Abstract
Despite the availability of licensed vaccines, influenza causes considerable morbidity and mortality worldwide. Current influenza vaccines elicit an immune response that primarily targets the head domain of the viral glycoprotein hemagglutinin (HA). Influenza viruses, however, readily evade this response by acquiring mutations in the head domain. While vaccines that target the more conserved HA stalk may circumvent this problem, low levels of anti-stalk antibodies are elicited by vaccination, possibly due to the poor accessibility of the stalk domain to B cell receptors. In this work, we demonstrated that nanoparticles presenting HA in an inverted orientation generated ten-fold higher anti-stalk antibody titers after a prime immunization and five-fold higher anti-stalk titers after a boost than nanoparticles displaying HA in its regular orientation. Moreover, nanoparticles presenting HA in an inverted orientation elicited a broader anti-stalk response that reduced mice weight loss and improved survival after challenge to a greater extent than nanoparticles displaying HA in a regular orientation. Refocusing the antibody response towards conserved epitopes by controlling antigen orientation may enable the design of broadly protective nanovaccines targeting influenza and other pathogens with pandemic potential.
Keywords: influenza, hemagglutinin, vaccine
Table of Contents Entry:

The immune system generally targets the variable head domain of the influenza protein hemagglutinin (HA) rather than its conserved stalk domain, which results in a strain-specific response. This phenomenon may be a result of HA orientation. In this work, nanoparticles that displayed HA in an inverted orientation were developed and found to elicit an enhanced stalk-directed antibody response.
1. Introduction
Influenza represents a major global health problem. Seasonal influenza virus epidemics have been estimated to cause between 290,000 and 650,000 deaths annually worldwide before the coronavirus disease 2019 (COVID-19) pandemic.[1] The spread of seasonal influenza viruses can be prevented through vaccination. Such immunizations, as well as natural infection, typically induce an immune response that primarily targets the globular head domain of the viral hemagglutinin (HA).[2–4] The HA head domain rapidly acquires mutations – a phenomenon referred to as antigenic drift – which enables the virus to escape pre-existing immunity. For that reason, the influenza virus vaccine must be reformulated and readministered annually. However, there can be a mismatch between the strains selected for incorporation into seasonal vaccines and circulating viruses, resulting in a decrease in vaccine efficacy.[5] Another serious concern is that commercially available vaccines would provide little protection against potentially pandemic influenza virus strains, such as highly pathogenic H5N1 or H7N9 viruses, and the consequences could be devastating. As an example, the influenza pandemic of 1918 has been estimated to have claimed tens of millions of lives.[6, 7]
In contrast to the highly plastic HA head domain,[8] the stalk domain is conserved and has a lower tolerance for mutations.[9, 10] The conserved nature of the stalk domain makes it a prime target for broadly neutralizing antibodies. In fact, stalk-directed antibodies isolated from mice and humans are capable of neutralizing across different influenza virus strains and subtypes.[11–13] Moreover, monoclonal antibodies that target the stalk domain have proven to be cross-protective in animal models.[11, 12, 14–17] Although these broadly protective stalk-specific antibodies are not commonly elicited by current influenza virus vaccines, different vaccines with broadly neutralizing potential that target the HA stalk domain are under development.[18] Examples include an HA molecule with a hyperglycosylated head domain that prevents B cells from accessing the HA head through steric shielding,[19] stalk-only HA constructs in which the HA head domain has been removed,[20–24] and chimeric HA molecules with variable heads and a conserved stalk domain that boost the stalk-directed response upon sequential immunizations.[16, 25, 26]
This work was motivated by a fundamental unresolved question: Why do natural infection and current vaccines elicit an immune response predominately targeting the HA head? Overcoming the natural immunodominance of the HA head may have important implications for the development of an effective broadly protective influenza virus vaccine. While several possible explanations for the immunodominance of the HA head exist,[27–29] one intriguing hypothesis suggests that the natural immunodominance of the HA head may result from its accessibility on the virion. Specifically, the HA is positioned on the viral particle in a way that its globular head is displayed as its outermost domain and is therefore easily accessible to head-specific B cell receptors.[30, 31] In contrast, the HA stalk is membrane-proximal and its interaction with stalk-specific B cells may be sterically hindered. The HA would have a similar orientation in live or inactivated viral vaccines, as well as in currently available subunit vaccines where HA molecules form rosettes clustering around the stalk and present the HA head as the outermost portion of the vaccine.[32] Therefore, in the case of both natural infection and vaccination, the accessibility of the stalk domain to stalk-specific B-cell receptors may be limited. This limited accessibility might inhibit the effective induction of broadly protective anti-stalk antibodies.
We hypothesized that inverting the orientation of the HA on virus-like particles (VLPs) would make the HA stalk domain more accessible to stalk-specific B cells, and enhance the induction of broadly protective anti-stalk antibodies. To test this hypothesis, we created a VLP that displays multiple copies of an antigen binding fragment (Fab) which recognizes the apex of the HA head; the subsequent binding of HA would result in its presentation in an inverted orientation. To investigate the effect of the orientation of the HA on VLPs and the protective anti-HA antibody response induced following vaccination, we immunized mice with either a construct displaying multiple copies of the HA in the inverted orientation or a construct in which the HA is in the natural orientation as on the surface of the virus. The inverted HA construct generated higher antibody titers against the HA stalk. Furthermore, mice immunized with the inverted HA construct were better protected against an influenza virus displaying a chimeric hemagglutinin (cHA) made up of the same stalk used for immunization but a completely different head domain. This indicates that the inverted HA construct generated enhanced stalk-based protection, suggesting that the orientation of HA affects the generation of anti-stalk antibodies. These results may inform the design of broadly protective influenza vaccines, as antibodies targeting the conserved stalk domain of HA have been found to be broadly protective. Furthermore, controlling antigen presentation using Fabs conjugated to VLPs may be a generally applicable strategy to enhance the immune response against immunosubdominant viral epitopes.
2. Results
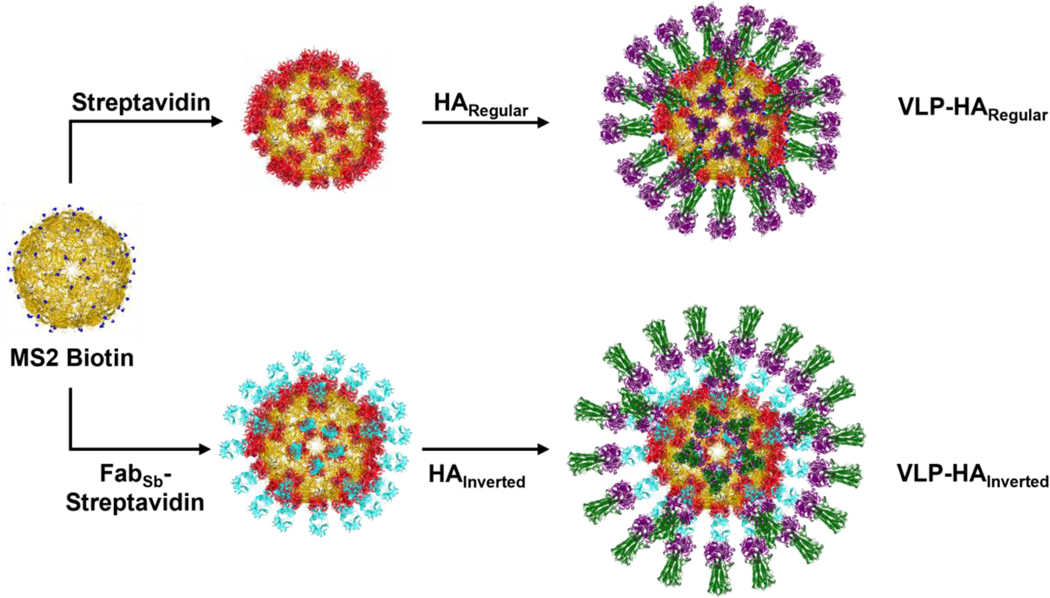
To develop a virion-like scaffold that could be used to present the HA in varying orientations (Figure 1), we generated streptavidin-coated VLPs that can display biotinylated protein by exploiting the strong interaction between biotin and streptavidin (SA).[33, 34] Specifically, we used BL21(DE3) competent Escherichia coli (E. coli) to express a single chain dimer of the MS2 bacteriophage coat protein[35, 36] with an inserted AviTag,[33, 34] which allowed for the site-specific biotinylation of the VLPs. MS2 VLPs have been shown to be stable for 12 months at 4 °C [37] and the inserted AviTag was placed in a surface loop that had previously been shown to tolerate a peptide insertion such that the MS2 VLP retains a relatively stable structure.[38, 39] After the MS2-AviTag VLPs had been expressed, they were purified using both Capto Core 700 resin and size exclusion chromatography (SEC). Once pure, the VLPs were biotinylated in vitro and subsequently separated from biotinylation reagents using SEC. Next, the resulting biotin-coated VLPs were added dropwise to an excess of streptavidin, which had been expressed in E. coli as inclusion bodies, refolded, and purified using iminobiotin affinity chromatography. The mixture was then run on an SEC column, which resulted in streptavidin-coated VLPs that could be used to display biotinylated proteins such as hemagglutinin.
Figure 1.
Assembly of VLP-HARegular and VLP-HAInverted. VLP-HARegular was assembled by first mixing MS2 Biotin (yellow; PDB: 2MS2) with streptavidin (red; PDB: 3RY2) to create MS2-SA VLP. Then the MS2-SA VLP was mixed with HARegular, which is biotinylated at its C-terminus (head domain purple, stalk domain green; PDB: 1RU7). VLP-HAInverted was assembled by first mixing MS2 Biotin with a FabSb (cyan; PDB: 7FAB – representative Fab) - Streptavidin conjugate. Then the resulting VLP displaying the FabSb was mixed with HAInverted (head domain purple, stalk domain green; PDB: 1RU7).
To display the hemagglutinin on the streptavidin-coated MS2 VLPs in the orientation that the HA naturally appears on the influenza virion, we appended an AviTag to the C-terminus HA ectodomain from the A/Puerto Rico/8/1934 H1N1 influenza virus strain. The biotinylation of this AviTag allowed conjugation of the HA to the streptavidin-coated VLPs such that the HA head was presented as the outermost domain of the construct, while the stalk was proximal to the surface. The HA was expressed using the baculovirus-insect cell expression system, purified via a C-terminal hexa-histidine tag using immobilized metal affinity chromatography (IMAC), and biotinylated. The biotinylated HA trimer was then separated from biotinylation reagents by using SEC. This purified, biotinylated HA was suitable for display on the streptavidin-coated MS2 VLPs. The approximate stoichiometric ratio of the biotinylated HA to MS2-SA VLP was determined by mixing the two constructs in varying ratios and running the resulting mixtures on a Sodium Dodecyl Sulfate Polyacrylamide Gel Electrophoresis (SDS-PAGE) gel to characterize the amount of excess protein in each mixture. Once the appropriate ratio was determined, the MS2-SA VLPs were added to a slight excess of biotinylated HA. This mixture was run on an SEC column to purify the resulting VLPs displaying densely packed HA oriented in the same manner that HA would naturally be oriented on the influenza virus (Figure 1).
To present the HA in the inverted orientation, the Fab region of the H28-D14 monoclonal antibody was first displayed on VLPs. This Fab is specific to the Sb antigenic site of the HA such that it binds the apex of the HA head,[40] resulting in an inverted display of the HA on the surface of VLPs with the HA stalk as the outermost portion of the construct (Figure 1). The H28-D14 Fab with a C-terminal AviTag and hexa-histidine tag on the heavy chain was expressed in mammalian cells, purified via Immobilized Metal ion Affinity Chromatography (IMAC), biotinylated in vitro, and separated from biotinylation reagents by using SEC. The biotinylated H28-D14 Fab was then added dropwise to excess streptavidin, and SEC was used to purify the resulting SA-Fab conjugate. The SA-Fab conjugate could then be attached to biotin-coated MS2 VLPs by way of the streptavidin’s unoccupied biotin binding sites. To determine the appropriate stoichiometric ratio of SA-Fab conjugate to biotinylated MS2 VLP, varying ratios of the two constructs were mixed and run on an SDS-PAGE gel. The resulting gel was then analyzed to determine the ratio at which little to no excess protein remained. After the appropriate stoichiometry had been determined, the biotinylated MS2 VLPs and SA-Fab conjugate were mixed in the established ratio. After a short incubation, the VLPs displaying H28-D14 Fab were added to a large excess of HA. This final mixture was run through an SEC column to purify the resulting VLPs displaying HA in the inverted orientation.
The constituent proteins and the final constructs displaying HA in both the regular and inverted orientations were characterized in vitro using a variety of bioanalytical methods. The proteins used to make both VLP constructs were run on an SDS-PAGE gel, which confirmed that all proteins were pure and ran as expected according to their molecular weights (Figure 2a). To ensure that the HA proteins retained their conformation, we conducted an enzyme-linked immunosorbent assay (ELISA) and confirmed that the anti-head antibody H28-D14[40] and the anti-stalk antibody CR6261[14] recognized both hemagglutinin proteins, HARegular and HAInverted (Figure 2b).
Figure 2.
Characterization of proteins used to assemble VLP-based vaccines. a) SDS-PAGE characterization of proteins used to assemble VLP-based vaccines. b) Characterization of hemagglutinin proteins, HARegular and HAInverted, by ELISA where gray bars represent binding of anti-stalk antibody CR6261 and white bars represent binding of anti-head antibody H28-D14 (mean ± SD; n=6).
Dynamic light scattering (DLS) was used to determine the approximate size of the VLP constructs (Figure 3a). The sizes of all the constructs are consistent with their expected sizes based on the theoretical sizes of the individual proteins. The regularly oriented HA construct is approximately 42.4 nm in radius, while the inverted HA construct is approximately 47.8 nm in radius – the difference likely being due to the Fab present in the inverted HA construct. We also imaged VLPs presenting HA in the regular and inverted orientations using cryo-electron microscopy (Figure 3b). As seen in the cryo-electron micrographs, the HA molecules are oriented on VLP-HARegular in a manner that positions the globular head as the outermost distal domain away from the particle center. In contrast, the VLP-HAInverted resembles a sea urchin, with the HA stalks being the most distal from the particle center. We performed a 3D reconstruction for the VLP-HAInverted construct and obtained a low resolution structure of the nanoparticle which indicates the different layers – MS2, streptavidin, anti-HA-head H28-D14 Fab (FabSb) and the HA head – as shown in Figs. 3c and 3d. We were not able to resolve the entire HA molecules due to their flexibility.
Figure 3.
In vitro characterization of VLP-HARegular and VLP-HAInverted. a) Characterization of (i) intermediate VLPs, (ii) VLP-HARegular, and (iii) VLP-HAInverted by dynamic light scattering (mean ± SD; n=3). b) Representative cryoelectron micrographs of (i) VLP-HARegular and (ii) VLP-HAInverted. Arrowheads (white) indicate oriented HA proteins on the VLP surface. The white scale bars correspond to 100 nm. c) Representative 2D class averages of VLP-HAInverted and the corresponding 3D model (d) with segmented volume of the putative MS2 shell (yellow), streptavidin (red), anti-HA-head H28-D14 Fab (FabSb; cyan), and HA head moiety (purple).
We conducted two additional assays to characterize the orientation of the HA on the surface of VLPs (Figure 4). The first assay was an “aggregation assay” in which we used DLS to measure the size of the oriented HA constructs in solution after mixing with VLPs displaying the H28-D14 Fab (Figure 4a and 4b). We first mixed VLPs displaying H28-D14 Fab with the HA regularly oriented on VLPs. We would expect that when the HA was regularly oriented, the VLPs displaying the H28-D14 Fab would bind the HA and induce aggregation. Characterization of the resulting mixture by DLS confirmed the occurrence of this aggregation, consistent with the HA being oriented on VLPs as it would be on the virus. Next, we mixed VLPs displaying H28-D14 Fab with VLPs presenting HA in the inverted orientation. In this case, we would not expect aggregation because access to the apex of the HA head would be sterically hindered. DLS confirmed that no large-scale aggregation had occurred. These results are consistent with the VLPs presenting the HAs in the desired orientation – regular or inverted. To further probe the orientation of the HAs on the VLPs, we conducted a hemagglutination assay (Figure 4c). Here we examined whether each construct was capable of agglutinating red blood cells, which contain sialic acid on their surface. HA binds to sialic acid near the apex of its head domain. Therefore, we would expect the regularly oriented HA, which presents the HA head as the outermost portion of the construct, to interact with and agglutinate the red blood cells. On the other hand, we would not expect the HA in the inverted orientation to interact with the sialic acid on the red blood cells. The results of the hemagglutination assay (Figure 4c) were consistent with these expectations. Therefore, we concluded that both the regularly oriented HA and the inverted HA are displayed on the VLPs as desired.
Figure 4.
Characterization of HA orientation on VLP. a) Cartoon and corresponding DLS measurement of (i) VLP-HARegular alone and (ii) VLP-HARegular mixed with VLP-FabSb. b) Cartoon and corresponding DLS measurement of (i) VLP-HAInverted alone and (ii) VLP-HAInverted mixed with VLP-FabSb. c) Characterization of HA orientation on VLP by hemagglutination assay.
We next evaluated the in vivo immunogenicity of the constructs. We immunized mice with either the regularly oriented HA (positive control), the inverted HA, or MS2-SA VLPs alone (negative control) (Figure 5a). Mice immunized with the HA constructs each received 2.5 μg of HA per dose. Mice immunized with VLPs alone received the same amount of MS2-SA VLPs as is in a dose of the regularly oriented HA. Post-prime sera were collected on day 21. After 28 days the mice were boosted. Post-boost sera were collected on day 42. An ELISA was conducted using the post-prime and post-boost sera to determine antibody titers against cH6/1, a chimeric hemagglutinin made up of an H6 head and an H1 stalk. As the mice were immunized with H1 HA, the resulting antibodies should only be capable of binding to the H1 stalk of the cH6/1 protein but not to its H6 head domain. Therefore, antibody titers against cH6/1 are a direct measure of anti-stalk antibody titers. We found that sera from mice immunized with the inverted HA construct had significantly higher stalk antibody titers than did sera from mice immunized with the regularly oriented HA construct, where titers are expressed as endpoint titers. A single dose of the inverted HA construct generated a geometric mean anti-stalk antibody titer of 2.10 × 104, whereas a single dose of the regularly oriented HA construct generated an anti-stalk titer of only 2.01 × 103 – a 10-fold difference (Figure 5b). Likewise, boosting with the inverted HA construct resulted in a geometric mean anti-stalk antibody titer of 9.77 × 104, whereas the regularly oriented HA construct generated a post-boost titer of 1.95 × 104 – a 5-fold difference (Figure 5b). Importantly, antibodies elicited in mice vaccinated with the inverted HA construct also displayed significantly broader reactivity against both influenza A and influenza B HAs (Figure 5c). The breadth of the reactivity was assessed by a cell-based ELISA using virus-infected MDCK cells displaying a natural conformation of the HA on the cell surface. Sera from mice vaccinated with the inverted HA construct showed reactivity against HA from H1, H5, and H9 viruses (group 1), H3 and H14 viruses (group 2), and even against the HA from viruses of the B/Victoria (B/Washington/02/2019) and B/ Yamagata (B/Phuket/3073/2013) lineages. By contrast, the reactivity for sera from mice vaccinated with the regularly oriented HA construct was lower, except against H5, which requires further investigation. Overall, these results indicate that the inverted HA construct elicits an enhanced and broadly reactive stalk-directed immune response. These results are exciting yet expected, as the HA in the inverted orientation presents the stalk such that it is accessible to stalk specific B cells, while the stalk of regularly oriented HA is membrane proximal, shielded by the globular HA head, and likely difficult for B cells to access.
Figure 5.
Antibody response to VLP-HARegular and VLP-HAInverted. a) Schedule for vaccination of mice, serum collection, and viral challenge. b) Endpoint antibody titers against cH6/1 HA generated by the priming dose and boosting dose of VLP-HARegular, VLP-HAInverted, and VLP. Endpoint titers using serially 3-fold diluted sera were expressed as the reciprocal of the largest dilution with an optical density at 490 nm cutoff value > 0.2. † - Endpoint titer is less than the reciprocal of the smallest dilution (< 100). (geometric mean with geometric standard deviation; n = 15 mice immunized with oriented HA constructs; n=12 mice immunized with VLP). ****P < 0.0001 determined by a Mann-Whitney test (α = 0.05). c) Reactivity of sera from vaccinated mice by cell-based ELISA. Heatmap intensity is based on the geometric mean of area under the curve (AUC) values of every group of mice against the indicated HA (n = 7 mice immunized with oriented HA constructs; n=6 mice immunized with VLP).
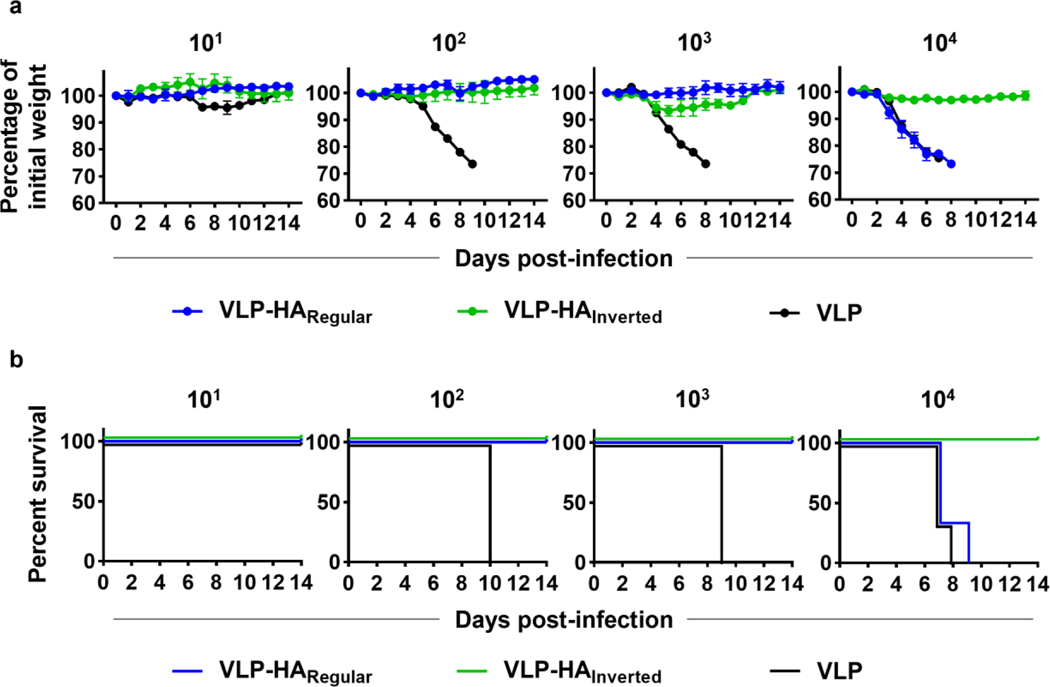
After this encouraging result, the vaccinated mice were challenged with a cH6/1N5 influenza virus at four different doses (101, 102, 103, or 104 plaque-forming units (PFU)/mouse ~ 0.3–300 LD50) to assess the degree of stalk-based protection generated by the HA constructs and the MS2-SA VLP control. As expected, the MS2-SA VLP control provided negligible protection against infection. Mice immunized with the MS2-SA VLP displayed rapid weight loss and only survived at the lowest administered dose of cH6/1N5 (Figs. 6a and 6b). Both the regularly oriented and inverted HA constructs provided a protective immune response against the three lowest doses of the cH6/1N5 virus, as all mice immunized with these constructs lost little bodyweight (Figure 6a) and survived the duration of these challenges (Figure 6b). However, the protection offered by the oriented HA constructs differed drastically against the highest challenge dose of the cH6/1N5 virus (~300 LD50). While mice immunized with the inverted HA lost very little body weight (Figure 6a) and survived the duration of the challenge (Figure 6b), mice immunized with the regularly oriented HA rapidly lost bodyweight (Figure 6a) and died by day 9 of the challenge (Figure 6b). These results indicate that the inverted HA generated a more protective stalk-directed response than did the regularly oriented HA. These results are consistent with initial expectations as the stalk should be more accessible to B cells when the HA is inverted. Therefore, we have concluded that HA orientation influences the generation of anti-stalk antibodies.
Figure 6.
Protective efficacy of oriented HA constructs. a) Weight loss of mice immunized with either VLP-HARegular, VLP-HAInverted, or VLP and subsequently infected with cH6/1N5 virus at four different doses (101, 102, 103, or 104 pfu/mouse; mean ± SEM; n=3). b) Survival of mice immunized with either VLP-HARegular, VLP-HAInverted, or VLP and subsequently infected with cH6/1N5 virus at four different doses (101, 102, 103, or 104 pfu/mouse; n=3).
3. Conclusion
In this work, we have developed and characterized VLPs presenting HA in an inverted orientation. Inverting the orientation of HA is expected to make the conserved HA stalk more accessible to B cells than it would be either in traditional vaccine formulations or on the influenza virus. Accordingly, these constructs that display HA in an inverted orientation were found to generate a broader and more robust anti-stalk response that reduced mice weight loss and improved survival after challenge to a greater extent than constructs displaying HA in its regular orientation. Additional refinement of the inverted HA construct could further enhance its efficacy. Efforts to enhance the durability of the immune response would be advantageous. It may also be interesting to include the inverted HA construct in a cocktail with an engineered HA antigen that directs the immune response to conserved sites in the HA head. While additional work may be required to fully optimize the inverted HA construct, we are encouraged by the success of VLP-based vaccines in clinical trials – several of which have been authorized for use in humans.[41]
Overall, the effect of HA orientation on the generation of anti-stalk antibodies is a fundamental result that may have applications in the development of broadly protective influenza vaccines. Furthermore, the approach of controlling antigen orientation using Fabs conjugated to VLPs can likely be applied more generally to enhance the immune response against conserved immunosubdominant epitopes of other antigens.
4. Methods
Expression and purification of HA:
DNA encoding both BirA and the ectodomain of HA (A/Puerto Rico/8/1934, H1N1) with a C-terminal T4 fibritin trimerization domain, AviTag, and hexa-histidine tag were cloned into pFastBac Dual (Gibco) to create the HARegular DNA construct. Similarly, the ectodomain of HA (A/Puerto Rico/8/1934) with a C-terminal T4 fibritin trimerization domain and Strep-Tag II was cloned into pFastBac Dual (Gibco) to create the HAInverted DNA construct. These plasmids were then transformed into DH10Bac cells (Gibco) according to the manufacturer’s instructions to generate recombinant bacmid. The transformation was plated on Luria Bertani (LB) agar plates and a blue/white screen was used to identify colonies containing the recombinant bacmid. A colony containing the recombinant bacmid was grown in LB media (10 mL), which was incubated overnight at 37°C. The following morning, the cells of the culture were pelleted and bacmid was midiprepped using the PureLink HiPure Plasmid DNA Miniprep Kit (Invitrogen) and its associated protocol. The bacmid was then transfected into Sf9 cells using Cellfectin II (Gibco) to create P1 baculovirus stock. The baculovirus was amplified until at least a P3 baculovirus stock was made as described by Margine et al.[42] In brief, Sf9 cells were plated in T175 flasks at a density of 2 × 105 cells/cm2. After a 20-minute incubation to allow for the cells to adhere to the flask, the media in the T175 flask was replaced with Grace’s Insect Medium (Gibco) supplemented with fetal bovine serum (3%; FBS), Pen-Strep (1%; 100 U ml−1 penicillin and 100 μg ml−1 streptomycin) and Pluronic F68 (0.1%). Then the P1 baculovirus stock (400 μL) was added to the T175 flask. The flask was incubated at 28°C for 6 days, after which the media was harvested by centrifugation at 2,000xg for 5 minutes to give the P2 baculovirus stock. This procedure was repeated using the P2 stock to further amplify the baculovirus and create subsequent stocks (P3, P4, P5 etc.). Ultimately, these amplified stocks were used to express the HA constructs using a procedure similar to that described by Margine et al.[42] For a 100 mL expression, 30 million cells were centrifuged at 100xg for 7 minutes. The media was discarded, and the cell pellet was resuspended in a baculovirus suspension (9 mL). After a 20-minute incubation, the baculovirus-cell suspension was added to a 250 mL Erlenmeyer flask containing Express Five Serum Free Media (90 mL; Gibco) supplemented with L-glutamine (Gibco) to a concentration of 18 mM. This culture was incubated for 3–4 days, after which the cells were pelleted at 5,500xg for 20 minutes. Resulting media containing secreted HA with a C-terminal Strep-Tag II was purified using a 5 mL StrepTrap HP column (Cytiva), which was operated according to the manufacturer’s instructions. The eluate was concentrated to approximately 1 mL using a 10 kDa molecular weight cut-off (MWCO) centrifugal filter (Millipore Sigma) and run on a Superdex 200 Increase 10/300 column (Cytiva) to separate HA trimer from aggregate. Media containing secreted HA with a C-terminal hexa-histidine tag was run through a gravity flow column (G-Biosciences) containing HisPur Ni2+-nitriloacetic acid (Ni-NTA) resin (1 mL; Thermo Scientific) that had been washed with DI water (60 column volumes) and pre-equilibrated with phosphate buffered saline (30 column volumes; PBS). The column was then washed with wash buffer (90 column volumes; 42 mm sodium bicarbonate, 8 mm sodium carbonate, 300 mm NaCl, 20 mm imidazole) and the hexa-histidine tagged protein was eluted by incubating the column in elution buffer (3 mL; 42 mm sodium bicarbonate, 8 mm sodium carbonate, 300 mm NaCl, 300 mm imidazole) for 5 minutes before collecting the eluate. This elution procedure was repeated three times to generate a total of 9 mL of eluate, which was buffer exchanged to 20 mm Tris, 20 mm NaCl, pH 8.0 to allow for in vitro biotinylation. The HA was then quantified using the bicinchoninic acid (BCA) assay (Thermo Scientific).
Expression and purification of MS2:
GenScript Biotech Corporation (Piscataway, NJ) synthesized DNA encoding an MS2 coat protein single chain dimer with an AviTag insertion made between the fourteenth and fifteenth amino acids of the second coat protein monomer. GenScript then cloned this DNA into pET-28b between the NdeI and XhoI restriction sites. This construct was expressed and purified as previously described.[34] The plasmid was co-transformed into BL21(DE3) E. coli (New England BioLabs) with a plasmid encoding a biotin ligase (pBirAcm, Avidity L.L.C.). The transformed cells were added to 2xYT (5 mL) supplemented with kanamycin and chloramphenicol, and this culture was incubated overnight at 37°C. The next morning, the 5 mL culture was added to 2xYT (1L) supplemented with kanamycin and chloramphenicol. This larger culture was incubated at 37°C until reaching an optical density (OD) of 0.6. At this point, expression of MS2 and BirA was induced with isopropyl β-d-1-thiogalactopyranoside (IPTG) (1 mM; GoldBio), biotin (12.5 mg per L of culture) was added, and the incubator temperature was reduced to 30°C. After overnight incubation, the culture was centrifuged at 7000xg for 7 minutes to pellet the cells. The resulting media was discarded and the cell pellet was resuspended in 20 mm Tris base, pH 9.0, (25 mL) supplemented with lysozyme (0.5 mg/mL; Alfa Aesar), a protease inhibitor tablet (Sigma-Aldrich), and benzonase (125 units; EMD Millipore). This cell suspension was kept on ice and mixed occasionally for 20 minutes, after which sodium deoxycholate (Alfa Aesar) was added to a concentration of 0.1% (w/v). The cells were then sonicated on ice for 3 minutes with 3 second pulses at an amplitude of 35% (Sonifier S-450, Branson Ultrasonics). The lysate was allowed to cool on ice for 2 minutes and then the sonication was repeated. The lysate was then centrifuged for 30 minutes at 27,000xg, and the resulting supernatant was centrifuged again for 15 minutes at 12,000xg. The supernatant resulting from the second centrifugation was then diluted 3-fold with 20 mm Tris, pH 8.0, and filtered using a 0.45 μm bottle-top filter. The MS2 was then purified from this diluted lysate using four HiScreen Capto Core 700 columns (Cytiva) in series according to the manufacturer’s operating instructions. Fractions resulting from the purification were run on an SDS-PAGE gel to determine purity and recovery of MS2. The desired fractions were then pooled, concentrated to approximately 1 mL using a 10 kDa MWCO centrifugal filter (Millipore Sigma), and further purified using a Superdex 200 increase 10/300 column (Cytiva). The purified MS2 was then buffer exchanged into 20 mm Tris, 20 mm NaCl, pH 8.0, in preparation for in vitro biotinylation. The MS2 was quantified using the BCA assay (Thermo Scientific).
Expression, refolding, and purification of streptavidin (SA):
Streptavidin was generally expressed, refolded, and purified as described previously.[43, 44] Plasmid pET21-Streptavidin-Glutamate_Tag was a gift from Mark Howarth (Addgene plasmid # 4636741; http://n2t.net/addgene:46367; RRID: Addgene_46367). This plasmid encoding streptavidin was transformed into BL21(DE3) E. coli and added to four culture tubes each containing 2xYT (5 mL per culture tube) with ampicillin. These cultures were incubated overnight at 37°C. Then, each of the 5 mL cultures were added to shake flasks containing 2xYT (1L) and ampicillin. The 1L cultures were incubated at 37°C until the cultures reached an OD of 0.6. At this point, expression of inclusion bodies was induced with IPTG (1M; GoldBio) and the temperature of the incubator was reduced to 30°C. After overnight incubation, the cultures were centrifuged at 7,000xg for 7 minutes such that the cultures resulted in two cell pellets. The supernatant was discarded, and each cell pellet was homogenized into resuspension buffer (50 mL; 50 mm Tris, 100 mm NaCl, pH 8.0) supplemented with lysozyme (1 mg/mL; Alfa Aesar) and benzonase (500 units; EMD Millipore). The resuspended cells were incubated at 4°C for an hour. The cells were further homogenized before adding sodium deoxycholate (Alfa Aesar) to a total concentration of 0.1% (w/v) and sonicating (Sonifier S-450, Branson Ultrasonics) for 3 minutes with 3 second pulses at 35% amplitude. The lysed cells were then centrifuged at 27,000xg for 15 minutes and the supernatant was discarded. This lysis procedure was repeated, but with a shorter 15-minute incubation at 4°C prior to sonication. The result was two inclusion body pellets that were then each suspended in wash buffer (50 mL; 50 mm Tris, 100 mm NaCl, 100 mm ethylenediamine tetraacetic acid (EDTA), 0.5% (v/v) Triton X-100, pH 8.0), homogenized, sonicated for 30 seconds at an amplitude of 35%, and centrifuged at 27,000xg for 15 minutes. This step was repeated two more times. The resulting washed inclusion body pellets were then suspended in a second wash buffer (50 mm Tris, 10 mm EDTA, pH 8.0), homogenized, sonicated for 30 seconds at an amplitude of 35%, and centrifuged at 15,000xg for 15 minutes. This second wash step was repeated once more. The final washed inclusion body pellets were then unfolded by resuspension and homogenization in a guanidine hydrochloride solution (7.12 m; 10 mL), which was stirred vigorously at room temperature for an hour. The unfolded streptavidin was then centrifuged at 12,000xg for 10 minutes, and the supernatant was added dropwise at a rate of 30 mL/h to chilled PBS (1L) being stirred vigorously to refold the streptavidin. After stirring overnight at 4°C, the refolded streptavidin in PBS was centrifuged at 7,000xg for 10 minutes to remove insoluble protein. The supernatant was filtered with a 0.45 μm bottle-top filter, and ammonium sulfate was slowly added to the filtered solution to a concentration of 1.9 m in order to precipitate out impurities. The resulting solution was stirred at 4°C for at least three hours. After three hours, the streptavidin solution with the precipitated impurities was centrifuged at 7,000xg for 10 minutes and the supernatant was filtered using an 0.45 μm bottle-top filter. Ammonium sulfate was added to the filtrate to bring the total concentration of ammonium sulfate to 3.68 M, which caused the precipitation of streptavidin. The resulting solution was stirred at 4°C for at least three hours, and was then centrifuged at 7,000xg for 20 minutes. The pellet was resuspended in Iminobiotin Affinity Chromatography (IBAC) binding buffer (20 mL; 50 mm sodium borate, 300 mm NaCl, pH 11.0) and allowed to flow through a gravity flow column (G-Biosciences) with Pierce Iminobiotin Agarose (5 mL; Thermo Scientific) that had been washed with deionized (DI) water and equilibrated with IBAC binding buffer. The column was then washed with IBAC binding buffer (20 column volumes) and the streptavidin was eluted with elution buffer (6 column volumes; 20 mm KH2PO4, pH 2.2). The eluate was dialyzed into PBS and concentrated using a 10 kDa MWCO centrifugal filter (Millipore Sigma). Streptavidin was quantified by measuring the UV absorption at 280 nm.
Expression and purification of H28-D14 Fab:
DNA encoding the variable heavy and light chains of H28-D14 (sequence kindly provided by Dr. Jonathan Yewdell, National Institutes of Health[40]) were cloned into TGEX-FH and TGEX-LC (Antibody Design Labs), respectively, by Gene Universal, Inc. (Newark, DE). The TGEX-FH vector was modified to include a C-terminal AviTag immediately upstream of the hexa-histidine tag. These plasmids were transfected into Expi293F cells in a 2:1 light chain to heavy chain ratio using the ExpiFectamine 293 Transfection Kit (Gibco) and associated protocol. Four days after transfection, the culture was centrifuged at 5,500xg for 20 minutes and the supernatant containing secreted H28-D14 Fab was dialyzed into PBS. The resulting H28-D14 in PBS was passed through a gravity flow column (G-Biosciences) with HisPur Ni-NTA resin (1 mL; Thermo Scientific) that had been washed with DI water (60 column volumes) and equilibrated with PBS (30 mL). The column was then washed with wash buffer (90 column volumes; 42 mm sodium bicarbonate, 8 mm sodium carbonate, 300 mm NaCl, 20 mm imidazole). To elute the Fab, the column was incubated in elution buffer (3 mL; 42 mm sodium bicarbonate, 8 mm sodium carbonate, 300 mm NaCl, 300 mm imidazole), which was collected after five minutes. This was repeated twice to create 9 mL of eluate. The eluate was buffer exchanged into 20 mm Tris, 20 mm NaCl, pH 8.0 to allow for in vitro biotinylation. H28-D14 Fab was then quantified by BCA assay (Thermo Scientific).
Expression and purification of H28-D14 and CR6261 antibodies:
The DNA encoding the variable heavy chains and the variable light chains of the H28-D14 antibody[40] (sequence kindly provided by Dr. Jonathan Yewdell, National Institutes of Health) and the CR6261 antibody[17] was cloned into TGEX-HC and TGEX-LC (Antibody Design Labs), respectively, by Gene Universal, Inc. (Newark, DE). The resulting plasmids were transfected into Expi293F cells in a 2:1 light chain to heavy chain ratio using the ExpiFectamine 293 Transfection Kit (Gibco) and protocol. After 5 days, cells were pelleted by centrifugation for 20 minutes at 5,500xg. The supernatant containing secreted antibody was diluted in PBS and purified using a MabSelect SuRe (Cytiva) column according to the manufacturer’s protocol.
In vitro biotinylation of AviTagged HA, MS2, and H28-D14 Fab:
AviTagged proteins were biotinylated in vitro using the BirA500 kit (Avidity LLC) essentially according to the manufacturer’s instructions. In brief, AviTagged protein was buffer exchanged into 20 mm Tris, 20 mm NaCl, pH 8.0 and its concentration was adjusted to 45 μM. BirA and Biomix B (a proprietary solution containing biotin, adenosine triphosphate (ATP), and magnesium acetate) were added to the protein solution as specified in the instructions. This solution was then shaken vigorously at 37°C for two hours. After the two-hour incubation, more Biomix B was added to the solution – the same volume that had been added previously – before incubating on a nutator overnight at 4°C. The biotinylated proteins were then purified using a Superdex 200 Increase 10/300 column (Cytiva) and quantified using the BCA assay (Thermo Scientific).
Assembly of VLP-HARegular:
As reported previously[34], biotinylation of the AviTagged MS2 resulted in near 100% biotinylation. Biotinylated MS2 (1 mL; ~0.7 mg/mL) was added dropwise to a streptavidin solution (~50 mg/mL) that was in approximately 20 times molar excess and being stirred vigorously in a 5 mL glass vial. The solution was stirred for 30 minutes before purifying the MS2-SA from excess streptavidin using a Superdex 200 Increase 10/300 column (Cytiva). Characterization of the purified MS2-SA suggests that each particle contains approximately 72 streptavidin molecules.[34] The MS2-SA was concentrated using a 10 kDa MWCO centrifugal filter (Millipore Sigma), and the amount of streptavidin in solution was quantified by running a boiled MS2-SA sample on an SDS-PAGE gel with lanes that included boiled streptavidin standards previously quantified by measuring the UV absorption at 280 nm. To determine appropriate stoichiometry between MS2-SA and biotinylated HARegular, small-scale mixtures of MS2-SA and biotinylated HARegular were run on an SDS-PAGE gel at 4°C in varying ratios. Once the appropriate stoichiometry had been determined, MS2-SA was added to a slight stoichiometric excess of biotinylated HARegular. After incubating the mixture on a nutator at room temperature for an hour, the resulting VLP-HARegular construct was separated from excess biotinylated HARegular using a Superdex 200 Increase 10/300 column (Cytiva). The amount of HA in the VLP-HARegular solution was quantified by running a boiled VLP-HARegular sample on an SDS-PAGE gel with bovine serum albumin (BSA) standards. This gel was also used to determine that each VLP-HARegular particle contains approximately 70 HA trimers.
Assembly of VLP-HAInverted:
Biotinylated H28-D14 Fab (~1mg; ~1 mg/mL) was added 5 μL at a time to a four-fold molar excess of streptavidin (~50 mg/mL) in a 1.5 mL Eppendorf tube. The mixture was vortexed between every 5 μL addition of biotinylated H28-D14 Fab. The streptavidin-Fab conjugate was purified from both excess streptavidin and Fab-streptavidin-Fab conjugates using a HiLoad Superdex 200 column (Cytiva). To determine the appropriate stoichiometry between MS2 biotin and the streptavidin-Fab conjugate, small scale mixtures of biotinylated MS2 and streptavidin-Fab conjugate were run on an SDS-PAGE gel at 4°C in varying ratios. Biotinylated MS2 was then added to the streptavidin-Fab conjugate in the appropriate stoichiometry and allowed to incubate on a nutator at room temperature for 45 minutes. Biotin in 10-fold molar excess of streptavidin was then added to occupy any biotin-binding sites that had not been occupied by the biotinylated MS2 or Fab. This mixture was incubated on a nutator at room temperature for 15 minutes before being added to a large excess of HAInverted to maximize the number of antigens per particle in this orientation as well. This final mixture was incubated on a nutator at room temperature for an hour. The VLP-HAInverted was then purified using a Superdex 200 Increase 10/300 column (Cytiva). The amount of HA in the VLP-HAInverted solution was quantified by running a heated sample of VLP-HAInverted on an SDS-PAGE gel with BSA standards. This gel was also used to determine that each VLP-HAInverted particle contains approximately 24 HA trimers.
SDS-PAGE:
PageRuler Plus Prestained Protein Ladder (Thermo Scientific) and protein samples mixed with Nu-PAGE lithium dodecyl sulfate (5 μL; LDS) sample buffer (Invitrogen) were pipetted into the wells of a 4–12% Bis-Tris gel (Invitrogen). Gels were cooled at 4°C while running in 2-(N-morpholino)ethanesulfonic acid-SDS (MES-SDS) buffer for 60 minutes at 110 V. The gel was then stained with SimplyBlue SafeStain (Invitrogen), subsequently destained, and imaged using the ChemiDoc MP imaging system (Bio-Rad).
DLS:
Protein was diluted in PBS to 100 μL and pipetted into a UVette (Eppendorf), which was loaded into a DynaPro NanoStar Dynamic Light Scattering detector (Wyatt Technology). Dynamics software (Wyatt Technology) allowed the temperature to equilibrate to 25°C before each measurement. Each measurement was the result of 10 acquisitions and was output as % intensity vs. radius. Radius is presented as the mean ± standard deviations (SD) of three measurements, while the plotted size distribution is the result of a single representative measurement.
Animal experiments:
Mouse experiments were performed in line with the protocols for Dr. Florian Krammer approved by the Icahn School of Medicine at Mount Sinai Institutional Animal Care and Use Committee (protocol # IACUC-2014–0254). We have complied with all relevant ethical regulations. Blood samples were obtained by submandibular bleeding. 6- to 8-week-old mice were immunized with the regularly oriented HA, the inverted HA, or MS2-SA VLPs. Mice receiving the HA constructs were immunized with 2.5 μg of HA per dose. Mice receiving VLPs alone got an equal amount of MS2-SA VLPs to the regularly oriented HA VLPs. Mice were boosted at day 28 after the first vaccine dose administration. Blood samples were collected at day 21 and day 42 after the first vaccine dose administration. For viral challenge, mice were anesthetized with 0.15 mg of ketamine/kg and 0.03 mg of xylazine/kg of body weight in a volume of 0.1 ml/mouse intraperitoneally. Mice were challenged 42 days after the first vaccine dose administration. For this, 50 μl of PBS containing 101 – 104 PFU/mouse of cH6/1N5 virus were administered intranasally. For ELISAs against cH6/1, sera were analyzed individually for every mouse, and antibody endpoint titer values were reported. For cell based ELISAs, sera were analyzed individually for every mouse, and the area under the curve (AUC) values were reported.
ELISA to Characterize Antigens:
Antigen (0.1 μg HA in 100 μL of PBS per well) was coated onto Nunc MaxiSorp 96-well flat-bottom plates (Invitrogen) and allowed to incubate for an hour. The antigen solution was then discarded and 5% BSA (Millipore) in PBST (200 μL/well; PBS with 0.05% Tween-20) was added to each well. The plate was incubated for 45 minutes before the wells of the plate were emptied and washed three times with PBST (200 μL/well). Then, CR6261 or H28-D14 antibody in 1% BSA in PBST (100 μL/well) was added to the appropriate wells such that the molar ratio of antibody to hemagglutinin was 1:1. These antibody solutions were incubated with the antigen for an hour before the wells were emptied and washed three times. Next, horseradish peroxidase (HRP)-conjugated anti-human IgG Fc fragment antibody (MP Biomedicals; 1:5,000 dilution) in 1% BSA in PBST (100 μL/well) was added to each well. After an hour incubation, the secondary antibody solution was discarded, and the plate was washed three times. To develop the plates, TMB substrate solution (100 μL/well; Millipore) was added to each well and was stopped after 3 minutes using sulfuric acid (0.16 m; 100 μL/well). The absorbance of the wells at 450 nm was read using a Spectramax i3x plate reader (Molecular Devices).
ELISA for Characterizing Antibody Titers:
Antigen (0.1 μg HA in 50 μL of PBS per well) was coated onto 96-well microtiter plates (Corning) and allowed to incubate for an hour. The antigen solution was then discarded and blocking solution consisting of 0.5% milk powder (American Bio), 3% bovine serum albumin (millipore) in PBST (200 μL/well; PBS with 0.1% Tween-20) was added. Plates were incubated for 1 hour at room temperature (RT) and washed three times with PBST (200 μL/well). Samples were initially diluted 1:100 and serially diluted (3-fold) in blocking solution. Diluted samples (100 μL/well) were transferred to blocked ELISA plates. Plates were incubated for two hours at RT and washed three times with PBST. A goat anti-mouse IgG horseradish peroxidase (HRP)-conjugated antibody (Rockland) was added at a 1:3000 dilution in blocking solution. After one hour incubation, the secondary antibody solution was removed, and the plate was washed three times with PBST. To develop the plates, Sigmafast o-phenylenediamine dihydrochloride (Sigmafast OPD, Sigma-Aldrich) was added (100 μL/well). Plates were incubated for 10 min and the reaction was stopped using hydrochloric acid (3m; 50 μL/well; Thermo Fisher). The optical density (OD) was measured at 490 nm wavelength with a microplate reader (BioTek, SYNERGY H1). Antibody endpoint titers were expressed as the reciprocal of the largest dilution with an optical density at 490 nm cutoff value > 0.2. Values were plotted using Prism 9 software (GraphPad).
Cell based ELISA:
To assess the breadth of reactivity of antibodies contained in sera from immunized mice, binding to the HA expressed on the surface of virus-infected Madin-Darby Canine Kidney (MDCK) cells was assessed. On day one, 96-well cell culture plates were seeded with 2×104 cells/well. On day two, confluent cells (~90%) were infected with influenza viruses carrying the HA of group 1 ((A/Michigan/45/2015(H1N1), A/Vietnam/1203/04(H5N1), A/Hong Kong/33982/2009(H9N2)), group 2 (A/Hong Kong/4801/2014(H3N2), A/mallard duck/Astrakhan/263/1982(H14N5)) or influenza B (B/Phuket/3073/2013, B/Washington/02/2019) viruses at a multiplicity of infection (MOI) of 2. On day three (16 hours post-infection), cells were washed 1x with phosphate buffer solution (PBS) and fixed with a 10% paraformaldehyde (PFA) solution (100μL/well) for 1 hour at RT. Unspecific binding was blocked with a 3% milk powder (American Bio) solution (200μL/well) for 1 hour at RT. Serum samples were pre-diluted to a 1:100 initial dilution, followed by 3-fold serial dilutions in PBS 1% milk powder. Blocking solution was removed and serial dilutions of antibodies were added. As a positive control, the broadly reactive stalk antibody CR9114 (produced in house) was prepared at an initial concentration of 30μg/ml and serially diluted 3-fold. Samples and positive control were incubated for 2 hours at room temperature and were removed by aspiration. Plates were washed 2x with PBS and an HRP-labeled goat anti-mouse IgG antibody (Rockland) diluted 1:3,000 were added (50μL/well). Plates were washed 2x with PBS and TMB (3,3’,5,5’-Tetramethylbenzidine) peroxidase substrate (Bio-Rad) were added (100μL/well) for 20 min at RT. Reaction was stopped by addition of sulfuric acid (4N; 100μL/well; Fisher Scientific). Plates were read at 450 nm using a SYNERGY H1 microplate reader (BioTek). The area under the curve (AUC) values were calculated and plotted using Prism 9 software (GraphPad).
Cryo-EM sample preparation and data collection:
Aliquots (3 μl) of the VLP-HAInverted and VLP-HARegular nanoparticles were applied to UltrAuFoil gold R2/2 grids and subsequently blotted for 3 seconds at blot force 3 at 20°C and 100% humidity, then plunge-frozen in liquid ethane using an FEI Vitrobot Mark IV. Grids were imaged on a Glacios microscope operated at 200 kV and a Gatan K3 Summit direct electron detector. Movies were collected in counting mode with a total dose of 65.15 e–/Å2. Images were collected at a magnification of 120,000, corresponding to a calibrated pixel size of 1.24 Å/pixel with a defocus of –2.5μm.
Aggregation assay:
VLP-HA (1 μg of HA) was diluted in PBS to 100 μL and its size was measured by DLS. Next, biotinylated MS2 was mixed with streptavidin-Fab conjugate in the appropriate stoichiometric ratio that was determined by running small scale mixtures of each in varying ratios on an SDS-PAGE gel to create VLP-Fab. The VLP-Fab was diluted in PBS to 50 μL and mixed with VLP-HA (1 μg of HA) diluted in PBS to 50 μL such that Fab was in 1.5x molar excess to HA. The 100 μL mixture was then pipetted into a UVette (Eppendorf) and DLS measurements were made using the DynaPro NanoStar Dynamic Light Scattering detector (Wyatt Technology) and Dynamics software (Wyatt Technology).
Hemagglutination assay:
A solution of Turkey Red Blood Cells (5%; 1 mL; Lampire) was pipetted into a 15 mL tube and mixed with PBS (14 mL). The red blood cells were washed by centrifuging for 7 minutes at 100xg, decanting the PBS, and resuspending in fresh PBS. This washing procedure was repeated twice, and the washed red blood cell pellet was resuspended in PBS (10 mL) to create an 0.5% red blood cell solution. Next, wells of a 96 well plate were filled with antigen solution (VLP-HARegular, VLP-HAInverted, MS2-SA, inactivated virus (A/California/07/2009 H1N1)) that would be tested for the capability to agglutinate the red blood cells. The first well of each row was loaded with antigen (100 μL) containing 1.5 μg of HA, or an amount of MS2-SA VLP alone (the negative control) equivalent to the amount of MS2-SA VLP in the VLP-HARegular sample. The remaining wells in each row were filled with PBS (50 μL). Then, antigen solution (50 μL) was taken from the first well of each row and 2-fold serial dilutions were made across the row. Each well was then mixed with the 0.5% red blood cell solution (50 μL), and the plate was incubated at room temperature for at least an hour. The plate was imaged using the ChemiDoc MP imaging system (Bio-Rad).
Statistical Analysis:
For all plotted data, the sample size, measures of center and spread, and statistical testing methods (if relevant) are described in the corresponding figure caption as well as the following text. The ELISA data in Figure 2b are presented as mean ± SD with n=6. In Figure 3a, radius values are presented as the mean ± SD of three measurements, while plotted size distributions are the result of a single representative measurement. Figure 5b includes a plot of endpoint antibody titers elicited in mice immunized with the oriented constructs (n=15 per construct) and mice immunized with VLP (n=12). This data in Figure 5b is presented as geometric mean with geometric standard deviation and significance (****P < 0.0001) was determined by a Mann-Whitney test (α = 0.05). Figure 5c presents reactivity of sera from vaccinated mice by cell-based ELISA in the form of a heatmap, where heatmap intensity is based on the geometric mean of area under the curve (AUC) values of every group of mice against the indicated HA (n = 7 mice immunized with oriented HA constructs; n=6 mice immunized with VLP). In Figure 6a, data are presented as mean ± SEM with n=3. Survival data presented in Figure 6b are representative of three mice (n=3) per dose. All statistical analysis was carried out using GraphPad Prism 9.
Acknowledgements
RSK acknowledges support from the Garry Betty/V Foundation Chair Fund. This work was partially funded by the NIAID Collaborative Influenza Vaccine Innovation Centers (CIVIC) contract 75N93019C00051, the Centers of Excellence for Influenza Research and Response (CEIRR) contract 75N93021C00014, and a Public Health Service Institutional Research Training Award AI07647. Some of this work was performed at the National Center for CryoEM Access and Training (NCCAT) and the Simons Electron Microscopy Center located at the New York Structural Biology Center, supported by the NIH Common Fund Transformative High Resolution Cryo-Electron Microscopy program (U24 GM129539) and by grants from the Simons Foundation (SF349247) and New York State Assembly.
Footnotes
Conflict of Interest
The Icahn School of Medicine at Mount Sinai has filed patent applications relating to influenza virus vaccines which list Florian Krammer as co-inventor. Mount Sinai has spun out a company, Kantaro, to market serological tests for SARS-CoV-2. Florian Krammer has consulted for Merck and Pfizer (before 2020), and is currently consulting for Pfizer, Seqirus, 3rd Rock Ventures, Merck and Avimex. The Krammer laboratory is also collaborating with Pfizer on animal models of SARS-CoV-2 and with Dynavax on universal influenza virus vaccines.
Contributor Information
Steven J. Frey, School of Chemical & Biomolecular Engineering, Georgia Institute of Technology, Atlanta, Georgia, 30332, USA.
Juan Manuel Carreño, Department of Microbiology, Icahn School of Medicine at Mount Sinai, New York, New York, 10029, USA; Center for Vaccine Research and Pandemic Preparedness (C-VARPP), Icahn School of Medicine at Mount Sinai, New York, NY, USA.
Dominika Bielak, Department of Microbiology, Icahn School of Medicine at Mount Sinai, New York, New York, 10029, USA; Center for Vaccine Research and Pandemic Preparedness (C-VARPP), Icahn School of Medicine at Mount Sinai, New York, NY, USA.
Ammar Arsiwala, School of Chemical & Biomolecular Engineering, Georgia Institute of Technology, Atlanta, Georgia, 30332, USA.
Clara Altomare, Department of Microbiology, Icahn School of Medicine at Mount Sinai, New York, New York, 10029, USA.
Chad Varner, School of Chemical & Biomolecular Engineering, Georgia Institute of Technology, Atlanta, Georgia, 30332, USA.
Tania Rosen-Cheriyan, School of Chemical & Biomolecular Engineering, Georgia Institute of Technology, Atlanta, Georgia, 30332, USA.
Goran Bajic, Department of Microbiology, Icahn School of Medicine at Mount Sinai, New York, New York, 10029, USA.
Florian Krammer, Department of Microbiology, Icahn School of Medicine at Mount Sinai, New York, New York, 10029, USA; Center for Vaccine Research and Pandemic Preparedness (C-VARPP), Icahn School of Medicine at Mount Sinai, New York, NY, USA; Department of Pathology, Molecular and Cell Based Medicine, Icahn School of Medicine at Mount Sinai, New York, NY, USA.
Ravi S. Kane, School of Chemical & Biomolecular Engineering, Georgia Institute of Technology, Atlanta, Georgia, 30332, USA Wallace H. Coulter Department of Biomedical Engineering, Georgia Institute of Technology, Atlanta, Georgia, 30332, USA.
Data Availability Statement
The data needed to support the conclusions of this study have been included in the paper. The data that support the plots within the paper are available from the corresponding authors upon reasonable request.
References
- [1].Krammer F, Palese P, Nat Rev Drug Discov 2015, 14, 167. [DOI] [PubMed] [Google Scholar]
- [2].Moody MA, Zhang R, Walter EB, Woods CW, Ginsburg GS, McClain MT, Denny TN, Chen X, Munshaw S, Marshall DJ, Whitesides JF, Drinker MS, Amos JD, Gurley TC, Eudailey JA, Foulger A, DeRosa KR, Parks R, Meyerhoff RR, Yu JS, Kozink DM, Barefoot BE, Ramsburg EA, Khurana S, Golding H, Vandergrift NA, Alam SM, Tomaras GD, Kepler TB, Kelsoe G, Liao HX, Haynes BF, PLoS One 2011, 6, e25797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Wrammert J, Smith K, Miller J, Langley WA, Kokko K, Larsen C, Zheng NY, Mays I, Garman L, Helms C, James J, Air GM, Capra JD, Ahmed R, Wilson PC, Nature 2008, 453, 667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Margine I, Hai R, Albrecht RA, Obermoser G, Harrod AC, Banchereau J, Palucka K, García-Sastre A, Palese P, Treanor JJ, Krammer F, J Virol 2013, 87, 4728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Krammer F, Palese P, Nat Immunol 2014, 15, 3. [DOI] [PubMed] [Google Scholar]
- [6].Palese P, Nat Med 2004, 10, S82. [DOI] [PubMed] [Google Scholar]
- [7].Johnson NP, Mueller J, Bull Hist Med 2002, 76, 105. [DOI] [PubMed] [Google Scholar]
- [8].Heaton NS, Sachs D, Chen CJ, Hai R, Palese P, Proc Natl Acad Sci U S A 2013, 110, 20248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Roubidoux EK, Carreño JM, McMahon M, Jiang K, van Bakel H, Wilson P, Krammer F, mBio 2021, 12, [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Kirkpatrick E, Qiu X, Wilson PC, Bahl J, Krammer F, Sci Rep 2018, 8, 10432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Ekiert DC, Wilson IA, Curr Opin Virol 2012, 2, 134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Tan GS, Krammer F, Eggink D, Kongchanagul A, Moran TM, Palese P, J Virol 2012, 86, 6179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Wang TT, Tan GS, Hai R, Pica N, Petersen E, Moran TM, Palese P, PLoS Pathog 2010, 6, e1000796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Ekiert DC, Bhabha G, Elsliger MA, Friesen RH, Jongeneelen M, Throsby M, Goudsmit J, Wilson IA, Science 2009, 324, 246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Ekiert DC, Friesen RH, Bhabha G, Kwaks T, Jongeneelen M, Yu W, Ophorst C, Cox F, Korse HJ, Brandenburg B, Vogels R, Brakenhoff JP, Kompier R, Koldijk MH, Cornelissen LA, Poon LL, Peiris M, Koudstaal W, Wilson IA, Goudsmit J, Science 2011, 333, 843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Krammer F, Pica N, Hai R, Margine I, Palese P, J Virol 2013, 87, 6542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Throsby M, van den Brink E, Jongeneelen M, Poon LL, Alard P, Cornelissen L, Bakker A, Cox F, van Deventer E, Guan Y, Cinatl J, ter Meulen J, Lasters I, Carsetti R, Peiris M, de Kruif J, Goudsmit J, PLoS One 2008, 3, e3942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Krammer F, Curr Opin Virol 2016, 17, 95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Eggink D, Goff PH, Palese P, J Virol 2014, 88, 699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Deng L, Mohan T, Chang TZ, Gonzalez GX, Wang Y, Kwon YM, Kang SM, Compans RW, Champion JA, Wang BZ, Nat Commun 2018, 9, 359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Impagliazzo A, Milder F, Kuipers H, Wagner MV, Zhu X, Hoffman RM, van Meersbergen R, Huizingh J, Wanningen P, Verspuij J, de Man M, Ding Z, Apetri A, Kukrer B, Sneekes-Vriese E, Tomkiewicz D, Laursen NS, Lee PS, Zakrzewska A, Dekking L, Tolboom J, Tettero L, van Meerten S, Yu W, Koudstaal W, Goudsmit J, Ward AB, Meijberg W, Wilson IA, Radosevic K, Science 2015, 349, 1301. [DOI] [PubMed] [Google Scholar]
- [22].Lu Y, Welsh JP, Swartz JR, Proc Natl Acad Sci U S A 2014, 111, 125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Mallajosyula VV, Citron M, Ferrara F, Lu X, Callahan C, Heidecker GJ, Sarma SP, Flynn JA, Temperton NJ, Liang X, Varadarajan R, Proc Natl Acad Sci U S A 2014, 111, E2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Yassine HM, Boyington JC, McTamney PM, Wei CJ, Kanekiyo M, Kong WP, Gallagher JR, Wang L, Zhang Y, Joyce MG, Lingwood D, Moin SM, Andersen H, Okuno Y, Rao SS, Harris AK, Kwong PD, Mascola JR, Nabel GJ, Graham BS, Nat Med 2015, 21, 1065. [DOI] [PubMed] [Google Scholar]
- [25].Bernstein DI, Guptill J, Naficy A, Nachbagauer R, Berlanda-Scorza F, Feser J, Wilson PC, Solórzano A, Van der Wielen M, Walter EB, Albrecht RA, Buschle KN, Chen Y.-q., Claeys C, Dickey M, Dugan HL, Ermler ME, Freeman D, Gao M, Gast C, Guthmiller JJ, Hai R, Henry C, Lan LY-L, McNeal M, Palm A-KE, Shaw DG, Stamper CT, Sun W, Sutton V, Tepora ME, Wahid R, Wenzel H, Wohlbold TJ, Innis BL, García-Sastre A, Palese P, Krammer F, The Lancet Infectious Diseases 2020, 20, 80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Liu WC, Nachbagauer R, Stadlbauer D, Solorzano A, Berlanda-Scorza F, Garcia-Sastre A, Palese P, Krammer F, Albrecht RA, Front Immunol 2019, 10, 756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Knight M, Changrob S, Li L, Wilson PC, Immunol Rev 2020, 296, 191. [DOI] [PubMed] [Google Scholar]
- [28].Jegaskanda S, Andrews SF, Wheatley AK, Yewdell JW, McDermott AB, Subbarao K, JCI Insight 2019, 4, [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Angeletti D, Kosik I, Santos JJS, Yewdell WT, Boudreau CM, Mallajosyula VVA, Mankowski MC, Chambers M, Prabhakaran M, Hickman HD, McDermott AB, Alter G, Chaudhuri J, Yewdell JW, Proc Natl Acad Sci U S A 2019, 116, 13474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Andrews SF, Huang Y, Kaur K, Popova LI, Ho IY, Pauli NT, Henry Dunand CJ, Taylor WM, Lim S, Huang M, Qu X, Lee JH, Salgado-Ferrer M, Krammer F, Palese P, Wrammert J, Ahmed R, Wilson PC, Sci Transl Med 2015, 7, 316ra192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Avnir Y, Tallarico AS, Zhu Q, Bennett AS, Connelly G, Sheehan J, Sui J, Fahmy A, Huang CY, Cadwell G, Bankston LA, McGuire AT, Stamatatos L, Wagner G, Liddington RC, Marasco WA, PLoS Pathog 2014, 10, e1004103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Buckland B, Boulanger R, Fino M, Srivastava I, Holtz K, Khramtsov N, McPherson C, Meghrous J, Kubera P, Cox MM, Vaccine 2014, 32, 5496. [DOI] [PubMed] [Google Scholar]
- [33].Castro A, Carreno JM, Duehr J, Krammer F, Kane RS, Adv Healthc Mater 2021, e2002140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Chiba S, Frey SJ, Halfmann PJ, Kuroda M, Maemura T, Yang JE, Wright ER, Kawaoka Y, Kane RS, Communications Biology 2021, 4, [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Frietze KM, Peabody DS, Chackerian B, Curr Opin Virol 2016, 18, 44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Valegard K, Murray JB, Stockley PG, Stonehouse NJ, Liljas L, Nature 1994, 371, 623. [DOI] [PubMed] [Google Scholar]
- [37].Hashemi K, Ghahramani Seno MM, Ahmadian MR, Malaekeh-Nikouei B, Bassami MR, Dehghani H, Afkhami-Goli A, Sci Rep 2021, 11, 19851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Peabody DS, Manifold-Wheeler B, Medford A, Jordan SK, do Carmo Caldeira J, Chackerian B, J Mol Biol 2008, 380, 252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Caldeira JC, Peabody DS, J Nanobiotechnology 2011, 9, 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Angeletti D, Gibbs JS, Angel M, Kosik I, Hickman HD, Frank GM, Das SR, Wheatley AK, Prabhakaran M, Leggat DJ, McDermott AB, Yewdell JW, Nat Immunol 2017, 18, 456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Mohsen MO, Bachmann MF, Cell Mol Immunol 2022, 19, 993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Margine I, Palese P, Krammer F, J Vis Exp 2013, e51112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Fairhead M, Krndija D, Lowe ED, Howarth M, J Mol Biol 2014, 426, 199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Howarth M, Ting AY, Nat Protoc 2008, 3, 534. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data needed to support the conclusions of this study have been included in the paper. The data that support the plots within the paper are available from the corresponding authors upon reasonable request.