Abstract
Iron is an essential element involved in various metabolic processes. The ferritin family of proteins forms nanocage assembly and is involved in iron oxidation, storage, and mineralization. Although several structures of human ferritins and bacterioferritins have been solved, there is still no complete structure that shows both the trapped Fe-biomineral cluster and the nanocage. Furthermore, whereas the mechanism of iron trafficking has been explained using various approaches, structural details on the biomineralization process (i.e. the formation of the mineral itself) are generally lacking. Here, we report the cryo-electron microscopy (cryo-EM) structures of apoform and biomineral bound form (holoforms) of the Streptomyces coelicolor bacterioferritin (ScBfr) nanocage and the subunit crystal structure. The holoforms show different stages of Fe-biomineral accumulation inside the nanocage, in which the connections exist in two of the fourfold channels of the nanocage between the C-terminal of the ScBfr monomers and the Fe-biomineral cluster. The mutation and truncation of the bacterioferritin residues involved in these connections significantly reduced the iron and phosphate binding in comparison with those of the wild type and together explain the underlying mechanism. Collectively, our results represent a prototype for the bacterioferritin nanocage, which reveals insight into its biomineralization and the potential channel for bacterioferritin-associated iron trafficking.
Keywords: ferritin-like molecules, Fe cluster, nanocage, Streptomyces coelicolor, cryo-EM structure
Significance Statement.
Iron is an essential element for all living organisms, and it is involved in various metabolic processes. The ferritin family of proteins forms nanocage assembly and is involved in iron oxidation, storage, and mineralization. Although several structures of human ferritin and bacterioferritin subunits have been resolved, there is still no complete structure that shows the trapped iron cluster along with the nanocage. While the mechanism of iron trafficking was explained previously using various approaches, structural details on the biomineralization process are generally lacking. Here, we report four cryo-EM structures of the bacterioferritin that shows different stages of Fe-biomineral accumulation inside the nanocage.
Introduction
Iron is involved in various physiological processes, including DNA synthesis, gene regulation, respiration, and oxygen transport (1). However, free iron is toxic to cells and is linked with the formation of free radicals from reactive oxygen species (ROS). Thus, iron levels are carefully regulated to prevent iron overload and oxidative stress. The ferritin (Ftn) family of proteins play an important role in stress management by sequestering excess iron (Fe2+-substrate) and storing it as insoluble Fe3+ (product) clusters within a nanocage assembly. When required, these iron stores can be reduced and transported out of the cavity for cellular processes (2–4).
Ferritin-like molecules are present in archaea, bacteria, and eukarya, and their structures are well conserved despite poor sequence identity (5). There are three types of ferritin-like molecules found in bacteria: bacterioferritin (Bfr), Ftn, and DNA-binding proteins from starved cells (Dps) (6). Bfr and Ftn nanocages are composed of 24 subunits and can store around 2,500 Fe ions (7–9), whereas Dps nanocages comprise 12 subunits and able to store up to 500 Fe ions in the cavity (5). Bfr proteins bind heme between two monomers at the dimeric interface, with the nanocage capable of binding 12 heme molecules (10). Fe2+ ions enter into the ferroxidase center situated in the helical bundle of Bfr monomer through the B-pore (11–13). Recent studies have shown that Bfr ferroxidase center reacts faster with H2O2 than with O2 suggesting a prominent role in ROS detoxification (14).
Several crystal structures of animal Ftn and Bfr have been resolved (15–17) (Table S2), and cryo-EM structures of mammalian Ftn nanocages have been reported, along with their functional studies (18–20). The biomineral core formation in Ftn is also reported in various studies (21–23). However, to date, there is no cryo-electron microscopy (cryo-EM) nanocage structure of Bfr. Moreover, none of the reported structures shows the 3D structure of the nanocage cavity with the trapped Fe-biomineral cluster. Although the mechanism of iron trafficking has been detailed previously, structural details on the biomineralization process is generally lacking.
Streptomyces coelicolor is a model organism used for studying Mycobacterium tuberculosis and Corynebacterium diphtheriae due to its similarity in core genome (24). A recent study demonstrated that iron promotes the intrinsic resistance of aerobic actinomycetes S. coelicolor and Mycobacterium smegmatis against bactericidal antibiotics (25). The Streptomyces genus is responsible for producing a majority of the antibiotics in use today, as well as some immunosuppressants and antitumor agents (26).
Here, we report the high-resolution cryo-EM structures of the apo (empty) and holo (containing Fe-biomineral) states of the S. coelicolor Bfr (ScBfr) nanocage, along with the crystal structure of the ScBfr subunit. The holo-ScBfr structures illustrate the various stages of mineralization of the Fe cluster sequestered inside the nanocage cavity. Notably, the holoform-II maintains connections between the protein shell inner surface and the Fe-biomineral cluster suggestive of potential pathways for iron accumulation and release. Taken together, this study helps to clarify the multiple functions of Ftn, particularly the molecular mechanism of iron transport.
Results
Cryo-EM structures of ScBfr nanocages
The 3D reconstructions of the ScBfr images show four major species of nanocages: the apo-ScBfr (no Fe-biomineral cluster) form, and three forms of holo-ScBfr—form-I, form-II, and form-III with Fe-biomineral clusters accumulated/trapped inside the nanocage (Figs. 1 and 2 and Table S1).
Fig. 1.
The cryo-EM structures of A) apo-ScBfr, B) holo-ScBfr-form-I, and C) holo-ScBfr-form-III refined with O symmetry. The trans-sectional view of the model shown below each form. The Fe-mineral accumulated in the nanocage cavity is shown in yellow color in holo-form-I and holoform-III. The apo-ScBfr structure was solved at 2.7-Å resolution while the holo-form-I was solved at 3.6 and holoform-III at 3.2-Å resolution.
Fig. 2.
The cryo-EM structures of holoform-II models. Holoform-II model with O symmetry, A) closed model and B) Clipped model showing the biomineral core. Holoform-II model with C1 symmetry, C) Closed model and D) Clipped model showing biomineral core. Holoform-II model refined after subtraction, E) Clipped model of shell refined with O symmetry after signal subtraction, F) Biomineral core refined with C1 symmetry after signal subtraction and G) Merged view.
Structure of apo-ScBfr nanocage
The apo-ScBfr nanocage has an external diameter of 125 Å and an internal diameter of 82 Å with an empty cavity (Fig. 1A). The subunits in the nanocage are arranged in an antiparallel fashion, forming a dimer (Fig. S1A and B). In total, 12 dimeric units assemble to form the nanocage. There are six symmetric fourfold channels, eight symmetric threefold channels, and 24 B-pores formed by the subunits of the nanocage structure (Fig. S1A). The fourfold channels are formed by the C-terminal regions (146 to 157 aa) of the four monomers (one monomer from each dimeric unit), which protrudes toward the nanocage cavity (Fig. S1A).
The cryo-EM map of apo-ScBfr (Figs. 1A and 3A) shows density for all residues of the 24 chains of the ScBfr nanocage, except the last 10 residues in the C-terminus. The densities for the heme molecules are well defined in the dimeric interface of ScBfr (Fig. 3B), and a total of 12 heme molecules are modeled in the nanocage. Furthermore, the presence of heme was verified using the UV-Vis absorbance spectrum (Fig. S9). The Met52 residues from two monomers form coordination bonds with the heme molecule (Fig. 3B), and residues Arg45, Phe49, and Glu56 from both monomers make hydrogen bonding contacts with heme. The density of the Fe3+ ion located in the ferroxidase center is also well defined, and residues Glu18, Glu51, His54, and Glu127 form coordination bonds with the Fe3+ ion (Fig. 3C). Further, self-assembly of the subunits in the nanocage is predominantly maintained by electrostatic interactions (Fig. S1C and D).
Fig. 3.
A) The representative cryo-EM density map of apo-ScBfr nanocage to show the quality of the map. B) The density map of apo-ScBfr nanocage showing the heme molecule and the adjacent residues. The heme molecule is sandwiched between the residues Met52 from both monomers of the dimer; for clarity, only Met52 from 1 monomer is shown in this figure. C) The density map of apo-ScBfr nanocage (cryo-EM model) showing Fe ion in the ferroxidase center and the coordinating residues His54, Glu18, Glu51, and Glu127. These figures are prepared using the EM density for the apo-ScBfr model.
Structure of holo-ScBfr nanocages
The Fe-biomineral–trapped holo-ScBfr nanocage structures have same internal diameter and external diameter as the apo-ScBfr nanocage. The three different holoforms (holoform-I, holoform-II, and holoform-III) identified in this data set showed different amount of Fe trapped in the nanocage (Figs. 1, 2, and S4). The number of particles with these holoforms were very fewer (1–2% of the total number of particles) than the apoform. Due to the limitations in the number of particles, the holoform-I and holoform-III were refined with O symmetry that gave a meaningful resolution, while the holoform-II was refined with C1 symmetry and O symmetry to a higher resolution (Table S1). It should be noted that the symmetry averaged reconstructions by imposing O symmetry averaging results in the loss of unique features, which would be overwhelmed by signal of the dominant features. The holoform-II showed the presence of biomineral in both models (Fig. 2), but the biomineral core was asymmetric in the C1 symmetry refined model (Fig. 2D). While the O symmetry model showed connections between C-terminal region of ScBfr and biomineral core in all fourfold channels (Fig. 2B), only two of these connections were present in the C1 symmetry model (Fig. 2).
The refinement of all three holoform models with O symmetry showed different levels of Fe-biomineral accumulation: holoform-II showed larger amount of biomineral core compared to other two forms (Figs. 1 and 2A–D). The higher number of particles in the holoform-II data allowed us to subtract the signal for the biomineral core and refined the nanocage with O symmetry and the biomineral core with C1 symmetry. This refinement of only biomineral core yielded a resolution of only 5.7 Å (Fig. 2F), while the nanocage refined with O symmetry gave a resolution of 2.9 Å. The combined model (merged model in which shell at O symmetry and biomineral with C1 symmetry; Fig. 2G) shows good EM density for both nanocage and the biomineral core. The Fe-biomineral core does not fit to any symmetry, which is consistent with other reports. Previous studies (5, 9, 27, 28) suggested that the chemical content and structure of iron inside the core is polyphasic or heterogenous and may not be packed regularly.
Insights into the pathway of Fe ion movement
Although various pathways (5, 15, 29, 30) have been proposed to explain the passage of iron in and out of the nanocage cavity, the exact mechanism remains unclear. The iron pathways have been identified with different level of detail in different Ftns, and animal Ftns are studied in detail when compared to other Ftns. In this context, our structures of the holoforms of the ScBfr nanocages reveal the following intriguing features.
The holoform-I refined with octahedral symmetry showed small Fe-biomineral assemblies located close to the fourfold channels in the inner surface of the shell (Fig. 1B). Holoform-III is an intermediate between holoform-I and holoform-II, in which no connections observed to the inner surface of the nanocage may be due to the significantly lower density than the formed mineral or due to the subtraction (averaging out) of weak signal in the octahedral symmetry (Fig. 1C). Notably, the holoform-II had more particles, which allowed us to refine this model in three different ways (refer section 2.1.2 for details). In this form, the cryo-EM map shows connections between the trapped Fe-biomineral inside the nanocage cavity and C-terminal in models refined with O symmetry and C1 symmetry. We observed two connections maintained between the fourfold channel and biomineral core in the model refined with C1 symmetry. Collectively these models suggest that the Fe-biomineralization process is asymmetric, and it starts from the inner surface of the Ftn nanocage, and the fourfold channel serves as the starting point (seed) for the nucleation. Further it suggests that the Fe-biomineral cluster grows toward the center of the nanocage and fill the cavity. Moreover, the connections in holoform-II suggest a continuous pathway spanning 38 Å between the ferroxidase center in each monomer and the nanocage cavity (Fig. 4A and B).
Fig. 4.
A) Overall view of the ScBfr nanocage with the trapped Fe-biomineral cluster. B) The density map of the nanocage showing the connections between the trapped Fe-biomineral cluster inside the nanocage cavity and the ScBfr protein shell. B) Right panel: the Thr157 in the C-terminal of each monomer is connected to the Fe-core in the nanocage cavity and thus forms a continuous pathway spanning 38 Å between the ferroxidase center located in each monomer and the nanocage cavity (shown as brown arrow). B) Left panel: The 12 residues possibly involved in the movement of Fe3+ from the ferroxidase center to the nanocage cavity in ScBfr are shown as stick model and labeled. Fe atoms are shown as orange spheres. C) The heme molecule sandwiched between the dimeric interfaces is also having connections with the Fe-core through the adjacent residues Met52 and Phe49. D) Residues such as Lys42 and Tyr43 structurally located near the C-terminal region of ScBfr, along with Tyr133 and Asp132 connect the B-pore at the surface to the Fe cluster in the nanocage cavity.
These connections represent one possible route for Fe3+ ion transport from the ferroxidase center into the nanocage cavity. Turano et al. (31) proposed a similar pathway based on NMR studies of the Lithobates catesbeiana M-ferritin subunit. Superposition of the monomeric ScBfr (crystal structure; see section 2.3) with the L. catesbeiana M-ferritin subunit showed similarity in at least 12 residues (Ala21, Tyr25, His28, Glu47, Val83, Phe87, Asp90, Leu134, Leu138, Glu146, Glu155, and Thr157) involved in the movement of Fe3+ from the ferroxidase center to the nanocage cavity (Fig. 4B). Notably, most of these residues are highly conserved (around 40% residues) among Bfr proteins (Fig. S2A), and we also observed a continuous negative patch between the ferroxidase center and the C-terminal (Fig. S1C).
To confirm the role of these residues, in particular the C-terminal region, in iron transport, we generated a C-terminal truncated version of Bfr (ScBfr1–152: residues 153–167 removed) and examined Fe binding compared with the wild type. Inductively coupled plasma-optical emission spectrometry (ICP-OES) was used for determining the amount of Fe ion in ScBfr wild type and the C-terminal truncated mutant. We observed significantly reduced Fe ion binding in the C-terminal truncated mutant as compared with the wild type (0.04 ppm vs. 0.27 ppm), suggesting an important role for the C-terminal region in Fe-biomineral accumulation. Moreover, we performed an inorganic phosphate assay to determine the amount of phosphate which showed substantial reduction in the phosphate concentration in the C-terminal truncated mutant as compared with the wild type (0.015 µm vs 0.42µm). In Bfr wild type (which consists of both apoform and holoform), the calculated iron:phosphate ratio is 8:1, while the average protein:phosphate ratio is 1:0.37. In addition, the dynamic light scattering (DLS) experiment indicated that the C-terminal truncated mutant also has hydrodynamic radius and the calculated molecular weight corresponding to 24-mer assembly, similar to the wild type (Fig. S8A). Furthermore, we verified the C-terminal truncated mutant using negative-stained electron microscope images that also confirmed the presence of spherical assembly similar to the wild-type Bfr (Fig. S10).
In the holoform-II model refined with C1 and O symmetry, we observed connections between the Fe-biomineral and the heme binding region, involving residues Met52 and Phe49 (Fig. 4C). Whereas Met52 is highly conserved in Bfr proteins, Phe49 is conserved only in S. coelicolor and M. tuberculosis. Heme plays an important role in the release of iron from the central cavity of the Bfr nanocage by facilitating electron transfer (32). In the present study, the observed connections in holoform-II with or without symmetry suggest the possibility that the electrons transferred by heme enter the nanocage cavity to facilitate the release of Fe2+ from the cavity (Fig. 4C). A similar heme-mediated electron transfer process has been previously reported (16, 33).
We also observed a continuous connection (in holoform-II model refined with both O and C1 symmetry) between the B-pore at the surface and the Fe-biomineral in the nanocage cavity in holoform-II, spanning residues Lys42 and Tyr43, (which are structurally located near the C-terminal) and residues Tyr133 and Asp132. Residue Lys42 is connected to the Fe-biomineral core, whereas Asp132 is connected to the B-pore in the surface of the nanocage (Fig. 4D). We speculate that this connection might play a role in electron transfer into the nanocage cavity as well as Fe ion transfer. Indeed, mutating this commonly found Lys42 to Ala (ScBfr K42A) reduces the Fe ion binding as compared with the wild type (0.06 ppm vs. 0.27 ppm).
Crystal structure of ScBfr
The crystal structure of ScBfr was determined at 2.6-Å resolution (Table S2). The monomeric ScBfr adopts an α-helical structure (Fig. S2B), similar to its homologs (34). Four glutamic acids (Glu18, Glu51, Glu127, and Glu94-water mediated) and His 130 and His54 from three helices are involved in the Fe coordination in the ferroxidase center (Fig. S2B). Previous reports of Bfr crystal structures suggest that Bfr can form various crystal packing depending on the space group (i.e. 1 molecule to 48 molecules in the asymmetric unit) (Table S3); yet, no Fe-biomineral has been observed in any of these asymmetric unit oligomers.
A structural homology search for ScBfr identified conservation among several Ftns and Dps proteins (Table S4). The Bfr homologs are presents in several species, including pathogenic M. tuberculosis and C. diphtheriae. A comparison of the M. tuberculosis Bfr structure showed an overall similarity with the ScBfr structure (Fig S2C). The C-terminal regions of human H-ferritin (HuHF), human L-ferritin (HuLF), and bacterial Ftn (PDB codes: 2FHA, 2FFX, and 4XGS) are well ordered with an α-helical secondary structure that folds back to the core protein (Fig. S2D). In contrast, the C-terminal tail of ScBfr is flexible (5 residues in the C-terminal missing in the crystal structure) (Fig. S2D). The flexible C-terminal region of S. coelicolor might increase iron chelation into the core of the nanocage. A flexible/extended C-terminus has previously been implicated in ferroxidase activity and iron release in M. tuberculosis BfrB (34). Indeed, C-terminal truncation of Bfr significantly reduced Fe and phosphate binding as compared with wild-type Bfr (see section 2.2). Similarly, Bfr and Dps proteins have high overall structural similarity and low sequence identity (Fig. S3A and B).
The superposition of the nanocage which was generated using our ScBfr crystal structure with the cryo-EM apo/holo ScBfr nanocages indicates that the rmsd was around 1 Å for the core structure except in the N-terminal and C-terminal regions that showed a maximum difference of up to 3.5 Å and indicated that both N-terminal and C-terminal regions are flexible (Fig. S2C). Moreover, comparing the human Ftn (HuHF-HuLF) nanocage cryo-EM structures with that of ScBfr shows a difference of ∼3 to 5 Å in various regions (Fig. S2E). Notably, the C-terminal tail at the entrance of the nanocage cavity shows more flexibility for ScBfr than human Ftn (HuHF-HuLF).
Discussion
The iron-storage function of Ftn plays a central role in iron homeostasis in the cell (31). Despite the wealth of structural and functional data, the mechanism of Fe-biomineral accumulation inside the Ftn nanocage and its subsequent release for cellular activities remains unclear. In Bfr, Fe ions can be found inside the ferroxidase center, bound to heme in between monomers (i.e. at the dimeric interface) and in Fe-biomineral cluster inside the nanocage cavity. In our cryo-EM structures of the ScBfr nanocage, we show Fe ions in all these locations. Notably, we show a physiologically relevant structure of a 24-mer bacterioferritin nanocage with trapped Fe-biomineral. In the holoform-II cryo-EM structure, the C-terminus of ScBfr is linked to the Fe-biomineral within the nanocage cavity (Figs. 2 and S7). This observation is consistent with the previous studies that suggested the Fe-biomineral that grows from the inner surface of the nanocage to the center of the cavity (21, 22, 35). Using scanning transmission electron microscopy images from human liver samples, Pan et al. (21) proposed that the ferric oxide biomineral can grow from the inner surface of the nanocage toward the center of the core. Similarly, by using NMR studies, Pozzi et al. (21) reported the growth of an initial iron cluster on the inner surface of HuLF. Ciambellotti et al. (36) recently reported the expansion of the iron cluster precursor in HuLF.
Further we observed that the flexible C-terminal region of ScBfr may extend toward the center of the nanocage cavity (Fig. 4B). The role of the C-terminal in the Fe oxidation and reduction process is well documented in Bfr (34). Here we show that, C-terminal truncation of ScBfr significantly reduced iron and phosphate binding as compared with wild-type. Notably the Fe-biomineral accumulations are observed near the fourfold channel where the C-terminal regions from 4 monomers are clustered (Fig. 1B).
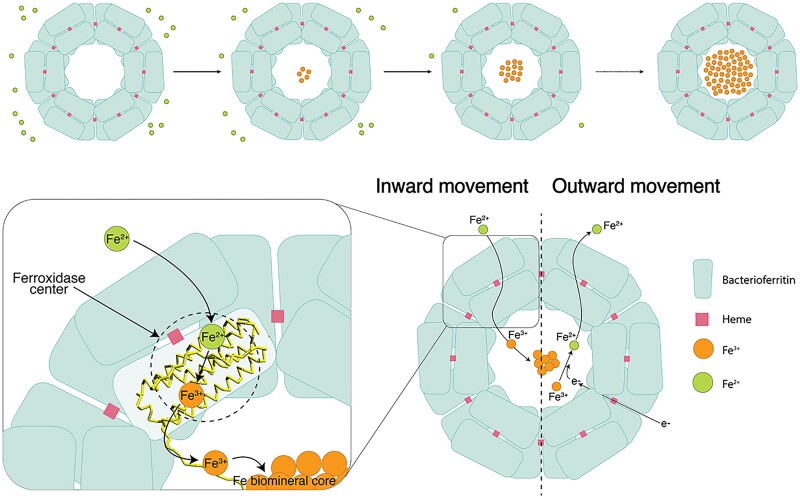
Combining our observations with previous literature (30–32, 37, 38), we propose the following mechanism to explain Fe trafficking in and out of the nanocage cavity (Fig. 5). Fe2+ ions enter through the B-pore and move into the ferroxidase center, where Fe2+ is converted to Fe3+ (38). These Fe3+ ions then move through the four-helical bundle channel to reach the C-terminal helix of ScBfr (Fig. 4B) and undergo the processes of olation/oxolation that give rise to the ferrihydrite mineral core close to the fourfold channel which further grows into the nanocage cavity (Figs. 1B, 1C and 2) (5, 39, 40). For outward trafficking, we propose that heme is involved in transferring electrons into the nanocage cavity (Fig. 4C). Using these electrons, Fe3+ is reduced to Fe2+, with Fe2+ then exiting either through the B-pore or via attachment to a chelating agent (9, 32).
Fig. 5.
Schematic representation showing the inward and outward movement of iron in Bfr nanocage. The Fe2+ atom enters into the Bfr shell through the B-pore and move to the ferroxidase center where it gets oxidized to form Fe3+. Fe3+ moves through the four-helix bundle into the C-terminal region of the Bfr and initiate the nucleation process resulting in the formation of Fe-biomineral cluster. Based on our cryo-EM structures of the ScBfr nanocage, we propose that the biomineralization of the nanocage starts from the inner surface of the nanocage near the fourfold channel. Similarly, heme molecule will help to transport electron (e-) into the nanocage cavity, once the Fe3+ is reduced it is converted to Fe2+ that can move out of the Bfr shell.
In holoform-II, a continuous connection is observed between the B-pore at the surface and the Fe-biomineral in the nanocage cavity, spanning residues Lys42 (connected to Fe-biomineral core) and Tyr43 and residues Tyr133 and Asp132 (connected to the B-pore in the surface of the nanocage) (Fig. 4D). We speculate that this connection might play a role in electron transfer into the nanocage cavity as well as Fe ion transfer. Lys42 to Ala (ScBfr K42A) reduces the Fe ion binding as compared with the wild type (0.06 ppm vs. 0.27 ppm). Notably, in Escherichia coli Bfr, Tyr25 has been reported to act as a molecular capacitor/electron donor for facilitating Fe oxidation (30, 37), and Trp133 facilitates electron transfer (30). Collectively, these findings suggest that Tyr43 and Tyr133 in ScBfr might also contribute to electron transfer.
Since the Fe-biomineral accumulation is a dynamic process (41), we believe that there are various intermediate forms of the nanocage structures other than these four reported structures, and it is supported by the observation of several intermediates in the 2D classes (Fig. S4). Notably the holoforms represent only 1 to 2% of the total number of particles used in our analysis. We identified and separated this small percentage of particles from the large data set and refined to a reasonable resolution where the biomineral clusters inside the nanocage cavity were observed. Hitherto, no biomineral core was reported in the naturally purified Bfr (without the addition of extra iron atoms). A limitation of this study is the application of octahedral symmetry to the whole model (that might average out/cancel weak signals), and we acknowledge that all the fourfold channels might not be having the connection which is evident from the structure determined without any symmetry (holoform-II), in which we observed connections only in two fourfold channels of the nanocage. However, the overall mechanism will be same albeit the difference in the saturation level at different fourfold channels (Figs. 1B and 2).
In summary, here we report the apo and holo states of ScBfr nanocages with Fe ions, heme molecules, and trapped Fe-biomineral clusters. The holoforms show different stages of Fe-biomineral accumulation inside the nanocage cavity. In particular, the holoform-II cryo-EM density shows various connections between the Fe-biomineral cluster and several residues of the protein shell. The holoform-II refined with C1 symmetry (no symmetry) shows biomineral cluster formation near two of the fourfold channels. This model shows the asymmetric nature of biomineralization of nanocage. The residues involved in these connections are critical for the movement of Fe ions in and out of the nanocage cavity. This study provides insight into the biomineralization process in Bfr. These ScBfr nanocage structures are the representative structures of Bfr family of proteins with Fe-biomineral trapped nanocage. Taken together, this study aids to clarify the multiple functions of Bfr, particularly the molecular mechanism of iron mineralization and transport.
Materials and methods
Cloning expression and protein purification
Bfr from S. coelicolor (ScBfr) was cloned into expression vector pET22b (GenScript, USA) between the Bamh1 and Xho1 restriction sites without a stop codon, and the recombinant plasmid was transformed into E. coli BL21 competent cells for protein overexpression. The transformed BL21 cells were used to inoculate LB medium (1 L) containing (100 mg/L) ampicillin, and the culture was grown at 37°C. The medium was not supplemented with iron salts. The Fe-biominerals are naturally trapped in this cell culture condition. After reaching the OD at A600nm of 0.6–0.8, the culture was induced by the addition of 0.3 mm isopropyl β-D-1-thiogalactopyranoside followed by incubation at 28°C for 7 h. Cells were harvested by centrifugation at 6000 rpm using a JLA8.1 rotor in a Beckman Coulter centrifuge for 30 min at 4°C. The harvested cells were resuspended in 30 mL of lysis buffer (30 mm Tris pH 7.0, 200 mm NaCl, 1 mm phenylmethylsulfonyl fluoride, 5% glycerol, and 0.5% Triton × 100) and lysed in sonicator. The resulting lysate was centrifuged at 18,000 rpm (JA20 rotor, Beckman Coulter) for 30 min at 4°C. The supernatant was added to Ni-agarose affinity column equilibrated with same buffer. The unbound proteins were washed off with the same buffer thrice. The protein was then eluted twice with increased imidazole concentration of 250 and 350 mm. The eluted protein samples were injected into Superdex S-75 column equilibrated with gel filtration buffer (30 mm Tris pH 7.0, 200 mm NaCl, and 5% glycerol). The peak fractions were pooled and concentrated, and the same samples were used for both cryo-EM and crystallography experiments (Fig. S8C). For crystallization screening, the protein was concentrated to 10 mg/mL, while for Cryo-EM experiments, the protein was used at 1.8 mg/mL concentration. The purity and homogeneity of ScBfr were verified by using DLS experiments. DLS experiment indicated a polydispersity index of 0.30. Moreover, using the above protocol, we have expressed and purified 2 mutant versions such as ScBfr K42A and ScBfr1–152 for the negative stain imaging, ICP-OES experiment and inorganic phosphate assay. The DLS results show that the truncated protein behaves similar to the wild-type protein (Fig. S8A and B).
Sample preparation and cryo-EM data acquisition
The ScBfr sample at concentration of 1.8 mg/mL was blotted (3.5 µL per blot) onto Au 300 mesh Quantifoil 1.2/1.3 grids (Quantifoil Micro Tools) and immediately plunge frozen using a Vitrobot (FEI). The data set containing 10,060 movies were collected using a Titan Krios microscope (FEI), operating at 300 kV, and equipped with a K3 camera (Gatan) in two batches. Fifty-frame movies were collected at 81,000× magnification (set on microscope) in super-resolution mode with an image pixel size of 0.858 Å/pixel. The dose rate was 6.8 electrons/pixel/second, with a total exposure time of 12 s. The defocus values ranged from −0.70 to −2.5 µm.
Image processing
Movies were motion-corrected to compensate for the beam-induced motion using MotionCor2 (42) with a B-factor of 150 pixels. The data processing was performed using the cryoSPARC program (43) (see Fig. S6) and Relion (44). The defocus values of the motion-corrected micrographs were estimated using CTFFIND4 (45). The particle picking was done using auto-picking method using Relion-Autopick, and approximately, 2 million particles were picked from the 10,060 micrographs. The auto-picked particles that were subjected to multiple rounds of 2D classification to remove suboptimal particles and finally 600,000 particles were selected and used for further 3D classification to obtain the four models (Figs. 1 and 2). An initial 3D model was generated from our present ScBfr crystal structure and low-pass filtered to 8 Å and used as the initial reference for auto-refine. The apo form was refined with half million particles and yielded a resolution of 2.7 Å after postprocessing. For holoform-I and holoform-III, there were 5,200 and 8,174 particles in these two classes and were subjected to 3D auto-refinement and postprocessing to yield the final models with final resolution of 3.6 and 3.2 Å (see Fig. S5A, B, and D). The holo-ScBfr form-II contained 11,617 particles and further subjected to 3D auto-refinement and sharpen 3D to yield the final model. 3D reconstruction was carried out after imposing O symmetry without masking out the inner cavity/Fe-biomineral core. The map has an overall resolution of 3.27 Å for holo-ScBfr form-II (see Figs. S5C and S6). The volume of the Fe-biomineral is 5% of the total volume of the nanocage cavity in the holo-ScBfr form-III but 70% in form-II. The Fe-biomineral cores inside the nanocages of the holo-ScBfr form-I, form-II, and form-III do not fit in any symmetry. Moreover, we refined the holo-ScBfr form-II without imposing symmetry to confirm the presence of the Fe-biomineral in the absence of symmetry. Although the resolution was poor (around 4.1 Å), the map clearly showed the presence of Fe-biomineral inside the nanocage (Figs. 2 and S7). Moreover, we carried out particle subtraction for the holoform-II particles and refined the nanocage and biomineral separately using O symmetry and C1 symmetry, respectively (Fig. 2).
ScBfr model building
The ScBfr crystal structure (see next section) was used as an initial model and aligned to the ScBfr cryo-EM map calculated with Relion. Phenix dock-in-map (46) was used for initial model building followed by manual model building in COOT (47) using the cryo-EM map. After initial model building, the model was refined against the EM-derived maps using the phenix.real_space_refinement tool from the PHENIX software package (46), employing rigid body, local grid, non-crystallographic symmetry, and gradient minimization. This model was then subjected to additional rounds of manual model-fitting and refinement which resulted in a final model-to-map cross-correlation coefficient of 0.83, 0.88, 0.80, and 0.86 for apo-ScBfr and holo-ScBfr (form-I, form-II, and form-III), respectively. Stereochemical properties of the model were evaluated by Molprobity (48).
Additionally, the holoform-II model was subjected to particle subtraction, the nanocage was refined using the subtracted particles with O symmetry, and the biomineral core was refined using C1 symmetry (Fig. 2E–G). We also refined the holoform-II with C1 symmetry to compare the difference in the models in the presence and absence of symmetry (Fig. 2C and D). All figures were prepared using Pymol (49) and ChimeraX (50).
Crystallization and structure determination
Crystallization screening was carried out at room temperature (24°C) using 1 µL of protein (10 mg/mL) mixed with equal volume of reservoir solution. Screens were set up manually by hanging-drop vapor diffusion method. All commercially available screens, viz. Hampton kits, Qiagen kits, and Emerald BioSystems kits were used for initial screening. The initial conditions were obtained from Wizard screens (Rigaku Reagents, USA). The better-quality crystals were obtained with the condition PEG3000, 0.1 m HEPES pH 7.5 and 0.2 m NaCl through the grid screen optimization process. Crystals were cryo-protected in the reservoir solution supplemented with 25% glycerol and flash cooled at 100 K. The data were collected NSRRC beamline BL13B1 using ADSC Quantum 315r CCD detector at 1-Å wavelength. Data processing was done with HKL2000 program (51). The Matthews coefficient was 2.71 Å3/Da (52), corresponding to a solvent content of 55% with six molecules in the asymmetric unit.
The structure was determined by molecular replacement method using Phenix-Phaser program (53) using the structure of Bfr from M. smegmatis (PDB code: 3BKN) as a search model. Initial model building was carried out using the AutoBuild program (54) followed by several rounds of manual model building using COOT program (47). The structure refinement was carried out using Phenix-Refine (55). The model had good stereochemistry, with 99.2% residues falling within the allowed regions of the Ramachandran plot. The density for modeling the second Fe ion was not present in the ferroxidase center, and hence, we modeled only one Fe ion in the ferroxidase center. Similarly, the current model did not have well-defined electron density map to build the heme molecule; instead, we modelled Fe ion in the dimeric interface.
Inductively coupled plasma-optical emission spectrometry
ICP-OES was used to estimate the amount of Fe in the samples. Perkin Elmer Avio 500 ICP-OES was used for this study. The purified wild-type and Bfr mutants (ScBfr1–152 and ScBfr K42A) at a concentration of 0.01 mg/ml were subsequently treated with 0.1 m HNO3 to prepare the samples. Samples were analyzed on the ICP-OES along with a blank with only buffer and 0.1 m HNO3. Fe was not detected in the blank samples.
Inorganic phosphate assay
Both the WT and mutant samples were purified, and pure samples were diluted to a final concentration of 50, 75, 100, and 125 µmol. Phosphate concentration was determined using the phosphate assay kit (catalog number: MAK308). Standards and samples were prepared, and 100 µL of malachite green dye was added and incubated at room temperature for 30 min. The standard graph was plotted using the standards’ concentrations against their absorbance at 620 nm. Using linear regression analysis, the free phosphates in the WT and mutant samples were determined. The absorbance was measured using Tecan Infinite 200 Pro and was controlled by the Tecan i-control software.
Detection of the presence of heme using Uv-Vis measurement
UV/Vis electronic absorption spectra were measured on BioTek Epoch 2 microplate spectrophotometer. The measurements were carried out at 25°C in a 96-well plate and performed in the range of 350–680 nm. Two-hundred microliters of buffer alone, bovine serum albumin (1 and 2 mg/mL), and Bfr WT (1 and 2 mg/mL) were prepared in 30 mm Tris pH 7.0, 200 mm NaCl, and 5% glycerol and loaded to the 96-well plate. The presence of heme was detected at the wavelength of 410 nm.
Negative stain imaging
Five microliters of apoferritin C-terminal truncated Bfr protein (Δ152) at 0.64 mg/mL was incubated on glow-discharged Lacey carbon grid for a minute. The excess sample was removed by a gentle blot with filter paper at the edge of the grid. The grid was then rinsed with 5µL of UranyLess EM stain and blotted gently at the edge of the grid. Then, the grid was incubated with 5µL of UranyLess EM stain for a minute and blotted again with filter paper. The grid was left to air-dry for about 5 min. The grids were screened in an FEI Tecnai T-12 electron microscope, and images were captured using Gatan UltraScan CCD camera.
Supplementary Material
Acknowledgments
We acknowledge the NSRRC, Taiwan beamline 13B1 for crystallographic data collection and National University of Singapore (NUS) CBIS for cryo-EM data collection. We thank Dr Kang Wei Tan, Department of Chemistry, NUS, and Dr Matthew Belousoff, Monash University, Australia, for the discussions on EM experiments and analysis. Preliminary version of this manuscript was on a preprint: https://www.biorxiv.org/content/10.1101/2021.02.04.429857v1. We also acknowledge the support from Mr Chung-Han Tsai from Centre for Advanced Microscopy, Australian National University, Australia. The computational resources from National Computational Infrastructure, Canberra, Australia was used for final refinement of Holoform -II model. Molecular graphics and analyses were performed with UCSF ChimeraX, developed by the Resource for Biocomputing, Visualization, and Informatics at the University of California, San Francisco.
Contributor Information
Chacko Jobichen, Department of Biological Sciences, National University of Singapore, Singapore 117543, Singapore.
Tan Ying Chong, Department of Biological Sciences, National University of Singapore, Singapore 117543, Singapore.
Rajesh Rattinam, Department of Biological Sciences, National University of Singapore, Singapore 117543, Singapore; Department of Microbial Biotechnology, Bharathiar University, Coimbatore 641046, Tamil Nadu, India.
Sandip Basak, Department of Physiology and Biophysics, Case Western Reserve University, Cleveland, OH 44106, USA.
Mahalashmi Srinivasan, Department of Biological Sciences, National University of Singapore, Singapore 117543, Singapore.
Yeu Khai Choong, Department of Biological Sciences, National University of Singapore, Singapore 117543, Singapore.
Kannu Priya Pandey, Department of Biological Sciences, National University of Singapore, Singapore 117543, Singapore.
Tran Bich Ngoc, Department of Biological Sciences, National University of Singapore, Singapore 117543, Singapore.
Jian Shi, Department of Biological Sciences, National University of Singapore, Singapore 117543, Singapore.
Jayaraman Angayarkanni, Department of Biological Sciences, National University of Singapore, Singapore 117543, Singapore; Department of Microbial Biotechnology, Bharathiar University, Coimbatore 641046, Tamil Nadu, India.
J Sivaraman, Department of Biological Sciences, National University of Singapore, Singapore 117543, Singapore.
Supplementary Material
Supplementary material is available at PNAS Nexus online.
Funding
This work was partially supported by Ministry of Education, Singapore (MoE Tier-2) grant (R18 154-000-B03-112) and R154-000-A72-114 (AcRF Tier 1 grant) respectively.
Author contributions
J.S. and J.A. conceived the project; C.J., and J.S. designed the experiments; C.J., T.Y.C., R.R., M.S., Y.K.C., K.P. performed research; T.B.N. and S.J. assisted in cryo-EM data collection; C.J., S.B., and J.S. analyzed the data; and C.J. and J.S. wrote the paper with input from other authors.
Data availability
The coordinates of the ScBfr crystal structure was deposited in wwPDB under the accession number 5XX9. The coordinates of the ScBfr nanocage structures and the cryo-EM density map are deposited in wwPDB and EMDB under the following accession numbers PDB ID 7Y6F, EMD-33639 (Apoform), PDB ID 7Y6G, EMD-33640 (holoform-I), PDB ID 7Y6P, EMD-33645 (holoform-III), PDB ID 8JAX, EMD-36137 (holoform-II), PDB ID 8JB0, EMD-36139 (holoform-II in C1 symmetry), and EMD-36140 (biomineral core only in C1 symmetry).
References
- 1. Papanikolaou G, Pantopoulos K. 2005. Iron metabolism and toxicity. Toxicol Appl Pharmacol. 202:199–211. [DOI] [PubMed] [Google Scholar]
- 2. Simão M, Leite RB, Leonor Cancela M. 2020. Expression of four new ferritins from grooved carpet shell clam Ruditapes decussatus challenged with Perkinsus olseni and metals (Cd, Cu and Zn). Aquat Toxicol. 229:105675. [DOI] [PubMed] [Google Scholar]
- 3. Galaris D, Barbouti A, Pantopoulos K. 2019. Iron homeostasis and oxidative stress: an intimate relationship. Biochim Biophys Acta Mol Cell Res. 1866(12):118535. [DOI] [PubMed] [Google Scholar]
- 4. Hintze KJ, Theil EC. 2006. Cellular regulation and molecular interactions of the ferritins. Cell Mol Life Sci. 63:591–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bradley JM, Le Brun NE, Moore GR. 2016. Ferritins: Furnishing proteins with iron. J Biol Inorg Chem. 21:13–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Smith JL. 2004. The physiological role of ferritin-like compounds in bacteria. Crit Rev Microbiol. 30:173–185. [DOI] [PubMed] [Google Scholar]
- 7. Wang W, Knovich MA, Coffman LG, Torti FM, Torti SV. 2010. Serum ferritin: past, present and future. Biochim Biophys Acta. 1800(8):760–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Arosio P, Adelman TG, Drysdale JW. 1978. On ferritin heterogeneity. Further evidence for heteropolymers. J Biol Chem. 253(12):4451–4458. [PubMed] [Google Scholar]
- 9. Theil EC, Tosha T, Behera RK. 2016. Solving biology's iron chemistry problem with ferritin protein nanocages. Acc Chem Res. 49(5):784–791. [DOI] [PubMed] [Google Scholar]
- 10. Dautant A, Meyer JB, Yariv J, Sweet RM, Frolow F. 1998. Structure of a monoclinic crystal form of cytochrome b1 (bacterioferritin) from E. coli. Acta Crystallogr D Biol Crystallogr. 54(1):16–24. [DOI] [PubMed] [Google Scholar]
- 11. Tosha T, Ng HL, Bhattasali O, Alber T, Theil EC. 2010. Moving metal ions through ferritin-protein nanocages from three-fold pores to catalytic sites. J Am Chem Soc. 132:14562–14569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lawson DM, et al. 1989. Identification of the ferroxidase centre in ferritin. FEBS Lett. 254:207–210. [DOI] [PubMed] [Google Scholar]
- 13. Wang W, Zhang Y, Zhao G, Wang H. 2021. Ferritin with atypical ferroxidase centers takes B-channels as the pathway for Fe(2+) uptake from mycoplasma. Inorg Chem. 60:7207–7216. [DOI] [PubMed] [Google Scholar]
- 14. Pullin J, et al. 2021. Iron oxidation in Escherichia coli bacterioferritin ferroxidase centre, a site designed to react rapidly with H2 O2 but slowly with O2. Angew Chem Int Ed Engl. 60:8361–8369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Janowski R, Auerbach-Nevo T, Weiss MS. 2008. Bacterioferritin from Mycobacterium smegmatis contains zinc in its di-nuclear site. Protein Sci. 17:1138–1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Yao H, et al. 2012. The structure of the BfrB-Bfd complex reveals protein-protein interactions enabling iron release from bacterioferritin. J Am Chem Soc. 134:13470–13481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Granier T, et al. 2003. Structural description of the active sites of mouse L-chain ferritin at 1.2 Å resolution. J Biol Inorg Chem. 8:105–111. [DOI] [PubMed] [Google Scholar]
- 18. Ahn B, et al. 2018. Four-fold channel-nicked human ferritin nanocages for active drug loading and pH-responsive drug release. Angew Chem Int Ed Engl. 57:2909–2913. [DOI] [PubMed] [Google Scholar]
- 19. Russo CJ, Passmore LA. 2014. Electron microscopy: ultrastable gold substrates for electron cryomicroscopy. Science 346:1377–1380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Montemiglio LC, et al. 2019. Cryo-EM structure of the human ferritin-transferrin receptor 1 complex. Nat Commun. 10:1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Pozzi C, et al. 2017. Chemistry at the protein-mineral interface in L-ferritin assists the assembly of a functional (mu(3)-oxo)Tris[(mu(2)-peroxo)] triiron(III) cluster. Proc Natl Acad Sci U S A. 114:2580–2585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Pan YH, et al. 2009. 3D Morphology of the human hepatic ferritin mineral core: new evidence for a subunit structure revealed by single particle analysis of HAADF-STEM images. J Struct Biol. 166:22–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. López-Castro JD, et al. 2012. A new approach to the ferritin iron core growth: influence of the H/L ratio on the core shape. Dalt Trans. 41:1320–1324. [DOI] [PubMed] [Google Scholar]
- 24. Thompson CJ, Fink D, Nguyen LD. 2002. Principles of microbial alchemy: insights from the Streptomyces coelicolor genome sequence. Genome Biol. 3(7):REVIEWS1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Choi JS, Seok YJ, Cho YH, Roe JH. 2022. Iron-induced respiration promotes antibiotic resistance in actinomycete bacteria. mBio 13(2):e0042522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Alam K, et al. 2022. Streptomyces: The biofactory of secondary metabolites. Front Microbiol. 13:968053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Crabb E, Moore E. 2010. Metals and life. Cambridge: Royal Society of Chemistry. [Google Scholar]
- 28. Quintana C, Cowley JM, Marhic C. 2004. Electron nanodiffraction and high-resolution electron microscopy studies of the structure and composition of physiological and pathological ferritin. J Struct Biol. 147:166–178. [DOI] [PubMed] [Google Scholar]
- 29. Bradley JM, Moore GR, Le Brun NE. 2017. Diversity of Fe(2+) entry and oxidation in ferritins. Curr Opin Chem Biol. 37:122–128. [DOI] [PubMed] [Google Scholar]
- 30. Bradley JM, Svistunenko DA, Moore GR, Le Brun NE. 2017. Tyr25, Tyr58 and Trp133 of Escherichia coli bacterioferritin transfer electrons between iron in the central cavity and the ferroxidase centre. Metallomics 9:1421–1428. [DOI] [PubMed] [Google Scholar]
- 31. Turano P, Lalli D, Felli IC, Theil EC, Bertini I. 2010. NMR reveals pathway for ferric mineral precursors to the central cavity of ferritin. Proc Natl Acad Sci U S A. 107:545–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Yasmin S, Andrews SC, Moore GR, Le Brun NE. 2011. A new role for heme, facilitating release of iron from the bacterioferritin iron biomineral. J Biol Chem. 286:3473–3483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Pullin J, et al. 2021. Electron transfer from haem to the di-iron ferroxidase centre in bacterioferritin. Angew Chem Int Ed Engl. 60:8376–8379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Khare G, et al. 2011. Ferritin structure from Mycobacterium tuberculosis: comparative study with homologues identifies extended C-terminus involved in ferroxidase activity. PLoS One 6:e18570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Zeth K, Offermann S, Essen L-O, Oesterhelt D. 2004. Iron-oxo clusters biomineralizing on protein surfaces: structural analysis of Halobacterium salinarum DpsA in its low- and high-iron states. Proc Natl Acad Sci U S A. 101:13780–13785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ciambellotti S, Pozzi C, Mangani S, Turano P. 2020. Iron biomineral growth from the initial nucleation seed in L-ferritin. Chemistry 26:5770–5773. [DOI] [PubMed] [Google Scholar]
- 37. Ebrahimi KH, Hagedoorn PL, Hagen WR. 2013. A conserved tyrosine in ferritin is a molecular capacitor. Chembiochem. 14:1123–1133. [DOI] [PubMed] [Google Scholar]
- 38. Rui H, Rivera M, Im W. 2012. Protein dynamics and ion traffic in bacterioferritin. Biochemistry 51:9900–9910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Pettersen EF, et al. 2004. UCSF Chimera—a visualization system for exploratory research and analysis. J Comput Chem. 25:1605–1612. [DOI] [PubMed] [Google Scholar]
- 40. Rivera M. 2017. Bacterioferritin: structure, dynamics, and protein-protein interactions at play in iron storage and mobilization. Acc Chem Res. 50:331–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Ueno T, et al. 2009. Process of accumulation of metal ions on the interior surface of apo-ferritin: crystal structures of a series of apo-ferritins containing variable quantities of Pd(II) ions. J Am Chem Soc. 131:5094–5100. [DOI] [PubMed] [Google Scholar]
- 42. Zheng SQ, et al. 2017. Motioncor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat Methods. 14:331–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Punjani A, Rubinstein J, Fleet DJ, Brubaker MA. 2017. cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat Methods. 14:290–296. [DOI] [PubMed] [Google Scholar]
- 44. Zivanov J, et al. 2018. New tools for automated high-resolution cryo-EM structure determination in RELION-3. Elife 7:e42166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Rohou A, Grigorieff N. 2015. CTFFIND4: fast and accurate defocus estimation from electron micrographs. J Struct Biol. 192:216–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Adams PD, et al. 2010. PHENIX: a comprehensive python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 66:213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Emsley P, Cowtan K. 2004. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 60:2126–2132. [DOI] [PubMed] [Google Scholar]
- 48. Chen VB, et al. 2010. Molprobity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr. 66:12–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. The PyMOL Molecular Graphics System . Version 1.8. Schrödinger, LLC.
- 50. Pettersen EF, et al. 2021. UCSF ChimeraX: structure visualization for researchers, educators, and developers. Protein Sci. 30(1):70–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Otwinowski Z, Minor W. 1997. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzym. 276:307–326. [DOI] [PubMed] [Google Scholar]
- 52. Matthews BW. 1968. Solvent content of protein crystals. J Mol Biol. 33:491–497. [DOI] [PubMed] [Google Scholar]
- 53. McCoy AJ. 2007. Solving structures of protein complexes by molecular replacement with Phaser. Acta Crystallogr D Biol Crystallogr. 63:32–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Terwilliger TC, et al. 2008. Iterative model building, structure refinement and density modification with the PHENIX AutoBuild wizard. Acta Crystallogr D Biol Crystallogr. 64:61–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Afonine PV, et al. 2010. Joint X-ray and neutron refinement with phenix.refine. Acta Crystallogr D Biol Crystallogr. 66:1153–1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The coordinates of the ScBfr crystal structure was deposited in wwPDB under the accession number 5XX9. The coordinates of the ScBfr nanocage structures and the cryo-EM density map are deposited in wwPDB and EMDB under the following accession numbers PDB ID 7Y6F, EMD-33639 (Apoform), PDB ID 7Y6G, EMD-33640 (holoform-I), PDB ID 7Y6P, EMD-33645 (holoform-III), PDB ID 8JAX, EMD-36137 (holoform-II), PDB ID 8JB0, EMD-36139 (holoform-II in C1 symmetry), and EMD-36140 (biomineral core only in C1 symmetry).