Abstract
Nonsense mutations are involved in multiple peripheral neuropathies. These mutations induce the presence of a premature termination codon (PTC) at the mRNA level. As a result, a dysfunctional or truncated protein is synthesized, or even absent linked to nonsense-mediated mRNA degradation (NMD) system activation. Readthrough molecules or NMD inhibitors could be innovative therapies in these hereditary neuropathies, particularly molecules harboring the dual activity as amlexanox. Charcot–Marie–Tooth (CMT) is the most common inherited pathology of the peripheral nervous system, affecting 1 in 2500 people worldwide. Nonsense mutations in the GDAP1 gene have been associated with a severe form of CMT, prompting us to investigate the effect of readthrough and NMD inhibitor molecules. Although not clearly defined, GDAP1 could be involved in mitochondrial functions, such as mitophagy. We focused on the homozygous c.581C>G (p.Ser194*) mutation inducing CMT2H using patient human induced pluripotent stem cell (hiPSC)-derived neuronal cells. Treatment during 20 h with 100 µM of amlexanox on this cell model stabilized GDAP1 mRNAs carrying UGA-PTC and induced a restoration of the mitochondrial morphology. These results highlight the potential of readthrough molecules associated to NMD inhibitors for the treatment of genetic alterations in CMT, opening the way for future investigations and a potential therapy.
Keywords: GDAP1, CMT disease, nonsense mutation, nonsense-mediated decay (NMD), readthrough molecules, premature termination codon (PTC)
1. Introduction
Nonsense mutations cause 11% of genetic diseases [1] and are mainly induced by single-nucleotide changes in the DNA-coding region. These mutations can lead to a premature termination codon (PTC) in the mRNA, stopping the translation process before the natural termination codon (NTC). The presence of such PTCs in the mRNA sequence has two main consequences. First, the PTC can induce the synthesis of an aberrant, even nonfunctional, truncated protein. Additionally, the corresponding PTC-containing mRNA could be recognized and degraded by the nonsense-mediated mRNA decay (NMD) system, leading to the lack of protein expression, and preventing the production of dysfunctional or nonfunctional proteins [2].
Recent therapeutic strategies, based on pharmacological approaches, can be applied to limit or inhibit nonsense mutations consequences. The first approach consists in using readthrough (RT) molecules, that are able to bypass the PTC. These molecules allow, by the incorporation of a near cognate amino acid at the PTC position, to restore the full protein expression, sometimes potentially modified [3,4]. The second conceivable therapeutic approach, which is commonly associated to the first one, involves molecules able to inhibit the NMD system in order to prevent mRNA degradation [5].
Both types of molecules have been previously identified and characterized, and, it has been shown that, sometimes, they even combine both mechanical approaches. The first class, the RT molecules, is organized in aminoglycosides and non-aminoglycosides compounds [6]. Aminoglycosides, such as G418 and gentamicin, induce translational readthrough by binding to the decoding center (A-site) of the rRNA, which is located in the small ribosomal subunit. This binding influences the ribosome translation fidelity [7]. Despite their high efficiency to bypass nonsense mutations, these molecules are unfortunately known to induce ototoxicity and nephrotoxicity [7,8,9]. To overcome this pitfall, new molecules, referred as aminoglycosides derivatives, were engineered. These molecules have been tested in several in vitro and in vivo models, and few of them have even reached clinical trials [10,11,12]. NMD inhibitors represent the second type of molecules, interfering to prevent PTC detection and the PTC-containing mRNA degradation by NMD. Moreover, some molecules, such as amlexanox, are known to harboring both activities, offering an interesting and comprehensive strategy [5]. Initially, amlexanox, was used to treat aphthous ulcers and asthma [13], currently it is employed in clinical trial for the treatment of type 2 diabetes and obesity (NCT01842282).
In neurodegenerative disorders, the readthrough strategy has been displayed only twice in the literature, both times regarding the central nervous system [14,15]. For disorders affecting the peripheral nervous system, such as peripheral neuropathies such as Charcot–Marie–Tooth (CMT) disease, one study has been reported [16]. CMT is the most common inherited pathology of the peripheral nervous system, affecting 1 in 2500 people worldwide. CMT disease is a heterogeneous group of sensory-motor disorders transmitted in either autosomal or X-linked mode of inheritance, in dominant or recessive forms. Clinical manifestations include muscular weakness and atrophy, foot deformities, such as pes cavus, and sometimes sensory loss and balance issues [17]. Electrophysiological studies help to distinguish demyelinating forms characterized by reduced nerve conduction velocity (NCV) and axonal CMT forms, with preserved NCV values. More than 80 genes are currently known to be responsible for CMT when mutated. While the complete duplication of PMP22 gene (Peripheral Myelin Protein 22) remains the main genetic cause of this pathology, PTCs have been detected in numerous CMT genes and we estimated that they represent around 10% of the detected mutations (personal data). Among the genes involved in CMT disease, GDAP1 (ganglioside-induced differentiation associated protein 1) appears to be one of the top 10 [18].
GDAP1 gene, located on chromosome 8 (8q13-q21) of the human genome, encodes a 358 aa protein [19], which is part of the glutathione S-transferases (GST) family as it possesses two GST domains (GST N and GST C) [19]. GSTs are involved in several cellular functions including cellular detoxification, regulation of cellular glutathione (GSH) levels, hormone biosynthesis and intracellular signaling [20]. A part from these two domains, GDAP1 protein presents two alpha helices (α4-α5 loop), a hydrophobic domain (HD1), and the transmembrane domain (TMD).
This protein is highly expressed in the outer mitochondrial membrane of neuronal cells [21,22] such as motor neurons (MNs) and neuronal progenitors (NPs). Today, the roles of GDAP1 are not completely solved and some of them remain to be explored. Recent studies have shown that GDAP1 would be involved in Golgi function [23], autophagy [24], and morphology of peroxisomes [25]. Nevertheless, most of its functions would be related to mitochondria activities, such as mitochondrial dynamics [26], oxidative stress [27], calcium homeostasis [28], mitochondria-endoplasmic reticulum [24] and mitochondria-lysosome membrane contact sites [29].
Although mitochondria are known as the powerhouse of ATP production, they are also responsible for metabolism, apoptosis and cell signaling [30]. They are essential for the survival and the proper function of neurons, and they play a key role in axonal transport [31]. At the structural level, mitochondria possess a double phospholipid membrane: the outer mitochondrial membrane (OMM) is in direct contact with the cytoplasm, while the inner mitochondrial membrane (IMM) differs from the OMM by its composition and the presence of lamellar invaginations called cristae [32]. The mitochondrial cristae extend into the matrix, where mitochondrial DNA (mtDNA) lies.
Correct architectural structure of cristae reflects a crucial mitochondria functioning [33]. It is well known that the cristae membrane constitute the site of oxidative phosphorylation, with the presence of a wide range of proteins, such as the electron transport chain (ETC) complexes and the ATP synthase [33,34]. Tregón et al. demonstrated that the loss of GDAP1 in mouse embryonic motor neurons (eMNs), induces the alteration of cristae morphology and deregulation of mitochondrial bioenergetic [35]. This ultrastructural abnormalities in mitochondrial cristae have been revealed also in motor neurons from CMT patients, even if no deficit in ATP production has been demonstrated [27].
More than 100 mutations in GDAP1 have been described as related to CMT [23] including, as the vast majority of them, autosomal recessive traits. In a general way, GDAP1 mutations can present an autosomal recessive (AR-GDAP1) or an autosomal dominant (AD-GDAP1) mode of inheritance, and they can be involved in axonal forms (CMT2K), or demyelinating forms (CMT4A), which makes their classification complicated. The clinical characteristics of patients with AR-GDAP1 mutations are more severe than patients with AD-GDAP1 and they develop first symptoms at a very early age in childhood [36,37,38]. In AR-GDAP1 CMT, PTC is present in almost 100% of the cases either in the homozygous state or associated with other kind of mutations such as missense mutations or frameshift mutations.
Therefore, in view of the lack of efficient therapeutic approaches, assessing readthrough molecules for patients displaying nonsense mutation in this gene represents a major challenge. However, these therapeutic developments are often limited by the lack of pertinent in vitro models. Our team developed few years ago a human induced pluripotent stem cell (hiPSC) in vitro model from CMT2H patient fibroblasts carrying the nonsense mutation c.581C>G in GDAP1 [27,39]. which corresponded to the first functional study of motor neurons mutated on GDAP1 and derived from human iPS cells. In this study, RT-PCR analysis showed that, in controls, GDAP1 mRNA is mainly expressed in neuronal cells (PNs and MNs) compared to other cell types (fibroblasts, iPSC). In the motor neurons of the patient, this level of GDAP1 mRNA is drastically reduced, probably degraded by the NMD system [27]. Morphological and functional studies have revealed in CMT patient motor neurons a decrease in cell viability associated with lipid dysfunction and the development of oxidative stress [27].
To our knowledge, no curative and efficient treatment has ever been proposed for CMT caused by nonsense mutations in GDAP1 gene. In an original way, this study propose to assess the effect of amlexanox as readthrough molecule combined to NMD inhibitors activity, on neuronal progenitors derived from CMT patients’ hiPSCs, harboring the homozygous c.581C>G nonsense mutation in GDAP1 (p.Ser194*). We demonstrated the rescue effects of this cellular treatment at molecular and protein levels, but also investigating their impact on mitochondrial morphology.
2. Results
2.1. Generation and Characterization of Neuronal Progenitors (NPs) from hiPSCs
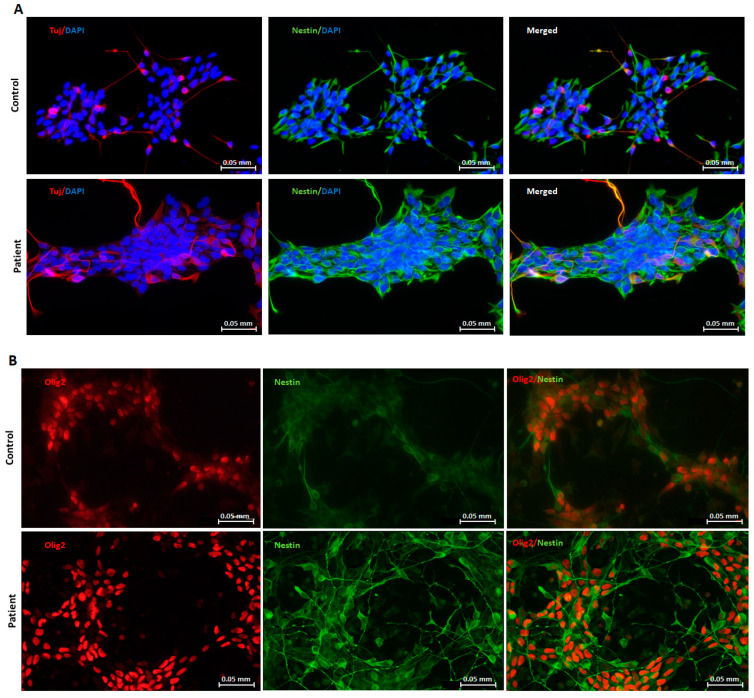
Two human induced pluripotent stem cell (hiPSC) clones, previously reprogrammed by our team [27], were used in this study: one harboring the homozygous GDAP1 nonsense mutation (c.581C>G, p.Ser194*) from a CMT patient, and the other used as control, free from any mutations in this same gene. To generate neuronal progenitors, we used a protocol adapted from Hörner et al. [40]. NPs were obtained in 10 days. Both control and patient clones expressed the typical neuronal progenitors’ markers Tuj1 and Nestin (Figure 1A,B). These patients and control cells also expressed Olig 2, a motor-neuron precursors’ marker. All together, these proteins’ expressions validate the cells’ orientation towards the motor neuron pathway.
Figure 1.
Characterization of NPs in control and patient clones at day 10 assessed by neural markers: (A) Nestin (green), Tuj1 (red), DAPI counterstain (blue). (B) Olig 2 (red) and Nestin (green). Scale bar = 0.05 mm.
2.2. Restoration of GDAP1 mRNA Expression in NPs
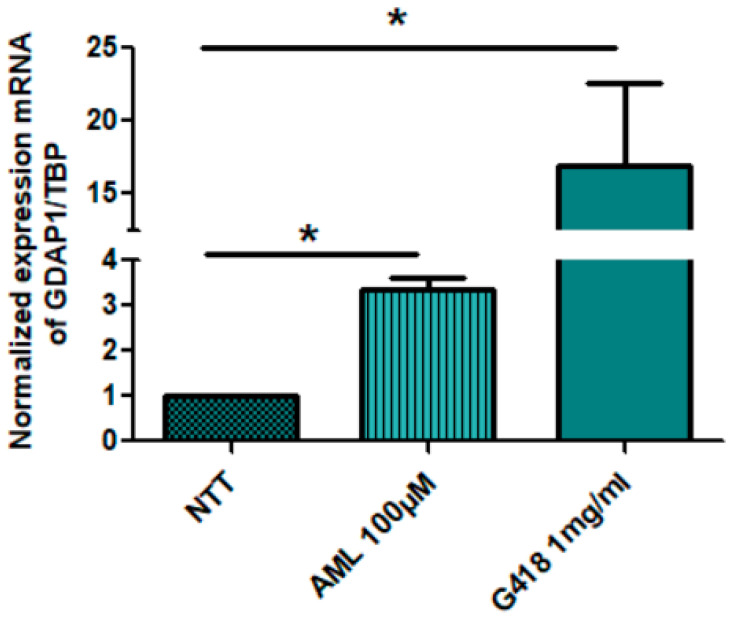
We have previously observed, by RT-qPCR, in neuronal cells (NPs and MNs) that patient mRNA levels of GDAP1 represented only 10 to 20% of mRNA levels in the control, suggesting that PTC-containing GDAP1 mRNA could be degraded by NMD [27]. To investigate the effect of drugs molecules on PTC-mRNA levels, patient NPs cells were treated for 20 h with 100 µM amlexanox, as recommended by Bordonaro et al. in 2019 and Cunha et al. in 2022 [41,42], or 1 mg/mL G418, a positive control, at day 4 after differentiation. With real-time PCR (RT-qPCR) (Figure 2), we highlighted that amlexanox, known as NMD inhibitor, led to a significant increase in GDAP1 mRNA to 3.35 fold compared to untreated NPs. As expected, G418 also allowed a significant increase in GDAP1 mRNA, but it cannot be used as a treatment for patients because of its toxicity. Our data supported that this mRNA carrying PTC is a substrate of the NMD system.
Figure 2.
Amlexanox restores GDAP1 mRNA expression in NPs carrying nonsense mutation (c.581C>G, p.Ser194*). The NPs of patient were treated with amlexanox (100 µM), or G418 (1 mg/mL), G418 was used as positive control. After 20 h of treatment, total RNAs were extracted. Reverse transcription and PCR was performed to measure the level of GDAP1 mRNA. To normalize the amount of PTC-containing mRNA, TBP was used as the reference gene. NTT: non treated cells; AML: amlexanox. The nonparametric Mann–Whitney test was performed; * p < 0.05. n = 3.
2.3. Restoration of GDAP1 Protein Expression in NPs
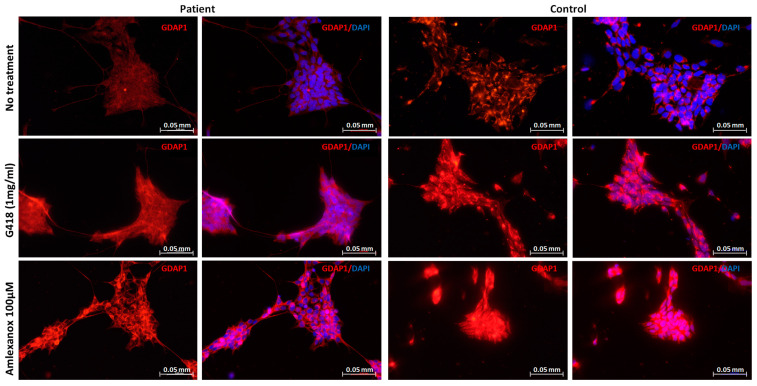
To determine whether amlexanox allows GDAP1 full-length protein re-expression, immunofluorescence staining was performed. NPs were incubated for 20 h with amlexanox (100 µM), or G418 (1 mg/mL). In the presence of amlexanox that display readthrough and NMD inhibitor activities, the protein expression was restored (Figure 3), such as in the presence of G418, the positive control.
Figure 3.
Amlexanox restores GDAP1 protein expression. NPs of patient and control were treated with amlexanox (100 µM); G418 was used as positive control. After 20 h of treatment, cells were fixed and stained with GDAP1 antibody (red), and DAPI (blue) for nuclei staining. Scale bar = 0.05 mm.
2.4. Amlexanox Effects on Mitochondrial Morphology in NPs
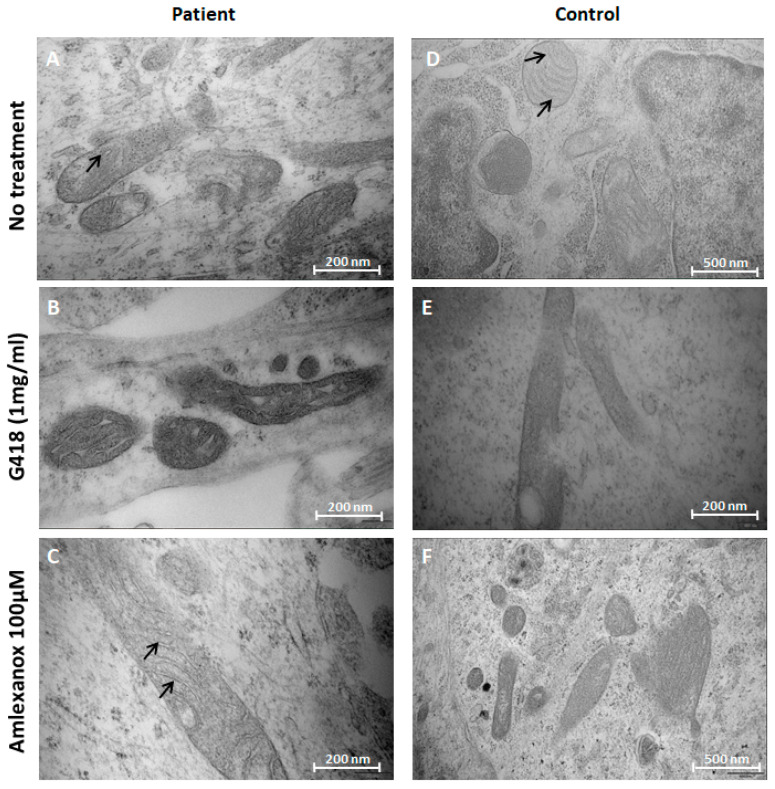
According to previous results on mitochondrial morphology alteration observed in patient MNs [27], we investigated the effect of amlexanox treatment on the structure of mitochondria using electron microscopy (EM) in NPs.
At mitochondrial level, no structural modifications emerged in control cells, in un-treated and treated conditions (Figure 4D–F). In contrast, in untreated patient NPs, we observed that mitochondrial cristae were disorganized and morphologically altered (Figure 4A). When treated by amlexanox, restoration of the inner mitochondrial morphology was observed, with regular cristae, in patient NPs (Figure 4C), suggesting that the re-expression of GDAP1 observed in Figure 3A would allow this restoration. However, G418 seemed to have a toxic effect on mitochondrial morphology, in both patient and control cells (Figure 4B,E) potentially explaining its in vivo toxicity already reported [7,8,9].
Figure 4.
Morphological analysis performed by EM, on mitochondria of NPs of patient on the left, and control on the right, in untreated, conditions (A,D). Untreated control and restoration of cristae, in patient PNs, are indicated by black arrows (C,D). Scale bar = 200 nm (A–C,E); scale bar = 500 nm (D,F).
3. Discussion
Cellular models allow screening a high number of molecules simultaneously. Unfortunately, a pertinent cellular model suitable for the evaluation of specific nonsense mutation is not always available. Cells of interest could be isolated from tissues of patients with genetic diseases, but, sometimes, the cell purification is difficult or impossible (such as neurons), leading to use the induced pluripotent stem cell (iPSC) strategy. Indeed, iPSCs, obtained from adult somatic cells (often fibroblasts), can be differentiated into all cell types including neurons. In our lab, we previously created hiPSCs from CMT patient fibroblasts carrying the nonsense mutation c.581C>G to characterize cells injuries and mechanisms involved in the pathogenicity [27]. In this study, we examine whether readthrough and NMD inhibitor molecules could rescue, in NPs, the native full-length GDAP1 protein and its associated regular mitochondrial phenotype, overcoming the effects of PTC (UGA) mutation in GDAP1 gene.
Our analysis has shown that, in NPs carrying the nonsense mutations in GDAP1, amlexanox (2-amino-7-isopropyl-5-oxo-5H-chromeno [2,3-b]pyridine-3-carboxylic acid) exhibited a promising beneficial effect, without any apparent cytotoxicity [5]. Thanks to its bivalent activity, amlexanox is known to both stabilize mRNA-containing nonsense mutation and promote PTC readthrough [5]. These activities have previously reported in several cellular models derived from patients with Duchenne Muscular Dystrophy (DMD), cancer and Cystic Fibrosis (CF) [5]. However, the mechanism combining both activities in cells remains unclear. Here, amlexanox has been shown to stabilize the mRNA of GDAP1 and to restore the synthesis of the GDAP1 protein. This molecule also induced a morphological improvement in mitochondria inner organization, evaluated by electron microscopy. These encouraging results indicate that amlexanox is able to display efficient readthrough inside neuronal cells, suggesting that it could be used as a strategy to treat neurological diseases induced by PTCs. This dual property has also been observed for the aminoglycoside G418 used as a positive control in our study. Unfortunately, this molecule exhibits toxic side effects that forced it to be withdrawn from clinical use in this indication. In order to reduce this toxicity, several G418 derivatives have been developed, such as ELX02 [43] which unfortunately, did not achieve statistical significance in a Phase 2 clinical trial for the treatment of CF.
In NPs, we demonstrated that both G418 and amlexanox offer the double property to stabilize mRNA, countering the involvement of the NMD system, and to induce readthrough activity. Previous studies are consistent with our results [5,44]. On the other hand, a recent study demonstrated that amlexanox was not able to increase the expression of the NEFL (Neurofilament light) protein in iPSC-derived motor neurons presenting a nonsense mutation in the related gene, which is involved in a CMT axonal form [16]. The efficiency discrepancy observed could be explained by the difference in dose used in treatment. Sainio et al. employed low concentration of the molecule (25 µM). Additionally, in the same study, they observed that this concentration was toxic for neuronal cells. In our model, no side effects on neuronal development were detected even with higher concentration (100 µM). Moreover, we decided to carry out the treatment on NPs, not directly on MNs which are more mature and specialized cells. The choice of 100 µM of amlexanox was also conducted by the positive results of other groups such as Bordonaro et al. in 2019 and Cunha et al. in 2022 who used it successfully on HCT-116 Colorectal Cancer Cells and on iPSC-derived ocular tissue models, respectively.
Our results show that amlexanox and G418 have an activity to bypass the UGA-PTC on the GDAP1 gene. Further investigations with adapted models, potentially created by the CRISPR-Cas9 technology, will be necessary to determine whether amlexanox displays the ability to bypass all three PTC on NPs. Indeed, amlexanox, was first characterized as an NMDI and its ability to correct the three forms of PTC has been highlighted on three different cellular models (DMD, CF, and cancers) [5]. Moreover, it has been previously reported that the success of readthrough strategy depends on the PTC nature but also on the nucleotide context in the vicinity of the PTC [45,46]. Indeed, a better readthrough efficiency of amlexanox was shown with an UGA-PTC followed by a C base compared to an UGA-PTC followed by a G base [45]. In our model, the UGA-PTC is followed by an A suggesting a relative activity. Furthermore, following readthrough activity, it is essential to inquire which amino acid is incorporated at the PTC position. Previous studies highlighted that, in case of readthrough, cysteine, arginine, and tryptophan were preferentially inserted at UGA-PTC [47]. Among these three amino acids, none can help, in our model, to fully restore the serine of the wild-type GDAP1 protein. However, the effect of amlexanox, in patient NPs, seems to maintain mitochondrial morphology, suggesting that the amino acid incorporated could induce a beneficial function.
In conclusion, these results underlined the proof of concept that this strategy combining NMD inhibitor and readthrough activities could be applied to CMT patients exhibited nonsense mutations in GDAP1, offering a restored, or a less severe, phenotype. However, it will be important to conduct further analysis to investigate the activity of amlexanox in other genes carrying PTC and mediated CMT disease.
4. Materials and Methods
4.1. Culture of Human iPS Cell Lines
The human induced pluripotent stem cells (hiPSCs) were obtained from dermal fibroblasts of a CMT patient and a control subject. The patient was a male child carrying the homozygous c.581C>G (p.Ser194*) mutation in GDAP1. He presented a severe axonal peripheral neuropathy and a polyvisceral disorder, which led to his death at the age of three. The control was a healthy 24-year-old male donor. Previous investigations allowed us to determine that the healthy individual did not show any signs of peripheral neuropathy, and did not carry any mutations in GDAP1 hiPSCs clones of both subjects were obtained as previously described [27]. hiPSCs were feeder-free cultured on a coating of diluted Geltrex (STEMCELL Technologies, Grenoble, France) and in mTeSR™1 medium (STEMCELL Technologies), at 37 °C, 5% CO2. The culture medium was changed daily. Passages were carried out every 5 to 8 days using StemMACS Passaging solution XF (Miltenyi Biotec, Bergisch Gladbach, Germany).
4.2. Differentiation of hiPSCs into Neuronal Progenitors (NPs)
The differentiation protocol was adapted from Hörner et al. [40]. Briefly, when hiPSCs reached 75% confluence, mTeSR™1 medium was replaced with differentiation medium composed by Neurobasal medium and Ko-DMEMF12 (Thermo Fisher SCIEN-TIFIC) (1:1) complemented with 1× B27 (Thermo Fisher SCIENTIFIC, Waltham, MA, USA), 1× N2 (Thermo Fisher SCIENTIFIC), 1% (non-essential amino acids(Thermo Fisher SCIENTIFIC), 1% Glutamax (Thermo Fisher SCIENTIFIC), extemporaneously supplemented with 2 µM SB-431542 (Tocris, Bioscience, Minneapolis, MN, USA), 2 µM dorsomorphin (Sigma-Aldrich, Merck, Saint-Louis, MO, USA), 3 µM CHIR 99,021 (Sigma-Aldrich, Merck), and 0.1 mM µM Ascorbic Acid (Tocris, Bio-Techne). The medium was changed daily for 5 days. To obtain NPs, cells were then dissociated using Accutase (STEMCELL Technologies, Saint Égrève, France) and plated at 1:6 ratio on Geltrex-coated 6-well plates. The culture medium was then supplemented with 2 µM SB-431542, 2 µM dorsomorphin, 1 µM CHIR 99021, 0.1 µM Ascorbic Acid, 0.1 µM retinoic acid (Sigma-Aldrich, Merck), 0.5 µM purmorphamin (Abcam, Waltham, MA, USA). The medium was changed daily for 5 days.
4.3. Amlexanox and G418 Preparation
Amlexanox was purchased from Sigma-Aldrich (ref: SML0517), and dissolved at 25 mM in DMSO. G418 was purchased from Life technologies, Waltham, MA, USA (ref: 11811031), and dissolved at 100 mg/mL in water.
4.4. Treatment of NPs
At day 10, NPs were seeded at a density of 500,000 cells in a 12-well plate, previously coated with Geltrex. Culture medium completed with small molecules (previously described in Section 4.2) was changed every day. Amlexanox and G418 were diluted in culture medium at 100 µM and 1 mg/mL, respectively, and added to cells (day 14). After 20 h, treatments were stopped by performing RNA extraction or slide fixation for Immunocytochemistry and Electron microscopy analysis.
4.5. RNA Extraction and Analysis
RNAs were extracted from patient and control NPs following the instructions of the RNeasy Plus Mini Kit (QIAGEN, Hilden, Germany). Reverse transcription of 1 μg of total RNA into cDNA was performed using the QuantiTect Reverse Transcription kit (QIAGEN) and following the manufacturer’s protocol. The analysis of gene expression was carried out by real-time quantitative PCR (qPCR). Primers were designed spanning the exon/exon boundaries, of exons 5 and 6 of the GDAP1 gene and exons 7 and 8 of the TATA box binding protein (TBP) gene, chosen as the reference gene. All primers used are reported in Table 1. Rotor-Gene SYBR Green PCR Kit (400) (QIAGEN) was used following the standard protocol. Reactions were performed on the Corbett Rotor-Gene 6000 Machine (QIAGEN). The Ct values of each real-time reaction were normalized, using TBP as endogenous control gene.
Table 1.
Primers of GDAP1 and TBP used.
| Gene | Primer Sequence | Direction | Exon |
|---|---|---|---|
| GDAP1 | GCTGCTTGATCATGACAATGT | F | 5–6 |
| CCTCTTCTGGGGTTTCTTCA | R | 5–6 | |
| TBP | ACAGGTGCTAAAGTCAGAGC | F | 7–8 |
| GAGGCAAGGGTACATGAGAG | R | 7–8 |
4.6. Immunocytochemistry
Expression of GDAP1 protein has been assessed by immunofluorescence (IF). NPs were seeded on glass slides coated with diluted Geltrex. On day 4, cells were treated with DMSO, or molecules, using preset concentrations according to literature. After 20 h of treatment, cells were fixed for 10 min with 4% paraformaldehyde (PFA) (Sigma-Aldrich, Merck), and then permeabilized with 0.1% Triton X-100 (Sigma-Aldrich, Merck) for one hour. Cells were incubated overnight at 4 °C with the primary antibody, prepared in 3% BSA (bovine serum albumin)/DPBS (Thermo Fisher SCIENTIFIC), then the next day with the secondary antibody (Alexa FluorTM 488 or Alexa FluorTM 594; Molecular Probes, Thermo-Fisher SCIENTIFIC, 1:2000 dilution), for 1 h at room temperature. Nuclei were stained with 2 µg/mL 4′,6′-diamidino-2-phénylindole dihydrochloride (DAPI) (Sigma-Aldrich, Merck). Images captures were acquired using a fluorescence microscope (Leica Microsystems, Wetzlar, Germany). All antibodies’ dilutions and references are reported in Supplementary Table S1. We chose to use Nestin as neural progenitors marker, Tuj-1 as a neuronal marker, and Olig 2, which can highlight motor-neuron precursors but also oligodendrocytes. When present together, these markers highlight only motor-neuron precursors.
4.7. Electron Microscopy
After fixation with 2.5% glutaraldehyde, NPs, were incubated in 2% OsO4 for 30 min (Euromedex, Souffelweyer-sheim, France), at room temperature. NPs, were than rinsed with distilled water. Dehydration was performed by a series of ethanol dilutions: 30%, 50%, 70%, 95%, and 100%., cells were embedded in Epon 812. Selected thick blocks were stained with uranyl acetate and lead citrate, and finally analyzed by Jeol 1011 electron microscope. Experiments for electron microscopy, were conducted in Neurology and Anatomic Pathology departments at University Hospital of Limoges.
4.8. Statistical Analysis
Statistical analyses were performed using the GraphPad Prism 8 software (GraphPad Software, Inc., San Diego, CA, USA). Data were presented as the mean ± SEM (Standard Error of the Mean). Data were compared using the nonparametric Mann–Whitney test (p < 0.05 was considered significant).
5. Conclusions
In conclusion, this study demonstrates for the first time the efficiency of amlexanox to restore the synthesis of GDAP1 protein and mitochondrial morphology in a NP model exhibiting an UGA-PTC mutation in the GDAP1 gene, mimicking the condition of patients with CMT. These results underline that combining readthrough and NMD inhibitors activities could be used in these genetic alterations involved in CMT, opening the way for future investigations and a potential therapy in patients without efficient treatment.
Acknowledgments
The authors thank “Agence Nationale de la Recherche”, “Region Nouvelle Aquitaine”, “Club 41-098”, “Lions Club St Yrieix La Perche”, “Rotary Club”, “Mairie de St Yrieix La Perche”, and the University Hospital of Limoges for their support. We thank the I-Stem institute (INSERM/UEVE UMR 861, AFM, Genopole, Evry, France) and the AFM-Téléthon institute for the training they offered to help us to generate hiPSCs, and. We would like to thank Paco Derouault, in the Bioinformatic platform at Limoges CHU for his informatic support.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/ph16071034/s1, Table S1: Antibodies dilutions and references.
Author Contributions
Conceptualization, F.F. and A.-S.L.; methodology, N.B., F.M., C.L., L.R., I.P. and P.-A.F.; validation, N.B., F.M., L.R., P.-A.F., F.F., F.L. and A.-S.L.; formal analysis, N.B., F.M., L.R., F.F. and A.-S.L.; investigation, N.B., F.M., C.L., L.R., A.N., I.P., P.-A.F., F.F., F.L. and A.-S.L.; resources, L.R. and F.L.; data curation, N.B., F.M., C.L., I.P., F.F. and A.-S.L.; writing—original draft preparation, N.B., F.M., C.L., F.F. and A.-S.L.; writing—review and editing, F.M., L.R., P.-A.F., F.F., F.L. and A.-S.L.; visualization, N.B., F.M., F.F., F.L. and A.-S.L.; supervision, F.F. and A.-S.L.; project administration, F.F. and A.-S.L.; funding acquisition, F.F. and A.-S.L. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
This study was conducted according to the guidelines of the Declaration of Helsinki, and approved by the Ethics Committee of Limoges University Hospital (N°384-2020-40, 10/07/2020).
Informed Consent Statement
Informed consent was obtained from all subjects involved in this study.
Data Availability Statement
The data presented in this study are available on request from the corresponding author.
Conflicts of Interest
The authors declare no conflict of interest.
Funding Statement
This research was funded by Agence Nationale de la Recherche, grant number “ANR-20-CE17-0026” and Région Nouvelle Aquitaine, grant number “AAP NA 2021 MEXICOS”.
Footnotes
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
References
- 1.Mort M., Ivanov D., Cooper D.N., Chuzhanova N.A. A Meta-Analysis of Nonsense Mutations Causing Human Genetic Disease. Hum. Mutat. 2008;29:1037–1047. doi: 10.1002/humu.20763. [DOI] [PubMed] [Google Scholar]
- 2.Guissart C., Mouzat K., Kantar J., Louveau B., Vilquin P., Polge A., Raoul C., Lumbroso S. Premature Termination Codons in SOD1 Causing Amyotrophic Lateral Sclerosis Are Predicted to Escape the Nonsense-Mediated MRNA Decay. Sci. Rep. 2020;10:20738. doi: 10.1038/s41598-020-77716-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Martins-Dias P., Romão L. Nonsense Suppression Therapies in Human Genetic Diseases. Cell. Mol. Life Sci. 2021;78:4677–4701. doi: 10.1007/s00018-021-03809-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Palma M., Lejeune F. Deciphering the Molecular Mechanism of Stop Codon Readthrough. Biol. Rev. 2021;96:310–329. doi: 10.1111/brv.12657. [DOI] [PubMed] [Google Scholar]
- 5.Gonzalez-Hilarion S., Beghyn T., Jia J., Debreuck N., Berte G., Mamchaoui K., Mouly V., Gruenert D.C., Déprez B., Lejeune F. Rescue of Nonsense Mutations by Amlexanox in Human Cells. Orphanet J. Rare Dis. 2012;7:58. doi: 10.1186/1750-1172-7-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gómez-Grau M., Garrido E., Cozar M., Rodriguez-Sureda V., Domínguez C., Arenas C., Gatti R.A., Cormand B., Grinberg D., Vilageliu L. Evaluation of Aminoglycoside and Non-Aminoglycoside Compounds for Stop-Codon Readthrough Therapy in Four Lysosomal Storage Diseases. PLoS ONE. 2015;10:e0135873. doi: 10.1371/journal.pone.0135873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Greenwood G.J. Neomycin Ototoxicity; Report of a Case. AMA Arch. Otolaryngol. 1959;69:390–397. doi: 10.1001/archotol.1959.00730030400002. [DOI] [PubMed] [Google Scholar]
- 8.Hock R., Anderson R.J. Prevention of Drug-Induced Nephrotoxicity in the Intensive Care Unit. J. Crit. Care. 1995;10:33–43. doi: 10.1016/0883-9441(95)90029-2. [DOI] [PubMed] [Google Scholar]
- 9.McWilliam S.J., Antoine D.J., Smyth R.L., Pirmohamed M. Aminoglycoside-Induced Nephrotoxicity in Children. Pediatr. Nephrol. Berl. Ger. 2017;32:2015–2025. doi: 10.1007/s00467-016-3533-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lai C.-H., Chun H.H., Nahas S.A., Mitui M., Gamo K.M., Du L., Gatti R.A. Correction of ATM Gene Function by Aminoglycoside-Induced Read-through of Premature Termination Codons. Proc. Natl. Acad. Sci. USA. 2004;101:15676–15681. doi: 10.1073/pnas.0405155101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Du M., Jones J.R., Lanier J., Keeling K.M., Lindsey J.R., Tousson A., Bebök Z., Whitsett J.A., Dey C.R., Colledge W.H., et al. Aminoglycoside Suppression of a Premature Stop Mutation in a Cftr-/- Mouse Carrying a Human CFTR-G542X Transgene. J. Mol. Med. Berl. Ger. 2002;80:595–604. doi: 10.1007/s00109-002-0363-1. [DOI] [PubMed] [Google Scholar]
- 12.Dunant P., Walter M.C., Karpati G., Lochmüller H. Gentamicin Fails to Increase Dystrophin Expression in Dystrophin-Deficient Muscle. Muscle Nerve. 2003;27:624–627. doi: 10.1002/mus.10341. [DOI] [PubMed] [Google Scholar]
- 13.Meng W., Dong Y., Liu J., Wang Z., Zhong X., Chen R., Zhou H., Lin M., Jiang L., Gao F., et al. A Clinical Evaluation of Amlexanox Oral Adhesive Pellicles in the Treatment of Recurrent Aphthous Stomatitis and Comparison with Amlexanox Oral Tablets: A Randomized, Placebo Controlled, Blinded, Multicenter Clinical Trial. Trials. 2009;10:30. doi: 10.1186/1745-6215-10-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Frew J., Baradaran-Heravi A., Balgi A.D., Wu X., Yan T.D., Arns S., Shidmoossavee F.S., Tan J., Jaquith J.B., Jansen-West K.R., et al. Premature Termination Codon Readthrough Upregulates Progranulin Expression and Improves Lysosomal Function in Preclinical Models of GRN Deficiency. Mol. Neurodegener. 2020;15:21. doi: 10.1186/s13024-020-00369-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee P., Martin N.T., Nakamura K., Azghadi S., Amiri M., Ben-David U., Perlman S., Gatti R.A., Hu H., Lowry W.E. SMRT Compounds Abrogate Cellular Phenotypes of Ataxia Telangiectasia in Neural Derivatives of Patient-Specific HiPSCs. Nat. Commun. 2013;4:1824. doi: 10.1038/ncomms2824. [DOI] [PubMed] [Google Scholar]
- 16.Sainio M.T., Rasila T., Molchanova S.M., Järvilehto J., Torregrosa-Muñumer R., Harjuhaahto S., Pennonen J., Huber N., Herukka S.-K., Haapasalo A., et al. Neurofilament Light Regulates Axon Caliber, Synaptic Activity, and Organelle Trafficking in Cultured Human Motor Neurons. Front. Cell Dev. Biol. 2021;9:820105. doi: 10.3389/fcell.2021.820105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bird T.D. Charcot-Marie-Tooth Hereditary Neuropathy Overview. In: Adam M.P., Mirzaa G.M., Pagon R.A., Wallace S.E., Bean L.J., Gripp K.W., Amemiya A., editors. GeneReviews®. University of Washington; Seattle, WA, USA: 1993. [Google Scholar]
- 18.Yoshimura A., Yuan J.-H., Hashiguchi A., Ando M., Higuchi Y., Nakamura T., Okamoto Y., Nakagawa M., Takashima H. Genetic Profile and Onset Features of 1005 Patients with Charcot-Marie-Tooth Disease in Japan. J. Neurol. Neurosurg. Psychiatry. 2019;90:195–202. doi: 10.1136/jnnp-2018-318839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Marco A., Cuesta A., Pedrola L., Palau F., Marín I. Evolutionary and Structural Analyses of GDAP1, Involved in Charcot-Marie-Tooth Disease, Characterize a Novel Class of Glutathione Transferase-Related Genes. Mol. Biol. Evol. 2004;21:176–187. doi: 10.1093/molbev/msh013. [DOI] [PubMed] [Google Scholar]
- 20.Hayes J.D., Flanagan J.U., Jowsey I.R. Glutathione Transferases. Annu. Rev. Pharmacol. Toxicol. 2005;45:51–88. doi: 10.1146/annurev.pharmtox.45.120403.095857. [DOI] [PubMed] [Google Scholar]
- 21.Niemann A., Huber N., Wagner K.M., Somandin C., Horn M., Lebrun-Julien F., Angst B., Pereira J.A., Halfter H., Welzl H., et al. The Gdap1 Knockout Mouse Mechanistically Links Redox Control to Charcot-Marie-Tooth Disease. Pt 3Brain J. Neurol. 2014;137:668–682. doi: 10.1093/brain/awt371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pedrola L., Espert A., Wu X., Claramunt R., Shy M.E., Palau F. GDAP1, the Protein Causing Charcot-Marie-Tooth Disease Type 4A, Is Expressed in Neurons and Is Associated with Mitochondria. Hum. Mol. Genet. 2005;14:1087–1094. doi: 10.1093/hmg/ddi121. [DOI] [PubMed] [Google Scholar]
- 23.Binięda K., Rzepnikowska W., Kolakowski D., Kaminska J., Szczepankiewicz A.A., Nieznańska H., Kochański A., Kabzińska D. Mutations in GDAP1 Influence Structure and Function of the Trans-Golgi Network. Int. J. Mol. Sci. 2021;22:914. doi: 10.3390/ijms22020914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cantarero L., Juárez-Escoto E., Civera-Tregón A., Rodríguez-Sanz M., Roldán M., Benítez R., Hoenicka J., Palau F. Mitochondria-Lysosome Membrane Contacts Are Defective in GDAP1-Related Charcot-Marie-Tooth Disease. Hum. Mol. Genet. 2021;29:3589–3605. doi: 10.1093/hmg/ddaa243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huber N., Guimaraes S., Schrader M., Suter U., Niemann A. Charcot-Marie-Tooth Disease-Associated Mutants of GDAP1 Dissociate Its Roles in Peroxisomal and Mitochondrial Fission. EMBO Rep. 2013;14:545–552. doi: 10.1038/embor.2013.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Niemann A., Wagner K.M., Ruegg M., Suter U. GDAP1 Mutations Differ in Their Effects on Mitochondrial Dynamics and Apoptosis Depending on the Mode of Inheritance. Neurobiol. Dis. 2009;36:509–520. doi: 10.1016/j.nbd.2009.09.011. [DOI] [PubMed] [Google Scholar]
- 27.Miressi F., Benslimane N., Favreau F., Rassat M., Richard L., Bourthoumieu S., Laroche C., Magy L., Magdelaine C., Sturtz F., et al. GDAP1 Involvement in Mitochondrial Function and Oxidative Stress, Investigated in a Charcot-Marie-Tooth Model of HiPSCs-Derived Motor Neurons. Biomedicines. 2021;9:945. doi: 10.3390/biomedicines9080945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pla-Martín D., Rueda C.B., Estela A., Sánchez-Piris M., González-Sánchez P., Traba J., de la Fuente S., Scorrano L., Renau-Piqueras J., Alvarez J., et al. Silencing of the Charcot-Marie-Tooth Disease-Associated Gene GDAP1 Induces Abnormal Mitochondrial Distribution and Affects Ca2+ Homeostasis by Reducing Store-Operated Ca2+ Entry. Neurobiol. Dis. 2013;55:140–151. doi: 10.1016/j.nbd.2013.03.010. [DOI] [PubMed] [Google Scholar]
- 29.Cantarero L., García-Vargas G., Hoenicka J., Palau F. Differential Effects of Mendelian GDAP1 Clinical Variants on Mitochondria-Lysosome Membrane Contacts Sites. Biol. Open. 2023;12:bio059707. doi: 10.1242/bio.059707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Labbé K., Murley A., Nunnari J. Determinants and Functions of Mitochondrial Behavior. Annu. Rev. Cell Dev. Biol. 2014;30:357–391. doi: 10.1146/annurev-cellbio-101011-155756. [DOI] [PubMed] [Google Scholar]
- 31.Li S., Xiong G.-J., Huang N., Sheng Z.-H. The Crosstalk of Energy Sensing and Mitochondrial Anchoring Sustains Synaptic Efficacy by Maintaining Presynaptic Metabolism. Nat. Metab. 2020;2:1077–1095. doi: 10.1038/s42255-020-00289-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mannella C.A. Structure and Dynamics of the Mitochondrial Inner Membrane Cristae. Biochim. Biophys. Acta BBA—Mol. Cell Res. 2006;1763:542–548. doi: 10.1016/j.bbamcr.2006.04.006. [DOI] [PubMed] [Google Scholar]
- 33.Wiedemann N., Pfanner N. Mitochondrial Machineries for Protein Import and Assembly. Annu. Rev. Biochem. 2017;86:685–714. doi: 10.1146/annurev-biochem-060815-014352. [DOI] [PubMed] [Google Scholar]
- 34.Gilkerson R.W., Selker J.M.L., Capaldi R.A. The Cristal Membrane of Mitochondria Is the Principal Site of Oxidative Phosphorylation. FEBS Lett. 2003;546:355–358. doi: 10.1016/S0014-5793(03)00633-1. [DOI] [PubMed] [Google Scholar]
- 35.Civera-Tregón A., Domínguez L., Martínez-Valero P., Serrano C., Vallmitjana A., Benítez R., Hoenicka J., Satrústegui J., Palau F. Mitochondria and Calcium Defects Correlate with Axonal Dysfunction in GDAP1-Related Charcot-Marie-Tooth Mouse Model. Neurobiol. Dis. 2021;152:105300. doi: 10.1016/j.nbd.2021.105300. [DOI] [PubMed] [Google Scholar]
- 36.Baxter R.V., Ben Othmane K., Rochelle J.M., Stajich J.E., Hulette C., Dew-Knight S., Hentati F., Ben Hamida M., Bel S., Stenger J.E., et al. Ganglioside-Induced Differentiation-Associated Protein-1 Is Mutant in Charcot-Marie-Tooth Disease Type 4A/8q21. Nat. Genet. 2002;30:21–22. doi: 10.1038/ng796. [DOI] [PubMed] [Google Scholar]
- 37.Cuesta A., Pedrola L., Sevilla T., García-Planells J., Chumillas M.J., Mayordomo F., LeGuern E., Marín I., Vílchez J.J., Palau F. The Gene Encoding Ganglioside-Induced Differentiation-Associated Protein 1 Is Mutated in Axonal Charcot-Marie-Tooth Type 4A Disease. Nat. Genet. 2002;30:22–25. doi: 10.1038/ng798. [DOI] [PubMed] [Google Scholar]
- 38.Sevilla T., Jaijo T., Nauffal D., Collado D., Chumillas M.J., Vilchez J.J., Muelas N., Bataller L., Domenech R., Espinós C., et al. Vocal Cord Paresis and Diaphragmatic Dysfunction Are Severe and Frequent Symptoms of GDAP1-Associated Neuropathy. Brain. 2008;131:3051–3061. doi: 10.1093/brain/awn228. [DOI] [PubMed] [Google Scholar]
- 39.Benhabiles H., Gonzalez-Hilarion S., Amand S., Bailly C., Prévotat A., Reix P., Hubert D., Adriaenssens E., Rebuffat S., Tulasne D., et al. Optimized Approach for the Identification of Highly Efficient Correctors of Nonsense Mutations in Human Diseases. PLoS ONE. 2017;12:e0187930. doi: 10.1371/journal.pone.0187930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hörner S.J., Couturier N., Bruch R., Koch P., Hafner M., Rudolf R. HiPSC-Derived Schwann Cells Influence Myogenic Differentiation in Neuromuscular Cocultures. Cells. 2021;10:3292. doi: 10.3390/cells10123292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bordonaro M., Lazarova D. Amlexanox and UPF1 Modulate Wnt Signaling and Apoptosis in HCT-116 Colorectal Cancer Cells. J. Cancer. 2019;10:287–292. doi: 10.7150/jca.28331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cunha D.L., Eintracht J., Harding P., Zhou J.H., Moosajee M. Restoration of Functional PAX6 in Aniridia Patient IPSC-Derived Ocular Tissue Models Using Repurposed Nonsense Suppression Drugs. bioRxiv. 2022 doi: 10.1101/2022.10.12.511600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.de Poel E., Spelier S., Suen S.W.F., Kruisselbrink E., Graeber S.Y., Mall M.A., Weersink E.J.M., van der Eerden M.M., Koppelman G.H., van der Ent C.K., et al. Functional Restoration of CFTR Nonsense Mutations in Intestinal Organoids. J. Cyst. Fibros. 2022;21:246–253. doi: 10.1016/j.jcf.2021.09.020. [DOI] [PubMed] [Google Scholar]
- 44.Correa-Cerro L.S. DHCR7 Nonsense Mutations and Characterisation of MRNA Nonsense Mediated Decay in Smith-Lemli-Opitz Syndrome. J. Med. Genet. 2005;42:350–357. doi: 10.1136/jmg.2004.022749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Atanasova V.S., Jiang Q., Prisco M., Gruber C., Piñón Hofbauer J., Chen M., Has C., Bruckner-Tuderman L., McGrath J.A., Uitto J., et al. Amlexanox Enhances Premature Termination Codon Read-Through in COL7A1 and Expression of Full Length Type VII Collagen: Potential Therapy for Recessive Dystrophic Epidermolysis Bullosa. J. Investig. Dermatol. 2017;137:1842–1849. doi: 10.1016/j.jid.2017.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Manuvakhova M., Keeling K., Bedwell D.M. Aminoglycoside Antibiotics Mediate Context-Dependent Suppression of Termination Codons in a Mammalian Translation System. RNA. 2000;6:1044–1055. doi: 10.1017/S1355838200000716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Roy B., Friesen W.J., Tomizawa Y., Leszyk J.D., Zhuo J., Johnson B., Dakka J., Trotta C.R., Xue X., Mutyam V., et al. Ataluren Stimulates Ribosomal Selection of Near-Cognate TRNAs to Promote Nonsense Suppression. Proc. Natl. Acad. Sci. USA. 2016;113:12508–12513. doi: 10.1073/pnas.1605336113. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data presented in this study are available on request from the corresponding author.