Abstract
The second gene in the 3′-to-5′ gene order in respiratory syncytial virus (RSV) encodes the nonstructural protein NS2, for which there is no assigned function. To study the function of NS2, we have used a recently developed reverse genetics system to ablate expression of NS2 in recombinant RSV. A full-length cDNA copy of the antigenome of RSV A2 strain under the control of a T7 promoter was modified by introduction of tandem termination codons within the NS2 open reading frame (NS2stop) or by deletion of the entire NS2 gene (ΔNS2). The NS2 knockout antigenomic cDNAs were cotransfected with plasmids encoding the N, P, L, and M2-1 proteins of RSV, each controlled by the T7 promoter, into cells infected with a vaccinia virus recombinant expressing T7 RNA polymerase. Recombinant NS2stop and ΔNS2 RSVs were recovered and characterized. Both types of NS2 knockout virus displayed pinpoint plaque morphology and grew more slowly than wild-type RSV. The expression of monocistronic mRNAs for the five genes examined (NS1, NS2, N, F, and L) was unchanged in cells infected with either type of NS2 knockout virus, except that no NS2 mRNA was detected with the ΔNS2 virus. Synthesis of readthrough mRNAs was affected only for the ΔNS2 virus, where the NS1-NS2, NS2-N, and NS1-NS2-N mRNAs were replaced with the predicted novel NS1-N mRNA. Upon passage, the NS2stop virus stock rapidly developed revertants which expressed NS2 protein and grew with similar plaque morphology and kinetics wild-type RSV. Sequence analysis confirmed that the termination codons had reverted to sense, albeit not the wild-type assignments, and provided evidence consistent with biased hypermutation. No revertants were recovered from recombinant ΔNS2 RSV. These results show that the NS2 protein is not essential for RSV replication, although its presence greatly improves virus growth in cell culture. The attenuated phenotype of these mutant viruses, coupled with the expected genetic stability associated with gene deletions, suggests that the ΔNS2 RSV is a candidate for vaccine development.
Human respiratory syncytial virus (RSV) is one of the most important etiologic agents of pediatric respiratory disease worldwide (9). RSV is the prototype member of the genus Pneumovirus of the family Paramyxoviridae. Its genome consists of a single, negative-sense RNA of 15,222 nucleotides (for strain A2) encoding 10 major subgenomic mRNAs and 11 viral proteins. Three of these proteins form the minimal viral polymerase: the nucleocapsid (N) protein, the phosphoprotein (P protein), and the large polymerase (L) protein. The 22-kDa M2-1 protein encoded by the 5′-proximal open reading frame (ORF) of the M2 mRNA has been shown to be a transcription elongation/antitermination factor and probably is associated with the viral nucleocapsid (8, 18). The attachment (G) and fusion (F) proteins are viral transmembrane glycoproteins involved in virus assembly, budding, and entry. The M protein is thought to be the RSV counterpart of the paramyxovirus matrix (M) protein. However, several RSV proteins lack assigned functions. These are the small hydrophobic (SH) transmembrane glycoprotein, the M2-2 protein (encoded by the second ORF of the M2 RNA), and the nonstructural (NS1 and NS2) proteins. The SH gene has counterparts in the rubulaviruses SV5 and mumps virus but is dispensable for RSV growth in cell culture (2). The M2-2 and NS1 proteins downregulate RSV transcription and RNA replication in a minigenome model system, although the significance of these activities in the viral replicative cycle is unclear (1, 8).
The NS2 protein is encoded by the second gene in the 3′-to-5′ gene order. Due to the promoter-proximal location of its gene, the NS2 mRNA is one of the most abundant of the 10 RSV transcripts and its protein is expressed early in infection (10, 11). The NS2 gene of strain A2 is 503 nucleotides (nt) long and encodes a basic, 124-amino-acid (aa) protein with a predicted molecular mass of 14.7 kDa. Sequence analysis provided no obvious clues to its function or significance. Expression of the NS2 ORF inhibited viral transcription and RNA replication in the minigenome system, although the effect was small compared to that of the NS1 and M2-2 ORFs and occurred only at high levels of NS2 protein expression (references 1 and 8 and unpublished data). In addition, the NS2 protein may interact with the cytoskeleton of infected cells (16, 27). However, the role of the NS2 protein in RSV replication and pathogenesis is not known.
To examine more closely the function of the NS2 protein, we used a recently described reverse genetics system for recovering recombinant RSV (rRSV) from cDNA. This system involves the intracellular coexpression of the N, P, L, and M2-1 proteins and RSV antigenome, under the control of the T7 promoter, in cells concomitantly infected with a vaccinia virus recombinant expressing the T7 RNA polymerase (7). Here, we ablated expression of the NS2 protein by either introducing termination codons into its ORF or excising the complete gene from the antigenome cDNA clone. rRSV which no longer expresses the NS2 protein was recovered but displayed altered growth characteristics in cell culture.
MATERIALS AND METHODS
Plasmid construction.
Construction of a cDNA copy of the RSV antigenome under the control of a T7 promoter has been described previously (7). To introduce tandem termination codons into the NS2 ORF for the construction of NS2stop, an AatII-HindIII fragment from a plasmid containing the 3′ end of the antigenome through the SH gene end signal (D51) (Fig. 1a) was subcloned into pGEM 7Z(−) (Promega) to facilitate the production of single-stranded DNA for site-directed mutagenesis (21). The oligonucleotide 5′-TAT GGT CTC GAG TTA CTA CGG TCT CAT-3′ (corresponding to nucleotides 705 to 679 of the antigenome) was used for the mutagenesis reaction, where the mutations (underlined) and an XhoI site (italics) were simultaneously introduced. The AatII-AflII fragment of the mutagenized plasmid was excised, and reintroduced into D51 (Fig. 1a), and sequenced completely. The PacI-BamHI fragment of a plasmid containing the G, F, and M2 ORFs (GFM2) was inserted into the PacI-BamHI window of D51, reconstructing the previously described D50 plasmid (2). The full-length antigenome clone (NS2stop) was then assembled by inserting the BamHI-MluI fragment of D39 (2) into the mutant D50 plasmid.
FIG. 1.
Mutation of the NS2 gene to ablate expression of its encoded protein. (a) Site-directed mutagenesis was used to alter codons 21 and 22 in the NS2 ORF of the antigenome cDNA (sequences in positive sense). D51 is a cDNA containing nt 1 to 4623 of the RSV antigenome, which was an intermediate in the engineering (see Materials and Methods). Mutated bases are shown above the wild-type sequence. A novel XhoI restriction site (underlined) was added as a marker for mutants. Restriction enzyme sites used in DNA manipulation are shown (see Materials and Methods). (b) The entire NS2 gene was deleted by PCR, as described in Materials and Methods, such that the NS1 gene end signal was fused to the NS2-N intergenic region. The sequence is shown in positive sense, and the point of deletion is indicated by an open triangle.
To delete the NS2 gene to make ΔNS2 (Fig. 1b), D51 was subjected to PCR with Vent DNA polymerase (New England Biolabs) by the method of Byrappa et al. (3). Primers were designed such that they would amplify the entire D51 plasmid except the NS2 gene. The 5′-phosphorylated oligonucleotides used were as follows: forward primer, 5′-TTA AGG AGA GAT ATA AGA TAG AAG ATG-3′ (nt 1100 to 1126), and reverse primer, 5′-GTT TTA TAT TAA CTA ATG GTG TTA GTG-3′ (nt 577 to 551). The forward primer is located in the NS2-N intergenic region, and the reverse primer begins with the NS1 gene end signal (underlined). The PCR product was isolated by agarose gel electrophoresis, self-ligated, and transformed into DH10B competent cells (Life Technologies). The deletion was confirmed by restriction digestion and sequencing. The AatII-AvrII fragment of the resulting D51/ΔNS2 was inserted into a second version of the wild-type antigenome clone, which contains six translationally silent restriction site markers in the L gene as well as two coding changes in the F gene (D53/HEKsites [28]), resulting in rHEKsites/ΔNS2. Thus, the genetic backgrounds of the NS2stop and ΔNS2 viruses differ by two amino acid changes in F and six translationally silent restriction sites in L. The background of the NS2stop viruses is the original rRSV (7), while that of the ΔNS2 virus is one which our laboratory is using for construction of live-attenuated recombinant RSV as vaccine candidates (28). Importantly, these differences were phenotypically silent in vitro; therefore, the two versions of wild-type rRSV are equivalent for the purposes of the present study.
Recovery of rRSV.
Transfections were performed essentially as described previously (7). Briefly, monolayers of HEp-2 cells in six-well dishes were simultaneously infected with 3 focus-forming units per cell of a recombinant vaccinia virus (MVA strain) expressing T7 RNA polymerase (MVA-T7) (29) and transfected with a mixture of plasmids encoding the RSV N, P, L, and M2 (ORF1) proteins and the antigenome (wild-type or mutant) RNA, each under the control of the T7 promoter (with the amounts of input plasmid per well being 0.4, 0.3, 0.2, 0.1, and 1.0 μg, respectively), by using LipofectACE (Life Technologies). The transfection-infection mixture was removed after 18 h of incubation at 32°C and replaced with fresh medium (OptiMEM supplemented with 4% fetal bovine serum). At 48 h later, the clarified supernatants were passaged onto fresh HEp-2 cells and incubated at 37°C; they were harvested 7 days later.
Viral titers were determined by plaque assay for 7 days under 0.8% methylcellulose followed by fixation with 80% methanol. Plaques were visualized by incubation with a cocktail of three murine anti-RSV F monoclonal antibodies followed by horseradish peroxidase-coupled goat anti-mouse IgG antibodies and 4CN substrate (Kirkegaard & Perry Laboratories) as described previously (22).
Northern blot analysis.
Total cellular RNA from infected cells was isolated with Trizol reagent (Life Technologies). RNA (10 μg per sample) was electrophoresed in 1.5% agarose gels containing 0.44 M formaldehyde, transferred to supported nitrocellulose (Optitran, 0.2 μm; Schleicher & Schuell) with a Turboblotter apparatus (Schleicher & Schuell), and hybridized with 32P-labeled DNA probes prepared by random priming (Mega-prime; Amersham) from the indicated RSV ORFs. The blots were washed with 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)–0.1% sodium dodecyl sulfate (SDS) twice at room temperature and twice at 65°C and then subjected to autoradiography with Bio-Max MR film (Kodak).
Western blot analysis.
Cell pellets from infected cells were disrupted by addition of 2× sample buffer (100 mM Tris-Cl [pH 6.8], 4% SDS, 20% glycerol, 0.2% bromophenol blue, 200 mM dithiothreitol) and centrifugation through Qiashredders (Qiagen). Approximately 1.5 × 105 cell equivalents of each infected cell extract was subjected to electrophoresis on SDS–8 to 16% polyacrylamide gels (for the RSV structural proteins) or SDS–4 to 20% polyacrylamide gels (for NS1 and NS2) (Novex) and transferred to a polyvinylidene difluoride membrane (Novex). The blots were incubated with rabbit antiserum raised against either purified RSV (2) or a C-terminal peptide of NS2 (1). Viral proteins were visualized by secondary incubation with either horseradish peroxidase-coupled goat anti-rabbit IgG antibodies followed by chemiluminescence (Boehringer Mannheim) (see Fig. 5) or alkaline phosphatase-coupled goat anti-rabbit IgG antibodies followed by Western Blue substrate (Promega) (see Fig. 7).
FIG. 5.
Expression of RSV proteins by NS2stop, ΔNS2, and NS2stop revertant viruses. Total-cell extracts of HEp-2 cells were harvested 48 h postinfection and subjected to Western blot analysis with an antiserum directed against purified virions (a) or an antiserum against a C-terminal peptide of NS2 which cross-reacts with NS1 (b). Major viral proteins are indicated on the right (NS1 and NS2 run as a doublet).
FIG. 7.
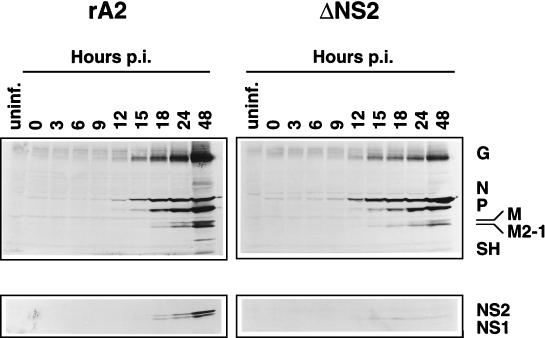
Time course of RSV protein expression by recombinant wild-type and ΔNS2 viruses. HEp-2 cells were infected with rA2 (left) or ΔNS2 (right) virus at a MOI of 3. Total-cell extracts were harvested at the indicated times postinfection (p.i.) and subjected to Western blot analysis with an antiserum against purified RSV (top) or an antiserum against an NS2 peptide (bottom), as in Fig. 5. Major viral proteins are indicated on the right.
RT-PCR and sequencing.
Total cellular RNA from infected cells (1 μg) was used as the template for reverse transcription (RT) with Superscript II Moloney murine leukemia virus reverse transcriptase (Life Technologies) to amplify the NS2 ORF from NS2-containing mRNA. The negative-sense oligonucleotide 5′-GCC AAT GCA TTC TAA GAA CCC-3′ corresponding to nt 948 to 928 in the RSV genome was used to prime cDNA synthesis. One-tenth of the RT reaction mixture was subjected to PCR (Gene Amp; Perkin-Elmer) with the above primer and a second positive-sense oligonucleotide 5′-CCA TGG ACA CAA CCC ACA ATG-3′, corresponding to nt 626 to 646 of the genome. PCR was performed by 35 cycles of denaturation (94°C for 30 s), annealing (42°C for 30 s), and extension (72°C for 1 min) followed by a 10-min extension at 72°C. The amplified region encompasses all but the last 16 aa of NS2. PCR fragments were isolated by agarose gel electrophoresis and cloned into pCR II (Invitrogen). Plasmids from six individual colonies per virus were sequenced in both directions by using Sequenase (USB) and primers which hybridized to the sequences flanking the insert.
RESULTS
Construction of cDNAs encoding rRSV antigenomes containing a mutant or deleted NS2 ORF.
We have previously described the construction of a full-length cDNA clone of the RSV antigenome under the control of a T7 promoter, which was used to rescue wild-type recombinant virus (7). We used two strategies to ablate expression of the NS2 ORF. First, we mutated codons 21 and 22 of the 124-codon NS2 ORF to encode translation terminations by site-directed mutagenesis (NS2stop [Fig. 1a]; see Materials and Methods). The NS2 ORF has methionyl codons at positions 1, 14, and 18; therefore, the two nonsense codons would terminate translation initiating at any of the three. The next in-frame AUG is at codon 67, more than halfway down the ORF. In addition, an adjacent XhoI restriction site was introduced for screening purposes. These mutations were expected to allow normal transcription of the NS2 gene and thus not disrupt the gradient of transcription attenuation down the genome. A polypeptide encompassing the N-terminal 20 aa of NS2 would result from translation of the mutant mRNA. The genetic background of this virus is identical to that in the first reported recovery of rRSV (7).
The second strategy we used was to excise completely the NS2 gene, including the transcription signals (ΔNS2 [Fig. 1b]; see Materials and Methods). For this, we used a PCR-based system whereby the NS1 gene end signal was fused to the NS2-N intergenic region. This mutation removed 523 nt from the antigenomic cDNA, resulting in a genome of 14,700 nt (the wild-type rRSV is 15,223 nt long, 1 nt longer than biologically derived wild-type RSV). The ΔNS2 virus would encode only 9 monocistronic mRNAs, compared to the 10 encoded by wild-type RSV, resulting in an alteration in the pattern of readthrough mRNAs from this region, as described below.
The genetic background of the ΔNS2 virus is a second version of wild-type rRSV, called rHEKsites (28), which contains six translationally silent restriction sites in the L gene and two amino acid substitutions in the F gene which are phenotypically silent in cell culture. The two changes in F make the encoded rRSV identical at the amino acid level to the wild-type A2/HEK strain, which was the progenitor of a series of vaccine candidate viruses, so that direct comparisons can be made in future evaluations of the ΔNS2 virus as a vaccine candidate. For the purposes of the present study, these differences are silent. Nonetheless, the ΔNS2 virus also will be compared with its matched wild-type virus, rHEKsites.
Recovery of rRSV which does not express NS2 protein.
The recovery of infectious rRSV from cDNA has been described in detail previously (7). Intracellular coexpression in HEp-2 cells of the N, P, M2 (ORF1), and L proteins in conjunction with the antigenomic RNA, each under the control of a T7 promoter, was driven by T7 RNA polymerase expressed from a recombinant vaccinia virus (MVA strain). Supernatants from these cells were clarified and passaged onto fresh HEp-2 monolayers. The MVA strain is severely host range restricted in most mammalian cells, including HEp-2, and is lost quickly during passage. NS2stop and ΔNS2 rRSV were recovered and detected by plaque assay under methylcellulose. Wild-type (rA2) and modified wild-type (rHEKsites) antigenome clones were used as controls for the NS2stop and ΔNS2 viruses, respectively.
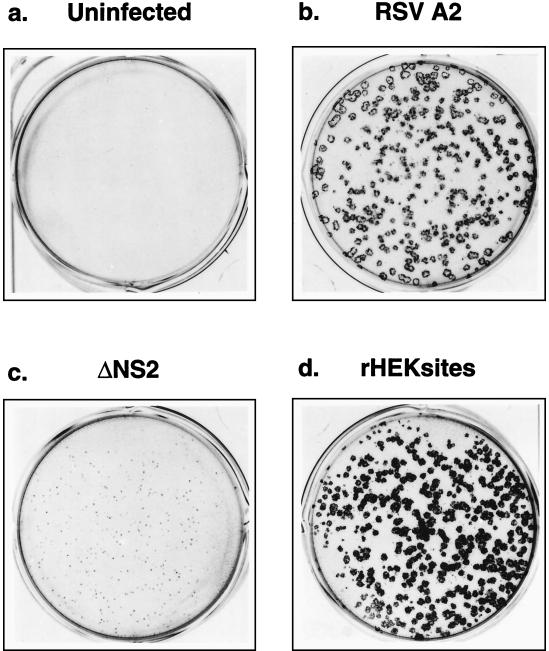
Both types of NS2 knockout viruses displayed a pinpoint plaque morphology even after incubation for 6 days (ΔNS2 is shown in Fig. 2c). These plaques could be visualized only by staining with monoclonal antibodies. In contrast, wild-type rHEKsites formed larger plaques (Fig. 2d), which were clearly visible under light microscopy without antibody staining 5 days postinfection. After six serial passages, a small number of larger, wild-type-like plaques emerged in the NS2stop virus plaque assays against the background of pinpoint plaques (results not shown). Four clonal populations of these large-plaque, putative revertant viruses (R1 to R4) were isolated and characterized as discussed below.
FIG. 2.
Photographs of recombinant RSV plaques. Monolayers of HEp-2 cells were uninfected (a) or infected with biologically derived wild-type A2 (b), ΔNS2 rRSV (c), or wild-type rHEKsites RSV (d) and incubated under methylcellulose for 6 days. The plaques were visualized as described in Materials and Methods and photographed.
Growth of NS2 knockout viruses in vitro.
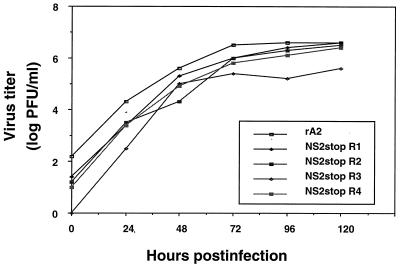
Since the NS2 knockout viruses displayed pinpoint plaque morphology, we investigated whether they exhibited slower growth than wild-type RSV. To do this, triplicate monolayer cultures were infected with NS2stop, ΔNS2, or wild-type RSV at a multiplicity of infection (MOI) of 1. Supernatant samples were harvested, and viral titers were determined by plaque assay (Fig. 3). Both the NS2stop and ΔNS2 viruses showed delayed growth kinetics and lower final viral titers compared to their corresponding wild-type RSV (Fig. 3). After 7 days, the titers of each NS2 knockout virus were approximately 10- to 50-fold lower than those of wild-type RSV. In contrast, the large-plaque revertants of NS2stop (R1 to R4) all displayed growth characteristics nearly equal to those of the wild type (Fig. 4). Both the growth kinetics and the final viral titers of these revertants were similar to those of wild-type RSV.
FIG. 3.
Growth curves of NS2 knockout viruses. NS2stop (a), ΔNS2 (b), or their respective wild-type control viruses were used to infect triplicate monolayers of HEp-2 cells at a MOI of 1. Supernatants were harvested at the indicated time points and assayed for viral titers. The two NS2stop viruses shown in panel a were derived from independent antigenome cDNAs.
FIG. 4.
Growth curves of NS2stop revertant viruses. The growth of four clonal pools of NS2stop revertant viruses (R1 to R4) was compared to that of wild-type rA2.
Western blot analysis of viral proteins.
To confirm that the mutations incorporated into the antigenome cDNA indeed ablated expression of the NS2 protein, we next examined the expression of viral proteins in cells infected with the recombinant viruses. Cells were infected with NS2stop, NS2stop revertant R1 or R2, ΔNS2, or wild-type rRSV at a MOI of 1. At 48 h postinfection, the cells were harvested and divided into two aliquots. One aliquot was processed for total-protein analysis, while the other was processed for RNA analysis as described below. Proteins were separated by SDS-polyacrylamide gel electrophoresis and subjected to Western blot analysis with rabbit antiserum directed against either purified virions or a carboxy-terminal peptide of NS2. As previously described, the anti-NS2 peptide antiserum cross-reacts weakly with NS1, most probably due to the identity of the four C-terminal residues in these two proteins (1).
Several major RSV structural proteins (G, N, P, M, M2-1, and SH) were detected in all infected-cell extracts by Western blotting with antibodies against RSV virions (Fig. 5a). While the overall level of the viral structural proteins was slightly lower in cells infected with the NS2 knockout viruses (lanes 2 to 4 compared to lanes 5 to 9), the relative molar amounts of the major proteins were consistent among all viruses. Analysis of the nonstructural proteins with the anti-NS2 peptide antiserum showed that expression of NS2 had been ablated by both the termination codons and deletion of the ORF (Fig. 5b, lanes 2 to 4). As expected, the NS2stop revertants had regained expression of NS2 (lanes 5 and 6), indicating that the presence of NS2 was responsible for the change in growth characteristics.
Northern blot analysis of viral RNAs.
We next investigated whether the lack of NS2 expression or the excision of the NS2 gene (and concomitant decrease in mRNA number and genome length) had an effect on the steady-state levels of viral RNAs. Total cellular RNA from infected cells from the experiment in Fig. 5 was isolated and used in a Northern blot analysis with DNA probes representing the NS1, NS2, N, F, and L genes (Fig. 6). The NS1 probe showed that the accumulation of monocistronic NS1 mRNA was essentially the same for all of the viruses tested, namely, the ΔNS2, NS2stop, revertant, and wild-type rRSVs (Fig. 6a). The NS2 probe confirmed the absence of expression of the NS2 mRNA by the ΔNS2 virus, but the remaining viruses expressed similar amounts of monocistronic NS2 mRNA (Fig. 6b). The N probe showed that the accumulation of monocistronic N mRNA was essentially the same for all tested viruses (Fig. 6c). Thus, loss of expression of the NS2 protein or deletion of the NS2 gene did not have a dramatic effect on the relative level of accumulation of this mRNA.
FIG. 6.
RSV RNA expression by NS2stop, ΔNS2, and NS2stop revertant viruses. Total-cell RNA was isolated from HEp-2 cells 48 h postinfection from the same experiment as in Fig. 5 and subjected to Northern blot analysis with probes directed against the NS1, NS2, N, F, and L ORFs. Monocistronic and readthrough mRNA species are indicated on the right. The novel NS1-N transcript present in the ΔNS2 virus is indicated in parentheses. G/AG, genome-antigenome.
The patterns of readthrough mRNAs containing NS1, NS2, or N sequence were similar for all of the viruses except for the ΔNS2 virus, for which the NS1-NS2, NS2-N, and NS1-NS2-N mRNAs were replaced by the novel NS1-N RNA, which is the expected result of the NS2 gene deletion. The novel NS1-N mRNA was readily apparent with the NS1 probe, whereas with the N probe it migrated at the same position as the NS2-N RNA due to the similarity in predicted size (1,761 and 1,732 nt, respectively [nonpolyadenylated]). The pattern of monocistronic and readthrough mRNAs containing F sequence was essentially the same for all tested viruses (Fig. 6d). The relative amounts of L mRNA genome and antigenome showed some variability among viruses in the experiment in Fig. 6e. However, these apparent differences were not consistent among multiple experiments (results not shown) and presumably reflect variability in the efficiency of recovery and transfer of these large RNAs. Thus, overall, the relative levels of L mRNA genome and antigenome did not appear to be affected by the lack of expression of the NS2 protein.
Time course of RSV protein and RNA synthesis.
While the steady-state levels of protein and RNA were not dramatically different between the NS2 knockout and wild-type viruses, it was possible that the rate of RSV transcription and protein synthesis was altered by the absence of NS2. Therefore, we infected cells with recombinant wild-type or ΔNS2 RSV at a MOI of 3 and harvested total protein and total cellular RNA at several time points. Western blot analysis of the protein extracts showed that the accumulation of viral proteins was detectable under these conditions beginning between 9 and 12 h postinfection for both viruses (Fig. 7). As expected, the NS2 protein was not detected in extracts from cells infected with ΔNS2 rRSV (Fig. 7, bottom right). The accumulation of viral proteins in ΔNS2 rRSV-infected cells appeared to proceed with the same kinetics but at a somewhat lower magnitude than in wild-type-rRSV-infected cells. By 48 h postinfection, the level of viral proteins was modestly higher in cells infected with rA2 (Fig. 7).
We also examined whether the absence of NS2 had an effect on the accumulation of RSV mRNAs over time. Total cellular RNA derived from the samples in Fig. 7 was subjected to Northern analysis with a probe for the N ORF (Fig. 8). The N-containing monocistronic and readthrough mRNAs appeared slightly sooner in cells infected with the ΔNS2 virus (Fig. 8, right) than in those infected with the wild type (left), but the level of accumulation was lower.
FIG. 8.
Time course of N mRNA expression by recombinant wild-type and ΔNS2 viruses. Total-cell RNA was isolated at the indicated times postinfection (p.i.) from the same experiment as in Fig. 7. Northern blot analysis was performed as in Fig. 6 with a probe directed against the N ORF. Monocistronic N mRNA and readthrough mRNA species are indicated on the right. The NS2-N (rA2 [left]) and NS1-N (ΔNS2 [right]) mRNAs comigrate.
Sequence analysis of NS2 mRNA from revertant viruses.
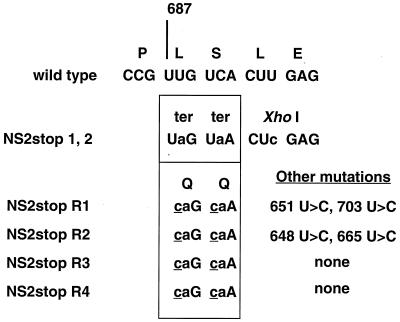
Since the NS2stop revertants had regained expression of the NS2 protein, we wanted to determine which sequence changes had occurred to allow translation of the NS2 ORF. Total RNA from cells infected with NS2stop or NS2stop revertants (R1 to R4) was subjected to RT-PCR with primers which specifically amplified positive-sense NS2 RNA (mRNA or antigenome). The PCR fragments, corresponding to positions 626 to 948 of the antigenome, were cloned into a TA vector, and six clones from each virus were sequenced in both directions. All clones derived from a single virus displayed identical sequences. The two independent NS2stop viruses which were examined contained the engineered changes as well as the XhoI site (Fig. 9), but contained no other mutations within the 324-base region, encompassing codons 1 to 108 of the NS2 ORF. In contrast, all four revertants showed second-site mutations in the two engineered translation termination codons, changing the U in the first position of each to C (Fig. 9). These changes led to the insertion of glutamine residues in place of the nonsense assignments. In addition, two of the revertants had two other U-to-C transitions each. The two additional mutations in NS2stop R1 (at nt 651 and 703, corresponding to aa 8 and 25, respectively) both coded for isoleucine-to-threonine changes. The change at nt 665 (aa 13) in NS2stop R2 encoded a leucine-to-proline change, while the mutation at nt 648 (aa 25) was silent. The other two revertants did not display any other mutations within the region sequenced besides the second-site reversions.
FIG. 9.
Sequence changes in NS2 mRNA from NS2stop revertant viruses. RT-PCR was performed on NS2 mRNA derived from cells infected with either of two independent NS2stop viruses or four independent revertant viruses. PCR fragments were cloned into a TA cloning vector. Six clones from each virus were sequenced; no differences were found within each virus preparation or between the two NS2stop viruses. Differences from wild type are shown in lowercase type; second-site substitutions are underlined. Nucleotide numbering is from the 3′ end of the genome.
DISCUSSION
We have shown in this study that rRSV mutants which do not express the nonstructural NS2 protein are viable in cell culture. However, these viruses are moderately debilitated compared to the wild type. The NS2 knockout viruses form pinpoint plaques and grow more slowly and to a lower final titer than does wild-type RSV. These data suggest that the NS2 protein performs an accessory function which, although not essential, improves virus growth.
The pinpoint plaque phenotype of the NS2 knockout RSVs, evidence of attenuation in vitro, suggests that these viruses might also be attenuated in the host. We are currently evaluating the ability of these viruses to infect and induce effective anti-RSV immunity in rodents and primates. In this instance, it is necessary to use the NS2 deletion mutant to preclude the emergence of revertant viruses. If NS2 knockout viruses are indeed attenuated in vivo, they may be potential candidates for a vaccine, since expression of the major protective antigens is not dramatically impaired. In this regard, we note that despite the phenotype of impaired growth, we have been able to produce large stocks of ΔNS2 virus with an infectivity titer of 2 × 107 PFU/ml. Since the NS2 protein does not appear to be a significant neutralization or protective antigen, its absence in a vaccine virus would not compromise its immunogenicity. The G and F glycoproteins are the major antigens involved in neutralization and resistance to reinfection. The NS2 protein was an antigen for cytotoxic T cells in a subgroup of human adult volunteers (6). However, studies with rodents showed that immunization with antigens which induce RSV-specific cytotoxic T cells provide only a short-lived resistance to RSV challenge, and a vaccinia virus recombinant expressing NS2 did not induce neutralizing antibody or significant protection from reinfection (12).
NS2stop viruses readily regained NS2 expression by reversion, consistent with the idea that NS2 fulfills an important function in RSV biology. The revertants display plaque morphology and growth rates similar to those of wild-type RSV. Interestingly, the mutations in the revertant viruses were second-site reversions within the termination codons, resulting in glutamine at both positions 21 and 22 compared with leucine-serine in the wild-type virus. Two observations suggest that the reversion involved biased hypermutation, which has been described for several RNA viruses including measles virus, hepatitis delta virus, and RSV (5). First, this mechanism involves A-to-G changes in the genome, which is negative sense. This would explain why the reversion to sense involved second-site mutation at the first nucleotide (A) in both codons rather than same-site mutation at the second nucleotide (U), which was originally altered from the wild type. Since the revertant viruses grew slightly less well than the wild-type virus, it might be that the reversions were suboptimal but occurred because they were the only changes possible by this mechanism. Second, biased hypermutation can introduce multiple changes simultaneously, allowing one-step reversion to sense at both codons. In addition, the nearby additional mutations in two of the four revertants analyzed were all A to G changes in negative sense. As expected, we have not detected revertants from RSV which had the NS2 gene deleted.
From the results with the NS2stop viruses, we can deduce that the first 20 aa of the protein are not sufficient to carry out its function. On the other hand, the protein can sustain substitutions at position 8 (Ile to Thr), 13 (Leu to Pro), 21 (Leu to Gln), 22 (Leu to Gln), or 25 (Ile to Thr). NS2 might positively regulate transcription and/or translation of RSV genes, which would lead to the slower accumulation of viral RNA and proteins seen with the NS2 knockout viruses. Slightly lower expression of RSV proteins in NS2 knockout viruses, particularly of the viral nucleocapsid, might then lead to a more pronounced decrease in virion production and plaque size. Alternatively, NS2 may function directly in virion morphogenesis to enhance the transport of viral nucleocapsids to the plasma membrane. NS2 associates with the cytoskeletal fraction of infected cells and colocalizes with N and P (27). However, the formation of infectious helper-dependent particles was not affected by the presence or absence of NS2 protein in a packaging and passage assay (26), suggesting that NS2 plays little or no role in this process. Further studies to characterize more extensively the function of NS2 both in cell culture and in animals are in progress.
While the studies presented here suggest an important though nonessential role for NS2 in RSV replication, its exact function remains unclear. Nonstructural proteins, arising from alternative translation initiation or editing of the P mRNA, are also present in other paramyxoviruses (13). The Sendai virus C protein, which is encoded by the P mRNA, specifically inhibits RNA synthesis from the genomic promoter (4, 25). In addition, abrogation of C protein expression from the plasmid P gene is necessary for recovery of recombinant Sendai virus (17). Studies of recombinant Sendai viruses which are unable to produce the transframe V protein have shown that these viruses are viable and grow similarly to wild-type Sendai in eggs and cell culture (14, 19). However, V appears to be an important determinant in Sendai virus pathogenesis (15, 19, 20). For measles virus, ablation of expression of either V or C in recombinant viruses also does not noticeably affect viral growth in culture (23, 24). Interestingly, recombinant measles viruses overexpressing the V protein form smaller syncytia, which lyse 1 to 2 days later than those formed by the wild type (24). Thus, NS2 is one of several paramyxovirus proteins which are nonessential and whose interaction with viral or host factors remains to be characterized.
ACKNOWLEDGMENTS
We thank Ena Camargo for growing virus stocks, Stephen Whitehead for D53/HEKsites, and Rachel Fearns, Brian Murphy, and Robert Chanock for critical readings of the manuscript.
REFERENCES
- 1.Atreya P L, Peeples M E, Collins P L. The NS1 protein of human respiratory syncytial virus is a potent inhibitor of minigenome transcription and RNA replication. J Virol. 1998;72:1452–1461. doi: 10.1128/jvi.72.2.1452-1461.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bukreyev A, Whitehead S S, Murphy B R, Collins P L. Recombinant respiratory syncytial virus from which the entire SH gene has been deleted grows efficiently in cell culture and exhibits site-specific attenuation in the respiratory tract of the mouse. J Virol. 1997;71:8973–8982. doi: 10.1128/jvi.71.12.8973-8982.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Byrappa S, Gavin D K, Gupta K C. A highly efficient procedure for site-specific mutagenesis of full-length plasmids using Vent DNA polymerase. Genome Res. 1995;5:404–407. doi: 10.1101/gr.5.4.404. [DOI] [PubMed] [Google Scholar]
- 4.Cadd T, Garcin D, Tapparel C, Itoh M, Homma M, Roux L, Curran J, Kolakofsky D. The Sendai paramyxovirus accessory C proteins inhibit viral genome amplification in a promoter-specific fashion. J Virol. 1996;70:5067–5074. doi: 10.1128/jvi.70.8.5067-5074.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cattaneo R. Biased (A→I) hypermutation of animal RNA virus genomes. Curr Opin Genet Dev. 1994;4:895–900. doi: 10.1016/0959-437x(94)90076-0. [DOI] [PubMed] [Google Scholar]
- 6.Cherrie A H, Anderson K, Wertz G W, Openshaw P J. Human cytotoxic T cells stimulated by antigen on dendritic cells recognize the N, SH, F, M, 22K, and 1b proteins of respiratory syncytial virus. J Virol. 1992;66:2102–2110. doi: 10.1128/jvi.66.4.2102-2110.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Collins P L, Hill M G, Camargo E, Grosfeld H, Chanock R M, Murphy B R. Production of infectious human respiratory syncytial virus from cloned cDNA confirms an essential role for the transcription elongation factor from the 5′ proximal open reading frame of the M2 mRNA in gene expression and provides a capability for vaccine development. Proc Natl Acad Sci USA. 1995;92:11563–11567. doi: 10.1073/pnas.92.25.11563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Collins P L, Hill M G, Cristina J, Grosfeld H. Transcription elongation factor of respiratory syncytial virus, a nonsegmented negative-strand RNA virus. Proc Natl Acad Sci USA. 1996;93:81–85. doi: 10.1073/pnas.93.1.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Collins P L, McIntosh K, Chanock R M. Respiratory syncytial virus. In: Fields B N, Knipe D M, Howley P M, Chanock R M, Melnick J L, Monath T P, Roizman B, Straus S E, editors. Fields virology. 3rd ed. Vol. 2. Philadelphia, Pa: Lippincott-Raven; 1996. pp. 1313–1352. [Google Scholar]
- 10.Collins P L, Wertz G W. cDNA cloning and transcriptional mapping of nine polyadenylylated RNAs encoded by the genome of human respiratory syncytial virus. Proc Natl Acad Sci USA. 1983;80:3208–3212. doi: 10.1073/pnas.80.11.3208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Collins P L, Wertz G W. Nucleotide sequences of the 1B and 1C nonstructural protein mRNAs of human respiratory syncytial virus. Virology. 1985;143:442–451. doi: 10.1016/0042-6822(85)90384-8. [DOI] [PubMed] [Google Scholar]
- 12.Connors M, Collins P L, Firestone C Y, Murphy B R. Respiratory syncytial virus (RSV) F, G, M2 (22K), and N proteins each induce resistance to RSV challenge, but resistance induced by M2 and N proteins is relatively short-lived. J Virol. 1991;65:1634–1637. doi: 10.1128/jvi.65.3.1634-1637.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Curran J, Kolakofsky D. Sendai virus P gene produces multiple proteins from overlapping open reading frames. Enzyme. 1990;44:244–249. doi: 10.1159/000468762. [DOI] [PubMed] [Google Scholar]
- 14.Delenda C, Hausmann S, Garcin D, Kolakofsky D. Normal cellular replication of Sendai virus without the trans-frame, nonstructural V protein. Virology. 1997;228:55–62. doi: 10.1006/viro.1996.8354. [DOI] [PubMed] [Google Scholar]
- 15.Delenda C, Taylor G, Hausmann S, Garcin D, Kolakofsky D. Sendai viruses with altered P, V, and W protein expression. Virology. 1998;242:327–337. doi: 10.1006/viro.1998.9027. [DOI] [PubMed] [Google Scholar]
- 16.Evans J E, Cane P A, Pringle C R. Expression and characterisation of the NS1 and NS2 proteins of respiratory syncytial virus. Virus Res. 1996;43:155–161. doi: 10.1016/0168-1702(96)01327-5. [DOI] [PubMed] [Google Scholar]
- 17.Garcin D, Pelet T, Calain P, Roux L, Curran J, Kolakofsky D. A highly recombinogenic system for the recovery of infectious Sendai paramyxovirus from cDNA: generation of a novel copy-back nondefective interfering virus. EMBO J. 1995;14:6087–6094. doi: 10.1002/j.1460-2075.1995.tb00299.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hardy R W, Wertz G W. The product of the respiratory syncytial virus M2 gene ORF1 enhances readthrough of intergenic junctions during viral transcription. J Virol. 1998;72:520–526. doi: 10.1128/jvi.72.1.520-526.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kato A, Kiyotani K, Sakai Y, Yoshida T, Nagai Y. The paramyxovirus, Sendai virus, V protein encodes a luxury function required for viral pathogenesis. EMBO J. 1997;16:578–587. doi: 10.1093/emboj/16.3.578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kato A, Kiyotani K, Sakai Y, Yoshida T, Shioda T, Nagai Y. Importance of the cysteine-rich carboxyl-terminal half of V protein for Sendai virus pathogenesis. J Virol. 1997;71:7266–7272. doi: 10.1128/jvi.71.10.7266-7272.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kunkel T A, Roberts J D, Zakour R A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Methods Enzymol. 1987;154:367–382. doi: 10.1016/0076-6879(87)54085-x. [DOI] [PubMed] [Google Scholar]
- 22.Murphy B R, Sotnikov A V, Lawrence L A, Banks S M, Prince G A. Enhanced pulmonary histopathology is observed in cotton rats immunized with formalin-inactivated respiratory syncytial virus (RSV) or purified F glycoprotein and challenged with RSV 3–6 months after immunization. Vaccine. 1990;8:497–502. doi: 10.1016/0264-410x(90)90253-i. [DOI] [PubMed] [Google Scholar]
- 23.Radecke F, Billeter M A. The nonstructural C protein is not essential for multiplication of Edmonston B strain measles virus in cultured cells. Virology. 1996;217:418–421. doi: 10.1006/viro.1996.0134. [DOI] [PubMed] [Google Scholar]
- 24.Schneider H, Kaelin K, Billeter M A. Recombinant measles viruses defective for RNA editing and V protein synthesis are viable in cultured cells. Virology. 1997;227:314–322. doi: 10.1006/viro.1996.8339. [DOI] [PubMed] [Google Scholar]
- 25.Tapparel C, Hausmann S, Pelet T, Curran J, Kolakofsky D, Roux L. Inhibition of Sendai virus genome replication due to promoter-increased selectivity: a possible role for the accessory C proteins. J Virol. 1997;71:9588–9599. doi: 10.1128/jvi.71.12.9588-9599.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Teng M N, Collins P L. Identification of the respiratory syncytial virus proteins required for formation and passage of helper-dependent infectious particles. J Virol. 1998;72:5707–5716. doi: 10.1128/jvi.72.7.5707-5716.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Weber E, Humbert B, Streckert H J, Werchau H. Nonstructural protein 2 (NS2) of respiratory syncytial virus (RSV) detected by an antipeptide serum. Respiration. 1995;62:27–33. doi: 10.1159/000196385. [DOI] [PubMed] [Google Scholar]
- 28.Whitehead S S, Juhasz K, Firestone C Y, Collins P L, Murphy B R. Recombinant respiratory syncytial virus (RSV) bearing a set of mutations from cold-passaged RSV is attenuated in chimpanzees. J Virol. 1998;72:4467–4471. doi: 10.1128/jvi.72.5.4467-4471.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wyatt L S, Moss B, Rozenblatt S. Replication-deficient vaccinia virus encoding bacteriophage T7 RNA polymerase for transient gene expression in mammalian cells. Virology. 1995;210:202–205. doi: 10.1006/viro.1995.1332. [DOI] [PubMed] [Google Scholar]