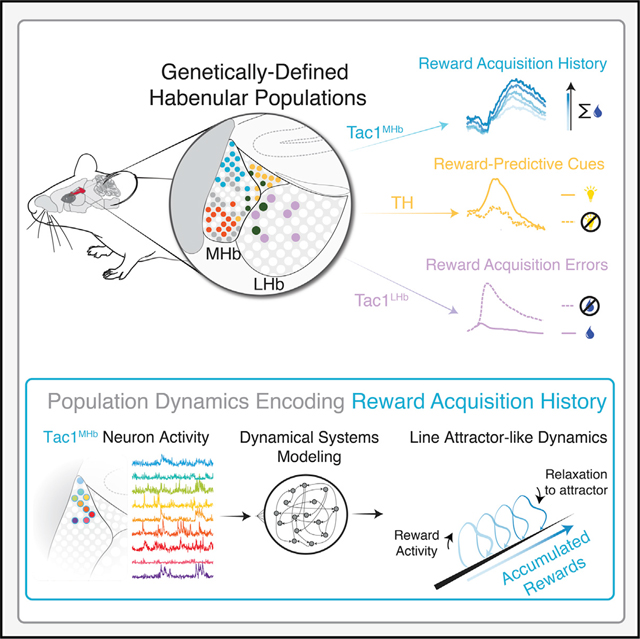
SUMMARY
Computational analysis of cellular activity has developed largely independently of modern transcriptomic cell typology, but integrating these approaches may be essential for full insight into cellular-level mechanisms underlying brain function and dysfunction. Applying this approach to the habenula (a structure with diverse, intermingled molecular, anatomical, and computational features), we identified encoding of reward-predictive cues and reward outcomes in distinct genetically defined neural populations, including TH+ cells and Tac1+ cells. Data from genetically targeted recordings were used to train an optimized nonlinear dynamical systems model and revealed activity dynamics consistent with a line attractor. High-density, cell-type-specific electrophysiological recordings and optogenetic perturbation provided supporting evidence for this model. Reverse-engineering predicted how Tac1+ cells might integrate reward history, which was complemented by in vivo experimentation. This integrated approach describes a process by which data-driven computational models of population activity can generate and frame actionable hypotheses for cell-type-specific investigation in biological systems.
In brief
Data-driven computational models of large-scale neural recordings predict celltype-specific probability dependence of reward response.
Graphical Abstract
INTRODUCTION
Across the animal kingdom, genetic targeting of cells within intermingled populations has enabled neuroscientists to identify specific cell types that can encode and drive precise brain states and behaviors (Adamantidis et al., 2007; Allen et al., 2019; Andalman et al., 2019; Buchholtz et al., 2020; Deisseroth, 2017; Füzesi et al., 2016; Jennings et al., 2019; Kim et al., 2013; Kohl et al., 2018; Krashes et al., 2014; Lovett-Barron et al., 2017, 2020; Sternson, 2020; Vesuna et al., 2020; Yu et al., 2009). Functional cell typology discoveries have generally not incorporated advances in neural population dynamics (in part because these analyses often use electrophysiology data, from which cell type identification is challenging to obtain). Yet understanding how molecularly defined populations are involved in brain and behavioral states may benefit from, or even require, integration of modern computational approaches.
Even in highly trained and well-performed behavior, cellular resolution brain activity exhibits substantial trial-to-trial variability that is often not well described by traditional concepts such as trial-averaged representations (Afshar et al., 2011; Pandarinath et al., 2018a; Yu et al., 2009; Mazzucato, 2022); similarly, under identical stimulus conditions, neural perturbations can elicit diverse neural and behavioral effects (Hallam, 1981; Pandarinath et al., 2018a; Remington et al., 2018; Shenoy et al., 2013; Sohn et al., 2019). Key conceptual advances have resulted from applying dynamical systems theory (e.g., Churchland et al., 2010; Shenoy et al., 2013); this approach, compared to the trial-averaged representational approach, may be of particular relevance for insight into neural and behavioral processes for which trial-to-trial variability itself has structure (for example, neural activity increasing as animals gather rewards during foraging in a time-varying environment).
To integrate genetic methods with computational methods leveraging population activity, we chose to study the transcriptionally diverse habenula, an epithalamic structure containing two main subdivisions with distinct gene expression, cytoarchitecture, and connectivity (Aizawa et al., 2012; Hikosaka, 2010; Namboodiri et al., 2016; Wagner et al., 2016). The lateral habenula (LHb) receives input from basal forebrain, prefrontal cortex, hypothalamus, and basal ganglia (Herkenham and Nauta, 1977; Hong and Hikosaka, 2008; Lazaridis et al., 2019; Stamatakis et al., 2013; Zhang et al., 2018) and sends projections to the ventral tegmental area (VTA/RMTg) and the dorsal raphe (Hong et al., 2011; Morales and Margolis, 2017; Quina et al., 2015). In contrast, the medial habenula (MHb) receives most of its input from the septum and nucleus of the diagonal band (Qin and Luo, 2009; Sutherland, 1982) and projects almost exclusively to the interpeduncular nucleus (IPN) (Qin and Luo, 2009; Quina et al., 2015). The relative simplicity of this anatomy belies its molecular diversity (Aizawa et al., 2012; Wagner et al., 2016; Díaz et al., 2011; Pandey et al., 2018). Classical habenula studies have focused on two mutually exclusive populations: dorsal tachykinin 1-expressing (Tac1, peptidergic) and ventral choline acetyltransferase-expressing (ChAT, cholinergic) neurons (Contestabile and Fonnum, 1983). Studies leveraging these genes have provided evidence for molecular-functional mapping, including the role of ChAT+ neurons in anxiety, nicotine sensitivity, and fear, as well as recent work identifying a role for Tac1+ neurons in spatial learning and motivated behavior (Cho et al., 2019; Seigneur et al., 2018; Yamaguchi et al., 2013; Pang et al., 2016; Frahm et al., 2011; Zhao-Shea et al., 2015; Hsu et al., 2014). Identification of additional cell type markers might further refine this mapping of cell type to function, particularly when extending the definition of function to the dynamical regime.
Here, we integrate spatially resolved transcript amplicon readout mapping (STARmap) (Kebschull et al., 2020; Wang et al., 2018) with genetically resolved optical and electrical recording to link specific cellular populations to behavioral elements of reward seeking. We show that habenular tyrosine hydroxylase-expressing (TH+) neurons learn and encode reward-predicting cues, LHb Tac1+ neurons encode negative reward outcomes, and MHb Tac1+ neurons integrate rewards with accumulation dynamics that are well described by a line attractor. Using an approach custom-modified for Ca2+ signals, we demonstrate nonlinear dynamical systems modeling of in silico behavioral sessions to computationally test alternative reward contingencies, and finally compare the model’s activity dynamics to experimentally measured dynamics in mice. Together, this approach illustrates the combination of spatial and genetic cell typology information with dynamical systems computational modeling for elucidating the functional significance of neural populations in behaving animals.
RESULTS
Molecular characterization of the habenula
We simultaneously assessed expression and spatial distribution of 15 genes (including neuromodulator receptors and neuropeptide-encoding genes) via tissue sequencing enabled by hydrogel-tissue chemistry (STARmap; Wang et al., 2018 Figures 1A–1C). Consistent with previous reports (Contestabile and Fonnum, 1983; Wagner et al., 2016), these data highlight the non-overlapping expression of Chat and Tac1 populations (Figures 1D and S1A). We identified six transcriptionally defined clusters (Figures 1E and 1F), which occupied different spatial subregions in the MHb (Figures 1G and S1B), in line with recent observations (Hashikawa et al., 2020). One cluster (Hb1) mapped onto dorsal MHb cells that expressed Tac1 at high levels. Two clusters (Hb2, Hb3) were enriched for Chat. Another cluster, Hb4, was enriched for Th, Calb1, Cartpt, and Htr5B, with cells spatially intermingled in the stria medullaris and Region X (HbX), a suggested region between the MHb and LHb (Seigneur et al., 2018; Wagner et al., 2016; Wallace et al., 2020). Cluster Hb5 was defined by high Gpr151 expression and the remaining cluster (Hb6) by Gad2+ neurons located in the LHb (Figure 1G), as described previously (Quina et al., 2020; Wallace et al., 2020).
Figure 1. Molecular and anatomical characterization of medial habenula cell types.
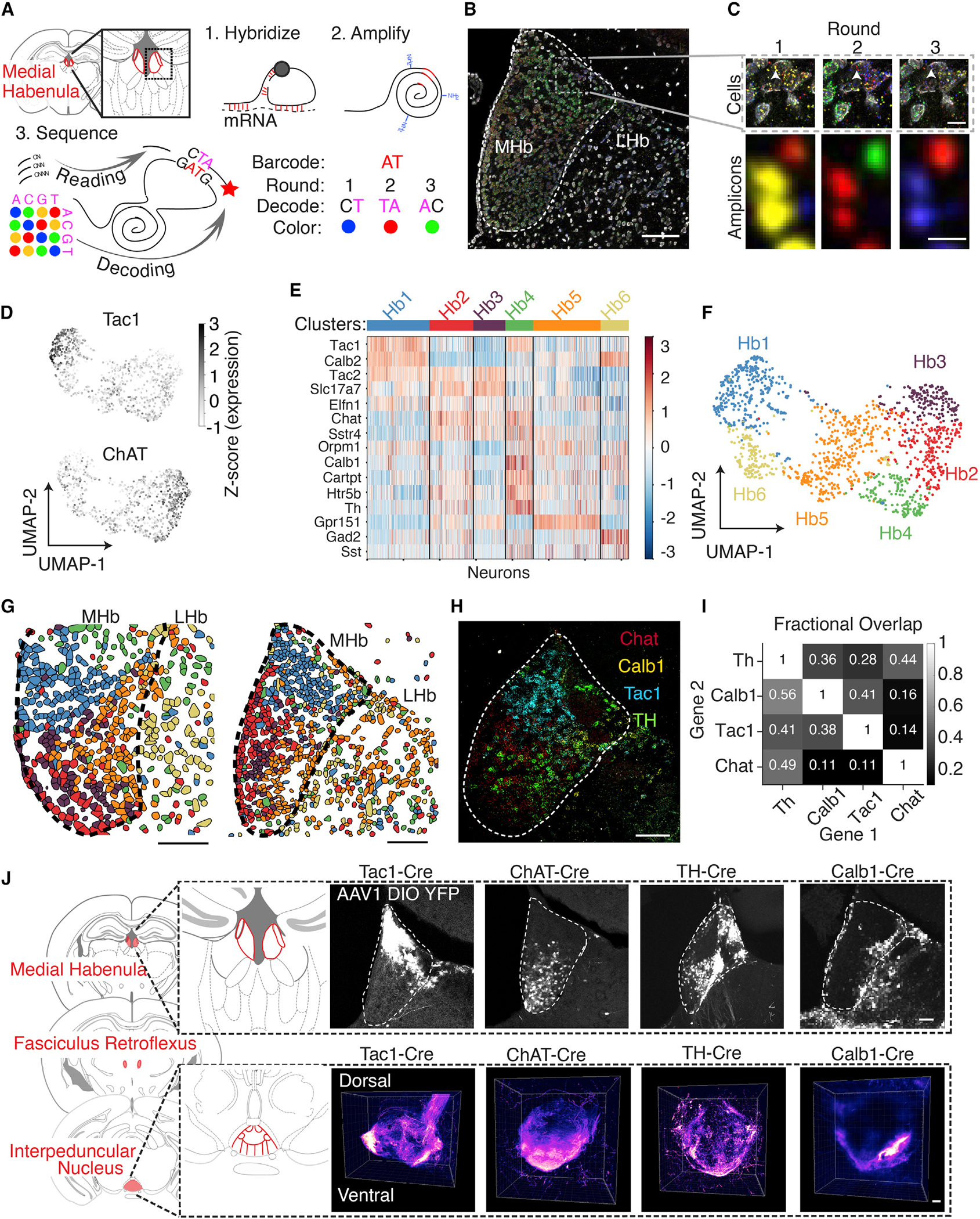
(A) Experimental design for 3 rounds of STARmap in situ sequencing of 15 genes in the habenula (Hb). Barcoded probes hybridize to mRNA targets and undergo rolling circle amplification. Sequential hybridization decodes each base on two adjacent rounds (STAR Methods).
(B) Deconvolved image from one round of in situ sequencing of Hb tissue. Dashed line indicates MHb boundaries. Box indicates ROI in (C). Scalebar: 100 μm.
(C) Top, magnified view of dotted box in (B) across 3 rounds of imaging. Scale bar: 10 μm. Bottom, magnified view indicated by arrow in top panels. Scale bar: 1 μm.
(D) Uniform manifold approximation projection (UMAP) of the expression of 15 genes for 1440 segmented Hb neurons from 2 biological replicates. Grayscale indicates the Z scored expression of Tac1 and Chat.
(E) Heatmap of expression levels of each gene (row) for each cell (column), color bar indicates Z score for each gene across all clusters.
(F) UMAP projection of all neurons. Color indicates cluster identity.
(G) Clusters identified in (F) are mapped onto the position of each cell in the Hb for two biological replicates. Scale bar: 100 μm.
(H) Quadruple in situ hybridization of Tyrosine Hydroxylase (Th), Tachykinin1 (Tac1), Choline Acetyltransferase (Chat), and Calbindin1 (Calb1) mRNA. Scale bar: 100 μm.
(I) Quantification of overlap in (H). Grayscale indicates the proportion of cells expressing Gene 1 that also express Gene 2. Fractional overlap listed inside each box. n = 3639 neurons.
(J) Left, coronal sections from mouse atlas showing the axonal projections from the medial Hb to the interpeduncular nucleus (IPN) (Konsman, 2001). Top right, neurons expressing AAV1-DIO-EYFP in X-Cre animals in the Hb, with α-GFP immunostaining. Bottom right, 3D rendering of YFP+ IPN axons of X-Cre:DIOYFP animals after tissue clearing (~3-mm-thick sections, pseudocolored for YFP intensity).
Scale bars: 100 μm.
See also Figure S1.
With the goal of linking molecular markers to functional features, we chose four candidate genes from the STARmap analysis (Tac1, Chat, Th, and Calb1), as these genes largely defined transcriptionally and spatially distinct neural populations, including dorsal MHb, ventral MHb, and HbX. Expression patterns were confirmed by quadruple in situ hybridization (Figures 1H and 1I and S1C and S1D), and injecting a virus driving Cre-dependent expression of yellow fluorescent protein (AAV-Ef1α-DIO-YFP) into transgenic Cre driver lines resulted in YFP expression consistent with the in situ data (Figure 1J, top row). Volumetric imaging of the YFP+ axon termination fields in the IPN revealed cell type-specific innervation patterns. Tac1+ axons terminated laterally, ChAT+ axons crisscrossed the midline (Video S1) (Ables et al., 2017; Hsu et al., 2013), and both TH+ and Calb1+ axons localized to the ventral IPN as a thin sheet (Figure 1J). Taken together, our STARmap sequencing screen identified genetically defined populations with distinct transcriptional, spatial, and axonal-projection properties.
Cell-type-specific neural dynamics in a visuospatial task
To directly compare the role of these cell types in behavior, we recorded population activity using fiber photometry from each MHb cell type and one pan-neuronal LHb population in the three-port serial reaction time task (3CSRTT), an operant task that involves learning, visuospatial attention, behavioral inhibition, motivation, and reward expectation (Figures 2A–2C and S2A–S2C) (Bari et al., 2008; Kim et al., 2016). Briefly, animals learned to nose poke to a brief visual cue in one of three ports; upon a correct response, a reward light was illuminated on the opposite wall with 4% sucrose in water delivered below. As mice progressed in training, they developed an attentive scanning behavior (Video S2) and premature pokes shifted to the end of the delay period, indicating cue light anticipation (Figure S2D). We injected AAV1-Ef1α-DIO-GCaMP6f into the MHb of Cre-expressing transgenic mice, or AAVdj-hSyn-GCaMP6m into the LHb of non-transgenic C57BL/6 mice, and recorded photometry signals during the 3CSRTT (Figures 2D–2K and S2E and S2F). Trial-aligned photometry showed distinctly structured activity patterns (Figure 2E). To determine if particular cell types showed activity dynamics correlated with attentional state (cue), behavioral inhibition (nose poke), or rewarded outcomes (reward port entry), we aligned photometry signals to each behavioral epoch and separated neural traces by trial outcome (Figures 2F–2H).
Figure 2. Habenular cell types show distinct reward-related activity.
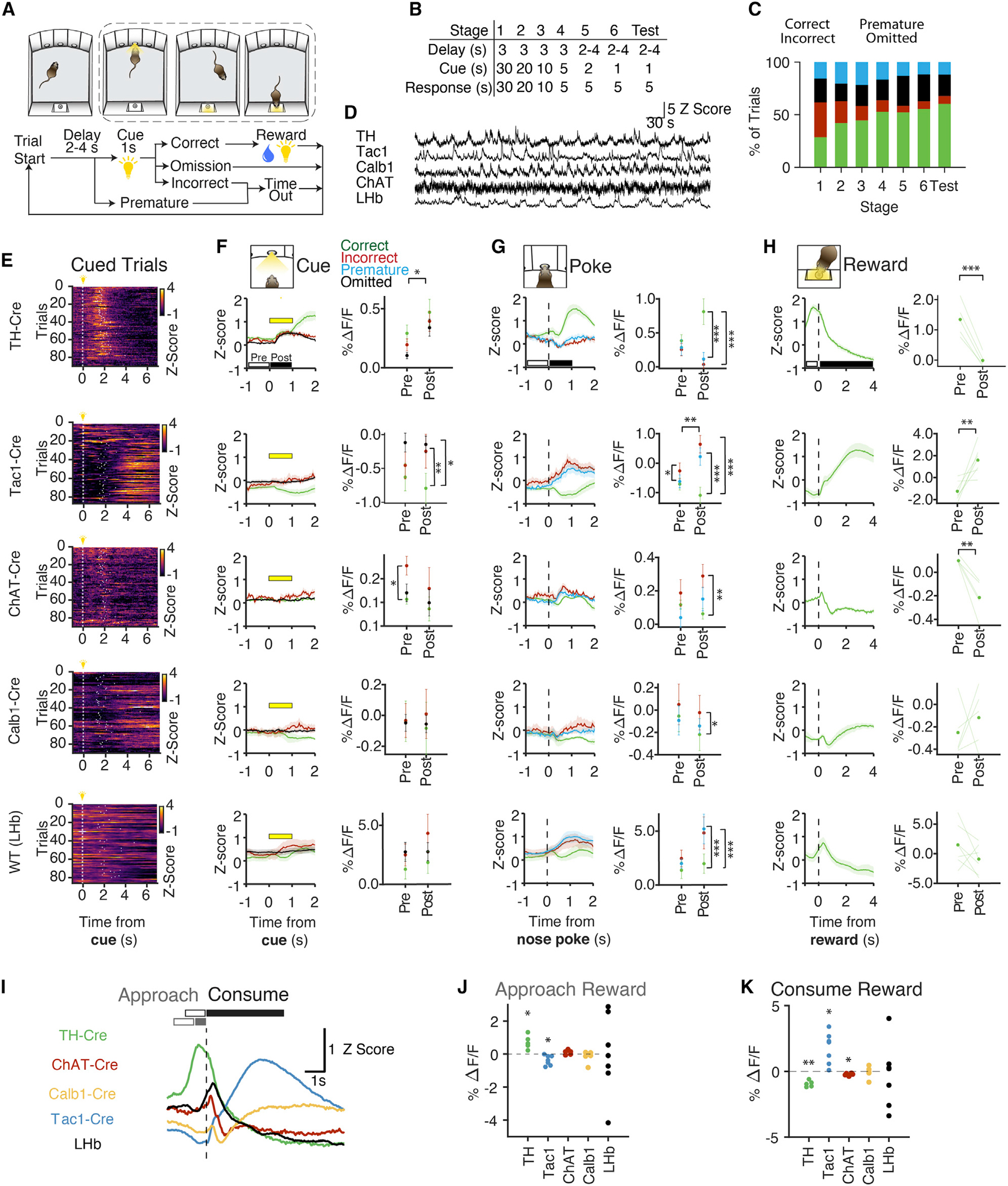
(A) Trial structure of 3-Choice Serial Reaction Time Task. After a variable delay, a cue light appears in one of three nose pokes for 1 s. Nose pokes into the lit port result in delivery of sucrose water at the reward port on the opposite wall. Premature and incorrect trials result in a 5 s time out.
(B) Training consists of 6 stages of progressively shorter cue durations and the introduction of a variable delay.
(C) Behavioral performance across training. Percentage of correct (green), incorrect (red), omitted (black), and premature responses (blue) for all animals (n = 29 mice).
(D) Example traces from photometry recording during behavior, Z scored across each session.
(E) Example photometry recordings from one behavioral session per genotype. Each row represents a single trial where t = 0 is cue onset. Color: Z scored fluorescence. Data from premature and omitted trials are not displayed.
(F–H) Left panels, photometry time series normalized to 405 nm control, Z scored across each session, and aligned to cue onset, nose poke, or reward port entry. Data is separated by behavioral outcome: correct (green), incorrect (red), omitted (black), and premature (blue). Error bars indicate SEM. Right panels, %ΔF/F calculated before and after the cue (F) or nose poke (G). Two-way ANOVA with repeated measures correction. Cue effect: Th+, p < 0.05. Nose poke effect: Tac1+, p <0 .01. Trial outcome vs cue: Tac1+, p < 0.05. Trial outcome versus nose poke: Th+, p <0 .01; Tac1+, p <0 .001; ChAT+, p < 0.05. See Table S1 for multiple comparisons. For reward (H), Th+, p <0 .001, Tac1+, p <0 .01, ChAT+, p <0 .01 by paired t test.
(I) Summary of mean Z scores for each genotype in (H), aligned to reward port entry.
(J and K) Quantification of the change in GCaMP fluorescence at reward approach or reward consumption with a one-sample t test with FDR correction. For reward approach: Th+, p < 0.05 and Tac1+, p < 0.05. For reward consumption, Th+, p < 0.01, Tac1+, p < 0.05, ChAT+, p < 0.05. In all panels, black bars indicate the time periods analyzed before (open) and after (filled) behavioral events. Data represents average across mice: Th-Cre, n = 5 mice; Tac1-Cre, n = 7 mice, ChAT-Cre, n = 5 mice, Calb1-Cre, n = 5 animals; LHb, n = 7 animals.
Error bars: SEM.
The 3CSRTT requires the animal to attend to the cue panel in anticipation of the cue light, allowing this paradigm to be used to assess visuospatial attention (Robbins, 2002). Only ChAT neurons showed pre-cue activity predictive of trial outcome, suggesting a possible role in the attentional component of the task (Figure 2F). Cue illumination significantly increased activity in TH+ neurons for all trial types, indicating that TH neurons may encode salient cues in the environment. We next aligned the data to nose pokes to compare activity before pokes on different trial outcomes (premature pokes are a suggested metric for impulsivity and are altered in MHb-lesioned mice (Kobayashi et al., 2013; Sanchez-Roige et al., 2011). We found only a small difference in correct versus incorrect trials for Tac1+ neurons (Figure 2G). In contrast, the post-nose poke period revealed trial outcome-dependent activity in TH+, Tac1+, ChAT+, and LHb populations, with prominent differences between rewarded (correct) versus unrewarded (incorrect or premature) trials. When we aligned data to the reward port entry, TH+, Tac1+, and ChAT+ neural signals were all significantly modulated by reward, but with distinct dynamics (Figure 2H). TH+ activity peaked as mice approached the reward port and then diminished quickly. Tac1+ activity gradually increased during reward consumption whereas ChAT+ activity decreased during this time period. Calb1+ and LHb populations were not significantly modulated by reward.
Separating reward approach from reward consumption, we found that TH+ neurons were active during reward approach, whereas Tac1+ neurons were active during reward consumption (Figures 2I–2K). To rule out movement-related signals, we compared these responses to other (unrewarded) head entries into the reward port. Tac1+ neurons increased activity during both rewarded and unrewarded head entries, but with different temporal dynamics (Figures S2G and S2H); unrewarded head entries triggered a subsecond-onset/offset signal whereas rewarded head entries elicited slower dynamics (τon = 0.22 s unrewarded versus 2.29 s rewarded, τoff = 1.32 s unrewarded versus 2.26 s rewarded). In other cell types, there was no activity change to unrewarded entries. For ChAT+ signals, there was little modulation in the 3CSRTT overall, but we observed behaviorally correlated activity in the same cohort of animals while in the elevated plus maze, consistent with a role in anxiety (McLaughlin et al., 2017; Figures S2I and S2J). Together, these data revealed diverse functional properties of transcriptionally defined neural populations in the habenula.
Habenular cell types respond to reward in a learned and expectation-based manner
We next sought to better understand the contingencies of these reward-related signals. We introduced variants of the task in which the reward-predictive cue (reward port light) and reward (sucrose) were unlinked in some trials (Figures 3A–3G), or in which we varied reward size (Figures S3A–S3D). We found that mice entered the reward port more quickly when the port light was on, indicating learned association between the port light and reward. Photometry signals from TH+ neurons during the reward period were not significantly different between rewarded and unrewarded trials but were significantly higher when the reward-port light was on (Figure 3C). Video tracking of head angle showed that TH+ signals were time-locked to orienting movements toward the reward port (Figures S3E and S3F), but reward-independent changes in luminance (e.g., house light) did not alter TH+ activity (Figure S3G). To rule out activity driven by the preceding nose poke, we delivered free sucrose rewards to 3CSRTT-trained mice outside of the trial structure (Figures S3H–S3J). TH+ signals increased only when free rewards were cued by a reward port light (Figure S3H), and doubling the reward size had no effect on TH+ activity (Figure S3B).
Figure 3. Cell-type-specific segregation of cue-related and outcome-related reward activity.
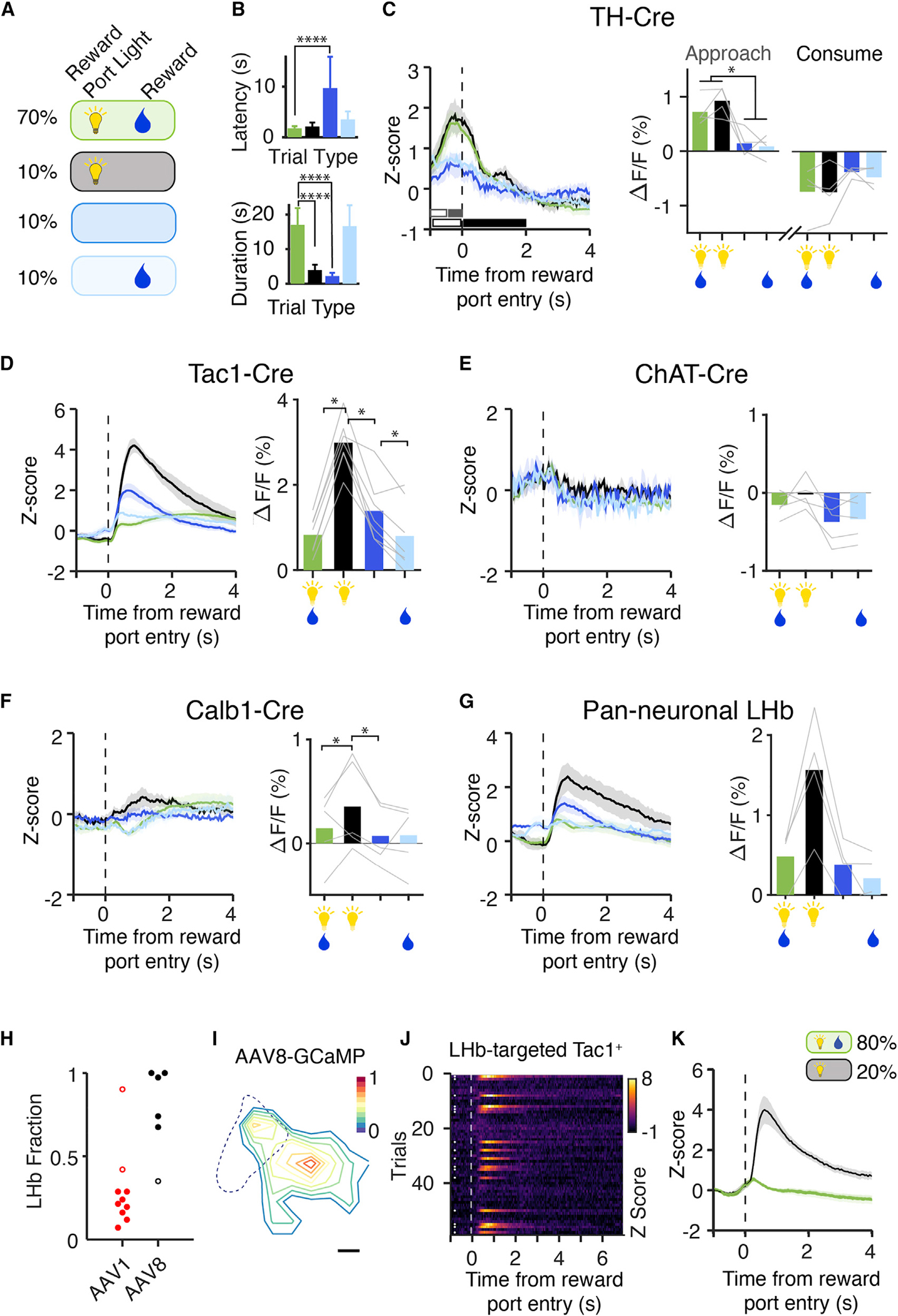
(A) A variant of the task where reward-predicting cues and reward probabilities were modified. In 70% of correct trials, rewards were cued and delivered (green). In the remaining correct trials, the reward was not cued (light blue), the reward was not delivered (black, 10%), or the reward was neither cued nor delivered (navy, 10%).
(B) Latency to retrieve reward on correct trials, and the duration in the reward port consuming the reward. Two-way ANOVA with repeated measures, corrected for FRD. For reward latency: cue effect, p < 0.01, reward effect, p < 0.01, Interaction, p < 0.01. For reward delivery: reward effect, p < 0.0001. n = 25 animals. Error bars: SEM.
(C–G) Reward-related activity in Hb cell types to predictive cues and reward delivery. Left panels, mean Z scored photometry data aligned to reward port entry. Black and gray bars indicate pre- (open bar) and post- (closed bar) comparison. Color indicates trial type in (A). Right panels, %ΔF/F at reward approach and consumption (C) or consumption (D–G). Gray lines represent individual animals, bars indicate the mean for each reward contingency: TH-Cre, n = 4 mice; Tac1-Cre, n = 7 mice (MHb-Targeted, 82% of neurons in the MHb); ChAT-Cre, n = 5 mice; Calb1-Cre, n = 5 animals; LHb, n = 4 animals. One-way ANOVA with repeated measures and FDR correction, *p < 0.05.
(H) Serotype tropism for LHb neurons. The ratio of GCaMP+ LHb neurons to all GCaMP+ neurons (see STAR Methods) was calculated for AAV1 and AAV8 injections. AAV1, n = 10 animals; AAV8, n = 5 animals. For photometry experiments in Figures 2 and 3, animals with confirmed fiber placement and >70% MHb neurons were included in the analysis in order to assess the activity of MHb Tac1 neurons (the majority of Tac1 neurons in the Hb). Animals excluded from analysis are indicated by open circles.
(I) Spatial distribution of LHb-targeted Tac1+ neurons. All GCaMP+ neurons in the LHb were counted and registered to a common coordinate system. Contour lines: the deciles of normalized cell density.
(J) Tac1+ neurons in the LHb were targeted for fiber photometry recording using Tac1-Cre mice (88% of neurons in the LHb) and lateral injection of AAV8-DIO-GCaMP6f. Example from one animal in a session where 20% of rewards were withheld, showing all correct trials aligned to the reward port entry. White dots: withheld trials.
(K) Mean Z score for LHb-targeted Tac1+ neurons at reward port entry for rewarded (green) and withheld (black) trials. n = 7 animals.
See also Figure S3.
These data suggested that the TH+ population can represent a learned reward-predicting cue. To test this hypothesis, we performed a reversal paradigm in trained mice in which the reward-port light was paired with incorrect (unrewarded) trials instead of correct (rewarded) trials. In this paradigm, the cue light was still a salient stimulus, but no longer associated with a reward. We found that the TH+ light response decreased across five reversal sessions (Figures S3K–S3M). Together, these experiments strongly suggested that the TH+ population can signal a learned encoding of reward-predicting cues. In contrast, in the withheld reward task, Tac1+ neurons demonstrated rapid and robust activity on unrewarded trials, particularly those with a reward cue (Figure 3D, black), while ChAT+ activity showed no dependence on these reward contingencies, and Calb1+ neurons showed only modest differences among trial conditions (Figures 3E and 3F). In summary, we found that TH+ and Tac1+ neurons signaled distinct task-related features of reward-guided behavior: TH+ neurons encoded learned reward-predicting cues and Tac1+ dynamics were linked to reward outcome.
Opposing Tac1+ signals resolved by topography
Tac1+ photometry signals increased in both rewarded and unrewarded trials. Why would they signal both acquired and missed rewards? The differences in the temporal dynamics of the photometry signal suggested that they might originate from two distinct Tac1+ populations. We had targeted our recordings to the MHb, but the LHb also contains Tac1+ neurons. Based on previous work, we next hypothesized that LHb Tac1+ neurons signaled negative outcomes, whereas MHb Tac1+ cells signaled acquired rewards (Figure 3G) (Bromberg-Martin and Hikosaka, 2011; Hsu et al., 2014, 2016; Matsumoto and Hikosaka, 2007).
We implemented a viral tropism targeting strategy to record more specifically from LHb Tac1+ neurons (hereafter Tac1LHb; Figures 3H, 3I and S2F). We found that Tac1LHb neurons responded reliably to withheld rewards (Figures 3J and 3K), with dynamics matching those in our previous recordings (Figure 3D). Conversely, Tac1LHb recordings did not recapitulate the reward-associated activity seen in the MHb-enriched Tac1+ population (Figures 3J and 3K). Together, these data strongly suggested that the opposing valence signals arise from anatomically distinct Tac1+ populations.
Tac1+ MHb cells exhibit long-timescale activity dynamics
The trial-averaged data from habenular cell types had revealed interesting differences in reward responses, but we also noticed longer timescale dynamics across behavioral sessions: rewards in early trials triggered little activity in Tac1+ neurons, whereas later rewards showed more reliable and robust responses (Figure 4A). In addition, Tac1+ reward responses were higher if the previous trial had been rewarded (Figures S4A–S4E), together suggesting a sensitivity to reward history. Tac1LHb, TH+, and ChAT+ populations lacked this ramping activity (Figures 4B and 4C). This pattern was of interest because it suggested that Tac1MHb populations might be important for integrating past reward outcomes and updating behavioral strategies. Testing the causal role of these neurons in reward-guided decision-making required more specific viral targeting of Tac1MHb neurons to yield an interpretable result—particularly because Tac1MHb and Tac1LHb neurons encode opposing valence events. To this end, we developed a method leveraging viral tropism and INTRSECT gene targeting (Fenno et al., 2014, 2020) to express the inhibitory opsin eNpHR3.0 in Tac1MHb. We trained Tac1MHb-eNpHR+ mice on a head-fixed, reward-guided decision-making task (Figures 4D–4G) in which mice were presented with two lick spouts: one with a high reward probability (90%) and one with a low reward probability (10%). The spout with the high reward port was alternated every 15–20 rewarded trials. Block switches were not signaled; thus, the task required mice to sample both lick spouts and integrate information about recent reward history to optimize reward seeking (Figure 4H).
Figure 4. Long-timescale activity dynamics and behavioral significance of habenular cell types.
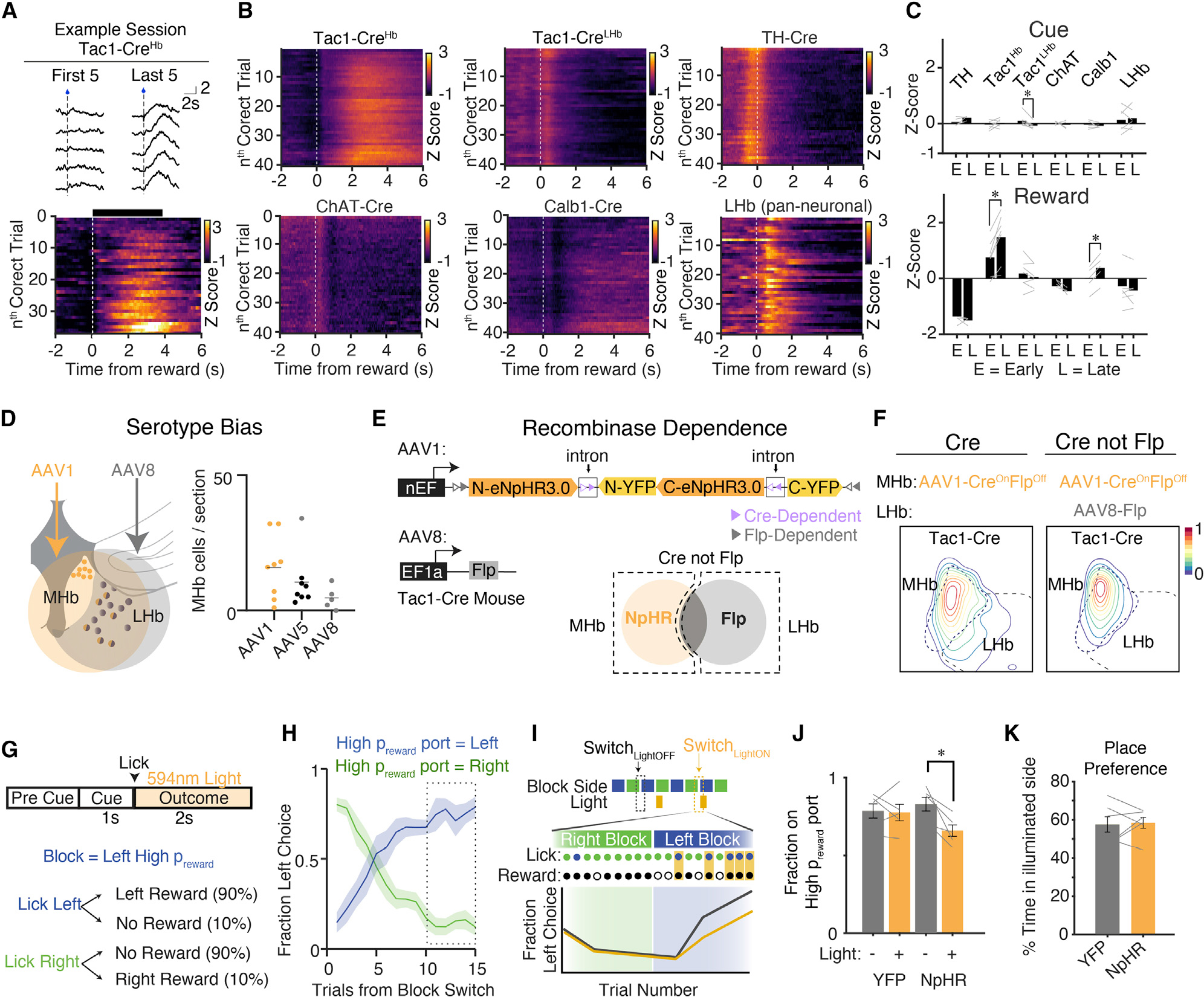
(A) Top, example traces of photometry signal for a Tac1-Cre mouse for the first 5 rewards and last 5 rewards of a behavioral session. Dotted line indicates reward delivery. Bottom, reward responses for correct trials in an example session from one mouse, sorted by nth correct trial.
(B) Average reward response for each genotype, Z scored across each session and sorted by nth correct trial. Only rewarded trials are displayed.
(C) Quantification of fluorescence changes over a behavioral session 1 s after cue onset and 4 s after head entry into the reward port. To look at changes across the session, the mean Z score of the first 5 correct trials were compared to 5 late trials (correct trials #36–40). *p < 0.05 by paired t test. Error bars indicate SEM All p values are FDR-corrected. Tac1Hb animals include AAV1 injections with fiber placements in the MHb (82% of GCaMP+ neurons in the MHb). Tac1LHb animals include AAV8 injections with fiber placements in the LHb (88% GCaMP+ neurons in the MHb). Gray lines: individual animals.
(D) Bias for AAV1-YFP, AAV5-YFP, and AAV8-YFP to infect MHb neurons. Injection locations at MHb/LHb boundary were verified with simultaneous fluorosphere injection. Each dot represents one animal.
(E) Intersectional strategy to target MHb Tac1+ neurons for optogenetic silencing. AAV1 CreOnFlpOff eNpHR3.0 injected in the MHb to turn on expression of eNpHR3.0 in all Tac1 Hb neurons. AAV8-Flp preferentially infects LHb neurons to turn off eNpHR3.0 expression laterally.
(F) Distribution of virally infected neurons in AAV1 CreONFlpOff alone versus AAV1 CreONFlpOff plus AAV8-Flp. Contour lines represent the deciles of normalized cell density. n = 5 animals/condition.
(G) Behavioral paradigm for head-fixed reward-guided decision-making task. Animals are presented with two lick spouts. In a block trial structure, one spout hasa high probability of reward (0.9) and the other a low probability of reward (0.1). After a pre-cue period in which premature licks terminate the trial, a 1 s cue light is illuminated. During this time, licks result in delivery of a water reward according to the reward contingencies defined for that block. Optogenetic inhibition is restricted to rewarded trials on the high probability lick spout.
(H) Fraction of left side choices on right-to-left block switches (blue) and left-to-right block switches (green). Error bars indicate SEM. Dotted box: trials quantified in (J).
(I) Schematic of stimulation paradigm at block switches and prediction of the behavioral response. On a subset of block switches, rewarded licks on trials 0–15 after the block switch trigger 2 s of 594 nm light to activate eNpHR3.0.
(J) Mean fraction of high probability port choices for each animal across trials 10–15 after the block switch for control and light inhibited trials (indicated by dotted box in [H]). n = 5 animals.
(K) Time spent in yellow light stimulated side of a two-chamber real time place preference assay. n = 6 animals.
See also Figure S4.
We delivered precisely timed optogenetic inhibition for 2 s after the first lick on rewarded trials, coinciding with reward-elicited Tac1MHb activity as seen in photometry experiments (Figure 4I). Silencing during this time period degraded adaptive transitions and behavioral performance (Figures 4 and S4F–S4I), though no changes were observed in a real-time place preference assay, suggesting the effect was not due to an aversion related to Tac1MHb inhibition (Figure 4K). Thus, Tac1MHb neuron activity exhibits a functional role in guiding future explore/exploit decisions.
Individual Tac1+ MHb cells ramp up as rewards are gathered
To investigate these reward signals at single-cell resolution, we used endoscopic two-photon Ca2+ imaging of Tac1+ or TH+ MHb neurons expressing H2B-GCaMP6f. We adapted a simple head-fixed cue-reward association task (Figure 5A), in which a visual cue signaled availability of water reward, which mice obtained by licking a spout (rewarded). In 15% of trials, no reward was delivered (unrewarded). Trial-averaged activity traces revealed that ~40% of Tac1+ cells and ~25% of TH+ cells were task modulated (Figures 5C and 5D). Averaging the activity of individual neurons revealed cell-type-specific long-timescale activity dynamics as in the photometry recordings (Figures 5C and 4B). In particular, the population-averaged activity of Tac1+ (but not TH+) neurons displayed the striking ramp-up across trials (Figures 5E–5H), and individual Tac1+ neurons also showed clear ramping across trials (Figure S5A).
Figure 5. Cell-type-specific imaging and electrophysiology reveals distinct long-timescale dynamics at single-neuron and population level.
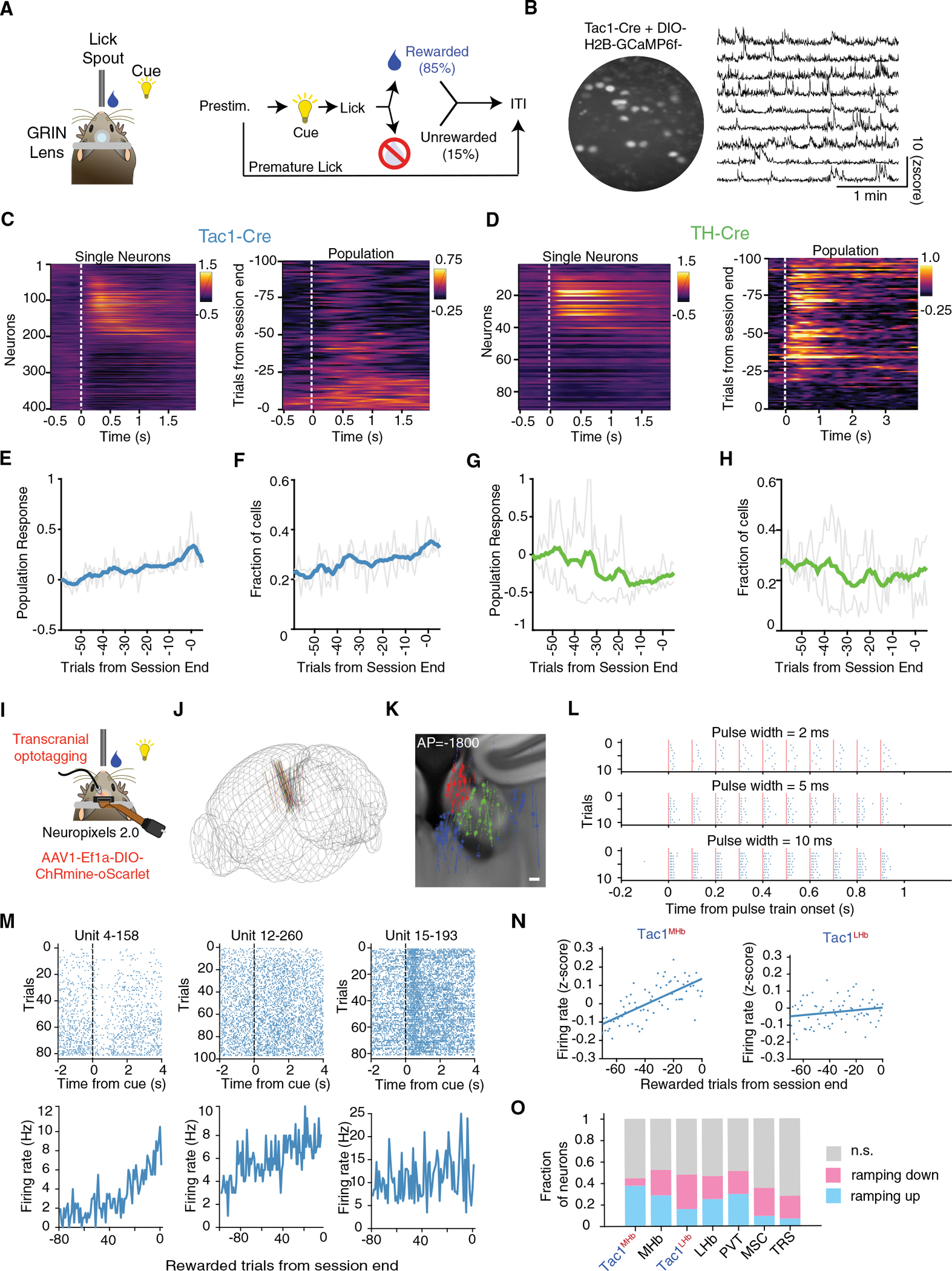
(A) A simplified reward task for head-fixed calcium imaging and electrophysiology. During a 0.5 s prestimulus period, animals must withhold licking. During a 1 s cue period, licks are rewarded with sucrose water delivery. In 15% of trials, earned rewards are withheld. Trials are separated by a variable ITI.
(B) Example 2p image of Tac1MHb neurons expressing H2B-GCaMP6f, imaged through a 600 μm GRIN lens. Right, example traces from Tac1MHb neurons.
(C–D) Heatmap showing (left) Z scored, trial-averaged activity from single Tac1MHb (C) or TH+ (D) neurons on rewarded trials. Neurons from 2 mice are included in each panel. (right) Heat maps from one example animal showing population activity (sum of fluorescence across all neurons) for each trial in one session (right). (E and G) Activity of reward-responsive neurons during the reward period over a behavioral session in Tac1MHb and TH+ neurons, respectively. Data normalized to the first trial shown. Mean (bold line) and individual mice (gray lines). Across the session, Tac1MHb populations showed stereotyped and statistically significant increasing population activity (p = 1.7×10−22) and TH+ populations showed more variable and slightly decreasing population activity (p = 3.6×10−5); p values for the null hypothesis that the slope of linear regression is zero.
(F and H) The proportion of reward-responsive cells over the session in Tac1MHb and TH+ mice, respectively. A responding cell is defined as a cell with a Z score greater than 0.25 for the 1 s reward period. Across the session, Tac1MHb neurons show a statistically significant increase (p = 6.4×10−15) and TH+ neurons show a statistically significant decrease (p = 5.8×10−4) in the fraction of active neurons.
(I) Experimental configuration for Neuropixels 2.0 recording. A 4-shank probe was approached at 10 from the midline. A 637 nm laser was illuminated above the skull.
(J) Summary of Neuropixels probe insertions targeting MHb. n = 7 animals, 18 behavioral sessions.
(K) Spatial position of recorded single neurons registered to the Allen Brain Atlas. Red, MHb; green, LHb; blue, others; *, optotagged. See Figure S5C for more complete visualization. n = 6099 Hb neurons, including 29 optotagged Tac1MHb and 25 optotagged Tac1LHb.
(L) Spike raster plot for an example optotagged Tac1MHb neuron at 2, 5, 10 ms pulsewidth.
(M) Spike raster plot and firing rate across trials, for three example optotagged Tac1MHb neurons.
(N) Left, population-averaged baseline firing rate across trials for Tac1MHb neurons (p = 4.6 × 10−16); right, baseline firing rate across trials for Tac1LHb neurons (p = 0.10).
(O) Fraction of neurons in each brain area showing significant ramping up or ramping down across a behavioral session. See STAR Methods for statistical criteria for classifying ramping characteristics.
See also Figures S4 and S5.
We complemented these single-cell findings with electrophysiological recordings in the same task. We used the 4-shank Neuropixels 2.0 probes (Steinmetz et al., 2021) and established a reliable workflow to access MHb neurons (Figures 5I–5K). We conferred cell type specificity through deep-brain transcranial optotagging with the opsin ChRmine (Chen et al., 2021; Marshel et al., 2019, Kishi et al., 2022) (Figure 5L). Our recordings spanned the habenular area and the septum, a major input to MHb (Figures S5B–S5E). We again observed ramping in single neurons and populations of the Tac1MHb, but not Tac1LHb, cell type (Figures 5M and N). All regions had mixtures of ramping-up and ramping-down neurons; however, in the Tac1MHb population, the ramping-up population greatly outnumbered the ramping-down population (Figure 5O).
Data-driven dynamical systems modeling: Tac1MHb line-attractor dynamics integrate reward history
To leverage and learn from our cellular-resolution and cell-type-specific experimental datasets (Figure 5), we developed a Ca2+ imaging-compatible data-driven dynamical systems approach for single trials based on latent factor analysis via dynamical systems (LFADS), a computational technique for inferring single-trial neural population dynamics (Pandarinath et al., 2018a), which we term type-resolved LFADS (trLFADS). Though trLFADS is applicable to both cell-type-specific electrophysiology and Ca2+ imaging datasets in principle, here we focus on imaging to harness the largest number of relevant simultaneously recorded neurons, and in so doing, enable LFADS for cell-type-specific activity data for the first time. In essence, trLFADS trains a recurrent neural network (RNN) that regenerates experimentally observed single-trial neural population activity (Figure 6A) by approximating the underlying neural dynamical system . If and represent population state and external input, respectively, where time is , then trLFADS learns a model consistent with the data. The state is a point in a high-dimensional state space of dimensionality set by the relevant neural latent factors, whose projection corresponds to the activity of individual neurons (Vyas et al., 2020; Pandarinath et al., 2018b). Thus, a single trial can be represented in the neural state space as a trajectory with time evolution perturbed by the external input (, which is inferred by the model). Here, we modified the network architecture and cost function from previous work (which could only handle spike counts) to enable direct processing of Ca2+ transients (STAR Methods). Applying trLFADS to imaging data enabled access both to denoised single-trial neural trajectories and dynamical systems models that generate trajectories.
Figure 6. Data-driven modeling of cellular-resolution population activity identifies cell-type-specific attractor dynamics.
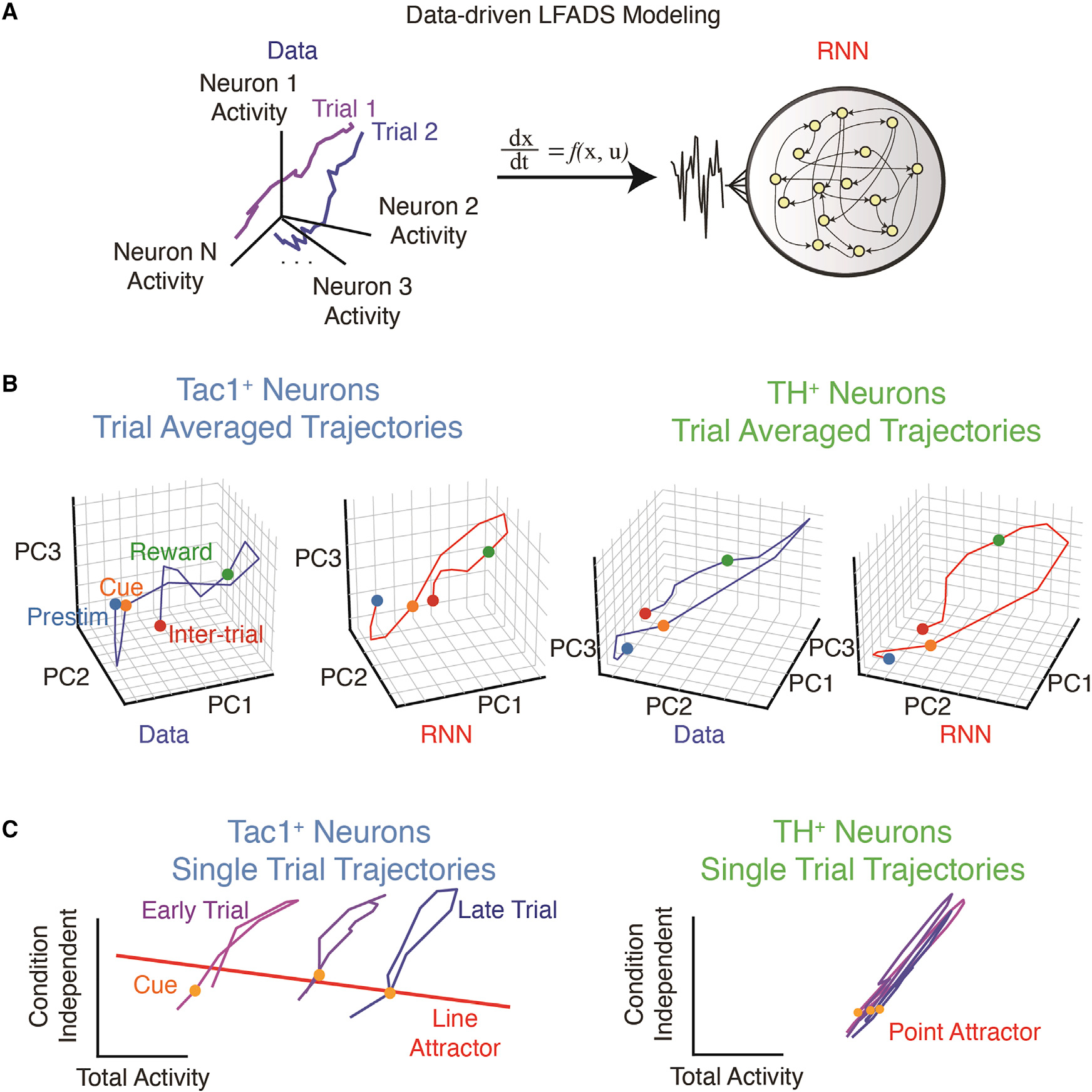
(A) LFADS modeling of neural population activity. The dynamics of Tac1MHb or TH+ neurons measured by two-photon microscopy can be described as trajectories in the neural state space (left). These trajectories can be generated by an RNN, which approximates the underlying neural dynamical system, at the single-trial level (right).
(B) The trial-averaged neural trajectory of Tac1MHb (left) and TH+ (right) neurons in the raw data and the RNN, demonstrating consistent epoch-dependent dynamics demarcated by the dots.
(C) Single-trial trajectories and the underlying attractor manifolds identified by fixed point analysis of the trLFADS generator RNN. In the Tac1MHb neurons (left), the line attractor integrates the external inputs over time, resulting in the trajectories shift along the line attractor as the session progress. Note the alignment of the line attractor and the total activity mode for Tac1MHb neurons, implying the progressive increase of total activity. The straight colored line is the top principal component of the identified fixed points. In the TH+ neurons (right), the discrete point attractor confines the trajectories, resulting in no change in total activity over time. The orange dots represent the cue onsets.
See also Figure S6.
We trained separate trLFADS models for the separately recorded Tac1MHb and TH+ populations. While the trial-averaged trajectories were characterized by a major loop time-locked to cue onset for both Tac1MHb and TH+ populations (Figure 6B), denoised single-trial trajectories revealed cell-type-specific structure across the session—a progressive shift across trials in Tac1MHb, but not TH+, models (Figures 6C and S6A and S6B). Here, population states were shown in two targeted orthogonal dimensions relevant to behavior (which also explained majority of the variance; Figure S6C), with the horizontal axis corresponding to population-averaged total neuronal activity. Within single trials, both Tac1MHb and TH+ models revealed a tilted loop trajectory, consistent with the transient increase of total activity after cue onset. Across trials, we found a horizontal procession corresponding to an overall increase in activity, which was specific to the Tac1MHb models (Figure 6C).
To study the dynamical structures governing these trajectories, we next reverse-engineered the learned dynamical systems (generator RNNs in trLFADS) using fixed point analysis (Sussillo and Barak, 2013; STAR Methods). In Tac1MHb models, we identified a continuous line attractor with a large total activity mode projection (Figures 6C and S6A). In contrast, TH+ model dynamics were governed by a discrete point attractor (Figures 6C and S6B). Neural attractors represent stable population activation patterns and thus their geometric arrangements provide insights into the logic of the neural dynamics storing information over long timescales. A line attractor integrates external inputs and stores this value as a state, remembering the sum of external stimuli (Mante et al., 2013; Seung, 1996)—as might be expected for the representation of reward accumulation over time.
We next explored the types of external inputs integrated by the Tac1MHb dynamics. We resolved single-trial dynamics into two parts: internal states and external inputs. We quantified condition-averaged inferred external inputs and the corresponding initial state shifts, finding distinct effects between the rewarded and unrewarded trial types (Figures S6D–S6F). Single-trial initial states (i.e., the states before the cue onset) shifted along the line attractor consistent with integration of trial-type-dependent external inputs (i.e., the activity of neural populations upstream to the experimentally observed population)—thus explicitly representing the animal’s reward history, consistent with our findings from the optogenetic reward-guided decision-making experiments (Figure 4).
Transient optogenetic perturbation and recovery
The identification of line attractor dynamics in the Tac1MHb dynamical systems models generated a series of experimentally testable theoretical predictions. For internal states, we considered that a transiently perturbed neural internal state would be predicted to smoothly and steadily return to the attractor; importantly, this transition would not be instantaneous as it would likely involve multiple cycles of synaptic communication typical of neural attractors. A second prediction was that the projection of external inputs onto the “selection vector” of the line attractor ought to determine the extent of neural state update or integration (Mante et al., 2013); if we pushed the population state in a random direction (nearly orthogonal to a fixed selection vector in high-dimensional neural state space), this transient perturbation would result in nearly zero integration along the line attractor, unlike the well-aligned “reward delivery” input described above (Figure 7A).
Figure 7. Transient optogenetic perturbation and reward history modulation experiments support the line attractor dynamics model.
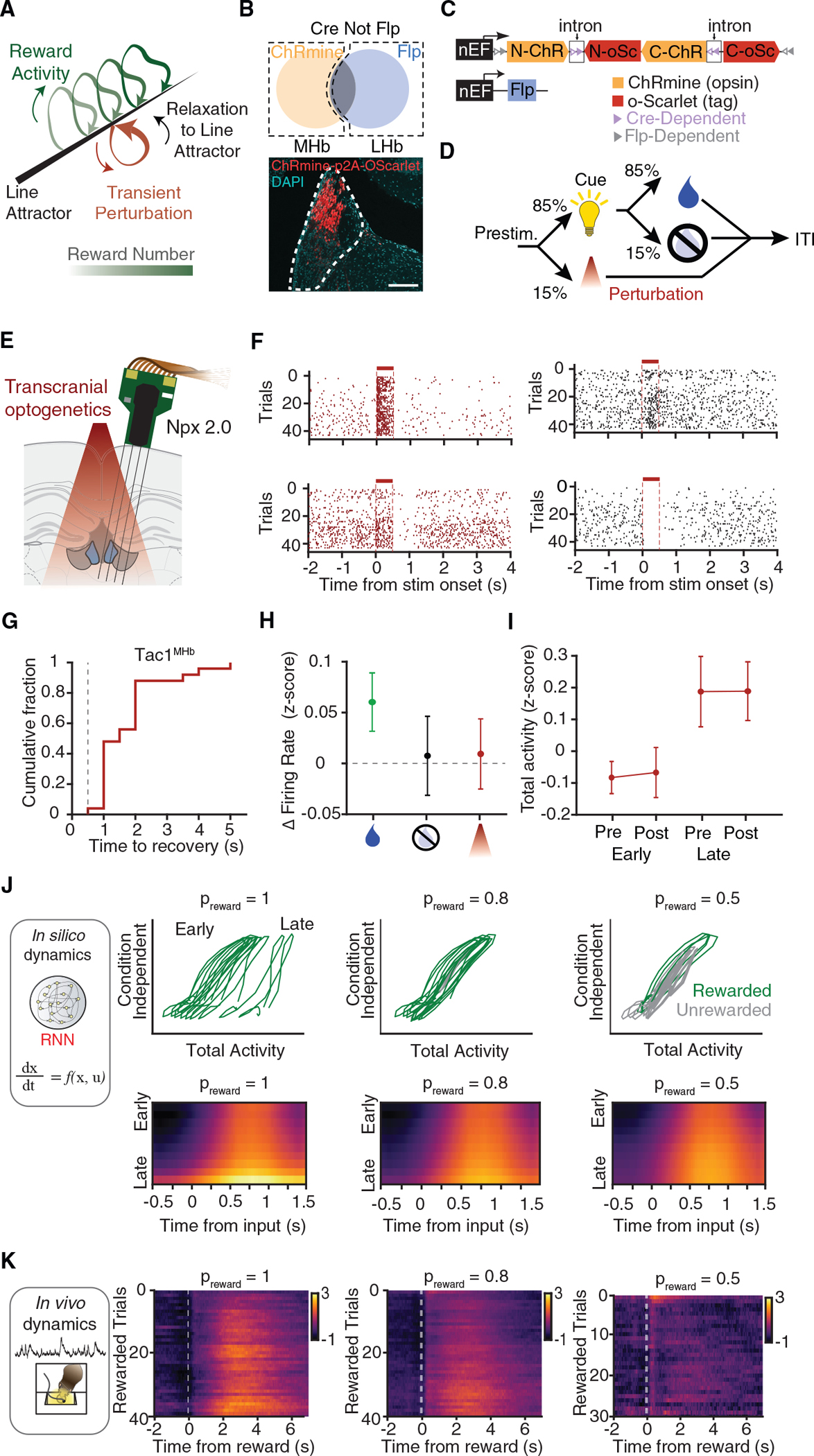
(A) Schematic representation of transient optogenetic perturbation of the line attractor dynamics.
(B) Intersectional gene targeting approach. AAV1 CreOnFlpOff ChRmine-p2A-oScarlet is delivered to MHb and LHb neurons of Tac1-Cre mice. AAV8-Flp is delivered to the LHb to turn off ChRmine expression in Tac1LHb neurons.
(C) Constructs used for INTRSECT implementation as described in (B).
(D) Trial structure for the perturbation experiment.
(E) Experimental configuration for transcranial optogenetic stimulation and neural recording using Neuropixels 2.0 probes.
(F) Spike raster plots and firing rate changes for example validated optotagged MHb Tac1 neurons (left) or nearby modulated MHb neurons (right), which were all simultaneously recorded.
(G) Time to baseline recovery after perturbation for MHb neurons.
(H) Average firing rate changes in rewarded (green), unrewarded (black), and perturbation (red) trials. Curves: mean; error bar: SEM from hierarchical bootstrap. n = 1,078 neurons, 4 sessions, 2 mice.
(I) Within-trial and across-trial firing rate changes for optogenetic perturbation of MHb Tac1 neurons. Trials were split into early/late halves for concise visualization.
(J) Simulation of reward signal accumulation with varying reward probability (1.0, 0.8, and 0.5). Top, example state space trajectories show generated single sessions, initialized at the identical initial state. Bottom, model predictions on changes in total population activity (fiber photometry signal) across rewarded trials. For each case, 1,000 simulated sessions with random initial states were averaged.
(K) Fiber photometry recordings in 3CSRTT at 3 different reward probabilities: preward = 1, preward = 0.8, and preward = 0.5. Mean fluorescence (%ΔF/F) during reward as a function of nth correct trial in a session.
See also Figure S7.
Accordingly, we performed brief Tac1MHb-specific optogenetic stimulation and electrophysiological recording during the behavior. We used INTRSECT targeting to express ChRmine in Tac1MHb (but not Tac1LHb) neurons (Figures 7B and 7C), which enabled transcranial optogenetic perturbation during the head-fixed reward-guided task (Figures 7D and 7E). Transient perturbation robustly triggered diverse types of MHb single-neuron responses (Figure 7F). Optotagged neurons were directly driven by the ChRmine stimulation and thus showed transient excitation. Other modulated units showed both excitation and inhibition, and a substation fraction had seconds-long return-to-baseline dynamics (Figure 7G). These prolonged relaxation kinetics were consistent with network effects predicted by attractor models (Mante et al., 2013).
We included perturbation trials in a range of the baseline states and found that the long-timescale ramping-up dynamics proceeded as expected, independent of the transient perturbations. Because stochastic opsin expression will set an arbitrary multidimensional direction for the transient population-level perturbations (which in this case are not designed to provide naturalistic single-cell-resolution dynamics), guided by the above theoretical predictions we hypothesized that the transient stimulation itself would not contribute to ramping. Quantification of within-trial firing rate changes found that only rewarded trials, but not unrewarded trials or perturbation trials, contributed to the ramping (Figures 7H and S7A and S7B). In summary, consistent with theoretical predictions on transient, unaligned perturbation of line attractor dynamics models, we observed within-trial relaxation onto the pre-stimulation baseline in the Tac1MHb population without significant integration, in contrast to across-trial movement of the baseline by interleaved rewarded trials (Figure 7H).
Reward history integration in silico and in vivo
We finally asked if trial-type-dependent external inputs would be sufficient to generate the long-timescale dynamics via cell-type-specific attractors. A crucial feature of data-driven system identification with trLFADS is the possibility for in silico neural dynamics experiments, by simulating temporal evolution of neural trajectories across trials while injecting trial-type-dependent external inputs into the learned dynamical systems; the state evolution of these dynamical systems then generates de novo neural trajectories (Figure 7J and STAR Methods). We discovered that injecting only rewarded-trial external inputs shifted the neural state along the line attractor, strikingly consistent with our photometry, imaging, and electrophysiology results. In contrast, the same type of simulation with TH+ models showed no activity increase over time, consistent instead with operation of a discrete point attractor (Figure S7C).
Finally, we considered how different reward probabilities might modulate these dynamics. Computational titration of reward delivery probability was carried out by varying the ratio of rewarded- and unrewarded-trial external inputs; these in silico experiments assessed reward probabilities not yet experimentally tested (preward = 0.5, 0.8, or 1.0). Differential slopes of activity accumulation were predicted in Tac1MHb but not TH+ simulations (Figures 7J and S7C). We compared the simulated dynamics to experimentally measured neural dynamics from photometry signals in freely moving mice. We found that ramping developed more steeply at higher reward-probabilities (Figure 7K), consistent with the trLFADS prediction and supporting the model that Tac1MHb population dynamics can serve as a line attractor that integrates reward history.
In summary, by computationally reverse-engineering the trained trLFADS models to generate testable hypotheses, we predicted dynamics consistent with a cell-type-specific line attractor system for Tac1MHb neurons. This outcome may thus reveal a computationally defined cell type marker (line-attractor integration of reward history), which in this case maps onto a specific genetically defined and regionally localized cell type.
DISCUSSION
Here, we measured neural population activity from multiple habenular cell types during rewarded behavioral tasks. We began by providing the initial report of temporal structure and probability-context dependence of habenular Tac1+ neuron responses and showed TH+ cells learn to encode reward-predictive cues (Figures 2 and S3E–S3G) (consistent with previous work suggesting separate roles for cholinergic and peptidergic MHb cell types; Seigneur et al., 2018). Next, through dynamical system modeling we identified a cell-type-specific line attractor (Tac1+ but not TH+) underlying long-timescale reward history integration, a computation important for establishing the value of actions over many behaviorally relevant timescales, particularly in environments with sparse or dynamic rewards (Hattori et al., 2019; Sugrue et al., 2004; Hwang et al., 2017; Kawai et al., 2015; Morcos and Harvey, 2016; Scott et al., 2017; Sul et al., 2010; Zalocusky et al., 2016; Bayer and Glimcher, 2005; Bromberg-Martin et al., 2010; Lazaridis et al., 2019; Ottenheimer et al., 2018; Tsutsui et al., 2016). These results show how the population activity of a genetically defined cell type can operate as a discrete computational element subserving complex, behaviorally important brain functions.
TH+: Cell-type-specific segregation of predictive valence
Unexpected rewards are known to activate dopaminergic neurons, but after repeated reward exposures, neural activity shifts back in time to cues that precede and predict rewards (Schultz et al., 1997)—a milestone discovery validating prior models of reinforcement learning (Sutton and Barto, 1990). TH, an obligatory enzyme for dopamine synthesis, is often used to mark dopaminergic neurons; however, our TH+ neurons are likely “dopaergic” rather than dopaminergic, since they lack other molecular machinery for dopamine processing and release aromatic acid acid decarboxylase (AADC) and vesicular monoamine transporter (VMAT2); Weihe et al., 2006) and may produce little of the TH enzyme itself (Lammel et al., 2015). Here, we find TH gene expression marks a group of neurons activated by reward-predicting cues. In the future, it will be interesting to explore how TH+ neuron activity in MHb/HbX develops before and during training—and perhaps is modulated by how reliably the cue predicts reward availability—akin to neurons in the LHb encoding the information content of a stimulus (Bromberg-Martin and Hikosaka, 2011).
Habenular pathways of reward and aversion
The habenula has been described as an anti-reward center, though this characterization has been predominantly informed by the activity of LHb neurons because of their response to aversive stimuli and negative reward prediction error (Hikosaka, 2010; Proulx et al., 2014) and to a lesser extent the ventral MHb ChAT+ neurons, which have been implicated in anxiety (Cho et al., 2019; Mathuru et al., 2013; Seigneur et al., 2018; Yamaguchi et al., 2013) and aversion (Buolos et al., 2020; Frahm et al., 2011; McLaughlin et al., 2017; Morton et al., 2018; Choi et al., 2021). Evidence of a parallel positive reward pathway in Hb had been suggested by observations that Hb lesions can block intracranial self-stimulation (Morissette and Boye, 2008), and more recently by dorsal MHb (predominantly Tac1+) optogenetic manipulations demonstrating a role in primary reinforcement and hedonic value (Hsu et al., 2014, 2016). Our data show opposing valence signals for MHb and LHb Tac1+ neurons and provide additional evidence of parallel, anatomically distinct reward and anti-reward pathways in the habenula.
The integration of reward history is a computation important for establishing value of actions over a range of behaviorally relevant timescales, particularly in environments with sparse or dynamic rewards. Recent-history effects have been observed in several rewarded tasks and species (Hattori et al., 2019; Sugrue et al., 2004; Hwang et al., 2017; Kawai et al., 2015; Morcos and Harvey, 2016; Scott et al., 2017; Sul et al., 2010; Zalocusky et al., 2016; Bayer and Glimcher, 2005; Tsutsui et al., 2016), including in the LHb (Lazaridis et al., 2019; Bromberg-Martin et al., 2010); further work is needed to test how reward history computations in upstream brain regions might influence Tac1MHb activity. Moreover, the functional impact of Tac1MHb ramping on downstream targets remains an open question. The gene Tac1 encodes preprotachykinin, the peptide precursor of Neurokinin A and Substance P; the latter is typically associated with pain and itch (Davidson and Giesler, 2010), but more recent work has shed light on the role of these peptidergic neurons in encoding hedonic value (Hsu et al., 2014) and novelty (Molas et al., 2017). Substance P is released from dense core vesicles upon high frequency stimulation, and it will be interesting to determine if ramping activity may be sufficient to push Tac1+ neurons into a peptidergic regime, which has been shown to promote plasticity at MHb-IPN synapses (Melani et al., 2019).
Cell-type-specific attractor dynamics revealed by data-driven modeling
While it is not trivial for short single-neuron time constants to specifically influence persistent population activity on timescales relevant for adaptive behavioral changes, attractor dynamics implemented in neural systems could in theory serve this bridging role and thus implement aspects of phenomena such as cognitive integration (Robinson, 1989; Seung, 1996), foraging (Hattori et al., 2019), and associative memory (Hopfield, 1982). Experimental observations have identified putative attractor dynamics in certain brain circuits (Finkelstein et al., 2021; Inagaki et al., 2019; Kim et al., 2017; Mante et al., 2013; Miri et al., 2011); however, previous work has not implicated specific cell types in attractor implementation, nor provided direct correspondence to single-trial data crucial for determining if the attractor dynamics could guide individual choices made by behaving animals. Here, we have carried out data-driven identification of the dynamical systems capable of regenerating the relevant experimental neural data (Pandarinath et al., 2018b; Yu et al., 2009; Zhao and Park, 2017; Zoltowski et al., 2020) and directly mapped the underlying attractor dynamics in the resulting models. We were able to quantitatively define cell-type-specific single-trial attractor dynamics, successfully test theoretical predictions describing neural attractor dynamics responding to perturbation, and use in silico experiments to simulatevarying reward probabilities.
Many opportunities for exploration remain, especially in the use of optogenetics to further probe these long-timescale flexible changes in behavior. Although we observed nearly zero integration along the putative Tac1MHb line attractor upon recovery from transient one-photon optogenetic perturbation (a perturbation that by design does not match naturally occurring ensemble dynamics), we anticipate that in vivo cellular-resolution two-photon optogenetic perturbation (Prakash et al., 2012) of multiple individually defined single cells during behavior (Jennings et al., 2019; Marshel et al., 2019; Carrillo-Reid et al., 2019) precisely aligned to the putative line attractor’s selection vector might be sufficient to result in nonzero integration, further advancing population activity level cellular-resolution causal neuroscience. Such ensemble-optogenetics intervention may be designed to elicit effects on the system that are either matched or mismatched to effects of natural inputs (e.g., from upstream structures such as septum). These future experiments may provide insight into the robustness and uniqueness of the putative line attractor (e.g., testing the possibility that MHb activity may represent readout of upstream populations implementing the identified computations). In addition, although here we studied a single cell type at a time (equivalent to observing the neural states projected onto a specific neural subspace), simultaneous access to multiple cell types may provide insight into higher-order cell type markers and the computations arising from interactions between cell types.
While we and others have previously described the brain-spanning activity arising from even simple behaviors, using electrophysiological recordings without cell type specificity (Allen et al., 2019; Steinmetz et al., 2019), our current results reveal logic linking specific neural population dynamics (often implicated in higher-level computation) to discrete cell types. Identifying specific cell types involved in integration of reward history is important for understanding the design, adaptation, and function of value computation over behaviorally relevant timescales (Roy et al., 2021). Laying groundwork for this direction of investigation, we show how dynamical systems modeling of specific cell types generates testable predictions in the form of computational cell type descriptors; this approach may be broadly useful in elucidating how principles of nervous system function arise from properties of constituent cellular elements.
Limitations of the study
A common assumption for dynamical systems modeling is that the neural circuit is “fixed” during the behavioral session. This invariance in time is the basis of widely used trial averaging and provides the inductive bias to identify underlying dynamical systems. The fixed-circuit assumption has historically been justified by the fact that animals are typically heavily trained to tasks before neural recordings, with stable performance metrics attained (training was also completed in the present study). However, structural changes due to plasticity may still occur, which may have multiple timescales and mechanisms. For example, synaptic potentiation or cell-autonomous excitability changes might also be able to implement reward history computation, by Tac1MHb or other populations, in the absence of causally relevant line attractor dynamics. A promising approach to test this possibility would involve all-optical electrophysiology (Fan et al., 2020) using voltage sensors for direct synaptic strength testing and excitability measurements in vivo. Although application of these emerging optical approaches to deep brain regions such as the habenula will be technically challenging, this approach will be useful even beyond plasticity studies (for example, by revealing mechanistic details of biological substrates underlying the line attractor dynamics, such as local versus multi-regional synaptic contributions).
Our analyses rely on single genes (or a combination of anatomical and genetic properties) to access cell types, but further molecular refinement will be possible or even likely. Our markers captured differences in activity of habenular neurons, but some cell types showed mixed responses (Calb1+), and our cellular resolution imaging indicated that not all monogenetically defined neurons exhibit specific reward- or cue-related responses. Recent transcriptional analyses in the habenula have noted that individual genes may be insufficient to unambiguously identify transcriptional cell type (Hashikawa et al., 2020; Wallace et al., 2020; Pandey et al., 2018), and intersectional viral targeting methods (Fenno et al., 2014, 2020) may further refine transcriptome-to-function mapping.
STAR★METHODS
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Karl Deisseroth (deissero@stanford.edu).
Materials availability
All unique/stable reagents generated in this study are freely available from the lead contact with a completed Materials Transfer Agreement.
Data and code availability
All code is available on GitHub. Modeling code is available at https://github.com/google-research/computation-thru-dynamics. Photometry code is available on https://github.com/SylwestrakLab/HbRewardCellTypes. In vivo electrophysiology code is available on https://github.com/youngju-jo. STARmap data is available on the NeMO Archive (NeMO: 9ACQ8G1, https://assets.nemoarchive.org/dat-9ACQ8G2) and electrophysiology data is availbe on the DANDI archive (DANDI: 000302, https://dandiarchive.org/dandiset/000302).
Any additional information required to reanalyze the data reported in this paper is available from the lead contact (KD) upon request.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Animal husbandry and all aspects of animal care and euthanasia as described were in accordance with guidelines from the NIH and have been approved by members of the Stanford Institutional Animal Care and Use Committee and University of Oregon Institutional Animal Care and Use Committee. For in situ sequencing, in situ hybridization, and serotype testing, C57/Bl6 mice aged 10w-16w were obtained from Jackson Laboratories (#000664). This study includes data from both male and female mice. For transgenic experiments Tac1-Cre (B6; 129S-Tac1tm1.1(cre)Hze; Jax #021877), Th-Cre (Ththm1(cre)Te; MGI:3056580), ChAT-Cre (Tg(Chat-cre) GM24Gsat/Mmucd; MMRRC #017269-UCD), and Calb1-Cre (B6.Cg-Calb1tm1.1(folA/cre)Hze/J; Jax #023531) mice were used.
METHOD DETAILS
STARmap in situ sequencing
The STARmap experiment was conducted as previously described (Wang et al., 2018). In brief, glass-bottom 12-well plates were treated by methacryloxypropyltrimethoxysilane (Bind-Silane) and poly-L-lysine (Sigma) following manufacturer’s instructions. Freshly harvested mouse brains were immediately embedded in O.C.T., snap-frozen by liquid nitrogen, and cut on a cryostat to 16-μmm slices. Slices containing habenula regions were mounted in the pretreated glass-bottom plates. Brain slices were fixed with 4% PFA in PBS at 22°C for 10 min, permeabilized with −20°C methanol, and then placed at −80°C for 15 min before hybridization. SNAIL probes were dissolved at 100 mM in ultrapure RNase-free water and pooled. The samples were taken from −80°C and equilibrated to r.t. for 5 min, washed by PBSTR (0.1% Tween 20, 0.1 U/μL SUPERase·In in PBS) for 2–5 min and incubated in 1 × hybridization buffer (2X SSC, 10% formamide, 1% Tween 20, 20 mM RVC, 0.1 mg/mL salmon sperm DNA and pooled SNAIL probes at 100 nM per oligo) in 40°C humidified oven with gentle shaking overnight. The samples were then washed for 20 min twice with PBSTR, followed by one 20 min wash in 4X SSC dissolved in PBSTR at 37°C.Finally, the sample was briefly rinsed with PBSTR once atr.t. The samples were then incubated for 2 h with T4 DNA ligation mixture (1:50 dilution of T4 DNA ligase supplemented with 1X BSA and 0.2 U/μL of SUPERase-In) at room temperature with gentle agitation. Then samples were washed twice with PBSTR, incubated with RCA mixture (1:50 dilution of Phi29 DNA polymerase, 250 mM dNTP, 1X BSA and and 20 μM 5-(3-aminoallyl)-dUTP) at 30°C for 2 h under agitation. The samples were next washed twice in PBST (PBSTR omitting SUPERase·In) and treated with 20 mM Acrylic acid NHS ester in PBST for 2 h at r.t. The samples were briefly washed with PBST once, then incubated with monomer buffer (4% acrylamide, 0.2% bis-acrylamide, 2X SSC) for 30 min at RT. The buffer was aspirated and 10 μL of polymerization mixture (0.2% ammonium persulfate, 0.2% tetramethylethylenediamine dissolved in monomer buffer) was added to the center of the sample, which was immediately covered by Gel Slick coated coverslip and incubated for 1 h at r.t., then washed by PBST twice for 5 min each. The tissue-gel hybrids were digested with Proteinase K mixture (0.2 mg/mL Proteinase K, 20 mM Tris = 7.5, 100 nM NaCl, 1%SDS) at 37°C for 2 h, then washed with PBST three times (5 min each). For in situ sequencing, each sequencing cycle began with treating the sample with stripping buffer (60% formamide, 0.1% Triton X-100) at r.t. for 10 min twice, followed by three PBST washes, 5 min each, then sequencing ligation mixture (Wang et al., 2018). Images were acquired using Leica TCS SP8 confocal microscopy with a white light laser, 40× oil-immersed objective (NA 1.3), with a voxel size of 78 × 78 × 315 nm.
In situ hybridization
Procedure
To prepare tissue for in situ hybridization, C57/Bl6 mice aged 10–12 weeks were anesthetized with isoflurane and rapidly decapitated. Brain tissue was immediately removed, embedded in OCT and flash frozen in liquid nitrogen. Tissue was equilibrated to −20°C and sectioned on a microtome (Leica CM500) to a thickness of 20μm and mounted on superfrost plus slides (Fisher Scientific). Tissue was promptly fixed in 4% PFA for 10 min, then transfer to prechilled methanol (−20°C) and incubated at −80°C for 1 h. Tissue was washed two times in 5xSSC, then incubated in hybridization buffer for 30 min. Probes targeting Tachykinin1, Tyrosine hydroxylase, Choline Acetyltransferase, and Calbindin1 mRNA were designed by Molecular Technologies and diluted to 2nM final concentration in hybridization buffer. Hybridization buffer with probes was added to the slides, covered with Hybrislips, and incubated overnight in a humidified chamber at 37°C. Using a Coplan jar, two washes of 30% formamide in 5xSSC were performed, followed by 2 washes in 5xSSCT. Slides were equilibrated in amplification buffer for 30 min. During this time, fluorophore-labeled hairpins were heated to 95°C for 90 s, then cooled to room temperature at bench top for 30 min. Cooled hairpins were added to amplification buffer and the resulting solution was added to tissue sections and coverslipped. Amplification reaction was run overnight at room temperature, protected from light exposure. Amplified sections were washed 4 times with 5xSSCT in a Coplan jar and then coverslipped for confocal imaging.
| 30% probe hybridization buffer | For 40 mL of solution |
|---|---|
| 30% formamide | 12 mL formamide |
| 5× sodium chloride sodium citrate (SSC) | 10 mL of 20× SSC |
| 9 mM citric acid (pH 6.0) | 360 μL 1 M citric acid, pH 6.0 |
| 0.1% Tween 20 | 400 μL of 10% Tween 20 |
| 50 μg/mL heparin | 200 μL of 10 mg/mL heparin |
| 1× Denhardt’s solution | 800 mL of 50× Denhardt’s solution |
| 10% dextran sulfate | 8 mL of 50% dextran sulfate |
| 30% probe wash buffer | For 40 mL of solution |
|---|---|
| Fill up to 40 mL with ultrapure H2O | |
| 30% formamide | 12 mL formamide |
| 5× sodium chloride sodium citrate (SSC) | 10 mL of 20× SSC |
| 9 mM citric acid (pH 6.0) | 360 μL 1 M citric acid, pH 6.0 |
| 0.1% Tween 20 | 400 μL of 10% Tween 20 |
| 50 μg/mL heparin | 200 μL of 10 mg/mL heparin |
| Amplification buffer | For 40 mL of solution |
|---|---|
| Fill up to 40 mL with ultrapure H2O | |
| 5× sodium chloride sodium citrate (SSC) | 10 mL of 20× SSC |
| 0.1% Tween 20 | 400 μL of 10% Tween 20 |
| 10% dextran sulfate | 8 mL of 50% dextran sulfate |
| 5×SSCT | For 40 mL of solution |
|---|---|
| Fill up to 40 mL with ultrapure H2O | |
| 5× sodium chloride sodium citrate (SSC) | 10 mL of 20× SSC |
| 0.1% Tween 20 | 400 μL of 10% Tween 20 |
| Fill up to 40 mL with ultrapure H2O | |
Viral injections
Mice were anesthetized with isoflurane and stereotaxic injections were used to deliver 200–300nL of virus to the medial habenula (AP: −1.4mm, ML +/− 0.3mm, DV, −3.05mm) at a rate of 10 nL/min. For serotype quantification experiments, fluorescent beads (Lumafluor, diluted 1:10) were added to determine center of injection location to verify targeting to the MHb/LHb boundary. Viruses used in the study were generated at the Stanford Viral Vector Core and are listed in the Key Resources Table
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
|
| ||
| Antibodies | ||
|
| ||
| anti-GFP antibody | Thermo Fisher Scientific | Cat# A-21311; RRID: AB_221477 |
| anti-GFP antibody | Thermo Fisher Scientific | Cat# A-31852; RRID: AB_162553 |
|
| ||
| Bacterial and virus strains | ||
|
| ||
| AAV1-EF1a-DIO-GCaMP6f | Stanford Viral Vector Core | custom prep |
| AAV1-EF1a-DIO-YFP | Stanford Viral Vector Core | custom prep |
| AAV1-hSyn-YFP | Stanford Viral Vector Core | custom prep |
| AAV5-hSyn-YFP | UNC Viral Vector Core | custom prep |
| AAV8-hSyn-YFP | Stanford Viral Vector Core | custom prep |
| AAVdj-hSyn-YFP | Stanford Viral Vector Core | custom prep |
| AAVdj-hSyn-GCaMP6m | Stanford Viral Vector Core | custom prep |
| AAV8-EF1a-DIO-GCaMP6m | Stanford Viral Vector Core | custom prep |
| AAV1-EF1a-DIO-H2B-GCaMP6f | Stanford Viral Vector Core | custom prep |
| AAV1-Ef1a-DIO-ChRmine-oScarlet | Stanford Viral Vector Core | custom prep |
| AAV1-nEF-CreOnFlpOff-ChRmine-p2A-oScarlet | Stanford Viral Vector Core | custom prep |
| AAV1-nEF-CreOnFlpOff-eNpHR3.0-YFP | Stanford Viral Vector Core | custom prep |
| AAV8-Ef1a-FlpO | Stanford Viral Vector Core | custom prep |
|
| ||
| Chemicals, peptides, and recombinant proteins | ||
|
| ||
| methacryloxypropyltrimethoxysilane | GE Healthcare | 17-1330-01 |
| poly-L-lysine | Sigma-Aldrich | P6407 |
| Glass Bottom Plates | MatTekq | P12G-1.5-14-F |
| Micro coverglass | EMS | 72,226-01 |
| OTC | Fisher | 23-730-571 |
| 16% PFA | EMS | 15710-S |
| PBS | Gibco | 70,011–044 |
| Triton X-100 | Sigma-Aldrich | 93,443 |
| OminiPur Formamide | Calbiochem | 75-12-7 |
| 20×SSC buffer | Sigma-Aldrich | S6639 |
| dNTP mix | Invitrogen | 100,004,893 |
| Tween 20 | Calbiochem | 655,206 |
| SUPERase | Invitrogen | AM2696 |
| RVC | New England Biolabs | S1402S |
| Salmon Sperm | Invitrogen | AM9680 |
| T4 DNA ligase | Thermo Scientific | EL0011 |
| Phi29 DNA polymerase | Thermo Fisher Scientific | EP0094 |
| BSA | New England Biolabs | EP0094 |
| BSPEG9 | Thermo Fisher Scientific | 21,582 |
| Phi29 DNA polymerase | Thermo Fisher Scientific | EP0094 |
| Acrylic acid NHS ester | Sigma-Aldrich | A8060 |
| Methacrylic acid NHS ester | Sigma-Aldrich | 730,300 |
| DMSO | Sigma-Aldrich | D12345 |
| Acrylamide, 40% | BioRad | 161-0140 |
| Bis-acrylamide, 2% | BioRad | 161-0142 |
| Ammonium persulfate | Sigma-Aldrich | A3678 |
| Tetramethylethylenediamine | Sigma-Aldrich | T9281 |
| OminiPur SDS | Calbiochem | 7991 |
| Shrimp Alkaline Phosphatase | New England Biolabs | M0371L |
| DAPI | Molecular Probes | D1306 |
| NeuroTrace Nissl Stain | Molecular Probes | N-21480 |
| Gel Slick | Lonza | 50,640 |
| Proteinase K | Invitrogen | 25,530,049 |
| superfrost plus slides | Fisher Scientific | 22-037-246 |
| Hybrislips | EMS | 7,032,962 |
| Trimethroprim (TMP) | Sigma-Aldrich | 92,131-1G |
| Heparin | Sigma-Aldrich | H4784-250MG |
| 50× Denhardt’s solution | Sigma-Aldrich | D2532-5ML |
| Dextran Sulfate | Sigma-Aldrich | D6001 |
| Fluorescent beads | Lumafluor | Red Retrobeads |
| Alexa 514-Labeled Hairpins | Molecular Technologies | B5-H1/H2-514 |
| Alexa 555-Labeled Hairpins | Molecular Technologies | B2-H1/H2-555 |
| Alexa 593-Labeled Hairpins | Molecular Technologies | B4-H1/H2-593 |
| Alexa 647-Labeled Hairpins | Molecular Technologies | B1-H1/H2-647 |
| Methanol | Sigma-Aldrich | 322,415 |
| Dichloromethane | Sigma-Aldrich | 270,997 |
| Ethyl cinnamate | Sigma-Aldrich | 112,372 |
| Trypsin | Sigma-Aldrich | 59427C |
| CM-DiI | Thermo Fisher Scientific | V22888 |
| DiD | Thermo Fisher Scientific | D7757 |
| DID-DS | Thermo Fisher Scientific | D12730 |
|
| ||
| Experimental models: Organisms/strains | ||
|
| ||
| Th-Cre (Ththm1(cre)Te | Lindeberg | MGI:3056580 |
| B6; 129S-Tac1tm1.1(cre)Hze | Jax | 021,877 |
| Tg(Chat-cre)GM24Gsat/Mmucd | MMRRC | #017269-UCD |
| B6.Cg-Calb1tm1.1(folA/cre)Hze/J | Jax | #023531 |
| C57/Bl6 | Jax | #000664 |
|
| ||
| Oligonucleotides | ||
|
| ||
| Calb1_01 | IDT (for STARmap) | /5Phos/ACATTAGCCAACTCTACAATTCCTATAATTATTAATGAAACATACACTAAAGATA |
| Calb1_02 | IDT (for STARmap) | /5Phos/ACATTATCAGCGTCGAAATGAAGCCAGAATTATTAATGAAACATACACTAAAGATA |
| Calb1_03 | IDT (for STARmap) | /5Phos/ACATTAGGAAATTTTCCTGCACTGGTAATTATTAATGAAACATACACTAAAGATA |
| Calb1_04 | IDT (for STARmap) | /5Phos/ACATTAATTTCATTTCCGGTGATAGCTAATTATTAATGAAACATACACTAAAGATA |
| Calb1_11 | IDT (for STARmap) | TCTTCTGTGGGTAAGACGTGTAATGTTATCTT |
| Calb1_12 | IDT (for STARmap) | CTTCCAGGTAACCACTTCCGTAATGTTATCTT |
| Calb1_13 | IDT (for STARmap) | TGATTCCCTGGAATTTAAGATAATGTTATCTT |
| Calb1_14 | IDT (for STARmap) | CTGTCCATATTGATCCACAAATAATGTTATCTT |
| Calb2_01 | IDT (for STARmap) | /5Phos/ACATTAGAGCACAATCTCCAGGTCCTAATTATTACTGAAACATACACTAAAGATA |
| Calb2_02 | IDT (for STARmap) | /5Phos/ACATTAGGTGGTGAGCTGTTGGATGTAATTATTACTGAAACATACACTAAAGATA |
| Calb2_03 | IDT (for STARmap) | /5Phos/ACATTAATCCGTAGTATGGTCTGGGTGTAATTATTACTGAAACATACACTAAAGATA |
| Calb2_04 | IDT (for STARmap) | /5Phos/ACATTAAGCCCACGTGCTGCCTGAAGCAAATTATTACTGAAACATACACTAAAGATA |
| Calb2_11 | IDT (for STARmap) | TTACACGGGGGGCTCACTGCTAATGTTATCTT |
| Calb2_12 | IDT (for STARmap) | AGGACATGACACTCTTCCTGTAATGTTATCTT |
| Calb2_13 | IDT (for STARmap) | TGCCATCTCCATTTAAGTCAAATAATGTTATCTT |
| Calb2_14 | IDT (for STARmap) | AAGCCTCCATAAACTCAGCGCTTAATGTTATCTT |
| Cartpt_01 | IDT (for STARmap) | /5Phos/ACATTACCTTTCCTCACTGCGCACTGAATTATTAGTGAAACATACACTAAAGATA |
| Cartpt_02 | IDT (for STARmap) | /5Phos/ACATTAAAGTTGCCGCCTTGGCAGCTAATTATTAGTGAAACATACACTAAAGATA |
| Cartpt_03 | IDT (for STARmap) | /5Phos/ACATTAGCGTTTACTCTTGAGCTTCTAATTATTAGTGAAACATACACTAAAGATA |
| Cartpt_04 | IDT (for STARmap) | /5Phos/ACATTAAGTAGCAGCAGGGCGGCGCCCAATTATTAGTGAAACATACACTAAAGATA |
| Cartpt_11 | IDT (for STARmap) | ACAGCTTCCCGATCCTGGCTAATGTTATCTT |
| Cartpt_12 | IDT (for STARmap) | AACATAGCGCCGGGAGCCCTAATGTTATCTT |
| Cartpt_13 | IDT (for STARmap) | ACTTCTTCTCGTAGATCGGATAATGTTATCTT |
| Cartpt_14 | IDT (for STARmap) | ACGGGCACCCAGCAAAGGTATAATGTTATCTT |
| Chat_01 | IDT (for STARmap) | /5Phos/ACATTATTCGCTCCATTCAAGCTGCAAATTATTATTGAAACATACACTAAAGATA |
| Chat_02 | IDT (for STARmap) | /5Phos/ACATTAGGACGCCATTTTGACTATCTAATTATTATTGAAACATACACTAAAGATA |
| Chat_03 | IDT (for STARmap) | /5Phos/ACATTATCTCTCATGTCAACAAGGCTAATTATTATTGAAACATACACTAAAGATA |
| Chat_04 | IDT (for STARmap) | /5Phos/ACATTAATTAATGACAACATCCAAGACAAATTATTATTGAAACATACACTAAAGATA |
| Chat_11 | IDT (for STARmap) | GGGACTTGTCATACCAACGTAATGTTATCTT |
| Chat_12 | IDT (for STARmap) | GGCAGGCGTTCATCCTCGTTAATGTTATCTT |
| Chat_13 | IDT (for STARmap) | AGGCTGCCTCGAACTACAGATAATGTTATCTT |
| Chat_14 | IDT (for STARmap) | TCACCCTCACTGAGACGGCGGTAATGTTATCTT |
| Elfn1_01 | IDT (for STARmap) | /5Phos/ACATTAAGTCTGAGACGCTCCCAGATAATTATTAATGAACCATACACTAAAGATA |
| Elfn1_02 | IDT (for STARmap) | /5Phos/ACATTACGTCGATGCAGTTGTTAATGAATTATTAATGAACCATACACTAAAGATA |
| Elfn1_03 | IDT (for STARmap) | /5Phos/ACATTATACAGGTACTCGAGCTTGCTAATTATTAATGAACCATACACTAAAGATA |
| Elfn1_04 | IDT (for STARmap) | /5Phos/ACATTACGTACTGGCGACTCTTGTCGGCAATTATTAATGAACCATACACTAAAGATA |
| Elfn1_11 | IDT (for STARmap) | CCTTGTGTCTCCGTCGGCTTAATGTTATCTT |
| Elfn1_12 | IDT (for STARmap) | GGTGGACTCGGACTTGAGGTAATGTTATCTT |
| Elfn1_13 | IDT (for STARmap) | CGATGAGGTTGGCCTGTAGTAATGTTATCTT |
| Elfn1_14 | IDT (for STARmap) | GGGGTATGAGTGCCGATGCTCTAATGTTATCTT |
| GAD2_01 | IDT (for STARmap) | /5Phos/ACATTAGGACATCAGTAACCCTCCACAATTATTACTGAACCATACACTAAAGATA |
| GAD2_02 | IDT (for STARmap) | /5Phos/ACATTACTCTAACCAGGAGAGCTGAAAATTATTACTGAACCATACACTAAAGATA |
| GAD2_03 | IDT (for STARmap) | /5Phos/ACATTAAACGCGTAGTTGACATCCCCTTAATTATTACTGAACCATACACTAAAGATA |
| GAD2_04 | IDT (for STARmap) | /5Phos/ACATTACTCCAGATTTTGCGGTTGGTCTAATTATTACTGAACCATACACTAAAGATA |
| GAD2_11 | IDT (for STARmap) | AGCTTCCACTTGTGTTTCCTAATGTTATCTT |
| GAD2_12 | IDT (for STARmap) | AGCTCTGCATCAGTCCCTCCTAATGTTATCTT |
| GAD2_13 | IDT (for STARmap) | AGCAGGTCTGTTGCGTGCATAATGTTATCTT |
| GAD2_14 | IDT (for STARmap) | TGTTTGGCAATGCGTCAAAATTTAATGTTATCTT |
| Gpr151_01 | IDT (for STARmap) | /5Phos/ACATTAAACAAATGTCAAGCTCTTGGAATTATTAGTGAACCATACACTAAAGATA |
| Gpr151_02 | IDT (for STARmap) | /5Phos/ACATTAATTGTCTTGTGCTGAAGGGAAATTATTAGTGAACCATACACTAAAGATA |
| Gpr151_03 | IDT (for STARmap) | /5Phos/ACATTATAGGATGCATGTTTGGGTCTAATTATTAGTGAACCATACACTAAAGATA |
| Gpr151_04 | IDT (for STARmap) | /5Phos/ACATTAGTTGCTGGAATTGGTGTCGGAATTATTAGTGAACCATACACTAAAGATA |
| Gpr151_11 | IDT (for STARmap) | AAGCATGCTTTGGCTACCATAATGTTATCTT |
| Gpr151_12 | IDT (for STARmap) | TGTTCCCAGGGTATAGGGTTAATGTTATCTT |
| Gpr151_13 | IDT (for STARmap) | AGACCCACGTCCGTCTGTGTTAATGTTATCTT |
| Gpr151_14 | IDT (for STARmap) | GCGAGCAAACGACTCGTTCATAATGTTATCTT |
| Htr5b_01 | IDT (for STARmap) | /5Phos/ACATTACACGCCAATCAAGATCCCGAAATTATTATTGAACCATACACTAAAGATA |
| Htr5b_02 | IDT (for STARmap) | /5Phos/ACATTAAAGGTTACTGTTGCTCGGCGAATTATTATTGAACCATACACTAAAGATA |
| Htr5b_03 | IDT (for STARmap) | /5Phos/ACATTAGGACAACTCGCTCACCAGGCTAATTATTATTGAACCATACACTAAAGATA |
| Htr5b_04 | IDT (for STARmap) | /5Phos/ACATTAATTCCATAAGAAAGTGGCAACGAATTATTATTGAACCATACACTAAAGATA |
| Htr5b_11 | IDT (for STARmap) | GGGGATCCAACAAAGCACAATAATGTTATCTT |
| Htr5b_12 | IDT (for STARmap) | CCACGAGTCTCCGCTTGTCTTAATGTTATCTT |
| Htr5b_13 | IDT (for STARmap) | CCTAGCTGCCAACGTCGCCCATAATGTTATCTT |
| Htr5b_14 | IDT (for STARmap) | AGGATAGTCACCAGAACTAGCTAATGTTATCTT |
| Oprm1_01 | IDT (for STARmap) | /5Phos/ACATTAGCATGATGAAGGCGAAGATGAATTATTAATGAAGCATACACTAAAGATA |
| Oprm1_02 | IDT (for STARmap) | /5Phos/ACATTACTTGAGTCGTAAGATCATCAGTAATTATTAATGAAGCATACACTAAAGATA |
| Oprm1_03 | IDT (for STARmap) | /5Phos/ACATTAACCCCTGCCTGTATTTTGTGGTAATTATTAATGAAGCATACACTAAAGATA |
| Oprm1_04 | IDT (for STARmap) | /5Phos/ACATTACCGTGGAGGGGTGTTCCCTAGTAATTATTAATGAAGCATACACTAAAGATA |
| Oprm1_11 | IDT (for STARmap) | ACACAGTGATGATGAGGACCTAATGTTATCTT |
| Oprm1_12 | IDT (for STARmap) | GGAGCCCGACAGCATGCGGACATAATGTTATCTT |
| Oprm1_13 | IDT (for STARmap) | AGAACGTGAGGGTGCAATCTATTAATGTTATCTT |
| Oprm1_14 | IDT (for STARmap) | GGTTAGTTCGATCCACTGTATTTAATGTTATCTT |
| Slc17a7_01 | IDT (for STARmap) | /5Phos/ACATTAGACGTAAAGAAGCGCCTCCAAATTATTACTGAAGCATACACTAAAGATA |
| Slc17a7_02 | IDT (for STARmap) | /5Phos/ACATTAACATTATGTGACGACTGCGCAATTATTACTGAAGCATACACTAAAGATA |
| Slc17a7_03 | IDT (for STARmap) | /5Phos/ACATTATTCAAAGTAGGCGGGCTGAGAGAATTATTACTGAAGCATACACTAAAGATA |
| Slc17a7_04 | IDT (for STARmap) | /5Phos/ACATTAAGGGTGGAGGTAGCCACAATGGAATTATTACTGAAGCATACACTAAAGATA |
| Slc17a7_11 | IDT (for STARmap) | ATGATGGCATAGACGGGCATTAATGTTATCTT |
| Slc17a7_12 | IDT (for STARmap) | AGCTTTCGCACGTTGGTAGTTAATGTTATCTT |
| Slc17a7_13 | IDT (for STARmap) | GCTGATCTCAAAGCCGAACACTTAATGTTATCTT |
| Slc17a7_14 | IDT (for STARmap) | GCTGCTGAAGGGATCAACATGTTAATGTTATCTT |
| Sst_01 | IDT (for STARmap) | /5Phos/ACATTACCATTGCTGGGTTCGAGTTGAATTATTAGTGAAGCATACACTAAAGATA |
| Sst_02 | IDT (for STARmap) | /5Phos/ACATTACCAGAAGAAGTTCTTGCAGCAATTATTAGTGAAGCATACACTAAAGATA |
| Sst_03 | IDT (for STARmap) | /5Phos/ACATTACCAGTTCCTGTTTCCCGGTGAATTATTAGTGAAGCATACACTAAAGATA |
| Sst_04 | IDT (for STARmap) | /5Phos/ACATTAGACGGAGTCTGGGGTCCGAGGGAATTATTAGTGAAGCATACACTAAAGATA |
| Sst_11 | IDT (for STARmap) | AGCTTTGCGTTCCCGGGGTTAATGTTATCTT |
| Sst_12 | IDT (for STARmap) | CTAACAGGATGTGAATGTCTTAATGTTATCTT |
| Sst_13 | IDT (for STARmap) | AGCTCTGCCAAGAAGTACTTTAATGTTATCTT |
| Sst_14 | IDT (for STARmap) | CGCCAGAGACTTCTGCAGAAATAATGTTATCTT |
| Sstr4_01 | IDT (for STARmap) | /5Phos/ACATTAAAGCACTGCGCGACACAGCAAATTATTATTGAAGCATACACTAAAGATA |
| Sstr4_02 | IDT (for STARmap) | /5Phos/ACATTAGCACGAGCCTAGTGATCTTCAATTATTATTGAAGCATACACTAAAGATA |
| Sstr4_03 | IDT (for STARmap) | /5Phos/ACATTAGATGACCAGGGCGTTTCCCAAATTATTATTGAAGCATACACTAAAGATA |
| Sstr4_04 | IDT (for STARmap) | /5Phos/ACATTAAGAACCGGGAGTAGGAAGCCCAAATTATTATTGAAGCATACACTAAAGATA |
| Sstr4_11 | IDT (for STARmap) | ATGTTCAGGCCGTCCACGCTAATGTTATCTT |
| Sstr4_12 | IDT (for STARmap) | AAAGACGGTCACCACCATTTAATGTTATCTT |
| Sstr4_13 | IDT (for STARmap) | TGGCATAGCGTAGGATCACGTAATGTTATCTT |
| Sstr4_14 | IDT (for STARmap) | AGCAGGTAGCATAATCCGATGGTAATGTTATCTT |
| Syt10_01 | IDT (for STARmap) | /5Phos/ACATTAGTCGTCTTTTCTGTTGCCCTAATTATTAATGAATCATACACTAAAGATA |
| Syt10_02 | IDT (for STARmap) | /5Phos/ACATTATTCGACACACGCCTATGACCTAATTATTAATGAATCATACACTAAAGATA |
| Syt10_03 | IDT (for STARmap) | /5Phos/ACATTATTTCCATACTGTGGCTTCCCTAATTATTAATGAATCATACACTAAAGATA |
| Syt10_04 | IDT (for STARmap) | /5Phos/ACATTACTCGTTCCAATGGTCTCTCCCAAATTATTAATGAATCATACACTAAAGATA |
| Syt10_11 | IDT (for STARmap) | AGTTTCCCACAGGTCTTGATAATGTTATCTT |
| Syt10_12 | IDT (for STARmap) | AGACCCTCAGCGTCTAGTCCTAATGTTATCTT |
| Syt10_13 | IDT (for STARmap) | TCTGTGGTAGCACAGTGGATATAATGTTATCTT |
| Syt10_14 | IDT (for STARmap) | TGGCTTTCGGTGATAGGCCAGCTAATGTTATCTT |
| Tac1_01 | IDT (for STARmap) | /5Phos/ACATTAGCGATTCTCTGCAGAAGATGAATTATTACTGAATCATACACTAAAGATA |
| Tac1_02 | IDT (for STARmap) | /5Phos/ACATTACAAAGAACTGCTGAG GCTTGAATTATTACTGAATCATACACTAAAGATA |
| Tac1_03 | IDT (for STARmap) | /5Phos/ACATTAATCGCGCTTCTTTCATAAGCAATTATTACTGAATCATACACTAAAGATA |
| Tac1_04 | IDT (for STARmap) | /5Phos/ACATTACCCATTAGTCCAACAAAGGAATAATTATTACTGAATCATACACTAAAGATA |
| Tac1_11 | IDT (for STARmap) | GCTGAGGCTTGGGTCTTCGTAATGTTATCTT |
| Tac1_12 | IDT (for STARmap) | ATCCCGCTTGCCCATTAATTAATGTTATCTT |
| Tac1_13 | IDT (for STARmap) | ACGTCTTCTTTCGTAGTTCTTAATGTTATCTT |
| Tac1_14 | IDT (for STARmap) | GCCACAGAATTTAAAGCTCTTTTAATGTTATCTT |
| Tac2_01 | IDT (for STARmap) | /5Phos/ACATTAAGCCGCAAACAGCATGGCGCAATTATTAGTGAATCATACACTAAAGATA |
| Tac2_02 | IDT (for STARmap) | /5Phos/ACATTACGGTGGGAGTGTCTGGTTGGCAATTATTAGTGAATCATACACTAAAGATA |
| Tac2_03 | IDT (for STARmap) | /5Phos/ACATTATGTCACGTTTCTGTGGAAGTGAATTATTAGTGAATCATACACTAAAGATA |
| Tac2_04 | IDT (for STARmap) | /5Phos/ACATTATCTCCGAAGCAGGGACGGAGGCAATTATTAGTGAATCATACACTAAAGATA |
| Tac2_11 | IDT (for STARmap) | AGCCAAGCTGAGGGCGAGGATAATGTTATCTT |
| Tac2_12 | IDT (for STARmap) | GGGGTGTTCTCTTCAACCACTAATGTTATCTT |
| Tac2_13 | IDT (for STARmap) | AGTCCCACAAAGAAGTCGTGTAATGTTATCTT |
| Tac2_14 | IDT (for STARmap) | AGAGACAGGGCGGCTGTCGTAGTAATGTTATCTT |
| Th_01 | IDT (for STARmap) | /5Phos/ACATTAGAAGTGAGACACATCCTCCAAATTATTATTGAATCATACACTAAAGATA |
| Th_02 | IDT (for STARmap) | /5Phos/ACATTACTCGAATACCACAGCCTCCAAATTATTATTGAATCATACACTAAAGATA |
| Th_03 | IDT (for STARmap) | /5Phos/ACATTAGGCTTCAAATGTCTCAAACACTAATTATTATTGAATCATACACTAAAGATA |
| Th_04 | IDT (for STARmap) | /5Phos/ACATTAGAGGCATAGTTCCTGAGCTTGTAATTATTATTGAATCATACACTAAAGATA |
| Th_11 | IDT (for STARmap) | AAGCCAGTCCGTTCCTTCATAATGTTATCTT |
| Th_12 | IDT (for STARmap) | ACAGCATTTCCATCCCTCTTAATGTTATCTT |
| Th_13 | IDT (for STARmap) | CCGGGTCTCTAAGTGGTGGATTTAATGTTATCTT |
| Th_14 | IDT (for STARmap) | ACAGAGAATGGGCGCTGGATACTAATGTTATCTT |
| Mus-Calb1-B4P1 | IDT (for ISH) | cactctcaaactagccgctgcaccacgatggcagaatcccacctgcagtcat |
| Mus-Calb1-B4P2 | IDT (for ISH) | agaaaagtagatatcgtgaatcattatagccgaaggactctaataaaaattt |
| Mus-Calb1-B4P3 | IDT (for ISH) | aatagtaccaagtttggcattcttactaatagtagtagaacaaatcctataa |
| Mus-Calb1-B4P4 | IDT (for ISH) | cgccgcgcccagctcagcctgcgcagccctctcgcccgaggttcgcgctccg |
| Mus-Calb1-B4P5 | IDT (for ISH) | aagactgtggatgatacaaaactagcagagtacacagacctcatgctgaaac |
| Mus-Calb1-B4P6 | IDT (for ISH) | ccagtgcaggaaaatttccttcttaaattccagggaatcaaaatgtgtggga |
| Mus-Calb1-B4P7 | IDT (for ISH) | ggagggaagctgtaccgaacagaccttgctcttattctttctgctggagaca |
| Mus-Calb1-B4P8 | IDT (for ISH) | tagagttggtgaccacaaccacttgctagtgatacattgtatctaaaaccat |
| Mus-Calb1-B4P9 | IDT (for ISH) | aaatatcaacagttaattatggctttattctgaaacgatctccctagagatt |
| Mus-Calb1-B4P10 | IDT (for ISH) | atgcctatatttccaagaagtctactgccagagagtatgaccatagcccatt |
| Mus-Calb1-B4P11 | IDT (for ISH) | ctaaattattttcatgtgttccagatgacaattattctagtaaactgctgtt |
| Mus-Calb1-B4P12 | IDT (for ISH) | tatttcatcaaaacttgtgtattctgtggattctatggttcatattgagatc |
| Mus-Calb1-B4P13 | IDT (for ISH) | aggtttgggtcaggttggatttaagcactttttttcaattgttgttcataaa |
| Mus-Calb1-B4P14 | IDT (for ISH) | atttgtttctaccccaaagtgttaattgtcatgtaatctgttatcaattagg |
| Mus-Calb1-B4P15 | IDT (for ISH) | gcgcgaaagaaggctggattggagctatcaccggaaatgaaatcctttgtgg |
| Mus-Calb1-B4P16 | IDT (for ISH) | caatatggacagagagatgatggaaaaataggaattgtagagttggctcacg |
| Mus-Calb1-B4P17 | IDT (for ISH) | ttacccacagaagagaatttcttgctgctctttcgatgccagcaactgaagt |
| Mus-Calb1-B4P18 | IDT (for ISH) | atcgaaaccgaggaacttaagaactttctaaaggacctactagagaaagcaa |
| Mus-Calb1-B4P19 | IDT (for ISH) | tttgattcaaataatgacggaaagctggaactgacagagatggccaggttac |
| Mus-Calb1-B4P20 | IDT (for ISH) | gagttcaataaggcttttgagttatatgatcaggatggcaacggatacatag |
| Mus-ChAT-B5P1 | IDT (for ISH) | gcgtccaacgaggatgaacgcctgcctccaatcggcctgctgacgtcagacg |
| Mus-ChAT-B5P2 | IDT (for ISH) | gctaggatgcctatcctggaaaaggtccccccaaagatgcctgtacaagctt |
| Mus-ChAT-B5P3 | IDT (for ISH) | gaatactggctgaatgacatgtatctaaacaaccgcctggccctgccagtca |
| Mus-ChAT-B5P4 | IDT (for ISH) | cgccgtctcagtgagggtgatctgttcactcagttgagaaagatagtcaaaa |
| Mus-ChAT-B5P5 | IDT (for ISH) | atgagagacctctgtagttcgaggcagcctgctgaaggcaagccaccaacag |
| Mus-ChAT-B5P6 | IDT (for ISH) | aaggaaagagctagaggcccaaccaagccaagcaatcttgactactcccact |
| Mus-ChAT-B5P7 | IDT (for ISH) | ggtctttagaaacttaacctttctgcttctttcccagcaacacccagtggtg |
| Mus-ChAT-B5P8 | IDT (for ISH) | gcgtaacagcccaggagagcaggtcggcagctctgctactctggattaagaa |
| Mus-ChAT-B5P9 | IDT (for ISH) | agctgtgaggaggtgctggacttacctaagttgccagtgcccccactgcagc |
| Mus-ChAT-B5P10 | IDT (for ISH) | aggaagagccaggccattgtgaagcggtttggggcccctggtggcctgggtg |
| Mus-ChAT-B5P11 | IDT (for ISH) | tctagccctgctgtgatctttgctcggcagcacttccaagacaccaatgacc |
| Mus-ChAT-B5P12 | IDT (for ISH) | ctaaggtttgcagccagcctcatctctggtgtgcttagctacaaggctctgc |
| Mus-ChAT-B5P13 | IDT (for ISH) | ggcactggagacctcagtgacacacacagggccctccagctccttcatggtg |
| Mus-ChAT-B5P14 | IDT (for ISH) | ggctgcagcttgaatggagcgaatcgttggtatgacaagtccctgcagtttg |
| Mus-ChAT-B5P15 | IDT (for ISH) | aagaagctcgtccgagctgactcagtgagtgaactccctgctcccagaaggc |
| Mus-ChAT-B5P16 | IDT (for ISH) | gccctccagctggcttactacaggctttaccagaggctggtgcccacctatg |
| Mus-ChAT-B5P17 | IDT (for ISH) | tacacagtcatggccataaccggcatggccattgacaaccatcttctggcac |
| Mus-ChAT-B5P18 | IDT (for ISH) | tataacccccagcctgaggccatcaccttctgcatctccagctttcacggct |
| Mus-ChAT-B5P19 | IDT (for ISH) | gcttgttgctgctcccctatccttgggggctcacatgaagctggcatgttaa |
| Mus-ChAT-B5P20 | IDT (for ISH) | atgagacccagcctggcttggaagcagcctgggtgggctgggagctccctct |
| Mus-Tac1-B2P1 | IDT (for ISH) | gtgcgcacctgcggagcatccccgcggtctgaccgcaaaatcgaacatgaaa |
| Mus-Tac1-B2P2 | IDT (for ISH) | aatcgatgccaacgatgatctaaattattggtccgactggtccgacagtgac |
| Mus-Tac1-B2P3 | IDT (for ISH) | gatcaaggaggcaatgccggagccctttgagcatcttctgcagagaatcgcc |
| Mus-Tac1-B2P4 | IDT (for ISH) | tttaaattctgtggcttatgaaagaagcgcgatgcagaactacgaaagaaga |
| Mus-Tac1-B2P5 | IDT (for ISH) | ataatgtactgagacttcggtatttgactctatttgtatcctagcagcatgt |
| Mus-Tac1-B2P6 | IDT (for ISH) | ctctcacaaaaggcataaaacagattcctttgttggactaatgggcaaaaga |
| Mus-Tac1-B2P7 | IDT (for ISH) | taaataaacccctgaacgcactatctattcatcttcatctgtgtcagtgagc |
| Mus-Tac1-B2P8 | IDT (for ISH) | gtaatttcagcaaagcacagtgatgaaggagctgtccaagcttggcagtgac |
| Mus-Tac1-B2P9 | IDT (for ISH) | gcagagactcctgtgcgtctctctcacgctacccctggttctgctttcatgc |
| Mus-Tac1-B2P10 | IDT (for ISH) | cctgtttcgtgactatatagagatgttttgaaaaagtttcaatgtaattctc |
| Mus-Tac1-B2P11 | IDT (for ISH) | aagacccaagcctcagcagttctttggattaatgggcaagcgggatgctgat |
| Mus-Tac1-B2P12 | IDT (for ISH) | cggacctgctccgctcctgcaccgcggccaaggagagcaaagagcgcccagc |
| Mus-Tac1-B2P13 | IDT (for ISH) | cctcgtggccgtggcggtcttttttctcgtttccactcaactgtttgcagag |
| Mus-Tac1-B2P14 | IDT (for ISH) | ctcagttgaaaaacaagtggccctgttaaaggctctttatggacatggccag |
| Mus-Tac1-B2P15 | IDT (for ISH) | tgaacggtaaaataaaatgtgcgctatgaggaatgattatttatttaataac |
| Mus-Tac1-B2P16 | IDT (for ISH) | ccaataagccttgtaattctaatgtggtgacctccccagaagtagaaattag |
| Mus-Tac1-B2P17 | IDT (for ISH) | atgttgttgtgagtgaaaaactcaaaaaagaagtgtttattttttcatattg |
| Mus-Tac1-B2P18 | IDT (for ISH) | agtctccaaagaaaggacccttctgtgagccagcgcaggcagctgctgctgg |
| Mus-Tac1-B2P19 | IDT (for ISH) | gtcttcagtcattgtatgatgtgttgtgatagctaccattttaaataaaaga |
| Mus-Th-B1P1 | IDT (for ISH) | ctcctcagttctgtgcgtcgggtgtctgacgatgtgcgcagtgccagagagg |
| Mus-Th-B1P2 | IDT (for ISH) | gtactggacagtcctcacaccatccggcgctccttagagggggtccaggatg |
| Mus-Th-B1P3 | IDT (for ISH) | tcagagcaggataccaagcaggccgaggctgtcacgtccccaaggttcattg |
| Mus-Th-B1P4 | IDT (for ISH) | cggcggcagagtctcatcgaggatgcccgcaaggagcgggaggcagcagcag |
| Mus-Th-B1P5 | IDT (for ISH) | gcagcagcggctgcggtagcctccgcggaacctgggaacccattggaggctg |
| Mus-Th-B1P6 | IDT (for ISH) | gtattcgaggagagggatggaaatgctgttctcaacctgctcttctccttga |
| Mus-Th-B1P7 | IDT (for ISH) | gccaaaatccaccacttagagacccggcctgcccagaggccactggcaggaa |
| Mus-Th-B1P8 | IDT (for ISH) | ccccacctggagtactttgtgcgcttcgaggtgcccagtggcgacctggctg |
| Mus-Th-B1P9 | IDT (for ISH) | aaggttccctggttcccaaggaaagtgtcagagttggataagtgtcaccacc |
| Mus-Th-B1P10 | IDT (for ISH) | gaggtatacgccacgctgaagggcctctatgctacccatgcctgccgggaac |
| Mus-Th-B1P11 | IDT (for ISH) | cgacccgtggccggtctactgtctgcccgtgattttctggccagtctggcct |
| Mus-Th-B1P12 | IDT (for ISH) | cgtgtgtttcagtgcacacagtacatccgtcatgcctcctcacctatgcact |
| Mus-Th-B1P13 | IDT (for ISH) | tatggagagctcctgcactccctgtcagaggagcccgaggtccgggcctttg |
| Mus-Th-B1P14 | IDT (for ISH) | gtcaccaagtttgaccctgacctggacctggaccatccgggcttctctgacc |
| Mus-Th-B1P15 | IDT (for ISH) | gcgtatcgccagcgccggaagctgattgcagagattgccttccaatacaagc |
| Mus-Th-B1P16 | IDT (for ISH) | ctggaggctttccagcttctggaacggtactgtggctaccgagaggacagca |
| Mus-Th-B1P17 | IDT (for ISH) | Cccgagccagactgctgccacgagctgctgggacacgtacccatgttggctg |
| Mus-Th-B1P18 | IDT (for ISH) | ctgtgtaaacagaatggggagctgaaggcttacggtgcagggctgctgtctt |
| Mus-Th-B1P19 | IDT (for ISH) | ccagacacagcagccgtgcagccctaccaagatcaaacctaccagccggtgt |
| Mus-Th-B1P20 | IDT (for ISH) | cgtatccagcgcccattctctgtgaagtttgacccgtacaccctggccattg |
|
| ||
| Software and algorithms | ||
|
| ||
| MedPC-IV | MedAssociates | https://www.med-associates.com/med-pc-v/ |
| Viewer3 | BioObserve | http://www.biobserve.com/behavioralresearch/ |
| FIJI | OpenSource | https://ImageJ.net/ImageJ |
| MATLAB | Mathworks | https://www.mathworks.com/products/matlab.html |
| DeepLabCut | Mathis et al. (2018) | https://github.com/DeepLabCut |
| CNMF | Pnevmatikakis et al. (2016) | https://github.com/epnev/ca_source_extraction |
| SpikeGLX (and associated command-line tools: CatGT, TPrime, and C_Waves) | Bill Karsh (Janelia Research Campus) | https://billkarsh.github.io/SpikeGLX |
| Extracellular electrophysiology data processing pipeline | Jennifer Colonell (Janelia Research Campus) | https://github.com/jenniferColonell/ecephys_spike_sorting |
| Kilosort 2.5 | Pachitariu et al., (2016) | https://github.com/MouseLand/Kilosort |
| Phy | Cyrille Rossant (International Brain Laboratory) | https://github.com/cortex-lab/phy |
| Shield-2018 | Swaney et al., (2019) | https://hub.docker.com/r/chunglabmit/shield-2018 https://github.com/chunglabmit/shield-2018 |
| Elastix | Klein et al., (2010) | https://elastix.lumc.nl |
| Lasagna | OpenSerialSection (University College London) | https://github.com/SainsburyWellcomeCentre/lasagna |
| Atlas Electrophysiology | International Brain Laboratory; Liu et al., (2021) | https://github.com/int-brain-lab/iblapps/tree/master/atlaselectrophysiology |
| AllenCCF | University College London | https://github.com/cortex-lab/allenCCF |
| Computation-through-dynamics (LFADS and fixed point analysis) | Pandarinath et al. (2018b); Sussillo and Barak (2013) | https://github.com/google-research/computation-thru-dynamics |
|
| ||
| Deposited data | ||
|
| ||
| STARmap data | NeMO Archive | asset.nemoarchive.org/dat-9ACQ8G2 |
| Electrophysiology data | DANDI Archive | N/A |
TMP injections
For Calb1-Cre animals, Cre was under the control of trimethroprim (TMP). To induce Cre, animals were given an i.p. injection of TMP (15 mg/kg). To prepare TMP, a stock solution of 100 mg/ml in DMSO was diluted 1:10 in sterile saline and immediately injected into the animal. All TMP injections were made at least one week after surgery.
Tissue clearing
For hydrogel-based cleared tissue experiments, Tac1-Cre, TH-Cre, Calb1-Cre, or ChAT-Cre animals were injected with AAV1-DIO-YFP virus, allowed to recover for 3 weeks, deeply anesthetized with Beuthanasia, and transcardially perfused with 1% hydrogel solution. Brain tissue was removed and post fixed in hydrogel solution overnight at 4C. Dissolved oxygen was removed under vacuum and samples were incubated at 37C for 5 h in degassed hydrogel solution. Tissue was transferred to clearing solution and incubated at 37°C until clear (approx. 2–3 weeks). Clearing solution was refreshed daily for the first week, then every other day until clear. For refractive index matching, tissue was transferred to 25% glycerol overnight, then 50% glycerol for 1–3 h, followed by 65% glycerol until transparent (approx. 2 h). Images were acquired on a Leica SP5 Confocal microscope using a 10× objective. Image planes were acquired at 5 μm intervals. Three dimensional renderings were generated by Imaris Software.
CLARITY hydrogel solution
| Stock Solution | Volume | Final Concentration |
|---|---|---|
|
| ||
| Acrylamide (40%) | 10 mL | 1% |
| Bis (2%) | 2.5 mL | 0.00625% |
| VA-044 Initiator | 1g | 0.25% |
| 10X PBS | 40 mL | 1× |
| 16% PFA | 100 mL | 4% |
| H2O | 247.5 mL | – |
SDS clearing solution
| Stock Solution | Volume | Final Concentration |
|---|---|---|
|
| ||
| Borate Buffer (5M) | 1 L | 1M |
| SDS (20%) | 1 L | 4% |
| H2O | 3 L | – |
Animal behavior
In preparation for behavioral testing, animals were transferred to reverse light cycle rooms at least 1 week prior to testing. Animals were handled a minimum of 5 days prior to the onset of behavior training. Mice were acclimated to sucrose either by replacing cage water with a 4% sucrose solution, or by allowing animals already under water restriction temporary access to a 10% sucrose solution until animals consumed their day’s allotment of water. Animals were weighed and water restricted prior to the onset of training and weight was monitored to ensure animals did not lose >20% of body weight. For 3CSRTT training, operant conditioning boxes from Med Associates were fit with a custom 3D printed reward wall so as to minimize the contact between the fiber optic emanating from the animal’s skull and the wall of the chamber when then animal was consuming the water reward. The standard Med Associates Five-Choice Serial Reaction Task was modified to include a mid-box beam break to start trials since animals consumed the water reward for a variable duration. In addition to the measurements acquired in the standard protocol, premature poke latency and reward consumption duration were also recorded.
At the onset of training, animals were first acclimated to the operant box for 15 min on Day 1, during which time the house lights were extinguished and the nose pokes were covered. On Day 2, house lights were illuminated and free sucrose water rewards were delivered at variable intervals in the reward port and the reward port light was illuminated until 500 ms after head entry into the reward port. When animals retrieved water rewards with low latency (<5s), nose pokes were uncovered and all three nose poke cues were illuminated. Animals were required to poke into any lit nose poke to retrieve a reward in the reward port (nonspatial training). When a mouse completed 30 trials in 60 min, it was moved onto Stage 1 of the training regimen, from which point onward only one nose poke was active per trial. When animals met the criterion indicated below, they were moved to the next training stage. For most sessions, animals performed 100 trials, or 60 min, whichever came first. In cases with poor performance, additional trials were run to enable animals to reach their daily quota for water to maintain weight. If additional water was required to maintain weight, the remaining water was delivered >1 h after the end of the training session.
Stage parameters
| Stage | Cue Duration (s) | Limited Hold (s) | ITI |
|---|---|---|---|
|
| |||
| Stage 1 | 30 | 30 | 3 |
| Stage 2 | 20 | 20 | 3 |
| Stage 3 | 10 | 10 | 3 |
| Stage 4 | 5 | 5 | 3 |
| Stage 5 | 2 | 5 | 2,3,4 |
| Stage 6 | 1 | 5 | 2,3,4 |
Advancement criteria
| Stage | %Correct | % Accurate | % Omitted |
|---|---|---|---|
|
| |||
| Stage 1 | 30 | – | – |
| Stage 2 | 40 | – | – |
| Stage 3 | 50 | – | – |
| Stage 4 | 50 | 80 | – |
| Stage 5 | 50 | 80 | 20 |
| Stage 6 | 50 | 80 | 20 |
Elevated plus maze
Animals previously trained on the 3CSRTT were placed in an elevated plus maze, containing 4 arms 35cm in length and 5cm in width. Two arms contained walls 20cm high (closed arms). The entire apparatus was elevated 60cm off the floor. Animals were placed at the center of maze to begin and fiber photometry was recorded for 10 min while the animals explored the maze. Fiber photometry data and video (10 fps) were acquired with a TDT RZ5 using Synapse software.
Real time place preference
A custom clear acrylic arena (44 cm × 24 cm × 27cm) was divided into two chambers with unique flooring and wall patterns in each chamber. The arena was recorded from above at 20Hz by a commercially available USB webcam (https://www.elpcctv.com/ELP-USBFHD06H-SFV(2.8-12)). For optogenetic stimulation, mice with fiber optic implants (ThorLabs, CFM32L20) were connected to either a yellow (593nm) or blue (473nm) laser (OptoEngine LLC) via a patch cord (Thorlabs, TT200R5F1B and BFY32FL1). On the day prior to experimentation, mice were acclimated to the arena for 20 min while attached to the fiber optic rotary joint patch cord. The Track-Control toolkit, an open-source object detection and closed loop feedback toolbox run through Python (Zhang et al., 2019) was used to manually assign the stimulated chamber at the start of each experiment, and track the centroid of the mouse. When the centroid of the mouse crossed into the stimulated chamber, an Arduino microcontroller (www.arduino.cc) drove continuous yellow laser stimulation at 15mW or blue light stimulation at 20Hz (20% duty cycle), measured at 15mW when continuous. Pulsed blue light stimulation was achieved using a master 8 programmable pulse generator (A.M.P.I). Stimulation ceased when the animal returned to the non-stimulated chamber. Experiments were run without user interference and automatically terminated after 20 min.
Fiber photometry
Surgical procedures
For fiber placement for MHb-targeted fiber photometry experiments, AAV1-EF1a-DIO-GCaMP6f was injected in the MHb as described above and a fiber optic cannula (Doric Lenses, MFC_400/430–0.48_3.5mm_MF2.5(8 mm)_A45) was placed above the habenula (AP: −1.4, ML: +/−0.5, DV: 2.85–2.95). Tips of the angled fiber optics were positioned on the lateral side of the target location. Cannulas were cemented in place with Metabond (Parkell) and animals were allowed to recover for at least 2 weeks. Of note, we found some serotype tropism for medial or lateral subregions of the habenula, and use AAV1 for all MHb-targeted injections. (Figure 3H). For LHb-targeted injections, AAV8-EF1a-DIO-GCaMP6f was injected in the LHb at AP: −1.4, ML: +/− 0.45, DV: −3.05 and a fiber optic cannula (Doric Lenses, MFC_400/430–0.48_3.5mm_MF2.5(8 mm)_A45) was placed above the habenula (AP: −1.4, ML: +/− 0.55, DV: 2.85–2.95).
Recording
Fiber photometry signals were recorded with a fiber optic patch cord (Doric Lenses, MFP_400/430/1100–0.48_5m_FC-MF2.5) coupled to the animals with a ceramic or brass sleeve. GCaMP was illuminated with LEDs emitting 405 and 490nm light, modulated with a lock-in amplifier, using the optical setup described (Lerner et al., 2015). Fluorescence was captured by a photodetector and processed with an RZ50 digital acquisition system (TDT Instruments) and Synapse Software (TDT instruments). Photometry data was synced to behavioral data using TTL outputs from Med Associates operant boxes, including trial starts, nose pokes, and entry into the reward port. Photometry data was analyzed offline using MATLAB. We would like to note that due to the low frequency of withheld:-withheld trial types, data is pooled from stages 4–6 during training for Figure S4B–E.
Reward history-guided decision-making
Surgical procedure
Mice were anesthetized with isoflurane and stereotaxic injections were used to deliver 2 viruses bilaterally. In the medial habenula, (AP: −1.4mm, ML +/− 0.3mm, DV, −3.05mm) 200–300 nL of AAV1-nEF-CreONFlpOFF-eNpHR3.0-YFP at 5 × 1012 was delivered at a rate of 10 nL/min and 200nL of AAV8-Ef1a-FlpO at 7 × 1011 was delivered to the lateral habenula (AP: −1.4mm, ML +/− 0.55mm, DV, −3.05mm). A dual core fiber optic cannula (ThorLabs CFM32L20) was cut to 3.5mm in length and inserted at AP −1.4 to a depth of DV −2.7. The fiber optic cannula and a custom headbar were cemented in place and the animal was allowed to recover.
Behavioral training
Two weeks after surgery, animals were water restricted and acclimated to head fixation. Behavioral shaping included several stages. Animals first learned to lick to both ports to receive water and rewarded licks resulted in an auditory cue. Next, a 1s pre-cue period was imposed during which all licks resulted in a premature termination of the trial. Upon auditory cue onset, licks at either port resulted in a water reward. After premature licking subsided, animals were forced to alternate between both lick ports in a block structure to reduce lick side bias. To encourage switching between sides in animals will side biases, small (2ul) rewards were delivered at the “correct” port after an “incorrect” port lick. After behavioral shaping, animals were moved to a block structure task. In each block, one lick port had a reward probability of 0.9 and the other a reward probability of 0.1. Blocks switched when animals had performed a minimum of 15 +/− 3 “correct” licks at the high preward port. For the first 3 block switches in a session on a given day, a small reward was delivered on the new high preward port to prevent perseverative behavior toward one lick port. These first three block switches were not included in the analysis.
Optogenetic inhibition
Continuous 594 nm light was delivered bilaterally through a dual core patch cord (ThorLabs BFY32FL1) at 15mW combined power across both bilateral fibers. A subset of block switches were illuminated. On those blocks, any rewarded trials on trials 1–15 after the block switch were stimulated for 2s after the first rewarded lick.
Histology
Procedure
Following fiber photometry, optogenetic inhibition, and DIO-YFP experiments (Cre line validation and Serotype Testing), animals were deeply anesthetized, transcardially perfused with 4% PFA and postfixed overnight at 4C. Brains were sectioned at 50 μm thickness near the site of cannula placement, permeabilized in 0.1% TX100, and incubated overnight at 4C in anti-GFP antibody (ThermoFisher Cat # A-21311 or Cat# A-31852; 1:3000) to stain GCaMP expressing neurons. Sections were washed in PBS with 0.1% TX100 and imaged on a Leica SP5 Confocal Microscope.
Endoscopic two-photon calcium imaging
Surgery
Mice were anesthetized with 5% isoflurane and then maintained at surgical plane with 1–2% isoflurane. After betadine and ethanol cleaning, skin covering the dorsal aspect of the skull was made, and a 2 mm craniotomy was made above the injection side. After bleeding was stopped, a 36 gauge beveled needle attached to a nanoinjector Hamilton syringe was inserted into the medial habenula [+0.3 M/L, −1.4 A/P, −2.95 D/V]. 300 nL of AAV1-Ef1a-DIO-H2B-GCaMP6f was injected slowly (25 nL/min) (#3560 from Stanford Viral Vector core, 1.2 × 1012, undiluted). Ten minutes after injection ended, the syringe was removed. A 600 μm GRIN (Inscopix) was slowly (100 mm/min) inserted above the injection site [+0.3 M/L, −1.4 A/P, −2.60 D/V], and then cemented to the skull. A custom made head bar was cemented to the skull before covering the lens with wax paper and then Kwik-Cast (World Precision Instruments) for protection. Animals recovered in a warmed cage and then were individually housed. Two weeks later, animals that showed potential GCaMP expression were trained on this behavior.
Head-fixed behavior
A head-fixed behavioral setup was constructed using Bpod (Sanworks) and custom-fabricated parts. Each trial began with a 0.5 s pre cue period, in which a lick aborted the trial to the next ITI. If no premature licks occurred, a cue light was illuminated for 1 s, during which the animal could respond by licking an optical lickport (Sanworks). Timely response produced a 10–15 ul reward, which was followed by a 7–9 s ITI.
Imaging
Two photon imaging was conducted on a standard Neurolabware two photon microscope (8KHz resonance scanning, 512 lines per frame at 30 Hz/volume) tuned to 920 nm with a 16× Nikon dipping objective (NA = 0.8, WD – 3.0). Three planes were captured per imaging session. Imaging frames were aligned to trial start via TTL pulse.
Electrophysiology
Surgery and head-fixed behavior
The surgical procedure and the head-fixed behavior were mostly similar with those for two-photon calcium imaging experiments. For ChRmine expression, 300 nL of AAV1-nEF-DIO-ChRmine-oScarlet at 2 × 1012 vg/ml was injected to MHb [AP −1.4 mm, ML +/−0.3 mm, DV −2.95 mm]. For MHb-specific ChRmine expression, 300 nL of AAV1-nEF-CreOnFlpOff-ChRmine-oScarlet at 3 × 1012 vg/ml was injected to MHb, and 300 nL of AAV8-Ef1a-FlpO at 1 × 1012 vg/ml was injected to LHb [AP −1.4 mm, ML +/− 0.5 mm, DV −2.95 mm]. Injection speed was 25 nL/min and we waited at least 10 min post-injection before retracting syringes. A custom made headplate was cemented to the skull, and a reference electrode (ED1058-ND, Digi-key) was implanted away from the injection sites to just touch the brain surface. Then the rest of the skull was covered by “clear-skull cap”, i.e., layers consisting of cyanoacrylate adhesive (Krazy Glue) and clear nail polish (72,810, Electron Microscopy Sciences) (Zengcai Guo et al., Neuron, 2014; Allen, Kauvar et al. Neuron, 2017). Upon recovery for at least 3 days, the animals were water-restricted for at least 5 days before training onset. Then we trained the animals with 6 ul of plain water per rewarded trial. The durations of pre-cue, cue, and ITI epochs were 3, up to 2, and 4–6 s, respectively. Licking was detected by either optical (Sanworks) or electrical (comparator circuit, Janelia Research Campus) lickport.
In vivo electrophysiology
We used Neuropixels 2.0 probes (Steinmetz et al., 2021) lti-probe manipulator system (New Scale Technologies) and controlled by the SpikeGLX software (Janelia Research Campus), for the acute in vivo extracellular electrophysiological recordings acquired at 30 kHz. Prior to recording sessions, we performed craniotomy and durotomy, and the craniotomies were kept moist by frequent application of PBS and were covered by silicone elstomers (Qwik-Cast, World Precision Instruments) between recordings. For MHb-targeted recordings, a relatively large craniotomy (~2.5 mm diameter) was made on top of the midline at AP −1.4 mm. A 4-shank probe was inserted along an oblique (~10 deg) direction at slow speed (down to 200 μm/min near the target depth). While we typically targeted the probe tip to ~3500 μm from the brain surface, we decided the final depth on-the-fly by inferring the probe position relative to brain structures using the following heuristics. First, dentate gyrus shows high LFP signals and scarce spikes, and the spiking structure right below is habenula. Second, because MHb is right next to the third ventricle with no spikes, by examining overall spike rates of the individual shanks (separated ~250 μm from each other) approximate mediolateral position could be inferred. The probe was lowered ~100 μm past the final depth and then retracted to the final depth. Then we waited at least 15 min prior to recording for stabilization. The sites at the bottom ~700 μm of all shanks were used for recording. For septum-targeted recording, a small craniotomy (~1 mm) was made right next to the midline at AP +0.5 mm. Either a single- or 4-shank probe was inserted in a similar manner, typically targeting ~5000 μm from the brain surface. Bottom ~3000 μm of one shank was used for recording. The probes were cleaned with trypsin (Sigma-Aldrich) between recording sessions.
Transcranial optogenetics
For MHb recordings combined with transcranial optogenetics using ChRmine, we used a fiber-pigtailed 637 nm diode laser (OBIS, Coherent) driven by a voltage input (Bpod analog output module, Sanworks). The laser light was guided by a relayed 400 μm core, 0.39 NA optical fiber (M82L01, Thorlabs) positioned several millimeters above the skull by another manipulator arm. The space between the fiber tip and the skull was filled with PBS to improve coupling of stimulation light. Square pulses with ~800 mW/mm2 peak irradiance (at the fiber tip) and varying temporal parameters were used during or after behavioral recordings for perturbation and optotagging experiments, respectively. For transient perturbation, we used 10 ms width pulses at 30 Hz for 0.5 s per trial. For optotagging, we used 30 trials of 10 ms width pulses preceded by at least 1 s no-light periods.
QUANTIFICATION AND STATISTICAL ANALYSIS
In situ hybridization
To determine overlap of Tac1, Th, Calb1, and ChAT gene expression, confocal images were acquired on a Leica SP8 using Alexa 488, 514, 555, and 647 fluorophores attached to HCR hairpins, as well as DAPI staining to stain all cells. Using a FIJI macro, all DAPI labeled cells were hand-annotated, and then the corresponding fluorescence channels were scored for the presence or absence of each mRNA transcript.
Elevated plus maze
Positional data was analyzed with BioObserve software. Fiber photometry signals were downstampled to the frame rate of the camera (10 fps). To generate heat maps, xy positions were binned every 1/3 cm and the mean of Z-scored photometry data was calculated for each bin.
Behavioral video analysis
Animals were recorded while performing a 3-Choice Serial Reaction Time Task using commercially available USB web cameras. Frames were captured at 30 Hz with 1280 × 720 pixel resolution. Video analysis was performed with the open source software DeepLabCut (Mathis et al., 2018), post experimentation. Approximately 5 percent of frames per video were extracted to represent the diversity of animal poses during behavior, and manually labeled for nose, the center of implant, the back of implant, center of body mass, and tail base. Extracted and labeled frames were used to train a neural network for >100,000 iterations. The trained network was then used to analyze and estimate poses for the remaining frames. The x and y coordinates generated for the center and back of implant were used to create a vector indicating the head angle and movement of the animal. Head angle and movement were then aligned to fiber photometry data to fluorescence peaks correlated to behaviorally relevant periods.
Fiber photometry
Fiber photometry data was collected using Synapse software (TDT Instruments). Timestamps were acquired from Med Associates operant training boxes (sampled at 10ms) and synced with the fiber photometry data in the Synapse Software. Raw photodetector measurements were processed offline in MATLAB (Lerner et al., 2015). Briefly, data from the control 405nm channel was downsampled to 60Hz and low pass filtered, and upsampled to the original sampling frequency (1017Hz). The smoothed control signal was fit to the GCaMP signal, and then subtracted from the GCaMP signal. The resulting value is reported as ΔF. For ΔF/F measurements, baseline rig fluorescence was measured without the animal subject and subtracted from the mean fluorescence during a session to determine the denominator F. For Z-score measurements, Z-scores were calculated using the entire behavioral session, from the start of the first trial to 10 s after the end of the last trial. For comparisons of timeseries across animals and genotypes or for visualization, we use Z-score to reduce animal to animal variability in viral expression. However, when reward statistics change across sessions (e.g., Figure 7K), we use ΔF/F to avoid Z-scoring artifacts. For within-animal, trial-wise comparisons, we use ΔF/F measurements.
Reward history-guided decision-making
To reduce behavioral performance variability across days, only sessions where animals choice for the high reward port increased to >0.5 for trials on 10–15 after the block switch (across both stimulated and unstimulated transitions). To analyze block transitions (Figure S4F), the fraction of trials in which animals chose the new high preward port was calculated for light block and no light blocks. In light blocks, only rewarded licks in the new high preward port receive light illumination (0–2s after the first lick). The first lick to the high preward port can vary across animals and blocks, thus, the time of the first illumination also varies. To determine the effect of inhibition during reward at trialt on the choice at trialt+1, the fraction of “stay” choices was calculated by taking all rewarded trials during trialst=1–15 of block switches and looking at the fraction of high preward port choices on trailt+1 (choices to stay). This was analyzed separately for trialt = no light and trialt = light. The response rate was calculated for this same set of trialst=1–15 by calculating the number of trials where the animals licked to either port, divided by the total number of trials. There were very few omissions during this task.
Histology
Confocal images were analyzed in FIJI and quantified in MATLAB. Boundaries of the MHb and LHb were manually annotated for each section using a DAPI counterstain. In sections containing, or immediately adjacent to the fiberoptic cannula, cells expressing GCaMP were manually annotated. The small size, dense packing, and neuropil staining made automated cell counting unreliable in this format. For fiber photometry analyses, cells were determined to be in the collection cone of the fiber optic if they were <200 μm away from the face of the fiber, taking into account the NA of the fiber.
Endoscopic two-photon calcium imaging
Images were extracted to tiff stacks using custom software. Image stacks were motion corrected using the FIJI plug in moco, and then downsampled to 256×256 pixels. CNMF (Pnevmatikakis et al., 2016) was run on these processed stacks to extract spatial footprints and time series for individual cells.
Electrophysiology
Spike sorting
The electrophysiological traces were processed by a custom Python-based pipeline, with preprocessing (CatGT), spike sorting (Kilosort 2.5; ), waveform calculation (C_Waves), quality control metrics calculation (quality_metrics), and activity-behavior synchronization (TPrime) modules, initially written by the Allen Institute for Brain Science and Jennifer Colonell (Janelia Research Campus), and the sorted clusters were manually refined using Phy (Cyrille Rossant, International Brain Laboratory). We considered the units with <0.5 inter-spike interval violation metric (Hill et al., 2011; Siegle et al., 2021) single units, or “neurons”, and analyzed the neurons with >0.2 Hz firing rates.
Transcranial optogenetics
Prior to the spike sorting pipeline, light artifacts in the raw traces were computationally removed by subtracting trial-averaged traces for individual channels. To determine the optotagged single units, we used stimulus-associated spike latency test based on firstspike latency distributions (Kvitsiani et al., 2013; Kim et al., 2016).
Atlas registration
The imaging data were destripped and stitched by an accompanying software package (Swaney et al., 2019), obtaining full 3D volumes. Each autofluorescence volume was registered to a standard 3D mouse brain atlas (Allen Institute Common Coordinate Framework; CCFv3), and the corresponding transform was applied to all channels. For registration, we first used a series of automated rigid and non-rigid registration algorithms implemented by Elastix (elastix: a toolbox for intensity based medical image registration IEEE Transactions on Medical Imaging, Klein et al., 2010; Ye et al., 2016) and then manually adjusted the control points for further refinement (Swaney et al., 2019). For tracing the Neuropixels insertion tracks, we imported the registered volumes to Lasagna (OpenSerialSection) and then manually clicked on the visualized CM-DiI or DiD (spectrally and spatially distinguishable) tracks to obtain several coordinates along each track. These coordinates and the corresponding electrophysiological features (e.g., firing rates and LFPs over channels and time) were imported to a Python-based GUI for final registration. Specifically, several heuristics (e.g., no spikes in the ventricles, high LFPs in the dentate gyrus, or similar depths of individual shanks for the 4-shank probes) were utilized to find brain-region boundaries along the initial tracks, which determined to coordinates of the individual channels and thus of the single units recorded there. These coordinates determined the brain region label for each single unit based on the annotations in the CCFv3. Recorded brain regions were grouped based on CCFv3 hierarchy into the following 15 structures for downstream analyses: MHb (medial habenula), lateral habenula (LHb), LAT (lateral group of the dorsal thalamus), MED (medial group of the dorsal thalamus), MTN (midline group of the dorsal thalamus; includes PVT, paraventricular nucleus of the thalamus), ILM (intralaminar nuclei of the dorsal thalamus), DORsm (thalamus, sensorymotor cortex related), MB (midbrain), HIP (hippocampus), MSC (medial septal complex), TRS (triangular nucleus of septum), LSX (lateral septal complex), STR (striatum), BST (bed nuclei of the stria terminalis), and HY (Hypothalamus).
Data analysis
Task-modulated neurons were identified by paired, two-sided Wilcoxon signed rank test of average firing rates for 1 s pre-cue baseline and four sequential 1 s windows from cue onset, separately for each trial type. A neuron was deemed task-modulated if any of these windows showed significant changes from the baseline (p < 0.01 after FDR correction across all neurons and windows using the Benjamini-Hochberg correction).
Time to baseline recovery for individual neurons upon transient optogenetic perturbation was estimated by paired, two-sided Wilcoxon signed rank test of average firing rates for 2 s pre-perturbation baseline and sequential 0.5 s windows from perturbation onset. We defined the recovery time as the time point such that the three consecutive windows starting from this time point show no significant changes from the baseline (p < 0.01 after FDR correction).
Individual neurons or populations were classified by ramping characteristics based on linear regression from trial index to average firing rate for 2 s pre-cue baseline. Two-sided p value for the null hypothesis that the slope is zero was calculated by estimating confidence interval of the slope. The significance threshold was p < 0.05, and the ramping direction was determined by the sign of the slope.
For quantifying trial-type-dependent within-trial population dynamics and integration, we first subtracted average firing rate for 2 s baseline for each trial to remove the baseline changes across trials. Then hierarchical bootstrap was used to combine data from multiple levels (Saravanan, Berman, Sober. Application of the hierarchical bootstrap to multi-level data in neuroscience. Neuron Behav Data Anal Theory. 2020). Each resampled dataset was constructed by randomly selecting a session, randomly selecting a neuron in the selected session, and randomly selecting a trial for the selected neuron, all with replacement, matching the size of the original dataset. Mean firing rate dynamics was calculated for each resampled dataset. After 100 iterations of this procedure, mean and SEM were calculated as mean and SD of the resampled means, respectively. For integration, we considered the baseline-subtracted average firing rate for the final 2 s window (5–7 s from cue or perturbation onset; right before the next event onset for the shortest ITI). The one-sided p values for the null hypothesis that this firing rate change, or integration along the total activity mode, is zero were calculated as the fraction of resampled means that are larger or smaller than zero. The significance threshold was p < 0.05.
Dynamical systems modeling
We modified the original version of LFADS (Pandarinath et al., 2018a) to handle two-photon Ca2+ imaging data. Specifically, the Poisson distribution modeling discrete spikes was replaced by the Gaussian distribution modeling continuous fluorescence traces, and the corresponding likelihood function was changed accordingly. In addition, the processing of input data by the encoder RNN was set to be forward-directional in time only, and causal; that is, when decoding a time point t in a trial, the trLFADS generator only received information up to and including time point t. We made the architecture causal to aid interpretation of the trLFADS inferred inputs; this does not imply qualitative differences between the bidirectional versus unidirectional processing in time. For each session, the odd and even trials were split into training and validation sets respectively, to optimize the hyperparameters used in downstream analyses. We trained 4 total trLFADS models: one for each of two mice, and one for each Tac1+ and TH+ populations. Hyperparameters were as follows: number of units in the generator: 256; number of latent factors: 32; dimensionality of the inferred input: 1; number of units in the encoder: 256; number of units in the controller: 256; size of time bins: 200 ms; L2 penalty: 10−9 (Tac1+) or 10−3 (TH+); dropout keep rate: 0.98 (Tac1+) or 0.9 (TH+).
The fixed points were identified by minimizing of the trained generator RNN using gradient-based optimization (Barak et al., 2013). The input to the generator during optimization was the trial-averaged pre-cue input, which was close to zero. The squared speed thresholds were 0.0003 and 0.001 for Tac1+ and TH+, respectively.
The targeted dimensionality reduction (Mante et al., 2013) was based on three directions defined as follows. The total activity mode was defined as a vector of all 1s; the condition independent mode was defined as average activity difference between 1 s reward period and 1 s pre-cue period; the line attractor mode was defined as the first principal component of the relevant fixed points. The three vectors were orthonormalized using the above order using the Gram-Schmidt process before projection.
For the in silico dynamics experiments, we ran the trained generator RNN forward in time with trial-averaged inferred inputs (between −1 s before and 3 s after the cue onset) of either rewarded or unrewarded trials, sampled from a Bernoulli distribution of given reward probability. Between two consecutive trials 1 s relaxation time with zero effective input was given to address the inter-trial interval. We ran up to 10 rewarded trials per session and projected the trajectory to the total activity mode in order to predict fiber photometry. We simulated and averaged across 1,000 sessions, with random initial conditions on the line attractor, for each reward probability. All code was written in Python 3 with Jax for auto-differentiation (Frostig et al., 2018) and is available at https://github.com/google-research/computation-thru-dynamics.
Supplementary Material
Highlights.
TH+ neurons in medial habenula encode reward-predictive cues
Tac1+ neurons in medial habenula encode reward outcome and reward history
Population dynamics of Tac1+ neurons track reward history via a putative line attractor
Dynamical systems modeling predicts probability dependence of Tac1+ reward response
ACKNOWLEDGMENTS
We thank Niles Pierce (Caltech) and Harry Choi (Molecular Technologies) for sharing HCR v3 probes and protocols with us; Tom Davidson for providing the initial code for processing photometry data; Yiming Chen for input; Dan O’Shea, Felicity Gore, Alex Attinger, and Jennifer Colonell (Janelia Research Campus) for electrophysiology advice; Ed Friedrich (New Scales Technologies) and Josh Siegle (Allen Institute) for sharing electrode mounts; Misha Raffiee for advice on light sheet imaging; Surya Ganguli for suggestions on modeling; and Deisseroth lab members for discussion. This work was supported by the LIGHT-SPACE U19 of the NIH BRAIN initiative (K.D., K.V.S., D.S.), NIMH (K.D.), NIDA (K.D.), the NSF NeuroNex program (K.D.), the AE and Gatsby Foundations (K.D.), BBRF (E.L.S.), the Bio-X Bowes Fellowship (Y.J.), and the Asan Foundation Biomedical Science Scholarship (Y.J.).
Footnotes
DECLARATION OF INTERESTS
These tools, and all protocols, clones, and sequences, are freely available to nonprofit institutions and investigators. D.S. is employed as a research scientist by Meta (Meta Reality Labs); his work there is unrelated to this study. K.V.S. is a consultant to Neuralink Corp. and CTRL-Labs Inc. in the Reality Labs Division of Meta (formerly Facebook); he is also on the Scientific Advisory Boards of Inscopix Inc., Mind X Inc., and Heal Inc. S.V. is a consultant to Compass Therapeutics. X.W., W.E.A., and K.D. hold IP for the hydrogel-tissue chemistry (STARmap) methods; K.D. is also a member of the Cell advisory board and co-founded and advises Maplight Therapeutics. These entities did not support or influence this work.
SUPPLEMENTAL INFORMATION
Supplemental information can be found online at https://doi.org/10.1016/j.cell.2022.08.019.
REFERENCES
- Ables JL, Gorlich A, Antolin-Fontes B, Wang C, Lipford SM, Riad MH, Ren J, Hu F, Luo M, Kenny PJ, et al. (2017). Retrograde inhibition by a specific subset of interpeduncular α5 nicotinic neurons regulates nicotine preference. Proceedings of the National Academy of Sciences 114, 13012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adamantidis AR, Zhang F, Aravanis AM, Deisseroth K, and de Lecea L (2007). Neural substrates of awakening probed with optogenetic control of hypocretin neurons. Nature 450, 420–424. 10.1038/nature06310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Afshar A, Santhanam G, Yu B, Ryu S, Sahani M, and Shenoy K (2011). Single-trial neural correlates of arm movement preparation. Neuron 71, 555–564. 10.1016/j.neuron.2011.05.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aizawa H, Kobayashi M, Tanaka S, Fukai T, and Okamoto H (2012). Molecular characterization of the subnuclei in rat habenula. J. Comp. Neurol. 520, 4051. 10.1002/cne.23230. [DOI] [PubMed] [Google Scholar]
- Allen WE, Chen MZ, Pichamoorthy N, Tien RH, Pachitariu M, Luo L, and Deisseroth K (2019). Thirst regulates motivated behavior through modulation of brainwide neural population dynamics. Science 364, 253. 10.1126/science.aav3932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen WE, Chen MZ, Pichamoorthy N, Tien RH, Pachitariu M, Luo L, and Deisseroth K (2019). Thirst regulates motivated behavior through modulation of brainwide neural population dynamics. Science 364, 253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andalman AS, Burns VM, Lovett-Barron M, Broxton M, Poole B, Yang SJ, Grosenick L, Lerner TN, Chen R, Benster T, et al. (2019). Neuronal Dynamics Regulating Brain and Behavioral State Transitions. Cell 177, 970–985.e20. 10.1016/j.cell.2019.02.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barak O, Sussillo D, Romo R, Tsodyks M, and Abbott LF (2013). From fixed points to chaos: three models of delayed discrimination. Progress in Neurobiology 103, 214–222. 10.1016/j.pneurobio.2013.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bari A, Dalley JW, and Robbins TW (2008). The application of the 5-choice serial reaction time task for the assessment of visual attentional processes and impulse control in rats. Nat. Protoc. 3, 759–767. 10.1038/nprot.2008.41. [DOI] [PubMed] [Google Scholar]
- Bayer HM, and Glimcher PW (2005). Midbrain dopamine neurons encode a quantitative reward prediction error signal. Neuron 47, 129–141. 10.1016/j.neuron.2005.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bromberg-Martin ES, and Hikosaka O (2011). Lateral habenula neurons signal errors in the prediction of reward information. Nat. Neurosci. 14, 1209–1216. 10.1038/nn.2902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bromberg-Martin ES, Matsumoto M, Nakahara H, and Hikosaka O (2010). Multiple Timescales of Memory in Lateral Habenula and Dopamine Neurons. Neuron 67, 499–510. 10.1016/j.neuron.2010.06.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Buchholtz LJ, Lam RM, Emrick JJ, Chesler AT, and Ryba NJ (2020). Assigning transcriptomic class in the trigeminal ganglion using multiplex in situ hybridization and machine learning. Pain 161, 2212–2224. 10.1097/j.pain.0000000000001911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buolos LJ, Ben Hamida S, Bailly J, Maitra M, Ehrlich AT, Gavériaux-Ruff C, Darcq E, and Kieffer BL (2020). Mu opioid receptors in the medial habenula contribute to naloxone aversion. Neuropsychopharmacology 45, 247–255. 10.1038/s41386-019-0395-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrillo-Reid L, Han S, Yang W, Akrouh A, and Yuste R (2019). Controlling visually guided behavior by holographic recalling of cortical ensembles. Cell 178, 447–457. 10.1016/j.cell.2019.05.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen R, Gore F, Nguyen Q-A, Ramakrishnan C, Patel S, Kim SH, Raffiee M, Kim YS, Hsueh B, Krook-Magnusson E, et al. (2021). Deep brain optogenetics without intracranial surgery. Nat. Biotechnol. 39, 161–164. 10.1038/s41587-020-0679-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho CH, Lee S, Kim A, Yarishkin O, Ryoo K, Lee YS, Jung HG, Yang E, Lee DY, Lee B, et al. (2020). TMEM16A expression in cholinergic neurons of the medial habenula mediates anxiety-related behaviors. EMBO Rep. 21, e48097. 10.15252/embr.201948097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi J-H, Duboué ER, Macurak M, Chanchu J-M, and Halpern ME (2021). Specialized neurons in the right habenula mediate response to aversive olfactory cues. Elife 10, e72345. 10.7554/elife.72345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Churchland MM, Cunningham JP, Kaufman MT, Ryu SI, and Shenoy KV (2010). Cortical preparatory activity: representation of movement or first cog in a dynamical machine? Neuron 68, 387–400. 10.1016/j.neuron.2010.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Contestabile A, and Fonnum F (1983). Cholinergic and GABAergic forebrain projections to the habenula and nucleus interpeduncularis: surgical and kainic acid lesions. Brain Res. 275, 287–297. 10.1016/0006-8993(83)90989-7. [DOI] [PubMed] [Google Scholar]
- Díaz E, Bravo D, Rojas X, and Concha ML (2011). Morphologic and immunohistochemical organization of the human habenular complex. J. Comp. Neurol. 519, 3727–3747. 10.1002/cne.22687. [DOI] [PubMed] [Google Scholar]
- Davidson S, and Giesler GJ (2010). The multiple pathways for itch and their interactions with pain. Trends Neurosci 33, 550–558. 10.1016/j.tins.2010.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deisseroth K (2017). Optical and chemical discoveries recognized for impact on biology and psychiatry. EMBO Rep. 18, 859–860. 10.15252/embr.201744405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan LZ, Kheifets S, Böhm UL, Wu H, Piatkevich KD, Xie ME, Parot V, Ha Y, Evans KE, Boyden ES, et al. (2020). All-Optical Electrophysiology Reveals the Role of Lateral Inhibition in Sensory Processing in Cortical Layer 1. Cell 180, 521–535.e18. 10.1016/j.cell.2020.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenno LE, Mattis J, Ramakrishnan C, Hyun M, Lee SY, He M, Tucciarone J, Selimbeyoglu A, Berndt A, Grosenick L, et al. (2014). Targeting cells with single vectors using multiple-feature Boolean logic. Nat Meth. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenno LE, Ramakrishnan C, Kim YS, Evans KE, Lo M, Vesuna S, Inoue M, Cheung KY, Yuen E, Pichamoorthy N, et al. (2020). Comprehensive Dual- and Triple-Feature Intersectional Single-Vector Delivery of Diverse Functional Payloads to Cells of Behaving Mammals. Neuron 107, 836–853.e11. 10.1016/j.neuron.2020.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkelstein A, Fontolan L, Economo MN, Li N, Romani S, and Svoboda K (2021). Attractor dynamics gate cortical information flow during decision-making. Nat. Neurosci. 24, 843–850. 10.1038/s41593-021-00840-6. [DOI] [PubMed] [Google Scholar]
- Frahm S, Slimak M, Ferrarese L, Santos-Torres J, Antolin-Fontes B, Auer S, Filkin S, Pons S, Fontaine J-F, Tsetlin V, et al. (2011). Aversion to Nicotine Is Regulated by the Balanced Activity of beta-4 and alpha-5 Nicotinic Receptor Subunits in the Medial Habenula. Neuron 70, 522–535. 10.1016/j.neuron.2011.04.013. [DOI] [PubMed] [Google Scholar]
- Frostig R, Johnson M, and Leary C (2018). Compiling machine learning programs via high-level tracing. In Systems for Machine Learning. [Google Scholar]
- Füzesi T, Daviu N, Wamsteeker Cusulin JI, Bonin RP, and Bains JS (2016). Hypothalamic CRH neurons orchestrate complex behaviours after stress. Nat. Commun. 7, 11937. 10.1038/ncomms11937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallam TG (1981). David G. Luenberger: Introduction to Dynamic Systems, Theory, Models, and Applications1979 (New York: John Wiley & Sons; ). 446 pp. Behav. Sci. 26, 397–398. [Google Scholar]
- Hashikawa Y, Hashikawa K, Rossi MA, Basiri ML, Liu Y, Johnston NL, Ahmad OR, and Stuber GD (2020). Transcriptional and Spatial Resolution of Cell Types in the Mammalian Habenula. Neuron 106, 743–758.e5. 10.1016/j.neuron.2020.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hattori R, Danskin B, Babic Z, Mlynaryk N, and Komiyama T (2019). Area-Specificity and Plasticity of History-Dependent Value Coding During Learning. Cell 177, 1858–1872.e15. 10.1016/j.cell.2019.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herkenham M, and Nauta WJH (1977). Afferent connections of the habenular nuclei in the rat. A horseradish peroxidase study, with a note on the fiber-of-passage problem. J. Comp. Neurol. 173, 123–145. 10.1002/cne.901730107. [DOI] [PubMed] [Google Scholar]
- Hikosaka O (2010). The habenula: from stress evasion to value-based decision-making. Nat. Rev. Neurosci. 11, 503–513. 10.1038/nrn2866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill DN, Mehta SB, and Kleinfeld D (2011). Quality metrics to accompany spike sorting of extracellular signals. Journal of Neuroscience 31, 8699–8705. 10.1523/JNEUROSCI.0971-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong S, and Hikosaka O (2008). The globus pallidus sends reward-related signals to the lateral habenula. Neuron 60, 720–729. 10.1016/j.neuron.2008.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong S, Jhou TC, Smith M, Saleem KS, and Hikosaka O (2011). Negative reward signals from the lateral habenula to dopamine neurons are mediated by rostromedial tegmental nucleus in primates. J. Neurosci. 31, 11457–11471. 10.1523/jneurosci.1384-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopfield JJ (1982). Neural networks and physical systems with emergent collective computational abilities. Proc. Natl. Acad. Sci. USA 79, 2554–2558. 10.1073/pnas.79.8.2554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu YWA, Morton G, Guy EG, Wang SD, and Turner EE (2016). Dorsal Medial Habenula Regulation of Mood-Related Behaviors and Primary Reinforcement by Tachykinin-Expressing Habenula Neurons. eNeuro 3. ENEURO. 0109–16.2016. 10.1523/eneuro.0109-16.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu YWA, Tempest L, Quina LA, Wei AD, Zeng H, and Turner EE (2013). Medial Habenula Output Circuit Mediated by 5 Nicotinic Receptor-Expressing GABAergic Neurons in the Interpeduncular Nucleus. J. Neurosci. 33, 18022–18035. 10.1523/jneurosci.2927-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu Y-WA, Wang SD, Wang S, Morton G, Zariwala HA, de la Iglesia HO, Turner EE, and Turner EE (2014). Role of the dorsal medial habenula in the regulation of voluntary activity, motor function, hedonic state, and primary reinforcement. J. Neurosci. 34, 11366–11384. 10.1523/jneurosci.1861-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang EJ, Dahlen JE, Mukundan M, and Komiyama T (2017). History-based action selection bias in posterior parietal cortex. Nat. Commun. 8, 1242. 10.1038/s41467-017-01356-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inagaki HK, Fontolan L, Romani S, and Svoboda K (2019). Discrete attractor dynamics underlies persistent activity in the frontal cortex. Nature 566, 212–217. 10.1038/s41586-019-0919-7. [DOI] [PubMed] [Google Scholar]
- Jennings JH, Kim CK, Marshel JH, Raffiee M, Ye L, Quirin S, Pak S, Ramakrishnan C, and Deisseroth K (2019). Interacting neural ensembles in orbitofrontal cortex for social and feeding behaviour. Nature 565, 645–649. 10.1038/s41586-018-0866-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawai T, Yamada H, Sato N, Takada M, and Matsumoto M (2015). Roles of the Lateral Habenula and Anterior Cingulate Cortex in Negative Outcome Monitoring and Behavioral Adjustment in Nonhuman Primates. Neuron 88, 792–804. 10.1016/j.neuron.2015.09.030. [DOI] [PubMed] [Google Scholar]
- Kebschull JM, Richman EB, Ringach N, Friedmann D, Albarran E, Kolluru SS, Jones RC, Allen WE, Wang Y, Cho SW, et al. (2020). Cerebellar nuclei evolved by repeatedly duplicating a conserved cell-type set. Science 370, eabd5059. 10.1126/science.abd5059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SS, Rouault H, Druckmann S, and Jayaraman V (2017). Ring attractor dynamics in the Drosophila central brain. Science 356, 849–853. 10.1126/science.aal4835. [DOI] [PubMed] [Google Scholar]
- Kim S-Y, Adhikari A, Lee SY, Marshel JH, Kim CK, Mallory CS, Lo M, Pak S, Mattis J, Lim BK, et al. (2013). Diverging neural pathways assemble a behavioural state from separable features in anxiety. Nature 496, 219–223. 10.1038/nature12018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H, Ährlund-Richter S, Wang X, Deisseroth K, and Carlén M (2016). Prefrontal Parvalbumin Neurons in Control of Attention. Cell 164, 208–218. 10.1016/j.cell.2015.11.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kishi KE, Kim YS, Fukuda M, Inoue M, Kusakizako T, Wang PY, Ramakrishnan C, Byrne EFX, Thadhani E, Paggi JM, et al. (2022). Structural basis for channel conduction in the pump-like channelrhodopsin ChRmine. Cell 185, 672–689. 10.1016/j.cell.2022.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein S, Staring M, Murphy K, Viergever MA, and Pluim JPW (2010). elastix: a toolbox for intensity-based medical image registration. IEEE Transactions on Medical Imaging 29, 196–205. 10.1109/TMI.2009.2035616. [DOI] [PubMed] [Google Scholar]
- Kobayashi Y, Sano Y, Vannoni E, Goto H, Suzuki H, Oba A, Kawasaki H, Kanba S, Lipp H-P, Murphy NP, et al. (2013). Genetic dissection of medial habenula-interpeduncular nucleus pathway function in mice. Front. Behav. Neurosci. 7, 17. 10.3389/fnbeh.2013.00017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohl J, Babayan BM, Rubinstein ND, Autry AE, Marin-Rodriguez B, Kapoor V, Miyamishi K, Zweifel LS, Luo L, Uchida N, and Dulac C (2018). Functional circuit architecture underlying parental behaviour. Nature 556, 326–331. 10.1038/s41586-018-0027-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konsman J-P (2001). Paxinos and Franklin’s the Mouse Brain in Stereotaxic Coordinates (New York: Academic Press; ), pp. 827–828. [Google Scholar]
- Krashes MJ, Shah BP, Madara JC, Olson DP, Strochlic DE, Garfield AS, Vong L, Pei H, Watabe-Uchida M, Uchida N, et al. (2014). An excitatory paraventricular nucleus to AgRP neuron circuit that drives hunger. Nature 507, 238–242. 10.1038/nature12956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kvitsiani D, Ranade S, Hangya B, Taniguchi H, Huang JZ, and Kepecs A (2013). Distinct behavioural and network correlates of two interneuron types in prefrontal cortex. Nature 498, 363–366. 10.1038/nature12176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lammel S, Steinberg E, Földy C, Wall N, Beier K, Luo L, and Malenka R (2015). Diversity of Transgenic Mouse Models for Selective Targeting of Midbrain Dopamine Neurons. Neuron 85, 429–438. 10.1016/j.neuron.2014.12.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazaridis I, Tzortzi O, Weglage M, Märtin A, Xuan Y, Parent M, Johansson Y, Fuzik J, Fürth D, Fenno LE, et al. (2019). A hypothalamus-habenula circuit controls aversion. Mol Psychiatry 24, 1351–1368. 10.1038/s41380-019-0369-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerner TN, Shilyansky C, Davidson TJ, Evans KE, Beier KT, Zalocusky KA, Crow AK, Malenka RC, Luo L, Tomer R, and Deisseroth K (2015). Intact-Brain Analyses Reveal Distinct Information Carried by SNc Dopamine Subcircuits. Cell 162, 635–647. 10.1016/j.cell.2015.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu LD, Chen S, Hou H, West SJ, Faulkner M, Economo MN, Li N, and Svoboda K; International Brain Laboratory (2021). Accurate Localization of Linear Probe Electrode Arrays across Multiple Brains. eNeuro 8. 10.1523/ENEURO.0241-21.2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovett-Barron M, Andalman AS, Allen WE, Vesuna S, Kauvar I, Burns VM, and Deisseroth K (2017). Ancestral Circuits for the Coordinated Modulation of Brain State. Cell 171, 1411–1423.e17. e1417. 10.1016/j.cell.2017.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovett-Barron M, Chen R, Bradbury S, Andalman AS, Wagle M, Guo S, and Deisseroth K (2020). Multiple convergent hypothalamus-brainstem circuits drive defensive behavior. Nat. Neurosci. 23, 959–967. 10.1038/s41593-020-0655-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mante V, Sussillo D, Shenoy KV, and Newsome WT (2013). Context-dependent computation by recurrent dynamics in prefrontal cortex. Nature 503, 78–84. 10.1038/nature12742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshel JH, Kim YS, Machado TA, Quirin S, Benson B, Kadmon J, Raja C, Chibukhchyan A, Ramakrishnan C, Inoue M, et al. (2019). Cortical layer-specific critical dynamics triggering perception. Science 365, eaaw5202. eaaw5202–eaaw5214. 10.1126/science.aaw5202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathis A, Mamidanna P, Cury KM, Abe T, Murthy VN, Mathis MW, and Bethge M (2018). DeepLabCut: markerless pose estimation of user-defined body parts with deep learning. Nat. Neurosci. 21, 1281–1289. 10.1038/s41593-018-0209-y. [DOI] [PubMed] [Google Scholar]
- Mathuru AS, Jesuthasan S, and Jesuthasan S (2013). The medial habenula as a regulator of anxiety in adult zebrafish. Front Neural Circuits 7, 99. 10.3389/fncir.2013.00099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto M, and Hikosaka O (2007). Lateral habenula as a source of negative reward signals in dopamine neurons. Nature 447, 1111–1115. 10.1038/nature05860. [DOI] [PubMed] [Google Scholar]
- Mazzucato L (2022). Neural mechanisms underlying the temporal organization of naturalistic animal behavior. Elife 11, e76577. 10.7554/elife.76577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLaughlin I, Dani JA, and De Biasi M (2017). The medial habenula and interpeduncular nucleus circuitry is critical in addiction, anxiety, and mood regulation. J. Neurochem. 142 (Suppl 2), 130–143. 10.1111/jnc.14008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melani R, Von Itter R, Jing D, Koppensteiner P, and Ninan I (2019). Opposing effects of an atypical glycinergic and substance P transmission on interpeduncular nucleus plasticity. Neuropsychopharmacology 44, 1828–1836. 10.1038/s41386-019-0396-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miri A, Daie K, Arrenberg AB, Baier H, Aksay E, and Tank DW (2011). Spatial gradients and multidimensional dynamics in a neural integrator circuit. Nat. Neurosci. 14, 1150–1159. 10.1038/nn.2888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molas S, Zhao-Shea R, Liu L, DeGroot SR, Gardner PD, and Tapper AR (2017). A circuit-based mechanism underlying familiarity signaling and the preference for novelty. Nat. Neurosci. 20, 1260–1268. 10.1038/nn.4607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morales M, and Margolis EB (2017). Ventral tegmental area: cellular heterogeneity, connectivity and behaviour. Nature Publishing Group 18, 73–85. 10.1038/nrn.2016.165. [DOI] [PubMed] [Google Scholar]
- Morcos AS, and Harvey CD (2016). History-dependent variability in population dynamics during evidence accumulation in cortex. Nat. Neurosci. 19, 1672–1681. 10.1038/nn.4403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morissette M-C, and Boye SM (2008). Electrolytic lesions of the habenula attenuate brain stimulation reward. Behav. Brain Res. 187, 17–26. 10.1016/j.bbr.2007.08.021. [DOI] [PubMed] [Google Scholar]
- Morton G, Nasirova N, Sparks DW, Brodsky M, Sivakumaran S, Lambe EK, and Turner EE (2018). Chrna5-Expressing Neurons in the Interpeduncular Nucleus Mediate Aversion Primed by Prior Stimulation or Nicotine Exposure. J Neurosci 38, 6900–6920. 10.1523/JNEUROSCI.0023-18.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Namboodiri VMK, Rodriguez-Romaguera J, and Stuber GD (2016). The habenula. Curr. Biol. 26, R873–R877. 10.1016/j.cub.2016.08.051. [DOI] [PubMed] [Google Scholar]
- Ottenheimer D, Richard JM, and Janak PH (2018). Ventral pallidum encodes relative reward value earlier and more robustly than nucleus accumbens. Nat. Commun. 9, 4350. 10.1038/s41467-018-06849-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pachitariu M, Steinmetz N, Kadir S, Carandini M, and Kenneth D, H. (2016). Kilosort: realtime spike-sorting for extracellular electrophysiology with hundreds of channels. bioRxiv. 10.1101/061481. [DOI] [Google Scholar]
- Pandarinath C, Ames KC, Russo AA, Farshchian A, Miller LE, Dyer EL, and Kao JC (2018a). Latent Factors and Dynamics in Motor Cortex and Their Application to Brain-Machine Interfaces. J. Neurosci. 38, 9390–9401. 10.1523/jneurosci.1669-18.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandarinath C, O’Shea DJ, Collins J, Jozefowicz R, Stavisky SD, Kao JC, Trautmann EM, Kaufman MT, Ryu SI, Hochberg LR, et al. (2018b). Inferring single-trial neural population dynamics using sequential auto-encoders. Nat Meth 15, 805–815. 10.1038/s41592-018-0109-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang X, Liu L, Ngolab J, Zhao-Shea R, McIntosh JM, Gardner PD, and Tapper AR (2016). Habenula cholinergic neurons regulate anxiety during nicotine withdrawal via nicotinic acetylcholine receptors. Neuropharmacology 107, 294–304. 10.1016/j.neuropharm.2016.03.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey S, Shekhar K, Regev A, and Schier AF (2018). Comprehensive Identification and Spatial Mapping of Habenular Neuronal Types Using Single-Cell RNA- Seq. Curr. Biol. 28, 1052–1065.e7. e1057. 10.1016/j.cub.2018.02.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pnevmatikakis E, Soudry D, Gao Y, Machado TA, Merel J, Pfau D, Reardon T, Mu Y, Lacefield C, Yang W, et al. (2016). Simultaneous Denoising, Deconvolution, and Demixing of Calcium Imaging Data. Neuron 89, 285–299. 10.1016/j.neuron.2015.11.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prakash R, Yizhar O, Grewe B, Ramakrishnan C, Wang N, Goshen I, Packer AM, Peterka DS, Yuste R, Schnitzer MJ, and Deisseroth K (2012). Two-photon optogenetic toolbox for fast inhibition, excitation and bistable modulation. Nat Methods 9, 1171–1179. 10.1038/nmeth.2215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proulx CD, Hikosaka O, and Malinow R (2014). Reward processing by the lateral habenula in normal and depressive behaviors. Nature Publishing Group 17, 1146–1152. 10.1038/nn.3779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin C, and Luo M (2009). Neurochemical phenotypes of the afferent and efferent projections of the mouse medial habenula. Neuroscience 161, 827–837. 10.1016/j.neuroscience.2009.03.085. [DOI] [PubMed] [Google Scholar]
- Quina LA, Tempest L, Ng L, Harris JA, Ferguson S, Jhou TC, and Turner EE (2015). Efferent pathways of the mouse lateral habenula. J. Comp. Neurol. 523, 32–60. 10.1002/cne.23662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quina LA, Walker A, Morton G, Han V, and Turner EE (2020). GAD2-expression defines a class of excitatory lateral habenula neurons in mice that project to the raphe and pontine tegmentum. eNeuro ENEURO.0527 19, 2020–2043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remington ED, Egger SW, Narain D, Wang J, and Jazayeri M (2018). A Dynamical Systems Perspective on Flexible Motor Timing. Trends Cogn. Sci. (Regul. Ed.) 22, 938–952. 10.1016/j.tics.2018.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robbins TW (2002). The 5-choice serial reaction time task: behavioural pharmacology and functional neurochemistry. Psychopharmacology 163, 362–380. 10.1007/s00213-002-1154-7. [DOI] [PubMed] [Google Scholar]
- Robinson DA (1989). Integrating with Neurons. Annu. Rev. Neurosci. 12, 33–45. 10.1146/annurev.ne.12.030189.000341. [DOI] [PubMed] [Google Scholar]
- Roy NA, Bak JH, Akrami A, Brody CD, Pillow JW, and Pillow JW (2021). Extracting the dynamics of behavior in sensory decision-making experiments. Neuron 109, 597–610.e6. 10.1016/j.neuron.2020.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Roige S, Peña-Oliver Y, and Stephens DN (2011). Measuring impulsivity in mice: the five-choice serial reaction time task. Psychopharmacology 219, 253–270. 10.1007/s00213-011-2560-5. [DOI] [PubMed] [Google Scholar]
- Schultz W, Dayan P, and Montague PR (1997). A neural substrate of prediction and reward. Science 275, 1593–1599. 10.1126/science.275.5306.1593. [DOI] [PubMed] [Google Scholar]
- Scott BB, Constantinople CM, Akrami A, Hanks TD, Brody CD, and Tank DW (2017). Fronto-parietal Cortical Circuits Encode Accumulated Evidence with a Diversity of Timescales. Neuron 95, 385–398.e5. e385. 10.1016/j.neuron.2017.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seigneur E, Polepalli JS, and Südhof TC (2018). Cbln2 and Cbln4 are expressed in distinct medial habenula-interpeduncular projections and contribute to different behavioral outputs. Proc. Natl. Acad. Sci. USA 115, E10235–E10244. 10.1073/pnas.1811086115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seung HS (1996). How the brain keeps the eyes still. Proc. Natl. Acad. Sci. USA 93, 13339–13344. 10.1073/pnas.93.23.13339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shenoy KV, Sahani M, and Churchland MM (2013). Cortical control of arm movements: a dynamical systems perspective. Annu. Rev. Neurosci. 36, 337–359. 10.1146/annurev-neuro-062111-150509. [DOI] [PubMed] [Google Scholar]
- Siegle JH, Jia X, Durand S, Gale S, Bennett C, Graddis N, Heller G, Ramirez TK, Choi H, Luviano JA, et al. (2021). Survey of spiking in the mouse visual system reveals functional hierarchy. Nature 592, 86–92. 10.1038/s41586-020-03171-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sohn H, Narain D, Meirhaeghe N, and Jazayeri M (2019). Bayesian Computation through Cortical Latent Dynamics. Neuron 103, 934–947.e5. e935. 10.1016/j.neuron.2019.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A, Jennings J, Ung R, Blair G, Weinberg R, Neve R, Boyce F, Mattis J, Ramakrishnan C, Deisseroth K, and Stuber G (2013). A unique population of ventral tegmental area neurons inhibits the lateral habenula to promote reward. Neuron 80, 1039–1053. 10.1016/j.neuron.2013.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinmetz NA, Zatka-Haas P, Carandini M, and Harris KD (2019). Distributed coding of choice, action and engagement across the mouse brain. Nature 576, 266–273. 10.1038/s41586-019-1787-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinmetz NA, Aydin C, Lebedeva A, Okun M, Pachitariu M, Bauza M, Beau M, Bhagat J, Böhm C, Broux M, et al. (2021. Apr 16). Neuropixels 2.0: A miniaturized high-density probe for stable, long-term brain recordings. Science 372, eabf4588. 10.1126/science.abf4588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sternson SM (2020). Exploring internal state-coding across the rodent brain. Curr. Opin. Neurobiol. 65, 20–26. 10.1016/j.conb.2020.08.009. [DOI] [PubMed] [Google Scholar]
- Sugrue LP, Corrado GS, and Newsome WT (2004). Matching behavior and the representation of value in the parietal cortex. Science 304, 1782–1787. 10.1126/science.1094765. [DOI] [PubMed] [Google Scholar]
- Sul JH, Kim H, Huh N, Lee D, and Jung MW (2010). Distinct Roles of Rodent Orbitofrontal and Medial Prefrontal Cortex in Decision Making. Neuron 66, 449–460. 10.1016/j.neuron.2010.03.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sussillo D, and Barak O (2013). Opening the black box: low-dimensional dynamics in high-dimensional recurrent neural networks. Neural Comput. 25, 626–649. 10.1162/neco_a_00409. [DOI] [PubMed] [Google Scholar]
- Sutherland RJ (1982). The dorsal diencephalic conduction system: a review of the anatomy and functions of the habenular complex. Neurosci. Biobehav. Rev. 6, 1–13. 10.1016/0149-7634(82)90003-3. [DOI] [PubMed] [Google Scholar]
- Sutton RS, and Barto AG (1990). Time-derivative Models of Pavlovian Reinforcement. [Google Scholar]
- Swaney J, Kamentsky L, Evans NB, Xie K, Park Y-G, Drummond G, Yun DH, and Chung K (2019). Scalable image processing techniques for quantitative analysis of volumetric biological images from light-sheet microscopy. bioRxiv. 10.1101/576595. [DOI] [Google Scholar]
- Tsutsui K-I, Grabenhorst F, Kobayashi S, and Schultz W (2016). A dynamic code for economic object valuation in prefrontal cortex neurons. Nat. Commun. 7, 12554. 10.1038/ncomms12554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vesuna S, Kauvar IV, Richman E, Gore F, Oskotsky T, Sava-Segal C, Luo L, Malenka RC, Henderson JM, Nuyujukian P, et al. (2020). Deep posteromedial cortical rhythm in dissociation. Nature 586, 87–94. 10.1038/s41586-020-2731-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vyas S, Golub MD, Sussillo D, and Shenoy KV (2020). Computation Through Neural Population Dynamics. Annu. Rev. Neurosci. 43, 249–275. 10.1146/annurev-neuro-092619-094115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner F, French L, and Veh RW (2016). Transcriptomic-anatomic analysis of the mouse habenula uncovers a high molecular heterogeneity among neurons in the lateral complex, while gene expression in the medial complex largely obeys subnuclear boundaries. Brain Struct. Funct. 221, 39–58. 10.1007/s00429-014-0891-9. [DOI] [PubMed] [Google Scholar]
- Wallace ML, Huang KW, Hochbaum D, Hyun M, Radeljic G, and Sabatini BL (2020). Anatomical and single-cell transcriptional profiling of the murine habenular complex. Elife 9, e51271. 10.7554/elife.51271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Allen WE, Wright MA, Sylwestrak EL, Samusik N, Vesuna S, Evans K, Liu C, Ramakrishnan C, Liu J, et al. (2018). Three-dimensional intact-tissue sequencing of single-cell transcriptional states. Science 361, eaat5691. 10.1126/science.aat5691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weihe E, Depboylu C, Schütz B, Schäfer MKH, and Eiden LE (2006). Three Types of Tyrosine Hydroxylase-Positive CNS Neurons Distinguished by Dopa Decarboxylase and VMAT2 Co-Expression. Cell. Mol. Neurobiol. 26, 657–676. 10.1007/s10571-006-9053-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi T, Danjo T, Pastan I, Hikida T, and Nakanishi S (2013). Distinct Roles of Segregated Transmission of the Septo-Habenular Pathway in Anxiety and Fear. Neuron 78, 537–544. 10.1016/j.neuron.2013.02.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye L, Allen WE, Thompson KR, Tian Q, Hsueh B, Ramakrishnan C, Wang A-C, Jennings JH, Adhikari A, Halpern CH, et al. (2016). Wiring and molecular features of prefrontal ensembles representing distinct experiences. Cell 165, 1776–1778. 10.1016/j.cell.2016.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu BM, Cunningham JP, Santhanam G, Ryu SI, Shenoy KV, and Sahani M (2009). Gaussian-process factor analysis for low-dimensional single-trial analysis of neural population activity. J. Neurophysiol. 102, 614–635. 10.1152/jn.90941.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zalocusky KA, Ramakrishnan C, Lerner TN, Davidson TJ, Knutson B, and Deisseroth K (2016). Nucleus accumbens D2R cells signal prior outcomes and control risky decision-making. Nature 531, 642–646. 10.1038/nature17400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang G-W, Shen L, Zhong W, Xiong Y, Zhang LI, and Tao HW (2018). Transforming Sensory Cues into Aversive Emotion via SeptalHabenular Pathway. Neuron 99, 1016–1028.e5. 10.1016/j.neuron.2018.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang G-W, Shen L, Li Z, Tao HW, and Zhang LI (2019). Track-Control, an automatic video-based real-time closed-loop behavioral control toolbox. Preprint at bioRxiv. 12.11.873372. [Google Scholar]
- Zhao Y, and Park IM (2017). Variational Latent Gaussian Process for Recovering Single-Trial Dynamics from Population Spike Trains. Neural Comput. 29, 1293–1316. 10.1162/neco_a_00953. [DOI] [PubMed] [Google Scholar]
- Zhao-Shea R, DeGroot SR, Liu L, Vallaster M, Pang X, Su Q, Gao G, Rando OJ, Martin GE, George O, et al. (2015). Increased CRF signalling in a ventral tegmental area-interpeduncular nucleus-medial habenula circuit induces anxiety during nicotine withdrawal. Nat. Commun. 6, 6770. 10.1038/ncomms7770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoltowski D, Pillow J, and Linderman S (2020). A general recurrent state space framework for modeling neural dynamics during decision-making 119, 11680–11691. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All code is available on GitHub. Modeling code is available at https://github.com/google-research/computation-thru-dynamics. Photometry code is available on https://github.com/SylwestrakLab/HbRewardCellTypes. In vivo electrophysiology code is available on https://github.com/youngju-jo. STARmap data is available on the NeMO Archive (NeMO: 9ACQ8G1, https://assets.nemoarchive.org/dat-9ACQ8G2) and electrophysiology data is availbe on the DANDI archive (DANDI: 000302, https://dandiarchive.org/dandiset/000302).
Any additional information required to reanalyze the data reported in this paper is available from the lead contact (KD) upon request.


