Abstract
Interstitial pneumonia is a frequent and critical manifestation of human cytomegalovirus (CMV) disease in immunocompromised patients, in particular in recipients of bone marrow transplantation. Previous work in the murine CMV infection model has identified the lungs as a major organ site of CMV latency and recurrence. It was open to question whether the viral genome is transcriptionally silent or active during latency. Transcription could be latency associated and thus be part of the latency phenotype. Alternatively, transcriptional activity could reflect episodes of reactivation. We demonstrate here that transcription of the immediate-early (IE) transcription unit ie1-ie3 selectively generates ie1-specific transcripts during latency. Notably, while the latent viral DNA was found to be evenly distributed in the lungs, transcription was focal and randomly distributed. This finding indicates that IE transcription is not a feature inherent to murine CMV latency but rather reflects foci of primordial reactivation. However, this reactivation did not initiate productive infection, since ie3 gene mRNA specifying the essential transactivator IE3 of murine CMV early gene expression was not detectable. Accordingly, transcripts encoding gB were absent during latency. By contrast, during induced virus recurrence, IE-phase transcription switched from focal to generalized and ie3-specific transcripts were generated. These data imply that latency and recurrence are regulated not only at the IE promoter-enhancer and that there exists an additional checkpoint at the level of precursor RNA splicing. We propose that focal transcription reflects random episodes of nonproductive reactivation that get terminated before IE3 is expressed and ignites the productive cycle.
Latent infection with human cytomegalovirus (CMV) entails a risk of virus recurrence and consequent multiple-organ CMV disease in immunocompromised patients, in particular in patients who have undergone hematoablative conditioning for bone marrow (BM) transplantation (BMT) (8, 51). There is a dual risk. First, latent CMV present in the donor can be transmitted with the BM cells (BMC) transplanted or with blood products and can reactivate, or become reactivated, leading to productive infection in the recipient. This implies that BMC and/or blood cells are a source of latent CMV, and there is indeed a lot of evidence for human CMV latency in hematopoietic myeloid lineage progenitors (11, 23, 27, 29, 32) and mature progeny thereof, monocytes and macrophages in particular (45, 49, 50). In major histocompatibility complex (MHC)-mismatched BMT, a graft-versus-host response may facilitate human CMV recurrence from a particular type of macrophage that expresses dendritic cell markers (45). Second, latent CMV present in the recipient can give rise to recurrent infection within organs of the recipient. That this occurs became evident from the high incidence of human CMV infection in CMV-seropositive latent CMV carriers receiving a transplant from a CMV-negative donor (reviewed in reference 4). While current research on human CMV latency is focused on latency in hematopoietic cell types, there are good arguments for the existence of a second type of latency of CMV residing in stromal and/or parenchymal cells of various tissues. First, recurrence occurs in the carriers of latent CMV after the depletion of hematopoietic cell types. Second, latent CMV is transmitted most efficiently by solid organ transplantation (6, 10), actually more efficiently than by BMT (4).
The existence of at least two independent cellular sites of CMV latency is also suggested by findings in the model of murine CMV latency. Recent work by Pollock et al. has documented murine CMV latency in peritoneal exudate macrophages (35), thereby corroborating more indirect earlier conclusions (5). Whereas spleen tissue explant cultures are a long-established method of eliciting recurrence of murine CMV (17), the latently infected cell type in the spleen has remained enigmatic. Since the spleen is rich in macrophages, they are candidates for harboring latent murine CMV. However, the latent genome as well as the potential to reactivate in explant cultures was found to copurify with class II MHC-negative stromal cells (30, 37), most likely sinusoidal endothelial cells (30). Likewise, renal peritubular endothelial cells were discussed as a cellular site of latency in the kidney (21). More recently, Koffron et al. have presented histological data supporting the view that murine CMV may become latent in macrophages and endothelial cells (22).
The problem of the cellular sites of human CMV and murine CMV latency also affects the studies on potential transcriptional activity during latency, since the molecular state and activity of the latent viral genome are likely to be influenced by the cell type. Some of the discrepant data in the literature may thus not represent controversial findings but might rather relate to a heterogeneity in the latently infected cell types. Evidence for a latency-specific transcription of human CMV was provided by Kondo et al. for the example of latency in granulocyte-macrophage progenitors. Latency-associated transcripts from the regulatory immediate-early (IE) region differed from the IE transcripts of the productive phase. A class of sense transcripts was found to use alternative start sites in the human CMV IE1/IE2 promoter-enhancer, while a second class was transcribed in the antisense direction (23, 24).
Analogous transcripts are not known for murine CMV. The occasional detection of murine CMV ie1 gene transcript sequences in the reverse transcriptase (RT) PCR analysis of organ RNA derived from operationally-defined latent mice (12, 52) was not interpreted as due to latency-associated transcription but as an indication of a persisting productive infection at a level below the detection limit of conventional infectivity assays (52).
We have shown previously that latent murine CMV DNA is harbored in a series of organs, including the salivary glands, adrenal glands, spleen, heart, kidney, and lungs (40), with a particularly high load of latent viral genome in the lungs (3, 40, 47). Notably, this “tissue latency” of murine CMV was found several months after clearance of viral DNA from BM and blood (2, 3, 25). This fact made it evident that blood leukocytes were not the source of latent CMV during that advanced stage of latency and supported the hypothesis that a stromal and/or parenchymal site of latency must exist.
Here we have employed this model to study the transcriptional activity of murine CMV in the stromal-parenchymal type of latency in the lungs.
MATERIALS AND METHODS
Experimental BMT and infection.
For sex-mismatched male-into-female BMT, male and female mice of the inbred strain BALB/c (MHC haplotype H-2d) were used at the age of 8 weeks as BMC donors and recipients, respectively. Hematoablative conditioning of the recipients was performed by total-body γ-irradiation with a single, sublethal dose of 6 Gy from a γ-137Cs source (OB58; Buchler, Braunschweig, Germany). Cell suspensions of donor femoral and tibial BMC were obtained by flushing medium through the bone shafts, and contaminating vascular and sinusoidal CD8 T cells were depleted as described previously (47). BMT was performed by infusion of 5 × 106 donor BMC into the tail vein of each recipient at ca. 6 h after the irradiation. Infection with 105 PFU of purified murine CMV, strain Smith ATCC VR-194/1981, was performed by subcutaneous injection at the left hind footpad at ca. 2 h after BMT (25).
High-sensitivity assay of viral infectivity.
Viral latency in the lungs was verified, and virus recurrence was monitored, by the recently described RT PCR-based focus expansion assay (25). In essence, monolayers of permissive fetal mouse cells, previously often referred to as mouse “embryo fibroblasts,” were inoculated with 2 ml of freeze-thawed and homogenized tissue, representing a 1/18 aliquot of the whole lungs, under the influence of a 1,000 × g centrifugal force. Specifically, for each individual mouse tested, 2-ml homogenates were prepared from nine pieces derived from the left lung and the postcaval lobe. After a culture period of 72 h, which is equivalent to ca. three rounds of murine CMV replication in the indicator cells, poly(A)+ RNA was isolated and analyzed for the presence of viral IE1 transcripts by ie1 exon 3-exon 4 (exon 3/4)-specific RT PCR. As we have shown in great detail in a previous report (25), this assay revealed a genome-to-infectivity ratio of ca. 5:1 for purified murine CMV and consistently detected infectivity in lung homogenates supplemented with 0.05 PFU of murine CMV, which equals five infectious genomes. It is important to emphasize that all of the homogenate was plated in order to avoid a sampling error. Latency studies were performed at 1 year after primary infection, which was at least 6 months after the resolution of acute infection in organs, including the long-persisting productive infection in the salivary glands (2, 25, 40). Recurrence was monitored by the detection of infectious virus and of productive cycle IE3 transcripts on day 14 after immunoablative treatment by 6.5 Gy total-body γ-irradiation.
Isolation of DNA.
Total DNA, cellular and viral, was isolated from whole blood and from BMC.
(i) Isolation of DNA from blood.
Blood was taken by heart puncture (3) and was transferred immediately for DNA extraction into the extraction buffer of the NucleoSpin blood kit (Macherey & Nagel, Düren, Germany). The DNA isolation was performed according to the instructions in the supplier’s method manual. In brief, for each preparation, a 200-μl sample of whole blood was incubated for 10 min at 70°C with 200 μl of extraction buffer and 0.4 mg of freshly added proteinase K. DNA was precipitated with ethanol and separated from the soluble phase by centrifugation in NucleoSpin columns, followed by extensive washing with ethanol-containing buffers. The purified DNA was eluted from the NucleoSpin column matrix by incubation with 100 μl of elution buffer at 70°C for 10 min, followed by centrifugation.
(ii) Isolation of DNA from BMC.
Femoral and tibial BMC were resuspended in phosphate-buffered saline (5 × 106 cells in a total volume of 200 μl) and digested for 2 h at 56°C with proteinase K. After addition of 200 μl of extraction buffer, incubation was continued for 10 min at 70°C. DNA was extracted by using the NucleoSpin method described above.
Simultaneous isolation of DNA and poly(A)+ RNA from lung tissue.
Pieces of lung tissue, specifically from the superior, middle, and inferior lobes of the lungs, were dissected and were shock-frozen in liquid nitrogen. DNA as well as poly(A)+ RNA of individual pieces, each of which represented ca. 1/18 of the whole lungs, was isolated in parallel as described previously (25). Particular care was taken to reduce the time spent between sacrifice of latently infected mice and shock-freezing of tissue pieces in order to minimize the risk of postmortem transcriptional reactivation. Specifically, animals were sacrificed one-by-one, lungs were excised immediately and cut into pieces, and pieces were transferred without delay into vials containing liquid nitrogen. With some skill, this can be managed within 5 min, or within 10 min at the most. Nonetheless, it is understood that the argument that postmortem reactivation occurs can never be refuted with certainty. It is also understood that pieces cut so as to be equal in size are never absolutely equal. Therefore, after transfer of the frozen pieces into the extraction buffer, vials were weighed, and weight differences were later accounted for by appropriate adjustments of the DNA or RNA aliquots included in the assays. For the sake of clarity, we refer to a 1/18 fraction of the lungs as a “piece”; as the mouse lungs consist of ca. 60 × 106 tissue cells (25), one piece contains ca. 3.3 × 106 lung cells. The average yields of cellular DNA and poly(A)+ RNA per piece were found to be ca. 20 μg and ca. 2 μg, respectively.
Quantitation of viral DNA.
The amount of purified organ DNA was determined from the optical density measured at 260 nm. The cellular DNA in hematopoietic cells and their mature progeny, that is, in BMC and blood cells, was quantitated in the male-into-female BMT chimeras by a PCR specific for the male sex (testes)-determining gene tdy as described previously (48). The number of viral genomes present in total DNA was quantitated by diluting the DNA serially, followed by performing a PCR specific for a 363-bp sequence within exon 4 of the IE gene ie1 of murine CMV, essentially as described in previous reports (3, 47) but with further improvements. Specifically, PCR reactions were here performed in 96-well microplates in the GeneAmp PCR System 9700 thermocycler (Perkin-Elmer, Foster City, Calif.). Amplification was achieved in 30 cycles with denaturation at 95°C for 3 min in the first cycle and at 96°C for 30 s in the remaining 29 cycles, primer annealing at 58°C for 1 min, and primer extension at 72°C for 1 min with extension to 6 min in the last cycle. Cellular DNA derived from the respective specimens of uninfected mice was supplemented with defined copy numbers of plasmid pIE111 (31), which encompasses genes ie1 and ie3, and was subjected to the ie1 gene-specific PCR to serve as a standard in the quantification. Amplification products (20 μl thereof) were vacuum dot blotted onto nylon membrane, hybridized with a γ-32P-end-labeled internal oligonucleotide probe, and analyzed quantitatively by digital phosphorimaging as described previously (47).
Construction of recombinant plasmids for in vitro transcription.
The map locations of the synthetic RNAs are illustrated in Fig. 1. Recombinant plasmids containing coding sequences of the murine CMV genes ie1, ie3, and gB were constructed as described below. For construction of pSP64 Poly(A)-ie1, a 2.1-kb EcoRI fragment containing the complete ie1 open reading frame (19) was excised from plasmid p62/3 (7), and was cloned blunt end into the SmaI site of vector pSP64 Poly(A) (GenBank/EMBL accession no. X65328; cat. no. P1241, Promega, Madison, Wis.). The resulting plasmid, pSP64 Poly(A)-ie1, thus includes the complete ie1 open reading frame (exons 2, 3, and 4 of the ie1-ie3 transcription unit) starting at a synthetic EcoRI site located 21 bp upstream of the initiation codon and ending at the AvaI site at the beginning of exon 5 of ie3 (31). Synthetic RNA transcribed from this plasmid has a length of 2.17 kb, including a 30-nucleotide poly(A) tail. For construction of pBlue-ie3, the vector pBluescript (pBluescript II KS(+) phagemid vector, cat. no. 212207; Stratagene, Heidelberg, Germany) was digested with EcoRV and XbaI, and a part of the ie3 sequence was inserted as a 985-bp EcoRV-XbaI fragment derived from plasmid pie3 (31). EcoRV cleaves in intron 1, 58 nucleotides upstream of the ATG located in exon 2 of the ie1-ie3 transcription unit (19), and XbaI cleaves at position 534 of exon 5 (GenBank accession no. M77846). The resulting plasmid, pBlue-ie3, thus includes exon 2, intron 2, exon 3, and part of exon 5. Synthetic RNA transcribed from this plasmid has a length of 1.045 kb and lacks a poly(A) tail. For construction of pSP64 Poly(A)-gB, part of the open reading frame of the gB gene was amplified by PCR from plasmid pACYC-gB (38) by using oligonucleotides 5′-(CAGAATTC)AAGGACTACATCAACGATGC and 5′-(CAGAGCTC)GCGGAACGACTTGTAAAAAG as forward and reverse primers, respectively (38) (GenBank accession no. M86302). The amplification product was digested with SacI and inserted into the SacI site of vector pSP64 Poly(A). The resulting plasmid, pSP64 Poly(A)-gB, thus includes the gB sequence from position 1895 to position 3154 (38). Synthetic RNA transcribed from this plasmid has a length of 1.34 kb and includes a 30-nucleotide poly(A) tail.
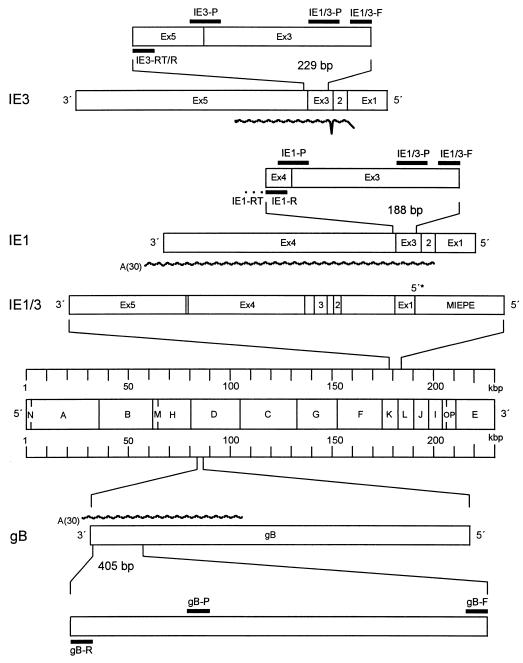
FIG. 1.
Map of the transcriptional analysis. The HindIII physical map of the murine CMV Smith strain genome is shown in the center. The gB gene is expanded below. The ie1-ie3 transcription unit (IE1/3) and the IE1 and IE3 splicing products derived thereof are resolved to greater detail at the top. IE1, IE3, and gB are drawn to scale. Boxes represent the genomic organization with exons (Ex) indicated. Ex1 is noncoding. The wavy lines illustrate the positions and lengths of in vitro-synthesized RNAs. The asterisk marks the 5′ transcriptional start site of the ie1-ie3 transcription unit. Locations of primers and probes are illustrated by black bars at the amplified sequences. The suffix -RT denotes a specific primer for reverse transcription. An oligo(dT) primer was used in the case of gB. The suffix -R denotes a reverse primer, -F denotes a forward primer, and -P denotes a hybridization probe. MIEPE, major IE promoter-enhancer.
In vitro synthesis of viral transcripts.
Synthetic transcripts of the murine CMV genes ie1, ie3, and gB were prepared according to the instructions in the Promega technical manual. Plasmids pSP64 Poly(A)-ie1 and pSP64 Poly(A)-gB were linearized with EcoRI, and transcription was performed with RNA polymerase SP6 (Promega). Plasmid pBlue-ie3 was cut with XbaI, and transcription was performed with RNA polymerase T3 (Promega). Specifically, after complete linearization, plasmid DNA was purified by phenol-chloroform extraction followed by ethanol precipitation. The in vitro transcription reactions were carried out at 37°C for 2 h in a total volume of 0.1 ml, containing 2 μg of plasmid DNA, 40 U of the corresponding RNA polymerase, 100 U of RNasin, 0.5 mM of each ribonucleoside triphosphate, 10 mM dithiothreitol, 40 mM Tris-HCl (pH 7.5), 6 mM MgCl2, 2 mM spermidine, and 10 mM NaCl. Transcripts were separated by vertical low-melting agarose (1.2%, wt/vol) gel electrophoresis. Bands were cut out, the gel was melted at 65°C, and RNA was isolated by phenol-chloroform extraction and precipitation with ethanol. The RNA was dissolved in water, and contaminating plasmid DNA was degraded by using RQ1 RNase-free DNase (M6101; Promega). The reaction was performed at 37°C for 2 h in a volume of 0.1 ml containing 4 U of the RQ1 DNase, 100 U of RNasin, 10 mM dithiothreitol, 40 mM Tris-HCl (pH 7.5), 6 mM MgCl2, 2 mM spermidine, and 10 mM NaCl. To remove trace amounts of residual DNA, extraction was performed twice with acidic phenol. In detail, 400 μl of distilled water, 50 μl of 2.5 M potassium acetate, and 500 μl of water-saturated phenol were added to the reaction volume. After the mixture was swirled with a vortex apparatus, 100 μl of chloroform was added and the mixture was incubated for 5 min on ice. After centrifugation, this procedure was repeated, and RNA was extracted with chloroform followed by precipitation with ethanol. Finally, the purified RNA was dissolved in water, and its concentration was determined.
Analysis and quantitation of transcripts.
Poly(A)+ RNA was isolated from the shock-frozen lung tissue pieces (see above) by an oligo(dT)-cellulose affinity method as described previously (1, 25). Transcripts of the viral genes ie1, ie3, and gB were detected by specific RT PCRs (for a map, see Fig. 1). The reactions were carried out as described previously except that they were performed in 96-well plates in a GeneAmp 5700 thermocycler (Perkin-Elmer) and that the cDNAs were amplified in 33 cycles. In the previously described ie1-specific RT PCR (25), the probe directed against the exon 3/4 splicing junction was elongated by six nucleotides downstream in order to improve the specificity (the new probe is referred to as IE1-P). The length of the IE1-specific amplificate is 188 bp. Primers and probe for an RT PCR specific for the cellular transcript of hypoxanthine phosphoribosyltransferase (HPRT), yielding a fragment of 163 bp, have also been specified in the previous report (25). For the detection of the ie3 gene transcript, the oligonucleotide (5′) nucleotide (nt) 3445 to 3426 (located in exon 5, with the counting beginning at the 5′ transcription start site of the ie1-ie3 transcription unit) (31) was used as the RT primer as well as the reverse primer in the PCR (IE3-RT/R), the oligonucleotide (5′) nt 1361 to 1380 (exon 3, shared by ie1 and ie3) served as the forward primer (IE1/3-F), and oligonucleotide (5′) nt 1511 to 1524 (exon 3) 3379 to 3396 (exon 5) served as the probe directed against the exon 3/5 splicing junction (probe IE3-P). The fragment length of the correct amplificate is 229 bp, while an amplificate from contaminating viral DNA can readily be identified by its length of 2,085 bp. In case of a comparative quantitation of IE1 and IE3 transcripts, the exon 3 specific oligonucleotide 5′nt 1392 to 1418 was used as a common probe (probe IE1/3-P) in order to minimize the variables. The gB gene transcript was reverse transcribed by using an oligo(dT) primer. The resulting cDNA was amplified by using the oligonucleotide (5′) nt 83010 to 83029 as the forward primer (gB-F), (5′) nt 83415 to 83396 as the reverse primer (gB-R), and (5′) nt 83127 to 83146 as the probe (gB-P) (39) (GenBank accession no. MCU68299 [complete genome]). Since gB lacks introns, amplificates derived from RNA and DNA cannot be distinguished by size. Reactions performed in the absence of the enzyme RT were therefore used to verify the specific detection of RNA. Amplification products were analyzed by gel electrophoresis, Southern blotting, and hybridization with the corresponding γ-32P-end-labeled internal probe, followed by autoradiography. The radioactivity was measured by digital phosphorimaging. Defined numbers of in vitro-synthesized transcripts served as standards for quantifications.
Frequency estimation of transcriptional foci in the lungs.
The frequency of transcriptional foci of murine CMV in latently infected lungs was estimated from the fraction of transcriptionally negative lung pieces, f(0), by using the Poisson distribution equation λ = −ln f(0). The fraction of pieces containing n (n = 0, 1, 2, 3, and so on) foci, F, was calculated by using the formula F(n) = λn × e−λ/n!. The estimate was made for 45 pieces derived from the superior, middle, and inferior lobes of five latently infected lungs and was then extrapolated to 18 pieces of a complete “statistical lung.” For the upper estimate, neighboring positive pieces were all regarded as being individually positive. For the lower estimate, neighboring positive pieces were counted as only one positive piece assuming a single transcriptional focus located at the border between the two pieces.
RESULTS
Rapid exhaustion of the bone marrow pool of murine CMV DNA.
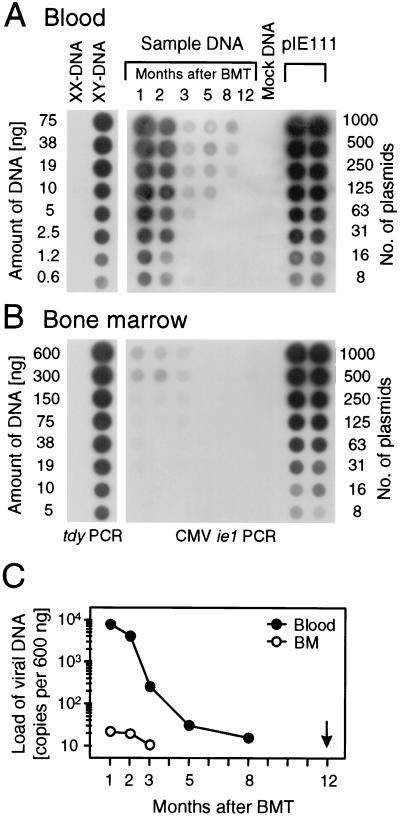
After experimental BMT and concurrent murine CMV infection, prevention of lethal multiple-organ CMV disease and clearance of productive infection depend on the efficacy of lymphohematopoietic CD8 T-cell reconstitution (33, 47). Under experimental conditions that permit survival, infectious virus is cleared from organs but viral genome is retained in a nonreplicative state referred to as molecular latency (25, 36). Based on site-specific differences in the efficacy of antiviral control, molecular latency can be established in a particular organ, while productive infection persists in other organs. Typically, in the model used here and introduced previously (25), molecular latency is established after 1 to 2 months in the spleen, after 3 to 4 months in the lungs, and after 5 to 6 months in the salivary glands. Thus, by 6 months after BMT and primary infection, productive infection is resolved and viral latency is established at all potential organ sites. As we have documented previously (25) and reproduced here, viral DNA is retained in blood cells for an extended period of time after the resolution of productive infection in organs, but the viral DNA load in the blood declines progressively and viral DNA is eventually also cleared (Fig. 2A). For human CMV, latently infected hematopoietic progenitor cells in the BM are currently discussed as a source of latently infected progeny in the blood (11, 45). Notably, for murine CMV, the viral genome load in the BM is much lower than is the load in the blood, and clearance from the BM precedes clearance from the blood by several months (Fig. 2B). It is pertinent to our work that all subsequent experiments and findings pertain to a parenchymal or stromal type of murine CMV latency present in the lungs many months after the clearance of viral DNA from BM and blood.
FIG. 2.
Kinetics of viral DNA clearance in hematopoietic cells and their progeny. Sex-mismatched BMT was performed with BMC derived from male (XY) BALB/c donors transplanted into female (XX) BALB/c recipients. DNA was isolated, at the indicated time points after BMT and infection, from blood and BMC pooled from three recipients per time point. (A) Quantitation of viral DNA in blood. (B) Quantitation of viral DNA in BM. On the left are shown cellular gene controls documenting equivalent efficacy of PCR for DNA preparations derived from blood and BM at 12 months after BMT and infection. The cellular sequence amplified was that from the tdy gene, which is located on the Y chromosome of the repopulating, donor-derived male cells. DNA isolated from uninfected female recipients of a sex-matched syngeneic BMT (XX-DNA/mock DNA) was used for the negative controls. Amplification of a 363-bp sequence within exon 4 of gene ie1 of murine CMV is shown on the right. Mock DNA derived from blood or BM of the uninfected female BMT recipients was supplemented with plasmid pIE111, which includes ie1, and was titrated in duplicate as a standard for the quantitation. Shown are the autoradiographs of the dot blots obtained after hybridization with the respective γ-32P-end-labeled internal oligonucleotide probes. (C) Comparison of the viral DNA loads in blood and BM. The radioactivity per dot was measured by phosphorimaging, and the number of viral ie1 genes per 600 ng of DNA, which is equivalent to the DNA content of 105 diploid cells, was calculated from the linear portions of the titrations. The arrow marks the time point for all subsequent analyses.
Mosaic approach to the analysis of pulmonary latency of murine CMV.
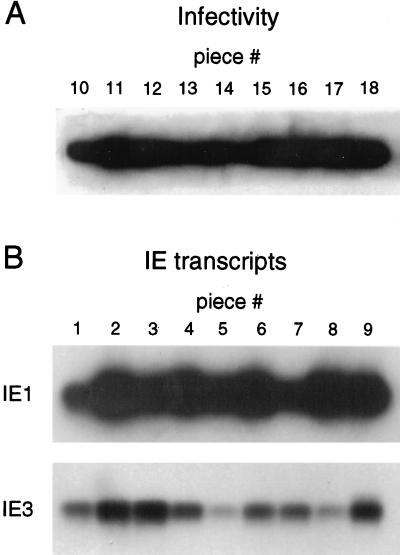
Assays for viral infectivity are technically not compatible with the molecular analysis of DNA load and in vivo viral transcriptional activity. We have therefore used the “mosaic approach” illustrated in Fig. 3. The superior lobe, middle lobe, and inferior lobe of the lungs were cut into nine pieces, designated piece #1 to piece #9, which were used for the simultaneous analysis of latent viral DNA load and viral transcription in each piece. The postcaval lobe and left lung were cut into nine pieces, designated piece #10 to piece #18, which were used to verify latency by the absence of infectious virus. This mosaic approach allows one to discuss results in the context of their topographical relations in the organ. Furthermore, a statistical evaluation is greatly facilitated by the lobular anatomy of the lungs that limits the number of neighboring pieces to only one “neighbor” in the case of 10 pieces and to two “neighbors” in the case of the remaining 8 pieces.
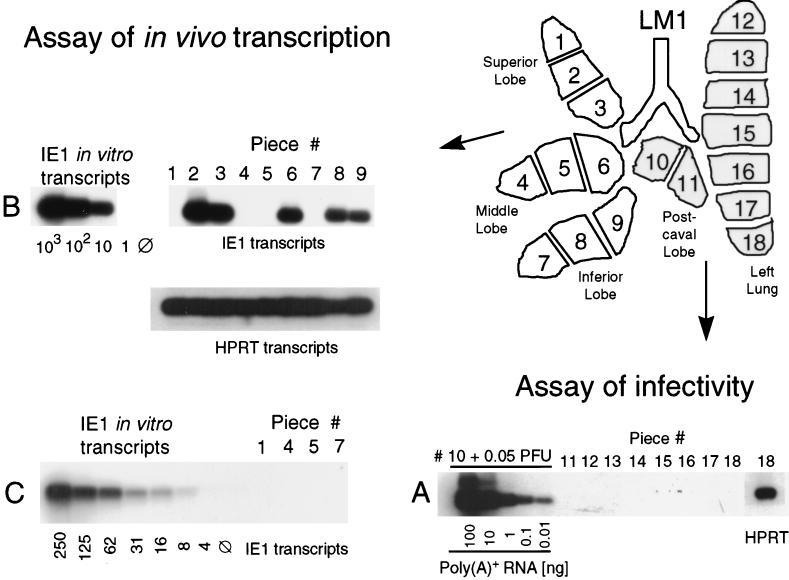
FIG. 3.
General outline of the approach. A scheme illustrating the lobular anatomy of the lungs in ventral view is shown at the upper right. To examine the results in their topographical relation, the lobes were cut into equal pieces designated on the lung map as #1 through #18. Minor size (weight) differences were compensated by adjustment of the aliquots included in the assays. (A) Verification of murine CMV latency. The absence of infectious virus was confirmed at 12 months after BMT and infection for latent mouse LM1 by using a high-sensitivity assay of infectivity, the RT PCR-based focus expansion assay. As a positive control, 0.05 PFU of purified murine CMV were added to the homogenate of piece #10 before the infection of an indicator culture of permissive mouse fetal cells. Poly(A)+ RNA derived from this culture after 72 h of viral replication was serially diluted as indicated, whereas for indicator cultures infected with homogenates of pieces #11 through #18 a constant amount (100 ng) of poly(A)+ RNA was subjected to an ie1 exon 3/4-specific RT PCR. Shown are the autoradiographs obtained after gel electrophoresis, Southern blotting, and hybridization with the γ-32P-end-labeled oligonucleotide probe IE1-P directed against the exon 3/4 splice junction. For the indicator culture corresponding to piece #18, the presence of RNA in the preparation is exemplarily demonstrated by RT PCR specific for the cellular housekeeping gene transcript HPRT. (B) Direct analysis of in vivo transcriptional activity. Poly(A)+ RNA isolated from pieces #1 through #9 (a 1/10 aliquot thereof [200 ng]) was subjected to the ie1 exon 3/4-specific RT PCR. As a positive control and standard, carrier poly(A)+ RNA derived from uninfected lung tissue was supplemented with the indicated numbers of in vitro-synthesized IE1 RNA molecules. HPRT-specific RT PCR served to document the presence of RNA in all samples. (C) For increasing the sensitivity of detection, pieces scored as negative in the analysis illustrated in panel B were retested by including a 7/10 aliquot of the poly(A)+ RNA yield in the ie1 exon 3/ 4-specific RT PCR.
Verification of murine CMV latency in the lungs.
All assays were performed at 12 months after BMT and infection, that is, at a time when productive primary infection was resolved in all organs (25) and when, as documented in Fig. 2, viral DNA was cleared from the BM and blood. However, these general precautions did not exclude a productive infection that may result from a spontaneous recurrence, possibly occurring by chance at the time of analysis, in an individual piece of lung tissue. We therefore tested for the presence of infectious virus in lung pieces #11 to #18. An example for an individual mouse designated LM1 (latent mouse 1) is shown in Fig. 3A, and all other latently infected mice were checked in the same manner. The previously described RT PCR-based focus expansion assay detects infectivity with very high sensitivity (25). This was documented again by supplementing the homogenate of piece #10 with 0.05 PFU of purified murine CMV, which equals ca. 25 viral genomes. As shown in detail previously, as few as 5 genomes are required for initiating infection in an indicator culture of permissive cells, but 25 genomes are needed to avoid negative results for the cultures due to the Poisson statistics of the sampling (25). Infection in this positive control culture was indicated by a large amount of amplificate obtained after ie1 exon 3/4-specific RT PCR performed with poly(A)+ RNA isolated from the indicator culture after 72 h of viral replication. By contrast, no signal was obtained from the indicator cultures inoculated with the homogenates of pieces #11 to #18. Mice LM2 to LM5 included (results not shown) 40 lung pieces tested altogether, and none contained infectious virus.
Focal transcriptional activity in latently infected lungs.
Based on the absence of infectious virus, the presence of productive-phase transcripts, such as the ie1 gene-derived IE1 transcript, was not expected, and transcriptional analysis was originally performed with the intention of documenting the absence of such transcripts as a further argument in favor of molecular murine CMV latency in the lungs. It was therefore a surprise to find IE1 transcripts among the poly(A)+ RNAs isolated directly from the nine pieces of the right lung lobes, as is documented for mouse LM1 in Fig. 3B. Notably, an on-or-off pattern was seen, in this particular example with five positive pieces and four virtually negative pieces. That RNA was present in all samples and in comparable amounts was verified by amplification of transcripts specified by the cellular gene coding for HPRT. The detection limit in this assay, as determined by titration of in vitro-synthesized IE1 transcripts, was ca. 10 molecules. Since an aliquot containing 1/10 of the poly(A)+ RNA yield from each piece of tissue, ca. 200 ng, was included in the RT PCR, positive pieces contained >100 IE1 transcripts in total. In order to improve the sensitivity and to lower the detection limit, negative pieces #1, #4, #5, and #7 were retested with 7/10 of the poly(A)+ RNA yield (Fig. 2C). Since these four samples were again negative without exception, the corresponding tissue pieces must have contained <10 transcripts. We infer from this finding that the difference between positive pieces and negative pieces may indeed be a qualitative difference. It is understood that the numbers of transcripts represent minimum estimates and depend upon the efficacy of the isolation procedure. After supplementing of lung tissue homogenate from uninfected mice with defined numbers of either in vitro-synthesized IE1 transcripts or natural IE1 transcripts derived from productively infected lungs, the retrieval after passage through the oligo(dT) column was found to be 50% or better in either case. Because a factor of two is not meaningful in this type of assay, calculated numbers of transcripts are reported herein with no correction for isolation efficacy. Thus, a positive score is given for >100 transcripts and a negative score is given for <10 transcripts calculated per piece, with each piece representing ca. 1/18 fraction of the lungs. While the possibility that transcription occurred at a level below the limit of detection can never be excluded, significant quantitative differences between the data for the lung tissue pieces were obvious. In conclusion, the observed mosaic pattern of transcription in the latently infected lungs suggests the existence of “foci” of IE1 transcription. For clarity it should be emphasized that the physical correlate of such a focus is yet to be defined. It could represent a single cell with a detectable number of transcripts or a group of cells collectively having a detectable number of transcripts. Thus, the term focus is used here to indicate a detectable transcriptional event.
Latent viral DNA is contained in all parts of the lungs.
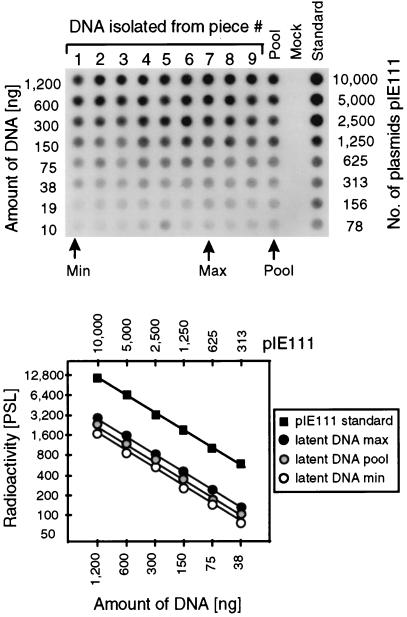
The question arose whether the focal transcriptional activity was related to an accordingly focal distribution of the latent viral genomes. Specifically, absence of latent viral DNA in a particular piece could be a trivial explanation for the absence of IE1 transcripts. Viral DNA was therefore quantitated in lung tissue pieces #1 to #9 of mouse LM1 (Fig. 4), that is, in the very same pieces for which the transcriptional activity had been determined (Fig. 3B). This was possible because DNA and RNA were isolated in parallel. All pieces contained latent viral DNA in comparable amounts. Specifically, the transcriptionally silent pieces, #1, #4, #5, and #7, did not contain less viral DNA than did the transcriptionally active pieces. In fact, the pieces with the lowest and highest DNA load, namely piece #1 and piece #7, respectively, both belonged to the group of transcriptionally silent pieces. Likewise, the high transcriptional activity found in piece #2 did not correspond to an accordingly high load of latent viral DNA. Quantitation by phosphorimaging revealed an average load of 7,500 viral genomes per 106 lung tissue cells (6 μg of cellular DNA), with a lower value of 6,000 viral genomes (for piece #1) and an upper value of 9,000 viral genomes (for piece #7). In essence, these differences in the latent viral DNA load between the pieces are minor, if at all significant, and we conclude that the total latent viral DNA load in a particular piece does not determine its transcriptional status. On the basis of the chosen resolution of the analysis, namely the mosaic of 18 pieces per lung, the latent viral DNA appeared to be evenly distributed. To avoid a misinterpretation, it should be emphasized that at higher resolution the latent viral DNA could still form hot spots critical for the initiation of transcription. The answer to this question awaits further analysis.
FIG. 4.
Determination of the viral DNA load during murine CMV latency in the lungs. In parallel with the isolation of the poly(A)+ RNA used for the transcriptional analyses shown in Fig. 3B and C, DNA was isolated from lung tissue pieces #1 through #9 derived from mouse LM1. The DNA from the individual pieces as well as a pool of the DNA from all nine pieces (Pool) were titrated and subjected to an ie1 exon 4-specific PCR. A negative control (Mock) was provided by DNA derived from uninfected lungs. As a standard for the quantitation, the mock DNA was supplemented with plasmid pIE111. (Top) Autoradiograph of the dot blot obtained after hybridization with a γ-32P-end-labeled internal oligonucleotide probe. (Bottom) Computed phosphorimaging results for the same blot. For the sake of clarity, the computations are depicted only for the pool, representing the average of all samples, as well as for the individual samples with the lowest load (Min) and the highest load (Max) among the nine samples tested. Log-log plots of radioactivity measured as phosphostimulated luminescence (PSL) units (ordinate) versus the amount of sample DNA (abscissa) are shown. The upper rule relates the amount of DNA to the number of plasmids in the pIE111 standard.
Foci of transcriptional activity are randomly distributed in latently infected lungs.
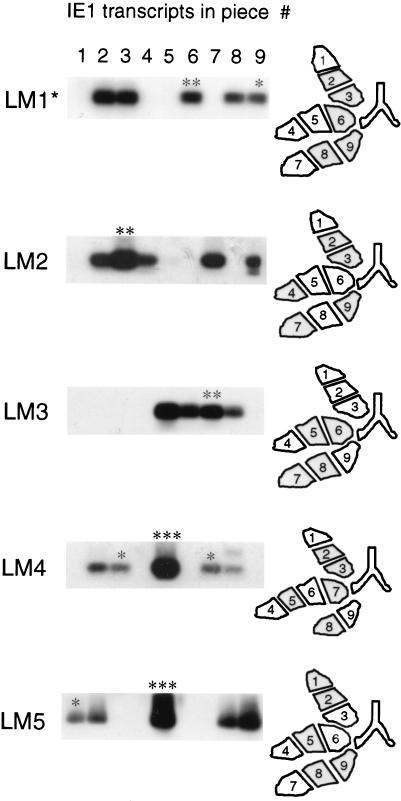
In order to verify that mouse LM1 was not peculiar with regard to transcription during latency, transcriptional activity was tested in the same manner in mice LM2 through LM5, and the analysis for mouse LM1 was repeated with the remaining 1/10 aliquot of the poly(A)+ RNA preparations (Fig. 5). In essence, the pattern was reproduced for mouse LM1, and on-or-off patterns were observed in like manner for all lungs analyzed. Topographical maps of the lung lobes illustrate the distributions of transcriptionally active and silent tissue pieces (Fig. 5, right). For mouse LM1, the particular distribution might have suggested a spread of transcriptional activity in the three lobes from the proximal portion to the distal portion relative to the bronchial tree. However, this interpretation was quickly proven wrong by the patterns observed in the other four mice. Actually, no rule can be recognized, and we hence conclude that viral transcriptional activity is randomly distributed in latently infected lungs.
FIG. 5.
Stochastic distribution of IE1 transcripts in latently infected lungs. Transcription from the ie1 gene was analyzed for mice LM1 through LM5 as outlined in Fig. 3B for mouse LM1. The analysis for mouse LM1 was repeated (LM1*) with the final aliquot of the poly(A)+ RNA preparation. The locations of positive and negative pieces in the three lobes of the lungs tested are illustrated by topographical maps, with transcriptionally active pieces being indicated by shading. Samples marked by one, two, and three asterisks were classified as samples with low, intermediate, and high transcriptional activity, respectively, and were used later for the quantitation of transcripts (see Fig. 7).
Frequency of transcriptional foci in a statistically generated prototypic latent lung.
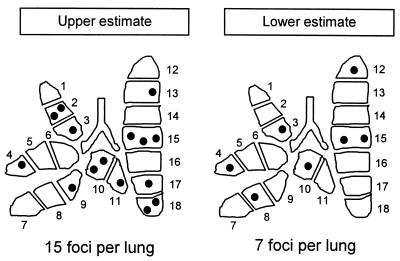
By definition, silent pieces do not harbor a transcriptional focus, whereas transcriptionally active pieces must harbor at least one focus but may also contain more than one focus. The Poisson distribution statistics can be employed to estimate the frequency of transcriptional foci from the fraction of silent pieces among all pieces and to calculate the fractions of pieces containing one, two, three, or more foci. The calculations were made for the 45 pieces of the five lungs as shown in Fig. 5, and the results were then extrapolated to 18 pieces of a complete “prototypic latent lung” (Fig. 6). A problem arises from the fact that a focus may be located by chance right at the experimental cut border between two pieces, which could lead to an overestimation of the number of transcriptionally positive pieces. We thus arrived at an upper estimate by assuming that neighboring positive pieces are always both positive (Fig. 6, left). The other extreme, and actually the less likely one in our opinion, is to assume that two neighboring positive pieces always have only one focus. This gives us the lower estimate (Fig. 6, right). In conclusion, the statistical calculations show us that latently infected lungs, under the specific conditions of our experimental setting, contain 7 to 15 foci of transcription and that the number of foci per positive piece is unlikely to exceed three.
FIG. 6.
Estimation of the average number of transcriptional foci in latently infected lungs. The statistical calculations were based on the patterns of transcription observed for 45 pieces derived from the superior, middle, and inferior lung lobes of five latently infected mice (see Fig. 5). The observed fraction, f(0), of transcriptionally negative lung pieces was used to calculate the Poisson distribution parameter λ (lambda). Based on λ, the fraction of pieces containing n (n = 0, 1, 2, and 3) foci, F(n), was calculated, and the results were extrapolated to a prototypic complete lung. The upper estimate was obtained by assuming that neighboring positive pieces are always both transcriptionally active. In that case, f(0) = 21/45. The lower estimate was obtained by assuming that neighboring positive pieces always share a transcriptional focus and count as a single positive piece. In that case, f(0) = 29/45. Shown are topographical maps of the statistically generated prototypic latent lungs, with pieces containing n (n = 0, 1, 2, and 3) foci being indicated by the corresponding number of dots.
Quantitation of ie1-specific transcripts in latently infected lungs.
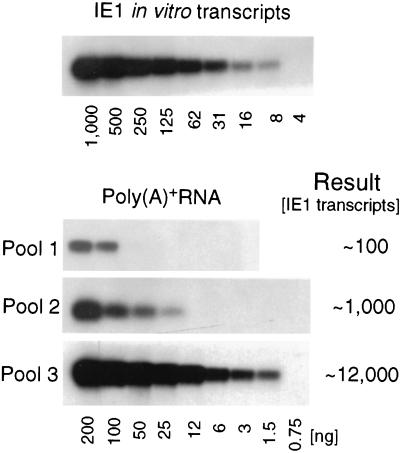
The signal intensities were found to differ markedly between transcriptionally active pieces of lung tissue (Fig. 5). This might reflect different numbers of foci per piece and/or different numbers of transcripts per focus. Since, as outlined above, the Poisson distribution statistics predicted an occupancy of positive pieces by one, two, or three but not by more foci of transcription, we classified positive pieces into those with low, intermediate, and high transcriptional activity (Fig. 5). Pools were formed for the three so defined classes, and the number of transcripts for each pool was measured by titration of poly(A)+ RNA followed by ie1 exon 3/4-specific RT PCR, specific hybridization, and phosphorimaging (Fig. 7). In vitro-synthesized IE1 transcripts were titrated to serve as a standard for the quantitation. If the amounts of transcripts were determined only by the number of foci per piece, we would expect proportions of 1:2:3 for the three pools. However, the actually observed proportions were on the order of ca. 1:10:100. As a consequence, we must then conclude that the amount of transcripts is primarily determined by the number of transcripts per focus, thus suggesting a heterogeneity in the transcriptional activity per focus.
FIG. 7.
Quantitation of IE1 transcripts in transcriptionally active lung tissue pieces. Positive pieces from mice LM1 through LM5 (see Fig. 5) were classified as having low, intermediate, or high transcriptional activity. Representative samples of the three so defined classes (indexed in Fig. 5 by the number of asterisks) were used to form poly(A)+ RNA pools 1, 2, and 3, respectively. Quantitation was performed by titration of the RNA pools, ie1 exon 3/ 4-specific RT PCR, and phosphorimaging. Shown are the autoradiographs. In vitro-synthesized IE1 transcripts provided the standard. The results are expressed as rounded-off numbers of IE1 transcripts per piece.
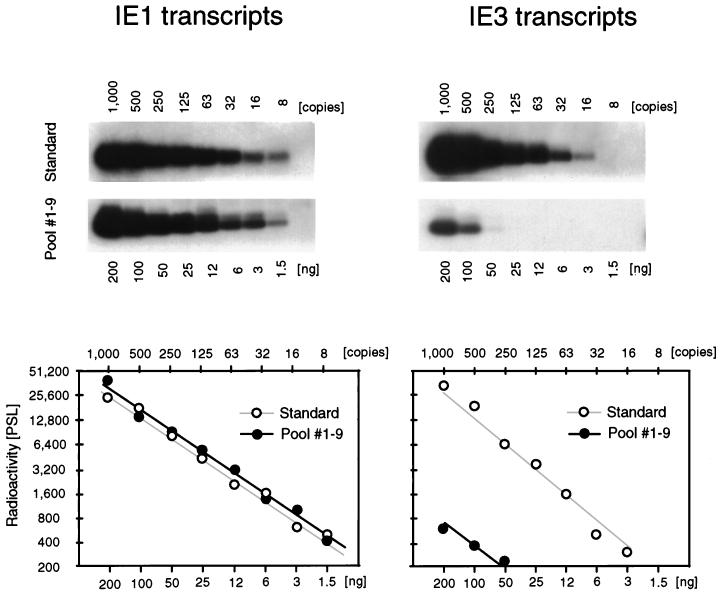
High ie1-specific transcriptional activity is not indicative of productive cycle reactivation.
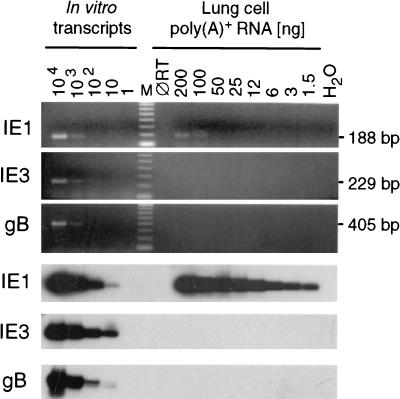
Even though all tissue pieces derived from the corresponding left lungs and postcaval lobes were negative in the high-sensitivity infectivity assay (Fig. 3), one might still surmise that transcriptional activity indicates a low-level persistent productive infection at a level below the detection limit of infectivity assays, an interpretation favored by other authors previously (52). The pieces containing more than 10,000 IE1 transcripts, that is, piece #5 of mouse LM4 and piece #5 of mouse LM5 (Fig. 5 and pool 3 in Fig. 7), are those which are most likely to be implicated in either persistent or recurrent productive infection. However, IE1 transcripts are not a proper indicator for initiation of the viral productive cycle. Whereas the IE1 protein contributes to the transactivation of early-phase genes, the main and essential transactivator of murine CMV is the 88-kDa protein IE3 (31), which corresponds to the 86-kDa IE2 protein of human CMV (13; for a review see reference 46). Accordingly, IE3 transcripts as well as transcripts specifying a virion structural protein, such as gB (38), should be present in persistent productive infection or intermittent recurrence. RT PCRs specific for IE1, IE3, and gB transcripts detected the corresponding in vitro-synthesized transcripts with comparable sensitivities. Specifically, in each case, 10 transcripts could clearly be visualized (Fig. 8, left). Whereas ca. 12,000 IE1 transcripts were again detected within the poly(A)+ RNA of the high-activity pool, pool 3, IE3 transcripts and gB transcripts were virtually absent (Fig. 8, right). To improve the sensitivity of detection, the remaining 1.4 μg of the pool 3 poly(A)+ RNA was subjected to the ie3-specific RT PCR, and it was found to be negative (not shown). Based on the detection limit of eight transcripts (in a log2 titration of the standard), we conclude that the number of IE3 transcripts, if they did exist at all, was less than the number of IE1 transcripts by at least a factor of 1,000. Altogether, sensitive analyses of IE3 and gB transcripts did not give any hint of or evidence for persistent or recurrent productive infection.
FIG. 8.
Survey of productive cycle transcription in latently infected lungs. Transcription from genes ie1 (exon 3/4 of the ie1-ie3 transcription unit), ie3 (exon 3/5 of the ie1-ie3 transcription unit), and gB was analyzed for the high-activity poly(A)+ RNA pool (pool 3; see Fig. 7) by the appropriate RT PCRs. (Top) Ethidium bromide-stained gels verifying the correct sizes of the amplificates. M, lane M contains the 100-bp size markers; ØRT, control reactions performed with 200-ng samples and all components except the RT. (Bottom) Corresponding Southern blot autoradiographs obtained after hybridization with the γ-32P-end-labeled oligonucleotide probes. For the IE1 and IE3 transcripts, the probes were directed against the exon 3-exon 4 (probe IE1-P) and exon 3-exon 5 (probe IE3-P) splicing junctions, respectively.
Experimental induction of recurrence is associated with detectable IE3 transcripts.
While the comparison between IE1 transcripts and IE3 transcripts did not provide evidence in favor of a productive cycle transcription, the observed great difference per se cannot be viewed as evidence against it. IE1 transcripts are known to be abundant and fairly stable during productive infection (18, 19). The virtual absence of IE3 transcripts during latency could thus result from an overall low level of transcriptional activity during latency in combination with a generally low abundance and/or stability of the IE3 mRNA. It is important to recall that the ie1 and ie3 genes are transcribed as a transcription unit and that mRNAs are generated from a 5.1-kb IE1/3 precursor transcript by differential splicing connecting exons 1 through 3 with exon 4 for IE1 mRNA and with exon 5 for IE3 mRNA (19, 31) (illustrated in Fig. 1). It was therefore of relevance to determine the IE1 transcript: IE3 transcript ratio during productive infection. Specifically, a switch from latent nonproductive infection to recurrent productive infection should involve a significant rate of production of IE3 mRNA specifying the essential transactivator IE3. As members of our group have documented repeatedly in previous reports, rapid recurrence of infectious virus from latently infected lungs can be detected after total-body γ-irradiation (3, 25, 40, 47), and recurrence of murine CMV after other regimens of immunosuppressive treatment is a long-recognized phenomenon in this field (reviewed in reference 16). We have therefore used our model of radiation-facilitated recurrence for analyzing the IE-phase transcription during reinitiation of productive infection (Fig. 9). Successful induction of recurrence is documented for a representative case by the RT PCR-based focus expansion assay of infectivity, performed on day 14 after γ-irradiation with a dose of 6.5 Gy (Fig. 9A). It is worth noting that infectious virus can be found in individual pieces as early as day 4 after the irradiation, which indicates an immediate entry of latent murine CMV into the productive cycle (not shown). By day 14, infectious virus was detected in all nine tissue pieces derived from the left lung and the postcaval lobe (i.e., piece #10 through piece #18). Since none of 40 lung tissue pieces collected from five mice had been found to be positive during latency (Fig. 3), the statistical significance of the recurrence is undebatable. Tissue pieces #1 through #9 of the remaining three lobes of the lungs were used to detect IE1 and IE3 transcripts (Fig. 9B). Regarding IE1, the latency-associated on-or-off pattern of focal transcriptional activity was now replaced by the presence of a large amount of transcripts in all nine pieces. Notably, IE3 transcripts became detectable under these conditions, and, as was the case for IE1 transcripts, all nine pieces were positive.
FIG. 9.
Analysis of reactivation and recurrent infection in the lungs. Recurrence of infectious with virus from latently infected lungs was induced at 12 months after BMT and infection by total-body γ-irradiation with a dose of 6.5 Gy. Data are shown for a particular case on day 14 after the irradiation. (A) Assay of infectivity. Infectious virus was detected in lung tissue pieces #10 through #18 (left lungs and postcaval lobe) by using the RT PCR-based focus expansion assay (details are as described in the legend for Fig. 3). (B) Detection of IE1 and IE3 transcripts. Poly(A)+ RNA was isolated from lung tissue pieces #1 through #9 (remaining three lobes) on day 14 after the irradiation, and 1/10 aliquots (ca. 200 ng) were subjected to the RT PCRs.
Recurrence is characterized by an increased rate of ie3-specific splicing.
Nonetheless, one could still surmise that detection of IE3 transcripts reflected merely an enhanced level of overall viral transcriptional activity during recurrence. We therefore pooled the poly(A)+ RNA of the nine pieces and quantitated IE1 and IE3 transcripts by direct comparison with the corresponding in vitro-synthesized RNAs that served as standards (Fig. 10). As one would expect, the total amount of IE1 transcripts in the lungs was increased during recurrence. Notably, however, the average number of IE1 transcripts per piece, ca. 15,000 molecules, did not significantly exceed the number of IE1 transcripts determined during latency in the high-activity pool; pool 3 (compare Fig. 7 and Fig. 10, left), in which IE3 transcripts were not detectable at all (Fig. 8). By contrast, during recurrence, the average number of IE3 transcripts was calculated to be ca. 300 molecules per piece of lung tissue (Fig. 10, right). In other words, the IE1 transcript: IE3 transcript ratio was ca. 50 during recurrence and >1,000 during latency, which clearly indicates a change in the program of IE1/3 precursor transcript splicing.
FIG. 10.
Quantitative comparison between IE1 transcripts and IE3 transcripts during recurrence. Aliquots of the poly(A)+ RNAs derived from lung tissue pieces #1 through #9 (see Fig. 9) were pooled. Aliquots of the pool (200 ng, which equals 1/10 of the yield of one piece) were serially diluted and subjected to the RT PCRs. The in vitro-synthesized transcripts were titrated in parallel as standards for the quantitation. (Top) Autoradiographs obtained after gel electrophoresis of the amplificates, Southern blotting, and hybridization with the γ-32P-end-labeled oligonucleotide probe IE1/3-P directed against the common exon 3. (Bottom) Computed phosphorimaging data for the same blots. Log-log plots of radioactivity measured as phosphostimulated luminescence (PSL) units (ordinate) versus the amount of poly(A)+ RNA (abscissa) are shown. The upper rule relates the amount of poly(A)+ RNA to the number of molecules in the standard.
In conclusion, murine CMV latency in the lungs is associated with a focal, randomly distributed generation of IE1 transcripts in the absence of productive cycle transcripts, while recurrence is associated with enhanced generation of IE3 transcripts.
DISCUSSION
While most current concepts focus on human CMV and murine CMV latency in hematopoietic cells of the myeloid differentiation lineage, our data on murine CMV latency have reconfirmed our previous conclusions regarding the significance of multiple-organ tissue latency (40), in particular of latency in the lungs (3, 25), a major organ site of CMV pathogenesis and immune control (14, 43). Specifically, at a time after acute infection of BMT recipients when the level of viral DNA in BM and blood had declined to below the detection limit (<100 copies per 106 cells), the lungs still harbored the latent viral genome, with a load of 6,000 to 9,000 copies per 106 lung cells. The maintenance of latent CMV in organs after exhaustion of the putative source in BM and blood does not formally exclude the possibility that the origin of latently infected cells in the lungs is hematopoietic lineage, but we must then postulate that the latently infected cells accumulate in organs, are extremely long-lived, and are not replaced during natural turnover by the uninfected cells delivered by the BM and circulating in the blood. While our data do not argue against the existence of a hematopoietic and leukocytic reservoir of latent CMV for a significant and most probably clinically meaningful period, we favor the interpretation that cells of the myeloid lineage are only a temporary site of CMV latency and that long-lived stromal cells are the cellular site of enduring latency in organs. The kinetics of viral DNA clearance from BM and blood indicates that the viral DNA load in the blood results primarily from the productive infection and not from exported progeny of latently infected hematopoietic progenitor cells residing in the BM. Throughout the course of the kinetics, the load in the BM was always far below the load in the blood. One may argue that a few hematopoietic progenitor cells in the BM can give rise to a huge number of progeny and that this will also lead to an amplification of the latent viral DNA. However, although true in an absolute sense, this argument ignores the fact that the uninfected hematopoietic progenitor cells also produce progeny. One might raise the interesting argument that DNA derived from the uninfected cells of the erythroid differentiation lineage may dilute the viral DNA signal in the BM, while latent viral DNA is relatively enriched in blood leukocytes due to the anuclear status of erythrocytes. However, although erythrocytes by far outnumber leukocytes in peripheral blood, the maintenance pool of erythroid-lineage cells in the BM is about the size of the myelomonocytic-lineage cell pool. We have shown this recently by a comparative histochemical quantitation of myelomonocytic and erythroid colony cells in repopulating BM after experimental BMT (48), and much earlier studies have determined a myeloid:erythroid ratio of ca. 1.5:1 for femoral BM of adult mice (26). DNA derived from erythroid-lineage cells may thus dilute the viral DNA in BM by twofold at the most, but this certainly cannot account for the observed viral load differences between BM and blood. Therefore, unless we assume that latently infected cells gain a growth advantage or that the copy number of latent viral DNA markedly increases during cell differentiation, although presently there is no evidence for this, the relative load in the blood cannot exceed the relative load in the putative BM source. In fact, our data do not exclude the possibility that the load determined for the BM resulted mainly, if not exclusively, from blood cells present in the BM capillaries and sinusoids. We have previously shown by immunohistological detection of viral IE1 protein as well as by in situ hybridization specific for viral DNA that murine CMV does indeed infect reticular stromal cells in the hematopoietic cord; however, even during acute, lethal infection of mice, the number of infected cells in the BM stroma was minute and most infected cells did not complete the productive cycle (28). The recently discussed idea that a persistent productive infection in the BM stroma may be the source from which myeloid progenitor cells or monocytes and macrophages acquire CMV (35, 45) is thus not supported by the data obtained in the murine-model system. Considering the high load of latent viral DNA in various organs, we rather propose that macrophages pick up the latent CMV genome in tissues by phagocytosis of latently infected, damaged or senescent stromal or parenchymal cells. We must admit, of course, that our arguments apply to murine CMV and that human CMV in general, or isolates with a particular cell tropism (44), may differ in this respect. For resolving this question, a quantitative and longitudinal analysis of the human CMV genome load is needed. Only a comparative study of the kinetics of the viral DNA load in the BM, blood, and organs of BMT patients will allow us to evaluate the relative contributions of myeloid-lineage cells and of tissue stromal cells to human CMV latency.
Transcriptional activity of latent human CMV has been analyzed in a model of latency established in cultures of in vitro-infected granulocyte-macrophage progenitors (23, 24). These cells do not support productive infection, but they do carry the viral DNA. Two classes of novel transcripts derived from the regulatory ie1-ie2 gene region, namely sense and antisense latency-associated transcripts, were detected in a small percentage of the cells. While infectious virus was recovered from these cultures after cocultivation with permissive cells, it remained open to question whether recurrence originated from the transcriptionally active minority or from the transcriptionally silent majority of the latently infected cells. Notably, the latency-associated transcripts could also be detected in myeloid-lineage-committed progenitor cells derived from latently infected, healthy individuals (11, 24).
In models of in vivo latency of murine CMV, ie1-specific sequences derived from spliced RNA were detected by RT PCR by some investigators (12, 52) but not by others (22). Absence of IE1 transcripts was taken as an argument in favor of molecular latency (22), whereas presence of IE1 transcripts was interpreted as being indicative of a persistent productive infection at a low level, one below the detection limit of then-available conventional infectivity assays (52). However, the assumption of low-level productive infection was not supported by any further evidence, and more recent studies employing much-improved assays for testing viral infectivity have corroborated the conclusion that murine CMV does establish molecular latency characterized by absence of infectivity (25, 36).
Data presented herein can explain these controversial previous findings. We have documented that latently infected lungs can be viewed as a mosaic, with transcriptionally active and silent pieces coexisting and randomly distributed in the very same organ at the very same time. Whereas the frequency of transcriptionally active foci was quite high, 7 to 15 foci per lung, infectious virus could not be detected with the previously described high-sensitivity focus expansion assay capable of detecting 0.01 PFU (25). Thus, apparently, IE1 transcripts are not necessarily indicative of productive infection. The IE1 protein pp89 of murine CMV (20) is not the transactivator of viral early gene expression, and its human CMV counterpart, the IE1 protein p72, is not essential for viral replication (9). Transcripts from the murine ie3 gene, which is the counterpart of the human CMV ie2 gene (13, 46) and encodes the IE3 transactivator (31), are therefore a much better marker for the initiation of the productive cycle. While IE3 transcripts were clearly detected during experimentally induced recurrent infection of the lungs, these transcripts, as well as transcripts specifying the structural glycoprotein gB, were not detectable during murine CMV latency in the lungs, not even in those individual pieces of tissue that contained an amount of IE1 transcripts comparable to the average amount present during recurrent infection. Taken together, these data demonstrated a switch in the transcriptional and splicing program from latency associated to recurrence associated and thus represent the strongest evidence presented to date against a low-level persistence of productive infection during murine CMV latency in organs. The fact that the ie1 and ie3 genes are transcribed as a transcription unit has a very important implication. While an on-position of the major IE promoter-enhancer allowing for IE1/3 precursor transcription is certainly the primary condition for recurrence to occur, a subsequent regulation at the level of splicing appears to be critical for the generation of the essential IE3 transactivator that starts the productive cycle.
So far, the small absolute amount of IE1 transcripts in the lungs has precluded their precise identification. Determination of whether the authentic, pp89-encoding 2.75-kb transcript (19) is produced at all must await further studies. An immunohistological analysis failed to detect intranuclear pp89 protein, even though hundreds of sections from latently infected lungs were screened (data not shown). We have evidence that a fraction of the ie1-specific transcripts is deficient in exon 2-encoded sequences (unpublished data). We are thus faced with a mixture of transcripts, a fact that further complicates their identification. It is worth noting that alternative start sites have not been identified upstream of the ie1-ie3 transcription start site of murine CMV. Therefore, analogs of the latency-associated sense transcripts of human CMV, which use alternative start sites in the analogous IE1/IE2 promoter-enhancer region of human CMV (24), are not to be expected for murine CMV.
A comment is needed on the definition of a latency-associated transcript. We have found that IE1-phase transcriptional activity is focal and randomly distributed in the lungs, whereas the latent viral DNA is evenly distributed. Thus, apparently, the observed transcription is not a feature inherent to latency. Rather, it appears that the latent viral genome can exist in a transcriptionally active and in a transcriptionally silent state. This is not peculiar to murine CMV but appears to apply also to human CMV latency in granulocyte-macrophage progenitors, since the latency-associated transcripts of human CMV were also not found in all cells that carried latent viral DNA but actually only in a small percentage at a given time point (23). Upon experimental induction of recurrence, we have found IE3 transcripts as well as infectious virus in all pieces of the lungs. Since the absolute amount of infectious virus is quite small early in recurrence, and since antiviral antibody prevents virus spread in seropositive mice (15, 40), we consider it unlikely that the observed generalized infection of the lungs resulted from virus dissemination. If we are right, this would imply that recurrence can occur in transcriptionally active and silent pieces.
However, one has to be aware of the fact that our data can provide only a snapshot of what happens in the lungs during latency. Thus, a piece of lung tissue that was found to contain transcriptionally silent latent viral DNA at the time of assay might have been transcriptionally active the day before and a transcriptionally active piece may return to quiescence. We have no immediate approach at hand to prove this, but let us nevertheless speculate a bit as to what it might mean. Randomly distributed IE1 transcripts could indicate episodes of spontaneous reactivation of viral gene expression that is terminated before the IE3 transactivator protein is expressed at a level sufficient to initiate the productive cycle. How could reactivation be terminated? Provided that IE1 transcripts lead to protein synthesis, not necessarily of the authentic pp89, and to the presentation of the immunodominant ie1 exon 4-encoded IE1 nonapeptide of murine CMV (7, 14, 42), IE1 nonapeptide-specific cytolytic T cells, which are in fact present during latency as memory cells (14, 41), could eliminate the reactivating cells and thereby prevent virus recurrence. The spontaneous recurrence observed in tissue explant cultures (reference 17 and reviewed in reference 16) as well as the frequent and rapid recurrence observed in organs of latently infected mice after various protocols of immunoablative treatment (34, 40) are in accordance with this hypothesis of an immune surveillance of CMV latency.
However, we doubt that the phenomena of CMV latency and reactivation are entirely explainable by, respectively, the presence and withdrawal of immune control. The observed switch in the IE transcript splicing program, from selective IE1 splicing during latency to IE1 and IE3 splicing during reactivation, may indicate a role for splicing regulation.
ACKNOWLEDGMENTS
This work will be part of the Ph.D thesis of S. K. Kurz at the Faculty of Biology of the Johannes Gutenberg-University. The previous scientific contributions of Angelika Ebeling-Keil, Günther Keil, and Martin Messerle have provided the essential basis of this work.
Support was provided by a grant to M.J.R. by the Bundesministerium für Bildung, Wissenschaft, Forschung und Technologie (BMBF), Collaborative Research Project on CMV, individual project 01Kl 9607/5. We thank all members of the collaborative project for helpful discussion.
REFERENCES
- 1.Badley J E, Bishop G A, St. John T, Frelinger J A. A simple, rapid method for the purification of poly A+ RNA. BioTechniques. 1988;6:114–116. [PubMed] [Google Scholar]
- 2.Balthesen M, Dreher L, Lucin P, Reddehase M J. The establishment of cytomegalovirus latency in organs is not linked to local virus production during primary infection. J Gen Virol. 1994;75:2329–2336. doi: 10.1099/0022-1317-75-9-2329. [DOI] [PubMed] [Google Scholar]
- 3.Balthesen M, Messerle M, Reddehase M J. Lungs are a major organ site of cytomegalovirus latency and recurrence. J Virol. 1993;67:5360–5366. doi: 10.1128/jvi.67.9.5360-5366.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bowden, R. A. 1991. Cytomegalovirus infections in transplant patients: methods of prevention of primary cytomegalovirus. Transplant. Proc. 23(Suppl. 3):136–138. [PubMed]
- 5.Brautigam A R, Dutko F J, Olding L B, Oldstone M B. Pathogenesis of murine cytomegalovirus infection: the macrophage as a permissive cell for cytomegalovirus infection, replication, and latency. J Gen Virol. 1979;44:349–359. doi: 10.1099/0022-1317-44-2-349. [DOI] [PubMed] [Google Scholar]
- 6.Chou S W. Acquisition of donor strains of cytomegalovirus by renal-transplant recipients. N Engl J Med. 1986;314:1418–1423. doi: 10.1056/NEJM198605293142205. [DOI] [PubMed] [Google Scholar]
- 7.Del Val M, Volkmer H, Rothbard J B, Jonjić S, Messerle M, Schickedanz J, Reddehase M J, Koszinowski U H. Molecular basis for cytolytic T-lymphocyte recognition of the murine cytomegalovirus immediate-early protein pp89. J Virol. 1988;62:3965–3972. doi: 10.1128/jvi.62.11.3965-3972.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Forman, S. J. 1991. Bone marrow transplantation. Transplant. Proc. 23(Suppl. 3):110–114. [PubMed]
- 9.Greaves R F, Mocarski E S. Defective growth correlates with reduced accumulation of a viral DNA replication protein after low-multiplicity infection by a human cytomegalovirus ie1 mutant. J Virol. 1998;72:366–379. doi: 10.1128/jvi.72.1.366-379.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Grundy J E, Super M, Lui S, Sweny P, Griffiths P D. The source of cytomegalovirus infection in seropositive renal allograft recipients is frequently the donor kidney. Transplant Proc. 1987;19:2126–2128. [PubMed] [Google Scholar]
- 11.Hahn G, Jores R, Mocarski E S. Cytomegalovirus remains latent in a common precursor of dendritic and myeloid cells. Proc Natl Acad Sci USA. 1998;95:3937–3942. doi: 10.1073/pnas.95.7.3937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Henry S C, Hamilton J D. Detection of murine cytomegalovirus immediate early 1 transcripts in the spleens of latently infected mice. J Infect Dis. 1993;167:950–954. doi: 10.1093/infdis/167.4.950. [DOI] [PubMed] [Google Scholar]
- 13.Hermiston T W, Malone C L, Witte P R, Stinski M F. Identification and characterization of the human cytomegalovirus immediate-early region 2 gene that stimulates gene expression from an inducible promoter. J Virol. 1987;61:3214–3221. doi: 10.1128/jvi.61.10.3214-3221.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Holtappels R, Podlech J, Geginat G, Steffens H-P, Thomas D, Reddehase M J. Control of murine cytomegalovirus in the lungs: relative but not absolute immunodominance of the immediate-early 1 nonapeptide during the antiviral cytolytic T-lymphocyte response in pulmonary infiltrates. J Virol. 1998;72:7201–7212. doi: 10.1128/jvi.72.9.7201-7212.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jonjic S, Pavic I, Polic B, Crnkovic I, Lucin P, Koszinowski U H. Antibodies are not essential for the resolution of primary cytomegalovirus infection but limit dissemination of recurrent virus. J Exp Med. 1994;179:1713–1717. doi: 10.1084/jem.179.5.1713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jordan M C. Latent infection and the elusive cytomegalovirus. Rev Infect Dis. 1983;5:205–215. doi: 10.1093/clinids/5.2.205. [DOI] [PubMed] [Google Scholar]
- 17.Jordan M C, Mar V L. Spontaneous activation of latent cytomegalovirus from murine spleen explants: role of lymphocytes and macrophages in release and replication of virus. J Clin Investig. 1982;70:762–768. doi: 10.1172/JCI110672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Keil G M, Ebeling-Keil A, Koszinowski U H. Temporal regulation of murine cytomegalovirus transcription and mapping of viral RNA synthesized at immediate early times after infection. J Virol. 1984;50:784–795. doi: 10.1128/jvi.50.3.784-795.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Keil G M, Ebeling-Keil A, Koszinowski U H. Sequence and structural organization of murine cytomegalovirus immediate-early gene 1. J Virol. 1987;61:1901–1908. doi: 10.1128/jvi.61.6.1901-1908.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Keil G M, Fibi M R, Koszinowski U H. Characterization of the major immediate-early polypeptides encoded by murine cytomegalovirus. J Virol. 1985;54:422–428. doi: 10.1128/jvi.54.2.422-428.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Klotman M E, Henry S C, Greene R C, Brazy P C, Klotman P E, Hamilton J D. Detection of mouse cytomegalovirus nucleic acid in latently infected mice by in vitro enzymatic amplification. J Infect Dis. 1990;161:220–225. doi: 10.1093/infdis/161.2.220. [DOI] [PubMed] [Google Scholar]
- 22.Koffron A J, Hummel M, Patterson B K, Yan S, Kaufman D B, Fryer J P, Stuart F P, Abecassis M I. Cellular localization of latent murine cytomegalovirus. J Virol. 1998;72:95–103. doi: 10.1128/jvi.72.1.95-103.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kondo K, Kaneshima H, Mocarski E S. Human cytomegalovirus latent infection of granulocyte-macrophage progenitors. Proc Natl Acad Sci USA. 1994;91:11879–11883. doi: 10.1073/pnas.91.25.11879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kondo K, Xu J, Mocarski E S. Human cytomegalovirus latent gene expression in granulocyte-macrophage progenitors in culture and in seropositive individuals. Proc Natl Acad Sci USA. 1996;93:11137–11142. doi: 10.1073/pnas.93.20.11137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kurz S, Steffens H-P, Mayer A, Harris J R, Reddehase M J. Latency versus persistence or intermittent recurrences: evidence for a latent state of murine cytomegalovirus in the lungs. J Virol. 1997;71:2980–2987. doi: 10.1128/jvi.71.4.2980-2987.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lawkowicz W, Czerski P. Comparative haematology of laboratory animals. (Selected aspects) Acta Haematol. 1966;36:13–25. doi: 10.1159/000209088. [DOI] [PubMed] [Google Scholar]
- 27.Maciejewski J P, Bruening E E, Donahue R E, Mocarski E S, Young N S, St. Jeor S C. Infection of hematopoietic progenitor cells by human cytomegalovirus. Blood. 1992;80:170–178. [PubMed] [Google Scholar]
- 28.Mayer A, Podlech J, Kurz S, Steffens H-P, Maiberger S, Thalmeier K, Angele P, Dreher L, Reddehase M J. Bone marrow failure by cytomegalovirus is associated with an in vivo deficiency in the expression of essential stromal hemopoietin genes. J Virol. 1997;71:4589–4598. doi: 10.1128/jvi.71.6.4589-4598.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mendelson M, Monard S, Sissons J G P, Sinclair J H. Detection of endogenous human cytomegalovirus in CD34+ bone marrow progenitors. J Gen Virol. 1996;77:3099–3102. doi: 10.1099/0022-1317-77-12-3099. [DOI] [PubMed] [Google Scholar]
- 30.Mercer J A, Wiley C A, Spector D H. Pathogenesis of murine cytomegalovirus infection: identification of infected cells in the spleen during acute and latent infections. J Virol. 1988;62:987–997. doi: 10.1128/jvi.62.3.987-997.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Messerle M, Bühler B, Keil G M, Koszinowski U H. Structural organization, expression, and functional characterization of the murine cytomegalovirus immediate-early gene 3. J Virol. 1992;66:27–36. doi: 10.1128/jvi.66.1.27-36.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Minton E J, Tysoe C, Sinclair J H, Sissons J G P. Human cytomegalovirus infection of the monocyte/macrophage lineage in bone marrow. J Virol. 1994;68:4017–4021. doi: 10.1128/jvi.68.6.4017-4021.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Podlech J, Holtappels R, Wirtz N, Steffens H-P, Reddehase M J. Reconstitution of CD8 T cells is essential for the prevention of multiple-organ cytomegalovirus histopathology after bone marrow transplantation. J Gen Virol. 1998;79:2099–2104. doi: 10.1099/0022-1317-79-9-2099. [DOI] [PubMed] [Google Scholar]
- 34.Polic B, Hengel H, Krmpotic A, Trgovcich J, Pavic I, Lucin P, Jonjic S, Koszinowski U H. Hierarchical and redundant lymphocyte subset control precludes cytomegalovirus replication during latent infection. J Exp Med. 1998;188:1047–1054. doi: 10.1084/jem.188.6.1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pollock J L, Presti R M, Paetzold S, Virgin H W., IV Latent murine cytomegalovirus infection in macrophages. Virology. 1997;227:168–179. doi: 10.1006/viro.1996.8303. [DOI] [PubMed] [Google Scholar]
- 36.Pollock J L, Virgin H W., IV Latency, without persistence, of murine cytomegalovirus in the spleen and kidney. J Virol. 1995;69:1762–1768. doi: 10.1128/jvi.69.3.1762-1768.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pomeroy C, Hilleren P J, Jordan M C. Latent murine cytomegalovirus DNA in splenic stromal cells of mice. J Virol. 1991;65:3330–3334. doi: 10.1128/jvi.65.6.3330-3334.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rapp M, Messerle M, Bühler B, Tannheimer M, Keil G M, Koszinowski U H. Identification of the murine cytomegalovirus glycoprotein B gene and its expression by recombinant vaccinia virus. J Virol. 1992;66:4399–4406. doi: 10.1128/jvi.66.7.4399-4406.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rawlinson W D, Farrell H E, Barrell B G. Analysis of the complete DNA sequence of murine cytomegalovirus. J Virol. 1996;70:8833–8849. doi: 10.1128/jvi.70.12.8833-8849.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Reddehase M J, Balthesen M, Rapp M, Jonjic S, Pavic I, Koszinowski U H. The conditions of primary infection define the load of latent viral genome in organs and the risk of recurrent cytomegalovirus disease. J Exp Med. 1994;179:185–193. doi: 10.1084/jem.179.1.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Reddehase M J, Keil G M, Koszinowski U H. The cytolytic T lymphocyte response to the murine cytomegalovirus. II. Detection of virus replication stage-specific antigens by separate populations of in vivo active cytolytic T lymphocyte precursors. Eur J Immunol. 1984;14:56–61. doi: 10.1002/eji.1830140111. [DOI] [PubMed] [Google Scholar]
- 42.Reddehase M J, Rothbard J B, Koszinowski U H. A pentapeptide as minimal antigenic determinant for MHC class I-restricted T lymphocytes. Nature (London) 1989;337:651–653. doi: 10.1038/337651a0. [DOI] [PubMed] [Google Scholar]
- 43.Reddehase M J, Weiland F, Münch K, Jonjic S, Lüske A, Koszinowski U H. Interstitial murine cytomegalovirus pneumonia after irradiation: characterization of cells that limit viral replication during established infection of the lungs. J Virol. 1985;55:264–273. doi: 10.1128/jvi.55.2.264-273.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Simmons P, Kaushansky K, Torok-Storb B. Mechanisms of cytomegalovirus-mediated myelosuppression: perturbation of stromal cell function versus direct infection of myeloid cells. Proc Natl Acad Sci USA. 1990;87:1386–1390. doi: 10.1073/pnas.87.4.1386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Soderberg-Naucler C, Fish K N, Nelson J A. Reactivation of latent human cytomegalovirus by allogeneic stimulation of blood cells from healthy donors. Cell. 1997;91:119–126. doi: 10.1016/s0092-8674(01)80014-3. [DOI] [PubMed] [Google Scholar]
- 46.Spector D H. Activation and regulation of human cytomegalovirus early genes. Intervirology. 1996;39:361–377. doi: 10.1159/000150507. [DOI] [PubMed] [Google Scholar]
- 47.Steffens H-P, Kurz S, Holtappels R, Reddehase M J. Preemptive CD8 T-cell immunotherapy of acute cytomegalovirus infection prevents lethal disease, limits the burden of latent viral genomes, and reduces the risk of virus recurrence. J Virol. 1998;72:1797–1804. doi: 10.1128/jvi.72.3.1797-1804.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Steffens H-P, Podlech J, Kurz S, Angele P, Dreis D, Reddehase M J. Cytomegalovirus inhibits the engraftment of donor bone marrow cells by downregulation of hemopoietin gene expression in recipient stroma. J Virol. 1998;72:5006–5015. doi: 10.1128/jvi.72.6.5006-5015.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Taylor-Wiedeman J, Sissons J G P, Borysiewicz L K, Sinclair J H. Monocytes are a major site of persistence of human cytomegalovirus in peripheral blood mononuclear cells. J Gen Virol. 1991;72:2059–2064. doi: 10.1099/0022-1317-72-9-2059. [DOI] [PubMed] [Google Scholar]
- 50.Taylor-Wiedeman J, Sissons J G P, Sinclair J H. Induction of endogenous human cytomegalovirus gene expression after differentiation of monocytes from healthy carriers. J Virol. 1994;68:1597–1604. doi: 10.1128/jvi.68.3.1597-1604.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Winston D J, Ho W G, Champlin R E. Cytomegalovirus infections after bone marrow transplantation. Rev Infect Dis. 1990;12:S776–S792. doi: 10.1093/clinids/12.supplement_7.s776. [DOI] [PubMed] [Google Scholar]
- 52.Yuhasz S A, Dissette V B, Cook M L, Stevens J G. Murine cytomegalovirus is present in both chronic active and latent states in persistently infected mice. Virology. 1994;202:272–280. doi: 10.1006/viro.1994.1343. [DOI] [PubMed] [Google Scholar]