Abstract
Purpose
To provide a holistic and complete comparison of the five most advanced AI algorithms in the augmentation of low dose 18F-FDG PET data over the entire dose reduction spectrum.
Methods
In this multicentre study, five AI algorithms were investigated for restoring low-count whole-body PET/MRI, covering convolutional benchmarks – U-Net, enhanced deep super-resolution network (EDSR), generative adversarial network (GAN) – and the most cutting-edge image reconstruction transformer models in computer vision to date – swin transformer image restoration network (SwinIR) and EDSR-ViT (vision transformer). The algorithms were evaluated against six groups of count levels representing the simulated 75%, 50%, 25%, 12.5%, 6.25%, and 1% (extremely ultra-low-count) of the clinical standard 3 MBq/kg 18F-FDG dose. The comparisons were performed upon two independent cohorts – (1) a primary cohort from Stanford University and (2) a cross-continental external validation cohort from Tübingen University – in order to ensure the findings are generalizable. 476 original count and simulated low-count whole-body PET/MRI scans were incorporated into this analysis.
Results
For low-count PET reconstruction on the primary cohort, the mean structural similarity index (SSIM) scores for dose 6.25% were 0.898 (95% CI, 0.887–0.910) for EDSR, 0.893 (0.881–0.905) for EDSR-ViT, 0.873 (0.859–0.887) for GAN, 0.885 (0.873–0.898) for U-Net, and 0.910 (0.900–0.920) for SwinIR. In continuation, SwinIR’s performance was also discreetly evaluated at each simulated radiotracer dose levels. Using the primary Stanford cohort, the mean SSIM scores were 0.970 (0.959–0.981) for dose 75%, 0.947 (0.931–0.962) for dose 50%, 0.935 (0.917–0.952) for dose 25%, 0.913 (0.890–0.934) for dose 12.5%, 0.914 (0.896–0.932) for dose 6.25%, and 0.848 (0.816–0.880) for dose 1%.
Conclusion
Swin transformer model outperforms the conventional convolutional neural network benchmarks, enabling 6.25% low-dose PET reconstruction with better structure fidelity and lesion-to-background contrast. A radiotracer dose reduction to 1% of the current clinical standard radiotracer dose is out of scope for current AI techniques.
Keywords: PET reconstruction, whole-body PET imaging, Transformer model, CNN, Deep learning
INTRODUCTION
The use of artificial intelligence (AI) technology for medical image reconstruction has accelerated rapidly in the past decade. AI-powered deep learning neural networks are increasingly being used to augment low-count medical images, such as those acquired by positron emission tomography (PET)[1]. PET has been considered as the gold standard for staging and treatment monitoring of patients with solid cancers[2–4]. However, the disadvantages of PET imaging as compared to magnetic resonance imaging (MRI) are its high cost and ionizing radiation exposure[5–7]. Reductions in radiotracer dosage could minimize radiation exposure, and reductions in scan time could enhance patient throughput and reduce scan costs. However, reductions in radiotracer dosage and scan times lower the detection of PET annihilation events, resulting in low-count PET scans with reduced diagnostic image quality (DIQ)[8]. Based on a comprehensive literature review, the reconstruction of a standard full-count PET imaging from this reduced DIQ cannot be achieved by simple postprocessing operations such as denoising, since lowering the number of coincidence events in the PET detector introduces both noise and local uptake value changes[9]. Hence, sophisticated AI-powered deep learning techniques for image reconstruction became increasingly more popularized to facilitate PET image reconstruction[10–12].
Multiple algorithms have emerged in recent years to enhance low-count PET scans[13–15], with some convolutional neural network (CNN) methods approved by the U.S. Food and Drug Administration (FDA)[16]. However, the FDA does not recommend which specific FDA-approved software should be used for a given medical problem. Most available AI-powered PET reconstruction publications feature a single AI algorithm. As such, the literature currently lacks an unbiased, systematic evaluation comparing multiple state-of-the-art AI algorithms in this context. Moreover, the rapid rate of progress in AI and deep learning research has given way to transformer-based models with innate global self-attention mechanisms capable of outperforming CNN-based benchmarks in a variety of imaging-related tasks including image reconstruction[17–20]. To our knowledge, transformers have not yet been well-adapted and utilized for PET reconstruction, nor have they been directly compared against the state-of-the-art CNNs. Thus, we herein seek to fulfill an unmet need by performing a comprehensive comparison of state-of-the-art AI algorithms for low-count whole-body (WB) PET imaging reconstruction.
Reducing the 18F-FDG dose increases image artifacts, because the image quality is proportional to the number of coincidence events in the PET detector following radiopharmaceutical positron annihilation[1]. Such significant artifacts and noise introduce challenges for the recovery of true radiotracer signal by AI models. Three recent studies have explored AI-based augmentation in WB PET images at 50%[16, 21], 25%[1], and 6.25%[22] of the clinical standard doses. To date, few efforts have been reported on conducting a comprehensive assessment across the dosage reduction spectrum[16]. There is also a lack of PET databases containing list-mode data that can be used to generate a wide array of dose-reduced images for direct comparison[23]. A key question that has not yet been addressed in low-count PET image augmentation is that of model limitation (i.e. what is the lowest reduction percentage that a given AI algorithm can enhance with acceptable clinical utility).
To close the gaps on the aforementioned challenges, our study aimed to compare five different AI-algorithms in the augmentation of low dose 18F-FDG PET data. Using two cross-continental independent PET/MRI datasets, we examined six PET dose level percentages ranging from 75% to 1% against the five most advanced algorithms – spanning the CNN and transformer categories. The five algorithms include three CNN benchmarks: U-Net[24], enhanced deep super-resolution network EDSR[22], generative adversarial network (GAN)[25], and two transformer models: SwinIR[17] and EDSR-ViT[18]. Notably, the recent advancement – Swin transformer – was leveraged for whole-body PET reconstruction for the first time in this study.
To integrate these AI-powered low-count PET reconstruction in a clinical setting, a comprehensive investigation is critical. Hence, we considered different anatomical regions for the training of our algorithm, which has been underexplored in previous studies. This study is pertinent for implementers developing AI models optimized for achieving PET imaging that preserves the best image quality with the lowest possible radiation exposure to patients. To promote the continued advancement of this domain, we have open-sourced the code underpinning the five AI algorithms tailored for PET/MRI reconstruction.
MATERIALS AND METHODS
Participants and Dose Reduction Spectrum
In this multicentre, restrospective evaluation of data from the Health Insurance Portability and Accountability (HIPAA)-compliant clinical trials, two participating centers (University of Tübingen, Germany, and Stanford University, CA, USA) obtained approval from their institutional review board (IRB). Written informed consent was obtained from all adult patients and parents of pediatric patients. Stanford cohort: Between July 2015 and June 2019, we collected 48 whole-body PET/MRI scans (Supplementary pp 1-2) from 22 children and young adults (13 females, 9 males) with lymphoma and a mean age (standard deviation; range) of 17 years (7; 6–30 years). Tumor histology consisted of 14 patients with Hodgkin lymphoma, six with non-Hodgkin lymphoma, and two patients with posttransplant lymphoproliferative disorder (PTLD). Tübingen cohort: 20 whole-body PET/MRI scans (Supplementary pp 1-2) from 10 patients (5 females, 5 males) with a mean age (standard deviation; range) of 14 years (5; 3–18 years) were collected. The distribution of tumor histologies was eight with Hodgkin lymphoma and two with non-Hodgkin lymphoma.
Radiotracer input data were used to generate images. Full-dose (3 MBq/kg) PET data were acquired in list mode, which helps detect coincidence events across the entire duration of the PET bed time (3 minutes 30 seconds). Low- dose PET images were retrospectively simulated by unlisting the PET list-mode data and reconstructing them based on the percentage of coincidence events[26]. List-mode PET input data were collected over time periods: the first block of 3 minutes 30 seconds, 2 minutes 38 seconds, 1 minute 45 seconds, 53 seconds, 26 seconds, 13 seconds, and 2 seconds. These were used to simulate 100%, 75%, 50%, 25%, 12.5%, 6.25%, and 1% 18F-FDG PET dose levels, respectively. This resulted in 476 original count standard-dose and simulated low-count PET/MRI images (336 from the Stanford cohort and 140 from the Tübingen cohort) included in this study.
Study Design
Five different AI-algorithms were trained and tested separately over six dose reduction percentages ranging from 75% to 1% (of the clinical standard-dose) on the primary Stanford PET/MRI images; the GAN model was only evaluated on 6.25% dose due to its underperformance relative to the other algorithms. This resulted in 25 AI models in total. All of the 25 AI models were further tested on the Tübingen external validation cohort. The Tübingen cohort was not included in the training of each algorithm, making it a true external test set. The same image pre-processing steps (Supplementary p 2) were applied to all PET/MRI images from each cohort. Using an approach which aimed to alleviate additional burden on the network learning methods to find patterns between images for final reconstruction, the top and bottom 0.1% of the pixels in PET images were clipped. This operation was critical for model convergence and training stability, as these pixels possessed high noise and were therefore outliers of the distribution.
The 3D whole-body volume was inferenced in a slice-by-slice fashion and the predicted 2D slices were stacked together to reconstruct the final 3D PET prediction. We adopted 2.5D input scheme to ensure vertical spatial consistency. Five consecutive axial slices from both PET and MRI modalities were fed into the model as combined inputs, resulting in ten input slices in total for one evaluation. Five-fold cross-validation was applied to ensure generalization in model performance. A combination of mean square error (MSE) and the structural similarity index measure (SSIM) loss was used to train the model (Supplementary pp 2-3).
Five AI Algorithms Evaluated
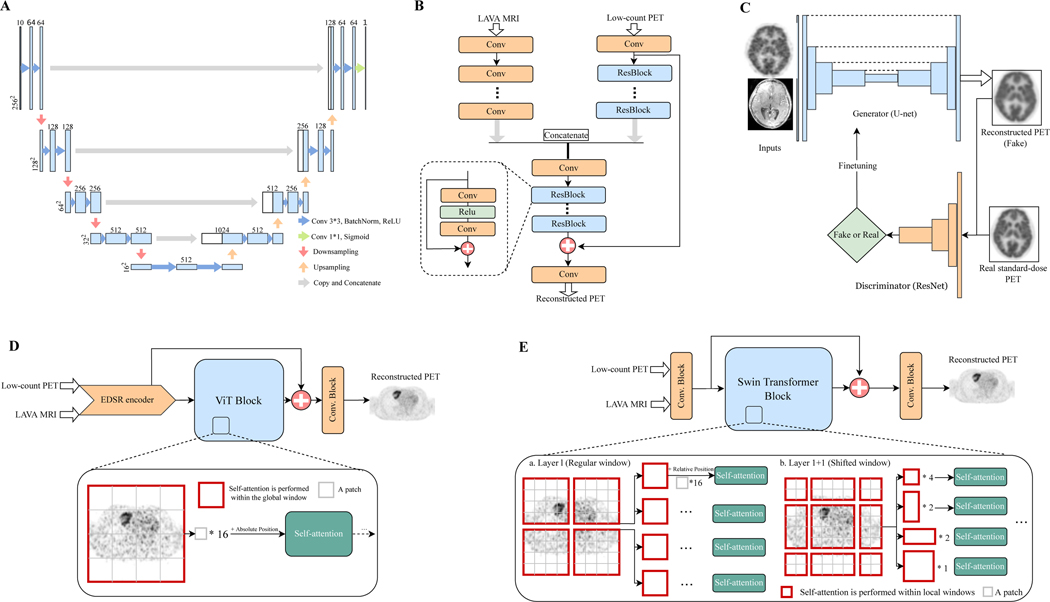
The framework illustrating the five AI algorithms in low-count PET reconstruction is shown in Figure 1. We investigated three CNN benchmarks (U-Net, EDSR, and GAN) and two transformer models (EDSR-ViT and SwinIR). Below, we detailed the algorithms and their algorithmic advantages.
Figure 1: Schematic overviews of AI algorithm frameworks for low-count PET reconstruction.
(A) The classic U-net model. (B) The adapted EDSR (enhanced deep super-resolution network) model. (C) The GAN (generative adversarial network) model. (D) The EDSR-ViT model. EDSR-ViT takes the feature encoder part from the adapted EDSR (B) directly, and makes use of the ViT (Visual transformer) block to obtain global self-attention within the image. (E) The SwinIR model, consisting of Swin transformer blocks. The main difference of Swin transformer and ViT transformer is where the self-attention operation applies. For Swin transformer block, the self-attention is applied within each of the local windows, including the regular window partitions (Layer l) and the following shifted-windows (Layer l+1, etc). For ViT, the self-attention is applied within the global image, which is equally partitioned into fixed-size patches.
U-Net
Proposed in 2015[24], the U-Net was first invented for biomedical image segmentation and has rapidly become the most well-recognized and classic AI model in the medical imaging community. Previous studies[1, 16] have utilized U-Net in low-count PET reconstruction. The name “U-Net” borrows intuitively from the U-shaped structure of the model diagram, as shown in Figure 1A. It consists of (1) the left side encoder, where convolution layers intercalate with max-pooling layers that gradually reduce the dimensions of the image, and (2) the right side decoder, where a set of convolution operations and upscaling brings the feature map back to the original dimensions. This architecture is well-suited for middle-level segmentation tasks, as the semantic information extracted from the encoder, along with the spatial information kept from the skip connection and decoder, provides almost everything needed for semantic segmentation in biomedical images.
EDSR
Investigated on 6.25%-low-count PET/MRI reconstruction in 2021[22], the adapted EDSR is inspired by the classic enhanced deep super-resolution network[27] model in computer vision. The main innovation of EDSR is the organization and optimization of the building block, with only two convolutions, a rectified linear unit (ReLU) activation in between, and an add residual – as shown in Figure 1B. The unnecessary modules – batch normalization and follow-up ReLU activation – in conventional residual networks, ResNet[28] and SRResNet[29], are removed.
GAN
First proposed in 2014[30] and now widely used in image generation, generative adversarial networks (GAN) originated from the notion of having two neural networks, a generator and a discriminator, pitted against one other as adversaries in order to generate new, synthetic instances of data that can pass for real data (Figure 1C); in short, the generator’s goal is to fool the system by trying to produce images that the discriminator cannot distinguish from real-world ones[31]. Several studies[25, 32] have explored GANs in PET reconstruction. However, most of the superior performance has been achieved by introducing additional clinical data – e.g. amyloid status within the brain[25] – which are not always available in real practice. Moreover, the longstanding challenges with GAN training, i.e. model collapse, non-convergence, and instability[33], preclude widespread use in the medical image community.
EDSR-ViT
Originally designed for sequence-to-sequence prediction in natural language processing (NLP)[34], transformer applications had been expanded to image processing very recently and soon became a game-changing technique in computer vision[35]. As opposed to FCN, where the receptive fields are gradually expanded through a series of convolution operations, the self-attention operations inherited in Transformers allow full coverage of the entire input space at the beginning, demonstrating exceptional representation power. Vision Transformer (ViT) – a transformer adapted for image processing – has shown impressive performance on high-level vision tasks[36, 37], but few efforts have been made to explore its role in image reconstruction. In order to examine its performance on PET/MRI reconstruction, we tailored the original ViT by adding an EDSR CNN encoder on top of the transformer block, as shown in Figure 1D. The rationale for this is that the global long-range dependency from ViT and the precise localization from CNN encoder are complimentary for low-level vision tasks[38].
SwinIR
Proposed in 2021[17], SwinIR is among the pioneering efforts in Transformer utilization for image restoration, showing surperior performance over a variety of state-of-the-art methods spanning image super-resolution, image denoising, and JPEG compression artifact reduction. The highlight of SwinIR is the adoption of Swin Transformer[19]. Swin Transformer is a hierarchical transformer whose representation is computed with shifted windows, reducing the border artifacts in ViT – as ViT usually divides the input image into patches with fixed size (e.g. ). This brings greater efficiency by limiting self-attention computation to these local shifted windows and allowing cross-window connection to capture global dependency (Figure 1E). According to a recent study[19], Swin Transformer outperformed ViT in high-level tasks including image classification, object detection, and semantic segmentation. In this study, we adopted the backbone of SwinIR[17], which consists of 24 Swin Transformer blocks for PET reconstruction.
Evaluation Framework
We adopted three quantitative metrics to measure the quality of the reconstructed PET images: SSIM (the structural similarity index), PSNR (peak signal-to-noise ratio), and VIF (Visual information fidelity). SSIM is the most widely used metric in radiology imaging reconstruction[39] (which are a combination of luminance, contrast, and structural comparison functions). Specifically, the SSIM score was derived by comparing the AI-reconstructed PET to the original standard-dose PET sequences and quantifying similarity on a scale of 0 (no similarity) to 1 (perfect similarity). PSNR is most commonly used to measure the reconstruction quality of a lossy transformation[40]. The higher the PSNR, the better the degraded image has been reconstructed to match the original image. SSIM and PSNR mainly focus on pixel-wise similarity; thus, we introduce VIF, which uses natural statistics models to evaluate psychovisual features of the human visual system[41]. The code for calculating the performance was written with Python using SciPy and Scikit-image toolkits (script; Supplementary p 3).
To investigate the utility of AI-reconstructed PET scans in providing quantitative measures of tumor metabolism required for clinical PET interpretations, we measured standardized uptake values (SUVs) for the tumors and used liver as an internal reference standard. SUVs are the most widely used metric in clinical oncologic imaging and play a germane role in assessing tumor glucose metabolism on FDG-PET[42, 43]. The SUVmax of target lesions and SUVmax of liver were measured by placing separate three-dimensional volumes of interest over tumor lesions and the liver. SUVs were measured using OsiriX version 12.5.1. (OsiriX software; Supplementary p 3). SUV values were calculated based on patient body weight and injected dose by using the equation in Supplementary p 3.
Statistical Analysis
We used Wilcoxon signed-rank t test as implemented in R software (V4.0.3) to assess the significance of the difference between two algorithms. We used a predefined P < 0.05 for significance. The performance tables show the mean, standard deviation (SD), and the first (25%) and third (75%) quartiles of the data. The evaluation metrics are provided with two-sided 95% confidence intervals (CIs). All algorithms were written in Python3, with model training and testing performed using the Pytorch package (version 1.10).
RESULTS
Both baseline and follow-up WB PET/MRI scans of 32 children and young adult lymphoma patients were collected and six dose levels (75%, 50%, 25%, 12.5%, 6.25%, and 1%) were simulated, resulting in 476 PET/MRI scans (336 from the primary Stanford cohort and 140 from the Tübingen external cohort). The cross-continental PET/MRI cohorts were used to examine the generalization of our findings. To the best of our knowledge, large pooled PET/MRI databases containing PET list-mode data amenable to simulate low-dose PET for AI model evaluation do not exist. As such, our collected cohort is unique in that it is among the first PET/MRI databases for AI-enabled dose reduction studies.
CNN Models on Low-Count PET Reconstruction
The classic U-Net[24] model, which is the most well-recognized approach in PET reconstruction, ranked 4 out of 5 in the SSIM metric on the 6.25%-low-count Stanford testing cohort (Table 1). The possible explanation is that the frequent sparse operations from U-Net – down-sampling and up-sampling – inevitably lose localization in the pixel space, thus rendering U-Net less suitable for precise low-level PET reconstruction task. EDSR, which has been investigated on 6.25%-low-count PET/MRI reconstruction in 2021[22], ranked 2 in SSIM on the 6.25%-low-count Stanford cohort. Unlike U-Net, no sparse operation is introduced in EDSR, ensuring spatial information integrity. Even so, EDSR with its limited receptive field and lack of global understanding of the input image might restrict its performance for low-count PET reconstruction. The last CNN approach we investigated was generative adversarial networks (GAN). In our experiment, GAN ranked the lowest on 6.25%-low-count PET reconstruction (Table 1). Comparisons of the five AI algorithms in low-count PET reconstruction is summarized in Table 2.
Table 1. Performance metrics of five state-of-the-art AI algorithms on 6.25% low-count PET reconstruction.
Measures of performance include structural similarity index (SSIM), peak signal-to-noise ratio (PSNR), and visual information fidelity (VIF). Performance is based on the Stanford primary cohort (32 scans from 21 patients; indicated by test) and the Tübingen external validation cohort (20 scans from 10 patients; indicated by external). All comparisons are calculated with the ground truth standard-count PET images.
| Model | SSIMtest | PSNRtest | VIFtest | SSIMexternal | PSNRenternal | VIEexternal |
|---|---|---|---|---|---|---|
| EDSR | ||||||
| Mean (SD) | 0.898 (0.033) | 39.7 (2.37) | 0.454 (0.048) | 0.949 (0.020) | 39.5 (1.14) | 0.466 (0.041) |
| Median (Q1.Q3) | 0.908 (0.878,0.923) | 40.3 (38.6,41.3) | 0.462 (0.415,0.481) | 0.952 (0.931,0.966) | 39.8 (38.8,40.2) | 0.478 (0.432,0.501) |
| EDSR-ViT | ||||||
| Mean (SD) | 0.893 (0.035) | 38.7(1.83) | 0.433 (0.051) | 0.947 (0.021) | 38.4 (1.00) | 0.436 (0.042) |
| Median (Q1,Q3) | 0.901 (0.864,0.921) |
38.9 (37.7, 39.9) | 0.438 (0.399,0.457) | 0.950 (0.925,0.964) | 38 5 (38.0, 38.9) | 0.449 (0.395,0.475) |
| GAN | ||||||
| Mean (SD) | 0.873 (0.040) | 37.4(2.14) | 0.417 (0.047) | 0.939 (0.022) | 35.7 (0.957) | 0.427 (0.039) |
| Median (Q1,Q3) | 0.875 (0.845,0.912) | 37.6 (36.2, 38.6) | 0.420 (0.386, 0.445) | 0.939 (0.921,0.957) | 35.7 (35.2,36.1) | 0.435 (0.385, 0.459) |
| U-net | ||||||
| Mean (SD) | 0.885 (0.036) | 39.1 (2.39) | 0.442 (0.048) | 0.947 (0.020) | 39.6 (1.29) | 0.454 (0.042) |
| Median (Q1,Q3) | 0.893 (0.859,0.919) | 39.5 (38.1,40.8) | 0.447 (0.410, 0.471) | 0.951 (0.928, 0.964) | 39.8 (38.7, 40.4) | 0.463 (0.413, 0.494) |
| SwinIR | ||||||
| Mean (SD) | 0.910 (0.029) | 39.9 (2.26) | 0.485 (0.046) | 0.950 (0.019) | 39.1 (1.08) | 0.483 (0.043) |
| Median (Q1,Q3) | 0.918 (0.889, 0.934) | 40.3 (38.5,41.5) | 0.492 (0.453,0.516) | 0.952 (0.933, 0.966) | 39 3 (38.5,39.7) | 0.491 (0.443, 0.524) |
| 6.25% low-count PET | ||||||
| Mean (SD) | 0.786 (0.047) | 35.0(2.42) | 0.263 (0.046) | 0.735 (0.030) | 34.9(1.43) | 0.257 (0.030) |
| Median (Q1,Q3) | 0.802 (0.749,0.816) | 35.4 (33.4, 36.6) | 0.261 (0.234, 0.289) | 0.730 (0.711, 0.751) | 35.1 (34.3,35.1) | 0.249 (0.230, 0.282) |
All P-values, calculated using Wilcoxon signed-rank test between the Al-reconstructed PET and the low-count PET, are below 0.001
Table 2. Comparions of five AI algorithms in low-count PET reconstruction.
The five advanced AI algorithms are compared from nine perspectives. 1) Number of parameters of the model. M = million; 2) Number of operations running the algorithm. Gflops = one billion floating point operations; 3) Time cost for training; 4) Inference time for one low-count PET/MRI scan; 5) Algorithm category – convolutional neural network or transformer category; 6) Model requirement for pretraining; 7) Overall pros of the model; 8) Overall cons of the model; 9) FDA approval status.
| Criteria | U-net | EDSR | EDSR-ViT | GAN | SwinIR |
|---|---|---|---|---|---|
| Number of Parameters (M) | 17.27 | 0.94 | 19.03 | 28.44 | 7.78 |
| Number of Operations (Gflops) | 40.38 | 63.34 | 51.72 | 42.66 | 504.48 |
| Training Time (min/epoch) | 7 | 4 | 4 | 9 | 129 |
| Inference Time (sec/subject) | 10 | 13 | 15 | 10 | 122 |
| Model Category | CNN | CNN | ViT Transformer | CNN | Swin Transformer |
| Pretraining Scheme | None | None | ImageNet2lk-ViT | I mageNet-ResNet | None |
| Pros of the model | Stable | Good performance | Retain more texture details than EDSR | ✗ | Superior performance |
| Cons of the model | Average Performance | Prone to over smooth image | Sensitive to training strategy | Difficult to train; Additional clinical information is needed | Huge number of operations |
| FDA-approved | ✓ | ✗ | ✗ | ✗ | ✗ |
Training is performed on 4 GeForce RTX 3090 GPUs; Inferencing is performed on 4 GeForce GTX 1080 Ti GPUs
Transformer-Based Models on Low-Count PET Reconstruction
The quantitative metrics of EDSR-ViT did not improve as compared with EDSR on the 6.25%-low-count Stanford testing cohort (Table 1), but EDSR-ViT – with the help of transformer addition – did recover more texture details and alleviate the over-smoothing issue of the conventional CNN approach (Figure 2D). One noteworthy limitation of ViT is that its use requires substantial computational resources, as it relies heavily on large-scale datasets – such as ImageNet-21k[44] and JFT-300M (which are not publically available) – for model pretraining[45].
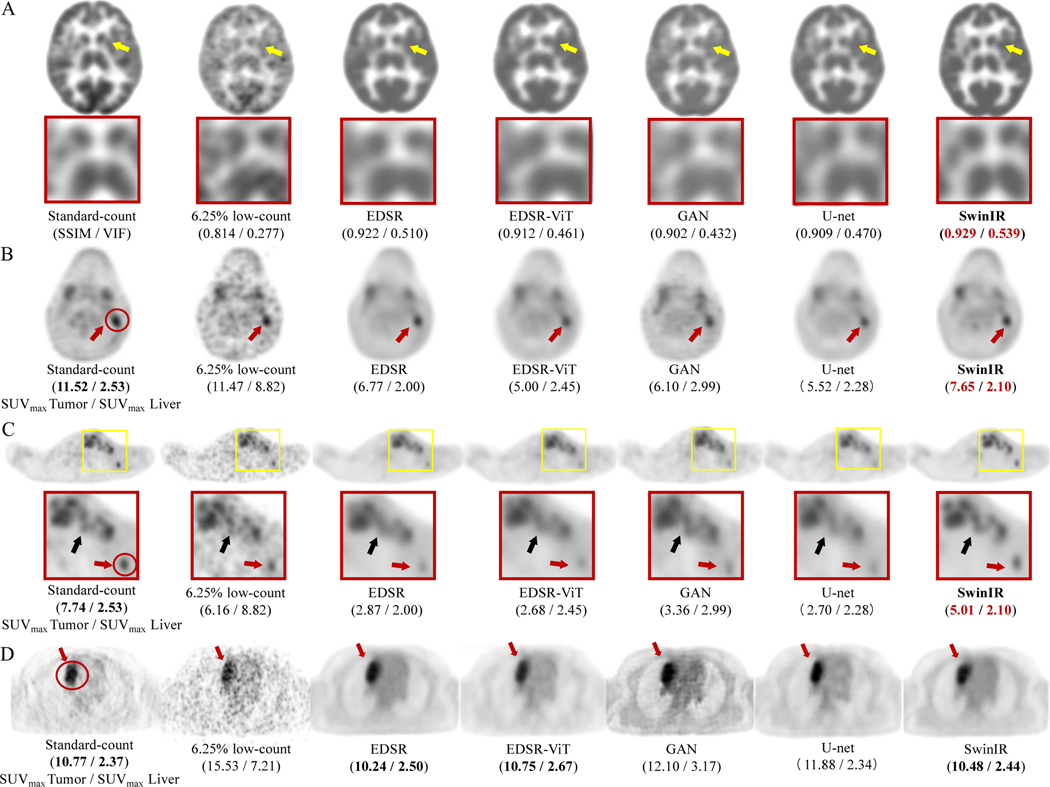
Figure 2: PET image comparison across five state-of-the-art AI algorithms on 6.25% low-count PET reconstruction.
(A) Representative 18F-FDG PET scan of a 29-year-old female patient with Hodgkin lymphoma (HL). The enlarged patches are shown on the second panel (yellow arrows: basal ganglia). The structural similarity index (SSIM) and visual information fidelity (VIF) metrics are presented under each PET image. (B) Representative 18F-FDG PET scan of a 14-year-old male patient with HL. The SUVmax of the lesion (delineated by red circle) and liver for this patient are shown under each PET image. (C) The same patient as (B). The small lesion (less than 1.5 cm3; 5mm < width <10mm; height > 10mm; red arrow ) is enhanced by SwinIR with the lesion-to-liver contrast of SUVmax retained. The lesions (black arrow) are also clearly depicted by SwinIR, in contrast with being blurred and mixed together by the other reconstructions. (D) Representative 18F-FDG PET scan of a 17-year-old female patient from the external Tübingen testing cohort. All AI algorithms successfully denoise the 6.25% low-count images and provide similar diagnostic conspicuity of the lesion (red circle; red arrows) as the standard-dose PET, demonstrating the model is generalizable across different institutions for all AI algorithms. SwinIR shows superiority in retaining lesion-to-liver contrast and structural fidelity.
SwinIR demonstrates significant advantages in low-count whole-body PET reconstruction, ranking one in all metrics on the 6.25%-low-count Stanford testing cohort (Table 1). It shows superiority in retaining lesion-to-background contrast and structural fidelity (Figure 2). Another advantage of SwinIR in low-count PET/MRI reconstruction is that no pre-training is needed according to our experiments, which saves computational resources and introduces flexibility for model tailoring. A major drawback, however, is the large number of operations required in SwinIR – resulting in training and testing times that were 10x longer compared to other state-of-the-art models (Table 2).
Five AI Algorithms on Six Dose Reduction Percentages
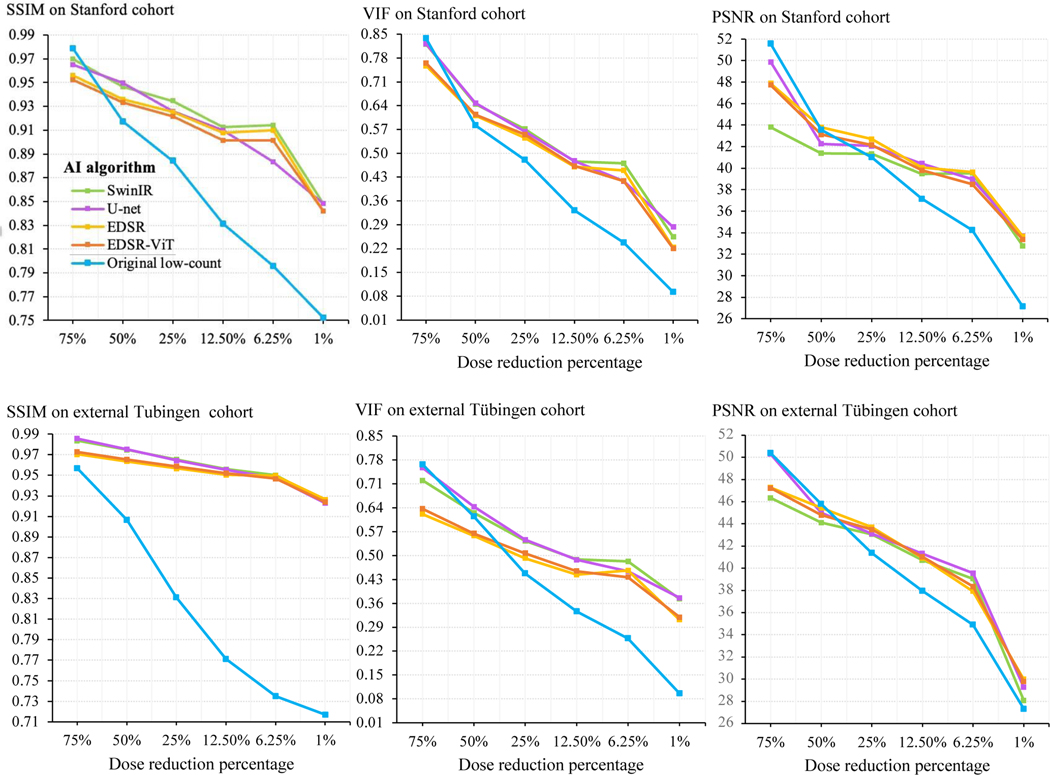
To provide a holistic comparison of the five AI algorithms, all algorithms were evaluated in the reconstruction of low-count whole-body PET images at six reduction percentages (75%, 50%, 25%, 12.5%, 6.25%, and 1% of the clinical standard 3 MBq/kg 18F-FDG dose). The quantitative performance metrics of all AI algorithms over the entire dose reduction spectrum are shown in Figure 3. Model comparisons at doses 25% and 12.5% revealed that SSIM scores were highest for SwinIR and lowest for EDSR-ViT on the Stanford internal test set. At dose 6.25%, SSIM scores were highest for SwinIR and lowest for U-Net. Differences in SSIM score became apparent between algorithms at dose 6.25%, ranging from 0.883 (U-Net) to 0.914 (SwinIR). At dose 1%, SSIM scores were highest for SwinIR and U-Net, and lowest for EDSR and EDSR-ViT. Differences in SSIM score between algorithms were the least appreciable at dose 1%, ranging from 0.842 (SwinIR and U-Net) to 0.848 (EDSR and EDSR-ViT). For the Tübingen cohort, SwinIR also achieved the best performance in the SSIM metric with doses below 50% (Figure 3). More detailed performance metrics for 6.25% low-count PET reconstruction are shown in Table 1. Dose 6.25% was the lowest dosage with around 40 dB PSNR for the AI reconstruction and thereby became our dose of choice for further investigation. The systematic evaluation presented herein is rendered in summary form, with mean and median quantitative values over the four-fold cross-validations on the two cohorts of interest (Table 1). SwinIR achieved the best quantitative results, with the highest SSIM score of 0.910 (95% CI 0.900–0.920), PSNR score of 39.9 (39.1–40.6), and VIF score of 0.485 (0.469–0.501) on the primary Stanford test set. It was also generalized to the external Tübingen test set with the highest SSIM score of 0.950 (0.942–0.958) and VIF score of 0.483 (0.464–0.502), demonstrating superior model generalization across different institutions and scanner types.
Figure 3: Quantitative metrics over the dose reduction spectrum.
The five AI algorithms were adapated for the low-count PET reconstruction task. The AI models were trained on 75%, 50%, 25%, 12.5%, 6.25%, and 1% of the clinical standard 18F-FDG dose PET/MRI images from the primary Stanford cohort. One round of cross-validation was adopted. The trained models were then evaluated on the corresponding low-count PET/MRI test set. The performance on the Stanford internal test set is shown on the top panel, and the performance on the external Tubingen test cohort is shown on the bottom panel. Measures of performance include structural similarity index (SSIM), peak signal-to-noise ratio (PSNR), and visual information fidelity (VIF). For all three metrics, higher represents better reconstruction. All comparisons are made against the ground-truth standard-count PET images. The blue line presents the original low-count PET images without AI enhancement and serves as the baseline for direct comparisons.
The qualitative comparisons between the five AI algorithms on 6.25% low-count reconstruction are shown in Figure 2. The PET images reconstructed from the SwinIR model were superior in reflecting the underlying anatomic patterns of the tracer uptake (the basal ganglia; Figure 2A) when compared to the images generated from the other four models. Meanwhile, though lesions could be detected on all AI-reconstructed scans (Figure 2B-D), lesion-to-background contrast and confidence for lesion detection were significantly improved on SwinIR – especially for small lesions (Figure 2C). Compared to the standard-dose 18F-FDG PET scans, the simulated 6.25% low-count PET images had significantly higher SUVmax values of the liver as a result of increased image noise. All five AI algorithms managed to recover SUVmax values of the liver similar to the values in standard-dose PET, demonstrating good denoising capability. All tumors had SUV values above that of the liver on all AI-reconstructed PET images. For small lesions less than 1.5 cm3, high lesion-to-liver SUV contrast (i.e. the commonly used standard employed by radiologists for lesion detection) was only retained by SwinIR (Figure 2B,C).
Reconstruction Across the Dose Reduction Spectrum
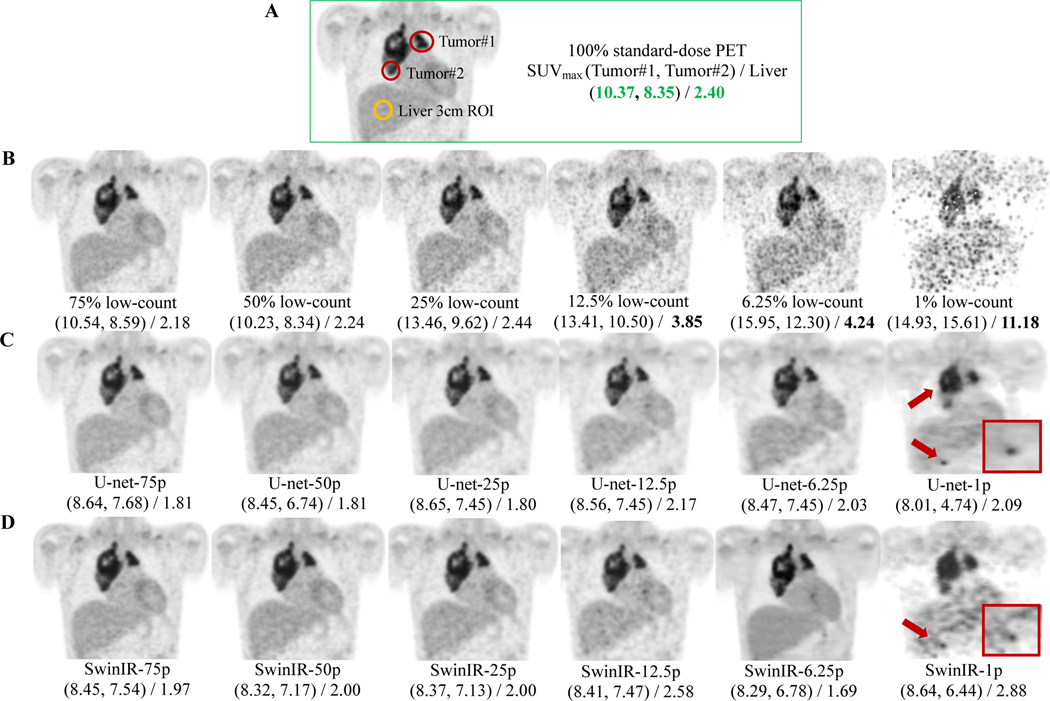
Next, we examine the AI-powered PET reconstruction through the lens of dose reduction spectrum. AI-reconstructed PET images consistently achieved improved SSIM, VIF, and PSNR over original low-count PET images at dose 25%, 12.5%, 6.25%, and 1% (Figure 3). Among the six dose reduction percentages, the improvement from AI reconstruction was largest at dose 6.25%. The average improvement scores for the five AI algorithms were 0.106 (95% CI 0.102–0.110) in SSIM, 3.97 dB (3.78–4.16) in PSNR, and 0.183 (0.178–0.188) in VIF on Stanford internal test cohort; 0.211 (0.208–0.215) in SSIM, 3.54 dB (3.20–3.88) in PSNR, and 0.196 (0.190–0.202) in VIF on Tübingen external test set. Pair-wise t-tests between the AI-reconstructed PET images and the low-count PET images revealed p-values consistently less than 0.001, suggesting that all AI algorithms possessed statistically significant capacities for reconstruction and generalization. Figure 4 provide the detailed qualitive PET image comparisons between different dosages. With reduction in simulated radiotracer dose, PET images exhibited higher noise and information loss, leading to markedly increased SUVmax values in the liver and tumors (Figure 4D). The AI models tested herein reduced artifacts for the low-count PET images and recovered the SUVmax values of liver and tumors to values commensurate with those derived from standard-dose PET (Figure 4C,D).
Figure 4: PET image comparisons across the dose reduction spectrum from 75% to 1% (of the clinical standard 3 MBq/kg 18F-FDG dose).
Representative 18F-FDG PET scan of 13-year-old male patient with Diffuse large B cell lymphoma (DLBCL). The SUVmax of two tumors and liver were measured for each PET image. SwinIR and U-Net are our demonstration models of choice, representing the transformer and CNN categories, respectively. (A) The coronal slice of the standard-dose PET, showing the chest region. SUVmax of two tumors and liver were measured for direct comparison. (B) The original low-count PET images with SUVmax measured under the same regions of tumors and liver as in (A). (C) U-Net reconstructed low-count PET images. The red arrows point to corrupted reconstruction in mediastinum and erroneous upstaging in liver. Red rectangle: enlargement of false upstaging in the liver area. U-Net-75p = U-Net reconstructed 75% low-count PET image. (D) SwinIR reconstructed low-count PET images. The red arrows point to the erroneous upstaging. Red rectangle: enlargement of the degraded reconstruction in liver. SwinIR-75p = SwinIR reconstructed 75% low-count PET image.
For doses 75% and 50%, there were discrepancies between quantitative metrics and visual appearances. All AI models have enhanced the 75% and 50% low-count PETs visually with reduced image noise (Figure 4), but the improvements were not reflected quantitatively (Figure 3). A possible explanation is that 75% and 50% low-count PET images are sufficiently similar to standard-dose PET. Their PSNR values are greater than the threshold – 40dB – which corresponds to nearly undiscernable differences, and thus passes the considerations for good image quality[46, 47]. Therefore, the quantitative metrics might not be able to reasonably depict improvements above this threshold.
In general, the quantative metrics – SSIM, VIF, and PSNR – of both original low-count PET and AI-reconstructed PET images decreased over the dose reduction spectrum. However, AI reconstructions (powered by SwinIR, EDSR, and EDSR-ViT) between doses 12.5% and 6.25% achieved similar performance in the three metrics (Figure 3). This is partly owing to the smoothing effect of 6.25% low-count reconstruction (the liver area in SwinIR-6.25p; Figure 4D). The AI models in 6.25% low-count reconstruction converged on an approach that smoothed particular regions with significantly decreased noise.
From doses ranging from 6.25% to 1%, there was a steep drop (Figure 3) in SSIM, PSNR, and VIF across both Stanford and Tübingen cohorts, indicating the challenge of extreme-low-count PET reconstruction. Indeed, AI reconstruction introduced hallucinated signals and erroneous upstaging in 1% low-count PET reconstruction (Figure 4C,D; far right column). Qualitatively, all of the AI reconstructed PET images in Figure 4 closely resemble the true standard-dose PET, except for dose 1%. The extreme-low-count scenario degraded PET images with substantial artifacts and information loss that were difficult for the current AI techniques to handle without the incorporation of additional information. Supplementary Figure 2 shows the whole-body PET reconstruction from the coronal view, across the dose reduction spectrum powered by SwinIR.
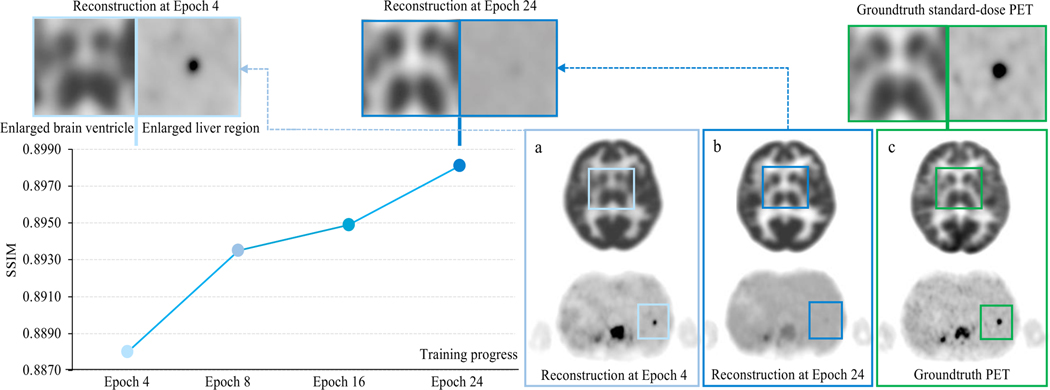
Model Training Strategy
Figure 5 demonstrates an interesting observation when training SwinIR in 6.25% low-count PET images. In epoch 24, the trained model was able to reconstruct the shape and contrast of the basal ganglia in the brain, but failed to clearly depict a small lesion (less than 1.5 cm3) in the liver. Meanwhile in epoch 4, the brain structure was not well-reconstructed, but the diagnostic conspicuity of the small lesion was preserved. Our experiment suggested that the discrepancies in reconstruction quality between different anatomical regions were agnostic to specific model architectures. The possible reasons may be two-fold: (1) the commonly used loss functions – mean square loss (MSE loss) and the structural similarity index loss (SSIM loss) – were originally proposed for natural image reconstruction and not specifically designed for diagnostic radiology images, thus limiting their ability to guide model training for these specific clinical needs; and (2) whole-body PET images have large intra-patient uptake variation. The metabolic activities of the brain and bladder are greater than other anatomical locations, shown as hyperintensities in PET images. As the training progresses, the focus of model optimization can shift to these hyperintense regions easily, as they can possess larger absolute loss penality values; this can in turn cause over-smoothing of other relatively low-intensity regions (e.g. the liver).
Figure 5: Representative discrepancy in reconstruction quality between different anatomical regions over the course of model training.
SwinIR is the model of choice for this demonstration. The performance is based on the primary Stanford PET/MRI cohort. The line chart shows the SSIM metric of the Stanford validation set over models at different training epochs. PET images illustrate cases from the Stanford testing set. The patches (top panel) are enlarged crops of a, b, and c, respectively. As the training progresses from epoch 4 to epoch 24, the structure of the basal ganglia within the brain becomes better reconstructed, while the small lesion (less than 1 cm3) within the liver gets over-smoothed.
DISCUSSION
In this study, we provide the first unbiased and comprehensive investigation of AI-enabled low-count whole-body PET reconstruction from two perspectives: the reconstruction algorithms and the dose reduction percentages. Six reduction percentages covering the entire dose spectrum – 75%, 50%, 25%, 12.5%, 6.25%, and 1% (extreme low count) of the clinical standard 18F-FDG dose – were investigated. In addition, we adapted five state-of-the-art AI algorithms for this task, including the classic CNN benchmarks and the most advanced transformer models. Two cross-continental PET/MRI cohorts were used to examine the generalization of our findings.
All five AI algorithms possess PET reconstruction capability. In particular, transformer models – especially Swin transformer – demonstrated superior performance in restoring structural details and lesion consipicuity in low-count PET reconstruction. The transformer approach complemented the conventional CNN approaches in that the innate global self-attention mechanisam provided long-range dependency that is otherwise lacking in CNNs due to the limited receptive field of convolution operations. The only work to date applying transformers on PET reconstruction is focused on 25% low-count brain images[48], whereas our investigation leverages WB scans over the complete dose reduction spectrum. Additionally, this is the first study utilizing Swin transformer – the most recent advancement in computer vision – for whole-body PET reconstruction. The improved Swin transformer model (SwinIR) with its shifted window mechanism further improved the depiction of structural details and small lesions that could be missed if the fixed partition operations of ViT transformer alone were used.
While AI deep learning architectures are essential in low-count PET reconstruction, equally important is the model training strategy, i.e. the procedure used to carry out the learning process; this includes specific considerations such as the loss function and when to stop training. To date, few efforts have been made to reconcile these considerations. We made an oberservation about the discrepancy in reconstruction quality among different anatomical regions on PET reconstruction over the course of training. This observation underscores the role of training strategy in building up the optimal model for low-count whole-body PET reconstruction. Our findings suggest that engaging radiologists in the model development loop is imperative so that the PET reconstruction training process can be effectively and efficiently guided by domain experts in a task-specific fashion. Another possible direction is region-based reconstruction that takes the regional differerence priors into consideration for effectively designing WB PET reconstruction models.
Another key contribution of this study is the examination of AI-powered PET reconstruction over six groups of count levels, representing 75%, 50%, 25%, 12.5%, 6.25%, to extremely ultra-low-count 1% (of the clinical standard 3 MBq/kg 18F-FDG dose). In order to perform a holistic assessment of low-count PET reconstruction, we adapted multiple AI algorithms – with the exception of GAN – upon the complete dose reduction spectrum. The GAN model was excluded due to its underperformance and difficulty of training relative to the other models. The most relevant work to our study, published in 2021[16], evaluated the FDA-approved U-Net software across various dosages. This commercially available software was trained only on 25% low-count PET images and was tested at other percentages. In contrast, our study takes the approach of training and testing images in a manner consistent with the relevant reduced dosage. To our best knowledge, this study is the first complete investigation of AI-powered PET reconstruction over the entire dose reducing spectrum. The most cutting-edge AI algorithms enabled low-count PET reconstruction of doses above 6.25%, while dose 1% without additional clincal information was out of scope for the AI techniques evaluated herein.
This study has the following limitations. Simulated low-dose PET images were used instead of injecting multiple different PET tracer doses in a single patient, considering ethically feasiblity. Though previous data have shown that simulated low-dose images have characteristics similar to those of actual low-dose images[49], evidence of AI-reconstruction in true injected low-dose cases is needed. In addition, this study only included patients scanned with FDG, due to its clinical prevalency. The use of the deep-learning approaches to reconstruct images obtained with non-FDG radiotracers may entail different performances dependent upon signal-to-noise ratios, and the uptake dynamics and locations.
In conclusion, the findings from this study hold important implications for implementers developing the optimal AI model in order to achieve PET imaging with the lowest radiation exposure to patients and non-inferior DIQ. Mitigation of ionizing radiation exposure from medical imaging procedures holds critically important potential for clinical impact, as reducing such exposure could minimize the potential risk of secondary cancer development later in life[5, 50, 51]. This is especially important for pediatric patients or patients receiving therapies that require repeat imaging with reoccurring radiation exposure. Toward further advancement of this domain, we open-sourced the five AI algorithms specifically tailored for low-count PET/MRI reconstruction. Of note, our code may easily be applied to other medical imaging modalities (e.g. MRI, CT) and could thereby potentially serve as a common foundation for medical image reconstruction.
Supplementary Material
ACKNOWLEDGMENTS
This study was supported by a grant from the National Cancer Institute of the US National Institutes of Health, grant number R01CA269231, and the Andrew McDonough B+ Foundation.
DISCLOSURES
The funders had no role in the study design, data collection and analysis, decision to publish, and preparation of the manuscript.
Footnotes
The authors declare no competing financial or non-financial interests.
REFERENCES
- 1.Chaudhari AS, Mittra E, Davidzon GA, Gulaka P, Gandhi H, Brown A, et al. Low-count whole-body PET with deep learning in a multicenter and externally validated study. NPJ digital medicine. 2021;4:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baum SH, Frühwald M, Rahbar K, Wessling J, Schober O, Weckesser M. Contribution of PET/CT to prediction of outcome in children and young adults with rhabdomyosarcoma. Journal of Nuclear Medicine. 2011;52:1535–40. [DOI] [PubMed] [Google Scholar]
- 3.Kleis M, Daldrup-Link H, Matthay K, Goldsby R, Lu Y, Schuster T, et al. Diagnostic value of PET/CT for the staging and restaging of pediatric tumors. European journal of nuclear medicine and molecular imaging. 2009;36:23–36. [DOI] [PubMed] [Google Scholar]
- 4.Baratto L, Hawk KE, Qi J, Gatidis S, Kiru L, Daldrup-Link HE. PET/MRI Improves Management of Children with Cancer. Journal of Nuclear Medicine. 2021;62:1334–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Huang B, Law MW-M, Khong P-L. Whole-body PET/CT scanning: estimation of radiation dose and cancer risk. Radiology. 2009;251:166–74. [DOI] [PubMed] [Google Scholar]
- 6.Meulepas JM, Ronckers CM, Smets AM, Nievelstein RA, Gradowska P, Lee C, et al. Radiation exposure from pediatric CT scans and subsequent cancer risk in the Netherlands. JNCI: Journal of the National Cancer Institute. 2019;111:256–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brenner DJ, Doll R, Goodhead DT, Hall EJ, Land CE, Little JB, et al. Cancer risks attributable to low doses of ionizing radiation: assessing what we really know. Proceedings of the National Academy of Sciences. 2003;100:13761–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Townsend D. Physical principles and technology of clinical PET imaging. Annals-Academy of Medicine Singapore. 2004;33:133–45. [PubMed] [Google Scholar]
- 9.Wang T, Lei Y, Fu Y, Curran WJ, Liu T, Nye JA, et al. Machine learning in quantitative PET: A review of attenuation correction and low-count image reconstruction methods. Physica Medica. 2020;76:294–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang G, Ye JC, Mueller K, Fessler JA. Image reconstruction is a new frontier of machine learning. IEEE transactions on medical imaging. 2018;37:1289–96. [DOI] [PubMed] [Google Scholar]
- 11.Reader AJ, Corda G, Mehranian A, da Costa-Luis C, Ellis S, Schnabel JA. Deep learning for PET image reconstruction. IEEE Transactions on Radiation and Plasma Medical Sciences. 2020;5:1–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Raj A, Bresler Y, Li B. Improving robustness of deep-learning-based image reconstruction. International Conference on Machine Learning. 2020:7932–42. [Google Scholar]
- 13.Häggström I, Schmidtlein CR, Campanella G, Fuchs TJ. DeepPET: A deep encoder–decoder network for directly solving the PET image reconstruction inverse problem. Medical image analysis. 2019;54:253–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gong K, Catana C, Qi J, Li Q. PET image reconstruction using deep image prior. IEEE transactions on medical imaging. 2018;38:1655–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xu J, Gong E, Pauly J, Zaharchuk G. 200x low-dose PET reconstruction using deep learning. arXiv preprint arXiv:171204119. 2017.
- 16.Theruvath AJ, Siedek F, Yerneni K, Muehe AM, Spunt SL, Pribnow A, et al. Validation of Deep Learning–based Augmentation for Reduced 18F-FDG Dose for PET/MRI in Children and Young Adults with Lymphoma. Radiology: Artificial Intelligence. 2021;3:e200232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liang J, Cao J, Sun G, Zhang K, Gool LV, Timofte R. SwinIR: Image Restoration Using Swin Transformer. IEEE/CVF International Conference on Computer Vision Workshops (ICCVW). 2021:1833–44. [Google Scholar]
- 18.Dosovitskiy A, Beyer L, Kolesnikov A, Weissenborn D, Zhai X, Unterthiner T, et al. An image is worth 16×16 words: Transformers for image recognition at scale. arXiv preprint arXiv:201011929. 2020.
- 19.Liu Z, Lin Y, Cao Y, Hu H, Wei Y, Zhang Z, et al. Swin transformer: Hierarchical vision transformer using shifted windows. arXiv preprint arXiv:210314030. 2021.
- 20.Han K, Wang Y, Chen H, Chen X, Guo J, Liu Z, et al. A survey on visual transformer. arXiv preprint arXiv:201212556. 2020.
- 21.Whiteley W, Luk WK, Gregor J. DirectPET: full-size neural network PET reconstruction from sinogram data. Journal of Medical Imaging. 2020;7:032503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang Y-RJ, Baratto L, Hawk KE, Theruvath AJ, Pribnow A, Thakor AS, et al. Artificial intelligence enables whole-body positron emission tomography scans with minimal radiation exposure. European Journal of Nuclear Medicine and Molecular Imaging. 2021:1–11. [DOI] [PMC free article] [PubMed]
- 23.Schramm G, Rigie D, Vahle T, Rezaei A, Van Laere K, Shepherd T, et al. Approximating anatomically-guided PET reconstruction in image space using a convolutional neural network. NeuroImage. 2021;224:117399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ronneberger O, Fischer P, Brox T. U-net: Convolutional networks for biomedical image segmentation. International Conference on Medical image computing and computer-assisted intervention. 2015:234–41. [Google Scholar]
- 25.Ouyang J, Chen KT, Gong E, Pauly J, Zaharchuk G. Ultra‐low‐dose PET reconstruction using generative adversarial network with feature matching and task‐specific perceptual loss. Medical physics. 2019;46:3555–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sekine T, Delso G, Zeimpekis KG, de Galiza Barbosa F, Ter Voert EE, Huellner M, et al. Reduction of 18F-FDG dose in clinical PET/MR imaging by using silicon photomultiplier detectors. Radiology. 2018;286:249–59. [DOI] [PubMed] [Google Scholar]
- 27.Lim B, Son S, Kim H, Nah S, Mu Lee K. Enhanced deep residual networks for single image super-resolution. Proceedings of the IEEE conference on computer vision and pattern recognition workshops. 2017:136–44. [Google Scholar]
- 28.He K, Zhang X, Ren S, Sun J. Deep residual learning for image recognition. Proceedings of the IEEE conference on computer vision and pattern recognition. 2016:770–8. [Google Scholar]
- 29.Ledig C, Theis L, Huszár F, Caballero J, Cunningham A, Acosta A, et al. Photo-realistic single image super-resolution using a generative adversarial network. Proceedings of the IEEE conference on computer vision and pattern recognition. 2017:4681–90. [Google Scholar]
- 30.Goodfellow I, Pouget-Abadie J, Mirza M, Xu B, Warde-Farley D, Ozair S, et al. Generative adversarial nets. Advances in neural information processing systems. 2014;27.
- 31.Lucas A, Iliadis M, Molina R, Katsaggelos AK. Using deep neural networks for inverse problems in imaging: beyond analytical methods. IEEE Signal Processing Magazine. 2018;35:20–36. [Google Scholar]
- 32.Islam J, Zhang Y. GAN-based synthetic brain PET image generation. Brain informatics. 2020;7:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liang H, Bao W, Shen X. Adaptive Weighting Feature Fusion Approach Based on Generative Adversarial Network for Hyperspectral Image Classification. Remote Sensing. 2021;13:198. [Google Scholar]
- 34.Wolf T, Chaumond J, Debut L, Sanh V, Delangue C, Moi A, et al. Transformers: State-of-the-art natural language processing. Proceedings of the 2020 Conference on Empirical Methods in Natural Language Processing: System Demonstrations. 2020:38–45. [Google Scholar]
- 35.Tang C, Zhao Y, Wang G, Luo C, Xie W, Zeng W. Sparse MLP for Image Recognition: Is Self-Attention Really Necessary? arXiv preprint arXiv:210905422. 2021.
- 36.Carion N, Massa F, Synnaeve G, Usunier N, Kirillov A, Zagoruyko S. End-to-end object detection with transformers. European conference on computer vision. 2020:213–29.
- 37.Touvron H, Cord M, Douze M, Massa F, Sablayrolles A, Jégou H. Training data-efficient image transformers & distillation through attention. International Conference on Machine Learning. 2021:10347–57. [Google Scholar]
- 38.Chen J, Lu Y, Yu Q, Luo X, Adeli E, Wang Y, et al. Transunet: Transformers make strong encoders for medical image segmentation. arXiv preprint arXiv:210204306. 2021.
- 39.Preetha CJ, Meredig H, Brugnara G, Mahmutoglu MA, Foltyn M, Isensee F, et al. Deep-learning-based synthesis of post-contrast T1-weighted MRI for tumour response assessment in neuro-oncology: a multicentre, retrospective cohort study. The Lancet Digital Health. 2021;3:e784–e94. [DOI] [PubMed] [Google Scholar]
- 40.Hore A, Ziou D. Image quality metrics: PSNR vs. SSIM. 20th international conference on pattern recognition. 2010:2366–9. [Google Scholar]
- 41.Sheikh HR, Bovik AC. Image information and visual quality. IEEE Transactions on image processing. 2006;15:430–44. [DOI] [PubMed] [Google Scholar]
- 42.Thie JA. Understanding the standardized uptake value, its methods, and implications for usage. Journal of Nuclear Medicine. 2004;45:1431–4. [PubMed] [Google Scholar]
- 43.Fletcher J, Kinahan P. PET/CT standardized uptake values (SUVs) in clinical practice and assessing response to therapy. NIH Public Access. 2010;31:496–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Deng J, Dong W, Socher R, Li L-J, Li K, Fei-Fei L. Imagenet: A large-scale hierarchical image database. 2009 IEEE conference on computer vision and pattern recognition. 2009:248–55. [Google Scholar]
- 45.Yuan L, Chen Y, Wang T, Yu W, Shi Y, Jiang Z, et al. Tokens-to-token vit: Training vision transformers from scratch on imagenet. arXiv preprint arXiv:210111986. 2021.
- 46.Yang Q, Tan K-H, Ahuja N. Real-time O (1) bilateral filtering. 2009 IEEE Conference on Computer Vision and Pattern Recognition. 2009:557–64. [Google Scholar]
- 47.Paris S, Durand F. A fast approximation of the bilateral filter using a signal processing approach. European conference on computer vision. 2006:568–80.
- 48.Luo Y, Wang Y, Zu C, Zhan B, Wu X, Zhou J, et al. 3D Transformer-GAN for High-Quality PET Reconstruction. International Conference on Medical Image Computing and Computer-Assisted Intervention: Springer; 2021. p. 276–85. [Google Scholar]
- 49.MD Dipl-math SG, Seith F, Schäfer JF, Christian la Fougère M, Nikolaou K, Schwenzer NF. Towards tracer dose reduction in PET studies: simulation of dose reduction by retrospective randomized undersampling of list-mode data. Hellenic Journal of Nuclear Medicine. 2016;19:15–8. [DOI] [PubMed] [Google Scholar]
- 50.Brenner DJ, Hall EJ. Computed tomography—an increasing source of radiation exposure. New England journal of medicine. 2007;357:2277–84. [DOI] [PubMed] [Google Scholar]
- 51.Chawla SC, Federman N, Zhang D, Nagata K, Nuthakki S, McNitt-Gray M, et al. Estimated cumulative radiation dose from PET/CT in children with malignancies: a 5-year retrospective review. Pediatric radiology. 2010;40:681–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.