Abstract
Herpes simplex virus type 1 (HSV-1) infection causes the active degradation of the catalytic subunit of DNA-dependent protein kinase (DNA-PKcs), and this process is reliant on the expression of the HSV-1 immediate-early protein Vmw110. In this study we investigated in more detail the mechanism by which the degradation occurs, the domains of Vmw110 which are required, and whether Vmw110 is by itself sufficient for the effect. We found that proteasome inhibitors prevented the degradation of DNA-PKcs, indicating the involvement of a proteasome pathway. Furthermore, the continued activity of DNA-PK during infection in the presence of these inhibitors indicated that Vmw110 does not directly alter the enzyme activity of DNA-PKcs prior to its degradation in a normal infection. Indeed, Vmw110 was found to bind to neither the catalytic nor Ku subunits of DNA-PK. Using mutant Vmw110 viruses we show that the RING finger domain of Vmw110 is essential for the induced degradation of DNA-PKcs but that the ability of Vmw110 to bind to a cellular ubiquitin-specific protease (HAUSP) is not required. When expressed in the absence of other viral proteins, Vmw110 was sufficient to cause the degradation of DNA-PKcs, indicating that the effect on the stability of DNA-PKcs was a direct consequence of Vmw110 activity and not an indirect Vmw110-dependent effect of virus infection. Finally, the Vmw110-induced degradation of DNA-PKcs and loss in DNA-PK activity appears to be beneficial to HSV-1 infection, as virus replication was more efficient in cells lacking DNA-PKcs, especially at low multiplicities of infection.
Herpes simplex virus type 1 (HSV-1) is a common human pathogen which establishes a life-long latent infection in sensory neurons after initiating a lytic infection in epithelia (reviewed in reference 41). The virus has a linear double-stranded DNA genome of approximately 152 kb, and the expression of the encoded genes is controlled by both viral factors and the host’s RNA polymerase II (RNAP II) transcription machinery (45). At least 76 genes are expressed during lytic infection (34), and this number of expressed genes differs dramatically from the situation in latency, when only one set of related viral transcripts can be detected (reviewed in reference 17). The lytic genes are expressed in a temporal cascade and can be classified as immediate-early, (IE), early, and late, depending on their time course of synthesis and requirements for prior viral gene expression and DNA replication (41, 51). Four of the five IE proteins, Vmw110 (ICP0), Vmw175 (ICP4), Vmw68 (ICP22), and Vmw63 (ICP27), have been shown to have roles in the regulation of gene expression during lytic infection (10, 33, 38–40, 44, 46).
Vmw110, a RING finger protein encoded by IE gene 1, is a strong and promiscuous activator of gene expression in transfection assays (reviewed in reference 10) and has been implicated in the regulation of both the lytic cycle and reactivation from latency. Several lines of evidence indicate that Vmw110 might play a specific role in the control of the balance between the latent and lytic states, such that in its presence the latter is favored (3, 6, 19, 30, 42, 48, 49, 54). It is likely that Vmw110 carries out its role in activation of transcription and reactivation from latency by interacting with cellular proteins. Consistent with this, Vmw110 has recently been found to interact with and stabilize cyclin D3 (25) and to bind strongly and specifically to a cellular protein which is a functional novel member of the ubiquitin-specific protease (USP) family (14, 35, 36). The latter protein has been referred to as HAUSP (herpesvirus-associated USP), and it has a molecular mass of approximately 130 kDa. In uninfected cells, HAUSP exhibits a micropunctate nuclear staining pattern with a limited number of larger discrete foci, some of which coincide with specific nuclear structures called ND10 domains, PML nuclear bodies, or PODs (14). These domains of unknown function are associated with the nuclear matrix and contain at least six cellular proteins, of which the most widely studied is PML (a protein implicated in promyelocytic leukemia) (1, 2, 7a, 14, 26, 50).
At early times of infection Vmw110 also localizes to ND10 (31), and its interaction with HAUSP leads to an increased proportion of ND10 containing this USP (14). The consequence of the localization of Vmw110 at ND10 domains is their disruption (13, 32), and it has recently been found that this disruption correlates with the virus-induced and Vmw110-dependent degradation of several high-molecular-weight isoforms of PML (15). Other recent studies have shown that these isoforms of PML are very likely to comprise covalent conjugates with the small ubiquitin-like protein PIC1 (also known as GMP1, SUMO-1, Sentrin, and UBL-1 [reviewed in references 21 and 43; see also references 15, 24, 37, and 47]). Further, it has also been observed that virus infection leads to the degradation of a large number of uncharacterized PIC1-conjugated proteins in a Vmw110-dependent manner (15).
Another cellular protein targeted for degradation during HSV-1 infection is the catalytic subunit of the DNA-dependent protein kinase (DNA-PK) (29), which is a complex nuclear protein composed of a large catalytic polypeptide of approximately 460 kDa (DNA-PKcs), and the heterodimeric Ku protein, which targets DNA-PK to DNA (reviewed in references 8 and 52). DNA-PK is required for DNA double-strand break repair and V(D)J recombination and may play additional roles in the cell, for example, in the regulation of transcription from both RNAP I and II promoters and also during apoptosis (reviewed in references 8 and 27).
The previous study of the effect of virus infection on DNA-PKcs clearly showed that the degradation was dependent on the expression of Vmw110; however, the mechanism by which this occurred and whether Vmw110 was sufficient were not determined. In view of the effects of Vmw110 on the stabilities of other cellular proteins, we have investigated this phenomenon in more detail. Possible models include a situation in which direct interactions between Vmw110 and DNA-PKcs induce the degradation of the latter, or one in which Vmw110 modulates the proteolytic pathways which control the amount of DNA-PKcs in the cell. With respect to the latter scenario, the interaction of Vmw110 with HAUSP is intriguing. USP enzymes remove ubiquitin adducts from substrate proteins, thereby protecting them from being targeted to the proteasome for rapid degradation (reviewed in references 20 and 53). Therefore, Vmw110 may control the stability of cellular and/or viral proteins which are ubiquitinated and subject to degradation by the ubiquitin-dependent proteasome pathway. For example, Vmw110 may transport HAUSP either to or from these proteins, enhancing or preventing, respectively, the removal of polyubiquitin chains and thus controlling the level of the protein in the cell. If DNA-PKcs is a normal substrate of HAUSP, inhibition of the latter’s activity by binding to Vmw110 might lead to DNA-PKcs degradation.
In this paper we have investigated the mechanism by which Vmw110 causes DNA-PKcs degradation during infection. The above hypotheses were tested with various mutant viruses which express Vmw110 proteins which either do or do not bind HAUSP, and the involvement of the proteasome pathway was determined by infecting in the presence or absence of proteasome inhibitors. We have also investigated whether Vmw110 physically interacts with either the DNA-PKcs or the Ku subunits of DNA-PK and have determined whether Vmw110 is sufficient in the absence of other viral proteins to cause DNA-PKcs degradation. Finally, the potential relevance of the degradation of DNA-PKcs to virus infection was assessed by comparing the levels of growth of the wild type and of Vmw110-deficient viruses on two human cell lines, MO59K and MO59J, that either do or do not express DNA-PKcs, respectively (28).
MATERIALS AND METHODS
Cells and viruses.
HeLa S3, a human epithelioid cervical carcinoma cell line, was grown in Dulbecco’s modified Eagle’s medium with 10% fetal calf serum (FCS), 100 U of penicillin per ml, and 100 μg of streptomycin per ml. MO59J and MO59K cells, derived from a human malignant glioma, were grown in Dulbecco’s modified Eagle’s medium NUT Mix F-12 (HAM) supplemented with 0.5 mM sodium pyruvate, nonessential amino acids, 10% FCS, and antibiotics as described previously (28). All viruses were grown and titrated in baby hamster kidney (BHK) cells propagated in Glasgow modified Eagle’s medium supplemented with 10% newborn calf serum, 10% tryptose phosphate broth, and antibiotics as described above. The wild-type HSV-1 strains 17syn+ and KOS1.1 were used as stated below. The Vmw110 mutant 17syn+ viruses dl1403, FXE, D12, and E52X were as described previously (9, 36, 49), and viruses E58X, A8X, and A78 had lesions as described in Table 1 and detailed elsewhere (16).
TABLE 1.
Properties of viral mutant Vmw110 proteins
| Virus | Vmw110a | HAUSP bindingb | Comments |
|---|---|---|---|
| 17syn+ | 1–775 | Yes | Wild type |
| dl1403 | 1–150 | No | Taken as null mutant |
| E52X | 1–594 | No | |
| E58X | 1–633 | No | |
| A8X | 1–646 | Minimal | |
| A78 | Δ592–646 | Minimal | |
| D12 | Δ594–633 | Minimal | |
| FXE | Δ106–149 | Yes | RING finger deletion |
Amino acids present or deleted (Δ) from the wild-type sequence.
As detected by coimmunoprecipitation. Trace amounts of HAUSP have sometimes been detected in experiments using A8X, D12, and A78, but these are very substantially reduced compared to the wild type. The data in this Table are presented in more detail elsewhere (16).
Plasmids.
The pCI expression plasmid pCI110 was based on the pCIneo vector (Promega). pCI110 was made by inserting the Vmw110 coding region into the cloning sites of pCIneo.
Transfections.
HeLa S3 cells at a density of 105 cells per well of a 24-well plate were transfected, using the reagent Tfx50 (Promega) according to the manufacturer’s instructions. The DNA-to-Tfx50 ratio was 4.5:1, and the DNA-medium-Tfx50 mix was applied to the cells for 40 min before being replaced with complete medium.
Antibodies.
Anti-Vmw110 monoclonal antibody (MAb) 11060, anti-Vmw175 MAb 10176, and polyclonal anti-HAUSP r201 have been described previously (11, 12, 14). Polyclonal anti-Vmw110 r190 was isolated after immunization of a rabbit with a glutathione S-transferase fusion protein containing residues 594 to 775 of Vmw110 (6a), and anti-HAUSP MAb 16613 was isolated after immunization of mice with a glutathione S-transferase fusion protein containing the N-terminal 193 amino acids of HAUSP (6a). Anti-DNA-PK rabbit serum DPK1 and anti-Ku rabbit serum 31 were as previously described (4). MAbs to DNA-PKcs (MAbs 42-27 and 18-2) were a gift from Tom Shenk (Princeton University).
Immunoprecipitations.
Extracts from 4 × 106 HeLa S3 cells were prepared from mock-infected or infected samples. The medium was removed, and the cells were scraped into 1× RIPA (5 mM Tris-HCl [pH 7.4], 75 mM NaCl, 5 mM KCl, 0.5 mM EDTA), pelleted, washed in 1× RIPA, and resuspended in 250 μl of 1× RIPA containing 0.5% Nonidet P-40, 5 mM N-ethylmaleimide, 0.1 mM phenylmethylsulfonyl fluoride, 5 μg of leupeptin per ml, and 5 μg of aprotinin per ml. Extracts were frozen in liquid nitrogen and stored at −70°C. For immunoprecipitations, extracts were sonicated, cell debris was pelleted at 3,000 × g for 10 min at 4°C, and the supernatants were used as the total immunoprecipitation sample. All subsequent steps were performed at 4°C. Samples were precleared by the addition of 40 μl of protein A-Sepharose or protein G-Sepharose (Sigma), which had been equilibrated in the immunoprecipitation buffer, and mixed for 20 min on a rotary shaker. The Sepharose beads were pelleted, the supernatant was split in two for addition of preimmune serum or immune serum, and the incubation continued for 2 h. A further 40 μl of Sepharose beads was added, and the incubation continued for a further 30 min. The Sepharose beads were pelleted, the supernatant containing unbound proteins was retained, and the beads were washed four times in the immunoprecipitation buffer and taken up in sodium dodecyl sulfate (SDS)-gel loading buffer. Samples were then analyzed by immunoblotting.
DNA-PK activity assays.
HeLa S3 cells were mock infected or infected at 10 PFU per cell in the presence of 1% dimethyl sulfoxide (DMSO) or 1% DMSO containing 10 μM lactacystin for 8 h. Cells were harvested and lysed, and the DNA-PK activity was assayed as described previously (29).
Western blot (immunoblot) analysis.
HeLa S3 cells at a density of 2 × 105 cells per well in 24-well plates were infected with virus at 10 PFU per cell. After a 1-h adsorption period, medium was added and the infections continued. After 24 h the medium was removed and any cells were pelleted. The cell monolayer was washed in phosphate-buffered saline (PBS), resuspended directly in SDS-gel loading buffer containing 5 mM N-ethylmaleimide, and added to cells pelleted from the medium. When proteasome inhibitors were used, medium containing 1% DMSO or 1% DMSO with lactacystin or MG132 to give a final concentration of 10 μM was added after the adsorption period. Proteins were analyzed by SDS-polyacrylamide gel electrophoresis and Western blotting by the enhanced-chemiluminescence (Amersham) method. Antibodies were stripped from membranes according to the Amersham protocol, and the membranes were reprobed as required.
Immunofluorescence.
HeLa cells were plated at a density of 105 cells per well on glass coverslips in 24-well plates. After infection or transfection the cells were fixed with formaldehyde (5% in PBS containing 2% sucrose) for 10 min at room temperature and permeabilized with acetone-methanol (70:30) for 5 min at −20°C. Samples were initially blocked for 45 min with PBS containing 10% FCS. Antibodies were diluted in PBS containing 1% FCS, and samples were stained with the primary antibody for 1 h, washed with PBS–1% FCS several times, and then stained with the secondary antibody for 30 min. Goat anti-mouse fluorescein isothiocyanate-labelled and goat anti-rabbit tetramethylrodamine isothiocyanate-labelled secondary antibodies (Sigma) were used at a dilution of 1/100. After additional washing, stained coverslips were mounted in Citifluor and examined on a Nikon Microphot-SA microscope with appropriate filters. Images were captured with a Digital Pixel CCD digital camera and prepared for printing by using Photoshop.
Confocal microscopy.
Samples for analysis by confocal microscopy were processed as described above except that goat anti-rabbit cy3 at a dilution of 1/2,000 (Amersham) was used instead of goat anti-rabbit tetramethylrodamine isothiocyanate-labelled secondary antibody. Stained cells were examined with a Zeiss LSM 510 confocal microscope system, with two lasers giving excitation lines at 488 nm (fluorescein isothiocyanate) and 543 nm (cy3), and a Zeiss Axioplan microscope, using a 63× oil immersion objective lens, NA 1.4. The data from the channels were collected simultaneously by using narrow-band filter settings built into the instrument, and channel overlap was not detected. Data were collected with eightfold averaging at a resolution of 1,024 by 1,024 pixels with optical slices between 0.5 and 1 μm. Data sets were processed with the LSM 510 software and then exported for preparation for printing using Photoshop.
Growth of virus on DNA-PK-positive and -negative cells.
MO59J and MO59K cells at 105 cells per well in 24-well plates were infected with virus at 0.05, 0.01, 0.005, or 0.001 PFU per cell, the cells were harvested into the medium 2 days postinfection (dpi), and the progeny virus was titrated on BHK cells.
RESULTS
The RING finger domain but not the HAUSP binding domain of Vmw110 is required for DNA-PKcs degradation.
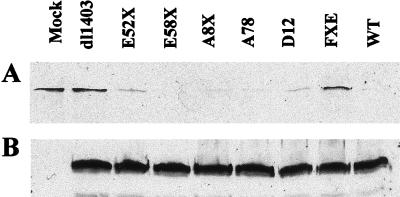
Because of its potential role in the control of protein stability, the interaction of Vmw110 with the cellular USP HAUSP may be important for HSV-1-induced degradation of DNA-PKcs in HeLa S3 cells. To investigate this hypothesis, and to determine which regions of Vmw110 are required for the degradation of DNA-PKcs to occur, HeLa S3 cells were mock infected or infected with wild-type strain 17syn+ virus and the derivative Vmw110 mutant viruses dl1403, E52X, E58X, A8X, A78, D12, and FXE. The properties of the expressed mutant proteins are summarized in Table 1. The cells were harvested 24 h postinfection (hpi) and analyzed by Western blotting (Fig. 1A). This period of infection is longer than that used previously with viruses based on the KOS1.1 wild-type strain (29) because we found that 17syn+ was not as efficient as KOS1.1 at inducing the degradation of DNA-PKcs (data not shown). However, using these conditions we confirmed that strain 17syn+ caused the degradation of DNA-PKcs, whereas the Vmw110 null mutant dl1403 did not (Fig. 1A). The results with the other mutant viruses indicated that there was no correlation between the ability of mutant Vmw110 to bind HAUSP and the effect of the viruses on DNA-PKcs stability; for example, viruses D12 and E52X, which express HAUSP-binding-deficient Vmw110, induced DNA-PKcs degradation while the mutant protein expressed by FXE, which binds HAUSP, did not. Therefore, these results indicate that there is no apparent connection linking the HAUSP-Vmw110 interaction with the degradation of DNA-PKcs during virus infection. However, the failure of mutant virus FXE to induce DNA-PKcs degradation indicates that the RING finger domain of Vmw110 is essential for this effect. In this regard, the results are similar to those concerning the degradation of the high-molecular-weight isoforms of PML, since in that case the RING finger of Vmw110 was also essential (15). As a control for the efficiency of infection in these experiments, the blots were reprobed to monitor the levels of Vmw175 (Fig. 1B) and Vmw110 (data not shown) that were expressed, and the results confirmed that the amounts of the viral proteins that were being expressed by the mutant viruses were comparable.
FIG. 1.
Effect of Vmw110 mutant viruses on DNA-PKcs protein levels. HeLa S3 cells were mock infected or infected with wild-type (WT) 17syn+ virus and Vmw110 mutant viruses as shown at 10 PFU per cell. At 24 hpi cells were harvested into Laemli buffer and samples were analyzed by Western blotting. (A) Blot probed with polyclonal anti-DNA-PKcs DPK1 at a dilution of 1/3,000; (B) same blot reprobed with MAb anti-Vmw175 10176 at a dilution of 1/2,000.
Virus-induced degradation of DNA-PKcs occurs via a proteasome pathway.
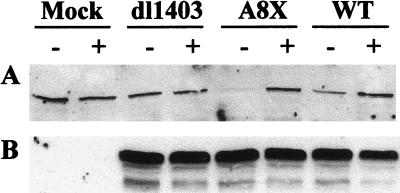
Despite the apparent lack of involvement of HAUSP, it remained possible that the degradation of DNA-PKcs occurs via a proteasome-dependent pathway. To investigate this possibility, cells were mock infected or infected with strain 17syn+ and the mutant viruses dl1403 and A8X in the presence or absence of the proteasome inhibitor MG132. Samples were taken 24 hpi, and analysis by Western blotting showed that virus-induced degradation of DNA-PKcs was prevented in the presence of the proteasome inhibitor (Fig. 2A), which did not of itself affect the level of DNA-PKcs in the cell. As an indication of the efficiency of virus infection, reprobing the blot for Vmw175 showed that, under the conditions employed, Vmw175 levels were unaffected by the inhibitor (Fig. 2B). Levels of Vmw110 expression were also found to be unaffected (data not shown). We conclude that the virus-induced degradation of DNA-PKcs occurs via a proteasome pathway. Similar results were obtained with the proteasome inhibitor lactacystin (data not shown).
FIG. 2.
Prevention of virus-induced DNA-PKcs degradation by the proteasome inhibitor MG132. HeLa S3 cells were mock infected or infected with the viruses as shown at 10 PFU per cell in the presence (+) or absence (−) of 10 μM MG132. Samples were harvested 24 hpi and analyzed by Western blotting. (A) Blot probed with polyclonal anti-DNA-PKcs DPK1 at a dilution of 1/3,000; (B) same blot reprobed with MAb anti-Vmw175 10176 at a dilution of 1/2,000. WT, wild type.
DNA-PK activity is not directly affected by Vmw110.
The decrease in the activity of DNA-PK upon infection (29) may be due directly to the degradation of DNA-PKcs or due to an initial modification of DNA-PKcs caused by Vmw110 prior to degradation. To test this possibility, HeLa S3 cells were mock infected or infected with wild-type strain 17syn+ or KOS1.1 virus in the presence or absence of the proteasome inhibitor lactacystin. Cells were harvested 8 hpi, and whole-cell extracts were prepared for DNA-PK assays and immunoblotting. This time point was selected first because of the experience of Lees-Miller et al. (29) and second because lactacystin is biochemically unstable in aqueous solutions, loosing its potency at later times (7).
In the presence of the proteasome inhibitor the decrease in DNA-PK enzyme activity was substantially less than that observed in its absence during virus infection (Table 2). The decrease in the presence of the inhibitor, especially for KOS1.1-infected cells, is probably due to the fact that lactacystin has to be converted into the active β-lactone form, leading to a delay in the action of the inhibitor (7). As KOS1.1 degrades DNA-PKcs faster than syn17+, this would explain the larger decrease in enzyme activity in the presence of the inhibitor in KOS1.1-infected cells. In the absence of the inhibitor, the decrease in enzyme activity occurred in parallel to the decrease in DNA-PKcs protein levels determined by immunoblotting (data not shown, but see reference 29). Therefore, the reduction in DNA-PK activity upon infection is due to the degradation of the catalytic subunit and not due to Vmw110 directly or indirectly altering the enzyme activity of DNA-PK.
TABLE 2.
DNA-PK activity in the presence of a proteasome inhibitor
| Samplea | Lactacystinb | Activityc | % of mockd |
|---|---|---|---|
| Mock | − | 22.6 | 100 |
| Mock | + | 19.6 | 86.9 |
| KOS1.1 | − | 2.6 | 11.3 |
| KOS1.1 | + | 15.9 | 68.7 |
| 17syn+ | − | 12.1 | 53.6 |
| 17syn+ | + | 21.6 | 93.4 |
Samples of cells infected with the viruses indicated or mock infected were taken 8 h pi.
Absence or presence of lactacystin at 10 μM from time of infection.
DNA-PK activity in units per milligram of protein (average of two independent determinations).
Percentage of level of activity in mock-infected cells without lactacystin.
Vmw110 does not physically interact with either subunit of DNA-PK.
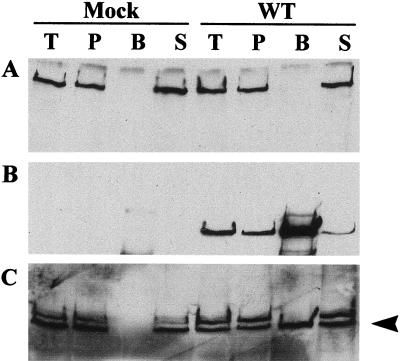
It is possible that the Vmw110-dependent degradation of DNA-PKcs during virus infection requires a direct interaction between the two proteins. We investigated this possibility by conducting anti-Vmw110 immunoprecipitation experiments using extracts from mock-infected cells and strain 17syn+-infected cells which had been harvested before the degradation of DNA-PKcs had been completed. The protein samples were immunoblotted, and probing for DNA-PKcs indicated that coprecipitation of the two proteins was not detectable (Fig. 3A). Reprobing for Vmw110 confirmed that the immunoprecipitation itself was successful (Fig. 3B), and further reprobing for HAUSP (a protein known to be coprecipitated with Vmw110 [35]) indicated that under the conditions of the experiment Vmw110-HAUSP coprecipitation occurred (Fig. 3C). The results were confirmed by performing the experiment in reverse with anti-DNA-PKcs rabbit serum to precipitate DNA-PK from virus-infected cell extracts; again, no coprecipitation of Vmw110 with DNA-PK was observed (data not shown). Similar experiments to investigate the interaction of Vmw110 and Ku subunits indicated that no coprecipitation of these DNA-PK subunits with Vmw110 occurred either (data not shown).
FIG. 3.
DNA-PKcs does not coimmunoprecipitate with Vmw110. HeLa S3 cells were mock infected or infected with wild-type (WT) virus at 10 PFU per cell, and extracts for immunoprecipitation with MAb anti-Vmw110 11060 were made 8 hpi. Proteins that were in the total cell extract (lanes T), that were bound to the preclear beads (lanes P), that immunoprecipitated (lanes B), and that remained in the supernatant after precipitation (lanes S) were analyzed by Western blotting. Blots were probed sequentially with polyclonal anti-DNA-PK DPK1 at a dilution of 1/3,000 (A), MAb anti-Vmw110 11060 at a dilution of 1/10,000 (B), and polyclonal anti-HAUSP r201 at a dilution of 1/1,000 (C). The arrowhead in panel C indicates the band corresponding to HAUSP.
Degradation of DNA-PKcs does not appear to be spatially regulated.
Immunoblot experiments monitor the amount of DNA-PKcs in a total cell population, but we considered it advantageous to be able to monitor the levels of DNA-PKcs in single cells. Therefore, we developed an indirect immunofluorescence assay to determine the fate of DNA-PKcs in individual cells. Staining of two human cell lines, MO59K and MO59J, in which DNA-PKcs is present and absent, respectively, showed that in MO59K cells DNA-PKcs is distributed throughout the nuclei but excluded from the nucleoli but that in MO59J cells no DNA-PKcs is evident (data not shown). This finding indicated that it is possible to use immunofluorescence to distinguish between cells which do and do not contain DNA-PKcs.
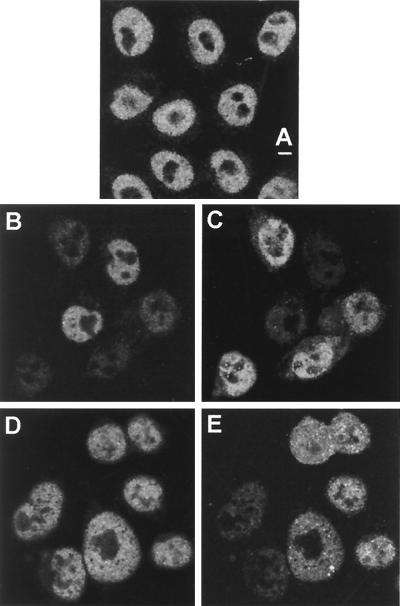
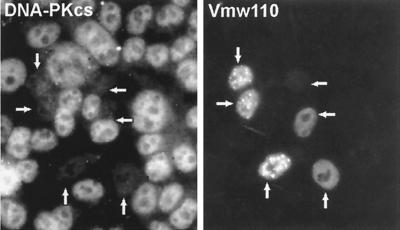
Early in infection Vmw110 localizes to ND10 domains and causes their disruption. As noted above, this effect correlates with the Vmw110-dependent proteasome degradation of a number of high-molecular-weight isoforms of PML and this degradation occurs most efficiently when Vmw110 is capable of colocalizing with PML at ND10 (15). Therefore, the induced degradation of the PML isoforms appears to be spatially regulated in that it occurs in the vicinity of local concentrations of Vmw110. We wished to determine if evidence of a similar effect on DNA-PKcs could be observed. As DNA-PKcs is present throughout the nucleus, it is possible that its degradation may occur in a manner related to the localization of Vmw110 early in infection. To investigate this possibility, cells at different stages of infection were observed by immunofluorescence. No difference in the staining patterns of DNA-PKcs in mock-infected and strain 17syn+-infected cells was observed before 6 hpi (data not shown). After this time, DNA-PKcs fluorescence was completely absent from the nuclei of a proportion of cells, the number of which increased upon infection, and there was no evidence of uneven degradation. For example, compare the degree of DNA-PKcs staining in the uninfected cells (Fig. 4B) with that of the infected cells expressing Vmw110, identified in Fig. 4C. In fact, at the time DNA-PKcs degradation was observed, Vmw110 was no longer present in a punctate, ND10-related pattern but it was diffuse throughout the nucleus (Fig. 4C).
FIG. 4.
Virus-induced degradation of DNA-PKcs seen by confocal microscopy. HeLa S3 cells were mock infected (A) or infected with wild-type syn17+ (B and C) or FXE (D and E) viruses at 10 PFU per cell. The cells were fixed at 8 hpi and costained with MAb anti-DNA-PKcs 18-2 at a dilution of 1/75 (A, B, and D) and polyclonal anti-Vmw110 r190 at a dilution of 1/1,000 (C and E). The bar indicates 5 μm.
The decrease in DNA-PKcs staining in infected cells was seen more rapidly and extensively in cells infected with the KOS1.1 strain than in those infected with the 17syn+ strain, decreasing throughout the nucleus from 2 h onwards. By 8 h there appeared to be two populations of infected cells, one containing DNA-PKcs and one in which DNA-PKcs was absent, the latter being in the majority (data not shown). In cells infected with the Vmw110 mutant FXE (Fig. 4E), infection had no effect on the staining of DNA-PKcs at 8 hpi (Fig. 4D), a time at which at least some loss of DNA-PKcs from cells infected with strain 17syn+ could be observed. This result is consistent with the results of Western blot analysis of DNA-PKcs during infection with this virus (Fig. 1A). The effect of the other mutant viruses on DNA-PKcs as assayed by immunofluorescence was also consistent with Western blot results (data not shown).
Vmw110 alone is sufficient to induce DNA-PKcs degradation.
The results presented above showed that the DNA-PKcs content in an individual cell could be assayed. Therefore, we were able to determine if Vmw110 expressed alone in the absence of other viral proteins was sufficient to cause the degradation. HeLa S3 cells were transfected with pCI110, a plasmid expressing Vmw110, or with pCIneo, a control plasmid expressing the neomycin gene, and analyzed by immunofluorescence microscopy a day later. Transfected cells expressing Vmw110 (Fig. 5) exhibited a clear reduction in DNA-PKcs, which was evident even in cells expressing very low levels of Vmw110. This result is particularly well illustrated by the rightmost two cells in Fig. 5. Cells could occasionally be found in which both Vmw110 and DNA-PKcs were present, but these were the exception. All cells transfected with the control plasmid expressed normal levels of DNA-PKcs, and although there was no means of determining which cells were transfected, these normal levels indicated that the transfection conditions per se did not affect DNA-PKcs (data not shown). This result clearly shows that Vmw110 is by itself sufficient to cause the loss of DNA-PKcs from the cell, and therefore this provides the first evidence that Vmw110 affects protein stability in the absence of other viral factors.
FIG. 5.
Vmw110 expressed from a plasmid is sufficient to induce DNA-PKcs degradation. HeLa S3 cells were transfected with plasmid pCI110 expressing Vmw110, processed for immunofluorescence after 24 h, and costained with polyclonal anti-DNA-PKcs DPK1 at a dilution of 1/75 and MAb anti-Vmw110 11060 at a dilution of 1/2,000. Arrows indicate transfected cells. Note that the upper right cell only weakly expressed Vmw110 but that, despite this, DNA-PKcs was still eliminated.
DNA-PK affects the replication of HSV-1 in mammalian cells.
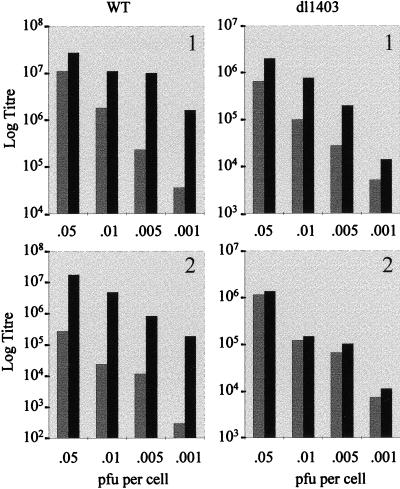
While the above results clearly show that Vmw110 by itself is able to induce the degradation of DNA-PKcs via a mechanism requiring the RING finger domain of Vmw110 and an active proteasome pathway, the significance of these observations for the outcome of virus infection remains to be clarified. The availability of the human cell lines MO59K and MO59J (see above) made it possible to address the issue of whether the presence of active DNA-PK influences the efficiency of viral replication. MO59K and MO59J cells were infected with 17syn+ and dl1403 viruses at a variety of input multiplicities, and the virus yield after 2 days was determined by titration in BHK cells. The results showed that virus yield from MO59J cells was greater than from MO59K cells for both viruses, and this difference became more pronounced and significant at low multiplicities of infection (MOI) (Table 3). Data comparing each individual virus in the two cell lines in two independent experiments are represented visually in Fig. 6. In the first experiment, up to 50-fold more strain 17syn+ virus was produced in MO59J cells than in MO59K cells, but this difference was only up to 8-fold more with dl1403. In the second experiment, the effect was more impressive, with MO59J cells producing up to 400-fold more wild-type virus but only up to 2-fold more dl1403 virus. Since the infections were performed at low multiplicity and progeny virus was harvested 2 dpi, the results represent the yields from successive rounds of virus infection. Therefore, these data indicate that the presence of DNA-PK has a moderate but reproducible inhibitory effect on the ability of the virus to replicate.
TABLE 3.
Virus growth on MO59K and MO59J cells
| Virus | MOI | Virus yield (PFU/ml)
|
|||
|---|---|---|---|---|---|
| Expt 1
|
Expt 2
|
||||
| MO59K | MO59J | MO59K | MO59J | ||
| 17syn+ | 0.05 | 1.1 × 107 | 2.7 × 107 | 2.7 × 105 | 1.7 × 107 |
| 0.01 | 1.8 × 106 | 1.1 × 107 | 2.3 × 104 | 4.8 × 106 | |
| 0.005 | 2.3 × 105 | 1.0 × 107 | 1.1 × 104 | 8.2 × 105 | |
| 0.001 | 3.6 × 104 | 1.6 × 106 | 3.0 × 102 | 1.8 × 105 | |
| dl1403 | 0.05 | 6.6 × 105 | 2.0 × 106 | 1.2 × 106 | 1.4 × 106 |
| 0.01 | 1.0 × 105 | 7.8 × 10105 | 1.2 × 105 | 1.5 × 105 | |
| 0.005 | 2.8 × 104 | 2.1 × 105 | 6.6 × 104 | 1.0 × 105 | |
| 0.001 | 5.2 × 103 | 1.4 × 104 | 7.4 × 103 | 1.1 × 104 | |
FIG. 6.
Replication of wild-type (WT) and dl1403 viruses in MO59K and MO59J cells. MO59K and MO59J cells were infected with wild-type or dl1403 virus at the MOI shown, and virus yields were titrated on BHK cells 2 days pi. Graphs show virus yields (log titers) versus input MOI (PFU per cell) for wild-type and dl1403 viruses in MO59K (gray bars) and MO59J (black bars) cells from two separate experiments. (Note that as the comparison is between the two cell lines and not between the two viruses, the scale range for log titers is different for each virus. In experiment 2 the effect of the MOI was greater on MO59K cells than in experiment 1, so the lower limit of the graph was reduced.)
Although the DNA-PKcs in MO59K cells will be degraded by the action of Vmw110 during wild-type virus infection, this takes several hours to complete, so the reduction in virus yield in these cells compared to that in MO59J cells is consistent with DNA-PK remaining active in the early stages of infection. For example, 54% of DNA-PKcs remained active in HeLa S3 cells after 8 h of infection with strain 17syn+ (Table 2). In both experiments, the differential between the virus yields from the two cell types increased by an order of magnitude over the input multiplicity range used, and this multiplicity dependence is consistent with the multiplicity-related requirement for Vmw110 during infection. The interpretation of the data obtained with Vmw110 null mutant dl1403 is not straightforward. On one hand, the presence of active DNA-PK in MO59K cells does not have an effect on the replication of the mutant virus. On the other hand, because the absence of Vmw110 results in an extremely high particle-to-PFU ratio for this virus, these infections are effectively being conducted at much higher multiplicities in terms of potentially active virus particles than those of the wild-type virus infections, and these higher MOI may overcome any inhibitory effect of the activity of DNA-PK.
DISCUSSION
This paper demonstrates that the expression of Vmw110 during HSV-1 infection, and by itself in transfected cells, leads to the active degradation of the catalytic subunit of DNA-PK, a nuclear enzyme which plays a role in double-strand DNA break repair, in V(D)J recombination, and possibly in the regulation of transcription and apoptosis. Vmw110 has also previously been shown to interact with and bind to a novel cellular USP named HAUSP. This fact together with the degradation of DNA-PKcs suggested a hypothesis in which Vmw110 could prevent HAUSP from removing polyubiquitin chains from DNA-PKcs, thus rendering it susceptible to degradation by the ubiquitin-dependent proteasome pathway. Using Vmw110 mutant viruses expressing Vmw110 proteins which do or do not bind HAUSP, we have shown that there is no apparent correlation between the binding of HAUSP by Vmw110 and the degradation of DNA-PKcs, thus disproving this hypothesis. However, the RING finger of Vmw110 was found to be essential for the degradation of DNA-PKcs, since virus FXE, which expresses a Vmw110 protein lacking this domain, failed to induce degradation. Infecting in the presence of proteasome inhibitors also prevented DNA-PKcs degradation, indicating that a proteasome pathway is involved. This pathway is not used as a result of Vmw110 modifying DNA-PK such that the altered form is recognized as an aberrant conformation, leading to targeting of DNA-PKcs for degradation, as DNA-PK was still active in the presence of proteasome inhibitors. The effect of Vmw110 on DNA-PKcs is probably indirect and may not be specific but rather the result of a more global effect, as discussed below. This idea is strengthened by the fact that no evidence of a direct interaction between the two proteins was found.
In view of the fact that a proteasome pathway is involved in the induced degradation of DNA-PKcs, it is surprising that HAUSP appears to play no role. It is certainly an obvious candidate for involvement, and although the significance of the interaction between Vmw110 and HAUSP is still unclear, it is likely to be of importance in some aspect of the virus life cycle. The requirement for the RING finger region of Vmw110 may indicate that this region interacts with other cellular proteins, leading to the activation of the degradation pathway. Despite the apparent lack of involvement of HAUSP, as degradation of DNA-PKcs occurs via a proteasome pathway, it is possible that DNA-PKcs is ubiquitinated. The ubiquitin-mediated protein degradation pathway is a common mechanism for the regulation of the level of a protein within a cell and can lead to very rapid elimination of the target protein; it involves the formation of a polyubiquitin chain on the protein, which targets the protein to the proteasome for degradation. Alternatively, we have found that HSV-1 infection results in the degradation of several high-molecular-weight isoforms of PML and a large number of uncharacterized PIC1-conjugated proteins. This process also requires the Vmw110 RING finger and active proteasomes (15), and the parallel between the fates of PML and DNA-PKcs suggests that a similar general mechanism may be operating in both cases. The PML isoforms that are targeted for degradation are likely themselves to be PIC1 conjugates, and therefore it is possible that the common feature is PIC1 conjugation and that DNA-PKcs is a natural PIC1-conjugated protein. Indeed, PIC1 has been found to be covalently conjugated to a large number of cellular proteins, including many nuclear proteins (23).
We attempted to test these possibilities by immunoprecipitation of DNA-PKcs and probing for the presence of ubiquitin or PIC1 in the precipitated protein by immunoblotting. To detect modified DNA-PKcs, immunoprecipitates were probed for endogenous ubiquitin or PIC1 with specific antibodies or for epitope-tagged versions of ubiquitin or PIC1 in extracts from transfected or electroporated cells. However, none of the approaches were successful, which suggests that any potential modification by ubiquitin or PIC1 of DNA-PKcs is difficult to detect. Similar problems of detecting endogenous PIC1-conjugated forms of PML with anti-PIC1 antibodies have been experienced (47), and unlike the case of PML, any higher-molecular-weight isoforms of DNA-PKcs resulting from its modification would be difficult to resolve on gels due to the large size of DNA-PKcs itself. For these reasons, we consider that current evidence is insufficient to provide confident conclusions on the possible modification of DNA-PKcs by ubiquitin or related polypeptides.
The degradation of certain selected proteins upon infection is probably a mechanism for aiding virus replication. This idea was strengthened by the finding that the virus replicated more efficiently in DNA-PKcs-negative cells (MO59J) than in DNA-PKcs-positive cells (MO59K), suggesting that the Vmw110-mediated elimination of DNA-PKcs might enhance virus replication. The improved virus growth in MO59J compared to that in MO59K cells was not particularly large, but it was reproducible over a number of experiments, and this finding poses the question of why the lack of DNA-PK activity may be beneficial to virus replication. A relevant consideration is that elimination of DNA-PK activity takes several hours to complete, whereas a major requirement for Vmw110 occurs in the earliest stages of infection, during commitment to the life cycle. One hypothesis is that since DNA-PK is required for nonhomologous end joining of DNA, it may interfere with the proper replication and dynamics of progeny viral genomes. The depletion of DNA-PK during infection may alter the recombination and repair mechanisms of the host cell to prevent this. Another possibility is that elimination of DNA-PK activity alters the modification of key cellular regulatory factors that promote transcription of the viral DNA or translation or the functions of the viral protein products. DNA-PK is known to inhibit transcription by RNAP II (18), so inhibition of DNA-PK activity may provide a mechanism for increasing the levels of RNAP II transcription on viral DNA. For example, it has been suggested that DNA-PK may interfere with the basal or activated levels of RNAP II transcription of viral genes, both when viral DNA first enters the nucleus and when viral DNA enters a quiescent latent state in neurons. DNA-PK may accomplish this repression by phosphorylating (and inactivating) transcription factors such as Oct-1 that enhance viral gene expression (5, 18, 27, 29), and this possibility is consistent with the finding that activation of gene expression by Vmw110 occurs at or before initiation of transcription (22). However, the time course of loss of DNA-PKcs from the cell is not compatible with this latter hypothesis, and we are more in favor of the view that enhanced replication in the absence of DNA-PKcs is due to its role in DNA mechanics.
ACKNOWLEDGMENTS
We thank Joan Turner for the human MO59J and MO59K cell lines and Duncan McGeoch for constructive criticism.
This research was supported by the Medical Research Council, by a grant from the Medical Research Council of Canada and the Natural Sciences and Engineering Research Council of Canada, and by a visiting-scientist award to Jane Parkinson from the Alberta Heritage Foundation for Medical Research.
REFERENCES
- 1.Ascoli C A, Maul G G. Identification of a novel nuclear domain. J Cell Biol. 1991;112:785–795. doi: 10.1083/jcb.112.5.785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boddy M N, Howe K, Etkin L D, Solomon E, Freemont P S. PIC1, a novel ubiquitin-like protein which interacts with the PML component of a multiprotein complex that is disrupted in acute promyelocytic leukaemia. Oncogene. 1996;13:971–982. [PubMed] [Google Scholar]
- 3.Cai W, Astor T D, Liptak L M, Cho C, Coen D, Schaffer P A. The herpes simplex virus type 1 regulatory protein ICP0 enhances replication during acute infection and reactivation from latency. J Virol. 1993;67:7501–7512. doi: 10.1128/jvi.67.12.7501-7512.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chan D W, Lees-Miller S P. The DNA-dependent protein kinase is inactivated by autophosphorylation of the catalytic subunit. J Biol Chem. 1996;271:8936–8941. doi: 10.1074/jbc.271.15.8936. [DOI] [PubMed] [Google Scholar]
- 5.Chibazakura T, Watanabe F, Kitajima S, Tsukada K, Yasukochi Y, Teraoka H. Phosphorylation of human general transcription factors TATA-binding protein and transcription factor IIB by DNA-dependent protein kinase—synergistic stimulation of RNA polymerase II basal transcription in vitro. Eur J Biochem. 1997;247:1166–1173. doi: 10.1111/j.1432-1033.1997.01166.x. [DOI] [PubMed] [Google Scholar]
- 6.Clements G B, Stow N D. A herpes simplex virus type 1 mutant containing a deletion within immediate-early gene 1 is latency competent in mice. J Virol. 1989;70:2501–2506. doi: 10.1099/0022-1317-70-9-2501. [DOI] [PubMed] [Google Scholar]
- 6a.Cross, A., and R. Everett. Unpublished data.
- 7.Dick L R, Cruikshank A A, Destree A T, Grenier L, McCormack T A, Melandri F D, Nunes S L, Palombella V J, Parent L A, Plamondon L, Stein R L. Mechanistic studies on inactivation of the proteasome by lactacystin in cultured cells. J Biol Chem. 1997;272:182–188. doi: 10.1074/jbc.272.1.182. [DOI] [PubMed] [Google Scholar]
- 7a.Dyck J A, Maul G G, Miller W H, Jr, Chen J D, Kakizuka A, Evans R M. A novel macromolecular structure is a target of the promyelocytic-retinoic acid receptor oncoprotein. Cell. 1994;76:333–343. doi: 10.1016/0092-8674(94)90340-9. [DOI] [PubMed] [Google Scholar]
- 8.Dynan W D, Yoo S. Interaction of Ku protein and DNA-dependent protein kinase catalytic subunit with nucleic acids. Nucleic Acids Res. 1998;26:1551–1559. doi: 10.1093/nar/26.7.1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Everett R D. Construction and characterisation of herpes simplex virus type 1 mutants with defined lesions in immediate-early gene 1. J Gen Virol. 1989;70:1185–1202. doi: 10.1099/0022-1317-70-5-1185. [DOI] [PubMed] [Google Scholar]
- 10.Everett R D, Preston C M, Stow N D. Functional and genetic analysis of the role of Vmw110 in herpes simplex virus replication. In: Wagner E K, editor. The control of herpes simplex virus gene expression. Boca Raton, Fla: CRC Press, Inc.; 1991. pp. 50–76. [Google Scholar]
- 11.Everett R D, Cross A, Tyler J K, Orr A. An epitope within the DNA binding domain of the herpes simplex virus immediate-early protein Vmw175 is conserved in the varicella-zoster virus gene 62 protein. J Gen Virol. 1993;74:1955–1958. doi: 10.1099/0022-1317-74-9-1955. [DOI] [PubMed] [Google Scholar]
- 12.Everett R D, Cross A, Orr A. A truncated form of herpes simplex virus type 1 immediate-early protein Vmw110 is expressed in a cell-type dependent manner. Virology. 1993;197:751–756. doi: 10.1006/viro.1993.1651. [DOI] [PubMed] [Google Scholar]
- 13.Everett R D, Maul G G. HSV-1 IE protein Vmw110 causes redistribution of PML. EMBO J. 1994;13:5062–5069. doi: 10.1002/j.1460-2075.1994.tb06835.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Everett R D, Meredith M R, Orr A, Cross A, Kathoria M, Parkinson J. A novel ubiquitin-specific protease is dynamically associated with the PML nuclear domain and binds to a herpesvirus regulatory protein. EMBO J. 1997;16:1519–1530. doi: 10.1093/emboj/16.7.1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Everett R D, Freemont P, Saitoh H, Dasso M, Orr A, Kathoria M, Parkinson J. The disruption of the ND10 during herpes simplex virus infection correlates with the Vmw110- and proteasome-dependent loss of several PML isoforms. J Virol. 1998;72:6581–6591. doi: 10.1128/jvi.72.8.6581-6591.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Everett R D, Meredith M, Orr A. The ability of herpes simplex virus type 1 immediate-early protein Vmw110 to bind to a ubiquitin-specific protease contributes to its role in the activation of gene expression and stimulation of virus replication. J Virol. 1999;73:417–426. doi: 10.1128/jvi.73.1.417-426.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fraser N W, Block T M, Spivack J G. The latency associated transcripts of herpes simplex virus: RNA in search of a function. Virology. 1992;191:1–8. doi: 10.1016/0042-6822(92)90160-q. [DOI] [PubMed] [Google Scholar]
- 18.Giffin W, Torrence H, Rodda D J, Prefontaine G G, Pope L, Hache R J G. Sequence-specific DNA binding by Ku autoantigen and its effects on transcription. Nature (London) 1996;380:265–268. doi: 10.1038/380265a0. [DOI] [PubMed] [Google Scholar]
- 19.Harris R A, Everett R D, Zhu X, Silverstein S, Preston C M. The HSV immediate early protein Vmw110 reactivates latent HSV type 2 in an in vitro latency system. J Virol. 1989;63:3513–3515. doi: 10.1128/jvi.63.8.3513-3515.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hochstrasser M. Ubiquitin, proteasomes, and the regulation of intracellular protein degradation. Curr Opin Cell Biol. 1995;7:215–223. doi: 10.1016/0955-0674(95)80031-x. [DOI] [PubMed] [Google Scholar]
- 21.Johnson P R, Hochstrasser M. SUMO-1: ubiquitin gains weight. Trends Cell Biol. 1997;7:408–413. doi: 10.1016/S0962-8924(97)01132-X. [DOI] [PubMed] [Google Scholar]
- 22.Jordan R, Schaffer P A. Activation of gene expression by herpes simplex virus type 1 ICP0 occurs at the level of mRNA synthesis. J Virol. 1997;71:6850–6862. doi: 10.1128/jvi.71.9.6850-6862.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kamitani T, Nguyen H P, Yeh E T H. Preferential modification of nuclear proteins by a novel ubiquitin-like molecule. J Biol Chem. 1997;272:14001–14004. doi: 10.1074/jbc.272.22.14001. [DOI] [PubMed] [Google Scholar]
- 24.Kamitani T, Nguyen H P, Kito K, FuKuda-Kamitani T, Yeh E T H. Covalent modification of PML by the Sentrin family of ubiquitin-like proteins. J Biol Chem. 1998;273:3117–3120. doi: 10.1074/jbc.273.6.3117. [DOI] [PubMed] [Google Scholar]
- 25.Kawaguchi Y, van Sant C, Roizman B. Herpes simplex virus regulatory protein ICP0 interacts with and stabilizes the cell cycle regulator cyclin D3. J Virol. 1997;71:7328–7336. doi: 10.1128/jvi.71.10.7328-7336.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Korioth F, Gieffers C, Maul G G, Frey J. Molecular characterisation of NDP52, a novel protein of nuclear domain 10 which is redistributed upon infection and interferon treatment. J Cell Biol. 1995;130:1–14. doi: 10.1083/jcb.130.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lees-Miller S P. The DNA-dependent protein kinase DNA-PK: 10 years and no ends in sight. Biochem Cell Biol. 1996;74:503–512. doi: 10.1139/o96-054. [DOI] [PubMed] [Google Scholar]
- 28.Lees-Miller S P, Godbout R, Chan D W, Weinfeld M W, Day III R S, Barron G M, Allalunis-Turner J. Absence of the p350 subunit of DNA-activated protein kinase from a radiosensitive human cell line. Science. 1995;267:1183–1185. doi: 10.1126/science.7855602. [DOI] [PubMed] [Google Scholar]
- 29.Lees-Miller S P, Long M C, Kilvert M A, Lam V, Rice S A, Spencer C A. Attenuation of DNA-dependent protein kinase activity and its catalytic subunit by the herpes simplex virus type 1 transactivator ICP0. J Virol. 1996;70:7471–7477. doi: 10.1128/jvi.70.11.7471-7477.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Leib D A, Coen D M, Bogard C L, Hicks K A, Yager D R, Knipe D M, Tyler K L, Schaffer P A. Immediate early regulatory gene mutants define different stages in the establishment and reactivation of herpes simplex virus latency. J Virol. 1989;63:759–768. doi: 10.1128/jvi.63.2.759-768.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Maul G G, Guldner H H, Spivack J G. Modification of discrete nuclear domains induced by herpes simplex virus type 1 immediate-early gene 1 product ICP0. J Gen Virol. 1993;74:2679–2690. doi: 10.1099/0022-1317-74-12-2679. [DOI] [PubMed] [Google Scholar]
- 32.Maul G G, Everett R D. The nuclear location of PML, a cellular member of the C3HC4 zinc binding domain protein family, is rearranged during herpes simplex virus infection by the C3HC4 viral protein ICP0. J Gen Virol. 1994;75:1223–1233. doi: 10.1099/0022-1317-75-6-1223. [DOI] [PubMed] [Google Scholar]
- 33.McCarthy A M, McMahan L, Schaffer P A. Herpes simplex virus type 1 ICP27 mutants exhibit altered patterns of transcription and are DNA deficient. J Virol. 1989;63:18–27. doi: 10.1128/jvi.63.1.18-27.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McGeoch D J, Barnett B C, MacLean C A. Emerging functions of alphaherpesvirus genes. Semin Virol. 1993;4:125–134. [Google Scholar]
- 35.Meredith M R, Orr A, Everett R D. Herpes simplex virus type 1 immediate-early protein Vmw110 binds strongly and specifically to a 135 kd cellular protein. Virology. 1994;200:457–469. doi: 10.1006/viro.1994.1209. [DOI] [PubMed] [Google Scholar]
- 36.Meredith M R, Orr A, Elliott M, Everett R D. Separation of the sequence requirements for HSV-1 Vmw110 multimerisation and interaction with a 135kD cellular protein. Virology. 1995;209:174–187. doi: 10.1006/viro.1995.1241. [DOI] [PubMed] [Google Scholar]
- 37.Muller S, Matunis M J, DeJean A. Conjugation of the ubiquitin-related modifier SUMO-1 regulates the partitioning of PML within the nucleus. EMBO J. 1998;17:61–70. doi: 10.1093/emboj/17.1.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Phelan A, Carmo-Fonseca M, McLaughlan J, Lamond A I, Clements J B. A herpes simplex virus 1 immediate-early gene product, IE63, regulates small nuclear ribonucleoprotein distribution. Proc Natl Acad Sci USA. 1993;90:9056–9060. doi: 10.1073/pnas.90.19.9056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Poffenberger K L, Raichlen P E, Herman R C. In vitro characterisation of a herpes simplex virus type 1 ICP22 deletion mutant. Virus Genes. 1993;7:171–186. doi: 10.1007/BF01702397. [DOI] [PubMed] [Google Scholar]
- 40.Rice S A, Long M L, Lam V, Schaffer P A, Spencer C A. Herpes simplex virus immediate-early protein ICP22 is required for viral modification of host RNA polymerase II and establishment of the normal viral transcription program. J Virol. 1995;69:5550–5559. doi: 10.1128/jvi.69.9.5550-5559.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Roizman B, Sears A E. Herpes simplex viruses and their replication. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology. 3rd ed. New York, N.Y: Raven Press; 1996. pp. 2231–2296. [Google Scholar]
- 42.Sacks W R, Schaffer P A. Deletion mutants in the gene encoding the herpes simplex virus type 1 immediate-early protein ICP0 exhibit impaired growth in cell culture. J Virol. 1987;61:829–839. doi: 10.1128/jvi.61.3.829-839.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Saitoh H, Pu R T, Dasso M. SUMO-1: wrestling with a new ubiquitin-related modifier. Trends Biochem Sci. 1997;22:374–376. doi: 10.1016/s0968-0004(97)01102-x. [DOI] [PubMed] [Google Scholar]
- 44.Sandri-Goldin R M, Mendoza G E. A herpes simplex virus regulatory protein appears to act post-transcriptionally during infection to regulate gene expression. Genes Dev. 1992;6:848–863. doi: 10.1101/gad.6.5.848. [DOI] [PubMed] [Google Scholar]
- 45.Smiley J R, Panning B, Smibert C A. Regulation of cellular genes by HSV products. In: Wagner E K, editor. Herpesvirus transcription and regulation. Boca Raton, Fla: CRC Press, Inc.; 1991. pp. 151–179. [Google Scholar]
- 46.Smith C A, Bates P, Rivera-Gonzalez R, Gu B, DeLuca N A. ICP4, the major transcriptional regulatory protein of herpes simplex virus type 1, forms a tripartite complex with TATA-binding protein and TFIIB. J Virol. 1993;67:4676–4687. doi: 10.1128/jvi.67.8.4676-4687.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sternsdorf T, Jensen K, Will H. Evidence for covalent modification of the nuclear dot-associated proteins PML and Sp100 by PIC1/SUMO-1. J Cell Biol. 1997;139:1621–1634. doi: 10.1083/jcb.139.7.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stow E C, Stow N D. Complementation of a herpes simplex virus type 1 deletion mutant by human cytomegalovirus. J Gen Virol. 1989;70:695–704. doi: 10.1099/0022-1317-70-3-695. [DOI] [PubMed] [Google Scholar]
- 49.Stow N D, Stow E C. Isolation and characterisation of a herpes simplex virus type 1 mutant containing a deletion within the gene encoding the immediate-early polypeptide Vmw110. J Gen Virol. 1986;67:2571–2585. doi: 10.1099/0022-1317-67-12-2571. [DOI] [PubMed] [Google Scholar]
- 50.Stuurman N, DeGraaf A, Josso A, Humbel B, DeYong L, van Driel R. A monoclonal antibody recognising nuclear matrix associated nuclear bodies. J Cell Sci. 1992;101:773–784. doi: 10.1242/jcs.101.4.773. [DOI] [PubMed] [Google Scholar]
- 51.Wagner E K. Herpesvirus transcription—general aspects. In: Wagner E K, editor. Herpesvirus transcription and regulation. Boca Raton, Fla: CRC Press, Inc.; 1991. pp. 1–17. [Google Scholar]
- 52.Weaver D T. Regulation and repair of double-strand DNA breaks. Crit Rev Eukaryot Gene Expr. 1996;6:345–375. doi: 10.1615/critreveukargeneexpr.v6.i4.20. [DOI] [PubMed] [Google Scholar]
- 53.Wilkinson K D. Roles of ubiquitination in proteolysis and cellular regulation. Annu Rev Nutr. 1995;15:161–189. doi: 10.1146/annurev.nu.15.070195.001113. [DOI] [PubMed] [Google Scholar]
- 54.Zhu X, Chen J, Young C S H, Silverstein S. Reactivation of latent herpes simplex virus by adenovirus recombinants encoding mutant IE-0 gene products. J Virol. 1990;64:4489–4498. doi: 10.1128/jvi.64.9.4489-4498.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]