Abstract
The family Poxviridae contains two subfamilies: the Entomopoxvirinae (poxviruses of insects) and the Chordopoxvirinae (poxviruses of vertebrates). Here we present the first characterization of the genome of an entomopoxvirus (EPV) which infects the North American migratory grasshopper Melanoplus sanguinipes and other important orthopteran pests. The 236-kbp M. sanguinipes EPV (MsEPV) genome consists of a central coding region bounded by 7-kbp inverted terminal repeats and contains 267 open reading frames (ORFs), of which 107 exhibit similarity to previously described genes. The presence of genes not previously described in poxviruses, and in some cases in any other known virus, suggests significant viral adaptation to the arthropod host and the external environment. Genes predicting interactions with host cellular mechanisms include homologues of the inhibitor of apoptosis protein, stress response protein phosphatase 2C, extracellular matrixin metalloproteases, ubiquitin, calcium binding EF-hand protein, glycosyltransferase, and a triacylglyceride lipase. MsEPV genes with putative functions in prevention and repair of DNA damage include a complete base excision repair pathway (uracil DNA glycosylase, AP endonuclease, DNA polymerase β, and an NAD+-dependent DNA ligase), a photoreactivation repair pathway (cyclobutane pyrimidine dimer photolyase), a LINE-type reverse transcriptase, and a mutT homologue. The presence of these specific repair pathways may represent viral adaptation for repair of environmentally induced DNA damage. The absence of previously described poxvirus enzymes involved in nucleotide metabolism and the presence of a novel thymidylate synthase homologue suggest that MsEPV is heavily reliant on host cell nucleotide pools and the de novo nucleotide biosynthesis pathway. MsEPV and lepidopteran genus B EPVs lack genome colinearity and exhibit a low level of amino acid identity among homologous genes (20 to 59%), perhaps reflecting a significant evolutionary distance between lepidopteran and orthopteran viruses. Divergence between MsEPV and the Chordopoxvirinae is indicated by the presence of only 49 identifiable chordopoxvirus homologues, low-level amino acid identity among these genes (20 to 48%), and the presence in MsEPV of 43 novel ORFs in five gene families. Genes common to both poxvirus subfamilies, which include those encoding enzymes involved in RNA transcription and modification, DNA replication, protein processing, virion assembly, and virion structural proteins, define the genetic core of the Poxviridae.
The Poxviridae family consists of large cytoplasmic double-stranded DNA viruses separated into two subfamilies: the Entomopoxvirinae (poxviruses of insects) and the Chordopoxvirinae (poxviruses of vertebrates) (130). The entomopoxvirus (EPV) subfamily is divided into three genera based primarily on differences in viral host range and virion morphology. Genus A viruses infect coleopterans, genus B viruses infect lepidopterans and orthopterans, and genus C viruses infect dipterans (7, 54). Insects are the only known hosts of EPVs, and observed viral host range is restricted to one or a few related species (7).
A detailed genetic comparison of the two subfamilies has been limited by the lack of information on EPV genomics. Restriction endonuclease analysis and DNA cross-hybridization studies have, however, suggested large genomic differences between lepidopteran group B EPVs and chordopoxviruses (ChPVs) (63, 102). Limited gene comparisons have also shown that at certain loci, lepidopteran EPV gene order is distinct from that of ChPVs (66, 175) and that the degree of amino acid similarity between EPV and ChPV enzymatic and structural proteins is low (6, 66, 175).
EPVs have been studied mainly because they are potential insect biocontrol agents and expression vectors (7, 41, 180). However, EPV genomic organization and molecular mechanisms of replication, pathogenesis, and host range are largely unknown. Few EPV genes have been characterized in detail, and additional information on the viral genome and virus-host interactions is necessary to further develop and improve these viruses as biocontrol agents (6, 175).
Melanoplus sanguinipes EPV (MsEPV) infects the North American migratory grasshopper M. sanguinipes, an agriculturally important insect pest, as well as two related grasshopper species (M. differentialis and M. packardii), the desert locust (Schistocerca gregaria) (179), and the African migratory locust (Locusta migratoria) (82, 108). MsEPV produces a large ellipsoid virion (250 to 300 nm in length) with a rectangular core. Grasshopper nymphs are infected by MsEPV after oral ingestion of virus-containing occlusion bodies. Presumably, the virus infects cells of the midgut prior to generalization of infection to the major target organ, the fat body (40, 72). Infection results in a slow and debilitating disease with high mortality, occurring 25 to 30 days postinfection. High titers of infectious spheroids, which can number up to 8 × 107 per grasshopper, are evident at 12 to 15 days postinfection (72, 127, 208).
Here, we present a genomic analysis of MsEPV. These data represent the first characterization of an EPV genome; further, they define the genetic core of the Poxviridae.
MATERIALS AND METHODS
MsEPV DNA isolation and cloning.
MsEPV genomic viral DNA was extracted from gradient-purified viral occlusion bodies obtained from the North American migratory grasshopper, M. sanguinipes, as previously described (102). Random DNA fragments were obtained by incomplete enzymatic digestion with Tsp509I endonuclease (New England Biolabs, Beverly, Mass.). DNA fragments of 1.5 to 2.5 kbp were isolated after separation on agarose gels, cloned into the dephosphorylated EcoRI site of plasmid pUC19, and grown in Escherichia coli DH10B cells (GIBCO BRL, Gaithersburg, Md.). Double-stranded plasmids pUC19 was purified by the rapid boiling method (162). DNA templates were sequenced from both ends with M13 forward and reverse primers, using dideoxy chain terminator sequencing chemistries (163) and an Applied Biosystems PRISM 377 automated DNA sequencer (Perkin-Elmer, Foster City, Calif.). Applied Biosystems sequence software (version 3.0) was used for lane tracking and trace extraction. Chromatogram traces were base called with Phred software (43), which also produced a quality file containing a predicted probability of error at each base position. The sequences were assembled with Phrap software (42), using the quality files and default settings to produce a consensus sequence. Subsequent manual editing was done with the Consed sequence editor (56). The final DNA consensus sequence represented on average an eightfold redundancy at each base position.
MsEPV genome organization was confirmed by comparing observed BamHI, HindIII, and ScaI restriction fragments to the consensus sequence data. Right and left ends of the genome were confirmed by using AluI, BglII, ClaI, NheI, PmlI, and Sau3AI restriction digests (New England Biolabs).
DNA sequence analysis.
Genome DNA composition, structure, repeats, and restriction enzyme patterns were analyzed with the Wisconsin Genetics Computer Group (GCG) programs (33). Open reading frames (ORFs) consisting of more than 60 amino acids, and with a methionine start codon and codon usage consistent with known EPV gene sequences from GenBank were considered to be protein encoding (176, 177). DNA and protein comparisons with entries in genetic databases were performed with FASTA (141) and BLAST programs (2). Multiple sequence alignments were performed with the CLUSTAL (74, 187), GCG Pileup (33), MSA (116), and Macaw (166) computer programs. Motif searches were done against the SBASE release 5 (44) and Prosite release 14 (9) databases. Other protein patterns were determined with the profile scanning programs PROBE (134), GIBBS, ASSET (133), and Hidden Markov Model (37, 38). Prediction of transmembrane domains was accomplished with MEMSAT (84, 85) and TopPred (24) software. Signal peptides were predicted with the program Sigseq (195). Physical descriptions of proteins were obtained by using SAPS software (19). Phylogenetic analysis was done with the Phylip computer programs (45) and Phylo_Win graphic tools (51). Gene families were identified by using the following criteria: (i) similarity based on BLAST scores (3) and pairwise clustering with CLUS (94); (ii) cluster profiles produced by PROBE (134); (iii) statistical significance, determined by using PRDF (142); and (iv) the presence of unique motifs, determined by using Pileup, GIBBS, and MACAW (33, 88, 133, 166).
Abbreviations.
Organisms have been abbreviated as follows: Amsacta moorei EPV, AmEPV; Autographa californica nuclear polyhedrosis virus, AcNPV; African swine fever virus, ASFV; Choristoneura biennis EPV, CbEPV; Choristoneura fumiferana EPV, CfEPV; cowpox virus, CPV; fowlpox virus, FPV; Heliothis armigera EPV, HaEPV; Melolontha melolontha EPV, MmEPV; Molluscum contagiosum virus, MCV; Orgyia pseudosugata NPV, OpNPV; rabbit fibroma virus, RFV; swinepox virus, SPV; and variola virus, VAR.
Nucleotide sequence accession number.
The MsEPV genome sequence has been deposited in GenBank under accession no. AF063866.
RESULTS AND DISCUSSION
Organization of the MsEPV genome.
The MsEPV genome was assembled into a contiguous sequence of 236,120 bp, similar in size to a previous estimate of 235 kbp (108). Because genomic termini were not sequenced, the left-most nucleotide of the assembled sequence was arbitrarily designated base no. 1.
The nucleotide composition is 81.7% A+T, as previously estimated for MsEPV (101), and is uniformly distributed over the entire length of the MsEPV genome. The total amino acid composition of all MsEPV ORFs reflects a bias for residues with A+T-rich codons. As previously noted in DNAs of other A+T-rich organisms (182), MsEPV preferentially encodes the 6 amino acids specified by codons exclusively composed of A and/or T (Lys, Asn, Ile, Leu, Tyr, and Phe). These amino acids represent the majority (61%) of all those encoded.
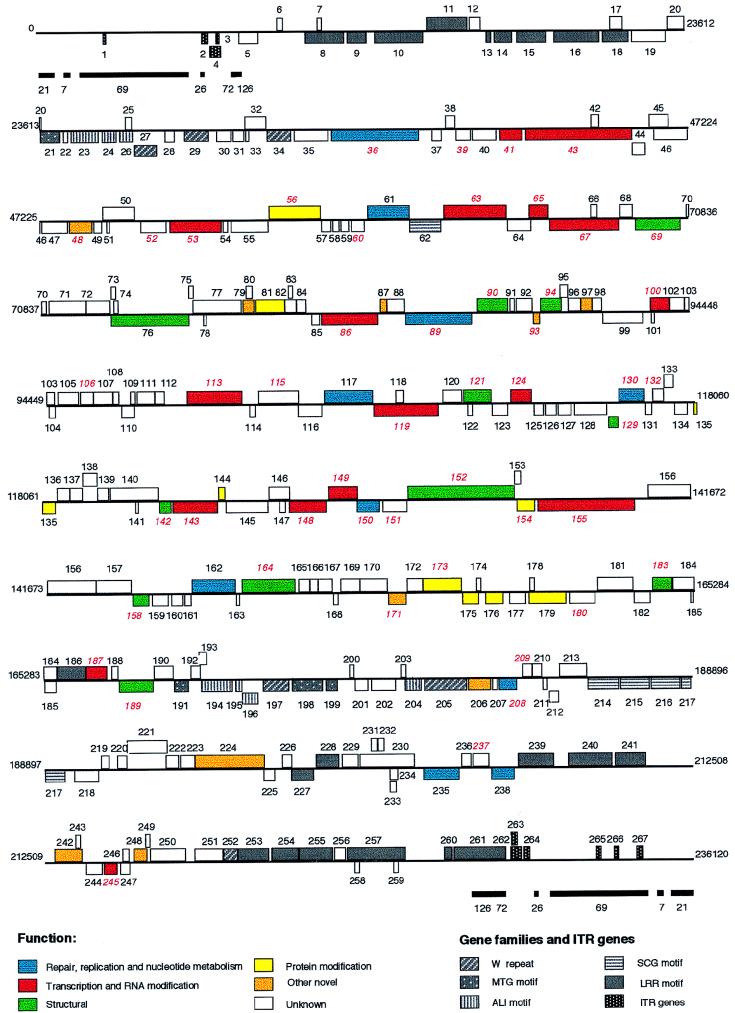
Two hundred and sixty-seven ORFs defined as methionine-initiated ORFs of greater than 60 amino acids are present (Fig. 1). The high A+T content, which results in a paucity of start codons (2.1%) and a large number of stop codons (14.5%), facilitates ORF identification. Predicted ORFs represent a 99% coding density, with an average ORF length of 854 nucleotides. Forty-four ORFs overlap other ORFs, and 28 smaller ORFs are completely contained within larger ORFs. Only 98 of the 267 MsEPV ORFs have been assigned a putative similarity or function based on homologies with other viral or cellular genes (Tables 1 and 2). Of the 155 most centrally located ORFs (MSV036 to MSV190), 45 (29%) are ChPV homologues. In contrast, of 112 ORFs in the terminal genomic regions (MSV001 to MS035 and MSV191 to MSV267), only 4 (3.5%) are identifiable ChPV homologues (Fig. 1).
FIG. 1.
Linear map of the MsEPV genome. ORFs are numbered from left to right based on initiation codon position. ORFs transcribed to the right are located above the horizontal lines; ORFs transcribed to the left are below. ChPV homologues are indicated with red italicized numbers. Genes with similar functions and members of gene families are colored according to the figure key. ITRs are represented as heavy black bars underneath the ORF map (numbers indicate sizes [in base pairs] of nucleotide repeats).
TABLE 1.
MsEPV ORFs
| ORF | Position (length, aa)a | Best matchb | BlastP score | % Identity | Length, aaa | Predicted structure and/ or functionc | Promoter typed |
|---|---|---|---|---|---|---|---|
| MSV001 | 2631–2446 (62) | ITR, 62 aa | |||||
| MSV002 | 6320–6054 (89) | ITR, 89 aa | |||||
| MSV003 | 6756–6577 (60) | ITR, 60 aa, TM | |||||
| MSV004 | 6814–6350 (155) | ITR, 155 aa | E | ||||
| MSV005 | 8171–7434 (246) | TM | |||||
| MSV006 | 8831–9055 (75) | ||||||
| MSV007 | 10282–10473 (64) | ||||||
| MSV008 | 11327–9828 (500) | [P28854, AmEPV ORF Q3] | 333 | 40 | 205 | LRR | E |
| MSV009 | 12138–11377 (254) | [P28854, AmEPV ORF Q3] | 286 | 38 | 204 | LRR | E |
| MSV010 | 14199–12367 (611) | [P28854, AmEPV ORF Q3] | 298 | 35 | 218 | LRR | E |
| MSV011 | 14293–15807 (505) | [P28854, AmEPV ORF Q3] | 298 | 39 | 226 | LRR | E |
| MSV012 | 15855–16244 (130) | E | |||||
| MSV013 | 16654–16439 (72) | [P28854, AmEPV ORF Q3] | 77 | 36 | 71 | LRR | E |
| MSV014 | 17462–16731 (244) | [P28854, AmEPV ORF Q3] | 256 | 42 | 171 | LRR, TM | E |
| MSV015 | 18692–17535 (386) | [P28854, AmEPV ORF Q3] | 325 | 38 | 207 | LRR | E |
| MSV016 | 20651–18936 (572) | [P28854, AmEPV ORF Q3] | 315 | 40 | 218 | LRR | |
| MSV017 | 20990–21430 (147) | ||||||
| MSV018 | 21717–20701 (339) | [P28854, AmEPV ORF Q3] | 315 | 37 | 213 | LRR | |
| MSV019 | 23074–21764 (437) | ||||||
| MSV020 | 23121–23645 (175) | E | |||||
| MSV021 | 24438–23659 (260) | AF003534, Chilo iridescent virus ORF 074R | 128 | 22 | 193 | MTG motif | E |
| MSV022 | 24696–24478 (73) | E | |||||
| MSV023 | 25850–24756 (365) | L44593, bacteriophage BK5-T ORF266 | 135 | 27 | 144 | ALI motif | E |
| MSV024 | 26483–25875 (203) | AF003534, Chilo iridescent virus OFR 011L | 239 | 37 | 154 | ALI motif | |
| MSV025 | 26794–26997 (68) | TM | |||||
| MSV026 | 27095–26526 (190) | AF003534, Chilo iridescent virus ORF 011L | 201 | 32 | 154 | ALI motif | |
| MSV027 | 27979–27089 (297) | M96361, AcNPV 41.6-kDa protein | 190 | 32 | 148 | Tryptophan repeat | E |
| MSV028 | 28594–28172 (141) | AF003534, Chilo iridescent virus ORF 011L | 125 | 28 | 98 | E | |
| MSV029 | 29846–28881 (322) | M96361, AcNPV 41.6-kDa protein | 133 | 29 | 167 | Tryptophan repeat | E |
| MSV030 | 30674–30081 (198) | E | |||||
| MSV031 | 31112–30690 (141) | TM | L | ||||
| MSV032 | 31140–31895 (252) | SP | |||||
| MSV033 | 31309–31112 (66) | TM, SP | |||||
| MSV034 | 32866–31925 (314) | M96361, AcNPV 41.6-kDa protein | 159 | 28 | 150 | Tryptophan repeat | E |
| MSV035 | 34198–32882 (439) | ||||||
| MSV036 | 37490–34254 (1079) | [P30319, CbEPV DNA polymerase] (E9L) | 1,668 | 40 | 958 | DNA polymerase | E |
| MSV037 | 38330–37944 (129) | E | |||||
| MSV038 | 38456–38821 (122) | L | |||||
| MSV039 | 39408–38830 (193) | [U60315, MCV MCV062R protein] (G6R) | 117 | 25 | 127 | L | |
| MSV040 | 40359–39412 (316) | E | |||||
| MSV041 | 41293–40409 (295) | [L22579, VAR poly(A) polymerase regulatory] (J3R) | 299 | 35 | 244 | Poly(A) polymerase (small subunit) PAPS | E |
| MSV042 | 43777–44037 (87) | ||||||
| MSV043 | 45280–41324 (1319) | [P20504, vaccinia virus RNA polymerase RPO147] (J6R) | 1,313 | 30 | 1,185 | RNA polymerase, RPO147 | E |
| MSV044 | 45770–45276 (165) | L | |||||
| MSV045 | 45881–46579 (233) | TM, SP | |||||
| MSV046 | 47237–45897 (447) | TM, SP | E | ||||
| MSV047 | 48296–47304 (331) | TM, SP | E | ||||
| MSV048 | 49188–48325 (288) | D12680, Rhizopus niveus lipase | 142 | 29 | 158 | Lipase, TM | L |
| MSV049 | 49565–49218 (116) | TM | L | ||||
| MSV050 | 49581–50717 (379) | TM | L | ||||
| MSV051 | 49855–49661 (65) | TM | |||||
| MSV052 | 51931–50897 (345) | [P20998, vaccinia virus A23R protein] (A23R) | 116 | 26 | 324 | ||
| MSV053 | 53902–51962 (647) | [P24486, CbEPV NPH-1] (D11L) | 1,997 | 58 | 648 | Nucleoside phosphohydrolase, NPH-1 | L |
| MSV054 | 54163–53918 (82) | ||||||
| MSV055 | 55645–54248 (466) | TM | L | ||||
| MSV056 | 55659–57545 (629) | [X76267, VAR (Garcia 66) F2L] (G1L) | 98 | 23 | 237 | Metalloprotease, TM | L |
| MSV057 | 57947–57552 (132) | L | |||||
| MSV058 | 58207–57947 (87) | L | |||||
| MSV059 | 58573–58241 (111) | E | |||||
| MSV060 | 59204–58623 (194) | [P20496, vaccinia virus H2 late protein] (H2R) | 316 | 37 | 183 | TM | L |
| MSV061 | 59208–60800 (531) | Z83109, Caenorhabditis elegans F44G3.3 gene product | 308 | 28 | 338 | RT | |
| MSV062 | 61995–60793 (401) | SCG motif, TM | L | ||||
| MSV063 | 62037–64316 (760) | [X76265, VAR 82-kDa subunit] (A7L) | 410 | 25 | 583 | Early transcription factor, VETFS | L |
| MSV064 | 65158–64319 (280) | E | |||||
| MSV065 | 65228–65881 (218) | [P07609, vaccinia virus late transactivator protein] (A2L) | 194 | 22 | 198 | Late transcription factor, VLTF-3 | L |
| MSV066 | 67390–67614 (75) | TM, SP | |||||
| MSV067 | 68454–65875 (860) | [P20979, vaccinia virus mRNA capping enzyme, large subunit] (D1R) | 639 | 28 | 750 | mRNA capping | L |
| MSV068 | 68466–68945 (160) | TM, SP | L | ||||
| MSV069 | 70676–68952 (575) | [U44841, HaEVP rifampicin resistance protein] (D13L) | 1,697 | 54 | 572 | Morphogenesis, rifampin resistance | L |
| MSV070 | 70701–71036 (112) | ||||||
| MSV071 | 71179–72504 (442) | ||||||
| MSV072 | 72504–73397 (298) | L | |||||
| MSV073 | 73510–73707 (66) | ||||||
| MSV074 | 73619–73843 (75) | ||||||
| MSV075 | 76302–76490 (63) | ||||||
| MSV076 | 76309–73400 (970) | [P29815, AmEPV spheroidin] | 439 | 25 | 707 | Spheroidin | L |
| MSV077 | 76411–78204 (598) | TM | E | ||||
| MSV078 | 76944–76744 (67) | TM | |||||
| MSV079 | 78257–78670 (138) | 70 | 26 | 88 | C2H2 zinc finger | L | |
| MSV080 | 78363–78590 (76) | TM | |||||
| MSV081 | 78693–79763 (357) | P36993, Mus musculus PP2C, beta isoform | 329 | 25 | 323 | Protein phosphatase, PP2C | L |
| MSV082 | 79775–80191 (139) | L | |||||
| MSV083 | 79878–80060 (61) | ||||||
| MSV084 | 80206–80574 (123) | E | |||||
| MSV085 | 81072–80719 (118) | TM | L | ||||
| MSV086 | 83232–81082 (717) | [P20502, vaccinia virus RNA helicase] (I8R) | 776 | 33 | 651 | RNA helicase, NPH-II | L |
| MSV087 | 83258–83485 (76) | D45892, Neurospora crassa thioredoxin | 60 | 26 | 67 | Thioredoxin | L |
| MSV088 | 83508–84122 (205) | L | |||||
| MSV089 | 86650–84149 (834) | [G41700, RFV C5 protein] (D5R) | 548 | 27 | 643 | Nucleic acid-dependent NTPase, TM | |
| MSV090 | 86786–87925 (380) | [U60315, MCV MC121L protein] (A16L) | 389 | 29 | 274 | Potential membrane protein, TM | |
| MSV091 | 87971–88189 (73) | E | |||||
| MSV092 | 88220–88807 (196) | L | |||||
| MSV093 | 89112–88792 (107) | [P33821, VAR E10R protein] (E10R) | 194 | 45 | 90 | Potential redox, yeast ERV1 | L |
| MSV094 | 89128–89850 (241) | [P24361, vaccinia virus F9 protein] (F9L) | 262 | 32 | 186 | Potential membrane protein, TM | L |
| MSV095 | 89843–90082 (80) | TM | L | ||||
| MSV096 | 90112–90564 (151) | E | |||||
| MSV097 | 90573–90992 (140) | PRF:1906390A, Atriplex nummularia caltractin-like protein | 120 | 28 | 128 | Calcium binding protein | L |
| MSV098 | 91014–91337 (108) | L | |||||
| MSV099 | 92883–91327 (519) | TM, SP | E | ||||
| MSV100 | 93123–93812 (230) | P33813, VAR RNA polymerase RPO19 (A5R) | 68 | 26 | 126 | RNA polymerase subunit, RPO19, TM | |
| MSV101 | 93298–93095 (68) | ||||||
| MSV102 | 93834–94364 (177) | E | |||||
| MSV103 | 94376–94828 (151) | ||||||
| MSV104 | 94827–94513 (105) | TM | |||||
| MSV105 | 94873–95634 (254) | TM | E | ||||
| MSV106 | 95701–96189 (163) | [P20997, vaccinia virus A22 protein] (A22R) | 134 | 29 | 154 | E, L | |
| MSV107 | 96207–96884 (226) | TM, SP | L | ||||
| MSV108 | 96891–97118 (76) | TM, SP | L | ||||
| MSV109 | 97531–97743 (71) | TM, SP | |||||
| MSV110 | 97709–97167 (181) | TM, SP | E | ||||
| MSV111 | 97802–98404 (201) | TM, SP | E | ||||
| MSV112 | 98404–98793 (130) | TM | |||||
| MSV113 | 99618–101639 (674) | [P04308 vaccinia virus 70 kDa subunit (D6R) | 734 | 44 | 379 | Early transcription factor, VETFS | L |
| MSV114 | 102136–101864 (91) | TM, SP | E | ||||
| MSV115 | 102182–103696 (505) | [J03399, vaccinia virus G5R protein] (G5R) | 189 | 28 | 270 | ||
| MSV116 | 104571–103621 (317) | TM | E | ||||
| MSV117 | 104621–106429 (603) | M13961, Rattus norvegicus DNA polymerase β | 185 | 31 | 186 | DNA polymerase β, SP | L |
| U40707, Caenorhabditis elegans AP endonuclease | 162 | 31 | 180 | AP endonuclease | |||
| MSV118 | 107228–107512 (95) | TM | |||||
| MSV119 | 108840–106420 (807) | [P33067, VAR RNA polymerase-associated protein] (H4L) | 564 | 26 | 797 | RNA polymerase-associated factor, RAP94, TM | L |
| MSV120 | 108922–109674 (251) | E | |||||
| MSV121 | 109692–110690 (333) | [P15909, FPV protein FP1] (G9R) | 380 | 30 | 317 | Potential membrane protein, TM | L |
| MSV122 | 110059–109847 (71) | SP | |||||
| MSV123 | 111375–110686 (230) | ||||||
| MSV124 | 111409–112209 (267) | [S42252, FPV mRNA capping enzyme, small subunit] (D12L) | 95 | 18 | 221 | mRNA capping, TM | E, L |
| MSV125 | 112651–112220 (144) | E, L | |||||
| MSV126 | 113119–112676 (148) | ||||||
| MSV127 | 113641–113126 (172) | ||||||
| MSV128 | 114960–113695 (422) | ||||||
| MSV129 | 115345–114947 (133) | [P07615, vaccinia virus L5R protein] (L5R) | 90 | 31 | 114 | TM | L |
| MSV130 | 115362–116345 (328) | [U80056, AmEPV DNA topoisomerase 1] (H6R) | 986 | 59 | 330 | Type I topoisomerase L | |
| MSV131 | 116598–116344 (85) | TM | |||||
| MSV132 | 116604–117029 (142) | [P29816, AmEPV G4R protein] (A28L) | 463 | 58 | 139 | AmEPV G4R, TM, SP | L |
| MSV133 | 117022–117405 (128) | L | |||||
| MSV134 | 117881–117408 (158) | TM | L | ||||
| MSV135 | 118619–117903 (239) | P40371, Schizosaccharomyces pombe PP2C | 128 | 22 | 246 | Protein phosphatase, PP2C | E, L |
| MSV136 | 118649–119098 (150) | L | |||||
| MSV137 | 119105–119551 (149) | ||||||
| MSV138 | 119548–120117 (190) | L | |||||
| MSV139 | 120121–120537 (139) | E, L | |||||
| MSV140 | 120541–122364 (608) | L | |||||
| MSV141 | 121630–121433 (66) | ||||||
| MSV142 | 122781–122365 (139) | [M17418, FPV 15.6-kDa protein] (J5L) | 171 | 33 | 132 | Potential membrane protein | L |
| MSV143 | 124506–122794 (571) | [P33809, VAR poly(A) polymerase catalytic] (E1L) | 249 | 29 | 322 | Poly(A) polymerase (PAPL), TM | |
| MSV144 | 124539–124778 (80) | U01220, Neurospora crassa ubiquitin | 356 | 86 | 80 | Ubiquitin | |
| MSV145 | 126345–124771 (525) | L | |||||
| MSV146 | 126360–127091 (244) | ||||||
| MSV147 | 127018–126704 (105) | TM | |||||
| MSV148 | 128490–127078 (471) | [P20534, vaccinia virus DNA helicase] (A18R) | 423 | 30 | 376 | DNA helicase, TM | L |
| MSV149 | 128526–129569 (348) | P21087, vaccinia virus RNA polymerase RP035 (A29L) | 90 | 20 | 213 | RNA polymerase subunit, RPO35 | L |
| MSV150 | 130430–129564 (289) | [P32817, FPV D10 protein] (D10R) | 186 | 31 | 200 | NTP pyrophosphohydrolase, MutT | |
| MSV151 | 131400–130462 (313) | [P33836, VAR A11R protein] (A11R) | 158 | 22 | 314 | L | |
| MSV152 | 131427–135344 (1306) | [P33817, VAR major core protein precursor P4a] (A10L) | 256 | 22 | 840 | Core protein, P4a | L |
| MSV153 | 135325–135537 (71) | TM | |||||
| MSV154 | 136112–135357 (252) | AB000449, Homo sapiens Ser/Thr protein kinase 1 (B1R) | 206 | 32 | 170 | Ser/Thr protein kinase, VRK1 | E |
| MSV155 | 139728–136159 (1190) | [P17474, CPV RNA polymerase RPO132] (A24R) | 925 | 30 | 770 | RNA polymerase, RPO132 | E, L |
| MSV156 | 140126–143506 (1127) | E, L | |||||
| MSV157 | 143533–144822 (430) | E | |||||
| MSV158 | 145485–144811 (225) | [X76267, VAR core protein VP8 precursor] (L4R) | 133 | 28 | 219 | Core protein, VP8 | L |
| MSV159 | 146202–145528 (225) | L | |||||
| MSV160 | 146701–146231 (157) | L | |||||
| MSV161 | 147002–146724 (93) | AFO019224, HaEPV ORF F2 protein | 154 | 39 | 78 | HaEPV F2, TM, SP | L |
| MSV162 | 147019–148584 (522) | P26996, Thermus thermophilus NAD+-DNA ligase | 178 | 25 | 356 | NAD+-dependent DNA ligase | |
| MSV163 | 148808–148593 (72) | TM | L | ||||
| MSV164 | 148850–150793 (648) | [P17355 FPV major core protein precursor P4b] (A3L) | 297 | 24 | 519 | Core protein, P4b, TM | L |
| MSV165 | 150915–151292 (126) | TM, SP | L | ||||
| MSV166 | 151313–151600 (96) | TM, SP | L | ||||
| MSV167 | 151629–152162 (178) | E | |||||
| MSV168 | 152391–152176 (72) | ||||||
| MSV169 | 152435–153124 (230) | TM | |||||
| MSV170 | 153165–154136 (324) | E | |||||
| MSV171 | 154863–154132 (244) | [P21055, vaccinia virus A32L protein] (A32L) | 168 | 30 | 204 | ATP/GTP binding motif | |
| MSV172 | 154891–155442 (184) | L | |||||
| MSV173 | 155472–156842 (457) | [P32216, SPV Ser/Thr protein kinase C20L] (F10L) | 247 | 26 | 375 | Ser/Thr protein kinase, KRF1 | L |
| MSV174 | 157386–157568 (61) | TM | |||||
| MSV175 | 157502–156840 (221) | U90931, Bacteroides fragilis metalloprotease toxin 2 | 85 | 29 | 48 | Metalloprotease, SP | |
| MSV176 | 158396–157683 (238) | X89576, Homo sapiens MT2-MMP protein | 103 | 29 | 130 | Metalloprotease, SP | |
| MSV177 | 159202–158573 (210) | TM, SP | E | ||||
| MSV178 | 159328–159507 (60) | ||||||
| MSV179 | 160698–159304 (465) | U82541, Xenopus laevis matrix metalloprotease | 163 | 25 | 179 | Metalloprotease, TM, SP | |
| MSV180 | 161788–160760 (343) | [P07614, vaccinia virus protein L3L protein] (L3L) | 188 | 32 | 162 | ||
| MSV181 | 161820–163121 (434) | TM, SP | |||||
| MSV182 | 163771–163121 (217) | ||||||
| MSV183 | 163810–164535 (242) | [U60315, MCV MC069R protein] (L1R) | 503 | 48 | 224 | Myristylated membrane protein, TM | |
| MSV184 | 164558–165802 (415) | TM | L | ||||
| MSV185 | 165438–165253 (62) | ||||||
| MSV186 | 165805–166824 (340) | [P28854, AmEPV ORF Q3] | 124 | 30 | 164 | LRR | |
| MSV187 | 166845–167627 (261) | [S42254, FPV transactivator protein] (A1L) | 98 | 30 | 106 | Late transcription factor, VLTF-2 | |
| MSV188 | 167805–168008 (68) | E, L | |||||
| MSV189 | 169331–168003 (443) | [P29817, AmEPV G1L protein] (I7L) | 1,074 | 48 | 452 | Core protein, I7L | |
| MSV190 | 169350–170030 (227) | [P29818 AmEPV G2R protein] | 268 | 31 | 217 | AmEPV G2R, TM | |
| MSV191 | 170628–170020 (203) | MTG motif | E | ||||
| MSV192 | 170671–171021 (117) | L | |||||
| MSV193 | 170978–171286 (103) | TM | |||||
| MSV194 | 172239–171013 (409) | L44593, bacteriophage BK5-T ORF 266 protein | 262 | 40 | 157 | ALI motif | E |
| MSV195 | 172550–172290 (87) | P24655, AcNPV 38-kDa protein, orf2 | 91 | 33 | 84 | ALI motif | |
| MSV196 | 173145–172540 (202) | AF003534, Chilo iridescent virus ORF 011L | 275 | 35 | 190 | ALI motif | E |
| MSV197 | 174289–173282 (336) | M96361, AcNPV 41.6-kDa protein | 166 | 29 | 162 | Tryptophan repeat | |
| MSV198 | 175515–174319 (399) | AF003534, Chilo iridescent virus ORF 074R | 150 | 23 | 240 | MTG motif | E |
| MSV199 | 176049–175576 (158) | AF003534, Chilo iridescent virus ORF 074R | 120 | 25 | 148 | MTG motif | E |
| MSV200 | 176444–176632 (63) | ||||||
| MSV201 | 177155–176625 (177) | ||||||
| MSV202 | 178207–177224 (328) | E | |||||
| MSV203 | 178373–178555 (61) | TM | |||||
| MSV204 | 179133–178468 (222) | AF003534, Chilo iridescent virus ORF 011L | 137 | 27 | 161 | ALI motif | E |
| MSV205 | 180752–179163 (530) | M96361, AcNPV 41.6-kDa Protein | 182 | 33 | 146 | Tryptophan repeat | E |
| MSV206 | 181634–180774 (287) | U94833, Haemophilus sommus lipooligosaccharide biosynthesis | 91 | 25 | 236 | Glycosyltransferase, TM | L |
| MSV207 | 181871–181638 (78) | ||||||
| MSV208 | 182595–181900 (232) | U20824, equine herpesvirus uracil DNA glycosylase (D4R) | 264 | 34 | 214 | UNG | E |
| MSV209 | 182790–183128 (113) | [P20996, vaccinia virus A21 protein] (A21L) | 69 | 24 | 115 | SP | |
| MSV210 | 183142–183474 (111) | ||||||
| MSV211 | 183743–183501 (81) | ||||||
| MSV212 | 184138–183740 (133) | L | |||||
| MSV213 | 184168–185160 (331) | L | |||||
| MSV214 | 186301–185144 (386) | SCG motif, TM | L | ||||
| MSV215 | 187434–186325 (370) | SCG motif, TM, SP | L | ||||
| MSV216 | 188543–187434 (370) | SCG motif, TM | L | ||||
| MSV217 | 189685–188543 (381) | SCG motif, TM | L | ||||
| MSV218 | 190939–189980 (320) | ||||||
| MSV219 | 190981–191469 (163) | ||||||
| MSV220 | 191566–191925 (120) | E | |||||
| MSV221 | 191912–193381 (490) | ||||||
| MSV222 | 193368–193838 (157) | TM | |||||
| MSV223 | 193896–194408 (171) | TM | E | ||||
| MSV224 | 194435–196933 (833) | 141 | 23 | 627 | NTPase/Helicase | E, L | |
| MSV225 | 197342–196917 (142) | E | |||||
| MSV226 | 197604–197930 (109) | E | |||||
| MSV227 | 198805–197888 (306) | [P28854, AmEPV ORF Q3] | 165 | 30 | 200 | LRR | E |
| MSV228 | 198841–199677 (279) | [P28854, AmEVP ORF Q3] | 148 | 32 | 167 | LRR | L |
| MSV229 | 199808–200353 (182) | AF003534 Chilo iridescent virus ORF 011L | 195 | 37 | 123 | E | |
| MSV230 | 200414–202423 (670) | E | |||||
| MSV231 | 200853–201053 (67) | ||||||
| MSV232 | 201060–201341 (94) | ||||||
| MSV233 | 201796–201521 (92) | TM | E | ||||
| MSV234 | 201830–201480 (117) | ||||||
| MSV235 | 204120–202723 (466) | D31902, Monodelphis domestica CPD photolyase | 1,377 | 57 | 429 | CPD photolyase | |
| MSV236 | 204132–204530 (133) | E | |||||
| MSV237 | 204558–205133 (192) | [P20999, vaccinia virus B2R protein (B2R) | 434 | 44 | 189 | E | |
| MSV238 | 206102–205227 (292) | L08594, Arabidopsis thaliana thymidylate synthase | 930 | 59 | 286 | Thymidylate synthase | E |
| MSV239 | 206210–207496 (429) | [P28854, AmEPV ORF Q3] | 341 | 41 | 217 | LRR | E, L |
| MSV240 | 208070–209650 (527) | [P28854, AmEPV ORF Q3] | 330 | 38 | 219 | LRR | |
| MSV241 | 209732–210892 (387) | [P28854 AmEPV ORF Q3] | 310 | 38 | 213 | LRR | E |
| MSV242 | 212889–213875 (329) | L22858 AcNPV apoptosis inhibitor protein | 129 | 25 | 132 | IAP, TM | |
| MSV243 | 213634–213822 (63) | ||||||
| MSV244 | 214649–214005 (215) | ||||||
| MSV245 | 215215–214658 (186) | [P33058, variola virus RNA polymerase RPO18] (D7R) | 93 | 23 | 156 | RNA polymerase, RPO18 | E |
| MSV246 | 215380–215583 (68) | ||||||
| MSV247 | 215660–215232 (143) | TM | E | ||||
| MSV248 | 215803–216252 (150) | U75930 OpNPV inhibitor of apoptosis protein | 217 | 29 | 144 | IAP | E |
| MSV249 | 216210–216398 (63) | TM | L | ||||
| MSV250 | 216350–217672 (441) | E | |||||
| MSV251 | 218047–219033 (329) | C3H2C3 RING finger | |||||
| MSV252 | 219048–219530 (161) | Tryptophan repeat | E | ||||
| MSV253 | 219557–220756 (400) | [P28854, AmEPV ORF Q3] | 275 | 36 | 213 | LRR | E |
| MSV254 | 220798–221796 (333) | [P28854, AmEPV ORF Q3] | 258 | 41 | 160 | LRR, TM | E |
| MSV255 | 221829–223037 (403) | [P28854, AmEPV ORF Q3] | 341 | 42 | 208 | LRR | E |
| MSV256 | 223107–223508 (134) | ||||||
| MSV257 | 223560–225680 (707) | [P28854, AmEPV ORF Q3] | 238 | 35 | 207 | LRR | E |
| MSV258 | 224029–223835 (65) | ||||||
| MSV259 | 225490–225224 (89) | SP | |||||
| MSV260 | 227136–227414 (93) | [P28854, AmEPV ORF Q3] | 116 | 38 | 81 | LRR | |
| MSV261 | 227470–229341 (624) | [P28854, AmEPV ORF Q3] | 341 | 42 | 219 | LRR | E |
| MSV262 | 229505–229969 (155) | ITR, 155 aa | E | ||||
| MSV263 | 229563–229742 (60) | ITR, 60 aa; TM | |||||
| MSV264 | 229999–230265 (89) | ITR, 89 aa | |||||
| MSV265 | 232644–232829 (62) | ITR, 62 aa | |||||
| MSV266 | 233334–233519 (62) | ITR, 62 aa | |||||
| MSV267 | 234162–234347 (62) | ITR, 62 aa |
aa, amino acids.
Accession numbers are from the GenBank or SwissProt database (unless otherwise indicated). Poxvirus data are in brackets; vaccinia virus data are in parentheses.
Function was deduced from the degree of amino acid similarity to either known genes or Prosite signatures. TM, Z score of ≥ 1.96 for the prediction of transmembrane domains by using the Memsat computer program; SP, Z score of ≥ 2.5 for the prediction of signal peptides by using Sigcleave.
Putative promoter type. E, early; L, late.
TABLE 2.
Chordopoxvirus homologues in MsEPV
| Function | MsEPV ORF | Vaccinia virus ORF | % Amino acid identity (length) | Poxvirus with highest degree of homology | % Amino acid identity (length) | Gene name and/ or function |
|---|---|---|---|---|---|---|
| Transcription/mRNA modification | ||||||
| RNA polymerase | ||||||
| MSV043 | J6R | 30 (1,185) | RPO147 | |||
| MSV100 | A5R | 25 (162) | VAR | 26 (162) | RPO19 | |
| MSV119 | H4L | 26 (797) | VAR | 26 (797) | RAP94 | |
| MSV149 | A29L | 20 (213) | RPO35 | |||
| MSV155 | A24R | 30 (770) | CPV | 30 (770) | RPO132 | |
| MSV245 | D7R | 23 (133) | VAR | 23 (156) | RPO18 | |
| Transcription factors | ||||||
| MSV063 | A7L | 25 (583) | VAR | 25 (583) | VETFL | |
| MSV065 | A2L | 22 (198) | VLTF-3 | |||
| MSV113 | D6R | 44 (379) | VETFS | |||
| MSV187 | A1L | 26 (78) | FPV | 30 (106) | VLTF-2 | |
| NTPase/helicase | ||||||
| MSV053 | D11L | 36 (633) | FPV | 37 (633) | NPH-I | |
| MSV086 | I8R | 33 (651) | RNA helicase/NPH-II | |||
| MSV148 | A18R | 30 (376) | DNA helicase | |||
| mRNA modification | ||||||
| MSV041 | J3R | 35 (244) | VAR | 35 (244) | PAPS | |
| MSV067 | D1R | 28 (750) | Capping enzyme, large subunit | |||
| MSV124 | D12L | 20 (177) | FPV | 18 (221) | Capping enzyme, small subunit | |
| MSV143 | E1L | 29 (322) | VAR | 29 (322) | PAPL | |
| DNA replication/repair | ||||||
| MSV036 | E9L | 29 (943) | FPV | 30 (926) | DNA polymerase | |
| MSV089 | D5R | 29 (560) | RFV | 27 (643) | NTPase | |
| MSV130 | H6R | 34 (315) | FPV | 37 (321) | Topoisomerase | |
| MSV150 | D10R | 35 (91) | FPV | 31 (200) | mutT motif | |
| MSV208 | D4R | 18 (199) | FPV | 30 (117) | UNG | |
| Structural | ||||||
| MSV069 | D13L | 28 (445) | Rifampicin resistance | |||
| MSV090 | A16L | 27 (368) | MCV | 29 (274) | Putative membrane protein | |
| MSV094 | F9L | 32 (186) | Putative membrane protein | |||
| MSV121 | G9R | 25 (335) | FPV | 30 (317) | Putative membrane protein | |
| MSV129 | L5R | 31 (114) | Putative membrane protein | |||
| MSV142 | J5L | 29 (131) | FPV | 33 (132) | Putative membrane protein | |
| MSV152 | A10L | 22 (718) | VAR | 22 (840) | Core protein, P4a | |
| MSV158 | L4R | 27 (219) | VAR | 28 (219) | Core protein, VP8 | |
| MSV164 | A3L | 23 (532) | FPV | 24 (519) | Core protein, P4b | |
| MSV183 | L1R | 38 (224) | MCV | 48 (224) | Membrane protein | |
| MSV189 | I7L | 26 (416) | FPV | 26 (427) | Core protein | |
| Enzymes | ||||||
| MSV048 | CPV | 25 (75) | Lipase | |||
| MSV093 | E10R | 42 (100) | VAR | 45 (90) | Potential redox, ERV1 | |
| MSV154 | B1R | 32 (161) | Protein kinase | |||
| MSV171 | A32L | 30 (204) | ATP/GTP binding motif | |||
| MSV173 | F10L | 24 (407) | SPV | 26 (375) | Protein kinase | |
| MSV056 | G1L | 23 (591) | VAR | 23 (591) | Metalloprotease | |
| Unknown | ||||||
| MSV039 | G6R | 21 (146) | MCV | 25 (127) | ||
| MSV052 | A23R | 26 (324) | ||||
| MSV060 | H2R | 37 (183) | ||||
| MSV106 | A22R | 29 (154) | ||||
| MSV115 | G5R | 28 (270) | ||||
| MSV132 | A28L | 33 (146) | MCV | 38 (141) | ||
| MSV151 | A11R | 22 (314) | VAR | 22 (314) | ||
| MSV180 | L3L | 32 (162) | ||||
| MSV209 | A21L | 24 (115) | ||||
| MSV237 | B2R | 44 (189) |
MsEPV has a genomic organization similar to that of other known chordopoxviruses (53, 122, 169, 170). There is no evidence for introns, both strands are protein encoding, and there are few overlapping ORFs. ORFs frequently occur in head-to-tail tandem arrays (Fig. 1). Within the terminal 50 kbp of the genome, most ORFs are transcriptionally oriented toward their respective termini. As seen in other poxviruses, the MsEPV genome contains a central coding region bounded by two inverted terminal repeat (ITR) regions of approximately 7 kbp each (Fig. 1). The first 126-bp repeat marks the boundary between the ITR and the central coding region (Fig. 1). There are also regions internal to the ITR containing additional tandem repeats and several gene families.
ITRs.
Although lacking sequence identity, MsEPV and ChPV ITR tandem repeats are similar (Fig. 1) (121, 190, 204). MsEPV ITRs contain a series of tandemly repeated sequences, 21-bp repeats (27 and 34 copies in the left and right ITRs, respectively) followed by blocks of 7-bp repeats (33 and 32 copies), 69-bp repeats (58 and 52 copies), 26-bp repeats (5 copies each), 72-bp repeats (2 copies each), and a 126-bp repeat (1 copy each). A variable number of incomplete 69-bp repeats (56 to 59 bp) separate the four terminal blocks of repeats. Nucleotide identity within sets of repeats is approximately 60% for the 21-bp repeats and 80 to 98% for the others. Comparison of 4-kbp noncoding ITR regions of vaccinia virus and MsEPV shows that the most numerous repeats, 69 or 70 bp long, are accompanied by less-abundant repeats of 125 to 126 bp and 26 bp and incomplete forms of the 69- to 70-bp repeats (54 to 58 bp). As in orthopoxviruses, there is a nonrepetitive spacer region within each ITR (53, 122). The coding capacity of MsEPV ITRs is limited. Four ORFs are present in the left ITR, and six ORFs are found in the right ITR. Within each ITR, four ORFs are present as single-copy genes. Three copies of the most-terminal ORF are present in the right ITR (MSV265, MSV266, and MSV267).
Gene expression regulatory elements.
DNA sequences upstream, within, and downstream of MsEPV ORFs exhibited similarity to promoter and regulatory elements described for other poxviruses, thus suggesting some degree of conservation of gene regulatory mechanisms. Of the 14 MsEPV ORFs with homology to known poxvirus early genes, 8 contain a TGAAAxxxxA motif in the region 5′ of the putative translational start codon (Table 1), and an additional four genes contain this motif with only a single nucleotide substitution (data not shown). This putative early-type promoter element resembles the early-promoter core consensus sequence found in other ChPVs (130, 170). This motif has previously been found upstream of the thymidine kinase (TK) genes of other EPVs (58, 119), and a similar motif (TGAATxxxxA) is found upstream of the CbEPV DNA polymerase gene (131). Interestingly, the upstream sequence of the MmEPV fusolin gene, which demonstrates early promoter activity (111), also contains the TGAAAxxxxA motif (52). The vaccinia virus consensus early transcriptional stop sequence (TTTTTxT), which has also been observed downstream of EPV TK and other EPV gene sequences (58, 110, 175, 216), is found within 100 bases of the 3′ ends of 49 of the 84 MsEPV ORFs preceded by the TGAAAxxxxA motif.
Twenty-four of 36 MsEPV homologues of late ChPV genes (170) contain the consensus poxvirus-late-promoter sequence (TAAATG) at the translational start site (15, 157, 198). This late-promoter sequence has been previously described for other EPV genes, including those encoding spheroidin, the nucleoside triphosphatase (NTPase) hydrolase I (NPH-I), and topoisomerase (64, 66, 110, 164, 175). Eleven of the 12 remaining putative MsEPV late genes contain upstream sequences that have been found at the start of poxvirus late genes. Such sequences include TAAAT upstream of the translational start site (eight ORFs) (158), TAAAAT (one ORF) (95), and TAATG (three ORFs) (157, 158). Similar to the fusolin genes of other EPVs, 33 MsEPV ORFs contain the TAATG motif at the translational start site (27, 140, 215).
Transcription and mRNA biogenesis.
MsEPV contains homologues of 18 of the 26 vaccinia virus genes thought to be involved in transcriptional processes (130) (Fig. 1; Table 2). This suggests the presence of conserved mechanisms for generating functional mRNA among the two poxvirus subfamilies. Vaccinia virus RNA polymerase is encoded by at least eight viral genes ranging in size from 7 to 147 kDa. MsEPV homologues of the two largest vaccinia virus subunits, RPO147 (J6R) and RPO132 (A24R), and the smaller subunits RPO35 (A29L), RPO19 (A5R), and RPO18 (D7R) are MSV043, MSV155, MSV149, MSV100, and MSV245, respectively (Table 2). Homologues of the three remaining vaccinia virus RNA polymerase subunits are not identifiable in MsEPV. Amino acid variability within homologous ChPV RPO subunits suggests that other MsEPV subunits may also be highly variable and, thus, undetectable by current computer search and analysis algorithms. In addition, MsEPV contains a homologue (MSV119) of the RNA polymerase-associated protein RAP94 (H4L), which is specifically required for transcription of early-promoter templates (Table 2) (130).
Four homologues of vaccinia virus transcription factors are encoded in MsEPV. MSV113 and MSV063 are homologues of the two subunits of the vaccinia virus early transcription factor, VETFS (D6R) and VETFL (A7L), respectively. MSV187 and MSV065 are homologues of the two late transcription factors VLTF-2 (A1L) and VLTF-3 (A2L) (Table 2). While MSV113 has 44% amino acid identity to vaccinia virus VETFS, the levels of amino acid identity to VETFL, VLTF-2, and VLTF-3 homologues are much lower (22 to 24%). The vaccinia virus late transcription factor VLTF-1 (G8R), VLTF-4 (H5R), and G2R gene product homologues are either absent from the MsEPV genome or unidentifiable. The absence of a VLTF-1 homologue is surprising, since this gene is essential for vaccinia virus replication and is conserved among ChPV genera (130, 170).
Four MsEPV ORFs, MSV053, MSV086, MSV113, and MSV148, are homologues of four NTPase-helicase genes found in vaccinia virus (Table 2). These include the NPH-I homologue (D11L), the RNA-DNA helicase (NPH-II) homologue (I8R), the small subunit of the early transcription factor VETFS (D6R), and the DNA helicase (A18R), respectively (Table 2). These MsEPV ORFs contain motifs conserved among the NTPase and helicase enzymes of the RNA and DNA helicase superfamily II (55, 93). Only NPH-I homologues have been previously described in other EPVs (65, 110, 175, 217). As expected, this gene (MSV053) has a higher level of amino acid identity to EPV genes (58%) than to orthopox-, molluscipox-, and leporipoxvirus homologues (33 to 37%). Another ORF (MSV224) also contains carboxy-terminal helicase and NTPase motifs in addition to a cysteine-rich amino terminus, but it lacks homology to other poxvirus genes.
MSV148 encodes a homologue of the vaccinia virus A18R gene. A18R encodes a late virion-associated DNA helicase that is essential for correct viral gene expression and productive infection (12, 174). The essential nature of A18R suggests a similar function for MSV148 in MsEPV.
MsEPV contains homologues of vaccinia virus genes involved in transcriptional termination, capping, and polyadenylation. MSV067 and MSV124 are homologues of the large and small subunits of the vaccinia virus capping enzyme, D1R and D12L, respectively (Table 2). MSV143 and MSV041 are homologues of the large and small polyadenylation polymerase (PAP) subunits, PAPL (E1L) and PAPS (J3R), respectively (Table 2).
Nucleotide metabolism.
MsEPV lacks all previously described poxvirus genes involved in nucleotide metabolism (130). Absent are genes encoding TK, thymidylate kinase, the large and small subunits of ribonucleotide reductase, dUTPase, glutaredoxin, and guanylate kinase and the cytidine kinase gene found in FPV (92). The absence of a TK gene in MsEPV is surprising given that TK genes have been identified in other group B EPVs (AmEPV, CbEPV, and CfEPV) (58, 119). This paucity of viral enzymes suggests that MsEPV replication is heavily dependent on host cell nucleotide biosynthesis. These differences in nucleotide metabolism must be of significance for viral cell and/or tissue tropism within the grasshopper host.
Interestingly, and unlike other known poxviruses, MSV238 encodes a thymidylate synthase (TSY) homologue. MSV238 is very similar to TSY genes from eukaryotes (52 to 59% amino acid identity). The 29-amino-acid TSY Prosite motif (PS00091), which contains the catalytic cysteine residue, is also conserved in MSV238 with the exception of a single conservative substitution (leucine to isoleucine) at position 170. Homodimeric TSY catalyzes the methylation of dUMP to the nucleotide precursor dTMP, thus representing an important part of the de novo pathway of pyrimidine biosynthesis (21). Despite its ubiquitous distribution in nature, a viral TSY gene has been observed only in a few herpesviruses and bacteriophages (13, 79, 80).
DNA replication.
MsEPV contains homologues of most vaccinia virus genes involved in DNA replication, including DNA polymerase (E9L), ATP-GTP binding protein (D5R), DNA topoisomerase (H6R), and replication essential protein kinase (B1R) (Table 2). However, it lacks the processivity factor (A20R) and an ATP-dependent DNA ligase (A50R). Notably, and unlike any other known virus, MsEPV encodes an NAD+-dependent DNA ligase homologue (Table 1).
The MsEPV DNA polymerase (MSV036) is homologous to family B replicative DNA polymerases found in CbEPV (40% identity over 958 amino acids) and other ChPVs (29 to 30% amino acid identity over 900 amino acids). MSV036 also exhibits similarity to the DNA polymerase genes of chlorella virus PBCV-1 (39% identity over 247 amino acids; GenBank accession no. S35209) and ASFV (21% identity over 452 amino acids; GenBank accession no. U27575). MSV036 identity to family B DNA polymerases includes the highly conserved region I, in which the Prosite family signature (PS00116) is 100% conserved (5, 207).
MSV089 is homologous to the vaccinia virus ATP-GTP binding protein D5R (29% identity over 560 amino acids). Although more divergent than other ChPV D5R homologues, MSV089 contains regions of similarity throughout the protein, including the extended I (A) type of nucleoside triphosphate binding motif (55). D5R is known to be essential for viral DNA replication and is involved in homologous recombination (159).
Like AmEPV, MSV130 encodes a eukaryotic type I DNA topoisomerase with homology to vaccinia virus H6R. In MsEPV, the type I DNA topoisomerase Prosite motif (PS00176) and the active site residue (Tyr-292) are 100% conserved (172). In addition, critical residues required for transesterification by H6R are also conserved (205). Of six critical DNA recognition motifs described in the vaccinia virus protein, three are conserved and two are conservatively substituted in MSV130 (168).
MSV162 exhibits similarity to bacterial NAD+-dependent DNA ligases (Fig. 2B). To our knowledge, this is the first NAD+-dependent DNA ligase found in a virus genome. Eukaryotic organisms and other DNA viruses, including all known poxviruses, encode ATP-dependent DNA ligases (26, 67, 89, 103, 115, 213). MSV162 is most similar to bacterial Thermus thermophilus ligase and includes 12 of the 16 Prosite signature residues (PS01055) and the active-site motif Lys-X-Asp-Gly (118). Residues essential for adenylation and deadenylation steps are conserved in MSV162 (Lys-112 and Asp-118, respectively) (118). In addition, MSV162 exhibits partial conservation of a second NAD+-ligase Prosite signature (PS01056) but lacks residues typically conserved in the carboxyl terminus. Given that NAD+-dependent DNA ligases have been found only in bacteria, the presence of this gene in a eukaryotic virus is surprising and suggests that MSV162 has a prokaryotic origin.
FIG. 2.
Multiple amino acid sequence alignments of MsEPV ORFs with DNA repair and replication enzymes. Boldfaced letters represent active site residues, asterisks mark residues that match Prosite signatures, and shaded residues represent amino acids with identity to those of the corresponding MsEPV ORF. Amino acid positions are indicated on the right. (A) Alignment of MSV235 with class 2 CPD photolyases; regions I and II represent class 2 CPD photolyase Prosite signatures PS01083 and PS01084, respectively. Abbreviations: Monodelph, Monodelphis domestica, accession no. D31902; Oryzias, Oryzias latipes, accession no. S52048; Drosoph, Drosophila melanogaster, accession no. S52047; Arab, Arabidopsis thaliana, accession no. X99301; Methano, Methanobacterium thermoautotrophicum, accession no. D30752. (B) Alignment of MSV162 with NAD+-dependent DNA ligases; regions I and II represent Prosite signatures PS01055 and PS01056 for NAD+-dependent DNA ligases, respectively. Abbreviations: Thermus, Thermus aquaticus, accession no. P26996; Ecoli, E. coli, accession no. P15042; Mycoplas, Mycoplasma pneumoniae, accession no. AE000047.
DNA repair.
MsEPV encodes at least seven genes with putative DNA repair functions (Table 1). These include homologues of genes encoding a uracil DNA glycosylase (UNG) (MSV208), DNA polymerase β (MSV117), AP endonuclease (MSV117), DNA helicase (MSV148), a ChPV mutT homologue (MSV150), cyclobutane pyrimidine dimer (CPD) photolyase (MSV235), and a LINE-type reverse transcriptase (RT) (MSV061). Genes for DNA polymerase β and AP endonuclease have not been previously described in poxviruses. Further, the CPD photolyase and the LINE-type RT have not been previously found in any virus genome. The presence of this complement of genes in MsEPV suggests that virally encoded DNA repair functions are important for virus survival in nature.
Although other large DNA viruses, such as vaccinia virus and ASFV, contain some genes of the base excision repair (BER) pathway (181, 191, 212), MsEPV provides the first example of a virus potentially encoding all genes required for BER. DNA damage in eukaryotic cells arises spontaneously from hydrolytic events, oxygen free-radical attack, or methylation of ring nitrogen by endogenous agents (114). This pathway is also essential for resistance to DNA damage inflicted by exogenous DNA-damaging agents such as ionizing radiation and other radical-inducing agents (167). UNG, AP endonuclease, DNA polymerase β, and DNA ligase act sequentially through the BER pathway to repair damaged DNA (30). UNG removes deaminated cytosine (uracil) to generate apurinic or apyrimidinic sites (AP sites), class II AP endonucleases remove AP sites after cleaving the DNA strand 5′ to the AP site, DNA polymerase β fills the gap by DNA repair synthesis, and DNA ligase finishes the repair process by closing the gap (20, 30, 124, 203).
MSV208 is most similar to the equine herpesvirus UNG (34% identity over 214 amino acids); it has significant identity to UNG in bacteria (Bacillus subtilis PIR accession no. S39712), lower eukaryotes (slime mold [Dictostelium sp.]; GenBank accession no. U32866), and higher eukaryotes (mouse [Mus musculus]; GenBank accession no. X99018) (29 to 34% identity over 195 to 216 amino acids). Surprisingly, MSV208 exhibits much less similarity to the poxvirus UNG homologues in vaccinia virus (19% identity over 78 amino acids) and FPV (30% identity over 117 amino acids). The MsEPV gene does, however, contain amino acid substitutions at the predicted UNG active site. Most notably, an aspartic acid residue has been replaced by an arginine at the N-glycosylic bond cleavage site (residue 60 in MSV208). The significance of these residue changes for protein function is not known. In vaccinia virus and human cytomegalovirus, UNG enzymes have been implicated in other functions, including establishing the correct temporal progression of DNA synthesis and viral replication (126, 146, 181). Thus, it is possible that MSV208 performs other functions unrelated to BER.
Homologues of both class II AP endonucleases and DNA polymerase β are encoded by MSV117. This gene has homology to eukaryotic and viral class II AP endonucleases at its amino terminus (amino acids 1 to 296) and homology to DNA polymerase β at its carboxy terminus (amino acids 296 to 607). MSV117 contains most residues of AP endonuclease class II Prosite signatures 2 and 3 (PS00730 and PS00731, respectively), which include conserved and potentially metal-binding cysteine and histidine residues. The degree of identity of MSV117 to eukaryotic and viral DNA polymerase β enzymes is highest at the catalytic region (31% identity, over 186 amino acids, to rat DNA polymerase β (GenBank accession no. M13961) and includes the DNA polymerase X Prosite signature (PS00522). Rat DNA polymerase β is a smaller protein (335 amino acids) consisting of two domains connected by a protease-sensitive region (96). The 31-kDa carboxyl-terminal domain contains the residues critical for catalytic activity as defined by the crystal structure (29). These residues are present in MsEPV (Arg-469, Asp-476, and Asp-478). ASFV, another cytoplasmic DNA virus with an arthropod host, encodes both an AP endonuclease and a DNA polymerase β in separate ORFs (212). The ASFV DNA polymerase β (174 amino acids) is the smallest functional DNA polymerase β enzyme that has been described (137). MSV117 has homology to both ASFV genes.
The fusion of the AP endonuclease and DNA polymerase β genes into one gene has not been previously described. This fusion is reasonable, however, since the activities of both enzymes are coordinately required for DNA BER (14). Although a common strategy for RNA viruses and retroviruses, polyprotein processing has been observed for only a few vaccinia virus and ASFV structural proteins (4, 130, 173). The absence of vaccinia virus and ASFV proteolytic cleavage consensus sequences (Ala-Gly/Ala-Ser and Gly-Gly-X, respectively) at the AP endonuclease-DNA polymerase β junction in MSV117 suggests that this gene product may have a dual enzymatic function.
A role in DNA repair or recombination is possible for MSV117. DNA polymerase β is the simplest naturally occurring DNA polymerase known, and it is thought to function in a variety of repair mechanisms, including mismatched base repair (203), AP lesion repair (123), and monofunctional adduct repair (34). DNA polymerase β also seems to be involved in a repair-type DNA synthesis associated with recombination (68, 77) and with replicative DNA synthesis (183).
MSV235 shares similarity to class II CPD photolyases from marsupials, fish, insects, plants, and bacteria (38 to 57% identity over 428 to 445 amino acids) (Fig. 2B). This gene represents the first photolyase homologue found in a viral genome. CPD photolyase is a photoreactive enzyme that mediates repair of UV-induced CPDs in DNA (71, 87). The predicted protein of 466 residues exhibits 119 of 141 conserved class II residues (214). Both class II Prosite signatures (PS01083 and PS01084) in the carboxyl-terminal region (residues 298 to 312 and 425 to 444, respectively) are present, except for a conservative arginine-to-lysine substitution at position 306. Eukaryotic photolyases possess a protruding amino terminus with three regions of clustered positively charged amino acids which have been proposed to contain sequences for nuclear or mitochondrial transport (214). Consistent with a cytoplasmic mode of replication, these regions are absent from MSV235.
The importance of light-dependent DNA repair mechanisms in maintaining virus populations in nature has recently been demonstrated. Host cell light-dependent repair mechanisms have been reported to restore infectivity in up to 52% of sunlight-damaged viruses in natural marine virus communities (197). The ubiquity of CPD enzymes in nature (they are found in bacteria, plants, and mammals), the efficiency of light energy to repair UV-induced DNA damage, the unienzymatic nature of the system (71), and the detrimental effects of UV damage on survival of insect DNA viruses (8) suggest that a photolyase gene might be found in an insect virus. A virus-encoded photorepair system may thus confer a selective advantage for MsEPV in nature, where long periods of environmental exposure may occur.
MSV150 is a homologue of vaccinia virus genes D9R and D10R (Table 2) (91). All three genes contain the Prosite signature (PS00893) for MutT proteins. The amino acid identity of MSV150 to Shope fibroma, vaccinia, and molluscum contagiosum virus D10R homologues is 28 to 35% over 87 to 91 residues. MSV150 colinearity to D10R is interrupted by two regions (amino acids 65 to 117 and 146 to 180) which are absent in the vaccinia virus homologue. Although the specific function of the vaccinia virus D9R and D10R homologues is unknown (91), bacterial mutT pyrophosphohydrolase genes help prevent DNA damage and assure fidelity of RNA transcription within the GO error avoidance system that is responsible for removing an oxidatively damaged form of guanine (8-hydroxyguanine or 7,8-dihydro-8-oxoguanine) from both DNA and the nucleotide pools (125, 184).
MSV061 has significant homology to LINE-type RTs. The seven conserved regions (domains I to VII) characteristic of diverse retroelements, which include the two critical RT-identifying motifs Asp-h-2X-Ala-Phe and Tyr-h-Asp-Asp-X-3h (where h is any hydrophobic amino acid and X is any amino acid), are also present in MSV061 (Fig. 3) (210). Domains II, III, V, and VII are perfectly conserved, while domains I, IV, and VI contain one, two, and one substituted residues, respectively (Fig. 3). Multiple alignments and phylogenetic trees generated by the neighbor-joining method with 1,000 bootstrap replicates (161) show that MSV061 is most closely related (98% bootstrap support) to the LINE-type transposable elements and is least closely related to RT from yeast introns and E. coli MsDNA (Fig. 3 and data not shown). Other distinctive features of LINE retrotransposons are missing in MSV061, suggesting that it may be the remnant of an old transposition or a truncated LINE. Genes normally adjacent to LINE RTs, such as ORF1 or zinc finger-containing ORFs, are not found adjacent to MSV061 (46, 152, 209). A triple 21-bp repeat located immediately 3′ of the MSV061 translational stop codon may be the remnant of a transpositional event. All available data suggest that MSV061 is a functional viral gene: critical RT motifs are conserved (Fig. 3), the gene shows normal MsEPV base composition with typical MsEPV codon usage (data not shown), and a potential late promoter (TAATG) is located at the translational start site of the ORF.
FIG. 3.
Multiple amino acid sequence alignments of MSV061 with RTs. The seven RT motifs are indicated with roman numbers I to VII (210). Boldfaced letters indicate invariant amino acids, shaded letters indicate amino acids that are identical to corresponding ones in MSV061, and consensus residues are indicated at the bottom as follows: h, hydrophobic; p, small polar; c, charged; and x, any amino acid. Uppercase letters indicate the one-letter amino acid code. Amino acid positions are indicated on the right. Abbreviations: LINE, LINE type of RT; Intron, group II intron; MsDNA, multicopy single-stranded DNA; Cele, Caenorhabditis elegans, accession no. U00063; Rat, Rattus norvegicus, accession no. X61294; Xenla, Xenopus laevis, accession no. P14381; Dictyo, Dictyostelium discoideum, accession no. X57031; Aedes, Aedes aegypti, accession no. M95171; Yeast, Saccharomyces cerevisiae, accession no. P21325; Ecoli, E. coli, accession no. V00694.
Roles for MSV061 in DNA repair, viral DNA replication, or possibly gene acquisition are all plausible. LINE RT-mediated repair of double-strand chromosomal breaks has recently been demonstrated (129, 185). RTs from human L1 or yeast Ty1 or from the trypanosomatid protozoan Crithidia sp. (CRE1 transposon) can repair double-strand breaks by the insertion of complementary DNA at the break site. In the absence of homologous recombination, RTs repair double-strand breaks by nonhomologous end joining with capture of DNA within the cleavage site (129, 185).
Long terminal repeat (LTR)-containing retrotransposons have been found integrated in other DNA viruses. Integration of LTR-type retrotransposons into baculovirus AcNPV DNA has been described previously (49). This retroelement is flanked by LTRs and contains three ORFs similar in size and location to the gag, pol, and env genes of retroviruses (107). Also, integrated sequences of avian reticuloendotheliosis virus have been recently identified in field and vaccine strains of FPV, thus demonstrating that retroviral genomes can be integrated into the DNA of large cytoplasmic viruses (73). MSV061 does not, however, resemble the RT found in either of these LTR-type transposable elements, and the MsEPV genome does not contain any other retroviral elements associated with LTR transposons, such as gag, pol, env, RNase H, integrase, or LTR DNA sequences.
Protein modification.
Active participation of MsEPV in protein modification is indicated by the presence of eight viral and cellular gene homologues. These homologues include two protein kinases (MSV154 and MSV173), two type 2C cellular protein phosphatases (MSV081 and MSV135), ubiquitin (MSV144), and three metalloproteases (MSV175, MSV176, and MSV179).
MSV154 and MSV173 are similar to the two vaccinia virion-associated serine/threonine protein kinases, VPK1 (B1R) and VPK2 (F10L), respectively (Table 2). Both ORFs contain the conserved catalytic region IV with an active-site motif of serine/threonine protein kinases (Prosite PS00108), and MSV173 has region I with a protein kinase ATP-binding signature (Prosite PS00107) (69). Although MSV154 lacks the glycine residues that are conserved in the ATP-binding region of other poxvirus and mammalian homologues, it does contain the lysine ATP-binding residue (Prosite PS00107) essential for the kinase activity found in vaccinia virus VPK1 (113). VPK1 is necessary for vaccinia virus DNA replication (151), and VPK2 also appears to be essential for virus viability (112).
MSV081 and MSV135 encode protein phosphatase 2C (PP2C) homologues which are similar to each other and to PP2Cs from a broad range of organisms (Fig. 4A). To our knowledge, this is the first report of a PP2C gene in a viral genome. PP2C is the prototypic member of a large family of Mg2+/Mn2+-dependent protein serine/threonine phosphatases (PPM family) present in both eukaryotes and prokaryotes (11). The six invariant metal-coordinating residues common to all PP2C amino-terminal catalytic domains are conserved in MSV081 and MSV135 (Fig. 4A) (11). In addition, MSV081 contains all 8 amino acids present in the PP2C Prosite signature (PS01032) while MSV135 has only a single substitution (Fig. 4A). The different sizes of MSV081 and MSV135 (357 and 239 residues, respectively) indicate that the two are isoforms. MSV081 contains a signal peptide and a cleavage site at the amino terminus, suggesting that it is a secreted protein. Neither ORF contains the 90-amino-acid carboxyl-terminal region characteristic of mammalian PP2Cs (28). Among other functions, PP2C reverses stress-activated protein kinase cascades in the fission yeast (171), inactivates cystic fibrosis transmembrane conductance regulation in humans (189), determines cell fate in bacteria (36), and promotes sex determination in Caenorhabditis elegans (22). Although the pleiotropic functions of this enzyme preclude predictions of specific roles during viral infection, a role in regulation of host intracellular signaling pathways is likely.
FIG. 4.
Multiple amino acid sequence alignments of MsEPV ORFs with protein modification enzymes. (A) Alignment of MSV175, MSV176, and MSV179 with the catalytic or zinc-binding regions of zinc-dependent proteases. Boldfaced letters represent amino acids which are either histidine zinc ligands or glutamic acid catalytic residues, and shaded residues represent amino acids with identity to the corresponding MsEPV ORF. The consensus for the metzincin (Metzn) subfamily (81) is exhibited underneath. (where b is any bulky hydrophobic amino acid and x is any amino acid). Abbreviations: Xenop, Xenopus laevis, accession no. L49412; Human, Homo sapiens, accession no. P39900; Mus, Mus musculus, accession no. L36244; Gmax, Glycine max, accession no. U63725; Bfrag, Bacteroides fragilis, accession no. U90931. (B) Alignment of MSV081 and MSV135 with eukaryotic PP2C proteins. Boldfaced letters represent metal-coordinating residues (28), asterisks mark highly conserved residues, shaded residues represent amino acids with identity to MsEPV, and overlined residues mark the Prosite signature (PS00142). Abbreviations: Human, Homo sapiens, accession no. P35813; Param, Paramecium tetraurelia, accession no. Z36985; Sacch, Saccharomyces cerevisiae, accession no. U72346; Arabid, Arabidopsis thaliana, accession no. U78721.
MSV144 encodes a ubiquitin homologue (Table 1). Ubiquitin is a highly conserved protein which forms covalent attachments to protein substrates and induces degradation of targeted proteins by the 26S proteasome complex (23). Amino acid identity between MSV144 and eukaryotic ubiquitin (83 to 88%) includes residues required for protein ubiquitination (data not shown). This percentage of identity is lower than that observed among eukaryotic ubiquitin genes (approximately 96%). Several baculoviruses also encode ubiquitin genes (v-ubi) which are among the most divergent known (76% amino acid identity to the mammalian ubiquitin consensus) (61, 160, 194).
The presence of ubiquitin and ubiquitin-conjugating enzymes in different arthropod viruses (59, 75, 150, 194) and the role of ubiquitination in insect development (60) suggest that MSV144 performs an insect-host-related function. ASFV, another cytoplasmic DNA virus with an arthropod host (144), encodes a ubiquitin-conjugating enzyme and incorporates ubiquitinated proteins into the virion (75, 76, 154). The baculovirus v-ubi product is a nonessential structural protein that affects viral growth in cell culture (150). Covalent attachment of cellular ubiquitin to specific targets and their subsequent degradation affect numerous processes, including regulation of gene expression, cell cycle, signal transduction, apoptosis, receptor-mediated endocytosis, and antigen processing (23, 199). Indeed, over 45 confirmed or putative cellular substrates for ubiquitination have been identified, and many are from independent cellular regulatory pathways (199).
MsEPV encodes homologues of two types of metalloproteases (Table 1). The first type, represented by MSV056, is a homologue of vaccinia virus G1L. Like G1L, this gene contains an amino-terminal His-2X-Glu-His inverted metalloprotease motif and downstream glutamate residues (201). The presence of a homologue for G1L, a protein known to be involved in virion core protein processing, and the presence of potential proteolytic cleavage sites in virion core protein homologues in MsEPV suggest conservation in poxvirus structural protein processing and morphogenesis (193, 200).
A second type of metalloprotease catalytic domain, His-Glu-2X-His (86), characterizes two of three similar ORFs (Fig. 4B). MSV176 and MSV179 each contain a perfect His-Glu-2X-His consensus, while MSV175 has a glutamic acid-to-glutamine substitution at the active-site residue (Fig. 4B). These three ORFs also contain residues, including a third histidine zinc ligand and Met turn region downstream of the core His-Glu-2X-His domain, which are consistent with the metzincin subfamily of zinc-dependent metalloproteases (16, 81). In addition, all three ORFs contain putative amino-terminal signal peptides which are common among extracellular metalloendopeptidases (149). The presence of the His-Glu-2X-His motif and potential signal peptide and the significant degree of similarity to matrixins (mammalian extracellular matrix metalloproteinases) suggest that at least MSV176 and MSV179 may function as extracellular metalloproteases.
Baculoviruses have been shown to encode a metalloprotease (enhancin) which enhances virulence during infection by digesting the proteins of the host midgut peritrophic membrane (106, 156). A protein with enhancin-like activity has also been reported in Pseudaletia separata EPV (211). Thus, MSV176 and MSV179 may perform a similar host-related function in MsEPV infection.
Cellular functions.
MSV048 has significant homology to triacylglyceride lipases found in fungi (Rhizopus spp.), eubacteria (Synechocystis spp.), protozoa (Plasmodium spp.), and higher plants (Ipomoea spp.) and similarity at the potential lipase active site of previously described cowpox and ectromelia virus ORFs (196) (Fig. 5D). MSV048 contains a potential catalytic triad (Ser-173, Asp-227, and His-265), the Prosite signature (PS00120), and a high overall degree of amino acid similarity to known lipases (Fig. 5D; Table 1) (18, 31, 32). The predicted size of MSV048 (288 amino acids) is similar to that of most fungal lipases (265 to 297 amino acids), and it exhibits 29% identity over 158 amino acids to the most closely related lipase (from Rhizopus niveous). The presence of a potential signal peptide suggests that the protein is secreted. Given that the grasshopper fat body is the major organ infected by MsEPV (72) and is also the main site of triacylglycerol storage (50, 188), this viral lipase could conceivably be involved in the hydrolysis of lipids, perhaps functioning as a virulence factor.
FIG. 5.
Multiple amino acid sequence alignments of MsEPV ORFs with cellular and viral homologues. Boldfaced letters represent active site residues, asterisks mark residues from a Prosite signature (when indicated) or those exhibiting ≥85% conservation, and shaded residues represents amino acids with identity to MsEPV. Amino acid positions are indicated on the right. (A) Alignment of MSV048 at the active site of triacylglycerol lipases. Abbreviations: Rhizopus, Rhizopus niveus, accession no. D12680; Synecho, Synechocystis sp., accession no. D64004; Celegans, Caenorhabditis elegans, accession no. U97001; Ipomoea, Ipomoea nill, accession no. U55867; and Cowpox, CPV putative lipase, accession no. X94355. The Prosite signature is PS00120. (B) Alignment of MSV097 with two EF-hand motifs from calcium-binding proteins. Abbreviations: Tvaginalis, Trichomonas vaginalis, accession no. U38786; Calbicans, Candida albicans, accession no. P23286; Atriplex, Atriplex nummularia, accession no. PRF: 1906390A; Smansoni, Schistosoma mansoni, accession no. P15845; Brassica, Brassica napus, accession no. D63152. The Prosite signature is PS00018. (C) Alignment of MSV206 with bacterial glycosyltransferase genes. Abbreviations: Nmening, Neisseria meningitidis, accession no. U65788; Hinfluen, Haemophilus influenzae, accession no. U36398; Hpylori, Helicobacter pylori, accession no. AE000592; Phaemol, Pasteurella haemolytica, accession no. U15958; Hsomnus, Haemophilus somnus, accession no. U94833; Hducreyi, Haemophilus ducreyi, accession no. U58147. (D) Alignment of MSV087 with thioredoxin genes. Abbreviations: Strept, Streptomyces aureofaciens, accession no. P33791; Coryne, Corynebacterium nephridii, accession no. P00275; Ecoli, E. coli, accession no. M54881; Eubact, Eubacterium acidaminophilum, accession no. P21610; Neuros, Neurospora crassa, accession no. D45892. The Prosite signature is PS00194.
MSV242 and MSV248 are similar to viral and cellular inhibitor of apoptosis genes (iap) (Table 1) (35, 186). Both predicted MsEPV IAP proteins contain an amino-terminal baculovirus IAP repeat motif (BIR motif; Prosite PS01282) and one C3HC4 RING finger motif (Prosite PS00518) at the carboxyl terminus. Like baculovirus genes, MSV242 contains two BIR motifs while the smaller gene, MSV248, contains only a single BIR. iap genes were initially described in baculoviruses, where they were shown to inhibit apoptosis of infected cells and to increase viral infectivity (25). iap-like genes have been identified in only three virus families, the Baculoviridae (25), Iridoviridae (GenBank accession no. P40629), and Asfarviridae (ASFV) (132), all of which have arthropod hosts. The presence of iap genes in these viruses suggests that an apoptotic cellular response to viral infection may be an important host defense mechanism in diverse arthropods.
MSV097 encodes a protein with homology to the EF-hand superfamily of calcium binding proteins (Fig. 5B). These include regulatory and structural proteins such as calmodulin and caltractin (135). The 12-residue EF-hand loop motif (Prosite PS00018) responsible for calcium binding is represented twice in the amino terminus of MSV097 (Fig. 5B). The level of conservation within these two EF-hand motifs indicates high-affinity calcium binding. The carboxyl terminus is, however, less similar to other calcium binding proteins. Because calcium binding proteins control multiple intracellular processes, a role for this gene in virus-cell interactions is likely.
MSV206 has similarity to bacterial glycosyltransferases involved in lipopolysaccharide capsule biosynthesis and pathogenicity (Fig. 5C). These enzymes, which transfer sugar residues to lipid moieties or other sugar residues, have not been described previously in poxviruses (83, 145, 178). The presence of a transmembrane domain at the carboxyl terminus of MSV206 (amino acids 252 to 276) suggests that the protein is membrane associated. In bacteria, sugar polymerization is catalyzed by an inner-membrane-bound transferase complex. In MsEPV-infected grasshoppers, changes in the distribution of cell membrane carbohydrates on hemocytes have been observed (72, 127). Thus, MSV206 may modify surface polysaccharides on infected cells. MSV206 has no similarity to the baculovirus UDP-glucosyltransferase, which interferes with normal molting of virus-infected larvae by catalyzing the transfer of glucose to ecdysteroids (138).
MSV087 and MSV093 contain conserved cysteine residues indicative of redox-active centers found in glutaredoxin and thioredoxin (Fig. 5A) (39, 78, 105). MSV087 has similarity to thioredoxin of the fungus Neurospora crassa (23% amino acid identity over 67 amino acids) and shows partial conservation at the thioredoxin Prosite signature PS00194 (Fig. 5A). Prolines 27 and 65, which are necessary for maintenance of the E. coli thioredoxin structure, are conserved in MSV087 (39). Thioredoxins are small proteins of approximately 100 amino acids which participate in redox reactions via reversible oxidation of a redox-active disulfide bond (78). These enzymes are multifunctional, performing roles in DNA replication, protein synthesis, protein folding, and photosynthesis (78).
MSV093 has significant homology to the vaccinia virus gene E10R (53) and a lower level of identity to potential E10R homologues found in other cytoplasmic DNA viruses and eukaryotes (62, 97, 117, 170, 212). The yeast ERV1 gene has been shown to function in oxidative phosphorylation and appears to function in eukaryotic cell growth (47, 117). All of these genes contain the pair of conserved cysteine residues typical of glutaredoxin and thioredoxin redox-active centers (78).
Structural proteins.
Four ChPV virion core protein homologues are present in MsEPV (Table 2). MSV152, MSV164, MSV158, and MSV189 are homologues of vaccinia virus A10L, A3L, L4R, and I7L, which encode the virion core precursor proteins P4a, P4b, and VP8 and the core-associated I7L protein, respectively (130). Interestingly, MSV152 and MSV158, like their vaccinia virus homologues, contain potential proteolytic cleavage sites (104, 192, 193), which suggests that aspects of structural protein processing may be conserved between MsEPV and ChPVs. The proteolysis of P4a, P4b, and VP8 precursor proteins is intimately associated with normal vaccinia virus morphogenesis and production of infectious virions (104, 130, 202).
The overall degree of amino acid similarity between MsEPV and ChPV core protein homologues is low (22 to 28% identity) compared to the similarity observed among ChPV homologues (45 to 65% amino acid identity). Additionally, homologues of the following vaccinia virus structural genes are not found in MsEPV, including A4L core protein, F17L and I3L DNA-binding phosphoproteins, and the G7L, D2L, D3R, and A12L proteins associated with internal parts of intracellular mature virions (130).
Of the 14 known membrane proteins in vaccinia virus (130), only L1R is conserved in MsEPV (MSV183), perhaps reflecting the closer relationship between these proteins and host-specific functions. The vaccinia virus L1R gene, which encodes a major myristylated membrane protein that is associated exclusively with the primary membrane surrounding the virion core, is involved in virion assembly (48, 147, 148).
Homologues of five genes representing two conserved ChPV gene families are present in MsEPV. Invariant cysteine residues and putative transmembrane domains unique to each family are conserved in these MsEPV ORFs (170). MSV183 and MSV094, homologues of vaccinia virus L1R and F9L, respectively, comprise one gene family. MSV090, MSV121, and MSV142 are homologues of the vaccinia virus genes A16L, G9R, and J5L and comprise the second gene family. Although most of the genes in these two ChPV gene families remain poorly characterized, G9R and A16L have been shown to be myristylated and potentially soluble proteins (120). J5L is thought to be an essential gene (218). The presence of these two gene families in both subfamilies of the Poxviridae suggests that they may provide highly conserved replicative or structural functions.
MSV069 is 25 to 28% identical to ChPV rifampin resistance proteins (vaccinia virus D13L) and 54% identical to the HaEPV D13L homologue (139). In vaccinia virus, this essential gene is associated with virion assembly and may direct the formation of Golgi complex-derived viral crescents, which are the first morphologically distinct structures observed during poxvirus assembly (130). Viral crescents have been observed in cells infected with MsEPV and other genus B EPVs (54, 57, 72, 98). Thus, a similar role for MSV069 in EPV morphogenesis is likely. Interestingly, homologues of other vaccinia virus genes associated with early events of virus morphogenesis, such as A14L and A17L (153, 155, 206), were not identified.
ChPV homologues of unknown function.
MsEPV also encodes homologues of 10 ChPV genes of unknown function (Table 2). Four of these homologues exhibit a high degree of amino acid identity to their ChPV counterparts. MSV237 and vaccinia virus B2R, which exhibit 44% amino acid identity, are located in the right termini of their respective genomes (53). MSV060, which contains a putative signal peptide, and MSV132, which contains a putative amino-terminal transmembrane and signal peptide, exhibit up to 37 and 44% amino acid identity with ChPV homologues of vaccinia virus H2R and A28L, respectively (Table 2). Although MSV115 and vaccinia virus G5R exhibit only 28% amino acid identity, they contain two prominent protein motifs (Asp-Ala-Glu-Phe-X-Met-Cys-2X-Ala and Trp-Pro-4X-Asp-Gln-Asp) which are also conserved in the MCV homologue MC060R.
Gene families of unknown function.
MsEPV contains 43 novel ORFs grouped into five gene families. Genes from these families are asymmetrically distributed in regions terminal to the conserved central part of the genome. Family members form tandem arrays occasionally interrupted by other genes (Fig. 1). The lack of similarity between gene family members and known ChPV genes and the presence of host range genes in similar genomic locations in ChPVs suggest that MsEPV gene families perform host range functions. Analysis of sequences upstream of the translational initiation sites suggests that four of the five gene families may be expressed early in infection (Table 1).
The leucine-rich repeat (LRR) family contains 21 ORFs which range in size from 72 to 707 amino acids. The distinguishing feature of this family is a 44-amino-acid repeat that contains regularly spaced leucine residues at 22-amino-acid intervals. A similar motif is present in the AmEPV Q3 ORF (58) and accounts for the homology between LRR family ORFs and the Q3 ORF (Table 1). MsEPV LRR regions are also similar to Listeria internalin proteins (SwissProt accession no. P25146), Trypanosoma adenylate cyclase regulatory protein (SwissProt accession no. P23799), and yeast protein phosphatase SDS22 regulatory protein (SwissProt accession no. P22194). Other known LRR domain-containing proteins participate in protein-protein interactions, and most are involved in signal transduction (90, 109, 136).
Seven MsEPV ORFs belong to the alanine-leucine-isoleucine (ALI) motif family. The distinguishing feature of this gene family is an amino-terminal motif which contains invariant alanine, leucine, and isoleucine residues. ORFs containing the ALI motif are divided into two subgroups. Subgroup I includes four ORFs (MSV024, MSV026, MSV196, and MSV204) which range in size from 190 to 222 amino acids and contain Ile-Ile-X-Cys-Phe and Ile-Asp-Leu-Try/Phe-Phe motifs between residues 100 and 150. Subgroup II includes two longer proteins (MSV023 and MSV194) which lack the carboxyl-terminal motifs of the first group and instead contain a different, highly conserved (55% over 365 amino acids) carboxyl terminus. An additional subgroup II ALI motif protein (MSV195) has only 87 amino acids. Both subgroups of ALI family ORFs demonstrated amino-terminal homology to motif regions in putative genes from Chilo iridescent virus (GenBank accession no. AF003534); AcNPV (GenBank accession no. L22858); bacteriophages BK5-T (GenBank accession no. L44593), N15 (GenBank accession no. AF064539), and A2 (GenBank accession no. Y12813); and the bacterium Haemophilus influenzae (GenBank accession no. U32821). While the Chilo iridescent virus ORF showed extensive homology over the length of subgroup I ALI family ORFs, subgroup II ORFs shared an extended amino-terminal motif with the invertebrate virus, bacteriophage, and bacterial ORFs (data not shown).
MsEPV ORFs MSV027, MSV029, MSV034, MSV197, MSV205, and MSV252 comprise the tryptophan (W) repeat family. The distinguishing feature of this family is the presence of a repetitive 23-amino-acid motif that contains tryptophan, leucine, and isoleucine residues. These ORFs contain 3 to 12 copies of the motif, with a tryptophan residue spaced every 23 amino acids and a leucine or isoleucine residue spaced every 11 or 12 amino acids. Three of these ORFs (MSV027, MSV197, and MSV205) contain a carboxyl-terminal C3H2C3 (RING-H2) variation of the C3HC4 RING finger motif (17, 165), and one ORF (MSV029) contains a partial RING-H2 motif. The similarity of MsEPV W-repeat ORFs to an uncharacterized 41.6-kDa protein from AcNPV (Table 1) is based primarily on the periodically repeated tryptophan residues present in both proteins.
The methionine-threonine-glycine (MTG) family contains four ORFs (MSV021, MSV191, MSV198, and MSV199) and is defined by a 50-amino-acid amino-terminal motif containing these three invariant residues and by an internal motif [SxWxI(5x)FK]. MTG motif ORFs range in size from 158 to 399 amino acid residues. Interestingly, MTG family ORFs demonstrate similarity to ORFs C72R and C74R from Chilo iridescent virus (Table 1).
The serine-cysteine-glycine (SCG) family contains five ORFs (MSV214, MSV215, MSV216, MSV217, and MSV062). The distinguishing feature of this family is a 37- to 38-amino-acid motif with invariant serine, cysteine, and glycine residues around position 170 and a predicted transmembrane domain. SCG ORFs are relatively uniform in length (386 to 401 amino acids) and exhibit 34 to 46% amino acid identity.
Relationship of MsEPV to other genus B EPVs.
MsEPV resembles other EPVs in genome size, DNA composition, and the presence of conserved genes (6, 7). However, the absence of some genes present in other group B EPVs, low levels of amino acid identity to gene homologues, and extensive rearrangements in gene order suggest that MsEPV is distantly related to other described genus B EPVs. Genomic differences among EPVs may reflect the diversity within the hexopod class of arthropods and the long evolutionary presence of insects.
The size of the MsEPV genome (236 kbp) is similar to those reported for AmEPV (225 kbp) and HaEPV (233 kbp) (63, 175), and the high A+T content of MsEPV (82%) is comparable to that of the AmEPV genome (81.5%) (99, 100). ITRs of approximately 7 kbp may also be present in HaEPV (175).
MsEPV contains 10 of 16 ORFs previously described in other EPVs. These include the following: AmEPV G1L (vaccinia virus I7L), G2R, G4R (vaccinia virus A28), G5R (spheroidin), and G6L (vaccinia virus NPH-1) (66) and their homologues in CfEPV, CbEPV, and HaEPV (110, 175, 217); CbEPV DNA polymerase (131); AmEPV topoisomerase (143); HaEPV rifampicin resistance protein (vaccinia virus D13L) (139; and the F2 ORF from HaEPV (175). In addition, the 21 ORFs comprising the MsEPV LRR family are similar to the Q3 ORF that is located adjacent to the AmEPV TK gene (58). The LRR gene family and the Q3 ORF contain similar repetitive leucine-rich sequences, and both are located in terminal genomic locations.
Known EPV inclusion proteins are divergent or absent in MsEPV. The MsEPV spheroidin protein exhibits only about 20% amino acid identity to spheroidins of genus A (MmEPV) and genus B (HaEPV, AmEPV, and CbEPV) viruses. This contrasts with the 76 to 92% amino acid identity observed among other spheroidins of genus B viruses (6). In addition, MsEPV spheroidin contains approximately one-half (20 residues) the number of cysteines present in AmEPV and CbEPV (10, 215). As might be expected given the lack of observed spindle-shaped inclusions in MsEPV-infected cells (72), MsEPV does not encode a homologue of the major spindle body protein (fusolin, spindolin) that has been identified in group B and group A EPVs (6, 52, 70, 128), nor does it encode a gene with similarity to the AmEPV filament-associated protein (1). MsEPV lacks a TK gene, which is present in AmEPV, CbEPV, and CfEPV (6).
The MsEPV genome is not colinear with other described genus B EPV genomes. Of six known genes that are grouped adjacently in the AmEPV genome, MsEPV contains five homologues that are spread over 115 kbp (58, 66). This includes the spheroidin and NPH-I homologues, which despite their juxtaposition in all known genus B EPVs are separated by approximately 20 kbp in MsEPV (66). Similarly, the HaEPV F2L ORF is only 3 kbp from the HaEPV spheroidin gene, while MsEPV homologues of these genes are separated by 70 kbp (175). The MsEPV topoisomerase is adjacent to the vaccinia virus A28L homologue, while in AmEPV these genes are separated by over 100 kbp (143). Thus, the conserved colinear core of genes that has been proposed to exist for EPV genomes (65, 175) may in fact be conserved only among the lepidopteran viruses. Given the above discussion, orthopteran and lepidopteran EPVs may represent two distinct genera of entomopoxviruses.
Conclusions.
The MsEPV DNA sequence provides the first view of EPV genomics. Comparison of MsEPV with ChPVs establishes the genetic core of the Poxviridae. EPV genome analysis provides basic knowledge of viral functions, including response to DNA damage, nucleotide metabolism, manipulation of cellular stress responses, and virulence, which underlie viral interactions with the arthropod host and the environment. An improved understanding of these interactions will lead to the design of insect biocontrol strategies with enhanced efficacy and versatility.
ACKNOWLEDGMENTS
We thank A. Ciupryk, S. Mireilles, and J. R. Emanuelli for excellent technical assistance; T. Lewis for helpful discussion and assistance; and F. Barany, K. Becker, J. D. Boeke, J. Cox, T. H. Eickbush, A. Gabriel, B. D. Hammock, R. Kretsinger, H. Krokan, S. Parikh, and A. Sancar for comments on EPV gene homologues.
REFERENCES
- 1.Alaoui-Ismaili M H, Richardson C D. Identification and characterization of a filament-associated protein encoded by Amsacta mooreientomopoxvirus. J Virol. 1996;70:2697–2705. doi: 10.1128/jvi.70.5.2697-2705.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 3.Altschul S F, Madden T L, Schaffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Andres G, Simon-Mateo C, Vinuela E. Characterization of two African swine fever virus 220-kDa proteins: a precursor of the major structural protein p150 and an oligomer of phosphoprotein p32. Virology. 1993;194:284–293. doi: 10.1006/viro.1993.1259. [DOI] [PubMed] [Google Scholar]
- 5.Argos P. A sequence motif in many polymerases. Nucleic Acids Res. 1988;16:9909–9916. doi: 10.1093/nar/16.21.9909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Arif B M. Recent advances in the molecular biology of entomopoxviruses. J Gen Virol. 1995;76:1–13. doi: 10.1099/0022-1317-76-1-1. [DOI] [PubMed] [Google Scholar]
- 7.Arif B M, Kutstak E. The entomopoxviruses. In: Kutstak E, editor. Viruses of invertebrates. New York, N.Y: Marcell Dekker, Inc.; 1991. pp. 179–195. [Google Scholar]
- 8.Bailey L. Control of invertebrates by viruses. In: Gibbs A J, editor. Viruses and invertebrates. Vol. 31. New York, N.Y: Elsevier Publishing Company; 1973. pp. 534–553. [Google Scholar]
- 9.Bairoch A, Bucher P, Hofmann K. The PROSITE database: its status in 1997. Nucleic Acids Res. 1997;25:217–221. doi: 10.1093/nar/25.1.217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Banville M, Dumas F, Trifiro S, Arif B, Richardson C. The predicted amino acid sequence of the spheroidin protein from Amsacta mooreientomopoxvirus: lack of homology between major occlusion body proteins of different poxviruses. J Gen Virol. 1992;73:559–566. doi: 10.1099/0022-1317-73-3-559. [DOI] [PubMed] [Google Scholar]
- 11.Barford D. Molecular mechanisms of the protein serine/threonine phosphatases. Trends Biochem Sci. 1996;21:407–412. doi: 10.1016/s0968-0004(96)10060-8. [DOI] [PubMed] [Google Scholar]
- 12.Bayliss C D, Condit R C. The vaccinia virus A18R gene product is a DNA-dependent ATPase. J Biol Chem. 1995;270:1550–1556. doi: 10.1074/jbc.270.4.1550. [DOI] [PubMed] [Google Scholar]
- 13.Belfort M, Moelleken A, Maley G F, Maley F. Purification and properties of T4 phage thymidylate synthetase produced by the cloned gene in an amplification vector. J Biol Chem. 1983;258:2045–2051. [PubMed] [Google Scholar]
- 14.Bennett R A O, Wilson III D M, Wong D, Demple B. Interaction of human apurinic endonuclease and DNA polymerase β in the base excision repair pathway. Proc Natl Acad Sci USA. 1997;94:7166–7169. doi: 10.1073/pnas.94.14.7166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bertholet C, Stocco P, Van Meir E, Wittek R. Functional analysis of the 5′ flanking sequence of a vaccinia virus late gene. EMBO J. 1986;5:1951–1957. doi: 10.1002/j.1460-2075.1986.tb04449.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bode W, Gomis-Ruth F X, Stockler W. Astacins, serralysins, snake venom and matrix metalloproteinases exhibit identical zinc-binding environments (HEXXHXXGXXH and Met-turn) and topologies and should be grouped into a common family, the ‘metzincins’. FEBS Lett. 1993;331:134–140. doi: 10.1016/0014-5793(93)80312-i. [DOI] [PubMed] [Google Scholar]
- 17.Borden K L B, Freemont P S. The RING finger domain: a recent example of a sequence-structure family. Curr Opin Struct Biol. 1996;6:395–401. doi: 10.1016/s0959-440x(96)80060-1. [DOI] [PubMed] [Google Scholar]
- 18.Brady L, Brzozowski A M, Derewenda Z S, Dodson E, Dodson G, Tolley S, Turkenburg J P, Christiansen L, Huge-Jensen B, Norskov L, Thim L, Menge U. A serine protease triad forms the catalytic centre of a triacylglycerol lipase. Nature. 1990;343:767–770. doi: 10.1038/343767a0. [DOI] [PubMed] [Google Scholar]
- 19.Brendel V, Bucher P, Nourbakhsh I R, Blaisdell B E, Karlin S. Methods and algorithms for statistical analysis of protein sequences. Proc Natl Acad Sci USA. 1992;89:2002–2006. doi: 10.1073/pnas.89.6.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cappelli E, Taylor R, Cevasco M, Abbondandolo A, Caldecott K, Frosina G. Involvement of XRCC1 and DNA ligase III gene products in DNA base excision repair. J Biol Chem. 1997;272:23970–23975. doi: 10.1074/jbc.272.38.23970. [DOI] [PubMed] [Google Scholar]
- 21.Carreras C W, Santi D V. The catalytic mechanism and structure of thymidylate synthase. Annu Rev Biochem. 1995;64:721–762. doi: 10.1146/annurev.bi.64.070195.003445. [DOI] [PubMed] [Google Scholar]
- 22.Chin-Sang I D, Spence A M. Caenorhabditis eleganssex-determining protein FEM-2 is a protein phosphatase that promotes male development and interacts directly with FEM-3. Genes Dev. 1996;10:2314–2325. doi: 10.1101/gad.10.18.2314. [DOI] [PubMed] [Google Scholar]
- 23.Ciechanover A, Schwartz A L. The ubiquitin-proteasome pathway: the complexity and myriad functions of proteins death. Proc Natl Acad Sci USA. 1998;95:2727–2730. doi: 10.1073/pnas.95.6.2727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Claros M G, von Heijne G. TopPred II: an improved software for membrane protein structure predictions. Comput Appl Biosci. 1994;10:685–686. doi: 10.1093/bioinformatics/10.6.685. [DOI] [PubMed] [Google Scholar]
- 25.Clem R J, Miller L K. Control of programmed cell death by the baculovirus genes p35 and iap. Mol Cell Biol. 1994;14:5212–5222. doi: 10.1128/mcb.14.8.5212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Colinas R J, Goebel S J, Davis S W, Johnson G P, Norton E K, Paoletti E. A DNA ligase gene in the Copenhagen strain of vaccinia virus is nonessential for viral replication and recombination. Virology. 1990;179:267–275. doi: 10.1016/0042-6822(90)90295-3. [DOI] [PubMed] [Google Scholar]
- 27.Dall D, Sriskantha A, Vera A, Lai-Fook J, Symonds T. A gene encoding a highly expressed spindle body protein of Heliothis armigeraentomopoxvirus. J Gen Virol. 1993;74:1811–1818. doi: 10.1099/0022-1317-74-9-1811. [DOI] [PubMed] [Google Scholar]
- 28.Das A K, Helps N R, Cohen P T W, Barford D. Crystal structure of the protein serine/threonine phosphatase 2C at 2.0 Å resolution. EMBO J. 1996;15:6798–6809. [PMC free article] [PubMed] [Google Scholar]
- 29.Davies J F I, Almassy R J, Hostomska Z, Ferre R A, Hostomsky Z. 2.3 Å crystal structure of the catalytic domain of DNA polymerase β. Cell. 1994;76:1123–1133. doi: 10.1016/0092-8674(94)90388-3. [DOI] [PubMed] [Google Scholar]
- 30.Demple B, Harrison L. Repair of oxidative damage to DNA: enzymology and biology. Annu Rev Biochem. 1994;63:915–948. doi: 10.1146/annurev.bi.63.070194.004411. [DOI] [PubMed] [Google Scholar]
- 31.Derewenda Z S, Derewenda U, Dodson G G. The crystal and molecular structure of the Rhizomucor mieheitriacylglyceride lipase at 1.9 Å resolution. J Mol Biol. 1992;227:818–839. doi: 10.1016/0022-2836(92)90225-9. [DOI] [PubMed] [Google Scholar]
- 32.Derewenda Z S, Sharp A M. News from the interface: the molecular structures of triacylglyceride lipases. Trends Biochem Sci. 1993;18:20–25. doi: 10.1016/0968-0004(93)90082-x. [DOI] [PubMed] [Google Scholar]
- 33.Devereux J, Haeberli P, Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984;12:387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dianov G, Price A, Lindahl T. Generation of single-nucleotide repair patches following excision of uracil residues from DNA. Mol Cell Biol. 1992;12:1605–1612. doi: 10.1128/mcb.12.4.1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Duckett C S, Nava V E, Gedrich R W, Clem R J, Van Dongen J L, Gilfillan M C, Shiels H, Hardwick J M, Thompson C B. A conserved family of cellular genes related to the baculovirus iapgene and encoding apoptosis inhibitors. EMBO J. 1996;15:2685–2694. [PMC free article] [PubMed] [Google Scholar]
- 36.Duncan L, Alper S, Arigoni F, Losick R, Stragier P. Activation of cell-specific transcription by a serine phophatase at the site of asymmetric division. Science. 1995;270:641–646. doi: 10.1126/science.270.5236.641. [DOI] [PubMed] [Google Scholar]
- 37.Eddy S R. Hidden Markov models. Curr Opin Struct Biol. 1996;6:361–365. doi: 10.1016/s0959-440x(96)80056-x. [DOI] [PubMed] [Google Scholar]
- 38.Eddy S R, Mitchison G, Durbin R. Maximum discrimination hidden Markov models of sequence consensus. J Comput Biol. 1995;2:9–23. doi: 10.1089/cmb.1995.2.9. [DOI] [PubMed] [Google Scholar]
- 39.Ellis L B M, Saurugger P, Woodward C. Identification of the three-dimensional thioredoxin motif: related structure in the ORF3 protein of the Staphylococcus aureus meroperon. Biochemistry. 1992;31:4882–4891. doi: 10.1021/bi00135a020. [DOI] [PubMed] [Google Scholar]
- 40.Erlandson M. Protease activity associated with occlusion body preparations of an entomopoxvirus from Melanoplus sanguinipes. J Invertebr Pathol. 1991;57:255–263. [Google Scholar]
- 41.Erlandson M A, Streett D A, Goettel M S, Johnson D L, editors. Memoirs of the Entomological Society of Canada. Vol. 171. Ottawa, Ontario, Canada: Entomological Society of Canada; 1997. Entomopoxviruses associated with grasshoppers and locusts: biochemical characterization; pp. 131–146. [Google Scholar]
- 42.Ewing B, Green P. Base-calling of automated sequencer traces using Phred. II. Error probabilities. Genome Res. 1998;8:186–194. [PubMed] [Google Scholar]
- 43.Ewing B, Hillier L, Wendl M C, Green P. Base-calling of automated sequencer traces using Phred. I. Accuracy assessment. Genome Res. 1998;8:175–185. doi: 10.1101/gr.8.3.175. [DOI] [PubMed] [Google Scholar]
- 44.Fabian P, Murvai J, Hatsagi Z, Vlahovicek K, Hegyi H, Pongor S. The SBASE protein domain library, release 5.0: a collection of annotated protein sequence segments. Nucleic Acids Res. 1997;25:240–243. doi: 10.1093/nar/25.1.240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Felsenstein J. PHYLIP—phylogeny inference package (version 3.2) Cladistics. 1989;5:164–166. [Google Scholar]
- 46.Feng Q, Moran J V, Kazazian H H, Jr, Boeke J D. Human L1 retrotransposon encodes a conserved endonuclease required for retrotransposition. Cell. 1996;87:905–916. doi: 10.1016/s0092-8674(00)81997-2. [DOI] [PubMed] [Google Scholar]
- 47.Francavilla A, Vujanovic N L, Polimeno L, Azzarone A, Iacobellis A, Deleo A, Hagiya M, Whiteside T L, Starzl T E. The in vivoeffect of hepatotrophic factors augmenter of liver regeneration, hepatocyte growth factor, and insulin-like growth factor-II on liver natural killer cell functions. Hepatology. 1997;25:411–415. doi: 10.1002/hep.510250225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Franke C A, Wilson E M, Hruby D E. Use of a cell-free system to identify the vaccinia virus L1R gene product as the major late myristylated virion protein M25. J Virol. 1990;64:5988–5996. doi: 10.1128/jvi.64.12.5988-5996.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Friesen P D, Nissen M S. Gene organization and transcription of TED, a lepidopteran retrotransposon integrated within the baculovirus genome. Mol Cell Biol. 1990;10:3067–3077. doi: 10.1128/mcb.10.6.3067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gade G, Beenakkers A M T. Adipokinetic hormone-induced lipid mobilization and cyclic AMP accumulation in the fat body of Locusta migratoriaduring development. Gen Comp Endocrinol. 1977;32:481–487. doi: 10.1016/0016-6480(77)90231-3. [DOI] [PubMed] [Google Scholar]
- 51.Galtier N, Gouy M, Gautier C. SEAVIEW and PHYLO_WIN: two graphic tools for sequence alignment and molecular phylogeny. CABIOS. 1996;12:543–554. doi: 10.1093/bioinformatics/12.6.543. [DOI] [PubMed] [Google Scholar]
- 52.Gauthier L, Cousserans F, Veyrunes J C, Bergoin M. The Melolontha melolontha entomopoxvirus (MmEPV) fusolin is related to the fusolins of lepidopteran EPVs and to the 37K baculovirus glycoprotein. Virology. 1995;208:427–436. doi: 10.1006/viro.1995.1173. [DOI] [PubMed] [Google Scholar]
- 53.Goebel S J, Johnson G P, Perkus M E, Davis S W, Winslow J P, Paoletti E. The complete DNA sequence of vaccinia virus. Virology. 1990;179:247–266. doi: 10.1016/0042-6822(90)90294-2. [DOI] [PubMed] [Google Scholar]
- 54.Goodwin R H, Milner R J, Beaton C D. Entomopoxvirinae. In: Adams J R, Bonami J R, editors. Atlas of invertebrate viruses. Vol. 8. Boca Raton, Fla: CRC Press, Inc.; 1991. pp. 259–285. [Google Scholar]
- 55.Gorbalenya A E, Koonin E V, Donchenko A P, Blinov V M. Two related superfamilies of putative helicases involved in replication, recombination, repair and expression of DNA and RNA genomes. Nucleic Acids Res. 1989;17:4713–4730. doi: 10.1093/nar/17.12.4713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gordon D, Abajian C, Green P. Consed: a graphical tool for sequence finishing. Genome Res. 1998;8:192–202. doi: 10.1101/gr.8.3.195. [DOI] [PubMed] [Google Scholar]
- 57.Granados R R, Roberts D W. Electron microscopy of a poxlike virus infecting an invetebrate host. Virology. 1970;40:230–243. doi: 10.1016/0042-6822(70)90398-3. [DOI] [PubMed] [Google Scholar]
- 58.Gruidl M E, Hall R L, Moyer R W. Mapping and molecular characterization of a functional thymidine kinase from Amsacta mooreientomopoxvirus. Virology. 1992;186:507–516. doi: 10.1016/0042-6822(92)90016-i. [DOI] [PubMed] [Google Scholar]
- 59.Guarino L A, Smith G, Dong W. Ubiquitin is attached to membranes of baculovirus particles by a novel type of phospholipid anchor. Cell. 1995;80:301–309. doi: 10.1016/0092-8674(95)90413-1. [DOI] [PubMed] [Google Scholar]
- 60.Haas A L, Baboshina O, Williams B, Schwartz L M. Coordinated induction of the ubiquitin conjugation pathway accompanies the developmentally programmed death of insect skeletal muscle. J Biol Chem. 1995;270:9407–9412. doi: 10.1074/jbc.270.16.9407. [DOI] [PubMed] [Google Scholar]
- 61.Haas A L, Katzung D J, Reback P M, Guarino L A. Functional characterization of the ubiquitin variant encoded by the baculovirus Autographa californica. Biochemistry. 1996;35:5385–5394. doi: 10.1021/bi9524981. [DOI] [PubMed] [Google Scholar]
- 62.Hagiya M, Francavilla A, Polimeno L, Ihara I, Sakai H, Seki T, Shimonishi M, Porter K A, Starzl T E. Cloning and sequence analysis of the rat augmenter of liver regeneration (ALR) gene: expression of biologically active recombinant ALR and demonstration of tissue distribution. Proc Natl Acad Sci USA. 1994;91:8142–8146. doi: 10.1073/pnas.91.17.8142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hall R L, Hink W F. Physical mapping and field inversion gel electrophoresis of Amsacta mooreientomopoxvirus DNA. Arch Virol. 1990;110:77–90. doi: 10.1007/BF01310704. [DOI] [PubMed] [Google Scholar]
- 64.Hall R L, Li Y, Feller J, Moyer R W. The Amsacta mooreientomopoxvirus spheroidin gene is improperly transcribed in vertebrate poxviruses. Virology. 1996;224:427–436. doi: 10.1006/viro.1996.0549. [DOI] [PubMed] [Google Scholar]
- 65.Hall R L, Moyer R W. Identification of an Amsacta spheroidin-like protein within the occlusion bodies of Choristoneuraentomopoxviruses. Virology. 1993;192:179–187. doi: 10.1006/viro.1993.1020. [DOI] [PubMed] [Google Scholar]
- 66.Hall R L, Moyer R W. Identification, cloning, and sequencing of a fragment of Amsacta mooreientomopoxvirus DNA containing the spheroidin gene and three vaccinia virus-related open reading frames. J Virol. 1991;65:6516–6527. doi: 10.1128/jvi.65.12.6516-6527.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hammond J M, Kerr S M, Smith G L, Dixon L K. An African swine fever virus gene with homology to DNA ligases. Nucleic Acids Res. 1992;20:2667–2671. doi: 10.1093/nar/20.11.2667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hammond R A, McClung J K, Miller M R. Effect of DNA polymerase inhibitors on DNA repair in intact and permeable human fibroblasts: evidence that DNA polymerase δ and β are involved in DNA repair synthesis induced by N-methyl-N′-nitro-N-nitrosoguanidine. Biochemistry. 1990;29:286–291. doi: 10.1021/bi00453a039. [DOI] [PubMed] [Google Scholar]
- 69.Hanks S K, Quinn A M, Hunter T. The protein kinase family: conserved features and deduced phylogeny of the catalytic domains. Science. 1988;241:42–52. doi: 10.1126/science.3291115. [DOI] [PubMed] [Google Scholar]
- 70.Hayakawa T, Xu J, Hukuhara T. Cloning and sequencing of the gene for an enhancing factor from Pseudaletia separataentomopoxvirus. Gene. 1996;177:269–270. doi: 10.1016/0378-1119(96)00297-1. [DOI] [PubMed] [Google Scholar]
- 71.Hearst J E. The structure of photolyase: using photon energy for DNA repair. Science. 1995;268:1858–1859. doi: 10.1126/science.7604259. [DOI] [PubMed] [Google Scholar]
- 72.Henry J E, Nelson B P, Jutila J W. Pathology and development of the grasshopper inclusion body virus in Melanoplus sanguinipes. J Virol. 1969;3:605–610. doi: 10.1128/jvi.3.6.605-610.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hertig C, Coupar B E H, Gould A R, Boyle D B. Field and vaccine strains of fowlpox virus carry integrated sequences from the avian retrovirus, reticuloendotheliosis virus. Virology. 1997;235:367–376. doi: 10.1006/viro.1997.8691. [DOI] [PubMed] [Google Scholar]
- 74.Higgins D G, Thompson J D, Gibson T J. Using CLUSTAL for multiple sequence alignments. Methods Enzymol. 1996;266:383–402. doi: 10.1016/s0076-6879(96)66024-8. [DOI] [PubMed] [Google Scholar]
- 75.Hingamp P M, Arnold J E, Mayer R J, Dixon L K. A ubiquitin conjugating enzyme encoded by African swine fever virus. EMBO J. 1992;11:361–366. doi: 10.1002/j.1460-2075.1992.tb05058.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hingamp P M, Leyland M L, Webb J, Twigger S, Mayer R J, Dixon L K. Characterization of a ubiquitinated protein which is externally located in African swine fever virions. J Virol. 1995;69:1785–1793. doi: 10.1128/jvi.69.3.1785-1793.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Hirose F, Hotta Y, Yamaguchi M, Matsukage A. Difference in the expression level of DNA polymerase β among mouse tissues: high expression in the pachytene spermatocyte. Exp Cell Res. 1989;181:169–180. doi: 10.1016/0014-4827(89)90191-2. [DOI] [PubMed] [Google Scholar]
- 78.Holmgren A. Thioredoxin. Annu Rev Biochem. 1985;54:237–271. doi: 10.1146/annurev.bi.54.070185.001321. [DOI] [PubMed] [Google Scholar]
- 79.Honess R W, Bodemer W, Cameron K R, Niller H-H, Fleckenstein B, Randall R E. The A+T-rich genome of Herpesvirus saimiricontains a highly conserved gene for thymidylate synthase. Proc Natl Acad Sci USA. 1986;83:3604–3608. doi: 10.1073/pnas.83.11.3604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Honess R W, Craxton M A, Williams L, Gompels U A. A comparative analysis of the sequence of the thymidine kinase gene of a gammaherpesvirus, herpesvirus saimiri. J Gen Virol. 1989;70:3003–3013. doi: 10.1099/0022-1317-70-11-3003. [DOI] [PubMed] [Google Scholar]
- 81.Hooper N M. Families of zinc metalloproteases. FEBS Lett. 1994;354:1–6. doi: 10.1016/0014-5793(94)01079-x. [DOI] [PubMed] [Google Scholar]
- 82.Jaeger B, Langridge W H R. Infection of Locusta migratoria with entomopoxviruses from Arphia conspersa and Melanoplus sanguinipesgrasshoppers. J Invert Pathol. 1984;43:374–382. [Google Scholar]
- 83.Jennings M P, Hood D W, Peak I R A, Virji M, Moxon R E. Molecular analysis of a locus for the biosynthesis and phase-variable expression of the lacto-N-neotetraose terminal lipopolysaccharide structure in Neisseria meningitidis. Mol Microbiol. 1995;18:729–740. doi: 10.1111/j.1365-2958.1995.mmi_18040729.x. [DOI] [PubMed] [Google Scholar]
- 84.Jones D T, Taylor W R, Thornton J M. A model recognition approach to the prediction of all-helical membrane protein structure and topology. Biochemistry. 1994;33:3038–3049. doi: 10.1021/bi00176a037. [DOI] [PubMed] [Google Scholar]
- 85.Jones D T, Taylor W R, Thornton J M. A mutation data matrix for transmembrane proteins. FEBS Lett. 1994;339:269–275. doi: 10.1016/0014-5793(94)80429-x. [DOI] [PubMed] [Google Scholar]
- 86.Jongeneel C V, Bouvier J, Bairoch A. A unique signature identifies a family of zinc-dependent metallopeptidases. FEBS Lett. 1989;242:211–214. doi: 10.1016/0014-5793(89)80471-5. [DOI] [PubMed] [Google Scholar]
- 87.Kanai S, Kikuno R, Toh H, Ryo H, Todo T. Molecular evolution of the photolyase-blue-light photoreceptor family. J Mol Evol. 1997;45:535–548. doi: 10.1007/pl00006258. [DOI] [PubMed] [Google Scholar]
- 88.Karlin S, Brendel V, Bucher P. Significant similarity and dissimilarity in homologous proteins. Mol Biol Evol. 1992;9:152–167. doi: 10.1093/oxfordjournals.molbev.a040704. [DOI] [PubMed] [Google Scholar]
- 89.Kerr S M, Smith G L. Vaccinia virus encodes a polypeptide with DNA ligase activity. Nucleic Acids Res. 1989;17:9039–9050. doi: 10.1093/nar/17.22.9039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kobe B, Deisenhofer J. Crystal structure of porcine ribonuclease inhibitor, a protein with leucine-rich repeats. Nature. 1993;366:751–756. doi: 10.1038/366751a0. [DOI] [PubMed] [Google Scholar]
- 91.Koonin E V. A highly conserved sequence motif defining the family of MutT-related proteins from eubacteria, eukaryotes and viruses. Nucleic Acids Res. 1993;21:4847. doi: 10.1093/nar/21.20.4847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Koonin E V, Senkevich T G. Fowlpox virus encodes a protein related to human deoxycytidine kinase: further evidence for independent acquisition of genes for enzymes of nucleotide metabolism by different viruses. Virus Genes. 1993;7:289–295. doi: 10.1007/BF01702589. [DOI] [PubMed] [Google Scholar]
- 93.Koonin E V, Senkevich T G. Vaccinia virus encodes four putative DNA and/or RNA helicases distantly related to each other. J Gen Virol. 1992;73:989–993. doi: 10.1099/0022-1317-73-4-989. [DOI] [PubMed] [Google Scholar]
- 94.Koonin E V, Tatusov R L, Rudd K E. Protein sequence comparison at genome scale. Methods Enzymol. 1996;266:295–322. doi: 10.1016/s0076-6879(96)66020-0. [DOI] [PubMed] [Google Scholar]
- 95.Kotwal G J, Hugin A W, Moss B. Mapping and insertional mutagenesis of a vaccinia virus gene encoding a 13,800-Da secreted protein. Virology. 1989;171:579–587. doi: 10.1016/0042-6822(89)90627-2. [DOI] [PubMed] [Google Scholar]
- 96.Kumar A, Widen S G, Williams K R, Kedar P, Karpel R L, Wilson S H. Studies of the domain structure of mammalian DNA polymerase β. J Biol Chem. 1990;265:2124–2131. [PubMed] [Google Scholar]
- 97.Kutish G F, Li Y, Lu Z, Furuta M, Rock D L, Van Etten J L. Analysis of 76 kb of the chlorella virus PBCV-1 330-kb genome: map positions 182 to 258. Virology. 1996;223:303–317. doi: 10.1006/viro.1996.0482. [DOI] [PubMed] [Google Scholar]
- 98.Lai-Fook J, Brooks E M, Dall D J. Intracellular structures associated with in vitro replication of Heliothis armigeraentomopoxvirus. J Invertebr Pathol. 1994;64:269–272. doi: 10.1006/jipa.1999.4905. [DOI] [PubMed] [Google Scholar]
- 99.Langridge W H R, Roberts D W. Molecular weight of DNA from four entomopoxviruses determined by electron microscopy. J Virol. 1977;21:301–308. doi: 10.1128/jvi.21.1.301-308.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Langridge W H R, Bozarth R F, Roberts D W. The base composition of entomopoxvirus DNA. Virology. 1977;76:616–620. doi: 10.1016/0042-6822(77)90243-4. [DOI] [PubMed] [Google Scholar]
- 101.Langridge W H R, Henry J E. Molecular weight and base composition of DNA isolated from Melanoplus sanguinipesentomopoxvirus. J Invertebr Pathol. 1981;37:34–37. [Google Scholar]
- 102.Langridge W H R, Oma E, Henry J E. Characterization of the DNA and structural proteins of entomopoxviruses from Melanoplus sanguinipes, Arphia conspirsa, and Phoetaliotes nebrascensis(Orthoptera) J Invertebr Pathol. 1983;42:327–333. [Google Scholar]
- 103.Lasko D D, Tomkinson A E, Lindahl T. Eukaryotic DNA ligases. Mutat Res. 1990;236:277–287. doi: 10.1016/0921-8777(90)90011-s. [DOI] [PubMed] [Google Scholar]
- 104.Lee P, Hruby D E. transprocessing of vaccinia virus core proteins. J Virol. 1993;67:4252–4263. doi: 10.1128/jvi.67.7.4252-4263.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.LeMaster D M, Springer P A, Unkefer C J. The role of the buried aspartate of Escherichia colithioredoxin in the activation of the mixed disulfide intermediate. J Biol Chem. 1997;272:29998–30001. doi: 10.1074/jbc.272.48.29998. [DOI] [PubMed] [Google Scholar]
- 106.Lepore L S, Roelvink P R, Granados R R. Enhancin, the granulosis virus protein that facilitates nucleopolyhedrovirus (NPV) infections, is a metalloprotease. J Invertebr Pathol. 1996;68:131–140. doi: 10.1006/jipa.1996.0070. [DOI] [PubMed] [Google Scholar]
- 107.Lerch R A, Friesen P D. The baculovirus-integrated retrotransposon TED encodes gag and polproteins that assemble into viruslike particles with reverse transcriptase. J Virol. 1992;66:1590–1601. doi: 10.1128/jvi.66.3.1590-1601.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Levin D B, Adachi D, Williams L L, Myles T G. Host specificity and molecular characterization of the entomopoxvirus of the lesser migratory grasshopper, Melanoplus sanguinipes. J Invertebr Pathol. 1993;62:241–247. [Google Scholar]
- 109.Li J, Chory J. A putative leucine-rich repeat receptor kinase involved in brassinosteroid signal transduction. Cell. 1997;90:929–938. doi: 10.1016/s0092-8674(00)80357-8. [DOI] [PubMed] [Google Scholar]
- 110.Li X, Barrett J W, Yuen L, Arif B M. Cloning, sequencing and transcriptional analysis of the Choristoneura fumiferanaentomopoxvirus spheroidin gene. Virus Res. 1997;47:143–154. doi: 10.1016/s0168-1702(96)01409-8. [DOI] [PubMed] [Google Scholar]
- 111.Li Y, Hall R L, Moyer R W. Transient, nonlethal expression of genes in vertebrate cells by recombinant entomopoxviruses. J Virol. 1997;71:9557–9562. doi: 10.1128/jvi.71.12.9557-9562.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Lin S, Broyles S S. Vaccinia protein kinase 2: a second essential serine/threonine protein kinase encoded by vaccinia virus. Proc Natl Acad Sci USA. 1994;91:7653–7657. doi: 10.1073/pnas.91.16.7653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Lin S, Chen W, Broyles S S. The vaccinia virus B1R gene product is a serine/threonine protein kinase. J Virol. 1992;66:2717–2723. doi: 10.1128/jvi.66.5.2717-2723.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Lindahl T. Instability and decay of the primary structure of DNA. Nature. 1993;362:709–715. doi: 10.1038/362709a0. [DOI] [PubMed] [Google Scholar]
- 115.Lindahl T, Barnes D E. Mammalian DNA ligases. Annu Rev Biochem. 1992;61:251–281. doi: 10.1146/annurev.bi.61.070192.001343. [DOI] [PubMed] [Google Scholar]
- 116.Lipman D J, Altschul S F, Kececioglu J D. A tool for multiple sequence alignment. Proc Natl Acad Sci USA. 1989;86:4412–4415. doi: 10.1073/pnas.86.12.4412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Lisowsky T. ERV1 is involved in the cell-division cycle and the maintenance of mitochondrial genomes in Saccharomyces cerevisiae. Curr Genet. 1994;26:15–20. doi: 10.1007/BF00326299. [DOI] [PubMed] [Google Scholar]
- 118.Luo J, Barany F. Identification of essential residues in Thermus thermophilusDNA ligase. Nucleic Acids Res. 1996;24:3079–3085. doi: 10.1093/nar/24.15.3079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Lytvyn V, Fortin Y, Banville M, Arif B, Richardson C. Comparison of the thymidine kinase genes from three entomopoxviruses. J Gen Virol. 1992;73:3235–3240. doi: 10.1099/0022-1317-73-12-3235. [DOI] [PubMed] [Google Scholar]
- 120.Martin K H, Grosenbach D W, Franke C A, Hruby D E. Identification and analysis of three myristylated vaccinia virus late proteins. J Virol. 1997;71:5218–5226. doi: 10.1128/jvi.71.7.5218-5226.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Massung R F, Knight J C, Esposito J J. Topography of variola smallpox virus inverted terminal repeats. Virology. 1995;211:350–355. doi: 10.1006/viro.1995.1416. [DOI] [PubMed] [Google Scholar]
- 122.Massung R F, Liu L, Qi J, Knight J C, Yuran T E, Kerlavage A R, Parsons J M, Venter J C, Esposito J J. Analysis of the complete genome of smallpox variola major virus strain Bangladesh-1975. Virology. 1994;201:215–240. doi: 10.1006/viro.1994.1288. [DOI] [PubMed] [Google Scholar]
- 123.Matsumoto Y, Bogenhagen D F. Repair of a synthetic abasic site in DNA in a Xenopus laevisoocyte extract. Mol Cell Biol. 1989;9:3750–3757. doi: 10.1128/mcb.9.9.3750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Matsumoto Y, Kim K. Excision of deoxyribose phosphate residues by DNA polymerase B during DNA repair. Science. 1995;269:699–702. doi: 10.1126/science.7624801. [DOI] [PubMed] [Google Scholar]
- 125.Michaels M L, Miller J H. The GO system protects organisms from the mutagenic effect of the spontaneous lesion 8-hydroxyguanine (7,8-dihydro-8-oxoguanine) J Bacteriol. 1992;174:6321–6325. doi: 10.1128/jb.174.20.6321-6325.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Millns A K, Carpenter M S, DeLange A M. The vaccinia virus-encoded uracil DNA glycosylase has an essential role in viral DNA replication. Virology. 1994;198:504–513. doi: 10.1006/viro.1994.1061. [DOI] [PubMed] [Google Scholar]
- 127.Miranpuri G S, Erlandson M A, Gillespie J P, Khachatourians G G. Changes in hemolymph of the migratory grasshopper, Melanoplus sanguinipes, infected with an entomopoxvirus. J Invertebr Pathol. 1992;60:274–282. [Google Scholar]
- 128.Mitsuhashi W, Saito H, Sato M. Complete nucleotide sequence of the fusolin gene from an entomopoxvirus in the cupreous chafer, Anomala cuprea Hope(Coleoptera: Scarabaeidae) Insect Biochem Mol Biol. 1997;27:869–876. doi: 10.1016/s0965-1748(97)00069-6. [DOI] [PubMed] [Google Scholar]
- 129.Moore J K, Haber J E. Capture of retrotransposon DNA at the sites of chromosomal double-strand breaks. Nature. 1996;383:644–646. doi: 10.1038/383644a0. [DOI] [PubMed] [Google Scholar]
- 130.Moss B. Poxviridae: the viruses and their replication. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology. Philadelphia, Pa: Lippincott-Raven; 1996. pp. 2637–2671. [Google Scholar]
- 131.Mustafa A, Yuen L. Identification and sequencing of the Choristoneura biennisentomopoxvirus DNA polymerase gene. J DNA Sequencing Mapping. 1991;2:39–45. doi: 10.3109/10425179109008437. [DOI] [PubMed] [Google Scholar]
- 132.Neilan J G, Lu Z, Kutish G F, Zsak L, Burrage T G, Borca M V, Carrillo C, Rock D L. A BIR motif containing gene of African swine fever virus, 4CL, is nonessential for growth in vitroand viral virulence. Virology. 1997;230:252–264. doi: 10.1006/viro.1997.8481. [DOI] [PubMed] [Google Scholar]
- 133.Neuwald A F, Green P. Detecting patterns in protein sequences. J Mol Biol. 1994;239:698–712. doi: 10.1006/jmbi.1994.1407. [DOI] [PubMed] [Google Scholar]
- 134.Neuwald A F, Liu J S, Lipman D J, Lawrence C E. Extracting protein alignment models from the sequence database. Nucleic Acids Res. 1997;25:1665–1677. doi: 10.1093/nar/25.9.1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Niki I, Yokokura H, Sudo T, Kato M, Hidaka H. Ca2+ signalling and intracellular Ca2+binding proteins. J Biochem. 1996;120:685–698. doi: 10.1093/oxfordjournals.jbchem.a021466. [DOI] [PubMed] [Google Scholar]
- 136.Ohkura H, Yanagida M. S. pombegene sds22+ essential for a midmitotic transition encodes a leucine-rich repeat protein that positively modulates protein phosphatase-1. Cell. 1991;64:149–157. doi: 10.1016/0092-8674(91)90216-l. [DOI] [PubMed] [Google Scholar]
- 137.Oliveros M, Yanez R J, Salas M L, Salas J, Vinuela E, Blanco L. Characterization of an African swine fever virus 20-kDa DNA polymerase involved in DNA repair. J Biol Chem. 1997;272:30899–30910. doi: 10.1074/jbc.272.49.30899. [DOI] [PubMed] [Google Scholar]
- 138.O’Reilly D R, Miller L K. A baculovirus blocks insect molting by producing ecdysteroid UDP-glucosyl transferase. Science. 1989;245:1110–1112. doi: 10.1126/science.2505387. [DOI] [PubMed] [Google Scholar]
- 139.Osborne R J, Symonds T M, Sriskantha A, Lai-Fook J, Fernon C A, Dall D J. An entomopoxvirus homologue of the vaccinia virus D13L-encoded ‘rifampicin resistance’ protein. J Gen Virol. 1996;77:839–846. doi: 10.1099/0022-1317-77-5-839. [DOI] [PubMed] [Google Scholar]
- 140.Pearson A, Richardson C, Yuen L. The 5′ noncoding region sequence of the Choristoneura biennisentomopoxvirus spheroidin gene functions as an efficient late promoter in the mammalian vaccinia expression system. Virology. 1991;180:561–566. doi: 10.1016/0042-6822(91)90070-r. [DOI] [PubMed] [Google Scholar]
- 141.Pearson W R. Rapid and sensitive sequence comparison with FASTP and FASTA. Methods Enzymol. 1990;183:63–98. doi: 10.1016/0076-6879(90)83007-v. [DOI] [PubMed] [Google Scholar]
- 142.Pearson W R, Lipman D J. Improved tools for biological sequence comparison. Proc Natl Acad Sci USA. 1988;85:2444–2448. doi: 10.1073/pnas.85.8.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Petersen B O, Hall R L, Moyer R W, Shuman S. Characterization of a DNA topoisomerase encoded by Amsacta mooreientomopoxvirus. Virology. 1997;230:197–206. doi: 10.1006/viro.1997.8495. [DOI] [PubMed] [Google Scholar]
- 144.Plowright W, Parker J, Peirce M A. African swine fever virus in ticks (Ornithodoros moubata, Murray) collected from animal burrows in Tanzania. Nature. 1969;221:1071–1073. doi: 10.1038/2211071a0. [DOI] [PubMed] [Google Scholar]
- 145.Potter M D, Lo R Y C. Cloning and characterization of the galE locus of Pasteurella haemolyticaA1. Infect Immun. 1996;64:855–860. doi: 10.1128/iai.64.3.855-860.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Prichard M N, Duke G M, Mocarski E S. Human cytomegalovirus uracil DNA glycosylase is required for the normal temporal regulation of both DNA synthesis and viral replication. J Virol. 1996;70:3018–3025. doi: 10.1128/jvi.70.5.3018-3025.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Ravanello M P, Hruby D E. Characterization of the vaccinia virus L1R myristylprotein as a component of the intracellular virion envelope. J Gen Virol. 1994;75:1479–1483. doi: 10.1099/0022-1317-75-6-1479. [DOI] [PubMed] [Google Scholar]
- 148.Ravanello M P, Hruby D E. Conditional lethal expression of the vaccinia virus L1R myristylated protein reveals a role in virion assembly. J Virol. 1994;68:6401–6410. doi: 10.1128/jvi.68.10.6401-6410.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Rawlings N D, Barrett A J. Evolutionary families of metallopeptidases. Methods Enzymol. 1995;248:183–228. doi: 10.1016/0076-6879(95)48015-3. [DOI] [PubMed] [Google Scholar]
- 150.Reilly L M, Guarino L A. The viral ubiquitin gene of Autographa californicanuclear polyhedrosis virus is not essential for viral replication. Virology. 1996;218:243–247. doi: 10.1006/viro.1996.0185. [DOI] [PubMed] [Google Scholar]
- 151.Rempel R E, Anderson M K, Evans E, Traktman P. Temperature-sensitive vaccinia virus mutants identify a gene with an essential role in viral replication. J Virol. 1990;64:574–583. doi: 10.1128/jvi.64.2.574-583.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Ring M, Pfeifer T A, Grigliatti T A. Identification of a 5′ truncated non-LTR-retrotransposon, YAKPs1, from the variegated cutworm, Peridroma saucia, using PCR. Insect Biochem Mol Biol. 1996;26:511–518. doi: 10.1016/0965-1748(96)00007-0. [DOI] [PubMed] [Google Scholar]
- 153.Rodríguez D, Esteban M, Rodríguez J R. Vaccinia virus A17L gene product is essential for an early step in virion morphogenesis. J Virol. 1995;69:4640–4648. doi: 10.1128/jvi.69.8.4640-4648.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Rodriguez J M, Salas M L, Vinuela E. Genes homologous to ubiquitin-conjugating proteins and eukaryotic transcription factor SII in African swine fever virus. Virology. 1992;186:40–52. doi: 10.1016/0042-6822(92)90059-x. [DOI] [PubMed] [Google Scholar]
- 155.Rodríguez J R, Risco C, Carrascosa J L, Esteban M, Rodríguez D. Vaccinia virus 15-kilodalton (A14L) protein is essential for assembly and attachment of viral crescents to virosomes. J Virol. 1998;72:1287–1296. doi: 10.1128/jvi.72.2.1287-1296.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Roelvink P W, Corsaro B G, Granados R R. Characterization of the Helicoverpa armigera and Pseudaletia unipunctagranulovirus enhancin genes. J Gen Virol. 1995;76:2693–2705. doi: 10.1099/0022-1317-76-11-2693. [DOI] [PubMed] [Google Scholar]
- 157.Rosel J, Moss B. Transcriptional and translational mapping and nucleotide sequence analysis of a vaccinia virus gene encoding the precursor of the major core polypeptide 4b. J Virol. 1985;56:830–838. doi: 10.1128/jvi.56.3.830-838.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Rosel J L, Earl P L, Weir J P, Moss B. Conserved TAAATG sequence at the transcriptional and translational initiation sites of vaccinia virus late genes deduced by structural and functional analysis of the HindIII H genome fragment. J Virol. 1986;60:436–449. doi: 10.1128/jvi.60.2.436-449.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Roseman N A, Hruby D E. Nucleotide sequence and transcript organization of a region of the vaccinia virus genome which encodes a constitutively expressed gene required for DNA replication. J Virol. 1987;61:1398–1406. doi: 10.1128/jvi.61.5.1398-1406.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Russell R L Q, Rohrmann G F. Nucleotide sequence of the ubiquitin-39K region from the Orgyia pseudotsugatamultinucleocapsid nuclear polyhedrosis virus genome. J Gen Virol. 1993;74:1191–1195. doi: 10.1099/0022-1317-74-6-1191. [DOI] [PubMed] [Google Scholar]
- 161.Saitou N, Nei M. The neighbor-joining method: a new method for recontructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 162.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 163.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Sanz P, Veyrunes J-C, Cousserans F, Bergoin M. Cloning and sequencing of the spherulin gene, the occlusion body major polypeptide of the Melolontha melolontha entomopoxvirus (MmEPV) Virology. 1994;202:449–457. doi: 10.1006/viro.1994.1361. [DOI] [PubMed] [Google Scholar]
- 165.Saurin A J, Borden K L B, Boddy M N, Freemont P S. Does this have a familiar RING? Trends Biochem Sci. 1996;21:208–214. [PubMed] [Google Scholar]
- 166.Schuler G D, Altschul S F, Lipman D J. A workbench for multiple alignment construction and analysis. Proteins. 1991;9:180–190. doi: 10.1002/prot.340090304. [DOI] [PubMed] [Google Scholar]
- 167.Seeberg E, Eide L, Bjoras M. The base excision repair pathway. Trends Biochem Sci. 1995;20:391–397. doi: 10.1016/s0968-0004(00)89086-6. [DOI] [PubMed] [Google Scholar]
- 168.Sekiguchi J, Shuman S. Mutational analysis of vaccinia virus topoisomerase identifies residues involved in DNA binding. Nucleic Acids Res. 1997;25:3649–3656. doi: 10.1093/nar/25.18.3649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Senkevich T G, Bugert J J, Sisler J R, Koonin E V, Darai G, Moss B. Genome sequence of a human tumorigenic poxvirus: prediction of specific host response-evasion genes. Science. 1996;273:813–816. doi: 10.1126/science.273.5276.813. [DOI] [PubMed] [Google Scholar]
- 170.Senkevich T G, Koonin E V, Bugert J J, Darai G, Moss B. The genome of Molluscum contagiosumvirus: analysis and comparison with other poxviruses. Virology. 1997;233:19–42. doi: 10.1006/viro.1997.8607. [DOI] [PubMed] [Google Scholar]
- 171.Shiozaki K, Russell P. Counteractive roles of protein phosphatase 2C (PP2C) and a MAP kinase kinase homolog in the osmoregulation of fission yeast. EMBO J. 1995;14:492–502. doi: 10.1002/j.1460-2075.1995.tb07025.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Shuman S, Kane E M, Morham S G. Mapping the active-site tyrosine of vaccinia virus DNA topoisomerase I. Proc Natl Acad Sci USA. 1989;86:9793–9797. doi: 10.1073/pnas.86.24.9793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Simon-Mateo C, Andres G, Vinuela E. Polyprotein processing in African swine fever virus: a novel gene expression strategy for a DNA virus. EMBO J. 1993;12:2977–2987. doi: 10.1002/j.1460-2075.1993.tb05960.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Simpson D A, Condit R C. Vaccinia virus gene A18R encodes an essential DNA helicase. J Virol. 1995;69:6131–6139. doi: 10.1128/jvi.69.10.6131-6139.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Sriskantha A, Osborne R J, Dall D J. Mapping of the Heliothis armigeraentomopoxirus (HaEPV) genome, and analysis of genes encoding the HaEPV spheroidin and nucleoside triphosphate phosphohydrolase 1 proteins. J Gen Virol. 1997;78:3115–3123. doi: 10.1099/0022-1317-78-12-3115. [DOI] [PubMed] [Google Scholar]
- 176.Staden A, McLachlan A D. Codon preference and its use in identifying protein coding regions in long DNA sequences. Nucleic Acids Res. 1982;10:141–156. doi: 10.1093/nar/10.1.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Staden R. An interactive graphics program for comparing and aligning nucleic acid and amino acid sequences. Nucleic Acids Res. 1982;10:2951–2961. doi: 10.1093/nar/10.9.2951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Stevens M K, Klesney-Tait J, Lumbley S, Walters K A, Joffe A M, Radolf J D, Hansen E J. Identification of tandem genes involved in lipooligosaccharide expression by Haemophilus ducreyi. Infect Immun. 1997;65:651–660. doi: 10.1128/iai.65.2.651-660.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Streett D A, Oma E A, Henry J E. Cross infection of three grasshopper species with the Melanoplus sanguinipesentomopoxvirus. J Invertebr Pathol. 1990;56:419–421. [Google Scholar]
- 180.Streett D A, Woods S A, Erlandson M A, Goettel M S, Johnson D L, editors. Memoirs of the Entomological Society of Canada. Vol. 171. Ottawa, Ontario, Canada: Entomological Society of Canada; 1997. Entomopoxviruses of grasshoppers and locusts: biological control potential; pp. 115–130. [Google Scholar]
- 181.Stuart D T, Upton C, Higman M A, Niles E G, McFadden G. A poxvirus-encoded uracil DNA glycosylase is essential for virus viability. J Virol. 1993;67:2503–2512. doi: 10.1128/jvi.67.5.2503-2512.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Sueoka N. Correlation between base composition of deoxyribonucleic acid and amino acid composition of protein. Biochemistry. 1961;47:1141–1149. doi: 10.1073/pnas.47.8.1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Sweasy J B, Loeb L A. Mammalian DNA polymerase β can substitute for DNA polymerase I during DNA replication in Escherichia coli. J Biol Chem. 1992;267:1407–1410. [PubMed] [Google Scholar]
- 184.Taddei F, Hayakawa H, Bouton M-F, Cirinesi A-M, Matic I, Sekiguchi M, Radman M. Counteraction by MutT protein of transcriptional errors caused by oxidative damage. Science. 1997;278:128–130. doi: 10.1126/science.278.5335.128. [DOI] [PubMed] [Google Scholar]
- 185.Teng S-C, Kim B, Gabriel A. Retrotransposon reverse-transcriptase-mediated repair of chromosomal breaks. Nature. 1996;383:641–644. doi: 10.1038/383641a0. [DOI] [PubMed] [Google Scholar]
- 186.Teodoro J G, Branton P E. Regulation of apoptosis by viral gene products. J Virol. 1997;71:1739–1746. doi: 10.1128/jvi.71.3.1739-1746.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Thompson J D, Higgins D G, Gibson T J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Tietz A, Weintraub H. Hydrolysis of glycerides by lipases of the fat body of the locust, Locusta migratoria. Insect Biochem. 1978;8:11–16. [Google Scholar]
- 189.Travis S M, Berger H A, Welsh M J. Protein phosphatase 2C dephosphorylates and inactivates crystic fibrosis transmembrane conductance regulator. Proc Natl Acad Sci USA. 1997;94:11055–11060. doi: 10.1073/pnas.94.20.11055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Upton C, McFadden G. DNA sequence homology between the terminal inverted repeats of Shope fibroma virus and an endogenous cellular plasmid species. Mol Cell Biol. 1986;6:265–276. doi: 10.1128/mcb.6.1.265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Upton C, Stuart D T, McFadden G. Identification of a poxvirus gene encoding a uracil DNA glycosylase. Proc Natl Acad Sci USA. 1993;90:4518–4522. doi: 10.1073/pnas.90.10.4518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.VanSlyke J K, Franke C A, Hruby D E. Proteolytic maturation of vaccinia virus core proteins: identification of a conserved motif at the N termini of the 4b and 25K virion proteins. J Gen Virol. 1991;72:411–416. doi: 10.1099/0022-1317-72-2-411. [DOI] [PubMed] [Google Scholar]
- 193.VanSlyke J K, Whitheead S S, Wilson E M, Hruby D E. The multistep proteolytic maturation pathway utilized by vaccinia virus P4a protein: a degenerate conserved cleavage motif within core proteins. Virology. 1991;183:467–478. doi: 10.1016/0042-6822(91)90976-i. [DOI] [PubMed] [Google Scholar]
- 194.van Strien E A, Jansen B J H, Mans R M W, Zuidema D, Vlak J M. Sequence and transcriptional analysis of the ubiquitin gene cluster in the genome of Spodoptera exiguanucleopolyhedrovirus. J Gen Virol. 1996;77:2311–2319. doi: 10.1099/0022-1317-77-9-2311. [DOI] [PubMed] [Google Scholar]
- 195.von Heijne G. A new method for predicting signal sequence cleavage sites. Nucleic Acids Res. 1986;14:4683–4690. doi: 10.1093/nar/14.11.4683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Wall E M, Cao J-X, Chen N, Buller R M L, Upton C. A novel poxvirus gene and its human homolog are similar to an E. colilysophospholipase. Virus Res. 1997;52:157–167. doi: 10.1016/s0168-1702(97)00122-6. [DOI] [PubMed] [Google Scholar]
- 197.Weinbauer M G, Wilhelm S W, Suttle C A, Garza D R. Photoreactivation compensates for UV damage and restores infectivity to natural marine virus communities. Appl Environ Microbiol. 1997;63:2200–2205. doi: 10.1128/aem.63.6.2200-2205.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Weir J P, Moss B. Regulation of expression and nucleotide sequence of a late vaccinia virus gene. J Virol. 1984;51:662–669. doi: 10.1128/jvi.51.3.662-669.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Weissman A M. Regulating protein degradation by ubiquitination. Immunol Today. 1997;18:189–198. doi: 10.1016/s0167-5699(97)84666-x. [DOI] [PubMed] [Google Scholar]
- 200.Whitheead S S, Hruby D E. Differential utilization of a conserved motif for the proteolytic maturation of vaccinia virus proteins. Virology. 1994;200:154–161. doi: 10.1006/viro.1994.1174. [DOI] [PubMed] [Google Scholar]
- 201.Whitehead S S, Hruby D E. A transcriptionally controlled trans-processing assay: putative identification of a vaccinia virus-encoded proteinase which cleaves precursor protein P25K. J Virol. 1994;68:7603–7608. doi: 10.1128/jvi.68.11.7603-7608.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Wilcock D, Smith G L. Vaccinia virus core protein VP8 is required for virus infectivity, but not for core protein processing or for INV and EEV formation. Virology. 1994;202:294–304. doi: 10.1006/viro.1994.1346. [DOI] [PubMed] [Google Scholar]
- 203.Wilson S H. Gene regulation and structure-function studies of mammalian DNA polymerase β. In: Strauss P, Wilson S H, editors. The eukaryotic nucleus. Vol. 1. Caldwell, N.J: Telford Press; 1990. pp. 199–234. [Google Scholar]
- 204.Wittek R, Menna A, Müller H K, Schümperli D, Boseley P G, Wyler R. Inverted terminal repeats in rabbit poxvirus and vaccinia virus DNA. J Virol. 1978;28:171–181. doi: 10.1128/jvi.28.1.171-181.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Wittschieben J, Shuman S. Mutational analysis of vaccinia DNA topoisomerase defines amino acid residues essential for covalent catalysis. J Biol Chem. 1994;269:29978–29983. [PubMed] [Google Scholar]
- 206.Wolffe E J, Moore D M, Peters P J, Moss B. Vaccinia virus A17L open reading frame encodes an essential component of nascent viral membranes that is required to initiate morphogenesis. J Virol. 1996;70:2797–2808. doi: 10.1128/jvi.70.5.2797-2808.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Wong S W, Wahl A F, Yuan P-M, Arai N, Pearson B E, Arai K, Korn D, Hunkapiller M W, Wang T-S. Human DNA polymerase α gene expression is cell proliferation dependent and its primary structure is similar to both prokaryotic and eukaryotic replicative DNA polymerases. EMBO J. 1988;7:37–47. doi: 10.1002/j.1460-2075.1988.tb02781.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Woods S A, Streett D A, Henry J E. Temporal patterns of mortality from an entomopoxvirus and strategies for control of the migratory grasshopper (Melanoplus sanguinipesF.) J Invertebr Pathol. 1992;60:33–39. [Google Scholar]
- 209.Wright D A, Ke N, Smalle J, Hauge B M, Goodman H M, Voytas D F. Multiple non-LTR retrotransposons in the genome of Arabidopsis thaliana. Genetics. 1996;142:569–578. doi: 10.1093/genetics/142.2.569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Xiong Y, Eickbush T H. Origin and evolution of retroelements based upon their reverse transcriptase sequences. EMBO J. 1990;9:3353–3362. doi: 10.1002/j.1460-2075.1990.tb07536.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Xu J, Hukuhara T. Biochemical properties of an enhancing factor of an entomopoxvirus. J Invertebr Pathol. 1994;63:14–18. [Google Scholar]
- 212.Yanez R J, Rodriguez J M, Nogal M L, Yuste L, Enriquez C, Rodriguez J F, Vinuela E. Analysis of the complete nucleotide sequence of African swine fever virus. Virology. 1995;208:249–278. doi: 10.1006/viro.1995.1149. [DOI] [PubMed] [Google Scholar]
- 213.Yanez R J, Vinuela E. African swine fever virus encodes a DNA ligase. Virology. 1993;193:531–536. doi: 10.1006/viro.1993.1161. [DOI] [PubMed] [Google Scholar]
- 214.Yasui A, Eker A P M, Yasuhira S, Yajima H, Kobayashi T, Takao M, Oikawa A. A new class of DNA photolyases present in various organisms including aplacental mammals. EMBO J. 1994;13:6143–6151. doi: 10.1002/j.1460-2075.1994.tb06961.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Yuen L, Dionne J, Arif B, Richardson C. Identification and sequencing of the spheroidin gene of Choristoneura biennisentomopoxvirus. Virology. 1990;175:427–433. doi: 10.1016/0042-6822(90)90427-s. [DOI] [PubMed] [Google Scholar]
- 216.Yuen L, Moss B. Oligonucleotide sequence signaling transcriptional termination of vaccinia virus early genes. Proc Natl Acad Sci USA. 1987;84:6417–6421. doi: 10.1073/pnas.84.18.6417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Yuen L, Noiseux M, Gomes M. DNA sequence of the nucleoside triphosphate phosphohydrolase I (NPH I) of the Choristoneura biennisentomopoxvirus. Virology. 1991;182:403–406. doi: 10.1016/0042-6822(91)90690-d. [DOI] [PubMed] [Google Scholar]
- 218.Zajac P, Spehner D, Drillien R. The vaccinia virus J5L open reading frame encodes a polypeptide expressed late during infection and required for viral multiplication. Virus Res. 1995;37:163–173. doi: 10.1016/0168-1702(95)00025-l. [DOI] [PubMed] [Google Scholar]