Abstract
Abstract
The biocatalysis of β-myrcene into value-added compounds, with enhanced organoleptic/therapeutic properties, may be performed by resorting to specialized enzymatic machinery of β-myrcene-biotransforming bacteria. Few β-myrcene-biotransforming bacteria have been studied, limiting the diversity of genetic modules/catabolic pathways available for biotechnological research. In our model Pseudomonas sp. strain M1, the β-myrcene catabolic core-code was identified in a 28-kb genomic island (GI). The lack of close homologs of this β-myrcene-associated genetic code prompted a bioprospection of cork oak and eucalyptus rhizospheres, from 4 geographic locations in Portugal, to evaluate the environmental diversity and dissemination of the β-myrcene-biotransforming genetic trait (Myr+).
Soil microbiomes were enriched in β-myrcene-supplemented cultures, from which β-myrcene-biotransforming bacteria were isolated, belonging to Alphaproteobacteria, Betaproteobacteria, Gammaproteobacteria, and Sphingobacteriia classes. From a panel of representative Myr+ isolates that included 7 bacterial genera, the production of β-myrcene derivatives previously reported in strain M1 was detected in Pseudomonas spp., Cupriavidus sp., Sphingobacterium sp., and Variovorax sp. A comparative genomics analysis against the genome of strain M1 found the M1-GI code in 11 new Pseudomonas genomes. Full nucleotide conservation of the β-myrcene core-code was observed throughout a 76-kb locus in strain M1 and all 11 Pseudomonas spp., resembling the structure of an integrative and conjugative element (ICE), despite being isolated from different niches.
Furthermore, the characterization of isolates not harboring the Myr+-related 76-kb locus suggested that they may biotransform β-myrcene via alternative catabolic loci, being thereby a novel source of enzymes and biomolecule catalogue for biotechnological exploitation.
Key points
• The isolation of 150 Myr+ bacteria hints the ubiquity of such trait in the rhizosphere.
• The Myr+ trait is spread across different bacterial taxonomic classes.
• The core-code for the Myr+ trait was detected in a novel ICE, only found in Pseudomonas spp.
Supplementary Information
The online version contains supplementary material available at 10.1007/s00253-023-12650-w.
Keywords: Cork oak rhizosphere, Eucalyptus rhizosphere, Monoterpene biotransformation, β-Myrcene, Pseudomonas, Integrative conjugative element
Introduction
Plant monoterpenes and their derivatives have been widely used as starting materials for the production of fine chemicals, because of their biotechnologically relevant organoleptic and therapeutic properties (Edris 2007; Bicas et al. 2009). The industrial exploitation of monoterpenes usually involves their chemical oxidation/hydroxylation and incorporation into aqueous formulations. Such strategy may be troublesome due to the high hydrophobicity and structural instability of monoterpenes, which often requires the utilization of surfactants and the presence of a second phase of organic solvents (Krings and Berger 1998; Schwab et al. 2013). These catalytic constrains may be overcome by resorting to the specialized enzymatic machinery of monoterpene-catabolizing microbes. They are able to quickly adapt to the hydrophobic character of monoterpenes, overcome the limited substrate availability, and modify the monoterpene backbone with high regio- and stereo-selectivity (Krings and Berger 1998). Nowadays, increasing research effort is being performed towards a more profitable bio-based transformation and catalysis, by prospecting ecological niches as source of novel cell factories and enzymes (Soares-Castro et al. 2021, and references therein).
The acyclic β-myrcene (7-methyl-3-methylene-1,6-octadiene) is one of the most promising monoterpenes, being used as starting material for the chemical synthesis of top selling flavors and fragrances (e.g., linalool, geraniol, citral, menthol), compounds with pharmacological potential (e.g., antimutagenics, analgesics, tyrosinase inhibitors) and also in the production of polymers, biodegradable surfactants, pheromones, and agrochemicals (Kauderer et al. 1991; Matsuura et al. 2006; Behr and Johnen 2009). Despite efforts to describe β-myrcene biotransformation in different models (Pseudomonas putida S4-2, Pseudomonas sp. strain M1, Pseudomonas aeruginosa PTCC 1074, Rhodococcus erythropolis MLT1, and Castellaniella defragrans 65Phen strains) (Narushima et al. 1981; Iurescia et al. 1999; Thompson et al. 2010; Esmaeili and Hashemi 2011; Lüddeke and Harder 2011; Soares-Castro et al. 2017), the majority of the studies lacked a holistic characterization of the β-myrcene catabolic pathways towards proper biotechnological exploitation. The knowledge regarding the repertoire of enzymes available in the environment for β-myrcene biotransformation is still scarce.
Pseudomonas sp. strain M1, isolated from soil sediments of the Rhine River (Netherlands), is able to use β-myrcene as sole carbon (myr+ trait) (Iurescia et al. 1999; Santos and Sá-Correia 2009; Soares-Castro and Santos 2015; Soares-Castro et al. 2017). The molecular basis of such trait was characterized through the combination of genome-wide, bioanalytical chemistry, molecular biology, and microbial physiology approaches which led to the description of a novel 28-kb genomic island (GI) associated with β-myrcene catabolism (Santos and Sá-Correia 2009; Soares-Castro and Santos 2015).
Moreover, a detailed phylogenetic analysis of the 28-kb GI showed (i) low homology levels to sequences available in public databases and (ii) the majority of genes comprised in the island shared significant identity with sequences from organisms that belong to a taxonomic class not related to Pseudomonas spp. In other words, the analysis suggested the 28-kb GI might have resulted from a modular-like assembly of DNA sequences acquired from different environmental sources, which were functionalized towards β-myrcene utilization in strain M1 (Soares-Castro and Santos 2015). These hints also suggested the existence of broader spectrum of bacteria able to metabolize/biotransform β-myrcene, when compared with the current knowledge. Such functional modules might hold biotechnological potential as targets for engineering strategies, by mimicking the transformations intended on the chemical catalysis of β-myrcene. Soares-Castro et al. (2017) have detected the production of myrcen-8-ol ((2E)-2-methyl-6-methylideneocta-2,7-dien-1-ol) and 4-methylhexanoic acid during the biotransformation of β-myrcene by M1 cells, which have been reported as insect pheromones and can be exploited as biocontrol agents, as well as the aroma compound 4-methyl-3-hexenoic acid (4-methylhex-3-enoic acid).
In nature, monoterpenes are mainly present in aerial organs of plants but have also been detected in the soil, derived from root exudates and accumulation of dead plant material (Wilt et al. 1993; Smolander et al. 2006; Lin et al. 2007). Previous studies report that accumulated soil monoterpenes might become available as carbon sources and modulate the dynamics of the soil microbiome and nutrient recycling (Misra and Pavlostathis 1997; Vokou et al. 2002; Smolander et al. 2006). Having this in mind, the soil can be considered a rich reservoir of novel gene products and metabolic pathways and a potential source of monoterpene biocatalysts for biotechnological research (Lee and Lee 2013).
The catalytic potential of soil microbial communities may be exploited by promoting the growth and enrichment of the microbial community of a soil sample towards the mineralization of a desired substrate (Steele and Stowers 1991). Despite the limitations of such strategies in underestimating the soil microbial diversity, due to singular growth requirements of the uncultured fraction, the cultivation-dependent approaches allow the physical isolation of strains presenting the desired metabolic trait, whose genetic traits can be further characterized.
Having in mind the limited knowledge regarding β-myrcene-biotransforming bacteria and the underlying genetic modules, in the present study, the bacterial communities of plant-associated soil samples, from 4 different geographic locations in Portugal, were enriched towards the biotransformation of β-myrcene. We aimed at the isolation of novel β-myrcene-catabolizing bacteria to broaden the current knowledge about the taxonomic context underlying the prokaryotic β-myrcene biotransformation, as well as to provide new insights regarding the environmental dissemination of the Myr+ trait, for future biotechnological exploitation.
Materials and methods
Soil collection and sample preparation
The soil sediments used in this study were collected in four different geographic locations in Portugal. Reis et al. (2019) provided soil samples derived from the rhizosphere of cork oak trees (Quercus suber L.) in Cabril (41°45′43.05″N 8°1′39.09″W) and Ermida (41°42′39.76″N 8°6′14.87″W), in Peneda-Gerês National Park (located in northern Portugal), as well as soil samples from the rhizosphere of cork oak trees in Grândola (38°11′32.37″N 8°37′11.41″W, located in southern Portugal). In Cabril, Ermida, and Grândola, five independent trees without signs of disease or infestation, separated at least 30 m from each other, were selected for soil sampling, under the middle of the cork oak canopy, resulting in 5 replicates per location (Reis et al. 2019). Soil samples were also collected in Guimarães (41°27′46.24″N 8°16′2.92″W, northern Portugal), from the rhizosphere of three eucalyptus trees (Eucalyptus globulus Labill.) separated by 10 m from each other (5 replicates).
Enrichment cultures of soil samples towards β-myrcene utilization
To perform the enrichment cultures, 10 g of soil replicates from each location were resuspended in 30 mL of 0.9% (w/v) NaCl, vortexed vigorously at 2500 × g for 10 min and the particles cleared by centrifuging at 1000 × g for 3 min. The cleared supernatants were centrifuged at 16,000 × g for 5 min to sediment microbial cells. The cells were resuspended in 1 mL of 0.9% (w/v) NaCl, and the replicates of each location were pooled into a single aliquot to inoculate the enrichment cultures (1 per each location). The microbial growth of soil suspensions was promoted during 48 h, in 50 mL of Mineral Medium (MM) (Soares-Castro and Santos 2015) in 250 mL flasks, supplemented with 0.05% (w/v) yeast extract, 40 mM glucose, and 40 mM pyruvate, at 30 °C and 200 rpm, to broaden the range of culturable bacteria retrieved from the soil suspensions (e.g., copiotrophs). Afterwards, 5 mL of the enrichment cultures were transferred to 50 mL of MM supplemented with 0.05% (w/v) yeast extract and with the saturating volume of 100 μL of β-myrcene [Acros Organics; CAS 123–35-3; log Kow: 4.17; water solubility at 25 °C of 5.6 mg L−1 or 41.1 μmol L−1 (Soares-Castro and Santos 2015); a volume of 100 μL corresponds to 130 mg L−1, 47.8 μmol in solution (Soares-Castro et al. 2017)], at 30 °C and 200 rpm, envisaging the enrichment and isolation of β-myrcene-biotransforming bacteria.
The amount of β-myrcene used in the experiments assured constant water phase saturation with the monoterpene to avoid carbon starvation effects. The flasks used with β-myrcene-supplemented cultures were sealed with rubber stoppers. During the enrichment approach, the optical density measured at 600 nm (OD600 nm) and pH values were monitored every 3 days, followed by supplementation of β-myrcene and correction of pH to values ranging from 6.5 to 7.0.
Isolation and identification of β-myrcene-biotransforming bacteria
Putative β-myrcene-biotransforming bacteria were isolated between 6 and 30 days of enrichment. Bacteria cells were plated on MM agar supplemented with 0.05% (w/v) yeast extract and saturating amount of β-myrcene placed on the plate lid. A set of colonies representing different types of morphologies were isolated, and their ability to use β-myrcene as carbon source (the Myr+ trait) was evaluated in culture tubes at 30 °C and 200 rpm, containing 10 mL liquid cultures of MM with 0.05% (w/v) yeast extract, supplemented with the saturating 50 μL of β-myrcene (65 mg L−1, 23.9 μmol in solution), and prepared with an initial OD600 nm value of 0.1. Cultures of MM + 0.05% (w/v) yeast extract, without supplementation with the monoterpene, were used as controls, to differentiate the β-myrcene-supported growth from residual growth supported by other constituents of the culture medium.
The taxonomic identification of the β-myrcene-biotransforming soil isolates was performed by Sanger sequencing of the 16S rRNA gene. Genomic DNA was extracted using the Wizard Genomic DNA Purification Kit (Promega), and the 16S rRNA gene was amplified by PCR, using the conserved Bacteria primer 8F (5′AGAGTTTGATCCTGGCTCAG 3′) (Gardener et al. 2000). Amplification was performed according to the following protocol: initial denaturation step at 96 °C for 3 min, 30 cycles of denaturation at 96 °C for 30 s, annealing at 50 °C for 30 s and extension at 72 °C for 1 min and 30 s, followed by a final extension step at 72 °C for 5 min. The PCR fragments were sequenced with an automatic ABI GeneAmp PCR System 9700 (GENEWIZ Ltd.) and the trimmed DNA sequences comprising the variable regions V2 to V4 (approximately 615 bp), used for taxonomic identification with the RDP server (Cole et al. 2013). The 16S rRNA gene sequences obtained from Sanger sequencing were aligned with MAFFT version 7 (Katoh and Standley 2013), and the maximum likelihood phylogenetic midpoint rooted tree was generated by using PhyML version 3.0 (Guindon et al. 2010), with 1000 bootstrap sets, the GTR (General Time Reversible) as the best-fit model of nucleotide substitution (evaluated by the W-IQ-Tree tool; Trifinopoulos et al. 2016), kappa estimated, 4 substitution rate categories, gamma distribution parameter estimated, BIONJ starting tree, with optimization of topology, branch lengths and rate parameter.
In this work, isolates obtained from Cabril, Ermida, Grândola, and Guimarães soils are designated as “UMC,” “UME,” “UMG,” and “UMA,” respectively.
Growth kinetics of selected Myr+ isolates
The ability to use β-myrcene as carbon source, by Myr+ isolates belonging to different bacterial genera and showing different phenotypic traits, was validated in 50 mL cultures of MM, supplemented with 0.05% (w/v) yeast extract and the saturating amount of 100 μL of β-myrcene as carbon source, in 250 mL flasks sealed with rubber stoppers, at 30 °C and 200 rpm.
The cultures were prepared to an initial OD600 nm value of 0.1. Similarly, control cultures of MM + 0.05% (w/v) yeast extract, without supplementation with β-myrcene, were used to distinguish the β-myrcene-supported growth (threshold of OD600 nm at 8 h and 24 h > 0.2) from residual growth. Pseudomonas sp. strain M1 was used as reference to evaluate and compare growth kinetic parameters of the isolates. The maximum specific growth rate of each isolate using β-myrcene as carbon source (μmyr) was estimated using the standard equation μmyr = ln(N2/N1)/t2-t1, in which “N1” and “N2” correspond to the variation of the cell abundance measured as OD600 nm, at two different time-points “t1” and “t2,” during the initial hours of the exponential phase. The pH values were monitored after 8 h and 24 h of growth to assess alterations of the chemical properties of the growth medium, as previously described in M1-grown cultures supplemented with β-myrcene as sole carbon source (Santos and Sá-Correia 2009; Soares-Castro et al. 2017).
The hierarchical clustering with growth parameters of each isolate (μmyr, OD600 nm registered at approximately 8 h and 24 h of growth, pH values registered at approximately 24 h of growth) was performed based on the calculated Canberra distances and UPGMA agglomeration method, with the hclust function from the core-package “stats,” version 4.0.3, of the R statistical software (R Core Team 2013).
Monitoring the activation of the promoter P5 of the β-myrcene hydroxylase MyrH of Pseudomonas sp. strain M1 in the Myr+ soil isolates
The GFP-based promoter-probe pSEVA637-P5 (GmR; pBBR1 broad-host origin of replication) was mobilized to selected Myr+ soil isolates to evaluate their ability to activate the promoter P5 of the gene coding for the β-myrcene hydroxylase MyrH of Pseudomonas sp. strain M1 (MRY70_06145) (Soares-Castro et al. 2017).
The isolates harboring the promoter-probe were cultured in 10 mL MM supplemented with 0.05% (w/v) yeast extract, 40 mM glucose, 40 mM pyruvate, and 30 μg mL−1 of gentamicin. The reporter system was induced when cells reached an OD600 nm of 0.3, by supplementing the culture medium with a saturating amount of 50 μL of β-myrcene (65 mg L−1, 23.9 μmol in solution). The β-myrcene-dependent activity of the promoter P5 was quantified at 24 h after β-myrcene supplementation, by fluorometric detection in a Qubit 3.0 fluorometer and expressed as the fold induction of GFP fluorescence normalized by the culture OD600 nm between β-myrcene-induced and non-induced cells.
Qualitative GC–MS analysis of β-myrcene-derivatives from cultures of representative Myr+ isolates
A panel of selected Myr+ isolates was cultured in 50 mL of MM with 0.05% (w/v) yeast extract, supplemented with the saturating amount of 100 μL of β-myrcene as sole carbon source. The cultures were prepared to an initial OD600 nm value of 0.1 and were collected in the mid-exponential growth phase (OD600 nm ranging from 0.4 to 0.5), for the following identification of the respective β-myrcene biotransformation products in the culture’s supernatant, according to Soares-Castro et al. (2017). Cultures of the wild-type strain of Pseudomonas sp. M1 were used as a reference.
A total of 50 mL of the supernatant (the whole culture) was collected, mixed with 10 mL ethyl acetate and vigorously shaken for 5 min. For each strain, 3 replicates of the extracts were produced for analysis. The ethyl acetate extracts were dried with nitrogen gas stream, dissolved in 1 mL of hexane and concentrated with a nitrogen gas stream to a final volume around 500 μL, for subsequent gas chromatography analysis coupled to mass spectrometry (Soares-Castro et al. 2017). Control metabolite profiles were prepared by using (i) pure β-myrcene standard in hexane; (ii) culture supernatants from cells grown with a mixture of 40 mM glucose and 40 mM pyruvate as sole carbon sources, without supplementation with terpenes; and (iii) abiotic control of MM with β-myrcene supplementation, without cells (Soares-Castro et al. 2017).
The gas chromatography analysis of the supernatants from biotransformation experiments was performed on a GC–MS Bruker Scion 436 with single quadrupole module, coupled to a CP8400 automated sampler and equipped with a Shimadzu RXi-5 Sil apolar capillary column (1,4-bis(dimethylsiloxy)phenylene dimethyl polysiloxane; 30 m × 0.25 mm I.D. × 0.25 μm film thickness). Helium was used as carrier gas at a flow rate of 1.0 mL min−1, and 1 uL of sample was injected in splitless mode. The column temperature was initially kept at 60 °C for 1 min and then gradually increased to 250 °C at a rate of 10 °C min−1, with a final hold time of 2 min (Soares-Castro et al. 2017). The mass spectra were acquired with an electron impact ionization voltage of 70 eV, scanned in the m/z range of 40–500 atomic mass unit. The baseline of the obtained chromatograms was defined with the statistics-sensitive non-linear iterative peak-clipping algorithm, which showed to be the best fit for the shape of different peak-free regions throughout the chromatogram (Soares-Castro et al. 2017).
Metabolite identification
The total ion chromatograms were deconvoluted with the AMDIS tool version 2.71 (Stein 1999) to generate the ion spectra for every integer m/z component in the data acquisition range of atomic mass units. The deconvoluted m/z spectra whose intensity values increase, maximize, and decrease together are derived from the same chemical species, thus comprise its ion profile used for compound identification. The ion profile of each chemical species (here defined as the query peak) was obtained by considering the ion intensity at the query peak maximum and subtracting the intensity of the m/z components from the region of the GC–MS spectrum at the beginning and end of the query peak (to remove any influence derived from background ions of the matrix, and/or from adjacent peaks on either side of the query peak). Similarly, the ion profile of overlapped peaks, corresponding to co-eluted compounds, was manually filtered by subtracting the intensity of the m/z components of the contaminant peak from the ion profile of the query peak.
The identification of terpene-derivatives was performed based on (i) the NIST MS Search tool version 2.0 libraries (mainlib comprising 163,198 spectra and Wiley comprising 228,996 spectra) and (ii) by comparing calculated retention indices (RI) with those reported in the literature and/or from the National Institute of Standards and Technology (NIST) Mass Spec Data Center (Wallace 2023). Retention indices of each query compound were determined by sampling a mixture of n-alkanes (C10–C28) under the same GC–MS conditions (van Den Dool and Kratz 1963).
Genome drafts of β-myrcene-biotransforming isolates
β-Myrcene-biotransforming soil isolates were selected for whole-genome sequencing, to determine their β-myrcene-associated genetic trait. The genomic DNA of the isolates was processed according to Illumina instructions to generate Nextera XT paired-end libraries. Isolates UMC13, UMC65, UMC76, UME9, UME65, UME77, and UME83 were sequenced using the Illumina MiSeq platform (2 × 250 bp), and isolates UMC631, UMC3103, UMC3106, UMC3129, UMA601, UMA603, and UMA643 were sequenced using the Illumina NextSeq500 platform (2 × 150 bp), at Instituto Gulbenkian de Ciência (IGC).
All raw paired-reads obtained from high throughput sequencing were initially corrected for sequencing errors with the k-spectrum-based BLUE tool set (Greenfield et al. 2014). Corrected read datasets were then filtered based on Phred quality scores (Ewing and Green 1998), and eventual adapter contamination, low quality, and ambiguous nucleotides were trimmed off by using the fastq-mcf tool (Aronesty 2011).
The derived filtered reads were then de novo assembled with the Unicycler pipeline (Wick et al. 2017), based on the SPAdes assembler, with default parameterization. Genome annotation was performed using the NCBI’s Prokaryotic Genomes Annotation Pipeline version 4.8 (Angiuoli et al. 2008).
Comparative genomics between the 14 sequenced isolates and reference strains
The genome-based taxonomy was assigned to the sequenced isolates with the Bacsort toolset (https://github.com/rrwick/Bacsort), by estimating the average nucleotide identity (ANI) with FastANI version 1.33 (Jain et al. 2018), to determine the most suitable phylogenetically closer reference strains for the following species-level comparative genomics analyses. The following reference genomes were retrieved from NCBI using the Bacsort toolset and based on the ANI obtained: Cupriavidus basilensis strain DSM 11853 (GenBank accession GCF_008801925.2), Pseudomonas protegens strain CHA0 (GenBank accession NC_021237.1), Sphingobacterium paramultivorum strain BIGb0170 (GenBank accession CP058555) and Variovorax ginsengisoli strain S09.D (GenBank accession GCF_006438845.1).
Assessing the conservation of the 28-kb β-myrcene core-code of strain M1 in 11 Pseudomonas isolates
The structural organization and nucleotide conservation of the genetic modules of M1-GI in the 11 isolates UMC65, UMC76, UMC631, UMC3103, UMC3106, UMC3129, UME65, UME83, UMA601, UMA603, and UMA643 was assessed by aligning the nucleotide sequence of the homologous loci in the CLC Genomics Workbench software (version 22.0.2, QIAGEN) with default parameters. The BLASTP-based homology search of the genes surrounding the 28-kb GI from M1 strain, putatively involved in DNA mobilization processes, was performed by using sequences described in the literature or retrieved from the ICEberg database (Bi et al. 2011).
Results
The isolation of bacterial strains harboring the Myr+ trait in the 4 Portuguese soils
In the present study, we prospected soil samples from the rhizosphere of cork oak trees and eucalyptus trees, of 4 different geographic locations in Portugal (UMC—Cabril; UME—Ermida; UMA—Guimarães; UMG – Grândola; Table S1), to isolate novel β-myrcene-catabolizing bacteria with biotechnological potential for β-myrcene biotransformation. The bacterial communities from the different soils were enriched during 30 days, in minimal medium supplemented with saturating amounts of β-myrcene as carbon-source, and the screenings for putative β-myrcene-biotransforming bacteria were carried out at different enrichment timepoints (between 6 and 30 days). The taxonomic identification was performed by Sanger sequencing of the 16S rRNA gene (Table S2). A total of 150 Myr+ Gram-negative bacteria were isolated and included members of the classes Alphaproteobacteria (2 Agrobacterium sp.), Betaproteobacteria (4 Achromobacter sp., 5 Burkholderia sp., 11 Cupriavidus sp., 2 Delftia sp., 2 Variovorax sp.), Gammaproteobacteria (1 Acinetobacter sp., 2 Citrobacter sp., 10 Enterobacter sp., 6 Lelliottia sp., 63 Pseudomonas sp., 3 Raoultella sp., 31 Serratia sp., 4 Shigella sp.), and Sphingobacteriia (4 Sphingobacterium sp.), as highlighted in Table 1 (the corresponding phylogenetic contextualization is shown in Fig. S1, Fig. S2 and Table S2).
Table 1.
Overview of the bacterial taxa of the 150 soil isolates obtained from the enrichment cultures with β-myrcene selection. Their associated geographic location, taxonomic identification and phenotypic information (growth yield and pH at 24 h of culture in β-myrcene-supplemented medium) are listed in Table S2. Their phylogenetic clustering is shown in Fig. S1 and Fig. S2. Soil location: C, Cabril; E, Ermida; G, Grândola; A, Guimarães
| Bacterial classes | Bacterial genera | Soil location | No. of isolates |
|---|---|---|---|
| Alphaproteobacteria | Agrobacterium-Rhizobium group | A | 2 |
| Betaproteobacteria | Achromobacter sp. | C, E, G | 4 |
| Burkholderia sp. | C, G | 5 | |
| Cupriavidus sp. | C, E | 11 | |
| Delftia sp. | E | 2 | |
| Variovorax sp. | C | 2 | |
| Gammaproteobacteria | Acinetobacter sp. | G | 1 |
| Citrobacter sp. | G | 2 | |
| Enterobacter sp. | C, E, G, A | 10 | |
| Lelliottia sp. | G, A | 6 | |
| Pseudomonas sp. | C, E, G, A | 63 | |
| Raoultella sp. | G | 3 | |
| Serratia sp. | C, E, G, A | 31 | |
| Shigella sp. | G | 4 | |
| Sphingobacteriia | Sphingobacterium sp. | C, E | 4 |
| Total | 150 | ||
From the selected colonies, Pseudomonas spp. comprised 42.8% of all isolated bacteria, accounting for approximately 16%, 15%, 10%, and 4% of the retrieved isolates from Cabril, Ermida, Grândola, and Guimarães, respectively. The taxonomic classification of the 63 Pseudomonas spp. resulted in 3 distinct phylogenetic groups: 19 P. putida-like isolates, a second group of 12 P. protegens-like isolates, and a third group comprised by 26 isolates clustered with Pseudomonas sp. strain M1.
Members of the genera Serratia and Enterobacter were also isolated from all 4 locations, although with relative frequencies less than 10% and 3%, respectively, for each soil. On the contrary, the isolated Variovorax spp. were only retrieved from Cabril, and Delftia spp. were only retrieved from Ermida. Members of the genera Acinetobacter, Citrobacter, Raoultella, and Shigella were isolated from Grândola forests. Agrobacterium isolates were obtained from the eucalyptus rhizosphere of Guimarães.
Pseudomonas isolates showed higher μmyr in β-myrcene supplemented cultures
We validated the ability to use β-myrcene as carbon source in 43 Myr+ isolates, with different phenotypic traits (e.g., growth yield) and different genomic backgrounds, carried out in 50 mL batch cultures, to evaluate and compare their growth kinetics with the reference strain M1.
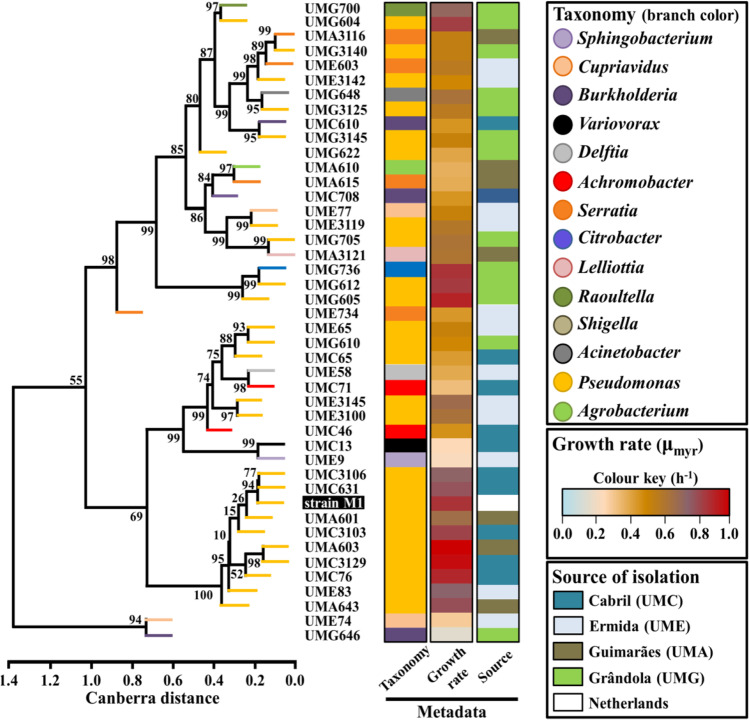
As shown in Fig. 1 and Table S3, the reference strain M1 registered a maximum specific growth rate (μmyr) of 0.85 ± 0.06 h−1 and OD600 nm values > 1.0 in the culture conditions used, being only outperformed by the phylogenetically close Pseudomonas isolates UMA603 (0.98 ± 0.09 h−1), UMC3129 (0.95 ± 0.01 h−1), and UMC76 (0.88 ± 0.03 h−1).
Fig. 1.
Clustering of 43 Myr+ isolates from the 4 geographic locations, based on the physiological characterization. The hierarchical clustering was performed with the calculated Canberra distances and UPGMA agglomeration, based on the estimated maximum specific growth rate (μmyr), OD600 nm at 8 h and 24 h of growth, and pH at 24 h of growth. Isolate codes: “UMC,” Cabril; “UME,” Ermida; “UMG,” Grândola; “UMA,” Guimarães. Bootstrap values presented as percentage are shown in the tree branches. Colored columns summarize the metadata associated with each isolate: (i) taxonomy based on 16S rRNA gene Sanger sequencing, (ii) ranges of μmyr in MM supplemented with 0.05% (w/v) yeast extract and β-myrcene, and (iii) source of isolation. Information about the 43 Myr+ isolates is detailed in Table S3
The remaining 6 Pseudomonas isolates clustered with the strain M1 (clade G13 in Fig. S2) and reached similarly high OD600 nm values after 8 h of growth, with μmyr ranging from 0.65 ± 0.05 h−1 in isolate UMA601 to 0.81 ± 0.04 h−1 in isolate UMC3103. Based on the measured growth kinetics parameters (estimated μmyr, OD600 nm, and pH), the clustering profile obtained for strain M1 and the isolates UMA601, UMA603, UMA643, UMC76, UMC631, UMC3103, UMC3106, UMC3129, and UME83 (Fig. 1) was similar to the phylogenetic clustering from Fig. S2 (shared 100% nucleotide homology of the sequenced 16S rRNA gene fragment).
The Pseudomonas sp. isolate UMG605 and the Citrobacter sp. isolate UMG736 also registered maximum specific growth rates comparable to the μmyr of strain M1 (0.89 ± 0.14 h−1 and 0.85 ± 0.03 h−1, respectively), however, associated with the production of less biomass. Whereas members of the Pseudomonas genus seemed to registered higher μmyr, values below 0.4 h−1 were estimated for Delftia sp. isolate UME58 (0.39 ± 0.05 h−1), Serratia sp. isolate UMA615 (0.38 ± 0.05 h−1), Agrobacterium sp. isolate UMA610 (0.37 ± 0.02 h−1), Achromobacter sp. isolate UMC71 (0.33 ± 0.04 h−1), Cupriavidus sp. isolate UME74 (0.29 ± 0.04 h−1), Variovorax sp. isolate UMC13 (0.25 ± 0.01 h−1), Sphingobacterium sp. isolate UME9 (0.23 ± 0.02 h−1), and Burkholderia sp. isolate UMG646 (0.15 ± 0.07 h−1).
Moreover, the characteristic acidification of the culture medium reported for strain M1, during growth using β-myrcene as sole carbon-source (Santos and Sá-Correia 2009; Soares-Castro et al. 2017) was detected for isolates clustered with this strain (UMA601, UMA603, UMA643, UMC76, UMC631, UMC3103, UMC3106, UMC3129, and UME83), as well as for the Pseudomonas sp. isolates UMC65 and UME65 (Table S3).
The β-myrcene biotransformation of several soil isolates produced β-myrcene derivatives previously reported in strain M1
A panel of 17 Myr+ isolates was selected to identify the diversity of β-myrcene derivatives produced in biotransformation experiments, by GC–MS. The panel comprised representatives of different species, to which the β-myrcene supplementation supported considerable growth and biomass production (OD600 nm > 1.0): Achromobacter sp. isolate UMC46; Cupriavidus sp. isolate UME77; Delftia sp. isolate UME58; Pseudomonas sp. UMC76, UMC631, UMC3103, UMC3106, UMC3129, UME83, UMA601, UMA603, UMA643, UME65, and UME3145; Serratia sp. isolate UME734; Sphingobacterium sp. isolate UME9; and Variovorax sp. isolate UMC13 (Table S3).
The analysis of culture’s supernatants of the strain M1 detected all key β-myrcene derivatives reported previously in the biotransformation of the monoterpene substrate (Table 2; Soares-Castro et al. 2017): myrcen-8-ol (C10H16O; compound 4), myrcenal (C10H14O; compound 3; (2E)-2-methyl-6-methylideneocta-2,7-dien-1-al), myrcenoic acid (C10H14O2; compound 5; (2E)-2-methyl-6-methylideneocta-2,7-dienoic acid), 4-methyl-3-hexenoic acid (C7H12O2; 4-Me-3-HA; compound 2), and 4-methylhexanoic acid (C7H14O2; 4-MeHA; compound 1).
Table 2.
Metabolites successfully identified in β-myrcene (C10H16) biotransformation experiments with the selected representatives of the panel of Myr+ soil isolates, which registered considerable growth and biomass production (OD600 nm > 1.0) in β-myrcene-supplemented cultures. The Pseudomonas sp. strain M1 was used as reference
| Metabolite identification | ||||||
|---|---|---|---|---|---|---|
|
Predicted metabolite ID (formula) (compound no.) |
4-MeHA C7H14O2 (1) |
4-Me-3-HA C7H12O2 (2) |
Myrcenal C10H14O (3) |
Myrcen-8-ol C10H16O (4) |
Myrcenoic acid C10H14O2 (5) |
|
| ID method | MS, RI | MS, RI | MS | MS, RI | MS | |
| Retention index | 1039 | 1079 | 1217 | 1241 | 1393 | |
| GC peaksa | ||||||
| Reference strain | M1 | 0.004 ± 2.8E − 4 | 0.077 ± 3.6E − 2 | 0.081 ± 9.8E − 3 | 0.215 ± 2.6E − 2 | 0.560 ± 1.0E − 1 |
| Pseudomonas spp. | UMA601 | 0.003 | 0.037 ± 6.0E − 3 | 0.071 ± 2.0E − 2 | 0.282 ± 3.5E − 2 | 0.355 ± 6.2E − 2 |
| (M1-like) | UMA603 | 0.004 ± 2.3E − 5 | 0.123 ± 1.1E − 2 | ND | 0.032 | 1.041 ± 1.3E − 2 |
| UMA643 | 0.003 | 0.032 ± 3.7E − 3 | 0.098 | 0.312 | 0.180 ± 3.1E − 2 | |
| UMC3103 | 0.013 ± 3.4E − 3 | 0.308 ± 1.3E − 1 | 0.034 | 0.125 | 0.506 ± 2.9E − 1 | |
| UMC3106 | 0.003 ± 3.4E − 4 | 0.048 ± 9.1E − 3 | 0.020 ± 2.8E − 3 | 0.036 ± 2.6E − 2 | 0.222 ± 2.1E − 2 | |
| UMC3129 | 0.010 ± 1.9E − 3 | 0.092 ± 1.6E − 2 | 0.056 ± 6.2E − 3 | 0.425 ± 1.1E − 1 | 1.179 ± 1.2E − 1 | |
| UMC631 | 0.020 ± 7.2E − 3 | 0.202 ± 4.1E − 2 | ND | 0.070 | 0.349 ± 4.4E − 2 | |
| UMC76 | 0.002 ± 2.5E − 4 | 0.050 ± 2.6E − 2 | 0.025 ± 7.1E − 3 | 0.162 ± 1.7E − 2 | 0.874 ± 3.0E − 1 | |
| UME83 | 0.004 | 0.057 ± 3.9E − 2 | ND | 0.053 | 0.366 ± 1.2E − 1 | |
| Pseudomonas sp. | UME65 | 0.018 ± 7.2E − 3 | 0.436 ± 7.4E − 2 | 0.071 ± 1.9E − 2 | 0.183 ± 6.5E − 2 | 9.821 ± 2.3 |
| Sphingobacterium sp. | UME9 | 0.012 ± 4.2E − 3 | 0.324 ± 8.5E − 2 | ND | 0.022 ± 8.9E − 3 | 11.027 ± 4.8 |
| Variovorax sp. | UMC13 | 0.008 ± 1.3E − 3 | 0.171 ± 6.5E − 3 | ND | 0.016 | 1.201 ± 2.1E − 1 |
| Cupriavidus sp. | UME77 | ND | ND | ND | 0.002 ± 1.0E − 3 | ND |
aPeak areas of the metabolites identified in the supernatants of each strain, normalized as a percentage of the total area of the chromatogram, shown as average values ± standard deviation when replicates were available, at mid-exponential phase of β-myrcene (7-methyl-3-methylideneocta-1,6-diene) biotransformation. 4-MeHA, 4-Methylhexanoic acid; 4-Me-3-HA, 4-Methyl-3-hexenoic acid; Myrcenal ((2E)-2-methyl-6-methylideneocta-2,7-dien-1-al); Myrcen-8-ol ((2E)-2-methyl-6-methylideneocta-2,7-dien-1-ol); Myrcenoic acid ((2E)-2-methyl-6-methylideneocta-2,7-dienoic acid); MS, mass spectrum search; RI, retention index; ND, not detected in any replicate
Significantly, myrcen-8-ol (compound 4), myrcenoic acid (compound 5), 4-methyl-3-hexenoic acid (compound 2) and 4-methylhexanoic acid (compound 1) were also detected in supernatants of the Pseudomonas sp. isolates UMC76, UMC631, UMC3103, UMC3106, UMC3129, UME83, UMA601, UMA603, UMA643, and UME65, as well as in supernatants of the Sphingobacterium sp. isolate UME9 and Variovorax sp. isolate UMC13. The initial oxidation product of β-myrcene biotransformation, myrcen-8-ol (compound 4), was also detected in cultures of Cupriavidus sp. isolate UME77.
Moreover, we were not able to identify β-myrcene derivatives, including any of the metabolites reported in the literature, in the analysis of the supernatants of Achromobacter sp. isolate UMC46, Delftia sp. isolate UME58, Pseudomonas sp. isolate UME3145, and Serratia sp. isolate UME734.
Whole-genome sequencing of 14 bacteria harboring the Myr+ trait
The isolates Cupriavidus sp. UME77; Pseudomonas sp. UMC65, UME65, UMA601, UMA603, UMA643, UMC76, UMC631, UMC3103, UMC3106, UMC3129, and UME83; Sphingobacterium sp. UME9; and Variovorax sp. UMC13, for which β-myrcene derivatives were successfully identified, were selected for whole-genome sequencing and characterization of the respective genomic background underlying the Myr+ phenotype. Even though the draft genomes were deposited in 2020, the assemblies were confirmed with recent versions of the tools, without significative changes in the outcome (contiguity, size, etc.). Assembly metrics of the final draft genomes are shown in Table S3.
The estimation of the average nucleotide identity (ANI) against reference genomes deposit in NCBI allowed to determine phylogenetic closer strains with available sequenced genomes, for the validation of the taxonomic assignment and the following comparative genomics analysis. The phylogenetic relationships between Myr+ bacteria and the selected references were assessed by the 16S rRNA gene alignment (Fig. 2a) and validated by whole-genome pairwise BLAST-based alignment (Fig. 2b).
Fig. 2.
Phylogeny of the 14 Myr+ isolates and phylogenetically closer reference strains. Panel a shows the phylogenetic tree with midpoint rooting based on the alignment of the 16S rRNA gene sequence extracted from the assembled and reference genomes. Bootstrap values presented as percentage are shown in the tree branches. Panel b shows the hierarchical clustering based on the pairwise distances estimated with BLAST alignments between all strains. Two identical genomes would result in a distance value of 0. The distances were estimated according to the ratio of nucleotide identity per total length of the compared genomes. Branches are colored according to the bacterial genera. Isolate codes: “UMC,” Cabril; “UME,” Ermida; “UMA,” Guimarães; “UMG,” Grândola. The estimated ANI values are available in Table S4
The draft genome of the isolates Cupriavidus sp. UME77 showed 96.7% of average nucleotide identity (ANI) against the genome of Cupriavidus basilensis strain DSM 11853, whereas the draft genomes of Sphingobacterium sp. UME9 and Variovorax sp. UMC13 showed 97.7% and 85.9% to the genomes of the closest references Sphingobacterium paramultivorum strain BIGb0170 and Variovorax ginsengisoli strain S09.D, respectively (Table S4). Moreover, the pairwise genomic distances also evidenced a high degree of genome similarity between several isolates retrieved from different locations, such as the pair UMC65 and UME65, which showed 98.9% of ANI to the type strain Pseudomonas protegens CHA0 (Fig. 2b and Table S4). These 2 isolates shared 99.9% of nucleotide identity among them, even though their sampling source was separated by 8.5 km. The isolates UMA601, UMA603, UMA643, UMC76, UMC631, UMC3103, UMC3106, UMC3129, and UME83, also retrieved from 3 different locations, shared around 98.1% of ANI with the Pseudomonas citronellolis and around 99.9% of ANI with the strain M1 (GenBank accession CP094343). Having the high percentage of ANI similarity, we hypothesize that M1 strain (and related isolates) belongs to Pseudomonas citronellolis species. This hypothesis is corroborated by a genome-based taxonomy comparative analysis using TYGS (https://tygs.dsmz.de/) which indicated that Pseudomonas sp. M1 and related isolates belong to Pseudomonas citronellolis species. However, further experimental evidences are required to confirm such hypothesis.
Even though β-myrcene biotransformations with Cupriavidus sp. UME77, Sphingobacterium sp. UME9, and Variovorax sp. UMC13 generated β-myrcene-derivatives reported in strain M1, the genome mining of their 3 genomes did not detect close homologs of the gene products of the M1 28-kb β-myrcene core-code (BLASTP analysis showed < 70% identity at the protein level, whose hits were not organized in genetic clusters; Table S6). The genome analysis of these 3 isolates, thereby, hinted the existence of alternative functional modules underlying their Myr+ trait and the detected metabolic profile. On the contrary, the activation of the reporter system in the isolates UMA601, UMA603, UMA643, UMC76, UMC631, UMC3103, UMC3106, UMC3129, UME83, UMC65, and UME65 (Table S3), together with the similar metabolic footprint of β-myrcene-derivatives as reported for strain M1, suggested that their genomes might harbor the full or partial 28-kb locus responsible for the M1 Myr+ trait.
The 28-kb β-myrcene core-code is located within a 76-kb genomic locus conserved in strain M1 and 11 other Pseudomonas isolates
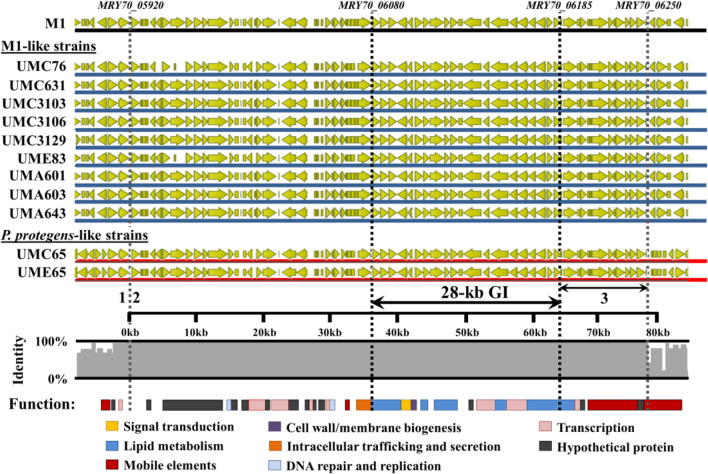
A closer inspection of the draft genomes of Pseudomonas sp. isolates UMC76, UMC631, UMC3103, UMC3106, UMC3129, UME83, UMA601, UMA603, and UMA643 revealed the presence of the 22 genes of the 28-kb GI from strain M1 (MRY70_06080-MRY70_06185), showing 100% nucleotide identity and alignment coverage (Table S5), displaying a similar structural organization, in the 9 M1-like Pseudomonas isolates (Fig. 3), as supported by the GC–MS analysis and the screening carried out the reporter system.
Fig. 3.
Nucleotide conservation of a 76-kb genomic region among 11 Pseudomonas spp., comprising the β-myrcene-responsive locus characterized in Pseudomonas sp. M1. The DNA backbone sequence is colored in blue for M1-like isolates and in red for P. protegens-like isolates: M1, Pseudomonas sp. M1 (MRY70_05920-MRY70_06250); UMC76 (A9979_06855-A9979_07180); UMC631 (FCN13_00270-FCN13_00620); UMC3103 (FCN10_11380-FCN10_11730); UMC3106 (FCJ51_00270-FCJ51_00620); UMC3129 (FCN12_00270-FCN12_00620); UME83 (A9972_23140-A9972_23470); UMA601 (FCJ53_19215-FCJ53_19415 and FCJ53_19910-FCJ53_20055); UMA603 (FCJ55_16630-FCJ55_16830 and FCJ55_04610-FCJ55_04755); UMA643 (FCJ56_13360-FCJ56_13560 and FCJ56_04410-FCJ56_04555); UMC65 (A9978_04360-A9978_04690); UME65 (A9971_23150-A9971_23480). The identity per base of the nucleotide alignment is represented in the grey plot. The grey vertical dotted lines depict the flanks of the genomic 76-kb locus fully conserved in the five Pseudomonas strains, whereas the black vertical dotted lines depict the flanks of the β-myrcene 28-kb core-code in Pseudomonas sp. M1. CDS within this 76-kb conserved locus are colored by function, based on COG classes. The category “Mobile elements” includes integrases, transposases, conjugative elements and plasmid-associated proteins. Using the genome of M1 strain as reference, the “1” represents the local of insertion downstream the GMP synthase (guaA) gene MRY70_05915 and corresponding homologs; “2” indicates the upstream flank starting with the MRY70_05920 gene, which codes for a putative integrase; “3” represents the downstream flank, ending with a cluster coding putative conjugal transfer proteins from Tra and Trb family (MRY70_06190-MRY70_06250)
Furthermore, the comparative genomics analysis between the strain M1 and the other sequenced Pseudomonas isolates evidenced the presence of the fully conserved 28-kb in the P. protegens-like isolates UMC65 and UME65. Strikingly, a whole-genome alignment between these Pseudomonas sp. isolates showed that the full nucleotide conservation of the β-myrcene core-code was not restricted to the 28-kb GI and included, in fact, a larger genomic region of approximately 76-kb (Fig. 3).
This 76-kb conserved region was located downstream the housekeeping gene guaA (MRY70_05915), coding for a glutamine-hydrolyzing GMP synthase, in the 12 aligned genomes (Fig. 3, indicated with “1”), flanked by a gene coding for a putative integrase (MRY70_05920 in strain M1; Fig. 3, indicated with “2”) and a cluster coding putative conjugal transfer proteins (MRY70_06190- MRY70_06250) in strain M1; Fig. 3, indicated with “3”). The integrase MRY70_05920 is homologous to the tyrosine site-specific P4 integrase of P. mendocina ymp located in the genomic island ympGIguaA (Pmen_3484: 91.6% a.a. identity and 99% of sequence coverage) and of Acidovorax avenae subsp. citrulli AAC00-1 located in the genomic island AAC00-1GIguaA (Aave_3366: 85% a.a. identity covering 99% of aligned sequence) strains (Table S6). The conjugational cluster located downstream of the β-myrcene core-code encodes a putative trbBCDEJLFGI operon and a traG gene, described as homologous to the conjugation system of IncP1 plasmids (RP4/Ti-like) (Lessl et al. 1992). Although homology for several genes located in the upstream flank of the 76-kb locus was not found in the ICEberg database, the conjugational cluster showed a.a. identity ranging from 67 to 89% with the Tn4371 ICE from Cupriavidus oxalatica A5 and from 37 to 89% with the ICE-KKS from Acidovorax sp. KKS102 (Table S6), both belonging to a family of ICEs (the Tn4371 family) only reported in Gamma-proteobacteria and Beta-proteobacteria (Springael et al. 2001; Toussaint et al. 2003). Besides the β-myrcene 28-kb catabolic modules, genes comprised within the integrative locus code for putative proteins involved in DNA integration and replication, transcription, and a major fraction encode proteins with unknown function (Fig. 3).
Discussion
The enrichment protocol and culture-dependent approach used in the present study allowed to isolate 150 β-myrcene-biotransforming bacteria living in the rhizosphere of cork oak trees and eucalyptus trees, from 4 different geographic sources in Portugal: from Cabril, Ermida, and Guimarães in the north, as well as from Grândola in the south. The ability to biotransform β-myrcene was, thus, widespread in the sampled soil communities of cork oak and eucalyptus forests. Hence, the soil surrounding these forests may comprise a valuable reservoir of potential β-myrcene (and other monoterpenes) catalysts.
The library of isolates extended the current taxonomy associated with the aerobic β-myrcene catabolism to 15 bacterial genera, ubiquitous in the environment and reported to establish biological interactions with plants. In fact, members of the genera Achromobacter, Acinetobacter, Burkholderia, Delftia, Enterobacter, Pseudomonas, Raoultella, Serratia, Sphingobacterium and Variovorax reported in this study were also found in the soil microbial communities of Q. suber (Bevivino et al. 2014). Pseudomonas spp. were also detected in the rhizosphere of several eucalyptus trees (Berendsen et al. 2015; da Silva Fonseca et al. 2018). Furthermore, the analysis of the microbiome from the pinewood nematode Bursaphelenchus xylophilus resulted in the identification of genes putatively associated with the catabolism of α-pinene, limonene, and geraniol, in Pseudomonas, Achromobacter, and Agrobacterium (Cheng et al. 2013). Similarly, the microbiome of bark beetles seems to comprise monoterpene-metabolizing bacteria from the genera Pseudomonas, Rahnella, Serratia, and Stenotrophomonas (Adams et al. 2011). In 2014, the metatranscriptome analysis of the resin-tolerant microbial community associated with pine tree galls, formed by the moth Retinia resinella, detected expression of genes associated with α-pinene catabolism and the acyclic terpene utilization pathway (atu pathway), widespread in more than 40 bacterial genera, from which Pseudomonas was the most abundant bacterial genus (Vilanova et al. 2014). Although functional characterization is required to assess the catalytic novelty of the reported genes, these metagenomics studies support the taxonomic knowledge acquired in the present study associating a broader taxonomic context and genomic background with the monoterpene-biotransforming trait.
The growth kinetics of the isolated bacteria was evaluated with representative isolates belonging to the different genera (Fig. 1; Table S3), in cultures supplemented with β-myrcene as carbon source. All assayed isolates phylogenetically closer to our reference Pseudomonas sp. strain M1 reached OD600 nm values higher than 1.5 at 24 h of growth and μmyr higher than 0.7 h−1 (with the exception of UMA601). This outstanding catabolic performance of the M1-like isolates was paralleled by other Pseudomonas spp. from the P. putida clade (strains UMG604, UMG605 and UMG612 from clade G11 in Fig. S2, with μmyr > 0.80 h−1), the Citrobacter isolate UMG736 (0.85 ± 0.02 h−1) and the Raoultella isolate UMG700 (0.70 ± 0.02 h−1), despite registering lower biomass yield. To our knowledge, Achromobacter spp., Acinetobacter spp., Burkholderia spp., Lelliottia spp., Raoultella spp., and Sphingobacterium spp. have never been experimentally associated to the ability of catalyzing catabolic transformations in the monoterpene backbone, whereas some strains of several Pseudomonas species (e.g., P. putida, P. aeruginosa, P. citronellolis, P. protegens, among others), Serratia marcescens, and Citrobacter braakii have been studied by their monoterpene-biotransforming enzymatic repertoire (Soares-Castro et al. 2021, and references therein).
The detection of several β-myrcene derivatives, previously reported for the β-myrcene catabolism of strain M1, such as myrcen-8-ol, myrcenoic acid, 4-methyl-3-hexenoic acid and 4-methylhexanoic acid, in Pseudomonas sp. isolates UMC76, UMC631, UMC3103, UMC3106, UMC3129, UME83, UMA601, UMA603, UMA643, and UME65, evidenced that their β-myrcene biotransformation pathways follow similar enzymatic steps, towards the channeling of the β-myrcene derivatives to the central metabolism. Additionally, the GFP fluorescence of the M1 β-myrcene hydroxylase MyrH promoter-probe was only induced by the supplementation of β-myrcene in M1-like isolates and in the P. protegens-like isolates UMC65 and UME65, thereby indicating the presence of M1-GI related modules in the genome of these 11 Pseudomonas spp. Evidence of the dissemination of the 28-kb GI in the isolates UMC76, UMC631, UMC3103, UMC3106, UMC3129, UME83, UMA601, UMA603, UMA643, UMC65, and UME65 was obtained by high-throughput genome sequencing, which revealed the full nucleotide and structural conservation of the 28-kb β-myrcene core code found in strain M1 (Fig. 3). Most importantly, the complete conservation of the nucleotide sequence was extended to a region spanning 76-kb, whose genetic content resembled the guaA-associated ICEs (Song et al. 2012).
As self-transmissible elements, ICEs encode the machinery required to control their excision and mediate conjugation (Wozniak and Waldor 2010). In M1 cells grown in the presence of β-myrcene, gene expression levels quantified by RNA-seq showed the induction of several CDS associated with the mobility of integrative and conjugative elements (Soares-Castro and Santos 2015), namely the upregulation of the integrase-coding gene (MRY70_05920) by 39-fold, the putative transcriptional regulator of the AlpA family (MRY70_05935) by fourfold, and the MRY70_06190-MRY70_06250 cluster, putatively coding conjugative transfer Tra and Trb proteins, by levels ranging from 1.7- to 3.5-fold. In particular, the putative β-myrcene-induced AlpA regulator (98% identity and 99% of alignment coverage with the putative AlpA-family regulator from P. stutzeri; accession WP_044316124) is reported to be the positive regulatory factor of P4 integrases (Kirby et al. 1994; Trempy et al. 1994). The induction of these CDS suggested the presence of an active regulatory cascade targeting the mobilization systems (or remnants of them) upon β-myrcene supplementation. These activation mechanisms could result in the excision of the 76-kb locus, albeit experimental evidences of a free excised circular replicon were never obtained. Such mobilization events may have resulted in the acquisition of the 76-kb ICE by the P. protegens-like strains UMC65 and UME65, since M1-like bacteria were also isolated in both Cabril (UMC76, UMC631, UMC3103, UMC3106, UMC3129) and Ermida (UME83) soils. Evidence of the existence of multiple replicons in strain M1 was not detected during assemblies with different types of datasets. The transference frequency and ability to transfer a mobile genetic element, such as genomic islands, even between closely related strains, have been described to be dependent on the host background (Minoia et al. 2008; Miyazaki et al. 2012): in part resulting from the integration of the GI-borne regulatory system within the cell global regulatory network and potential cross-talks between conserved regulatory signals of different mobile elements co-existing in the host, which can lead to the repression or induction of the island. Furthermore, the genetic structure and nucleotide composition of the 76-kb locus containing the β-myrcene core-code was conserved in all 12 strains of our library, even though they were isolated from geographically and chronologically distinct sources. Pseudomonas sp. strain M1 was isolated from sediments of the Rhine river (Netherlands) in 1997–1998, Pseudomonas sp. strains UMC76, UME83, UMC65, and UME65 were isolated from soil rhizosphere collected in 2015, and Pseudomonas sp. strains UMC631, UMC3103, UMC3106, UMC3129, UMA601, UMA603, and UMA643 were isolated from soil rhizosphere collected in 2017, from forests in the north of Portugal. Therefore, following experimental evidence is required to understand if the 76-kb locus containing the β-myrcene core-code is stably integrated in the genome or being actively mobilized, even if at low frequencies in the population, as described for the ICEclc in P. knackmussii strain B13 (Minoia et al. 2008; Miyazaki et al. 2012). Following work will also be focused on the characterization of factors modulating a potential excision (e.g., growth phase, efficiency of excision, and conjugation), putative interplay between the catabolic modules and the mobility-related elements, and the range of hosts, currently only observed in Pseudomonas spp.
The comparative genomics between Pseudomonas sp. strain M1 and the newly sequenced Pseudomonas sp. strains UMC76, UMC631, UMC3103, UMC3106, UMC3129, UME83, UMA601, UMA603, and UMA643 showed 99.9% of genomic similarity, according to the estimated ANI (Table S4). The high similarity detected at the genomic level between the 10 strains may suggest that they belong to the same species, resulting from unique evolutionary track of each strain, probably driven by niche-specific circumstances. Strikingly, the full conservation of the 76-kb locus hints the existence of molecular mechanisms maintaining the integrity and stability of this catabolic locus, which may exert a tight control of its transcription (e.g., DNA methylation associated to DNA repair systems) (Camacho and Casadesús 2002; Casadesús and Low 2006). Although we did not find clear evidence in the literature linking DNA methylation patterns to the conservation of specific DNA sequences and bacteria evolution, several reports associate N6-adenine methylation through Dam methyltransferase with the effectiveness of DNA repair systems. In Escherichia coli strain K12, mutants of the Dam methyltransferase resulted in an increase spontaneous mutation frequency up to 46-fold when compared to the Dam+ strain (Marinus and Morris 1975). Similar effects were also described for Dam– mutants on Serratia typhimurium (Torreblanca and Casadesús 1996). Moreover, the review of Wion and Casadesús (2006) reported that Dam-mediated methylation can also repress the mobility of the bacterial transposons IS10, IS50, Tn5, and Tn903 in Escherichia coli. Dam-mediated methylation of GATC sites in the promoter of both transposons hampered the binding of RNA polymerase and inhibited the transcription of the transposase required for transposition to occur. Additionally, methylation of GATC sites at the inner terminus of both IS10 and IS50 transposons inhibited the transposase activity, thus contributing to the host genome stability (Wion and Casadesús 2006).
Furthermore, the performed physiological, molecular, and analytical screenings suggested that the other Myr+ bacteria did not harbor a genomic context compatible with the activation of the promoter of the β-myrcene hydroxylase MyrH of the strain M1. The genomes of the sequenced isolates not harboring the 76-kb ICE (Sphingobacterium sp. isolate UME9, Variovorax sp. isolate UMC13, and Cupriavidus sp. isolate UME77) were further screened in silico for homologs of known monoterpene-catabolizing enzymes, involved in (i) the atu pathway from Pseudomonas citronellolis (gene products from atuRABCDEFGH) (Förster-Fromme et al. 2006); (ii) oxidation of linallool/geraniol by Castellaniella defragrans 65Phen (GeoA, GeoB and Ldi) (Lüddeke et al. 2012) and Pseudomonas putida (LinC) (Ropp et al. 1993); (iii) catabolism of limonene and carveol by Rhodococcus erythropolis DCL14 (LimABC and Mlhb) (van der Werf et al. 1999; van der Werf and Boot 2000) and Geobacillus stearothermophilus BR388 (gene product of pOT435 gene) (Cheong and Oriel 2000); (iv) catabolism of camphor by Pseudomonas putida NCIMB10007 (gene products of camABCDEFG) (Poulos et al. 1985; Bell et al. 2010) and Rhodococcus sp. (camK) (Roberts et al. 2004); (v) catabolism of p-cymene by P. putida F1 (gene products of cymBCAaAbDE and cmtABCDEFGHI) (Eaton 1996, 1997); and (vi) oxidation of 1,8-cineole by Citrobacter braakii (CinA) (Hawkes et al. 2002). This BLASTP analysis did not detect close homologs of the queried enzymes (BLASTP showed < 70% identity at the protein level, not organized in an operon-like structure; Table S5), and, thereby, candidate clusters coding for putative β-myrcene-biotransforming modules could not be considered. Having this in mind, the production of myrcen-8-ol, myrcenoic acid, 4-methyl-3-hexenoic acid, and 4-methylhexanoic acid by Sphingobacterium sp. isolate UME9 and Variovorax sp. isolate UMC13 supported the existence of β-myrcene-biotransforming modules with similar catalytic properties to those found in strain M1, even though corresponding to a distinct Myr+ genetic trait. The detection of myrcen-8-ol in β-myrcene-supplemented cultures of Cupriavidus sp. isolate UME77 suggested, at least, a common initial oxidation step with strain M1, but also derived from a different, and yet to be defined, Myr+ genetic trait. On the contrary, the lack of identification of known β-myrcene derivatives (or other monoterpene-related isomers, alcohols, aldehydes, or acids) in β-myrcene-growing cultures of Achromobacter sp. UMC46, Delftia sp. UME58, Pseudomonas sp. UME3145, and Serratia sp. UME734 may have derived from the fact that standard metabolite databases do not comprise a large number of mass spectra of β-myrcene-derived compounds (Soares-Castro et al. 2017), suggesting different metabolites may be generated by novel catabolic machinery. This also reflects the lack of knowledge available regarding β-myrcene catabolic pathways, which in turn impairs the successful metabolite identification (Soares-Castro et al. 2021). The missed detection of the initial β-myrcene derivatives could also have been influenced by strain-specific physiological traits (e.g., the rate of biotransformation) coupled to the single time-point analysis carried out at mid-exponential phase, even though saturating amounts of the monoterpene were used to supplement the cultures and ensure an unlimited feed of the substrate in a aqueous-gaseous equilibrium.
Nevertheless, the Myr+ genetic trait underlying the β-myrcene catabolism in the isolates not harboring the M1-related ICE remains to be identified and validated with functional approaches (e.g., RNA-seq) in following work, which will greatly increase the knowledge regarding the β-myrcene biotransformation in these strains.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
The authors would like to acknowledge to Jörg Becker and João Sobral for the sequencing services provided at the Instituto Gulbenkian de Ciência.
Author contribution
F.R. and T.L.-N. collected and processed the soil samples, as well as the respective metadata. P.S.-C. and F.S. carried-out the microbiological experiments. P.S.-C. and P.M.S. performed the in silico analyses. P.S.-C., F.S. and P.M.S. drafted the manuscript. P.S.-C. created the illustrations and tables. P.S.-C., F.R., T.L.-N., and P.M.S. reviewed and edited the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
Open access funding provided by FCT|FCCN (b-on). This work was supported by the project VALEU (PTDC/EAM-AMB/30488/2017), by the strategic program UID/BIA/04050/2013 (POCI-01–0145-FEDER-007569), by the GenomePT project (POCI-01–0145-FEDER-022184), supported by COMPETE 2020—Operational Programme for Competitiveness and Internationalisation (POCI), Lisboa Portugal Regional Operational Programme (Lisboa2020), Algarve Portugal Regional Operational Programme (CRESC Algarve2020), under the Portugal 2020 Partnership Agreement, funded by national funds through the Fundação para a Ciência e a Tecnologia (FCT) I.P. and the European Regional Development Fund (ERDF), through the COMPETE2020-Programa Operacional Competitividade e Internacionalização (POCI). The work was supported as well through the Ph.D. grants to P.S-C (grant number SFRH/BD/76894/2011) and to F.S. (grant number PD/BD/146184/2019).
Data availability
The Whole Genome Shotgun projects generated in this work have been deposited at DDBJ/EMBL/GenBank under the following accession numbers: Cupriavidus sp. strain UME77 (LZZU00000000), Pseudomonas sp. strain UMC65 (LZZX00000000), Pseudomonas sp. strain UME65 (LZZQ00000000), Pseudomonas sp. strain UMC76 (LZZY00000000), Pseudomonas sp. strain UMC631 (SUQJ00000000), Pseudomonas sp. strain UMC3103 (SWJU00000000), Pseudomonas sp. strain UMC3106 (SUQK00000000), Pseudomonas sp. strain UMC3129 (SWJT00000000), Pseudomonas sp. strain UME83 (LZZR00000000), Pseudomonas sp. strain UMA601 (SUQI00000000), Pseudomonas sp. strain UMA603 (SUQS00000000), Pseudomonas sp. strain UMA643 (SUQL00000000), Sphingobacterium sp. strain UME9 (LZZP00000000), Variovorax sp. strain UMC13 (LZZW00000000).
Strain availability
The reference strain, Pseudomonas sp. M1, and the representative isolates harboring the myr + Pseudomonas sp. UMA601, Pseudomonas sp. UMAC76, Pseudomonas sp. UME83 have been deposited at the University of Coimbra Bacteria Culture Collection (UCCCB) under the accession numbers UCCCB 159, UCCCB 160, UCCCB 161, and UCCCB 162, respectively, and at the Spanish Type Culture Collection (CECT) under the accession numbers CECT 20780, CECT 20781, CECT 20782, and CECT 20783, respectively.
Declarations
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Conflict of interest
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- Adams AS, Boone CK, Bohlmann J, Raffa KF. Responses of bark beetle-associated bacteria to host monoterpenes and their relationship to insect life histories. J Chem Ecol. 2011;37:808–817. doi: 10.1007/s10886-011-9992-6. [DOI] [PubMed] [Google Scholar]
- Angiuoli SV, Gussman A, Klimke W, Cochrane G, Field D, Garrity GM, Kodira CD, Kyrpides N, Madupu R, Markowitz V, Tatusova T, Thomson N, White O. Toward an online repository of standard operating procedures (SOPs) for (meta) genomic annotation. OMICS. 2008;12:137–141. doi: 10.1089/omi.2008.0017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aronesty E (2011) ea-utils: command-line tools for processing biological sequencing data. Available at https://github.com/ExpressionAnalysis/ea-utils
- Behr A, Johnen L. Myrcene as a natural base chemical in sustainable chemistry: a critical review. Chemsuschem. 2009;2:1072–1095. doi: 10.1002/cssc.200900186. [DOI] [PubMed] [Google Scholar]
- Bell SG, Dale A, Rees NH, Wong L-L. A cytochrome P450 class I electron transfer system from Novosphingobium aromaticivorans. Appl Microbiol Biotechnol. 2010;86:163–175. doi: 10.1007/s00253-009-2234-y. [DOI] [PubMed] [Google Scholar]
- Berendsen RL, van Verk MC, Stringlis IA, Zamioudis C, Tommassen J, Pieterse CMJ, Bakker PAHM. Unearthing the genomes of plant-beneficial Pseudomonas model strains WCS358, WCS374 and WCS417. BMC Genomics. 2015;16:539. doi: 10.1186/s12864-015-1632-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bevivino A, Paganin P, Bacci G, Florio A, Pellicer MS, Papaleo MC, Mengoni A, Ledda L, Fani R, Benedetti A. Soil bacterial community response to differences in agricultural management along with seasonal changes in a Mediterranean region. PLoS One. 2014;9:e105515. doi: 10.1371/journal.pone.0105515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi D, Xu Z, Harrison EM, Tai C, Wei Y, He X, Jia S, Deng Z, Rajakumar K, Ou H-Y. ICEberg: a web-based resource for integrative and conjugative elements found in Bacteria. Nucleic Acids Res. 2011;40(Database issue):D621–6. doi: 10.1093/nar/gkr846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bicas JL, Dionísio AP, Pastore GM. Bio-oxidation of terpenes: an approach for the flavor industry. Chem Rev. 2009;109:4518–4531. doi: 10.1021/cr800190y. [DOI] [PubMed] [Google Scholar]
- Camacho EM, Casadesús J. Conjugal transfer of the virulence plasmid of Salmonella enterica is regulated by the leucine-responsive regulatory protein and DNA adenine methylation. Mol Microbiol. 2002;44:1589–1598. doi: 10.1046/j.1365-2958.2002.02981.x. [DOI] [PubMed] [Google Scholar]
- Casadesús J, Low D. Epigenetic gene regulation in the bacterial world. Microbiol Mol Biol Rev. 2006;70:830–856. doi: 10.1128/MMBR.00016-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng X-Y, Tian X-L, Wang Y-S, Lin R-M, Mao Z-C, Chen N, Xie B-Y. Metagenomic analysis of the pinewood nematode microbiome reveals a symbiotic relationship critical for xenobiotics degradation. Sci Rep. 2013;3:1869. doi: 10.1038/srep01869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheong TK, Oriel PJ. Cloning and expression of the limonene hydroxylase of Bacillus stearothermophilus BR388 and utilization in two-phase limonene conversions. Appl Biochem Biotechnol. 2000;84–86:903–915. doi: 10.1385/abab:84-86:1-9:903. [DOI] [PubMed] [Google Scholar]
- Cole JR, Wang Q, Fish JA, Chai B, McGarrell DM, Sun Y, Brown CT, Porras-Alfaro A, Kuske CR, Tiedje JM. Ribosomal database project: data and tools for high throughput rRNA analysis. Nucleic Acids Res. 2013;42(Database issue):D633–42. doi: 10.1093/nar/gkt1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- da Silva FE, Peixoto RS, Rosado AS, de Carvalho BF, Tiedje JM, da Costa Rachid CTC. The microbiome of Eucalyptus roots under different management conditions and its potential for biological nitrogen fixation. Microb Ecol. 2018;75:183–191. doi: 10.1007/s00248-017-1014-y. [DOI] [PubMed] [Google Scholar]
- Eaton RW. p-Cumate catabolic pathway in Pseudomonas putida Fl: cloning and characterization of DNA carrying the cmt operon. J Bacteriol. 1996;179:3171–3180. doi: 10.1128/jb.179.10.3171-3180.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eaton RW. p-Cymene catabolic pathway in Pseudomonas putida F1: cloning and characterization of DNA encoding conversion of p-cymene to p-cumate. J Bacteriol. 1997;178:1351–1362. doi: 10.1128/jb.178.5.1351-1362.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edris AE. Pharmaceutical and therapeutic potentials of essential oils and their individual volatile constituents: a review. Phyther Res. 2007;21:308–323. doi: 10.1002/ptr.2072. [DOI] [PubMed] [Google Scholar]
- Esmaeili A, Hashemi E. Biotransformation of myrcene by Pseudomonas aeruginosa. Chem Cent J. 2011;5:26. doi: 10.1186/1752-153X-5-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewing B, Green P. Base-calling of automated sequencer traces using Phred. II. Error probabilities. Genome Res. 1998;8:186–194. doi: 10.1101/gr.8.3.175. [DOI] [PubMed] [Google Scholar]
- Förster-Fromme K, Höschle B, Mack C, Bott M, Armbruster W, Jendrossek D. Identification of genes and proteins necessary for catabolism of acyclic terpenes and leucine/isovalerate in Pseudomonas aeruginosa. Appl Environ Microbiol. 2006;72:4819–4828. doi: 10.1128/AEM.00853-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardener BBM, Schroeder KL, Kalloger SE, Raaijmakers JM, Thomashow LS, Weller DM. Genotypic and phenotypic diversity of phlD-containing Pseudomonas strains isolated from the rhizosphere of wheat. Appl Environ Microbiol. 2000;66:1939–1946. doi: 10.1128/aem.66.5.1939-1946.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenfield P, Duesing K, Papanicolaou A, Bauer DC. Blue: correcting sequencing errors using consensus and context. Bioinformatics. 2014;30:2723–2732. doi: 10.1093/bioinformatics/btu368. [DOI] [PubMed] [Google Scholar]
- Guindon S, Dufayard J-F, Lefort V, Anisimova M, Hordijk W, Gascuel O. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol. 2010;59:307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- Hawkes DB, Adams GW, Burlingame AL, de Montellano PRO, De Voss JJ. Cytochrome P450cin (CYP176A), isolation, expression, and characterization. J Biol Chem. 2002;277:27725–27732. doi: 10.1074/jbc.M203382200. [DOI] [PubMed] [Google Scholar]
- Iurescia S, Marconi AM, Tofani D, Gambacorta A, Paternò A, Devirgiliis C, van der Werf MJ, Zennaro E. Identification and sequencing of β-myrcene catabolism genes from Pseudomonas sp. strain M1. Appl Environ Microbiol. 1999;65:2871–2876. doi: 10.1128/AEM.65.7.2871-2876.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain C, Rodriguez-R LM, Phillippy AM, Konstantinidis KT, Aluru S. High throughput ANI analysis of 90K prokaryotic genomes reveals clear species boundaries. Nat Commun. 2018;9:5114. doi: 10.1038/s41467-018-07641-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kauderer B, Zamith H, Paumgartten FJR, Speit G, Holden HE. Evaluation of the mutagenicity of β-myrcene in mammalian cells in vitro. Environ Mol Mutagen. 1991;18:28–34. doi: 10.1002/em.2850180106. [DOI] [PubMed] [Google Scholar]
- Kirby JE, Trempy JE, Gottesman S. Excision of a P4-like cryptic prophage leads to Alp protease expression in Escherichia coli. J Bacteriol. 1994;176:2068–2081. doi: 10.1128/jb.176.7.2068-2081.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krings U, Berger GR. Biotechnological production of flavours and fragrances. Appl Microbiol Biotechnol. 1998;49:1–8. doi: 10.1007/s002530051129. [DOI] [PubMed] [Google Scholar]
- Lee MH, Lee SW. Bioprospecting potential of the soil metagenome: novel enzymes and bioactivities. Genom Inform. 2013;11:114–120. doi: 10.5808/GI.2013.11.3.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lessl M, Balzer D, Pansegrau W, Lanka E. Sequence similarities between the RP4 Tra2 and the Ti VirB region strongly support the conjugation model for T-DNA transfer. J Biol Chem. 1992;267:20471–20480. doi: 10.1016/S0021-9258(19)88726-4. [DOI] [PubMed] [Google Scholar]
- Lin C, Owen SM, Peñuelas J. Volatile organic compounds in the roots and rhizosphere of Pinus spp. Soil Biol Biochem. 2007;39:951–960. doi: 10.1016/j.soilbio.2006.11.007. [DOI] [Google Scholar]
- Lüddeke F, Harder J. Enantiospecific (S)-(+)-linalool formation from beta-myrcene by linalool dehydratase-isomerase. Z Naturforsch C. 2011;66:409–412. doi: 10.1515/znc-2011-7-813. [DOI] [PubMed] [Google Scholar]
- Lüddeke F, Wülfing A, Timke M, Germer F, Weber J, Dikfidan A, Rahnfeld T, Linder D, Meyerdierks A, Harder J. Geraniol and geranial dehydrogenases induced in anaerobic monoterpene degradation by Castellaniella defragrans. Appl Environ Microbiol. 2012;78:2128–2136. doi: 10.1128/AEM.07226-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marinus MG, Morris NR. Pleiotropic effects of a DNA adenine methylation mutation (dam-3) in Escherichia coli K12. Mutat Res Mol Mech Mutagen. 1975;28:15–26. doi: 10.1016/0027-5107(75)90309-7. [DOI] [PubMed] [Google Scholar]
- Matsuura R, Ukeda H, Sawamura M. Tyrosinase inhibitory activity of citrus essential oils. J Agric Food Chem. 2006;54:2309–2313. doi: 10.1021/jf051682i. [DOI] [PubMed] [Google Scholar]
- Minoia M, Gaillard M, Reinhard F, Stojanov M, Sentchilo V, van der Meer JR. Stochasticity and bistability in horizontal transfer control of a genomic island in Pseudomonas. Proc Natl Acad Sci. 2008;105:20792–20797. doi: 10.1073/pnas.0806164106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misra G, Pavlostathis SG. Biodegradation kinetics of monoterpenes in liquid and soil-slurry systems. Appl Microbiol Biotechnol. 1997;47:572–577. doi: 10.1007/s002530050975. [DOI] [Google Scholar]
- Miyazaki R, Minoia M, Pradervand N, Sulser S, Reinhard F, van der Meer JR. Cellular variability of RpoS expression underlies subpopulation activation of an integrative and conjugative element. PLoS Genet. 2012;8:e1002818. doi: 10.1371/journal.pgen.1002818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narushima H, Omori T, Minoda Y. Microbial oxidation of β-myrcene. In: Vezina C, Singh K, editors. Advances in Biotechnology. Oxford: Pergamon Press; 1981. pp. 525–531. [Google Scholar]
- Poulos TL, Finzel BC, Gunsalus IC, Wagner GC, Kraut J. The 2.6-A crystal structure of Pseudomonas putida cytochrome P-450. J Biol Chem. 1985;260:16122–16130. doi: 10.1016/S0021-9258(17)36209-9. [DOI] [PubMed] [Google Scholar]
- R Core Team (2013) R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. Available online at https://www.R-project.org/. Accessed Feb 2023
- Reis F, Soares-Castro P, Costa D, Tavares RM, Baptista P, Santos PM, Lino-Neto T. Climatic impacts on the bacterial community profiles of cork oak soils. Appl Soil Ecol. 2019;143:89–97. doi: 10.1016/j.apsoil.2019.05.031. [DOI] [Google Scholar]
- Roberts GA, Grogan G, Turner NJ, Flitsch SL. Nucleotide sequence of a portion of the camphor-degrading gene cluster from Rhodococcus sp. NCIMB 9784. DNA Seq. 2004;15:96–103. doi: 10.1080/10425170310001656765. [DOI] [PubMed] [Google Scholar]
- Ropp JD, Gunsalus IC, Sligar SG. Cloning and expression of a member of a new cytochrome P-450 family: cytochrome P-450lin (CYP111) from Pseudomonas incognita. J Bacteriol. 1993;175:6028–6037. doi: 10.1128/jb.175.18.6028-6037.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos PM, Sá-Correia I. Adaptation to β-myrcene catabolism in Pseudomonas sp. M1: An expression proteomics analysis. Proteomics. 2009;9:5101–5111. doi: 10.1002/pmic.200900325. [DOI] [PubMed] [Google Scholar]
- Schwab W, Fuchs C, Huang F-C. Transformation of terpenes into fine chemicals. Eur J Lipid Sci Technol. 2013;115:3–8. doi: 10.1002/ejlt.201200157. [DOI] [Google Scholar]
- Smolander A, Ketola RA, Kotiaho T, Kanerva S, Suominen K, Kitunen V. Volatile monoterpenes in soil atmosphere under birch and conifers: effects on soil N transformations. Soil Biol Biochem. 2006;38:3436–3442. doi: 10.1016/j.soilbio.2006.05.019. [DOI] [Google Scholar]
- Soares-Castro P, Santos PM. Deciphering the genome repertoire of Pseudomonas sp. M1 toward β-myrcene biotransformation. Genome Biol Evol. 2015;7:1–17. doi: 10.1093/gbe/evu254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soares-Castro P, Montenegro-Silva P, Heipieper HJ, Santos PM. Functional characterization of a 28-kilobase catabolic island from Pseudomonas sp. strain M1 involved in biotransformation of β-myrcene and related plant-derived volatiles. Appl Environ Microbiol. 2017;83:e03112–e3116. doi: 10.1128/AEM.03112-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soares-Castro P, Soares F, Santos PM. Current advances in the bacterial toolbox for the biotechnological production of monoterpene-based aroma compounds. Molecules. 2021;26:91. doi: 10.3390/molecules26010091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song L, Pan Y, Chen S, Zhang X. Structural characteristics of genomic islands associated with GMP synthases as integration hotspot among sequenced microbial genomes. Comput Biol Chem. 2012;36:62–70. doi: 10.1016/j.compbiolchem.2012.01.001. [DOI] [PubMed] [Google Scholar]
- Springael D, Ryngaert A, Merlin C, Toussaint A, Mergeay M. Occurrence of Tn4371-related mobile elements and sequences in (chloro) biphenyl-degrading bacteria. Appl Environ Microbiol. 2001;67:42–50. doi: 10.1128/AEM.67.1.42-50.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steele DB, Stowers MD. Techniques for selection of industrially important microorganisms. Annu Rev Microbiol. 1991;45:89–106. doi: 10.1146/annurev.mi.45.100191.000513. [DOI] [PubMed] [Google Scholar]
- Stein SE. An integrated method for spectrum extraction and compound identification from gas chromatography/mass spectrometry data. J Am Soc Mass Spectrom. 1999;10:770–781. doi: 10.1016/S1044-0305(99)00047-1. [DOI] [Google Scholar]
- Thompson ML, Marriott R, Dowle A, Grogan G. Biotransformation of β-myrcene to geraniol by a strain of Rhodococcus erythropolis isolated by selective enrichment from hop plants. Appl Microbiol Biotechnol. 2010;85:721–730. doi: 10.1007/s00253-009-2182-6. [DOI] [PubMed] [Google Scholar]
- Torreblanca J, Casadesús J. DNA adenine methylase mutants of Salmonella typhimurium and a novel Dam-regulated locus. Genetics. 1996;144:15–26. doi: 10.1093/genetics/144.1.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toussaint A, Merlin C, Monchy S, Benotmane MA, Leplae R, Mergeay M, Springael D. The biphenyl-and 4-chlorobiphenyl-catabolic transposon Tn4371, a member of a new family of genomic islands related to IncP and Ti plasmids. Appl Environ Microbiol. 2003;69:4837–4845. doi: 10.1128/AEM.69.8.4837-4845.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trempy JE, Kirby JE, Gottesman S. Alp suppression of Lon: dependence on the slpA gene. J Bacteriol. 1994;176:2061–2067. doi: 10.1128/jb.176.7.2061-2067.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trifinopoulos J, Nguyen L-T, von Haeseler A, Minh BQ. W-IQ-TREE: a fast online phylogenetic tool for maximum likelihood analysis. Nucleic Acids Res. 2016;44(W1):W232–W235. doi: 10.1093/nar/gkw256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Den Dool H, Kratz P. A generalization of the retention index system including linear temperature programmed gas—liquid partition chromatography. J Chromatogr A. 1963;11:463–471. doi: 10.1016/S0021-9673(01)80947-X. [DOI] [PubMed] [Google Scholar]
- van der Werf MJ, Boot AM. Metabolism of carveol and dihydrocarveol in Rhodococcus erythropolis DCL14. Microbiology. 2000;146:1129–1141. doi: 10.1099/00221287-146-5-1129. [DOI] [PubMed] [Google Scholar]
- van der Werf MJ, Swarts HJ, de Bont JAM. Rhodococcus erythropolis DCL14 contains a novel degradation pathway for limonene. Appl Environ Microbiol. 1999;65:2092–2102. doi: 10.1128/AEM.65.5.2092-2102.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilanova C, Marín M, Baixeras J, Latorre A, Porcar M. Selecting microbial strains from pine tree resin: biotechnological applications from a terpene world. PLoS One. 2014;9:e100740. doi: 10.1371/journal.pone.0100740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vokou D, Chalkos D, Karamanlidou G, Yiangou M. Activation of soil respiration and shift of the microbial population balance in soil as a response to Lavandula stoechas essential oil. J Chem Ecol. 2002;28:755–768. doi: 10.1023/A:1015236709767. [DOI] [PubMed] [Google Scholar]
- Wallace WE (2023). Retention indices in NIST Chemistry WebBook. In: Linstrom PJ, Mallard WG (eds) NIST standard reference database number 69. National Institute of Standards and Technology, Gaithersburg MD, p 20899. 10.18434/T4D303
- Wick RR, Judd LM, Gorrie CL, Holt KE. Unicycler: resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput Biol. 2017;13:e1005595. doi: 10.1371/journal.pcbi.1005595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilt FM, Miller GC, Everett RL, Hackett M. Monoterpene concentrations in fresh, senescent, and decaying foliage of singleleaf pinyon (Pinus monophylla Torr. & Frem.: Pinaceae) from the western Great Basin. J Chem Ecol. 1993;19:185–194. doi: 10.1007/BF00993688. [DOI] [PubMed] [Google Scholar]
- Wion D, Casadesús J. N6-methyl-adenine: an epigenetic signal for DNA–protein interactions. Nat Rev Microbiol. 2006;4:183. doi: 10.1038/nrmicro1350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wozniak RAF, Waldor MK. Integrative and conjugative elements: mosaic mobile genetic elements enabling dynamic lateral gene flow. Nat Rev Microbiol. 2010;8:552–563. doi: 10.1038/nrmicro2382. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The Whole Genome Shotgun projects generated in this work have been deposited at DDBJ/EMBL/GenBank under the following accession numbers: Cupriavidus sp. strain UME77 (LZZU00000000), Pseudomonas sp. strain UMC65 (LZZX00000000), Pseudomonas sp. strain UME65 (LZZQ00000000), Pseudomonas sp. strain UMC76 (LZZY00000000), Pseudomonas sp. strain UMC631 (SUQJ00000000), Pseudomonas sp. strain UMC3103 (SWJU00000000), Pseudomonas sp. strain UMC3106 (SUQK00000000), Pseudomonas sp. strain UMC3129 (SWJT00000000), Pseudomonas sp. strain UME83 (LZZR00000000), Pseudomonas sp. strain UMA601 (SUQI00000000), Pseudomonas sp. strain UMA603 (SUQS00000000), Pseudomonas sp. strain UMA643 (SUQL00000000), Sphingobacterium sp. strain UME9 (LZZP00000000), Variovorax sp. strain UMC13 (LZZW00000000).
The reference strain, Pseudomonas sp. M1, and the representative isolates harboring the myr + Pseudomonas sp. UMA601, Pseudomonas sp. UMAC76, Pseudomonas sp. UME83 have been deposited at the University of Coimbra Bacteria Culture Collection (UCCCB) under the accession numbers UCCCB 159, UCCCB 160, UCCCB 161, and UCCCB 162, respectively, and at the Spanish Type Culture Collection (CECT) under the accession numbers CECT 20780, CECT 20781, CECT 20782, and CECT 20783, respectively.