Abstract
Introduction
A genetic variant explaining a part of the exposure of many kinase inhibitors (KIs) is the single nucleotide polymorphism (SNP) CYP3A4*22, resulting in less CYP3A4 enzyme activity. The primary aim of this study was to investigate if the systemic exposure is non-inferior after a dose reduction of KIs metabolized by CYP3A4 in CYP3A4*22 carriers compared to patients without this SNP (i.e., wildtype patients) receiving the standard dose.
Methods
In this multicenter, prospective, non-inferiority study, patients were screened for the presence of CYP3A4*22. Patients with the CYP3A4*22 SNP received a 20–33% dose reduction. At steady state, a pharmacokinetic (PK) analysis was performed and compared to the PK results from wildtype patients treated with the registered dose using a two-stage individual patient data meta-analysis approach.
Results
In total, 207 patients were included in the final analysis. The CYP3A4*22 SNP was found in 16% of the patients in the final analysis (n = 34). Most of the included patients received imatinib (37%) or pazopanib (22%) treatment. The overall geometric mean ratio (GMR) comparing the exposure of the CYP3A4*22 carriers to the exposure of the wildtype CYP3A4 patients was 0.89 (90% confidence interval: 0.77–1.03).
Conclusion
Non-inferiority could not be proven for dose reduction of KIs metabolized by CYP3A4 in CYP3A4*22 carriers compared to the registered dose in wildtype patients. Therefore, an up-front dose reduction based upon the CYP3A4*22 SNP for all KIs does not seem an eligible new way of personalized therapy.
Trial Registration
International Clinical Trials Registry Platform Search Portal; number NL7514; registered 11/02/2019.
Supplementary Information
The online version contains supplementary material available at 10.1007/s40262-023-01260-4.
Key Points
| The SNP CYP3A4*22 results in a decreased CYP3A4 enzyme activity, which is involved in the metabolism of several drugs. |
| CYP3A4*22 carriers receiving an up-front 20–33% dose reduction of a CYP3A4 metabolized KI did not have a non-inferior exposure compared to wildtype patients receiving the registered dose. |
| CYP3A4*22 genotype-guided dosing does not seem a suitable dosing strategy for all CYP3A4 metabolized KIs. |
Introduction
Kinase inhibitors (KIs) are important in various cancer treatments. Kinase inhibitors can inhibit kinases, which are responsible for many cell functions, including cell signaling, growth, and division. Although KI treatment has improved the survival of many cancer types over the last decades, side effects are common and severe toxicity still raises the concern of patients and their treating physicians.
For many KIs, the incidence and severity of the toxicity are related to drug exposure [1]. For example, higher imatinib exposure has been associated with anemia, neutropenia, leukopenia, and thrombocytopenia [2], while a higher pazopanib exposure has been associated with hypertension [3], and a high sunitinib exposure has been associated with more dose-limiting and grade ≥ 3 toxicities [4, 5]. Besides, KI exposure has a high inter- and intra-patient variability [6, 7]. This high variability is caused by several factors. Variability in the rate of drug metabolism is one of the factors [8].
Most KIs are predominantly metabolized by the CYP3A4 enzyme [9]. The CYP3A4 iso-enzyme forms a major part of the cytochrome P450 enzyme family [10]. The activity of CYP3A4 is highly variable between patients [11, 12]. A genetic variant explaining a part of the variability in the CYP3A4 activity is the CYP3A4 intron 6 single nucleotide polymorphism (SNP) (rs35599367C >T or CYP3A4*22) [13, 14]. Due to this SNP, the formation of a non-functional splice variant of CYP3A4 is increased and consequently CYP3A4 protein levels and enzyme activity are decreased. The minor allelic frequency (MAF) for CYP3A4*22 is 5.0% in Europeans, 2.6% in the admixed American population and < 1% for the Asian and African population [15]. The phenotypic prevalence of CYP3A4*22 carriers in Europeans is thereby estimated to be around 10%.
As a result of the reduced CYP3A4 enzyme activity, the drug exposure of CYP3A4*22 carriers who are treated with a CYP3A4 metabolized KI is higher compared to patients without this SNP (i.e., wildtype patients). This was acknowledged in several previous studies. For example, in a retrospective study including 114 patients treated with sunitinib, a 22.5% decrease in clearance of sunitinib was found for CYP3A4*22 carriers [16]. Furthermore, in a population-pharmacokinetic model with data from 97 patients treated with pazopanib, a substantial lower clearance of 35% was found for CYP3A4*22 carriers [17]. Moreover, the performed simulations estimated that trough concentrations at steady state are 50% higher in CYP3A4*22 carriers [17]. A higher KI exposure caused by CYP3A4*22 could possibly lead to more toxicity given the frequently observed exposure-toxicity relationships in anticancer drugs with a narrow therapeutic index [18].
The aim of the current study was to demonstrate that a dose reduction of KIs metabolized by CYP3A4 in CYP3A4*22 carriers does not result in a lower exposure compared to wildtype patients receiving the standard dose. The secondary aim was to compare the incidence of toxicity and dose modifications after pharmacokinetic assessment between CYP3A4*22 carriers and wildtype patients.
Methods
Study Drugs
Oral targeted anti-cancer drugs were included in the study protocol if drugs were primarily metabolized by CYP3A4, defined as an increase of at least 25% on area under the curve (AUC) when the KI was co-administered with a strong CYP3A4 inhibitor, like ketoconazole or itraconazole, according to the Summary of Product Characteristics (SmPC) of the drug. Eventually, 19 drugs were included in this study (Table 1).
Table 1.
Overview of all study drugs and number of included patients per drug
| Study drug | Total patients | CYP3A4*22 carriers |
|---|---|---|
| Brigatinib | 1 | 0 |
| Cabozantinib | 15 | 3 |
| Cobimetinib | 4 | 0 |
| Dabrafenib | 40 | 5 |
| Dasatinib | 0 | 0 |
| Encorafenib | 5 | 0 |
| Erlotinib | 0 | 0 |
| Everolimus | 17 | 0 |
| Imatinib (400 mg starting dose only) | 92 | 11 |
| Lapatinib | 0 | 0 |
| Nilotinib | 0 | 0 |
| Olaparib | 45 | 1 |
| Osimertinib | 22 | 3 |
| Palbociclib | 3 | 0 |
| Pazopanib | 58 | 12 |
| Ponatinib | 0 | 0 |
| Regorafenib | 7 | 0 |
| Ruxolitinib | 1 | 0 |
| Sunitinib | 41 | 3 |
| Total | 351 | 38 |
Patient Selection and Study Procedures
All adult cancer patients with an indication to start treatment (or have started treatment less than 7 days ago) with one of the study drugs described in Table 1 with a WHO performance status ≤ 2, not using medication or (herbal) supplements, which are known or suspected to strongly inhibit or induct the CYP3A4 enzymes, and who were able and willing to undergo blood sampling for pharmacokinetic (PK) and pharmacogenetic (PG) analysis, met the study selection criteria.
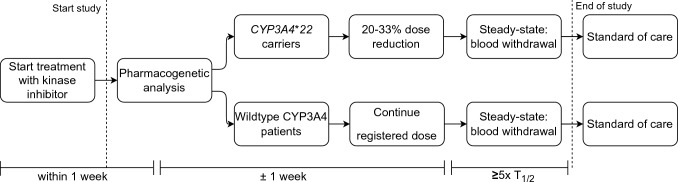
The study procedures are depicted in Fig. 1. All included patients were screened for the presence of the CYP3A4*22 SNP using PG analysis. Due to the available formulations, it was not possible to apply a uniform dose reduction for all drugs. Therefore, depending on available tablet/capsule forms, a dose reduction between 20 and 33% was chosen per drug. Within the group of CYP3A4*22 carriers treated with the same drug the same dose reduction was applied. Since no in vivo data regarding differences in CYP3A4 activity between heterozygous and homozygous CYP3A4*22 carriers is available, the same dose reduction was applied for these groups. The drug dose was not adjusted based on other CYP3A4 SNPs. The dose reduction lasted for 2–4 weeks (or at least five times the drug elimination half-life) after reporting of the SNP status to guarantee a steady-state exposure. Wildtype patients were treated with the standard dose in accordance with the SmPC of the European Medicine Agency (EMA). At steady state, a blood sample was collected for the PK analysis. Patients were instructed to obtain a PK sample directly prior to drug intake (i.e., Ctrough) at steady state. If withdrawal of the PK sample at Ctrough was impossible for logistical reasons, a blood sample for PK analysis was collected after reaching the time of maximum drug concentration (i.e., Tmax). Samples taken after Tmax were extrapolated to a calculated Ctrough using the time after dose, drug elimination half-life reported in the SmPC, and dose interval, by log-linear extrapolation [19]. Patient characteristics, toxicity data, and laboratory results were collected at baseline and during the study period. The study ended after withdrawal of blood for the PK analysis and patients continued treatment with the standard dose or reduced dose according to physicians’ choice. The 4 participating centers were Erasmus MC Cancer Institute, the Netherlands Cancer Institute, Leiden University Medical Centre and ADRZ Goes. Ethical approval was obtained from the Medical Ethical Committee of the Erasmus Medical Center and the study protocol was also approved by the board of directors of each participating center. The trial was conducted in accordance with the International Conference on Harmonization Good Clinical Practice guidelines, applicable regulations, and the Declaration of Helsinki. The trial was registered in the Netherlands Trial Register (International Clinical Trials Registry Platform Search Portal [trialsearch.who.int]; number NL7514). All patients provided written informed consent.
Fig. 1.
Timeline of the study procedures
Pharmacogenetic and Pharmacokinetic Analysis
All PG analyses were executed at the Erasmus University Medical Center. A blood sample of 4 mL was collected and isolated with the Nucleic Acid Isolation Kit I on the MagNA Pure Compact Instrument (Roche Diagnostics, Almere, The Netherlands). Genotyping was performed with the INFINITI® CYP450 3A4-3A5 Assay on the INFINITI® High Throughput System (HTS) (AutoGenomics Inc., Vista, CA USA). For confirmation, a 5′ nuclease assay qPCR (TaqMan™) using a Real-Time Quantstudio PCR System (ThermoFisher, Waltham, MA, US) was executed.
For the PK analysis, a 4 mL blood sample was collected after reaching steady state. Pharmacokinetic samples were processed to plasma or serum depending on the bioanalytical assay and stored at T < − 70 °C until the time of analysis. Samples were analyzed in the local laboratory or transferred to the laboratory of another study site, depending on the study drug and availability of a validated liquid chromatography tandem mass spectrometry (LC-MS/MS) assay [20–24]. All bioanalytical methods were validated according to US FDA guidelines [25]. Pharmacokinetic measurements were performed in the laboratory of Translational Pharmacology, Department of Medical Oncology at the Erasmus Medical Center, the laboratory of Clinical Pharmacy and Toxicology at the Leiden University Medical Centre or the Bioanalytical Laboratory of the Pharmacy at the Netherlands Cancer Institute. Inter-laboratory comparisons were performed to guarantee interchangeability of the PK measurements.
Sample Size and Statistical Analyses
The aim of the study was to demonstrate that the trough level of CYP3A4*22 carriers with a reduced dose is non-inferior to the trough level of non-carriers with a standard dose. The trough level was assumed to follow a log-normal distribution. With a non-inferiority margin of − 20% in relative difference (RD) (i.e., a difference of − 0.2231 on log-transformed data) and an average coefficient of variation of 40% (translated to a standard deviation [SD] of approximately 0.40 on the log-scale), at least 198 patients were required for this study of whom at least 33 had to be a CYP3A4*22 carrier (power of 90% and one-sided alpha of 5%) [26]. Only study drugs with at least two CYP3A4*22 carriers per drug were included in the final analysis. Inclusion of patients stopped when all these three criteria were met (i.e., at least 33 CYP3A4*22 carriers, at least two CYP3A4 carriers per drug, at least 165 CYP3A4 wildtype patients).
The two-stage individual patient data (IPD) meta-analysis was applied considering that several drugs were included. Each study drug was seen as a separate study within this analysis. Hereto, the ‘ipdmetan’ package in Stata was used [27]. Non-inferiority of the trough level was evaluated by looking at whether the lower bound of the two-sided 90% confidence interval (CI) for difference between carriers and non-carriers was higher than the non-inferiority boundary of −0.2231 on the log-scale (this boundary is equivalent to a − 20% relative difference and a geometric mean ratio of 0.80).
Non-inferiority regarding drug exposure was also analyzed for the two study drugs with the most CYP3A4*22 carriers separately (i.e., pazopanib and imatinib). Pharmacokinetic data were log normalized before performing an independent sample t-test. Geometric mean ratio and corresponding 90% CI, interpreted as the RD in percentage, were calculated by exponentiation of the mean difference and 90% CI of this difference derived from the independent sample t-test. Exposure was regarded as non-inferior if the lower boundary of the CI of the geometric mean ratio was higher than 0.8 (i.e., equivalent to a −20% relative difference). For these compounds, the incidence of patients with an exposure above TDM targets proposed by Verheijen et al were compared between the CYP3A4*22 carriers and the wildtype patients by the Fisher’s exact test [28].
For the Hardy-Weinberg equilibrium, the distribution of genotypes was tested by a chi-squared test. Severe or medically significant toxicities (i.e., Grade ≥ 3 based on Common Terminology Criteria for Adverse Events [CTCAE] version 4.03) were described with numbers of patients and percentages for CYP3A4*22 carriers and wildtype patients separately. Incidence of dose modifications at the end of study period was described using descriptive statistics.
Results
Patient Characteristics
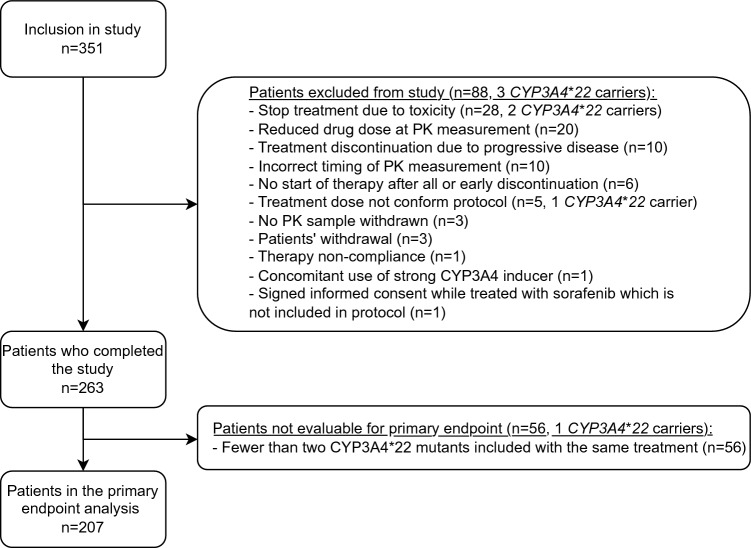
In total, 351 patients were included in the study (Fig. 2), 88 of whom were excluded from the analysis. The most common reasons for exclusion were preliminary stop with the study drug due to toxicity (n = 28: 26 wildtype patients, two CYP3A4*22 carriers) or progressive disease (n = 10; 9 wild-type patients, one unknown), a dose reduction not according to protocol (n = 20; 20 wildtype patients) and incorrect timing of the blood draw for PK analysis (n = 10). Furthermore, another 56 patients did complete the study successfully but were not evaluable for the primary endpoint because fewer than two CYP3A4*22 carriers treated with the same drug were included in the study group (i.e., brigatinib, cobimetinib, encorafenib, everolimus, olaparib, palbociclib, regorafenib, ruxolitinib). Therefore, 207 patients were eventually included in the final analysis.
Fig. 2.
Study flowchart
The median age of the patients included in the final analysis (n = 207) was 64 years and 54% were male (Table 2). Most patients had a WHO performance status of 0 (46%) or 1 (43%), 72% had metastatic disease, and 58% had not received systemic treatment before. More than half of the patients included in the final analysis received imatinib (37%) or pazopanib (22%), followed by dabrafenib (15%), sunitinib (12%), osimertinib (10%) and cabozantinib (4%). The majority of the CYP3A4*22 carriers were treated with pazopanib (32%) or imatinib (29%). Table 2 depicts an extensive summary of the baseline characteristics.
Table 2.
Baseline characteristics of patients included in final analysis
| Characteristic | All patients (n = 207) | Wildtype CYP3A4 patients (n = 173) | CYP3A4*22 carriers (n = 34) |
|---|---|---|---|
| Sex | |||
| Female | 95 (46%) | 75 (43%) | 20 (59%) |
| Male | 112 (54%) | 98 (57%) | 14 (41%) |
| Ethnic origin | |||
| White | 188 (91%) | 155 (90%) | 33 (97%) |
| Asian | 6 (3%) | 6 (3%) | – |
| Arabic | 6 (3%) | 6 (3%) | – |
| Black | 3 (2%) | 3 (2%) | – |
| Other | 4 (2%)a | 3 (2%)b | 1 (3%)c |
| Age (at inclusion) | |||
| Median [IQR] in years | 64 [55–73] | 65 [56–73] | 62 [55–72] |
| WHO performance score | |||
| 0 | 96 (46%) | 79 (46%) | 17 (50%) |
| 1 | 89 (43%) | 75 (43%) | 14 (41%) |
| 2 | 22 (11%) | 19 (11%) | 3 (9%) |
| CYP3A4*22 status | |||
| No CYP3A4 *22 carrier | 173 (84%) | 173 (100%) | NA |
| Heterozygote CYP3A4 *22 carrier | 31 (15%) | NA | 31 (91%) |
| Homozygote CYP3A4 *22 carrier | 3 (1%) | NA | 3 (9%) |
| Other CYP3A4 SNPs | |||
| CYP3A4 *1/*1B | 14 (6.8%) | 14 (8%) | – |
| CYP3A4 *1B/*1B | 1 (0.5%) | 1 (0.6%) | – |
| CYP3A4 *1/*3 | 1 (0.5%) | 1 (0.6%) | – |
| CYP3A4 *1/*12 | 1 (0.5%) | 1 (0.6%) | – |
| CYP3A4 *3/*22 | 5 (2.4%) | NA | 5 (15%) |
| No other SNPs | 185 (89%) | 156 (90%) | 29 (85%) |
| Drug | |||
| Cabozantinib | 9 (4%) | 6 (4%) | 3 (9%) |
| Dabrafenib | 30 (15%) | 26 (15%) | 4 (12%) |
| Imatinib (400 mg starting dose only) | 76 (37%) | 66 (38%) | 10 (29%) |
| Osimertinib | 21 (10%) | 18 (10%) | 3 (9%) |
| Pazopanib | 46 (22%) | 35 (20%) | 11 (32%) |
| Sunitinib | 25 (12%) | 22 (13%) | 3 (9%) |
| Primary malignancy | |||
| GIST | 85 (41%) | 75 (43%) | 10 (29%) |
| Sarcoma | 46 (22%) | 35 (20%) | 11 (32%) |
| Melanoma | 26 (13%) | 23 (13%) | 3 (9%) |
| Lung cancer | 25 (12%) | 21 (12%) | 4 (12%) |
| Renal cell carcinoma | 24 (12%) | 18 (10%) | 6 (18%) |
| Neuroendocrine tumor | 1 (1%) | 1 (1%) | 0 (0%) |
| Metastatic disease | |||
| Yes | 148 (71%) | 125 (72%) | 23 (68%) |
| No | 59 (29%) | 48 (28%) | 11 (32%) |
| Prior systemic treatment | |||
| Yes | 87 (42%) | 71 (41%) | 16 (47%) |
| No | 120 (58%) | 102 (59%) | 18 (53%) |
GIST gastro-intestinal stromal tumor, IQR interquartile range, NA not applicable, SNP single nucleotide polymorphism, WHO World Health Organization
aMixed racial parentage (n = 2), latino (n = 1), unknown ethnic origin (n = 1)
bMixed racial parentage (n = 1), latino (n = 1), unknown ethnic origin (n = 1)
cMixed racial parentage (n = 1)
Pharmacogenetics
From all 351 patients who were included in the study, 38 CYP3A4*22 carriers were identified (11%) (Table 1). The CYP3A4*22 SNP was in Hardy–Weinberg equilibrium (p = 0.08). The MAF of the SNP in all these patients was 6%. From the 207 patients included in the final analysis, 34 patients were CYP3A4*22 carrier (16%) (Table 2). Most of these patients had a heterozygote CYP3A4*22 genotype (15%), but three patients were homozygote for the SNP CYP3A4*22 (1%) who were treated with a dose conform the heterozygote CYP3A4*22 carriers. Exact genotypes observed in the patients included in the primary analysis are depicted in Table 2. Notably, from the nine included cabozantinib patients, three were CYP3A4*22 carrier (33%) and from the 46 pazopanib patients, 11 were CYP3A4*22 carrier (24%), both of which are considerably higher proportions than those reported in the other study drugs (osimertinib 3/21 [14%], dabrafenib 4/30 [13%], imatinib 10/76 [13%], sunitinib 3/25 [12%]).
Results of Pharmacokinetic Analysis
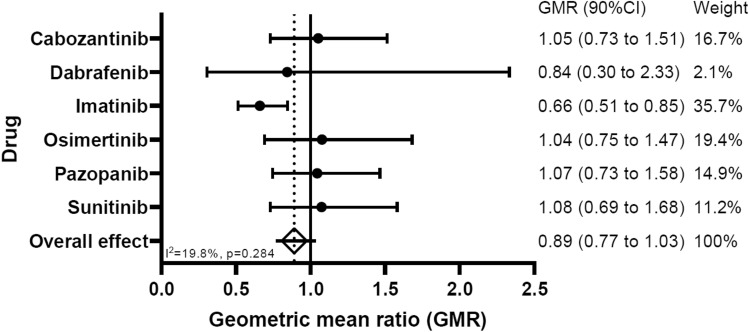
True Ctrough measurements were taken in 35% of all PK samples. For the other 65% of the samples, the Ctrough was estimated [19, 29]. An overview of the geometric mean Ctrough and the 95% CI of the geometric mean per drug is given in Table 3. The overall geometric mean ratio comparing the exposure of the CYP3A4*22 carriers to the exposure of the wildtype CYP3A4 patients is 0.89 (90% CI 0.77–1.03). This is equal to a RD in Ctrough exposure (CYP3A4*22 vs wildtype CYP3A4) of − 11.0% (90% CI RD: − 23 to 3.3%). Figure 3 represents a forest plot of the two-stage IPD meta-analysis approach with the effect sizes of the individual drugs (i.e., geometric mean ratios of Ctrough and their 90% CIs), the weight of each drug on the overall effect and the overall effect size.
Table 3.
Overview of the geometric mean Ctrough and 95% CI of the GM per drug
| Drug | Wildtype CYP3A4 | CYP3A4*22 carriers | ||
|---|---|---|---|---|
| GM Ctrough | 95% CI GM | GM Ctrough | 95% CI GM | |
| Cabozantinib | 1063 ng/mL | 728–1550 | 1,117 ng/mL | 796–1568 |
| Dabrafenib | 120 ng/mL | 74–195 | 101 ng/mL | 39–262 |
| Imatinib | 1200 ng/mL | 1072–1345 | 792 ng/mL | 636–985 |
| Osimertinib | 219 ng/mL | 177–270 | 235 ng/mL | 72–766 |
| Pazopanib | 27 mg/L | 22–34 | 28 mg/L | 21–39 |
| Sunitinib | 61 ng/mL | 51–73 | 66 ng/mL | 40–107 |
CI confidence interval, GM geometric mean
Fig. 3.
Forest plot of the two-stage individual patient data meta-analysis
Pharmacokinetics of Pazopanib
In total, 46 patients treated with pazopanib were included in the final analysis (22%). Ten patients were genotyped CYP3A4*1/*22 while one patient was CYP3A4*22/*22. The other 35 patients were wildtype patients.
The geometric mean Ctrough of the CYP3A4*22 carriers treated with 600 mg once daily (OD) was 28.3 mg/L (95% C: 20.6–39.0 mg/L) while the geometric mean Ctrough of the wildtype patients treated with 800 mg OD was 27.1 mg/L (95% CI 21.9–33.6 mg/L). The geometric mean ratio comparing the Ctrough of CYP3A4*22 carriers versus wildtype patients was 1.05 (90% CI 0.74–1.48).
In the group of CYP3A4*22 carriers, four patients (36%) had a Ctrough exposure below the earlier proposed efficacy threshold of 20.5 mg/L [28], compared to 34% (n = 12) of the wildtype pazopanib patients (p = 1.00).
Pharmacokinetics of Imatinib
In total, 76 patients treated with imatinib were included in the final analysis (37%). Ten patients were genotyped as heterozygote CYP3A4*22 carriers and 66 as wildtype patients.
The geometric mean Ctrough of the CYP3A4*22 carriers treated with imatinib 300 mg OD was 792 ng/mL (95% CI 636–986 ng/mL). The geometric mean Ctrough of the wildtype patients treated with imatinib 400 mg OD was 1201 ng/mL (95% CI 1071–1345 ng/mL). The geometric mean ratio comparing the Ctrough of CYP3A4*22 carriers versus wildtype patients was 0.66 (90% CI 0.51–0.85).
In the group of CYP3A4*22 carriers, nine out of ten patients (90%) had a Ctrough exposure below the adopted efficacy threshold of 1100 ng/mL [28], while the incidence of patients with a exposure below the target was 45% (n = 30) in the wildtype patients (p = 0.014).
Toxicity and Incidence of Dose Modifications
Overall, during the study period (i.e., after the baseline screening until the end of study) eight (24%) CYP3A4*22 carriers presented with grade ≥ 3 adverse events compared to 30 (17%) patients with grade ≥ 3 adverse events in the wildtype group. An overview of the grade ≥ 3 adverse events occurring during the study period (i.e., after the baseline screening until the end of study) is presented in Table 4.
Table 4.
Overview of grade ≥3 toxicity
| Grade ≥ 3 toxicity | Wildtype CYP3A4 patients (n = 173) | CYP3A4*22 carriers (n = 34) | All patients (n = 207) |
|---|---|---|---|
| Hyperglycemia | 3 (1.7%) | 1 (2.9%) | 4 (1.9%) |
| eGFR decrease | 1 (0.6%) | – | 1 (0.5%) |
| Hyponatremia | 4 (2.3%) | 1 (2.9%) | 5 (2.4%) |
| Hypomagnesemia | 1 (0.6%) | – | 1 (0.5%) |
| Hypoalbuminemia | 1 (0.6%) | – | 1 (0.5%) |
| AST increase | 3 (1.7%) | – | 3 (1.4%) |
| ALT increase | 3 (1.7%) | – | 3 (1.4%) |
| GGT increase | 6 (3.5%) | 1 (2.9%) | 7 (3.4%) |
| ALP increase | 3 (1.7%) | – | 3 (1.4%) |
| Bilirubin increase | 2 (1.2%) | 1 (2.9%) | 3 (1.4%) |
| Anemia | 2 (1.2%) | – | 2 (1%) |
| Thrombocytopenia | – | 1 (2.9%) | 1 (0.5%) |
| Leukocytopenia | 1 (0.6%) | – | 1 (0.5%) |
| Neutropenia | 1 (0.6%) | – | 1 (0.5%) |
| Nausea | 1 (0.6%) | – | 1 (0.5%) |
| Vomiting | 1 (0.6%) | – | 1 (0.5%) |
| Mucositis | 1 (0.6%) | – | 1 (0.5%) |
| Diarrhea | 1 (0.6%) | – | 1 (0.5%) |
| Sensory neuropathy | 1 (0.6%) | 1 (2.9%) | 2 (1.0%) |
| Motoric neuropathy | – | 1 (2.9%) | 1 (0.5%) |
| Fatigue | 1 (0.6%) | – | 1 (0.5%) |
| Fever | 1 (0.6%) | – | 1 (0.5%) |
| Rash | 1 (0.6%) | – | 1 (0.5%) |
| Pain | 2 (1.2%) | 1 (2.9%) | 3 (1.4%) |
| Hypertension | 6 (3.5%) | 3 (8.8%) | 9 (4.3%) |
| Vasculitis | 1 (0.6%) | – | 1 (0.5%) |
ALP alkaline phosphatase, ALT alanine transaminase, AST aspartate transaminase, eGFR estimated glomerular filtration rate, GGT gamma glutamyl transferase
For the patients treated with imatinib, grade ≥3 adverse events were observed in 12% (n = 8) of the wildtype patients compared to 10% (n = 1) of the CYP3A4*22 carriers. Of the wildtype patients treated with pazopanib, 29% (n = 10) reported grade ≥3 adverse events while 36% (n = 4) of the CYP3A4*22 carriers reported grade ≥3 adverse events. Supplementary Tables 1 and 2 summarize the grade ≥3 adverse events observed during the study period (i.e., after the baseline screening until the end of study) of the patients treated with imatinib and pazopanib, respectively.
Of the patients enrolled in the study (n = 351), 10 patients (9 wildtype patients, one unknown) discontinued treatment prematurely due to progressive disease. Furthermore, 48 patients (2 CYP3A4*22 carriers and 46 wildtype patients) received a dose reduction or discontinued treatment due to toxicity prior to PK assessment. These patients were excluded from the final analysis (Fig. 2). The two CYP3A4*22 carriers who stopped their treatment due to toxicity, stopped their treatment before their CYP3A4 genotype was known and were therefore treated with the full dose at the time of toxicity/end of therapy. From the patients included in the final analysis (n = 207), the dose was lowered due to toxicity after the pharmacokinetic analysis in three CYP3A4*22 carriers (9%) and 30 wildtype patients (17%). A dose increase was performed in 14 CYP3A4*22 carriers (41%) after the PK analysis in the study: ten based on Ctrough (imatinib n = 8, pazopanib n = 2), three preference patient/physician (osimertinib n = 2, pazopanib n = 1) and one because of disappointing efficacy (dabrafenib n = 1). In 16 wildtype patients (9%) the dose was increased due to low Ctrough and for one patient the dose was increased because of no toxicity.
Discussion
In this prospective study, CYP3A4*22 carriers receiving an up-front 20–33% dose reduction of a CYP3A4 metabolized KI did not have a non-inferior exposure compared to wildtype patients receiving the registered dose. Although a trend towards comparable exposure was seen for patients treated with cabozantinib, pazopanib, sunitinib and osimertinib, the exposure of imatinib was significantly lower in CYP3A4*22 carriers treated with the reduced dose, exposing them to a risk of under-dosing. In terms of toxicity, wildtype patients more often received a dose reduction due to toxicity, but the incidence of grade ≥ 3 adverse events was comparable between CYP3A4*22 carriers and wildtype patients. Based on the results from the current study, standard dose reductions based on CYP3A4*22 genotyping for patients treated with CYP3A4 metabolized KI should not be recommended.
The rationale of performing a dose reduction for CYP3A4*22 carriers was based on results from previous literature. Due to the CYP3A4*22 SNP, the formation of a nonfunctional CYP3A4 alternative splice variant was 2-fold higher, while the CYP3A4*22 genotype resulted in a CYP3A4 mRNA level and CYP3A4 protein level of 58% and 28% of the levels produced with a wildtype CYP3A4 genotype [13, 14, 30]. In CYP3A4*22 carriers the clearance of erythromycin (a CYP3A4 probe drug), sunitinib, and pazopanib, was reduced by 40%, 22.5%, and 35%, respectively [16, 17, 31]. There are several possible explanations why the hypothesis of the study could not be confirmed. First, the current study included many different KIs introducing statistical challenges. It could be that up-front genotyping of CYP3A4 is useful for specific agents, but not for all KIs that are primarily metabolized by CYP3A4. In the current study, the non-inferiority requirements were not met for the entire study group nor for the individual drugs. Imatinib exposure was even significantly lower after dose reduction in CYP3A4*22 carriers in the current study, so initial dose reduction for this drug should not be recommend. The weight of the imatinib results is 35% and thereby has a considerable influence on the overall effect. However, looking at the results from cabozantinib, osimertinib, pazopanib and sunitinib, the lower boundary of the CI of the GMR for these drugs was just below 0.8 and the GMR point estimates above 1.0 (Fig. 3). Although conclusions for individual drugs based on the current analysis are underpowered, these results suggest that a non-inferior exposure might be found for cabozantinib, osimertinib, pazopanib, and sunitinib in a study with a larger sample size. Future studies are warranted to investigate this. Another possible explanation of why the hypothesis was not confirmed in the current study is that all included drugs are predominantly, but not exclusively, metabolized by CYP3A4. For instance, cabozantinib is also partly metabolized by CYP2C9 and dabrafenib and pazopanib by CYP2C8, neither of which were measured or genotyped in this study [32–34]. Drug transporters such as P-glycoprotein or Breast Cancer Resistance Protein can also influence drug exposure of pazopanib and dabrafenib [33, 34]. Variation in activity of other CYP enzymes and drug transporters were not tested in our study but could have influenced the observed drug exposure. In previous studies, the exposure of pazopanib after 600 mg and 800 mg doses was almost comparable despite CYP3A4 genotype [35], which can be attributed to its complex non-linear pharmacokinetics with saturable absorption plateauing at doses above 600 mg. Of note, removal of the patients treated with pazopanib from our final analysis did not change the overall results of our study (results not shown). Contrary to our expectations and results from previous literature, the exposure of imatinib was significantly lower for CYP3A4*22 carriers. Imatinib is a CYP3A4 inhibitor by itself. Due to this auto-inhibition of CYP3A4, the metabolism of imatinib might not solely rely on CYP3A4 but relies on other metabolism routes at steady-state [36]. Therefore the influence of CYP3A4*22 on the exposure of imatinib might be lower than expected.
Surprisingly, the incidence of grade ≥ 3 toxicity did not seem higher in the wildtype patients compared to CYP3A4*22 carriers, but dose reductions due to toxicity after the PK analysis were more often performed in this group. This might partly be explained given that many patients who experienced toxicity received a dose reduction early in the study and were therefore excluded from the final analysis. Adverse events experienced by the excluded patients were not accounted for in the current toxicity analysis, which could have biased the overview of the toxicity in both groups.
This is the first prospective study investigating up-front CYP3A4*22 genotyping and dosing of KIs in cancer patients. The prospective non-inferiority design of the study and the inclusion of multiple KIs are strengths of this study. At the same time, the inclusion of several KIs means that the study was underpowered for analysis of individual drugs. Therefore, it is hard to make statements about the effect of CYP3A4*22 genotyping for each individual KI. Since we considered it very complicated to include a sufficient number of patients for each drug, a two-stage IPD meta-analysis approach was chosen. Besides, patients were not randomized in the current study because of the challenge to include a sufficient number of patients. Furthermore, comparing the toxicity between CYP3A4*22 carriers and wildtype patients introduced a challenge. Every study drug has its own toxicity profile and a different number of patients was included per study drug, making it hard to extrapolate the toxicity data to the entire group. Finally, the proportion of CYP3A4*22 carriers in some of the study drugs was remarkably high (e.g., 33% of the cabozantinib patients was CYP3A4*22 carrier and 24% of the pazopanib patients) compared to the MAF of 5.0% in Europeans reported in previous literature [15].
Although CYP3A4*22 genotype-guided initial dosing does not seem a suitable dosing strategy for all CYP3A4 metabolized KIs based on this study, it still has potential for specific scenarios. For example, for KIs with an established exposure-toxicity relationship but without exposure-response relationship, CYP3A4*22 genotype-guided dosing seems an interesting approach to prevent (unnecessary) toxicity without the risk to clinically relevantly under-dose CYP3A4*22 carriers. Furthermore, it could be useful to determine CYP3A4*22 status for patients who experienced severe toxicity on a previous CYP3A4 metabolized cancer drug, since that might increase the a priori chance of being a CYP3A4*22 carrier with a high exposure on previous therapy. Finally, future studies should focus on the role of CYP3A4*22 genotype-guided dosing for cabozantinib, osimertinib and sunitinib, since the results of these individual drugs in the current study were promising and the non-inferiority requirements might be met for these drugs in future studies with a larger sample size.
Supplementary Information
Below is the link to the electronic supplementary material.
Declarations
Funding
This study was financially supported by unrestricted research grants from de Merel Stichting.
Conflicts of Interests
The authors declare no potential conflicts of interest.
Availability of Data and Material
The datasets generated and/or analysed during the current study are available from the corresponding author on reasonable request.
Ethics Approval
Ethical approval was obtained from the Medical Ethical Committee of the Erasmus Medical Center and the study protocol was also approved by the board of directors of each participating center. The trial was conducted in accordance with the International Conference on Harmonization Good Clinical Practice guidelines, applicable regulations, and the Declaration of Helsinki. The trial was registered in the Netherlands Trial Register (International Clinical Trials Registry Platform Search Portal (trialsearch.who.int); number NL7514).
Consent to Participate
All patients provided written informed consent.
Consent for Publication
Not applicable
Code Availability
Not applicable
Authors' Contributions
Wrote Manuscript: RvE, NIJ, EO, RM, SK. Designed Research: RvE, NIJ, EO, NS, RM, SK. Performed Research: RvE, NIJ, MvM, EO, NG, AV, MM, RvS, PdB, DM, PJ, HG, AH, NS, RM, SK. Analyzed Data: RvE, NIJ, EO. Contributed New Reagents/Analytical Tools: NA.
Footnotes
Ruben A. G. van Eerden, Nikki S. IJzerman have contributed equally.
References
- 1.Mueller-Schoell A, Groenland SL, Scherf-Clavel O, van Dyk M, Huisinga W, Michelet R, et al. Therapeutic drug monitoring of oral targeted antineoplastic drugs. Eur J Clin Pharmacol. 2021;77(4):441–464. doi: 10.1007/s00228-020-03014-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Guilhot F, Hughes TP, Cortes J, Druker BJ, Baccarani M, Gathmann I, et al. Plasma exposure of imatinib and its correlation with clinical response in the Tyrosine Kinase Inhibitor Optimization and Selectivity Trial. Haematologica. 2012;97(5):731–738. doi: 10.3324/haematol.2011.045666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Suttle AB, Ball HA, Molimard M, Hutson TE, Carpenter C, Rajagopalan D, et al. Relationships between pazopanib exposure and clinical safety and efficacy in patients with advanced renal cell carcinoma. Br J Cancer. 2014;111(10):1909–1916. doi: 10.1038/bjc.2014.503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Noda S, Otsuji T, Baba M, Yoshida T, Kageyama S, Okamoto K, et al. Assessment of sunitinib-induced toxicities and clinical outcomes based on therapeutic drug monitoring of sunitinib for patients with renal cell carcinoma. Clin Genitourin Cancer. 2015;13(4):350–358. doi: 10.1016/j.clgc.2015.01.007. [DOI] [PubMed] [Google Scholar]
- 5.Westerdijk K, Krens SD, van der Graaf WTA, Mulder SF, van Herpen CML, Smilde T, et al. The relationship between sunitinib exposure and both efficacy and toxicity in real-world patients with renal cell carcinoma and gastrointestinal stromal tumour. Br J Clin Pharmacol. 2021;87(2):326–335. doi: 10.1111/bcp.14332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Klümpen HJ, Samer CF, Mathijssen RH, Schellens JH, Gurney H. Moving towards dose individualization of tyrosine kinase inhibitors. Cancer Treat Rev. 2011;37(4):251–260. doi: 10.1016/j.ctrv.2010.08.006. [DOI] [PubMed] [Google Scholar]
- 7.Groenland SL, van Eerden RAG, Westerdijk K, Meertens M, Koolen SLW, Moes DJAR, et al. Therapeutic drug monitoring based precision dosing of oral targeted therapies in oncology: a prospective multicentre study. Ann Oncol. 2022;33:1071–1082. doi: 10.1016/j.annonc.2022.06.010. [DOI] [PubMed] [Google Scholar]
- 8.Thummel KE, Lin YS. Sources of interindividual variability. Methods Mol Biol. 2014;1113:363–415. doi: 10.1007/978-1-62703-758-7_17. [DOI] [PubMed] [Google Scholar]
- 9.Duckett DR, Cameron MD. Metabolism considerations for kinase inhibitors in cancer treatment. Expert Opin Drug Metab Toxicol. 2010;6(10):1175–1193. doi: 10.1517/17425255.2010.506873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Danielson PB. The cytochrome P450 superfamily: biochemistry, evolution and drug metabolism in humans. Curr Drug Metab. 2002;3(6):561–597. doi: 10.2174/1389200023337054. [DOI] [PubMed] [Google Scholar]
- 11.Shimada T, Yamazaki H, Mimura M, Inui Y, Guengerich FP. Interindividual variations in human liver cytochrome P-450 enzymes involved in the oxidation of drugs, carcinogens and toxic chemicals: studies with liver microsomes of 30 Japanese and 30 Caucasians. J Pharmacol Exp Ther. 1994;270(1):414–423. [PubMed] [Google Scholar]
- 12.Westlind-Johnsson A, Malmebo S, Johansson A, Otter C, Andersson TB, Johansson I, et al. Comparative analysis of CYP3A expression in human liver suggests only a minor role for CYP3A5 in drug metabolism. Drug Metab Dispos. 2003;31(6):755–761. doi: 10.1124/dmd.31.6.755. [DOI] [PubMed] [Google Scholar]
- 13.Wang D, Guo Y, Wrighton SA, Cooke GE, Sadee W. Intronic polymorphism in CYP3A4 affects hepatic expression and response to statin drugs. Pharmacogenomics J. 2011;11(4):274–286. doi: 10.1038/tpj.2010.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Okubo M, Murayama N, Shimizu M, Shimada T, Guengerich FP, Yamazaki H. CYP3A4 intron 6 C>T polymorphism (CYP3A4*22) is associated with reduced CYP3A4 protein level and function in human liver microsomes. J Toxicol Sci. 2013;38(3):349–354. doi: 10.2131/jts.38.349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhou Y, Ingelman-Sundberg M, Lauschke VM. Worldwide distribution of cytochrome P450 ALLELES: A META-ANALYSIS OF POPULATION-SCALE SEQUENCING PROJECTs. Clin Pharmacol Ther. 2017;102(4):688–700. doi: 10.1002/cpt.690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Diekstra MH, Klümpen HJ, Lolkema MP, Yu H, Kloth JS, Gelderblom H, et al. Association analysis of genetic polymorphisms in genes related to sunitinib pharmacokinetics, specifically clearance of sunitinib and SU12662. Clin Pharmacol Ther. 2014;96(1):81–89. doi: 10.1038/clpt.2014.47. [DOI] [PubMed] [Google Scholar]
- 17.Bins S, Huitema ADR, Laven P, Bouazzaoui SE, Yu H, van Erp N, et al. Impact of CYP3A4*22 on pazopanib pharmacokinetics in cancer patients. Clin Pharmacokinet. 2019;58(5):651–658. doi: 10.1007/s40262-018-0719-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mathijssen RH, Sparreboom A, Verweij J. Determining the optimal dose in the development of anticancer agents. Nat Rev Clin Oncol. 2014;11(5):272–281. doi: 10.1038/nrclinonc.2014.40. [DOI] [PubMed] [Google Scholar]
- 19.van Eerden RAG, Oomen-de Hoop E, Noordam A, Mathijssen RHJ, Koolen SLW. Feasibility of extrapolating randomly taken plasma samples to trough levels for therapeutic drug monitoring purposes of small molecule kinase inhibitors. Pharmaceuticals (Basel) 2021;14(2):119. doi: 10.3390/ph14020119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.van Erp NP, de Wit D, Guchelaar HJ, Gelderblom H, Hessing TJ, Hartigh J. A validated assay for the simultaneous quantification of six tyrosine kinase inhibitors and two active metabolites in human serum using liquid chromatography coupled with tandem mass spectrometry. J Chromatogr B Analyt Technol Biomed Life Sci. 2013;15(937):33–43. doi: 10.1016/j.jchromb.2013.08.013. [DOI] [PubMed] [Google Scholar]
- 21.Herbrink M, de Vries N, Rosing H, Huitema AD, Nuijen B, Schellens JH, et al. Quantification of 11 therapeutic kinase inhibitors in human plasma for therapeutic drug monitoring using liquid chromatography coupled with tandem mass spectrometry. Ther Drug Monit. 2016;38(6):649–656. doi: 10.1097/FTD.0000000000000349. [DOI] [PubMed] [Google Scholar]
- 22.Herbrink M, de Vries N, Rosing H, Huitema ADR, Nuijen B, Schellens JHM, et al. Development and validation of a liquid chromatography-tandem mass spectrometry analytical method for the therapeutic drug monitoring of eight novel anticancer drugs. Biomed Chromatogr. 2018;32(4):e4147. doi: 10.1002/bmc.4147. [DOI] [PubMed] [Google Scholar]
- 23.Janssen JM, de Vries N, Venekamp N, Rosing H, Huitema ADR, Beijnen JH. Development and validation of a liquid chromatography-tandem mass spectrometry assay for nine oral anticancer drugs in human plasma. J Pharm Biomed Anal. 2019;10(174):561–566. doi: 10.1016/j.jpba.2019.06.034. [DOI] [PubMed] [Google Scholar]
- 24.Bruin MAC, de Vries N, Lucas L, Rosing H, Huitema ADR, Beijnen JH. Development and validation of an integrated LC-MS/MS assay for therapeutic drug monitoring of five PARP-inhibitors. J Chromatogr B Analyt Technol Biomed Life Sci. 2020;1(1138):121925. doi: 10.1016/j.jchromb.2019.121925. [DOI] [PubMed] [Google Scholar]
- 25.Center for Drug Evaluation and Research Center for Veterinary Medicine, Bioanalytical Method Validation Guidance for Industry, Food and Drug Administration (FDA), FDA-2013-D-1020 . 2018.
- 26.HyLown Consulting LLC. Compare 2 Means: 2-Sample Non-Inferiority or Superiority. [cited 2019. http://powerandsamplesize.com/Calculators/Compare-2-Means/2-Sample-Non-Inferiority-or-Superiority. Accessed Oct 2018.
- 27.Fisher D. Two-stage individual participant data meta-analysis and generalized forest plots. Stata J. 2015;15(2):369–396. doi: 10.1177/1536867X1501500203. [DOI] [Google Scholar]
- 28.Verheijen RB, Yu H, Schellens JHM, Beijnen JH, Steeghs N, Huitema ADR. Practical recommendations for therapeutic drug monitoring of kinase inhibitors in oncology. Clin Pharmacol Ther. 2017;102(5):765–776. doi: 10.1002/cpt.787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang Y, Chia YL, Nedelman J, Schran H, Mahon FX, Molimard M. A therapeutic drug monitoring algorithm for refining the imatinib trough level obtained at different sampling times. Ther Drug Monit. 2009;31(5):579–584. doi: 10.1097/FTD.0b013e3181b2c8cf. [DOI] [PubMed] [Google Scholar]
- 30.Wang D, Sadee W. CYP3A4 intronic SNP rs35599367 (CYP3A4*22) alters RNA splicing. Pharmacogenet Genomics. 2016;26(1):40–43. doi: 10.1097/FPC.0000000000000183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Elens L, Nieuweboer A, Clarke SJ, Charles KA, de Graan AJ, Haufroid V, et al. CYP3A4 intron 6 C>T SNP (CYP3A4*22) encodes lower CYP3A4 activity in cancer patients, as measured with probes midazolam and erythromycin. Pharmacogenomics. 2013;14(2):137–149. doi: 10.2217/pgs.12.202. [DOI] [PubMed] [Google Scholar]
- 32.European Medicines Agency. Summary of product characteristics cabozantinib.
- 33.European Medicines Agency. Summary of product characteristics dabrafenib.
- 34.European Medicines Agency. Summary of product characteristics pazopanib.
- 35.Lubberman FJE, Gelderblom H, Hamberg P, Vervenne WL, Mulder SF, Jansman FGA, et al. The Effect of Using Pazopanib With Food vs. Fasted on Pharmacokinetics, Patient Safety, and Preference (DIET Study) Clin Pharmacol Ther. 2019;106(5):1076–1082. doi: 10.1002/cpt.1515. [DOI] [PubMed] [Google Scholar]
- 36.van Erp NP, Gelderblom H, Karlsson MO, Li J, Zhao M, Ouwerkerk J, et al. Influence of CYP3A4 inhibition on the steady-state pharmacokinetics of imatinib. Clin Cancer Res. 2007;13(24):7394–7400. doi: 10.1158/1078-0432.CCR-07-0346. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.