Summary
Sox transcription factors are crucial for vertebrate nervous system development. In zebrafish embryo, sox1 genes are expressed in neural progenitor cells and neurons of ventral spinal cord. Our recent study revealed that the loss of sox1a and sox1b function results in a significant decline of V2 subtype neurons (V2s). Using single-cell RNA sequencing, we analyzed the transcriptome of sox1a lineage progenitors and neurons in the zebrafish spinal cord at four time points during embryonic development, employing the Tg(sox1a:eGFP) line. In addition to previously characterized sox1a-expressing neurons, we discovered the expression of sox1a in late-developing intraspinal serotonergic neurons (ISNs). Developmental trajectory analysis suggests that ISNs arise from lateral floor plate (LFP) progenitor cells. Pharmacological inhibition of the Notch signaling pathway revealed its role in negatively regulating LFP progenitor cell differentiation into ISNs. Our findings highlight the zebrafish LFP as a progenitor domain for ISNs, alongside known Kolmer-Agduhr (KA) and V3 interneurons.
Subject areas: Neuroscience, Cell biology, Omics, Transcriptomics
Graphical abstract

Highlights
-
•
Sox1a is expressed in ISNs
-
•
Lateral floor plate gives rise to ISNs
-
•
Notch signaling inhibits ISN differentiation
Neuroscience; Cell biology; Omics; Transcriptomics.
Introduction
Differential expression and combinatorial action of distinct transcription factors (TFs) play crucial roles during embryonic development by establishing unique transcriptional expression programs that determine tissue-specific cell fate.1,2 In the vertebrate spinal cord, the coordinated expression of TF combinations in neural progenitor domains along the dorsoventral (DV) axis is triggered by dorsal and ventral antagonistic signals.3,4,5 This process leads to the generation of diverse postmitotic neurons at specific DV positions.6,7,8 For instance, in the ventral spinal cord, the concentration of the morphogen Sonic hedgehog (Shh) released from the notochord and the medial floor plate specifies the development of motor neurons (MNs) and ventral interneurons (IN).9,10 Close to the source of Shh, KA", and V3 interneurons are generated from progenitor cells of the lateral floor plate (LFP).11,12,13,14 Subsequently, the MN, KA', V2, V1, and V0 interneurons are produced from the progenitor domains pMN, p2, p1, and p0, respectively.3,14,15,16 Additionally, certain populations of neural progenitor cells in the spinal cord differentiate into distinct neuronal subtypes.17 For example, in the mouse and zebrafish embryo, the p2 progenitor domain located one level below p1 gives rise to V2 neurons. These neurons generate at least three different subtypes of V2 interneurons, called V2a, V2b, and V2c (V2s in zebrafish).18,19,20,21,22,23,24
In a whole-mount in situ hybridization gene expression screen for TFs expressed in the zebrafish spinal cord, we identified two orthologs of the mammalian sox1 gene.25 We found that these genes were expressed in the p2 progenitor domain, KA″ and KA' interneurons, as well as in a subset of V2 interneurons called V2s.19 Furthermore, a transgenic reporter line, sox1a:eGFP, exhibited specific expression patterns in KA" neurons, KA' neurons, and V2s interneurons in the zebrafish spinal cord.19
To gain further insights into the transcriptome of sox1a lineage neural progenitors and neurons, we employed single-cell RNA sequencing (scRNA-seq) at 1-, 2-, 3-, and 5-day post fertilization (dpf) during zebrafish spinal cord development. Our data not only confirmed the previously described expression patterns of sox1a in p2, KA, and V2s interneurons but also revealed its presence in intraspinal serotonergic neurons (ISNs).
ISNs in zebrafish are characterized as fev- (also known as pet1) and tph2-positive neurons located ventrally in the spinal cord. Unlike many other ventral spinal cord neurons that develop earlier, ISNs emerge after 2 dpf.26,27 They possess ovoid/round soma and extend a single process toward the dorsal and lateral regions of the motor spine.26,28,29,30 Pharmacological and optogenetic studies have demonstrated that ISNs contribute to the generation and modulation of locomotor activity, with serotonin released from ISNs reducing spine-generated locomotor activity.26 Additionally, ISNs play a crucial role in axonal regrowth and restoration of spinal cord function following injury.31,32,33,34 Despite these findings, the precise progenitor cells responsible for generating ISNs and the gene regulatory networks governing their development and specification remain incompletely understood in the zebrafish.
Considering the close proximity of the LFP and ISNs in the ventral spinal cord and the identification of the LFP as a domain of neural progenitors giving rise to KA" and V311,12,13,14 interneurons, we embarked on investigating whether ISNs also originate from LFP progenitor cells using our scRNA-seq data. By reconstructing the trajectory of LFP progenitor cells, ISN precursors (ISN-pre), and ISNs, we confirmed that ISNs originate from the most ventral progenitor domain of the spinal cord, namely LFP. Furthermore, pharmacological inhibition of the Notch signaling pathway between 2 and 3 dpf indicates that this pathway is required to negatively regulate the development of LFP progenitor cells into ISN populations.
Results
Besides KA and V2s interneurons, sox1a is also expressed in ISNs and multiple neural progenitor populations in the zebrafish spinal cord
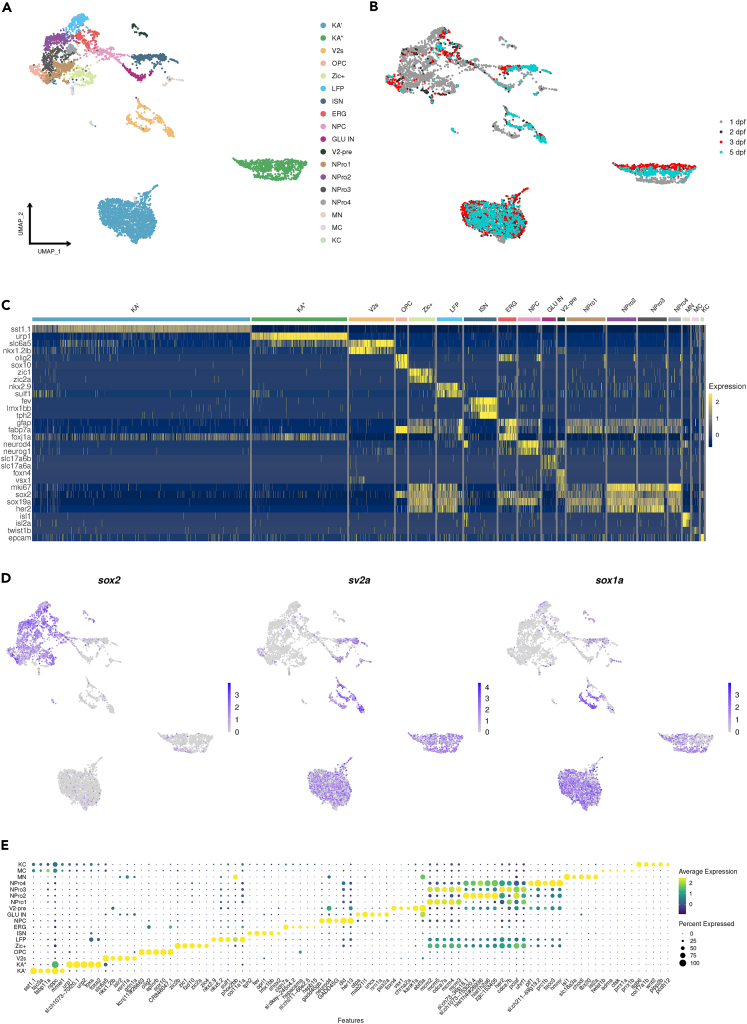
To obtain an exhaustive overview of all cell populations in the zebrafish spinal cord expressing the High-Mobility Group Box sox1a gene and to analyze the temporal changes in the transcriptional profile of different sox1a-positive neural progenitors and neuronal populations, the body (trunk and tail) of the transgenic sox1a:eGFP zebrafish line at 1, 2, 3, and 5 dpf was manually dissected, cells were dissociated and sorted GFP-positive (GFP+) cells were subjected to scRNA-seq (Figure 1A). Unsupervised cell clustering by Seurat35 on the integrated data from all four developmental stages revealed 25 and 40 unique cell clusters using the resolutions 0.5 and 1.5, respectively (Figures 1B and S1A–S1E), and the characteristic marker genes expressed in the major cell types are displayed by heatmap (Figure 1C). Since we used the Tg(sox1a:eGFP) line to sort the spinal cord cells, we checked for co-expression of the gfp and sox1a genes in each cluster and found that gfp mRNA was highly expressed in sox1a-positive cells (Figure 1D), especially in cells corresponding to the lateral line and neuronal/interneuronal progenitors (Figure 1D). This co-expression of gfp and sox1a confirmed the enrichment of sox1a cells in the zebrafish spinal cord. In addition to these cell populations, we identified other groups, including mesoderm, fin fold, hematopoietic system, cardiomyocytes, fast skeletal muscle, and vascular cells (Figures 1B and 1C; Table S1). The main objective of our study was to obtain a comprehensive overview of all neural progenitors and neurons expressing sox1a in the zebrafish spinal cord. To achieve this, we excluded cell populations that did not belong to these two categories, as well as the lateral line cell cluster, which was not relevant to our study. The resulting single-cell sequencing data were re-analyzed using only the dataset corresponding to the neural progenitor/interneuron cell populations (Figures 2A, S2A, and S2B; Table S2). Using the remaining cell populations, we identified distinct cell clusters corresponding to various progenitor cells, such as LFP (nkx2.9+) and V2 precursor (V2-pre; vsx1+ and foxn4+) (Figures 2A–2E). Additionally, we identified several neural progenitor groups of cells (NPro) that expressed markers of proliferation (mki67), as well as neural progenitors (sox2 and sox19a), but as we could not conclusively link them to a known progenitor domain of the spinal cord, these groups were designated as NPro1 to 4 (Figures 2A–2E). Furthermore, we identified another neural progenitor cell cluster (mki67, sox2, and sox19a) that exhibited enriched expression of different zic genes (zic1 and zic2a) and designated it as zic+. Among several other clusters, we also identified a glial population consisting of ependymal radial glial cells (ERG; GFAP+, foxja1+, fabp7+), which correspond to neural stem cells of the spinal cord (Figures 2A–2E). We also detected clusters of sox1a+ cells corresponding to KA", KA', and V2s neurons (Figures 2A and 2D) in agreement with the previous studies,18,19 thus validating our approach and findings. Interestingly, one new sox1a cluster representing the ISNs (tph2+ and fev+)26,27 was also discovered (Figures 2A and 2C–2E; Table S2; Figures 3A–3C).
Figure 1.
Integration of all single-cell datasets and annotation of main cell types
(A) Schematic workflow of scRNA-seq (part of it was created with BioRender.com).
(B) UMAP shows the main cell types annotated based on the top marker and known marker genes.
(C) A heatmap showing the representative marker genes expressed in main cell types.
(D) Feature Plots showing marker genes used to sort in-silico neural progenitor and neuronal cell populations. Note: cldnb is used to exclude lateral line cells, which are gfp+.
Figure 2.
Re-analyses of the neural progenitor population and neurons identify several cell subtypes in the spinal cord
(A) UMAP shows the neural progenitors and neuronal sub-clusters.
(B) The distribution of cells is based on the developmental stages.
(C) A heatmap showing the representative marker genes specifically expressed in each neural progenitor and neuronal subtype.
(D) Expression of sox2 (progenitor marker), sv2a (neuronal marker) and sox1a on Feature Plots.
(E) Dot Plot showing the top 5 marker genes for each cluster.
Figure 3.
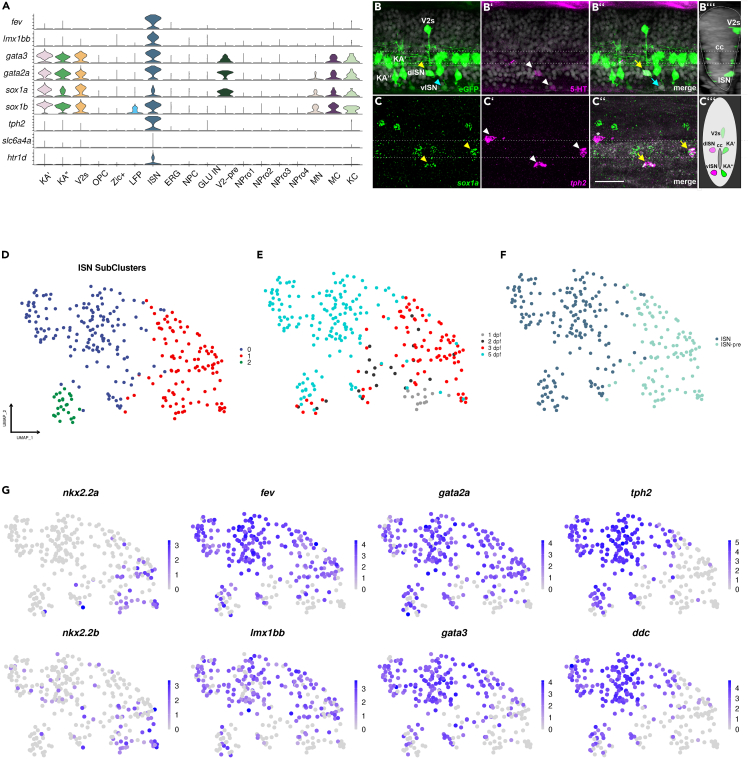
sox1a is expressed in several neuronal populations including ISNs in the zebrafish spinal cord
(A) Violin plots showing key marker genes of ISNs co-expressed with sox1a/b. (B-B″) Co-localization of 5-HT antibody staining with eGFP gene expression in the zebrafish spinal cord at 3 dpf.
(B) Lateral view of the spinal cord showing GFP+ V2s, KA', KA", more dorsal ISN (dISN) with strong eGFP expression (yellow arrowhead), and more ventral ISN (vISN) with weak eGFP expression (blue arrowhead).
(B′) The white arrowhead indicates two 5-HT neurons.
(B″) Co-expression of 5-HT with eGFP (yellow arrow: strong eGFP expression, blue arrow: weak eGFP expression).
(B‴) Optical transverse section showing a V2s neuron dorsal to the central canal (CC) and an ISN ventral to the CC.
(C–C″) Co-expression of sox1a and tph2 mRNA in the zebrafish spinal cord at 3 dpf using HCR-RNA FISH. (C) Yellow arrowheads indicate sox1a+ cells. (C′) White arrowheads indicate three tph2+ neurons. (C″) Yellow arrows indicate neurons that are tph2+ and sox1a+ while white asterisks show a tph2+ and sox1a-neuron.
(C‴) Schematic showing the position of all sox1a+ neurons in the zebrafish spinal cord. (D-E) ISN cluster is constituted of diverse cell populations.
(D) UMAP shows the sub-clustering of the ISN cluster into three sub-clusters. (E) Distribution of cells based on developmental stages.
(F) The majority of the ISN cluster consists of fev+ and tph2- (ISN-pre) or fev+ and tph2+ cells (ISNs).
(G) Feature plots showing key TFs specifically or co-expressed in ISN-pre and ISNs. In embryos (B) and (C), the dorsal side is facing upwards, while the ventral side is facing downwards. The anterior side is on the left, and the posterior side is on the right. Scale bar: 25 μm.
The ISN cluster is made of precursors and mature neurons
To characterize the newly identified ISN cell type, we determined the genes expressed in the corresponding cell cluster (Figure 3A; Table S2). This cell cluster is characterized by the expression of the TF genes fev, lmx1bb, gata3, gata2a, sox1a, and sox1b as well as genes involved in the synthesis, transport, and reception of the neurotransmitter serotonin, namely tph2 (tryptophan hydroxylase 2), slc6a4a (solute carrier family 6 member 4) and htr1d (5-hydroxytryptamine (serotonin) receptor 1D, G protein-coupled) (Figure 3A; Table S2;36). Analysis of the tph2+ cells in the ISN cluster indicated that 39% of the tph2+ neurons also expressed sox1a+ (68 sox1a+ in 172 tph2+ cells). Further analysis using hybridization chain reaction RNA-fluorescent in situ hybridization (HCR RNA-FISH) on Tg(sox1a:eGFP) embryos showed that some of the serotonergic (5-HT) neurons in close proximity to KA interneurons co-expressed GFP and 5-HT, as seen in Figure 3B. To support these findings, HCR RNA-FISH was conducted on 3 dpf zebrafish embryos using sox1a and tph2 probes, and it confirmed that 46% of ventrally located serotonergic neurons in the zebrafish spinal cord co-expressed sox1a with tph2 mRNA (out of 39 tph2+ cells counted in 7 embryos 18 cells were double-positive), as illustrated in Figure 3C. Temporal analysis of sequencing data indicated that the ISN cell population begins to emerge between 2 and 3 dpf (Figures S3A and S3B). Further analysis of the data corresponding to the ISN cluster led us to subdivide this group into 3 subgroups with different and overlapping gene expression profiles (Figures 3D–3G). A cluster (cluster 1) mainly composed of cells from time points 2 and 3 dpf showed expression of genes characteristic of ISN-pre (Figures 3D–3G; Table S3).37,38 Cells in this subcluster were positive for fev, gata3, gata2a, nkx2.2a and nkx2.2b, but negative for tph2 and ddc (Figure 3G). The other two subclusters were both positive for fev and tph2 (Figure 3G). The largest cluster (cluster 0) is composed of cells exclusively from 5 dpf, while the smallest cluster (2) is composed of cells from 2, 3, and 5 dpf (Figures 3D and 3E). This result demonstrates that the ISN cluster is a diverse group of cells composed of a committed progenitor cluster (fev+ and tph2- ISN-pre) and a mature cluster (fev+ and tph2+ ISN) (Figures 3F and 3G).
ISNs develop from lateral floor plate progenitor cells
Previous cell lineage tracing and genetic data indicated that LFP progenitor cells give rise to KA" and V3 interneurons in a sequential manner.11,12,14,39 In this sequence of events, KA" cells are the first to be generated at about 16 hpf (hours post fertilization), followed by V3 cells at approximately 24 hpf. The specification of LFP progenitor cells into different interneuron types depends on the duration of Notch signaling, where LFP cells with high Notch signaling remain in a progenitor state and do not differentiate. Inhibition of this pathway leads to the differentiation of progenitors into postmitotic interneurons.11,12,39
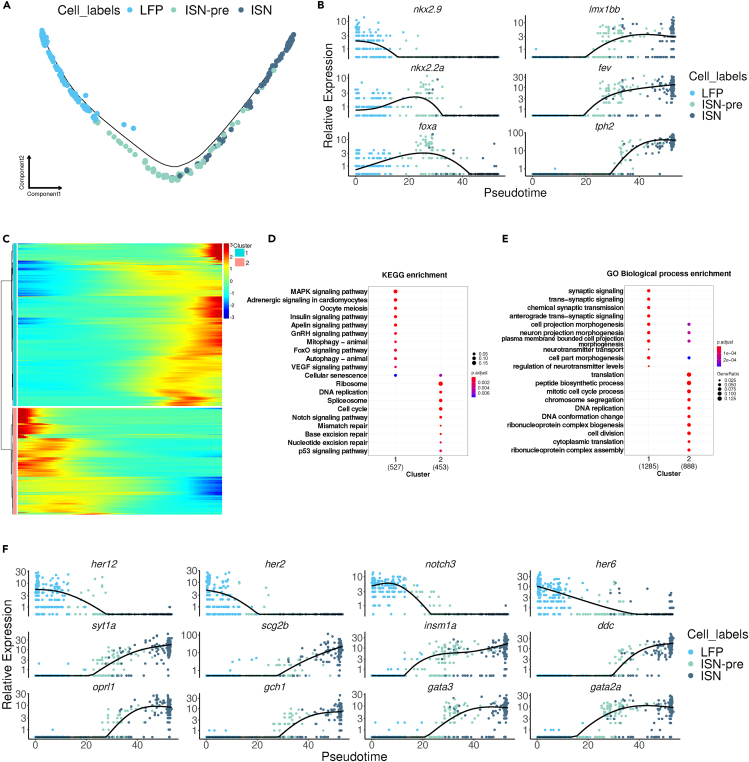
Since ISNs are located ventrally in the spinal cord, their precursors share the expression of several TF genes with LFP cells such as foxa, nkx2.2a, and nkx2.2b (Figure S4A; Table S4). In mice, some serotonergic neurons are generated from the ventral progenitor domain, which is positive for NKX2.2 and adjacent to the floor plate.40 Based on these results, we hypothesized that ISN-pre cells in the zebrafish spinal cord likely originate from the LFP and are generated by inhibiting the Notch signaling pathway, similar to KA" and V3 cells. To test this hypothesis, we revisited the integrated single-cell data from LFP, ISN-pre, and ISN clusters. Using pseudotime analysis of gene expression in these clusters, with LFP serving as the starting point, we predicted a single lineage trajectory that begins at LFP, passes through ISN-pre, and ends at ISN (Figures 4A and S4B). These data also clearly show that LFP cells originate from 1 dpf development stage (Figure S4C). The ISN-pre cells are composed of 2 and 3 dpf cells while the ISNs are at the beginning of their trajectory composed of 2 and 3 dpf embryonic stage cells but at the end of the trajectory, all ISNs are from the 5 dpf developmental stage (Figure S4C). Gene expression analysis over pseudotime shows that nkx2.9 is exclusively expressed in LFP cells (Figure 4B; Figures S4D and S4E; Table S4). nkx2.2a and b are expressed in the LFP and ISN-pre cells (Figures 4B, S4D, and S4E; Table S4). foxa is expressed in LFP and ISN-pre cells and some ISNs (Figures 4B, S4D, and S4E; Table S4). lmx1bb and fev marks ISN-pre and ISNs while tph2 is expressed mainly in ISNs (Figures 4B, S4A, and S4E). In line with these observations, analysis of the expression level of the nkx2.9, nkx2.2a, foxa, lmx1bb, fev and tph2 genes along the developmental trajectory confirmed that nkx2.9 is highly expressed in most LFP cells while tph2 is strongly expressed in ISNs. fev and nkx2.2a have a broader and shallower pattern of cellular expression, both of which are co-expressed in some of the ISN-pre cells in addition to ISNs (fev) and the LFP (nkx2.2a) (Figure S4E).
Figure 4.
LFP is the precursor of ISNs
(A) Pseudotime cell trajectory of LFP, ISN-pre, and ISN cells.
(B) The expression of key transcription markers for LFP and ISN is shown on pseudotime.
(C) Pseudotime heatmap showing pseudotime-dependent differentially expressed genes between clusters 1 and 2. GO and KEGG analyses of pseudotime-dependent differentially expressed genes between clusters 1 and 2.
(D) KEGG pathway analyses.
(E) GO analyses for Biological Processes.
(F) The expression of key transcription markers for LFP and ISN is shown on pseudotime.
To identify the dynamics of gene expression during the specification of LFP to ISN cells, we used monocle to identify gene modules and clusterProfiler for gene ontology (biological process) and pathway (KEGG) analyses of the module-specific genes, (Figures 4C–4E; Tables S5, S6, and S7). This analysis indicated that LFP cells (cluster 2) are enriched in marker genes belonging to the Notch signaling pathway, as her6, notch3, her12 and her2 were among the top 25 genes of cluster 2 (Figures 4C, 4D, and 4F; Tables S5, S6, and S7). Other genes in the top 25 were id1, sox3 and mki67, which implies that these cells are neural progenitors (NPro) (Tables S5, S6, and S7). In agreement with this result, undifferentiated LFP cells showed strong expression of Notch signaling and a decrease in this pathway triggered the differentiation of progenitors toward neuronal fate (Figure 4F). As expected, the top 25 genes of cluster 1 (ISN, ISN-pre) were genes with a role in synaptic signaling, neurotransmitter transport or synthesis, and axonogenesis such as synaptotagmin Ia (syt1a), dopa decarboxylase (ddc), tph2, fev, insm1a, opiate receptor-like 1 (oprl1), GTP cyclohydrolase 1 (gch1), gata3 and gata2a (Figures 4C, 4E, and 4F; Tables S5, S6, and S7).
The integrated single-cell RNA sequencing dataset from olig2:eGFP+ and sox1a:eGFP+ spinal cord cells confirmed the common origin of V3 interneurons and ISNs
Recent work has indicated that LFP progenitors generate KA" interneurons first, followed by V3 interneurons later.12 Our findings suggest that ISNs, which are generated even later than V3 neurons, also develop from LFP progenitors.
To further reinforce our results and provide evidence that both V3 and ISNs originate from LFP cells, we decided in addition to our data to analyze the scRNA-seq data published by Scott et al., (2021; Figures S5A and S5B).41 These sequencing data are particularly relevant to our study because they were obtained from olig2:eGFP+ cells in the zebrafish spinal cord. Since olig2 and sox1a are co-expressed in many ventral cells of the spinal cord, both sequencing sets include a significant number of cell clusters corresponding to neural progenitors and neurons, such as LFP, V2-pre, OPC, radial glia cells, KA" and KA' interneurons. Notably, the single-cell sequencing data of Scott et al. (2021)41 included two cell clusters corresponding to V3-pre (precursors) and V3 cell populations (Figure S5A), which are crucial for demonstrating that V3 interneurons are generated from the LFP and these two clusters were not present in our sequencing data. Therefore, we had to integrate a selected group of cells from the Scott et al. (2021) data into our sequencing results to analyze the common origin of ISNs and V3 cells. Our analysis began with examining the sequencing data of olig2+ cells at 48 hpf to identify cells that potentially corresponded to ISNs. Interestingly, we were able to identify a small group of cells among those annotated as V3-pre that were positive for fev, lmx1bb, and gata2a but negative for tph2. These findings suggested that these cells were ISN-pre cells (Figures S5A and S5B). We performed a careful analysis of the cell cluster annotated as V3 in the olig2+ sequencing data and observed that not all cells in this cluster were sim1a+ (Figure S5C). Thus, we removed all sim1a-cell sub-clusters from this cluster, which corresponded mainly to the lhx5+ group of cells (Figure S5C). Next, we pooled all sim1a+ cells, including V3-pre and V3 cells, into the LFP cluster cells, and re-analyzed the integrated data (Figures 5A and 5B; Figure S5D). This analysis revealed six distinct clusters, which corresponded to LFP, MN, NPC (neural precursor cell), ISN-pre, V3-pre, and V3 cells (Figure 5A). The expression of genes specific to each of these clusters is shown in Figures 5B and S5D, and Table S8. We then integrated these cell clusters into the previously identified LFP, ISN-pre, and ISN clusters from our own sox1a+ sequencing results and analyzed the distribution and time points of origin of these clusters, as well as their corresponding gene lists (Figures 5C, S5E, and S5F, Table S9). We observed that the first population to be generated was V3-pre, followed by V3, ISN-pre, and ISN (Figures 5C, 5D, and S5E). The developmental trajectory of the cells over pseudotime highlighted the LFP cells as the origin of both neuronal populations (Figure 5D). This trajectory from the LFP cells transit over NPC further split into two branches. The lower branches of the younger cell population correspond to V3-pre and V3, and the upper branch of the older cell population contains the ISN-pre and ISN cells (Figures 5D, S5H, and S5I). To analyze the marker genes involved in this transition, we identified four gene clusters using monocle based on pseudotime (Figure 5E).
Figure 5.
LFP is the precursor of both V3 and ISNs
(A) UMAP shows the integration of LFP, V3, and ISN-pre cells and their subtypes (publicly available dataset; Tg(olig2:eGFP).41.
(B) Feature plots of marker genes for LFP (nkx2.9), V3 (sim1a), and ISN-pre (fev) cells.
(C) UMAP shows the integration of LFP, V3, and ISN cells and their subtypes (current study: Tg(sox1a:eGFP)) integrated with the same cells from Tg(olig2:eGFP).
(D) Cell trajectory showing LFP as the precursor of V3 and ISN.
(E) Heatmap showing pseudotime-dependent differentially expressed genes between LFP and V3/ISN cells.
(F) The expression of key TFs for LFP and ISN/V3 is shown in pseudotime. Note that in (C), LFP-1 corresponds to progenitor cells in proliferation originating from 24 hpf and 36 hpf, while LFP-2 represents older non-proliferative LFP cells that correspond to cells coming from 2, 3, and 5 dpf. For this reason, LFP-2 is not included in the pseudotime analysis in (D).
According to the heatmap, cluster 1 genes are highly expressed in ISNs, and their cell type-specific markers are unique to differentiated ISNs (e.g., fev, gata3, Figure 5F; Table S10). Cluster 2 genes are significantly expressed in V3 cells (e.g., sim1a, nos1, Figure 5F; Table S10). Cluster 3 genes are strongly expressed in LFP cells and ISNs during the transition to V3 and ISNs (e.g., foxp4, fosab, Figure 5F; Table S10). Cluster 4 genes are specifically expressed in LFP and V3 cells during the transition to V3 and ISNs (e.g., nkx2.2a, pcdh19, Figure 5F; Table S10).
Notch signaling controls the differentiation of LFP cells into ISNs
Our analysis of LFP cells using scRNA-seq revealed the expression of several genes associated with neural progenitor cells in proliferation, such as sox3, sox2, sox19a, and sox19b, along with cell cycle genes including mki67, mcm5, and mcm2 (Table S2). Additionally, these cells exhibited a robust expression of genes involved in Notch signaling, including notch3 receptor and downstream targets such as her12, her2, her9, her6, her8a, and her4 (Table S2). Previous studies have shown that the differentiation of KA" and V3 interneurons from LFP progenitors requires downregulation of the Notch signaling pathway.12,39 To investigate whether the development of ISNs from LFP cells is also influenced by the downregulation of the Notch signaling pathway, we treated wild-type (WT) embryos with the γ-secretase inhibitor LY411575, which inhibits Notch processing after ligand binding.42 By doing so, we aimed to determine whether the downregulation of Notch signaling is necessary for the development of ISNs from LFP cells.
To carry out the experiment, we treated WT zebrafish embryos with LY411575 from 2 to 3 dpf. We specifically chose this time frame because it is when the first ISN-pre and ISN cells are observed in the ventral position of the spinal cord, while KA" and V3 neurons are already specified. We previously found that treating embryos between 1 and 2 dpf had no effect on the number of tph+ cells (Figures S6C and S6C′).
To assess the impact of LY411575 treatment, we examined the number of tph2+ neurons in the trunk of zebrafish embryos at 3 dpf (Figure 6)). Our results revealed a significant increase in the number of tph2+ cells in embryos treated with LY411575 compared to those treated with DMSO alone (Figures 6A–6C and S6F and S6F′). However, we did not observe any noticeable changes in the number of cells expressing urp1 (a KA" marker) regardless of the treatment time window (Figures S6A, S6A’, S6D, and S6D’). We observed an increase in GABAergic neurons (Figures S6B, S6B’, S6E, and S6E’) that cannot be attributed to the increase in KA" cells (since urp1 expression was not changed), but rather to other GABAergic neurons in the spinal cord. These results suggest that the specification of ISN cells from 2 dpf onwards relies on a reduction of Notch signaling, as previously established for KA" and V3 interneurons prior to 1 dpf and from 1 dpf, respectively.12
Figure 6.
Inhibition of the Notch signaling pathway leads to an increase in tph2+ neurons
(A and B) HCR RNA-FISH against tph2 mRNA. Arrowheads indicate tph2-expressing neurons. (A) Control embryo treated with DMSO. (B) Embryo treated with the Notch inhibitor LY.
(C) Quantification of tph2+ neurons. Embryos were treated from 2 to 3 dpf. The dorsal side is oriented upwards, and the anterior side is oriented toward the left. Statistical significance was assessed using Welch’s t-test, and the level of significance is indicated by the number of asterisks: ∗∗ for p < 0.01. Scale bar: 20 μm.
Discussion
Our sequencing data uncovered previously unknown expression domains for sox1a. In addition to KA and V2 neurons, we discovered that sox1a is also expressed in certain ISNs located in the ventral spinal cord of zebrafish. Our findings suggest that, like KA" and V3 interneurons, ISNs develop from LFP progenitor cells through latent specification after 48 hpf, following the development of other interneuronal subtypes. The specification of ISNs is dependent on the downregulation of the Notch signaling pathway, similar to KA" and V3 interneurons.
How do LFP progenitor cells give rise to three molecularly and functionally distinct populations of ventral neurons?
One question that arises is how LFP progenitor cells give rise to three molecularly and functionally distinct populations of ventral neurons. The generation of different types of neurons, such as GABAergic KA" cells, glutamatergic V3 interneurons, and serotonergic ISNs, from the same pool of LFP cells, remains unclear. One crucial factor to consider is that the three neuronal populations are generated at different stages of embryonic development. Consequently, the progenitor pool they originate from may not be exposed to the same cellular environment or signaling pathways and their cues. This could lead to the activation of different gene regulatory networks in each neuronal population, ultimately resulting in the specification of distinct neuron types. During spinal cord development, the amount and duration of exposure to the signaling protein Shh play a critical role in activating specific genes in neural progenitor cells along the DV axis of the spinal cord.4,43 Similarly, the distinct progenitor cells in the LFP may be exposed to different concentrations and durations of Shh, leading to the activation of different genes in each of the three progenitor populations and ultimately resulting in the development of KA", V3, and ISNs. Furthermore, the temporal and spatial expression pattern of Shh, which is initially limited to the medial floor plate until 36 hpf and later extends to the LFP44 (Figure S5G), suggests that the progenitor cells generating KA" neurons (developing before 1 dpf) and ISNs (developing after 1 dpf) are exposed to different concentrations of Shh. It is plausible that the duration of exposure to the Shh signal significantly varies for each group of progenitor cells. Differences in the concentration and duration of Shh exposure for distinct progenitor populations in the LFP, as observed in different DV progenitor domains in the spinal cord, could be a possible mechanism involved in the generation of the three different neuronal populations from a common pool of progenitor cells.
Gene regulatory networks governing the development of serotoninergic neurons are conserved during evolution
In mice, the signaling cascade involved in the specification of 5-HT neurons has been extensively studied in the ventral hindbrain. Cells adjacent to the floor plate and positive for NKX2.2 develop into serotonergic neurons,40 with NKX2.2 and FOXA2 acting upstream of the genes encoding for Gata2, Gata3, and Insm1.36 Activation of these TFs induces the expression of Lmx1b and Pet1.45 In postmitotic precursor neurons, Pet1, Lmx1b, and Gata3 activate the expression of genes necessary for the synthesis of serotonin, such as Tph2, Ddc, and Dch1.36,46,47 Consistent with this gene expression cascade, the binding sites of Gata and Pet1 on the promoter regions of Pet1 and Tph2, respectively, have been identified and found to be essential for the expression of these genes in serotonergic neurons.48,49,50 Genetic evidence points to a pivotal role of GATA2 in the generation of 5-HT neurons.45
In zebrafish ISN-pre cells appear slightly earlier than ISNs during spinal cord development (Figure S4B), these cells are postmitotic (elavl3+) and express the same combination of TFs expressed in mouse serotonin precursor neurons, such as gata2a, gata3, lmx1bb and fev (Figures 4B and S4B). Additionally, ISNs in zebrafish also express several genes characteristic of serotonergic neurons in mice, including tph2, ddc, and htr1d. This suggests that the gene regulatory network during the specification of ISNs in zebrafish is conserved during evolution, involving genes and anatomical regions that are almost identical to those in mouse embryos.
However, further experiments are necessary to characterize the gene regulatory network involved in the development of serotonergic neurons in zebrafish. Several mutant lines in zebrafish for genes with potentially relevant roles in ISN specification, such as gata3, gata2a, lmx1ba, and lmx1bb, are available.18,51 Detailed characterization of their phenotypes could provide insights into the possible role of these factors in ISN development. Furthermore, the temporal and spatial contributions of specific TFs in ISN development could be investigated using zebrafish transgenic lines with TF-specific Cre drivers. Simultaneously, studying the transcriptional regulatory logic involved in the regulation of fev and tph2 expression in serotonergic neurons would be interesting. A knockdown approach using morpholino suggests that nkx2.9 is the most upstream TF gene in the genetic program leading to ISN specification (Köhler and Rastegar, unpublished). Generating a stable gene editing line through the CRISPR/Cas9 approach for nkx2.9 could confirm the phenotype observed with the nkx2.9 morphant and provide a better understanding of the genetic program required for upstream ISN development.
Although KA", V3, and ISNs are derived from LFP progenitor cells, they do not share a common genetic program
The expression and combinatorial action of specific TFs determine the developmental fate and functions of distinct cell populations. When comparing the expression of key TFs in KA", V3, and ISNs, we found only a few common TFs between these cells, most of which are related to their shared geographic position in the spinal cord. For example, these neurons express genes such as insm1a and several hox genes including hoxb9a, hoxb10a, hoxc10a, and hoxc9a.
Interestingly, we observed more shared TFs between KA" cells and ISNs, even though KA" cells develop at an earlier embryonic stage than ISNs. Thus, KA" cells and ISNs share the expression of gata2a, gata3, sox1a, sox1b, insm1a, and insm1b among other TFs.
In mice, Gata2, Gata3, and Insm1 are part of the gene regulatory network of 5-HT neurons. GATA2 has been shown to control Pet1 transcription in serotonergic neurons by directly binding to an upstream regulatory sequence.48,49,50 Furthermore, GATA2 plays an important role in the generation of 5-HT neurons.45
Previous work has partially unraveled the gene regulatory network of KA" specification in zebrafish embryos.14,18 Interestingly, in KA" interneurons, Gata2a function is required for the specification of GABAergic KA" neurons and inhibition of V3 fate. Data from mouse ISNs (serotonergic neurons) and zebrafish KAs (GABAergic neurons) suggest an important role for gata2 in their specification. However, further work is needed to better understand the role of the gata2a gene in the specification of ISNs in zebrafish and to gain a comprehensive understanding of the gene regulatory network involved in ISN development. It should be noted that the expression of a common set of TFs in two cell populations is not sufficient to conclude the existence of a shared gene regulatory network. For example, we found that the same transcription genes expressed in closely related GABAergic neurons KA" and KA' have different functions in each of these cells.14
Do ISNs consist of a heterogeneous subpopulation of tph2+ neurons?
scRNA-seq analysis of the sox1a:eGFP transgenic line revealed that ISNs in the zebrafish spinal cord originate from the LFP. Interestingly, our preliminary studies on ISN clusters demonstrated that not all ISNs exhibit the same pattern of TF co-expression. This finding suggests that the ISN population may consist of heterogeneous subpopulations with molecularly distinct identities. Specifically, we observed that only 39% of tph2+ neurons were positive for sox1a, while 88.5% of these neurons were sox1b+ and 36.6% were triple positive (tph2, sox1a, and sox1b). The differential expression of sox1a and sox1b in ISNs raises the question of whether these distinct subpopulations have unique functions or distinct functions, as has been shown for the V2a population in mice and zebrafish, where distinct types of spinal V2a interneurons activate different motor neuron pools during fast locomotion, such as escape behavior in zebrafish.52,53,54 To fully understand the significance of these molecular differences, further investigations are required. It is essential to analyze the morphology of axonal processes and synaptic connections in molecularly distinct ISNs, as well as establish the relationship between marker gene expression, neuronal morphology, and connectivity. These comprehensive analyses will provide valuable insights into the existence of different ISN subpopulations and their potential distinct functions within the zebrafish spinal cord.
Limitations of the study
The limitations of this study are as follows: First, the study focuses exclusively on the expression of sox1a in a specific subgroup of ISNs, which may not represent all subtypes or populations of ISNs. Second, the investigation only examines the origin of ISNs from LFP progenitor cells at 2 days dpf, and other time frames of specification remain unexplored. Further research is needed to investigate additional developmental stages. Third, while the study highlights the importance of inhibiting the Notch signaling pathway for the differentiation of LFP progenitor cells into ISNs, it is possible that other downstream mechanisms are involved, requiring further investigation. Furthermore, the study does not delve into the molecular interactions that govern the precise mechanisms involved in the sequential generation of GABAergic KA interneurons, glutamatergic V3 interneurons, and ISNs from LFP progenitor cells. Future studies should address these fine-tuning mechanisms.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| ChromoTek GFP-Booster ATTO488 | Proteintech | Cat#gba488; LOT:90918037AF1-01; RRID: AB_2631386 |
| Anti-Serotonin antibody | Sigma-Aldrich | Cat#S5545-100UL; RRID: AB_477522 |
| Bacterial and virus strains | ||
| XL1-Blue Competent Cells | Agilent Technologies | Cat#200249 |
| Chemicals, peptides, and recombinant proteins | ||
| LY411575 Notch inhibitor | Sigma-Aldrich | Cat#HY-50752 |
| Critical commercial assays | ||
| Neural Tissue Dissociation Kit (P) | Miltenyi | Cat#130-092-628 |
| 10x Chromium Kit | 10x Genomics | Cat#1000092 |
| HCR RNA-FISH | Molecular Instruments, Inc. | https://www.molecularinstruments.com/ |
| NGS Fragment Kit | Agilent Technologies | Cat#DNF-473-0500 |
| Chromium Next GEM Chip G Single Cell Kit, 16 rxns | 10x Genomics | Cat#PN-1000127 |
| Chromium Next GEM Single Cell 3ʹ Kit v3.1, 16 rxns | 10x Genomics | Cat#PN-1000268 |
| Chromium Single Cell 3′ Library & Gel Bead Kit v2, 16 rxns | 10x Genomics | Cat#PN-120237 |
| NovaSeq 6000 Reagent Kits | Illumina | Cat#20028312 |
| Deposited data | ||
| Single cell transcriptome | this study | GSE226494 |
| Single cell transcriptome | Scott et al.41 | GSE173350 |
| Ensembl 104 | Ensembl | https://www.ensembl.org/; RRID:SCR_002344 |
| Zebrafish reference genome NCBI, GRCz11 | Genome Reference Consortium | https://www.ncbi.nlm.nih.gov/grc/zebrafish; RRID:SCR_006472 |
| Experimental models: Organisms/strains | ||
| Zebrafish: Tg(sox1a:eGFP) | Gerber et al.19 | ZDB-FISH-200408-5 |
| Zebrafish: Tg(pet1:eGFP) | Lillesaar et al.55 | ZDB-FISH-150901-18516 |
| Oligonucleotides | ||
| tph2 HCR® probe set (B5) | Molecular Instruments, Inc. | LOT: RTD209 |
| sox1a HCR® probe set (B3) | Molecular Instruments, Inc. | NM_001002483.1; LOT: PRB115 |
| GAD1b HCR® probe set (B1) | Molecular Instruments, Inc. | NM_194419.1; LOT: PRA299 |
| urp1 HCR® probe set (B5) | Molecular Instruments, Inc. | LOT: PRO979 |
| B5-546 HCR® amplifier | Molecular Instruments, Inc. | N/A; LOT: S018425; LOT: S013725 |
| B3-647 HCR® amplifier | Molecular Instruments, Inc. | N/A; LOT: S020325; LOT: S022025 |
| B3-488 HCR® amplifier | Molecular Instruments, Inc. | N/A; LOT: S017624; LOT: S042523 |
| B1-546 HCR® amplifier | Molecular Instruments, Inc. | N/A; LOT: S019224; LOT: S030423 |
| Software and algorithms | ||
| R v4.1.0 | R project | RRID:SCR_001905 |
| Monocle2 | Qiu et al.56 | RRID:SCR_016339 |
| Seurat | Hao et al.35 | RRID:SCR_016341 |
| ImageJ | Schneider et al.57 | https://imagej.nih.gov/ij/; RRID:SCR_003070 |
| clusterProfiler | Wu et al.58 | https://bioconductor.org/packages/release/bioc/html/clusterProfiler.html; RRID:SCR_016884 |
| Cell Ranger v6.1.2 | 10x Genomics | RRID:SCR_017344 |
| Primer-BLAST | NCBI. Ye et al.59 | https://www.ncbi.nlm.nih.gov/tools/primer-blast/; RRID:SCR_003095 |
| GraphPad Prism 9 | GraphPad | https://www.graphpad.com/; RRID:SCR_002798 |
| Custom Scripts | this study | https://github.com/Fushun-Chen/Sox1a_scRNA.git |
| Other | ||
| BD FACSARIA III cell sorter | BD | RRID:SCR_016695 |
| Confocal microscope TCS SP8 STED | Leica | N/A |
| 5200 Fragment Analyzer System | Agilent Technologies | Cat#M5310AA; RRID:SCR_019417 |
| SPRIselect | Beckman Coulter Life Sciences | Cat# B23319 |
| Illumina NovaSeq 6000 | Illumina | Cat#20012850; RRID:SCR_016387 |
| HC PL APO 93×/1.30 GLYC CORR STED WHITE objective |
Leica | Cat#11506417 |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Sepand Rastegar (sepand.rastegar@kit.edu).
Materials availability
All zebrafish lines utilized in this study have been duly deposited at the European Zebrafish Resource Center (EZRC). For access to the remaining materials, please contact the lead researcher, Sepand Rastegar.
Experimental model and study participant details
Fish stocks and embryo collection
WT zebrafish were obtained from the ABO line, an inbred line initially derived from an intercross between the AB and OX lines (European Zebrafish Resource Center). WT and transgenic zebrafish sox1a:eGFP,19 pet1:eGFP55 were maintained on a 14 h/10 h light-dark cycle at 28.5°C in a recirculation system (Schwarz) and fed commercial food and in-house hatched brine shrimp as described previously.62 Embryos were cultured in an embryo medium and staged according to.63
For the scRNA-seq, Tg(sox1a:eGFP) and WT-AB fish were crossed and embryos were collected within 15 min to minimize the age difference between embryos. Dead and unfertilized eggs were removed at 4 hpf. Batches of 40 embryos were collected and kept in 30 mL of E3 zebrafish medium with methylene blue at 28°C and in a dark incubator until they reached the desired developmental stage.
Method details
Cell dissociation
Cell dissociation was performed as described.64,65 A total of 60 embryos at developmental stages 1, 2, 3 and 5 dpf were kept on ice for 10 min, then the trunk was separated from the head and the yolk removed. The bodies were placed directly into the dissociation buffer and kept on ice. Dissociation was done at 28°C by rotating the cells and trituration was done every 10 min until complete dissociation. Cells were filtered through 40 μm filters (Falcon, Cat#431750) and centrifuged at 300 g in 2% BSA and resuspended in 4% BSA. A total of 22,000 cells were sorted based on the GFP signal and the cells were subsequently treated for encapsulation.
Single-cell transcriptomics
Single-cell transcriptome sequencing was performed based on the 10x Genomics Single-cell transcriptome workflow.66 Specifically, 22,000 FACS-sorted cells were recovered in BSA-coated tubes containing 5 μL PBS. Accurate cell numbers were obtained using a Neubauer Hemocytometer. Cell samples were then carefully mixed with Reverse Transcription Reagent (Chromium Single Cell NextGem 3′ Library Kit v3.1) and loaded onto a Chromium Single Cell G Chip to reach a recovery of up to 10,000 cells per sample. The samples were processed further following the 10x Genomics user manual guidelines for single cell 3′ RNA-seq v3.1. In short, the droplets were directly subjected to reverse transcription, the emulsion was broken, and cDNA was purified using silane beads (Chromium Single Cell 3′ Gel Bead Kit v2). After amplification of cDNA with 11 cycles, samples were purified with 0.6x volume of SPRI select beads to deplete DNA fragments smaller than 400 bp and cDNA quality was monitored using the Agilent FragmentAnalyzer 5200 (NGS Fragment Kit). 10 μL of the resulting cDNA was used to prepare scRNA-seq libraries - involving fragmentation, dA-Tailing, adapter ligation and 10–13 cycles of indexing PCR following manufacturers guidelines. After quantification, libraries were sequenced on multiple Illumina NovaSeq 6000 S4 flowcells in 100 bp paired-end mode, aiming for 30,000 fragments per cell.
The raw sequencing data were processed with the ‘count’ command of the Cell Ranger software (v6.1.2) provided by 10X Genomics. To build the reference, the zebrafish genome (GRCz11), as well as gene annotation (Ensembl 104), were downloaded from Ensembl and the annotation was filtered with the ‘mkgtf’ command of Cell Ranger to include gene of the following types: ‘protein_coding’, ‘lincRNA’, ‘antisense’, ‘IG_LV_gene’, ‘IG_V_gene’, ‘IG_V_pseudogene’, ‘IG_D_gene’, ‘IG_J_gene’, ‘IG_J_pseudogene’, ‘IG_C_gene’, ‘IG_C_pseudogene’, ‘TR_V_gene’, ‘TR_V_pseudogene’, ‘TR_D_gene’, ‘TR_J_gene’, ‘TR_J_pseudogene’, ‘TR_C_gene’. Sequence and annotation of the GFP construct were included manually. Genome sequence and filtered annotation were then used as input to the ‘mkref’ command of Cell Ranger to build the appropriate Cell Ranger Reference.
Seurat clustering and integration of datasets
We used Seurat (version 4)35 and the following common steps for each round of analysis. Cells (i) with more than 20% mitochondrial RNA genes, (ii) with less than 500 total reads (nCount_RNA), and 200 unique genes (nFeature_RNA) were removed from analyses. Genes found in less than three cells were also removed from analyses. The remaining cells were converted to Seurat objects, data was normalized (NormalizeData), the top 2000 variable genes were identified (FindVariableFeatures), and all genes were used to scale data (ScaleData) by regressing out nCount_RNA. The top 30 Principal Component Analyses (PCAs) (RunPCA) were calculated and these PCAs were used for dimensional reduction (RunUMAP), finding cell clusters (FindNeighbors, FindClusters) with resolution = 0.5. To integrate all datasets, we used Seurat objects and the top 2000 variable genes identified above. First, the anchors were identified (FindIntegrationAnchors), objects were integrated (IntegrateData), and all genes were used for scaling data (ScaleData) by regressing out nCount_RNA. The top 30 PCAs (RunPCA) was calculated and these PCAs were used for dimensional reduction (RunUMAP), finding cell clusters (FindNeighbors, FindClusters) with resolutions 0.5, 1, 1.5 and 2. Unless indicated the above steps and settings were used for sub-setting cells for iterative analyses.
Preliminary quality control
A preliminary analysis of 1 dpf embryos showed a cell population that is highly expressing hbb genes (e.g., hbbe1.1). We used scaled data from 1 dpf embryos Seurat object and labeled cells that are expressing the hbbe1.1 gene more than 0.0. These cells were removed from 1 dpf embryos and another round of clustering was performed as above for 1 dpf raw counts (hbb cells removed). The remaining hbb cells, which were clustered together, were removed as well. After quality control and removing hbb expressing cells from the 1 dpf dataset, developmental stages 1, 2, 3 and 5 dpf datasets were converted to a Seurat object, and the above settings were used for all datasets.
Identifying main cell types
After integration and clustering, we identified 25 and 40 clusters of cells using the resolutions 0.5 and 1.5, respectively. We identified the marker genes using the FindAllMarkers function. We annotated the main cell types using the top 20 marker genes and known marker genes from the literature. For example, we used fabp7a/sox2/gfap for neural progenitors67,68 (NPro), elavl3/sv2a for neurons,67 cldnb/epcam/sox2 for lateral lines,69,70 meox1 for mesoderm,71 vcanb for fin fold,72 ccl25b for the hematopoietic system,73 pitx2 for cardiomyocytes,74 mylpfa for skeletal muscle75 and fli1a for the vascular system.76 The remaining cells without clear markers were labeled as "unclassified" cells.
Iterative clustering of neurons and neural progenitors
To further analyze neurons and neural progenitors, we subset the cells annotated as NP and neurons and generated new Seurat objects and integrated objects as described above. We annotated the cells based on the following markers: sst1.1 (KA'), urp1 (KA"), slc6a5/nkx1.2lb (V2s), olig2/sox10 (oligodendrocyte precursor cells, OPC), zic1/zic2a (zic+), nkx2.9/sulf1 (lateral floor plate, LFP), fev/lmx1bb/tph2 (intraspinal serotonergic neurons, ISN), fabp7a/gfap/foxj1a (ependymoradial glia, ERG), neurod4/neurog1 (neural precursor cells, NPC), slc17a6a/slc17a6b (glutamatergic inhibitory neurons, GLU IN), foxn4/vsx1 (V2 precursor cells, V2-pre), mki67/sox2/sox19a/her4 (Neural Progenitors, NP), isl1/isl2a (motor neurons, MN), twist1b (mesodermal cells, MC) and epcam (keratinocyte cells, KC). The top markers genes and top TFs (obtained from AnimalTFDB (v3.0))77 were identified from markers genes identified by FindAllMarkers function.
Similarly, we subset ISNs identified above and re-analyzed and integrated them as described above. Then, we identified ISN precursor cells (fev and lmx1bb) and ISNs (fev and tph2).
Temporal analyses (pseudotime) of LFP, ISN and V3 cells
To generate the cell trajectory for LFP and ISN cells or V3 cells, we used monocle (version 2).78 The counts and metadata from Seurat objects were used to generate input for monocle, estimateSizefactors, estimateDispersions, detectGenes, differentialGeneTest functions were used sequentially for normalization, estimating dispersion, and selecting genes for ordering. Then, dimensional reduction (reduceDimension) and cell orderings (orderCells) were done. We used a heatmap to visualize the changed genes (qval <0.00001) based on pseudotime and the num_clusters = 2.
To perform cell trajectory between LFP and V3 cells, we used LFP and V3 cells and removed LFP_2 (late stage) and MN clusters. We performed the same analyses using monocle. Additionally, we used a publicly available dataset41 (GEO Accession: GSE173350) for LFP and V3 cells. In brief, we performed the same analyses above using Seurat and annotated cells and subset LFP, V3-pre, and V3, using nkx2.2a/sim1a to select V3. Then, we used monocle to generate cell trajectory and pseudotime as described above.
GO and KEGG pathway analyses
We used clusterProfiler58 package and gene list identified by Seurat or monocle to find enriched GO and KEGGs. Terms with a q-value <0.05 were considered statistically significant.
Hybridization chain reaction RNA fluorescent in situ hybridization (HCR RNA-FISH)
HCR RNA-FISH was performed according to the protocol for whole-mount zebrafish embryos and larvae (Molecular Instruments, Inc.). The tph2 HCR probe set (B5) for detecting ISNs, the sox1a HCR probe set (B3), the urp1 HCR probe set (B5) and GAD1b HCR probe set (B1) were used at a concentration of 16 nM, while the HCR amplifiers (B5-546, B3-647, B3-488, B1-546) were used at a concentration of 6 nM.
The GFP-Booster ATTO488 (1:500, ChromoTek & Proteintech) was used to amplify the fluorescence signal in GFP transgenic larvae.
The head of stained embryos and larvae was cut off manually, and the embryos were embedded laterally in Aqua-Poly/Mount (Polysciences).
Immunohistochemistry
All incubation steps were performed at room temperature unless stated otherwise. Embryos are rehydrated with gradual methanol in PBST (1x PBS containing 0.1% Tween 20) series for 5 min each. Then, embryos are incubated in pre-warmed 150 mM Tris-HCI pH 9 for 15 min. After the incubation, embryos are washed two times in PBST for 5 min and treated with acetone at −20°C for 20 min to increase penetration. Embryos are incubated in a blocking buffer for 4 h, before incubation with primary antibodies in a blocking buffer at 4°C overnight. The next day, embryos are washed four times with PBT (1x PBS containing 0,8 Triton X-100) for 30 min, and then secondary antibody incubation is performed for 2 h. After staining, spinal cords are mounted using Aqua-Poly/Mount.
LY411575 treatment
LY411575 (42; Sigma Aldrich, St. Louis, MO, USA) was reconstituted with dimethyl sulfoxide (DMSO) to a stock concentration of 25 mM. Embryos were incubated in embryonic medium containing 10 μM LY411575, 0.04% DMSO or 0.04% DMSO alone from 24 to 48 hpf or 48 to 72 hpf. and then washed with embryonic medium containing 0.04% DMSO.19 After treatment, the embryos were fixed with 4% PFA for 2 h at room temperature and transferred to 100% methanol at −20°C.
Imaging
For obtaining single-cell resolution images in both the control and LY411175 treated groups, an SP8 inverted confocal microscope (Leica) with an HC PL APO 93×/1.30 GLYC CORR STED WHITE objective was utilized. Fiji/ImageJ software was employed for image processing, such as generating Z-projects and adjusting brightness or contrast.
Quantification and statistical analysis
Cell counting
To examine the effect of Notch inhibition on the number of cells expressing tph2+ and urp1+, the stained cells were counted over the span of five somites above the yolk extension at 2 and 3 dpf.
Statistical analysis
To compare the number of cells expressing tph2 between control and treated larvae, the Welch’s t-test was performed using GraphPad Prism 9. Significance was set at the level of 5%, meaning that a p value higher or equal to 0.05 represents no significance (n.s.), whereas p values smaller than 0.05 show significant results. The significance level is graded with the number of asterisks: ∗ for p < 0.05, ∗∗ for p < 0.01, ∗∗∗ for p < 0.001 and ∗∗∗∗ for p < 0.0001.
Acknowledgments
We would like to express our gratitude to T. Beil, A. Schröck, and M. Sobucki for their valuable technical support throughout this project. We are thankful to C. Lillesaar for generously sharing the zebrafish Tg(pet1:eGFP) line.55 We would also like to acknowledge S. Pfitsch for proofreading our manuscript. We would like to thank DRESDEN-Concept Genome Center deep sequencing facility and CMCB Flow Cytometry Core Facility for their assistance and resources. The research in Sepand Rastegar’s lab is supported by the Helmholtz Association BioInterfaces in Technology and Medicine and Natural, Artificial, and Cognitive Information Processing (NACIP) Programs and by project grants of the German Research Foundation (Deutsche Forschungsgemeinschaft (DFG)) GRK2039, STR 439/17-1, and RA 3469/5-1.
The permit for the operation of animal husbandry has been obtained according to §11 TSchG (Regierungspräsidium Karlsruhe, Aktenzeichen 35–9185.64/BH KIT). The animals are kept according to the recommendations of the European Society for Fish Models in Biology and Medicine (EuFishBioMed) of the Karlsruhe Institute of Technology (KIT), and according to the respective white paper.60 All experiments with zebrafish embryos were done in developmental stages between 24 hpf and 120 hpf, zebrafish embryos are not protected during this period.61
Author contributions
Conceptualization: S.R., Methodology: F.C., M.K., G.C., M.T., and M.I.C.; Software: F.C. and M.I.C.; Validation: F.C., M.K., G.C., M.T., M.I.C., and S.R.; Formal analysis: F.C., M.K., G.C., M.T., M.I.C., and S.R.; Investigation: F.C., M.K., G.C., M.T., and M.I.C.; Resources: S.R., M.I.C., and C.K.; Data curation: S.R., F.C., and M.I.C.; Writing - original draft: S.R., M.I.C., and C.K.; Writing - review and editing: S.R. and C.K.; Visualization: F.C., M.K., G.C., M.T., and M.I.C.; Supervision: S.R. Project administration: S.R. Funding acquisition: S.R.
Declaration of interests
C.K. serves as an advisor to Neuron D GmbH, which had no influence or financial support on the design and execution of this study. The authors declare no competing or financial interests.
Inclusion and diversity
We support inclusive, diverse, and equitable conduct of research.
Published: July 11, 2023
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2023.107342.
Supplemental information
Using the main cell types as Idents, the top marker genes were identified via FindAllMarkers from Seurat. These markers are relevant to the findings presented in Figures 1B and 1C.
Using the annotated cell clusters as Idents, the top marker genes were identified via FindAllMarkers from Seurat. These markers are relevant to the findings presented in Figures 2A, 2B, and 2C.
Using the sub-clusters from the ISN population as Idents, the top marker genes were identified via FindAllMarkers from Seurat. These markers are relevant to the findings presented in Figure 3D.
Using the clusters from the LFP, ISN-pre, and ISN populations as Idents, the top marker genes were identified via FindAllMarkers from Seurat. These markers are relevant to the findings presented in Figure 3F.
DifferentialGeneTest from Monocle was used to identify marker genes, which were then utilized to create a pseudotime-dependent heatmap. This heatmap is relevant to the findings presented in Figure 4C.
ClusterProfiler identified the Gene Ontology and Biological Process enriched using genes from Table S5, which is relevant to the findings in Figure 4E.
ClusterProfiler identified the KEGG enriched using genes from Table S5, which is relevant to the findings in Figure 4D.
Identification of Top Marker Genes using FindAllMarkers from Seurat, using the annotated cell clusters as Idents. These clusters are LFP, ISN-pre, V3, V3-pre, MN and NPC.
Identification of Top Marker Genes using FindAllMarkers from Seurat. These results are related to findings presented in Figure 5C. The gene lists correspond to the clusters LFP-1 (progenitor cells), LFP-2 (non-proliferating), ISN-pre, ISN, V3, V3-pre, MN and NPC.
The Branch Expression Analysis Model (BEAM) in monocle2 was used to reveal the marker genes, which were subsequently utilized to generate a heatmap finding the changes in gene expression over pseudotime. This is related to Figure 5E.
Data and code availability
-
•
sox1a Single-cell RNA-seq data have been deposited at NCBI GEO and are publicly available with accession number GSE226494. olig2 Single-cell RNA-seq data can be accessed with access number GSE173350.
-
•
The code generated in this work has been deposited on Github and is publicly available at https://github.com/Fushun-Chen/Sox1a_scRNA.git
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- 1.Ferg M., Armant O., Yang L., Dickmeis T., Rastegar S., Strähle U. Gene transcription in the zebrafish embryo: regulators and networks. Brief. Funct. Genomics. 2014;13:131–143. doi: 10.1093/bfgp/elt044. [DOI] [PubMed] [Google Scholar]
- 2.Reiter F., Wienerroither S., Stark A. Combinatorial function of transcription factors and cofactors. Curr. Opin. Genet. Dev. 2017;43:73–81. doi: 10.1016/j.gde.2016.12.007. [DOI] [PubMed] [Google Scholar]
- 3.Dessaud E., McMahon A.P., Briscoe J. Pattern formation in the vertebrate neural tube: a sonic hedgehog morphogen-regulated transcriptional network. Development. 2008;135:2489–2503. doi: 10.1242/dev.009324. [DOI] [PubMed] [Google Scholar]
- 4.Sagner A., Briscoe J. Establishing Neuronal Diversity in the Spinal Cord: A Time and a Place. Development. 2019;146 doi: 10.1242/dev.182154. [DOI] [PubMed] [Google Scholar]
- 5.Le Dréau G., Martí E. Dorsal-ventral patterning of the neural tube: a tale of three signals. Dev. Neurobiol. 2012;72:1471–1481. doi: 10.1002/dneu.22015. [DOI] [PubMed] [Google Scholar]
- 6.Alaynick W.A., Jessell T.M., Pfaff S.L. SnapShot: spinal cord development. Cell. 2011;146:178–178.e1. doi: 10.1016/j.cell.2011.06.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lu D.C., Niu T., Alaynick W.A. Molecular and cellular development of spinal cord locomotor circuitry. Front. Mol. Neurosci. 2015;8:25. doi: 10.3389/fnmol.2015.00025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lai H.C., Seal R.P., Johnson J.E. Making sense out of spinal cord somatosensory development. Development. 2016;143:3434–3448. doi: 10.1242/dev.139592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Goulding M. Circuits controlling vertebrate locomotion: moving in a new direction. Nat. Rev. Neurosci. 2009;10:507–518. doi: 10.1038/nrn2608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McMahon A.P. Neural patterning: the role of Nkx genes in the ventral spinal cord. Genes Dev. 2000;14:2261–2264. doi: 10.1101/gad.840800. [DOI] [PubMed] [Google Scholar]
- 11.Huang P., Xiong F., Megason S.G., Schier A.F. Attenuation of Notch and Hedgehog signaling is required for fate specification in the spinal cord. PLoS Genet. 2012;8 doi: 10.1371/journal.pgen.1002762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jacobs C.T., Kejriwal A., Kocha K.M., Jin K.Y., Huang P. Temporal cell fate determination in the spinal cord is mediated by the duration of Notch signalling. Dev. Biol. 2022;489:1–13. doi: 10.1016/j.ydbio.2022.05.010. [DOI] [PubMed] [Google Scholar]
- 13.Schäfer M., Kinzel D., Winkler C. Discontinuous organization and specification of the lateral floor plate in zebrafish. Dev. Biol. 2007;301:117–129. doi: 10.1016/j.ydbio.2006.09.018. [DOI] [PubMed] [Google Scholar]
- 14.Yang L., Rastegar S., Strähle U. Regulatory interactions specifying Kolmer-Agduhr interneurons. Development. 2010;137:2713–2722. doi: 10.1242/dev.048470. [DOI] [PubMed] [Google Scholar]
- 15.Andrews M.G., Kong J., Novitch B.G., Butler S.J. New perspectives on the mechanisms establishing the dorsal-ventral axis of the spinal cord. Curr. Top. Dev. Biol. 2019;132:417–450. doi: 10.1016/bs.ctdb.2018.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang L., Wang F., Strähle U. The Genetic Programs Specifying Kolmer-Agduhr Interneurons. Front. Neurosci. 2020;14 doi: 10.3389/fnins.2020.577879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Delile J., Rayon T., Melchionda M., Edwards A., Briscoe J., Sagner A. Single Cell Transcriptomics Reveals Spatial and Temporal Dynamics of Gene Expression in the Developing Mouse Spinal Cord. Development. 2019;146 doi: 10.1242/dev.173807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Andrzejczuk L.A., Banerjee S., England S.J., Voufo C., Kamara K., Lewis K.E. Tal1, Gata2a, and Gata3 Have Distinct Functions in the Development of V2b and Cerebrospinal Fluid-Contacting KA Spinal Neurons. Front. Neurosci. 2018;12:170. doi: 10.3389/fnins.2018.00170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gerber V., Yang L., Takamiya M., Ribes V., Gourain V., Peravali R., Stegmaier J., Mikut R., Reischl M., Ferg M., et al. The HMG box transcription factors Sox1a and Sox1b specify a new class of glycinergic interneuron in the spinal cord of zebrafish embryos. Development. 2019;146 doi: 10.1242/dev.172510. [DOI] [PubMed] [Google Scholar]
- 20.Karunaratne A., Hargrave M., Poh A., Yamada T. GATA proteins identify a novel ventral interneuron subclass in the developing chick spinal cord. Dev. Biol. 2002;249:30–43. doi: 10.1006/dbio.2002.0754. [DOI] [PubMed] [Google Scholar]
- 21.Li S., Misra K., Matise M.P., Xiang M. Foxn4 acts synergistically with Mash1 to specify subtype identity of V2 interneurons in the spinal cord. Proc. Natl. Acad. Sci. USA. 2005;102:10688–10693. doi: 10.1073/pnas.0504799102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Panayi H., Panayiotou E., Orford M., Genethliou N., Mean R., Lapathitis G., Li S., Xiang M., Kessaris N., Richardson W.D., Malas S. Sox1 is required for the specification of a novel p2-derived interneuron subtype in the mouse ventral spinal cord. J. Neurosci. 2010;30:12274–12280. doi: 10.1523/JNEUROSCI.2402-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Smith E., Hargrave M., Yamada T., Begley C.G., Little M.H. Coexpression of SCL and GATA3 in the V2 interneurons of the developing mouse spinal cord. Dev. Dyn. 2002;224:231–237. doi: 10.1002/dvdy.10093. [DOI] [PubMed] [Google Scholar]
- 24.Zhou Y., Yamamoto M., Engel J.D. GATA2 is required for the generation of V2 interneurons. Development. 2000;127:3829–3838. doi: 10.1242/dev.127.17.3829. [DOI] [PubMed] [Google Scholar]
- 25.Armant O., März M., Schmidt R., Ferg M., Diotel N., Ertzer R., Bryne J.C., Yang L., Baader I., Reischl M., et al. Genome-wide, whole mount in situ analysis of transcriptional regulators in zebrafish embryos. Dev. Biol. 2013;380:351–362. doi: 10.1016/j.ydbio.2013.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Montgomery J.E., Wahlstrom-Helgren S., Wiggin T.D., Corwin B.M., Lillesaar C., Masino M.A. Intraspinal serotonergic signaling suppresses locomotor activity in larval zebrafish. Dev. Neurobiol. 2018;78:807–827. doi: 10.1002/dneu.22606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Montgomery J.E., Wiggin T.D., Rivera-Perez L.M., Lillesaar C., Masino M.A. Intraspinal serotonergic neurons consist of two, temporally distinct populations in developing zebrafish. Dev. Neurobiol. 2016;76:673–687. doi: 10.1002/dneu.22352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McLean D.L., Fetcho J.R. Ontogeny and innervation patterns of dopaminergic, noradrenergic, and serotonergic neurons in larval zebrafish. J. Comp. Neurol. 2004;480:38–56. doi: 10.1002/cne.20280. [DOI] [PubMed] [Google Scholar]
- 29.McLean D.L., Fetcho J.R. Relationship of tyrosine hydroxylase and serotonin immunoreactivity to sensorimotor circuitry in larval zebrafish. J. Comp. Neurol. 2004;480:57–71. doi: 10.1002/cne.20281. [DOI] [PubMed] [Google Scholar]
- 30.McLean D.L., Fetcho J.R. Spinal interneurons differentiate sequentially from those driving the fastest swimming movements in larval zebrafish to those driving the slowest ones. J. Neurosci. 2009;29:13566–13577. doi: 10.1523/JNEUROSCI.3277-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Barreiro-Iglesias A., Mysiak K.S., Scott A.L., Reimer M.M., Yang Y., Becker C.G., Becker T. Serotonin Promotes Development and Regeneration of Spinal Motor Neurons in Zebrafish. Cell Rep. 2015;13:924–932. doi: 10.1016/j.celrep.2015.09.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gabriel J.P., Mahmood R., Kyriakatos A., Söll I., Hauptmann G., Calabrese R.L., El Manira A. Serotonergic modulation of locomotion in zebrafish: endogenous release and synaptic mechanisms. J. Neurosci. 2009;29:10387–10395. doi: 10.1523/JNEUROSCI.1978-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Huang C.X., Zhao Y., Mao J., Wang Z., Xu L., Cheng J., Guan N.N., Song J. An injury-induced serotonergic neuron subpopulation contributes to axon regrowth and function restoration after spinal cord injury in zebrafish. Nat. Commun. 2021;12:7093. doi: 10.1038/s41467-021-27419-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kuscha V., Frazer S.L., Dias T.B., Hibi M., Becker T., Becker C.G. Lesion-induced generation of interneuron cell types in specific dorsoventral domains in the spinal cord of adult zebrafish. J. Comp. Neurol. 2012;520:3604–3616. doi: 10.1002/cne.23115. [DOI] [PubMed] [Google Scholar]
- 35.Hao Y., Hao S., Andersen-Nissen E., Mauck W.M., 3rd, Zheng S., Butler A., Lee M.J., Wilk A.J., Darby C., Zager M., et al. Integrated analysis of multimodal single-cell data. Cell. 2021;184:3573–3587.e29. doi: 10.1016/j.cell.2021.04.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Spencer W.C., Deneris E.S. Regulatory Mechanisms Controlling Maturation of Serotonin Neuron Identity and Function. Front. Cell. Neurosci. 2017;11:215. doi: 10.3389/fncel.2017.00215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Deneris E., Gaspar P. Serotonin neuron development: shaping molecular and structural identities. Wiley Interdiscip Rev Dev Biol. 2018;7 doi: 10.1002/wdev.301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Deneris E.S., Wyler S.C. Serotonergic transcriptional networks and potential importance to mental health. Nat. Neurosci. 2012;15:519–527. doi: 10.1038/nn.3039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jacobs C.T., Huang P. Notch signalling maintains Hedgehog responsiveness via a Gli-dependent mechanism during spinal cord patterning in zebrafish. Elife. 2019;8 doi: 10.7554/eLife.49252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pattyn A., Vallstedt A., Dias J.M., Samad O.A., Krumlauf R., Rijli F.M., Brunet J.F., Ericson J. Coordinated temporal and spatial control of motor neuron and serotonergic neuron generation from a common pool of CNS progenitors. Genes Dev. 2003;17:729–737. doi: 10.1101/gad.255803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Scott K., O'Rourke R., Winkler C.C., Kearns C.A., Appel B. Temporal single-cell transcriptomes of zebrafish spinal cord pMN progenitors reveal distinct neuronal and glial progenitor populations. Dev. Biol. 2021;479:37–50. doi: 10.1016/j.ydbio.2021.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fauq A.H., Simpson K., Maharvi G.M., Golde T., Das P. A multigram chemical synthesis of the gamma-secretase inhibitor LY411575 and its diastereoisomers. Bioorg. Med. Chem. Lett. 2007;17:6392–6395. doi: 10.1016/j.bmcl.2007.07.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sagner A., Briscoe J. Morphogen interpretation: concentration, time, competence, and signaling dynamics. Wiley Interdiscip Rev Dev Biol. 2017;6 doi: 10.1002/wdev.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Al Oustah A., Danesin C., Khouri-Farah N., Farreny M.A., Escalas N., Cochard P., Glise B., Soula C. Dynamics of sonic hedgehog signaling in the ventral spinal cord are controlled by intrinsic changes in source cells requiring sulfatase 1. Development. 2014;141:1392–1403. doi: 10.1242/dev.101717. [DOI] [PubMed] [Google Scholar]
- 45.Craven S.E., Lim K.C., Ye W., Engel J.D., de Sauvage F., Rosenthal A. Gata2 specifies serotonergic neurons downstream of sonic hedgehog. Development. 2004;131:1165–1173. doi: 10.1242/dev.01024. [DOI] [PubMed] [Google Scholar]
- 46.Hendricks T., Francis N., Fyodorov D., Deneris E.S. The ETS domain factor Pet-1 is an early and precise marker of central serotonin neurons and interacts with a conserved element in serotonergic genes. J. Neurosci. 1999;19:10348–10356. doi: 10.1523/JNEUROSCI.19-23-10348.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hendricks T.J., Fyodorov D.V., Wegman L.J., Lelutiu N.B., Pehek E.A., Yamamoto B., Silver J., Weeber E.J., Sweatt J.D., Deneris E.S. Pet-1 ETS gene plays a critical role in 5-HT neuron development and is required for normal anxiety-like and aggressive behavior. Neuron. 2003;37:233–247. doi: 10.1016/s0896-6273(02)01167-4. [DOI] [PubMed] [Google Scholar]
- 48.Krueger K.C., Deneris E.S. Serotonergic transcription of human FEV reveals direct GATA factor interactions and fate of Pet-1-deficient serotonin neuron precursors. J. Neurosci. 2008;28:12748–12758. doi: 10.1523/JNEUROSCI.4349-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Scott M.M., Krueger K.C., Deneris E.S. A differentially autoregulated Pet-1 enhancer region is a critical target of the transcriptional cascade that governs serotonin neuron development. J. Neurosci. 2005;25:2628–2636. doi: 10.1523/JNEUROSCI.4979-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wyler S.C., Spencer W.C., Green N.H., Rood B.D., Crawford L., Craige C., Gresch P., McMahon D.G., Beck S.G., Deneris E. Pet-1 Switches Transcriptional Targets Postnatally to Regulate Maturation of Serotonin Neuron Excitability. J. Neurosci. 2016;36:1758–1774. doi: 10.1523/JNEUROSCI.3798-15.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hilinski W.C., Bostrom J.R., England S.J., Juárez-Morales J.L., de Jager S., Armant O., Legradi J., Strähle U., Link B.A., Lewis K.E. Lmx1b is required for the glutamatergic fates of a subset of spinal cord neurons. Neural Dev. 2016;11:16. doi: 10.1186/s13064-016-0070-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.El Manira A. Dynamics and plasticity of spinal locomotor circuits. Curr. Opin. Neurobiol. 2014;29:133–141. doi: 10.1016/j.conb.2014.06.016. [DOI] [PubMed] [Google Scholar]
- 53.Menelaou E., McLean D.L. Hierarchical control of locomotion by distinct types of spinal V2a interneurons in zebrafish. Nat. Commun. 2019;10:4197. doi: 10.1038/s41467-019-12240-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Song J., Dahlberg E., El Manira A. V2a interneuron diversity tailors spinal circuit organization to control the vigor of locomotor movements. Nat. Commun. 2018;9:3370. doi: 10.1038/s41467-018-05827-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lillesaar C., Tannhäuser B., Stigloher C., Kremmer E., Bally-Cuif L. The serotonergic phenotype is acquired by converging genetic mechanisms within the zebrafish central nervous system. Dev. Dyn. 2007;236:1072–1084. doi: 10.1002/dvdy.21095. [DOI] [PubMed] [Google Scholar]
- 56.Qiu X., Mao Q., Tang Y., Wang L., Chawla R., Pliner H.A., Trapnell C. Reversed graph embedding resolves complex single-cell trajectories. Nat. Methods. 2017;14:979–982. doi: 10.1038/nmeth.4402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Schneider C.A., Rasband W.S., Eliceiri K.W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wu T., Hu E., Xu S., Chen M., Guo P., Dai Z., Feng T., Zhou L., Tang W., Zhan L., et al. clusterProfiler 4.0: A universal enrichment tool for interpreting omics data. Innovation. 2021;2 doi: 10.1016/j.xinn.2021.100141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ye J., Coulouris G., Zaretskaya I., Cutcutache I., Rozen S., Madden T.L. Primer-BLAST: a tool to design target-specific primers for polymerase chain reaction. BMC Bioinf. 2012;13:134. doi: 10.1186/1471-2105-13-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Aleström P., D'Angelo L., Midtlyng P.J., Schorderet D.F., Schulte-Merker S., Sohm F., Warner S. Zebrafish: Housing and husbandry recommendations. Lab. Anim. 2020;54:213–224. doi: 10.1177/0023677219869037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Strähle U., Scholz S., Geisler R., Greiner P., Hollert H., Rastegar S., Schumacher A., Selderslaghs I., Weiss C., Witters H., Braunbeck T. Zebrafish embryos as an alternative to animal experiments--a commentary on the definition of the onset of protected life stages in animal welfare regulations. Reprod. Toxicol. 2012;33:128–132. doi: 10.1016/j.reprotox.2011.06.121. [DOI] [PubMed] [Google Scholar]
- 62.Westerfield M. University of Oregon Press; 2000. The Zebrafish Book: A Guide for the Laboratory Use of Zebrafish (Danio Rerio) [Google Scholar]
- 63.Kimmel C.B., Ballard W.W., Kimmel S.R., Ullmann B., Schilling T.F. Stages of embryonic development of the zebrafish. Dev. Dyn. 1995;203:253–310. doi: 10.1002/aja.1002030302. [DOI] [PubMed] [Google Scholar]
- 64.Cosacak M.I., Bhattarai P., Kizil C. Protocol for Dissection and Dissociation of Zebrafish Telencephalon for Single-Cell Sequencing. STAR Protoc. 2020;1 doi: 10.1016/j.xpro.2020.100042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Cosacak M.I., Bhattarai P., Reinhardt S., Petzold A., Dahl A., Zhang Y., Kizil C. Single-Cell Transcriptomics Analyses of Neural Stem Cell Heterogeneity and Contextual Plasticity in a Zebrafish Brain Model of Amyloid Toxicity. Cell Rep. 2019;27:1307–1318.e3. doi: 10.1016/j.celrep.2019.03.090. [DOI] [PubMed] [Google Scholar]
- 66.Zheng G.X.Y., Terry J.M., Belgrader P., Ryvkin P., Bent Z.W., Wilson R., Ziraldo S.B., Wheeler T.D., McDermott G.P., Zhu J., et al. Massively parallel digital transcriptional profiling of single cells. Nat. Commun. 2017;8 doi: 10.1038/ncomms14049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lange C., Rost F., Machate A., Reinhardt S., Lesche M., Weber A., Kuscha V., Dahl A., Rulands S., Brand M. Single cell sequencing of radial glia progeny reveals the diversity of newborn neurons in the adult zebrafish brain. Development. 2020;147 doi: 10.1242/dev.185595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.März M., Chapouton P., Diotel N., Vaillant C., Hesl B., Takamiya M., Lam C.S., Kah O., Bally-Cuif L., Strähle U. Heterogeneity in progenitor cell subtypes in the ventricular zone of the zebrafish adult telencephalon. Glia. 2010;58:870–888. doi: 10.1002/glia.20971. [DOI] [PubMed] [Google Scholar]
- 69.Behra M., Gallardo V.E., Bradsher J., Torrado A., Elkahloun A., Idol J., Sheehy J., Zonies S., Xu L., Shaw K.M., et al. Transcriptional signature of accessory cells in the lateral line, using the Tnk1bp1:EGFP transgenic zebrafish line. BMC Dev. Biol. 2012;12:6. doi: 10.1186/1471-213X-12-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lush M.E., Piotrowski T. Sensory hair cell regeneration in the zebrafish lateral line. Dev. Dyn. 2014;243:1187–1202. doi: 10.1002/dvdy.24167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Row R.H., Pegg A., Kinney B.A., Farr G.H., 3rd, Maves L., Lowell S., Wilson V., Martin B.L. BMP and FGF signaling interact to pattern mesoderm by controlling basic helix-loop-helix transcription factor activity. Elife. 2018;7 doi: 10.7554/eLife.31018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Rabinowitz J.S., Robitaille A.M., Wang Y., Ray C.A., Thummel R., Gu H., Djukovic D., Raftery D., Berndt J.D., Moon R.T. Transcriptomic, proteomic, and metabolomic landscape of positional memory in the caudal fin of zebrafish. Proc. Natl. Acad. Sci. USA. 2017;114:E717–E726. doi: 10.1073/pnas.1620755114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wattrus S.J., Zon L.I. Stem cell safe harbor: the hematopoietic stem cell niche in zebrafish. Blood Adv. 2018;2:3063–3069. doi: 10.1182/bloodadvances.2018021725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Franco D., Sedmera D., Lozano-Velasco E. Multiple Roles of Pitx2 in Cardiac Development and Disease. J. Cardiovasc. Dev. Dis. 2017;4 doi: 10.3390/jcdd4040016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zhang W., Roy S. The zebrafish fast myosin light chain mylpfa:H2B-GFP transgene is a useful tool for in vivo imaging of myocyte fusion in the vertebrate embryo. Gene Expr. Patterns. 2016;20:106–110. doi: 10.1016/j.gep.2016.02.001. [DOI] [PubMed] [Google Scholar]
- 76.Lawson N.D., Weinstein B.M. In vivo imaging of embryonic vascular development using transgenic zebrafish. Dev. Biol. 2002;248:307–318. doi: 10.1006/dbio.2002.0711. [DOI] [PubMed] [Google Scholar]
- 77.Hu H., Miao Y.R., Jia L.H., Yu Q.Y., Zhang Q., Guo A.Y. AnimalTFDB 3.0: a comprehensive resource for annotation and prediction of animal transcription factors. Nucleic Acids Res. 2019;47:D33–D38. doi: 10.1093/nar/gky822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Cao J., Wang Z., Likhtman A.E., Huang X., Ibrahim D.M., Hill A.J., Zhang F., Mundlos S., Christiansen L., Steemers F.J., et al. The single-cell transcriptional landscape of mammalian organogenesis. Nature. 2019;11:496–502. doi: 10.1038/s41586-019-0969-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Using the main cell types as Idents, the top marker genes were identified via FindAllMarkers from Seurat. These markers are relevant to the findings presented in Figures 1B and 1C.
Using the annotated cell clusters as Idents, the top marker genes were identified via FindAllMarkers from Seurat. These markers are relevant to the findings presented in Figures 2A, 2B, and 2C.
Using the sub-clusters from the ISN population as Idents, the top marker genes were identified via FindAllMarkers from Seurat. These markers are relevant to the findings presented in Figure 3D.
Using the clusters from the LFP, ISN-pre, and ISN populations as Idents, the top marker genes were identified via FindAllMarkers from Seurat. These markers are relevant to the findings presented in Figure 3F.
DifferentialGeneTest from Monocle was used to identify marker genes, which were then utilized to create a pseudotime-dependent heatmap. This heatmap is relevant to the findings presented in Figure 4C.
ClusterProfiler identified the Gene Ontology and Biological Process enriched using genes from Table S5, which is relevant to the findings in Figure 4E.
ClusterProfiler identified the KEGG enriched using genes from Table S5, which is relevant to the findings in Figure 4D.
Identification of Top Marker Genes using FindAllMarkers from Seurat, using the annotated cell clusters as Idents. These clusters are LFP, ISN-pre, V3, V3-pre, MN and NPC.
Identification of Top Marker Genes using FindAllMarkers from Seurat. These results are related to findings presented in Figure 5C. The gene lists correspond to the clusters LFP-1 (progenitor cells), LFP-2 (non-proliferating), ISN-pre, ISN, V3, V3-pre, MN and NPC.
The Branch Expression Analysis Model (BEAM) in monocle2 was used to reveal the marker genes, which were subsequently utilized to generate a heatmap finding the changes in gene expression over pseudotime. This is related to Figure 5E.
Data Availability Statement
-
•
sox1a Single-cell RNA-seq data have been deposited at NCBI GEO and are publicly available with accession number GSE226494. olig2 Single-cell RNA-seq data can be accessed with access number GSE173350.
-
•
The code generated in this work has been deposited on Github and is publicly available at https://github.com/Fushun-Chen/Sox1a_scRNA.git
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.