Abstract
Earlier studies have revealed that human cytomegalovirus rapidly inhibits the growth of fibroblasts, blocking cell cycle progression at multiple points, including the G1-to-S-phase transition. The present study demonstrates that the UL69 protein, a virus-encoded constituent of the virion, is able to arrest cell cycle progression when introduced into uninfected cells. Expression of the UL69 protein causes U2 OS cells and primary human fibroblasts to accumulate within the G1 compartment of the cell cycle, and serum fails to induce the progression of quiescent human fibroblasts into the S phase when the protein is present. Therefore, the UL69 protein is at least partially responsible for the cell cycle block that is instituted after infection of permissive cells with human cytomegalovirus.
Human cytomegalovirus (HCMV), a beta-herpesvirus, is a major cause of morbidity and mortality in immunocompromised individuals (reviewed in reference 7) and the leading viral cause of birth defects (2). To develop strategies to prevent and treat HCMV infection, it is crucial to understand the early interactions between the virus and its host cell that lead to the establishment and progression of the virus replication cycle.
We have previously reported (30) that HCMV infection rapidly inhibits the growth of human primary fibroblasts. This growth inhibition results from a block in host cell cycle progression (5, 17, 22, 30). The arrest occurs at multiple phases of the cell cycle, including the transition from G1 to S. A recent report by Salvant et al. (38) showed that infection of cells in the S phase leads to a delay in the expression of immediate-early viral genes, further emphasizing the importance of cell cycle regulation to HCMV replication.
Other herpesviruses also appear to inhibit cell cycle progression. De Bruyn Kops and Knipe (14) have shown that cells fail to progress to the S phase when cultures in the G1 compartment are infected with herpes simplex virus, an alpha-herpesvirus. A mutant virus lacking the ICP27 gene fails to institute the cell cycle block (29), suggesting that the ICP27 protein plays a role in the process. Epstein-Barr virus, a gamma-herpesvirus, also inhibits cell cycle progression during lytic infection. The immediate-early BZLF1 viral protein is responsible for the arrest and appears to function at least in part by inducing the accumulation of the p53 tumor suppressor protein, which, in turn, induces cell cycle arrest in G1 (8). Since herpesviruses encode many of their own DNA replication and nucleotide metabolism enzymes, a cell cycle block in late G1 might be a general strategy by which this family of viruses creates a favorable environment for viral DNA replication while preventing cellular DNA synthesis (reviewed in reference 13).
Here we demonstrate that expression of the UL69 protein of HCMV inhibits cell cycle progression. U-2 OS cells transfected with a plasmid that expresses UL69 protein and primary human fibroblast (HF) cells infected with a recombinant retrovirus expressing the HCMV protein accumulate in the G1 compartment of the cell cycle. Further, serum fails to induce quiescent cells to progress to the S phase of the cell cycle in the presence of the viral protein. The UL69 protein, which is a constituent of the HCMV virion (45) and is related in its sequence to the herpes simplex virus ICP27 protein (10), is at least partially responsible for the ability of HCMV to institute a cell cycle arrest within infected fibroblasts.
MATERIALS AND METHODS
Cells and viruses.
The maintenance of primary HF cells and their infection with HCMV strain AD169 infection were as described previously (30). U-2 OS cells were obtained from the American Type Culture Collection. To inactivate HCMV by treatment with UV light (46), 0.3 ml of an HCMV stock obtained by infection at a multiplicity of 0.01 PFU/cell was placed in a six-well dish and irradiated at 2 J/m2/s for 10 min with mixing every 2 min. UV-treated virus stocks failed to express detectable IE1 and IE2 proteins when examined by immunofluorescent staining 12 h after infection.
Plasmids.
pHM160 (44) contains the UL69 open reading frame from HCMV strain AD169 under control of the HCMV major immediate-early promoter in the expression vector pCB6 (43). pCGN65 contains the pp65 open reading frame with its amino terminus fused to a 9-amino-acid epitope derived from the influenza virus hemagglutinin protein (3). pRc/CMVhp53 contains the human p53 cDNA controlled by the HCMV major immediate-early promoter (28). pBB14 contains the enhanced green-fluorescence protein (EGFP) open reading frame fused to sequences encoding the C terminus of the pseudorabies virus Us9 protein (6). pGFP-spectrin contains a pleckstrin homology domain from spectrin fused to the C terminus of EGFP (23). pRetro-EBNA (O. Petrenko, Princeton) was modified from LZRSpBMN-Z (26) by removing the lacZ gene. pRetro-GFP (provided by O. Petrenko, Princeton University) contains the EGFP coding region in the vector pRetro-EBNA.
pflipUL69 was constructed by digesting pHM160 with MluI, blunting the DNA ends with Klenow DNA polymerase, and digesting with EcoRI to liberate bases 412 to 2235 of the UL69 open reading frame. This fragment was then joined, using DNA ligase, to pCB6(+) that had been digested with EcoRI and EcoRV. Transcription of this construct is controlled by the HCMV major immediate-early promoter, which directs the synthesis of a transcript that is antisense in its orientation to UL69 mRNA.
pRetro-UL69 was constructed in three steps. First, the 5′ portion of the UL69 open reading frame (the ATG codon to the MluI site) was amplified from HCMV strain AD169 DNA by PCR with the PFU enzyme (Stratagene) and 5′-CGGGATCCCCATGGAGCTGC-3′ and 5′CGGAATTCCACGCGTCGCTTGAAAG-3′ primers, which introduced BamHI and EcoRI recognition sites into the 5′ and 3′ ends, respectively, of the amplified DNA. The product was digested with BamHI plus EcoRI and joined by ligation to BamHI-EcoRI-digested pKS-Bluescript (Stratagene), generating pKS-RUL69-5. The cloned product was sequenced to confirm the fidelity of the PCR amplification. Second, the 3′ portion of the UL69 open reading frame was liberated from pHM160 by digestion with MluI-HindIII and joined by ligation to MluI-HindIII-digested pKS-RUL69-5, thereby inserting the full-length UL69 open reading frame into pKS-RUL69. Third, the full-length UL69 open reading frame was removed from pKS-RUL69 by digestion with BamHI-EcoRI and joined by ligation to the BamHI-EcoRI-digested pRetro-EBNA vector. This plasmid, with UL69 expression under control of the long terminal repeat (LTR) was termed pRetro-UL69.
The UL69 open reading frame from the Towne strain was amplified by PCR from HCMV genomic DNA with 5′-GCGGATCCCCATGGAGCTGCACTCACGCG-3′ and 5′-CGCTCGAGGACAGCAATCATCACGCACAAC-3′ primers. The amplified product was digested with BamHI-XhoI and joined by ligation to BamHI-XhoI-digested pRetro-EBNA. This plasmid, with Towne UL69 expression under control of the LTR, was called pRetro-Towne-UL69. Three clones of the Towne UL69 open reading frame were obtained from independent PCRs and inserted into pRetro-EBNA. The retro-Towne-UL69 viruses, generated after transfection of these independent clones into packaging cells, all induced a G1 arrest in HF cells.
Transient expression of UL69 in U-2 OS cells.
The cell cycle analysis in U-2 OS cells transfected with pHM160 or pCB6 and control plasmids was performed as described by Kalejta et al. (23). U-2 OS cells (3.5 × 106) were transfected with a test plasmid (20 μg) together with pBB14 (2 μg). At 24 h after transfection, 40 ng of nocodazole per ml was added to the culture medium; at 24 h after the addition of nocodazole, the cells were harvested, subjected to fluorescence-activated cell sorter (FACS) analysis with a FACScan (Becton Dickinson), and data analysis was performed with CellQuest software (Becton Dickinson). For analysis of UL69 expression, cells were lysed at 48 h after transfection in buffer containing 0.05 M Tris (pH 6.8), 2% sodium dodecyl sulfate (SDS), 10% glycerol, 0.025% (wt/vol) bromophenol blue, and 1% (vol/vol) β-mercaptoethanol and subjected to electrophoresis to separate the three forms of UL69 (21, 45). After electrophoresis, the proteins were transferred overnight at 30 V or for 2 h at 70 V to a nitrocellulose membrane (Schleicher & Schuell) in buffer containing 0.025 M Tris (pH 8), 0.192 M glycine, 20% methanol, and 0.01% (wt/vol) SDS. After the transfer, the membrane was blocked in TBST buffer (10 mM Tris [pH 8], 150 mM NaCl, 0.05% Tween 20)–10% nonfat milk at room temperature for 1 h; incubated with a monoclonal antibody specific for UL69 protein at a 1:10 dilution in TBST–1% nonfat milk for 1 h; washed three times with TBST; incubated with horseradish peroxidase-conjugated goat anti-mouse immunoglobulin G (Amersham) at a 1:10,000 dilution in TBST–1% nonfat milk at room temperature for 45 min; washed three times with TBST; and developed with the Renaissance system (NEN).
Expression of UL69 in HF cells by using a recombinant retrovirus.
To prepare recombinant retrovirus stocks, pRetro-UL69 or pRetro-GFP was transfected into Phoenix-A cells (26) with Lipofectamine (GIBCO-BRL). At 48 h after transfection, a culture supernatant was obtained, from which cell debris was removed. The supernatant, containing the retrovirus, was stored at −80°C.
To quantify the amount of infectious recombinant virus in supernatants, HF cells were infected with dilutions of virus stocks in 4 μg of Polybrene per ml (26). At 48 h after infection, the titer of retro-GFP was calculated from the number of EGFP-positive cells and the titer of retro-UL69 was calculated from the number of UL69 protein-positive cells determined by immunofluorescent staining. Retro-GFP and retro-UL69 virus stocks each contained approximately 0.8 × 106 to 2.5 × 106 fluorescence units per ml. Western blot analysis was used to quantify UL69 protein expression following retro-UL69 infection (21, 45).
To analyze the effect of proteins expressed by recombinant retroviruses on cell cycle progression, HF cells were infected with retro-UL69 or retro-GFP at a multiplicity of 5 fluorescence units per cell. At 48 or 60 h after infection, the cells were removed from culture dishes by trypsinization and fixed with 70% ethanol at −20°C for at least 3 h. They were then resuspended in phosphate-buffered saline containing 50 μg of propidium iodide per ml and 100 μg of RNase A per ml, and the DNA content was measured by FACS analysis. To induce a G2/M arrest, nocodazole (50 ng/ml) was added to the culture medium at 12 h before the cells were harvested. When infections were performed at a multiplicity of 0.5 fluorescence unit per cell, retro-GFP-infected cells were identified by their green fluorescent signal and retro-UL69-infected cells were identified by immunofluorescent staining with an antibody against UL69. In this case, fixed cells were washed with PBS containing 1% serum, permeabilized by incubation in PBSBT buffer (PBS with 0.5% bovine serum albumin and 0.05% Tween 20)–10% serum at room temperature for 30 min, and then reacted with a UL69-specific antibody at a dilution of 1:10 in PBSBT at 37°C for 1 h. After being washed with PBSBT, the cells were reacted with fluorescein isothiocyanate-conjugated goat anti-mouse immunoglobulin G (Amersham) at a dilution of 1:1,000 in PBSBT at 37°C for 45 min. After another wash with PBSBT, the cells were suspended in PBS containing 50 μg of propidium iodide per ml and 100 μg of RNase A per ml to measure their DNA content.
To assay for S-phase entry of quiescent cells, HF cells were infected with retro-UL69 or retro-GFP at a multiplicity of 5 fluorescence units per cell. From 24 to 72 h after infection, the cells were synchronized in G0/G1 by maintenance in Dulbecco’s modified Eagle’s medium containing 0.2% serum. At 72 h after infection, the cells were fed with DMEM containing 10% serum to induce cell cycle progression; 24 h later (96 h after infection), the cells were harvested and subjected to FACS analysis.
RESULTS
UV-inactivated HCMV inhibits cell growth.
HCMV inhibits the growth of primary HF cells in the early stage of infection, independently of viral DNA replication (30). The growth-inhibitory effect could result from a signal generated when the virus interacts with the cell surface, from the action of a virion protein within the cell, or from a newly synthesized viral gene product. To distinguish between an effect on cell growth mediated by the virus particle and an effect of newly synthesized viral products, we examined the ability of HCMV inactivated with UV light to block cell growth. We have shown previously that UV treatment does not block internalization of the virus particle or movement of virion tegument proteins to the nucleus but does prevent detectable expression of virus-encoded mRNAs (46).
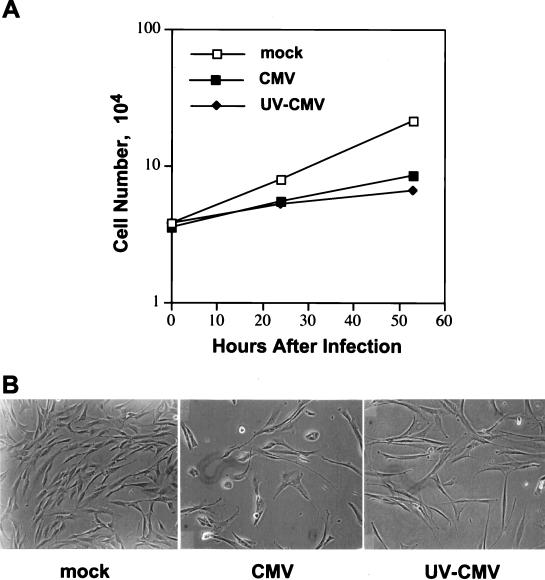
We confirmed that no virus-encoded IE1 mRNA could be detected at 12 h after infection with UV-inactivated HCMV (data not shown), but this stock of virus retained its ability to inhibit the growth of HF cells by 24 h after infection (Fig. 1A). Cytopathic effect was evident by 48 h after infection with the competent virus but not after infection with the UV-inactivated virus (Fig. 1B). Nevertheless, substantially fewer cells accumulated after infection with the UV-inactivated virus in comparison to mock-infected cells (Fig. 1), consistent with the view that cells did not die after infection but failed to divide. It is unlikely that this growth arrest was induced by release of the damaged viral DNA into cells, since UV-inactivated virus does not induce mRNAs encoding DNA repair enzymes (47). We conclude that the initial interaction of the virus particle with the cell surface, the process of internalization, or the action of a virion protein within the cell is responsible for blocking cell growth.
FIG. 1.
Cell growth experiments demonstrating the growth-inhibitory effect of UV-inactivated HCMV. Cultures of preconfluent HF cells were mock infected, infected with HCMV at a multiplicity of 5 PFU/cell, or infected with a quantity of UV-inactivated HCMV equivalent to 5 PFU/cell prior to its inactivation. (A) At different times after infection, cells were collected by trypsinization and counted with a hemacytometer. The av mean of triplicate determinations is presented. (B) Preconfluent HF cells were mock infected or infected as described above and photographed at 53 h after infection.
UL69 protein induces G1 arrest in U-2 OS cells.
The possibility that a virion protein was responsible for the effect on cell growth drew our attention to pUL69, which has recently been shown to be a constituent of the tegument domain of the virus particle (45). As noted above, UL69 is a homologue of the herpes simplex virus ICP27 gene that is needed for the induction of a cell cycle block by herpes simplex virus (29). We initially tested the effect of UL69 protein on cell cycle progression in U-2 OS cells.
pHM160 (44), a plasmid expressing the UL69 gene under the control of the HCMV immediate-early promoter, was transfected into U-2 OS cells together with pBB14, a plasmid expressing a Golgi membrane-localized EGFP-containing fusion protein (6). EGFP served as a marker to distinguish transfected (Fig. 2A, R3 region) from untransfected cells (R2 region), so that the cell cycle distribution of the two cell populations could be compared. Since asynchronous U-2 OS cells include a large G1 population, it is somewhat difficult to observe and quantify a G1 arrest (23). Therefore, nocodazole was added to the culture medium at 24 h before cell harvest to induce a G2/M block. As a result, cells will accumulate in the G2/M compartment, unless they are transfected with a plasmid expressing a gene product that induces a G1 arrest.
FIG. 2.
FACS analysis demonstrating that UL69 induces a G1 arrest in U-2 OS cells. (A) pHM160, which expresses UL69, was transfected into U2OS cells together with pBB14, which expresses an EGFP-containing fusion protein. EGFP serves as a marker to distinguish the highly transfected population (R3) from the untransfected population (R2) of cells. (B) Following a nocodazole block, untransfected (R2) and transfected (R3) cells were analyzed for DNA content by FACS. (C) The ratio of the G1 population in transfected cells to that in untransfected cells was calculated based on three independent FACS determinations. Standard deviations were too small to be seen on the scale used for this presentation.
Following the nocodozole block, UL69-transfected cultures (Fig. 2B) contained a much larger G1 population (11.3%) than was observed in nontransfected cells (3.4%) or in cells transfected with the vector plasmid containing no insert (3.3% [data not shown]). In multiple, independent experiments, pUL69 more efficiently blocked cells in G1 than did overexpression of the p53 tumor suppressor protein (Fig. 2C), a prototypical inducer of G1 arrest (reviewed in reference 27). Neither pflipUL69, expressing an antisense UL69 transcript, nor pCGN65, expressing the abundant pp65 HCMV tegument protein, induced a G1 arrest in this assay (Fig. 2C). Therefore, we conclude that UL69 induces a G1 cell cycle arrest when transiently expressed in U-2 OS cells.
UL69 induces a G1 arrest in HF cells.
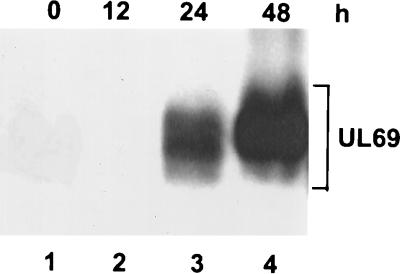
Since U-2 OS cells are nonpermissive for HCMV infection, we next asked whether UL69 protein induces a G1 arrest in HF cells which are permissive for HCMV infection. Due to the low transfection efficiency of HF cells (0.1 to 1%), it is difficult to analyze the effect of UL69 on cell cycle distribution by using the experimental design devised for U-2 OS cells. Therefore, a retrovirus vector (26) was used to express UL69 in HF cells. The retrovirus vector is referred to as retro-UL69 and expresses the UL69 coding region from the retroviral LTR. The expression of UL69 following infection with the recombinant retrovirus at a multiplicity of 5 fluorescence units per cell was monitored by Western blotting. UL69 protein was first detected at 24 h, and it accumulated to a high level at 48 h after infection (Fig. 3). The expression kinetics of UL69 was similar to that of EGFP following infection with a control recombinant retrovirus, retro-GFP (data not shown). Furthermore, UL69 expressed by retro-UL69 was resolved into a broad band after electrophoresis, presumably reflecting its accumulation as multiple phosphoforms.
FIG. 3.
Analysis of UL69 expression in HF cells from a recombinant retrovirus by Western blotting with a UL69-specific monoclonal antibody. Samples are from cells that were infected with retro-UL69 at a multiplicity of 5 fluorescence units per cell and harvested at 0 h (lane 1), 12 h (lane 2), 24 h (lane 3) and 48 h (lane 4) after infection. The broad smear includes bands corresponding to the multiple UL69 phosphoforms.
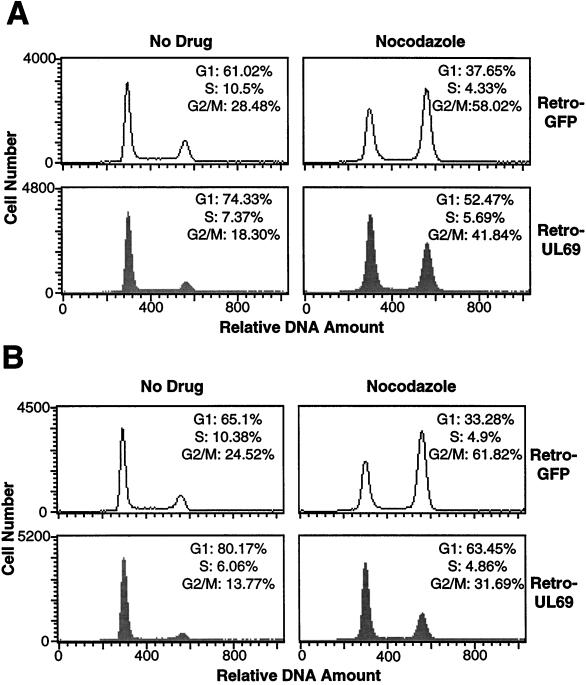
By using the same conditions for retro-UL69 infection as described above, where all the cells in the culture were infected, the cell cycle distribution of HF cells was analyzed by FACS analysis. At 24 h after infection, there was no difference in the cell cycle distribution of retro-UL69-infected and that of retro-GFP-infected cells (data not shown). At 48 h after infection (Fig. 4A, No Drug), a larger G1 population was observed in the retro-UL69-infected culture (74.3%) than in the retro-GFP-infected culture (61.0%); and at 60 h after infection (Fig. 4B, No Drug), the accumulation of cells in the G1 compartment continued to be higher for the retro-UL69-infected culture (80.2%) than for the retro-GFP-infected culture (65.1%). The G1 arrest following retro-UL69 infection was also evident when nocodazole was added to the culture medium at 12 h before cell harvest. At 60 h after infection, 63.5% of cells infected with retro-UL69 and 33.3% of cells infected with retro-GFP were in the G1 compartment (Fig. 4B, Nocodazole). Presumably, the proportion of cells reaching the drug-induced block at G2/M would be even greater for the retro-GFP-infected culture if the cells were maintained in the drug for a longer period. The increased number of cells in the G1 compartment (Fig. 4) correlated well with the accumulation of UL69 protein after infection with retro-UL69 (Fig. 3). Clearly, HF cells accumulate in the G1 compartment following infection with retro-UL69.
FIG. 4.
FACS analysis of HF cells infected with retroviruses expressing the UL69 protein or EGFP at a multiplicity of 5 fluorescence units per cell. At 48 h (A) or 60 h (B) after infection (left panels), the cells were subjected to FACS analysis. Nocodazole was added at 12 h before harvest to the samples analyzed in the right panels. The data displayed are from a single experiment representative of three independent experiments that produced similar results.
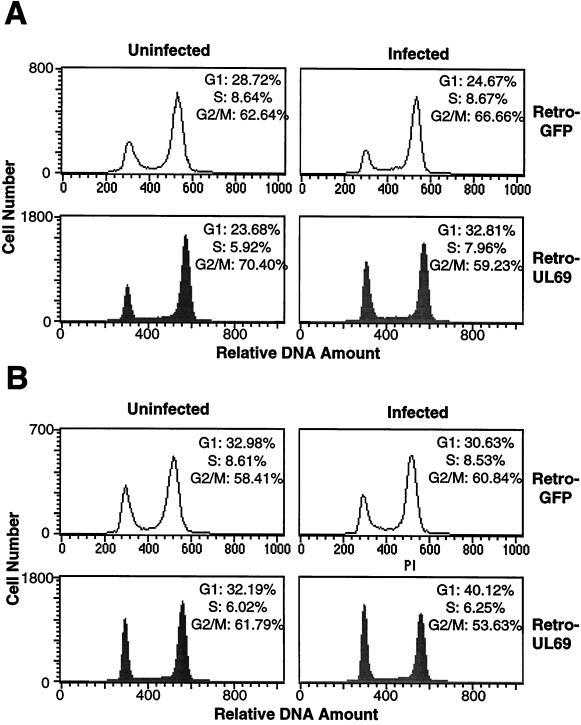
Although unlikely, it was conceivable that the G1 cell cycle arrest observed following retro-UL69 infection was induced by a nonviral contaminant in the retro-UL69 inoculum rather than by the UL69 protein. To exclude this possibility, FACS analysis was performed after infection at a multiplicity of 0.5 fluorescence unit per cell, where approximately 50% of the cells in the HF culture were infected by retro-UL69 or the control retro-GFP. Retro-UL69-infected cells were identified by immunofluorescent staining with a UL69-specific antibody, while retro-GFP-infected cells were identified by their green fluorescent signal. At 48 h after infection, 32.8% of retro-UL69-infected cells were in G1 while only 23.7% of uninfected cells in the same culture resided in this compartment of the cell cycle after a nocodazole block (Fig. 5A). In contrast, no increase in the portion of cells residing in the G1 phase was observed for the retro-GFP-infected cells compared to uninfected cells in the same culture (Fig. 5A). Similar results were obtained when the analysis was performed at 60 h after infection (Fig. 5B), arguing that the altered cell cycle distribution results from expression of UL69 protein rather than from a contaminant in the viral stocks. The increase in the proportion of cells in the G1 compartment was smaller in the experiment performed at a multiplicity of 0.5 fluorescence unit per cell (Fig. 5) than in the experiment performed with 5 fluorescent units per cell (Fig. 4). We suspect that this difference results from a reduced accumulation of UL69 protein in the cells infected at the lower multiplicity of infection, but we have not yet tested this possibility.
FIG. 5.
FACS analysis of HF cells infected with retroviruses expressing the UL69 protein or EGFP at a multiplicity of 0.5 fluorescence unit per cell. At 36 and 48 h after infection, nocodazole was added to the culture medium; 12 h later, at 48 (A) and 60 h after infection (B), the cells were subjected to FACS analysis. Retro-UL69-infected cells were identified by immunofluorescent staining with an antibody to UL69, while the retro-GFP-infected cells were identified by their green fluorescent signal. The data shown are from a single experiment representative of two independent experiments with similar results.
Taken together, the experiments described above demonstrate that UL69 expression results in a G1 cell cycle arrest in HF cells, which are permissive for HCMV infection.
UL69 expression blocks S-phase entry upon serum stimulation of quiescent HF cells.
HCMV infection was observed to induce a G1 cell cycle arrest and block S-phase entry (5, 17, 30, 38). To further explore the role that UL69 plays in HCMV-induced cell cycle arrest, we tested its ability to block the G0/G1-to-S transition after stimulation of quiescent cells with serum. Immunofluorescence analysis confirmed that UL69 or EGFP continued to be expressed for at least 1 week after infection with recombinant retroviruses (data not shown), and this long-term expression allowed us to investigate the effect of UL69 expression on the transition from G0/G1 to the S phase.
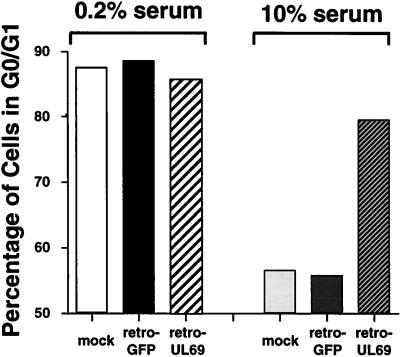
HF cells were infected with retro-UL69 or retro-GFP at a multiplicity of 5 fluorescence units per cell, and 24 h later the cultures were fed with medium containing 0.2% serum. After an additional 48 h, during which cells accumulated in the G0/G1 compartment, the cultures were fed with medium containing 10% serum to reverse the G0/G1 block and induce cell cycle progression. FACS analysis revealed that by 24 h after serum addition (96 h after infection), the portions of mock-infected and retro-GFP-infected cells in the G0/G1 compartment both decreased from nearly 90% to less than 60% (Fig. 6), indicating that these cells moved from the quiescent state and progressed into the S compartment. In contrast, S-phase entry was substantially suppressed in the retro-UL69-infected cells, which still maintained a G1 population of 79.5% (Fig. 6). We suspect that the small decrease in the portion of retro-UL69-infected cells in the G0/G1 phase after serum stimulation (85.8 to 79.5%, Fig. 6) might be due to a decreased steady-state level of UL69 protein, although we have not made quantitative estimates of protein levels.
FIG. 6.
FACS analysis demonstrating that UL69 blocks S-phase entry. HF cells were mock infected or infected at a multiplicity of 5 fluorescence units per cell with retro-GFP or retro-UL69. At 24 h after infection, the cells were synchronized in the G0/G1 compartment by serum starvation; 48 h later (72 h after infection), the cells were fed with serum-containing medium. To analyze cell cycle progression before and after serum stimulation, the cells were subjected to FACS analysis at 72 h after infection, before serum stimulation (0.2% serum), or at 96 h after infection, i.e., 24 h after serum stimulation (10% serum). The percentage of cells in the G0/G1 phase of each infected population was determined. The data shown are from a single experiment representative of two independent experiments with similar results.
These observations demonstrate that UL69 expression blocks S-phase entry upon stimulation of quiescent cells with serum; in this regard, expression of UL69 from the retrovirus vector mimics the effect on cell cycle progression observed after infection with HCMV. However, whereas infection with HCMV also induces a block to cell cycle progression in the G2 compartment (22, 30), UL69-expressing cells did not accumulate to any greater extent in the G2 compartment than did control cells (data not shown). This suggests that another protein coded for or induced by the virus is needed to institute the G2 arrest.
The experiments described above were performed with the UL69 gene product from the AD169 strain of HCMV to modulate cell cycle progression. When the protein from the Towne strain of HCMV was expressed in HF cells with the same retrovirus-derived system, a G1 arrest was observed (data not shown), confirming that the cell cycle effect is a general feature of the protein encoded by different strains of HCMV.
DISCUSSION
The HCMV UL69 protein can arrest cell cycle progression in the G1 compartment (Fig. 2, 4, and 5) and inhibit the transition of quiescent cells into the S compartment (Fig. 6). The UL69 protein is a constituent of the virion (45), and as a result, it is available to act soon after infection to mediate the rapid effects on cell growth and cell cycle progression that we have observed (30). The fact that UL69 protein is delivered to cells within virus particles is also consistent with the ability of UV-inactivated HCMV to inhibit cell growth (Fig. 1), even though it is not able to express its genome. Therefore, we propose that UL69 is at least partially responsible for the G1 block observed in HCMV-infected cells (5, 17, 30), and we are currently constructing a mutant virus lacking the UL69 open reading frame to determine whether additional viral gene products contribute to the G1 block.
HCMV infection arrests progression at multiple points within the cell cycle (30), whereas UL69 blocks only within G1 (Fig. 6). Consequently, other HCMV gene products in addition to UL69 must influence the cell cycle. UL69 might cooperate with other viral proteins to arrest the cell cycle outside of G1; alternatively, other viral products might influence the cell cycle independently of UL69.
As noted above, the HCMV UL69 protein exhibits homology to the herpes simplex virus ICP27 protein, and a mutant virus unable to express ICP27 fails to induce a cell cycle block (29). Therefore, it appears likely that these two proteins share a function, blocking cell cycle progression. It is important to note, however, that the two proteins are not functionally homologous at all levels; UL69 cannot correct the growth defect of an ICP27 mutant (44). Sequence homologues of UL69 and ICP27 have been identified in other members of the herpesvirus family, including ORF4 of varicella-zoster virus (20), UL3 of equine herpesvirus 1 (42), BMLF1 of Epstein-Barr virus (11, 12), and IE52k of herpesvirus saimiri (1, 32). ICP27 (31, 35, 41), UL69 (3, 36, 44), and several other members of this protein family (BMLF1 [24] and ORF4 [15, 16]) have been shown to be transcriptional regulatory proteins. Perhaps they will prove to share cell cycle regulatory functions as well.
How does UL69 protein block cell cycle progression? Since it can influence transcription (44), it might act indirectly by influencing the expression of a cellular protein that regulates cell cycle progression. Inhibition of either cyclin E or cyclin A blocks S-phase entry and cellular DNA synthesis (18, 33, 34), and the level of cyclin A and its associated kinase activity have been shown to be substantially reduced following HCMV infection (5, 22, 38). UL69 protein might act to inhibit cyclin A expression. Perhaps it interferes with posttranscriptional processing of host cell mRNAs, as has been shown for its homologue, ICP27 (19, 39). It is also possible that UL69 induces a G1 cdk inhibitor, such as an INK4 family member (reviewed in reference 40), although two other G1 cdk inhibitors, p21 and p27, are expressed at reduced levels within HCMV-infected cells (5).
Active (30) or UV-inactivated (Fig. 1) HCMV can inhibit cell proliferation, and we have recently reported that active or UV-inactivated virus also can induce interferon-responsive genes (46). Interferon can inhibit the growth of a variety of cell types (reviewed in references 25 and 37), including human embryo lung fibroblasts (4), whose growth is blocked following HCMV infection (5). Similarly to HCMV infection (5, 17, 30), interferon can inhibit the transition from G0/G1 to the S phase, and in some cell types interferon can delay progression at many points in the cell cycle (9). It is conceivable, therefore, that HCMV induces interferon-responsive genes that then mediate the inhibitory effects on cell growth and cell cycle progression. Work is in progress to determine whether UL69 influences the expression of interferon-responsive genes.
ACKNOWLEDGMENTS
We are grateful to J. Liu, D. Knipe, and H. Zhu for sharing their unpublished results. We thank J. Baldick, B. Banfield, A. Brideau, R. Kalejta, J. Lin, O. Petrenko, G. Nolan, T. Stamminger, and M. Stinski for plasmids and cells. We also thank A. Beavis for assistance with FACS analysis, and we thank R. Kalejta and S. Hayashi for helpful discussions and comments on the manuscript.
M.L. was supported by a fellowship from the American Heart Association, and T.S. is an Investigator of the Howard Hughes Medical Institute and an American Cancer Society Professor.
REFERENCES
- 1.Albrecht J-C, Nicholas J, Biller D, Cameron K R, Biesinger R, Newman C, Wittmann S, Craxton M A, Coleman H, Fleckenstein B, Honess R W. Primary structure of the herpesvirus saimiri genome. J Virol. 1992;66:5047–5058. doi: 10.1128/jvi.66.8.5047-5058.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alford C A, Stagno S, Pass R F, Britt W. Congenital and perinatal cytomegalovirus infections. Rev Infect Dis. 1990;12:S745–753. doi: 10.1093/clinids/12.supplement_7.s745. [DOI] [PubMed] [Google Scholar]
- 3.Baldick C J, Marchini A, Patterson C E, Shenk T. Human cytomegalovirus tegument protein pp71 (ppUL82) enhances the infectivity of viral DNA and accelerates the infectious cycle. J Virol. 1997;71:4400–4408. doi: 10.1128/jvi.71.6.4400-4408.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Borden E C, Hogan T F, Voelkel J G. Comparative antiproliferative activity in vitro of natural interferons alpha and beta for diploid and transformed human cells. Cancer Res. 1982;42:4948–4953. [PubMed] [Google Scholar]
- 5.Bresnahan W A, Boldogh I, Thompson E A, Albrecht T. Human cytomegalovirus inhibits cellular DNA synthesis and arrests productively infected cells in late G1. Virology. 1996;224:150–160. doi: 10.1006/viro.1996.0516. [DOI] [PubMed] [Google Scholar]
- 6.Brideau A D, Banfield B W, Enquist L W. The Us9 gene product of pseudorabies virus, an alphaherpesvirus, is a phosphorylated, tail-anchored type II membrane protein. J Virol. 1998;72:4560–4570. doi: 10.1128/jvi.72.6.4560-4570.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Britt W J, Alford C A. Cytomegalovirus. In: Fields B N, Knipe D M, Howley P M, et al., editors. Fields virology. Philadelphia, Pa: Lippincott-Raven Publishers; 1996. pp. 2493–2523. [Google Scholar]
- 8.Cayrol C, Flemington E K. The Epstein-Barr virus bZIP transcription factor Zta causes G0/G1 cell cycle arrest through induction of cyclin-dependent kinase inhibitors. EMBO J. 1996;15:2748–2759. [PMC free article] [PubMed] [Google Scholar]
- 9.Chang A Y, Keng P C. Inhibition of cell growth in synchronous human hypernephroma cells by recombinant interferon alpha-D and irradiation. J Interferon Res. 1983;3:379–385. doi: 10.1089/jir.1983.3.379. [DOI] [PubMed] [Google Scholar]
- 10.Chee M S, Bankier A T, Beck S, Bohni R, Brown C M, Cerny R, Horsnell T, Hatchison C A I, Kouzarides T, Martignetti J A, et al. Analysis of the protein-coding content of the sequence of human cytomegalovirus strain AD169. Curr Top Microbiol Immunol. 1990;154:125–170. doi: 10.1007/978-3-642-74980-3_6. [DOI] [PubMed] [Google Scholar]
- 11.Chevallier-Greco A, Manet E, Chavrier P, Mosnier C, Daillie J, Sergeant A. Both Epstein-Barr virus (EBV)-encoded trans-acting factors, EB1 and EB2, are required to activate transcription from an EBV early promoter. EMBO J. 1986;5:3243–3249. doi: 10.1002/j.1460-2075.1986.tb04635.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cho M-S, Milman G, Hayward S D. A second Epstein-Barr virus early antigen gene in BamHI fragment M encodes a 48- to 50-kilodalton nuclear protein. J Virol. 1985;56:860–866. doi: 10.1128/jvi.56.3.860-866.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.de Beeck A O, Caillet-Fauquet P. Viruses and the cell cycle. Prog Cell Cycle Res. 1997;3:1–19. doi: 10.1007/978-1-4615-5371-7_1. [DOI] [PubMed] [Google Scholar]
- 14.de Bruyn Kops A, Knipe D M. Formation of DNA replication structures in herpesvirus-infected cells requires a viral DNA-binding protein. Cell. 1988;55:857–868. doi: 10.1016/0092-8674(88)90141-9. [DOI] [PubMed] [Google Scholar]
- 15.Defechereux P, Melen L, Baudoux L, Merville-Louis M P, Rentier B, Piette J. Characterization of the regulatory functions of varicella-zoster virus open reading frame 4 gene product. J Virol. 1993;67:4379–4385. doi: 10.1128/jvi.67.7.4379-4385.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Defechereux P, Debrus S, Baudoux L, Rentier B, Piette J. Varicella-zoster virus open reading frame 4 encodes an immediate-early protein with posttranscriptional regulatory properties. J Virol. 1997;71:7073–7079. doi: 10.1128/jvi.71.9.7073-7079.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dittmer D, Mocarski E S. Human cytomegalovirus infection inhibits G1/S transition. J Virol. 1997;71:1629–1634. doi: 10.1128/jvi.71.2.1629-1634.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Girard F, Strausfeld U, Fernandez A, Lamb N J. Cyclin A is required for the onset of DNA replication in mammalian fibroblasts. Cell. 1991;67:1169–1179. doi: 10.1016/0092-8674(91)90293-8. [DOI] [PubMed] [Google Scholar]
- 19.Hardy W R, Sandri-Goldin R M. Herpes simplex virus inhibits host cell splicing, and regulatory protein ICP27 is required for this effect. J Virol. 1994;68:7790–7799. doi: 10.1128/jvi.68.12.7790-7799.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Inchauspe G, Nagpal S, Ostrove J M. Mapping of two varicella-zoster virus-encoded genes that activate the expression of viral early and late genes. Virology. 1989;173:700–709. doi: 10.1016/0042-6822(89)90583-7. [DOI] [PubMed] [Google Scholar]
- 21.Irmiere A, Gibson W. Isolation and characterization of a noninfectious virion-like particle released from cells infected with human strains of cytomegalovirus. Virology. 1983;130:118–133. doi: 10.1016/0042-6822(83)90122-8. [DOI] [PubMed] [Google Scholar]
- 22.Jault F M, Jault J M, Ruchti F, Fortunato E A, Clark C, Corbeil J, Richman D D, Spector D H. Cytomegalovirus infection induces high levels of cyclins, phosphorylated Rb, and p53, leading to cell cycle arrest. J Virol. 1995;69:6697–6704. doi: 10.1128/jvi.69.11.6697-6704.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kalejta R F, Shenk T, Beavis A J. Use of membrane-localized green fluorescent protein allows simultaneous identification of transfected cells and cell cycle analysis by flow cytometry. Cytometry. 1997;29:286–291. doi: 10.1002/(sici)1097-0320(19971201)29:4<286::aid-cyto4>3.0.co;2-8. [DOI] [PubMed] [Google Scholar]
- 24.Kenney S, Kamire J, Holley-Guthrie E, Mar E C, Lin J C, Markovitz D, Pagano J. The Epstein-Barr virus immediate-early gene product, BMLF1, acts in trans by a posttranscriptional mechanism which is reporter gene independent. J Virol. 1989;63:3870–3877. doi: 10.1128/jvi.63.9.3870-3877.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kimchi A. Cytokine triggered molecular pathways that control cell cycle arrest. J Cell Biochem. 1992;50:1–9. doi: 10.1002/jcb.240500102. [DOI] [PubMed] [Google Scholar]
- 26.Kinsella T M, Nolan G P. Episomal vectors rapidly and stably produce high-titre recombinant retrovirus. Hum Gene Ther. 1996;7:1405–1413. doi: 10.1089/hum.1996.7.12-1405. [DOI] [PubMed] [Google Scholar]
- 27.Levine A J. p53, the cellular gatekeeper for growth and division. Cell. 1997;88:323–331. doi: 10.1016/s0092-8674(00)81871-1. [DOI] [PubMed] [Google Scholar]
- 28.Lin J, Chen J, Ellenbaas B, Levine A J. Several hydrophobic amino acids in the p53 amino-terminal domain are required for transcriptional activation, binding to mdm-2 and the adenovirus 5 E1B 55-kD protein. Genes Dev. 1994;8:1235–1246. doi: 10.1101/gad.8.10.1235. [DOI] [PubMed] [Google Scholar]
- 29.Liu, J. J., and D. M. Knipe. Personal communication.
- 30.Lu M, Shenk T. Human cytomegalovirus infection inhibits cell cycle progression at multiple points, including the transition from G1 to S. J Virol. 1996;70:8850–8857. doi: 10.1128/jvi.70.12.8850-8857.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McMahan L, Shaffer P A. The repressing and enhancing functions of the herpes simplex virus regulatory protein ICP27 map to C-terminal regions and are required to modulate viral gene expression very early in infection. J Virol. 1990;64:3471–3485. doi: 10.1128/jvi.64.7.3471-3485.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nicholas J, Cameron K R, Coleman H, Newman C, Honess R W. Analysis of nucleotide sequence of the rightmost 43 kbp of herpesvirus saimiri (HVS) L-DNA: general conservation of genetic organization between HVS and Epstein-Barr virus. Virology. 1992;188:296–310. doi: 10.1016/0042-6822(92)90759-i. [DOI] [PubMed] [Google Scholar]
- 33.Ohtsubo M, Theodoras A M, Schumacher J, Roberts J M, Pagano M. Human cyclin E, a nuclear protein essential for the G1-to-S phase transition. Mol Cell Biol. 1995;15:2612–2624. doi: 10.1128/mcb.15.5.2612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pagano M, Pepperkok R, Verde F, Ansorge W, Draetta G. Cyclin A is required at two points in the human cell cycle. EMBO J. 1992;11:961–971. doi: 10.1002/j.1460-2075.1992.tb05135.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rice S A, Knipe D M. Gene-specific transactivation by herpes simplex virus type 1 alpha protein ICP27. J Virol. 1988;62:3814–3823. doi: 10.1128/jvi.62.10.3814-3823.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Romanowski M J, Shenk T. pIRS1 and pTRS1 are present in human cytomegalovirus virions. J Virol. 1997;71:5703–5705. doi: 10.1128/jvi.71.7.5703-5705.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Romeo G, Fiorucci G, Rossi G B. Interferons in cell growth and development. Trends Genet. 1989;5:19–24. doi: 10.1016/0168-9525(89)90007-3. [DOI] [PubMed] [Google Scholar]
- 38.Salvant B S, Fortunato E A, Spector D H. Cell cycle dysregulation by human cytomegalovirus: influence of the cell cycle phase at the time of infection and effects on cyclin transcription. J Virol. 1998;72:3729–3741. doi: 10.1128/jvi.72.5.3729-3741.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sandri-Goldin R M, Mendoza G E. A herpes virus regulatory protein appears to act posttranscriptionally affecting mRNA splicing. Genes Dev. 1992;6:848–863. doi: 10.1101/gad.6.5.848. [DOI] [PubMed] [Google Scholar]
- 40.Sherr C J, Roberts J M. Inhibitors of mammalian G1 cyclin-dependent kinases. Genes Dev. 1995;9:1149–1163. doi: 10.1101/gad.9.10.1149. [DOI] [PubMed] [Google Scholar]
- 41.Sekulovich R E, Leary K, Sandri-Goldin R M. The herpes simplex virus type 1 α protein ICP27 can act as a trans-repressor or trans-activator in combination with CP4 and ICP0. J Virol. 1988;62:4510–4522. doi: 10.1128/jvi.62.12.4510-4522.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Telford E A, Watson M S, McBride K, Davison A J. The DNA sequence of equine herpesvirus-1. Virology. 1992;189:304–316. doi: 10.1016/0042-6822(92)90706-u. [DOI] [PubMed] [Google Scholar]
- 43.Thomsen D R, Stenberg R M, Goins W F, Stinski M F. Promoter-regulatory region of the major immediate early gene of human cytomegalovirus. Proc Natl Acad Sci USA. 1984;81:659–663. doi: 10.1073/pnas.81.3.659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Winkler M, Rice S A, Stamminger T. UL69 of human cytomegalovirus, an open reading frame with homology to ICP27 of herpes simplex virus, encodes a transactivator of gene expression. J Virol. 1994;68:3943–3954. doi: 10.1128/jvi.68.6.3943-3954.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Winkler M, Stamminger T. A specific subform of the human cytomegalovirus transactivator protein pUL69 is contained within the tegument of virus particles. J Virol. 1996;70:8984–8987. doi: 10.1128/jvi.70.12.8984-8987.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhu H, Cong J-P, Shenk T. Use of differential display analysis to assess the effect of human cytomegalovirus infection on the accumulation of cellular RNAs: Induction of interferon-responsive RNAs. Proc Natl Acad Sci USA. 1997;94:13985–13990. doi: 10.1073/pnas.94.25.13985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhu, H., and T. Shenk. Unpublished data.