Abstract
Vpr and Vpx proteins from human and simian immunodeficiency viruses (HIV and SIV) are incorporated into virions in quantities equivalent to those of the viral Gag proteins. We demonstrate here that Vpr and Vpx proteins from distinct lineages of primate lentiviruses were able to bind to their respective Gag precursors. The capacity of HIV type 1 (HIV-1) Vpr mutants to bind to Pr55Gag was correlated with their incorporation into virions. Molecular analysis of these interactions revealed that they required the C-terminal p6 domain of the Gag precursors. While the signal for HIV-1 Vpr binding lies in the leucine triplet repeat region of the p6 domain reported to be essential for incorporation, SIVsm Gag lacking the equivalent region still bound to SIVsm Vpr and Vpx, indicating that the determinants for Gag binding are located upstream of this region of the p6 domain. Binding to Gag cleavage products showed that HIV-1 Vpr interacted directly with the nucleocapsid protein (NC), whereas SIVsm Vpr and Vpx did not interact with NC but with the p6 protein. These results (i) reveal differences between HIV-1 and SIVsm for the p6 determinants required for Vpr and Vpx binding to Gag and (ii) suggest that HIV-1 Vpr and SIVsm Vpr and Vpx interact with distinct cleavage products of the precursor following proteolytic processing in the virions.
The genomes of human and simian immunodeficiency viruses (HIV and SIV) contain several genes encoding auxiliary proteins which are not required for viral growth in vitro but are essential for viral replication and pathogenesis in vivo (41). Two of these proteins, Vpr and Vpx, play an important role in vivo, since rhesus monkeys infected with a vpr-vpx doubly defective SIV had a low virus burden and did not develop immunodeficiency disease (14). The two vpr and vpx genes are not found in all primate lentivirus genomes. Members of the HIV type 2 (HIV-2)–SIVsm group contain both genes, while other primate lentivirus lineages (HIV-1–SIVcpz, SIVagm, SIVmnd, and SIVsyk) have only the vpr gene.
Vpr and Vpx are two small proteins (14 to 16 kDa) that associate with viral particles and then accumulate in the nuclei of infected cells (7, 19, 26, 44). Although the molecular mechanism supporting the biological roles of Vpr and Vpx in vivo has not been elucidated, at least two separate functions of HIV-1 Vpr have been defined in vitro. Vpr is essential for the infection of terminally differentiated macrophages by HIV-1 (1, 4, 8, 17) and induces an arrest of the cell cycle in the G2 phase (16, 18, 34, 36, 37). In HIV-2–SIVsm, these two functions are segregated between the Vpr and Vpx proteins: Vpr is able to induce a cell cycle arrest but is dispensable for the infection of macrophages, while Vpx has no effect on the cell cycle but is required for virus replication in macrophages (11, 34).
Unlike other auxiliary proteins, Vpr and Vpx are specifically packaged into virions in quantities similar to those of the viral Gag proteins (7, 19, 33). Several studies have shown that HIV-1 Vpr is incorporated into virions formed in the absence of the pol and env gene products and is independent of viral RNA encapsidation (23, 33). These results indicate that expression of the HIV-1 Gag precursor (Pr55Gag) is sufficient to mediate the incorporation of Vpr into virions. The C-terminal p6 region of Pr55Gag is essential for this process since Vpr is not incorporated when the p6 domain is deleted (23, 26, 33), and Vpr is efficiently incorporated into particles formed by chimeric Rous sarcoma virus (RSV) or murine leukemia virus (MLV) Gag polyproteins containing the HIV-1 p6 sequence fused to their C termini (21, 25). The minimal region of HIV-1 Pr55Gag required for Vpr incorporation has been defined within a domain located near the C terminus of the p6 and containing a repeated leucine triplet sequence (LXX)4 (20, 25). The packaging signal for incorporation of the Vpx protein from HIV-2 also lies in the p6 part of the HIV-2 Gag precursor (32, 42).
We have explored the molecular mechanism mediating the incorporation of the HIV and SIV Vpr and Vpx proteins into virions by investigating the ability of these auxiliary proteins to interact physically with their homologous Gag precursor. We used the yeast two-hybrid system and an in vitro binding assay to demonstrate that Vpr and Vpx could bind directly to the precursors and that the C-terminal p6 domain was required for these interactions. While the (LXX)4 region of the p6 domain of the HIV-1 precursor was essential for Vpr binding, the minimal sequence required for SIVsm Vpr and Vpx binding was found upstream of the equivalent (LXX)3 region of the SIVsm p6 domain. Binding analysis performed with the cleavage products of the HIV-1 and SIVsm Gag precursors also revealed substantial differences between these two distinct groups of primate lentiviruses. HIV-1 Vpr interacted directly with the NCp7 protein, whereas SIVsm Vpr and Vpx interacted strongly with the SIVsm p6 protein, suggesting that HIV-1 Vpr and SIVsm Vpr and Vpx interact with distinct cleavage products of the precursor after proteolytic processing in the mature virions.
MATERIALS AND METHODS
Plasmid construction. (i) Yeast two-hybrid expression vectors.
The construction of the vectors for expression of HIV-1 (Lai isolate), SIVsm (Pbj1.9 isolate), and SIVagm (vervet isolate SM9063) Vpr and Vpx fused to the LexA DNA binding domain (LexABD) has been previously described (39). Vectors for expression of HIV-2 Vpr and Vpx fused to the Gal4 activation domain (Gal4AD) were constructed by PCR amplification of the vpr and vpx genes from the HIV-2 Rod isolate (30), which were then inserted in frame with the Gal4AD in the pGad1318 plasmid. Except for the HIV-2 Rod Gag precursor, which was expressed in fusion with the LexABD, the Gag precursors of HIV-1, SIVsm, and SIVagm, were fused to the Gal4AD. Briefly, the various gag genes were amplified by PCR using specific primers and then subcloned in frame with the LexABD or Gal4AD into the pLex10 or pGad1318 plasmid (39). HIV-2 gag was cloned into the EcoRI-SalI restriction sites of pLex10. HIV-1 and SIVagm gag were cloned into the BamHI-XhoI sites of pGad1318, while SIVsm gag was cloned into the EcoRI-XhoI sites of the same plasmid. The deletion mutants of HIV-1 Gag (GagΔp6) and SIVsm Gag (GagΔp6, GagΔL1, GagΔL2, and GagΔL3) (see Fig. 4) were generated by introducing premature termination codons into the HIV-1 and SIVsm gag genes by PCR (22). We used site-directed mutagenesis to generate single-point mutants of HIV-1 (GagL44A) and SIVsm (GagL54A) Gag by substituting Ala for the last Leu residue of the HIV-1 and SIVsm Gagp6 coding sequences located at positions 44 (HIV-1) and 54 (SIVsm) of p6 (30). Vectors for expression of the various HIV-1 and SIVsm Gag cleavage products fused to Gal4AD were generated by PCR amplification with specific primers of the matrix (MA), capsid (CA), nucleocapsid (NC), and p6 coding sequences from the HIV-1 Lai and SIVsmPbj1.9 gag genes (30). All the mutants and cleavage products of SIVsm Gag were subcloned at the EcoRI-XhoI sites of pGad1318, while those of HIV-1 Pr55Gag were subcloned at the BamHI-XhoI sites of the same plasmid. HIV-1 Pr55GagΔD1, NCp7ΔD1, and NCp7ΔD2 mutants were constructed by PCR with the pGad1318 plasmid as previously described (31).
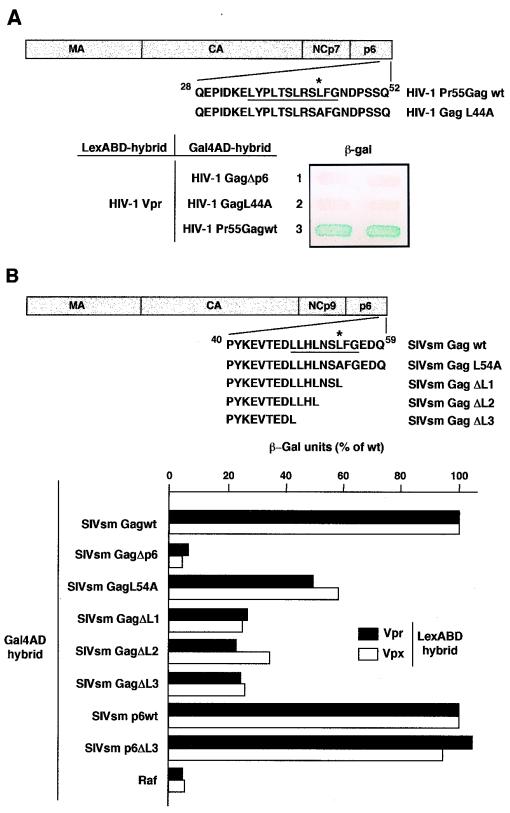
FIG. 4.
Role of the LXX repeats of the HIV-1 and SIVsm Gag p6 domain in Vpr and Vpx binding. (A) Mutation within the p6 (LXX)4 region disrupts HIV-1 Vpr binding to Pr55Gag. A diagram shows the HIV-1 wt Pr55Gag and GagL44A mutant. The p6 (LXX)4 repeats are underlined, and the Leu residue replaced by Ala in the GagL44A mutant is indicated by an asterisk. Numbering refers to the HIV-1 Lai p6 domain (30). L40 expressing HIV-1 Vpr fused to the LexABD and either GagΔp6 (lane 1), GagL44A (lane 2), or Pr55Gag wt (lane 3) fused to the Gal4AD was analyzed for β-gal activity. The filter assay was carried out overnight. (B) Quantitative β-gal assay of SIVsm Vpr and Vpx binding to Gag and p6 mutants. A diagram shows the SIVsm Gag wt and GagL54A, GagΔL1, GagΔL2, and GagΔL3 mutants. The p6 (LXX)3 repeats are underlined, and the Leu residue replaced by Ala in the GagL54A mutant is indicated by an asterisk. Numbering refers to the SIVsmPbj1.9 p6 domain (30). L40 expressing either the SIVsm Vpr (solid bars) or Vpx (white bars) LexABD hybrid in combination with each of the Gal4AD hybrids indicated was assayed for β-gal activity in a liquid culture assay. The results are expressed as the percentages of the β-gal activity determined for each Gag or p6 mutant relative to the activity obtained with the wt Gag and p6, respectively. The background level is approximately 2 U and corresponds to L40 expressing either the SIVsm Vpr- or Vpx-LexABD hybrid and the Gal4AD-Raf hybrid.
(ii) Bacterial expression vector.
The vector for expression in Escherichia coli of Pr55Gag fused to the glutathione S-transferase (GST) was constructed by subcloning the HIV-1 Lai gag gene, amplified by PCR, into the BamHI-EcoRI sites of pGEX-2T (Pharmacia).
(iii) Mammalian expression vectors.
Vectors for expression of the wild type (wt) or mutated HIV-1 Vpr proteins were constructed in the pAS1B plasmid. This vector contains an initiation codon followed by the nucleotide sequence encoding the nine-amino-acid epitope tag from the influenza virus hemagglutinin (HA) and thus allows expression, driven by the cytomegalovirus promoter, of Vpr molecules fused at their N termini to HA (HA-tagged Vpr). The vectors pCMVΔR9.1, pHR′-CMVLacZ, and pMD.G used for viral production in the Vpr packaging assay were kindly provided by D. Trono (Geneva, Switzerland) (45).
Two-hybrid assay.
The L40 yeast reporter strain containing the two LexA-inducible genes, HIS3 and LacZ, was cotransformed with the indicated LexABD and Gal4AD hybrid expression vectors and plated on selective medium lacking tryptophan and leucine (39). Double transformants were patched on the same medium and replica plated on selective medium lacking tryptophan, leucine, and histidine for histidine auxotrophy analysis and on Whatman 40 filters for β-galactosidase (β-gal) activity assay. This latter qualitative assay was monitored by incubation for from 1 h to overnight at 30°C, and the reaction was then stopped with 1 M Na2CO3 (39). The liquid culture assay for quantitative β-gal activity was performed in triplicate as described previously (39). The construction of the HIV-1 Vpr mutant library and the two-hybrid screening procedure of this library have been described previously (39). Briefly, the vpr gene from the HIV-1 Lai isolate was amplified by error-prone PCR as previously described (5), and the fragments were inserted into pLex10. Library plasmid DNA from about 7 × 103 independent E. coli clones, representing the complexity of the vpr potential mutant library, was prepared and used to transform the L40-MATa yeast strain. About 1,500 yeast clones were then screened to select Vpr mutants defective for binding to Pr55Gag by mating with the AMR70-MATα yeast strain previously transformed with the Gal4AD-Pr55Gag expression vector. Plasmids from mutants unable to interact with Pr55Gag were rescued, and their insert was completely sequenced. Two mutants (Vpr*E25K and Vpr*H33L), each containing a single point mutation, were isolated. Two other Vpr mutants (Vpr*A and Vpr*W18R) giving a strong β-gal activity in the filter assay were also selected.
In vitro binding study.
The GST-Pr55Gag fusion protein was expressed in E. coli and immobilized on glutathione (GSH)-agarose beads as described previously (2). One microgram of GST-Pr55Gag or GST immobilized on GSH-agarose beads was incubated overnight at 4°C with 2 μg of HIV-1 Lai Vpr obtained by chemical synthesis (a gift from B. Roques, Paris, France) (9), in phosphate-buffered saline containing 5 μg of bovine serum albumin/ml and 0.05% Tween. The beads were washed four times with a buffer containing 50 mM Tris-HCl (pH 7.4), 1 mM EDTA, 300 mM NaCl, 10% glycerol, and 1% Nonidet P-40 and then subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). Vpr binding was analyzed by Western blotting with rabbit anti-Vpr antibodies (a gift from E. Cohen, Montreal, Canada) and the enhanced chemiluminescence system (Amersham).
Cell culture and transfection.
293T cells were cultured in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal calf serum, and 25 mM HEPES was added during virus production. About 2 × 106 cells were cotransfected with 10 μg of pCMVΔR8.91, 5 μg of pMD.G, 20 μg of pHR′-CMVLacZ (45), and 5 μg of pAS1B-Vpr wt or mutated, using the procedure described by Boussif et al. (3) with the 22-kDa polyethylenimine (Euromedex).
Viral protein analysis.
Culture supernatants were collected 48 h after transfection and filtered through 0.45-μm-pore-size filters, and an aliquot was assayed for CAp24 Gag antigen by enzyme-linked immunosorbent assay (DuPont). Virions were collected by centrifugation for 1 h at 100,000 × g and suspended in 100 μl of ice-cold lysis buffer (10 mM Tris [pH 7.6], 150 mM NaCl, 2 mM EDTA, 0.5% Triton X-100) as described previously (10). For preparation of cell lysates, transfected cells were trypsinized, collected by centrifugation at 450 × g for 5 min, and suspended in 300 μl of ice-cold lysis buffer. They were then incubated on ice for 5 min and clarified by centrifugation for 5 min at 5,000 rpm in an Eppendorf model 5415C microcentrifuge. The supernatant was transferred to a fresh tube, and the protein concentration of the cell lysates was measured (Bio-Rad). Proteins from cell (50 μg of total proteins) and virion (50 ng of CAp24) lysates were separated by SDS-PAGE and analyzed by Western blotting with anti-HA 12CA5 (Boehringer) or anti-CAp24 monoclonal antibodies and the enhanced chemiluminescence system (Amersham). Anti-CAp24 monoclonal antibody was obtained from the National Institutes of Health AIDS Research Program (40).
RESULTS
Direct interaction between HIV and SIV Vpr and Vpx proteins and the Gag precursors.
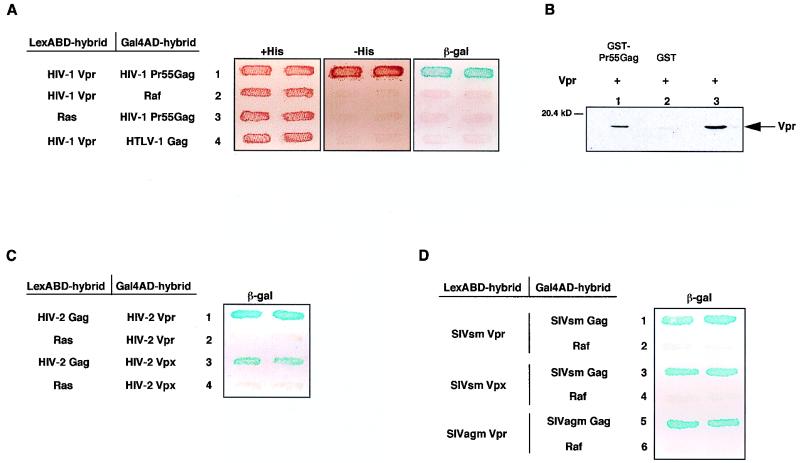
Virological studies have shown that expression of the HIV-1 Pr55Gag is sufficient to mediate Vpr incorporation into virions (20, 23, 33). We therefore analyzed whether HIV-1 Vpr bound to Pr55Gag, using the yeast two-hybrid system. The Vpr protein from the HIV-1 Lai isolate was fused to the LexABD and analyzed for interaction with the Pr55Gag fused to the Gal4AD in the L40 yeast strain containing the two LexA-inducible reporter genes, LacZ and HIS3. Interaction between HIV-1 Vpr and Pr55Gag was indicated by the ability of the L40 strain cotransformed with the hybrid expression plasmids to express β-gal activity and to grow in the absence of histidine (Fig. 1A, lane 1). This interaction was specific, since there was no transcriptional activation of the reporter genes in yeast cells transformed with the LexABD Vpr and Gal4AD-Pr55Gag expression plasmids alone (not shown) or in combination with irrelevant hybrid proteins fused to the Gal4AD and LexABD, Gal4AD-Raf, and LexABD Ras, respectively (Fig. 1A, lanes 2 and 3). In addition, HIV-1 Vpr did not interact with the Gag precursor of another retrovirus, human T-cell leukemia virus type 1 (Fig. 1A, lane 4), indicating that the interaction with HIV-1 Pr55Gag involved specific determinants within this precursor. We confirmed the interaction between HIV-1 Vpr and Pr55Gag by an in vitro binding assay, using recombinant Pr55Gag expressed in E. coli as a GST fusion protein and Vpr obtained by chemical synthesis. As shown in Fig. 1B, Vpr bound specifically to GST-Pr55Gag (lane 1) but not to GST (lane 2). This in vitro binding study demonstrates that Vpr and Pr55Gag are capable of direct physical interaction independent of any yeast intermediate protein.
FIG. 1.
Interactions of Vpr and Vpx proteins from primate lentiviruses with their homologous Gag precursor. (A) Binding of HIV-1 Vpr to Pr55Gag in the two-hybrid system. The L40 yeast reporter strain expressing the pairs of indicated hybrids fused to the LexABD and Gal4AD was analyzed for histidine auxotrophy and β-gal activity. Double transformants were patched on selective medium with histidine (+His) (left panel) and then replica plated on medium without histidine (−His) (middle panel) and on Whatman filters for β-gal assay (right panel). Growth in the absence of histidine and expression of β-gal activity indicate interaction between hybrid proteins. The β-gal filter assay was carried out overnight. (B) Vpr-Pr55Gag interaction in vitro. Chemically synthesized Vpr (lane 3) was incubated with equal amounts of GST-Pr55Gag (lane 1) or GST (lane 2) immobilized on GSH-agarose beads. Bound Vpr was then analyzed by Western blotting with a rabbit anti-Vpr antiserum. (C and D) Binding of HIV-2, SIVsm, and SIVagm Vpr and Vpx to homologous Gag precursors. L40 expressing the pairs of indicated hybrids fused to the LexABD and Gal4AD was analyzed for β-gal activity. The filter assay was incubated for 3 h. Each patch represents an independent transformant.
Since Vpr and Vpx from other primate lentiviruses are also virion-associated proteins (6, 11, 17, 42, 44), we assayed the ability of Vpr and Vpx from the HIV-2–SIVsm lineage and of Vpr from SIVagm to interact with their homologous Gag precursor. The Vpr and Vpx proteins from SIVsm and SIVagm were expressed in fusion with the LexABD, while HIV-2 Rod Vpr and Vpx were fused to the Gal4AD because they gave some transcriptional background in the L40 strain when they were expressed in fusion with the LexABD. As shown in Fig. 1C and D, the Vpr and Vpx proteins from HIV-2, SIVsm, and SIVagm also interacted with their homologous Gag precursor in this two-hybrid assay. Monitoring of the β-gal activities by filter assay revealed stronger interactions with the HIV-2 and SIV proteins than with HIV-1 proteins, since the blue color was detected in less than 3 h (Fig. 1C and D) compared to the overnight incubation required to detect the HIV-1 Vpr-Pr55Gag interaction in the same assay (Fig. 1A). Direct binding to the Gag precursor appeared to be a general property of Vpr and Vpx proteins from the distinct lineages of primate lentiviruses.
Binding of HIV-1 Vpr to Pr55Gag and incorporation into virions.
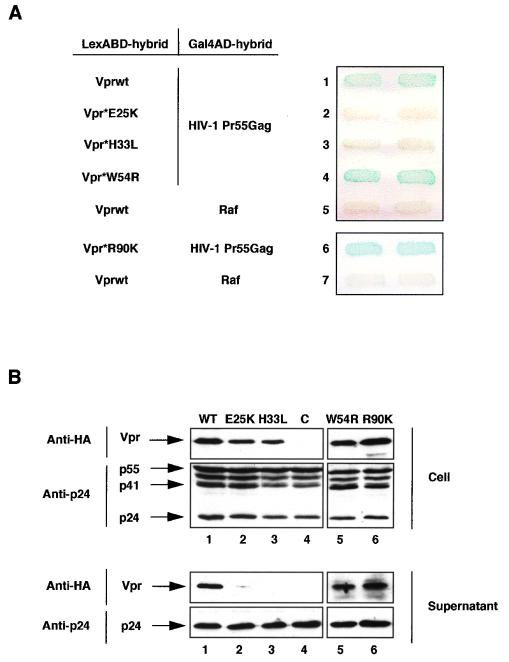
We then analyzed the relationship between the binding of Vpr to the Gag precursor and the virion incorporation by selecting HIV-1 Vpr mutants unable to interact with Pr55Gag in the two-hybrid system. A library of Vpr mutants fused to the LexABD was generated by random mutagenesis and expressed in combination with the Gal4AD-Pr55Gag hybrid in the L40 strain. We isolated two Vpr point mutants (Vpr*E25K and Vpr*H33L) that were unable to bind to Pr55Gag as indicated by the absence of β-gal expression (Fig. 2A, lanes 2 and 3). Both mutants were correctly expressed in yeast, as evidenced by Western blot analysis of yeast cell lysates (not shown). The Glu25 and His33 residues are located within the predicted N-terminal alpha-helical structure of Vpr, which is important for efficient Vpr incorporation (10, 27, 33, 43). Two other mutants (Vpr*W54R and Vpr*R90K) (39) containing mutations in the C-terminal part of Vpr retained, like the wt protein (Fig. 2A, lane 1), the ability to interact with Pr55Gag (lanes 4 and 6).
FIG. 2.
HIV-1 Vpr binding to Pr55Gag and virion incorporation. (A) Selection of Vpr mutants deficient for binding to Pr55Gag. L40 strain expressing the pairs of indicated hybrids fused to the LexABD and Gal4AD was analyzed for β-gal activity. Vpr*E25K (lane 2) and Vpr*H33L (lane 3) mutants were selected from a random Vpr mutant library, while Vpr*W54R and Vpr*R90K have been described previously (39). (B) Incorporation of HIV-1 Vpr mutants into virions. 293T cells were cotransfected with the pCMVΔR8.91, pMD.G, and pHR′-CMVLacZ plasmids and either a wt (lane 1) or a mutated (lanes 2, 3, 5, and 6) HA-tagged Vpr expression vector. Lysates from transfected cells (Cell) and virions (Supernatant) released into the culture medium and adjusted to contain similar amounts of CAp24 were separated by SDS-PAGE and analyzed by Western blotting with anti-HA (Cell and Supernatant upper panels) or anti-CAp24 (Cell and Supernatant lower panels) monoclonal antibodies. C (lane 4), cell and virion lysates from cells transfected with the pCMVΔR8.91, pMD.G, and pHR′-CMVLacZ vectors and the pAS1B plasmid without an insert.
These Vpr mutants were then analyzed for their ability to be incorporated into virions. We used a transient Vpr packaging assay in which Vpr fused at its N terminus to the HA epitope (HA-tagged Vpr) is expressed in trans (33) and incorporated into HIV-1 virions pseudotyped by the G protein of vesicular stomatitis virus (45). 293T cells were transfected with the HA-tagged Vpr (wt or mutated) expression vector in combination with an HIV-1-based vector lacking env and all of the auxiliary genes from HIV-1 and a VSV G protein expression vector. The virions released into the culture medium were collected 2 days later and lysed, and the virion lysates were adjusted to contain similar amounts of CAp24 (10). Cell lysates were also prepared from transfected cells. The virions produced from each transfection had retained full infectivity since they infected 293T cells with a similar efficiency (data not shown). The amount of virion- and cell-associated wt or mutated Vpr was then assessed by Western blot analysis using an anti-HA monoclonal antibody (Fig. 2B). The Vpr*W54R and Vpr*R90K mutants, which still bound to Pr55Gag in the two-hybrid assay, were incorporated into virions as efficiently as the wt protein, since a 14-kDa protein corresponding to HA-tagged Vpr was detected in the supernatants of transfected cells (lanes 1, 5, and 6, lower panel). In contrast, no Vpr was detected in the supernatants of cells expressing either the Vpr*E25K or Vpr*H33L mutant (lanes 2 and 3, lower panel), despite detectable levels of the protein in transfected cell lysates (lanes 2 and 3, upper panel). We also verified that similar amounts of virions had been produced in each transfection by probing the same blots with an anti-CAp24 monoclonal antibody (Fig. 2B). This striking correlation between the ability of Vpr to bind to Pr55Gag and to be packaged into virions indicates that Vpr and Vpx proteins are incorporated into virions through direct interaction with the Gag precursor. These results also indicate that residues in the N-terminal domain of HIV-1 Vpr are critical for interaction with Pr55Gag and for efficient packaging into virions.
Vpr and Vpx binding to the cleavage products of the Gag precursors.
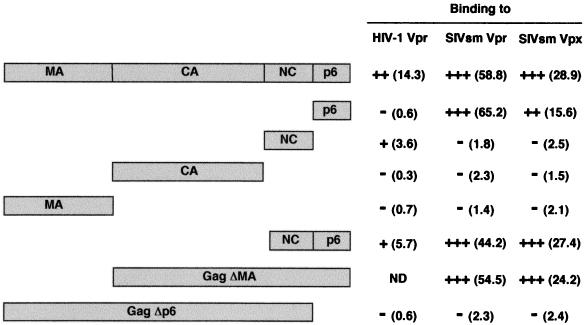
The binding of HIV-1 Vpr to the cleavage products of Pr55Gag was then analyzed. The coding sequences of the MA, CA, NCp7, and p6 proteins were fused to the Gal4AD and assayed for interaction with the LexABD-Vpr hybrid in the L40 strain. Only the NCp7 protein interacted weakly with Vpr in this two-hybrid assay (Fig. 3). The NCp15, covering the NCp7 and p6 coding sequences of Pr55Gag, also bound Vpr weakly. By contrast, no interaction between Vpr and the MA, CA, or p6 protein was detectable, although the hybrid proteins were correctly expressed in yeast cells as judged by Western blot analysis of yeast extracts (not shown). Since virological studies showed that HIV-1 Vpr packaging into virions is disrupted by deletion of the p6 C-terminal region of Pr55Gag (23, 26, 33), we investigated whether the p6 region was also essential for Vpr binding. Deletion of the last 52 C-terminal amino acids of Pr55Gag, corresponding to the p6 sequence (HIV-1 GagΔp6), completely abolished the interaction of the precursor with Vpr (Fig. 3), as indicated by the absence of β-gal activity in yeast cells expressing the LexABD-Vpr and Gal4AD-GagΔp6 hybrids (Fig. 4A, lane 1). These results suggest that the determinants mediating HIV-1 Vpr binding are located within the C-terminal region of Pr55Gag, corresponding to the NCp7 and p6 domains.
FIG. 3.
Binding of HIV-1 and SIVsm Vpr and Vpx to the cleavage products of the Gag precursors. L40 strain expressing either HIV-1 Vpr or SIVsm Vpr or Vpx fused to the LexABD in combination with each of the Gal4AD hybrids indicated on the left was analyzed for histidine auxotrophy and β-gal activity. The interactions between hybrid proteins were scored as follows: +++, cell growth on medium without histidine and development of a blue color in 3 h by β-gal filter assay; ++, cell growth on medium without histidine and development of a blue color after overnight incubation; +, cell growth on medium without histidine and development of a light blue color after overnight incubation; −, no growth on medium without histidine and development of a white color after overnight incubation of the β-gal assay. Quantitative β-gal activities expressed in β-gal units and determined by liquid culture assay are given in parentheses. The background level is approximately 1 to 2 U and corresponds to L40 expressing either the HIV-1 Vpr- or SIVsm Vpr- or Vpx-LexABD hybrid and the Gal4AD-Raf hybrid. ND, not done.
A similar analysis was then performed with SIVsm Vpr and Vpx and the cleavage products of the SIVsm Gag precursor (Fig. 3). The p6 protein of SIVsm Gag interacted with Vpr and Vpx as efficiently as the whole SIVsm precursor, but no binding was detected with the MA, CA, and particularly the NCp9 protein. As for HIV-1, deletion of the p6 domain (SIVsm GagΔp6) prevented interactions of both Vpr and Vpx with the Gag precursor (Fig. 4B). By contrast, deletions of the MA (GagΔMA) or the MA and CA (NCp9-p6) coding sequences did not impair interactions with Vpr and Vpx (Fig. 3). Hence, the C-terminal p6 domain contains the specific determinants required for binding of Vpr and Vpx to the SIVsm Gag, whereas the other domains of the precursor do not seem to be involved in these interactions. They also suggest that HIV-1 Vpr and SIVsm Vpr and Vpx interact with distinct maturation products after proteolytic processing of the Gag precursor.
Role of the LXX repeat sequences of the HIV-1 and SIVsm Gag p6 domain in Vpr and Vpx binding.
Previous studies have shown that a region at the C terminus of the HIV-1 p6 domain of Pr55Gag (residues 35 to 46 [Fig. 4A]) contains a leucine triplet repeat sequence (LXX)4 and is required for Vpr incorporation into HIV-1 virions (20, 25). A single point mutation of the Leu residue at position 44 within the fourth LXX motif completely prevented Vpr incorporation (20). We therefore analyzed the effect of this point mutation on Vpr binding. The Leu residue of Pr55Gag was replaced by an Ala to generate the HIV-1 GagL44A point mutant fused to Gal4AD. This mutant did not interact with the LexABD-Vpr hybrid in the L40 strain (Fig. 4A, lane 2), despite an expression level comparable to that of the wt Pr55Gag (data not shown). This result demonstrates that this Leu residue is essential for both Vpr binding and Vpr incorporation into virions.
Since there is an equivalent (LXX)3 repeat sequence in the p6 domain of SIVsm Gag (30), the corresponding Leu residue at position 54 was mutated to Ala to generate the GagL54A mutant (Fig. 4B). Quantitative two-hybrid analysis revealed that this mutant still interacted with Vpr and Vpx but did so less strongly than the wt precursor, giving β-gal activities about 50 and 60%, respectively, of those obtained with the wt SIVsm Gag. The importance of the (LXX)3 region for SIVsm Vpr and Vpx binding was further examined by using deletion mutants to sequentially remove the LXX repeats. Three SIVsm Gag mutants (GagΔL1, GagΔL2, and GagΔL3) were generated by removal of 5, 8, and 11 amino acids, respectively, from the C terminus of the precursor (Fig. 4B). Western blot analysis showed that these three deletion mutants and the GagL54A point mutant were expressed in yeast at levels comparable to that of the wt SIVsm Gag precursor (results not shown). The qualitative filter assay indicated that deletion of the three LXX repeats did not disrupt the binding of Vpr or Vpx to the Gag precursor (not shown). However, quantitative analysis of β-gal activities showed that the SIVsm GagΔL1, GagΔL2, and GagΔL3 mutants were severely impaired in their ability to bind Vpr and Vpx (Fig. 4B), suggesting that the (LXX)3 C-terminal region of the p6 domain participates in Vpr and Vpx binding in the context of the whole SIVsm Gag precursor. By contrast, the p6 lacking this region (p6ΔL3) interacted with Vpr and Vpx as efficiently as the wt p6 (Fig. 4B), indicating that the region upstream of the (LXX)3 motif contains the minimal sequence required for Vpr and Vpx binding. In addition, this result suggests that the (LXX)3 region has no influence on Vpr and Vpx binding in the context of the SIVsm mature p6 protein after proteolytic processing of the precursor.
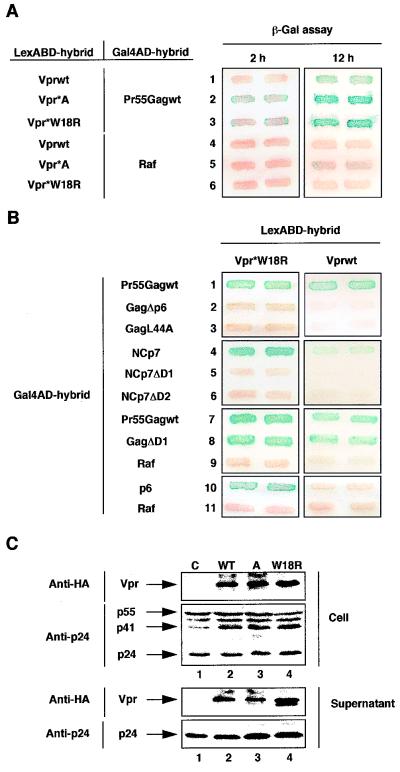
Characterization of an HIV-1 Vpr mutant displaying a strong avidity for Pr55Gag.
The two-hybrid screening of the random Vpr mutant library also revealed two mutants (Vpr*A and Vpr*W18R) that interacted more strongly with Pr55Gag than the wt Vpr protein. Interactions between Pr55Gag and these mutants were detected in the filter assay after 2 h of incubation (Fig. 5A, lanes 2 and 3, left panel), while overnight incubation was required to detect interaction with the wt protein (lane 1, right panel). Binding of Vpr*W18R to Pr55Gag was completely abolished by deletion of the p6 domain or a mutation in the fourth LXX motif of p6 (Fig. 5B, lanes 2 and 3), confirming the importance of the (LXX)4 region of the p6 domain in the binding of Vpr to Pr55Gag. While the Vpr*W18R mutant contained a single point mutation, Vpr*A contained multiple mutations, including the same W18R substitution, indicating that this mutation is sufficient to increase, at least in the two-hybrid system, the ability of HIV-1 Vpr to interact with Pr55Gag.
FIG. 5.
Characterization of an HIV-1 Vpr mutant displaying a higher avidity for Pr55Gag in the two-hybrid system. (A) Binding to Pr55Gag of Vpr*A and Vpr*W18R mutants. The L40 strain expressing either the Gal4AD-Pr55Gag (lanes 1 to 3) or the irrelevant Gal4AD-Raf (lanes 4 to 6) hybrid in combination with each of the LexABD hybrids indicated was analyzed for β-gal activity. Double transformants were patched on selective medium and then replica plated twice on Whatman filters for β-gal assay. The filter assays were incubated for 2 h (left panel) or 12 h (right panel). Vpr*A (lanes 2 and 5) and Vpr*W18R (lanes 3 and 6) mutants were selected from a random Vpr mutant library. (B) Binding of Vpr*W18R to Pr55Gag cleavage products. The L40 strain expressing either the wt Vpr (right panels) or Vpr*W18R (left panels) LexABD hybrid in combination with each of the Gal4AD hybrids indicated was analyzed for β-gal activity. The filter assays were incubated for 2 h (lanes 1 to 9, left panel) or 12 h (right panels, and lanes 10 to 11, left panel). (C) Incorporation of Vpr*A and Vpr*W18R into virions. Virion packaging was analyzed as described in the legend to Fig. 2. Lysates from transfected cells (Cell) and from virions (Supernatant) released into the culture medium and adjusted for similar amounts of CAp24 were separated by SDS-PAGE and analyzed by Western blotting with anti-HA (Cell and Supernatant, upper panels) or anti-CAp24 (Cell and Supernatant, lower panels) monoclonal antibodies. HA-tagged Vpr protein has appeared as a doublet (lane 4, Supernatant, upper panel) in some experiments (35). C (lane 1), cell, and virion lysates from cells transfected with the pCMVΔR8.91, pMD.G, and pHR′-CMVLacZ vectors and the pAS1B plasmid without an insert.
We then analyzed the binding of Vpr*W18R to the cleavage products of Pr55Gag. While the wt protein interacted weakly with NCp7 (Fig. 5B, lane 4, right panel), Vpr*W18R interacted strongly with NCp7, as indicated by the high β-gal activity detected in less than 1 h in the filter assay (lane 4, left panel). This Vpr mutant also interacted with the p6 protein, but with a lower efficiency, since β-gal activity was revealed only after overnight incubation (lane 10, left panel), suggesting that HIV-1 Vpr has a higher affinity for the mature NCp7 than for the mature p6 protein. The strong binding of Vpr*W18R to NCp7 was dependent on the zinc finger motifs in NCp7, since deletion mutants lacking the first or second zinc finger motif (NCp7ΔD1 and NCp7ΔD2) did not interact with Vpr*W18R (lanes 5 and 6, left panel). By contrast, deletion of the first motif of NCp7 within the context of the Pr55Gag precursor (GagΔD1) did not affect binding of either Vpr*W18R or wt Vpr (lane 8), indicating that the NCp7 zinc finger motifs are probably not required for direct binding of HIV-1 Vpr to the whole precursor. Last, using the same virion packaging assay described above (Fig. 5C), we determined that the Vpr*W18R and Vpr*A mutants were efficiently incorporated into virus particles. However, these two mutants did not display a higher efficiency of incorporation into virions despite their higher ability to interact with Pr55Gag (Fig. 5A). Altogether, these results confirm that the interaction of HIV-1 Vpr with Pr55Gag is mediated by the p6 domain and suggest that Vpr can then interact with the mature NCp7 protein after proteolytic processing of the precursor.
DISCUSSION
Vpr and Vpx are the only auxiliary proteins of primate lentiviruses incorporated in large amounts into virus particles budding at the surface of infected cells (7, 19, 26, 33, 44). We have used the yeast two-hybrid system to demonstrate that Vpr and Vpx proteins from distinct lineages of HIV and SIV can all interact with their homologous Gag precursor, suggesting that they are packaged into virions through direct interaction with the Gag precursor. This conclusion is supported by the ability of HIV-1 Vpr to interact directly with the Pr55Gag precursor in an in vitro binding assay. The incorporation of HIV-1 Vpr mutants is well correlated with binding to Pr55Gag. Two single-point mutants which do not interact with Pr55Gag are also not packaged into virions. Each of these mutants contains an amino acid substitution (Glu25 and His33) in the N-terminal putative alpha-helical structure, extending from residues 17 to 34 of HIV-1 Vpr (27, 28, 43). Several groups have shown that this Vpr domain must remain intact for efficient packaging into virions (10, 28, 43), and our results indicate that it is also involved in Vpr binding to Pr55Gag. Two other mutants, with substitutions in the C-terminal part of Vpr, interact with Pr55Gag and are efficiently incorporated into virions. Our data therefore strongly support the correlation between Vpr binding to Pr55Gag and packaging into virions.
Our results suggest that the p6 C-terminal domain of HIV-1 and SIVsm Gag contains the major determinants for efficient interactions with Vpr and Vpx, since deletions of this domain totally disrupt Vpr and Vpx binding in the two-hybrid assay. By contrast, deletion of the first zinc finger motif of the NCp7 region does not affect Vpr binding to HIV-1 Pr55Gag, suggesting that, in the context of the full-length precursor, the NCp7 region is not directly involved in Vpr binding. However, we cannot exclude the possibility that deletion of the p6 region could affect the global conformation of HIV-1 Gag and thus occlude the Vpr binding site on another domain of the precursor, in particular in the NCp7 region. The N-terminal MA region is also probably not involved in Vpr and Vpx binding, since SIVsm Gag lacking the MA region interacts with both proteins as well as the wt precursor.
Our findings agree well with studies indicating that the p6 C-terminal region of Gag is essential for the packaging of Vpr and Vpx into virions (23, 26, 32, 33, 42). Deletion of the p6 region of Pr55Gag abrogates Vpr incorporation (25, 33), while disruption of the NCp7 zinc finger structures has no deleterious effect on Vpr packaging (23, 33). However, recent results suggest that the NCp7 region could cooperate with the p6 domain to promote Vpr incorporation (9). Finally, the transfer of the HIV-1 p6 domain to the Gag precursors of RSV or MLV is sufficient to transfer the ability to incorporate Vpr into heterologous virus particles (21, 25), demonstrating that the Gag p6 region contains the major determinants for HIV-1 Vpr incorporation. Similarly, the determinants of the HIV-2 Gag precursor required for Vpx binding and incorporation into HIV-2 virions are exclusively located within the p6 region (32, 42), since deletion of this domain abrogates Vpx incorporation while deletion of the NC domain does not (42).
The p6 region of the Gag precursors exhibits considerable sequence variability among the different groups of primate lentiviruses. But two highly conserved motifs can be revealed within the HIV-1 p6 sequence (30). One motif lies near the N terminus of the p6 domain and is a proline-rich region required for efficient virus particle release (15) but not for Vpr incorporation into virions (20). The second region lies at the C terminus of p6 and consists of four repeats of a leucine triplet sequence (LXX). This motif is not important for virus release (15) but is directly involved in Vpr incorporation, since transfer of residues 35 to 47 of p6 encompassing the four LXX repeats to a heterologous RSV or MLV Gag led to Vpr incorporation levels comparable to those obtained with HIV-1 Pr55Gag (20, 25). Mutation of the Leu residue within the fourth LXX repeat prevented Vpr packaging into virions (20). This result is consistent with our finding that the same mutation (Pr55GagL44A) disrupts the interaction with Vpr, suggesting that the p6 (LXX)4 motif is also directly involved in the binding of HIV-1 Vpr to Pr55Gag.
There is an equivalent region containing three LXX repeats at the C terminus of the SIVsm p6 domain (30). However, this region does not seem to be critical for SIVsm Vpr and Vpx binding, since deletion of the three repeats affects but does not abolish binding to the Gag precursor. Thus, the p6 domain contains other determinants, located upstream of the C-terminal (LXX)3 region, that are involved in SIVsm Vpr and Vpx binding to Gag. Consistent with these results, the Vpx protein from HIV-2, a virus closely related to SIVsm, is efficiently incorporated into HIV-2 particles formed by the Gag precursor lacking the (LXX)3 region (32, 42). These data reveal significant differences between the distinct lineages of primate lentiviruses concerning the location of the Vpr and Vpx packaging signal within the Gag p6 domain. While the (LXX)4 region of HIV-1 Pr55Gag is critical for Vpr binding and packaging into HIV-1 virions, the equivalent (LXX)3 region of HIV-2–SIVsm Gag is not critical for binding, but could stabilize the interactions that lead to Vpr and Vpx incorporation into HIV-2 and SIVsm virions. Alternatively, the (LXX)3 region of HIV-2–SIVsm Gag could play an indirect role in maintaining the correct conformation of the p6 domain required for Vpr and Vpx binding in the context of the whole precursor. By contrast, this domain has no influence on the conformation required for Vpr and Vpx binding to the mature p6 protein after proteolytic cleavage of the precursor, since the SIVsm p6 protein with the three repeats deleted interacts with Vpr and Vpx as efficiently as the wt p6.
Analysis of Vpr and Vpx binding to the cleavage products of the HIV-1 and SIVsm Gag precursors also reveals substantial differences between these two groups of primate lentiviruses. We find a moderate interaction between the wt HIV-1 Vpr and the NCp7 protein, whereas the SIVsm Vpr and Vpx do not interact with the NCp9 protein but bind strongly to the p6 protein. Unlike some results showing a direct association of HIV-1 Vpr with the MAp17 protein in the two-hybrid system (38), we found no interaction between HIV-1 or SIVsm Vpr and Vpx and their homologous MA. Moreover, SIVsm Gag lacking the MA sequence binds to Vpr and Vpx as well as the wt precursor, suggesting that the MA domain is not required for these interactions, at least in SIVsm.
The capacity of HIV-1 Vpr to interact directly with NCp7 has been documented by in vitro binding assay using chemically synthesized polypeptides (9, 24) and is confirmed by our results obtained with the Vpr*W18R mutant. This mutant has a higher avidity than the wt Vpr for both Pr55Gag and NCp7 in the two-hybrid system and reveals the capacity of HIV-1 Vpr to interact with the p6 domain. Prediction of the amphipathic profile of the N-terminal helical structure of Vpr indicates that the Trp18 lies on the hydrophilic face of the helix (28, 43). Therefore, replacement of a Trp with a positively charged residue, such an Arg, will increase the amphipathic character of the helix. The Vpr*W18R mutant has retained the biological activities of the wt protein, since it is efficiently incorporated into virions, and this mutation does not affect the ability of the protein to induce an arrest of the cell cycle in the G2 phase (39). These findings suggest that the overall structure of the protein is probably maintained in the Vpr*W18R mutant. Interaction of Vpr with NCp7 depends on the two zinc finger motifs, since binding of Vpr*W18R or wt Vpr is abolished by deletion of these motifs. Others have reported similar results demonstrating the requirement of the NCp7 zinc finger motifs for direct association with HIV-1 Vpr (9). Conversely, deletion of the first zinc finger does not affect binding to Pr55Gag, suggesting that these motifs are not required for Vpr binding to the whole precursor. After assembly and proteolytic processing of Pr55Gag, Vpr could then switch from the p6 region of the precursor to the mature NCp7 protein. This model which assumes that the NCp7-Vpr interaction takes place after processing of Pr55Gag could account for the presence of HIV-1 Vpr in the viral preintegration complex after virus entry into the cell (13, 17), since NCp7 is an integral constituent of this complex (13). However, the results obtained with the cleavage products of the SIVsm Gag precursor indicate that neither Vpr nor Vpx has any significant affinity for the SIVsm NCp9 protein. Although interactions with mature NCp9 may take place in vivo, the binding of both Vpr and Vpx to the p6 suggests that these two proteins may use a mechanism distinct from that of HIV-1 Vpr to access the core of SIVsm virions after processing of the precursor.
The incorporation of Vpr and Vpx into HIV and SIV virions suggests that these proteins are required during the early stages of the viral life cycle. At least two biological functions have been attributed to Vpr and Vpx prior to de novo synthesis of viral proteins in newly infected cells. The best-documented function is linked to the involvement of Vpr and Vpx proteins in the import of the viral preintegration complex to the nuclei of nondividing cells, such as macrophages (4, 11, 12, 17). The second function indicates that Vpr might play another role during the first steps of the viral cycle, namely, increasing the accuracy of the reverse transcription process (29). The mutation rate of the HIV-1 reverse transcriptase is as much as fourfold higher in the absence of Vpr than in its presence. Incorporation into virions is required for this function, since introduction of a point mutation in the vpr gene, which prevents subsequent Vpr incorporation, gives rise to a mutation frequency similar to that of a vpr-defective HIV-1 (29).
In conclusion, the Vpr and Vpx proteins from primate lentiviruses are incorporated into virus particles through a direct physical interaction with the Gag precursor. Since incorporation of Vpr and Vpx into virions seems to be required for efficient virus replication in newly infected cells, the molecular interactions between viral proteins described in the present study could lead to novel antiviral strategies based on the inhibition of these interactions.
ACKNOWLEDGMENTS
We thank M. Douté, F. Letourneur, and E. Gomas for technical assistance; D. Trono, F. Barré-Sinoussi, E. Cohen, D. Lener, and B. Roques and the National Institutes of Health AIDS Research Program for the kind gifts of various reagents; P. Benaroch for critical reading of the manuscript; and O. Parkes for editing the manuscript.
L.S. and L.X.L. are fellows of the Agence Nationale de Recherche sur le SIDA and of the Fondation pour la Recherche Médicale. This work was supported by grants from the Agence Nationale de Recherche sur le SIDA and from SIDACTION.
REFERENCES
- 1.Balotta C, Lusso P, Crowley R, Gallo R C, Franchini G. Antisense phosphorothioate oligodeoxynucleotides targeted to the vpr gene inhibit human immunodeficiency virus type 1 replication in primary human macrophages. J Virol. 1993;67:4409–4414. doi: 10.1128/jvi.67.7.4409-4414.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Benichou S, Bomsel M, Bodéus M, Durand H, Douté M, Letourneur F, Camonis J, Benarous R. Physical interaction of the HIV-1 Nef protein with β-COP, a component of non-clathrin-coated vesicles essential for membrane traffic. J Biol Chem. 1994;269:30073–30076. [PubMed] [Google Scholar]
- 3.Boussif O, Lezoualc’h F, Zanta M A, Mergny M D, Scherman D, Demeneix B, Behr J P. A versatile vector for gene and oligonucleotide transfer into cells in culture and in vivo: polyethylenimine. Proc Natl Acad Sci USA. 1995;92:7297–7301. doi: 10.1073/pnas.92.16.7297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bukrinsky M I, Haggerty S, Dempsey M P, Sharova N, Adzhubel A, Spitz L, Lewis P, Goldfarb D, Emerman M, Stevenson M. A nuclear localization signal within HIV-1 matrix protein that governs infection of non-dividing cells. Nature. 1993;365:666–669. doi: 10.1038/365666a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cadwell R C, Joyce G F. Randomization of genes by PCR mutagenesis. PCR Methods Appl. 1992;2:28–33. doi: 10.1101/gr.2.1.28. [DOI] [PubMed] [Google Scholar]
- 6.Campbell B J, Hirsch V M. Vpr of simian immunodeficiency virus of African green monkeys is required for replication in macaque macrophages and lymphocytes. J Virol. 1997;71:5593–5602. doi: 10.1128/jvi.71.7.5593-5602.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cohen E A, Dehni G, Sodroski J G, Haseltine W A. Human immunodeficiency virus vpr product is a virion-associated regulatory protein. J Virol. 1990;64:3097–3099. doi: 10.1128/jvi.64.6.3097-3099.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Connor R I, Chen B K, Choe S, Landau N R. Vpr is required for efficient replication of human immunodeficiency virus type-1 in mononuclear phagocytes. Virology. 1995;206:935–944. doi: 10.1006/viro.1995.1016. [DOI] [PubMed] [Google Scholar]
- 9.De Rocquigny H, Petitjean P, Tanchou V, Decimo D, Drouot L, Delaunay T, Darlix J L, Roques B P. The zinc fingers of HIV nucleocapsid protein NCp7 direct interactions with the viral regulatory protein Vpr. J Biol Chem. 1997;272:30753–30759. doi: 10.1074/jbc.272.49.30753. [DOI] [PubMed] [Google Scholar]
- 10.Di Marzio P, Choe S, Ebright M, Knoblauch R, Landau N R. Mutational analysis of cell cycle arrest, nuclear localization, and virion packaging of human immunodeficiency virus type 1 Vpr. J Virol. 1995;69:7909–7916. doi: 10.1128/jvi.69.12.7909-7916.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fletcher T R, Brichacek B, Sharova N, Newman M A, Stivahtis G, Sharp P M, Emerman M, Hahn B H, Stevenson M. Nuclear import and cell cycle arrest functions of the HIV-1 Vpr protein are encoded by two separate genes in HIV-2/SIV(SM) EMBO J. 1996;15:6155–6165. [PMC free article] [PubMed] [Google Scholar]
- 12.Gallay P, Hope T, Chin D, Trono D. HIV-1 infection of nondividing cells through the recognition of integrase by the importin/karyopherin pathway. Proc Natl Acad Sci USA. 1997;94:9825–9830. doi: 10.1073/pnas.94.18.9825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gallay P, Swingler S, Song J, Bushman F, Trono D. HIV nuclear import is governed by the phosphotyrosine-mediated binding of matrix to the core domain of integrase. Cell. 1995;83:569–576. doi: 10.1016/0092-8674(95)90097-7. [DOI] [PubMed] [Google Scholar]
- 14.Gibbs J S, Lackner A A, Lang S M, Simon M A, Sehgal P K, Daniel M D, Desrosiers R C. Progression to AIDS in the absence of a gene for vpr or vpx. J Virol. 1995;69:2378–2383. doi: 10.1128/jvi.69.4.2378-2383.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Göttlinger H G, Dorfman T, Sodroski J G, Haseltine W A. Effect of mutations affecting the p6 gag protein on human immunodeficiency virus particle release. Proc Natl Acad Sci USA. 1991;88:3195–3199. doi: 10.1073/pnas.88.8.3195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.He J, Choe S, Walker R, Di Marzio P, Morgan D O, Landau N R. Human immunodeficiency virus type 1 viral protein R (Vpr) arrests cells in the G2 phase of the cell cycle by inhibiting p34cdc2 activity. J Virol. 1995;69:6705–6711. doi: 10.1128/jvi.69.11.6705-6711.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Heinzinger N K, Bukinsky M I, Haggerty S A, Ragland A M, Kewalramani V, Lee M A, Gendelman H E, Ratner L, Stevenson M, Emerman M. The Vpr protein of human immunodeficiency virus type 1 influences nuclear localization of viral nucleic acids in nondividing host cells. Proc Natl Acad Sci USA. 1994;91:7311–7315. doi: 10.1073/pnas.91.15.7311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jowett J B M, Planelles V, Poon B, Shah N P, Chen M-L, Chen I S Y. The human immunodeficiency virus type 1 vpr gene arrests infected T cells in the G2 + M phase of the cell cycle. J Virol. 1995;69:6304–6313. doi: 10.1128/jvi.69.10.6304-6313.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kappes J C, Parkin J S, Conway J A, Kim J, Brouillette C G, Shaw G M, Hahn B H. Intracellular transport and virion incorporation of vpx requires interaction with other virus type-specific components. Virology. 1993;193:222–233. doi: 10.1006/viro.1993.1118. [DOI] [PubMed] [Google Scholar]
- 20.Kondo E, Göttlinger H G. A conserved LXXLF sequence is the major determinant in p6gag required for the incorporation of human immunodeficiency virus type 1 Vpr. J Virol. 1996;70:159–164. doi: 10.1128/jvi.70.1.159-164.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kondo E, Mammano F, Cohen E A, Göttlinger H G. The p6gag domain of human immunodeficiency virus type 1 is sufficient for the incorporation of Vpr into heterologous viral particles. J Virol. 1995;69:2759–2764. doi: 10.1128/jvi.69.5.2759-2764.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kunkel T A, Roberts J D, Zakour R A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Methods Enzymol. 1987;154:367–382. doi: 10.1016/0076-6879(87)54085-x. [DOI] [PubMed] [Google Scholar]
- 23.Lavallée C, Yao X J, Ladha A, Göttlinger H, Haseltine W A, Cohen E A. Requirement of the Pr55gag precursor for incorporation of the Vpr product into human immunodeficiency virus type 1 viral particles. J Virol. 1994;68:1926–1934. doi: 10.1128/jvi.68.3.1926-1934.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li M S, Garcia A G, Bhattacharyya U, Mascagni P, Austen B M, Roberts M M. The Vpr protein of human immunodeficiency virus type 1 binds to nucleocapsid protein p7 in vitro. Biochem Biophys Res Commun. 1996;218:352–355. doi: 10.1006/bbrc.1996.0061. [DOI] [PubMed] [Google Scholar]
- 25.Lu Y-L, Bennett R P, Wills J W, Gorelick R, Ratner L. A leucine triplet repeat sequence (LXX)4 in p6gag is important for Vpr incorporation into human immunodeficiency virus type 1 particles. J Virol. 1995;69:6873–6879. doi: 10.1128/jvi.69.11.6873-6879.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lu Y-L, Spearman P, Ratner L. Human immunodeficiency virus type 1 viral protein R localization in infected cells and virions. J Virol. 1993;67:6542–6550. doi: 10.1128/jvi.67.11.6542-6550.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mahalingam S, Khan S A, Jabbar M A, Monken C E, Collman R G, Srinivasan A. Identification of residues in the N-terminal acidic domain of HIV-1 Vpr essential for virion incorporation. Virology. 1995;207:297–302. doi: 10.1006/viro.1995.1081. [DOI] [PubMed] [Google Scholar]
- 28.Mahalingam S, Khan S A, Murali R, Jabbar M A, Monken C E, Collman R G, Srinivasan A. Mutagenesis of the putative alpha-helical domain of the Vpr protein of human immunodeficiency virus type 1: effect on stability and virion incorporation. Proc Natl Acad Sci USA. 1995;92:3794–3798. doi: 10.1073/pnas.92.9.3794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mansky L M. The mutation rate of human immunodeficiency virus type 1 is influenced by the vpr gene. Virology. 1996;222:391–400. doi: 10.1006/viro.1996.0436. [DOI] [PubMed] [Google Scholar]
- 30.Myers G, Korber B, Berzofsky J A, Smith R F, Pavlakis G N. Human retroviruses and AIDS: a compilation and analysis of nucleic acid and amino acid sequences. Los Alamos, N.Mex: Los Alamos National Laboratory; 1992. [Google Scholar]
- 31.Ottmann M, Gabus C, Darlix J-L. The central globular domain of the nucleocapsid protein of human immunodeficiency virus type 1 is critical for virion structure and infectivity. J Virol. 1995;69:1778–1784. doi: 10.1128/jvi.69.3.1778-1784.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pancio H A, Ratner L. Human immunodeficiency virus type 2 Vpx-Gag interaction. J Virol. 1998;72:5271–5275. doi: 10.1128/jvi.72.6.5271-5275.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Paxton W, Connor R I, Landau N R. Incorporation of Vpr into human immunodeficiency virus type 1 virions: requirement for the p6 region of gag and mutational analysis. J Virol. 1993;67:7229–7237. doi: 10.1128/jvi.67.12.7229-7237.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Planelles V, Jowett J B, Li Q X, Xie Y, Hahn B, Chen I S. Vpr-induced cell cycle arrest is conserved among primate lentiviruses. J Virol. 1996;70:2516–2524. doi: 10.1128/jvi.70.4.2516-2524.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Poon B, Grovit-Ferbas K, Stewart S A, Chen I S Y. Cell cycle arrest by Vpr in HIV-1 virions and insensitivity to antiretroviral agents. Science. 1998;281:266–269. doi: 10.1126/science.281.5374.266. [DOI] [PubMed] [Google Scholar]
- 36.Re F, Braaten D, Franke E K, Luban J. Human immunodeficiency virus type 1 Vpr arrests the cell cycle in G2 by inhibiting the activation of p34cdc2-cyclin B. J Virol. 1995;69:6859–6864. doi: 10.1128/jvi.69.11.6859-6864.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rogel M E, Wu L I, Emerman M. The human immunodeficiency virus type 1 vpr gene prevents cell proliferation during chronic infection. J Virol. 1995;69:882–888. doi: 10.1128/jvi.69.2.882-888.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sato A, Yoshimoto J, Isaka Y, Miki S, Suyama A, Adachi A, Hayami M, Fujiwara T, Yoshie O. Evidence for direct association of Vpr and matrix protein p17 within the HIV-1 virion. Virology. 1996;220:208–212. doi: 10.1006/viro.1996.0302. [DOI] [PubMed] [Google Scholar]
- 39.Selig L, Benichou S, Rogel M E, Wu L I, Vodicka M A, Sire J, Benarous R, Emerman M. Uracil DNA glycosylase specifically interacts with Vpr of both human immunodeficiency virus type 1 and simian immunodeficiency virus of sooty mangabeys, but binding does not correlate with cell cycle arrest. J Virol. 1997;71:4842–4846. doi: 10.1128/jvi.71.6.4842-4846.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Steimer K S, Puma J P, Power M D, Powers M A, George N C, Stephans J C, Levy J A, Sanchez P R, Luciw P A, Barr P J, et al. Differential antibody responses of individuals infected with AIDS-associated retroviruses surveyed using the viral core antigen p25gag expressed in bacteria. Virology. 1986;150:283–290. doi: 10.1016/0042-6822(86)90289-8. [DOI] [PubMed] [Google Scholar]
- 41.Trono D. HIV accessory proteins: leading roles for the supporting cast. Cell. 1995;82:189–192. doi: 10.1016/0092-8674(95)90306-2. [DOI] [PubMed] [Google Scholar]
- 42.Wu X, Conway J A, Kim J, Kappes J C. Localization of the Vpx packaging signal within the C terminus of the human immunodeficiency virus type 2 Gag precursor protein. J Virol. 1994;68:6161–6169. doi: 10.1128/jvi.68.10.6161-6169.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yao X J, Subbramanian R A, Rougeau N, Boisvert F, Bergeron D, Cohen E A. Mutagenic analysis of human immunodeficiency virus type 1 Vpr: role of a predicted N-terminal alpha-helical structure in Vpr nuclear localization and virion incorporation. J Virol. 1995;69:7032–7044. doi: 10.1128/jvi.69.11.7032-7044.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yu X F, Ito S, Essex M, Lee T H. A naturally immunogenic virion-associated protein specific for HIV-2 and SIV. Nature. 1988;335:262–265. doi: 10.1038/335262a0. [DOI] [PubMed] [Google Scholar]
- 45.Zufferey R, Nagy D, Mandel R J, Naldini L, Trono D. Multiply attenuated lentiviral vector achieves efficient gene delivery in vivo. Nat Biotechnol. 1997;15:871–875. doi: 10.1038/nbt0997-871. [DOI] [PubMed] [Google Scholar]