Abstract
To improve the efficacy of lenvatinib in combination with programmed death-1 (PD-1) blockade therapy for hepatocellular carcinoma (HCC), we screened the suppressive metabolic enzymes that sensitize HCC to lenvatinib and PD-1 blockade, thus impeding HCC progression. After analysis of the CRISPR‒Cas9 screen, phosphatidylinositol-glycan biosynthesis class L (PIGL) ranked first in the positive selection list. PIGL depletion had no effect on tumor cell growth in vitro but reprogrammed the tumor microenvironment (TME) in vivo to support tumor cell survival. Specifically, nuclear PIGL disrupted the interaction between cMyc/BRD4 on the distant promoter of target genes and thus decreased the expression of CCL2 and CCL20, which are involved in shaping the immunosuppressive TME by recruiting macrophages and regulatory T cells. PIGL phosphorylation at Y81 by FGFR2 abolished the interaction of PIGL with importin α/β1, thus retaining PIGL in the cytosol and facilitating tumor evasion by releasing CCL2 and CCL20. Clinically, elevated nuclear PIGL predicts a better prognosis for HCC patients and presents a positive correlation with CD8 + T-cell enrichment in tumors. Clinically, our findings highlight that the nuclear PIGL intensity or the change in PIGL-Y81 phosphorylation should be used as a biomarker to guide lenvatinib with PD-1 blockade therapy.
Keywords: Nuclear PIGL, cMyc, PD-1 antibody, CCL2/20, Tumor immune evasion.
Subject terms: Cancer microenvironment, Immune evasion
Introduction
The high incidence and recurrence of hepatocellular carcinoma (HCC) have perplexed experts in the field. The currently available first-line therapeutic drugs, such as the tyrosine receptor kinase inhibitor lenvatinib and immunotherapies targeting the programmed death-1 (PD-1)/PD-L1 axis, are still insufficient to significantly improve the five-year survival rate of HCC patients [1, 2], reflecting the need to sensitize liver cancer to this combination treatment and explore HCC patients who are suitable for this treatment.
The immunosuppressive environment in tumor tissue, which is regulated by chemokine-directed pro-oncogenic immune cells, is considered a major obstacle to tumor therapy. For example, CCL2 secreted by tumor tissues, including tumor cells and macrophages, recruits tumor-associated macrophages into tumor tissue. CCL20 secreted by dendritic cells, macrophages or tumor cells recruits regulatory T cells that counteract the expansion and activation of cytotoxic T lymphocytes. The cell types described above create an immunosuppressive microenvironment in tumor tissues [1, 3, 4] that facilitates tumor cell evasion from immunosurveillance. We aimed to elucidate whether lenvatinib affected the tumor immune microenvironment and discovered the metabolic enzyme phosphatidylinositol-glycan biosynthesis class L (PIGL). PIGL is a member of the phosphatidylinositol-glycan biosynthesis (PIG) family and is involved in the second step of GPI biosynthesis by removing an acetyl group from N-acetyl-glucosaminyl-phosphatidylinositol (GPI) [5]. However, in this study, the function of PIGL depended on nuclear localization to inhibit the tumor immune-suppressive microenvironment rather than its enzyme activity. Although most metabolic enzymes are localized in the cytosol, particularly in the mitochondria and endoplasmic reticulum, some metabolic enzymes, such as UGDH, PKM2, and PGK1, exhibit obvious nuclear localization and function in the nucleus [6–8].
cMyc controls global gene expression and regulates cell proliferation, cell differentiation, the cell cycle, metabolism and apoptosis. According to some estimates, Myc, including cMyc and nMyc, is dysregulated in approximately 70% of human cancers, and strong evidence implicates aberrantly expressed Myc in both tumor initiation and maintenance [9, 10]. cMyc cooperates with its partners, such as bromodomain-containing (BRD) family proteins, to efficiently regulate gene expression and specifically and robustly upregulate its target genes. BRD4 is a chromatin reader protein that recognizes and binds acetylated histones, and BRD4 also presents acetyl-transferase activity [11]. BRD4 and cMyc are broadly cobound to the regulatory regions of active genes in promoters or distant promoters. The closeness of cMyc to BRD4 determines the expression levels of some cMyc target genes [12, 13]. However, cMyc is considered an undruggable target because of the peculiar characteristics of the shallow binding pockets on the cMyc protein [14–16]. Thus, disruption of the cMyc/BRD4 complex may be a potential strategy to delay cancer progression.
Here, we used C57BL/6 wild-type and C57BL/6 TCRα−/− mice in which T cells were depleted to screen the metabolic enzyme PIGL, which connects lenvatinib treatment to enhance the efficiency of subsequent PD-1 immunotherapy.
Results
PIGL synthetically induces HCC cell death with the combination of lenvatinib plus PD-1 blockade therapy
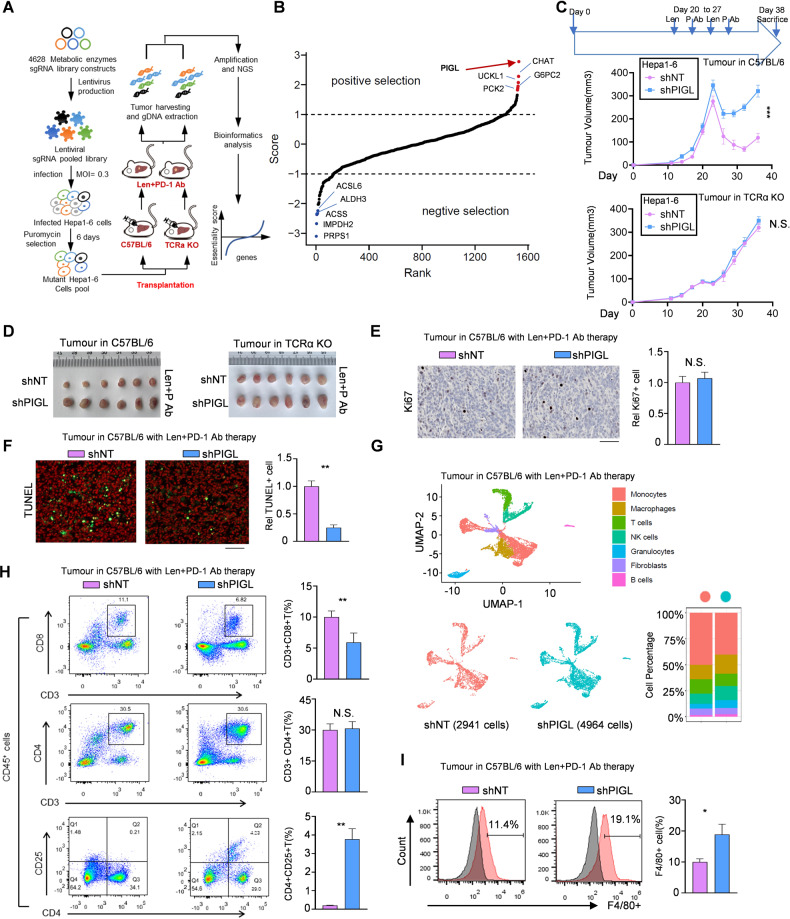
Hepa1-6 cells were infected with a lentivirus containing the metabolic enzyme sgRNA sublibrary and then orthotopically transplanted into C57BL/6 wild-type and C57BL/6 TCRα−/− mice. Next, we applied lenvatinib plus an anti-PD-1 antibody to discover the metabolic enzymes that synthetically induce HCC cell death upon exposure to lenvatinib plus immunotherapy. (Fig. 1A). Compared with C57BL/6 TCRα−/− mice, in C57BL/6 wild-type mice, all three sgRNAs targeting metabolic enzyme PIGL were significantly upregulated 8–17-fold after multiple-testing corrections, and we found that PIGL ranked first in the positive target list (Fig. 1B and S1A). We measured whether PIGL affected cell viability upon lenvatinib treatment and excluded the possibility that PIGL depletion exerted a significant effect on cell viability upon drug treatment (Fig. S1B, C).
Fig. 1.
PIGL synthetically induces HCC cell death with the combination of lenvatinib plus PD-1 blockade therapy. A, B The strategy using a sublibrary of metabolic enzyme sgRNA to screen the suppressive metabolic enzymes that improve the treatment efficiency of lenvatinib and PD-1 immunotherapy in liver cancer (A). The enrichment of positive and negative candidates in the sgRNA screen (B). C, D Subcutaneous transplantation of Hepa1-6 cells with the indicated genetic manipulation into C57BL/6 mice (n = 6) or C57BL/6 TCRα−/− mice (n = 6). P, PD-1 antibody. E, F Cell proliferation (E) and cell death (F) were measured in vivo by Ki67 and TUNEL staining, respectively. Scale bar, 200 μm. G The tumors were subjected to single-cell sequencing. The subtypes of immune cells, macrophages (M), CD8+ T cells, CD4+ T cells, fibroblasts (Fb) and dendritic cells (DC) are shown. H, I The proportions of the T-cell subsets helper T-cell (CD3+CD4+), cytotoxic T lymphocytes (CD3+CD8+), regulatory T cells (CD4+CD25+) (H) and F4/80+ macrophages (I) were analyzed using flow cytometry (FCM). Len, lenvatinib; PD-1 Ab, antibody against PD-1; +, positive. C, one-way ANOVA; E-F and H-I, two-tailed Student’s t test. (N.S., not significant; *p < 0.05; **p < 0.01; ***p < 0.001)
We observed that PIGL depletion in Hepa1-6 cells slightly drove tumor growth and induced significant resistance to the combination of lenvatinib and anti-PD-1 therapy compared with control cells. However, these phenotypes were not observed in subcutaneously transplanted Hepa1-6 cells in C57BL/6 TCRα−/− mice (Fig. 1C, D). We also used in vitro and in vivo competitive growth experiments to identify the survival advantage of PIGL-deficient tumor cells under immune conditions (Fig. S1D), implying the potential role of PIGL in the TIME. The in vivo cell proliferation and cell death assays showed that PIGL depletion had no effect on cell proliferation but upregulated the survival rate of Hepa1-6 cells forming tumors in WT-C57BL/6 mice (Fig. 1E, F) but not in C57BL/6 TCRα−/− mice (Fig. S1E, F). Therefore, we suspect that PIGL modulates TIME remodeling. After PD-1 blockade, single-cell RNA sequencing data indicated that the proportion of macrophages increased while the T-cell proportion decreased in PIGL-depleted tumors (Fig. 1G). Next, we performed flow cytometry to further detect the variation in T-cell subpopulations. As shown, PIGL depletion decreased the proportion of cytotoxic T lymphocytes (CD3+CD8+), and the overall percentage of helper T cells (CD3+CD4+) showed no significant change, while the percentage of regulatory T cells (CD4+CD25+) increased (Fig. S1G, H and Fig. 1H). In addition to the complicated regulatory circuit between T cells and macrophages, we also detected the proportion of macrophages in the solid tumor immune microenvironment by performing F4/80 staining and found that PIGL depletion increased the percentage of F4/80+ macrophages in solid tumors (Fig. 1I).
Taken together, the in vivo CRISPR screen and animal studies indicated that a member of the PIG family, PIGL, repressed the reprogramming of the solid TIME.
Nuclear PIGL was negatively correlated with the prognosis of HCC patients
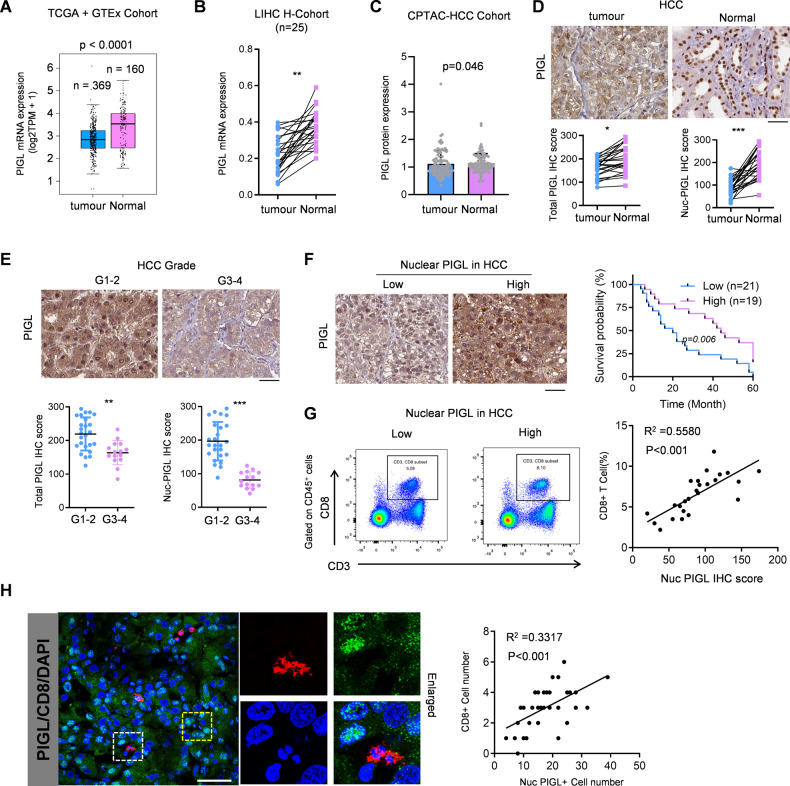
To explore the clinical significance of PIGL in HCC, we extracted information on PIGL mRNA expression in normal and tumor tissues from TCGA and GTEx cohorts. As shown in Fig. 2A, PIGL mRNA levels were downregulated in HCC tumor samples. Consistently, PIGL mRNA expression was also downregulated in our own collection of paired tumor and adjacent normal tissue samples (Fig. 2B). Consistent with the trend in mRNA levels, PIGL protein levels in tumor tissue were also decreased, with a slight difference (Fig. 2C). The subcellular localization of metabolic enzymes is often tightly associated with their functions and clinical significance. We found that in HCC samples, the PIGL protein was enriched in the nucleus in normal tissue but mainly distributed in the cytosol in tumor tissue. Notably, the total PIGL protein level in normal tissue was also slightly higher than that in tumor tissue (Fig. 2D). With increasing tumor grade, PIGL also showed a trend of a disappearance of nuclear localization and an overall decrease in expression (Fig. 2E).
Fig. 2.
Nuclear PIGL was negatively correlated with HCC patient prognosis. A. PIGL mRNA was extracted from the TCGA-HCC database and analyzed. B PIGL mRNA levels from HCC patients (n = 25) were measured using qRT‒PCR. C PIGL protein levels in HCC samples were analyzed. D IHC staining for total PIGL and nuclear PIGL was obtained and analyzed. E The HCC patients were grouped into high or low grades according to pathological diagnosis. Representative images are shown, and the IHC scores for total PIGL and nuclear PIGL were calculated and analyzed. F The survival curves were plotted using the best cutoff of the IHC score. G, H CD8+ T-cell infiltration was measured using FCM and immunofluorescence (IF) and analyzed by calculating the Pearson correlation coefficient. D–F Scale bar, 200 μm; (H), scale bar, 50 μm; A–E, one-way ANOVA; F, two-way ANOVA; G–H Pearson correlation coefficient test (*p < 0.05; **p < 0.01; ***p < 0.001)
We separated the HCC patients into two groups termed high and low, according to the level of total (Fig. S2A) or nuclear localized (Fig. 2F) proteins based on IHC scores, and then performed a Kaplan‒Meier survival analysis to discover whether the PIGL total protein levels or nuclear enrichment predicted the patients’ prognosis. Patients with higher levels of total PIGL protein or nuclear localization had better outcomes, especially patients with greater nuclear localization (Figs. S2A and 2F). Similar results were observed in the survival analysis of nuclear PIGL in TCGA-HCC samples (Fig. S2B). Importantly, nuclear PIGL expression was positively associated with CD8+ T-cell enrichment or recruitment in tumor tissues (Fig. 2G, H), further supporting the speculation that PIGL functions as a tumor suppressor in HCC.
PIGL hampers liver tumor growth independent of its enzyme activity, likely by suppressing CCL2/20 expression
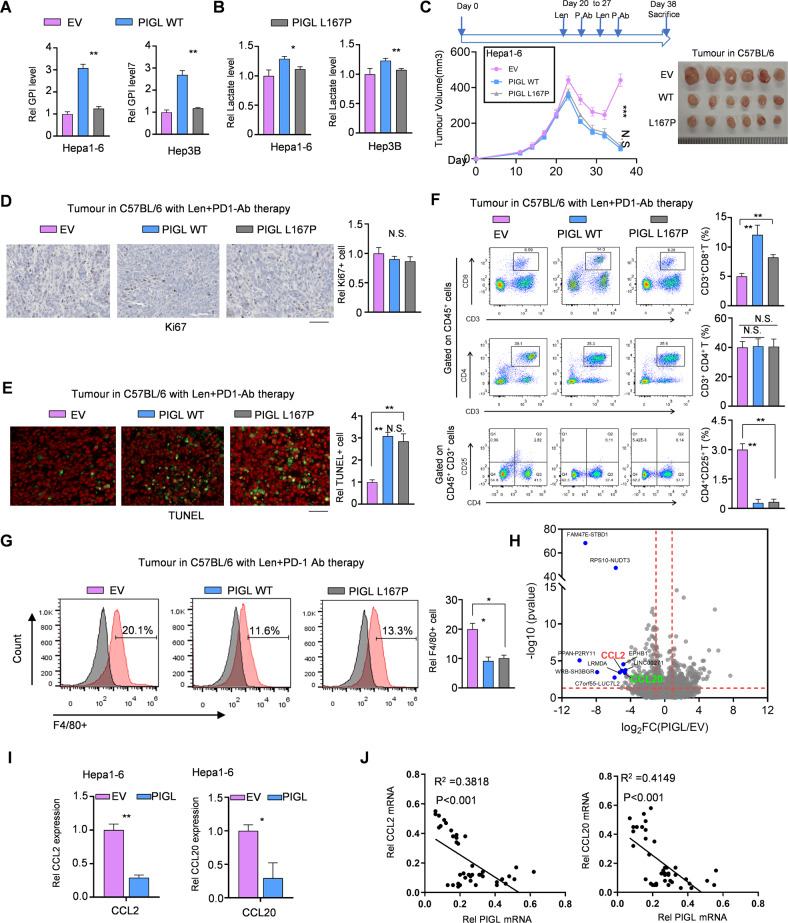
PIGL acts in the second step of GPI biosynthesis, and many eukaryotic proteins bind to membranes using a glycosylphosphatidylinositol (GPI) anchor, which contributes to TME remodeling. We constructed the mutant L167P-PIGL, which substantially impaired the enzyme activity (Fig. S3A) in driving GPI biosynthesis and glycolysis. Hence, we measured GPI and lactate levels in inactive PIGL-overexpressing cells and found that, compared with the WT group, GPI levels in the L167P group were considerably decreased and lactate levels were slightly decreased (Fig. 3A, B). Upon lenvatinib treatment in vitro, the cells forcibly expressing WT- or L167P-PIGL showed a similar trend of cell viability as the cells expressing the empty vector control (Fig. S3B), while the in vivo tumor growth assay showed that the cells forcibly expressing WT- or L167P-PIGL formed smaller solid tumors than the cells expressing the empty vector control. Notably, the tumor volume was not obviously different between the WT- and L167P-mutant PIGL groups (Fig. 3C). Compared with the empty vector group, the in vivo cell proliferation and cell death assays presented similar proliferation rates but lower survival probabilities in the WT- and L167P-PIGL groups. Of note, the tumor cells in both the WT- and L167P-PIGL groups showed the same proliferation and survival probability (Fig. 3D, E). We also detected the proportion of T-cell subsets, namely, helper T cells (CD3+CD4+), cytotoxic T lymphocytes (CD3+CD8+), and regulatory T cells (CD4+CD25+), in the solid tumors formed by the indicated Hepa1-6 cells in C57BL/6 mice. Compared with the empty vector control, overexpression of both WT-PIGL and L167P-PIGL in Hepa1-6 cells efficiently increased CD8+ T-cell populations but repressed regulatory T-cell populations in solid tumors (Figs. S3C and 3F). Additionally, the proportion of F4/80+ macrophages decreased in solid tumors formed by Hepa1-6 cells overexpressing WT- or L167P-PIGL (Fig. 3G).
Fig. 3.
PIGL hampers liver tumor growth independent of its activity, likely by suppressing CCL2/20 expression. A, B GPI and lactate levels were measured in Hepa1-6 cells overexpressing control, WT-PIGL or L167P-PIGL. C Subcutaneous transplantation of Hepa1-6 cells with the indicated genetic manipulation into C57BL/6 mice (n = 6). D, E In the solid tumors shown in (C), in vivo cell proliferation (D) and cell death (E) were measured by performing Ki67 and TUNEL staining, respectively. Scale bar, 200 μm. F, G The proportions of the T-cell subsets helper T-cell (F) and F4/80+ macrophages (G) were analyzed using flow cytometry. The gray peaks in (G) are negative isotype controls. H–J Using Hepa1-6 cells overexpressing WT-PIGL or EV, RNA profiling was performed. The differentially expressed genes (DEGs) were analyzed, and the top 10 DEGs are presented (H). qRT‒PCR was performed using specific primers against CCL2/20 mRNAs (I). The correlation between PIGL mRNA expression and CCL2 mRNA or CCL20 mRNA expression was analyzed by calculating the Pearson correlation coefficient (J). A, B, D–H Two-tailed Student’s t-test.; C Two-way ANOVA; J Pearson correlation coefficient test (N.S., not significant; *p < 0.05; **p < 0.01)
To explore the mechanism by which PIGL disturbs the immunosuppressive TME in solid tumors, we profiled the total RNA in PIGL-overexpressing and control tumor cells. CCL2 and CCL20 were the only two chemokines among the top 10 downregulated genes. qRT‒PCR confirmed the RNA sequencing results (Fig. 3I) obtained using PIGL-overexpressing and control cells. The levels of the secreted CCL2/20 proteins, as measured using ELISA, also showed a consistent trend with that of the mRNA level (Fig. S3D). Furthermore, the mRNA levels of both CCL2 and CCL20 were negatively correlated with PIGL mRNA expression in our own clinical HCC samples (Fig. 3J). Taken together, we proposed that independent of its activity, PIGL constrained tumor immune microenvironment (TIME) remodeling, probably by suppressing CCL2/20 expression.
The combination of CCL2/20 neutralization plus PD-1 blockade immunotherapy blocked PIGL depletion-driven liver cancer progression
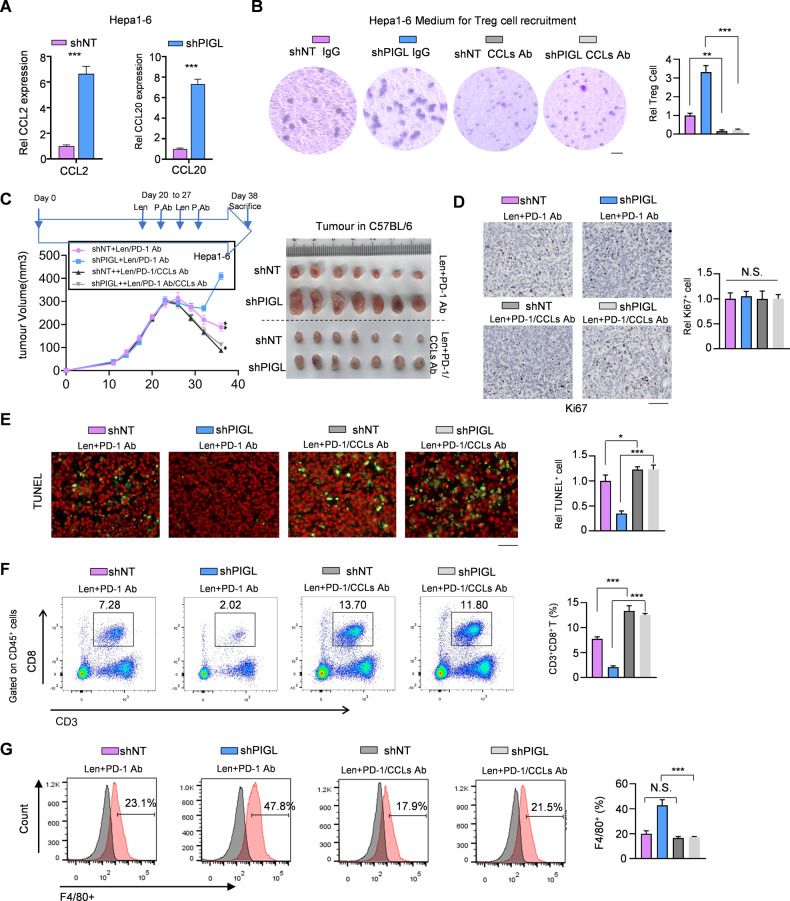
We primarily determined that PIGL depletion in Hepa1-6 cells increased CCL2/20 expression (Fig. 4A) and increased the levels of the secreted soluble CCL2/20 protein (Fig. S4A). Secreted CCL2 and CCL20 in tumor tissues mainly facilitate the recruitment of TAMs and regulatory T cells, respectively [17]. We observed that the medium from cultured PIGL-depleted Hepa1-6 cells recruited more regulatory T cells and macrophages, while the addition of antibodies neutralizing CCL2/20 impaired this recruitment (Figs. 4B and S4B, C).
Fig. 4.
The combination of CCL2/20 neutralization plus anti-PD-1 therapy blocked PIGL depletion-driven liver cancer progression. A CCL2/20 mRNA levels were detected in Hepa1-6 cells with or without PIGL depletion using qRT‒PCR. B After treatment with or without CCL2/20 neutralizing antibodies, the effect of conditioned medium (CM) from the indicated genetically manipulated Hepa1-6 cells on regulatory T-cell recruitment was detected by performing a transwell migration assay. Scale bar, 20 μm. C Subcutaneous transplantation of Hepa1-6 cells with the indicated genetic manipulation into C57BL/6 mice (n = 6). The treatment was performed as described in the supplementary methods. D, E In the solid tumors shown in (C), in vivo cell proliferation (D) and cell death (E) were measured by performing Ki67 and TUNEL staining, respectively. Scale bar, 200 μm. F, G In the solid tumors shown in (C), the proportions of the T-cell subsets (F) and F4/80+ macrophages (G) were analyzed using FCM. C One-way ANOVA; A, B, D–G Two-tailed Student’s t-test. (N.S., not significant; *p < 0.05; **p < 0.01; ***p < 0.001)
We injected the CCL2/20 antibody into the tail veins of mice to neutralize CCL2/20 in vivo, and ELISA showed that neutralization was successfully achieved (Fig. S4D). The recruitment of TAMs and regulatory T cells reflected an immunosuppressive microenvironment that drove cancer progression, especially by facilitating tumor cell evasion from immune checkpoint blockade. As expected, Hepa1-6 cells with PIGL depletion formed the largest tumors and resisted PD-1 blockade immunotherapy, while the addition of CCL2/20 antibody reversed this resistance and effectively improved the efficiency of PD-1 antibody treatment in PIGL depletion-driven liver cancer (Fig. 4C). Ki67 and TUNEL staining of tumor samples showed a consistent result that the addition of CCL2/20 antibody enhanced the efficiency of PD-1 antibody-induced cytotoxic T lymphocyte expansion, which exerted no effect on tumor cell proliferation but promoted tumor cell death (Fig. 4D, E). Flow cytometry showed that the addition of the CCL2/20 antibody increased the cytotoxic T lymphocyte population but blocked the recruitment of TAMs (Fig. 4F, G).
In summary, the administration of the CCL2/20 antibody rescued PIGL deficiency by enhancing cytotoxic T lymphocyte expansion, thereby improving the efficacy of immune checkpoint PD-1 blocking therapy.
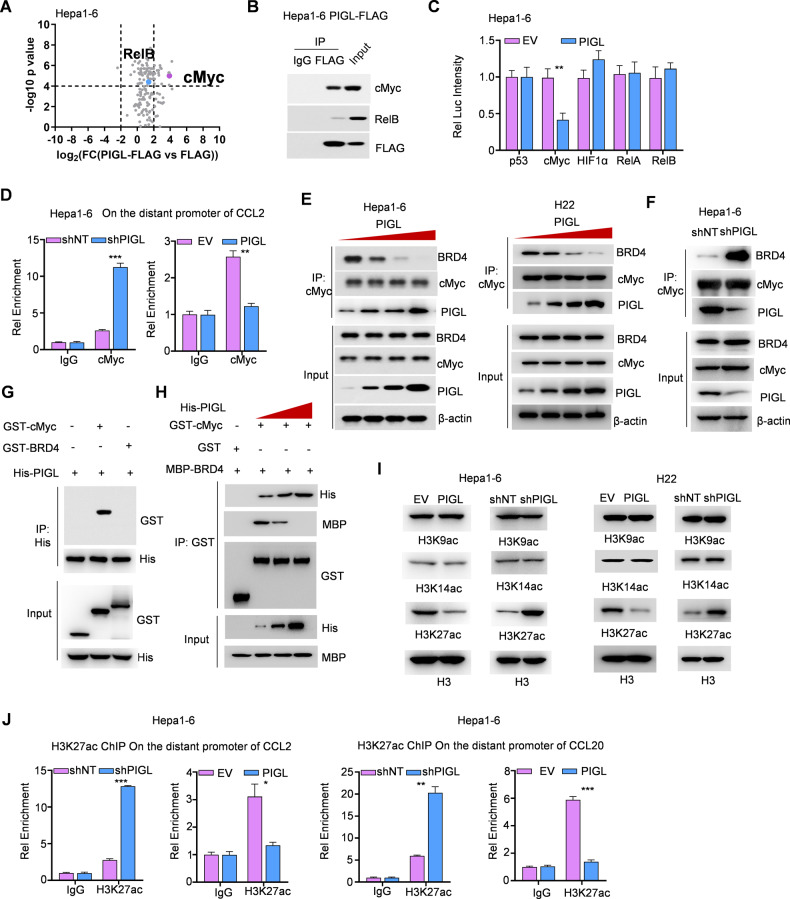
PIGL targeted cMyc to disrupt the cMyc/BRD4 axis and then reduced H3K27ac levels on the distant CCL2/20 promoter
The mechanism by which PIGL regulates CCL2/20 expression in tumor cells remains elusive. We enriched PIGL and its associated proteins in Hepa1-6 cells and subjected them to LC‒MS analysis to explore the mechanism by which PIGL modulates CCL2/20 expression. As shown in Fig. 5A, two transcription factors, cMyc and RelB, which directly regulate gene expression, were discovered to potentially interact with PIGL (Fig. 5A). Forward co-IP confirmed the interaction between PIGL and cMyc or RelB to different degrees (Fig. 5B). Luciferase assays showed that PIGL overexpression inhibited cMyc but not RelB/A, p53 or HIF1α transactivation (Fig. 5C). Conversely, PIGL depletion upregulated cMyc transactivation in Hepa1-6 cells (Fig. S5A).
Fig. 5.
PIGL targeted cMyc to disrupt the cMyc/BRD4 axis and then reduced H3K27ac levels on the distant CCL2/20 promoter. A, B FLAG-PIGL-associated proteins were identified using LC‒MS/MS (A) and ascertained by Co-IP (B). C Dual luciferase reporter assay. Five major transcription factors, p53, cMyc, HIF1α and RelA/B, were cotransfected with pGL3-response element and Renilla internal control plasmids into Hepa1-6 cells with or without PIGL overexpression. D ChIP‒qPCR assay. In Hepa1-6 cells with PIGL depletion or overexpression, an antibody against cMyc was used to enrich the corresponding DNA fragment. E, F Co-IP assay. In Hepa1-6 or H22 cells, PIGL was gradually expressed (E), and PIGL was depleted or overexpressed (F). An antibody against cMyc was used to enrich its associated complex. G In vitro pulldown assay. GST-tagged BRD4, cMyc and His-tagged PIGL were purified from E. coli and incubated together. Antibodies against His were used to enrich PIGL, and antibodies against GST were used to test the direct interaction between PIGL and cMyc or BRD4. H In vitro pulldown assay. GST-tagged cMyc, MBP-tagged BRD4 and His-tagged PIGL with gradually increasing levels were incubated together. Antibodies against GST were used to enrich cMyc, and antibodies against MBP or His were used to test the direct interaction between cMyc and BRD4 or PIGL. I In Hepa1-6 or H22 cells with PIGL depletion or overexpression, histone H3 acetylation at K9, K14 and K27 was detected. J ChIP‒qPCR assay. In Hepa1-6 or H22 cells with PIGL depletion or overexpression, an antibody against H3K27ac was used to enrich the corresponding DNA fragment. C One-way ANOVA; E, F, H, I Two-tailed Student’s t-test. (N.S., not significant; *p < 0.05; **p < 0.01; ***p < 0.001)
Then, Ch-IP using an antibody against cMyc profiled the region to which cMyc bound to the CCL2/20 promoters (Fig. S5B, C). Upon PIGL depletion, cMyc targeted the distant promoters of CCL2/20, while PIGL overexpression abolished this targeting (Fig. 5D and S5D, E), indicating that the interaction of cMyc with the distant promoters of CCL2/20 genes depends on the nuclear PIGL level. cMyc forms a complex with BRD4 at the promoters of target genes; thus, BRD4 and cMyc are broadly bound to the regulatory regions of active genes [13]. We suspected that PIGL enters the nucleus and disrupts the cMyc/BRD4 complex. We gradually overexpressed PIGL in both Hepa1-6 and H22 cells to test this hypothesis and observed that cMyc gradually lost its association with BRD4, accompanied by increased PIGL expression (Fig. 5E). Conversely, in Hepa1-6 cells with PIGL depletion, the interaction of cMyc with BRD4 was reinforced (Fig. 5F). In an in vitro binding assay, GST pulldown showed that PIGL directly interacted with cMyc but not BRD4 (Fig. 5G). The gradual increase in PIGL physically extruded BRD4 from cMyc (Fig. 5H).
BRD4 is a chromatin reader protein that recognizes and binds acetylated histones, and BRD4 presents acetyl-transferase activity. Hence, we tested the acetylation levels at the following sites: H3K9, H3K14 and H3K27. As shown in Fig. 5I, loss of PIGL increased H3K27 acetylation (H3K27ac), while overexpression of PIGL moderately blocked acetylation of this site (Fig. 5I). H3K27ac levels on the distant promoters of CCL2/20 were inversely regulated by PIGL levels in both Hepa1-6 and H22 cells (Fig. 5J and S5F).
Taken together, we ascertained that cMyc physically interacts with PIGL, the level of which determines the closeness between cMyc and BRD4.
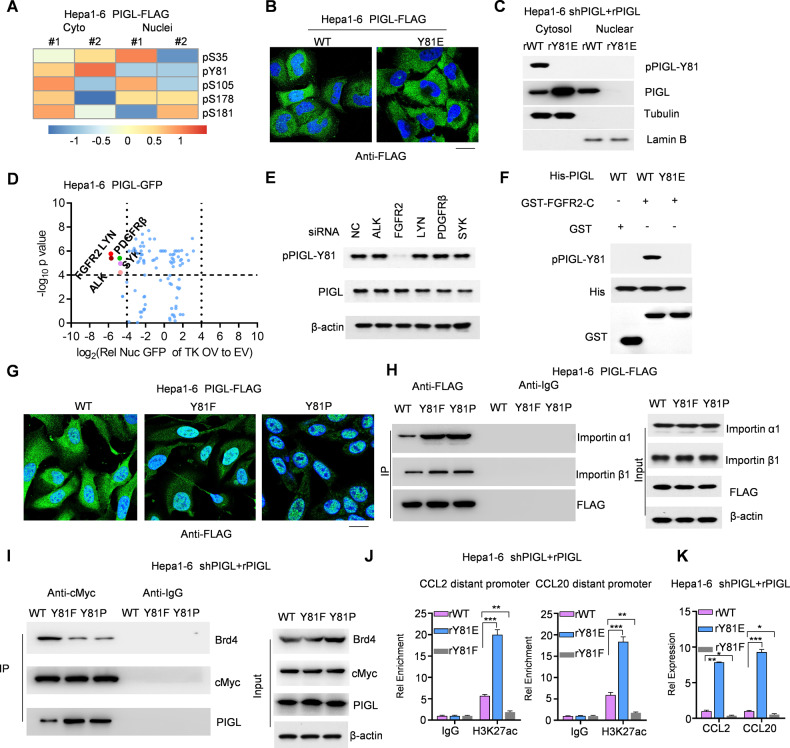
FGFR2-mediated PIGL-Y81 phosphorylation promoted PIGL retention in the cytosol and reinforced the cMyc/BRD4 axis
Previous data indicate that PIGL disrupts the cMyc/BRD4 complex on the distant promoters of CCL2/20 genes. We enriched cytosolic and nuclear PIGL and subjected them to LC‒MS analysis to explore the mechanism by which PIGL enters the nucleus. As shown, cytosolic PIGL had a much stronger intensity of phosphorylation at Y81, while other phosphorylated sites showed no significant differences in intensity in cytosolic and nuclear PIGL (Fig. 6A). We mimicked Y81 phosphorylation as Y81E mutant PIGL, which was mainly located in the cytosol, while WT-PIGL was located in both the cytosol and nucleus (Fig. 6B). We restored the expression of WT- or Y81E-PIGL in Hepa1-6 and H22 cells with PIGL depletion (Fig. S6A). Cytosol and nucleus fractionation showed that Y81 phosphorylation mainly occurred in the cytosol, which was tested using a specific antibody against phosphorylated Y81 (Fig. 6C and S6B).
Fig. 6.
FGFR2-mediated PIGL-Y81 phosphorylation promoted PIGL retention in the cytosol and reinforced the cMyc/BRD4 axis. A PIGL-FLAG from the cytosol and nucleus was subjected to LC‒MS to identify posttranslational modifications. The detected modifications with strong intensity are summarized in the heatmap. B WT- or Y81E-PIGL-FLAG was expressed in Hepa1-6 cells and stained with an antibody against FLAG. C The cytosol and nucleus of Hepa1-6 cells expressing restored WT-PIGL or Y81E-PIGL were fractioned. D Hepa1-6 cells expressing GFP-tagged PIGL were transfected with a tyrosine kinase (TK) overexpression vector (OV), and nuclear GFP intensity was captured. The ratio of GFP intensity in the TK OV group to that in the EV group was determined and analysed. E siRNAs targeting five TK coding genes were transiently transfected into Hepa1-6 cells. F The GST-tagged intracellular C-terminus of FGFR2 was incubated with His-tagged PIGL in the kinase reaction system. G, H WT-, Y81F- or Y81P-PIGL-FLAG was stably overexpressed in Hepa1-6 cells. An antibody against FLAG was used to stain PIGL-FLAG in these cells (G), and the association of WT- and PIGL-FLAG with importin α1/β1 was ascertained using Co-IP with FLAG antibody or IgG as a blank control (H). I–K Using Hepa1-6 cells with restored expression of WT-, Y81F- or Y81P-PIGL, Co-IP with cMyc antibody or IgG as a blank control was performed to test the association of cMyc with BRD4 and PIGL (I). J, K In the indicated Hepa1-6 cells, ChIP‒qPCR using an anti-H3K27ac antibody was performed to test H3K27ac enrichment on the distant promoters of CCL2/20 (J). Readout gene expression was detected using qRT‒PCR (K). C, D, J Two-tailed Student’s t-test. (N.S., not significant; *p < 0.05; **p < 0.01; ***p < 0.001)
As the number of tyrosine phosphorylation kinases is currently limited, by analyzing nuclear PIGL-GFP levels with tyrosine kinase overexpression, we observed that the five indicated tyrosine kinases should have the potential to phosphorylate PIGL at Y81 (Fig. 6D). Using siRNAs to deplete the respective tyrosine kinases (Fig. S6C), we primarily verified that FGFR2, a target of lenvatinib, may contribute to Y81 phosphorylation (Fig. 6E). An in vitro kinase assay ascertained that the FGFR2 C-terminus presented tyrosine kinase activity for PIGL-Y81 (Fig. 6F).
We mimicked dephosphorylation at Y81 by mutating Y81 to F81 or P81, and immunofluorescence (IF) staining showed that the mimicked dephosphorylation mutant PIGL was almost completely retained in the cytosol (Fig. 6G), which might be explained by the results from the Co-IP assay between importin α/β1 and the mimicked dephosphorylation mutant PIGL (Fig. 6H). We also restored the expression of WT-, Y81F-, and Y81P-PIGL in Hepa1-6 and H22 cells with PIGL depletion (Fig. S6D). After mimicking dephosphorylation at Y81, the interaction between cMyc and PIGL was substantially increased, and the cMyc/BRD4 axis was heavily impaired (Figs. 6I and S6E). Consequently, the intensity of the H3K27ac signal on the distant promoters was decreased upon mimicking dephosphorylation of Y81 in PIGL (Figs. 6J and S6F), and CCL2/20 expression was also downregulated (Fig. 6K and S6G, H).
Overall, FGFR2-mediated PIGL-Y81 phosphorylation retains PIGL in the cytosol by blocking the interaction of importin α/β1 with Y81-phosphorylated PIGL and reinforces the formation of the cMyc/BRD4 complex to promote CCL2/20 expression.
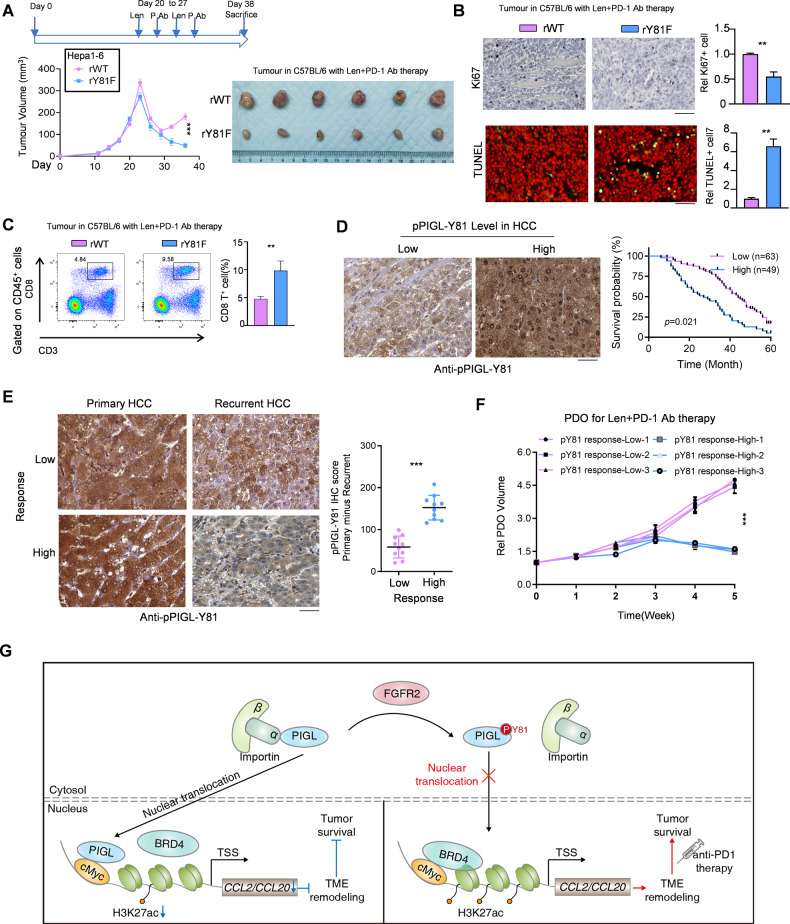
The alteration of PIGL-Y81 phosphorylation upon lenvatinib treatment predicts resistance to PD-1 blockade therapy
A clear mechanism or molecular marker for determining HCC patients who are suitable for combination therapy with lenvatinib and PD-1 blockade is still unavailable. We established a xenograft model to test whether PIGL-Y81 phosphorylation affects tumor growth following anti-PD-1 antibody treatment. As shown in Fig. 7A, compared with tumors formed by control Hepa1-6 cells, tumors formed by Hepa1-6 cells expressing the Y81-dephosphorylated PIGL mimic were much more sensitive to anti-PD-1 antibody treatment (Fig. 7A). Consistent with this finding, in vivo assays of cell proliferation and cell death, as measured by Ki67 and TUNEL staining, respectively, showed that the tumors formed by Hepa1-6 cells expressing the Y81-dephosphorylation mimic PIGL presented a similar proliferation rate but higher cell death (Fig. 7B). The proportion of intratumor cytotoxic T lymphocytes was increased when PIGL-Y81 in Hepa1-6 cells was not phosphorylated (Fig. 7C). Additionally, ELISAs showed that less CCL2/20 was secreted in the tumors formed by Hepa1-6 cells expressing the Y81-dephosphorylation mimic PIGL (Fig. S7A).
Fig. 7.
PIGL-Y81 phosphorylation predicts potential resistance to combined lenvatinib plus anti-PD-1 therapy. A Subcutaneous transplantation of Hepa1-6 cells with the indicated genetic manipulation into C57BL/6 mice (n = 6). B Cell proliferation (B, upper panel) and cell death (B, lower panel) were measured in vivo by performing Ki67 and TUNEL staining, respectively. Scale bar, 200 μm. C The CD8+CD3+ T-cell proportion in solid tumors shown in (A) was determined using FCM. D A total of 112 HCC patients were divided into two groups, indicated as high and low according to the IHC score for pPIGL-Y81 in HCC tissues. The survival curve was plotted with the best cutoff for the aforementioned IHC scores. Scale bar, 200 μm. E Twenty HCC patients who received lenvatinib treatment were separated into two groups by the median cutoff according to the change in IHC intensity in pPIGL-Y81 after lenvatinib treatment. A representative IHC image is shown (E, left panel). The change in the IHC score for HCC pPIGL-Y81 was analyzed (E, right panel). Scale bar, 200 μm. F Patient-derived organoids (PDOs) from three HCC samples with lower changes in pPIGL-Y81 and three HCC samples with higher changes in pPIGL-Y81 were prepared, and ectopic transplantation was performed for the combined treatment of lenvatinib and anti-PD-1 antibody with the indicated dosage described in the Methods section. G Schematic model of the mechanism by which FGFR2/PIGL axis-mediated CCL2/20 expression determines the efficiency of lenvatinib plus PD-1 therapy. A, D One-way ANOVA; B-C and E, two-tailed Student’s t-test; F Two-way ANOVA (**p < 0.01; ***p < 0.001)
We further explored the clinical significance of PIGL-Y81 phosphorylation in determining HCC patient prognosis and treatment strategies. We grouped HCC patients into two groups according to their PIGL-Y81 phosphorylation level, which was detected by a specific antibody (Fig. S7B). The survival curves were plotted by the best cutoff of the PIGL-Y81 phosphorylation level in the aforementioned HCC patients, and a lower PIGL-Y81 phosphorylation level predicted a better prognosis (Fig. 7D). Additionally, PIGL-Y81 phosphorylation was increased in tumor tissue compared with paired normal tissues (Fig. S7C). Lenvatinib constrained FGFR2 activation and further determined PIGL-Y81 phosphorylation, which should be a marker for HCC responding to PD1 blockade therapy. We collected 20 paired HCC samples from patients treated with lenvatinib and grouped them into two groups based on the change in PIGL-Y81 phosphorylation in the HCC samples before (collected as primary HCC samples) and after (collected as recurrent HCC samples) lenvatinib treatment (Fig. 7E). The survival curves of the two groups showed that a lower change in Y81 phosphorylation predicted a worse prognosis (Fig. S7D), indicating that HCC patients with a lower change in Y81 phosphorylation are not suitable for PD-1 blockade therapy. This result was further confirmed using a patient-derived organoid (PDO) model (Fig. 7F).
Overall, we explored the molecular symptoms of HCC patients who are suitable for the combination of lenvatinib and PD-1 immunotherapy in the clinic.
Discussion
In summary, we ascertained that PIGL, a member of the PIG family, functionally suppressed tumor immune evasion. After lenvatinib treatment, the alterations in PIGL phosphorylation at Y81 determined whether HCC patients were suitable for subsequent PD-1 blockade immunotherapy. In detail, FGFR2 phosphorylates PIGL at Y81 and retains PIGL in the cytosol. Once lenvatinib treatment is executed, the tyrosine kinase activity of FGFR2 is constrained, and PIGL is dephosphorylated at Y81, probably exposing areas for importin α/β1 binding. The import of PIGL into the nucleus indicates a new role for PIGL that is enzyme activity-independent. In detail, nuclear PIGL physically interacts with cMyc and disrupts the cMyc/BRD4 complex, in turn suppressing the expression of cMyc downstream genes, such as CCL2/20. CCL2 and CCL20 are secreted by tumor tissues and generally recruit TAMs and regulatory T cells, respectively, which remodel the TME and aid in the evasion of tumor cells from the immune checkpoint (Fig. 7G). Therefore, our study guided lenvatinib treatment in advanced HCC patients. Lenvatinib is required to drive nuclear PIGL accumulation in HCC cells with high FGFR2 activity. For HCC cells with low FGFR2 activity, lenvatinib may not be required to present the function of nuclear PIGL. After summarizing the significance of this study, we explored the implications of our findings.
The immunosuppressive TME is an obstacle to tumor immunotherapy. In recent years, numerous basic and clinical studies have explored strategies to improve the efficacy of tumor immunotherapy, such as developing immune checkpoint inhibitors, which are represented by PD-1/PD-L1 antibodies. These strategies are based on the study of resistance mechanisms of immune checkpoint inhibitors combined with targeted therapy to overcome resistance and promote the effects of immunotherapy. For example, the combination of the anti-PD-L1 antibody atezolizumab and BRAF/MEK inhibitor in patients with BRAF-mutated melanoma significantly improved progression-free survival (PFS) [18]. Moreover, the high rate of angiogenesis in the TME and the resulting abnormal vasculature and high interstitial pressure within the tumor can impair the infiltration of immune cells and checkpoint inhibitors. The combined treatment strategy exerted a significant effect on enhancing the antitumor immune response [4]. In addition to the intrinsic characteristics of tumors that lead to immunotherapy resistance, immune cells in the TME also regulate the effect of immunotherapy. Some studies have shown that inhibiting TAM activity enhances the response to immunotherapy and antitumor therapeutic effects because TAMs express anti-inflammatory cytokines and chemokines, inhibit cytotoxic T lymphocyte expansion and activation, promote the recruitment of regulatory T cells and contribute to the immune escape of tumor cells [19, 20].
In this study, an in vivo CRISPR-based screen of genes encoding metabolic enzymes that increased the sensitivity of liver tumors to anti-PD-1 antibody treatment was executed. A positive screen was performed to select the potential metabolic enzymes improving PD-1 antibody treatment. We selected PIGL, which ranked first among the positive candidate list. PIGL is a member of the PIG family, which generally localizes to the endoplasmic reticulum and regulates GPI biosynthesis [5]. GPI-anchored proteins were enriched in the membrane and mainly involved in regulating the TME. Interestingly, in this study, PIGL did not hamper TME remodeling through its enzyme activity but adjusted its function by changing its subcellular localization to the nucleus. In fact, many metabolic enzymes that are localized in the nucleus play important regulatory roles through their enzymatic activities [21, 22].
Another important finding from this study is that PIGL physically interacts with cMyc. The physical interaction between PIGL and cMyc indicates that PIGL might regulate the expression of cMyc target genes. cMyc induces gene expression at two levels: basal expression through the promoter and robust expression from enhancer regulation, which activates the expression of a series of genes that promote tumor cell survival under stress conditions. Epigenetically, cMyc competes with AR for coactivators, and AR/cMyc binding sites have markedly higher levels of H3K27ac, indicating the enrichment of AR/cMyc sites in functional distant promoters. Additionally, cMyc recruits histone acetyltransferases to hyperacetylate chromatin, facilitating the binding of RNA polII [23]. The ability of distant promoters to regulate gene expression depends on their epigenetic status: active distant promoters are enriched in H3K27ac, whereas a loss of H3K27ac and gain of H3K27me3 results in poised or repressed distant promoters [24]. Since changes in H3K27ac levels correlate with enhancer activity and gene expression [25, 26], cMyc overexpression consistently results in global transcriptional amplification with widespread increases in transcript levels, combined with elevated H3K27ac levels and enhancer activation [23].
In this study, the interaction between PIGL and cMyc prevented cMyc from forming a complex with BRD4 at the promoter regions of CCL2/20 and decreased CCL2/20 expression. BRD4 functions not only as a reader but also as a HAT enzyme because BRD4 contains two bromo domains in the N-terminus and one histone acetyltransferase domain (HAT) that presents HAT activity in the C-terminus. Additionally, BRD4 was tightly associated with the acetyltransferase complex p300/CBP and affected its acetyltransferase activity [11, 27]. Hence, BRD4 probably affects H3K27ac levels by integrating p300/CBP or exerts histone acetyltransferase activity in itself. ChIP sequencing would be more efficient in selecting the regulatory region where PIGL disrupts the cMyc/BRD4 complex. However, we screened the cMyc binding region upstream of the transcriptional start sites of CCL2/20 using ChIP‒qPCR and observed that cMyc bound the distant promoters of CCL2/20. As a result, the constrained expression of CCL2/20 was not sufficient to recruit large numbers of TAMs and regulatory T cells to reshape the TIME. The TIME promotes the survival of tumor cells during anti-PD-1 antibody treatment, especially in HCC [1, 3]. Therefore, tumor cells actively respond to anti-PD-1 antibody treatment, and weakening or inhibiting this response of tumor cells may substantially improve the effect of anti-PD-1 antibodies.
Chemokines are a family of small, secreted proteins with pleiotropic roles in inflammation-related pathological diseases, including cancer [28]. The chemokine CCL2 is a multifunctional factor involved in various aspects of liver pathogenesis, including acute liver injury, chronic HBV/HCV infection, cirrhosis and tumorigenesis [29, 30]. In the liver, CCL2 is secreted by hepatic stellate cells, hepatocytes, Kupffer cells and biliary epithelial cells [29]. CCR2, the only known receptor for CCL2, is expressed on monocytes and macrophages within the liver. When CCL2 and CCR2 are upregulated in the liver, they promote macrophage accumulation, inflammation, fibrosis and steatosis [29]. Hence, the CCL2/CCR2 axis influences tumor cell growth, angiogenesis, invasion and metastasis [31, 32]. In some preclinical cancer models, targeting CCL2 is an effective therapeutic approach. For example, carlumab, a neutralizing antibody against CCL2, has entered clinical trials for the treatment of prostate cancer [33]. CCL20 also plays an important role in tumor progression. Hepatitis C virus-induced CCL20 is a direct proangiogenic factor that binds to endothelial CCR6, suggesting that the CCL20/CCR6 axis contributes to hepatic angiogenesis and potential hepatocellular carcinoma [34]. CCL20 signaling promotes tumor growth, invasiveness, and chemoresistance by recruiting regulatory T cells [35, 36]. Disruption of the CCL20/CCR6 axis relieves T-cell exhaustion and extends survival. Some tumor-infiltrating cells expressing CCL20/CCR6 have previously been suggested to act as tumor-promoting cells by recruiting regulatory T cells or exhausting killer T cells [37]. In this study, according to the well-established roles of different immune cells, we selected TAMs, CD3+CD8+ T cells, CD3+CD4+ T cells and CD4+CD25+ T cells as candidate immune cells that are likely affected by CCL2/20.
Conclusively, we screened PIGL, which forms an axis with cMyc and impedes CCL2/20 expression upon lenvatinib treatment by epigenetically remodeling the state of the CCL2/20 promoters. FGFR2, one of the targets of lenvatinib, phosphorylates PIGL at Y81 to retain PIGL in the cytosol and then releases CCL2/20. Tumor cell-secreted CCL2/20 builds the TIME and further dampens the efficiency of PD-1 immunotherapy. Hence, the response to lenvatinib indicated by PIGL-Y81 phosphorylation is a potential clinical marker for the applicability of PD-1/PD-L1 blockade.
Materials and Methods
HCC patients and tissue specimens
Tumor tissues and their matched adjacent tissues were collected from HCC patients who were pathologically diagnosed at The Third Affiliated Hospital of Sun Yat-sen University from September 2018 to July 2019. All patients received standardized treatment for HCC. All patients signed the informed consent form and agreed to participate in the experiment. The research content and implementation plan of this experiment strictly complied with the Declaration of Helsinki and was approved by the ethics committee of The Third Affiliated Hospital of Sun Yat-sen University (Medical Ethics 2022-02-333-01).
In vivo CRISPR knockout screening of metabolic enzymes
The in vivo CRISPR screen was performed as previously described [38]. Briefly, the library containing 1516 mouse metabolic enzyme-encoding genes targeting sgRNAs (each gene has been targeted by 3 sgRNAs, and a very small number of genes with only 2 sgRNAs) and 91 nontargeting sgRNAs were also cloned into the LentiCRISPR-v2 vector. Lentiviruses were produced in 293 T cells using a pooled library, psPAX2, and pMD2.G at a ratio of 4:3:1 (m/m/m). Polycation polyethylenimine (PEI) was used as a transfection reagent with a ratio of 3:1 (m/m) to total plasmids. After lentivirus packaging, concentration, and titre determination, the target cells expressing luciferase were infected at an MOI = 0.3 and kept at a coverage of > 500 cells expressing each sgRNA. After 6 days of puromycin selection, positive cells were collected for orthotopic transplantation into the mouse liver, with 6 mice in each group. Seven days after transplantation, the mice were injected with drugs as indicated. After 2 weeks, the mouse liver tumors were collected and snap-frozen in liquid nitrogen for further manipulation. Each tumor’s genomic DNA was extracted and pooled together as templates for PCR amplification and next-generation sequencing. The screening data analysis was conducted using the well-established method MAGeCKFlute that includes read mapping, normalization, quality check, hit identification and functional analysis, as previously reported [39].
Mouse tumor model and treatments
A mouse liver tumor orthotropic transplantation model was established using the mouse HCC cell line Hepa1-6 or patient-derived organoids (PDOs) as previously described [40]. Briefly, 6- to 8-week-old C57BL/6 WT or TCRa−/− mice were anesthetized by isoflurane and injected in the subcapsular region of the liver with 50 μl of cell/Matrigel solution (containing 1 × 106 Hepa1-6 cells) using a syringe with a 33 G needle. PDOs were digested by trypsin for a short time and mixed with Matrigel. Six- to eight-week-old humanized huHSC-NCG mice were purchased from GemPharmatech, anesthetized by isoflurane, and then injected into the subcapsular region of the liver with 50 μl of cell/Matrigel solution (containing 25 μl of PDOs). After surgery, mice were incubated at 37 °C until awakening. A subcutaneous tumor transplantation model was established using Hepa1-6 cells. A total of 2 × 106 cells were injected subcutaneously into 6- to 8-week-old C57BL/6 WT or TCRa−/− mice. Mice of different sexes were used randomly in the experiment. All mice were maintained on a 12/12 h day/night cycle and allowed free access to food and water. All mouse experiments were approved by the Research Ethics Committee of Guangzhou University.
Lenvatinib and mouse or human PD-1 or CCL2/20 antibody treatment were executed at Day 20 when the tumor size exceeded 250 mm3 by intraperitoneal injection twice a week as indicated in the related figures. The dosages of drugs or antibodies were 0.2 μg lenvatinib/ml in mouse serum, 10 μg anti-PD-1 antibody/ml in mouse serum and 2 μg anti-CCL2 or anti-CCL20 antibody/ml in mouse serum.
Tumor-infiltrating immune cell isolation and flow cytometry
The mouse tumor was perfused using cold PBS, dissociated with 2 mg/mL collagenase A (Sigma) and 1× DNase I (Sigma) in RPMI-1640 medium at 37 °C for 30 min, and then filtered through a 70 μm strainer. Cells were centrifuged at 600 × g for 8 min, and the cell pellet was collected. Percoll cell separation medium (37.5%) was prepared. The cell pellet was resuspended in 10 ml Percoll in a 15 mL tube. After centrifugation at 500 × g for 30 min at 22 °C, the supernatant was removed, and the cell pellet was collected. One milliliter of erythrocyte lysis solution was added to the cell pellet, pipetted evenly, and placed on ice for 1–3 min to lyse the erythrocytes. The lysis was terminated by adding 9 ml of PBS and centrifuged at 600 × g for 8 min, and the cell pellet was collected.
For antibody staining, the cell pellet was resuspended in 250 µl PBS + 2% FBS solution. Before staining, single-cell suspensions were blocked with anti-mouse CD16/CD32 antibodies (Thermo Fisher) for 15 min. Then, the fluorophore-conjugated antibody mixture was added and stained on ice for 30 min in the dark. Isotype controls were used as negative controls to determine the specification of the primary antibodies. The flow cytometry antibodies used in this study were anti-mouse CD45-Brilliant Violet 412 (1:200, Biolegend, #103134), anti-mouse CD3-APC (1:200, Biolegend, #100312), anti-mouse CD8-Alexa Fluor 700 (1:200, eBioscience, #56-0081-82), anti-mouse CD4-PE-Cyanine7 (1:200, Biolegend, #100528), anti-mouse CD25-PerCP-Cyanine5.5 (1:100, Biolegend, #101912), and anti-mouse F4/80-FITC (1:200, Biolegend, #123107).
Flow cytometry analysis was then performed using BD FACSAria™ III. FlowJo v.10.4.2 was used for further analysis.
Single-cell RNA sequencing
The shNT or shPIGL fresh tumor single cell count was approximately 1 × 106 cells/mL, and cell viability greater than 95% was used for library preparation. The library preparation procedure was performed according to the standard manual of the 10X Genomics system. After library quantification and quality-checking using a DNA 1000 chip (Agilent Technologies), samples were diluted and loaded onto a HiSeq 4000 (Illumina). Next, raw data demultiplexing, barcode processing, alignment, filtering, and UMI counting were performed using the Cell Ranger analysis pipeline (v2.0). Then, the count data were used for downstream analysis in R 4.2.1 using Seurat (v.4.2.0). Cells with fewer than 200 features or higher than 5% mitochondrial gene content were removed prior to further analysis. The data were integrated using Harmony v0.1.0 [41] and then normalized using Seurat. Principal component analysis was performed using the top 3,000 variable genes to identify the number of significant components before clustering. Clustering was performed by calculating a shared nearest neighbor graph using a resolution of 0.8, which is shown as a UMAP. Subsetting into different cell types and cell type annotations were performed using the singleR package [42]. The percentage of each cell type was calculated and shown as a stacked bar plot using a set of packages in tidyverse v1.3.2.
RNA sequencing
Total RNA was extracted for RNA sequencing using an Illumina HiSeq 2500 system. Sequencing data analysis and management were performed with BaseSpace Sequence Hub. The KEGG pathway analysis was performed using the software clusterProfiler [43], and the plot was generated using ggplot2 in R (3.5.0). GSEA was conducted using GSEA software (v4.1.0), as described in a previous report [44].
Mass spectrometry analysis
Immunoprecipitated FLAG-PIGL from the cytosol or nuclei of Hepa1-6 cells was boiled with 40 μl of 1× loading buffer at 95 °C for 8 min and then loaded onto an SDS‒PAGE gel. The gel samples were processed through a series of routine flows, such as reductive alkylation, trypsin digestion and peptide extraction. The peptides were analyzed by liquid chromatography‒mass spectrometry (LC‒MS/MS) on a Q Exactive™ mass spectrometer (Thermo Fisher Scientific, Waltham, MA). Proteins were identified by a database search of the fragment spectra against the National SwissProt protein database (EBI) using Mascot Server 2.4 (Matrix Science, London, UK). After database searching, the intensity of each identified protein was used for further analysis. The data were normalized by log2 transformation, and the log2-fold fold change and p-value were calculated between the Vec-FLAG and PIGL-FLAG groups using a two-tailed Student’s t test. Phosphopeptide matches were analyzed by using MaxQuant v1.5.2.8 implemented in Proteome Discoverer and manually curated. The heatmap plot was constructed using the “pheatmap” package in R 4.2.1, and the color bar represents the normalized expression level of each site.
Statistical analysis
GraphPad Prism 9.0 software was used to analyze the experimental data. Each experiment was independently repeated at least three times, and the results are represented as the means ± S.D.s or means ± S.E.M.s. Comparisons were analyzed using an unpaired two-tailed Student’s t-test or ANOVA. Paired two-tailed Student’s t-tests were used to compare tumors and adjacent tissues. The p values are indicated in the related figures; *p < 0.05, **p < 0.01, ***p < 0.001 and NS, not significant (p > 0.05).
Supplementary information
Acknowledgements
The authors thank the Centre for Precise Gene Editing, Guangzhou University for technical support.
Author contributions
HY, TZS, LLY, and DWX designed, performed, and analyzed the experiments and prepared the figures. WL and QX provided technical and clinical assistance. XJW and YFD conceived this study and wrote the manuscript, and XJW, WL, and QX revised the manuscript.
Funding
This study was supported mainly by the Guangdong Natural Science Foundation of Guangdong Province of China 2021B1515020016 and in part by NSFC grants No. 82000616, 82272714, 82173149 and Science and Technology Program of Guangdong Province No. 2020B1212060019.
Competing interests
The authors declared no competing interests.
Ethical approval
The research content and implementation plan of this experiment strictly complied with the Declaration of Helsinki and was approved by the ethics committee of The Third Affiliated Hospital of Sun Yat-sen University (Medical Ethics 2022-02-333-01).
Footnotes
These authors contributed equally: Hua Yu, Tiezhu Shi, Linli Yao, Dongwei Xu.
Contributor Information
Wei Liu, Email: lwei6@mail.sysu.edu.cn.
Xiongjun Wang, Email: wangxiongjun@gzhu.edu.cn.
Supplementary information
The online version contains supplementary material available at 10.1038/s41423-023-01048-3.
References
- 1.Llovet JM, Castet F, Heikenwalder M, Maini MK, Mazzaferro V, Pinato DJ, et al. Immunotherapies for hepatocellular carcinoma. Nat Rev Clin Oncol. 2022;19:151–72. doi: 10.1038/s41571-021-00573-2. [DOI] [PubMed] [Google Scholar]
- 2.Makker V, Colombo N, Casado Herraez A, Santin AD, Colomba E, Miller DS, et al. Lenvatinib plus Pembrolizumab for advanced endometrial cancer. N Engl J Med. 2022;386:437–48. doi: 10.1056/NEJMoa2108330. [DOI] [PubMed] [Google Scholar]
- 3.Labani-Motlagh A, Ashja-Mahdavi M, Loskog A. The tumor microenvironment: A Milieu hindering and obstructing antitumor immune responses. Front Immunol. 2020;11:940. doi: 10.3389/fimmu.2020.00940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fukumura D, Kloepper J, Amoozgar Z, Duda DG, Jain RK. Enhancing cancer immunotherapy using antiangiogenics: opportunities and challenges. Nat Rev Clin Oncol. 2018;15:325–40. doi: 10.1038/nrclinonc.2018.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pottekat A, Menon AK. Subcellular localization and targeting of N-acetylglucosaminyl phosphatidylinositol de-N-acetylase, the second enzyme in the glycosylphosphatidylinositol biosynthetic pathway. J Biol Chem. 2004;279:15743–51. doi: 10.1074/jbc.M313537200. [DOI] [PubMed] [Google Scholar]
- 6.Wang X, Liu R, Zhu W, Chu H, Yu H, Wei P, et al. UDP-glucose accelerates SNAI1 mRNA decay and impairs lung cancer metastasis. Nature. 2019;571:127–31. doi: 10.1038/s41586-019-1340-y. [DOI] [PubMed] [Google Scholar]
- 7.Yang W, Xia Y, Ji H, Zheng Y, Liang J, Huang W, et al. Nuclear PKM2 regulates beta-catenin transactivation upon EGFR activation. Nature. 2011;480:118–22. doi: 10.1038/nature10598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liang C, Shi S, Qin Y, Meng Q, Hua J, Hu Q, et al. Localisation of PGK1 determines metabolic phenotype to balance metastasis and proliferation in patients with SMAD4-negative pancreatic cancer. Gut. 2020;69:888–900. doi: 10.1136/gutjnl-2018-317163. [DOI] [PubMed] [Google Scholar]
- 9.Dang CV. MYC on the path to cancer. Cell. 2012;149:22–35. doi: 10.1016/j.cell.2012.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dang CV. c-Myc target genes involved in cell growth, apoptosis, and metabolism. Mol Cell Biol. 1999;19:1–11. doi: 10.1128/MCB.19.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Morrison-Smith CD, Knox TM, Filic I, Soroko KM, Eschle BK, Wilkens MK, et al. Combined targeting of the BRD4-NUT-p300 axis in NUT Midline carcinoma by dual selective Bromodomain inhibitor, NEO2734. Mol Cancer Ther. 2020;19:1406–14. doi: 10.1158/1535-7163.MCT-20-0087. [DOI] [PubMed] [Google Scholar]
- 12.Patel MC, Debrosse M, Smith M, Dey A, Huynh W, Sarai N, et al. BRD4 coordinates recruitment of pause release factor P-TEFb and the pausing complex NELF/DSIF to regulate transcription elongation of interferon-stimulated genes. Mol Cell Biol. 2013;33:2497–507. doi: 10.1128/MCB.01180-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Muhar M, Ebert A, Neumann T, Umkehrer C, Jude J, Wieshofer C, et al. SLAM-seq defines direct gene-regulatory functions of the BRD4-MYC axis. Science. 2018;360:800–5. doi: 10.1126/science.aao2793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dang CV, Reddy EP, Shokat KM, Soucek L. Drugging the ‘undruggable’ cancer targets. Nat Rev Cancer. 2017;17:502–8. doi: 10.1038/nrc.2017.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen H, Liu H, Qing G. Targeting oncogenic Myc as a strategy for cancer treatment. Signal Transduct Target Ther. 2018;3:5. doi: 10.1038/s41392-018-0008-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Llombart V, Mansour MR. Therapeutic targeting of “undruggable” MYC. EBioMedicine. 2022;75:103756. doi: 10.1016/j.ebiom.2021.103756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Scharping NE, Menk AV, Moreci RS, Whetstone RD, Dadey RE, Watkins SC, et al. The tumor microenvironment represses T cell mitochondrial biogenesis to drive intratumoral T Cell metabolic insufficiency and dysfunction. Immunity. 2016;45:374–88. doi: 10.1016/j.immuni.2016.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gutzmer R, Stroyakovskiy D, Gogas H, Robert C, Lewis K, Protsenko S, et al. Atezolizumab, vemurafenib, and cobimetinib as first-line treatment for unresectable advanced BRAF(V600) mutation-positive melanoma (IMspire150): Primary analysis of the randomised, double-blind, placebo-controlled, phase 3 trial. Lancet. 2020;395:1835–44. doi: 10.1016/S0140-6736(20)30934-X. [DOI] [PubMed] [Google Scholar]
- 19.Rodell CB, Arlauckas SP, Cuccarese MF, Garris CS, Li R, Ahmed MS, et al. TLR7/8-agonist-loaded nanoparticles promote the polarization of tumour-associated macrophages to enhance cancer immunotherapy. Nat Biomed Eng. 2018;2:578–88. doi: 10.1038/s41551-018-0236-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhu Y, Yang J, Xu D, Gao XM, Zhang Z, Hsu JL, et al. Disruption of tumour-associated macrophage trafficking by the osteopontin-induced colony-stimulating factor-1 signalling sensitises hepatocellular carcinoma to anti-PD-L1 blockade. Gut. 2019;68:1653–66. doi: 10.1136/gutjnl-2019-318419. [DOI] [PubMed] [Google Scholar]
- 21.Xu D, Shao F, Bian X, Meng Y, Liang T, Lu Z. The evolving landscape of noncanonical functions of metabolic enzymes in cancer and other pathologies. Cell Metab. 2021;33:33–50. doi: 10.1016/j.cmet.2020.12.015. [DOI] [PubMed] [Google Scholar]
- 22.Luo W, Hu H, Chang R, Zhong J, Knabel M, O'Meally R, et al. Pyruvate kinase M2 is a PHD3-stimulated coactivator for hypoxia-inducible factor 1. Cell. 2011;145:732–44. doi: 10.1016/j.cell.2011.03.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rahl PB, Young RA. MYC and transcription elongation. Cold Spring Harb Perspect Med. 2014;4:a020990. doi: 10.1101/cshperspect.a020990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Durbin AD, Zimmerman MW, Dharia NV, Abraham BJ, Iniguez AB, Weichert-Leahey N, et al. Selective gene dependencies in MYCN-amplified neuroblastoma include the core transcriptional regulatory circuitry. Nat Genet. 2018;50:1240–6. doi: 10.1038/s41588-018-0191-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schoenfelder S, Fraser P. Long-range enhancer-promoter contacts in gene expression control. Nat Rev Genet. 2019;20:437–55. doi: 10.1038/s41576-019-0128-0. [DOI] [PubMed] [Google Scholar]
- 26.Morton AR, Dogan-Artun N, Faber ZJ, MacLeod G, Bartels CF, Piazza MS, et al. Functional enhancers shape extrachromosomal oncogene amplifications. Cell. 2019;179:1330–1341 e1313. doi: 10.1016/j.cell.2019.10.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Alekseyenko AA, Walsh EM, Wang X, Grayson AR, Hsi PT, Kharchenko PV, et al. The oncogenic BRD4-NUT chromatin regulator drives aberrant transcription within large topological domains. Genes Dev. 2015;29:1507–23. doi: 10.1101/gad.267583.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Roussos ET, Condeelis JS, Patsialou A. Chemotaxis in cancer. Nat Rev Cancer. 2011;11:573–87. doi: 10.1038/nrc3078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sahin H, Trautwein C, Wasmuth HE. Functional role of chemokines in liver disease models. Nat Rev Gastroenterol Hepatol. 2010;7:682–90. doi: 10.1038/nrgastro.2010.168. [DOI] [PubMed] [Google Scholar]
- 30.Mandrekar P, Ambade A, Lim A, Szabo G, Catalano D. An essential role for monocyte chemoattractant protein-1 in alcoholic liver injury: regulation of proinflammatory cytokines and hepatic steatosis in mice. Hepatology. 2011;54:2185–97. doi: 10.1002/hep.24599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang J, Patel L, Pienta KJ. Targeting chemokine (C-C motif) ligand 2 (CCL2) as an example of translation of cancer molecular biology to the clinic. Prog Mol Biol Transl Sci. 2010;95:31–53. doi: 10.1016/B978-0-12-385071-3.00003-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Qian BZ, Li J, Zhang H, Kitamura T, Zhang J, Campion LR, et al. CCL2 recruits inflammatory monocytes to facilitate breast-tumour metastasis. Nature. 2011;475:222–5. doi: 10.1038/nature10138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pienta KJ, Machiels JP, Schrijvers D, Alekseev B, Shkolnik M, Crabb SJ, et al. Phase 2 study of carlumab (CNTO 888), a human monoclonal antibody against CC-chemokine ligand 2 (CCL2), in metastatic castration-resistant prostate cancer. Invest N Drugs. 2013;31:760–8. doi: 10.1007/s10637-012-9869-8. [DOI] [PubMed] [Google Scholar]
- 34.Benkheil M, Van Haele M, Roskams T, Laporte M, Noppen S, Abbasi K, et al. CCL20, a direct-acting pro-angiogenic chemokine induced by hepatitis C virus (HCV): Potential role in HCV-related liver cancer. Exp Cell Res. 2018;372:168–77. doi: 10.1016/j.yexcr.2018.09.023. [DOI] [PubMed] [Google Scholar]
- 35.Cook KW, Letley DP, Ingram RJ, Staples E, Skjoldmose H, Atherton JC, et al. CCL20/CCR6-mediated migration of regulatory T cells to the Helicobacter pylori-infected human gastric mucosa. Gut. 2014;63:1550–9. doi: 10.1136/gutjnl-2013-306253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee B, Namkoong H, Yang Y, Huang H, Heller D, Szot GL, et al. Single-cell sequencing unveils distinct immune microenvironments with CCR6-CCL20 crosstalk in human chronic pancreatitis. Gut. 2022;71:1831–42. doi: 10.1136/gutjnl-2021-324546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kawanabe-Matsuda H, Takeda K, Nakamura M, Makino S, Karasaki T, Kakimi K, et al. Dietary Lactobacillus-derived exopolysaccharide enhances immune-checkpoint blockade therapy. Cancer Discov. 2022;12:1336–55. doi: 10.1158/2159-8290.CD-21-0929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Joung J, Konermann S, Gootenberg JS, Abudayyeh OO, Platt RJ, Brigham MD, et al. Genome-scale CRISPR-Cas9 knockout and transcriptional activation screening. Nat Protoc. 2017;12:828–63. doi: 10.1038/nprot.2017.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang B, Wang M, Zhang W, Xiao T, Chen CH, Wu A, et al. Integrative analysis of pooled CRISPR genetic screens using MAGeCKFlute. Nat Protoc. 2019;14:756–80. doi: 10.1038/s41596-018-0113-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen P, Luo X, Dai G, Jiang Y, Luo Y, Peng S, et al. Dexmedetomidine promotes the progression of hepatocellular carcinoma through hepatic stellate cell activation. Exp Mol Med. 2020;52:1062–74. doi: 10.1038/s12276-020-0461-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Korsunsky I, Millard N, Fan J, Slowikowski K, Zhang F, Wei K, et al. Fast, sensitive and accurate integration of single-cell data with Harmony. Nat Methods. 2019;16:1289–96. doi: 10.1038/s41592-019-0619-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Aran D, Looney AP, Liu L, Wu E, Fong V, Hsu A, et al. Reference-based analysis of lung single-cell sequencing reveals a transitional profibrotic macrophage. Nat Immunol. 2019;20:163–72. doi: 10.1038/s41590-018-0276-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yu G, Wang LG, Han Y, He QY. clusterProfiler: An R package for comparing biological themes among gene clusters. OMICS. 2012;16:284–7. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–50. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.