Abstract
Introduction
The success rate of in vitro fertilisation (IVF) treatment for couples with infertility remains low due to lack of a reliable tool in selecting euploid embryos for transfer. This study aims to compare the efficacy in embryo selection based on morphology alone compared with non-invasive preimplantation genetic testing for aneuploidy (niPGT-A) and morphology in infertile women undergoing IVF.
Methods and analysis
This is a randomised double-blind controlled trial conducted in two tertiary assisted reproduction centres. A total of 500 infertile women will be recruited and undergo IVF as indicated. They will be randomly assigned on day 6 after oocyte retrieval into two groups: the intervention group using morphology and niPGT-A and the control group based on morphology alone. In the control group, blastocysts with the best quality morphology will be replaced first. In the intervention group, blastocysts with the best morphology and euploid result of spent culture medium will be replaced first. The primary outcome is a live birth per the first embryo transfer. The statistical analysis will be performed with the intention to treat and per protocol.
Ethics and dissemination
Ethics approval was sought from the institutional review board of the two participating units. All participants will provide written informed consent before joining the study. The results of the study will be submitted to scientific conferences and peer-reviewed journals.
Trial registration number
Keywords: subfertility, reproductive medicine, genetics
STRENGTHS AND LIMITATIONS OF THIS STUDY.
This is a randomised double-blind controlled clinical trial.
The recruitment takes place in a large representative stakeholder group from the field of reproductive medicine in Hong Kong.
The intervention group of this study includes both morphology and non-invasive preimplantation genetic testing for aneuploidy (niPGT-A), as a result investigating the sole effect of addition of niPGT-A on embryo prioritisation alongside with morphology assessment.
One limitation is that our study is not designed to detect small differences in the live birth rates.
Introduction
One in seven couples experience difficulty in conceiving. Many of them will require in vitro fertilisation (IVF) treatment.1 IVF involves hormone injections to stimulate a woman’s ovaries to produce a number of oocytes which are collected by a minor operation and then mixed with sperm to form embryos in the laboratory. Usually, one or two embryos are transferred to the uterus 2–3 days (cleavage stage embryo transfer) or 5 days (blastocyst transfer) after oocyte retrieval. Despite advances in ovarian stimulation, culture medium and laboratory conditions, the pregnancy and birth rates remain 35% and 25% per transfer in Europe in 2014.2 The corresponding rates in the USA in 2016 were 45% and 36% per transfer, respectively (https://www.cdc.gov/art/reports/2016/national-summary.html).
The success of IVF depends on selection of the most competent embryos for transfer, which is still based on morphological criteria by examining the appearance of the embryos under a microscope. But it is well known that many women fail to achieve a pregnancy even after transfer of what are perceived to be good quality embryos. Therefore, some clinics replace multiple embryos in order to maximise pregnancy rates, a strategy which is associated with a high risk of multiple pregnancy.
Chromosome aneuploidy is an error in cell division that results in the ‘daughter’ cells having the wrong number of chromosomes. In some cases, there is a missing chromosome, while in others an extra one. It is a major reason for failure of pregnancy, miscarriage and congenital anomalies following both natural conception and IVF pregnancies and increases exponentially with maternal age.3–5
Our inability to assess embryo quality and select those with the highest potential for implantation on the basis of morphology has led to the use of preimplantation genetic testing for aneuploidy (PGT-A). PGT-A involves biopsy of a few cells from an embryo and assessment of the chromosome copy numbers. While PGT-A cannot create a healthy embryo or improve the quality of an embryo, it provides a method of selecting embryos with a normal number of chromosomes for transfer. This in turn has the potential to increase the chance of having a healthy live birth and reduce the risk of miscarriage or an abnormal fetus caused by an abnormal number of chromosomes.
Fluorescent in situ hybridisation (FISH) was first used in PGT-A studies but can only screen at most 9–12 chromosomes in multiple rounds of FISH with decreasing accuracy. A systematic review of nine randomised controlled trials failed to demonstrate the benefit of the use of PGT-A with FISH.6
Aneuploidy screening of all chromosomes is necessary to determine whether an embryo is chromosomally normal, that is, comprehensive chromosome screening. Several small randomised controlled trials showed significantly higher pregnancy or live birth rate and lower miscarriage rate following the use of using chromosomal microarray analysis.7–9 The emergence of more advanced genome sequencing such as next generation sequencing (NGS) provides a reliable, high throughput approach for PGT-A.10 The turnaround time of PGT-A with NGS is about a week, thus it is not possible to transfer blastocyst in the stimulated cycle. All blastocysts are frozen post biopsy and the blastocysts with normal genetic makeup are thawed and replaced in a subsequent menstrual cycle. Cryopreservation of blastocysts and replacing the frozen blastocysts after thawing in subsequent cycles become a common practice with vitrification as the cryopreservation method.11
A systematic review of the clinical utility of PGT-A with comprehensive chromosome screening found that three small randomised controlled trials demonstrated benefit in young and good prognosis patients in terms of clinical pregnancy rates and the use of single embryo transfer.12 However, a recent large randomised controlled trial of 661 women comparing PGT-A using NGS versus morphology showed PGT-A did not improve overall pregnancy outcomes in all women aged 25–40 years with at least two blastocysts that could be biopsied.13 It is possible that there is a detrimental effect of the biopsy of blastocysts on the embryo viability that nullifies the benefit of PGT-A.
The traditional PGT involves biopsy of a few cells from trophectoderm of a blastocyst, which requires skilful laboratory staff and additional instrumentation such as laser equipment. The trophectoderm biopsy is an invasive procedure and may lead to reduction in implantation potential, although the implantation potential is less affected when compared with blastomere biopsy from cleavage stage embryos.14 A non-invasive approach to PGT-A is definitely needed.
The demonstration of release of cell-free DNA from human embryos into the surrounding environment opens up the possibility of non-invasive PGT for aneuploidy (niPGT-A).15 Collection of spent culture medium (SCM) requires no specialised training and imposes negligible risk to the embryo. SCM may be more representative of the whole blastocyst as embryonic DNA is released from both trophectoderm and inner cell mass while the invasive trophectoderm biopsy obtains embryonic DNA from trophectoderm only. Multiple recent studies have demonstrated the ability to detect, extract and amplify cell-free DNA from SCM at the cleavage and blastocyst stages.16–23 It was shown that 24–48 hours of contact with the embryo was sufficient to collect cell-free DNA from SCM.21 The origin of cell-free DNA can be embryonic or parental. It is proposed that the cell-free DNA is derived from cells discarded by the embryos as a corrective mechanism for aneuploidies.16 24 However, the amount of cell-free DNA was not significantly greater in SCM from aneuploid versus euploid embryos, ruling out this possibility.20 Maternal and paternal contamination in SCM can be minimised by performing thorough oocyte striping and intracytoplasmic sperm injection (ICSI).25
In a recent review, the amplification success rate using sequential culture media for SCM ranged between 90% and 100% while the concordance rate in general ploidy, that is, euploid versus aneuploidy between SCM and blastocyst biopsy can be as high as 100% with an average of 75%.26 The difference in the general ploidy between SCM and trophectoderm biopsy can be due to mosaicism, which can be revealed in trophectoderm biopsy but not in SCM due to the nature of the DNA source and relatively low embryonic DNA fraction.
All relevant studies on niPGT-A focus the amplification success and concordance rates between SCM and trophectoderm biopsy. A large clinical randomised trial is urgently needed to confirm its efficacy in embryo selection during IVF in terms of live birth and miscarriage rates.
Objective and hypothesis
This randomised double-blind controlled trial aims to compare the efficacy in embryo selection based on morphology alone compared with niPGT-A and morphology in infertile women undergoing IVF.
Hypothesis to be tested include:
The embryo selection based on niPGT-A and morphology results in a higher live birth rate in IVF as compared with that based on morphology alone.
The embryo selection based on niPGT-A and morphology results in a lower miscarriage rate in IVF as compared with that based on morphology alone.
Methods and analysis
Trial design
This is a randomised double-blind controlled trial. Eligible women seeking fertility treatment in the Assisted Reproduction Units in Department of Obstetrics and Gynaecology in Kwong Wah Hospital (KWH) and Queen Mary Hospital (QMH) will be recruited for the study and informed written consent will be obtained after counselling.
Ethics approval was sought from the Institutional Review Board of the University of Hong Kong/Hospital Authority Hong Kong West Cluster (IRB number: UW 20-248) and Research Ethics Committee, Kowloon Central/Kowloon East (IRB number: KC/KE-20-0098/FR-2). The study was registered in Clinical Trials Registry (Identifier NCT04474522). Written informed consent will be sought before joining the study.
Selection and withdrawal of subjects
The population for trial will be infertile women undergoing IVF.
Inclusion criteria
Women admitted to the study will fulfil all of the following criteria:
Age less than 43 years at the time of ovarian stimulation.
Having at least two blastocysts suitable for freezing on day 6 after oocyte retrieval.
Exclusion criteria
Women should not be recruited in any of the following conditions:
Having less than two blastocysts suitable for freezing on day 6 after oocyte retrieval.
Undergoing PGT for monogenic diseases or structural rearrangement of chromosomes.
Using donor oocytes.
Having hydrosalpinx shown on pelvic scanning and not surgically treated.
Withdrawal criteria
Participation in the study is totally voluntary. Recruited women can withdraw from the study at any time without giving any reasons and subsequently they will receive standard medical care.
Treatment of subjects
Methods
Eligible women will be recruited for the study and informed written consent (see online supplemental material) will be obtained after counselling on the day of oocyte retrieval by research nurse or clinicians.
bmjopen-2023-072557supp001.pdf (107.8KB, pdf)
IVF protocol
Infertile women will undergo IVF as clinically indicated. They will receive ovarian stimulation as in standard operation procedure. Ovarian stimulation with gonadotropin injections (150–300 IU daily depending on the antral follicle count) will be given. Medroxyprogesterone acetate 10 mg daily will be started on the day 2 of ovarian stimulation or GnRH antagonist (cetrorelix) 0.25 mg daily will be started on the day 6 of ovarian stimulation to prevent premature ovulation. Ultrasound monitoring will be performed to monitor the growth of follicles. When three follicles reach >17 mm in diameter, human chorionic gonadotrophin (Ovidrel 0.25 mg) or GnRH agonist (Decapeptyl 0.3 mg) will be administered. Oocyte retrieval will be scheduled 36 hours after the trigger under transvaginal ultrasound guidance.
Oocytes will be fertilised conventionally or by ICSI depending on the semen parameters in accordance with the standard operating procedures and normal fertilisation will be assessed and confirmed by the presence of two pronuclei. Embryos will be grown individually to the blastocyst stage, up to day 6 after oocyte retrieval, in a monophasic medium. On day 3, embryos will be rinsed briefly in fresh culture medium. The culture medium will be replenished and culture will be continued at 37°C and 6% CO2 in reduced oxygen tension (5%). No fresh transfer of blastocysts will be performed in the stimulated cycle.
Grading of blastocyst by morphology
Blastocysts are graded according to Gardner’s classification.27 Each blastocyst will be cryopreserved on day 6 by vitrification individually and its SCM (~8 µL) will be frozen at −80°C separately and individually. The embryologist will grade the morphology of blastocysts according to Gardner’s criteria.
Then, on day 6 after oocyte retrieval, women will then be randomly assigned by a PGT laboratory staff into one of the following two groups according to a computer-generated randomisation list with a 1:1 ratio and a block size of 10. The randomisation list will be prepared by a research nurse, who is not involved in the clinical care of these women.
The intervention group using morphology and niPGT-A.
The control group based on morphology alone.
The women, clinicians and embryologists in the IVF laboratory will be blinded to the treatment groups they are assigned. Only the laboratory staff in the PGT laboratory will be aware of the group assignment.
niPGT-A of SCM
In the intervention group, comprehensive chromosome screening using NGS will be performed according to the recommendations of the company in all SCM samples. In the control group, the measurement will be done retrospectively after a live birth or when all blastocysts are replaced without a live birth.
A commercially available NI-PGT kit (PG-Seq Rapid Non-Invasive PGT kit, PerkinElmer) will be used to analyse the SCM samples. The protocol has been previously optimised with non-invasive samples from 15 laboratories around the world. The kit follows a single tube workflow, two-step PCR to whole genome amplification of the DNA in SCM and then attaches indexes and sequence-specific adapters to template DNA, resulting in sequencing ready samples.
After purification, equal molar concentration of indexed DNA from each sample will be pooled (96 samples) and then sequenced on a MiSeq system (Illumina) at 1×75 bp read length. On-board secondary analysis will be performed automatically by the MiSeq Reporter (Illumina), followed by the PG-Find Software (V.1.0, PerkinElmer). Reads aligning to anomalous, unstructured and highly repetitive sequences will be filtered from the analysis. A target bin size of 1000 kb will be used, giving a minimum resolution of 10 Mb. All genomic positions will be referred to the human genome build NCBI 37.
According to the default setting of the PG-Find software, classification of aneuploidy is determined by CNV (copy number variation) value. CNV value >2.7 is considered as gain while CNV value <1.3 is considered as loss. Sample will be concluded as non-euploid when one or more of the chromosomes show gain/loss.
The niPGT-A result of the SCM sample can be euploid, non-euploid and non-informative. It will be used only to prioritise the sequence of embryo transfer. Blastocysts with non-euploid result in the niPGT-A report will not be discarded and will be transferred with lower priority.
Blinding
The embryologist will grade the morphology of blastocysts according to Gardner’s criteria stated above and the grading of blastocysts will be entered into an online database, which will be managed by an IT technician. The laboratory staff in the PGT laboratory will enter the PGT result into an online database when the niPGT-A results are available. The IT technician will merge the data online to compile the sequence of embryo transfer according to a predetermined algorithm which depends on the day of blastocyst development (day 5 better than day 6), blastocyst morphology and niPGT-A result of the intervention group. The IT technician will issue the sequence of embryo transfer which does not contain information on the grading of the blastocyst and the NIPGT result to the embryologists in the IVF laboratory. Therefore, the subjects recruited, the clinicians and the embryologists will be blinded to the group allocation.
A pilot study
A pilot study was conducted on 82 SCM from February to September 2020. Media cultured in parallel but without contact with embryos were collected as controls (n=8). Amplification was successful in 80 SCM (97.6%, 80/82) and 72 SCM resulted in conclusive result (90.0%, 72/80). All controls showed no amplification.
In this cohort, 40 SCM with conclusive results were collected from PGT cycles in which trophectoderm biopsies were also performed. Overall, 85.0% (34/40) of samples showed concordance results between trophectoderm biopsy and SCM.
Frozen embryo transfer
Blastocysts will be replaced in the natural or hormonal replacement cycles, depending whether the women have regular menstrual cycles or not. Only one blastocyst will be transferred each time. The embryologists in the IVF laboratory will thaw and transfer the blastocyst according to the sequence of embryo transfer generated and issued by the IT technician. In the control group, blastocysts which develop on day 5 after the oocyte retrieval and have the best grading will be replaced first. In the intervention group, blastocysts which develop on day 5 and have the best grading and euploid result will be replaced first. If no blastocysts have euploid result, those with non-informative followed by non-euploid results will be replaced.
Pregnancy
A urine pregnancy test will be performed 14 days after the transfer. If the pregnancy test is positive, transvaginal ultrasound will be performed 2 weeks later to locate the pregnancy and confirm foetal viability and the number of fetuses. Subsequent management will be the same as other women with early pregnancy. They will be referred for antenatal care when the ongoing pregnancy is 8–10 weeks.
Follow-up
Written consent regarding retrieval of pregnancy and delivery data will be sought from the recruited women at the time of study. The women will be contacted after delivery by phone to retrieve the information of the pregnancy outcomes. The outcome of the pregnancy (delivery, miscarriage), number of babies born, birth weights and obstetrics complications will be recorded.
Women in both groups will continue to have blastocyst transfer until all the cryopreserved blastocysts are used up or they become pregnant within 6 months after randomisation. Cumulative live birth rate will be calculated (the number of live birth per couple within the study period). The pregnancy complication and congenital abnormalities of the pregnancies in the two groups will be traced through hospital records or patient contact by mail or phone of mail and compared.
Assessment of outcomes
The primary outcome is live birth beyond 22 weeks of gestation per the first frozen embryo transfer.
Secondary outcomes include:
Cumulative live birth rate: the number of pregnancies leading to live birth within 6 months of randomisation.
Time to pregnancy.
Positive urine pregnancy test.
Clinical pregnancy defined as the presence of intrauterine gestational sac on scanning at gestational week 6.
Ongoing pregnancy as the presence of a fetal pole with pulsation at 8–10 weeks of gestation.
Miscarriage defined as a clinically recognised pregnancy loss before the 22 weeks of pregnancy and whose denominator is the clinical pregnancy.
Multiple pregnancy: presence of more than one intrauterine sac at 6 weeks of gestation.
Ectopic pregnancy.
Pregnancy outcomes including preterm delivery, pre-eclampsia, gestational diabetes, congenital anomaly, perinatal mortality, Apgar score and birth weight of newborn.
Consistency of case management
The same standardised study protocol will be adopted in the two study centres. The clinicians who manage the women in KWH have all been trained at QMH, and are now adopting the same clinical management protocols in their respective unit. Regular and frequent communication among the two participating centres will also ensure dissemination of updated information on recruitment and safety issues.
Patient and public involvement
Patients or the public were not involved in the design, conduct, reporting or dissemination plans of this study.
Statistics
Sample size calculation
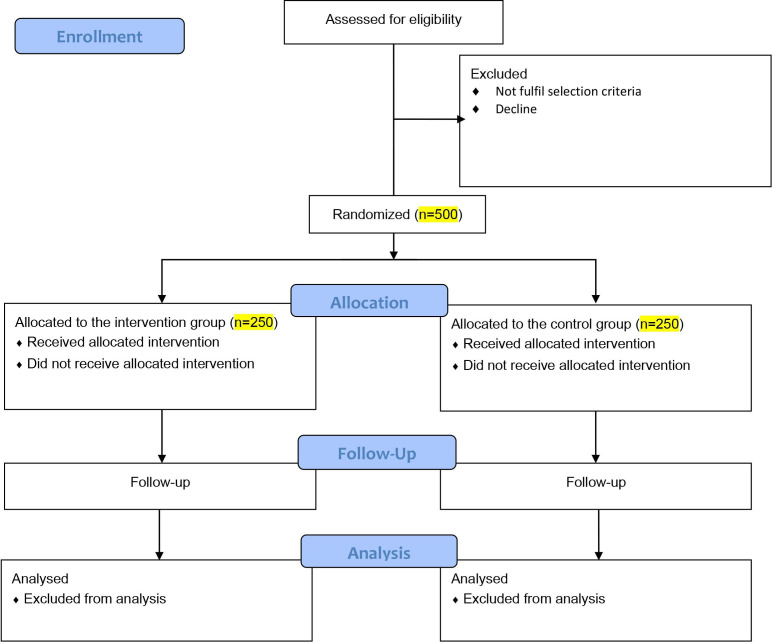
From the data in QMH and KWH, the live birth rate following transfer of one blastocyst based on morphology was about 35% in 2018 and 2019. We anticipate following blastocyst morphology and NIPGT-A, the live birth rate will increase from 35% to 50%, that is, 15% increase. The 50% live birth rate is based on the live birth rate observed following conventional PGT-A in the centre. The calculated sample size is 224 women in each group to give a power of 0.9 and type I error of 0.05. Assuming a 10% drop-out rate, the total sample size to be 500, 250 subjects in each group (figure 1).
Figure 1.
CONSORT 2010 flow diagram. CONSORT, Consolidated Standards of Reporting Trials.
A prespecified subgroup analysis will be performed: women aged <35 years vs ≥35 years.
Data analysis
Demographic features of women in the two groups will be compared. Comparison of quantitative variables will be performed using Student’s t-test or Mann-whitney U-test where appropriate, while categorical variables will be compared using a χ2 analysis, multivariable logistic regression or the one-way analysis of variance test if more than two categories will be compared. All statistical analyses of the data will be performed with the intention to treat and per protocol using the SPSS programme V.26.0 (SPSS), and a p<0.05 will be considered statistically significant.
The sensitivity, specificity, likelihood ratios and OR of the niPGT-A result will be calculated.
Ethics and dissemination
Ethics approval was sought from the Institutional Review Board of the University of Hong Kong/Hospital Authority Hong Kong West Cluster (IRB number: UW 20-248) and Research Ethics Committee, Kowloon Central/Kowloon East (IRB number: KC/KE-20-0098/FR-2). All participants will provide written informed consent before randomisation. The research findings from this study will be submitted to scientific conferences and peer-reviewed journals for publication so as to disseminate the results to other researchers and clinicians working in the field.
Discussion
The success rate of IVF has long been limited due to the inability to assess the implantation potential of each embryo accurately. Assessment by morphology using Gardner’s classification could not reflect chromosomal abnormalities in embryos, which is the reason for failure of implantation in most of the cases. NGS offers several advantages over chromosomal microarray analysis by (1) reduced DNA sequencing cost by high throughput sequencing technologies and a high number of samples can be simultaneously sequenced in a single testing; (2) enhanced detection of partial or segmental aneuploidies as a result of the increase in chromosomal analysis resolution to a few mega bases; (3) increased dynamic range enabling enhanced detection of mosaicism in multicellular samples and (4) automation of the sequencing library preparation and automation of the PGT-A diagnostic procedure. However, the beneficial effect of PGT-A has been nullified by the detrimental effect of embryo biopsy. If niPGT-A can be demonstrated to be a better blastocyst evaluation tool than the traditional morphology assessment during IVF treatment, it can potentially shorten the time-to-pregnancy in women with infertility.
Currently, all relevant studies on niPGT-A focused on the amplification success and concordance rates between SCM and trophectoderm biopsy but not on assessing its ability as a screening tool for blastocyst transfer prioritisation. This study could provide valuable information on the potential novel use of niPGT-A as an adjunct for morphological assessment of blastocysts.
Trial status
The first subject was recruited on 1 July 2021 and 400 women were recruited up to the writing of this protocol paper.
Supplementary Material
Footnotes
Contributors: All authors have substantial contribution to the protocol paper. HYHC: drafting the manuscript and revising it critically for important intellectual content, responsible for seeking ethics approval and registration on ClinicalTrials.gov. JFCC: substantial contribution to designing the work flow and logistics of niPGT-A, responsible for generating niPGT-A report. SFL: responsible for the recruitment of subjects and seeking ethics approval at the Kwong Wah Hospital. KKWL: responsible for collaborating with the PGT laboratory regarding logistics of collecting SCM and transferral. Responsible for IVF laboratory. EHYN: Substantial contributions to the conception and design of the whole study, and final approval of the version to be published. WSBY: Responsible for the design and worked on logistics of the study, and final approval of the version to be published.
Funding: The study is supported by the Health and Medical Research Fund (Project number 08192196).
Competing interests: None declared.
Patient and public involvement: Patients and/or the public were not involved in the design, or conduct, or reporting, or dissemination plans of this research.
Provenance and peer review: Not commissioned; externally peer reviewed.
Supplemental material: This content has been supplied by the author(s). It has not been vetted by BMJ Publishing Group Limited (BMJ) and may not have been peer-reviewed. Any opinions or recommendations discussed are solely those of the author(s) and are not endorsed by BMJ. BMJ disclaims all liability and responsibility arising from any reliance placed on the content. Where the content includes any translated material, BMJ does not warrant the accuracy and reliability of the translations (including but not limited to local regulations, clinical guidelines, terminology, drug names and drug dosages), and is not responsible for any error and/or omissions arising from translation and adaptation or otherwise.
Ethics statements
Patient consent for publication
Not applicable.
References
- 1.NICE . Fertility: assessment and treatment for people with fertility problems. NICE Guideline CG11 2014. [Google Scholar]
- 2.De C, Calhaz-Jorge C, Kupka MS, et al. European IVF-monitoring consortium (EIM) for the European society of human reproduction and Embryology (ESHRE). ART in Europe, 2014: results generated from European registries by ESHRE: the European IVF-monitoring consortium (EIM) for the European society of human reproduction and Embryology (ESHRE). Hum Reprod 2018;33:1586–601. [DOI] [PubMed] [Google Scholar]
- 3.Spandorfer SD, Davis OK, Barmat LI, et al. Relationship between maternal age and Aneuploidy in in vitro fertilization pregnancy loss. Fertil Steril 2004;81:1265–9. 10.1016/j.fertnstert.2003.09.057 [DOI] [PubMed] [Google Scholar]
- 4.Hassold T, Hall H, Hunt P. The origin of human Aneuploidy: where we have been, where we are going. Hum Mol Genet 2007;16 Spec No. 2:R203–8. 10.1093/hmg/ddm243 [DOI] [PubMed] [Google Scholar]
- 5.Andersen A-M, Wohlfahrt J, Christens P, et al. Maternal age and fetal loss: population based register linkage study. BMJ 2000;320:1708–12. 10.1136/bmj.320.7251.1708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mastenbroek S, Twisk M, van der Veen F, et al. Preimplantation genetic screening: a systematic review and meta-analysis of Rcts. Hum Reprod Update 2011;17:454–66. 10.1093/humupd/dmr003 [DOI] [PubMed] [Google Scholar]
- 7.Yang Z, Liu J, Collins GS, et al. Selection of single Blastocysts for fresh transfer via standard morphology assessment alone and with array CGH for good prognosis IVF patients: results from a randomized pilot study. Mol Cytogenet 2012;5:24. 10.1186/1755-8166-5-24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Scott RT, Upham KM, Forman EJ, et al. Blastocyst biopsy with comprehensive Chromosome screening and fresh embryo transfer significantly increases in vitro fertilization implantation and delivery rates: a randomized controlled trial. Fertil Steril 2013;100:697–703. 10.1016/j.fertnstert.2013.04.035 [DOI] [PubMed] [Google Scholar]
- 9.Forman EJ, Hong KH, Ferry KM, et al. In vitro fertilization with single Euploid Blastocyst transfer: a randomized controlled trial. Fertil Steril 2013;100:100–7. 10.1016/j.fertnstert.2013.02.056 [DOI] [PubMed] [Google Scholar]
- 10.Fiorentino F, Biricik A, Bono S, et al. Development and validation of a next-generation sequencing-based protocol for 24-Chromosome Aneuploidy screening of embryos. Fertil Steril 2014;101:1375–82. 10.1016/j.fertnstert.2014.01.051 [DOI] [PubMed] [Google Scholar]
- 11.Evans J, Hannan NJ, Edgell TA, et al. Fresh versus frozen embryo transfer: backing clinical decisions with scientific and clinical evidence. Hum Reprod Update 2014;20:808–21. 10.1093/humupd/dmu027 [DOI] [PubMed] [Google Scholar]
- 12.Lee E, Illingworth P, Wilton L, et al. The clinical effectiveness of Preimplantation genetic diagnosis for Aneuploidy in all 24 Chromosomes (PGD-A): systematic review. Hum Reprod 2015;30:473–83. 10.1093/humrep/deu303 [DOI] [PubMed] [Google Scholar]
- 13.Munné S, Kaplan B, Frattarelli JL, et al. Preimplantation genetic testing for Aneuploidy versus morphology as selection criteria for single frozen-thawed embryo transfer in good-prognosis patients: a multicenter randomized clinical trial. Fertil Steril 2019;112:1071–9. 10.1016/j.fertnstert.2019.07.1346 [DOI] [PubMed] [Google Scholar]
- 14.Scott RT, Upham KM, Forman EJ, et al. Cleavage-stage biopsy significantly impairs human embryonic implantation potential while Blastocyst biopsy does not: a randomized and paired clinical trial. Fertil Steril 2013;100:624–30. 10.1016/j.fertnstert.2013.04.039 [DOI] [PubMed] [Google Scholar]
- 15.Assou S, Aït-Ahmed O, El Messaoudi S, et al. Non-invasive pre-implantation genetic diagnosis of X-linked disorders. Med Hypotheses 2014;83:506–8. 10.1016/j.mehy.2014.08.019 [DOI] [PubMed] [Google Scholar]
- 16.Hammond ER, McGillivray BC, Wicker SM, et al. Characterizing nuclear and mitochondrial DNA in spent embryo culture media: genetic contamination identified. Fertil Steril 2017;107:220–8. 10.1016/j.fertnstert.2016.10.015 [DOI] [PubMed] [Google Scholar]
- 17.Xu J, Fang R, Chen L, et al. Noninvasive Chromosome screening of human embryos by genome sequencing of embryo culture medium for in vitro fertilization. Proc Natl Acad Sci U S A 2016;113:11907–12. 10.1073/pnas.1613294113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shamonki MI, Jin H, Haimowitz Z, et al. Proof of concept: Preimplantation genetic screening without embryo biopsy through analysis of cell-free DNA in spent embryo culture media. Fertil Steril 2016;106:1312–8. 10.1016/j.fertnstert.2016.07.1112 [DOI] [PubMed] [Google Scholar]
- 19.Feichtinger M, Vaccari E, Carli L, et al. Non-invasive Preimplantation genetic screening using array comparative Genomic hybridization on spent culture media: a proof-of concept pilot study. Reprod Biomed Online 2017;34:583–9. 10.1016/j.rbmo.2017.03.015 [DOI] [PubMed] [Google Scholar]
- 20.Vera-Rodriguez M, Diez-Juan A, Jimenez-Almazan J, et al. Origin and composition of cell-free DNA in spent medium from human embryo culture during Preimplantation development. Hum Reprod 2018;33:745–56. 10.1093/humrep/dey028 [DOI] [PubMed] [Google Scholar]
- 21.Kuznyetsov V, Madjunkova S, Antes R, et al. Evaluation of a novel non-invasive Preimplantation genetic screening approach. PLoS One 2018;13:e0197262. 10.1371/journal.pone.0197262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ho JR, Arrach N, Rhodes-Long K, et al. Pushing the limits of detection: investigation of cell-free DNA for Aneuploidy screening in embryos. Fertility and Sterility 2018;110:467–475. 10.1016/j.fertnstert.2018.03.036 [DOI] [PubMed] [Google Scholar]
- 23.Capalbo A, Romanelli V, Patassini C, et al. Diagnostic efficacy of Blastocoel fluid and spent media as sources of DNA for Preimplantation genetic testing in standard clinical conditions. Fertil Steril 2018;110:870–9. 10.1016/j.fertnstert.2018.05.031 [DOI] [PubMed] [Google Scholar]
- 24.Magli MC, Albanese C, Crippa A, et al. Deoxyribonucleic acid detection in Blastocoelic fluid: a new Predictor of embryo Ploidy and viable pregnancy. Fertil Steril 2019;111:77–85. 10.1016/j.fertnstert.2018.09.016 [DOI] [PubMed] [Google Scholar]
- 25.Tsai N-C, Chang Y-C, Su Y-R, et al. Validation of non-invasive Preimplantation genetic screening using a routine IVF laboratory Workflow[J]. Biomedicines 2022;10:1386. 10.3390/biomedicines10061386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Belandres D, Shamonki M, Arrach N. Current status of spent embryo media research for Preimplantation genetic testing. J Assist Reprod Genet 2019;36:819–26. 10.1007/s10815-019-01437-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gardner D, Schoolcraft W, Jansen R, et al. Towards reproductive certainty: infertility and genetics beyond. vitro culture of human blastocysts. Carnforth: Parthenon Press, 1999. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
bmjopen-2023-072557supp001.pdf (107.8KB, pdf)