Abstract
Jembrana disease virus (JDV) is a newly identified bovine lentivirus that is closely related to the bovine immunodeficiency virus (BIV). JDV contains a tat gene, encoded by two exons, which has potent transactivation activity. Cotransfection of the JDV tat expression plasmid with the JDV promoter chloramphenicol acetyltransferase (CAT) construct pJDV-U3R resulted in a substantial increase in the level of CAT mRNA transcribed from the JDV long terminal repeat (LTR) and a dramatic increase in the CAT protein level. Deletion analysis of the LTR sequences showed that sequences spanning nucleotides −68 to +53, including the TATA box and the predicted first stem-loop structure of the predicted Tat response element (TAR), were required for efficient transactivation. The results, derived from site-directed mutagenesis experiments, suggested that the base pairing in the stem of the first stem-loop structure in the TAR region was important for JDV Tat-mediated transactivation; in contrast, nucleotide substitutions in the loop region of JDV TAR had less effect. For the JDV LTR, upstream sequences, from nucleotide −196 and beyond, as well as the predicted secondary structures in the R region, may have a negative effect on basal JDV promoter activity. Deletion of these regions resulted in a four- to fivefold increase in basal expression. The JDV Tat is also a potent transactivator of other animal and primate lentivirus promoters. It transactivated BIV and human immunodeficiency virus type 1 (HIV-1) LTRs to levels similar to those with their homologous Tat proteins. In contrast, HIV-1 Tat has minimal effects on JDV LTR expression, whereas BIV Tat moderately transactivated the JDV LTR. Our study suggests that JDV may use a mechanism of transactivation similar but not identical to those of other animal and primate lentiviruses.
Jembrana disease was first recognized in 1964 as an acute and infectious disease affecting Bali cattle in the Jembrana district of Bali in Indonesia (5, 40). The virus that causes the disease was recently characterized (9, 10). The morphogenesis, protein structure, antigenic reactivity, and sequence analysis suggested that this virus is a lentivirus related to the bovine immunodeficiency virus (BIV) (9, 10, 40). The most noticeable difference between Jembrana disease virus (JDV) and BIV is the disease induced by each virus in cattle. JDV causes an acute disease in Bali cattle (Bos javanicus) and is endemic in parts of Indonesia (5, 40). In experimentally inoculated cattle, the incubation period varied from 4.5 to 12 days before the onset of clinical symptoms, which included fever, lethargy, anorexia, and lymphadenopathy; the mortality rate was about 17%. In recovered animals there was no recurrence of disease (35, 40). In contrast, experimental BIV infection results in only a subclinical disease syndrome, with transient lymphocytosis and possibly lymphadenopathy associated with follicular hyperplasia, and with no obvious clinical disease (6, 36). Another difference between BIV and JDV is the high titer of JDV (108 50% cattle infective doses per ml) in plasma during the febrile period of the disease; this has not been observed for BIV (6, 35, 36, 40).
JDV is closely related to BIV on the basis of nucleotide sequence analysis. The complete RNA genome of JDV is 7,732 bp (9). It is 750 bp shorter than the genome of BIV 127 (19). Like BIV and other lentiviruses, the JDV genome contains flanking long terminal repeats (LTRs) and the structural genes gag, pol, and env, which are characteristics of all retroviruses. On the basis of sequence analysis, a number of accessory genes represented by small open reading frames (ORFs) exist in the central and 3′-terminal regions of the JDV genome. In particular, the homologous sequence for the regulatory gene tat, which codes for the important trans-acting regulatory protein found in all lentiviruses characterized to date, was identified (9).
The putative Tat protein of JDV was predicted to be expressed from a multiply spliced transcript that includes two coding exons derived from separate ORFs in the central and 3′-terminal regions of the genome (9). In the first coding exon there was a cysteine-rich region, a core region, and a downstream basic domain that was found in the Tat proteins of most lentiviruses, including BIV 127 (19). These domains have been suggested to be important for their nucleic acid-binding and transactivation properties (8, 24). The presence of a BIV homologous tat sequence and the presence of a Tat response element (TAR)-like element in the extreme 5′ end of the JDV RNA strongly suggest that viral transactivation may occur and that it is mediated through an RNA stem-loop structure similar to those found in BIV, equine infectious anemia virus (EIAV), and primate lentiviruses (7, 8, 24).
To study the regulation of JDV gene expression, whether there is a functional Tat protein, and whether active JDV transcription and transactivation are responsible for high-titer JDV expression in infected animals, we characterized the JDV promoter and its ability to be transactivated by its homologous and heterologous Tat proteins. The JDV tat exon 1 coding region, based on sequence analysis, was cloned into a eukaryotic expression vector that contains the Rous sarcoma virus (RSV) promoter. The promoter activities of the intact JDV promoter, a series of 5′ and 3′ JDV LTR deletion mutants, and several site-directed mutants were then studied. Our studies showed that JDV Tat encoded by exon 1 possessed strong transactivation activities and that the predicted JDV TAR region was important for the transactivation. The JDV Tat is a ubiquitous and potent transactivator that activated other lentivirus promoters tested in a variety of cell types.
MATERIALS AND METHODS
Cell culture.
The CV-1 cell line (ATCC CCL70) and primary fetal bovine lung (FBL) cells (36) were cultured in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum (FBS) and penicillin-streptomycin. All FBL cells used for transient transfection were cultured in vitro for no more than six passages.
Construction of plasmids.
The various plasmids that were used in the study, pBIV-CAT, pBTATC, pHIV-CAT, pRSV-HTAT, pRSV-CAT, pHTLV-CAT, pSIV-CAT, and pHIV-2-CAT, have been described previously (25, 27).
To generate the Tat eukaryotic expression plasmid, the putative JDV tat exon 1 coding sequences were PCR amplified from JDV clone 147 (nucleotides [nt] 5000 to 7732) (9). By using the forward primer 5′ CAG ATA TGC CTG GTC CCT GG 3′ and the reverse primer 5′ TCC AGG ATC CAA CGA TCT AGT 3′, the 321-bp fragment from nt 5005 to nt 5326 was amplified. The PCR product was then cloned into the pGEM-T vector (Promega). To generate the Tat expression clone, the tat insert was released from the pGEM-T vector by digestion with NcoI, blunt ended by treatment with Klenow fragment, and then cut with BamHI. The vector plasmid pRSV-CAT (25) was cut with HindIII and filled in with Klenow fragment, followed by digestion with BamHI to release the chloramphenicol acetyltransferase (CAT) gene. The tat fragment was then ligated to the vector downstream of the RSV LTR promoter. This JDV tat expression plasmid was designated pRSV-JTAT.
To construct the JDV LTR clone from JDV clone 147, an EcoRI fragment (nt 6798 to 7732) which contained the U3 and R regions was subcloned into vector pUC18 to generate plasmid pUC18-U3R. To generate a clone with the entire LTR segment, pUC18-U3R and another JDV plasmid, 139, which contained a JDV gene fragment from nt 19 to nt 2881, were used as templates for overlapping PCR. The primers used were pUC18 primer 40 (5′ GTT TTC CCA GTC ACG AC 3′), which is upstream of the U3 region, and JDV primer U5 (5′ GCG CAA GCT TTT GGG TGG TTC T 3′, mutated at nt 159 to engineer a HindIII site), which ends at the boundary of JDV U5 and the noncoding region of the gag gene. Because the two template plasmids overlapped by 110 bp at the R region, the PCR product covered the entire JDV LTR. The PCR product was then inserted into vector pGEM-T to generate plasmid pGEM-JLTR. The JDV LTR fragment was released from pGEM-JLTR by cutting with HindIII and was then ligated into pUC18-CAT at the HindIII site to generate plasmid pJLTR-CAT. This JDV LTR clone was confirmed by DNA sequencing.
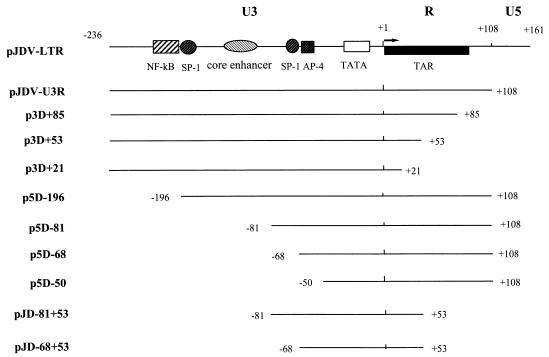
To construct various LTR 3′-end deletion clones, PCR was performed with a single forward upstream primer, JDV-U3.1 (nt 7324 to 7347; 5′ GTC CTC CTA GTT CGG ATC CTT T 3′, mutated to generate a BamHI site) and various LTR downstream deletion primers: JDV-R (nt 7732 to 7713; 5′ TGC CGA AAG CCA AAC GAC CT 3′), JDV-R2 (nt 7709 to 7690; 5′ TTC ACC TCG GCC GGG CTA CC 3′), JDV-R3 (nt 7677 to 7658; 5′ TGC CTT ACA GGG TAC CAG CT 3′, mutated to generate a KpnI site), and JDV-R4 (nt 7651 to 7628; 5′ TCG AAG CTT CAG CTA TCC AGA GC 3′, mutated to generate a HindIII site). The PCR products were then cloned into the pGEM-T vector before insertion into a CAT expression vector. These 3′ deletion clones were designated pJDV-U3R, p3D+85, p3D+53, and p3D+21, according to their positions in the LTR (Fig. 1).
FIG. 1.
Schematic representation of the JDV LTR promoter construct and the various derived deletion clones used for transient transfection analysis. The locations of the various promoter elements, U3, R, U5, the TATA box, and the putative TAR region, are indicated. Several predicted regulatory factor binding sites, for NF-κB, SP-1, AP-4, and the core enhancer element, are also shown. Solid lines represent the sequences retained in the LTR deletion plasmids. The 5′ and 3′ ends for each deletion plasmid are numbered with respect to the transcription start site (+1).
To construct 5′ deletion clones, similar strategies were used, and the 5′ deletion fragments were PCR amplified by using plasmid pJD+108 as a template. A single downstream reverse primer, CAT-R1 (5′ GTC TTT CAT TGC CAT ACG GA 3′), located in the CAT gene, and various upstream forward primers, U3-2 (nt 7431 to 7450; 5′ CCC GGA GCT CGA AAT ATC TGA 3′), U3-3 (nt 7543 to 7562; 5′ CAC GTA GCT TGG AGG ATC AG 3′), U3-4 (nt 7554 to 7573; 5′ GAG GAT CCG CTG ATA CCT AA 3′), and U3-5 (nt 7575 to 7598; 5′ AAT AGT AGT TCC CTT TTG CAT GCT 3′) were used for PCR. All the amplified fragments were cut with EcoRI at a site located within the reverse primer CAT-R1 and then ligated into vector pJLTR-CAT, which was cut with HindIII, blunt ended, and then cut with EcoRI. These 5′ deletion clones were designated p5D-196, p5D-81, p5D-68, and p5D-50. Likewise, two internal deletion clones, designated pJD-81+53 and pJD-68+53, were constructed by using plasmid p3D+53 as a template for PCR (Fig. 1).
Site-directed mutagenesis.
All mutants were generated by overlapping PCR methodology as described elsewhere (1). PCR products were cloned into vector pGCAT-A upstream of the CAT gene. The primers used in the mutagenesis studies are listed below (Table 1).
TABLE 1.
Oligonucleotide primers used for site-directed mutagenesis in the JDV TAR region
| Oligonucleotide | Sequencea | Positionsb | Polarityc |
|---|---|---|---|
| JMR1d | 5′CAACGGGTACCTGGATAGCTGACAGCTC3′ | −4 to +24 | + |
| JMR2 | 5′CTATCCAGGTACCCGTTGCAGAATGCTC3′ | +14 to −14 | − |
| JMR3 | 5′CTCTGATCAGCTGACAGCTCCGAGCC3′ | +5 to +30 | + |
| JMR4 | 5′AGCTGATCAGAGCCCCGTTGCAGAATG3′ | +16 to −11 | − |
| JMR5 | 5′CTCTGGATAGCGCTCAGCTCCGAGCC3′ | +5 to +30 | + |
| JMR6 | 5′CTCGGAGCTGAGCGCTATCCAGAGCC3′ | +28 to +3 | − |
| JMR7 | 5′GCTGACTAGTCCGAGCCCCAGCTGGTA3′ | +14 to +41 | + |
| JMR8 | 5′GCTCGGACTAGTCAGCTATCCAGA3′ | +29 to +6 | − |
Mutated nucleotides are underlined.
The transcription start site is referred to as +1.
+, sense orientation; −, antisense orientation.
JMR1 and JMR2 were used to generate mutant pJD+53M; JMR3 and JMR4 were used to generate mutant pJD+53M2; JMR5 and JMR6 were used to generate mutant pJD+53M3; JMR7 and JMR8 were used to generate mutant pJD+53M4 (see Fig. 4B).
Northern blotting.
Northern blotting was carried out as described previously (1, 42). Briefly, RNA samples were purified with the RNeasy Mini kit (Qiagen, Hilden, Germany), and 10 μg of each RNA sample was separated in a 1.2% formaldehyde agarose gel and then transferred to a supported nitrocellulose membrane. The membrane was baked at 80°C for 2 h, prehybridized (25 mM KPO4 [pH 7.4], 5× SSC [1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate], 5× Denhardt’s solution, 50 μg of salmon sperm DNA/ml, 50% formamide) for 2 to 5 h, then hybridized with a 32P-labeled DNA probe in hybridization solution (prehybridization solution containing 10% dextran sulfate). After hybridization, the membrane was washed twice, each time with 2× SSC–0.1% sodium dodecyl sulfate (SDS) and 0.25× SSC–0.1% SDS solutions at 65°C, and then exposed to Kodak XAR-5 film with an intensifying screen at −80°C overnight. The probes used were labeled by the random-primed labeling method with the NEBlot kit (New England Biolabs). The CAT DNA fragment used for a probe was cut out from plasmid pGCAT-A (Promega), and the glyceraldehyde-3-phosphate dehydrogenase (GAPDH) gene probe was used as an internal control. The GAPDH DNA fragment was amplified by PCR with primers GAPDH1 (5′ CCA TGG AGA AGG CTG GG 3′) and GAPDH2 (5′ CAA AGT TGT CAT GGA TGA CC 3′).
Transfection and CAT assay.
Transfection was carried out with CV-1 and FBL cells. About 2 × 105 cells were plated into each 60-mm plate with 5 ml of DMEM containing penicillin-streptomycin and 10% FBS. About 24 h after plating, the cells were transfected with plasmid DNA with Lipofectamine (GIBCO-BRL). Depending on the experiments, 0.25 to 1 μg of CAT reporter plasmid DNA, with various amounts (0 to 2.5 μg) of tat plasmid DNA, was mixed with 10 μl of Lipofectamine reagent in 500 μl of DMEM. The DNA mixture was then added to the cells, which had been washed twice with DMEM without FBS. Fresh DMEM with 20% FBS was added to the transfected cells 12 h after transfection. At 48 h after transfection, cells were lysed in 1 ml of lysis buffer (CAT ELISA kit; Boehringer Mannheim).
The protein concentration of each lysate was determined by the bicinchoninic acid protein assay (Pierce), and the amount of CAT enzyme in 10 μg of total cellular protein was determined by the CAT enzyme-linked immunosorbent assay (ELISA) using protocols described by the manufacturer (Boehringer Mannheim). In some cases, the β-galactosidase (β-Gal) expression plasmid was cotransfected into cells to normalize the transfection efficiency. The relative amounts of CAT were first determined by the CAT ELISA and then standardized to the experimental β-Gal units. The β-Gal activities were determined by a standard assay as described in the molecular cloning manual of Sambrook et al. (33). Each transfection was repeated three times, and the data were averaged.
Sequence analysis.
The secondary structure of RNA was determined by using the RNA folding program of the Genetics Computer Group (GCG) (Wisconsin package). Protein sequences were aligned with the GCG Pileup program. The tat sequences from the JDV genome (GenBank accession no. U21603), BIV genome (GenBank accession no. M32690), and human immunodeficiency virus type 1 (HIV-1) genome (GenBank locus, hivhxb2cg) were translated into amino acid sequences with the GCG Translate program. The HIV-2 Tat peptide sequence (GenBank accession no. p04605) was obtained from SwissProt.
RESULTS
JDV Tat encoded by exon 1 can activate LTR to high levels.
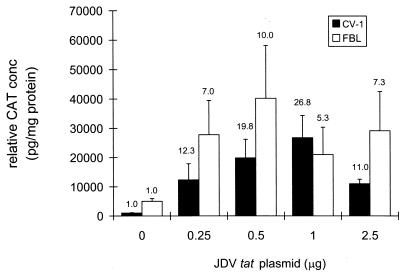
To test the ability of JDV Tat to transactivate the JDV LTR, the reporter plasmid pJDV-U3R was transfected into CV-1 and FBL cells with or without plasmid pRSV-JTAT, and the expression of the LTR was then determined by CAT analysis. The basal expression levels of the JDV LTR were higher in FBL cells than in CV-1 cells (data not shown). In CV-1 cells, when 1 μg of pJDV-U3R was cotransfected with different amounts of pRSV-JTAT (0, 0.25, 0.5, 1.0, and 2.5 μg), JDV Tat was found to transactivate LTR expression 27-fold with the addition of 1 μg of plasmid pRSV-JTAT (Fig. 2). However, high concentrations of the tat plasmid suppressed transactivation. In FBL cells, the highest transactivation level (10-fold) was achieved when 0.5 μg of the tat plasmid was added (Fig. 2). Even though overall CAT activity in the presence of Tat was much higher in FBL cells than in CV-1 cells, the relative activation by Tat was still much lower in FBL cells. The lower transactivation was likely due to the high basal expression levels of the JDV LTR in FBL cells.
FIG. 2.
Cotransfection of the JDV LTR with various amounts of JDV tat (pRSV-JTAT). pJDV-U3R was transfected into either FBL (0.5 μg of DNA) or CV-1 (1 μg of DNA) cells with varying amounts of pRSV-JTAT (0, 0.25, 0.5, 1.0, and 2.5 μg of DNA) by using Lipofectamine. The total amount of DNA used was kept constant by adjusting with pUC18 DNA. The fold activation, calculated as the average concentration of CAT protein (picograms of CAT protein per milligram of total cellular protein) in the presence of tat divided by the average concentration of CAT protein in the absence of tat, is shown above each bar.
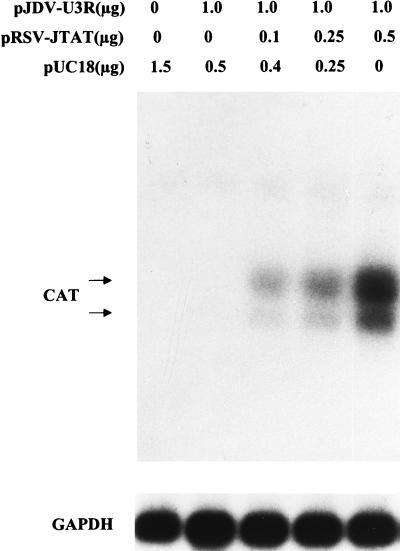
To further verify that JDV Tat encoded by exon 1 is a potent transactivator which acts on the JDV LTR at the transcriptional level, Northern blot analysis experiments were performed. When 1 μg of pJDV-U3R was cotransfected into CV-1 cells with different concentrations of the JDV tat plasmid, higher levels of CAT mRNA were detected in the presence of tat (Fig. 3). This is consistent with the increase in CAT protein levels in the presence of the JDV tat (Fig. 2). It remains to be determined whether the observed transactivation reflects direct interaction of the putative JDV Tat with the LTR or whether it is indirectly mediated by Tat through cellular factors.
FIG. 3.
Northern blot analysis for CAT-specific transcripts. The analysis was carried out with a 32P-labeled CAT-specific probe. The two CAT-specific RNA species indicated by arrows are the spliced and unspliced forms of the CAT mRNA (2). A GAPDH probe was used as an internal control to normalize the amount of RNA loaded onto each lane.
Localization of regulatory elements important for basal and transactivation activity in JDV LTR by deletion analysis.
Sequence analysis showed that the U3 region of the JDV LTR contained many cis-regulatory factor binding sites, such as those for NF-κB, SP-1, and AP-4 (Fig. 1) (9). The R region of JDV was predicted to contain the putative TAR region, corresponding to the TAR of BIV (19–21), and the JDV R region can potentially form three stem-loop secondary structures similar to those of BIV (Fig. 4A) (9). To define the cis- and trans-acting sequences in the LTR necessary for basal expression as well as the TARs on the JDV LTR, we constructed a series of 5′ deletion mutants in which we deleted one or more of the regulatory elements NF-κB, SP-1, core enhancer, and AP-4 (clones p5D-196, p5D-81, p5D-68, and p5D-50 [Fig. 1]). To map the potential TAR sites on the LTR, a set of 3′ deletion mutants was also generated. These mutants were designed in such a way as to delete the U5 region alone or the U5 region as well as one or more of the three predicted stem-loop structures in the putative TAR region in the LTR (clones pJDV-U3R, p3D+85, p3D+53, and p3D+21 [Fig. 1]). Two other internal deletion mutants, pJD-81+53 and pJD-68+53, were constructed to further narrow down the region in the JDV LTR that was required for the promoter activity and transactivation function. Transfection of these LTR deletion plasmids in the absence or presence of JDV tat (pRSV-JTAT) was performed in order to determine the effects of each of the predicted elements in viral transcription and transactivation.
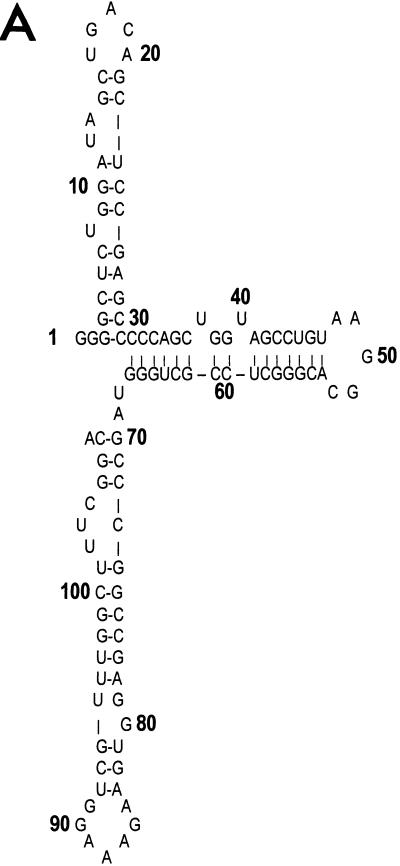
FIG. 4.
(A) The predicted secondary structure of the putative JDV TAR region. RNA folding was performed by the RNAFOLD program in the GCG Wisconsin package. The entire R region of the JDV LTR was included in the analysis. The free energy for the folded structure is −44.8 kJ/mol. (B) Effects of site-directed mutagenesis within the first stem-loop structure of JDV TAR on tat transactivation. Based on the deletion clone p3D+53, we constructed a set of mutation constructs, each of which bears three nucleotide substitutions. The mutated nucleotides are boldfaced, and their corresponding positions in the stem-loop structure are shown. In each experiment, 0.5 μg of the wild-type plasmid (p3D+53) or 0.5 μg of each mutant plasmid was cotransfected into CV-1 cells with the JDV tat plasmid. At 48 h posttransfection, cells were lysed and the lysates were subjected to a CAT ELISA as described in Materials and Methods. The average fold activation, calculated as described in the legend to Fig. 2 and derived from three independent experiments, is presented for each plasmid.
Like the wild-type JDV LTR, most LTR deletion mutants had higher basal activities in FBL cells than in CV-1 cells (Table 2). The JDV LTR construct, pJDV-LTR, which contained the entire U3, R, and U5 regions of the LTR, had activity similar to that of the promoter construct, pJDV-U3R, which contained only the U3 and R regions. This suggested that the U5 region does not play an important role in basal promoter functions, as seen in other retroviruses (7). It was interesting that the basal promoter activity of pJD-196, which had a 5′ deletion including the NF-κB site, was much higher than that of the intact LTR. Its activity was about fivefold higher in CV-1 cells and fourfold higher in FBL cells. It is possible that a JDV negative regulatory element is located between nt −236 and −196, as was found previously in a number of other lentivirus LTRs, including those of BIV, HIV, and EIAV (7, 15, 31). Deletion of sequences between nt −196 and −50, represented by clones p5D-81, p5D-68, and p5D-50, reduced the promoter activity relative to clone p5D-196 in both FBL and CV-1 cells. Clone p5D-50, in which all regulatory elements except the TATA box were deleted, had the lowest basal activity. The predicted regulatory elements, such as NF-κB, SP-1, core enhancer, and AP-4, may therefore play an important positive role in the basal expression of the JDV LTR. Deletion of the LTR from the 3′ end was not expected to affect basal promoter activity. The deletion of the U5 in pJDV-U3R had no effect, and deletion of part of the R region up to position +53 in p3D+53 also had little effect. Interestingly, deletion of part of the R region, including the predicted TAR stem-loops, in clone p3D+21, resulted in much-elevated basal expression levels both in CV-1 and in FBL cells (Table 2), suggesting that the presence of the secondary structure in the R region or other factors targeting the R region may be involved in affecting basal expression of the LTR. Two other deletion clones, pJD-81+53 and pJD-68+53, which retained all the 5′ (SP1, AP-1, and TATA) and 3′ elements essential for basal promoter activity, have activities slightly lower than, but not significantly different from, that of the intact promoter (Table 2).
TABLE 2.
Effects of JDV LTR deletions on basal expression and transactivation by JDV Tat
| Promoter plasmida | JDV LTR expression in:
|
|||||
|---|---|---|---|---|---|---|
| CV-1 cells
|
FBL cells
|
|||||
| Basalb | With JDV Tatb | Fold activationc | Basal | With JDV-Tat | Fold activation | |
| pJDV-LTR | 650 ± 187 | 12,500 ± 410 | 19.3 | 2,120 ± 960 | 17,320 ± 820 | 8.2 |
| pJDV-U3R | 626 ± 125 | 14,460 ± 2,579 | 23.1 | 1,800 ± 617 | 22,680 ± 1,776 | 12.6 |
| p3D+85 | 836 ± 276 | 27,560 ± 1,001 | 28.3 | 3,080 ± 556 | 17,960 ± 5,940 | 5.8 |
| p3D+53 | 676 ± 126 | 21,440 ± 4,667 | 31.7 | 1,994 ± 113 | 11,780 ± 4,861 | 5.9 |
| p3D+21 | 2,780 ± 607 | 5,060 ± 364 | 1.8 | 7,260 ± 1,735 | 4,640 ± 3,666 | 0.6 |
| p5D−196 | 3,180 ± 413 | 48,000 ± 17,224 | 15.1 | 8,280 ± 1,887 | 40,900 ± 10,252 | 5.1 |
| p5D−81 | 930 ± 185 | 14,700 ± 817 | 15.8 | 920 ± 54 | 8,520 ± 3,488 | 9.3 |
| p5D−68 | 650 ± 256 | 20,800 ± 1,258 | 32.1 | 1,060 ± 202 | 11,380 ± 2,920 | 10.8 |
| p5D−50 | 180 ± 98 | 220 ± 106 | 1.2 | 140 ± 15 | 180 ± 21 | 1.2 |
| pJD-81+53 | 320 ± 55 | 6,120 ± 113 | 19.2 | 580 ± 253 | 6,840 ± 1,548 | 11.8 |
| pJD-68+53 | 500 ± 101 | 12,020 ± 947 | 24.1 | 1,130 ± 204 | 11,740 ± 3,408 | 10.4 |
CV-1 cells were transfected with 1 μg of each promoter construct and 1 μg of pRSV-JTAT. FBL cells were transfected with 0.5 μg of promoter plasmid and 1 μg of pRSV-JTAT. The total amount of DNA used for each experiment was kept constant by adjusting with pUC19 plasmid DNA.
Expressed as picograms of CAT protein per milligram of total cellular protein. The relative CAT enzyme concentration was determined by extrapolation from the CAT ELISA, and the absorbance was converted to the amount of CAT enzyme per milligram of total cellular protein by using the CAT standard curve.
Calculated by dividing the CAT enzyme concentration in the presence of pRSV-JTAT by the concentration in the absence of pRSV-JTAT.
In the presence of JDV Tat, all the LTR 5′ deletion clones except p5D-50 were transactivated about 15- to 32-fold in CV-1 cells and about 5- to 10-fold in FBL cells (Table 2). Although basal promoter activity was retained in the deletion construct p5D-50, it was not transactivated by JDV Tat. This suggests that the upstream elements in the U3 region, such as AP-4, may be required for effective transactivation by Tat. Among the three active 5′ deletion clones (p5D-196, p5D-81, and p5D-68), p5D-196 was least responsive to Tat transactivation; the lower level of transactivation observed in pJD-196 could be due to the increased basal expression level. In the 3′ deletion mutants, deletion of U5 in pJDV-U3R and deletion of the third and of both the second and the third stem-loop of the TAR region in p3D+85 and p3D+53, respectively, seemed to have no marked effect on transactivation. Therefore, the second and third stem-loops of the TAR structure located in the R region of the LTR seem to be dispensable for transactivation by Tat. The most significant change in Tat transactivation was observed in pJD+21. This clone had lost its ability to be stimulated by Tat in both CV-1 and FBL cells (Table 2). This suggests that the putative stem-loop 1 of the TAR region formed by nt 1 to 30 was critical for Tat transactivation. It was also of interest that the transactivation by Tat in FBL cells, for most of the promoter constructs tested, seemed to be much lower than that induced in CV-1 cells. This was most likely due to the much higher basal expression levels of the promoter constructs in FBL cells. Despite these differences, the transactivation patterns of these promoters in the two different cell types were very similar.
The stem-loop structure is required for transactivation by JDV Tat.
The deletion analysis described above suggests that JDV Tat can strongly activate the JDV LTR through binding to the TAR region located downstream of the transcription start site. The TAR RNA could assume an extensive secondary structure that contains three stem-loop structures (Fig. 4A). Deletion to position +53 from the 3′ end of the TAR structure was predicted to have no effects on the formation of the first stem-loop structure and has no effects on transactivation (Table 2). To further test whether the first stem-loop secondary structure is required for transactivation by JDV Tat, a set of JDV LTR mutants, each of which differed from the deletion clone p3D+53 by 3 nt, was generated (Fig. 4B). Mutagenesis of the stem structure in clones pJD+53M1, pJD+53M2, and pJD+53M4 was predicted to perturb the proposed secondary structure within the first stem-loop region, whereas for clone pJD+53M3 it was predicted to have no effects. Our transactivation results supported the prediction. Clone pJD+53M3 still responded to Tat transactivation, even though the levels were slightly lower than those for the wild-type construct (p3D+53) (Fig. 4B). In contrast, mutants pJD+53M1, pJD+53M2, and pJD+53M4, which have mutations in the stem structure, lost their ability to be stimulated by Tat (Fig. 4B). Our results indicated that JDV Tat-mediated transactivation was quite sensitive to changes in RNA secondary structure in the stem structure of the first stem-loop but was relatively unaffected by sequence substitutions in the loop region.
Activation of the JDV LTR by BIV Tat and HIV Tat.
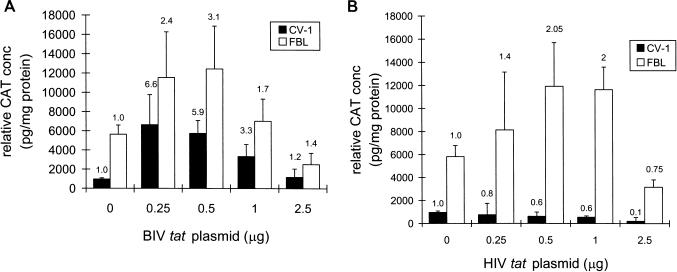
Given the similarities of JDV Tat and the JDV TARs with those of BIV and other primate lentiviruses, we determined if BIV and HIV Tat could stimulate the JDV LTR. The presence of BIV tat stimulated JDV LTR expression in both CV-1 and FBL cells (Fig. 5A). The transactivation levels seemed to be much lower than those observed with JDV tat (Fig. 2). As observed with pRSV-JTAT, the levels of transactivation by BIV Tat seemed to be lower in FBL cells than in CV-1 cells, probably due to the higher basal level of JDV LTR expression in bovine cells.
FIG. 5.
(A) Transactivation of the JDV LTR by BIV Tat in CV-1 and FBL cells. The pJDV-U3R plasmid (1 μg of pJDV-U3R for CV-1 cells and 0.5 μg for FBL cells) and varying amounts of the BIV tat plasmid (0, 0.25, 0.5, 1.0, and 2.5 μg) were cotransfected into cells by using Lipofectamine as described in Materials and Methods. (B) Effects of HIV Tat on JDV LTR expression in CV-1 and FBL cells. The pJDV-U3R plasmid (1 μg for CV-1 cells and 0.5 μg for FBL cells) and varying amounts of the HIV tat plasmid (0, 0.25, 0.5, 1.0, and 2.5 μg) were transfected by using Lipofectamine, and the CAT protein concentration were determined. The fold activation is shown above each bar.
Previous studies with HIV Tat and the BIV LTR have shown that HIV Tat can stimulate BIV expression, but only a two- to threefold increase in transactivation was observed in FBL cells (27). When different concentrations of HIV tat were cotransfected with JDV LTR into FBL cells (Fig. 5B), a maximum of about twofold transactivation was observed. Interestingly, no activation was observed with the largest amount of HIV tat used. In contrast to FBL cells, no transactivation of the JDV LTR by HIV tat was observed in CV-1 cells, suggesting that HIV Tat cannot activate JDV LTR effectively and that cellular factors may also play a role in the expression and transactivation of the JDV LTR.
Transactivation of BIV and HIV LTRs by JDV Tat.
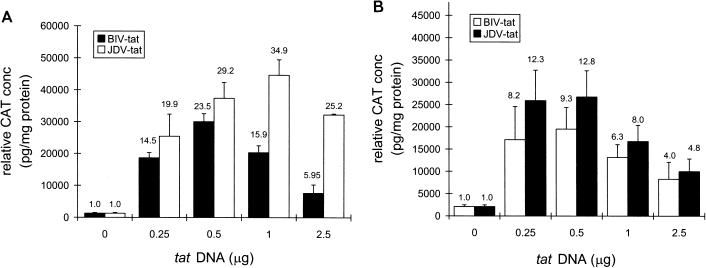
The JDV Tat protein not only has sequence homology with BIV Tat but is also structurally quite homologous to the HIV Tat protein, containing the cysteine, core, and basic functional domains (24). Therefore, we were interested in determining whether JDV Tat would stimulate the BIV and HIV LTRs. pRSV-JTAT was cotransfected with the BIV and HIV promoters, and the expression levels were determined. Cotransfection of pRSV-JTAT and pBIV-LTR-CAT into either CV-1 cells (Fig. 6A) or FBL cells (Fig. 6B) showed very strong transactivation of the BIV LTR by JDV Tat. JDV Tat was at least as active as BIV Tat in stimulating BIV LTR expression. It activated the BIV LTR in CV-1 cells about 29-fold when 0.5 μg of the plasmid DNA was added. In contrast, similar amounts of BIV Tat activated the BIV LTR only about 24-fold. Similar patterns of transactivation were observed in FBL cells. JDV Tat transactivated the BIV LTR about 13-fold, while BIV Tat activated the BIV LTR about 9-fold.
FIG. 6.
Comparison of the effects of JDV Tat and BIV Tat on BIV LTR expression in either CV-1 (A) or FBL (B) cells. Cells were transfected with 0.5 μg of pBIV-LTR-CAT and varying amounts of the JDV tat plasmid (0, 0.25, 0.5, 1.0, and 2.5 μg) by using Lipofectamine. The fold transactivation is shown above each bar.
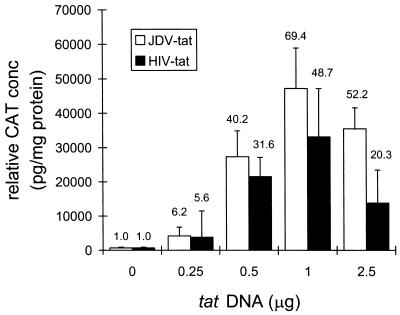
Since it has been demonstrated previously that BIV Tat can transactivate the HIV LTR (27), and given the similarity between BIV and JDV Tat, JDV Tat was tested for its ability to activate the HIV LTR. Either JDV tat (pRSV-JTAT) or HIV tat (pRSV-HTAT) DNA was cotransfected with pHIV-LTR-CAT into CV-1 cells at varying concentrations, and the levels of CAT expression were measured (Fig. 7). The presence of JDV tat transactivated HIV LTR expression very strongly. The presence of 1 μg of JDV tat plasmid stimulated HIV expression about 69-fold; a similar amount of HIV tat plasmid stimulated HIV LTR expression about 49-fold. These results suggest that JDV Tat is a potent transactivator for the HIV LTR and is as active as HIV Tat itself.
FIG. 7.
Comparison of the effects of JDV Tat and HIV Tat on HIV LTR expression. CV-1 cells were transfected with 0.1 μg of pHIV-LTR-CAT and varying amounts of the JDV or HIV tat plasmid (0, 0.25, 0.5, 1.0, and 2.5 μg) by using Lipofectamine. The fold transactivation is shown above each bar.
JDV Tat can transactivate several other retrovirus LTRs.
Since JDV Tat possesses potent transactivating functions on heterologous BIV and HIV-1 promoters, it may be a ubiquitous transactivator that can act in a TAR-dependent or TAR-independent manner. To test this possibility, JDV Tat was tested for its activity on several other lentivirus and nonlentivirus retrovirus promoters, such as the HIV-2, simian immunodeficiency virus (SIV), human T-lymphotropic virus type 1 (HTLV-1), and RSV promoters (Table 3). These promoter CAT constructs were cotransfected with JDV tat (pRSV-JTAT) into either CV-1 or FBL cells, and the transactivation levels were determined. As expected, JDV Tat activated both HIV-2 and SIV lentivirus promoters. The levels of activation were comparable to that observed with the JDV LTR, ranging from about 17- to 22-fold in CV-1 cells and from about 5- to 9-fold in FBL cells. The lower activation levels observed in FBL cells were probably due to higher basal expression levels of these promoters in FBL cells. Interestingly, JDV Tat activated the two nonlentivirus promoters HTLV-1 and RSV. Even though the levels were much lower with these promoters, they were significant and consistent. The HTLV-1 promoter was transactivated by JDV Tat about threefold in CV-1 cells and twofold in FBL cells; the RSV promoter was activated about five- and threefold in CV-1 and FBL cells, respectively.
TABLE 3.
Effects of JDV Tat on different viral promoters in CV-1 and FBL cells
| Promoter constructa | Fold activationb in:
|
|
|---|---|---|
| CV-1 cells | FBL cells | |
| pJDV-U3R | 22.0 ± 2.3 | 8.0 ± 2.7 |
| pHIV2-LTR-CAT | 16.7 ± 4.9 | 9.4 ± 0.6 |
| pSIV-LTR-CAT | 20.5 ± 7.8 | 5.4 ± 0.7 |
| pHTLV-LTR-CAT | 3.2 ± 1.1 | 1.8 ± 0.4 |
| pRSV-LTR-CAT | 5.1 ± 1.2 | 2.9 ± 0.3 |
For each transfection, 0.5 μg of promoter plasmid and 1 μg of JDV tat plasmid were used.
Fold activation was calculated as described in the legend to Fig. 2.
DISCUSSION
Our study is the first to demonstrate that JDV contains a functional tat gene and that exon 1 alone can transactivate the JDV LTR-directed gene expression to high levels. JDV Tat not only transactivated every lentivirus LTR tested but also weakly activated other, nonlentivirus promoters, probably via a TAR-independent mechanism. This suggested that JDV Tat is a potent and ubiquitous transactivator. This strong transactivation function of JDV Tat may be responsible for the ability of the virus to replicate to high titers in infected animals. Virus titers of 108 50% infective doses per ml in blood and plasma of infected animals have been reported (35, 40). The ability of JDV Tat to transactivate BIV, HIV, and other lentivirus promoters suggests that it may involve similar mechanisms of transactivation (8, 11, 12, 14, 15, 17, 28).
Alignment of JDV, BIV, HIV-1, and HIV-2 Tat proteins showed that JDV Tat has conserved cysteine-rich and core transactivation regions, like BIV or HIV (Fig. 8). This may explain why JDV Tat can transactivate BIV, HIV, and other lentivirus LTRs. The basic region and the amino terminus have high homology with BIV Tat but not with HIV Tat. The basic region is responsible for the binding of Tat to TAR. This suggests that JDV Tat may have a TAR recognition domain similar to that of BIV Tat and not to that of HIV Tat. Differences in TAR recognition domains may explain why BIV Tat can transactivate the JDV LTR and HIV Tat cannot. This speculation is further supported by previous studies demonstrating that BIV Tat can bind to its TAR site with high affinity and specificity. Unlike HIV Tat, BIV Tat does not appear to use cellular proteins to stabilize RNA binding in vivo (11). It turns out that the BIV TAR recognition domain simultaneously recognizes the bulge and stem regions of BIV TAR, which adopts an unusual structure, whereas HIV Tat proteins use a single arginine residue within a short region of basic amino acids to recognize a bulge region in TAR (11). Our results suggest that JDV Tat may involve a similar BIV-like TAR recognition domain. The JDV and BIV Tat proteins are much closer phylogenetically and may have evolved to use similar mechanisms to recognize their RNA targets.
FIG. 8.
Alignment of amino acid sequences of JDV, BIV, HIV-2, and HIV-1 Tat exon 1. Peptide sequences were aligned with the GCG Pileup program. The putative N-terminal (N-TERM), cysteine-rich (CYS-RICH), core region, basic, and C-terminal (C-TERM) domains of Tat are underlined and labeled (24). Asterisks mark amino acids that are conserved in JDV and other Tat sequences.
Previous studies on transactivation have focused mainly on the HIV tat gene. Cellular proteins termed cyclin T and cyclin-dependent kinase 9 (CDK9) have recently been found to be involved in transcription activation of the HIV-1 LTR by Tat (41, 43). HIV Tat is predicted to interact directly with cyclin T, which, in turn, specifically binds to CDK9 to form a Tat-cyclin T-CDK9 complex (13, 39). Tat appears to bind the cyclin-T complex and to recruit the complex to the HIV-1 promoter, a process that requires the binding of Tat to the TAR bulge and of cyclin T to the TAR loop. The cyclin-T-associated CDK9 can then phosphorylate the C-terminal domain of RNA polymerase II, leading to a transition from nonpossessive to possessive transcription (13). It is possible that in transactivation of the JDV LTR, JDV Tat adopts a mechanism similar to that of HIV. Several lines of evidence suggest that this might be the case. First, sequence analysis indicated that JDV Tat had some sequence homology with HIV Tat and contained similar highly conserved domains. Our data also showed that JDV Tat could strongly activate the HIV LTR, although HIV Tat cannot activate the JDV LTR. Second, the RNA sequence downstream of the transcription start site in JDV could assume stem-loop structures similar to those of the HIV TAR. Our deletion mapping and mutagenesis data presented in this study showed that the first TAR stem-loop structure was critical for transactivation by JDV Tat. Transactivation of the pJD+85 and pJD+53 deletion clones demonstrated wild-type levels of Tat transactivation, suggesting that the second and third stem-loop structures between +35 and +108 may be dispensable for transactivation. However, we cannot rule out the possibility that the distal stem-loop structures play a role in the regulation of JDV LTR transcriptional activation in vivo. Detailed analysis of stem-loops 2 and 3 of the JDV LTR is required in order to further elucidate their roles in transactivation. The roles of the additional stem-loop structures in transactivation have not been well characterized, although they are also present in other lentivirus LTRs, such as those of BIV, HIV-2, and SIV (3, 7, 16, 38). Previous reports of deletion analysis of the HIV-2 LTR have shown that in addition to the first stem-loop, the second stem-loop was also needed for optimal activity (3). Elimination of the second stem-loop resulted in a two- to threefold reduction in transactivation. In contrast, deletion of the third stem-loop had no effect on transactivation (16).
Like BIV and HIV, JDV Tat is encoded by two exons. This study focused only on exon 1 and indicated that the protein encoded by exon 1 is a strong transactivator analogous to HIV Tat. However, at this time we do not know what role exon 2 may play and whether it is dispensable for transactivation by JDV Tat in vivo. Since no infectious JDV cDNA clone is available, it is difficult at present to elucidate the function of JDV tat exon 2. For HIV, mutagenesis studies based on transfection assays indicated that the second coding exon is dispensable for transactivation (22, 26, 30, 32, 34). However, other studies have also shown that HIV-1 tat exon 2 plays a role in the optimal activation of integrated LTRs but not of unintegrated LTRs (23).
Besides the virus-encoded Tat, many cellular factors were reported to be involved in basal promoter expression and transactivation. For the JDV LTR, a deletion of the upstream sequence from nt −236 to nt −196 seemed to increase basal promoter activity about fivefold in CV-1 cells and fourfold in FBL cells, suggesting the possibility that a negative regulatory element exists, located between nt −196 and −236. Negative elements have indeed been found or suggested in a number of other lentiviruses, including EIAV, BIV, feline immunodeficiency virus, and HIV-1 (7, 15, 31). Previous studies on the HIV LTR indicate that the U3 region plays a very important role in TAR-dependent transactivation (4). In the JDV LTR, several enhancer motifs, such as NF-κB, the core enhancer element, Sp-1, and AP-4, were identified by sequence analysis (9). Our set of U3 deletion clones containing NF-κB, SP-1, core enhancer, and AP-4 deletions have different basal promoter activities, but their abilities to be transactivated by Tat were not affected. However, even though the deletion clone pJD-50, in which the last predicted cellular binding site (AP-4) was deleted, was still competent for basal expression, it was no longer transactivated by Tat. This suggests that besides TAR, additional elements, such as AP-4, may be required for efficient transactivation by Tat. Additional studies are required in order to elucidate the exact function of various elements in the U3 region that may participate in Tat transactivation.
We conclude that JDV, a newly characterized member of the lentivirus family, carries a potent tat gene and uses the common transactivation mechanisms adopted by other lentiviruses, including BIV and HIV. The ability of JDV Tat to transactivate its promoter expression strongly may play an important role in its pathogenesis. The disease course during JDV infection must involve a complex interaction between the virus and the host immune response, as well as an interplay between viral and cellular regulatory factors (18, 29, 34, 37). Therefore, whether a potent and ubiquitous transactivator like JDV Tat plays a role in viral pathogenesis and acute disease manifestation requires further investigation. Further understanding of the regulatory mechanisms of JDV and BIV may provide useful insights into the pathogenesis of lentiviruses.
ACKNOWLEDGMENTS
This work was supported in part by PHS grants TW00493, CA62810, A130356, CA76958 to C.W.
We thank K. Alexander-Nielsen for help in preparation of the manuscript.
REFERENCES
- 1.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: Green Publishing Associates; 1989. [Google Scholar]
- 2.Berkhout B, Silverman R H, Jeang K T. Tat trans-activates the human immunodeficiency virus through a nascent RNA target. Cell. 1989;59:273–282. doi: 10.1016/0092-8674(89)90289-4. [DOI] [PubMed] [Google Scholar]
- 3.Berkhout B, Gatignol A, Silver J, Jeang K T. Efficient trans-activation by the HIV-2 Tat protein requires a duplicated TAR RNA structure. Nucleic Acids Res. 1990;18:1839–1846. doi: 10.1093/nar/18.7.1839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berkhout B, Gatignol A, Rabson A B, Jeang K T. TAR-independent activation of the HIV-1 LTR: evidence that tat requires specific regions of the promoter. Cell. 1990;62:757–767. doi: 10.1016/0092-8674(90)90120-4. [DOI] [PubMed] [Google Scholar]
- 5.Budiarso I T, Hardjosworo S. Jembrana disease in Bali cattle. Aust Vet J. 1976;52:97. doi: 10.1111/j.1751-0813.1976.tb13867.x. [DOI] [PubMed] [Google Scholar]
- 6.Carpenter S, Miller L D, Alexandersen S, Whetstone C A, Van Der Maaten M J, Viuff B, Wannemuehler Y, Miller J M, Roth J A. Characterization of early pathogenic effects after experimental infection of calves with bovine immunodeficiency-like virus. J Virol. 1992;66:1074–1083. doi: 10.1128/jvi.66.2.1074-1083.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carpenter S, Nadin-Davis S A, Wannemuehler Y, Roth J A. Identification of transactivation-response sequences in the long terminal repeat of bovine immunodeficiency-like virus. J Virol. 1993;67:4399–4403. doi: 10.1128/jvi.67.7.4399-4403.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carvalho M, Derse D. Mutational analysis of the equine infectious anemia virus Tat-responsive element. J Virol. 1991;65:3468–3474. doi: 10.1128/jvi.65.7.3468-3474.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chadwick B J, Coelen R J, Sammels L M, Kertayadnya G, Wilcox G E. Genomic sequence analysis identifies Jembrana disease virus as a new bovine lentivirus. J Gen Virol. 1995;76:189–192. doi: 10.1099/0022-1317-76-1-189. [DOI] [PubMed] [Google Scholar]
- 10.Chadwick B J, Coelen R J, Wilcox G E, Sammels L M, Kertayadnya G. Nucleotide sequence analysis of Jembrana disease virus: a bovine lentivirus associated with an acute disease syndrome. J Gen Virol. 1995;76:1637–1650. doi: 10.1099/0022-1317-76-7-1637. [DOI] [PubMed] [Google Scholar]
- 11.Chen L, Frankel A D. An RNA-binding peptide from bovine immunodeficiency virus Tat protein recognizes an unusual RNA structure. Biochemistry. 1994;33:2708–2715. doi: 10.1021/bi00175a046. [DOI] [PubMed] [Google Scholar]
- 12.Cullen B R. Mechanism of action of regulatory proteins encoded by complex retroviruses. Microbiol Rev. 1992;56:375–394. doi: 10.1128/mr.56.3.375-394.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cullen B R. HIV-1 auxiliary proteins: making connections in a dying cell. Cell. 1998;93:685–692. doi: 10.1016/s0092-8674(00)81431-2. [DOI] [PubMed] [Google Scholar]
- 14.Cullen Davis J L, Clements J E. Characterization of a cDNA clone encoding the visna virus transactivating protein. Proc Natl Acad Sci USA. 1989;86:414–418. doi: 10.1073/pnas.86.2.414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dorn P L, Derse D. cis- and trans-acting regulation of gene expression of equine infectious anemia virus. J Virol. 1988;62:3522–3526. doi: 10.1128/jvi.62.9.3522-3526.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Emerman M, Guyader M, Montagnier L, Baltimore D, Muesing M A. The specificity of the human immunodeficiency virus type 2 transactivator is different from that of human immunodeficiency virus type 1. EMBO J. 1987;6:3755–3760. doi: 10.1002/j.1460-2075.1987.tb02710.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Feng S, Holland E C. HIV-1 Tat trans-activation requires the loop sequence within TAR. Nature (London) 1988;334:165. doi: 10.1038/334165a0. [DOI] [PubMed] [Google Scholar]
- 18.Fultz P N. Replication of an acutely lethal simian immunodeficiency virus activates and induces proliferation of lymphocytes. J Virol. 1991;65:4902–4909. doi: 10.1128/jvi.65.9.4902-4909.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Garvey K J, Oberste M S, Elser J E, Braunn M J, Gonda M A. Nucleotide sequence and genome organization of biologically active proviruses of the bovine immunodeficiency-like virus. Virology. 1990;175:391–409. doi: 10.1016/0042-6822(90)90424-p. [DOI] [PubMed] [Google Scholar]
- 20.Gonda M A, Luther D G, Fong S E, Tobin G J. Bovine immunodeficiency virus: molecular biology and virus-host interactions. Virus Res. 1994;32:155–181. doi: 10.1016/0168-1702(94)90040-x. [DOI] [PubMed] [Google Scholar]
- 21.Gonda M A. Bovine immunodeficiency virus. AIDS. 1992;6:759–776. doi: 10.1097/00002030-199208000-00001. [DOI] [PubMed] [Google Scholar]
- 22.Hauber J, Malim M H, Cullen B R. Mutational analysis of the conserved basic domain of human immunodeficiency virus Tat protein. J Virol. 1989;63:1181–1187. doi: 10.1128/jvi.63.3.1181-1187.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jeang K T, Berkhout B, Dropulic B. Effects of integration and replication on transcription of the HIV-1 long terminal repeat. J Biol Chem. 1993;268:24940–24949. [PubMed] [Google Scholar]
- 24.Jones K A, Peterlin B M. Control of RNA initiation and elongation at the HIV-1 promoter. Annu Rev Biochem. 1994;63:717–743. doi: 10.1146/annurev.bi.63.070194.003441. [DOI] [PubMed] [Google Scholar]
- 25.Jung M, Wood C. Activation of retroviral promoters by trans-acting factors. Virol (Life Sci Adv) 1991;10:77–88. [Google Scholar]
- 26.Kuppuswamy M, Subramanian T, Srinivasan A, Chinnadurai G. Multiple functional domains of Tat, the trans-activator of HIV-1, defined by mutational analysis. Nucleic Acids Res. 1989;17:3551–3561. doi: 10.1093/nar/17.9.3551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu Z-Q, Sheridan D, Wood C. Identification and characterization of the bovine immunodeficiency-like virus tat gene. J Virol. 1992;66:5137–5140. doi: 10.1128/jvi.66.8.5137-5140.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Muesing M A, Smith D H, Capon D J. Regulation of mRNA accumulation by a human immunodeficiency virus trans-activator protein. Cell. 1987;48:691–701. doi: 10.1016/0092-8674(87)90247-9. [DOI] [PubMed] [Google Scholar]
- 29.Novembre F J, Johnson P R, Lewis M G, Anderson D C, Klumpp S, McClure H M, Hirsch V M. Multiple viral determinants contribute to pathogenicity of the acutely lethal simian immunodeficiency virus SIVsmmPBj variant. J Virol. 1993;67:2466–2474. doi: 10.1128/jvi.67.5.2466-2474.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rhim H, Rice A P. Exon 2 of HIV-2 Tat contributes to transactivation of the HIV-2 LTR by increasing binding affinity to HIV-2 TAR RNA. Nucleic Acids Res. 1994;21:4405–4413. doi: 10.1093/nar/22.21.4405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rosen C A, Sodroski J G, Haseltine W A. The location of cis-acting regulatory sequences in the human T cell lymphotropic virus type III (HTLV-III/LAV) long terminal repeat. Cell. 1985;41:813–823. doi: 10.1016/s0092-8674(85)80062-3. [DOI] [PubMed] [Google Scholar]
- 32.Ruben S, Perkins A, Purcell R, Joung K, Sia R, Burghoff R, Haseltine W A, Rosen C A. Structural and functional characterization of human immunodeficiency virus Tat protein. J Virol. 1989;63:1–8. doi: 10.1128/jvi.63.1.1-8.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 34.Seigel L J, Ratner L, Josephs S F, Derse D, Feinberg M B, Reyes G R, O’Brien S J, Wong-Staal F. Transactivation induced by human T-lymphotropic virus type III (HTLV III) maps to a viral sequence encoding 58 amino acids and lacks tissue specificity. Virology. 1986;148:226–231. doi: 10.1016/0042-6822(86)90419-8. [DOI] [PubMed] [Google Scholar]
- 35.Soeharsono S, Hartaningsih N, Soetrisno M, Kertayadnya G, Wilcox G E. Studies of experimental Jembrana disease in Bali cattle. I. Transmission and persistence of the infectious agent in ruminants and pigs, and resistance of recovered cattle to re-infection. J Comp Pathol. 1990;103:49–59. doi: 10.1016/s0021-9975(08)80134-x. [DOI] [PubMed] [Google Scholar]
- 36.Suarez D L, Van Der Maaten M J, Wood C, Whetstone C A. Isolation and characterization of new wild-type isolates of bovine lentivirus. J Virol. 1993;67:5051–5055. doi: 10.1128/jvi.67.8.5051-5055.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Suarez D L, Whetstone C A. Identification of hypervariable and conserved regions in the surface envelope gene in the bovine lentivirus. Virology. 1995;212:728–733. doi: 10.1006/viro.1995.1532. [DOI] [PubMed] [Google Scholar]
- 38.Viglianti G A, Rubinstein E P, Graves K L. Role of TAR RNA splicing in translational regulation of simian immunodeficiency virus from rhesus macaques. J Virol. 1992;66:4824–4833. doi: 10.1128/jvi.66.8.4824-4833.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wei P, Garber M E, Fang S M, Fischer W H, Jones K A. A novel CDK9-associated C-type cyclin interacts directly with HIV-1 Tat and mediates its high-affinity, loop-specific binding to TAR RNA. Cell. 1998;92:451–462. doi: 10.1016/s0092-8674(00)80939-3. [DOI] [PubMed] [Google Scholar]
- 40.Wilcox G E, Soeharsono S, Dharma D M N, Copland J W. Jembrana disease and the bovine lentiviruses. ACIAR Proc. 1997;75:10–75. [Google Scholar]
- 41.Yang X, Herrmann C H, Rice A P. The human immunodeficiency virus Tat proteins specifically associate with TAK in vivo and require the carboxyl-terminal domain of RNA polymerase II for function. J Virol. 1996;70:4576–4584. doi: 10.1128/jvi.70.7.4576-4584.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhou Y, Chandran B, Wood C. Transcriptional patterns of the pCD41 (U27) locus of human herpesvirus 6. J Virol. 1997;71:3420–3430. doi: 10.1128/jvi.71.5.3420-3430.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhu Y, Pe’ery T, Peng J, Ramanathan Y, Marshall N, Marshall T, Amendt B, Mathews M B, Price D H. Transcription elongation factor P-TEFb is required for HIV-1 tat transactivation in vitro. Genes Dev. 1997;11:2622–2632. doi: 10.1101/gad.11.20.2622. [DOI] [PMC free article] [PubMed] [Google Scholar]