Abstract
The entry of the viral genomic DNA of cauliflower mosaic virus into the nucleus is a critical step of viral infection. We have shown by transient expression in plant protoplasts that the viral coat protein (CP), which is processed from the product of open reading frame IV, contains an N-terminal nuclear localization signal (NLS). The NLS is exposed on the surface of the virion and is thus available for interaction with a putative NLS receptor. Phosphorylation of the matured CP did not influence the nuclear localization of the protein but improved protein stability. Mutation of the NLS completely abolished viral infectivity, thus indicating its importance in the virus life cycle. The NLS seems to be regulated by the N terminus of the precapsid, which inhibits its nuclear targeting. This regulation could be important in allowing virus assembly in the cytoplasm.
Cauliflower mosaic virus (CaMV), the type member of the caulimovirus group (57), has a circular genomic DNA of 8 kbp with seven major open reading frames (ORF), six of which encode proteins that have been detected in vivo (25, 44). The virion is an icosahedral particle with a diameter of 53.8 nm made of 420 subunits of the viral coat protein (CP) (9). The N terminus of CP is believed to be exposed on the surface of the virion (9, 32).
Early in the replication cycle, CaMV delivers its genomic DNA to the nucleus, where it is assembled into a minichromosome by association with host proteins from the infected plant (48). Viral transcripts are then produced and used as mRNAs for the production of viral proteins or as templates for reverse transcription (50). Covey and Turner (12) observed that viral genomes, probably coming from mature virions in the cytosol, enter the nucleus to increase the pool of minichromosomes when protoplasts are prepared from CaMV-infected leaves. It is reasonable to assume that a virion-associated protein directs the DNA to the nucleus. The viral DNA alone is probably too large to easily enter the nucleus, as shown with mammalian cells (7, 28). Since CP is the most abundant viral protein in the virion, we hypothesized that it could participate in transporting viral DNA to the nucleus.
CaMV is a pararetrovirus and uses reverse transcriptase as part of the replicative cycle (53). An important feature that distinguishes the pararetroviruses from the retroviruses is the ability of the DNA proviral form of the retroviruses to be integrated into the host chromosome (4). The DNA of the pararetroviruses accumulates within the nucleus as multiple copies of circular minichromosomes (45, 49, 59).
Many of the genes of pararetroviruses are homologous in sequence and in function to those of retroviruses. Furthermore, the relative locations of some functions within the genome are conserved between the two groups (53). After entry into the cell, retroviruses disassemble in the cytoplasm and reverse transcribe the genomic RNAs into DNA. The postentry viral nucleoprotein complex, also called the preintegration complex (PIC), needs to harbor a signal to target the reverse-transcribed DNA into the nucleus. Retroviruses can be divided into two groups based on the ability of the PICs to be actively imported into the nucleus during interphase (6). Murine leukemia virus is an example of a retrovirus in which replication is restricted to dividing cells (52), in contrast to human immunodeficiency virus type 1 (HIV-1), which infects nonproliferating cells. Given the size of the PICs (15), it seems reasonable that one or more components of the PICs of the second group of retroviruses should harbor a nuclear localization signal (NLS) to mediate the transport of this complex into the nucleus.
The HIV-1 matrix protein (MA) has been implicated in directing the PICs to the nucleus in HIV-1, via an NLS (6). Gallay et al. (20, 21) proposed that phosphorylation of 1% of MA on a C-terminal Tyr was required to reverse the membrane binding of MA and promote an association between MA and the integrase, thus enabling MA with its NLS to direct the PIC to the nucleus. A mutation in the NLS of MA disabled the virus, abrogating infection of nondividing cells (6). However, those results are controversial, since recent evidence suggests that MA does not harbor an NLS (18). Furthermore, the blocking of Tyr phosphorylation of MA did not have detectable effect on virus infectivity of cells in a nondividing stage (19), contrary to earlier reports (20, 21). These results imply that other components of the PIC, namely, integrase, reverse transcriptase, nucleocapsid, Vpr, or cellular factors, must supply the NLS(s) (18). There is evidence that the Vpr protein of HIV-1 or the related protein Vpx in simian immunodeficiency virus, which are assembled in the virions, could contribute to nuclear targeting of the PICs (17, 29, 51). Another class of retroviruses, the foamy viruses, are often found in the nuclei of infected cells, and an NLS on the CP was shown to be responsible for this localization (55).
Pararetroviruses do not have PICs because reverse transcription is not necessary in the early stage of infection, since the viral genome is made of DNA. The targeting of human hepatitis B virus (HBV), a pararetrovirus, has been well documented. The core protein of HBV possesses a multifunctional domain at its C terminus that is important for DNA binding and packaging. An NLS (14, 63) and phosphorylation sites (36) were mapped in the same region. It was shown that in HepG2.2.15 cells, which constitutively produce HBV Gag, the core protein was firmly attached to nuclear pores (2). The affinity of the core protein for the pore is an important step in bringing genomic DNA to the nucleus (37). It has recently been suggested that the HBV polymerase transports the genomic DNA through the pore after disassembly of the particles at the nuclear envelope (37). It might be possible that CP has a similar function for CaMV.
In the present study, we have mapped a region of CaMV CP important for nuclear targeting that is exposed at the surface of the virion and present on the three processed forms of CP. In contrast to the case for HBV, phosphorylation of CP did not influence nuclear targeting, although the removal of the phosphorylation targets improved CP stability. This report is the first description of an NLS in a plant pararetrovirus.
MATERIALS AND METHODS
Plasmids.
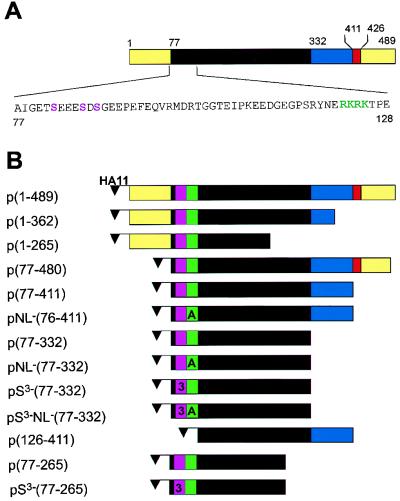
The human tenascin tag was removed from the plasmid pTTO (46) and replaced with the 11-amino-acid hemagglutinin epitope MYPYDVPDYAA (3). An NcoI site and a BglII site were added downstream of the tag to allow N-terminal fusions with different CP constructs. The resulting vector (pTAGNLS) contains a duplicated CaMV promoter and a CaMV poly(A) signal separated by the sequence encoding the tag and the cloning sites. CaMV CP constructs were generated by PCR. The sense oligonucleotides 5′TCAGAATTCCCATGGCAATAG3′ (9021), 5′GCTCCAGCACCATGGCCGAATC3′ (9483), 5′GAGAGAAAGACCATGGCCCCGGAGG3′ (13720), and 5′GAATTCCCCATGGC AATAGGAGGAACAGCTGAAGAAGAAAGCGATGCAGGAGGAG3′ (14986) and the antisense oligonucleotides 5′ATCCAGGATCCTCAATCTTTCT3′ (9482), 5′CATCGGATCCTCAGTCTGAGTCT3′ (9484), 5′CTTTTCGGATCCTCATGTAAATTC3′ (9485), 5′TTCGGATCCTTTCAGGATAAGTC3′ (10137), and 5′CTTGGGATCCTTTCATGTGGATG3′ (10138) were used for the amplifications. The combinations of oligonucleotides for the PCRs were 9483 and 9484 for p(1-489), 9483 and 10138 for p(1-362), 9021 and 9484 for p(77-480), 9021 and 9482 for p(77-411), 9021 and 10137 for p(77-332), 14986 and 10137 for pS3−(77-332), 9021 and 9485 for p(77-265), 14986 and 10137 for pS3−(77-265), 9021 and 9482 for p(77-411), and 13720 and 9482 for p(126-411). The clone pCa37 (39), containing the whole genome of CaMV in the SalI site of pBR322, was used as the substrate for the amplification. The construct p(77-480) stops at codon 480 because a mutation was incorporated which introduced a BamHI site 9 amino acids before the end of ORF IV. All of the PCRs were performed as follows: 94°C for 1 min, 50°C for 1 min, and 72°C for 2 min for 30 cycles. PCR products were cloned in the pTAGNLS or pET3d vector (Novagen) after digestion with NcoI and BamHI. The entire sequence of each clone was verified by DNA sequencing. A methionine codon was added as an initiation codon to clones starting at codon 76 or 125.
The mutations in the NLSs of p(77-332), pNL−(77-411), and pS3−(77-332) were introduced by site-directed mutagenesis (Amersham oligonucleotide-directed in vitro mutagenesis system, version II) with the oligonucleotide 5′GG AAAGTACCGGTCCTCCGGGGTTGCGGCCGCTGCCTCATTGTATCTA GATGGTCCTTC3′ to generate pNL−(77-332), pNL−(77-411), and pS3−NL−(77-332).
Plant and viruses.
Brassica rapa plants (turnip ’Just Right’) were grown at 22°C with a 16-h photoperiod as described previously (1). The Strasbourg strain of CaMV cloned in the SalI site of pBR322 (pCa37) was used for the infection test (39). The mutants pNL−(77-332) and pS3−(77-265) were transferred to the CaMV genome of pCa37 by exchanging an XbaI-AgeI fragment of pCa37 with the corresponding fragment from pNL−(77-332) or pS3−(77-265). Prior to inoculation, the plasmids harboring the wild-type (wt) and mutated versions of CaMV were linearized with SalI. Leaves of 4-week-old turnip plants were inoculated mechanically. For each experiment, two plants were inoculated with 20 μg of the wt CaMV. Six plants were inoculated with the mutant; three were inoculated with 5 μg and three were inoculated with 20 μg of viral DNA (1 μg/μl). The experiment was repeated three times under the same conditions. The virus was grown in B. rapa and purified as described previously (1).
Cloning and expression in Escherichia coli.
Expression plasmids were named after the positions of their start and stop codons within the CaMV ORF IV. Constructs were introduced into E. coli BL21(DE3). Liquid cultures were grown to an optical density at 600 nm of 0.6 and induced with 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) for 3 h at 37°C.
Plant protoplasts and analysis by direct immunofluorescence.
Mesophyll protoplasts of Nicotiana plumbaginifolia were transfected by the polyethylene glycol method as described by Goodall et al. (24), using 20 μg of plasmid per transfection (3 × 105 protoplasts). B. rapa protoplasts were made by the same protocol with modifications. The leaf pieces were digested in enzyme solution (1% cellulase R10, 0.2% Macerozyme R-10 (Yakult Pharmaceutical, Tokyo, Japan), 0.4 M mannitol, and 8 mM CaCl2) for 18 h at 25°C. The protoplasts were filtered, washed twice with washing solution (0.2 M CaCl2, 0.05% MES [morpholineethanesulfonic acid], pH 5.8), and resuspended in EP solution (10 mM HEPES, 150 mM NaCl2, 5 mM CaCl2, and 0.2 M mannitol) prior to transfection. Optimal expression of protein was found to be at 8 h after transfection. Protoplasts were collected and treated for indirect immunofluorescence (46). The samples were examined under oil with a Leitz microscope equipped with a Leitz Fluotar 40× objective and epifluorescence filters or with a confocal Leica (Heidelberg, Germany) DMIRBE microscope equipped with a Leitz 40× objective and Leica Scanware.
Extraction of proteins, SDS-PAGE, and immunoblotting.
The proteins were extracted with phenol from the transfected protoplasts or from the inoculated plants (33). Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) of protein samples was performed as described by Laemmli (38). Proteins were electroblotted from SDS-polyacrylamide gels to nitrocellulose membranes at 30 V (constant voltage) overnight. The membrane was treated as described previously (31).
The bacterial pellets containing the overexpressed constructs were resuspended in 1× Laemmli buffer, and 0.5 volume of loading dye (50% glycerol, 10 mM Tris [pH 8], 3% SDS, and 5 mM dithiothreitol) was added to the suspension before heating at 95°C for 3 to 5 min. Twenty micrograms of each sample was loaded on an SDS-polyacrylamide gel as described above.
Electron microscopy and antibodies.
The purified virus preparation was resuspended in 0.05 M Tris (pH 7.5) and adsorbed on electron microscope grids covered with carbon-coated colladium films after treatment by glow discharge and was negatively stained with 0.75% uranyl formate. For immunogold labeling, carbon-coated grids loaded with viruses were covered with anti-NLS antibodies, washed several times, and covered with 10-nm gold–antirabbit antibodies (Aurion). Samples were analyzed with a Zeiss EM 910 transmission electron microscope operated at 80 kV. The data were photographed on Kodak SO-163 films.
The peptide SRYNERKRKTPEDR, containing the NLS of CaMV Gag, was synthesized, conjugated to a heterologous protein, and injected into a rabbit to raise antibodies (Ready System AG, Bad Zurzach, Switzerland). The serum was purified and is referred to in this paper as NLS-immunoglobulin G (NLS-IgG). The antibodies raised against CP were already available in the laboratory and are referred to in this paper as CP-IgG (43). The tag antibody was obtained from Babco (Richmond, Calif.), and the goat antirabbit antibodies were obtained from Bio-Rad.
Dot blot hybridization.
The inoculated and upper inoculated leaves on the infected plants were chosen for sampling. One gram of fresh tissue was homogenized in 1 ml of 100 mM Tris (pH 8)–1 mM EDTA–50 mM NaCl. The homogenate was diluted 10×, 100×, and 1,000× with extraction buffer and spoted on nitrocellulose. The following spots were 10× dilutions of the previous one. The hybridization was performed as described previously (40).
Phosphorylation assay.
Inclusion bodies containing the recombinant proteins p(77-265) and p3S−(77-265) were isolated from E. coli. The proteins of the inclusion bodies were denatured in SDS and separated by SDS-PAGE (31). The protein of interest was visualized by incubation in 1 M KCl, electroeluted in SDS buffer according to the specifications of the manufacturer (Bio-Rad model 422), and dialyzed against 10 mM Tris-HCl, pH 7.5. Phosphorylation reactions with mixtures (30 μl) containing 0.25 μg of recombinant protein and 15 μg of CaMV particles were done as described previously (43).
RESULTS
Nuclear localization of CaMV CP in plant protoplasts.
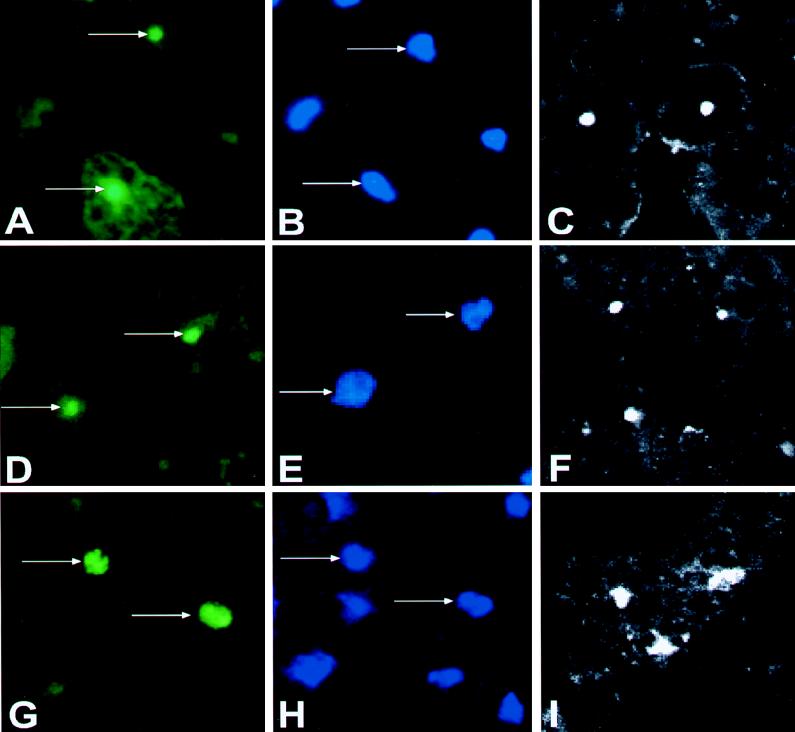
The CaMV ORF IV encodes a 57-kDa protein. The viral subunits correspond to three related capsid proteins (called p44, p39, and p37) that are derived from cleavage of the ORF IV product by a virus-encoded protease (58). In the case of p44, proteolytic cleavage occurs 75 amino acids from the N terminus of the ORF IV protein precursor (43). A construct starting at residue 77 of ORF IV, corresponding to the first amino acid of p44 (43), and ending at amino acid 480 (Fig. 1) was transiently expressed in N. plumbaginifolia protoplasts. The hemagglutinin epitope (MCYPYDVPDYASLA) (3) was fused to the N terminus of p44 as a tag to facilitate the detection of the protein with a commercial antibody. The expressed protein was detected by indirect immunofluorescence (46) with a rabbit antibody against the tag followed by a fluorescein isothiocyanate (FITC)-conjugated goat antirabbit antibody as a secondary antibody. Nuclei were stained with DAPI (4′,6-diamidino-2-phenylindole). The FITC signal of p44 (Fig. 2A) was coincident with the nuclear DAPI signal (Fig. 2B) but smaller, suggesting a sublocalization of the expressed protein within the nucleus, most probably in the nucleolus. p44 with a deletion of 43 amino acids at the C terminus [p(77-411)] showed the same sublocalization (Fig. 2D). However, when a deletion of 122 amino acids was made at the C terminus of p44 [p(77-332)], removing the lysine-rich domain of the protein, the fluorescent signal of this protein (Fig. 2G) became perfectly coincident with the DAPI signal (Fig. 2H).
FIG. 1.
CaMV CP constructs. (A) Schematic representation of CaMV CP. The yellow regions are rich in acidic amino acids. The black region is defined as the jelly roll (53) and is believed to be involved in protein-protein interaction between CP subunits in viral assembly (9). The blue region is the lysine-rich domain which is important for interaction with nucleic acids (9). The red region is the zinc finger believed to be involved in RNA binding. The amino acid sequence of the N terminus of p44 is shown below the scheme. The phosphorylation targets are marked in violet, and the NLS is in green. (B) Different forms of the CaMV CP used in the experiments. The small violet box represents the position of the phosphorylation target. The number 3 appears in the box when three of the serines are mutated to alanines. The letter A appears in the green box when the wt NLS sequence RKRK is mutated to AAAA. Each construct is named based on the numbering of the full-length ORF IV construct. The flag HA11 is fused to the N termini of all of the constructs to facilitate the immunodetection of these proteins.
FIG. 2.
Analysis by indirect immunofluorescence of the intracellular localizations of different forms of CaMV CP after transient expression in N. plumbaginifolia protoplasts. (A, D, and G) p(77-480), p(77-411), and p(77-332) were detected in the nuclei of transfected cells. (B, E, and H) The same field of view was stained with DAPI to visualize the nuclei. (C, F, and I) Confocal pictures of different cells from the same transfections.
Figure 2A, B, D, E, G, and H were taken with a conventional fluorescence microscope, while Fig. 2C, F, and I were taken with a confocal microscope and show different cells than do the adjacent pairs of FITC and DAPI pictures. Similar results were obtained when the constructs were expressed in protoplasts of the host B. rapa (data not shown). Based on these results, we conclude that p(77-332) contains all of the information necessary to allow nuclear localization of CP and that the C terminus of p44 influences sublocalization within the nucleus.
Mapping of the NLS of CaMV CP.
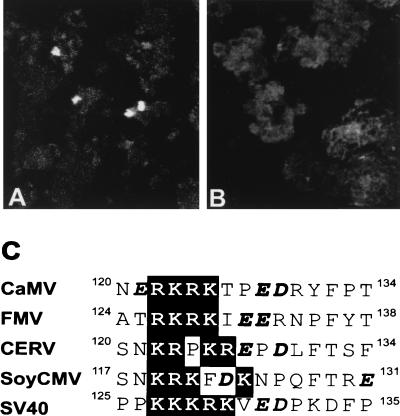
Many of the NLSs characterized to date are made essentially of positively charged amino acids arranged in one (monopartite) or two (bipartite) clusters (for a review, see reference 13). Inspection of the 255 amino acids of p(77-332) reveals a cluster of four positively charged residues at positions 122 to 125 (Fig. 1A). This motif (RKRK) resembles the monopartite simian virus 40 (SV40) T-antigen NLS (PKKKRKV) (13) (Fig. 3C). It is also conserved in sequence and position among other caulimoviruses (Fig. 3C). We introduced mutations in the basic cluster of p(77-332) to see the effect on nuclear targeting of CP after transient expression in N. plumbaginifolia protoplasts. The expressed proteins were immunodetected as described above. When all four positively charged amino acids were mutated to alanines in pNL−(77-332), no signal was detected in the nucleus (Fig. 3B), whereas expression of the wt sequence p(77-332) showed strong nuclear signals (Fig. 3A). Deletion of amino acids 77 to 125 [p(126-411)] also abolished nuclear targeting (data not shown). Since p(126-411) still contains most of the C-terminal lysine-rich domain of CP, this result suggests that the lysine-rich domain of CP does not harbor an NLS.
FIG. 3.
Mapping of the NLS of CP. (A and B) Confocal pictures showing indirect immunofluorescence of the intracellular localizations of different forms of CaMV CP after transient expression in N. plumbaginifolia protoplasts. (A) p(77-332), containing the wt sequence. (B) pNL−(77-332), with the four positively charged amino acids of RKRK replaced by AAAA. (C) Alignment of CPs of four caulimoviruses in the NLS region. CERV, carnation etched ring virus; FMV, figwort mosaic virus; SoyCMV, soybean chlorotic mottle virus. The SV40 T-antigen NLS is also shown for comparison. The basic clusters are highlighted in the black box. The acidic amino acids are shown in boldface italics.
We conclude that the basic cluster formed by amino acids 122 to 125 of CP is important for nuclear import of the protein. This sequence can be considered either an NLS itself or part of a bigger signal.
The NLS of CP is exposed on the surface of the virus.
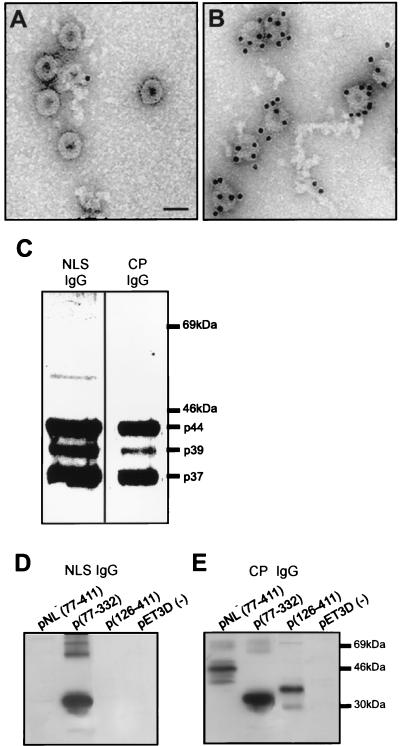
An NLS (SRYNERKRKTPEDR)-specific antiserum (NLS-IgG) was prepared and used to decorate the surface of the purified virus. Protein A-conjugated gold particles were added to the grid for the detection of NLS-IgG. Many gold particles could be detected at the virion surface, showing that this peptide is exposed on virus particles (Fig. 4B). This result further suggests that the sequence recognized by the antibodies is available for interaction with a putative plant receptor. The preimmune serum did not show any specific interaction with the virus particles (Fig. 4A).
FIG. 4.
The NLS of CaMV CP is exposed outside the virion and is present in the three processed forms of CP in the purified virion. (A and B) Gold labeling of partially purified CaMV particles with preimmune serum (A) or NLS-IgG (B). Bar, 50 nm. (C) Western blot of proteins from the purified virus probed with NLS-IgG or CP-IgG. (D) Western blot of p(126-411), pNL−(77-411), and p(77-332) expressed in E. coli and probed with NLS-IgG. (E) Same as panel D but probed with CP-IgG.
In Western blots, NLS-IgG reacted specifically with the three major forms of CP found in the purified virus (Fig. 4C). This result suggests that all of the processed forms of CP in the purified virus contain the NLS. CP-IgG also recognized the different forms of CP in the purified virus (Fig. 4C). The mutants pNL−(77-411) and p(126-411) and the wt p(77-332) were expressed in E. coli, and the recombinant proteins were analyzed by Western blotting with NLS-IgG or CP-IgG (Fig. 4D and E). The mutants pNL−(77-411) and p(126-411) reacted only with CP-IgG, while p(77-332) was recognized by both antibodies (Fig. 4D and E), indicating that the peptide antiserum is very specific for the wt basic cluster.
Does phosphorylation play a role in nuclear targeting of CP?
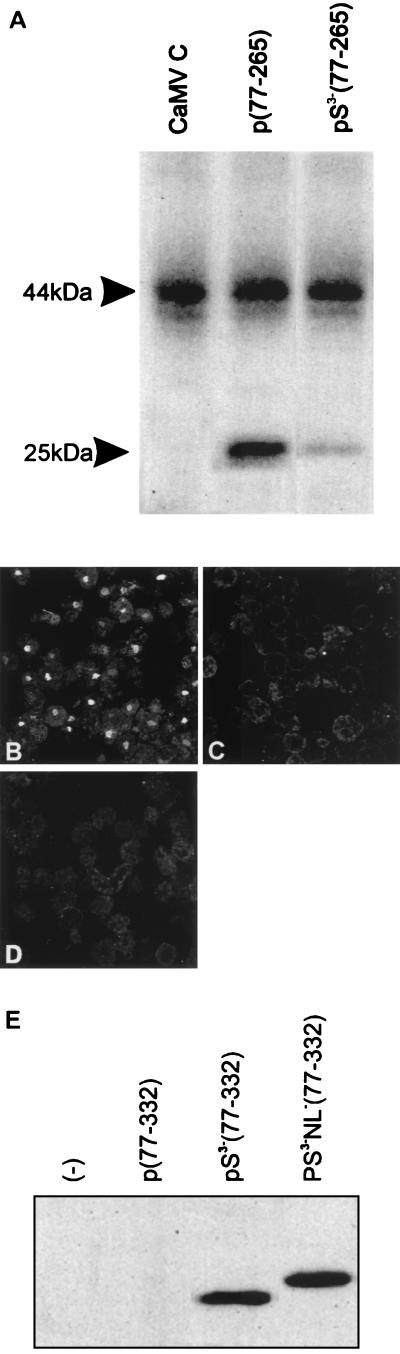
P44 is phosphorylated in vitro when the virions are incubated with [γ-32P]ATP (43). A host casein kinase II activity was shown to be associated with purified preparations of virions (43). Complete acid hydrolysis of labeled p44 isolated from viral particles followed by separation of the products by two-dimensional electrophoresis on cellulose thin-layer plates confirmed that all of the radioactivity was present as phosphoserine (43). To determine the positions of the phosphorylated serines, we have expressed and purified from E. coli p(77-265) and pS3−(77-265), in which the serines at positions 82, 86, and 88 were mutated to alanines. The protein pS3−(77-265) was shown to be only weakly phosphorylated by the virus-associated casein kinase activity compared to the wt protein p(77-265) (Fig. 5A). This result suggests that those three serines are major phosphorylation targets in the CaMV CP; the residual labeling might be due to phosphorylation of threonine residues found at positions 102 and 105 (43). The intensity of the phosphorylation of 50 ng of p(77-265) is comparable to the signal obtained by using 1 μg of p44 originating from virus particles (Fig. 5A). This observation is consistent with the results of Martinez-Izquierdo and Hohn (43), who calculated that only one radioactive phosphate per 650 molecules of p44 in the purified virions was incorporated in the in vitro assay. Furthermore, pretreatment of the purified virions with calf intestine alkaline phosphatase prior to incubation with [γ-32P]ATP led to a more extensive phosphorylation of p44, suggesting that most p44 is phosphorylated in vivo in the infected plants (43).
FIG. 5.
Influence of phosphorylation and the N-terminal acidic region of CP on nuclear targeting of CaMV CP. (A) In vitro phosphorylation of CaMV CP. Serines 82, 86, and 88 were mutated to alanines in pS3−(77-265). (B to D) Analysis by indirect immunofluorescence of the intracellular localizations of different forms of CaMV CP after transient expression in N. plumbaginifolia protoplasts. (B) pS3−(77-332), in which serines 82, 86, and 88 were mutated to alanine. (C) pS3−NL−(77-332), in which serines 82, 86, and 88 and all of the basic amino acids of the NLS were mutated to alanine. (D) p(1-362), with the first 76 amino acids of the ORF IV product. (E) Western blot of transfected protoplasts expressing wt p(77-332), pS3−(77-332), or pS3−NL−(77-332) probed with tag antibody.
Because of the proximity of the phosphorylated serines to the NLS, we wanted to test their influence on nuclear targeting of p(77-332). The three serines were mutated to alanines in the context of p(77-332) [pS3−(77-332); Fig. 1B], and this mutation was also combined with the NLS-negative mutant [pS3−NL−(77-332)].
After transient expression in N. plumbaginifolia protoplasts, the localizations of the proteins were detected by indirect immunofluorescence. Surprisingly, about five times more nuclei were stained with pS3−(77-332) (Fig. 5B) than with the wt protein p(77-332) (Fig. 3A). No nuclear localization could be detected with construct pS3−NL−(77-332) (Fig. 5C). When a Western blot was made by using tag or specific CaMV CP IgG with samples of the transfected protoplasts, we could show that the transiently expressed protein was detectable only when three of the serines were mutated to alanines (Fig. 5E). The wt protein was undetectable under these conditions. This suggested that the mutations improved the stability of CP. The increased abundance of nuclear signals observed with the mutants could thus be explained by an increased protein stability that leads to a greater accumulation of the protein, rather than by improved nuclear targeting. The change in the migration of pS3−NL−(77-332) in SDS-PAGE (Fig. 5E) is presumably caused by the exchange of four basic residues, which affects the net charge of the protein.
Viral mutants carrying mutations in the NLS or in the phosphorylation target sites affect the CaMV viral life cycle.
To examine the importance of the NLS for viral replication, we studied the effect of mutations in this region on viral viability. We tested the mutant in which the four basic amino acids RKRK were replaced with AAAA (Table 1). Viral symptoms were observed on control plants at 7 days postinoculation. However, more than 3 months after inoculation, the plants treated with viral DNA mutated in the NLS were still symptomless. To detect traces of viral replication, we performed dot blot hybridization on extracts made from leaves collected from plants inoculated with the mutant and wt viral DNAs. None of the samples infected with the mutant DNA showed any sign of viral replication. However, the samples infected with the wt viral DNA showed strong signals even after a 1,000-fold dilution of the plant extract (Table 1). PCR performed on the same samples could not amplify the expected viral fragment on the samples infected with the mutant, in contrast to the wt viral DNA, which was amplified as anticipated (Table 1). We concluded that the mutations completely abolished viral replication and that integrity of this region is essential for the life cycle of the virus.
TABLE 1.
Infection of B. rapa plants with CaMV clones harboring mutations in the NLS or in the phosphorylation target sites of CP
| CaMV | Symptoms at postinoculation day:
|
Nuclear targeting | Dot blot hybridization | PCR amplification | ||
|---|---|---|---|---|---|---|
| 7 | 10 | 90 | ||||
| RKRK | + | ++ | ++ | + | + | + |
| AAAA | − | − | − | − | − | − |
| S3− | − | − | − | + | ||
A similar experiment was also performed with a viral mutant in which the serines at positions 82, 86, and 88 were mutated to alanines (CaMV S3−). The plants inoculated with this mutant were symptomless even 3 months after inoculation, while the plants infected with the wt viral DNA showed symptoms at 7 days postinoculation (Table 1). A PCR product could be obtained from this mutant, suggesting that CaMV could replicate at a very low level (Table 1). We concluded that this mutation had a significant effect on the level of replication of CaMV.
Effect of the N terminus of the ORF IV product on the nuclear targeting of CP.
The CP present in the virus particles does not contain the first 76 amino acids of the pre-CP (58). To investigate the influence of this region on nuclear targeting of CP, we transfected N. plumbaginifolia protoplasts with the full-length ORF IV [p(1-489)], and versions truncated at the C terminus, namely, p(1-362) and p(1-265). No nuclear signal was detected in protoplasts transfected with p(1-362) and p(1-265) even when as much as 50 μg of DNA was used for transfection [see Fig. 5D for p(1-362)]. Thus, the presence of the first 76 amino acids of the ORF IV product may prevent nuclear targeting of CP. Similar results were observed when the serines at position 82, 86, and 88 were mutated to alanine residues in the context of the full-length ORF IV [p(1-489)] or p(1-362) (data not shown). The expression of p(1-489), p(1-362), and p(1-265) could not be detected by Western blotting with the human tenascin tag or CP IgG. We suggest that the presence of the first 76 amino acids reduces the stability of the protein, making its detection difficult with conventional techniques.
DISCUSSION
As a first step in infection, many DNA viruses import their genomic DNA into the nuclei of infected cells, where transcripts that lead to viral protein synthesis are made. The genomic DNA is too large to easily enter the nucleus, and for many viruses, such as HBV, influenza virus, adenovirus, SV40, and HIV, it has been shown that one or several viral proteins are used to mediate nuclear DNA import (5, 17, 20, 21, 27, 29, 37, 60, 61, 63).
In this study, we analyzed whether the CP of CaMV could facilitate the transit of the viral genome to the nucleus by investigating the nuclear targeting of this protein. We characterized a motif (amino acids 122 to 125) containing two lysines and two arginines that is important for targeting CP to the nuclei of transfected protoplasts. Mutation of these basic amino acids to alanines completely abolished the nuclear accumulation of the protein. Alignment of this region with the equivalent regions of three other caulimoviruses shows that the basic cluster is conserved. It is also notable that in all cases acidic residues are detected in the neighborhood of the basic cluster (Fig. 3C). Acidic residues have been shown to play an important role in several NLSs (42), and they might also be part of the signal in CaMV CP. Cloned CaMV harboring mutations in the basic cluster was not infectious, and revertants did not appear even 3 months after inoculation of the plants (Table 1). This result indicates the importance of this sequence for the virus life cycle.
The phosphorylation state of a protein can influence nuclear targeting. In HBV, the core protein (C) has phosphorylation sites positioned in close proximity to the NLS in the arginine-rich domain (41). The level of phosphorylation of the protein influences its capacity to accumulate in the nucleus (36). It was also suggested that only the phosphorylated form of C can adopt a structure in the virion that will expose the NLS (36). Similarly, it was shown for the SV40 T antigen that the negative charge provided by phosphorylation is an enhancer of the nuclear import (35). Protoplasts transfected with CaMV CP mutant constructs that cannot be phosphorylated apparently showed improved nuclear targeting (Fig. 5B). The frequency and the intensity of the signals were stronger than those with the wt CP. However, this mutation appeared to improve protein stability rather than nuclear targeting activity (Fig. 5E). It is important to mention that the proteins harboring wt sequences transiently expressed in plant protoplasts were not detected by Western blotting. By mutation of serines 82, 86, and 88 to alanine residues, which resulted in changes in phosphorylation, the proteins were stabilized, showing the importance of the basic cluster (residues 122 to 125) in nuclear targeting of CP. The NLS mutant pS3−NL−(77-332) accumulates in transfected protoplasts to a level similar to that of pS3−(77-332) (Fig. 5E). However, in contrast to the pS3−(77-332) construct, the mutant pS3−NL−(77-332) was not detected in the nucleus (Fig. 5C). Although phosphorylation seemed to be important for virus infectivity (Table 1) in our experiments, its role in the CaMV life cycle remains to be determined.
Transfection of protoplasts with constructs containing the very acidic N-terminal region of CP never showed detectable quantities of protein in the nucleus (Fig. 5D). We suggest that the acidic region can mask the basic cluster, as shown for human heat shock factor 2 (56). Nuclear targeting of CP would then be inhibited. This could be an important feature for the virus life cycle, since CP must be kept in the cytoplasm for viral assembly. We suggest that after assembly of the virus in the inclusion bodies in the cytoplasm, the first 75 amino acids of CP are removed by the viral protease to generate p44 (58). After this processing, the NLS becomes available for interaction with a plant receptor, such as importin-α (13, 54), that directs the complex to the nuclear pore. It is also possible that the pre-CP is simply less stable, making it undetectable by conventional techniques. We are presently trying to introduce mutations that will stabilize the pre-CP to evaluate its influence on nuclear targeting.
This work shows clearly that CaMV exposes NLSs at the surface of the virion that could be used for targeting the virus to the nucleus. The nuclear targeting of CP is not directly regulated by phosphorylation but may be influenced by the presence of the N-terminal acidic region that is removed by the viral protease (58). It is unclear how the viral DNA enters the nucleus. CaMV particles are not normally found in the nuclei of the infected plants. Two reports describe the presence of CaMV particles in the nuclei of infected plants (22, 26). These examples were reported as being unusual, because the CaMV isolates used do not infect natural hosts of the virus. However, we cannot rule out that CaMV particles enter the host nucleus through the nuclear pore complexes in a normal infection, as suggested for SV40 (10). SV40 virions are made of three structural proteins, VP1, VP2, and VP3. Each of these proteins contains an NLS that is involved in the nuclear localization of the virus (8, 11, 23, 34, 47, 60, 61). Soon after entry of SV40 into the nucleus, expression of the large T antigen can be detected (10, 62). The expression of this viral protein positively affects the rate of nuclear import by enlarging the size of the nuclear pores (16). Like for SV40, it is possible that CaMV, a 50-nm virion (similar in size to SV40), can pass through a 26-nm pore (30), if the pore is elastic. It might be possible that CaMV virions are rarely seen in the nuclei of infected plants because the rate of disassembly of the virus nucleus is very high.
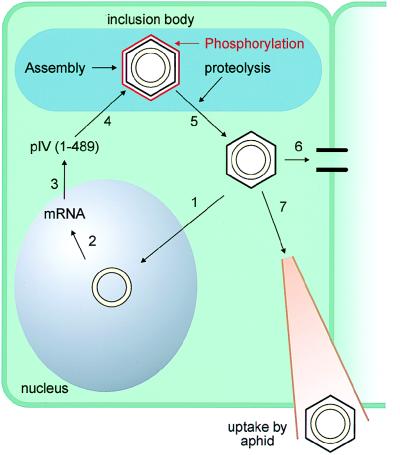
In summary, these results suggest that CaMV belongs to the group of viruses that are able to infect nonproliferating cells. This hypothesis is supported by the observation that CaMV is able to infect a plant if a fully developed leaf is inoculated. This allows us to suggest the following model (Fig. 6). CaMV virions infecting a new cell will be targeted to the nucleus of the plant cell via the NLS of the CP (step 1). It is not clear at this point if the virus is able to pass through the nuclear pore or if the virus disassembles at the nuclear membrane. Once the virion is in the nucleus, the viral genome is transcribed and mRNAs are transported to the cytoplasm for translation and for reverse transcription (step 2). The full-length CP (amino acids 1 to 489) is produced. The presence of the first 76 amino acids of CP keeps the protein in the cytoplasm, where it can be targeted to the inclusion body for viral assembly (step 3). Once the virus is assembled, the viral protease will cleave off the first 75 amino acids (step 4). This proteolysis will expose the NLS of the virus so that it can now be transported to the nucleus (step 5). The mature virus can also be transported to a neighboring cell via plasmodesmata modified by the viral movement protein (step 6) or can be captured by a feeding aphid that will transmit the virus to another plant (step 7).
FIG. 6.
Model describing the involvement of CaMV CP in the nuclear import of the CaMV genomic DNA to the nucleus.
ACKNOWLEDGMENTS
We thank M. Müller for the preparation of protoplasts and Andreas Hefti for assistance with the electron microscope. We also thank H. Rothnie for her critical reading of the manuscript.
This work was supported by a postdoctoral NSERC fellowship and by Friedrich Miescher fellowships.
REFERENCES
- 1.Blanc S, Cerutti M, Usmany M, Vlak J M, Hull R. Biological activity of cauliflower mosaic virus aphid transmission factor expressed in a heterologous system. Virology. 1993;192:643–650. doi: 10.1006/viro.1993.1080. [DOI] [PubMed] [Google Scholar]
- 2.Bock C T, Schwinn S, Schroder C H, Velhagen I, Zentgraf H. Localization of hepatitis B virus core protein and viral DNA at the nuclear membrane. Virus Genes. 1996;12:53–63. doi: 10.1007/BF00370001. [DOI] [PubMed] [Google Scholar]
- 3.Bosshart H, Humphrey J, Deignan E, Davidson J, Drazba J, Yuan L, Oarchchot V, Bonifacio J S, Peters P. The cytoplasmic domain mediates localisation of furin to the trans-golgi network en route to the endosomal/lysosomal system. J Cell Biol. 1984;126:1157–1172. doi: 10.1083/jcb.126.5.1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brown P H, Tiley L S, Cullen B R. Efficient polyadenylation within the human immunodeficiency virus type 1 long terminal repeat requires flanking U3-specific sequences. J Virol. 1991;65:3340–3343. doi: 10.1128/jvi.65.6.3340-3343.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bui M, Whittaker G, Helenius A. Effect of M1 protein and low pH on nuclear transport of influenza virus ribonucleoproteins. J Virol. 1996;70:8391–8401. doi: 10.1128/jvi.70.12.8391-8401.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bukrinsky M I, Haggerty S, Dempsey M P, Sharova N, Adzhubel A, Spitz L, Lewis P, Goldfarb D, Emerman M, Stevenson M. A nuclear localization signal within HIV-1 matrix protein that governs infection of non-dividing cells. Nature. 1993;365:666–669. doi: 10.1038/365666a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Capecchi M R. High efficiency transformation by direct microinjection of DNA into cultured mammalian cells. Cell. 1980;22:479–488. doi: 10.1016/0092-8674(80)90358-x. [DOI] [PubMed] [Google Scholar]
- 8.Chang D, Haynes J I, Brady J N, Consigli R A. Identification of a nuclear localization sequence in the polyomavirus capsid protein VP2. Virology. 1992;191:978–983. doi: 10.1016/0042-6822(92)90276-u. [DOI] [PubMed] [Google Scholar]
- 9.Cheng R H, Olson N H, Baker T S. Cauliflower mosaic virus: a 420 subunit (T=7), multilayer structure. Virology. 1992;186:655–668. doi: 10.1016/0042-6822(92)90032-k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Clever J, Yamada M, Kasamatsu H. Import of simian virus 40 virions through nuclear pore complexes. Proc Natl Acad Sci USA. 1991;88:7333–7337. doi: 10.1073/pnas.88.16.7333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Clever J, Kasamatsu H. Simian virus 40 Vp2/3 small structural proteins harbor their own nuclear transport signal. Virology. 1991;181:78–90. doi: 10.1016/0042-6822(91)90472-n. [DOI] [PubMed] [Google Scholar]
- 12.Covey S N, Turner D S. Changes in populations of cauliflower mosaic virus DNA and RNA forms during turnip callus proliferation. J Gen Virol. 1993;74:1887–1893. doi: 10.1099/0022-1317-74-9-1887. [DOI] [PubMed] [Google Scholar]
- 13.Dingwall C, Laskey R N. Nuclear targeting sequences: a consensus? Trends Biochem Sci. 1991;16:478–481. doi: 10.1016/0968-0004(91)90184-w. [DOI] [PubMed] [Google Scholar]
- 14.Eckhardt S G, Milich D R, McLachlan A. Hepatitis B virus core antigen has two nuclear localization signal sequences in the arginine-rich carboxyl terminus. J Virol. 1991;65:575–582. doi: 10.1128/jvi.65.2.575-582.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Farnet C M, Haseltine W A. Integration of human immunodeficiency virus type one DNA in vitro. Proc Natl Acad Sci USA. 1990;87:4164–4168. doi: 10.1073/pnas.87.11.4164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Feldher C, Cole C, Lanford R E, Akin D. The effects of SV40 large T-antigen and p53 on nuclear transport capacity in BALB/c 3T3 cells. Exp Cell Res. 1994;213:164–171. doi: 10.1006/excr.1994.1186. [DOI] [PubMed] [Google Scholar]
- 17.Fletcher T M, Brichacek B, Sharova N, Newman M A, Stivahtis G, Sharp P M, Emerman M, Hahn B H, Stevenson M. Nuclear import and cell cycle arrest functions of the HIV-1 Vpr protein are encoded by two separate genes in HIV-2/SIVSM. EMBO J. 1996;15:6155–6165. [PMC free article] [PubMed] [Google Scholar]
- 18.Fouchier R A M, Meyer B E, Simon J H M, Fisher U, Malim M H. HIV-1 infection of non-dividing cells: evidence that the amino-terminal basic region of the viral matrix protein is important for CP processing but not for post-entry nuclear import. EMBO J. 1997;16:4531–4539. doi: 10.1093/emboj/16.15.4531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Freed E O, Englund G, Maldarelli F, Martin M A. Phosphorylation of residue 131 of HIV-1 matrix is not required for macrophage infection. Cell. 1997;88:171–174. doi: 10.1016/s0092-8674(00)81836-x. [DOI] [PubMed] [Google Scholar]
- 20.Gallay P, Swingler S, Aiken C, Trono D. HIV-1 infection of non-dividing cells: C-terminal tyrosine phosphorylation of the viral matrix protein is a key regulator. Cell. 1995;80:379–388. doi: 10.1016/0092-8674(95)90488-3. [DOI] [PubMed] [Google Scholar]
- 21.Gallay P, Swingler S, Song J, Bushman F, Trono D. HIV nuclear import is governed by the phosphotyrosine-mediated binding of matrix to the core domain of integrase. Cell. 1995;83:569–576. doi: 10.1016/0092-8674(95)90097-7. [DOI] [PubMed] [Google Scholar]
- 22.Garbaczewska G, Kerlan C. Localization of cauliflower mosaic virus in the cell nucleus of Brassica pekinensis L. Res Virol. 1992;143:285–295. doi: 10.1016/s0923-2516(06)80117-2. [DOI] [PubMed] [Google Scholar]
- 23.Gharakhanian E, Takahashi J, Kasamatsu H. The carboxyl 35 amino acids of SV40 VP3 are essential for its nuclear accumulation. Virology. 1987;157:440–448. doi: 10.1016/0042-6822(87)90286-8. [DOI] [PubMed] [Google Scholar]
- 24.Goodall G J, Wiebauer K, Filipowicz W. Analysis of pre mRNA processing in transfected plant protoplasts. Methods Enzymol. 1990;181:148–161. doi: 10.1016/0076-6879(90)81117-d. [DOI] [PubMed] [Google Scholar]
- 25.Gordon K, Pfeiffer P, Futterer J, Hohn T. In vitro expression of cauliflower mosaic virus genes. EMBO J. 1988;7:309–317. doi: 10.1002/j.1460-2075.1988.tb02814.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gracia O, Shepherd R J. Cauliflower mosaic virus in the nucleus of Nicotiana. Virology. 1985;146:141–145. doi: 10.1016/0042-6822(85)90061-3. [DOI] [PubMed] [Google Scholar]
- 27.Greber U F, Webster P, Weber J, Helenius A. The role of the adenovirus protease in virus entry into cells. EMBO J. 1996;15:1766–1777. [PMC free article] [PubMed] [Google Scholar]
- 28.Hagstrom J E, Ludtke J J, Bassik M C, Sebestyén M G, Adam S A, Wolff J A. Nuclear import of DNA in digitonin-permeabilised cells. J Cell Sci. 1997;110:2323–2331. doi: 10.1242/jcs.110.18.2323. [DOI] [PubMed] [Google Scholar]
- 29.Heinzinger N K, Bukrinsky M I, Haggerty S A, Ragland A M, Kewalramani V, Lee M A, Gendelman H E, Ratner L, Stevenson M, Emerman M. The Vpr protein of human immunodeficiency virus type 1 influences nuclear localization of viral nucleic acids in non-dividing host cells. Proc Natl Acad Sci USA. 1994;91:7311–7315. doi: 10.1073/pnas.91.15.7311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hicks G R, Raikhel N. Protein import into the nucleus: an integrated review. Annu Rev Cell Dev Biol. 1995;11:155–188. doi: 10.1146/annurev.cb.11.110195.001103. [DOI] [PubMed] [Google Scholar]
- 31.Himmelbach A, Chapdelaine Y, Hohn T. Interaction between cauliflower mosaic virus inclusion body protein and capsid protein: implications for viral assembly. Virology. 1996;217:147–157. doi: 10.1006/viro.1996.0102. [DOI] [PubMed] [Google Scholar]
- 32.Hull R, Covey S N, Maule A J. Structure and replication of caulimovirus genomes. J Cell Sci. 1987;7:213–229. doi: 10.1242/jcs.1987.supplement_7.16. [DOI] [PubMed] [Google Scholar]
- 33.Hurkman W J, Tanaka C K. Solubilization of membrane proteins for analysis by two-dimensional gel electrophoresis. Plant Physiol. 1986;81:802–806. doi: 10.1104/pp.81.3.802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ishii N, Minami N, Chen E Y, Medina A L, Chico M M, Kasamatsu H. Analysis of a nuclear localization signal of simian virus 40 major capsid protein Vp1. J Virol. 1996;70:1317–1322. doi: 10.1128/jvi.70.2.1317-1322.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jans D A, Jans P. Negative charge at the casein kinase II site flanking the nuclear localization signal of the SV-40 large T-antigen is mechanistically important for enhanced nuclear import. Oncogene. 1994;9:2961–2968. [PubMed] [Google Scholar]
- 36.Kann M, Gerlich W H. Effect of core protein phosphorylation by protein kinase C on encapsidation of RNA within core particles of hepatitis B virus. J Virol. 1994;68:7993–8000. doi: 10.1128/jvi.68.12.7993-8000.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kann M, Bischof A, Gerlich W. In vitro model for the nuclear transport of the hepadnavirus genome. J Virol. 1997;71:1310–1316. doi: 10.1128/jvi.71.2.1310-1316.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 39.Lebeurier G, Hirth L, Hohn T, Hohn B. Infectivities of native and cloned cauliflower mosaic virus DNA. Gene. 1980;12:139–146. doi: 10.1016/0378-1119(80)90024-4. [DOI] [PubMed] [Google Scholar]
- 40.Leclerc D, Eweida M, Sing R P, Abouhaidar M. Biotinylated DNA probes for detecting potato virus Y and aucuba mosaic virus in leaves and dormant tubers of potato. Potato Res. 1992;35:173–182. [Google Scholar]
- 41.Liao W, Ou J-H. Phosphorylation and nuclear localization of the hepatitis B virus core protein: significance of serine in the three repeated SPRRR motifs. J Virol. 1995;69:1025–1029. doi: 10.1128/jvi.69.2.1025-1029.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Makkerh J P S, Dingwall C, Laskey R A. Comparative mutagenesis of nuclear localization signals reveals the importance of neutral and acidic amino acids. Curr Biol. 1996;6:1025–1027. doi: 10.1016/s0960-9822(02)00648-6. [DOI] [PubMed] [Google Scholar]
- 43.Martinez-Izquierdo J, Hohn T. Cauliflower mosaic virus coat protein is phosphorylated in vitro by a virion-associated protein kinase. Proc Natl Acad Sci USA. 1987;84:1824–1828. doi: 10.1073/pnas.84.7.1824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Maule A J, Harker C L, Wilson I J. The pattern of accumulation of cauliflower mosaic virus specific products in infected turnips. Virology. 1989;169:436–446. doi: 10.1016/0042-6822(89)90169-4. [DOI] [PubMed] [Google Scholar]
- 45.Ménissier J, de Murcia G, Lebeurier G, Hirth L. Electron microscopic studies of the different topological forms of the cauliflower mosaic virus DNA: knotted encapsidated DNA and nuclear minichromosome. EMBO J. 1983;2:1067–1071. doi: 10.1002/j.1460-2075.1983.tb01547.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mieszczak M, Klahre U, Levy J H, Gooddall G J, Filipowicz W. Multiple plant RNA binding proteins identified by PCR: expression of cDNAs encoding RNA binding proteins targeted to chloroplasts in Nicotiana plumbaginifolia. Mol Gen Genet. 1992;234:390–400. doi: 10.1007/BF00538698. [DOI] [PubMed] [Google Scholar]
- 47.Moreland R B, Garcea R L. Characterization of a nuclear localization sequence in the polyomavirus capsid protein VP1. Virology. 1991;185:513–518. doi: 10.1016/0042-6822(91)90811-o. [DOI] [PubMed] [Google Scholar]
- 48.Olszewski N, Hagen G, Guilfoyle T J. A transcriptionally active covalently closed minichromosome of cauliflower mosaic virus DNA isolated from infected turnip leaves. Cell. 1982;29:395–402. doi: 10.1016/0092-8674(82)90156-8. [DOI] [PubMed] [Google Scholar]
- 49.Olszewski N E, Guilfoyle T L. Nuclei purified from cauliflower mosaic virus infected turnip leaves contain subgenomic, covalently closed circular cauliflower mosaic virus DNAs. Nucleic Acids Res. 1983;11:8901–8914. doi: 10.1093/nar/11.24.8901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pfeiffer P, Hohn T. Involvement of reverse transcription in the replication of cauliflower mosaic virus: a detailed model and test of some aspects. Cell. 1983;33:781–789. doi: 10.1016/0092-8674(83)90020-x. [DOI] [PubMed] [Google Scholar]
- 51.Popov S, Rexach M, Zybarth G, Reiling N, Lee M-A, Ratner L, Lane C M, Shannon Moore M, Blobel G, Burinsky M. Viral protein R regulates nuclear import of the HIV-1 pre-integration complex. EMBO J. 1998;17:909–917. doi: 10.1093/emboj/17.4.909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Roe T Y, Reynolds T C, Yu G, Brown P O. Integration of murine leukemia virus DNA depends on mitosis. EMBO J. 1993;12:2099–2108. doi: 10.1002/j.1460-2075.1993.tb05858.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rothnie H M, Chapdelaine Y, Hohn T. Pararetroviruses and retroviruses: a comparative review of viral structure and gene expression strategies. Adv Virus Res. 1994;44:1–67. doi: 10.1016/s0065-3527(08)60327-9. [DOI] [PubMed] [Google Scholar]
- 54.Schledz M, Leclerc D, Neuhaus G, Merkle T. Characterization of four cDNAs ( Y14615, Y14616, Y15224, Y15225) encoding different importin alpha homologues from Arabidopsis thaliana designated AtIMPal-4. Plant Physiol. 1998;116:868. [Google Scholar]
- 55.Schliephake A W, Rethwilm A. Nuclear localization of foamy virus CP precursor protein. J Virol. 1994;68:4946–4954. doi: 10.1128/jvi.68.8.4946-4954.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sheldon L A, Kingston R E. Hydrophobic coiled-coil domains regulate the subcellular localization of human heat shock factor 2. Genes Dev. 1993;7:1549–1558. doi: 10.1101/gad.7.8.1549. [DOI] [PubMed] [Google Scholar]
- 57.Shepherd R J, Bruening G E, Wakeman R J. Double stranded DNA from cauliflower mosaic virus. Virology. 1970;41:339–347. doi: 10.1016/0042-6822(70)90086-3. [DOI] [PubMed] [Google Scholar]
- 58.Torruella M, Gordon K, Hohn T. Cauliflower mosaic virus produces an aspartic proteinase to cleave its polyproteins. EMBO J. 1989;8:2819–2825. doi: 10.1002/j.1460-2075.1989.tb08428.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tuttleman J S, Pourcel C, Karpas A, Summers J. Formation of the pool of covalently closed circular viral DNA in hepadnavirus-infected cells. Cell. 1986;47:451–460. doi: 10.1016/0092-8674(86)90602-1. [DOI] [PubMed] [Google Scholar]
- 60.Wychowski C, Benichou D, Girard M. A domain of SV40 capsid polypeptide VP1 that specifies migration into the cell nucleus. EMBO J. 1986;5:2569–2576. doi: 10.1002/j.1460-2075.1986.tb04536.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wychowski C, Benichou D, Girard M. The intranuclear localization of simian virus 40 polypeptides Vp2 and Vp3 depends on a specific amino acid sequence. J Virol. 1987;61:3862–3869. doi: 10.1128/jvi.61.12.3862-3869.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yamada M, Kasamatsu H. Role of nuclear pore complex in simian virus 40 nuclear targeting. J Virol. 1993;67:119–130. doi: 10.1128/jvi.67.1.119-130.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yeh C T, Liaw Y F, Ou J H. The arginine-rich domain of hepatitis B virus precore and core proteins contains a signal for nuclear transport. J Virol. 1990;64:6141–6147. doi: 10.1128/jvi.64.12.6141-6147.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]