Abstract
Neurocognitive impairment continues to be common comorbidity for people living with HIV (PLWH). Given the chronic nature of HIV disease, identifying reliable biomarkers of these impairments is essential to advance our understanding of the underlying neural foundation and facilitate screening and diagnosis in clinical care. While neuroimaging provides immense potential for such biomarkers, to date, investigations in PLWH have been mostly limited to either univariate mass techniques or a single neuroimaging modality. In the present study, connectome-based predictive modeling (CPM) was proposed to predict individual differences of cognitive functioning in PLWH, using resting-state functional connectivity (FC), white matter structural connectivity (SC), and clinical relevant measures. We also adopted an efficient feature selection approach to identify the most predictive features, which achieved an optimal prediction accuracy of r = 0.61 in the discovery dataset (n = 102) and r = 0.45 in an independent validation HIV cohort (n = 88). Two brain templates and nine distinct prediction models were also tested for better modeling generalizability. Results show that combining multimodal FC and SC features enabled higher prediction accuracy of cognitive scores in PLWH, while adding clinical and demographic metrics may further improve the prediction by introducing complementary information, which may help better evaluate the individual-level cognitive performance in PLWH.
Keywords: Individualized prediction, NeuroHIV, Neurocognitive impairment, Functional connectivity, Structural connectivity
I. Introduction
Widespread use of combination antiretroviral therapy (cART) has greatly extended the lifespan of people living with HIV (PLWH), but prevalence rates of neurocognitive impairment remain high, although deficits are usually mild to moderate [1]–[3]. The persistence of HIV-associated neurocognitive disorders (HAND) in PLWH may result from an irreversible central nervous system injury before the initiation of treatment [4], as well as a process of immune activation which persists after treatment [5]. HIV-associated neurocognitive deficits manifest in various areas, such as executive function, motor control, attention/working memory, and visuospatial processing [6], [7]. These deficiencies can impact the daily activities of PLWH, significantly decreasing the quality of their life [8]–[10], and they are also associated with HIV disease progression and increased risk of mortality [11]–[13]. Tracking changes in cognitive performance following HIV infection allows for early diagnosis to optimize long-term neurocognitive outcomes [14], [15].
While neuropsychological (NP) assessments are essential in diagnosing HAND among PLWH, they have several potential flaws. Specifically, NP batteries need to be completed and evaluated by a professional neuropsychologist and advised by an infectious disease clinician, which is not readily available in many communities. The tests can be sensitive to practice effects, limiting the ability to administer across short intermissions repeatedly. In addition, linguistic and cultural differences between PLWH around the world may affect the effectiveness of standardized tests, requiring additional validation and norms appropriate for each region [16], [17]. Meanwhile, some neuroimaging studies of HIV disease have shown that magnetic resonance imaging (MRI) may be sensitive in detecting brain abnormalities associated with neurocognitive impairments [18]–[20]. Given the relative objectivity of MRI results, neuroimaging has excellent potential to provide a readily operable and clinically relevant biomarker of HAND. Many studies have consistently suggested that MRI can efficiently identify subtle brain changes that occur prior to the clinical onset of overt behavioral deficits [21], which can improve the accuracy and diagnostic efficiency of HAND and help understand the mechanisms underlying neurocognitive impairment.
While both functional and structural MRI have independently revealed a neurobiological basis of neuroHIV, leveraging multimodal imaging has the potential to provide complementary information and offer new insights into HIV disease. Specifically, diffusion-weighted tractography has been used to explore microstructure variances in white matter (WM) tracts [22], [23]. PLWH typically display WM impairment across diffuse projection, association, and commissural tracts relative to healthy controls (HC) [24]. While deficits in the corpus callosum have been most frequently observed [25], the uncinate fasciculus and forceps minor are also commonly affected in HIV disease [26]–[28]. Resting-state functional MRI (rs-fMRI) analyses have identified HIV-related impairments in functional connectivity (FC) patterns involving the striatal, dorsolateral prefrontal cortex, parietal regions, and default mode network (DMN) [29]–[31]. DMN and frontoparietal network as well as DMN and sensorimotor network have demonstrated high connectivity, while inherent hypo-connectivity also have been found between the salience network and executive network [32], [33]. Both diffusion and functional imaging have been associated with impaired cognitive function in PLWH [18], [34], [35]. However, few studies have investigated the neurobiological substrates of HIV using multimodal neuroimaging features.
Despite the well-documented association between cognitive function and neuroimaging measures [36], most neuroHIV studies utilized univariate analysis methods (e. g., t-test and ANOVA), which primarily provide information about cognitive impairment at the group level. In addition, recent neuroHIV studies used neuroimaging features to classify persons into cognitively normal versus cognitively impaired using machine learning approaches [37], [38]. However, cognitive dysfunction is a continuous degenerative process, and forward-looking prediction of cognitive ability at the individual level is critical to determine appropriate remediation strategies before severe cognitive impairment occurs. As a popular machine learning method, connectome-based predictive modeling (CPM) can make full use of structural connectivity (SC) and/or FC measurements to quantitatively predict individuals’ cognitive performance [39], and has been used effectively to predict various cognitive behaviors from brain connectivity patterns, such as personality traits [40], [41], attention [42], creative capability [43], [44], and fluid intelligence [45], [46]. To date, one study applied CPM to predict global cognitive scores of 102 PLWH based on FC, with a predictive accuracy of r = 0.578 for the training set and r = 0.359 for the testing set [47]. Although some studies have successfully established correlations between FC and SC characteristics and cognitive behavior using prediction models [48], other studies have come to different or even opposite conclusions. For example, some researchers have used biological measures, SC, and FC signatures to better predict cognitive performance [49]–[51]. However, Mellem, et al. [52] developed prediction models that included a clinical scale assessment and both SC and FC features and found that structural features did not substantially contribute to the prediction strength of these models. Since white matter degradation is a characteristic feature of neuroHIV [53], we hypothesized that utilizing multimodal connectivity data, especially white matter SC and FC with CPM would improve the prediction accuracy of cognitive function in PLWH.
This study aimed to investigate the neural substrate in PLWH of cognitive dysfunction in the context of individualized prediction using multimodal MRI data. Specifically, we incorporated SC and FC as features into the CPM pipeline. In addition to incorporating demographics into the model, the impact of HIV characteristics and substance use was also taken into account. Considering that HIV clinical features and substance use are independently predictive of cognition in PLWH [38], [54], [55], we believed combining these characteristics with neuroimaging data may substantially improve estimation of global neurocognitive impairment.
II. MATERIALS AND METHODS
A. Participants and Study Procedures
Through advertisements in local infectious disease clinics, communities, and the internet, we recruited PLWH from the Raleigh-Durham area of North Carolina, USA. All participants had a confirmed diagnosis of HIV and were linked to HIV care. The discovery dataset included 102 participants recruited from 2010 to 2018, as described in [38]. The validation dataset was an independent cohort of eighty-eight participants recruited using similar strategies from 2017 to 2020. The parent studies examined the effects of substance use (including alcohol, marijuana, and/or cocaine) combined with HIV on the brain. The age range of all participants was 21–56. The exclusion criteria included any illicit drug use other than marijuana and cocaine for two days or more, less than eight years of education or severe learning disability, lack of fluency or illiteracy in English, unresolved neurological disorders or neuroinfectious, severe traumatic brain injury, severe mental illness or impaired mental status, and any MRI contraindications.
After screening interviews, including clinical interviews, questionnaires, and urine drug tests, eligible participants completed NP testing and the MRI scan on a subsequent day. Participants also signed release forms to allow staff to review their medical records, check for any exclusionary conditions, and abstract relevant clinical variables. Participants provided written informed consent, and all procedures were approved by the Duke University Health System Institutional Review Board.
B. NP Testing and Clinical Features
Cognitive function was assessed across 7 domains: (1) executive function (Trail Making Test Part B [56] and Stroop Interference [57]); (2) processing speed (Trail Making Test Part A [56] and Stroop Color Naming [57]); (3) attention/working memory (Paced Auditory Serial Addition Task-50 [58] and NAB Digit Span: forward & backward [59]); (4) verbal fluency (FAS letter fluency and category fluency – animals [60]); (5) learning (Hopkins Verbal Learning Test-Revised (HVLT-R), immediate trials [61]); (6) motor functioning (Grooved Pegboard Test, dominant and non-dominant hands [62]); (7) memory (HVLT-R, delayed trial [61]). The test battery was the same in the discovery and validation datasets, except that the validation dataset did not include the NAB Digit Span scores. Demographically corrected T-scores (mean ± standard deviation = 50 ± 10) were derived from raw scores for each test [58], [59], [63], [64]. Domain T-scores were calculated by averaging all tests in a domain, and global T-scores were calculated by averaging seven domain T-scores.
HIV clinical data were abstracted from participant medical records, including HIV disease duration, current and nadir CD4+ T-cell count, and the recent HIV viral load. Participants’ self-reported demographic characteristics such as age, gender, race, and education. The Addiction Severity Index-Lite assessed substance uses in the past 30 days, including alcohol, cigarettes, marijuana, and cocaine [65].
C. Image Acquisition and Preprocessing
All scans were collected on the same 3T GE Discovery MR750 imaging system with an 8-channel head coil at Duke University Hospital. The discovery dataset were acquired across three protocols and the potential effects were controlled using the ComBat method [66].
High-resolution T1-weighted images were collected using a spoiled echo sequence. The sequence parameters included: repetition time (TR) = 8.16/8.10 ms, echo time (TE) = 3.18 ms, voxel size = 1 mm × 1 mm × 1 mm, field of view (FOV) = 256 mm2, 256 × 256 matrix, 12° flip, 166/168 slices. For the validation dataset, the sequence parameters included: TR = 8.16 ms, TE = 3.18 ms, voxel size = 1 mm × 1 mm × 1 mm, FOV = 256 mm2, 256 × 256 matrix, 12° flip, 168 slices.
Diffusion-weighted imaging (DWI) was acquired in the axial plane using a single shot spin-echoplanar imaging (EPI) sequence (FOV = 256 mm2, voxel size = 2 mm × 2 mm × 2 mm, 128 × 128 matrix, 90° flip, 2 mm thickness). TE was set to use the minimum signal-to-noise ratio, and minimum TE ranged from 77.9 to 86.7. Additional parameters differed slightly between protocol (b-values = 900 or 800 s/mm2, TR = 10,000/8,000 ms, 73/67 slices, 30/64 directions). For validation dataset, DWI were collected using 64 directions, the sequence parameters included: FOV = 256 mm2, voxel size = 2 mm × 2 mm × 2 mm, 256 × 256 matrix, 90° flip, 2 mm thickness, b-value = 1,000 s/mm2, TR = 8,000 ms, minimum TE = 80.8 ms, 2 mm thickness, 67 slices. Using MATLAB dot() function (inner product) to downsample 64 direction data which identified the diffusion coding directions that were most similar to those in the protocol with 30 directions [67].
Diffusion-weighted imaging (DWI) was acquired in the axial plane using a single shot spin-echoplanar imaging (EPI) sequence (FOV = 256 mm2, voxel size = 2 mm3, 128 × 128 matrix, 90° flip, 2 mm thickness). TE was set to use the minimum signal-to-noise ratio, and minimum TE ranged from 77.9 to 86.7. Additional parameters differed slightly between protocol (b-values = 900 or 800 s/mm2, TR = 10,000/8,000 ms, 73/67 slices, 30/64 directions). For validation dataset, DWI were collected using 64 directions, the sequence parameters included: FOV = 256 mm2, voxel size = 2 mm3, 256 × 256 matrix, 90° flip, 2 mm thickness, b-value = 1,000 s/mm2, TR = 8,000 ms, minimum TE = 80.8 ms, 2 mm thickness, 67 slices. Data harmonization consisted of using a dot() function (inner product) to downsample 64 direction data which identified the diffusion coding directions that were most similar to those in the protocol with 30 directions [67]. Whole-brain, open-eye resting-state BOLD images were collected using T2*-weighted EPI (TR = 2,000 ms, voxel size =3.75 mm × 3.75 mm × 3.8 mm, FOV = 240 mm2, 64 × 64 matrix, interleaved data acquisition). For the discovery dataset, additional parameters varied slightly between one protocol (TE = 27 ms, 77° flip, 39 slices) and the other two protocols (TE = 25 ms, 90° flip, 35 slices). For validation dataset, additional parameters included: TE = 25 ms, 90° flip, 35 slices. The analysis included the first 150 volumes for consistency.
rs-fMRI data were oriented to LAS, and the first six volumes were excluded to ensure steady state. The processing pipeline was run using FSL v6.0.3. It included the following steps: slice-timing correction using Slicetimer, motion correction using MCFLIRT [68], rigid-body transformation, calculation of the affine registration of the functional data to the anatomical data, and non-linear registration of the anatomical data to MNI 2mm space using ANTs [18], [69] intensity normalization (scaling median intensity of each subject’s voxel data within the brain mask to 10,000), ICA-AROMA [70] to remove additional noise confounds, estimating and removing cerebrospinal fluid and WM signal components, high-pass temporal Gaussian filtering at 0.01 Hz, spatial smoothing using a Gaussian kernel with 6 mm full width at half maximum, and finally applying the previously-calculated registration to bring the data into common MNI space. All included scans had a relative motion cutoff of < 0.3 mm, measured using MCFLIRT with the middle volume as the reference.
Preprocessing of DWI data included denoising using dwide-noise [71]. For every denoised B0, a ”synthetic” undistorted B0 was created using Synb0-DisCo [72]. We then used FSL [73] and MRtrix3 [74] tools to process the denoised diffusion data. FSL’s topup [75] estimated and corrected the susceptibility-induced off-resonance field using the original and synthetic B0 images and applied the correction to the full diffusion data. The data were then run through FSL’s eddy [76], MRtrix’s dwi2mask [77], and FSL’s BET [78] to create a consensus mask, and MRtrix’s dwibiascorrect (for B1 field inhomogeneity correction). Diffusion images were registered to the MNI-space standard brain using ANTs [69], [79], with a rigid-body registration from the distortion-corrected B0 to the same subject’s T1-weighted images and a non-linear registration from the T1-weighted images to the FSL standard brain. Deterministic tractography was performed using MRtrix3’s tckgen, using the anatomically-constrained (ACT) algorithm with a five-tissue-type (5TT) segmentation of the T1 image using the Freesurfer-based algorithm in 5ttgen [73]. 100,000 tracks were generated from random seed points in 0.2 mm steps in the mask image, limited to a maximum angle of 60° between steps, excluding tracks with a length beyond the range of 4 mm-200 mm. 100,000 tracks were generated from random seed points in 0.2 mm steps in the mask image, limited to a maximum angle of 60° between steps, excluding tracks with a length beyond the range of 4 mm–200 mm [80].
D. Regression Modeling
The DWI data was parcellated into 170 ROIs according to the Automated Anatomical Labeling (AAL) template [81], the streamline count between every pair of nodes was extracted into weighted, undirected matrices with dimensions of 170 × 170 using tck2connectome [74]. We excluded edges consisting of fewer than three streamlines. In addition, to eliminate the effect of region size, the number of streamlines was normalized by the average of the two region sizes connected by each edge [80]. SC was defined as the number of streamlines/regions connecting each pair of nodes. rs-fMRI data was parcellated into 264 ROIs according to the Power atlas [82]. We obtained the timecourse of brain activity from each region across 144-time points and computed correlations between all possible pairs of regions to create the respective correlation matrices with dimensions of 264 × 264 for each subject, and the Fisher transform was performed on correlation matrices. Correlations between nodes less than 20mm apart were not calculated, as close physical proximity may artificially inflate the correlation values [83].
After removing the diagonal elements, we extracted the upper triangle part of the SC and FC matrices. Overall, the SC features were reshaped in a vector of (170 × 169)/2 = 14,365, while the FC features were reshaped in a vector of (264 × 263)/2 = 34,716.
The CPM was implemented to estimate global T-scores using FC and SC. Fig. 1 summarizes the analysis flowchart. Leave-one-out cross-validation (LOOCV) strategy was utilized for global T-score prediction in the discovery set. On each iteration of cross-validation, the model was trained on data from 101 participants and tested on the left-out individual. 102 rounds of cross-validation were conducted such that each subject was sequentially used as the testing sample [47] Each iteration included feature selection, model construction, and model application for the left-out individual. For details, see Algorithm 1.
Fig. 1. The prediction and validation flowchart incorporating feature selection and regression analysis.
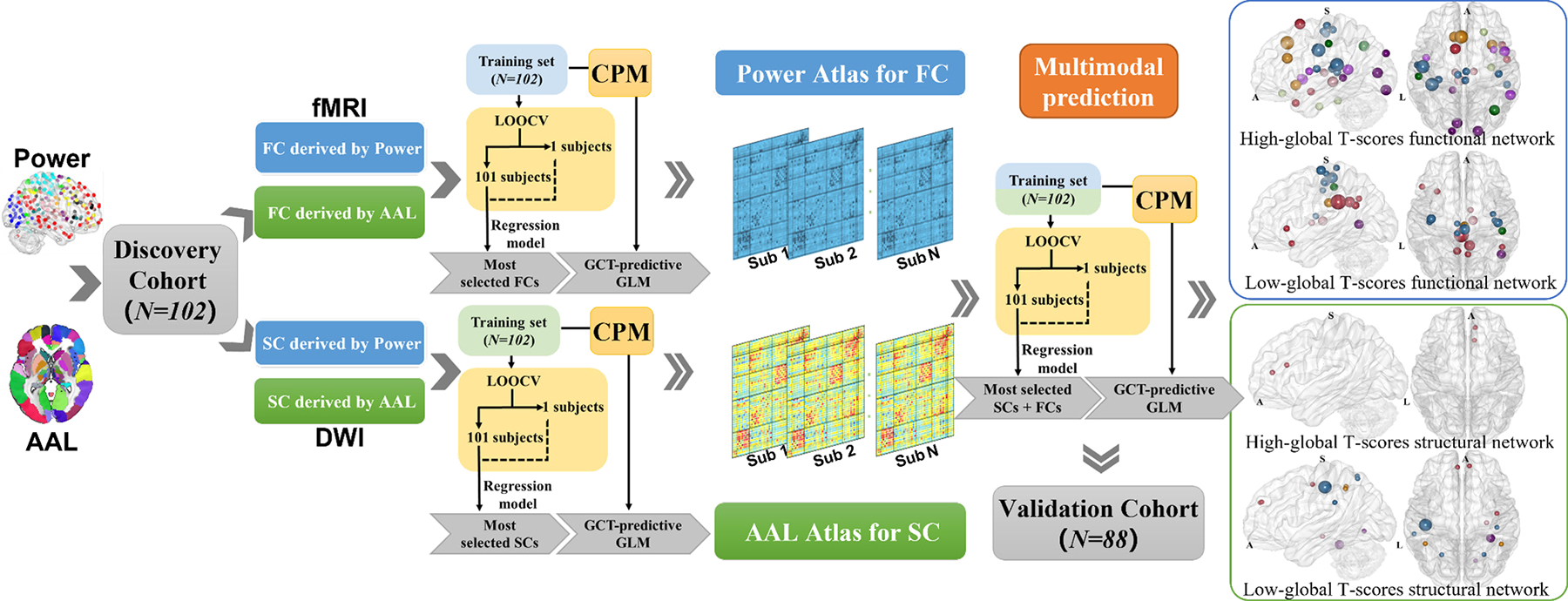
In the study, we estimate individuals’ global T-scores using functional, structural features or both in the connectome-based predictive models. Significantly, integrating both functional and structural features improved prediction accuracy. The rigorous validated analysis showed that the structural connectivities in sensorimotor and default mode network and functional connectivities linking subcortical, limbic, and dorsal attention networks are the most predictive connectome features.
Algorithm 1:
The Training and the validation strategy for connectome-based predictive modeling
| Require: global T-scores, functional and structural connectivity, and biological measures | |
| 1: | for subject i = 1 : N do |
| 2: | for threshold= 0 : 0.001 : 0.05 do |
| 3: | N-1 subject as training set and the ith as testing set; |
| 4: | All edges that were positively or negatively correlated with global T-scores and p < threshold constitute the positive and negative network; |
| 5: | The positive and negative network strength was calculated by summing the correlation coefficients of each edge in the positive and negative network; |
| 6: | Make predictions using simple or multiple linear regression models; |
| 7: | end for |
| 8: | Use models to predict global T-scores of ith subject; |
| 9: | end for |
| 10: | Apply predictive models trained in the entire N subjects using consensus network to the external validation set. |
In the feature selection step, we calculated the Pearson correlation between global T-scores and edges of FC/SC in the training set (after regressing out age, sex, education, and race). To maximize predictive accuracy using optimal p-value thresholds determined by the data, we selected edges obtained from significant correlation s [44], [45]. Edges that were positively correlated with global T-scores with a p < positive threshold (0 : 0.001 : 0.05) made up the positive network or high cognitive network, in which positive network strength was calculated by summing the correlation values of all edges in this network; similarly, edges negatively correlated with global T-scores with a p < negative threshold (0 : 0.001 : 0.05) made up the negative network or low cognitive network, and the negative network strength was calculated the same way. Positive network strength was calculated by summing the correlation coefficients of edges in the positive network, negative network strength was calculated by summing the correlation coefficients of edges in the negative network. Network strength was utilized as the imaging feature for model construction.
27 combinations (3 combinations of imaging features [SC, FC, SC + FC] × 3 combinations of strength features [positive network strength, negative network strength, positive and negative network strength] × 3 combinations of non-imaging characteristics [imaging only, demographic characteristics, demographic characteristics + clinical measures]) were treated as input features of the simple/multiple linear regression models to estimate the relationship between network strengths and the true global T-scores. Considering our aim is to explore whether integrating more modalities and clinical information could yield higher prediction accuracy, we did not test the prediction performance of two modalities combined HIV measure.
We quantified model performance by calculating Spearman rank correlation between observed and predicted global T-scores, and the coefficient of determination prediction R2. We determined the significance of any successful models based on a p-value calculated from 5,000 permutation tests. The specificity of the predictive model was determined by calculating the partial correlation between the predicted global T-scores and the true global T-scores when controlling for covariates, such as in Cui, et al. [84].
To assess whether prediction performance would be affected by different prediction models, we also probed performance with a variety of modeling algorithms for comparison. We used four prediction algorithms, which included three regularized general linear model regression algorithm, least absolute shrinkage, and selection operator (LASSO), partial least square regression (PLSR), and elastic net, and one nonlinear regression model algorithm, support vector regression (SVR) with RBF (nonlinear) kernels for the modeling. Positive and negative edges were selected the same way as in CPM (i.e., by thresholding the correlation with behavior in the training set using a statistical threshold) and both positive and negative edges were input as imaging features. All models were validated using LOOCV in the discovery set, and the model that performed best in the discovery set was used for evaluating the generalizability in the validation set.
E. Predictive Structural and Functional Networks
To simplify the characterization of the biological substrates, the edges within the CPM were further grouped into nine macroscopic brain regions functionally defined by the Yeo atlas [85], along with a cerebellar network (CB) and subcortical network (SUB) [86], [87]. Next, we employed a calculation to index the relative contribution of each of the nine networks [88]. That is, the contribution of edges was normalized for network size, such that a value of 1 indicates a proportionate contribution and larger values indicate a greater contribution of that network pair to the model than would be expected for its size.
F. External Validation
As selected features in each iteration were not wholly the same because of the LOOCV strategies, we then constructed a final model. Final predictive FC/SC edges applied to the validation dataset were determined by putting features that appeared at least 82 times among 102 loops (> 80% identification rate in all cross-validation) together. We defined these as the consensus FCs/SCs network. The external validation dataset was neither used to train the models nor to select parameters. Predictive models trained in the entire discovery dataset using consensus FCs/SCs network were applied to the external validation dataset. Spearman rank correlation between observed and predicted global T-scores and the coefficient of determination prediction R2 were calculated to evaluate predictive power. We determined the significance of any successful models based on a p-value calculated from 5,000 permutation tests. As a reference, we also examined the partial correlations between the global T-scores, controlling for demographic correlates.
III. RESULTS
A. Sample characteristics
Detailed information of sample characteristics of the discovery cohort (n = 102) and the validation cohort (n = 88) are summarized in Table I.
TABLE I.
Characteristics for Discovery and Validation Datasets
|
| |||
|---|---|---|---|
| Variables | Discovery set (N=102) | Validation set (N=88) | Statistic (p) |
|
| |||
| Demographic Variables | |||
|
| |||
| Age, years | 41.5±9.49 | 43.4±8.81 | t188=1.43(0.16) |
| Female | 74(72.55) | 70(79.55) | x21=0.93(0.34) |
| African-American race | 84(82.35) | 70(79.55) | x21=0.31(0.58) |
| Education, years | 13.4±2.40 | 13.8±2.61 | t188=1(0.32) |
|
| |||
| Substance Use | |||
|
| |||
| Daily cigarette smoking | 47(46.08) | 31(35.23) | x21=0.16(0.69) |
| aAlcohol | 66(64.71) | 66(75) | x21=0.88(0.35) |
| aCocaine | 33(32.35) | 24(27.27) | x21=0.22(0.64) |
| aMarijuana | 39(38.23) | 43(48.86) | x21=0.08(0.78) |
|
| |||
| HIV Clinical Measure | |||
|
| |||
| HIV Disease Duration, years | 10.9±7.95 | 13.4±7.01 | t188=2.25*(2.6e-2) |
| bCurrent CD4+ (cells/μL) | 615(346.5,854) | 759.5(464.5,970.5) | S=8610**(8.7e-3) |
| bNadir CD4+ (cells/μL) | 236(100,437) | 199(65.5,353) | S=10292(0.14) |
| cViral load | 23(22.77) | 6(6.82) | x21=16.84**(9.1e-4) |
|
| |||
Values are n(%) or mean ± standard deviation unless indicated. HIV, human immunodeficiency virus;
Any substance use in the past 30 days;
median (interquartile range);
< 50 copies/μL;
p < 0.05;
p < 0.01.
We analyzed the distribution differences of the characteristics between the two cohorts using two-sample t-tests for continuous demographic variables (age, education, and duration of HIV disease) and chi-square tests for categorical variables (gender, ethnicity, HIV suppression, and recent substance use). Wilcoxon rank-sum test was used to compare cohort differences between non-normally distributed demographic variables (current and nadir CD4+ T-cells counts). The significance threshold was two-sided, p <. No significant differences were found between the two datasets regarding demographic variables and recent substance use. However, participants in the validation dataset were infected with HIV longer, were more likely to be virally suppressed, and had higher current CD4+ T-cells count.
B. Individualized Prediction of global T-scores
CPM achieved appreciable estimations of global T-scores, using SC, FC, or a combination of both. For SC, the predictive accuracy ranged from 0.28 to 0.40, permutation test p <1.0e-3. For FC, the predictive accuracy ranged from 0.35 to 0.54, permutation test p < 1.0e-5.
C. Predictive Structural Networks and Functional Networks
Fig 2. Performance of predictive models in discovery dataset. The 27 Connectome-based predictive models (3 combinations of imaging features [SC, FC, SC + FC] × 3 combinations of strength features [positive network strength, negative network strength, positive and negative network strength] × 3 combinations of no-imaging characteristics [imaging only, demographic characteristics, demographic characteristics + clinical measures]), and 36 comparison models (3 combinations of imaging features [SC edge, FC edge, SC and FC edge] × 3 combinations of no-imaging characteristics [imaging only, demographic characteristics, demographic characteristics and clinical measures] × 4 prediction algorithms [LASSO, PLSR, elastic net, and SVR]) were assessed by correlating predicted and observed individual scores. Since models may consistently outperform others, we identified the consensus edges that appeared in more than 80% of all LOOCV loops. These edges are visualized on 3D glass brains in Fig. 3a–b. All SC and FC nodes belong to consensus edges, and their degrees are listed in Supplementary Table I and Supplementary Table II.
Fig. 2. Performance of predictive models in discovery dataset.
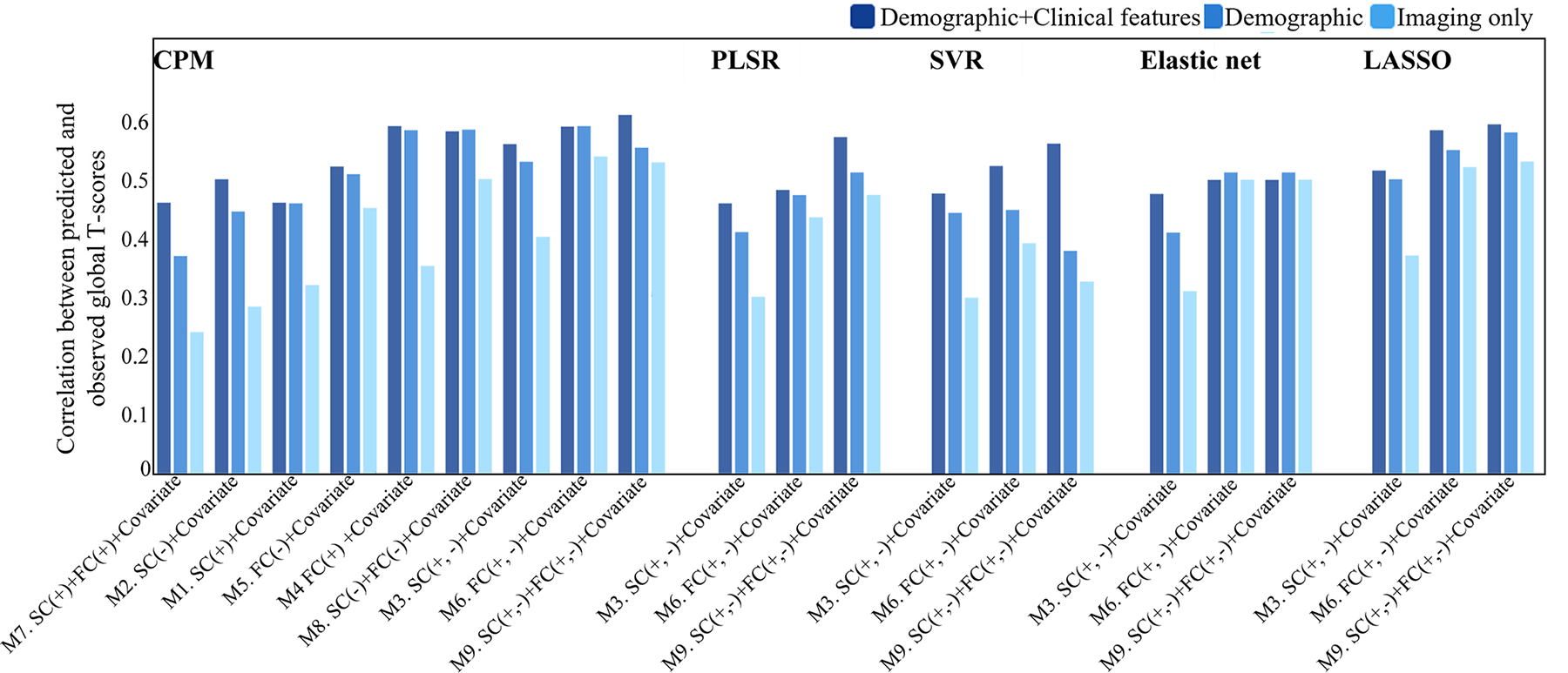
The 27 Connectome-based predictive models (3 combinations of imaging features [SC, FC, SC + FC] × 3 combinations of strength features [positive network strength, negative network strength, positive and negative network strength] × 3 combinations of no-imaging characteristics [imaging only, demographic characteristics, demographic characteristics + clinical measures]), and 36 comparison models (3 combinations of imaging features [SC edge, FC edge, SC and FC edge] × 3 combinations of no-imaging characteristics [imaging only, demographic characteristics, demographic characteristics and clinical measures] × 4 prediction algorithms [LASSO, PLSR, elastic net, and SVR]) were assessed by correlating predicted and observed individual scores.
Fig. 3. Distribution of the predictive functional and structural connections.
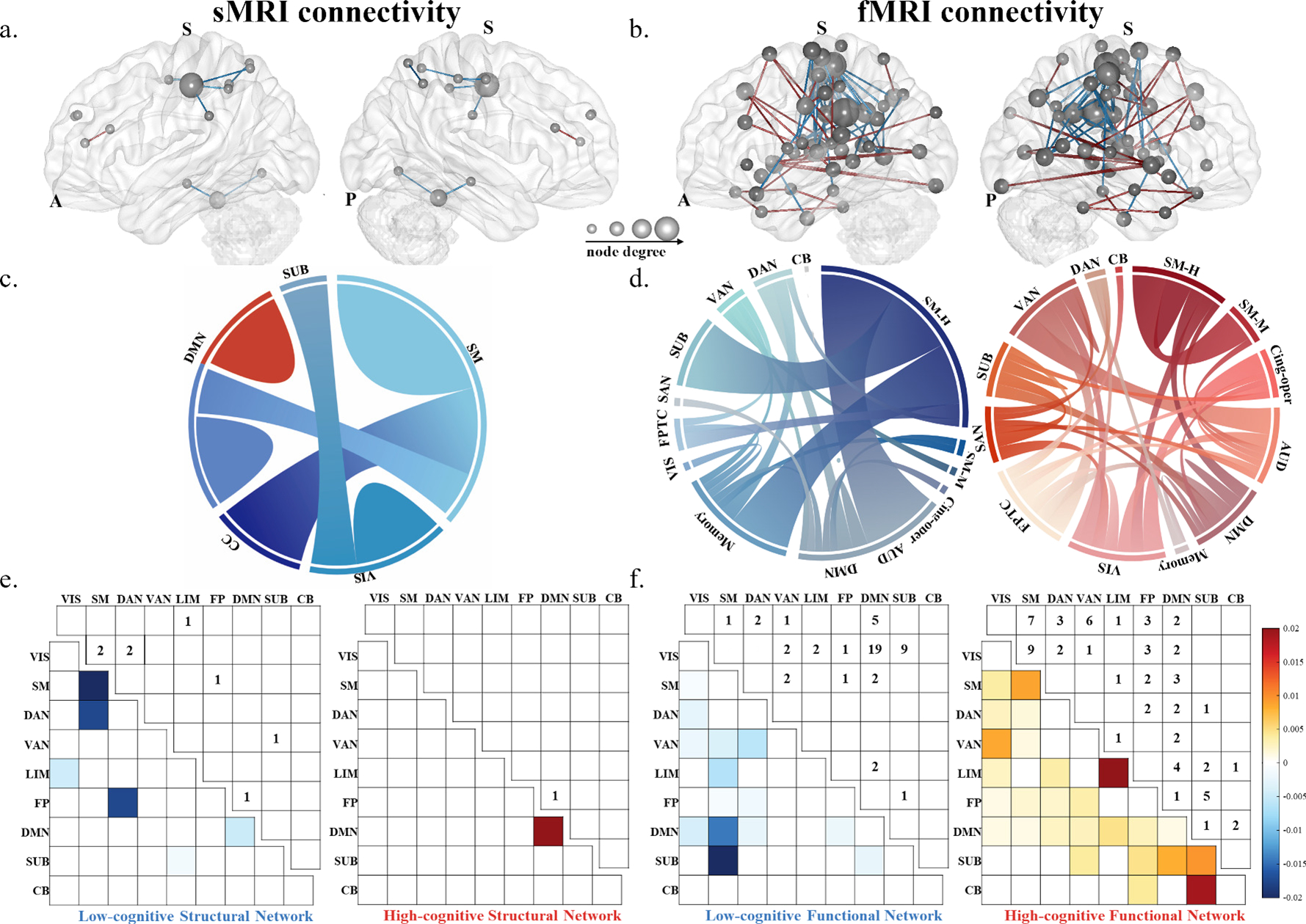
In brain figures, edges belonging to the high-cognitive network are in red and the edges belonging to the low-cognitive network are in blue in structural connections (a) and functional connections (b). The size of nodes represent higher degree (i.e., more connections). Ring plots illustrate intra- and inter- region connections for the high-cognitive (red) and low-cognitive (blue) structural networks (c) and functional networks (d),ribbon size is proportional to the number of edges. Matrices present the contributions (lower triangular) and raw number of edges (upper triangular) for each macroscale brain region for the high-cognitive (red) and the low-cognitive (blue) structural (e) and functional networks (f). Colored bars depict contributions. Region acronyms: SM-H = sensory/somatomotor hand; SM-M = sensory/somatomotor mouth; Cing-oper = cingulo-opercular task control; AUD = auditory; DMN = default mode; Memory = memory retrieval; VIS = visual; FPTC = frontal-partial task control; SAN = salience; SUB = subcortical; VAN = ventral attention; FP=frontal-partial; DAN = dorsal attention; CB = cerebellar; CC = cognitive control; SM = somatomotor; LIM = limbic.
For SC, the low-cognitive (negative) network included eight edges (~0.06% of the 14,365 total structural edges). Nodes with the highest k degrees (the number of edges identified in the final model) were the postcentral gyrus (k = 3) within the somatomotor network (SM). Greater SC in these edges was associated with lower global T-scores. In contrast, the high-cognitive (positive) SC network included one edge with 1 degree in the right anterior cingulate cortex (ACC), a hub region of the DMN, suggesting increased connectivity within the DWM was associated with higher global T-scores.
The consensus edges for the FC were more significant than the SC, which had 59 edges for the high-cognitive network (~0.17% of the 34,716 total functional edges) and 60 edges for the low-cognitive network (~0.17% of the 34,716 total functional edges) (Fig. 3b). In the low-cognitive network, the high degree nodes (k ≥ 3) were primarily in the sensory/somatomotor hand network (SM-H) and DMN, with greater connectivity associated with lower global T-scores. In the high-cognitive network, the high degree nodes were primarily in the sensory/somatomotor mouth network (SM-M), and frontal-partial network (FPTC), with greater connectivity associated with higher global T-scores. The FC positive network expressed a more substantial number of edges than the FC negative network. The distribution of SC nodes belonging to five macroscale brain regions defined by the AAL atlas are presented in Fig. 3c, while the distribution of FC nodes belonging to thirteen macroscale brain regions defined by the Power atlas are presented in Fig. 3d. To compare SC and FC at a homologous scale, we divided FC and SC nodes into nine macroscopic brain networks defined by the Yeo atlas along with CB and SUB. Matrices in Fig. 3e–f indicted edges’ contributions and the number of edges for each macroscopic brain region. The macroscopic networks showed that worse cognitive function was correlated with stronger SC within the SM, between the DAN and the frontal-partial network (FP), and between the DAN and SM. Higher cognitive function was related to increased SC within the DMN. Furthermore, worse cognitive function was associated with stronger FC between SM and DMN, and between the SM and SUB. Higher cognitive function was related to stronger FC within the SM, limbic network, and SUB, between the SUB and CB, between the SUB and DMN, and between the ventral attention and visual network.
D. External Validation
To evaluate the generalizability of the CPM built from the discovery dataset, we utilized the 9 SCs, and 119 FCs to predict the global T-scores for an independent validation cohort. As shown in Table III, the model successfully predicted the global T-scores in most (but not all) iterations with the validation dataset. The models combined with demographic characteristics and clinical features outperformed the imaging only models in the validation dataset. Specifically, the highest predictive accuracy was obtained with the negative FC networks (r = 0.47, p = 1.3e-5, R2 = 0.21), followed by the model with both positive and negative SC and FC (r = 0.45, p = 2.5e-5, R2 = 0.18) (Fig. 4). Even after controlling for demographic characteristics, the model taking clinical features into account was still significant (Supplementary Table V).
TABLE III.
External Validation Results of Connectome-Based Models
|
| |||
|---|---|---|---|
| Added Covariant | A+B+Imaging | A+Imaging | Imaging only |
|
| |||
| M1. SC(+) + Covariant | 0.38 (4.4e-4) | 0.27 (1.5e-2) | 0.22 (0.05) |
| M2. SC(−) + Covariant | 0.44 (5.3e-5) | 0.35 (1.0e-3) | 0.28 (0.01) |
| M3. SC(+, −) + Covariant | 0.43 (8.7e-5) | 0.34 (2.0e-3) | 0.25 (0.02) |
| M4. FC(+) + Covariant | 0.18 (8.1e-2) | 0.17(8.8e-2) | - |
| M5. FC(−) + Covariant | 0.47 (1.3e-5) | 0.39 (3.2e-4) | 0.33 (2.5e-3) |
| M6. FC(+, −) + Covariant | 0.20 (5.0e-2) | 0.17 (1.0e-1) | 0.02 (8.6e-1) |
| M7. SC(+) + FC(+) + Covariant | 0.17 (0.1) | 0.17 (0.1) | - |
| M8. SC(−) + FC(-) + Covariant | 0.46 (1.5e-5) | 0.39 (3.1e-4) | 0.33 (0.03) |
| M9. SC(+, −) + FC(+, −) + Covariant | 0.45 (2.5e-5) | 0.38 (4.9e-4) | 0.30 (3.0e-3) |
|
| |||
Correlation between predicted and observed scores (corresponding p–value). Bolded values correspond to the highest correlation in models. A – Demographic; B – HIV Measure; + = positive; − = negative; SC = structural connectivity; FC = functional connectivity
Fig. 4. Scatter plot for best predictive models in the discovery and the external validation dataset.
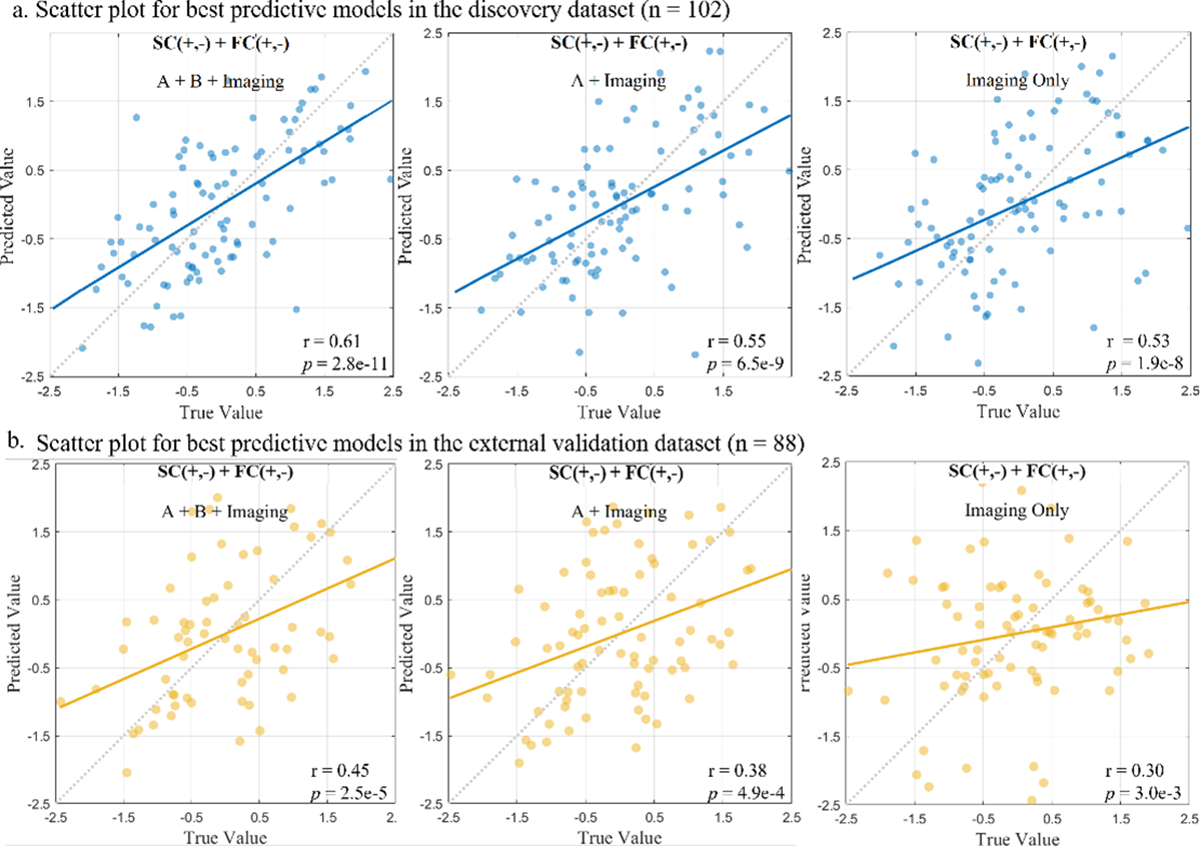
The accomplished robust prediction of global T-score using connectome-based prediction modeling in the sample of 102 PLWH (a), and the identified neuromarkers were generalized to validation cohort (n = 88) (b). Each dot in the scatter plot, represents a single participant from the dataset and their measured global T-scores score and predicted score.The dashed diagonal line represents a perfect 1-to-1 linear relationship between measured and predicted values. Values in the x-axis and y-axis were standardized to z-scores for visualization.
IV. DISCUSSION
The current study successfully implemented a modified CPM approach that integrated structural and functional MRI features to predict global cognitive function in PLWH. Here are some key points we found. First, we noticed that features from SC and FC could make complementary contributions to global T-score prediction, demonstrating the added value of incorporating multimodal MRI features into CPM. Second, we found that adding clinical characteristics, notable markers of HIV disease, and substance use improved model accuracy. We obtained fairly accurate prediction models relative to similar CPM studies. Importantly, we also investigated the generalizability of these connection-based biomarkers in a validation dataset comprising an independent cohort. The results of this study revealed that the combination of SC and FC features could yield sensitive markers of cognitive function in PLWH. Connectomes are particularly suited to uncover neural network deficits in neuroHIV. By incorporating nodes across the brain, we can take a holistic approach to study how cognitive function is associated with structural and functional connectivity within and between brain networks. Our approach is not limited to univariate mass techniques or a single neuroimaging modality, so it can uncover networks related to cognitive function in PLWH that might not typically be considered.
While CPM has been applied to successfully predict cognitive and behavioral dysfunction across multiple neural disorders such as Parkinson’s disease [89], cognitive impairment [90], and multiple sclerosis [91], only one study to date has used CPM to investigate neuroHIV [47]. Our results match the study by Yang, et al. [47] in which higher cognitive function was associated with increased FC between CB, SUB, and FP. However, we also found many additional functional networks associated with cognitive function. Importantly, our study builds upon this by incorporating both FC and SC into the CPM, allowing for an expanded investigation into how brain connectivity relates to neurocognitive function.
A. Contributions of Structural and Functional Networks to Models
Using a multimodal analytic model, we can identify complementary information between SC and FC that informs possible neural linkages. We observed higher within-network FC in the SM associated with low cognitive functioning. In contrast, higher within-network SC in SM was associated with high cognitive functioning. SM is associated with motor function, a prevalent cognitive dysfunction in PLWH [92]. Reduced within-network FC in SM has been observed in PLWH relative to HC [93]. Since SC indicates connections between nodes, FC represents the strength of nodes functionally co-activating. Therefore, higher SC is not necessarily linear with FC.
Greater within FC of the SUB was associated with higher cognitive function. HIV is commonly associated with disrupted functional connectivity in the SUB [94]. Other studies using seed-based approaches have also shown that increased FC in the SUB was associated with higher cognitive function [30], [95], [96]. Meanwhile, we found that higher cognitive function was correlated with greater connectivity between the SUB with CB and DMN. PLWH have been shown to have decreased connectivity between regions of SUB and DMN relative to HC [97].
Stronger FC between CB and SUB was associated with high cognitive function. As stated above, this effect was also observed in Yang, et al. [47]. Deficits in FC between the CB and SUB have been implicated in other diseases characterized by neurocognitive dysfunction [98]. While the CB is commonly associated with motor function, it has been shown that this region is heavily implicated in multiple facets of cognitive function [99]. FC in CB appears to be an important, albeit overlooked, function of neuroHIV and should be included in future cognitive models of this disease.
B. Limitations and Future Directions
Although this study obtained a high prediction accuracy for our multimodal CPM, which was replicated in an independent dataset, some limitations should be considered. The prediction performance of the external validation set is not as high as the prediction accuracy of the discovery dataset. This difference in prediction accuracy may be due to heterogeneity across samples. For example, participants in the validation cohort were more likely to have HIV viral suppression and had higher CD4+ T-cells counts. Moreover, differences in the measurement of the test battery between the discovery and validation cohorts can also be a potential reason. Further studies can include more heterogeneous subjects in both discovery and validation cohorts, which could expand the range of observed behavioral performance and add statistical power to the analysis. Furthermore, the model’s prediction accuracy using SC and FC features on the validation dataset is slightly lower than the model using only FC. Considering that changes in SC may occur over longer timescales than FC, it is likely to explain legacy neurobiological factors, such as HIV disease duration and nadir CD4+ T-cells count. However, the external validation prediction performance was still higher than that found in Yang, et al. [47], demonstrating the importance of multimodal data in predicting cognitive function in PLWH.
It should be noted that we found more FC features associated with cognition relative to SC. It may be that FC displays more sensitivity towards cognitive function relative to SC, which displays more specificity relative to associations with cognitive function. Since our diffusion images contained only thirty diffusion directions, a result could be less detailed mapping of SC with lower sensitivity to detect correlations with neurocognitive impairment. Future studies could use high angular resolution diffusion images to build upon our findings using denser structural connectomes. Additionally, due to the relative brevity of our NP battery, we focused on global cognition. Further research is needed to identify network models predictive of deficits in specific cognitive domains. Considering the interpretability of models and the limitation of samples, neural network modeling may be used in future studies. While the sample size is relatively small compared with previous CPM studies, it is sizable for the neuroHIV field. It provides a significant step forward in understanding neural biotypes of neurocognitive impairment in PLWH.
V. Conclusion
In this study, we integrated multimodal neuroimaging data into the CPM method. We established an adjusted global T-scores prediction model based on CPM and LOOCV strategies, which can be seen as a substantial improvement over the current cognitive prediction work in PLWH. Moreover, the proposed model can be generalized to an independent, demographically matched PLWH cohort and achieved appreciable estimation accuracy. We demonstrated that FC and SC provide complementary information. When combined with non-imaging behavioral data, these features can produce a set of potential biomarkers for global cognition. Such biomarkers are able to predict the global T-scores of novel individuals. In summary, this work helps us better understand the association between neuroimaging, HIV disease characteristics, and cognitive impairment in PLWH, which could improve neurocognitive impairment screening accuracy and diagnostic efficiency before manifesting behavioral deficits in PLWH. While promising, our CPM model requires additional refinement with additional validation evidence before being applied in clinical settings. In particular, research is needed to evaluate the predictive accuracy of CPM models on cognitive performance over time. Given our success in implementing multimodal CPM, Future studies could consider other morphological connectivity features, including gray matter structural connections, combined with longitudinal data to explore the longitudinal changes between SC, FC, and cognitive impairment.
Supplementary Material
TABLE II.
Prediction Performances of Prediction Methods in Discovery Dataset
|
| ||||
|---|---|---|---|---|
| Model | Added Covariate | A+B+Imaging | A+Imaging | Imaging only |
|
| ||||
| CPM | M1. SC(+) + Covariate | 0.46 (2.0e-6) | 0.46 (1.6e-6) | 0.32 (1.3e-3) |
| M2. SC(-) + Covariate | 0.50 (2.1e-7) | 0.44 (7.0e-6) | 0.28 (5.2e-3) | |
| M3. SC(+, -) + Covariate | 0.56 (2.7e-9) | 0.53 (2.9e-8) | 0.40 (4.7e-5) | |
| M4. FC(+) + Covariate | 0.59 (3.5e-10) | 0.58 (6.5e-10) | 0.35 (4.8e-4) | |
| M5. FC(-) + Covariate | 0.52 (4.6e-8) | 0.51 (1.3e-7) | 0.45 (3.5e-6) | |
| M6. FC(+, -) + Covariate | 0.59 (2.3e-10) | 0.59 (3.9e-10) | 0.54 (1.6e-8) | |
| M7. SC(+) + FC(+) + Covariate | 0.46 (2.0e-6) | 0.37 (1.9e-4) | 0.24 (1.8e-2) | |
| M8. SC(-) + FC(-) + Covariate | 0.58 (4.0e-10) | 0.58 (7.4e-10) | 0.50 (2.1e-7) | |
| M9. SC(+,-) + FC(+,-) + Covariate | 0.61 (2.8e-11) | 0.55 (6.5e-9) | 0.53 (1.9e-8) | |
|
| ||||
| SVR | M3. SC(+, -) + Covariate | 0.46 (1.2e-6) | 0.41 (2.3e-5) | 0.30 (1.8e-3) |
| M6. FC(+, -) + Covariate | 0.48 (4.7e-7) | 0.47 (5.0e-7) | 0.43 (7.5e-6) | |
| M9. SC(+,-) + FC(+,-) + Covariate | 0.57 (4.2e-10) | 0.51 (4.0e-8) | 0.47 (5.9e-7) | |
|
| ||||
| Elastic net | M3. SC(+, -) + Covariate | 0.47 (9.7e-4) | 0.44 (6.6e-6) | 0.30 (1.9e-3) |
| M6. FC(+, -) + Covariate | 0.52 (6.5e-8) | 0.45 (1.6e-6) | 0.39 (4.1e-5) | |
| M9. SC(+,-) + FC(+,-) + Covariate | 0.56 (4.0e-9) | 0.38 (NS) | 0.32 (8.9e-4) | |
|
| ||||
| LASSO | M3. SC(+, -) + Covariate | 0.47 (7.8e-7) | 0.41 (1.6e-5) | 0.31 (1.6e-3) |
| M6. FC(+, -) + Covariate | 0.50 (1.3e-7) | 0.51 (4.1e-8) | 0.50 (1.3e-7) | |
| M9. SC(+,-) +FC(+,-) + Covariate | 0.50 (1.3e-7) | 0.51 (4.0e-8) | 0.50 (1.2e-7) | |
|
| ||||
| PLSR | M3. SC(+, -) + Covariate | 0.51 (7.2e-8) | 0.50 (2.3e-7) | 0.37 (2.0e-4) |
| M6. FC(+, -) + Covariate | 0.58 (6.5e-11) | 0.55 (2.7e-11) | 0.52 (3.5e-8) | |
| M9. SC(+,-) + FC(+,-) + Covariate | 0.59 (6.4e-11) | 0.58 (2.0e-11) | 0.53 (2.9e-8) | |
|
| ||||
Correlation between predicted and observed scores (corresponding p-value). Bolded values correspond to the highest correlation in their respective models. Values in red correspond to the highest correlation when added the same covariate among five models. A – Demographic; B – HIV Measure; + = positive; - = negative; SC = structural connectivity; FC = functional connectivity; NS= not significant.
Acknowledgments
This work was supported by the National Institute on Drug Abuse at the National Institutes of Health NIH (R01-DA045565, R01-MH117107), and the National Science Foundation (2112455) (Corresponding author: Jing Sui, Christina S. Meade).
Contributor Information
Xiang Li, National Laboratory of Pattern Recognition, Institute of Automation, Chinese Academy of Sciences, Beijing, China, and School of Artificial Intelligence, University of Chinese Academy of Sciences, Beijing, China..
Sheri L. Towe, Department of Psychiatry & Behavioral Sciences, Duke University School of Medicine, Durham, NC, United States.
Ryan P. Bell, Department of Psychiatry & Behavioral Sciences, Duke University School of Medicine, Durham, NC, United States.
Rongtao Jiang, Department of Radiology and Biomedical Imaging, Yale School of Medicine, CT, United States..
Shana A. Hall, Department of Psychiatry & Behavioral Sciences, Duke University School of Medicine, Durham, NC, United States.
Vince D. Calhoun, Tri-Institutional Centre for Translational Research in Neuroimaging and Data Science (TReNDS), Georgia State University Georgia Institute of Technology, and Emory University, Atlanta, GA, United States..
Christina S. Meade, Department of Psychiatry & Behavioral Sciences, Duke University School of Medicine, Durham, NC, United States, and the Brain Imaging and Analysis Center, Duke University, Durham, NC, United States.
Jing Sui, Tri-Institutional Centre for Translational Research in Neuroimaging and Data Science (TReNDS), Georgia State University Georgia Institute of Technology, and Emory University, Atlanta, GA, United States..
References
- [1].Gisslen M et al. The definition of hiv-associated neurocognitive disorders: are we overestimating the real prevalence? BMC Infect Dis, 11:356, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Alford K and Vera JH. Cognitive impairment in people living with hiv in the art era: A review. Br Med Bull, 127(1):55–68, 2018. [DOI] [PubMed] [Google Scholar]
- [3].Kumar S et al. Prevalence of hiv associated neurocognitive disorder using modified mini mental state examination and its correlation with cd4 counts and anti-retroviral therapy. J Assoc Physicians India, 67(4):47–51, 2018. [PubMed] [Google Scholar]
- [4].Canestri A et al. Discordance between cerebral spinal fluid and plasma hiv replication in patients with neurological symptoms who are receiving suppressive antiretroviral therapy. Clin Infect Dis, 50(5):773–8, 2010. [DOI] [PubMed] [Google Scholar]
- [5].van Sighem A et al. Life expectancy of recently diagnosed asymptomatic hiv-infected patients approaches that of uninfected individuals. AIDS, 24(10):1527–1535, 2010. [DOI] [PubMed] [Google Scholar]
- [6].Woods SP et al. Cognitive neuropsychology of hiv-associated neurocognitive disorders. Neuropsychol Rev, 19(2):152–68, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Marquine MJ et al. A composite of multisystem injury and neurocognitive impairment in hiv infection: association with everyday functioning. J Neurovirol, 24(5):549–556, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Tozzi V et al. Neurocognitive impairment influences quality of life in hiv-infected patients receiving haart. Int J STD AIDS, 15(4):254–9, 2004. [DOI] [PubMed] [Google Scholar]
- [9].Scott JC et al. A neuropsychological investigation of multitasking in hiv infection: implications for everyday functioning. Neuropsychology, 25(4):511–9, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Clifford DB and Ances BM. Hiv-associated neurocognitive disorder. Lancet Infect Dis, 13(11):976–86, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Doyle K et al. Aging, prospective memory, and health-related quality of life in hiv infection. AIDS Behav, 16(8):2309–18, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Belete T et al. Prevalence of hiv associated neurocognitive deficit among hiv positive people in ethiopia: A cross sectional study at ayder referral hospital. Ethiop J Health Sci, 27(1):67–76, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Banerjee N et al. Impaired neurocognitive performance and mortality in hiv: Assessing the prognostic value of the hiv-dementia scale. AIDS Behav, 23(12):3482–3492, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].May M et al. Impact of late diagnosis and treatment on life expectancy in people with hiv-1: Uk collaborative hiv cohort (uk chic) study. BMJ, 343:d6016, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Brahmbhatt H et al. Impact of hiv and atiretroviral therapy on neurocognitive outcomes among school-aged children. J Acquir Immune Defic Syndr, 75(1):1–8, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Benloucif S et al. Morning or evening activity improves neuropsychological performance and subjective sleep quality in older adults. Sleep, 27(8):1542–51, 2004. [DOI] [PubMed] [Google Scholar]
- [17].Seelye A et al. The impact of sleep on neuropsychological performance in cognitively intact older adults using a novel in-home sensor-based sleep assessment approach. Clin Neuropsychol, 29(1):53–66, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Wang X et al. Abnormalities in resting-state functional connectivity in early human immunodeficiency virus infection. Brain Connect, 1(3):207–17, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Stubbe-Drger B et al. Early microstructural white matter changes in patients with hiv: a diffusion tensor imaging study. BMC Neurol, 12:23, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Ances BM and Hammoud DA. Neuroimaging of hiv-associated neurocognitive disorders (hand). Curr Opin HIV AIDS, 9(6):545–51, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Gonneaud J et al. Accelerated functional brain aging in pre-clinical familial alzheimer’s disease. Nat Commun, 12(1):5346, 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Sexton CE et al. A meta-analysis of diffusion tensor imaging in mild cognitive impairment and alzheimer’s disease. Neurobiol Aging, 32(12):2322 e5–18, 2011. [DOI] [PubMed] [Google Scholar]
- [23].Cysique LA et al. White matter measures are near normal in controlled hiv infection except in those with cognitive impairment and longer hiv duration. Journal of NeuroVirology, 23(4):539–547, 2017. [DOI] [PubMed] [Google Scholar]
- [24].Bell RP et al. Principal component analysis denoising improves sensitivity of mr diffusion to detect white matter injury in neurohiv. J Neuroimaging, 32(3):544–553, 2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].O’Connor EE et al. Brain structural changes following hiv infection: Meta-analysis. AJNR Am J Neuroradiol, 39(1):54–62, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Kuhn T et al. The joint effect of aging and hiv infection on microstructure of white matter bundles. Hum Brain Mapp, 40(15):4370–4380, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Chang K et al. Plasma inflammatory biomarkers link to diffusion tensor imaging metrics in virally suppressed hiv-infected individuals. AIDS, 34(2):203–213, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Zhao T et al. Diffusion tensor magnetic resonance imaging of white matter integrity in patients with hiv-associated neurocognitive disorders. Ann Transl Med, 8(20):1314, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Ipser JC et al. Hiv infection is associated with attenuated frontostriatal intrinsic connectivity: a preliminary study. J Int Neuropsychol Soc, 21(3):203–13, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Samboju V et al. Structural and functional brain imaging in acute hiv. Neuroimage Clin, 20:327–335, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Sui J et al. Structural and functional brain abnormalities in human immunodeficiency virus disease revealed by multimodal magnetic resonance imaging fusion: Association with cognitive function. Clin Infect Dis, 73(7):e2287–e2293, 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Chaganti JR et al. Functional connectivity in virally suppressed patients with hiv-associated neurocognitive disorder: A resting-state analysis. AJNR Am J Neuroradiol, 38(8):1623–1629, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Egbert AR et al. Hiv infection across aging: Synergistic effects on intrinsic functional connectivity of the brain. Prog Neuropsychopharmacol Biol Psychiatry, 88:19–30, 2019. [DOI] [PubMed] [Google Scholar]
- [34].Abidin AZ et al. Detecting cognitive impairment in hiv-infected individuals using mutual connectivity analysis of resting state functional mri. J Neurovirol, 26(2):188–200, 2020. [DOI] [PubMed] [Google Scholar]
- [35].Hall SA et al. Human immunodeficiency virus-related decreases in corpus callosal integrity and corresponding increases in functional connectivity. Hum Brain Mapp, 42(15):4958–4972, 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Jiang R et al. Interpreting brain biomarkers: Challenges and solutions in interpreting machine learning-based predictive neuroimaging. IEEE Signal Processing Magazine, 39(4):107–118, 2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Chockanathan U et al. Automated diagnosis of hiv-associated neurocognitive disorders using large-scale granger causality analysis of resting-state functional mri. Comput Biol Med, 106:24–30, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Xu Y et al. Machine learning prediction of neurocognitive impairment among people with hiv using clinical and multimodal magnetic resonance imaging data. J Neurovirol, 27(1):1–11, 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Shen X et al. Using connectome-based predictive modeling to predict individual behavior from brain connectivity. Nat Protoc, 12(3):506–518, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Hsu WT et al. Resting-state functional connectivity predicts neuroticism and extraversion in novel individuals. Soc Cogn Affect Neurosci, 13(2):224–232, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Jiang R et al. Connectome-based individualized prediction of temperament trait scores. Neuroimage, 183:366–374, 2018. [DOI] [PubMed] [Google Scholar]
- [42].Rosenberg MD et al. A neuromarker of sustained attention from whole-brain functional connectivity. Nat Neurosci, 19(1):165–71, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Beaty RE et al. Robust prediction of individual creative ability from brain functional connectivity. Proc Natl Acad Sci U S A, 115(5):1087–1092, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Liu Z et al. Neural and genetic determinants of creativity. Neuroimage, 174:164–176, 2021. [DOI] [PubMed] [Google Scholar]
- [45].Jiang R et al. Gender differences in connectome-based predictions of individualized intelligence quotient and sub-domain scores. Cereb Cortex, 30(3):888–900, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Jiang R et al. A neuroimaging signature of cognitive aging from whole-brain functional connectivity. Advanced Science, 9(24):2201621, 2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Yang FN et al. Connectome-based prediction of global cognitive performance in people with hiv. Neuroimage Clin, 30:102677, 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Jiang R et al. Multimodal data revealed different neurobiological correlates of intelligence between males and females. Brain imaging and behavior, 14(5):1979–1993, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Yu J et al. The individualized prediction of cognitive test scores in mild cognitive impairment using structural and functional connectivity features. NeuroImage, 223:117310, 2020. [DOI] [PubMed] [Google Scholar]
- [50].Cheng B et al. Semi-supervised multimodal relevance vector regression improves cognitive performance estimation from imaging and biological biomarkers. Neuroinformatics, 11(3):339–353, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Yu J et al. The functional and structural connectomes associated with geriatric depression and anxiety symptoms in mild cognitive impairment: Cross-syndrome overlap and generalization. Progress in NeuroPsychopharmacology and Biological Psychiatry, 110:110329, 2021. [DOI] [PubMed] [Google Scholar]
- [52].Mellem MS et al. Machine learning models identify multimodal measurements highly predictive of transdiagnostic symptom severity for mood, anhedonia, and anxiety. Biological Psychiatry: Cognitive Neuroscience and Neuroimaging, 5(1):56–67, 2020. [DOI] [PubMed] [Google Scholar]
- [53].Pomara N et al. White matter abnormalities in hiv-1 infection: A diffusion tensor imaging study. Psychiatry Research: Neuroimaging, 106(1):15–24, 2001. [DOI] [PubMed] [Google Scholar]
- [54].Sui J et al. Neuroimaging-based individualized prediction of cognition and behavior for mental disorders and health: Methods and promises. Biol Psychiatry, 88(11):818–828, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Prakash M et al. Quantitative longitudinal predictions of alzheimer’s disease by multi-modal predictive learning. J Alzheimers Dis, 79(4):1533–1546, 2021. [DOI] [PubMed] [Google Scholar]
- [56].Reitan RM and Wolfson D. The Halstead-Reitan neuropsychological test battery: Theory and clinical interpretation. Neuropsycholgy Press, Tucson, AZ, 2nd edition, 1993. [Google Scholar]
- [57].Golden CJ. Stroop Color and Word Test. Stoelting, Chicago, IL, 1978. [Google Scholar]
- [58].Diehr MC et al. The 50 and 100-item short forms of the paced auditory serial addition task (pasat): demographically corrected norms and comparisons with the full pasat in normal and clinical samples. J Clin Exp Neuropsychol, 25(4):571–85, 2003. [DOI] [PubMed] [Google Scholar]
- [59].Stern RA and White T. NAB Digits Forward/Digits Backward Test: Professional Manual. Psychological Assessment Resources, Inc. (PAR), Lutz, FL, 2009. [Google Scholar]
- [60].Benton A et al. Multilingual Aphasia Examination (3rd ed.). AJA Associates, Iowa City, IA, 1983. [Google Scholar]
- [61].Brandt J and Benedict RHB. Hopkins Verbal Learning Test – Revised professional manual. Psychological Assessment Resources, Inc, Lutz, FL, 2001. [Google Scholar]
- [62].Klove H. Grooved Pegboard Lafayette Instruments, Lafayette, IN, 1963. [Google Scholar]
- [63].Heaton RK. Revised comprehensive norms for an expanded Halstead-Reitan Battery: Demographically adjusted neuropsychological norms for African American and Caucasian adults, professional manual. Psychological Assessment Resources, 2004. [Google Scholar]
- [64].Golden CJ and Freshwater S. The Stroop Color and Word Test: A Manual for Clinical and Experimental Uses. Stoelting, Chicago, IL, 2002. [Google Scholar]
- [65].McLellan AT et al. The fifth edition of the addiction severity index. J Subst Abuse Treat, 9(3):199–213, 1992. [DOI] [PubMed] [Google Scholar]
- [66].Yu M et al. Statistical harmonization corrects site effects in functional connectivity measurements from multi-site fmri data. Human brain mapping, 39(11):4213–4227, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Chen NK et al. On the down-sampling of diffusion mri data along the angular dimension. Magn Reson Imaging, 82:104–110, 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Jenkinson M et al. Improved optimization for the robust and accurate linear registration and motion correction of brain images. Neuroimage, 17(2):825–41, 2002. [DOI] [PubMed] [Google Scholar]
- [69].Avants BB et al. A reproducible evaluation of ants similarity metric performance in brain image registration. Neuroimage, 54(3):2033–44, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Pruim RHR et al. Ica-aroma: A robust ica-based strategy for removing motion artifacts from fmri data. Neuroimage, 112:267–277, 2015. [DOI] [PubMed] [Google Scholar]
- [71].Veraart J et al. Diffusion mri noise mapping using random matrix theory. Magn Reson Med, 76(5):1582–1593, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Schilling KG et al. Synthesized b0 for diffusion distortion correction (synb0-disco). Magn Reson Imaging, 64:62–70, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Jenkinson M et al. Fsl. Neuroimage, 62(2):782–90, 2012. [DOI] [PubMed] [Google Scholar]
- [74].Tournier JD et al. Mrtrix3: A fast, flexible and open software framework for medical image processing and visualisation. Neuroimage, 202:116137, 2019. [DOI] [PubMed] [Google Scholar]
- [75].Andersson JLR et al. How to correct susceptibility distortions in spin-echo echo-planar images: application to diffusion tensor imaging. Neuroimage, 20(2):870–888, 2003. [DOI] [PubMed] [Google Scholar]
- [76].Andersson JLR and Sotiropoulos SN. An integrated approach to correction for off-resonance effects and subject movement in diffusion mr imaging. Neuroimage, 125:1063–1078, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Dhollander T and Connelly A. A novel iterative approach to reap the benefits of multi-tissue csd from just single-shell (+b=0) diffusion mri data, 2016.
- [78].Smith S. Fast robust automated brain extraction. Hum Brain Mapp, 2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Volkow ND et al. Addiction: beyond dopamine reward circuitry. Proc Natl Acad Sci U S A, 108(37):15037–42, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Hall SA et al. Strengthened and posterior-shifted structural rich-club organization in people who use cocaine. Drug Alcohol Depend, 235:109436, 2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Rolls ET et al. Automated anatomical labelling atlas 3. Neuroimage, 206:116189, 2020. [DOI] [PubMed] [Google Scholar]
- [82].Power JD et al. Functional network organization of the human brain. Neuron, 72(4):665–78, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Power JD et al. Spurious but systematic correlations in functional connectivity mri networks arise from subject motion. Neuroimage, 59(3):2142–54, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Cui Z et al. Individualized prediction of reading comprehension ability using gray matter volume. Cereb Cortex, 28(5):1656–1672, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Yeo BT et al. The organization of the human cerebral cortex estimated by intrinsic functional connectivity. J Neurophysiol, 106(3):1125–65, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Allen EA et al. A baseline for the multivariate comparison of resting-state networks. Front Syst Neurosci, 5:2, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Rashid B et al. Dynamic connectivity states estimated from resting fmri identify differences among schizophrenia, bipolar disorder, and healthy control subjects. Front Hum Neurosci, 8:897, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Greene AS et al. Task-induced brain state manipulation improves prediction of individual traits. Nature communications, 9(1):1–13, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Wang X et al. Antagonistic network signature of motor function in parkinson’s disease revealed by connectome-based predictive modeling. NPJ Parkinsons Dis, 8(1):49, 2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Zhu Y et al. Connectome-based model predicts episodic memory performance in individuals with subjective cognitive decline and amnestic mild cognitive impairment. Behav Brain Res, 411:113387, 2021. [DOI] [PubMed] [Google Scholar]
- [91].Manglani HR et al. Employing connectome-based models to predict working memory in multiple sclerosis. Brain Connect, 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Heaton R et al. Hiv-associated neurocognitive disorders persist in the era of potent antiretroviral therapy: Charter study. Neurology, 75(23):2087–2096, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Yang FN et al. The impacts of hiv infection, age, and education on functional brain networks in adults with hiv. J Neurovirol, 28(2):265–273, 2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Qiu W et al. A resting-state fmri study of patients with hiv infection based on regional homogeneity method. In 2011 7th Int.Conf. on Natural Computation, volume 2, pp. 997–1000, 2011. [Google Scholar]
- [95].Luckett PH et al. Modeling the effects of hiv and aging on resting-state networks using machine learning. JAIDS Journal of Acquired Immune Deficiency Syndromes, 88(4):414–419, 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].du Plessis L et al. Resting-state functional magnetic resonance imaging in clade c hiv: within-group association with neurocognitive function. J Neurovirol, 23(6):875–885, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Ortega M et al. Effects of hiv and combination antiretroviral therapy on cortico-striatal functional connectivity. AIDS, 29(6):703–12, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Guo CC et al. Network-selective vulnerability of the human cerebellum to alzheimer’s disease and frontotemporal dementia. Brain, 139(Pt 5):1527–38, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].Buckner RL. The cerebellum and cognitive function: 25 years of insight from anatomy and neuroimaging. Neuron, 80(3):807–15, 2013. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


