Abstract
Many enteroviruses, members of the family Picornaviridae, cause a rapid and drastic inhibition of host cell protein synthesis during infection, a process referred to as host cell shutoff. Poliovirus, one of the best-studied enteroviruses, causes marked inhibition of host cell translation while preferentially allowing translation of its own genomic mRNA. An abundance of experimental evidence has accumulated to indicate that cleavage of an essential translation initiation factor, eIF4G, during infection is responsible at least in part for this shutoff. However, evidence from inhibitors of viral replication suggests that an additional event is necessary for the complete translational shutoff observed during productive infection. This report examines the effect of poliovirus infection on a recently characterized 3′ end translational stimulatory protein, poly(A)-binding protein (PABP). PABP is involved in stimulating translation initiation in lower eukaryotes by its interaction with the poly(A) tail on mRNAs and has been proposed to facilitate 5′-end–3′-end interactions in the context of the closed-loop translational model. Here, we show that PABP is specifically degraded during poliovirus infection and that it is cleaved in vitro by both poliovirus 2A and 3C proteases and coxsackievirus B3 2A protease. Further, PABP cleavage by 2A protease is accompanied by concurrent loss of translational activity in an in vitro-translation assay. Similar loss of translational activity also occurs simultaneously with partial 3C protease-mediated cleavage of PABP in translation assays. Further, PABP is not degraded during infections in the presence of guanidine-HCl, which blocks the complete development of host translation shutoff. These results provide preliminary evidence that cleavage of PABP may contribute to inhibition of host translation in infected HeLa cells, and they are consistent with the hypothesis that PABP plays a role in facilitating translation initiation in higher eukaryotes.
Enteroviruses are members of the family Picornaviridae and are the etiologic agents responsible for many pathological syndromes. Since nearly all enteroviruses cause drastic inhibition of host cell translation (17, 25, 43), referred to as host cell shutoff, these viruses have been intensely studied in the last two decades as ideal systems to dissect the functionality and mechanisms of eukaryotic translation. One of the first insights into the mechanism of inhibition of capped cellular mRNA translation was gained when it was found that the eIF4G subunit (formerly called p220 or eIF-4γ) of eukaryotic initiation factor 4F (eIF4F; the cap-binding protein complex) was cleaved early in poliovirus infection, correlating well with the inhibition of host cell-derived, cap-dependent translation (19, 34). eIF4F is a complex of proteins containing eIF4G; eIF4A, an RNA helicase; and eIF4E, the cap-binding protein (42). The eIF4F complex plays a critical role in translation initiation by recruiting ribosomes to the mRNA-initiation factor precomplex via an interaction with 40S ribosome-associated eIF3 (for reviews on translation, see references 42, 43, and 55). Growing evidence suggests that eIF4G functions as a molecular bridge which serves to bring mRNA to the ribosome. The kinetics of eIF4G cleavage closely parallel the shutoff of host cell translation, though eIF4G cleavage has been noted to slightly precede the complete shutoff of host translation (19, 56). Though this cleavage separates the eIF4E and eIF3 binding domains on eIF4G and thus is thought to be essential for host cell shutoff to occur, several lines of evidence suggest that eIF4G cleavage is not in itself sufficient to cause the complete shutoff of host cell translation. First, studies of infected cells done with an inhibitor of poliovirus replication, guanidine, demonstrated that in the absence of detectable viral replication and viral proteins, translation of cellular protein was only inhibited by 40 to 60% in the presence of completely degraded eIF4G (6, 49). Further studies with other inhibitors of viral replication, such as monensin and nigericin, showed essentially the same result (30). In some instances, cellular translation continued at nearly normal levels while eIF4G had been completely degraded. Furthermore, a poliovirus mutant was described which did not cause cleavage of eIF4G but did cause global host translation inhibition (4), which indicated that other events were playing a role in the translation inhibition. Despite numerous efforts, cleavage or alteration of other translation initiation factors has not been found (16, 18, 64). eIF2α, which plays a role in forming the initiator Met-tRNA complex (61), has been shown to undergo phosphorylation during the latter part of infection (5, 47). However, this phosphorylation is thought to occur too late to be involved in host cell shutoff, and it is probably involved in the global inhibition of translation at the end of the infectious cycle. These studies have suggested that although eIF4G cleavage may be an integral part of host cell translation shutoff, another alteration(s) or event(s) must be occurring during infection to allow the complete switch from cellular cap-dependent mRNA translation to viral cap-independent translation.
Many recent studies have begun to uncover how translation initiation can occur by mechanisms dependent on the 3′ end of mRNA. There have been a variety of studies examining the effect of 3′ untranslated regions (UTRs) on translation regulation during development (14) and numerous studies showing that the polyadenylate tails present on the ends of most eukaryotic mRNAs act as translational initiation regulators (20, 28, 44). However, these data have been difficult to reconcile with current initiation models, which propose that the required steps in translation initiation took place on structures present at the 5′ ends of mRNAs via the 5′ mRNA cap structure. Recent studies of yeast and plant systems have shown that 5′ and 3′ regions of mRNAs are capable of associating via protein factors binding specifically to these regions. In particular, the poly(A)-binding protein (PABP) Pab1p, which interacts with the poly(A) tail present on most eukaryotic mRNAs, has been shown to interact with the eIF4G homologues in yeast, Tif4631p and Tif4632p (59). In plants, PABP interacts with eIF-iso4G and eIF4B (36) and increases PABP’s RNA-binding affinity. Studies of the translation efficiencies of mRNAs containing either a cap structure alone or a poly(A) tail alone show that both mRNAs are capable of undergoing translation but that the presence of both structures provides a synergistic stimulation of translation efficiency (20, 28). It has also been shown in yeast that the poly(A) tail is itself a translational promoter and that this structure together with the cap structure is able to promote efficient ribosome recruitment to the 5′ end of mRNA as well as to facilitate correct initiation codon choice (50). Thus, it has been proposed that in yeast, functional interactions occur between the 5′ and 3′ ends of mRNAs via Pab1p binding to eIF4G and possibly other initiation factors localized at the 5′ ends of RNAs (59). This has been termed the closed-loop model of initiation. In fact, it has recently been shown that yeast eIF4E, eIF4G, and Pab1p are sufficient to link 5′ and 3′ ends of mRNA in ring-like structures which can be visualized by atomic force microscopy (62). In the mammalian system, a newly identified protein called PAIP-1 (for PABP interacting protein 1), which shares homology with eIF4G and interacts with PABP and eIF4A, has been described (13). This protein is proposed to provide the physical link between the 5′ and 3′ ends of RNAs in mammalian cells, thus closing the loop, since evidence for other factor interactions is currently lacking. Further, recent findings suggest that PABP may also directly bind eIF4G via a segment near the amino terminus of eIF4G (29). The juxtaposing of the two ends of cellular mRNAs would then facilitate mechanisms encompassing regulation from both the 5′ and 3′ structures of RNAs and facilitate reinitiation of transiting ribosomes by a mechanism that is independent of intact eIF4G.
PABP has been cloned and sequenced from several divergent organisms and has been found to be a highly conserved protein containing four RNA-binding motifs and a less conserved proline-rich carboxyl-terminal domain (21, 24, 46). PABP is the founding member of the family of 68- to 72-kDa RNA-binding proteins containing conserved RNA recognition motifs (RRMs) (1, 52). The RRMs are composed of approximately 90 amino acids, with highly conserved hydrophobic cores (45). PABP from yeast (Pab1p) is essential for viability and is involved in translational regulation through interaction with eIF4G (33, 59). PABPs from both Xenopus laevis and humans have been characterized with regard to their RNA-binding affinities and their structures and functions (21, 46), though the domains responsible for high-affinity RNA binding in the two proteins differ. PABP is an important regulator of translation in development (63) and may be involved in RNA transport and stability by its binding to poly(A) tails (45, 51). These results imply an important role for PABP function in the regulation of translation, though evidence for this role has not yet been directly presented in a mammalian system.
Translation initiation can occur independently of the 5′ cap group by means of mRNA 3′ poly(A) tail-dependent mechanisms (36, 50, 60), which could thereby circumvent the loss of intact 4G in poliovirus infections in the presence of guanidine, allowing for continuing translation. Accordingly, we hypothesized that poliovirus infection may cause an alteration in a protein which interacts with the 3′ ends of cellular mRNAs. In this report, we examine the fate of PABP in human cells and show that PABP is specifically degraded during poliovirus infection, that is cleaved in vitro by poliovirus proteases 2A and 3C (2Apro and 3Cpro), as well as coxsackievirus B3 2Apro, and that its degradation is observed concurrently with a loss of in vitro-translation activity in extracts from uninfected HeLa cells and HeLa cells infected with poliovirus in the presence of guanidine.
MATERIALS AND METHODS
Cells and virus infection.
HeLa S10 cells were grown in spinner culture in Joklik’s minimal essential medium supplemented with 9% bovine calf serum–1% fetal calf serum and penicillin-streptomycin (Sigma). Poliovirus type 1 (Mahoney) was grown and purified as previously described (9). All virus infections were done at a multiplicity of infection of 100, including 2 mM guanidine-HCl in the media where poliovirus infections in the presence of guanidine were utilized. Serum was added to 5% at 30 min postinfection (p.i.), and the cells were harvested at the indicated time. For pulse labeling, an aliquot of cells was taken at the indicated time and pelleted. The cells were resuspended in 0.5 ml of methionine-free medium containing 30 μCi of 35S-TransLabel (ICN)/ml and labeled for 15 min at 37°C. The cells were then lysed in Nonidet P-40 (NP-40)–ribosome standard buffer (RSB) (1% NP-40, 10 mM NaCl, 10 mM Tris-HCl [pH 7.4] 1.5 mM MgCl2), and the cytoplasmic fraction was mixed with sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) sample buffer for gel analysis. Unlabeled infections were harvested, cells were lysed, and gel samples were made in an manner identical to that described above. For immunoprecipitations, the cells were pulse labeled in methionine-free medium containing 100 μCi of 35S-TransLabel/ml for 2 h, washed once with phosphate-buffered saline (PBS), resuspended in complete medium with or without additions, and then incubated as described in the figure legends.
PABP and protease purification.
Human PABP cloned into pET-11a vector (pET-PABP [21]) was transformed into Escherichia coli BLR(DE3) cells (Invitrogen) and induced with 0.4 mM IPTG (isopropyl-β-d-thiogalactopyranoside) for 3 h. Recombinant PABP was purified by standard purification techniques as follows. Cells were resuspended in 20 mM Tris (pH 7.0)–1 mM EDTA–50 mM KCl–50 mM NaCl–5% glycerol and sonicated to lyse them. The cell lysate was clarified by centrifugation at 10,000 × g for 15 min, and then the supernatant was loaded on a Sephacryl S300 (Pharmacia) gel filtration high-performance liquid chromatography column. The high-molecular-weight fractions were pooled and loaded on a Fast S Sepharose (Pharmacia) cation-exchange column at pH 6.0. Proteins were eluted on a 0.1 to 1.0 M NaCl gradient, and the fractions containing PABP were pooled. This pool was dialyzed against buffer containing 10 mM HEPES, pH 7.5, 100 mM KCl, 10 mM NaCl, 1 mM MgCl2, 0.1 mM EDTA, 10% glycerol and concentrated in a Centricon spin concentrator (Amicon) (10-kDa cutoff).
To make histidine-tagged PABP, the pET-PABP vector was used as a template for PCR amplification of the PABP insert. Primers were designed to facilitate cloning into the pTrcHisB vector (Invitrogen), using BamHI and KpnI sites to insert PABP. The primers were synthesized (Integrated DNA Technologies) as follows: forward primer, 5′ GGCGGATCCGATGAACCCCAGTGC 3′; reverse primer, 5′ GGCCACAAGGTTGACAAATTCCATGGCGC 3′. The 1.9-kb PCR product was ligated by TA cloning into pGEM-T (Promega), and then the insert was cut out with BamHI and KpnI and ligated into pTrcHisB. The construct was transformed into DH5α cells, grown, and induced in accordance with the manufacturer’s instructions. The growth of cells and induction of recombinant protein were done at 30°C. The His-PABP was purified on Talon resin (Clontech) under denaturing conditions, dialyzed into dialysis buffer (described above), and used for in vitro-cleavage assays.
For reactions with 35S-labeled PABP, the protein was synthesized in reticulocyte lysate (Promega) under standard translation conditions with the pET-PABP plasmid linearized with BamHI to express full-length PABP.
Poliovirus 2Apro was expressed in E. coli from pATH2A and used as a crude extract as previously described (7, 65). Poliovirus 3Cpro was expressed from pET3Chc plasmid (Thomas Pfister) as a C-terminal histidine-tagged protein in BLR(DE3) cells and purified to homogeneity on Talon resin under nondenaturing conditions.
Coxsackievirus B3 (CVB3) 2Apro was cloned by using the vector pCVB3-20 (11) as a template for PCR amplification of the 2Apro gene region. Primers were designed to facilitate cloning into the pET22b(+) vector (Novagen) by using NdeI and EcoRI sites and to introduce start and stop codons into the cDNA. The primers were synthesized (Integrated DNA Technologies) as follows: forward primer, 5′ GAGGATCCCATATGGGCGCATTTGGACAACAATC 3′; reverse primer, 5′ GGAATTCACTGTTCCATTGCATCATCTTCCAG 3′. The 0.46-kb PCR product was digested with NdeI and EcoRI and ligated into pET22b(+) previously digested with the same restriction endonucleases. The resulting construct, pET-Cx2A, was transformed into BLR(DE3) pLys S cells, and CVB3 2Apro was expressed at 34°C and purified to homogeneity by the procedure previously described for CVB4 2Apro (37).
PABP cleavage assays, immunoprecipitations, sequencing, and blotting.
In vitro-cleavage assays of PABP were performed with essentially the same conditions regardless of the source of the substrate. Ribosomal salt wash (RSW) was prepared as described previously (9) and used as a substrate in cleavage reactions with proteases in vitro. Reaction mixtures containing 2 to 4 μl of RSW, various amounts of protease, and 50 mM NaCl–5 mM MgCl2 were incubated at 37°C for 2 to 4 h (as indicated in the figure legends). Cleavage reactions with in vitro-translated PABP utilized 3 μl of reticulocyte translation reaction mixture mixed directly with protease. Reactions with purified recombinant PABP proteins contained purified protein to which protease was added and incubated at 37°C for the indicated time. All reactions were stopped by the addition of SDS-PAGE sample buffer and were analyzed on SDS-polyacrylamide gels by immunoblotting or staining with Coomassie blue. Data from the gels was scanned with an Artec Viewstation A6000C Plus scanner and imaged with Adobe Photoshop version 3.0. For sequencing of the PABP 2Apro cleavage product, a glutathione S-transferase–PABP fusion protein was generated and purified as previously described (3), and then cleavage assays with an equimolar ratio of substrate to protease were allowed to proceed overnight. The proteins were electrophoresed on a SDS–13% polyacrylamide gel and then transferred to polyvinylidene difluoride membranes. The Coomassie blue-stained protein band was subjected to microsequencing by the University of Oklahoma Health Sciences Center Molecular Biology Resource Facility.
For immunoblotting PABP, monoclonal antibody to PABP (10E10 [21]) was used at a 1:10,000 dilution in a solution of 5% dry milk containing 150 mM NaCl with overnight incubation at 4°C. Secondary antibody (goat anti-mouse horseradish peroxidase; Pierce) was used at 1:10,000 dilution in the same dry milk solution and incubated for 2 h at room temperature. The blots were washed thoroughly with PBS-T (PBS–0.05% Tween-20) and then developed with the SuperSignal enhanced chemiluminescence system (Pierce). Immunoblotting for eIF4G was done as previously described (38). Immunoprecipitations of PABP were done with monoclonal antibody 10E10 essentially as previously described (21), with the following exceptions: the cells were lysed in NP-40–RSB, the nuclei were removed, and then the clarified cell extract was brought to a volume of 0.5 ml with RSB containing 1% Empigen BB followed by incubation with 2 μl of ascites/reaction and 25 μl of protein A-agarose beads. The resulting autoradiographs were scanned with an Artec Viewstation A6000C plus scanner and imaged with Adobe Photoshop version 3.0.
Endogenous mRNA in vitro-translation assays.
Translation of endogenous mRNA in vitro was done essentially as described previously (39). Briefly, HeLa cells (either infected or mock-infected) were harvested by centrifugation and washed three times with ice-cold Earle’s balanced salt solution. The cells were resuspended in 1.5 cell volumes of lysis buffer [10 mM KCl, 2.5 mM dithiothreitol, 1.2 mM Mg(OAc)2, 20 mM HEPES-KOH, pH 7.4] and then incubated on ice for 10 min. The cells were lysed by a minimal number of Dounce homogenizer strokes (6 to 8), and then the lysate was clarified by centrifugation at 10,000 × g. The supernatant was removed, aliquoted, and stored at −80°C for translation reactions. The reaction mixtures contained 10 μl of cell extract, added exogenous protease where indicated, 20 μCi of 35S-TranSLabel, 5 μl of 5× translation cocktail [which delivered final concentrations of 1 mM Mg(OAc)2, 90 mM KOAc, 10 mM HEPES (pH 7.5), 0.1 mM amino acids (methionine minus), 1 mM dithiothreitol, 25 mM creatine phosphate, 5 μg of creatine kinase/ml, 0.5 mM GTP, and 1.0 mM ATP], and diethyl pyrocarbonate-treated H2O to a final volume of 25 μl. For preincubated reactions, all components but 5× translation cocktail and label were added and incubated for 60 min at 30°C. After the preincubation, the cocktail and label were added and the reaction was continued for 60 additional minutes at 37°C. At the end of the reaction, SDS-PAGE sample buffer was added, the samples were heated at 100°C for 3 to 4 min, and the reaction products were analyzed on 10% polyacrylamide gels. Half of the sample was used for autoradiography, and the other half was used for immunoblotting for PABP; the data was scanned with an Artec Viewstation A6000C Plus scanner and imaged with Adobe Photoshop version 3.0.
RESULTS
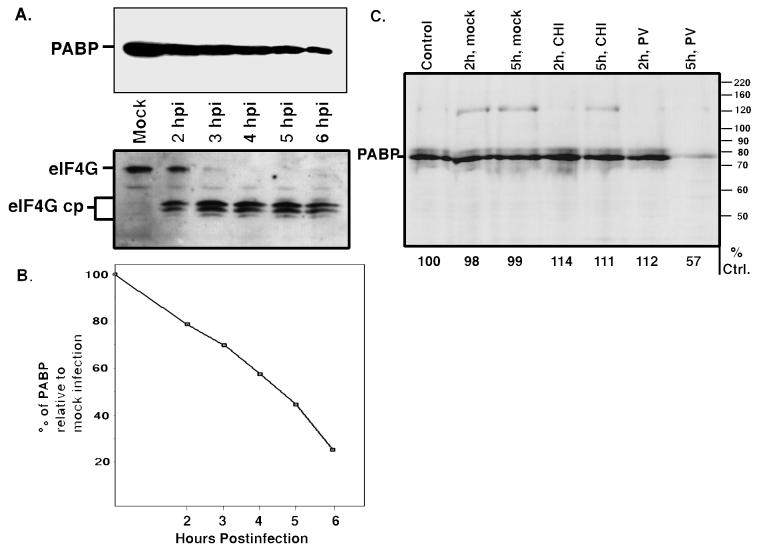
We first asked whether there was a detectable change in PABP during poliovirus infection, either in abundance or form. Figure 1A shows an immunoblot of cytoplasmic lysates from poliovirus-infected HeLa cells harvested at various times p.i. by using a monoclonal antibody to human PABP (21), while Fig. 1B shows the quantitation of this data. Figure 1A shows that during a typical infection, PABP is decreasing in abundance during the course of infection. Though we have only transiently observed cleavage products detectable by immunoblotting with this antibody (see Fig. 5), this decrease was most likely due to a proteolytic degradation. The extent of cleavage of PABP varies slightly from infection to infection and correlates with the multiplicity of infection and levels of viral protein expression (data not shown), but it has never been complete by 6 h p.i. Figure 1A also shows the corresponding cleavage of eIF4G in the same infection, demonstrating the typical complete cleavage which is manifest by approximately 3 h p.i. Most translation shutoff begins by 2 h p.i. and is complete by 4 h p.i. (reference 19 and data not shown). Therefore, over 50% of PABP is still intact in infected cells at times (3 to 4 h p.i.) when host translation inhibition is complete (Fig. 1B). In HeLa cells, PABP is normally a very stable and abundant protein, having a half-life greater than 8 h (21). To confirm this in our cells, we performed pulse-chase analysis of PABP stability in HeLa cells. Figure 1C shows that in both mock-treated and cycloheximide-treated cells, there is very little turnover of PABP. Therefore, PABP remains stable even during conditions of global translational inhibition. However, pulse-chase analysis confirmed a reduction in PABP of about 50% in infected cells, which agrees well with immunoblotting results. Taken together, these results suggest that PABP degradation is not likely due to increased turnover or lack of synthesis stemming from virus-induced translational inhibition but is consistent with degradation by a viral protease.
FIG. 1.
PABP is degraded during poliovirus infection. (A) HeLa spinner cells were mock infected (Mock) or infected with poliovirus, and samples were harvested at the indicated times p.i. (hpi). Aliquots of these samples were subjected to SDS-PAGE and immunoblotted with a monoclonal antibody to PABP (10E10) (upper left gel) or a polyclonal antibody to eIF4G (lower left gel). cp, cleavage product. (B) Densitometric images of three immunoblots prepared as for panel A were quantitated with an Alpha Innotech imager; the average of the data is shown. The data are represented as percentages of intact PABP levels relative to the amount in mock-infected cells. (C) Immunoprecipitations of PABP were done as described in Materials and Methods on extracts prepared from cells that were pulse labeled for 2 h and then harvested (Control) or subsequently chased for the indicated length of time with either complete medium (mock) or complete medium containing 10 μg of cycloheximide/ml (CHI) or infected with poliovirus for the indicated length of time. The number below each lane corresponds to the densitometric quantitation of the PABP band relative to the control lane (% Ctrl.).
FIG. 5.
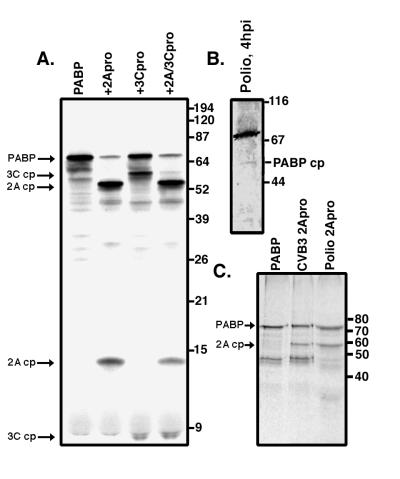
PABP cleavage products from in vitro cleavages correspond to the cleavage detected during poliovirus infection in vivo. (A) Reticulocyte lysate-translated PABP was used as a substrate for PABP cleavage reactions, as detailed in Materials and Methods. The cleavage reaction mixtures contained 1.0 μg of CVB3 2Apro, 1.5 μg of poliovirus 3Cpro, or a combination of both proteases and were incubated for 3 h at 37°C. The positions of the PABP cleavage products (cp) generated by each protease are indicated, as well as molecular mass marker positions. (B) In vivo cleavage product (PABP cp) detected by immunoblot analysis by PABP antibody in lysate derived from a poliovirus infection and harvested at 4 h. (C) Poliovirus 2Apro and CVB3 2Apro generate similar PABP cleavage products. Reticulocyte lysate-translated PABP was incubated alone or with 1.0 μg of CVB3 2Apro or 10 μl of poliovirus 2Apro extract and analyzed by gel electrophoresis and autoradiography. The position of the 57-kDa cleavage product (2A cp) is indicated.
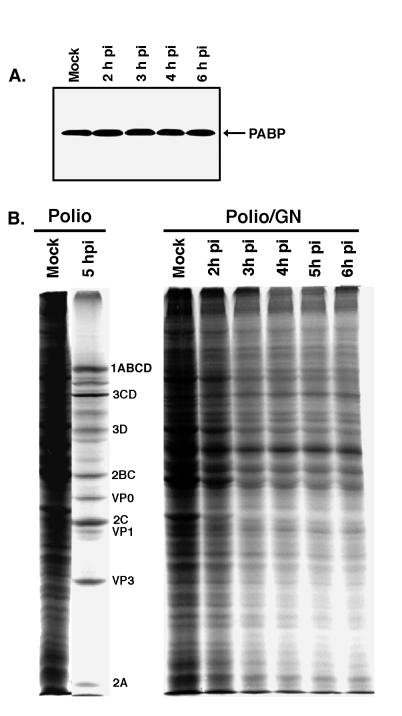
From these initial experiments, it became clear that PABP was degraded or cleaved during infection. Next, we were interested to know whether this cleavage still occurred during infection in the presence of guanidine-HCl, which inhibits viral replication but also blocks the development of complete host translation shutoff (6, 8). If PABP cleavage was required for complete host cell shutoff to occur, then PABP cleavage should be inhibited during infections in the presence of guanidine-HCl. The level of PABP was analyzed by immunoblotting at various time points during poliovirus infection in the presence of guanidine (Fig. 2A). It is readily apparent that the abundance of PABP does not change significantly during infection in the presence of guanidine, indicating that the cleavage observed during poliovirus infection requires efficient viral protein synthesis and is likely directly mediated by a viral protein. Quantitation of immunoblots showed that the PABP content of the cells varied less than 12% (data not shown) during this infection. Figure 2B shows the extent of translational inhibition under these conditions; as previously documented (6, 8, 49), the translation levels in poliovirus-guanidine-infected cells were only inhibited by approximately 50%, while eIF4G was completely cleaved (data not shown). Thus, the absence of PABP cleavage during poliovirus infections in the presence of guanidine is consistent with this alteration playing a role in the total translational shutoff induced by poliovirus infection.
FIG. 2.
PABP is not degraded during poliovirus infection in the presence of guanidine. (A) HeLa cells were mock-infected (Mock) or infected with poliovirus in the presence of 2 mM guanidine-HCl, and samples were harvested at the indicated times p.i. for immunoblot analysis with PABP antibody. (B) HeLa cells infected with poliovirus in the presence (Polio/GN) or absence (Polio) of guanidine were pulse labeled with [35S]Met at the indicated times p.i., and aliquots were analyzed by SDS-PAGE for radiolabeled proteins. The positions of the poliovirus proteins are indicated.
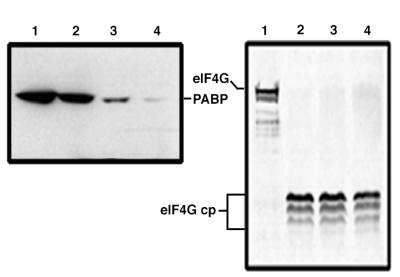
We were then interested in determining whether the poliovirus 2Apro was capable of cleaving PABP in vitro. 2Apro is involved in the cleavage of eIF4G (7, 34, 40) and appears to be the key viral component responsible for inducing host cell shutoff (15, 58) as well as enhancement of viral translation (26, 41, 48). In vitro-cleavage assays were performed with a crude translation initiation factor preparation (RSW) from uninfected HeLa cells as a PABP substrate. RSW was incubated with either no protease (Fig. 3, lanes 1) or increasing amounts of recombinant poliovirus 2Apro extract (Fig. 3, lanes 2 to 4). On the left of Fig. 3 is an immunoblot with PABP antibody, and on the right is a blot from the same samples with eIF4G antiserum. Clearly, at the highest level of added poliovirus 2Apro, nearly complete cleavage of PABP in the RSW sample was observed. By contrast, all concentrations of 2Apro caused complete cleavage of eIF4G substrate (Fig. 3, right blot, lanes 2 to 4). These cleavages in vitro reflect the highly efficient cleavage in vivo of eIF4G, which is completely degraded by 3 h p.i. (19). The incomplete cleavage of PABP in vitro and in vivo may reflect a less efficient cleavage reaction or the existence of variable substrate conformations or compartmentalized pools which inhibit cleavage, or it may simply be due to the abundance of PABP substrate in the cell.
FIG. 3.
Poliovirus 2Apro degrades PABP in vitro. In vitro-cleavage reaction mixtures were assembled as described in Materials and Methods, using 4 μl of RSW with 0 (lanes 1), 2 (lanes 2), 4 (lanes 3), or 10 μl (lanes 4) of recombinant poliovirus 2Apro bacterial extract. Reaction mixtures were incubated at 37°C for 3 h and then analyzed by gel electrophoresis and immunoblotting. The blots shown are from the same gel analyzed with two different antibodies, PABP (left) and eIF4G (right). The positions of eIF4G cleavage products (cp) are indicated.
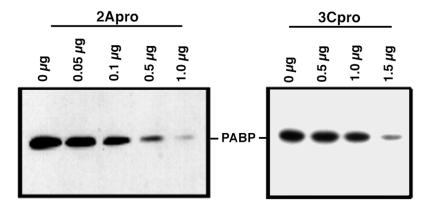
Next, we were interested in examining the cleavage of PABP by using purified proteases. To help exclude the possibility of cleavage artifacts, purified proteases were used for in vitro-cleavage assays. Poliovirus 2Apro is prone to aggregation, and stringent purification of active enzyme is very difficult (7). Therefore, we chose to utilize CVB3 2Apro since its substrate specificity and cleavage efficiency is nearly identical to those of poliovirus 2Apro but its biochemical properties enable it to be purified much more readily (7, 37). CVB3 and poliovirus 2Apros produced identical cleavage products of PABP in vitro (see Fig. 5); thus, purified CVB3 2Apro was substituted for poliovirus 2Apro in subsequent experiments. We examined poliovirus 3Cpro in vitro as well, since one other cellular protein, microtubule-associated protein 4 (MAP-4), is cleaved by 3Cpro during infection but not in the presence of guanidine (32), as is the case for PABP. For these assays, purified recombinant CVB3 2Apro or affinity-purified recombinant poliovirus 3Cpro with a C-terminal histidine tag was incubated with RSW substrate containing PABP. PABP was then analyzed by immunoblotting. Figure 4 (left blot) shows the same type of cleavage of PABP by CVB3 2Apro seen earlier, showing that at the highest concentration nearly complete cleavage or degradation of PABP occurred. As noted before, cleavage products which react with the monoclonal antibody were not detected. Other experiments have shown complete cleavage of PABP at this concentration, while all concentrations of purified 2Apro tested resulted in complete cleavage of eIF4G in the same samples (data not shown). Figure 4 (right blot) shows the result of incubating PABP with purified poliovirus 3Cpro. Though the protease concentrations necessary for comparable cleavage of PABP were much higher for 3Cpro than for 2Apro, it was readily apparent that both proteases were capable of cleaving PABP in vitro.
FIG. 4.
Purified 2Apro and 3Cpro cleave PABP in vitro. Cleavage reactions with RSW and purified recombinant 2Apro from CVB3 (left) or purified recombinant poliovirus 3Cpro (right) with the indicated quantities of protease were incubated for 3 h at 37°C and analyzed by gel electrophoresis and immunoblotting with PABP antibody.
Since we could demonstrate cleavage of PABP in vitro by both 2Apro and 3Cpro but could not detect cleavage products with the PABP monoclonal antibody, we next examined the cleavage pattern induced by incubating purified viral proteases with radiolabeled PABP translated in vitro in a reticulocyte lysate. Labeled PABP was incubated with purified proteases and analyzed by SDS-PAGE. The autoradiograph clearly shows the generation of major cleavage products for both proteases which are distinct from one another (Fig. 5A). Incubation of PABP with 2Apro results in the generation of cleavage products of approximately 57 and 14 kDa, the total of which correlates well with the apparent molecular mass for PABP (approximately 72 kDa). Cleavage by poliovirus 3Cpro results in the generation of cleavage products of approximately 61 and 8 kDa, again approximately the correct total molecular mass for PABP, suggesting that these are all primary cleavage products generated by the enzymatic activity of these proteases. It is possible that other, minor cleavage products are produced, but analysis is obscured by aberrant translation products in the sample (Fig. 5A, lane 1) previously noted to occur from translation of PABP in vitro (21, 24). Figure 5B shows a very faint immunoblot cleavage product band from a poliovirus-infected cell lysate of approximately the same size as the larger primary cleavage product generated by 2Apro in vitro. This band has only been observed sporadically by immunoblot analysis, and in very low abundance, but it is identical in size to those detected by cleavages in vitro with purified 2Apro, indicating that this cleavage is occurring during infection with poliovirus. It was also observed that the cleavage efficiency of 2Apro for PABP was greater than that of 3Cpro, since when the two proteases were incubated together (Fig. 5A), the observed cleavage products corresponding to 2Apro cleavage were much more abundant than those corresponding to 3Cpro cleavage. Interestingly, incubation of PABP with both proteases also demonstrated a lack of further processing when both proteases were present. The reason for this is unknown, but it could reflect differences in PABP conformation or complexes with other proteins which inhibit cleavage. Alternatively, mRNA binding may influence the susceptibility of PABP to cleavage. Figure 5C compares an in vitro cleavage assay of radiolabeled PABP incubated with CVB3 and poliovirus 2Apros. The 57-kDa cleavage product was again apparent and was identical in appearance for the two viral proteases, which indicated that the cleavage of PABP by poliovirus and CVB3 2Apros likely occurs at the same site.
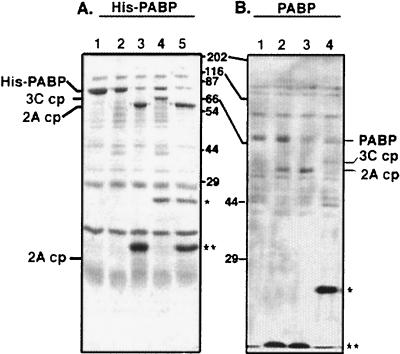
To help rule out the possibility of an indirect cleavage mechanism being involved in PABP cleavage, purified proteases were incubated with nearly purified PABP substrates (Fig. 6). As shown in Fig. 6B, affinity-purified His-PABP bearing an amino-terminal histidine tag was used for in vitro-cleavage assays. The same cleavage products (both slightly larger due to the histidine tag: approximately 62 and 66 kDa for 2A and 3C cleavage products, respectively) previously demonstrated for reticulocyte lysate-derived PABP were observed, indicating that both 2Apro and 3Cpro utilize PABP as a substrate. The smaller cleavage products at 14 and 8 kDa were difficult to observe in these experiments due to background E. coli proteins. Since there were still some bacterial contaminants present in the preparation, recombinant PABP was purified by means of conventional column chromatography and incubated in vitro with purified proteases (Fig. 6B). The same large (57- and 61-kDa) cleavage products were observed, suggesting that the cleavage of PABP by 2Apro and 3Cpro were the result of specific proteolytic cleavage by the proteases and not by contaminating proteins in the preparations.
FIG. 6.
Cleavage of purified PABP by purified 2Apro and 3Cpro. Purified recombinant histidine-tagged PABP (His-PABP) (left) or unmodified PABP (right) was incubated with purified CVB3 2Apro or poliovirus 3Cpro or both. (A) Lane 1, unincubated His-PABP; lane 2, His-PABP incubated alone; lane 3, His-PABP plus 1.0 μg of 2Apro; lane 4, His-PABP plus 1.5 μg of 3Cpro; lane 5, His-PABP plus 1.0 μg of 2Apro–1.5 μg of 3Cpro. (B) Lane 1, PABP incubated alone; lane 2, PABP plus 1.0 μg of 2Apro (37°C incubation); lane 3, PABP plus 1.0 μg of 2Apro (30°C incubation); lane 4, PABP plus 1.0 μg of 3Cpro (37°C incubation). The reactions shown in panel A were incubated for 16 h at 30°C, and those shown in panel B were incubated at the indicated temperature for 16 h and then analyzed by SDS-PAGE and Coomassie blue staining. ∗∗, 2Apro; ∗, 3Cpro.
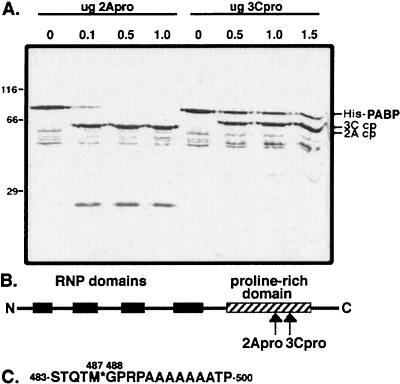
We next wanted to determine the approximate locations of the cleavage sites relative to the structural and functional domains of PABP. To do this, we used a commercially available monoclonal antibody to hexahistidine protein motifs and probed an immunoblot of cleavage reactions with amino-terminal-tagged His-PABP as a substrate (Fig. 7A). Lanes 1 and 5 contained purified His-PABP incubated without protease, while lanes 2 to 4 included increasing amounts of 2Apro and lanes 6 to 8 included increasing amounts of 3Cpro. The lower-molecular-mass proteins observed in the absence of protease may reflect minor degradation of the protein during purification. However, the bold cleavage products at approximately 62 kDa for 2Apro and 66 kDa for 3Cpro reflect the presence of the amino-terminal histidine tag on these cleavage products. This indicates that the cleavage sites for both proteases reside in the carboxyl-terminal domain of PABP. 2Apro may also generate a minor cleavage product (25 kDa) from a second cleavage site. These results also show that the primary cleavage sites for the two proteases are close to each other, since the masses of the cleavage products differ by only 4 to 5 kDa. The location of the primary cleavage sites of both poliovirus 2Apro and 3Cpro on PABP are indicated in the schematic diagram in Fig. 7B, which depicts the relative locations of the 4 RRMs for PABP responsible for poly(A) binding and indicates that both primary cleavage sites are located within the proline-rich region in the carboxyl terminus. Identification of the primary cleavage site for CVB3 2Apro was accomplished by microsequencing the small cleavage product (Fig. 7C). CVB3 2Apro cleaves the scissile bond between Met487 and Gly488 (numbering according to reference 24). While it is unusual for an enterovirus 2Apro to utilize a Met in the P1 position, the presence of Thr in the P2 and P4 positions, as well as the invariant Gly in the P1′ position, is a very favorable determinant of cleavage by 2Apro (27, 57).
FIG. 7.
PABP is cleaved in its carboxyl-terminal region by both 2Apro and 3Cpro. (A) Affinity-purified His-PABP was incubated with the indicated quantities of either CVB3 2Apro or poliovirus 3Cpro for 3 h at 37°C and then analyzed by immunoblotting with antibody to hexahistidine tag. The positions of the large PABP cleavage products (cp) generated by each protease are indicated. (B) Schematic diagram of PABP structure showing known functional domains and localization of the protease cleavage sites on PABP. (C) Sequence of the primary cleavage site (∗) for CVB3 2Apro on PABP.
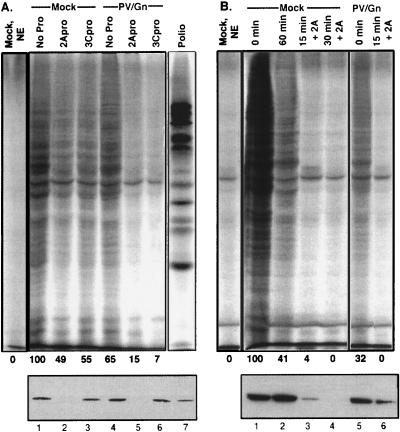
As a preliminary step to address the possibility that cleavage of PABP during poliovirus infection plays a role in host cell shutoff, we decided to examine the effect of the addition of purified proteases to an in vitro-translation assay of endogenous mRNA translation (Fig. 8). As mentioned previously, translation in poliovirus-guanidine-infected cells occurs at levels of 40 to 60% of normal rates, even though the eIF4G is completely cleaved (8). We reasoned that if cleavage of PABP were to cause a decrease in translation, this would be most evident in the extracts from infections in the presence of guanidine, due to the partial translation inhibition already present from prior in vivo cleavage of eIF4G. The translation assays utilized extracts prepared from mock-infected HeLa cells and cells infected in the presence or absence of guanidine-HCl, as indicated in Fig. 8. Lysates were preincubated without protease (Fig. 8A, lanes 1, 4, and 7), with purified 2Apro (Fig. 8A, lanes 2 and 5), or with 3Cpro (Fig. 8A, lanes 3 and 6) for 60 min to allow for protease enzymatic activity. After preincubation, radiolabel and translation cocktail were added and translation of endogenous mRNA was allowed to proceed for 60 min, after which the samples were processed for gel electrophoresis. Compared to the level of translation when the extracts were incubated in the absence of protease (Fig. 8A, lanes 1 and 4), the addition of 2Apro inhibited translation from both extracts, though the overall inhibition observed in the poliovirus-guanidine-infected extract was greater than that for the mock-infected lysates. These data indicated that 2Apro treatment of mock-infected extract inhibited translation approximately 50% while treatment of poliovirus-guanidine-infected extracts inhibited translation nearly completely (15 and 7% [Fig. 8A, lanes 5 and 6]). As shown in Fig. 8A, preincubation with 2Apro resulted in complete PABP cleavage concurrent with inhibition of translation of endogenous mRNA (lanes 2 and 5). Preincubation with 3Cpro resulted in a modest decrease in PABP, but surprisingly, it was also able to decrease translation rates in vitro. This inhibition of translation by 3Cpro alone was readily reproducible (data not shown), though typically not as extensive as that produced by 2Apro. An effect of 3Cpro on translation has not been reported previously, but it may require coexpression of 2Apro to be evident in vivo. Inhibition of translation upon addition of either protease was more nearly complete when eIF4G was previously cleaved in vivo (Fig. 8A, lane 6).
FIG. 8.
Inhibition of translation in vitro correlates with degradation of PABP by 2Apro and 3Cpro. (A) Extracts generated from mock-infected HeLa cells (Mock) or HeLa cells infected with poliovirus with (PV/Gn) or without (Polio) guanidine were incubated in translation reactions as described in Materials and Methods. The reactions were assembled with or without protease (1.0 μg/reaction) and preincubated for 60 min at 30°C. Radiolabel and cocktail were then added, and incubation was continued for 60 additional minutes at 37°C. Half of the sample was used for SDS-PAGE and autoradiography (top), while the other half is shown analyzed by immunoblotting with antibody to PABP (bottom). Lane 7 shows translation derived from poliovirus extracts without added protease, while the lane labeled Mock, NE, designates translation in a mock-infected extract reaction without an added energy source (creatine phosphate-creatine kinase). (B) Extracts from mock-infected HeLa cells (Mock) or poliovirus-guanidine-infected cells (PV/Gn) were assembled into reactions as described in Materials and Methods. All components except radiolabel were added initially and incubated at 30°C to allow endogenous translation. At the indicated time, radiolabel (20 μCi) was added to each reaction, and translation was allowed to continue for 60 min more at 37°C. The samples were processed as described for panel A. The Mock, NE, lane was prepared the same way as the Mock, 0 min, lane without creatine phosphate-creatine kinase. Below each lane, the percent translation rate compared to that of mock-infected lysate is shown as an average of two separate experiments (quantitated with National Institutes of Health image software).
In contrast to Fig. 8A, where extracts were preincubated with proteases before translation was initiated, Fig. 8B shows extracts in which protease incubation and translation were initiated concurrently. Translation lysates were pulse labeled at various times during the incubation to measure the effects on translation rates and PABP cleavage. As expected, preincubation of lysates alone for 60 min resulted in a 50% loss of translation efficiency (Fig. 8B, compare lanes 2 and 3) and reduced control translation to levels comparable to those observed for the lysates shown in Fig. 8A, which were also preincubated. Addition of 2Apro for only 15 min, however, resulted in the nearly complete abolishment of translation, which coincided with a 90% reduction in PABP levels. Thus, the rate of PABP cleavage appeared to increase as well, requiring just over 20 min for complete cleavage instead of the normal 45 to 60 min (data not shown). Similar results were observed in extracts from poliovirus-guanidine-infected cells, except that PABP degradation, though substantial, lagged slightly in comparison. If the extent of PABP cleavage is in some way linked to translation rates, less degradation of PABP may reflect the lower overall translation rate present in the poliovirus-guanidine-infected cells, which are partially shut off. In addition, similarly decreased translation rates were observed upon addition of 3Cpro, although the kinetics were significantly delayed compared to those in experiments with 2Apro addition (data not shown). Overall, these data point to a possible correlation between a loss of PABP in the translation lysates due to proteolytic cleavage and the degree of translation inhibition observed in vitro, though the exact role of PABP degradation in this inhibition has not been defined.
DISCUSSION
This report describes the identification of another cellular protein substrate, PABP, for poliovirus 2Apro and 3Cpro. Only one other cellular protein has been identified which is a target for both viral proteases during infection, the TATA-binding protein, TBP (66). TBP plays an integral role in promoting transcription from RNA polymerase II and III promoters, and its proteolysis plays a role in the transcriptional inhibition observed during poliovirus infection (12, 66). Similarly, PABP appears to play an integral role in another aspect of gene expression, poly(A) tail-mediated translational stimulation. Since PABP is cleaved in vitro by both poliovirus proteases and CVB3 2Apro (Fig. 4 to 7), it is likely that cleavage of PABP is important for some aspect of translation shutoff, viral translation, or viral replication. Though it is unknown whether both cleavage sites are utilized in infected cells, the fact that PABP can be targeted by both proteases and cleaved in the same region of the molecule by both implies a role for this cleavage during infection.
The data presented in Fig. 7 indicate that the cleavage of PABP by both 2Apro and 3Cpro occurs in the carboxyl-terminal domain of the protein. Several investigators have analyzed various structural domains of PABP from several species (10, 46, 54). These studies indicated that some RRM RNA-binding domains probably have a unique RNA-binding activity, which may be modulated when RRMs are analyzed singly versus in combinations. Human PABP appears to have RNA-binding activity in all its RRM domains, but the highest affinity for poly(A) resides in the first two domains together (10). Further, in yeast, the RRM2 region is involved in direct-binding interactions with eIF4G (33). Recent preliminary evidence suggests that this region is also involved in eIF4G binding in human cells (29). Thus, cleavage of PABP in the C-terminal domain indicates that interactions of PABP with eIF4G may not be grossly disrupted by 2Apro, since the large cleavage products still maintain the intact eIF4G binding site. The effect of cleavage on these protein interactions is currently being investigated.
Little information has been forthcoming about any specific function of the carboxyl region of PABP. Early results with yeast suggested a role in direct-binding interactions with the 60S ribosomal subunit (53), and others have suggested a role in dimerization or oligomerization of PABP (2, 35). Interestingly, the RNA-binding analyses of X. laevis PABP indicated an ancillary role for the carboxyl-terminal region in conferring specificity of binding to poly(A) (46). A large segment of the C-terminal domain 265 amino acids is proline rich, suggesting a loose or extended conformation. This 265-amino-acid domain contains a highly conserved segment at each end, separated by a poorly conserved region. In particular, the C-terminal 100 amino acids of PABP are highly conserved in eukaryotes. In the center of the nonconserved region, however, is a conserved polyalanine motif found in all eukaryotic PABPs. Interestingly, 2Apro cleaves adjacent to this motif (Fig. 7C), separating the polyalanine segment and the C-terminal conserved domain from the rest of the protein. It is possible that this region is involved in contacts with specific proteins, and these interactions are disrupted by cleavage during poliovirus infection. One candidate for this interaction is the recently described PAIP-1 (13). This protein has been shown to interact with PABP and with eIF4A in vitro and to enhance translation in vivo. The region responsible for this interaction with PABP was mapped to the C-terminal region of PAIP-1, but the corresponding domain on PABP which interacts with PAIP has not yet been reported. A recent study of chloroplasts derived from green algae also demonstrated that processing of a chloroplast-derived PABP modulated the function of the protein to allow it to stimulate translation of a specific subset of mRNAs (67). This modified PABP binds with high specificity to the 5′ UTRs of these mRNAs and activates translation of mRNAs containing this 5′ UTR. Cleavage of PABP during poliovirus infection could alter its RNA-binding specificity, thereby changing its function. Alternatively, cleavage of PABP may be important for another aspect of the viral life cycle, such as RNA replication or packaging. Definitive evidence of a function for PABP cleavage and/or cleavage products awaits further experimentation.
In this study, we have shown partial or complete cleavage of PABP concurrent with further inhibition of translation in vitro after eIF4G is cleaved (Fig. 8). Though the data presented in Fig. 8 do not show a complete abolishment of endogenous translation in the mock-infected extracts (Fig. 8A, lanes 1 to 3), there was an appreciable decrease in the level of translation. Previous studies with in vitro translation extracts and capped reporter constructs in the presence of purified 2Apros did not show a total inhibition of translation of these mRNAs as well, though the cleavage of 4G was complete under these conditions (37, 68). This lack of complete shutoff with these assays may reflect a “loosening” of the stringency of regulation concerning fidelity of translation in vitro. Such may be the case with the data from mock-infected extracts, though assays with poliovirus-guanidine-infected extracts did show essentially complete shutoff (Fig. 8A, lanes 5 and 6). Recently, Gradi et al. reported that the recently discovered eIF4GII (22) is incompletely cleaved in poliovirus-guanidine-infected cells and that the addition of 2Apro was able to hasten complete cleavage (23). This provides yet another protein factor whose cleavage by 2Apro may contribute to host translation shutoff. Interestingly, in our system 3Cpro was also able to affect translation, but it is not known if eIF4GII is cleaved by 3Cpro.
If PABP cleavage is indeed a player in the translational shutoff mechanism, it may be that cleavage of only a certain subset or pool of PABP may be necessary for this function, which would be reflected in vivo by incomplete cleavage of total cellular PABP during infection (Fig. 1). Certainly, PABP bound to messages in polyribosomes, or PABP complexed with certain cellular proteins (i.e., eIF4G, PAIP-1, or ribosomal proteins), could be envisioned to be a preferential target for cleavage by poliovirus proteases. Preliminary experiments indicate that cleavage of PABP by 2Apro may be less efficient when PABP is bound to poly(A); in contrast, cleavage by 3Cpro may be more efficient (data not shown), suggesting that mRNA binding and/or protein interactions may have a significant impact on PABP cleavage efficiency. Alternatively, incomplete cleavage of PABP may simply be a reflection of its abundance in the cell, since it is present at a high concentration of 4 μM, which gives a threefold excess of PABP over poly(A)-binding sites in the cytoplasm (21).
How does cleavage of PABP relate to the shutoff of translation during infection? Clearly, disruption of 5′ cap-dependent interactions by cleavage of eIF4G is insufficient to account for the complete inhibition of cellular translation observed in active infections, since infections in the presence of guanidine do not abolish translation of endogenous mRNA (6, 8, 49). However, cleavage of PABP may disrupt 3′ poly(A) tail-dependent translational activity, thereby allowing the complete translation shutoff to occur when the viral infection is productive. This hypothesis fits well with the closed-loop model (31), which proposes that interactions between the 5′ and 3′ ends of mRNAs made through protein-protein contacts can allow efficient initiation and also reinitiation of ribosomes, circumventing the need for 5′ cap-dependent initiation for each round of translation. During poliovirus infection, de novo 5′ cap-dependent interactions are abolished first through cleavage of eIF4G. It is not clear that eIF4G cleavage will open loops, since the N-terminal fragment of eIF4G released could still simultaneously bind PABP and eIF4E, thus linking both ends of mRNA. Furthermore, 5′-3′ interactions mediated by PAIP may not be disrupted by eIF4G cleavage. Thus, continued translation after eIF4G cleavage may represent reinitiation events on closed loops which still maintain 5′-end–3′-end contacts. Cleavage of PABP may disrupt (i) these 5′-3′ contacts, (ii) PABP cycling between various ligands or complexes, or (iii) interactions of PABP with 60S ribosomal subunits in a fashion which impairs ribosome rejoining during reinitiation. Proteolysis of other proteins, such as eIF4GII, may also be involved in mediating this inhibition, though the relative contributions of these events may be difficult to separate from one another. In conclusion, we have shown that PABP is proteolytically cleaved during poliovirus infection, that it is a substrate for both poliovirus and CVB3 2Apros and poliovirus 3Cpro in vitro, and that its degradation in translation assays was observed concurrently with an inhibition of endogenous cellular mRNA translation. These results indicate that PABP cleavage during poliovirus infection may contribute to the observed shutoff of host cell translation.
ACKNOWLEDGMENTS
We thank Thomas Pfister and Eckard Wimmer (State University of New York at Stony Brook) for the gift of the poliovirus His-tagged 3Cpro plasmid pET3Chc, Gideon Dreyfuss (Howard Hughes Medical Institute) for the generous donation of monoclonal antibody 10E10 to PABP and the pET-PABP expression plasmid, and J. Bag (University of Guelph, Ontario, Canada) for the glutathione S-transferase–PABP expression plasmid.
This work was supported by NIH grant AI27914 to R.E.L.
REFERENCES
- 1.Adam S A, Nakagawa T, Swanson M S, Woodruff T K, Dreyfuss G. mRNA polyadenylate-binding protein: gene isolation and sequencing and identification of a ribonucleoprotein consensus sequence. Mol Cell Biol. 1986;6:2932–2943. doi: 10.1128/mcb.6.8.2932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baer B W, Kornberg R D. The protein responsible for the repeating structure of cytoplasmic poly(A)-ribonucleoprotein. J Cell Biol. 1983;96:717–721. doi: 10.1083/jcb.96.3.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bag J, Wu J. Translational control of poly(A)-binding protein expression. Eur J Biochem. 1996;237:143–152. doi: 10.1111/j.1432-1033.1996.0143n.x. [DOI] [PubMed] [Google Scholar]
- 4.Bernstein H D, Sonenberg N, Baltimore D. Poliovirus mutant that does not selectively inhibit host cell protein synthesis. Mol Cell Biol. 1985;5:2913–2923. doi: 10.1128/mcb.5.11.2913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Black T L, Barber G N, Katze M G. Degradation of the interferon-induced 68,000-Mr protein kinase by poliovirus requires RNA. J Virol. 1993;67:791–800. doi: 10.1128/jvi.67.2.791-800.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bonneau A-M, Sonenberg N. Proteolysis of the p220 component of the cap-binding protein complex is not sufficient for complete inhibition of host cell protein synthesis after poliovirus infection. J Virol. 1987;61:986–991. doi: 10.1128/jvi.61.4.986-991.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bovee M L, Lamphear B, Rhoads R E, Lloyd R E. Direct cleavage of eIF4G by poliovirus 2A protease is inefficient in vitro. Virology. 1998;245:241–249. doi: 10.1006/viro.1998.9172. [DOI] [PubMed] [Google Scholar]
- 8.Bovee M L, Marissen W E, Zamora M, Lloyd R E. The predominant eIF4G-specific cleavage activity in poliovirus-infected HeLa cells is distinct from 2A protease. Virology. 1998;245:229–240. doi: 10.1006/viro.1998.9171. [DOI] [PubMed] [Google Scholar]
- 9.Brown B A, Ehrenfeld E. Translation of poliovirus RNA in vitro: changes in cleavage pattern and initiation sites by ribosomal salt wash. Virology. 1979;97:396–405. doi: 10.1016/0042-6822(79)90350-7. [DOI] [PubMed] [Google Scholar]
- 10.Burd C G, Matunis E L, Dreyfuss G. The multiple RNA-binding domains of the mRNA poly(A)-binding protein have different RNA-binding activities. Mol Cell Biol. 1991;11:3419–3424. doi: 10.1128/mcb.11.7.3419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chapman N M, Tu Z, Tracy S, Gauntt C J. An infectious cDNA copy of the genome of a non-cardiovirulent coxsackievirus B3 strain—its complete sequence analysis and comparison to the genomes of cardiovirulent coxsackieviruses. Arch Virol. 1994;135:115–130. doi: 10.1007/BF01309769. [DOI] [PubMed] [Google Scholar]
- 12.Clark M E, Lieberman P M, Berk A J, Dasgupta A. Direct cleavage of human TATA-binding protein by poliovirus protease 3C in vivo and in vitro. Mol Cell Biol. 1993;13:1232–1237. doi: 10.1128/mcb.13.2.1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Craig A W B, Haghighat A, Yu A T K, Sonenberg N. Interaction of polyadenylate-binding protein with the eIF4G homologue PAIP enhances translation. Nature. 1998;392:520–523. doi: 10.1038/33198. [DOI] [PubMed] [Google Scholar]
- 14.Curtis D, Lehmann R, Zamore P. Translational regulation in development. Cell. 1995;81:171–178. doi: 10.1016/0092-8674(95)90325-9. [DOI] [PubMed] [Google Scholar]
- 15.Davies M V, Pelletier J, Meerovitch K, Sonenberg N, Kaufman R J. The effect of poliovirus proteinase 2Apro expression on cellular metabolism. J Biol Chem. 1991;266:14714–14720. [PubMed] [Google Scholar]
- 16.Duncan R, Etchison D, Hershey J W B. Protein synthesis eukaryotic initiation factors 4A and 4B are not altered by poliovirus infection of HeLa cells. J Biol Chem. 1983;258:7236–7239. [PubMed] [Google Scholar]
- 17.Ehrenfeld E. Initiation of translation by picornavirus RNAs. In: Hershey J W B, Matthews M B, Sonenberg N, editors. Translational control. Cold Spring Harbor, N.Y: Cold Spring Harbor Press; 1996. pp. 549–573. [Google Scholar]
- 18.Etchison D, Hansen J, Ehrenfeld E, Edery I, Sonenberg N, Milburn S, Hershey J W B. Demonstration in vitro that eucaryotic initiation factor 3 is active but a cap-binding protein complex is inactive in poliovirus-infected HeLa cells. J Virol. 1984;51:832–837. doi: 10.1128/jvi.51.3.832-837.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Etchison D, Milburn S C, Edery I, Sonenberg N, Hershey J W B. Inhibition of HeLa cell protein synthesis following poliovirus infection correlates with the proteolysis of a 220,000-dalton polypeptide associated with eukaryotic initiation factor 3 and a cap binding protein complex. J Biol Chem. 1982;257:14806–14810. [PubMed] [Google Scholar]
- 20.Gallie D R. The cap and poly(A) tail function synergistically to regulate messenger RNA translational efficiency. Genes Dev. 1991;5:2108–2116. doi: 10.1101/gad.5.11.2108. [DOI] [PubMed] [Google Scholar]
- 21.Gorlach M, Burd C G, Dreyfuss G. The mRNA poly(A)-binding protein: localization, abundance, and RNA-binding specificity. Exp Cell Res. 1994;211:400–407. doi: 10.1006/excr.1994.1104. [DOI] [PubMed] [Google Scholar]
- 22.Gradi A, Imataka H, Svitkin Y V, Rom E, Raught B, Morino S, Sonenberg N. A novel functional human eukaryotic translation initiation factor 4G. Mol Cell Biol. 1998;18:334–342. doi: 10.1128/mcb.18.1.334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gradi A, Svitkin Y V, Imataka H, Sonenberg N. Proteolysis of human eIF4GII, but not eIF4GI, coincides with the shutoff of host protein synthesis after poliovirus infection. Proc Natl Acad Sci USA. 1998;95:11089–11094. doi: 10.1073/pnas.95.19.11089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Grange T, Martins de Sa C, Oddos J J, Pictet R. Human mRNA polyadenylate binding protein: evolutionary conservation of a nucleic acid binding motif. Nucleic Acids Res. 1987;15:4771–4787. doi: 10.1093/nar/15.12.4771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Haller A A, Semler B L. Translation and host cell shutoff. In: Rotbart H, editor. Human enterovirus infections. Washington, D.C: American Society for Microbiology; 1995. pp. 113–134. [Google Scholar]
- 26.Hambidge S J, Sarnow P. Translational enhancement of the poliovirus 5′ noncoding region mediated by virus-encoded polypeptide-2A. Proc Natl Acad Sci USA. 1992;89:10272–10276. doi: 10.1073/pnas.89.21.10272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hellen C U T, Lee C-K, Wimmer E. Determinants of substrate recognition by poliovirus 2A proteinase. J Virol. 1992;66:3330–3338. doi: 10.1128/jvi.66.6.3330-3338.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Iizuka N, Najita L, Franzusoff A, Sarnow P. Cap-dependent and cap-independent translation by internal initiation of mRNAs in cell extracts prepared from Saccharomyces cerevisiae. Mol Cell Biol. 1994;14:7322–7330. doi: 10.1128/mcb.14.11.7322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Imataka H, Gradi A, Sonenberg N. Abstracts from the 1998 Conference on Translational Control. Cold Spring Harbor, N.Y: Cold Spring Harbor Press; 1998. A newly identified N-terminal amino acid stretch of human eIF4G binds poly(A) binding protein and functions in poly(A) dependent translation; p. 147. [Google Scholar]
- 30.Irurzun A, Sánchez-Palomino S, Novoa I, Carrasco L. Monensin and nigericin prevent the inhibition of host translation by poliovirus, without affecting p220 cleavage. J Virol. 1995;69:7453–7460. doi: 10.1128/jvi.69.12.7453-7460.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jacobson A. Poly(A) metabolism and translation: the closed-loop model. In: Hershey J W B, Matthews M B, Sonenberg N, editors. Translational control. Cold Spring Harbor, N.Y: Cold Spring Harbor Press; 1996. pp. 451–479. [Google Scholar]
- 32.Joachims M, Harris K S, Etchison D. Poliovirus protease 3C mediates cleavage of microtubule-associated protein 4. Virology. 1995;211:451–461. doi: 10.1006/viro.1995.1427. [DOI] [PubMed] [Google Scholar]
- 33.Kessler S H, Sachs A B. RNA recognition motif 2 of yeast Pab1p is required for its functional interaction with eukaryotic translation initiation factor 4G. Mol Cell Biol. 1998;18:51–57. doi: 10.1128/mcb.18.1.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kräusslich H G, Nicklin M J H, Toyoda H, Etchison D, Wimmer E. Poliovirus proteinase 2A induces cleavage of eukaryotic initiation factor 4F polypeptide p220. J Virol. 1987;61:2711–2718. doi: 10.1128/jvi.61.9.2711-2718.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kuhn U, Pieler T. Xenopus poly(A)-binding protein: functional domains in RNA binding and protein-protein interaction. J Mol Biol. 1996;256:20–30. doi: 10.1006/jmbi.1996.0065. [DOI] [PubMed] [Google Scholar]
- 36.Le H, Tanguay R L, Balasta M L, Wei C-C, Browning K S, Metz A M, Goss D J, Gallie D R. Translation initiation factors eIF-iso4G and eIF-4B interact with the poly(A)-binding protein and increase its RNA binding activity. J Biol Chem. 1997;272:16247–16255. doi: 10.1074/jbc.272.26.16247. [DOI] [PubMed] [Google Scholar]
- 37.Liebig H D, Ziegler E, Yan R, Hartmuth K, Klump H, Kowalski H, Blaas D, Sommergruber W, Frasel L, Lamphear B, Rhoads R, Kuechler E, Skern T. Purification of two picornaviral 2A proteinases—interaction with eIF-4γ and influence on in vitro translation. Biochemistry. 1993;32:7581–7588. doi: 10.1021/bi00080a033. [DOI] [PubMed] [Google Scholar]
- 38.Lloyd R E, Bovee M. Persistent infection of human erythroblastoid cells by poliovirus. Virology. 1993;194:200–209. doi: 10.1006/viro.1993.1250. [DOI] [PubMed] [Google Scholar]
- 39.Lloyd R E, Etchison D, Ehrenfeld E. Poliovirus protease does not mediate cleavage of the 220,000-Da component of the cap binding protein complex. Proc Natl Acad Sci USA. 1985;82:2723–2727. doi: 10.1073/pnas.82.9.2723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lloyd R E, Grubman M J, Ehrenfeld E. Relationship of p220 cleavage during picornavirus infection to 2A proteinase sequences. J Virol. 1988;62:4216–4223. doi: 10.1128/jvi.62.11.4216-4223.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Macadam A J, Ferguson G, Fleming T, Stone D M, Almond J W, Minor P D. Role for poliovirus protease 2A in cap independent translation. EMBO J. 1994;13:924–927. doi: 10.1002/j.1460-2075.1994.tb06336.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Merrick W C, Hershey J W B. The pathway and mechanism of eukaryotic protein synthesis. In: Hershey J W B, Matthews M B, Sonenberg N, editors. Translational control. Cold Spring Harbor, N.Y: Cold Spring Harbor Press; 1996. pp. 31–69. [Google Scholar]
- 43.Morley S J, Curtis P S, Pain V M. eIF4G—translation’s mystery factor begins to yield its secrets. RNA. 1997;3:1085–1104. . (Review.) [PMC free article] [PubMed] [Google Scholar]
- 44.Munroe D, Jacobson A. Messenger RNA poly(A) tail, a 3′ enhancer of translational initiation. Mol Cell Biol. 1990;10:3441–3455. doi: 10.1128/mcb.10.7.3441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nagai K, Oubridge C, Ito N, Evans P. The RNP domain: a sequence-specific RNA-binding domain involved in processing and transport of RNA. Trends Biochem Sci. 1995;20:235–240. doi: 10.1016/s0968-0004(00)89024-6. [DOI] [PubMed] [Google Scholar]
- 46.Nietfeld W, Mentzel H, Pieler T. The Xenopus laevis poly(A) binding protein is composed of multiple functionally independent RNA binding domains. EMBO J. 1990;9:3699–3705. doi: 10.1002/j.1460-2075.1990.tb07582.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.O’Neill R E, Racaniello V R. Inhibition of translation in cells infected with a poliovirus 2Apro mutant correlates with phosphorylation of the alpha subunit of eucaryotic initiation factor 2. J Virol. 1989;63:5069–5075. doi: 10.1128/jvi.63.12.5069-5075.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Percy N, Belsham G J, Brangwyn J K, Sullivan M, Stone D M, Almond J W. Intracellular modifications induced by poliovirus reduce the requirement for structural motifs in the 5′ noncoding region of the genome involved in internal initiation of protein synthesis. J Virol. 1992;66:1695–1701. doi: 10.1128/jvi.66.3.1695-1701.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pérez L, Carrasco L. Lack of direct correlation between p220 cleavage and the shut-off of host translation after poliovirus infection. Virology. 1992;189:178–186. doi: 10.1016/0042-6822(92)90693-j. [DOI] [PubMed] [Google Scholar]
- 50.Preiss T, Hentze M W. Dual function of the messenger RNA cap structure in poly(a)-tail-promoted translation in yeast. Nature. 1998;392:516–520. doi: 10.1038/33192. [DOI] [PubMed] [Google Scholar]
- 51.Richter J D. Dynamics of poly(A) addition and removal during development. In: Hershey J W B, Matthews M B, Sonenberg N, editors. Translational control. Cold Spring Harbor, N.Y: Cold Spring Harbor Press; 1996. pp. 481–503. [Google Scholar]
- 52.Sachs A, Bond M W, Kornberg R D. A single gene from yeast for both nuclear and cytoplasmic polyadenylate-binding proteins: domain structure and expression. Cell. 1986;45:827–835. doi: 10.1016/0092-8674(86)90557-x. [DOI] [PubMed] [Google Scholar]
- 53.Sachs A B, Davis R W. The poly(A)-binding protein is required for poly(A) shortening and 60S ribosomal subunit dependent translation initiation. Cell. 1989;58:857–867. doi: 10.1016/0092-8674(89)90938-0. [DOI] [PubMed] [Google Scholar]
- 54.Sachs A B, Davis R W, Kornberg R D. A single domain of yeast poly(A)-binding protein is necessary and sufficient for RNA binding and cell viability. Mol Cell Biol. 1987;7:3268–3276. doi: 10.1128/mcb.7.9.3268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sachs A B, Sarnow P, Hentze A W. Starting at the beginning, middle, and end: translation initiation in eukaryotes. Cell. 1997;89:831–838. doi: 10.1016/s0092-8674(00)80268-8. [DOI] [PubMed] [Google Scholar]
- 56.Shatkin A J. mRNA cap binding proteins: essential factors for initiating translation. Cell. 1985;40:223–224. doi: 10.1016/0092-8674(85)90132-1. [DOI] [PubMed] [Google Scholar]
- 57.Sommergruber W, Ahorn H, Zophel A, Maurerfogy I, Fessl F, Schnorrenberg G, Liebig H D, Blaas D, Kuechler E, Skern T. Cleavage specificity on synthetic peptide substrates of human rhinovirus-2 proteinase-2A. J Biol Chem. 1992;267:22639–22644. [PubMed] [Google Scholar]
- 58.Sun X-H, Baltimore D. Human immunodeficiency virus tat-activated expression of poliovirus protein 2A inhibits mRNA translation. Proc Natl Acad Sci USA. 1989;86:2143–2146. doi: 10.1073/pnas.86.7.2143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tarun S Z, Sachs A B. Association of the yeast poly(A) tail binding protein with translation initiation factor eIF-4G. EMBO J. 1996;15:7168–7177. [PMC free article] [PubMed] [Google Scholar]
- 60.Tarun S Z, Jr, Wells S E, Deardorff J A, Sachs A B. Translation initiation factor eIF4G mediates in vitro poly(A) tail-dependent translation. Proc Natl Acad Sci USA. 1997;94:9046–9051. doi: 10.1073/pnas.94.17.9046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Trachsel H. Binding of initiator methionyl-tRNA to ribosomes. In: Hershey J W B, Matthews M B, Sonenberg N, editors. Translational control. Cold Spring Harbor, N.Y: Cold Spring Harbor Press; 1996. pp. 113–137. [Google Scholar]
- 62.Wells S E, Hillner P E, Vale R D, Sachs A B. Circularization of mRNA by eukaryotic translation initiation factors. Mol Cell. 1998;2:135–140. doi: 10.1016/s1097-2765(00)80122-7. [DOI] [PubMed] [Google Scholar]
- 63.Wickens M, Anderson P, Jackson R J. Life and death in the cytoplasm: messages from the 3′ end. Curr Opin Genet Dev. 1997;7:220–232. doi: 10.1016/s0959-437x(97)80132-3. [DOI] [PubMed] [Google Scholar]
- 64.Wyckoff E E, Lloyd R E, Ehrenfeld E. Relationship of eukaryotic initiation factor 3 to poliovirus-induced p220 cleavage activity. J Virol. 1992;66:2943–2951. doi: 10.1128/jvi.66.5.2943-2951.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wyckoff E E, Hershey J W B, Ehrenfeld E. Eukaryotic initiation factor-3 is required for poliovirus-2A protease-induced cleavage of the p220 component of eukaryotic initiation factor-4F. Proc Natl Acad Sci USA. 1990;87:9529–9533. doi: 10.1073/pnas.87.24.9529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yalamanchili P, Datta U, Dasgupta A. Inhibition of host cell transcription by poliovirus: cleavage of transcription factor CREB by poliovirus-encoded protease 3C(Pro) J Virol. 1997;71:1220–1226. doi: 10.1128/jvi.71.2.1220-1226.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yohn C B, Cohen A, Danon A, Mayfield S P. A poly(A)-binding protein functions in the chloroplast as a message-specific translation factor. Proc Natl Acad Sci USA. 1998;95:2238–2243. doi: 10.1073/pnas.95.5.2238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ziegler E, Borman A M, Deliat F G, Liebig H D, Jugovic D, Kean K M, Skern T, Kuechler E. Picornavirus 2A proteinase-mediated stimulation of internal initiation of translation is dependent on enzymatic activity and the cleavage products of cellular proteins. Virology. 1995;213:549–557. doi: 10.1016/s0042-6822(95)90001-2. [DOI] [PubMed] [Google Scholar]