Abstract
The long PCR technique was used to amplify the three size classes of viral mRNAs produced in cells infected by feline immunodeficiency virus (FIV). We identified in the env region a new splice acceptor site that generated two transcripts, each coding for an 11-kDa protein, p11rev, whose function is unknown. The small-size class of mRNAs included two bicistronic orf2/rev mRNAs and two rev-like mRNAs, consisting only of the second exon of rev and coding for a 15-kDa protein, p15rev. p15rev contained the minimal effector domain of Rev and was sufficient to mediate partial Rev activity. The bicistronic mRNAs encoded two distinct proteins, one of 23 kDa corresponding to Rev and a 9-kDa protein encoded by the orf2 gene. The orf2 gene product is a protein of 79 amino acids with characteristics similar to those of the Tat (transactivator) proteins of the ungulate lentiviruses. Transient expression assays, using the FIV long terminal repeat (LTR) to drive transcription of the bacterial gene for chloramphenicol acetyltransferase demonstrated that the orf2 gene transactivates gene expression an average of 14- to 20-fold above the basal level. Deletion mutants of the FIV LTR were generated to locate sequences responsive to transactivation mediated by the orf2 gene. A 5′ deletion mutant that removed the AP1 site resulted in residual low-level transactivation by orf2. Further experiments using LTR mutants with internal deletions identified three regions located between positions −126 and −47 relative to the cap site that were important for orf2-directed transactivation. These regions include the AP1 site, a C/EBP tandem repeat, and an ATF site.
Feline immunodeficiency virus (FIV) (25) is a lentivirus associated with a slow progressive disease in the domestic cat involving multiple organ systems (for a review, see reference 3). FIV has a disease pattern similar to that of human immunodeficiency virus (HIV), but its genomic organization is less complex and is more closely related to those of the ungulate lentiviruses visna virus and caprine arthritis encephalitis virus (CAEV) (6). Like with other lentiviruses, three size classes of viral mRNAs are produced in FIV-infected cells: full-length (9.2-kb), intermediate (5.2- and 4.4-kb), and small multiply spliced (1.7- and 1.4-kb) mRNAs which represent the gag/pol, vif/env, env, orf2/rev, and rev transcripts, respectively (27, 37). The multiply spliced mRNAs are produced shortly after infection and encode Rev and the orf2 gene product, which are essential for the viral life cycle. Rev functions through a specific cis-acting RNA target, the Rev-responsive element (RRE), and allows the nuclear export and cytoplasmic accumulation of unspliced (gag/pol) and singly spliced (vif/env and env) viral mRNAs. FIV Rev is a 23-kDa protein whose 80 N-terminal amino acids are encoded by the N terminus of env joined in frame to 73 residues from orfH, located at the 3′ end of the genome (27). Although FIV Rev does not share a leucine-rich effector domain found in HIV type 1 (HIV-1) Rev and the Rev-like proteins of many other retroviruses, mutational analyses showed that a short region in the second exon of FIV Rev was functionally interchangeable with the HIV-1 Rev effector domain (19). The effector domain of FIV Rev spanned a region of 27 amino acids downstream from a stretch of basic residues in the second exon of Rev. A short open reading frame (ORF), orf2, coincides by its size and location to the tat genes of visna virus and CAEV (7, 10, 14). The orf2 gene product has been shown to produce a three- to fivefold transactivation of the FIV long terminal repeat (LTR) (33, 35, 38). Whether this low-level transactivation is significant remains to be defined. Nonetheless, the orf2 gene product is an essential component for the virus life cycle. orf2-defective FIV failed to productively infect feline T cells and primary peripheral blood lymphocytes (PBLs) (36). Furthermore, the FIV 34TF10 infectious molecular clone, which contains a stop codon in the orf2 gene, replicated poorly in T cells (26). Substitution of the stop codon for a tryptophan codon allowed the efficient replication of the virus in T cells and primary PBLs (38), suggesting that an intact orf2 gene influences the host cell tropism of FIV.
Here, we report the amplification in a single PCR of the three different size classes of viral mRNAs produced in FIV-infected cells by using the long PCR technology. We have identified a new splice acceptor site located in the env region that generates two mRNAs producing a Rev-related protein, p11rev. In addition, we have characterized another Rev-related protein, p15rev, consisting only of the second exon of Rev. The latter protein displayed some degree of Rev activity, although less than the wild-type protein. Finally, using improved transfection protocols, we demonstrate that the orf2 gene encodes a potent transactivator of the FIV LTR. Deletional analysis of the FIV LTR demonstrated that a region located between positions −126 and −47, relative to the cap site, contains AP1, a C/EBP tandem repeat, and ATF sites that are important for the orf2 gene-mediated transactivation of the FIV LTR.
MATERIALS AND METHODS
Cells and virus.
Primary PBLs were obtained by Ficoll-Paque gradient purification from blood of a specific-pathogen-free cat. PBLs were maintained in RPMI 1640 medium supplemented with 15% heat-inactivated fetal bovine serum (Gemini Bioproducts, Calabasa, Calif.), 2 mM l-glutamine (Sigma, St. Louis, Mo.), 1 mM sodium pyruvate (Sigma), 10 mM HEPES buffer (Sigma), 1× nonessential amino acids (Sigma), 1× β-mercaptoethanol (Gibco BRL, Gaithersburg, Md.), 7.5 μg of concanavalin A (Sigma) per ml, 100 U of human recombinant interleukin-2 (a gift of Hoffmann-La Roche) per ml, and 50 μg of gentamicin (Gemini Bioproducts) per ml. Crandell feline kidney (CrFK) and HeLa cells, obtained from the American Type Culture Collection, were maintained in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal bovine serum. The San Diego isolate of FIV, PPR (26), was used for the analysis of the transcriptional pattern of FIV.
Synthetic oligonucleotide primers.
LA4 (5′-ACTTGAGAAGAGTGATTGAGGAAGTGAAGC-3′; nucleotides 373 to 403 [sense]), LA7 (5′-TAAGCAGCTGCTAGCGCTTTAACTATGAGTCATGTTCAGC-3′; nucleotides 9237 to 9198 [antisense]), and LA11 (5′-CAAAATGGATTCATATGACATATCTTCCTC-3′; nucleotides 8914 to 8885 [antisense]) were designed from the sequence of the PPR molecular clone (GenBank accession no. 36968 [26]).
RNA extraction and cDNA synthesis.
Total RNA was extracted from PBLs, 104-C1 cells, and MCH5-4 cells by using an RNeasy kit (Qiagen, Chatsworth, Calif.) as specified by the manufacturer. Briefly, total RNA (0.5 to 1 μg) was heated at 70°C for 5 min and cooled on ice. First-strand cDNA synthesis was carried out in a total volume of 50 μl containing 50 mM Tris-HCl (pH 8.3), 75 mM KCl, 3 mM MgCl2, 10 mM dithiothreitol, 50 mM deoxynucleoside triphosphates, 10 U of RNase inhibitor (Promega, Madison, Wis.) 50 U of Moloney murine leukemia virus reverse transcriptase (Stratagene, La Jolla, Calif.), and 0.3 μg of oligo(dT)15 primer (Promega). The reaction mixture was then incubated at 37°C for 2 h, and 1 to 5 μl was used for subsequent amplifications.
PCR amplification.
One to 5 μl of the cDNA reaction mixture was amplified by long PCR (2). Amplification was carried out in a total volume of 100 μl containing 20 mM Tris-HCl (pH 8.55 at 25°C), 16 mM (NH4)2SO4, 150 μg of bovine serum albumin per ml, 3.5 mM MgCl2, 250 μM each deoxynucleoside triphosphate, 100 mM Tris base, and 700 ng of each primer. The PCR mixture was overlaid with 75 μl of mineral oil (Promega); after 1 min of incubation at 94°C, 5 U of Taq (Promega) and 1:64 U of Deep Vent (New England Biolabs, Beverly, Mass.) polymerases were added, and the mixture was incubated at 94 and 68°C for 10 s and 7 min, respectively. The cycle was repeated 25 to 35 times in a Perkin-Elmer Cetus thermocycler.
Cloning and sequencing of amplified cDNAs.
The PCR-amplified cDNAs were cleaned up (Promega), gel purified, and cloned in the TA cloning vector (pCR3; Invitrogen, La Jolla, Calif.) under the control of the cytomegalovirus and T7 promoters. Sequences were determined by the dideoxy-chain termination procedure (30), using a Sequenase version 2.0 kit (United States Biochemical, Cleveland, Ohio).
In vitro transcription-translation and immunoprecipitations.
The orf2/rev and rev-like cDNAs cloned in pCR3 were transcribed and translated in vitro by using T7 polymerase and the coupled transcription-translation reticulocyte lysate system (Promega).
Construction of 5′ deletion and internal deletion mutants.
The 5′ deletion mutants have been previously described (35). Internal deletion mutations of the FIV LTR were generated by PCR-ligation-PCR (1). Briefly, two fragments of the FIV LTR, one corresponding to the 5′ end of U3 to the nucleotide immediately upstream of the site to be deleted and the second corresponding to the nucleotide immediately downstream of the site to be deleted to the 3′ end of U5, were amplified by using Deep-Vent DNA polymerase (New England Biolabs). The PCR products were gel purified, phosphorylated, and then ligated. The fusion gene was next amplified by using a primer pair specific for the 5′ end of U3 and the 3′ end of U5. The PCR product was then inserted in pFIVLTR-CAT deleted of the wild-type LTR insert. We generated five single internal deletion mutants corresponding to the first AP4 site, the AP1 site, the C/EBP tandem repeat, the NF1 site, and the ATF site.
Transfections and CAT assays.
DNAs were prepared by using Qiagen midi- and maxiprep kits or by the Merlin service offered by Bio101 (Vista, Calif.). The LTR-chloramphenicol acetyltransferase (CAT) and RRE-CAT constructs used in this study have been previously reported (27, 35). Constructs (1 μg of target, 5 or 10 μg of effector plasmids) were transfected in triplicate in CrFK cells by using a calcium phosphate precipitation procedure as previously described (38). For HeLa cells, 1 and 10 μg of LTR-CAT construct were used. Following transfection, cells were incubated for 40 h and lysed, and CAT activity was assayed on 20 μg of cell extract by phase extraction according to the protocol of Seed and Sheen (31). In our previous study (38), CAT activity was assayed on cell extracts that were normalized by a β-galactosidase assay. However, the orf2 gene has been shown to transactivate the Rous sarcoma virus promoter of our β-galactosidase construct (pRSV-βGal), which causes an error in normalizing cell extracts in the presence of orf2 cotransfection (39). Therefore, in the present study, CAT activity was assayed by using 20 μg of protein per cell extract.
RESULTS
Long PCR amplification of the different size classes of FIV transcripts.
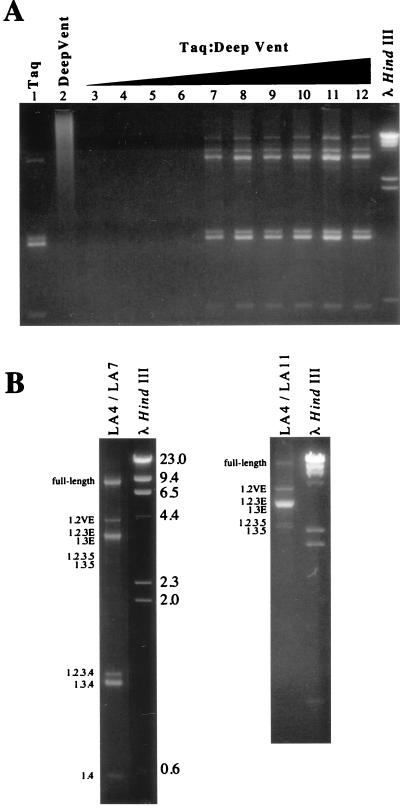
Long PCR has been recently described and shown to be an efficient method to amplify long DNA targets with high fidelity (2). Long PCR combines modifications of standard PCR buffers and thermal cycling profiles with a combination of two polymerases, providing both processivity and 3′-5′ proofreading exonuclease activity. Here, we used long PCR to investigate the gene expression of FIV. PBLs from a specific-pathogen-free cat were infected with FIV-PPR (26). At 15 days postinfection, cells were harvested and total RNA was extracted. One microgram of total RNA was reverse transcribed by using an RNase H-minus reverse transcriptase and oligo(dT)15 primer; 1 μl of the cDNA mixture was subsequently amplified with primers LA4 and LA7, located in exons 1 and 4 respectively, which are present in all viral transcripts (26, 37) (Fig. 1). We used the PCR buffer conditions described by Barnes (2) but a different combination of polymerases: the Taq DNA polymerase from Thermus aquaticus as a processive polymerase, and the Deep-Vent DNA polymerase from Thermoccocus litoralis as a proofreading polymerase. Long PCR amplification was carried out with Taq polymerase alone and in combination with various amounts of Deep-Vent polymerase. The use of Taq alone resulted in the amplification of four bands: 3 of 1.6, 1.5, and 0.5 kb, previously reported to correspond to exons 1.2.3.4, exons 1.3.4, and exons 1.4 (27, 37); and one band of about 3.5 kb (Fig. 2A, lane 1). No detectable amplicon was observed when Deep-Vent alone was used (Fig. 2A, lane 2). Only the 1.6-, 1.5-, and 0.5-kb products were observed when amplification with Taq or Deep-Vent was carried out under classic PCR buffer conditions (data not shown). Since long PCR amplification required a very low level of polymerase with 3′ exonuclease activity (2), we optimized the PCR conditions by combining 5 U of Taq with decreasing amounts of Deep-Vent, from 1/2 to 1/1,024 U. Long PCR using 1/2 to 1/16 U of Deep-Vent failed to amplify any detectable product (Fig. 2A, lanes 3 to 6). However, long PCR carried out with enzyme combinations using 1/32 to 1/1,024 U of Deep-Vent resulted in the amplication of a complex pattern of bands (Fig. 2A, lanes 7 to 12). These results are consistent with those previously reported (2). Three other major products of 9.4, 4.0, and 3.5 kb were observed under these conditions (Fig. 2A, lanes 7 to 12; Fig. 2B, left panel). Long PCR amplification with cDNAs from two FIV-infected T-cell lines and FIV-infected CrFK cells resulted in the same pattern of amplified products (data not shown). To verify that these three new amplified products represented the unspliced full-length mRNA and the singly spliced env mRNAs, we performed a long PCR with primers LA4 and LA11 (Fig. 1). LA11 is located 5′ of the splice acceptor at nucleotide 8944 of exon 4 (Fig. 1). This splice acceptor is used by all of the multiply spliced mRNAs to join exons 1, 2, and 3 to exon 4 (27, 37). Therefore, only unspliced or partially spliced mRNAs could be amplified with this primer set. Indeed, we observed three major products corresponding to the full-length mRNA and singly spliced mRNAs; a minor product, a doublet at 2.4 kb, was also amplified by this primer set (Fig. 2B, right panel).
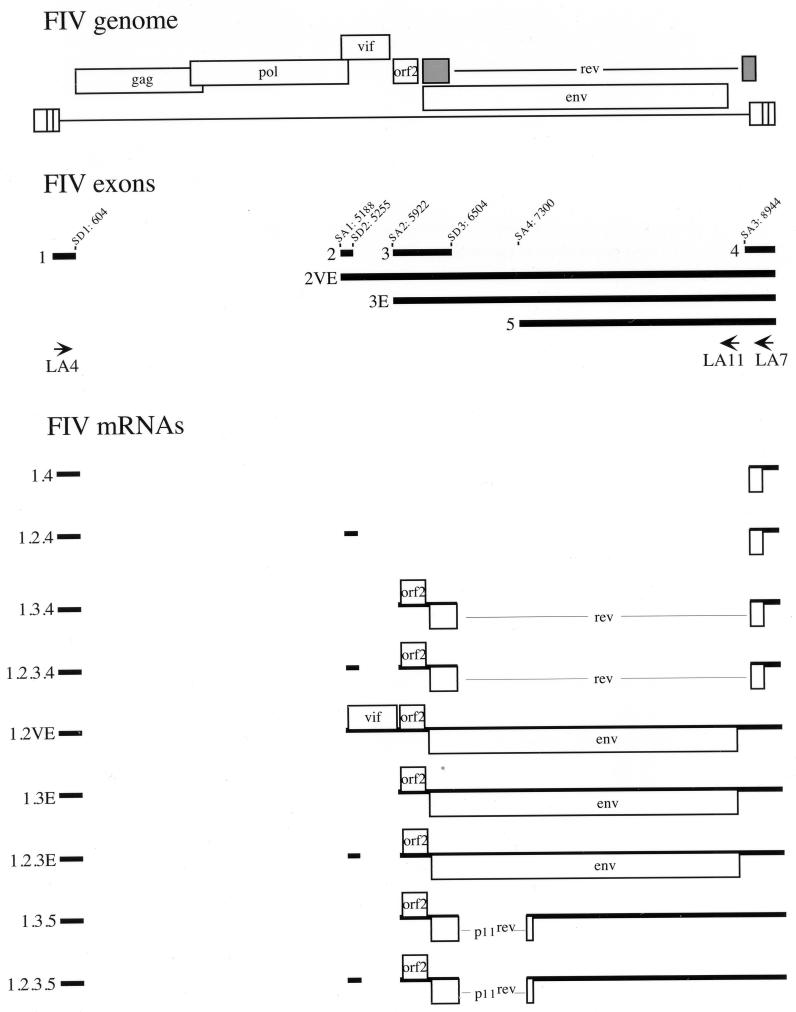
FIG. 1.
Transcriptional map of FIV. A schematic representation of the FIV genome is shown at the top. The exon structure and locations of splice donor (SD) and splice acceptor (SA) sites were determined by sequencing the PCR-amplified cDNAs. The arrows represent locations and orientations of the oligonucleotide primers used in this study. The viral transcripts are deduced from the sequence of the cDNA clones. Each transcript is designated by its exon composition. Transcripts 1.3.5 and 1.2.3.5 are newly described in this study.
FIG. 2.
Long PCR amplification of the different size classes of FIV transcripts. PCR products were resolved by electrophoresis on 1.3% agarose gel and stained with ethidium bromide. The sizes of the HindIII-cut λ DNA markers (outside lanes) are indicated in kilobases. (A) Optimization using different enzyme ratios. Taq and Deep-Vent polymerases were used alone (lanes 1 and 2) or in combination (lanes 3 to 12), where 5 U of Taq was mixed with serial twofold dilutions of Deep-Vent, starting with 0.5 U of Deep-Vent. (B) PCR amplification with the primer pairs LA4-LA7 (left panel) and LA4-LA11 (right panel). Numbers on the left indicate the exon composition of each PCR product, deduced from the sequence of the cDNA clones.
To exclude artifacts due to DNA contamination or to the use of a long annealing or extension time, the same PCRs with the primer sets LA4-LA7 and LA4-LA11 were performed on DNase-treated total RNA and genomic DNA extracted from FIV-infected cells. No detectable amplicon was observed after amplification on total RNA, and a single 9-kb band corresponding to the full-length proviral DNA was amplified when genomic DNA was used as a template (data not shown).
To verify that the amplified products represented FIV spliced transcripts, the PCR-amplified cDNAs were gel purified, cloned in the TA vector, and sequenced as described in Materials and Methods. A schematic representation of the different FIV exons and transcripts is depicted in Fig. 1. In addition of the spliced mRNAs previously reported for FIV (26, 27, 37), we identified two new transcripts. The 2.4-kb doublet observed when cDNAs were amplified with the primer pair LA4-LA11 consisted of two mRNAs generated by splicing of exon 1 directly, or via exon 2, to exon 3 and to a new exon, exon 5 (Fig. 1). These two mRNAs were named 1.3.5 and 1.2.3.5 (Fig. 1). The common splice donor site of exon 3 joined a new splice acceptor site at nucleotide 7300 of exon 5. Thus, in addition to the orf2 gene, a new ORF consisting of the first exon of rev spliced to a second exon of 19 amino acids was also present (Fig. 1).
Analysis of the coding potential of the orf2/rev and rev-like transcripts.
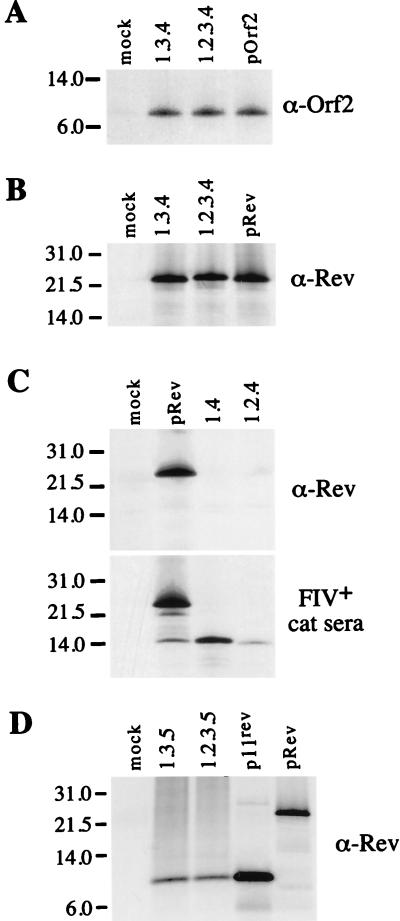
The coding potential of the different cDNAs was investigated in vitro by transcription-translation using a rabbit reticulocyte lysate system. The bicistronic orf2/rev transcripts (1.3.4 and 1.2.3.4) encoded two proteins of 9 and 23 kDa which were specifically recognized by anti-orf2 and anti-Rev rabbit sera, respectively (Fig. 3A and B). Two other small transcripts, 1.4 and 1.2.4, which contain only the second exon of rev, were analyzed in vitro for their coding potential. A potential initiator AUG is present at the beginning of the second exon of rev. Therefore, transcripts 1.4 and 1.2.4 might encode a protein corresponding to the second exon only. The anti-Rev rabbit serum failed to immunoprecipitate any product from the translation reaction with cDNAs 1.4. and 1.2.4 (Fig. 3C, top panel), while an FIV-positive cat serum known to react with Rev immunoprecipitated a product of 15 kDa (Fig. 3C, bottom panel). These results were not surprising, since the anti-Rev peptide serum was raised against an oligopeptide from the first exon of Rev (27). The 15-kDa product, which we refer to here as p15rev, was also immunoprecipitated in the pRev translation reaction (Fig. 3C, bottom panel), suggesting that a degree of internal initiation occurred from the AUG located in the second exon of rev. The coding potential of transcripts 1.3.5 and 1.2.3.5 was also investigated for the ability to direct the synthesis of the new ORF consisting of the first exon of rev joined in frame to 19 amino acid residues encoded by a 3′ ORF located in the env region. In vitro translation reactions performed with cDNAs 1.3.5 and 1.2.3.5 were immunoprecipitated with the anti-Rev peptide serum, and an 11-kDa product, termed p11rev, was detected (Fig. 3D). A product of similar size was also immunoprecipitated when the translation reaction was carried out with a cDNA clone encoding only the p11rev ORF (Fig. 3D).
FIG. 3.
Coding potential of the orf2/rev and rev-like cDNAs. The cDNA clones of the orf2/rev and rev-like transcripts were analyzed by an in vitro coupled transcription-translation system (Promega). 35S-labeled proteins were immunoprecipitated by an anti-orf2 rabbit serum (A), an anti-Rev rabbit serum (B to D), and an FIV-positive cat serum (C). Immune complexes were resolved by electrophoresis on a sodium dodecyl sulfate–10 to 20% polyacrylamide gel. Sizes of the protein molecular weight markers are indicated in kilodaltons on the left. The cDNA clones used as templates in the translation reactions are indicated above the lanes.
Analysis of the Rev activity of p11rev and p15rev.
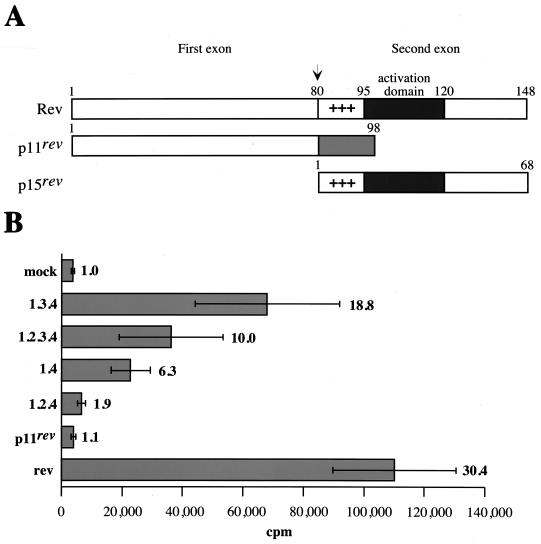
A schematic representation of Rev, p11rev, and p15rev is shown in Fig. 4A. p11rev contains the first exon of Rev, while p15rev contains the second exon of Rev. The effector domain of FIV Rev has been mapped to a region between amino acids 95 and 120, located immediately downstream of the basic domain of Rev (19). This region, which is unrelated to other retrovirus Rev effector domains, has been shown to be functionally interchangeable with the HIV-1 Rev effector domain (19). To determine whether p11rev and p15rev proteins also regulate virus protein expression, we used a reporter plasmid that contained the FIV RRE downstream of the CAT gene (27). The CAT gene and FIV RRE are flanked by splice donor and acceptor sites. In the absence of Rev, the CAT gene will be removed by splicing. Therefore, the functionality of Rev and Rev-like proteins can be assessed by the level of CAT activity in cells cotransfected with pCAT-RRE and a rev or rev-like construct. The different cDNAs encoding Rev and Rev-like proteins were introduced downstream of the cytomegalovirus promoter in the pCR3 vector. CrFK cells cotransfected with pCAT-RRE and pCR3 served as a mock control (Fig. 4B). As a positive control, pRev, which contains only the rev ORF, was used. Cotransfection of pRev increased the CAT activity 30-fold over the mock transfection control (Fig. 4B). Cotransfection with cDNAs 1.3.4 and 1.2.3.4 increased CAT activity 19- and 10-fold, respectively (Fig. 4B). These results demonstrate that cDNAs 1.3.4 and 1.2.3.4 encode a functional Rev protein. Six- and twofold increases in CAT activity were observed with cDNAs 1.4 and 1.2.4, respectively, which encode p15rev, while no detectable Rev activity was observed for p11rev (Fig. 4B). These results suggest that cDNAs 1.4 and 1.2.4 encode a Rev-like protein with partial Rev activity. The overall results are in agreement with the presence of an effector domain of Rev within the second exon (19) but also suggest that the first exon is necessary for full Rev activity.
FIG. 4.
Rev activity of the orf2/rev and rev-like transcripts. (A) Schematic structure of Rev, p11rev, and p15rev proteins. (B) Each rev and rev-like cDNA clone was cotransfected with a CAT-RRE vector in CrFK cells. Transfected cell lysates were assayed for CAT activity as described in Materials and Methods. Relative CAT activity is the mean value of two independent experiments performed in triplicate; standard deviations are shown as error bars. The relative increase of CAT activity compared to the mock control for each construct is shown on the right.
The orf2 gene encodes a transcriptional transactivator.
Since previous studies on FIV transactivation yielded conflicting results with regard to the ability of the orf2 gene product to mediate transactivation of the FIV LTR (22, 33, 35, 38), we wished to confirm that orf2 encodes the transactivating protein (Tat) of FIV. The orf2 gene coincides by its size and location to the tat (orfS) gene of visna virus (7, 10). Ungulate and feline ORFs encode 9- to 11-kDa products with domain similarities (4). In particular, acidic/hydrophobic, leucine-rich, and cysteine-rich domains are present in both products (Fig. 5A). To determine if orf2 encodes a transcriptional transactivator of the FIV LTR, we analyzed the effects of cDNAs 1.3.4 and 1.2.3.4 on expression of the CAT gene driven by the FIV LTR (pFIVLTR-CAT) in CrFK cells. Mock-cotransfected CrFK cells served as a background control for the assay. Cotransfection with cDNAs 1.3.4 and 1.2.3.4 resulted in average 6.5- and 5-fold increases in transactivation of the FIV LTR, respectively (Fig. 5B). Furthermore, transactivation of the FIV LTR in the presence of a vector expressing the orf2 gene of FIV-PPR was 17-fold above the basal level, while in the presence of a Rev-expressing vector, no increase in CAT activity was observed (Fig. 5B). These results demonstrate that the orf2 gene encodes a trans-acting factor that significantly increases gene expression directed by the FIV LTR.
FIG. 5.
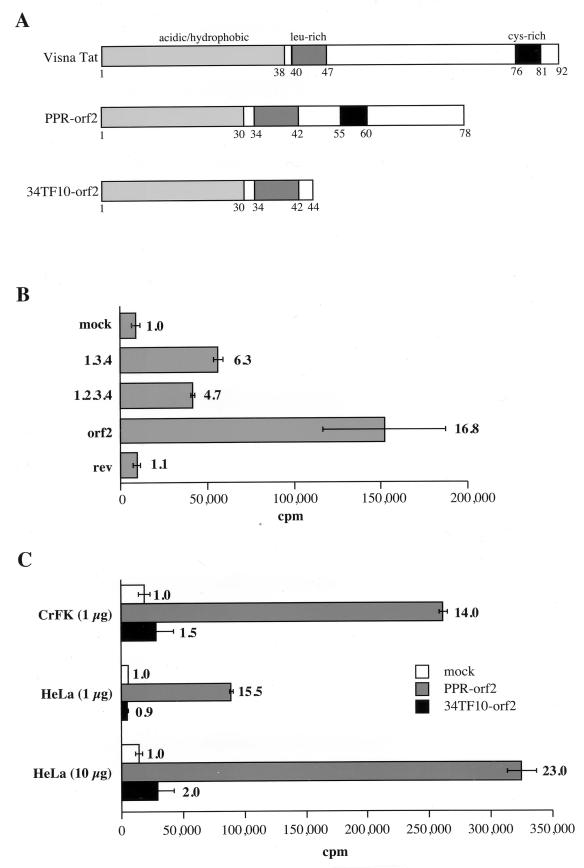
The orf2 gene encodes a transactivator of the FIV LTR. (A) Schematic organizations of the visna virus Tat protein and of the FIV-PPR and FIV-34TF10 orf2 gene products. (B) Each cDNA clone indicated on the left was cotransfected with an FIV LTR-CAT vector in CrFK cells. (C) Comparison of transactivation of the FIV LTR mediated by the PPR and 34TF10 orf2 genes in CrFK and HeLa cells. The amount of pFIV LTR-CAT vector is indicated in parentheses. The CAT activity of the transfected cells was assayed as described in Materials and Methods. The experiment were repeated at least twice, in triplicate; standard deviations are shown as error bars. The relative increase of CAT activity compared to the mock control for each construct is shown on the right.
We next investigated the ability of the 34TF10 orf2 gene to transactivate the FIV LTR. FIV-PPR contains an intact orf2, while FIV-34TF10 contains a truncated orf2 due to a termination codon at amino acid 44 (26, 34) (Fig. 5A). Cotransfection of PPR orf2 and 34TF10 orf2 resulted in average 14- and 1.5-fold increases in transactivation of the FIV LTR over the mock cotransfected control, respectively (Fig. 5C). The same experiment was also performed with HeLa cells. Since the basal activity of the FIV LTR is very low in HeLa cells compared to CrFK cells, the cotransfections were performed with either 1 or 10 μg of pFIVLTR-CAT. As shown in Fig. 5C, relative degrees of transactivation in HeLa cells were similar to those seen in CrFK cells (Fig. 5C). These results demonstrate that (i) the orf2 gene encodes the FIV Tat protein and (ii) an intact orf2 gene is necessary for transactivation of the FIV LTR.
Deletional analysis of LTR sequences required for transactivation.
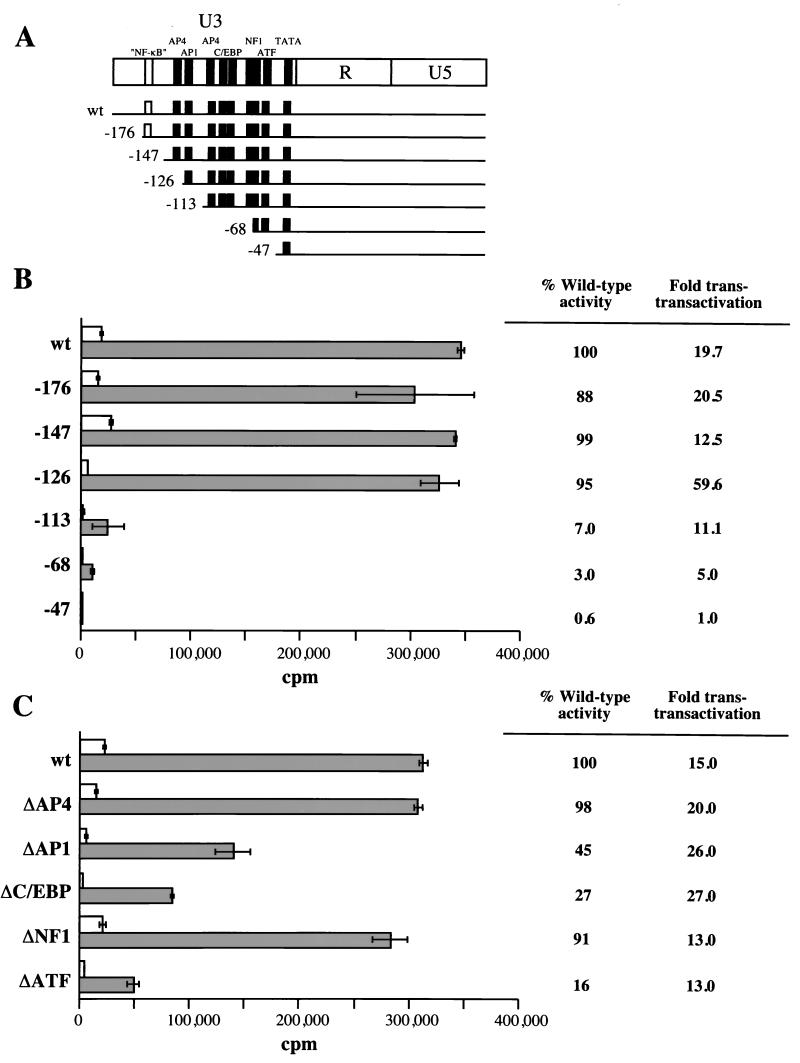
Several consensus sequences for known upstream enhancer-promoter elements, including AP4, AP1, C/EBP, NF1, and ATF, are present in the U3 region of the FIV LTR (21, 24, 26, 34). To map the LTR sequences required for transactivation, we used a panel of mutants with progressive 5′ deletions of the FIV-UK8 LTR (35). The full-length and deletion mutant LTR-CAT constructs were transfected in CrFK cells and tested for transactivation by cotransfection with the PPR orf2 plasmid (Fig. 6B). Mock cotransfections served to define the basal level activity for each construct. The mutants deleted in U3 between positions −176 and −126 responded to transactivation as efficiently as the wild-type LTR. However, deletion extending to positions −113, −68, and −47 led to a decrease both in basal promoter activity and in the magnitude of transactivation. The dramatic decrease in transactivation of the mutant deleted to position −113 suggested that a putative AP1 binding site located between positions −126 and −113 is involved in the transactivation process.
FIG. 6.
Deletion of the LTR sequences required for transactivation. (A) Deletion mutants of the FIV LTR promoter. The putative cis-acting regulatory elements contained in the U3 region of the LTR are indicated at the top. The panel of 5′ deletion mutants was previously described (35) and kindly provided by J. F. Thompson and colleagues. wt, wild type. (B and C) Transactivation of 5′ deletion mutants (B) and internal deletion mutants (C) of the FIV LTR by PPR orf2. Each deletion mutant was cotransfected in CrFK cells with the PPR orf2 vector (gray bars). Cotransfection of each mutant with pCR3 defined baseline activity (blank bars). CAT activity was assayed as described in Materials and Methods. The experiment was repeated twice, in triplicate; standard deviations are shown as error bars. The percentage of wild-type LTR activity of each deletion mutant in response to transactivation by orf2, as well as the relative increase of CAT activity compared to the basal level for each deletion mutant, is shown on the right.
The further reduction in transactivation of the mutants deleted to positions −68 and −47 may either reflect the deletion of important sequences responsive to orf2-mediated transactivation or be due to an underestimation of the level of transactivation caused by the decrease in the basal promoter activity. The region located between positions −113 and −47 contains putative regulatory elements, including an AP4 site, a C/EBP tandem repeat, and NF-1 and ATF sites (11, 17, 18, 20). Therefore these elements may play an important role in transactivation of the FIV LTR. To address this issue, we constructed a panel of FIV LTR mutants with internal deletions and analyzed their activity after cotransfection with the PPR orf2 plasmid in CrFK cells (Fig. 6C). While the ΔAP4 and ΔNF1 mutants responded as efficiently as the wild-type LTR, deletion of the AP1, the C/EBP tandem repeat, or the ATF site resulted in a reduced basal activity as well as a reduced response to orf2-mediated transactivation compared to the wild-type LTR (Fig. 6C). However, deletion of any one of these three sites was insufficient to abrogate the level of transactivation by orf2. This finding suggests that a mutation in any one site may be compensated for by the presence of the two other elements. Therefore, these sites may act synergistically to regulate transactivation of the FIV LTR by orf2.
DISCUSSION
The transcriptional map reported here was similar to those in previous studies (26, 27, 37), with two exceptions. First, we detected no monocistronic rev mRNA, a feature apparently unique to the TM1 strain of FIV (37). Second, we identified in the env region a new splice acceptor site which generates, in the presence or absence of exon 2, two transcripts with a coding capacity for an 11-kDa protein, p11rev. The p11rev transcripts are RRE-containing mRNAs, and their expression is probably dependent on the presence of Rev and occurs at a late stage during infection. Therefore, p11rev may constitute an accessory protein of FIV. p11rev and Rev have the same first env-encoded exon, but we detected no Rev activity for p11rev. Two others transcripts, 1.4 and 1.2.4, with a coding capacity for a Rev-related protein, were previously identified but not characterized (27). Similar transcripts were also reported for CAEV and equine infectious anemia virus (8, 28). Transcripts 1.4 and 1.2.4 contain the second rev ORF and encode a protein of 15 kDa, p15rev. An initiator AUG codon is present at the beginning of this ORF and is conserved only in two other FIV strains (23, 32). In CAEV and equine infectious anemia virus, this ORF lacks an initiator AUG but encodes a 16-kDa protein, suggesting that initiation occurs at a non-AUG codon (8, 28). This feature may also apply to FIV strains lacking this initiator AUG. Although p15rev lacks the env-encoded N-terminal domain of Rev, it contains two regions important for Rev function. The first is KRQRRRR, which is analogous to the arginine-rich RNA binding domain of the ungulate and primate lentivirus Rev proteins. The second is a polar effector domain, which is unrelated but functionally interchangeable to the leucine-rich effector domain of HIV-1 Rev protein (19). As expected, p15rev functioned via a mechanism similar to that for Rev. However, the Rev activity displayed by cDNA 1.4 was 20% of that observed for the full-length Rev-expressing clone (Fig. 4B). The difference in Rev activity may result from either a low expression of p15rev or the requirement of the N-terminal exon of Rev for full Rev function. We could not analyze the presence of p15rev in infected cells, since the FIV-positive cat serum used in this study also reacts with p15, the FIV matrix protein.
Comparison of the genomic organization of FIV, visna virus, and CAEV shows that the orf2 gene coincides in size and location to the tat ORFs of the ungulate viruses (7, 10, 14). Therefore, FIV orf2 has been postulated to encode a transcriptional transactivator (33). Transactivation of the FIV LTR by FIV proviruses has been shown to be weak and varied depending on the LTR, provirus, and cell type tested (22, 33, 35, 38). However, these studies provided evidence that FIV PPR strain might encode a weak transactivator, while the 34TF10 strain did not. The orf2 genes of FIV-PPR and FIV-34TF10 differ by the presence of a termination codon in FIV-34TF10 resulting in a truncated orf2 gene (26, 34). Truncation and mutation of the orf2 gene were also reported to impair the ability of FIV to efficiently replicate in PBLs and feline T cells (36, 38). Mutagenesis of the termination codon of 34TF10 orf2 to tryptophan codon resulted in an efficient replication of PBLs and feline T cells by the orf2-repaired 34TF10 virus (38). These findings were consistent with the interpretation that the orf2 gene (i) might encode a weak transactivator and (ii) is implicated in the viral replication efficiency.
Our study confirms that orf2 encodes the FIV Tat protein. Cotransfection experiments with the bicistronic orf2/rev cDNA clones resulted in a five- to sixfold increase in transactivation of the full-length FIV LTR (Fig. 5B). Transient expression assays resulted in an increase of gene expression from the full-length FIV LTR-CAT construct cotransfected with PPR orf2 in CrFK and HeLa cells. Average 14- to 17-fold increases in LTR activity were observed in CrFK cells cotransfected with the PPR orf2 gene, while no significant level of transactivation was detected with the truncated orf2 gene of FIV-34TF10 (Fig. 5C). orf2 has been previously reported to transactivate the FIV LTR three- to fivefold above the basal level in CrFK cells. A similar degree of transactivation has also been shown in HeLa cells, using 10 μg of pFIVLTR-CAT (38). In the present study, transactivation assays performed with HeLa cells and 1 and 10 μg of pFIVLTR-CAT resulted in average 15- and 23-fold increases in LTR activity, respectively (Fig. 5C). We attribute the differences in transactivation observed between the data presented here and those previously reported as reflecting better DNA transfection efficiency rather than simply lower basal LTR activity.
The orf2 gene product was not detected in infected cells by immunoprecipitation of labeled cells extracts. Difficulties in identifying visna virus Tat protein in infected cells have also been reported (7). However, the orf2 gene from cDNA clones 1.3.4, 1.2.3.4, and pOrf2 directed the in vitro synthesis of a 9-kDa polypeptide that was specifically immunoprecipitated by an anti-orf2 polypeptide serum. We are currently investigating the presence of orf2 gene product in infected cells by using a panel FIV-positive sera and monoclonal antibodies.
Several cis-acting regulatory elements, including AP4, AP1, C/EBP, NF1, and ATF, are present in the FIV LTR (21, 24, 26, 34). Site-directed and deletion mutagenesis have shown that the putative AP4/AP1 and ATF sites are required for full basal promoter activity (13, 15, 33, 35), and DNase I footprint analysis has identified three major binding domains covering the AP1, the C/EBP tandem repeat, and the ATF sites (35). To locate sequences in the U3 region of the FIV LTR that are important for promoter activity in basal and transactivation, we used a panel of 5′ deletion mutants of the FIV LTR. Our results demonstrated that a region located between positions −126 and −47 relative to the cap site was essential for LTR activity in response to transactivation by FIV orf2. Importantly, we showed that a 5′ deletion in the U3 region of the FIV LTR extending to position −113 that removed a putative AP1 site resulted in a dramatic reduction in promoter activity in response to transactivation by orf2. However, the present study indicates that although internal deletion of the AP1, C/EBP, or ATF motif resulted in reduced promoter activity, the level of transactivation of these mutants by orf2 was similar to that of the wild-type LTR. Deletion of these sites has been shown to reduce the basal activity of the FIV LTR (13, 16, 33), and deletion of both the AP1 and ATF sites resulted in a dramatic loss of basal LTR activity (13). These sites have also been identified as major protein binding domains by DNase footprint analysis and gel mobility shift assays (13, 16, 35). Gel supershift assays have shown that the AP1 and ATF sites were recognized by AP1- and ATF-like proteins in CrFK cells (13). Together with the results presented here, these data strongly suggest that the AP1, C/EBP, and ATF sites cooperate in transcriptional regulation of the FIV LTR by orf2.
There are interesting similarities among the activities of the visna virus, CAEV, and FIV LTRs. Secondary structure analysis of the R-U5 region of the visna virus, CAEV, and FIV LTRs revealed no stem-loop structure analogous to the HIV TAR region (9, 39). Also, experiments in our laboratory have failed to show direct binding of the orf2 gene product to the FIV LTR (5). Unlike the HIV LTR, which has very low basal activity, the visna virus, CAEV, and FIV LTRs have relatively high basal activity. Furthermore, visna virus and CAEV Tat proteins have been shown to mediate transactivation through an AP1/AP4 motif (12, 14), and here we demonstrated that an AP1 site is at least one of the targets of the FIV orf2-mediated transactivation. Finally, a cluster of cysteine residues contained within the carboxy-terminal domain of the visna virus and CAEV Tat proteins has been shown to be important for Tat function (29). There are also four cysteine residues in the orf2 gene product of FIV, and we demonstrated that a truncated orf2 gene (34TF10 orf2) that lacks this cluster of cysteine residues failed to transactivate the FIV LTR. These observations suggest that these proteins share a common transactivation mechanism by direct interaction with cellular transcription factors.
ACKNOWLEDGMENTS
We thank Udayan Chatterji, Ying-Chuan Lin, Laure Moutouh, and Huldrych Gunthard for careful reading and criticism of the manuscript, and we thank C. J. Kiser for assistance in preparation of the manuscript. We also thank James Neil for providing the LTR-CAT constructs and Tom Phillips for the RRE-CAT construct.
This work was supported in part by grants from the National Institute of Allergy and Infectious Diseases (AI 25825), the National Institute of Mental Health (MH 47680), and the National Institutes of Health (GM 48870).
REFERENCES
- 1.Ali S A, Steinkasserer A. PCR-ligation-PCR mutagenesis: a protocol for creating gene fusions and mutations. BioTechniques. 1995;18:746–750. [PubMed] [Google Scholar]
- 2.Barnes W M. PCR amplification of up to 35-kb DNA with high fidelity and high yield from λ-bacteriophage templates. Proc Natl Acad Sci USA. 1994;91:2216–2220. doi: 10.1073/pnas.91.6.2216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bendinelli M, Pistello M, Lombardi S, Poli A, Garzelli C, Matteucci D, Ceccherini-Nelli L, Malvaldi G, Tozzini F. Feline immunodeficiency virus: an interesting model for AIDS studies and an important cat pathogen. Clin Microbiol Rev. 1995;8:87–112. doi: 10.1128/cmr.8.1.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carruth L M, Hardwick J M, Morse B A, Clements J E. Visna virus Tat protein: a potent transcription factor with both activator and suppressor domains. J Virol. 1994;68:6137–6146. doi: 10.1128/jvi.68.10.6137-6146.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chatterji, U., and J. H. Elder. Unpublished observation.
- 6.Clements J E, Payne S L. Molecular basis of the pathobiology of lentiviruses. Virus Res. 1994;32:97–109. doi: 10.1016/0168-1702(94)90037-x. [DOI] [PubMed] [Google Scholar]
- 7.Davis J L, Clements J E. Characterization of a cDNA clone encoding the visna virus transactivating protein. Proc Natl Acad Sci USA. 1989;86:414–418. doi: 10.1073/pnas.86.2.414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gazit A, Mashiah P, Kalinski H, Gast A, Rosin-Abersfeld R, Tronick S R, Yaniv A. Two species of Rev proteins, with distinct N termini, are expressed by caprine arthritis encephaltis virus. J Virol. 1996;70:2674–2677. doi: 10.1128/jvi.70.4.2674-2677.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gdovin S L, Clements J E. Molecular mechanisms of visna virus Tat: identification of the targets for transcriptional activation and evidence for a post-transcriptional effect. Virology. 1992;188:438–450. doi: 10.1016/0042-6822(92)90497-d. [DOI] [PubMed] [Google Scholar]
- 10.Gourdou I, Mazarin V, Querat G, Sauze N, Vigne R. The open reading frame S of visna virus genome is a trans-activating gene. Virology. 1989;171:170–178. doi: 10.1016/0042-6822(89)90524-2. [DOI] [PubMed] [Google Scholar]
- 11.Gronostajski R M. Analysis of nuclear factor I binding to DNA using degenerate oligonucleatides. Nucleic Acids Res. 1986;14:9117–9132. doi: 10.1093/nar/14.22.9117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hess J L, Small J A, Clements J E. Sequences in the visna virus long terminal repeat that control transcriptional activity and respond to viral trans-activation: involvement of AP-1 sites in basal activity and trans-activation. J Virol. 1989;63:3001–3015. doi: 10.1128/jvi.63.7.3001-3015.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ikeda Y, Inoshima Y, Kawaguchi Y, Maeda K, Kohmoto M, Kai C, Miyazawa T, Mikami T. Protein-binding properties of the putative AP-1 and ATF sequences in the feline immunodeficiency virus long terminal repeat. J Gen Virol. 1998;79:95–99. doi: 10.1099/0022-1317-79-1-95. [DOI] [PubMed] [Google Scholar]
- 14.Kalinski H, Mashiah P, Rotem D, Orzech Y, Sherman L, Miki T, Yaniv A, Gazit A, Tronick S R. Characterization of cDNAs species encoding the Tat protein of caprine arthritis encephalitis virus. Virology. 1994;204:828–834. doi: 10.1006/viro.1994.1602. [DOI] [PubMed] [Google Scholar]
- 15.Kawaguchi Y, Norimine J, Miyazawa T, Kai C, Mikami T. Sequences within the feline immunodeficiency virus long terminal repeat that regulate gene expression and respond to activation by feline herpes virus type 1. Virology. 1992;190:465–468. doi: 10.1016/0042-6822(92)91235-m. [DOI] [PubMed] [Google Scholar]
- 16.Kawaguchi Y, Tomonaga K, Maeda K, Ono M, Miyazawa T, Kohmoto M, Tohya Y, Mikami T. The C/EBP site in the feline immunodeficiency virus (FIV) long terminal repeat (LTR) is necessary for its efficient replication and is also involved in the inhibition of FIV LTR-directed gene expression by pseudorabies virus ICP4. Virology. 1995;208:492–499. doi: 10.1006/viro.1995.1180. [DOI] [PubMed] [Google Scholar]
- 17.Landschultz W H, Johnson P F, Adashi E Y, Graves B J, McKnight S L. Isolation of a recombinant copy of the gene encoding C/EBP. Genes Dev. 1988;2:786–800. doi: 10.1101/gad.2.7.786. [DOI] [PubMed] [Google Scholar]
- 18.Lin Y-S, Green M R. Interaction of a common cellular transcription factor, ATF, with regulatory elements in both E1a- and cyclic AMP-inducible promotors. Proc Natl Acad Sci USA. 1988;85:414–417. doi: 10.1073/pnas.85.10.3396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mancuso V A, Hope T J, Zhu L, Derse D, Phillips T, Parslow T G. Posttranscriptional effector domains in the Rev proteins of feline immunodeficiency virus and equine infectious anemia virus. J Virol. 1994;68:1998–2001. doi: 10.1128/jvi.68.3.1998-2001.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mermod N, Williams T J, Tjian R. Enhancer binding factors AP-4 and AP-1 act in concert to activate SV40 late transcription in vitro. Nature (London) 1988;332:557–561. doi: 10.1038/332557a0. [DOI] [PubMed] [Google Scholar]
- 21.Miyazawa T, Fukasawa M, Hasegawa A, Maki N, Ikuta K, Takahashi E, Hayami M, Mikami T. Molecular cloning of a novel isolate of feline immunodeficiency virus biologically and genetically different from the original U.S. isolate. J Virol. 1991;65:1572–1577. doi: 10.1128/jvi.65.3.1572-1577.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Miyazawa T, Kohmoto M, Kawaguchi Y, Tomonaga K, Toyosaki T, Ikuta K, Adachi A, Mikami T. The AP-1 binding site in the feline immunodeficiency virus long terminal repeat is not required for virus replication in feline T lymphocytes. J Gen Virol. 1993;74:1573–1580. doi: 10.1099/0022-1317-74-8-1573. [DOI] [PubMed] [Google Scholar]
- 23.Morikawa S, Lutz H, Aubert A, Bishop D H L. Identification of conserved and variable regions in the envelope glycoprotein sequences of two feline immunodeficiency viruses isolated in Zurich, Switzerland. Virus Res. 1991;21:53–63. doi: 10.1016/0168-1702(91)90071-3. [DOI] [PubMed] [Google Scholar]
- 24.Olmsted R A, Barnes A K, Yamamoto J K, Hirsch V M, Purcell R H, Johnson P R. Molecular cloning of feline immunodeficiency virus. Proc Natl Acad Sci USA. 1989;86:2448–2452. doi: 10.1073/pnas.86.7.2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pedersen N C, Ho E H, Brown M L, Yamamoto J K. Isolation of a T-lymphotropic virus from domestic cats with an immunodeficiency-like syndrome. Science. 1987;235:790–793. doi: 10.1126/science.3643650. [DOI] [PubMed] [Google Scholar]
- 26.Phillips T R, Talbott R, Lamont C, Muir S, Lovelace K, Elder J H. Comparison of two host cell range variants of feline immunodeficiency virus. J Virol. 1990;64:4605–4613. doi: 10.1128/jvi.64.10.4605-4613.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Phillips T R, Vamont C, Konings D, Shacklett B, Hamson C, Luciw P, Elder J H. Identification of the Rev transactivation and Rev-responsive elements of feline immunodeficiency virus. J Virol. 1992;66:5464–5471. doi: 10.1128/jvi.66.9.5464-5471.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rosin-Arbesfeld R, Rivlin M, Noiman S, Mashiah P, Yaniv A, Miki T, Tronick S R, Gazit A. Structural and functional characterization of rev-like transcripts of equine anemia virus. J Virol. 1993;67:5640–5646. doi: 10.1128/jvi.67.9.5640-5646.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Saltarelli M J, Schoborg R, Gdovin S L, Clements J E. The CAEV tat gene trans-activates the viral LTR and is necessary for efficient viral replication. Virology. 1993;197:35–44. doi: 10.1006/viro.1993.1564. [DOI] [PubMed] [Google Scholar]
- 30.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Seed B, Sheen J-Y. A simple phase-extraction assay for chloramphenicol acetyltransferase activity. Genes. 1988;67:271–277. doi: 10.1016/0378-1119(88)90403-9. [DOI] [PubMed] [Google Scholar]
- 32.Sodora D L, Courcelle J, Brojatsch J, Berson A, Wang Y C, Dow S W, Hoover E A, Mullins J I. Analysis of a feline immunodeficiency virus provirus reveals patterns of gene sequence conservation distinct from human immunodeficiency virus type 1. AIDS Res Hum Retroviruses. 1995;11:531–533. doi: 10.1089/aid.1995.11.531. [DOI] [PubMed] [Google Scholar]
- 33.Sparger E, Shacklett B, Renshaw-Gegg L, Barry P, Pedersen N, Elder J, Luciw P. Regulation of gene expression directed by the long terminal repeat of the feline immunodeficiency virus. Virology. 1992;187:165–177. doi: 10.1016/0042-6822(92)90305-9. [DOI] [PubMed] [Google Scholar]
- 34.Talbott R L, Sparger E E, Lovelace K M, Fitch W M, Pedersen N C, Luciw P A, Elder J H. Nucleotide sequence and genomic organization of feline immunodeficiency virus. Proc Natl Acad Sci USA. 1989;86:5743–5747. doi: 10.1073/pnas.86.15.5743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Thompson J F, Elder J H, Neil J C. Cis- and trans-regulation of feline immunodeficiency virus: identification of functional binding sites in the long terminal repeat. J Gen Virol. 1994;75:545–554. doi: 10.1099/0022-1317-75-3-545. [DOI] [PubMed] [Google Scholar]
- 36.Tomonaga K, Miyazawa T, Sakuragi J-I, Mori T, Adachi A, Mikami T. The feline immunodeficiency virus ORF-A gene facilitates efficient viral replication in established T-cell lines and peripheral blood lymphocytes. J Virol. 1993;67:5889–5895. doi: 10.1128/jvi.67.10.5889-5895.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tomonaga K, Shin Y-S, Fukasawa M, Miyazawa T, Adachi A, Mikami T. Feline immunodeficiency virus gene expression: analysis of the RNA splicing pattern and the monocistronic rev mRNA. J Gen Virol. 1993;74:2409–2417. doi: 10.1099/0022-1317-74-11-2409. [DOI] [PubMed] [Google Scholar]
- 38.Waters A K, de Parseval A P, Lerner D L, Niels J C, Thompson F J, Elder J H. Influence of ORF2 on host cell tropism of feline immunodeficiency virus. Virology. 1996;215:10–16. doi: 10.1006/viro.1996.0002. [DOI] [PubMed] [Google Scholar]
- 39.Waters, A. K., and J. H. Elder. Unpublished observation.