino acid residues. These numbers are also used to refer to the corresponding mutants. Amino acid residues that are highly conserved among retroviruses and retroviruslike elements are indicated by an asterisk (10, 51). There was no clear basis upon which to distinguish the first and second histidines, which are each indicated by an asterisk.
Previous studies showed that Ty3 elements with mutations in the conserved D,D(35)E motif, as well as elements with carboxyl-terminal truncations of Ty3 IN, failed to complement transposition of a marked donor in a genetic helper-donor transposition assay (49). In this study, transposition of Ty3 IN mutants was monitored by using both quantitative and qualitative versions of a plasmid-based suppressor target assay (Fig. 2) (47). The assay is based on the expression of a transposition-competent Ty3 under control of the GAL1-10 promoter, carried on a URA3-marked donor plasmid, and its transposition into a HIS3-marked target plasmid. The target plasmid contains two divergent tRNA genes, sup2-o and tDNAVal. The tDNAVal gene acts to recruit Ty3 to the target site. The sup2-o gene is a transcriptionally inactive gene encoding an ochre suppressor tRNATyr. Expression of this suppressor is activated by Ty3 integration into the target site. Ty3 wild-type and IN mutant strains were grown on medium containing galactose and lacking histidine and uracil, to induce transposition and select for Ty3 and target plasmids, and then replica plated to minimal medium containing leucine and tryptophan. Under these conditions, cells must be Ade+ and Lys+ to grow. Transposition was scored as papillations on the latter medium, resulting from suppression of the ochre nonsense markers, ade2-101 lys2-1, in the host strain yTM443 (Fig. 2). A quantitative assay in which 106 cells were plated in duplicate or triplicate onto SD or SG medium lacking uracil and histidine was performed. The mutants were tested at least twice by using one or two separate transformants. These data are described in Fig. 2A. An example of the qualitative patch assay is shown in Fig. 2B; these results were in good agreement with those of the quantitative assay. The polypurine tract mutant 519A(1) was the least affected, with near wild-type levels of transposition. Ty3 IN mutants 11A(2) and 43A(2) retained 60 to 90% of the transposition activity relative to the wild type. Mutants 33A(2) and 69A(2) had 30 to 60% of the transposition activity compared to the wild type, and mutant 436A(2) had 10 to 30% of the transposition activity compared to the wild type. Transposition was indistinguishable from background for the majority (14 of 20) of the IN mutants. Thus, in addition to the previously characterized D,D(35)E motif, charged residues in the nonconserved amino- and carboxyl-terminal domains of Ty3 IN are required for Ty3 position-specific transposition.
FIG. 2.
Ty3 transposition assay. (A) Schematic diagram and transposition activity of charged-to-alanine scanning Ty3 IN mutants. The conserved HHCC zinc finger motif and the D,D(35)E motif are indicated. The positions of the mutations in the 536-amino-acid protein are indicated by lines. A quantitative plasmid-based suppressor target assay was used to score transposition events for each of the Ty3 IN mutants (see Materials and Methods). Transposition was scored by suppression of the ade2-101 lys2-1 markers of yeast strain yTM443 as papillations on minimal medium containing leucine and tryptophan. Quantitative transposition assays were performed at least twice with one or two independent transformants for wild-type Ty3 and IN mutants. A comparison of transposition levels between each mutant and the wild type (wt) is shown. (B) Qualitative assay for Ty3 transposition. Ty3 IN mutants were patched onto SD medium lacking uracil and histidine to select for the donor and target plasmids, respectively. The plate was incubated at 30°C for 24 h, and the cells were replica plated to minimal medium containing leucine and tryptophan (results not shown) and to SG lacking uracil and histidine to induce Ty3 transposition. After 48 h at 30°C on SG, the patches were replica plated onto minimal medium containing leucine and tryptophan and incubated at 30°C for 6 days. Transposition was scored as described in panel A.
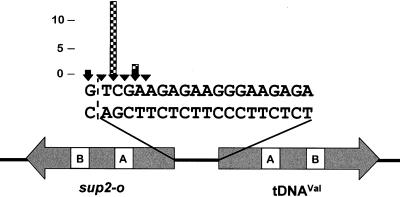
It is not known whether disruption of Ty3 integration specificity results in a default to no integration or relaxed specificity of integration. Therefore, it was of interest, even among this set of targeted integrants, to examine the accuracy of insertion. Defects in 3′-end processing of Ty3 DNA or targeting of Ty3 could potentially be detected by sequence analysis of the transposed element. Thus, a total of 16 independent Ty3 insertions (1 to 3 insertions per Ty3 element) into the target plasmid were recovered and mapped for two wild-type Ty3 elements and the six Ty3 IN mutants that retained transposition activity (Fig. 3). A previous study with the same target and wild-type Ty3 elements showed that insertions from positions −16 to −20 relative to the tDNAVal structural sequence could be detected, with the majority (59%) of insertions occurring at position −19 (47). In the present study, the two wild-type Ty3 insertions occurred at positions −17 and −21 relative to the tDNAVal structural sequence. Fourteen insertions of the mutant Ty3 elements occurred at positions −17 and −19 relative to the tDNAVal coding region; 13 of these were at position −19 and were thus similarly located to wild-type insertions in the previous study. Each of the 14 mutant insertions displayed the characteristic 5-bp target repeats. The sequence of these insertions suggests that IN mutant elements inserted into the target do not display relaxed targeting.
FIG. 3.
Positions of Ty3 insertions recovered after selection for suppressor tRNA gene expression. The structural coding sequences of the divergent tRNA genes of the target plasmid and the 19-bp sequence separating the genes are indicated. The positions and numbers of independent integration events that were isolated at those positions are represented by the arrowheads and height of the bars, respectively. Two wild-type Ty3 insertions and 14 IN mutant insertions into the target were rescued, with 1 to 3 insertions for each of the six transposition-competent IN mutants [11A(2), 33A(2), 43A(2), 69A(2), 436A(2), and 519A(1)]. Wild-type Ty3 sequence (solid bars) begins at positions −17 and −21 relative to tDNAVal, and IN mutant Ty3 sequence (checked bars) begins at positions −17 and −19. The dashed line demarcates the first base pair of the sup2-o structural sequence.
Mutations in IN do not affect GAG3 protein synthesis or assembly but can affect the stability and processing of the IN domain.
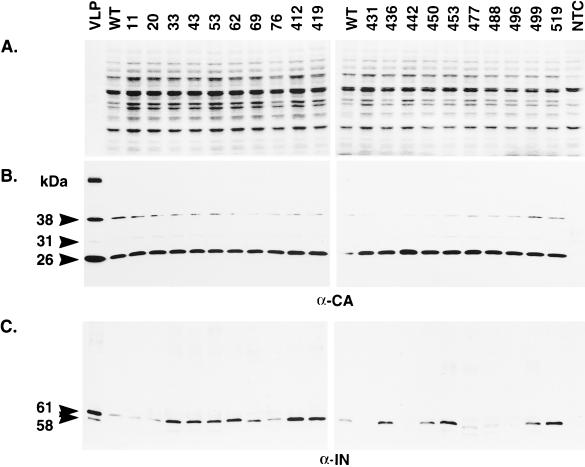
Ty3 particles are composed primarily of protein derived from the GAG3 reading frame. Nevertheless, stable particle formation is dependent on PR activity and so requires Gag3-Pol3p fusion protein in addition to mature CA (48). To determine whether polyprotein processing was affected in the IN mutants, whole-cell extract isolated from cells expressing Ty3 was separated on an SDS-polyacrylamide gel and either stained with Coomassie blue (Fig. 4A), or transferred to nitrocellulose membranes and subjected to immunoblot analysis with antibody to CA (Fig. 4B) and IN (Fig. 4C). All 20 IN mutants had wild-type levels of mature CA (26 kDa), indicating that polyprotein processing and VLP assembly are not grossly affected in these mutants. Mature Ty3 IN species (61 and 58 kDa) result from differential amino-terminal processing (48). These are both visible only in VLP preparations, but the 61-kDa IN species is the primary species detected in the whole-cell extract samples (unpublished data). Based on the results of several immunoblot analyses, none of the eight mutations in the amino-terminal domain of Ty3 IN, proximal to the processing sites, affected steady-state levels of mature 61 kDa IN protein. However, 3 of the 12 mutations in the carboxyl-terminal domain [431A(2), 442A(2), and 496A(2)] resulted in undetectable levels of mature IN and two [477A(2) and 488A(2)] displayed a species that was smaller than 61 kDa and also present at lower levels than wild-type IN. Thus, mutations in nonconserved regions of Ty3 IN can affect IN processing and/or stability.
FIG. 4.
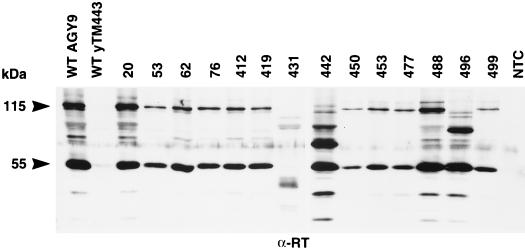
Immunoblot analysis of Ty3 proteins. (A) A 1-μg portion of wild-type Ty3 VLP protein or 10 μg of whole-cell extract isolated from yTM443 cells overexpressing wild-type Ty3 (WT) or IN mutant Ty3 or from nontransformed cells (NTC) was separated on a denaturing SDS–10% polyacrylamide gel and stained with Coomassie blue. (B and C) Identical samples were transferred to nitrocellulose (Hybond ECL; Amersham) and subjected to immunoblot analysis with a polyclonal rabbit anti-CA immunoglobulin G antibody (B) or a polyclonal rabbit anti-IN immunoglobulin G antibody (C). The positions of the structural proteins p38 (38 kDa), p31 (31 kDa), and CA (26 kDa) and the IN species (61 and 58 kDa [apparent only for VLP protein]) are indicated on the left. The 115-kDa RT-IN fusion protein was not detectable with anti-IN antibody on immunoblots of whole-cell extracts.
Mutations in IN severely affect the amount of reverse-transcribed DNA in vivo but do not completely disrupt RT activity in vitro.
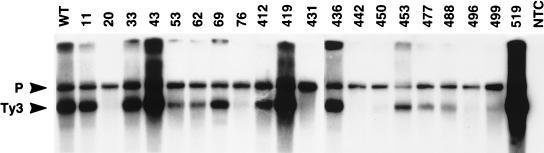
Reverse transcription of Ty3 DNA requires maturation of Ty3 RT. A previous study showed that even modest carboxyl-terminal truncations (27 amino acids) of Ty3 IN severely reduced the amount of Ty3 DNA present in vivo (49) but did not completely eliminate RT activity in vitro. The predominant Ty3 extrachromosomal DNA species in cells expressing wild-type or IN catalytic-site mutant Ty3 is the linear, full-length 5.4-kb species. To examine whether Ty3 extrachromosomal DNA was affected for the IN mutants in this study, nucleic acid was isolated from cells expressing Ty3, digested with BamHI to linearize the vector carrying the Ty3 element, and subjected to Southern analysis with a probe to the internal (non-LTR) portion of Ty3 DNA (Fig. 5). Mutations in both amino-terminal [20A(2) and 76A(2)] and carboxyl-terminal [431A(2), 442A(2), 450A(2), and 496A(2)] domains of Ty3 IN resulted in nearly undetectable amounts of reverse-transcribed DNA. Additional mutants [53A(2), 62A(2), 453A(2), 477A(2), 488A(2), and 499A(2)] had lower than wild-type levels of DNA. In contrast, mutants 43A(2), 419A(3), and 519A(1) consistently displayed higher than wild-type levels of DNA. Although it might be anticipated that Ty3 extrachromosomal DNA would accumulate in the presence of mutations blocking integration, this was not observed for the catalytic site D,D(35)E mutant (49). As anticipated, Ty3 IN mutants that were capable of transposition retained at least wild-type levels of reverse-transcribed DNA. However, the transposition-deficient mutants 412A(2) and 419A(3) also had at least wild-type levels of DNA, suggesting that in these mutants, and possibly in others, transposition was affected at a step after reverse transcription.
FIG. 5.
Southern blot analysis of nucleic acid from cells expressing Ty3 IN mutants. A 10-μg sample of total nucleic acid digested with BamHI was separated on a 1% agarose gel, transferred to nitrocellulose (Duralon UV; Stratagene) and probed with a 32P-labeled, BglII-digested DNA fragment containing the internal (non-LTR) region of Ty3. The position of the 5.4-kb replicated, full-length, extrachromosomal linear Ty3 DNA is indicated; the upper band represents the expression plasmid (P).
Decreased levels of VLP-associated Ty3 DNA could be attributable to incorrectly processed or unstable Ty3 RT protein or to inactive RT. Ty3 RT is not as reproducibly detectable as IN on immunoblots with whole-cell extracts. Therefore, to investigate whether the replication defect of the Ty3 IN mutants resulted from aberrant processing or gross instability of RT, VLPs were isolated from cells expressing Ty3, as described in Materials and Methods. VLP proteins were separated on an SDS-polyacrylamide gel and subjected to immunoblot analysis with antisera to RT (Fig. 6). Two forms of RT have previously been detected by immunoblot analysis, a 55-kDa species and a 115-kDa RT-IN fusion protein (49). All IN mutants, except 431A(2), had the mature 55-kDa species of RT. Based on this and other immunoblot analyses with antisera to RT, mutants 431A(2), 442A(2), and 496A(2) lacked the 115-kDa RT-IN fusion protein. Mature IN was also lacking in whole-cell extracts for each of these mutants (Fig. 4C). This suggests that the IN domain may have been destabilized by these mutations. VLPs from transposition-competent IN mutants, with normal levels of DNA, were not analyzed.
FIG. 6.
Immunoblot analysis of VLP proteins from cells expressing Ty3 IN mutants. VLPs were isolated from either AGY-9 or yTM443 cells expressing wild-type Ty3 (WT) or IN mutant Ty3, or fractions at the position of VLPs were taken from nontransformed cells (NTC). AGY-9 cells were used as the source of VLPs for mutants 20A(2), 431A(2), 442A(2), 488A(2), and 496A(2), as well as for the nontransformed cell control. YTM443 cells were used as the source of VLPs for 53A(2), 62A(2), 76A(2), 412A(2), 419A(3), 450A(2), 453A(2), 477A(2), and 499A(2). Equal amounts of protein (∼1.2 μg) were separated on a denaturing SDS–10% polyacrylamide gel, transferred to nitrocellulose (Hybond ECL; Amersham), and probed with a polyclonal rabbit anti-RT immunoglobulin G antibody. The positions of the 55-kDa RT and 115-kDa RT-IN fusion protein are indicated on the left.
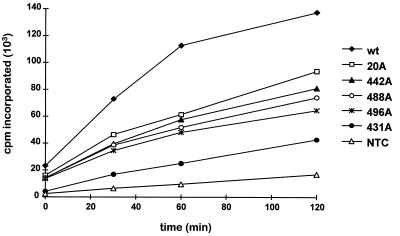
Ty3 VLPs represent a heterogeneous collection of particles at different stages of particle morphogenesis. Because of this, the VLP fraction contains both RNA and DNA and RT activity. To determine whether specific RT activity was affected by mutations in IN, VLPs were isolated from cells expressing Ty3 and used in a standard exogenous RT assay. RT activity was measured as the incorporation of [α-32P]dTTP, using a poly(rA) · p(dT)12–18 template-primer, as described in Materials and Methods. Incorporation was measured at 0, 30, 60, and 120 min. The wild-type VLP fraction displayed a linear increase in incorporation from 0 to 60 min. The RT activity of the IN mutant VLP fractions ranged from 22 to 55% of the activity of the wild-type VLP fraction (Fig. 7). Five IN mutants with reduced levels of VLP-associated DNA, as determined by Southern analysis of whole-cell extract DNA, were analyzed. Ty3 RT activity was severely diminished for mutant 431A(2). This was not surprising, since the 55-kDa RT protein was not detectable by immunoblot analysis (Fig. 6). Mutant 20A(2) displayed both IN and RT proteins (Fig. 4C and 6, respectively) and lacked replicated DNA in vivo. However, it retained considerable RT activity in vitro (Fig. 7). Mutants 442A(2), 488A(2), and 496A(2) had aberrant patterns of IN (Fig. 4C). The mutants had wild-type levels of 55-kDa RT protein, but 442A(2) and 496A(2) lacked the 115-kDa RT-IN fusion protein (Fig. 6). All three were replication defective in vivo yet retained significant RT activity in vitro (Fig. 7). Thus, residues in the nonconserved domains of Ty3 IN appear critical for reverse transcription in vivo and/or stability of full-length replicated Ty3 DNA within the VLP, even when the 55-kDa RT species is present. In contrast to the absence of DNA in these VLPs, the level of RT activity in vitro, with exogenous template and primer, was substantial.
FIG. 7.
In vitro RT activity of Ty3 IN mutants. VLPs were isolated from AGY-9 cells expressing wild-type Ty3 (wt) or IN mutant Ty3, or fractions at the position of VLPs were taken from non-transformed cells (NTC). A 10-μg portion of VLP protein was used for each sample. RT activity was measured as the incorporation of [α-32P]dTTP at 25°C, using a poly(rA) · p(dT)12–18 template-primer (Pharmacia). The cpm incorporated was plotted as a function of time. Points represent averages of duplicate samples differing at the most by 10%. This activity assay was performed three times; the plot represents the results of one such experiment.
Residues in the carboxyl-terminal domain of Ty3 IN are required for 3′-end processing of Ty3 DNA.
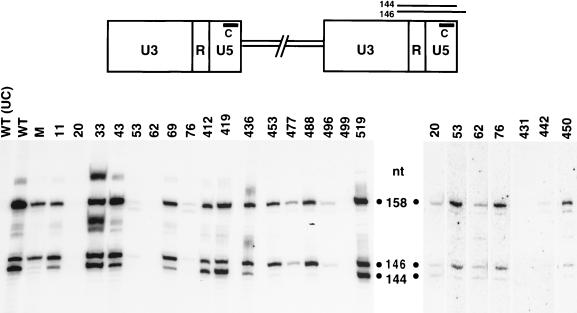
Full-length, replicated Ty3 DNA contains two bases at each 3′ end that are removed by IN in a reaction termed 3′-end processing. In a previous study, residues of the conserved D,D(35)E motif of Ty3 IN were shown to be absolutely required for 3′-end processing activity (49). To examine whether mutations in the nonconserved regions of Ty3 IN affected 3′-end processing of Ty3 DNA, nucleic acid was extracted from sucrose gradient fractions containing Ty3 VLPs and treated with RNase A and HinfI. The resulting fragments were separated on a denaturing polyacrylamide gel, as for sequencing, and transferred onto a nylon membrane for Southern analysis. Probe C, which is complementary to the U5 sequences at the 3′ end of the plus strand, was used to monitor the 3′-end processing event (Fig. 8, top). Previous analysis of the 3′-terminal structure of Ty3 plus-strand DNA revealed a hybridization pattern consisting of two distinct bands, representing the preprocessed (146-nucleotide [nt]) and the processed (144-nt) forms of Ty3 DNA at the U5 end of the LTR (49). A sequence ladder, representing the 3′-terminal region of Ty3 DNA, served as a molecular standard for determining the size of the 3′-end fragments (data not shown). In this study, VLP DNA from wild-type Ty3 served as a positive control for the pattern of cleaved and uncleaved plus-strand ends (Fig. 8, lane WT); DNA from a catalytic-site mutant (D225E, E261D), known to be defective for 3′-end processing activity (49), was used as a negative control (lane M). Uncut wild-type VLP DNA treated with RNase was used as a control for the detection of any Ty3 fragments that were not a result of HinfI digestion (lane UC). IN mutants with ample amounts of DNA [11A(2), 33A(2), 43A(2), 69A(2), 412A(2), 419A(3), 436A(2), and 519A(1)] were tested at least twice; mutants with small amounts of DNA [20A(2), 53A(2), 62A(2), 76A(2), 431A(2), 442A(2), 450A(2), 453A(2), 477A(2), 488A(2) and 496A(2)] were tested at least three times, with the exception of 499A(2), which was tested twice. Increasing amounts of VLP DNA were used in each additional experiment in an attempt to resolve whether 3′-end processing could occur in these mutants. DNA preparations from mutants with amino-terminal Ty3 IN mutations [11A(2), 20A(2), 33A(2), 43A(2), 53A(2), 62A(2), 69A(2), and 76A(2)], as well as mutants 412A(2), 419A(3), 436A(2), 450A(2), and 519A(1), showed two prominent bands (146 and 144 nt), similar to the wild-type pattern (Fig. 8). Thus, these IN mutants were capable of 3′-end processing. Of these mutants, 53A(2), 76A(2), and 450A(2), displayed a somewhat reduced ratio of processed to unprocessed DNA compared to that of the wild type. Although unprocessed DNA was detectable in VLPs from four mutants with carboxyl-terminal IN mutations [453A(2), 477A(2), 488A(2), and 496A(2)], very little or no processed Ty3 DNA was detected, indicating that these mutants are defective for 3′-end processing. Of these mutants, 453A(2) displayed correctly processed IN (Fig. 4C) and RT (Fig. 6) proteins, as well as the 115-kDa RT-IN species (Fig. 6). Because of the limited amount of DNA available for evaluating mutants 431A(2), 442A(2), and 499A(2), it was difficult to accurately determine the ratio of processed to unprocessed forms of Ty3 DNA. Although mutant 499A(2) produced low but detectable levels of DNA in vivo based on Southern analysis of whole-cell extracts (Fig. 5), it is interesting that VLPs prepared from this mutant, in contrast to several other mutants with low levels of DNA in that assay, contained virtually no detectable DNA.
FIG. 8.
Detection of 3′-end processing of Ty3 DNA by Southern analysis. A schematic diagram of Ty3 with the LTR regions U3, R, and U5 is shown at the top. VLP nucleic acid was isolated from cells expressing wild-type Ty3 (WT) or IN mutant Ty3. The catalytic-site double mutant (D225E, E261D) (lane M) was used as a control. VLP DNA, at 0.5 to 4 μg depending on the availability of DNA, was digested with HinfI or not digested for a WT DNA uncut control (UC), and separated on an 8% polyacrylamide gel containing 6 M urea. A sequence ladder (not shown) representing the 3′ terminus of Ty3 DNA was run in adjacent lanes to serve as a molecular standard for determining the size of the Ty3 DNA fragments. The fragments were transferred onto a nylon membrane (GeneScreen) for Southern analysis with a 32P-5′-end-labeled oligonucleotide, probe C, which is complementary to plus-strand sequences in the U5 region of the LTR, as indicated in the top panel. Predicted fragment sizes from HinfI-digested Ty3 DNA are shown in the top panel, and corresponding fragment sizes are indicated next to the blots, with the 146- and 144-nt species representing the 3′ preprocessed and processed Ty3 DNAs, respectively. The 158-nt species represents the HinfI fragment from the 3′ end of the plus-strand strong-stop DNA extended into the tRNA minus-strand primer. The composite of three blots shown on the left includes mutants for which sufficient amounts of VLP DNA were available for analysis. The composite blot on the right shows mutants with substantially smaller amounts of VLP DNA. This group of mutants was tested at least three times, with increasing amounts of DNA used in each experiment. The contrast of this blot was computer enhanced to display faint bands.
DISCUSSION
The catalytic functions of retroelement IN have been defined by in vitro assays of wild-type and mutant recombinant proteins. Despite the success of these in vitro studies of recombinant IN protein and its derivatives, understanding the multiple functions of IN in vivo in DNA replication, uncoating, nuclear entry, and target access is ongoing. In this respect, the Ty3 retrotransposon system offers certain advantages: IN protein function can be assessed without a requirement for infectivity; production of proteins is simple to assay; a linear form of the DNA can be directly analyzed to assess 3′-end processing in vivo; and, similar to HIV, nuclear targeting, presumably accompanied by uncoating, is required for integration. In addition, if Ty3 IN is directly involved in targeting, understanding Ty3 target specificity may help to discriminate the roles of IN domains in donor and target association.
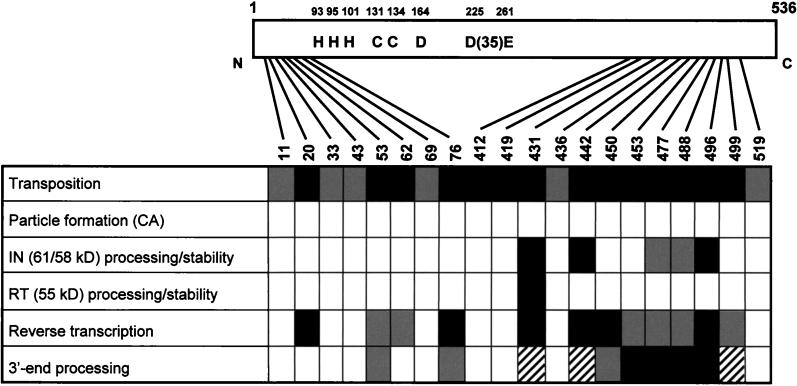
In the present study, the contributions of the nonconserved domains of Ty3 IN in vivo were investigated at the stages of protein maturation, particle assembly, DNA replication, 3′-end processing, and transposition. Charged-to-alanine scanning mutagenesis was used to dissect the functions of the nonconserved amino- and carboxyl-terminal domains of the protein. Twenty Ty3 IN mutants were assayed at multiple stages of the Ty3 life cycle. The results of this study are summarized in Fig. 9.
FIG. 9.
Summary of charged-to-alanine scanning Ty3 IN mutant phenotypes. Open boxes indicate Ty3 IN mutants with no detectable defect or presumed defect at the corresponding step of the life cycle. Shaded boxes indicate IN mutants with a slight defect. Solid boxes indicate IN mutants with a severe defect at the step of the Ty3 life cycle tested. Hatched boxes indicate IN mutants that could not be definitively analyzed in the corresponding assay.
Transposition, as assayed by a plasmid-based suppressor target assay, was abolished for 14 of the 20 IN mutants. This transposition assay allows the recovery of insertions within a small window upstream of the tRNA target gene. Ty3 insertions into the target plasmid were recovered and mapped for the six IN mutants that retained transposition activity. Although only a few integrants were assayed per mutant, these insertions were distributed similarly to insertions of wild-type elements. However, the suppressor target assay used in this study would not have detected transpositions where targeting was significantly relaxed or abolished. Therefore, mutants strongly affected in targeting would have a transposition-negative phenotype if they transposed nonspecifically.
Since the majority of the Ty3 IN mutants were severely affected for transposition, all the mutants were investigated to assess whether polyprotein processing and VLP assembly occurred. Immunoblot analysis of whole-cell extracts with antibodies against Ty3 CA showed that all the mutants were able to produce the mature, 26-kDa form of Ty3 CA protein at levels similar to the levels of CA in cells expressing wild-type Ty3. Unlike retroviruses, where accumulation of polyprotein precursors and formation of immature particles occurs in host cells and is observed in virions of PR mutants, only low levels of precursor polyproteins are present in Ty3 PR mutants, suggesting that immature particles are not stable (48). Thus, the normal amounts of mature protein observed suggested that VLP assembly is not grossly disturbed in the IN mutants and that Ty3 PR is active on properly displayed protein substrates. Therefore, the transposition defects of these mutants are more likely to involve downstream steps in Ty3 VLP assembly, such as specific steps in Gag3-Pol3p protein maturation, reverse transcription, 3′-end processing, uncoating, nuclear entry, or targeting of the preintegration complex to the target site.
Ty3 IN maturation was examined by performing immunoblot analysis on whole-cell extracts with antibodies to Ty3 IN. IN was correctly processed in the majority of mutants. No mutations in the amino-terminal region, proximal to the processing sites (48), affected the amount or mass of the IN species observed. Five mutants with changes in the carboxyl-terminal domain were defective for IN, as evidenced by an undetectable amount of mature IN or an aberrant pattern of IN-related protein. In four of these mutants, the 55-kDa RT species was produced, suggesting that amino-terminal processing of IN was probably not affected. The high proportion of mutations directed at predicted surface residues that apparently reduced IN stability suggests that the carboxyl-terminal IN domain mediates intermolecular protein or DNA interactions important for IN folding or changes in conformation concomitant with reverse transcription or uncoating.
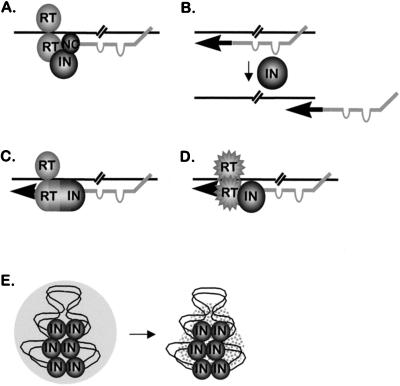
Previous studies showed that carboxyl-terminal deletions of Ty3 IN had severe effects on DNA replication in vivo (49). Similarly, the present study showed that point mutations in both amino- and carboxyl-terminal domains affected the amount of reverse-transcribed Ty3 DNA observed associated with VLPs. Data from previous studies suggested that Ty3 RT is a heterodimer, composed of RT-IN together with RT, such as the RT heterodimers of avian retroviruses. The amino terminus of the 55-kDa protein was determined by Edman degradation (48), and the size of this protein is consistent with a carboxyl-terminal end located close to the amino terminus of IN. This 55-kDa protein includes polymerase and RNase H domains (48). The 115-kDa protein observed in immunoblot analyses with antibody against RT and IN is of a size consistent with inclusion of both the 55-kDa RT and 61-kDa IN domains. In addition, mutations causing carboxyl-terminal truncation of IN resulted in similarly truncated species derived from the 115-kDa protein (49). These results, coupled with the low levels of DNA in Ty3 mutants truncated in the carboxyl-terminal domain of IN, led to the hypothesis that the Ty3 polymerase is an RT/RT-IN heterodimer. In the present study, abnormally small amounts of particle-associated DNA were also observed. Therefore, immunoblot analysis was performed with VLP protein from transposition-deficient IN mutants to examine whether the mutations in Ty3 IN affected the maturation of Ty3 RT and RT-IN. All mutants, except 431A(2), displayed mature 55-kDa Ty3 RT protein. Not surprisingly, the 115-kDa RT-IN fusion protein was absent in the three mutants [431A(2), 442A(2), and 496A(2)] that also lacked mature IN. Consistent with the heterodimer hypothesis, each mutant that lacked IN was deficient for DNA. Despite this apparent correlation, a reduction in the amount of DNA recovered was observed for roughly half of the IN mutants in our study, including several mutants that correctly processed both RT and IN. Formally, there are five possible explanations of the DNA defects produced by mutations in IN (Fig. 10). These are not mutually exclusive scenarios; thus, it is possible that the low DNA recovery in mutants stems from more than one cause. IN could (i) be required for assembly or activity of the reverse transcription initiation complex (Fig. 10A); (ii) affect strand transfer, for example by modulating RNase H activity (Fig. 10B); (iii) contribute a subdomain of a heterodimeric RT, composed of RT and RT-IN (Fig. 10C); (iv) facilitate correct folding of an RT-RT homodimer (Fig. 10D); or (v) stabilize Ty3 DNA within the VLP (Fig. 10E).
FIG. 10.
Hypothetical functions of Ty3 IN during in vivo replication. (A) Stabilization of the interaction between the initiator tRNAMet primer and primer binding site. (B) Mediation of primer/strong-stop strand transfer, possibly by modulation of RNase H activity. (C) Component of the subdomain of heterodimeric RT. (D) Chaperone for the RT-RT homodimer. (E) Stabilization of the full-length, replicated DNA within the VLP. The bipartite tRNAMet primer structure is shown schematically, according to the model for Ty3 reverse transcription initiation by Gabus et al. (27).
As indicated above, IN could be implicated at multiple stages in the retroelement life cycle, several of which could affect the presence of DNA in the preintegration complex. To determine whether the IN domain is essential for polymerization activity of Ty3 RT, a standard exogenous RT assay was performed. This showed that, except for mutant 431A(2), Ty3 IN mutants with severely diminished amounts of reverse-transcribed DNA in vivo displayed significant RT activity in vitro. Thus, the 115-kDa species, including the nonconserved domains of Ty3 IN, appeared not to be essential for RT activity on the exogenous primer and template. This finding argues against the models in Fig. 10C and D, where IN either is part of a heterodimeric RT or is necessary for folding RT. It is possible that the exogenous assay is relatively insensitive compared to in vivo replication, since the exogenous assay could reflect the cumulative activity of multiple polymerase molecules present in the VLP. The exogenous assay would fail to distinguish defects in the interaction between the tRNAMet primer and primer binding site (Fig. 10A) or at later steps in reverse transcription initiation, including RNase H degradation of the template RNA or primer/strong-stop strand transfer (Fig. 10B). These two roles for IN could be required for reverse transcription in vivo but are circumvented in an RT activity assay, where the template and primer are supplied exogenously. Because of the low level of activity of wild-type Ty3 VLPs in endogenous assays, it is difficult to directly test for defects in endogenous activity of the mutant particles. The fifth possibility involves IN, not in reverse transcription per se but in stabilizing full-length replicated DNA after the completion of reverse transcription (Fig. 10E). Because Ty3 DNA must be translocated into the nucleus, a transition from a relatively large particle composed primarily of Gag3-derived protein to a preintegration complex is deemed likely. By analogy to HIV-1, the preintegration complex may be composed primarily of IN, RT, NC, and certain host factors (7, 62). A structural role for IN in stabilizing the preintegration complex would be consistent with preservation of RT activity in the IN mutants and would also explain why VLPs of some mutants, in particular mutant 499A(2), appeared to contain very small or undetectable amounts of Ty3 DNA, although full-length DNA was detectable by Southern analysis of whole-cell extract nucleic acid. If IN is required to stabilize Ty3 DNA, it can be argued that mutants that lack IN completely, in particular 442A(2) and 496A(2), may be unable to form stable preintegration complexes. Thus, the low-DNA phenotype in these mutants would not be due to a defect in reverse transcription but, rather, would be due to degradation of Ty3 DNA released from a defective preintegration complex. This model could also explain the DNA-negative phenotype of HIV-1 IN deletion mutants that have RT activity (10, 54). Interestingly, a Moloney murine leukemia virus mutant with a portion of IN deleted was not affected for DNA levels (80). Because Moloney murine leukemia virus is not transported into the nucleus, IN may contribute less to the structure of the preintegration complex, and thus it is possible that the complex is correspondingly less sensitive to perturbations caused by defects in IN. In summary, it is quite likely that different explanations are appropriate for different Ty3 IN mutants with low levels of DNA. For example, mutants that process IN correctly but have almost no detectable DNA [20A(2), 76A(2), and 450A(2)] may assemble preintegration complexes correctly but be defective in reverse transcription. On the other hand, Ty3 IN mutants that do not have detectable IN would be candidates for defective preintegration complexes.
Still other mutants [11A(2), 33A(2), 43A(2), 69A(2), 412A(2), 419A(3), 436A(2), and 519A(1)] had levels of DNA indistinguishable from those in the wild type and thus were affected in transposition at stages downstream of DNA production. Candidate steps in the life cycle would include uncoating, 3′-end processing, nuclear localization, and integration. Of these mutants, 412A(2) and 419A(3) were the most dramatically affected, with no detectable transposition. Mutants 33A(2), 69A(2), and 436A(2) retained up to 60% of their transposition activity compared to the wild type. The majority of Ty3 IN mutants produced ample amounts of DNA for 3′-end analysis. Results from these experiments showed that mutants 453A(2), 477A(2), 488A(2), and 496A(2) were disrupted for 3′-end processing, producing very little or no processed DNA species. In addition, mutants with mutations in both amino- and carboxyl-terminal domains of Ty3 IN [53A(2), 76A(2), and 450A(2)], which had diminished levels of DNA, had reduced ratios of processed to preprocessed DNA species. Although the amount of Ty3 DNA is reduced in cells expressing these mutants, the amount of IN is similar to that in the wild type for mutants 53A(2), 76A(2), 450A(2), and 453A(2). Assuming that Ty3 particles contain one or a few DNA genomes, the concentration of DNA within a particle should not be affected in these mutants. Thus, the failure of processing to occur is likely to reflect a defect in IN rather than in the levels of IN or substrate. For mutants 431A(2), 442A(2), and 499A(2), the amount of DNA recovered from the VLPs was so small that it was difficult to reproducibly judge the ratios of processed to unprocessed DNA species. Recombinant retroviral IN core domain, with the amino- and carboxyl-terminal domains deleted, can perform disintegration but not 3′-end processing in vitro (8, 17). Mutations in the zinc finger domain of retroviral IN interfere with 3′-end processing and strand transfer (20, 55, 77, 78). Although the Ty3 IN zinc finger was not mutated, it is possible that one or more of these mutations, for example the mutation in 69A(2), affected its function. Based on the similarity of Ty3 and retroviral IN core domains, we infer that these mutations either cause the amino- or carboxyl-terminal region to interfere with catalysis or, more probably disrupt association with the LTR.
Previous studies have failed to produce Ty3 IN mutants that are defective for transposition but retain RT and IN 3′-end processing activity. In the present study, charged-to-alanine scanning mutagenesis of Ty3 IN was successfully used to create such mutants. The phenotypes of the IN mutants in this study are complex and suggest that IN participates in multiple aspects of the Ty3 life cycle (Fig. 9). Recent studies with Ty3 IN mutants 412A(2) and 419A(3), which are completely defective for transposition but display wild-type phenotypes at other stages of the Ty3 life cycle, showed that these mutations in the context of Ty3 IN fused to green fluorescent protein (GFP) disrupt the localization of IN-GFP to the nucleus (56). These data suggest that Ty3 IN contains an NLS located within the carboxyl-terminal domain of IN, which, at least in part, is composed of the mutated residues in IN mutants 412A(2) and 419A(3). Interestingly, these mutations did not affect 3′-end processing activity. These results do not rule out a role for this region in targeting, but they suggest for the first time that VLPs or their derivatives do not require association with the chromosomal target for 3′-end processing to occur. Instead, Ty3 IN, at least in this case, displayed catalytic activity in the cytoplasm, prior to nuclear entry of the preintegration complex. This is similar to what is observed for retroviruses (62, 68), but it appears to be different from the case of the site-specific element Tn7, which does not activate its transposase until it is in the presence of the target DNA complex (2). A subset of Ty3 IN mutants, in particular mutants 53A(2), 62A(2), and 69A(2) with mutations in the amino-terminal domain and mutant 436A(2) with a mutation in the carboxyl-terminal domain, produced and processed DNA, albeit at lower levels than in the wild type for mutants 53A(2) and 62A(2). The mutations in 69A(2) and 436A(2) lie in the vicinity of the Ty3 IN zinc finger and putative NLS, respectively, and could thus interfere with the proper folding or functions of these regions. Although the transposition deficiencies that we observed for these mutants could be attributable solely to the defects described above, it is possible that late steps of the Ty3 life cycle are affected as well. Because the region immediately surrounding mutations 412A(2) and 419A(3) appears sufficient to localize GFP to the yeast cell nucleus (56), it is possible that mutations outside this region affect a late step in the Ty3 life cycle other than nuclear entry. Although targeting specificity is a possible step that could have been affected, additional experiments are required to determine whether any of the above mutants are defective for targeting.
Studies of retroviral IN mutants in vivo have also shown pleiotropic effects on viral replication, including mutants with defects in virion morphology, polyprotein processing, and DNA synthesis and at post-DNA synthesis stages (10, 21, 54, 59, 67, 75, 81). More specifically, mutations in the zinc finger-like domain of HIV-1 IN were found to severely affect DNA synthesis in the mutant virions, as assayed by PCR, without affecting RT activity in vitro (54, 59). It is possible that these mutations affected the stability of the replicated DNA, similarly to what is proposed here for the Ty3 IN mutants. Analysis of effects of point mutations in the carboxyl-terminal region of HIV-1 identified two replication-defective mutants with normal levels of DNA and IN activity in vitro, suggesting that IN may be required for correct assembly and function of the preintegration complex in vivo (54). These mutations may be comparable to the late-acting mutations found in the present study.
In summary, the integrity of the Ty3 IN protein in the VLP is essential for DNA production and integration. Our results also suggest that IN plays a central role in the transition from an RNA to DNA genome and in the changes required for both nuclear access and target presentation. The mutants collected in this study can be used in epistasis studies to further define the order of IN functions in the retroelement life cycle and in suppression studies to identify cellular components that may interact directly with IN at later stages of the life cycle.
ACKNOWLEDGMENTS
We thank T. M. Menees and W. E. Robinson for helpful discussions.
This work was supported by Public Health Service grant GM33281 to S.B.S.
REFERENCES
- 1.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Current protocols in molecular biology. New York, N.Y: Greene Publishing Associates/Wiley-Interscience; 1998. [Google Scholar]
- 2.Bainton R J, Kubo K M, Feng J, Craig N L. Tn7 transposition: target DNA recognition is mediated by multiple Tn7-encoded proteins in a purified in vitro system. Cell. 1993;72:931–943. doi: 10.1016/0092-8674(93)90581-a. [DOI] [PubMed] [Google Scholar]
- 3.Baker T A, Luo L. Identification of residues in the Mu transposase essential for catalysis. Proc Natl Acad Sci USA. 1994;91:6654–6658. doi: 10.1073/pnas.91.14.6654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bass S H, Mulkerrin M G, Wells J A. A systematic mutational analysis of hormone-binding determinants in the human growth hormone receptor. Proc Natl Acad Sci USA. 1991;88:4498–4502. doi: 10.1073/pnas.88.10.4498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bradford M M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 6.Brown P O, Bowerman B, Varmus H E, Bishop J M. Retroviral integration: structure of the initial covalent product and its precursor, and a role for the viral IN protein. Proc Natl Acad Sci USA. 1989;86:2525–2529. doi: 10.1073/pnas.86.8.2525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bukrinsky M I, Sharova N, McDonald T L, Pushkarskaya T, Tarpley W G, Stevenson M. Association of integrase, matrix, and reverse transcriptase antigens of human immunodeficiency virus type 1 with viral nucleic acids following acute infection. Proc Natl Acad Sci USA. 1993;90:6125–6129. doi: 10.1073/pnas.90.13.6125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bushman F D, Engelman A, Palmer I, Wingfield P, Craigie R. Domains of the integrase protein of human immunodeficiency virus type 1 responsible for polynucleotidyl transfer and zinc binding. Proc Natl Acad Sci USA. 1993;90:3428–3432. doi: 10.1073/pnas.90.8.3428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bushman F D, Wang B. Rous sarcoma virus integrase protein: Mapping functions for catalysis and substrate binding. J Virol. 1994;68:2215–2223. doi: 10.1128/jvi.68.4.2215-2223.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cannon P M, Wilson W, Byles E, Kingsman S M, Kingsman A J. Human immunodeficiency virus type 1 integrase: effect on viral replication of mutations at highly conserved residues. J Virol. 1994;68:4768–4775. doi: 10.1128/jvi.68.8.4768-4775.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chalker D L, Sandmeyer S B. Ty3 integrates within the region of RNA polymerase III transcription initiation. Genes Dev. 1992;6:117–128. doi: 10.1101/gad.6.1.117. [DOI] [PubMed] [Google Scholar]
- 12.Chow S A, Vincent K A, Ellison V, Brown P O. Reversal of integration and DNA splicing mediated by integrase of human immunodeficiency virus. Science. 1992;255:723–726. doi: 10.1126/science.1738845. [DOI] [PubMed] [Google Scholar]
- 13.Copeland T D, Grandgenett D P, Oroszlan S. Amino acid sequence analysis of reverse transcriptase subunits from avian myeloblastosis virus. J Virol. 1980;36:115–119. doi: 10.1128/jvi.36.1.115-119.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Craigie R, Fujiwara T, Bushman F D. The IN protein of Moloney murine leukemia virus processes the viral DNA ends and accomplishes their integration in vitro. Cell. 1990;62:829–837. doi: 10.1016/0092-8674(90)90126-y. [DOI] [PubMed] [Google Scholar]
- 15.Doak T, Doerder F, Jahn C, Herrick G. A proposed superfamily of transposase genes: transposon-like elements in ciliated protozoa and a common “D35E” motif. Proc Natl Acad Sci USA. 1994;91:942–946. doi: 10.1073/pnas.91.3.942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Drelich M, Haenggi M, Mous J. Conserved residues Pro-109 and Asp-116 are required for interaction of the human immunodeficiency virus type 1 integrase protein with its viral DNA substrate. J Virol. 1993;67:5041–5044. doi: 10.1128/jvi.67.8.5041-5044.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Drelich M, Wilhelm R, Mous J. Identification of amino acid residues critical for endonuclease and integrase activities of HIV-1 IN protein in vitro. Virology. 1992;188:459–468. doi: 10.1016/0042-6822(92)90499-f. [DOI] [PubMed] [Google Scholar]
- 18.Ellison V, Gerton J, Vincent K A, Brown P O. An essential interaction between distinct domains of HIV-1 integrase mediates assembly of the active multimer. J Biol Chem. 1995;270:3320–3326. doi: 10.1074/jbc.270.7.3320. [DOI] [PubMed] [Google Scholar]
- 19.Engelman A, Bushman F D, Craigie R. Identification of discrete functional domains of HIV-1 integrase and their organization within an active multimeric complex. EMBO J. 1993;12:3269–3275. doi: 10.1002/j.1460-2075.1993.tb05996.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Engelman A, Craigie R. Identification of conserved amino acid residues critical for human immunodeficiency virus type 1 integrase function in vitro. J Virol. 1992;66:6361–6369. doi: 10.1128/jvi.66.11.6361-6369.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Engelman A, Englund G, Orenstein J M, Martin M A, Craigie R. Multiple effects of mutations in human immunodeficiency virus type 1 integrase on viral replication. J Virol. 1995;69:2729–2736. doi: 10.1128/jvi.69.5.2729-2736.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Engelman A, Hickman A B, Craigie R. The core and carboxyl-terminal domains of the integrase protein of human immunodeficiency virus type 1 each contribute to nonspecific DNA binding. J Virol. 1994;68:5911–5917. doi: 10.1128/jvi.68.9.5911-5917.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Engelman A, Mizuuchi K, Craigie R. HIV-1 DNA integration: mechanism of viral DNA cleavage and DNA strand transfer. Cell. 1991;67:1211–1221. doi: 10.1016/0092-8674(91)90297-c. [DOI] [PubMed] [Google Scholar]
- 24.Englund G, Theodore T S, Freed E O, Engelman A, Martin M A. Integration is required for productive infection of monocyte-derived macrophages by human immunodeficiency virus type 1. J Virol. 1995;69:3216–3219. doi: 10.1128/jvi.69.5.3216-3219.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fayet O, Ramond P, Polard P, Prere M F, Chandler M. Functional similarities between retroviruses and the IS3 family of bacterial insertion sequences? Mol Microbiol. 1990;4:1771–1777. doi: 10.1111/j.1365-2958.1990.tb00555.x. [DOI] [PubMed] [Google Scholar]
- 26.Fujiwara T, Mizuuchi K. Retroviral DNA integration: structure of an integration intermediate. Cell. 1988;54:497–504. doi: 10.1016/0092-8674(88)90071-2. [DOI] [PubMed] [Google Scholar]
- 27.Gabus C, Ficheux D, Rau M, Keith G, Sandmeyer S, Darlix J-L. The yeast Ty3 retrotransposon contains a 5′-3′ bipartite primer binding site and encodes nucleocapsid protein NCp9 functionally homologous to HIV-1 NCp7. EMBO J, 1998;17:4873–4880. doi: 10.1093/emboj/17.16.4873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gallay P, Hope T, Chin D, Trono D. HIV-1 infection of non-dividing cells through the recognition of integrase by the importin/karyopherin pathway. Proc Natl Acad Sci USA. 1997;94:9825–9830. doi: 10.1073/pnas.94.18.9825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gerard G F, Grandgenett D P. Retrovirus reverse transcriptase. New York, N.Y: Academic Press, Inc.; 1980. pp. 345–394. [Google Scholar]
- 30.Gerton J L, Brown P O. The core domain of HIV-1 integrase recognizes key features of its DNA substrates. J Biol Chem. 1997;272:25809–25815. doi: 10.1074/jbc.272.41.25809. [DOI] [PubMed] [Google Scholar]
- 31.Gibbs C S, Zoller M J. Rational scanning mutagenesis of a protein kinase identifies functional regions involved in catalysis and substrate interactions. J Biol Chem. 1991;266:8923–8931. [PubMed] [Google Scholar]
- 32.Grandgenett D, Quinn T, Hippenmeyer P J, Oroszlan S. Structural characterization of the avian retrovirus reverse transcriptase and endonuclease domains. J Biol Chem. 1985;260:8243–8249. [PubMed] [Google Scholar]
- 33.Hansen L J, Chalker D L, Orlinsky K J, Sandmeyer S B. Ty3 GAG3 and POL3 genes encode the components of intracellular particles. J Virol. 1992;66:1414–1424. doi: 10.1128/jvi.66.3.1414-1424.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hansen L J, Chalker D L, Sandmeyer S B. Ty3, a yeast retrotransposon associated with tRNA genes, has homology to animal retroviruses. Mol Cell Biol. 1988;8:5245–5256. doi: 10.1128/mcb.8.12.5245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hansen L J, Sandmeyer S B. Characterization of a transpositionally active Ty3 element and identification of the Ty3 integrase protein. J Virol. 1990;64:2599–2607. doi: 10.1128/jvi.64.6.2599-2607.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hoffman C S, Winston F. A ten-minute DNA preparation from yeast efficiently releases autonomous plasmids for transformation of Escherichia coli. Gene. 1987;57:267–272. doi: 10.1016/0378-1119(87)90131-4. [DOI] [PubMed] [Google Scholar]
- 37.Jenkins T M, Engelman A, Ghirlando R, Craigie R. A soluble active mutant of HIV-1 integrase: involvement of both the core and carboxyl-terminal domains in multimerization. J Biol Chem. 1996;271:7712–7718. doi: 10.1074/jbc.271.13.7712. [DOI] [PubMed] [Google Scholar]
- 38.Jenkins T M, Esposito D, Engelman A, Craigie R. Critical contacts between HIV-1 integrase and viral DNA identified by structure-based analysis and photo-crosslinking. EMBO J. 1997;16:6849–6859. doi: 10.1093/emboj/16.22.6849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Johnson M S, McClure M A, Feng D-F, Gray J, Doolittle R F. Computer analysis of retroviral pol genes: assignment of enzymatic functions to specific sequences and homologies with non-viral enzymes. Proc Natl Acad Sci USA. 1986;83:7648–7652. doi: 10.1073/pnas.83.20.7648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jonsson C B, Roth M J. Role of the His-Cys finger of Moloney murine leukemia virus integrase protein in integration and disintegration. J Virol. 1993;67:5562–5571. doi: 10.1128/jvi.67.9.5562-5571.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kalpana G V, Goff S P. Genetic analysis of homomeric interactions of human immunodeficiency virus type 1 integrase using the yeast two-hybrid system. Proc Natl Acad Sci USA. 1993;90:10593–10597. doi: 10.1073/pnas.90.22.10593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Katzman M, Katz R A, Skalka A M, Leis J. The avian retroviral integration protein cleaves the terminal sequences of linear viral DNA at the in vivo sites of integration. J Virol. 1989;63:5319–5327. doi: 10.1128/jvi.63.12.5319-5327.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Katzman M, Mack J P G, Skalka A M, Leis J. A covalent complex between retroviral integrase and nicked substrate DNA. Proc Natl Acad Sci USA. 1991;88:4695–4699. doi: 10.1073/pnas.88.11.4695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Katzman M, Sudol M. Mapping domains of retroviral integrase responsible for viral DNA specificity and target site selection by analysis of chimeras between human immunodeficiency virus type 1 and visna virus integrases. J Virol. 1995;69:5687–5696. doi: 10.1128/jvi.69.9.5687-5696.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kenna M A, Baker Brachmann C, Devine S E, Boeke J D. Invading the yeast nucleus: a nuclear localization signal at the C terminus of Ty1 integrase is required for transposition in vivo. Mol Cell Biol. 1998;18:1115–1124. doi: 10.1128/mcb.18.2.1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Khan E, Mack J P, Katz R A, Kulkosky J, Skalka A M. Retroviral integrase domains: DNA binding and the recognition of LTR sequences. Nucleic Acids Res. 1991;19:851–860. doi: 10.1093/nar/19.4.851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kinsey P, Sandmeyer S. Ty3 transposes in mating populations of yeast: a novel transposition assay for Ty3. Genetics. 1995;139:81–94. doi: 10.1093/genetics/139.1.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kirchner J, Sandmeyer S B. Proteolytic processing of Ty3 proteins is required for transposition. J Virol. 1993;67:19–28. doi: 10.1128/jvi.67.1.19-28.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kirchner J, Sandmeyer S B. Ty3 integrase mutants defective in reverse transcription or 3′ end processing of extrachromosomal Ty3 DNA. J Virol. 1996;70:4737–4747. doi: 10.1128/jvi.70.7.4737-4747.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kukolj G, Jones K S, Skalka A. Subcellular localization of avian sarcoma virus and human immunodeficiency virus type 1 integrases. J Virol. 1997;71:843–847. doi: 10.1128/jvi.71.1.843-847.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kulkosky J, Jones K S, Katz R A, Mack J P G, Skalka A M. Residues critical for retroviral integrative recombination in a region that is highly conserved among retroviral/retrotransposon integrases and bacterial insertion sequence transposases. Mol Cell Biol. 1992;12:2331–2338. doi: 10.1128/mcb.12.5.2331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kunkel T A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Proc Natl Acad Sci USA. 1985;82:488–492. doi: 10.1073/pnas.82.2.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.LaFemina R L, Schneider C L, Robbins H L, Callahan P L, LeGrow K, Roth E, Schleif W A, Emini E A. Requirement of active human immunodeficiency virus type 1 integrase enzyme for productive infection of human T-lymphoid cells. J Virol. 1992;66:7414–7419. doi: 10.1128/jvi.66.12.7414-7419.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Leavitt A D, Robles G, Alesandro N, Varmus H. Human immunodeficiency virus type 1 integrase mutants retain in vitro integrase activity yet fail to integrate viral DNA efficiently during infection. J Virol. 1996;70:721–728. doi: 10.1128/jvi.70.2.721-728.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Leavitt A D, Shiue L, Varmus H E. Site-directed mutagenesis of HIV-1 integrase demonstrates differential effects on integrase functions in vitro. J Biol Chem. 1993;268:2113–2119. [PubMed] [Google Scholar]
- 56.Lin, S. S., and S. B. Sandmeyer. Unpublished data.
- 57.Lodi P J, Ernst J A, Kuszewski J, Hickman A B, Engelman A, Craigie R, Clore G M, Gronenborn A M. Solution structure of the DNA binding domain of HIV-1 integrase. Biochemistry. 1995;34:9826–9833. doi: 10.1021/bi00031a002. [DOI] [PubMed] [Google Scholar]
- 58.Maniatis T, Fritsch E F, Sambrook J. Molecular cloning—a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 59.Masuda T, Planelles V, Krogstad P, Chen I S. Genetic analysis of human immunodeficiency virus type 1 integrase and the U3 att site: unusual phenotype of mutants in the zinc finger-like domain. J Virol. 1995;69:6687–6696. doi: 10.1128/jvi.69.11.6687-6696.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Menees, T. M. Personal communication.
- 61.Menees T M, Sandmeyer S B. Transposition of the yeast retroviruslike element Ty3 is dependent on the cell cycle. Mol Cell Biol. 1994;14:8229–8340. doi: 10.1128/mcb.14.12.8229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Miller M D, Farnet C M, Bushman F D. Human immunodeficiency virus type 1 preintegration complexes: studies of organization and composition. J Virol. 1997;71:5382–5390. doi: 10.1128/jvi.71.7.5382-5390.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Moore S P, Rinckel L A, Garfinkel D J. A Ty1 integrase nuclear localization signal required for retrotransposition. Mol Cell Biol. 1998;18:1105–1114. doi: 10.1128/mcb.18.2.1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mumm S R, Grandgenett D P. Defining nucleic acid-binding properties of avian retrovirus integrase by deletion analysis. J Virol. 1991;65:1160–1167. doi: 10.1128/jvi.65.3.1160-1167.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Pahl A, Flugel R M. Characterization of the human spuma retrovirus integrase by site-directed mutagenesis, by complementation analysis, and by swapping the zinc finger domain of HIV-1. J Biol Chem. 1995;270:2957–2966. doi: 10.1074/jbc.270.7.2957. [DOI] [PubMed] [Google Scholar]
- 66.Puras Lutzke R A, Vink C, Plasterk R H A. Characterization of the minimal DNA-binding domain of the HIV integrase protein. Nucleic Acids Res. 1994;22:4125–4131. doi: 10.1093/nar/22.20.4125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Reicin A S, Paik S, Berkowitz R D, Luban J, Lowy I, Goff S. Linker insertion mutations in the human immunodeficiency virus type 1 gag gene: effects on virion particle assembly, release, and infectivity. J Virol. 1995;69:642–650. doi: 10.1128/jvi.69.2.642-650.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Roe T, Chow S A, Brown P O. 3′-End processing and kinetics of 5′-end joining during retroviral integration in vivo. J Virol. 1997;71:1334–1340. doi: 10.1128/jvi.71.2.1334-1340.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Roth M J, Schwartzberg P L, Goff S P. Structure of the termini of DNA intermediates in the integration of retroviral DNA: dependence on IN function and terminal DNA sequence. Cell. 1989;58:47–54. doi: 10.1016/0092-8674(89)90401-7. [DOI] [PubMed] [Google Scholar]
- 70.Rowland S J, Dyke K G. Tn552, a novel transposable element from Staphylococcus aureus. Mol Microbiol. 1990;4:961–975. doi: 10.1111/j.1365-2958.1990.tb00669.x. [DOI] [PubMed] [Google Scholar]
- 71.Schauer M, Billich A. The N-terminal region of HIV-1 integrase is required for integration activity, but not for DNA binding. Biochem Biophys Res Commun. 1992;185:874–880. doi: 10.1016/0006-291x(92)91708-x. [DOI] [PubMed] [Google Scholar]
- 72.Schiff R D, Grandgenett D P. Virus-coded origin of a 32,000-dalton protein from avian retrovirus cores:structural relatedness of p32 and the beta polypeptide of the avian retrovirus DNA polymerase. J Virol. 1978;28:279–291. doi: 10.1128/jvi.28.1.279-291.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Shibagaki Y, Chow S A. Central core domain of retroviral integrase is responsible for target site selection. J Biol Chem. 1997;272:8361–8369. doi: 10.1074/jbc.272.13.8361. [DOI] [PubMed] [Google Scholar]
- 74.Shin C-G, Taddeo B, Haseltine W A, Farnet C M. Genetic analysis of the human immunodeficiency virus type 1 integrase protein. J Virol. 1994;68:1633–1642. doi: 10.1128/jvi.68.3.1633-1642.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Taddeo B, Haseltine W A, Farnet C M. Integrase mutants of human immunodeficiency virus type 1 with a specific defect in integration. J Virol. 1994;68:8401–8405. doi: 10.1128/jvi.68.12.8401-8405.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.van Gent D C, Elgersma Y, Bolk M W J, Vink C, Plasterk R H A. DNA binding properties of the integrase proteins of human immunodeficiency viruses types 1 and 2. Nucleic Acids Res. 1991;19:3821–3837. doi: 10.1093/nar/19.14.3821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.van Gent D C, Oude Groeneger A A M, Plasterk R H A. Mutational analysis of the integrase protein of human immunodeficiency virus type 2. Proc Natl Acad Sci USA. 1992;89:9598–9602. doi: 10.1073/pnas.89.20.9598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Vincent K A, Ellison V, Chow S A, Brown P O. Characterization of human immunodeficiency virus type 1 integrase expressed in Escherichia coli and analysis of variants with amino-terminal mutations. J Virol. 1993;67:425–437. doi: 10.1128/jvi.67.1.425-437.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Vink C, Oude Groeneger A A M, Plasterk R H A. Identification of the catalytic and DNA-binding region of the human immunodeficiency virus type 1 integrase protein. Nucleic Acids Res. 1993;21:1419–1425. doi: 10.1093/nar/21.6.1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wei S-Q, Mizuuchi K, Craigie R. A large nucleoprotein assembly at the ends of the viral DNA mediates retroviral DNA integration. EMBO J. 1997;16:7511–7520. doi: 10.1093/emboj/16.24.7511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wiskerchen M, Muesing M A. Human immunodeficiency virus type 1 integrase: effects of mutations on viral ability to integrate, direct viral gene expression from unintegrated viral DNA templates, and sustain viral propagation in primary cells. J Virol. 1995;69:376–386. doi: 10.1128/jvi.69.1.376-386.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wiskerchen M, Muesing M A. Identification and characterization of a temperature-sensitive mutant of human immunodeficiency virus type 1 by alanine scanning mutagenesis of the integrase gene. J Virol. 1995;69:597–601. doi: 10.1128/jvi.69.1.597-601.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Woerner A M, Klutch M, Levin J G, Marcus-Sekura C J. Localization of DNA binding activity of HIV-1 integrase to the C-terminal half of the protein. AIDS Res Hum Retroviruses. 1992;8:297–304. doi: 10.1089/aid.1992.8.297. [DOI] [PubMed] [Google Scholar]
- 84.Woerner A M, Marcus-Sekura C J. Characterization of a DNA binding domain in the C-terminus of HIV-1 integrase by deletion mutagenesis. Nucleic Acids Res. 1993;21:3507–3511. doi: 10.1093/nar/21.15.3507. [DOI] [PMC free article] [PubMed] [Google Scholar]