Abstract
Evodia rutaecarpa (Evodia) is a Chinese herbal medicine with analgesic and anti-neurodegenerative properties. However, whether Evodia compounds can be applied for the comorbid pain of Alzheimer's disease (AD) and the underlying mechanisms remain unclear. Herein, 137 common targets of Evodia between AD and pain were predicted from drug and disease target databases. Subsequently, protein-protein interaction (PPI) network, protein function module construction, and bioinformatics analyses were used to analyze the potential relationship among targets, pathways, and diseases. Evodia could simultaneously treat AD comorbid pain through multi-target, multi-component, and multi-pathway mechanisms, and inflammation was an important common phenotype of AD and pain. The relationship between important transcription factors such as RELA, NF-κB1, SP1, STAT3, and JUN on IL-17, TNF, and MAPK signaling pathways might be potential mechanisms of Evodia. Additionally, 10 candidate compounds were predicted, and evodiamine might be the effective active ingredient of Evodia in treating AD or pain. In summary, this study provided a reference for subsequent research and a novel understanding and direction for the clinical use of evodiamine to treat AD patients with comorbid pain.
Keywords: Evodia rutaecarpa, Alzheimer's disease, Pain, Evodiamine, Network pharmacology
1. Introduction
Alzheimer's disease (AD) is the most common neurodegenerative disease in the elderly. In 2019, about 13.14 million people were diagnosed with AD in China, accounting for 20% of the total number of AD cases worldwide [1]. Current investigations found that almost 50% of AD patients suffer from pain [[2], [3], [4]], and more than 75% experience pain during AD progression [5]. However, pain management in AD patients remains challenging due to the complex link between AD and pain [6,7]. Mild and moderate AD patients can still have pain. Nevertheless, patients lose almost all ability to express the pain in late AD stages, which makes the clinical diagnosis of pain more difficult [8]. Treatment is another clinical issue for AD patients besides evaluation and diagnosis. Non-steroidal anti-inflammatory drugs (NSAIDs), such as acetaminophen, are the first-line medicine for AD-related pain [9,10]. Except for the compliance and overdose risk of AD patients, NSAIDs might not present enough efficacy for chronic pain or neuralgia [11]. Other analgesics, such as opioids, still lack reliable clinical evidence and might increase adverse events when combined with psychotropic drugs to treat AD [12,13]. Meanwhile, the incidence of pain significantly increases with age, and pain level is positively correlated with the severity of cognitive impairment [14]. Therefore, the positive vicious cycle in AD patients with comorbid pain must be addressed.
Neuroinflammation is an important bridge linking pain and cognitive decline by regulating microglia function (e.g., pro-inflammatory factors, phagocytosis), synaptogenesis, and dysfunction in several brain areas (e.g., locus coeruleus, frontal cortex) [15,16]. Evodia rutaecarpa (Evodia) has been used to treat pain or AD patients in traditional Chinese medicine (TCM) [17,18]. Evodia and its compounds (e.g., evodiamine) can be anti-inflammation and anti-neurodegeneration candidate agents [18,19]. In a rat model of neuropathic pain, evodiamine administration significantly inhibited the elevation of inflammatory cytokines, ameliorated mitochondrial dysfunction, and maintained oxidative stress levels [20]. Rutaecarpine, an active compound of Evodia, can attenuate the pathological changes of AD-like mice, alleviate cognitive and memory impairments and enhance the plasticity of neural protrusions [21]. Despite the TCM emphasis on the comprehensive method to regulate homeostasis through the “sovereign (Jun), minister (Chen), assistant (Zuo) and guide (Shi)” herb medicines in the formula, further research is still necessary to determine the active compounds for reducing toxicity and side effects [22]. In recent years, network pharmacology has been used to explore potential targets among several diseases through a comprehensive network analysis to reveal the relationship among known TCM herbal targets, genes, and pathways from multiple dimensions [23,24]. Thus, network pharmacology is a convenient and feasible method to undermine potential active compounds and common targets of Evodia in AD patients with comorbid pain.
Therefore, we hypothesized that Evodia or its compounds might be used to develop treatments for AD patients with comorbid pain. Thus, we used network pharmacology to predict the common targets of Evodia for AD and pain. Then, we conducted bioinformatic analyses to determine the potential targets and candidate compounds, which supported our hypothesis and might facilitate future studies.
2. Methods
2.1. Collection of potential drug and disease targets
Different disease databases can be used for different research purposes, including studying the molecular basis of human diseases and their comorbidities and analyzing the properties of disease genes. First, we collected targets from multiple databases: Human Phenotype Ontology (HPO, http://www.human-phenotype-ontology.org) [25], DisGeNET (https://www.disgenet.org) [26], National Center for Biotechnology Information (NCBI, https://www.ncbi.nlm.nih.gov/gene), and Pharmacogenomics Knowledgebase (PharmGKB, https://www.pharmgkb.org) [27]. Subsequently, “Alzheimer's disease” and “pain” were used as search terms for AD and pain-related genes. Search results were collected from each database, duplicate values were removed, and the final results were the pathogenic targets of AD and pain.
The Traditional Chinese Medicine Systems Pharmacology and Analysis Platform (TCMSP, https://old.tcmsp-e.com/tcmsp.php) [28] contains the chemical composition, the action target, and related pharmacokinetic properties of natural compounds. We used “wuzhuyu" (Evodia pinyin name) as the keyword to obtain relevant drug targets and selected “Homo sapiens” as the species in the UniProt (https://www.uniprot.org) [29] database to standardize the retrieved target names into gene names. Next, we used Venny 2.1.0 (http://bioinfogp.cnb.csic.es/tools/Venny/index.html) to analyze the common targets of AD and pain, as well as the common targets of Evodia in AD and pain treatment. Then, gene names and inter-relationships of drug-treated diseases were entered into Cytoscape 3.9.1 (https://cytoscape.org) [30] to obtain the visual action network of Evodia in treating AD and pain.
2.2. Protein-protein interaction (PPI) network construction and network topology analysis
The PPI network comprises individual protein interactions, participating in signal transmission, gene expression regulation, energy, substance metabolism, cell cycle regulation, and other life processes. The STRING 11.5 platform (https://cn.string-db.org [31]) was used to determine protein-protein interactions between common targets. The species was selected as Homo sapiens, with the “highest confidence” set to 0.9 and the “Clustering Options” to “k-means clustering,” and the “number of clusters” to 3. Subsequently, the results of the PPI network analysis were imported into Cytoscape 3.9.1 software in the TSV format, and the Network Analyzer [32] plug-in was used for topology analysis to obtain the node degree and edge betweenness of each target gene to highlight their importance and eliminate targets with node degree of 0.
2.3. Enrichment analysis of the intersection of drug-disease targets
To understand the biological significance behind the targets, we used the Database for Annotation, Visualization, and Integrated Discovery (DAVID, https://david.ncifcrf.gov) [33,34] to perform Gene Ontology (GO) functional analysis of the intersection of drug-disease targets. GO consists of three ontologies: process (BP), cellular component (CC), and molecular function (MF). We also performed The Kyoto Encyclopedia of Genes and Genomes (KEGG, https://www.kegg.jp) [35] pathway enrichment analysis of these targets at the molecular level using the online bioinformatics tools (http://www.bioinformatics.com.cn). Then, we imported the intersection of these drug-disease targets into Metascape (http://Metascape.org) [36], and GO function and KEGG pathway enrichment analyses were performed again. The top 10 GO and top 20 KEGG pathway results were compared with DAVID and KEGG databases, respectively. At this time, the relationship between the top 20 targets with node degree in the PPI network and the intersection of KEGG pathways was input into Cytoscape 3.9.1 to obtain the network analysis diagram of the significant targets acting on the significant pathways.
2.4. Identification of the functional clusters of 107 intersection targets
Metascape is a platform that can cluster similar proteins and construct functional modules using the Molecular Complex Detection (MCODE) algorithm [37]. With 107 targets imported, MCODE analysis was used to identify protein relationships between molecular complexes and cluster them into a gene cluster where each target has the same or a similar gene function. Additionally, transcription factors that regulate other targets were further predicted, and information was obtained on target interactions.
2.5. Creation of the network diagram
Next, we used a Bioinformatics Analysis Tool for the Molecular Mechanism of Traditional Chinese Medicine (BATMAN-TCM, http://bionet.ncpsb.org/batman-tcm/) [38] to analyze Evodia. We used “wuzhuyu” as the search term and set the “Score cutoff” to 20, the “Adjusted P-value” to 0.05, and the species to “Homo sapiens.” The multi-component, multi-target, multi-pathway, and multi-disease network diagram was obtained for Evodia. The screening condition for disease enrichment results was set to p < 0.05, and the top ten results were ranked from large to small by target number. After the pain-related components of AD were obtained, they were entered into the TCMSP database for bioactivity prediction. The screening threshold conditions were: oral bioavailability (OB) ≥ 30%, similarity (DL) ≥ 0.18, and blood-brain barrier (BBB) ≥ 0.3. The relationship between eligible active ingredients and target diseases was plotted. Finally, GO and KEGG analyses were performed again for the targets of the component of Evodia.
3. Result
3.1. Common targets of Evodia for AD and pain
After retrieving and deleting duplicate genes from multiple disease databases, a total of 3507 AD-related targets were obtained, among which 200 were related to the drug target of Evodia. There were 2277 targets associated with pain, 157 of which were associated with Evodia. According to the Venn diagram intersection, 1101 common targets of AD and pain (Fig. 1A) and 137 overlapping targets of Evodia and AD-pain (Fig. 1B) were detected. Then, the visualization network of “AD targets”- “common targets of AD & Pain”- “Pain targets” indicated the target genes involved in AD and Pain (Fig. 1C).
Fig. 1.
The plot of intersection and interaction between drug and disease targets. A. Purple represents targets of AD, yellow represents targets of pain, and yellow-green represents targets of AD comorbid pain. B. Purple represents targets associated with Evodia in AD, yellow represents targets associated with Evodia in pain, and yellow-green represents targets associated with Evodia in comorbidity. C. Cytoscape 3.9.1 clearly showed the relationship of the targets between Evodia and AD comorbid pain. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
3.2. PPI network and topological importance of common targets of AD and pain
Then, we constructed a PPI network to systematically understand the mutual relationship between the expression and function of 137 overlapping targets. These targets were classified by “k-means clustering.” In the interaction network, a cluster comprises genes with similar functions (Fig. 2A). Thirty genes had no relationship with other genes and were mainly concentrated in the blue network. The red and green networks mainly comprised genes with more relationships and similar functions. Among them, the gene with the most relationships in the red network was AKT1, and JUN was related to 36 other genes in the green network. Then, the PPI network was imported into Cytoscape for topological analysis. We conducted the topological network analysis after removing 30 targets with node degree 0 (Fig. 2B). The connectivity between nodes, the node degree, represents the interaction between targets. Further, we selected the genes that were most related to each other. The node degree was reflected by node color and size, and the edge betweenness between target genes was reflected by the color depth and thickness of connecting lines. The top 10 targets are Jun proto-oncogene (JUN, node degree = 36), RELA proto-oncogene (RELA, node degree = 30), AKT Serine/Threonine kinase 1 (AKT1, node degree = 28), mitogen-activated protein kinase 1 (MAPK1, node degree = 27), interleukin 6 (IL6, node degree = 25), tumor necrosis factor (TNF, node degree = 25), catenin beta 1 (CTNNB1, node degree = 24), Fos proto-oncogene (FOS, node degree = 24), mitogen-activated protein kinase 14 (MAPK14, node degree = 24), and Tumor Protein P53 (TP53, node degree = 23). Larger nodes and darker colors suggest greater importance in the biological function of the disease.
Fig. 2.
PPI network. A. PPI network analysis was performed on overlapping targets and classified by K-means clustering. B. After removing out the targets with a node degree of 0 and performing a topological analysis of the PPI network, the weight of the targets is represented by the size and color depth of the nodes. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
3.3. GO and KEGG pathway enrichment analyses of common targets of AD and pain
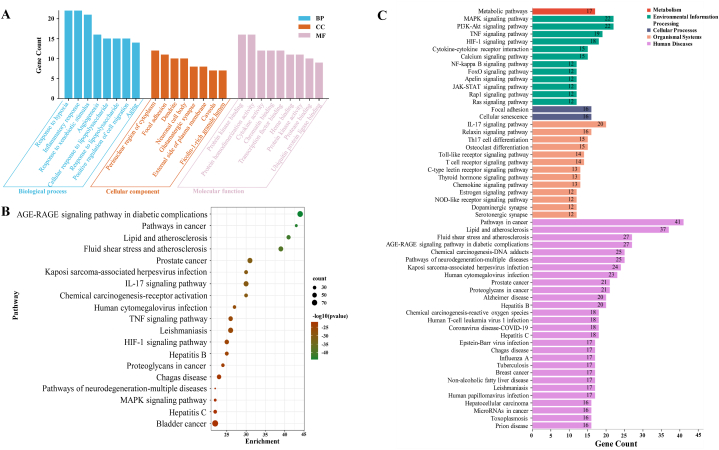
GO consists of three ontologies: biological process (BP), cellular component (CC), and molecular function (MF) of a gene. We used 107 targets with mutual interaction in the DAVID database and Metascape platform. Evodia was mainly enriched in response to hypoxia and inflammatory response in the BP category. For CC, Evodia was mainly enriched in the cytoplasm, dendrite, and neuronal cell body. For MF, it was mainly enriched in protein kinase binding, protein homodimerization activity, and cytokine activity (Fig. 3A). Additionally, 18 KEGG-enriched pathways were obtained after the intersection with Metascape (Fig. 3B, pathways arranged by p-value) and KEGG database (Fig. 3C, pathways arranged by classification and the number of enriched genes). Enriched pathways included pathways in cancer, the MAPK signaling pathway, the AGE-RAGE signaling pathway in diabetic complications, and the IL-17 signaling pathway.
Fig. 3.
GO and KEGG analysis. A. After GO and Metascape analysis of 107 key targets, the top 10 results of the two rankings were taken as the same. B. KEGG pathways enriched in Metascape platform (the size of the nodes indicates the number of enriched genes, and the color depth indicates the size of p-value). C. Results of KEGG enrichment analysis (different colors indicate the different systems of action, and the pathways in each system are arranged in descending order by the number of enriched genes). (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
Then, we used Cytoscape 3.9.1 to display the enrichment relationship between the top 20 node degree targets. The node degree of HIF1A, TGFB1, and VEGFA was 13, and 21 targets were in the top 20 in the PPI network. These 18 pathways are in the network graph (Fig. 4), which clarifies the genes' position in the pathway. In these pathways, genes mainly focused on inflammatory and oxidative stress-related responses to exert their biological significance.
Fig. 4.
Enrichment network diagram of the top 20 (21 in total) core targets in the PPI network and KEGG pathways (intersection of the top 20 in KEGG enrichment analysis and the top 20 in Metascape analysis).
3.4. MCODE analysis of common targets of AD and pain
Furthermore, we implemented the MCODE cluster analysis for the 107 targets using Metascape. We obtained five modules of Evodia for common targets of AD and pain (Fig. 5A). Among the top 20 node degree targets (21 in total) in the PPI network, 9 genes were enriched in MCODE1 (score = 4.86) (Fig. 5B, indicated by the gene name in orange), and 11 genes were enriched in MCODE2 (score = 6.38) (Fig. 5C, indicated by the gene name in orange). MCODE1 and MCODE2 are more relevant and closely related to Evodia treatment regarding protein relationships. The pathways enriched in MCODE1 were cancer, spinal cord injury, and IL-18 signaling pathways. The key genes were MAPK1 (degree = 18), AKT1 (degree = 13), CTNNB1 (degree = 13), and EGFR (degree = 13). The pathways enriched in MCODE2 were interleukins, PID AP1, and cytokine signaling pathways in the immune system. The key genes in MCODE2 were RELA (degree = 20), JUN (degree = 19), FOS (degree = 18), and TNF (degree = 17). Among the 21 genes, the targets regulated by the top 10 transcription factors with p-values (Fig. 5D) are represented by octagons in Fig. 5B and C, demonstrating the regulatory action of the top 10 transcription factors, except for MAPK1 and MAPK14.
Fig. 5.
MCODE cluster analysis and protein function module. A. MCODE cluster analysis was performed on 107 targets. B. Pathway enrichment results for MCODE1. Gene names in orange indicate genes belonging to the top 20 node degrees (21 in total) in the PPI network. C. Pathway enrichment results for MCODE2. D. Bar graph of transcription factors enrichment analysis (colored by p-values) (Genes of the top 20 node degrees (21 in total) regulated by the top 10 transcription factors are shown in B and C as an octagonal shape). (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
3.5. Construction of the component-target-pathway-disease network
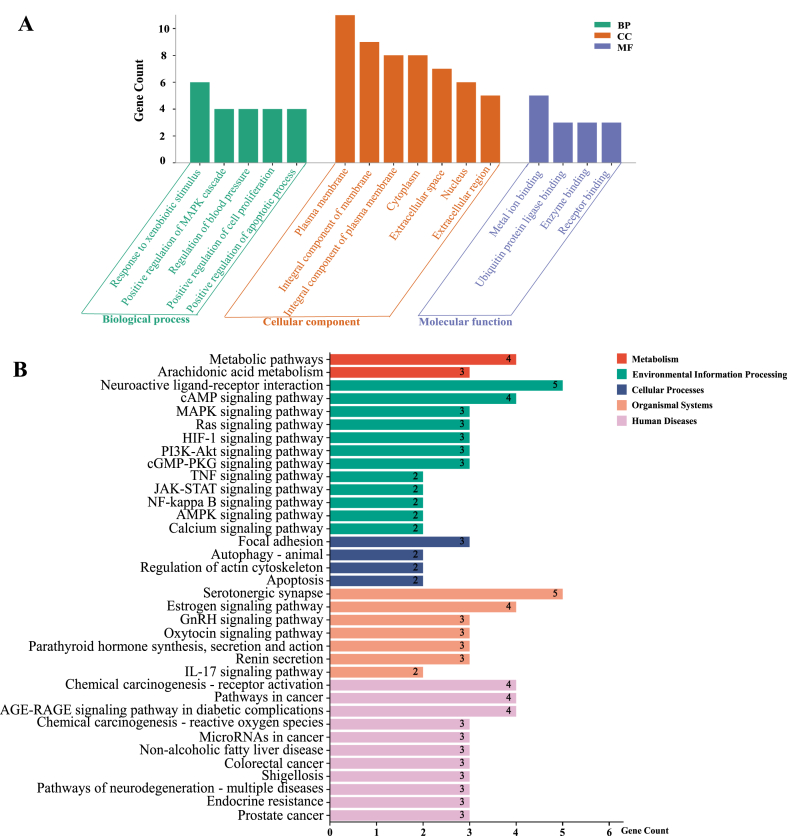
The BATMAN-TCM platform was used to enrich the drug network and disease phenotype of Evodia. We obtained a simplified network diagram of component-target-pathway-disease, in which the red squares indicated the main target diseases (Fig. 6A). The top 10 enriched phenotypes of Evodia-related diseases were mainly related to analgesics, with target diseases respectively ranked fourth (AD), fifth (pain, unspecified) and 10th (pain) (Fig. 6B). In total, we summarized 15 components involved in the occurrence and development of the target diseases, and their enrichment relationships with the four target diseases are shown in Fig. 6C. Finally, these results were input into the TCMSP database for biological activity prediction, and 10 eligible biological compounds were obtained (Table 1). Evodiamine had the highest OB (OB = 86.02%) and DL (DL = 0.64), followed by goshuyuamide I (OB = 83.19%) and evodiamide (OB = 73.77). After GO and KEGG analysis of the targets of evodiamine, which is the most likely bioactive component, it mainly concentrated in response to foreign stimuli in the BP category. For CC, evodiamine was mainly enriched in plasma membrane and cytoplasm. For MF, it was mainly enriched in ion binding and enzyme binding (Fig. 7A). In addition, 36 pathways are enriched in the KEGG database (Fig. 7B), including Metabolic pathways, Neuroactive ligand-receptor interaction, MAPK, TNF, NF-kappa B, JAK-STAT and IL-17 signaling pathways.
Fig. 6.
Drug network, disease phenotypes, and pathways of action associated with Evodia. A. Evodia-related components - targets – way – disease network diagram (purple heptagon nodes represent active ingredients of Evodia, blue star nodes represent target genes, yellow circle nodes represent enrichment pathways, green square nodes represent enrichments of the disease, in the middle of the four red squares represent the disease that would be discussed in this paper). B. Disease phenotypes associated with Evodia (arranged in descending order by the number of enriched genes). C. Relationship between target disease and corresponding active ingredient. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
Table 1.
Compounds related to Evodia that met the criteria in the BATMAN-TCM platform (OB-oral bioavailability, DL-drug likeness, BBB- blood-brain barrier).
| Mol ID | Molecule Name | OB (%) | DL | BBB |
|---|---|---|---|---|
| MOL003958 | Evodiamine | 86.02 | 0.64 | 0.85 |
| MOL004018 | Goshuyuamide I | 83.19 | 0.39 | 0.64 |
| MOL004014 | Evodiamide | 73.77 | 0.28 | 0.81 |
| MOL003974 | Evocarpine | 48.66 | 0.36 | 1.17 |
| MOL003950 | 1-Methyl-2-[(Z)-undec-6-enyl]-4-quinolone | 48.48 | 0.27 | 1.14 |
| MOL003947 | 1-Methyl-2-[(Z)-pentadec-10-enyl]-4-quinolone | 48.45 | 0.46 | 1.11 |
| MOL003972 | 1-Methyl-2-nonyl-4-quinolone | 48.42 | 0.2 | 1.21 |
| MOL003964 | 1-Methyl-2-undecyl-4-quinolone | 47.59 | 0.27 | 1.19 |
| MOL003957 | 1-Methyl-2-pentadecyl-4-quinolone | 44.52 | 0.46 | 1.05 |
| MOL002662 | Rutaecarpine | 40.3 | 0.6 | 0.71 |
Fig. 7.
GO and KEGG analysis. A. GO analysis results of the targets corresponding to evodiamine. B. Pathway enrichment results of the targets corresponding to evodiamine (different colors indicate the different systems of action, and the pathways in each system are arranged in descending order by the number of enriched genes). (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
4. Discussion
Evodia is a famous Chinese herbal medicine with analgesic and anti-neurodegeneration potential in chronic pain and AD patients, respectively. However, whether the compounds in Evodia can treat AD comorbid pain patients remains unknown. Herein, we predicted 137 common targets of AD and pain of Evodia using drug and disease databases. Then, 107 common targets of AD and pain were obtained in the PPI and topological network analysis. Then, the bioinformatic analysis suggested that the common targets of AD and pain of Evodia were enriched for inflammation, oxidative stress, and neurodegenerative functions and pathways. Furthermore, we predicted 10 candidate compounds for both target AD and pain-related diseases that might facilitate future studies to explore the potential mechanism and treatment for AD comorbid pain patients.
In the current study, we predicted and analyzed the 107 targets of 137 common targets of AD and pain by topological network analysis and GO and pathway enrichment analysis. Our topological and PPI analysis showed that the top 21 targets are associated with inflammatory and neurodegenerative pathways. Inflammatory-related biological processes (inflammatory response) and pathways (AGE-RAGE, IL-17, and TNF signaling pathways) were also enriched in the GO and pathway analysis. Previous studies have suggested that IL-17 could trigger neurocognitive deficits in early-stage AD patients [39] and is a potential target for chronic pain [40]. The therapeutic potential of TNF in AD [41] and chronic pain [42] have also been reported. These results suggested that inflammation was a vital common phenotype in AD and pain, consistent with our current results.
Moreover, to explore and understand the potential function and interaction of the 107 common targets, we used Metascape to provide more comprehensive results across multiple databases [43]. The top 21 common targets based on nodes degree were mainly enriched in MCODE1 and MCODE2. The enriched functions and pathways of MCODE1 and MCODE2 were related to spinal cord injury, interleukins, and cytokine in the immune system. Besides, RELA, NF-κB1, SP1, STAT3, and JUN are the top 5 enriched transcriptional factors among the 107 common targets.
RELA (NF-κB p65) and NF-κB1 (NF-κB p50), main members of the NF-κB family, form the p65/p50 complex to regulate the inflammatory response in the nervous system [44]. When pain hypersensitivity occurs, NF-κB signaling pathway activates downstream pro-inflammatory factors such as IL-1β and IL-6, resulting in significantly increased expression levels of inflammatory mediators [45]. Under oxidative stress, RELA and NF-κB1 are activated and promote the production of downstream inflammatory mediators, contributing to Aβ-induced neuronal apoptosis and aggravating the clinical symptoms of AD patients [46,47]. Kim et al. [48] knocked out IL-17, the upstream factor of the NF-κB signaling pathway, and then the mice showed a significant reduction of pain hypersensitivity, and the pain threshold recovered after the injection of IL-17. Similarly, researchers have found in AD mice that early silencing of IL-17 protects against cognitive deficits and synaptic dysfunction [49,50]. Thus, inhibition of IL-17 may have therapeutic effects in both pain mice and AD mice.
As a transcription factor, STAT3 is involved in the process of pain. IL-17 can activate STAT3 signal transduction through IL-6 induction and promote neutrophil aggregation [51]. The pro-inflammatory factor TNF-α also contributes to STAT3 activation and nerve excitability enhancement. Activation of the TNF-α/STAT3 pathway increases hypersensitivity and subsequently mediates injury-induced neuropathic pain [52]. Inhibition of the IL-6/STAT3 pathway in rats can also improve the development of pain and comorbidities [53]. In the pathological changes of AD, TNF signal transduction is involved in cell necrosis in neurodegeneration. STAT3 participates in Aβ-induced microglial activation and inflammatory cytokine release [54,55]. In contrast, in STAT3-null AD mice, the clearance pathway of Aβ is upregulated, neuronal apoptosis is inhibited, and mice eventually show improved learning and memory function [56,57]. The evidence is consistent with our current projections.
In neuropathic pain models, SP1 binds to its corresponding site in the promoter of transient receptor potential vanilloid type-1 (TRPV1), an important component of the mechanism of mechanical nociception associated with persistent pain conditions [58]. Previous in vivo and in vitro experiments have confirmed that knocking down SP1 can improve neuropathic pain [59,60]. Previous reviews have shown that SP1 also regulates Aβ and tau protein expression, especially under oxidative stress, affecting the survival of neuronal cells in AD patients [61,62]. In both in vivo and in vitro studies, activation of TRPV1 can induce clearance of Aβ by mediating autophagy, thereby alleviating cognitive decline [63].
Previous studies have shown that after the IL-17 signaling pathway is activated, IL-17 induces the downstream c-JUN and c-FOS to bind to the corresponding sites, significantly increases COX-2 levels, and leads to the occurrence and development of inflammatory responses [64] Under neuropathic pain, c-JUN on the MAPK pathway is significantly activated, the expression of downstream inflammatory cytokines IL-1β and IL-6 increases, and the plasticity of neurons changes [65] In the brain of AD patients, the cascade activation of c-JUN promotes the apoptosis of neuronal cells and the development of neurodegeneration [66] Our results suggested that Evodia and its active components might improve cognitive function and pain symptoms by regulating neuroinflammation, the neuroimmune system, and the synergistic effects of RELA, NF-κB1, SP1, STAT3, and JUN on IL-17, TNF, and MAPK signaling pathways, playing an important role in AD and chronic pain conditions.
Based on the bioinformatics analysis, Evodia exhibits potential pharmacological effects to alleviate the comorbid pain of AD patients. The pharmacological network analysis showed that several active compounds of Evodia have analgesia, anticancer and anti-neurodegenerative effects, consistent with previous studies [[67], [68], [69]]. The relevant components of Evodia were analyzed under various conditions to understand the major and key bioactive components. Oral availability reflects the proportion of herbal medicine taken into human circulation. Evodiamine, goshuyuamide I, evodiamide, and evocarpine were the four monomers with the highest OB. The GO and KEGG analysis of evodiamine also showed a similar enriched GO terms and KEGG pathways of Evodia. This indicates that evodiamine is likely to participate in and play an important role in the medicinal benefits of Evodia. Evodiamine significantly alleviates mechanical pain and acute visceral neuralgia in mice by reducing peripheral hypersensitivity, significantly affecting neuroprotection, anti-inflammation, and cardiac protection [70]. In clinical practice, evodiamine is often isolated or purified as an analgesic and anticancer agent [71]. The biological functions of goshuyuamide I and evodiamide are still poorly understood. Evocarpine has significant antibacterial activity against Mycobacterium tuberculosis, Helicobacter pylori, and methicillin-resistant Staphylococcus aureus [[72], [73], [74]]. However, its analgesic and anti-inflammatory effects still lack credible evidence. Therefore, evodiamine might be an important compound of Evodia in its main functions, such as anti-inflammation, pain relief, and improvement of neurodegeneration. Other active compounds, such as goshuyuamide I and evodiamide, still require further investigation.
5. Conclusion
In summary, Evodia can inhibit inflammation and oxidative stress via the IL-17, TNF, and MAPK signaling pathways, which might be a potential therapeutic mechanism against AD comorbid pain. Evodiamine is the most bioactive and therapeutic potential compound of Evodia. Although the mechanism of a single pathway or a single gene to treat AD and pain has been clarified, the therapeutic effect of evodiamine on AD comorbid pain might be multi-target, multi-pathway, and multi-mechanism, and our research on this network has limitations. Subsequent studies are needed to investigate further and prove the interrelationship of NF-κB (including RELA and NF-κB1), SP1, STAT3, and AP-1 (including JUN and FOS) on IL-17, TNF, and MAPK signaling pathways, as well as to determine the clinical potential of evodiamine. At present, no research has been found on the treatment of AD comorbid pain with Evodia or its component Evodiamine. We predict its mechanism through network pharmacology, which will be verified on the basis of constructing an animal model of AD comorbid pain in the future. Overall, this study provided some clues and evidence for the clinical application of evodiamine in treating AD comorbid pain, improving neurological prognosis, and relieving cognitive dysfunction and analgesia.
Author contribution statement
Tao Tao; Huiyi Jiang: Conceived and designed the experiments; Performed the experiments; Analyzed and interpreted the data; Wrote the paper.
Xin Deng; Danping Li: Contributed reagents, materials, analysis tools or data.
Jiamin Qiu: Conceived and designed the experiments; Performed the experiments; Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data; Wrote the paper.
Funding statement
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Data availability statement
Data included in article/supplementary material/referenced in article.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- 1.Ren R., Qi J., Lin S., et al. The China Alzheimer report 2022. Gen. Psychiatr. 2022;35(1) doi: 10.1136/gpsych-2022-100751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barry H.E., Parsons C., Passmore A.P., et al. Pain in care home residents with dementia: an exploration of frequency, prescribing and relatives' perspectives. Int. J. Geriatr. Psychiatr. 2015;30(1):55–63. doi: 10.1002/gps.4111. [DOI] [PubMed] [Google Scholar]
- 3.Zwakhalen S.M., Koopmans R.T., Geels P.J., et al. The prevalence of pain in nursing home residents with dementia measured using an observational pain scale. Eur. J. Pain. 2009;13(1):89–93. doi: 10.1016/j.ejpain.2008.02.009. [DOI] [PubMed] [Google Scholar]
- 4.van Kooten J., Binnekade T.T., van der Wouden J.C., et al. A review of pain prevalence in Alzheimer's, vascular, frontotemporal and lewy body dementias. Dement. Geriatr. Cogn. Disord. 2016;41:220–232. doi: 10.1159/000444791. [DOI] [PubMed] [Google Scholar]
- 5.de Tommaso M., Arendt-Nielsen L., Defrin R., et al. Pain in neurodegenerative disease: current knowledge and future perspectives. Behav. Neurol. 2016;2016 doi: 10.1155/2016/7576292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cao S., Fisher D.W., Yu T., et al. The link between chronic pain and Alzheimer's disease. J. Neuroinflammation. 2019;16(1):204. doi: 10.1186/s12974-019-1608-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Manietta C., Labonté V., Thiesemann R., et al. Algorithm-based pain management for people with dementia in nursing homes. Cochrane Database Syst. Rev. 2022;4(4):CD013339. doi: 10.1002/14651858.CD013339.pub2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Borg C., Sala E., Chainay H., et al. How do patients with Alzheimer's disease imagine their pain? Eur. J. Pain. 2021;25(2):466–472. doi: 10.1002/ejp.1685. [DOI] [PubMed] [Google Scholar]
- 9.Corbett A., Husebo B., Malcangio M., et al. Assessment and treatment of pain in people with dementia. Nat. Rev. Neurol. 2012;8(5):264–274. doi: 10.1038/nrneurol.2012.53. [DOI] [PubMed] [Google Scholar]
- 10.McLachlan A.J., Bath S., Naganathan V., et al. Clinical pharmacology of analgesic medicines in older people: impact of frailty and cognitive impairment. Br. J. Clin. Pharmacol. 2011;71(3):351–364. doi: 10.1111/j.1365-2125.2010.03847.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Baron R., Binder A., Attal N., et al. Neuropathic low back pain in clinical practice. Eur. J. Pain. 2016;20(6):861–873. doi: 10.1002/ejp.838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cravello L., Di Santo S., Varrassi G., et al. Chronic pain in the elderly with cognitive decline: a narrative review. Pain Ther. 2019;8(1):53–65. doi: 10.1007/s40122-019-0111-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hartikainen S., Mäntyselkä P., Louhivuori-Laako K., et al. Concomitant use of analgesics and psychotropics in home-dwelling elderly people-Kuopio 75 + study. Br. J. Clin. Pharmacol. 2005;60(3):306–310. doi: 10.1111/j.1365-2125.2005.02417.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rajkumar Anto P., Ballard Clive, Jane Fossey, et al. Epidemiology of pain in people with dementia living in care homes: longitudinal course, prevalence, and treatment implications. J. Am. Med. Dir. Assoc. 2017;18(5):453.e1–453.e6. doi: 10.1016/j.jamda.2017.01.024. [DOI] [PubMed] [Google Scholar]
- 15.Sommer C., Leinders M., Üçeyler N. Inflammation in the pathophysiology of neuropathic pain. Pain. 2018;159(3):595–602. doi: 10.1097/j.pain.0000000000001122. [DOI] [PubMed] [Google Scholar]
- 16.Zhang J., Ke K.F., Liu Z., et al. Th17 cell-mediated neuroinflammation is involved in neurodegeneration of aβ1-42-induced Alzheimer's disease model rats. PLoS One. 2013;8(10) doi: 10.1371/journal.pone.0075786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liao J.F., Chiou W.F., Shen Y.C., et al. Anti-inflammatory and anti-infectious effects of Evodia rutaecarpa (Wuzhuyu) and its major bioactive components. Chin. Med. 2011;6(1):6. doi: 10.1186/1749-8546-6-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fang Z., Tang Y., Ying J., et al. Traditional Chinese medicine for anti-Alzheimer's disease: berberine and evodiamine from Evodia rutaecarpa. Chin. Med. 2020;15:82. doi: 10.1186/s13020-020-00359-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sun Q., Xie L., Song J., et al. Evodiamine: a review of its pharmacology, toxicity, pharmacokinetics and preparation researches. J. Ethnopharmacol. 2020;262 doi: 10.1016/j.jep.2020.113164. [DOI] [PubMed] [Google Scholar]
- 20.Wu P., Chen Y. Evodiamine ameliorates paclitaxel-induced neuropathic pain by inhibiting inflammation and maintaining mitochondrial anti-oxidant functions. Hum. Cell. 2019;32(3):251–259. doi: 10.1007/s13577-019-00238-4. [DOI] [PubMed] [Google Scholar]
- 21.Zhao B., Wang Y., Liu R., et al. Rutaecarpine ameliorated high sucrose-induced Alzheimer's disease like pathological and cognitive impairments in mice. Rejuvenation Res. 2021;24(3):181–190. doi: 10.1089/rej.2020.2349. [DOI] [PubMed] [Google Scholar]
- 22.Huo M.Q., Peng S., Ren Y., et al. Discovery and application of traditional Chinese medicine efficacy markers based on systematic traditional Chinese medicine. Zhongguo Zhongyao Zazhi. 2020;45(14):3245–3250. doi: 10.19540/j.cnki.cjcmm.20200210.402. Chinese. [DOI] [PubMed] [Google Scholar]
- 23.Hopkins A.L. Network pharmacology. Nat. Biotechnol. 2007;25(10):1110–1111. doi: 10.1038/nbt1007-1110. [DOI] [PubMed] [Google Scholar]
- 24.Casas A.I., Hassan A.A., Larsen S.J., et al. From single drug targets to synergistic network pharmacology in ischemic stroke. Proc. Natl. Acad. Sci. U. S. A. 2019;116(14):7129–7136. doi: 10.1073/pnas.1820799116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Robinson Peter N., Köhler Sebastian, Bauer Sebastian, et al. The Human Phenotype Ontology: a tool for annotating and analyzing human hereditary disease. Am. J. Hum. Genet. 2008;83(5):610–615. doi: 10.1016/j.ajhg.2008.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Piñero Janet, Bravo Àlex, Queralt-Rosinach Núria, et al. DisGeNET: a comprehensive platform integrating information on human disease-associated genes and variants. Nucleic Acids Res. 2017;45(D1):D833–D839. doi: 10.1093/nar/gkw943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Whirl-Carrillo M., McDonagh E.M., et al. Pharmacogenomics knowledge for personalized medicine. Clin. Pharmacol. Ther. 2012;92(4):414–417. doi: 10.1038/clpt.2012.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ru Jinlong, Peng Li, Wang Jinan, et al. TCMSP: a database of systems pharmacology for drug discovery from herbal medicines. J. Cheminf. 2014;6:13. doi: 10.1186/1758-2946-6-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Consortium UniProt. UniProt: a worldwide hub of protein knowledge. Nucleic Acids Res. 2019 Jan 8;47(D1):D506–D515. doi: 10.1093/nar/gky1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shannon Paul, Markiel Andrew, Owen Ozier, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13(11):2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Damian Szklarczyk, Gable Annika L., Lyon David, et al. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019;47(D1):D607–D613. doi: 10.1093/nar/gky1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Assenov Y., Ramírez F., Schelhorn S.E., et al. Computing topological parameters of biological networks. Bioinformatics. 2008;24(2):282–284. doi: 10.1093/bioinformatics/btm554. [DOI] [PubMed] [Google Scholar]
- 33.Huang Da Wei, Sherman Brad T., Lempicki Richard A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009;4(1):44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 34.Sherman Brad T., Ming Hao, Qiu Ju, et al. DAVID: a web server for functional enrichment analysis and functional annotation of gene lists (2021 update) Nucleic Acids Res. 2022;50(W1):W216–W221. doi: 10.1093/nar/gkac194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kanehisa Minoru, Furumichi Miho, Sato Yoko, et al. KEGG: integrating viruses and cellular organisms. Nucleic Acids Res. 2021;49(D1):D545–D551. doi: 10.1093/nar/gkaa970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhou Yingyao, Zhou Bin, Pache Lars, et al. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat. Commun. 2019;10(1):1523. doi: 10.1038/s41467-019-09234-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bader G.D., Hogue C.W. An automated method for finding molecular complexes in large protein interaction networks. BMC Bioinf. 2003;4:2. doi: 10.1186/1471-2105-4-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu Zhongyang, Guo Feifei, Wang Yong, et al. BATMAN-TCM: a bioinformatics analysis tool for molecular apiens m of traditional Chinese medicine. Sci. Rep. 2016;6 doi: 10.1038/srep21146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Brigas H.C., Ribeiro M., Coelho J.E., et al. IL-17 triggers the onset of cognitive and synaptic deficits in early stages of Alzheimer's disease. Cell Rep. 2021;36(9) doi: 10.1016/j.celrep.2021.109574. [DOI] [PubMed] [Google Scholar]
- 40.Jiang X., Zhou R., Zhang Y., et al. Interleukin-17 as a potential therapeutic target for chronic pain. Front. Immunol. 2022;13 doi: 10.3389/fimmu.2022.999407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Torres-Acosta N., O'Keefe J.H., O'Keefe E.L., et al. Therapeutic potential of TNF-α inhibition for Alzheimer's disease prevention. J Alzheimers Dis. 2020;78(2):619–626. doi: 10.3233/JAD-200711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Leung L., Cahill C.M. TNF-alpha and neuropathic pain--a review. J. Neuroinflammation. 2010;7:27. doi: 10.1186/1742-2094-7-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhou Y., Zhou B., Pache L., et al. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat. Commun. 2019;10(1):1523. doi: 10.1038/s41467-019-09234-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Giridharan S., Srinivasan M. Mechanisms of NF-κB p65 and strategies for therapeutic manipulation. J. Inflamm. Res. 2018;11:407–419. doi: 10.2147/JIR.S140188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chen X., Zhang B., Li J., et al. Celastrol attenuates incision-induced inflammation and pain associated with inhibition of the NF-κB signalling pathway via SARM. Life Sci. 2018;205:136–144. doi: 10.1016/j.lfs.2018.05.020. [DOI] [PubMed] [Google Scholar]
- 46.Cristiano C., Volpicelli F., Lippiello P., et al. Neutralization of IL-17 rescues amyloid-β-induced neuroinflammation and memory impairment. Br. J. Pharmacol. 2019;176(18):3544–3557. doi: 10.1111/bph.14586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shi Z.M., Han Y.W., Han X.H., et al. Upstream regulators and downstream effectors of NF-κB in Alzheimer's disease. J. Neurol. Sci. 2016;366:127–134. doi: 10.1016/j.jns.2016.05.022. [DOI] [PubMed] [Google Scholar]
- 48.Kim C.F., Moalem-Taylor G. Interleukin-17 contributes to neuroinflammation and neuropathic pain following peripheral nerve injury in mice. J. Pain. 2011;12(3):370–383. doi: 10.1016/j.jpain.2010.08.003. [DOI] [PubMed] [Google Scholar]
- 49.Brigas H.C., Ribeiro M., Coelho J.E., et al. IL-17 triggers the onset of cognitive and synaptic deficits in early stages of Alzheimer's disease. Cell Rep. 2021;36(9) doi: 10.1016/j.celrep.2021.109574. [DOI] [PubMed] [Google Scholar]
- 50.Vellecco V., Saviano A., Raucci F., et al. Interleukin-17 (IL-17) triggers systemic inflammation, peripheral vascular dysfunction, and related prothrombotic state in a mouse model of Alzheimer's disease. Pharmacol. Res. 2023;187 doi: 10.1016/j.phrs.2022.106595. [DOI] [PubMed] [Google Scholar]
- 51.Wang L., Yi T., Kortylewski M., et al. IL-17 can promote tumor growth through an IL-6-Stat3 signaling pathway. J. Exp. Med. 2009;206(7):1457–1464. doi: 10.1084/jem.20090207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ding H.H., Zhang S.B., Lv Y.Y., et al. TNF-α/STAT3 pathway epigenetically upregulates Nav 1.6 expression in DRG and contributes to neuropathic pain induced by L5-VRT. J. Neuroinflammation. 2019;16(1):29. doi: 10.1186/s12974-019-1421-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhao Y.T., Deng J., Liu H.M., et al. Adaptation of prelimbic cortex mediated by IL-6/STAT3/Acp 5 pathway contributes to the comorbidity of neuropathic pain and depression in rats. J. Neuroinflammation. 2022;19(1):144. doi: 10.1186/s12974-022-02503-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jayaraman A., Htike T.T., James R., et al. TNF-mediated neuroinflammation is linked to neuronal necroptosis in Alzheimer's disease hippocampus. Acta Neuropathol Commun. 2021;9(1):159. doi: 10.1186/s40478-021-01264-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.An X.Q., Xi W., Gu C.Y., et al. Complement protein C5a enhances the β-amyloid-induced neuro-inflammatory response in microglia in Alzheimer's disease. Med. Sci. 2018:116–120. doi: 10.1051/medsci/201834f120. 34 Focus issue F1. [DOI] [PubMed] [Google Scholar]
- 56.Reichenbach N., Delekate A., Plescher M., et al. Inhibition of Stat3-mediated astrogliosis ameliorates pathology in an Alzheimer's disease model. EMBO Mol. Med. 2019;11(2) doi: 10.15252/emmm.201809665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chiba T., Yamada M., Aiso S. Targeting the JAK2/STAT3 axis in Alzheimer's disease. Expert Opin. Ther. Targets. 2009;13(10):1155–1167. doi: 10.1517/14728220903213426. [DOI] [PubMed] [Google Scholar]
- 58.Zavala K., Lee J., Chong J., et al. The anticancer antibiotic mithramycin-A inhibits TRPV1 expression in dorsal root ganglion neurons. Neurosci. Lett. 2014;578:211–216. doi: 10.1016/j.neulet.2014.01.021. [DOI] [PubMed] [Google Scholar]
- 59.Miao J., Chen Z., Wu Y., et al. Sp1 inhibits PGC-1α via HDAC2-catalyzed histone deacetylation in chronic constriction injury-induced neuropathic pain. ACS Chem. Neurosci. 2022;13(23):3438–3452. doi: 10.1021/acschemneuro.2c00440. [DOI] [PubMed] [Google Scholar]
- 60.Xie Y., Li Z., Xu H., et al. Downregulation of Sp1 inhibits the expression of HDAC1/SOX10 to alleviate neuropathic pain-like behaviors after spinal nerve ligation in mice. ACS Chem. Neurosci. 2022;13(9):1446–1455. doi: 10.1021/acschemneuro.2c00091. [DOI] [PubMed] [Google Scholar]
- 61.Santpere G., Nieto M., Puig B., et al. Abnormal Sp1 transcription factor expression in Alzheimer disease and tauopathies. Neurosci. Lett. 2006;397(1–2):30–34. doi: 10.1016/j.neulet.2005.11.062. [DOI] [PubMed] [Google Scholar]
- 62.Rossner S., Sastre M., Bourne K., et al. Transcriptional and translational regulation of BACE1 expression--implications for Alzheimer's disease. Prog. Neurobiol. 2006;79(2):95–111. doi: 10.1016/j.pneurobio.2006.06.001. [DOI] [PubMed] [Google Scholar]
- 63.Wang C., Huang W., Lu J., et al. TRPV1-Mediated microglial autophagy attenuates Alzheimer's disease-associated pathology and cognitive decline. Front. Pharmacol. 2022;12 doi: 10.3389/fphar.2021.763866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Li J.K., Nie L., Zhao Y.P., et al. IL-17 mediates inflammatory reactions via p38/c-Fos and JNK/c-Jun activation in an AP-1-dependent manner in human nucleus pulposus cells. J. Transl. Med. 2016;14:77. doi: 10.1186/s12967-016-0833-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ji R.R., Gereau R.W., 4th, Malcangio M., et al. MAP kinase and pain. Brain Res. Rev. 2009;60(1):135–148. doi: 10.1016/j.brainresrev.2008.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Okazawa H., Estus S. The JNK/c-Jun cascade and Alzheimer's disease. Am J Alzheimers Dis Other Demen. 2002;17(2):79–88. doi: 10.1177/153331750201700209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Matsuda H., Wu J.X., Tanaka T., et al. Antinociceptive activities of 70% methanol extract of evodiae fructus (fruit of Evodia rutaecarpa var. bodinieri) and its alkaloidal components. Biol. Pharm. Bull. 1997;20(3):243–248. doi: 10.1248/bpb.20.243. [DOI] [PubMed] [Google Scholar]
- 68.Liao C.H., Pan S.L., Guh J.H., et al. Antitumor mechanism of evodiamine, a constituent from Chinese herb Evodiae fructus, in human multiple-drug resistant breast cancer NCI/ADR-RES cells in vitro and in vivo. Carcinogenesis. 2005;26(5):968–975. doi: 10.1093/carcin/bgi041. [DOI] [PubMed] [Google Scholar]
- 69.Fang Z., Tang Y., Ying J., et al. Traditional Chinese medicine for anti-Alzheimer's disease: berberine and evodiamine from Evodia rutaecarpa. Chin. Med. 2020;15:82. doi: 10.1186/s13020-020-00359-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhang W.D., Chen X.Y., Wu C., et al. Evodiamine reduced peripheral hypersensitivity on the mouse with nerve injury or inflammation. Mol. Pain. 2020;16 doi: 10.1177/1744806920902563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tan Q., Zhang J. Evodiamine and its role in chronic diseases. Adv. Exp. Med. Biol. 2016;929:315–328. doi: 10.1007/978-3-319-41342-6_14. [DOI] [PubMed] [Google Scholar]
- 72.Hochfellner C., Evangelopoulos D., Zloh M., et al. Antagonistic effects of indoloquinazoline alkaloids on antimycobacterial activity of evocarpine. J. Appl. Microbiol. 2015;118(4):864–872. doi: 10.1111/jam.12753. [DOI] [PubMed] [Google Scholar]
- 73.Rho T.C., Bae E.A., Kim D.H., et al. Anti-Helicobacter pylori activity of quinolone alkaloids from Evodiae fructus. Biol. Pharm. Bull. 1999;22(10):1141–1143. doi: 10.1248/bpb.22.1141. [DOI] [PubMed] [Google Scholar]
- 74.Pan X., Bligh S.W., Smith E. Quinolone alkaloids from Fructus Euodiae show activity against methicillin-resistant Staphylococcus aureus. Phytother Res. 2014;28(2):305–307. doi: 10.1002/ptr.4987. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data included in article/supplementary material/referenced in article.