Abstract

In search of a potent small molecular PD-L1 inhibitor, we designed and synthesized a compound based on a 2-hydroxy-4-phenylthiophene-3-carbonitrile moiety. Ligand’s performance was tested in vitro and compared side-by-side with a known PD-L1 antagonist with a proven bioactivity BMS1166. Subsequently, we modified both compounds to allow 18F labeling that could be used for PET imaging. Radiolabeling, which is used in drug development and diagnosis, was applied to investigate the properties of those ligands and test them against tissue sections with diverse expression levels of PD-L1. We confirmed biological activity toward hPD-L1 for this inhibitor, comparable with BMS1166, while holding enhanced pharmacological properties.
Introduction
In recent decades, immunotherapy, mostly based on the PD-1/PD-L1 axis, emerged as a promising pharmacological approach in cancer treatment. Especially in lung cancer and melanoma, PD-1/PD-L1 targeting monoclonal antibodies are highly efficacious giving patients an improved chance to fight the disease.1 However, not every cancer patient responds to therapy and the possible side effects of antibody therapy vary from mild ones, such as skin rash and dry mouth, to more severe symptoms including hypothyroidism and autoimmunological diseases.2 Moreover, immunotherapy is expensive and the cost of nivolumab, a PD-1 antibody, is over 2500€ for 1 therapy cycle.3,4 Therefore, a search for effective treatment, which is preferably non-immunogenic, lacks adverse reactions, and is affordable is urgently needed. Those characteristics are addressed by small molecular weight inhibitors (SMIs), which are not only inexpensive to synthesize but also are usually characterized by good cell permeability and, unlike much larger antibodies, can be orally available.
The PD-1/PD-L1 axis emerged as one of the most effective immune checkpoint blockade (ICB) strategies in the last decade. Until now, only therapeutics targeting PD-1/PD-L1 and CTLA-4 have been approved by FDA. Recent studies on co-crystal structures of PD-L1 and antibodies complexes have shown that monoclonal antibodies [atezolizumab, durvalumab, or avelumab, ca. 150 kDa, binding affinity dissociation constant (pKd) of 9–10] bind by 5 complementarity-determining regions (CDRs) and those epitopes are conserved between those antibodies.5−8 Remarkably, smaller structures, such as PD-L1 nanobodies (KN035, ca. 15 kDa), bind just as efficiently with pKd of 8.5, but interact with only 2 CDRs. However, it was reported that only one CDR is in fact crucial for the high affinity toward PD-L1.8,9 This region represents a narrow binding area yielding a possibility for a “drug miniaturization,” which will be more specific. Small molecular ligands for PD-L1 (MW < 1 kDa) can either mimic antibody binding or stabilize PD-L1 dimer. The formation of this dimer results in the inability of PD-1 to bind to PD-L1, therefore, blocking PD-1 signaling. Compounds mimicking antibodies are macrocyclic peptides.10,11 Other ligands follow the biphenyl structure from Bristol-Myers Squibb (BMS) patented in 2015. They are characterized by low nanomolar IC50 values and have been recently found to restore T-cell cytolytic function to the extent comparable with nivolumab, as well as reduce tumor growth in vivo.11,12 The promising preclinical testing resulted in a few of the small molecular PD-L1 inhibitors being evaluated in clinical trials, such as MAX-10181 (Maxinovel) and GS-4224 (Gilead), both ligands based on the biphenyl core.13,14
To study the binding properties of novel small molecular PD-L1 inhibitors, different assays can be applied. Most commonly used are PD-1/PD-L1 blockade assays employing fluorescence measurements, either on a protein level such as the homogeneous time-resolved fluorescence (HTRF) technique or on a cellular level using cell lines equipped with a luciferase reporter. However, those assays do not display all the qualities and limitations that the PD-L1 antagonist can possess. Positron emission tomography (PET), as a non-invasive imaging technique, can be used not only as a diagnostic technique but also is very useful in a drug development process, as it allows for the tracer quantification with a high sensitivity, a monitoring of the tracer distribution, metabolism in living organisms, and a subsequent clinical translation. In the case of an isotopic exchange, the molecule structure and its binding properties are usually not affected, so its biodistribution and pharmacokinetics can be easily assessed by the gamma detector systems quantitatively.
Here, we present the design, synthesis, and biological evaluation of novel 2-hydroxy-4-phenylthiophene-3-carbonitrile-based PD-L1 inhibitors and compare them with the well-known biphenyl-based PD-L1 antagonist BMS1166.
Results
Molecular Design
Following our previously introduced pharmacophore model of PD-1/PD-L1 small molecular weight antagonists, we designed a general structure based on a central thiophene ring accessible by the three-component Gewald reaction (Gewald-3CR, Figure 1B).8,15,16 By examining the computational docking of a novel class of small molecular inhibitors to PD-L1 and comparing it to the BMS1166 compound, we anticipated that a phenyl–thiophene moiety (Figure 1B,C, block A) resembles the biphenyl moiety from the BMS compound inducing PD-L1 dimerization and, therefore, binding in a similar manner to a site located between PD-L1 dimers, a narrow, but extended hydrophobic cavity (Figure 2A). The 3-cyano moiety helps to twist the thiopheno phenyl moiety out of plane to introduce a preformed receptor binding conformation. In the majority of other reported small molecule PD-1/PD-L1 antagonists, conformational prefix is achieved by the introduction of bulky 2-halogen substituents in the biphenyl moiety.17,18 Additionally, the phenyl ring following the bicyclic structure through an ether bond enabled additional strong, positioning π–π interactions at the borderline of the pocket (Figure 1B,C, block B). Our co-crystal structures of PD-L1 and BMS1166 (PDB ID: 6R3K) revealed that the amine residue (Figure 1B,C, block C) does not contribute to the binding and, therefore, can be modified.19 We enhanced the ligand solubility by removing the proline residue and leaving a “naked” primary amine tail acting as both H-bond donor and acceptor. Furthermore, co-crystal structures have shown that benzonitrile moiety (Figure 1C, block D) is located outside of the binding cavity, which creates strong π–π interactions with Tyr123 that might stabilize ligand inside the active center of the PD-L1 dimer. Nevertheless, another known PD-L1 antagonist—BMS202 lacks the benzonitrile moiety, while maintaining a good affinity toward the target. In our design, we decided to abstain from introducing block D and focus on a minimal version of a small molecule complying with Lipinsky’s rule of five.
Figure 1.
Structural design of PD-L1 inhibitors. (A) Examples of potent PD-L1 dimerizer taken from patent literature. (B) Our design of novel inhibitor 2-((4-(aminomethyl)benzyl)oxy)-4-(4-fluorophenyl)thiophene-3-carbonitrile accessible by Gewald-3CR (4a) and (C) structural comparison with fluorine derivative of BMS1166 (9a). Similar parts of the scaffolds are indicated by the different colored boxes; synthetized BMS1166 was used as a reference compound and is referred in this work as 9b.
Figure 2.
Docking of PD-L1 antagonist. (A) Docking results of an example lead compound 4a (in green) to a PD-L1 dimer (from PDB ID: 6R3K). Blue and yellow represent two PD-L1 monomers (surface and ribbon models). (B,C) Comparison of binding modes of 4a, 9a, and 9b. (B) Superimposition of 9b from the co-crystal structure with an anticipated orientation of 9a from the molecular docking. (C) Superimposition of 9b from the co-crystal structure with an anticipated orientation of 4a from the molecular docking. Additional π–π stacking interactions and hydrogen bonds are highlighted as red dashes (distance ≤ 3.3 Å),20 transparent amino acids denote residues anticipated to change their conformation upon binding of a superimposed ligand (4a and 9a, respectively) to PD-L1 dimer. (D) Superimposition of ligand 4a into the pharmacophore model based on the BMS1166/PD-L1 co-crystal structure (from PDB ID: 6R3K). Ph4 model shows 3 aromatic rings (orange sphere), a hydrophobic group (green sphere), and a positive charged group (purple sphere).
Computational docking of novel ligands by the superposition onto the co-crystal structure of BMS1166 and PD-L1 dimer (PDB ID: 6R3K) as well as pharmacophore modelling of superimposed 4a and 9b confirmed similar binding modes of those two compounds (Figure 2). Although 2-((4-(aminomethyl)benzyl)oxy)-4-(4-fluorophenyl)thiophene-3-carbonitrile lacks a phenyl ring (Figure 1C, block D), which supports binding outside of the pocket through π–π interaction with Tyr123, its structure inside the pocket provides additional interactions. Block A of compound 4a is involved in a strong, stabilizing π–π interaction through its phenyl group with Tyr56 and hydrophobic contacts with all surrounding hydrophobic residues. Primary amine from block C, which lays outside of the binding pocket, is likely involved in the charge–charge contact with Asp122. Computational docking allowed us to explore additional modifications to the core molecule and their effect on binding to the target. The modification most interesting for us would be an addition of fluorine that would grant the radiolabeling with fluorine-18, useful for further biological evaluation. Substitution with fluorine at the phenyl-thiophene ring of 4a and the fluorine modification of 4-hydroxyproline of 9b did not alter the binding mode as no major changes of the conformation were observed. We continued with the synthesis of compound derivatives utilizing reactions shown in Schemes 1 and 2.
Scheme 1. Synthetic Route of Compounds 4a–b.

Reagents and conditions: (a) S8, DCM, 0 °C → RT, 24 h; (b) NaOEt, ethanol, reflux, 24 h; (c) K2CO3, ACN, 105 °C, 21 h; 17 h; (d) 7 N HCl in dioxane, RT, 4 h, (e) B2Pin2, Pd(dppf)Cl2·DCM, CH3COOK, dioxane, 95 °C; R = F, Br.
Scheme 2. Synthetic Route of Compounds 9a–b and 10.

Reagents and conditions: (a) Pd(dppf)Cl2, 80 °C, 12 h; (b) SOCl2, DCM, 0 °C → RT, 20 h; (c) 5-chloro-2,4-dihydroxybenzaldehyde, NaHCO3, KI, ACN, DMF, 65 °C, 20 h; (d) K2CO3, ACN, 75 °C; (e) (2S,4S)-4-fluoropyrrolidine-2-carboxylic acid (9a), (2S,4R)-4-hydroxypyrrolidine-2-carboxylic acid (9b) or methyl (2S,4R)-4-hydroxypyrrolidine-2-carboxylate (9c), NaCNBH3 or NaBH(OAc)3, DMF, DCE; and (f) TosCl, Et3N, DCM.
Ligands Synthesis
Our synthetic pathway toward the targeted compounds 4a–b, started with the Gewald three component reaction (G-3CR), which gives us access to the 4-phenyl substituted thiophene ring.21−25 Indeed, p-substituted acetophenones (R = F for 1a and Br for 1b) reacted with methyl cyanoacetate and elemental sulfur to generate compounds 1a–b, almost quantitatively, in 92 and 86% overall yields, respectively. Next, an rearrangement reaction was employed, and in the presence of sodium ethoxide, compounds 2a–b in 30 and 52% were yielded, respectively.21 A Williamson ether synthesis reaction was used to couple tert-butyl (4-(hydroxymethyl)benzyl)carbamate in 45 and 76% for 3a–b), followed by a quantitative Boc-deprotection using 4 M HCl in dioxane affording the fluorinated compound 4a. Finally, for bromine-derivative 3b, we opted to not perform a deprotection step, but performed a Miyaura borylation to obtain compound 4b in 60% yield.
Accessing compounds 9a–b and 10 was achieved following the procedure described below. First, a Suzuki coupling of (2,3-dihydrobenzo[b][1,4]dioxin-6-yl)boronic acid and 3-bromo-2-methylphenyl)methanol was accomplished to afford compound 5 in 90% yield. Then, a chlorination reaction of compound 5 was performed to afford compound 6 in quantitative yield. Subsequently, a nucleophilic substitution with 5-chloro-2,4-dihydroxybenzaldehyde was employed to obtain 7 (74% yield). An additional nucleophilic substitution involving compound 7 and 3-(chloromethyl)benzonitrile was carried out to provide quantitatively compound 8. The final step toward 9a–c was based on a reductive amination involving either (2R,4S)-4-fluoropyrrolidine-2-carboxylic acid (for 9a), (2R,4R)-4-hydroxypyrrolidine-2-carboxylic acid (for 9b), or methyl (2R,4R)-4-hydroxypyrrolidine-2-carboxylate (for 9c) affording 9a–c in 58–60% yield. An additional step was necessary involving compound 9c and 4-toluenesulfonyl chloride, attaining compound 10 for further 18F-labeling with a good yield of 56%.
Comparison of Ligands’ Biological Activity
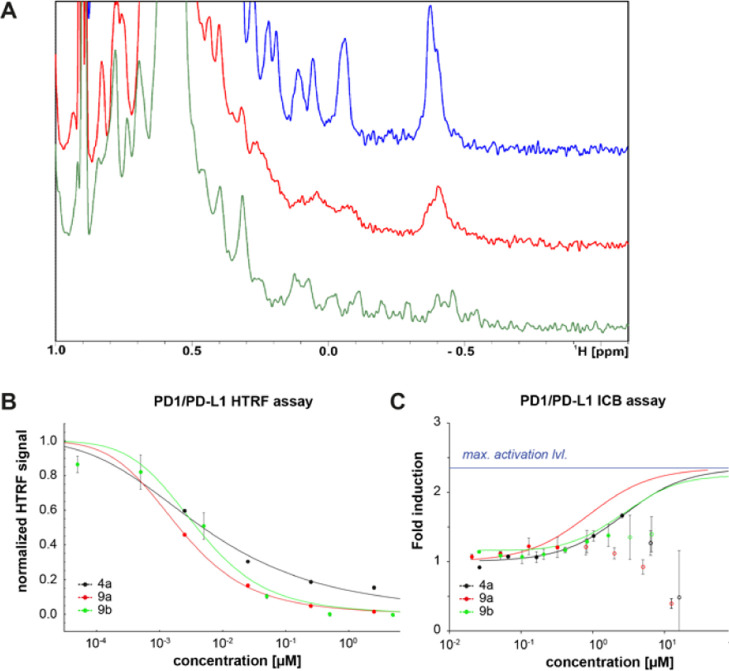
To investigate if our synthesized analogs are capable of disrupting PD-1/PD-L1 binding, we performed our previously described nuclear magnetic resonance (NMR) assay.16 We collected both 1D and 2D spectra of PD-L1 apo (blue) in the presence of 4a (red) and positive control BMS1166 (9b) (green) at molar ratio 1:1 (Figures 3A and S1A). Both 1H NMR and 1H–15N heteronuclear multiple quantum coherence (HMQC) signals show that the addition of 4a as well as reference compound BMS1166 induces dimerization of PD-L1 that was also previously observed for the biphenyl compounds.26 The PD-L1 dimerization causes peaks of the 1H aliphatic part PD-L1 spectrum to flatten (peak at −0.4 ppm) or disappear in the case of peaks from 1H–15N HMQC spectrum. Dimerization of PD-L1, in turn, prevents its binding to the PD-1.
Figure 3.
Biophysical characterization of the potency of tested compounds (A) aliphatic part of 1H NMR spectra of apo-PD-L1 (blue), PD-L1/4a (red), and PD-L1/9b (green) in the molar ratio 1:1. (B) IC50 determination using HTRF PD-1/PD-L1 assay. Measurements were done in two independent dilution series (C) PD-1/PD-L1 immune checkpoint blockade assay for 4a, 9a, and 9b. The fitted function was estimated from Hill’s equation and points without a filled circle were excluded from the fitting due to high cytotoxicity.
To determine the potency of synthesized inhibitors to disrupt the PD-1/PD-L1 complex, we used homogeneous time resolved fluorescence (HTRF), which is a well-established method for the inhibitory activity of PD-L1 assessment based on RT-FRET effect.10,11,26,27 In this binding assay, tagged PD-1 and PD-L1 are labelled with anti-tag reagent that generates FRET signal only when both proteins are bound (in spatial proximity). Presence of inhibitors (small molecules, proteins, and so forth) that disrupt the binding decreases the resulting signal allowing the assessment of the inhibitory activity of tested inhibitors via IC50 (the half maximal inhibitory concentration). Compared to the positive control BMS1166 (9b) IC50 of 3.78 nM, both tested compounds exhibit comparable inhibitory activities (9a 2.01 nM and 4a 4.97 nM) proving their potency in disrupting the PD-1/PD-L1 complex (Figure 3B). Additionally, we have tested compound 4a and 9a for their affinities to human PD-L1. Reported values showed that 4a binds to hPD-L1 with kD of ca. 27 nM, while 9a with ca. 8 nM, which is consistent with low nanomolar inhibitory activities of those compounds in HTRF assay (Figure S1C).
To determine if the in vitro results translate to the cell-based assay, we performed the cellular assay, in which coculturing Jurkat reporter T cells and artificial antigen-presenting cells (aAPCs) provided the binding between PD-1 and PD-L1. The disruption of the binding between PD-1 and PD-L1 via inhibitor enhances a TCR-mediated activation of the Jurkat cells leading to the increase in the coupled luciferase signal intensity. Therefore, reporting luciferase activity can be used to track the activation status of the Jurkat T cells. In our cellular assay, tested compounds gave EC50 values of 2.70 μM for 4a, 0.88 μM for 9a, and 1.57 μM for 9b (matching previously obtained value for the positive control).27 However, in higher concentrations, they exhibit cytotoxicity toward the Jurkat cell line, and the EC50 values, especially for 9a, should be approached as an estimation (Figures 3C and S1B).
Radiochemistry
For compounds 4a and 9a with a proven inhibitory activity toward PD-L1, we performed radiolabeling to explore other pharmacological properties, such as lipophilicity and short-term stability, as well as binding to PD-L1+/– tissue sections.
[18F]4a radiosynthesis was performed by the copper-mediated fluorination of boronate pinacol ester (4b) according to a procedure by Mossine et al. (Scheme 3).28 Optimal reaction time of 15 min was chosen based on TLC conversion measurements (54, 56, 57% at 15, 30, and 45 min, respectively, Figure S2A). Synthesis was accomplished with good radiochemical yield (up to 18% RCY) and very good molar activity (up to 138 669 GBq/mmol).
Scheme 3. Copper-Mediated Fluorination of [18F]4a from Boronic Pinacol Ester Precursor 4b.
Reagents and conditions: (a) [18F]F–, Et4N·HCO3, nBuOH, DMA, 110 °C, 15 min; (b) 6 N HClaq 110 °C, 10 min.
[18F]9a was accessed via nucleophilic substitution of 10 (tosylate) and subsequent esterolysis using 1 M LiOHaq (Scheme 4). The usage of acetonitrile, the most common solvent for nucleophilic substitution with 18F, resulted in a low conversion (<3%) and was replaced by DMSO. The optimal reaction time of 10 min was chosen based on TLC conversion measurements (33, 37, 36% at 10, 20, and 35 min, respectively, Figure S2B). Two routes were explored for this radiosynthesis, with purification in between two steps (method A) and subsequent two steps performed as a one-pot reaction (method B). Method A, although resulting in a much cleaner profile of hydrolysis reaction, took twice as long and resulted in a lower radiochemical yield (ca. 180 vs 85 min, the maximal value of RCY 2 vs 5% d. c., Figure S2C). [18F]9a synthesis was accomplished by method B with moderate radiochemical yield (up to 5% RCY) and very good molar activity (up to 144 836 GBq/mmol).
Scheme 4. Fluorination of [18F]9a from Tosylate Precursor 10.
Reagents and conditions: (a) [18F]F–, K2CO3, Kryptofix 222, DMSO, 100 °C, 10 min; (b) 1 M LiOHaq MeOH, 100 °C, 10 min.
Both tracers were obtained with very good radiochemical purity and molar activity as shown in Table 1 and were subjected to lipophilicity and stability measurements. Experimental lipophilicity listed in Table 1 matched predicted log P values (5.38 for [18F]9a and 3.7 for [18F]4a).29 The assay confirmed high lipophilicity of [18F]9a and moderate lipophilicity of [18F]4a. Stability in formulation solution after 4 h for both compounds maintained above 95% threshold, however, stability in human serum showed slightly higher degradation for [18F]9a than [18F]4a (minimal value 82 vs 93%, Figure S7).
Table 1. Comparison of properties of [18F]Labeled PD-L1 Tracers.
| tracer | [18F]4a | [18F]9a |
|---|---|---|
| radiochemical purity [%] | 99.8 ± 0.5b | 99.2 ± 1.1c |
| molar activity [GBq/mmol] | 78 876 ± 84 561b | 100 794 ± 48 716c |
| calculated log P (XLOGP329) | 3.70 | 5.38 |
| experimental log Pa | 3.6 ± 0.4 | 5.2 ± 0.6 |
| experimental log D7.4a | 3.9 ± 0.9 | 6.5 ± 0.2 |
| stability in formulation solution after 4 h [%]a | 97.5 ± 0.5 | 97.3 ± 1.1 |
| stability in serum after 4 h [%]a | 93.9 ± 0.5 | 88.7 ± 5.7 |
Experiment performed in triplicate.
n = 4.
n = 6.
Autoradiography with PDL1+/– Tissue Sections
Next, we explored the binding properties of the synthesized compounds in a human tissue setting. We selected available tumor tissue sections—ES2 (ovarian carcinoma) and H358 (lung adenocarcinoma). Tumors were prepared in two versions—wild type, expressing PD-L1 and PD-L1 knock-out, as shown in intra- and extracellular measurements of the above cell lines (Figure S8). We noted a 40–55% higher uptake of both, [18F]4a and [18F]9a compounds, in PD-L1+ H358 tumors than in wild-type counterparts. Uptake correlated with PD-L1 expression, which was confirmed by immunohistochemistry staining (Figure 4A). Interestingly, the uptake was localized mostly on the peripheral part of the tumor slide, similar to immunohistochemistry staining. For H358, both tracers performed similarly well (Figure 4B). Unfortunately, [18F]4a failed to discriminate between wild-type and knock-out for ES2 slides. Elevated, unspecific uptake in PD-L1– tumors could be explained by the high lipophilicity of compounds and nature of paraffin-embedded tissues, which often result in upregulated unspecific uptake. Afterward, to eliminate potential issues with paraffin embedding, we examined binding to snap-frozen human tonsils. Tonsils are known to overexpress PD-L1, especially in structures called germinal centers.30−32 We were able to achieve a similar imaging pattern with both of the tracers. Quantification confirmed no difference in binding between those two molecules (Figure 4C,D).
Figure 4.
Autoradiography with PD-L1+/– (H358 and ES2) tumor for [18F]4a and [18F]9a. (A) Images of tumor tissue slides incubated with [18F]4a and [18F]9a and comparison with immunohistochemistry staining with PD-L1 antibody. (B–D) Autoradiography quantification and tracer comparison (B). Autoradiography with human tonsils for [18F]4a and [18F]9a (C). Autoradiography quantification and tracer comparison (D); experiments performed in triplicate and groups compared with Mann–Whitney test (nonparametric, *p < 0.05, **p < 0.051), colorful scale signifies color coding from low to high tracer uptake.
Discussion and Conclusions
Immune checkpoint blockade, especially the PD-1/PD-L1 axis, has been widely popularized due to its phenomenal results in cancer treatment. Although the PD-1/PD-L1 role in tumor immunology and its downstream signaling has been extensively studied, the PD-L1 blockade and its effect on PD-1 are not fully comprehended. Particularly, the mode of action for small molecular, non-peptidic PD-L1 antagonists is not fully understood. Only recently, few studies investigated the effects of small molecular weight PD-1/PD-L1 antagonists.33−38 This research revealed how vastly different outcomes can be achieved for PD-L1 inhibitors using different biological assays. Notably, Aurigene-1 (a close derivative of clinical trial compound CA-170) exhibits nanomolar potency in the splenocyte recovery in an assay that was used as a scouting method for PD-1/PD-L1 inhibitors.33 However, Aurigene-1 does not show binding in SPR binding assay, nor does it show activity in ELISA biochemical assay, and it does not induce IL-2 production.35 Moreover, our research proved that CA-170 is not a direct binder to either human or murine PD-L1 by means of NMR, HTRF, and cellular assays.29 Additionally, BMS small molecular compounds, while being able to disrupt the PD-1/PD-L1 axis in Jurkat cells modified with NFAT promoter, display strong cytotoxicity at higher concentrations.35−38 On account of those conflicting characteristics, further understanding of the mode of action is necessary for a rational design and the discovery of novel PD-L1 inhibitors.
In this work, we report the design, synthesis, and biological evaluation of an exemplary ligand from the novel, 2-hydroxy-4-phenylthiophene-3-carbonitrile-based class of compounds. Those compounds act as PD-L1 antagonists, and their structural analysis based on molecular docking reveals similarities to small molecular PD-L1 inhibitors developed by Bristol-Myers Squibb. We confirmed their biological activity not only in computational docking and biophysical studies but also in various in vitro assays. We have proven that tested compounds structurally interact with hPD-L1 as evident by examining the shifts in both protein-based 1D and 2D NMR spectra upon compound addition. Furthermore, we assessed the ability of those compounds to disrupt PD-1/PD-L1 binding in HTRF assay to be low single digit nanomolar. Additionally, the cell-based PD-1/PD-L1 immune checkpoint blockade assay has confirmed the activity of all compounds. We observed similar binding properties of our novel scaffold to known and potent PD-L1 antagonist—BMS1166. We were able to radiolabel 2-hydroxy-4-phenylthiophene-3-carbonitrile and BMS1166-based inhibitors via copper-mediated fluorination of boronic pinacol esters and nucleophilic substitution. Tracer production was performed in moderate to good yields and afforded pure compounds with a molar activity. Radiolabeling with fluorine-18 allowed us to expand our knowledge of certain properties of those compounds, such as lipophilicity and short-term stability, as well as performed biological assays on tissues with various PD-L1 expressions. We established similar patterns of binding for the synthesized compounds to H358 cell lines (PDL1+/–) and human tonsils. [18F]4a was unable to bind specifically to ES2 cell lines (PDL1+/–), however, it showed lower background uptake than [18F]9a, which could be explained by lower lipophilicity. Additionally, better stability in human serum was established for [18F]4a. These findings suggest that [18F]4a might be a promising PD-1/PD-L1 antagonist candidate for diagnostics as a tracer or therapy as a non-radioactive compound 4a, however further evaluation needs to be performed. To prove utility of compound 4a, a comparative in vivo PET study with 9a must be performed utilizing immunocompromised mouse model inoculated with human tumor cell lines used in this work. This tracer study will reveal the optimal timeline for compound distribution, preferred targeted tumor cell line (characterized by different level of PD-L1 expression), and additional organ selectivity. Due to lipophilic nature of these compounds, a preferred pathway of degradation would be a biliary excretion. Therefore, an assessment of accumulation in the liver and hepatoxicity must be evaluated. Additionally, we observed toxicity in Jurkat cells at higher concentration and in vitro toxicity study would prove essential to further strategize usefulness of these compounds. Radiolabeling could be used not only to observe compound distribution in real time in a mouse model but also determine metabolic stability and formed metabolites. The design of this novel scaffold allows for several modifications that could further improve their binding, lipophilicity, and stability. The addition of a component representing block D (Figure 1) e.g., a phenyl, could provide increased affinity and we believe derivatization of this phenyl ring with hydrophilic moieties could lower the lipophilicity and increase compound’s water stability. We foresee that the synthesis and biological assessment of compounds based on 4a could be beneficial in finding effective PD-L1 inhibitors for diagnostic or therapeutic purposes.
Experimental Section
General Information
Reagents were acquired from commercial suppliers (Sigma-Aldrich, ABCR, Acros and AK Scientific) and used without any purification unless otherwise noted. Thin layer chromatography was performed on Fluka precoated silica gel plates (0.20 mm thick, particle size 25 μm). Flash chromatography was performed on a Teledyne ISCO Combiflash Rf, using RediSep Rf Normal-phase Silica Flash Columns (Silica Gel 60 Å, 230–400 mesh) and on a Reveleris X2 Flash Chromatography, using Grace Reveleris Silica flash cartridges (40, 24, 12, and 3 g). All HTRF experiments were performed using a Cisbio Bioassays Human PD1/PD-L1 biochemical binding assay. Nuclear magnetic resonance spectra were recorded on a Bruker Avance 500 spectrometer. Chemical shifts for 1H NMR were reported in ppm relative to TMS (δ = 0 ppm) or the corresponding solvent peak (CDCl3 δ = 7.26 ppm, DMSO-d6 δ = 2.50 ppm, and CD3OD δ = 3.31 ppm) and coupling constants were reported in hertz (Hz). The following abbreviations were used for spin multiplicity: s = singlet, bs = broad singlet, d = doublet, dd = doublet of doublets, t = triplet, ddt = doublet of doublet of triplets, q = quartet, and m = multiplet. Chemical shifts for 13C NMR were reported in ppm relative to the solvent peak (CDCl3 δ = 77.2 ppm, DMSO-d6 δ = 39.5 ppm, and CD3OD δ = 49.0 ppm). High resolution mass spectra (HRMS) were recorded using an Orbitrap-Velos Pro at a resolution of 60,000. All compounds are >95% pure by HPLC analysis.
Experimental Procedures and Analytical Data
Ethyl 2-Amino-4-(4-fluorophenyl)thiophene-3-carboxylate 1a
To a solution of 4-fluoroacetophenone (2.8 g, 2.4 mL, 20 mmol) in 100 mL dry DCM was added ethyl cyanoacetate (2.7 g, 2.6 mL, 24 mmol). The reaction mixture was cooled to 0 °C and stirred for 0.5 h. Subsequently, TiCl4 (7.6 g, 4.4 mL, 40 mmol) was added dropwise and the reaction mixture was stirred for 1 h at 0 °C, followed by addition of pyridine in two portions (5.4 mL, 67 mmol). The reaction mixture was stirred for 18 h at RT. Subsequently, the reaction mixture was poured into 100 mL 3 N HCl and extracted twice with 200 mL of DCM. The combined organic layer was washed with brine, filtered over MgSO4, and concentrated under reduced pressure. The resulting oil was dissolved in 19 mL THF, followed by addition of elemental sulfur (0.8 g, 25 mmol) and diethylamine (2.8 g, 3.9 mL, 38 mmol). Reaction mixture was stirred for 18 h at RT. The crude was diluted with diethyl ether, washed twice with water and brine, filtered over MgSO4, and concentrated under reduced pressure to provide the desired product as a yellow solid (4.9 g, 18.5 mmol, 92%). 1H NMR (500 MHz, DMSO-d6): δ 7.42 (s, 2H), 7.26 (ddd, J = 8.9, 5.6, 2.7 Hz, 2H), 7.13–7.07 (m, 2H), 6.16 (s, 1H), 3.95 (q, J = 7.1 Hz, 2H), 0.91 (t, J = 7.1 Hz, 3H); 13C NMR (126 MHz, DMSO-d6): δ 161.7 (d, J = 242.6 Hz), 135.1 (d, J = 3.2 Hz), 130.9 (d, J = 8.2 Hz), 114.4 (d, J = 21.2 Hz), 105.7 (d, J = 7.3 Hz), 59.1, 14.2; MS (ESI) m/z: calcd for C13H13FNO2S [M + H]+, 266.0646; found [M + H]+, 266.0645.
Ethyl 2-Amino-4-(4-bromophenyl)thiophene-3-carboxylate 1b
To a solution of the 4-bromoacetophenone (4.0 g, 20 mmol) in 100 mL dry DCM was added ethyl cyanoacetate (2.7 g, 2.6 mL, 24 mmol). The reaction mixture was cooled to 0 °C and stirred for 0.5 h. Subsequently, TiCl4 (7.6 g, 4.4 mL, 40 mmol) was added dropwise and the reaction mixture was stirred for 1 h at 0 °C, followed by addition of pyridine in two portions (5.4 mL, 67 mmol). Reaction mixture was stirred for 18 h at RT. The crude was poured into 100 mL 3 N HCl and extracted twice with 200 mL of DCM. Subsequently, the combined organic layer washed with brine, filtered over MgSO4, and concentrated under reduced pressure. The resulting oil was dissolved in 19 mL THF, followed by addition of elemental sulfur (0.8 g, 25 mmol) and diethylamine (2.8 g, 3.9 mL, 38 mmol). Reaction mixture was stirred for 18 h at RT. The crude was diluted with diethyl ether, washed twice with water and brine, filtered over MgSO4, and concentrated under reduced pressure. The crude mass was purified by silica gel flash chromatography using 10% EtOAc/PE as an eluent to provide the desired product as a yellow solid (5.6 g, 17 mmol, 86%). 1H NMR (500 MHz, CDCl3): δ 7.47–7.39 (m, 2H), 7.19–7.13 (m, 2H), 6.14 (s, 2H), 6.03 (s, 1H), 4.06 (q, J = 7.1 Hz, 2H), 0.98 (t, J = 7.1 Hz, 3H); 13C NMR (126 MHz, CDCl3): δ 165.6, 164.2, 140.3, 137.5, 130.7, 130.4, 120.9, 105.9, 105.7, 59.7, 13.9; HRMS (ESI) m/z: calcd for C13H13BrNO2S [M + H]+, 325.9845; found [M + H]+, 325.9842.
4-(4-Fluorophenyl)-2-hydroxythiophene-3-carbonitrile 2a
To a refluxing stirred solution of ethyl 2-amino-4-(4-fluorophenyl) thiophene-3-carboxylate (2.61 g, 9.83 mmol) in 20 mL of EtOH was added dropwise a solution consisting of sodium (452 mg, 19.7 mmol) in 20 mL EtOH. Reaction mixture was stirred for 4 h while refluxing. Subsequently, reaction mixture was allowed to cool to RT, and 80 mL water was added and stirred for 1 h at RT. The crude was filtered and the filtrate was acidified with 37% HClaq until a pH of 2 was reached. The resulting precipitate was filtered and air dried to provide the desired product as an off-white solid (647 mg, 2.95 mmol, 30%).1H NMR (500 MHz, DMSO-d6): δ 7.62–7.57 (m, 2H), 7.34–7.26 (m, 2H), 6.78 (s, 1H).
4-(4-Bromophenyl)-2-hydroxythiophene-3-carbonitrile 2b
To a refluxing stirred solution of ethyl 2-amino-4-(4-bromophenyl)thiophene-3-carboxylate (682 mg, 2.1 mmol) in 2.5 mL EtOH was added dropwise sodium ethoxide (285 mg, 4.2 mmol) in 2.5 mL EtOH. Reaction mixture was stirred for 4 h while refluxing. Subsequently, reaction mixture was allowed to cool to RT, and 20 mL water was added and stirred for 1.5 h at RT. The crude was filtered and the filtrate was acidified with 37% HClaq until a pH of 2 was reached. The resulting precipitate was filtered, air dried, and purified by silica gel flash chromatography using 0–4% MeOH/DCM as eluent to provide the desired product as an off-white solid (303 mg, 1.1 mmol, 52%).1H NMR (500 MHz, DMSO-d6): δ 7.64 (d, J = 8.0 Hz, 2H), 7.50 (d, J = 8.0 Hz, 2H), 6.82 (s, 1H); 13C NMR (126 MHz, DMSO-d6): δ 13C NMR (126 MHz, DMSO): δ 175.1, 136.0, 133.5, 132.0, 131.8, 129.1, 121.5, 115.4, 108.1, 87.7.
tert-Butyl (4-(((3-Cyano-4-(4-fluorophenyl)thiophen-2-yl)oxy)methyl)benzyl)carbamate 3a
A solution consisting of 4-(4-fluorophenyl)-2-hydroxythiophene-3-carbonitrile (647 mg, 2.95 mmol), tert-butyl (4-(bromomethyl)benzyl)carbamate (976 mg, 3.25 mmol), and potassium carbonate (450 mg, 3.25 mmol) in 10 mL ACN was stirred for 18 h at 80 °C. The reaction mixture was concentrated under reduced pressure. The crude mass was purified by silica gel flash chromatography with 0–100% EtOAc/PE to provide the desired product as a colorless oil (580 mg, 1.3 mmol, 45%). 1H NMR (500 MHz, CDCl3): δ 7.59–7.50 (m, 2H), 7.43 (d, J = 7.8 Hz, 2H), 7.33 (d, J = 7.8 Hz, 2H), 7.14–7.07 (m, 2H), 6.53 (s, 1H), 5.26 (s, 2H), 4.90 (s, 1H), 4.34 (d, J = 6.1 Hz, 2H), 1.46 (s, 10H); 13C NMR (126 MHz, CDCl3): δ 163.9, 138.7, 133.0, 130.0, 129.3, 129.2, 128.5, 127.9, 116.0, 115.8, 114.2, 107.7, 44.3, 28.4 ppm.
tert-Butyl (4-(((4-(4-Bromophenyl)-3-cyanothiophen-2-yl)oxy)methyl)benzyl)carbamate 3b
A solution consisting of 4-(4-bromophenyl)-2-hydroxythiophene-3-carbonitrile (380 mg, 1.73 mmol), tert-butyl (4-(bromomethyl)benzyl)carbamate (572 mg, 1.9 mmol), and potassium carbonate (263 mg, 1.9 mmol) in 10 mL ACN was stirred at 80 °C for 18 h. The reaction mixture was concentrated under reduced pressure. The crude mass was purified by silica gel flash chromatography with 10–20% EtOAc/PE to provide the desired product as a brown solid (654 mg, 0.35 mmol, 76%). 1H NMR (500 MHz, CDCl3): δ 7.55 (dd, J = 8.6 Hz, 2.5 Hz, 2H), 7.43 (dd, J = 8.4 Hz, 5.1 Hz, 4H), 7.33 (d, J = 10 Hz, 2H), 6.55 (s, 1H), 5.26 (s, 2H), 4.92 (s, 1H), 4.34 (s, 2H), 1.46 (s, 9H); 13C NMR (126 MHz, CDCl3): δ 140.3, 138.6, 133.1, 132.9, 132.2, 129.1, 128.6, 128,0, 122.9, 114.2, 108.3, 80.2, 77.0, 44.4, 28.5; HRMS (APCI) m/z: calcd for C24H24O3N2BrS [M – H]+, 499.0686; found [M + H]+, 499.0683.
2-((4-(Aminomethyl)benzyl)oxy)-4-(4-fluorophenyl)thiophene-3-carbonitrile Hydrochloride 4a
tert-Butyl (4-(((3-cyano-4-(4-fluorophenyl)thiophen-2-yl)oxy)methyl)benzyl)carbamate (68 mg, 0.16 mmol) was dissolved in 4 mL HCl (4 M in dioxane) and stirred at RT for 4 h. Reaction mixture was concentrated under reduced pressure to provide the desired product as a yellow solid (52 mg, 0.15 mmol, 99%).1H NMR (500 MHz, methanol-d4): δ 7.61 (tdd, J = 8.8, 5.6, 1.9 Hz, 4H), 7.56–7.51 (m, 2H), 6.91–6.86 (m, 1H), 5.41 (d, J = 2.6 Hz, 2H), 4.17–4.13 (m, 2H); 13C NMR (CDCl3, 126 MHz): 13C NMR (126 MHz, methanol-d4): δ 172.9, 162.3, 160.4, 136.4, 134.3, 132.4, 127.6, 127.5, 127.5, 127.3, 113.8, 113.7, 112.1, 107.2, 74.7, 41.0; HRMS (APCI) m/z: calcd for C19H16ON2FS [M – H]+: 339.0962; found [M – H]+: 339.0958.
tert-Butyl (4-(((3-Cyano-4-(4-(4,4,5,5-tetramethyl-1,3,2-dioxaborolan-2-yl)phenyl)thiophen-2-yl)oxy)methyl)benzyl)carbamate 4b
A solution of (4-(((4-(4-bromophenyl)-3-cyanothiophen-2-yl)oxy)methyl)benzyl)carbamate (554 mg, 1.1 mmol) in 10 mL anhydrous dioxane was placed under nitrogen and degassed for 10 min. Bispinacolato diboron (338 mg, 1.3 mmol), potassium acetate (327 mg, 3.3 mmol), and [1,1′-bis(diphenylphosphino)ferrocene]palladium(II) dichloride (73 mg, 0.1 mmol) was added. Reaction mixture was stirred at 95 °C for 17 h. The crude was extracted thrice with EtOAc. The combined organic layer was washed with brine, filtered over MgSO4, and concentrated under reduced pressure. Silica gel flash chromatography using 10–30% EtOAc/PE as eluent provided the desired product as a yellow solid (186 mg, 0.34 mmol, 42%).1H NMR (CDCl3, 500 MHz): δ 1H NMR (500 MHz, CDCl3): δ 7.87–7.84 (m, 2H), 7.59–7.56 (m, 2H), 7.44–7.41 (m, 2H), 7.32 (d, J = 7.9 Hz, 2H), 6.60 (s, 1H), 5.26 (s, 2H), 4.33 (d, J = 6.1 Hz, 2H), 1.46 (s, 9H), 1.35 (s, 12H), 1.23 (s, 8H).; 13C NMR (CDCl3, 126 MHz): δ 151.3 (Cq), 149.3 (CH), 123.7 (CH), 37.0 (CH2), 23.2 (CH2),13.5 (CH3) ppm; HRMS (APCI) m/z: calcd for C30H36O5N2BS [M – H]+, 546.2469; found [M + H]+, 546.2466.
(3-(2,3-Dihydrobenzo[b][1,4]dioxin-6-yl)-2-methylphenyl)methanol 5
A mixture of 3-bromo-2-methylphenyl)methanol (5.2 g, 25.9 mmol) and (2,3-dihydrobenzo[b][1,4]dioxin-6-yl)boronic acid in toluene/ethanol/sat. aq sodium bicarbonate solution (5:1:5, 0.3 M) was placed under nitrogen and degassed for 10 min. [1,1′-Bis(diphenylphosphino)ferrocene]palladium(II) chloride (95 mg, 0.1 mmol) was added, and the reaction mixture was heated to 85 °C for 12 h. Ethyl acetate and water were added to the reaction mixture. The organic phase was washed with 1 M NaOH solution and brine. The organic layer was filtered over MgSO4 and concentrated under reduced pressure. The crude product was purified by silica gel flash chromatography using 0–100% EtOAc/PE as eluent and provided the desired product as a colorless solid (6.0 g, 23.4 mmol, 90%); 1H NMR (CDCl3, 500 MHz): δ 7.35 (dd, J = 7.6, 1.6 Hz, 1H), 7.21 (t, J = 7.6 Hz, 1H), 7.16 (dd, J = 7.6, 1.6 Hz, 1H), 6.89 (d, J = 8.2 Hz, 1H), 6.80 (d, J = 2.1 Hz, 1H), 6.75 (dd, J = 8.2, 2.1 Hz, 1H), 4.73 (s, 2H), 4.28 (s, 4H), 2.24 (s, 3H); 13C NMR (CDCl3, 126 MHz): δ 143.1, 142.6, 142.3, 139.3, 135.5, 133.7, 129.6, 126.6, 125.6, 122.6, 118.3, 116.9, 64.5, 64.5, 64.0, 16.0; HRMS (APCI) m/z: calcd for C16H15O2 [M + H–H2O]+: 239.1067, found [M + H–H2O]+: 239.1066.
6-(3-(Chloromethyl)-2-methylphenyl)-2,3-dihydrobenzo[b][1,4]dioxine 6
3-(2,3-Dihydrobenzo[b][1,4]dioxin-6-yl)-2-methylphenyl)methanol (1.1 g, 4.3 mmol) was dissolved in 10 mL DCM and cooled to 0 °C. Thionyl chloride (3.1 mL, 42.5 mmol) was added dropwise and reaction mixture was allowed to warm to RT and stirred for 18 h. After reaction completion, crude was extracted thrice with EtOAc and sodium bicarbonate. Combined organic layer was filtered over MgSO4 and concentrated under reduced pressure to provide the desired product as an orange oil (1.2 g, 4.3 mmol, quant. Yield) 1H NMR (500 MHz, CDCl3): δ 7.30 (t, J = 4.6 Hz, 1H), 7.21–7.19 (m, 2H), 6.90 (d, J = 8.2 Hz, 1H), 6.81 (d, J = 2.1 Hz, 1H), 6.75 (dd, J = 8.2, 2.1 Hz, 1H), 4.67 (s, 2H), 4.29 (s, 4H), 2.33 (s, 3H); 13C NMR (126 MHz, CDCl3): δ 143.2, 142.8, 142.8, 136.2, 135.3, 135.1, 130.8, 129.0, 125.9, 122.6, 118.3, 117.0, 64.5, 45.7, 16.2; HRMS (APCI) m/z: calcd for C16H16ClO2 [M + H]+, 275.0833; found [M + H]+, 275.0832.
5-Chloro-4-((3-(2,3-dihydrobenzo[b][1,4]dioxin-6-yl)-2-methylbenzyl)oxy)-2-hydroxybenzaldehyde 7
A mixture of 6-(3-(chloromethyl)-2-methylphenyl)-2,3-dihydrobenzo[b][1,4]dioxine (2.7 g, 9.8 mmol), 5-chloro-2,4-dihydroxybenzaldehyde (1.7 g, 9.8 mmol), potassium iodide (1.6 g, 9.8 mmol), and sodium bicarbonate (0.8 g, 9.8 mmol) was dissolved in 40 mL ACN. The reaction mixture was stirred at 65 °C for 2 days. After reaction completion, reaction mixture was concentrated and crude was extracted thrice with DCM and water. Combined organic layer was concentrated under reduced pressure. Crude was suspended in THF, filtered, and washed with water to provide the desired product as a colorless solid (3.0 g, 7.3 mmol, 74%).1H NMR (500 MHz, CDCl3): δ 11.46 (s, 1H), 9.72 (s, 1H), 7.57 (s, 1H), 7.47 (dd, J = 6.6, 2.5 Hz, 1H), 7.32–7.23 (m, 3H), 6.94 (d, J = 8.2 Hz, 1H), 6.86 (d, J = 2.0 Hz, 1H), 6.81 (dd, J = 8.2, 2.0 Hz, 1H), 6.66 (s, 1H), 5.22 (s, 2H), 4.34 (s, 4H), 2.29 (s, 3H), 1.65 (s, 3H); 13C NMR (126 MHz, CDCl3): δ 193.9, 163.1, 161.1, 143.2, 142.8, 142.6, 135.2, 134.3, 134.2, 133.6, 130.6, 127.4, 125.8, 122.7, 118.4, 117.0, 115.1, 115.0, 101.7, 70.5, 64.6, 64.6, 25.7, 16.4; HRMS (ESI) m/z: calcd for C23H20ClO5 [M + H]+, 411.0994; found [M + H]+, 411.0993.
3-((4-Chloro-5-((3-(2,3-dihydrobenzo[b][1,4]dioxin-6-yl)-2-methylbenzyl)oxy)-2-formy)lphenoxy)methyl)benzonitrile 8
A mixture of 5-chloro-4-((3-(2,3-dihydrobenzo[b][1,4]dioxin-6-yl)-2-methylbenzyl)oxy)-2-hydroxybenzaldehyde (1.80 g, 4.39), 3-(chloromethyl)benzonitrile (729 mg, 4.81 mmol), and potassium carbonate (665 mg, 4.81 mmol) was dissolved in 30 mL ACN and 1 mL DMF. Reaction mixture was stirred at 80 °C for 16 h. After reaction completion, crude was extracted thrice with DCM and water. Combined organic layer was filtered over MgSO4 and concentrated under reduced pressure. The crude product was purified by silica gel flash chromatography using 0–100% EtOAC/PE) to provide the desired product as a pink solid (4.31 g, 4.32 mmol, 98%).1H NMR (500 MHz, CDCl3): δ 10.34 (s, 1H), 7.93 (s, 1H), 7.75 (d, J = 1.9 Hz, 1H), 7.70 (ddt, J = 7.8, 4.9, 1.9 Hz, 2H), 7.57 (t, J = 7.8 Hz, 1H), 7.42 (dd, J = 5.6, 3.5 Hz, 1H), 7.28 (dd, J = 5.8, 2.3 Hz, 2H), 6.94 (d, J = 8.2 Hz, 1H), 6.84 (d, J = 2.1 Hz, 1H), 6.79 (dd, J = 8.2, 2.1 Hz, 1H), 6.64 (s, 1H), 5.23 (s, 2H), 5.21 (s, 2H), 4.34 (s, 4H), 2.31 (s, 3H). 13C NMR (126 MHz, CDCl3): δ 186.8, 160.7, 160.1, 143.3, 142.9, 142.8, 137.3, 135.0, 134.4, 133.7, 132.3, 131.5, 130.8, 130.7, 130.4, 129.9, 127.5, 125.8, 122.6, 119.5, 118.4, 118.3, 117.2, 117.1, 113.3, 98.8, 70.6, 69.9, 64.6, 64.6, 16.5.; HRMS (APCI) m/z: calcd for C31H25ClNO5 [M + H]+, 526.1416; found [M + H]+, 526.1417.
(2R,4S)-1-(5-Chloro-2-((3-cyanobenzyl)oxy)-4-((3-(2,3-dihydrobenzo[b][1,4]dioxin-6-yl)-2-methylbenzyl)oxy)benzyl)-4-fluoropyrrolidine-2-carboxylic Acid 9a
To a stirred solution of 3-((4-chloro-5-((3-(2,3-dihydrobenzo[b][1,4]dioxin-6-yl)-2-methylbenzyl)oxy)-2-formylphenoxy)methyl)benzonitrile (150 mg, 0.3 mmol) and (2S,4S)-4-fluoropyrrolidine-2-carboxylic acid (193 mg, 1.5 mmol) in 3 mL DMF, 3 drops of acetic acid were added and stirring continued for 30 min at RT. Subsequently, sodium cyanoborohydride (105 mg, 1.7 mmol) was added and reaction mixture was heated to 80 °C for 3 h. The crude product was purified by silica gel flash chromatography using 0–12% MeOH/DCM to provide the desired product as a colorless oil (112 mg, 0.2 mmol, 60%).1H NMR (500 MHz, CDCl3): δ 7.68 (s, 1H), 7.62 (d, J = 7.4 Hz, 2H), 7.50 (t, J = 7.4 Hz, 1H), 7.40 (s, 1H), 7.35 (dd, J = 6.1, 3.0 Hz, 1H), 7.24–7.19 (m, 2H), 6.90 (d, J = 8.1 Hz, 1H), 6.80 (d, J = 1.9 Hz, 1H), 6.75 (dd, J = 8.1, 1.9 Hz, 1H), 6.57 (s, 1H), 5.14 (s, 2H), 5.06 (s, 2H), 4.30 (s, 4H), 4.23 (d, J = 12.8 Hz, 1H), 4.03 (d, J = 12.8 Hz, 1H), 3.93 (dd, J = 9.9, 7.7 Hz, 1H), 3.59 (ddd, J = 35.8, 13.5, 3.8 Hz, 1H), 3.20 (dd, J = 24.3, 13.5 Hz, 1H), 2.63 (ddd, J = 20.9, 15.1, 7.7 Hz, 1H), 2.24 (s, 3H),; 13C NMR (CDCl3, 126 MHz): δ 13C NMR (126 MHz, CDCl3): δ 172.3, 156.1, 155.6, 143.2, 142.8, 142.6, 137.6, 135.1, 134.3, 134.2, 133.1, 132.2, 131.7, 130.8, 130.5, 129.9, 127.4, 125.7, 122.7, 118.5, 118.3, 117.7, 115.7, 113.2, 100.1, 94.0, 92.6, 70.6, 69.8, 66.3, 64.6, 64.6, 58.9, 58.7, 54.5, 37.3, 37.2, 16.4.; HRMS (ESI) m/z: calcd for C36H33ClFN2O6 [M + H]+, 643.2006; found [M + H]+, 643.2004.
(2R,4S)-1-(5-Chloro-2-((3-cyanobenzyl)oxy)-4-((3-(2,3-dihydrobenzo[b][1,4]dioxin-6-yl)-2-methylbenzyl)oxy)benzyl)-4-hydroxypyrrolidine-2-carboxylic Acid 9b
To a stirred solution of 3-((4-chloro-5-((3-(2,3-dihydrobenzo[b][1,4]dioxin-6-yl)-2-methylbenzyl)oxy)-2-formylphenoxy)methyl)benzonitrile (350 mg, 0.7 mmol) and methyl (2S,4R)-4-hydroxypyrrolidine-2-carboxylic acid (292 mg, 2.0 mmol) in 4 mL DCE/DMF (3:1), 2 drops of acetic acid were added and stirring continued for 30 min at RT. Subsequently, reaction mixture was cooled to 0 °C, sodium triacetoxyborohydride (568 mg, 2.7 mmol) was added and reaction mixture was stirred for 12 h at RT. After reaction completion, the crude was extracted with EtOAc and sat. sodium bicarbonate aqueous solution. Organic layer was washed with brine, filtered over MgSO4, and concentrated under reduced pressure. Compound was purified by silica gel flash chromatography using 30–100% EtOAC/PE to provide the desired product as a colorless oil (246 mg, 0.4 mmol, 58%). 1H NMR (CDCl3, 500 MHz): δ 7.75 (s, 1H), 7.65 (ddt, J = 17.5, 7.8, 1.4 Hz, 2H), 7.51 (t, J = 7.8 Hz, 1H), 7.40 (dd, J = 5.6, 3.5 Hz, 1H), 7.34 (s, 1H), 7.22 (d, J = 2.0 Hz, 1H), 7.22 (s, 1H), 6.91 (d, J = 8.2 Hz, 1H), 6.81 (d, J = 2.0 Hz, 1H), 6.76 (dd, J = 8.2, 2.0 Hz, 1H), 6.56 (s, 1H), 5.10 (s, 2H), 5.06 (s, 2H), 4.30 (s, 4H), 4.27–4.22 (m, 1H), 3.82 (d, J = 13.3 Hz, 1H), 3.78 (d, J = 13.3 Hz, 1H), 3.34 (dd, J = 10.1, 4.0 Hz, 1H), 3.04 (dt, J = 9.8, 1.5 Hz, 1H), 2.66 (dd, J = 9.8, 4.0 Hz, 1H), 2.38 (ddd, J = 14.1, 10.1, 5.7 Hz, 1H), 2.28 (s, 3H), 1.95–1.88 (m, 1H). 13C NMR (H2O + D2O, 126 MHz): δ 158.1, 157.9, 144.6, 144.2, 143.9, 139.3, 136.4, 135.9, 135.5, 134.1, 133.9, 133.3, 132.7, 131.3, 131.2, 128.6, 126.6, 123.5, 119.6, 119.3, 118.0, 116.2, 113.9, 113.2, 101.1, 79.5, 79.2, 78.9, 71.5, 71.1, 70.0, 68.7, 65.7, 62.5, 54.7, 39.0, 30.8, 16.7; HRMS (ESI) m/z: calcd for C36H34ClN2O7 [M + H]+, 641.2049; found [M + H]+, 641.2045.
Methyl (2R,4R)-1-(5-Chloro-2-((3-cyanobenzyl)oxy)-4-((3-(2,3-dihydrobenzo[b][1,4]dioxin-6-yl)-2-methylbenzyl)oxy)benzyl)-4-hydroxypyrrolidine-2-carboxylate 9c
To a stirred solution of 3-((4-chloro-5-((3-(2,3-dihydrobenzo[b][1,4]dioxin-6-yl)-2-methylbenzyl)oxy)-2-formylphenoxy)methyl)benzonitrile (296 mg, 0.6 mmol) and methyl (2S,4R)-4-hydroxypyrrolidine-2-carboxylate (390 mg, 2.7 mmol) in 4 mL DCE/DMF (3:1), 2 drops of acetic acid were added and stirring continued for 30 min at RT. Subsequently, reaction mixture was cooled to 0 °C, sodium triacetoxyborohydride (475 mg, 1.7 mmol) was added and reaction mixture was stirred for 12 h at RT. After reaction completion, the crude was extracted with EtOAc and sat. sodium bicarbonate aqueous solution. Organic layer was washed with brine, filtered over MgSO4, and concentrated under reduced pressure. Compound was purified by silica gel flash chromatography using 30–100% EtOAC/PE to provide the desired product as a colorless oil (215 mg, 0.3 mmol, 58%). 1H NMR (CDCl3, 500 MHz): δ 7.75 (d, J = 1.8 Hz, 1H), 7.65 (ddt, J = 14.3, 7.7, 1.3 Hz, 2H), 7.51 (t, J = 7.7 Hz, 1H), 7.40 (dd, J = 5.6, 3.4 Hz, 1H), 7.34 (s, 1H), 7.25–7.19 (m, 2H), 6.91 (d, J = 8.2 Hz, 1H), 6.81 (d, J = 2.1 Hz, 1H), 6.77 (dd, J = 8.2, 2.1 Hz, 1H), 6.56 (s, 1H), 5.10 (s, 2H), 5.06 (s, 2H), 4.31 (s, 4H), 4.27–4.23 (m, 1H), 3.86–3.76 (m, 2H), 3.56 (s, 3H), 3.34 (dd, J = 10.1, 4.0 Hz, 1H), 3.05 (dt, J = 9.8, 1.5 Hz, 1H), 2.66 (dd, J = 9.8, 4.0 Hz, 1H), 2.38 (ddd, J = 14.2, 10.1, 5.8 Hz, 1H), 2.28 (s, 3H), 2.06 (d, J = 16.3 Hz, 1H), 1.92 (ddt, J = 14.2, 3.1, 1.5 Hz, 1H); 13C NMR (CDCl3, 126 MHz): δ 175.5, 155.6, 154.1, 143.1, 142.7, 142.4, 138.3, 135.1, 134.6, 134.2, 132.2, 131.7, 131.4, 130.6, 130.3, 129.5, 127.43, 125.6, 122.6, 120.4, 118.6, 118.2, 116.9, 115.5, 112.9, 100.5, 71.0, 70.6, 69.7, 64.6, 64.5, 63.2, 62.0, 52.0, 51.0, 39.3, 16.3; HRMS (ESI) m/z: calcd for C37H36ClN2O7 [M + H]+: 655.2206; found [M + H]+: 655.2204.
Methyl (2R,4R)-1-(5-Chloro-2-((3-cyanobenzyl)oxy)-4-((3-(2,3-dihydrobenzo[b][1,4]dioxin-6-yl)-2-methylbenzyl)oxy)benzyl)-4-(tosyloxy)pyrrolidine-2-carboxylate 10
Methyl (2R,4R)-1-(5-chloro-2-((3-cyanobenzyl)oxy)-4-((3-(2,3-dihydrobenzo[b][1,4]dioxin-6-yl)-2-methylbenzyl)oxy)benzyl)-4-hydroxypyrrolidine-2-carboxylate (215 mg, 0.3 mmol) was dissolved in 6 mL pyridine and cooled to 0 °C. 4-Toluenesulfonyl chloride (76 mg, 0.4 mmol) was added and after 1 h reaction time allowed to warm up to RT. The reaction mixture was stirred 2 days at RT. After reaction completion, crude was extracted twice with ethyl acetate and water. The combined organic layer was washed with brine, filtered over MgSO4, and concentrated under reduced pressure. Crude was purified by silica gel flash chromatography using 0–100% EtOAC/PE to provide the desired product as a colorless solid (148 mg, 0.2 mmol, 56%).1H NMR (CDCl3, 500 MHz): δ (d, J = 5.9 Hz, 2H), ppm; 13C NMR (CDCl3, 126 MHz): δ 7.75 (d, J = 8.4 Hz, 2H), 7.72 (d, J = 1.6 Hz, 1H), 7.66 (dt, J = 7.8, 1.4 Hz, 1H), 7.62 (dt, J = 7.8, 1.4 Hz, 1H), 7.51 (t, J = 7.7 Hz, 1H), 7.40 (dd, J = 5.5, 3.7 Hz, 1H), 7.32–7.28 (m, 3H), 7.24–7.21 (m, 2H), 6.91 (d, J = 8.2 Hz, 1H), 6.81 (d, J = 2.0 Hz, 1H), 6.77 (dd, J = 8.2, 2.0 Hz, 1H), 6.55 (s, 1H), 5.09 (s, 2H), 5.04 (d, J = 2.0 Hz, 2H), 4.99–4.95 (m, 1H), 4.31 (s, 4H), 3.85 (d, J = 13.6 Hz, 1H), 3.70–3.65 (m, 1H), 3.57 (s, 3H), 3.25 (t, J = 7.8 Hz, 1H), 3.15 (d, J = 11.4 Hz, 1H), 2.64 (dd, J = 11.4, 5.7 Hz, 1H), 2.52–2.44 (m, 1H), 2.44 (s, 3H), 2.28 (s, 3H), 2.24–2.15 (m, 1H); 13C NMR (CDCl3, 126 MHz): δ 172.7, 155.7, 154.3, 144.9, 143.2, 142.8, 142.5, 138.3, 135.2, 134.7, 134.3, 133.9, 132.2, 131.9, 131.7, 130.7, 130.4, 130.0, 129.9, 129.7, 127.9, 127.5, 125.7, 122.7, 118.7, 118.4, 117.00, 115.6, 112.9, 100.6, 78.7, 70.7, 69.8, 64.6, 64.5, 63.4, 58.4, 52.0, 50.5, 36.8, 21.8, 16.4; HRMS (ESI) m/z: calcd for C44H42ClN2O9S [M + H]+, 809.2294; found [M + H]+, 809.2294.
Tracer Production
Radiolabeling of [18F]4a was based on a recent publication (Mossine et al. 2015) that reports the usage of boronic acids or boronic esters via copper-mediated fluorination.2818F-fluorine was eluted from an activated QMA cartridge with 2.7 mg tetraethylammonium bicarbonate in 0.4 mL nBu-OH to a reaction vial containing precursor (5–15 mg, 9–28 μmol) and 13.6 mg Cu(OTf)2(py)4 (13.6 mg, 20 μmol) that were dissolved in 0.8 mL DMA. The reaction proceeded for 15 min at 110 °C with constant stirring. After completion, 100 μL 6 M HCl was added and left stirring for 10 min in the same conditions. The reaction was passed through an activated Sep-Pak C18 light SPE cartridge and purified further with RP HPLC (40% ACN/25 mM PBS; XBridge BEH Shield OBD Prep RP18 5 μm 10 × 250 mm). The formulation was performed by loading the product diluted in 60 mL of water on activated Sep-Pak C18 light SPE cartridge and elution with 0.8 mL EtOH. The organic solvent was diluted with 5 mL of PBS to achieve less than 14% of ethanol in the final solution.
Radiolabeling of [18F]9a was performed by standardly used nucleophilic fluorination. Briefly, [18F]F– from cyclotron was passed through a conditioned QMA light cartridge, dried with air, and eluted with 3.5 mg K2CO3 and 20 mg K2.2.2 in 0.7 mL ACN/0.2 mL water. [18F]F– was dried azeotropically (3 × 0.5 mL anhydrous ACN) and precursor was added in anhydrous ACN or DMSO (5 mg, 6.2 μmol, 0.3 mL). The reaction was monitored using TLC plates (eluent and purified further with RP HPLC (50% ACN/0.1 M sodium acetate; XBridge BEH Shield OBD Prep RP18 5 μm 10 × 250 mm). The formulation was performed by loading the product diluted in 60 mL of water on activated Sep-Pak C18 light SPE cartridge and elution with 0.8 mL EtOH. The organic solvent was diluted with 5 mL of water + 6 μL Tween-80 to achieve less than 14% of ethanol in the final solution. For lipophilicity, stability, and ex vivo study, synthesis of [18F]BMS1166 was performed in Synthera IBA automation module and purification was performed with a formulation in E&Z automation module.
The identity, purity, and purification of tracers were determined by high-performance liquid chromatography (HPLC). The Waters ACQUITY HPLC system was equipped with a dual wavelength absorbance detector, in-line radioactivity detector, and HPLC column: XBridge Prep Shield and XBridge BEH Shield OBD Prep column (both RP18 5 μm 10 × 250 mm, Waters, USA). Secondary quality control was performed using ultra performance liquid chromatography (UPLC). The Waters ACQUITY HPLC H-class UPLC system was equipped with a dual wavelength absorbance detector in line with LB513 radioactivity detector with an MX50-2 cell (Berthold) and UPLC column: BEH Shield and BEH Phenyl (both RP18 1.7 μm 3.0 × 50 mm, Acquity Waters, USA). For in vitro tracer stability, the radiochemical purity of labeled products was measured using thin-layer chromatography (TLC) (silica TLC plates on aluminum, MerckMilipore, USA). The TLCs were developed with an appropriate MeOH/DCM eluent system. The distribution of the radioactivity among the TLC plate was measured on GE Amersham Typhoon Scanner using phosphorus plates and ImageQuant TL 1D software for data processing (both GE Healthcare Life Science, USA).
The radioactivity was measured using a dose calibrator (VDC-505, Commecer, Netherlands) and 2480 Wizard Detector Gamma Counter (PerkinElmer, Netherlands).
In Vitro Stability
In vitro stability of [18F]4a and [18F]9a was evaluated in Tween-80 or PBS (original diluent) and serum. After purification, samples of tracers were mixed with equal volumes of the formulated tracer and solution in which stability was evaluated (100 μL tracer solution with 100 μL Tween-80/PBS) and measured every 30 min for a 4 h period. The radiochemical purity (RCP) of labeled products was measured using thin-layer chromatography (TLC) (silica TLC plates on aluminum, MerckMilipore, USA). The TLCs were developed with appropriate MeOH/DCM eluent system; 10% MeOH/DCM + NH3 for [18F]4a and 10% MeOH/DCM for [18F]9a. The distribution of the radioactivity among the TLC plate was measured on GE Amersham Typhoon Scanner using phosphorus plates and ImageQuant TL 1D software for data processing (both GE Healthcare Life Science, USA). The experiment was performed in triplicate.
Cell Lines
The human tumor cell line H358 (lung adenocarcinoma) was obtained from the American Type Culture Collection and ES2 (human ovarian clear cell carcinoma) cell line was a gift from Dr. Els Berns (Erasmus MC, The Netherlands). Tumor cells were cultured in RPMI 1640 medium (Gibco, Paisley, UK) supplemented with 10% fetal bovine serum (FBS, Bodinco BV, The Netherlands) and maintained in a 5% CO2 37 °C incubator. Cells were tested negative for mycoplasma contamination by PCR analysis and proven their authenticity in short tandem repeat profiling.
CHO-K1 artificial antigen-presenting cells (PD-L1+ aAPC/CHO-K1 cells, called aAPCs) overexpressing TCR ligand and PD-L1, and a modified Jurkat T cell line overexpressing PD-1 with a luciferase reporter under the control of NFAT promoter (PD-1 effector cells, called ECs) were obtained from Promega. PD-L1-expressing aAPC–CHO-K1 cells and PD-1 effector cells were cultured in RPMI 1640 medium (BioWest) supplemented with 10% FBS and 2 mM l-glutamine. Additionally, aAPCs and ECs were propagated in a constant presence of hygromycin B (50 μg/mL) and G418 (250 μg/mL) to provide a stable expression of the introduced genetic constructs. The two latter antibiotics were omitted in the experiments.
Flow Cytometry
To assess the extracellular expression of PD-L1 on used samples, cells were collected and stained with CD279 (PD-1)-APC and CD274 (PD-L1)-PE antibodies. After incubation at 4 °C for 30 min, cells were washed thrice with PBS + 2% FBS, and their surface expression was measured using BD Accuri c6 flow cytometer (BD Biosciences) and BD FACSVerse flow cytometer (BD Biosciences). Data analysis was performed with the use of Cytobank (www.cytobank.org).
Western Blotting
To assess the intracellular expression of PD-L1 in used samples, cells were collected and lysed in MPER buffer + 1% protease and 1% phosphatase inhibitor for 30 min on ice followed by pelleting of insoluble material by centrifugation. Sample concentrations were determined using Bradford assay (Bradford, 1976) and suitable amounts were heated to 100 °C in SDS sample buffer + 10% β-mercaptoethanol for 10 min.39 Lysates were separated by SDS–PAGE and transferred to the PVDF membrane (Immobilon P, Millipore). Membranes were blocked in 5% milk in TBS + 0.05% Tween-20, probed with the indicated antibodies, and reactive bands visualized using Image Lab software (Bio-Rad).
Generation of PD-L1 Knock-Out Cell Lines
PD-L1 knock-out cell line was constructed using the CRISPR-Cas9 engineering technique on H358 and ES2. The guide was prepared using primers CACCGTCTTTATATTCATGACCTAC (54 °C) and AAACGTAGGTCATGAATATAAAGAC (51 °C) reported in the literature (Liao et al., 2017).40 Guide duplex was ligated with thermocycler, and fused into pSpCas9(BB)-2A-GFP plasmid (pSpCas9(BB)-2A-GFP (PX458) was a gift from Feng Zhang; Addgene plasmid #48138; http://n2t.net/addgene:48138; RRID:Addgene_48138). The plasmid construct was subjected to bacterial transformation on agar Petri dishes containing ampicillin, lysed the next day, and purified using a DNA purification kit (Qiagen). As final validation of obtaining the right plasmid, samples were sent for sequencing to GATC Biotech. Transfection with the specific PD-L1-Cas9-GFP vector was performed using FuGENE HD Transfection Reagent (Promega) according to manufacturer protocol. For the experiment, cells were plated one day before transfection (6-well plate, 0.25–1 × 106 cells in 2 mL of growth medium without antibiotics, 80–90% confluent at the time of transfection). Cells were incubated at 37 °C in a CO2 incubator and visualized for positive transfection by green fluorescence positivity under a fluorescent microscope (EVOS FL imaging system, Thermo Fisher). After 4 days, GFP-positive cells were sorted into 96-well plates as single-cell clones using SH800S Sony sorter (Sony, USA). Expanded clones were analyzed by flow cytometry using CD274-PE antibody and selected ones characterized by Western blotting. The clones confirmed with a constitutive knockout of PD-L1 expression were used for the following functional assays.
Ex Vivo Tumor Cell Analysis
Formalin-fixed paraffin-embedded xenograft tumors (ES2 PD-L1+/– and H358 PD-L1+/–) were cut into 4 μm slices and placed on the glass slides. Before each immunohistochemistry and autoradiography experiment, tissue slides were deparaffinized by oven incubation (min 1 h at 60 °C) and xylene washes, then rehydrated through graded alcohol and distilled water. Additionally, heat-induced antigen retrieval with 10 mM citrate buffer pH = 6.0 was performed (15 min 400 W). Snap-frozen human tonsil tissue was a gift from the Pathology Department (UMCG). Before each experiment, they were incubated in cold acetone for 10 min.
For immunohistochemistry, antigen retrieval was followed by endogenous peroxidase block (S2003; Dako), endogenous IgG block (X0909; Dako), and incubation with anti-PD-L1 antibody (5 μg/mL, clone 28–8; Abcam) for 60 min. Next, sections were incubated with anti-rabbit Dako EnVision+ polymer for 30 min (K4010; Dako). The staining was visualized using 3,3′-diaminobenzidine + substrate (K3468; Dako) and counterstained using hematoxylin. The image was recorded on NanoZoomer Digital Slide Scanner and visualized by NDP View software (both Hamamatsu Photonics, Japan).
For autoradiography, antigen retrieval and acetone wash were followed by 30 min of incubation with a freshly formulated tracer at a concentration of 0.2 MBq/mL. After washing twice with cold PBS and once with water, slides were left to dry and exposed to phosphorus plates. The image was recorded on GE Amersham Typhoon Biomolecular Imager and analyzed with ImageQuant TL 1D software (both GE Healthcare Life Science, USA).
PD-L1 Expression
The PD-L1 protein (18–134) was expressed in the Escherichia coli BL21 (DE3) and purified as described previously (Zak et al., 2016).8 Protein expression was induced with 1 mM isopropyl β-d-1-thiogalactopyranosid (IPTG) at OD600 of 0.8 and the cells were cultured overnight at 37 °C. Afterward protein was refolded by drop-wise dilution into solution containing 0.1 M Tris pH 8.0, 1 M l-arginine hydrochloride, 0.25 mM oxidized glutathione, and 0.25 mM reduced glutathione. After refolding, protein was dialyzed 3 times against buffer containing 10 mM Tris pH 8.0 and 20 mM NaCl. Finally, PD-L1 was purified by SEC (size-exclusion chromatography) on HiLoad 26/600 Superdex 75 column (GE Healthcare) equilibrated with PBS pH 7.4.
NMR Binding Assay
For NMR measurements, the buffer was exchanged by gel filtration to PBS pH 7.4. 10% (v/v) of D2O was added to the samples to provide the lock signal. All spectra were recorded at 300 K using a Bruker Avance III 600 MHz spectrometer equipped with the nitrogen cryo-probe head.
Determination of binding of compounds to PD-L1 was carried out with the 1H NMR. The line width broadening in the proton NMR of PD-L1 suggests that the compounds induce protein oligomerization. In all the cases, the well-resolved narrow resonance peaks in the aliphatic region of 1H NMR spectrum of apo-PD-L1 exhibited significant broadening upon the addition of each compound indicating a significant increase in the molecular weight of the complex. The molecular weight of each complex was estimated from relaxation time analysis, which can only be explained by the compound-induced PD-L1 dimerization. No significant changes were observed upon the addition of the PD-1 with the tested compounds.
Microscale Thermophoresis Assay
PD-L1 was expressed as for NMR experiment. It was diluted to 10 μM and labelled with Protein Labeling Kit RED-NHS 2nd Generation from Nanotemper according to the manufacturer’s guidelines. The degree of labelling for PD-L1 was 0.74, which is with agreement with the protocol. Labelled PD-L1 at 100 nM concentration was then mixed with independent series of 4a and 9a compound dilution series (n = 4) and incubated for 30 min at RT. Next, samples were transferred to dedicated 384-well plate and measured on Diantus equipped with pico detector using auto-excitation and 5 s pulse (Nanotemper) for the affinity measurements. The results were exported, averaged, and fitted in Mathematica 12 using following equations:
where: FB—fraction bound, [A]—concentration of unlabeled titrated partner, [B]—concentration of fluorescent labeled partner that is fixed, [AB]—concentration of bound complex of A and B, Kd—equilibrium dissociation constant.
Homogeneous Time-Resolved Fluorescence Assay
The HTRF assay was performed using the certified Cis-Bio assay kit at 20 μL final volume using their standard protocol as described by Musielak et al.34 Measurements were performed on individual dilution series to determine the half maximal inhibitory concentration (IC50) of tested compounds. After mixing all components according to the Cis-Bio protocol, the plate was incubated for 2 h at RT. TR-FRET measurement was performed on the Tecan Spark 20 M. Collected data was background-subtracted on the negative control, normalized on the positive control, averaged, and fitted with normalized Hill’s equation to determine the IC50 value using Mathematica 12. For the compounds with IC50 values that were too high due to, e.g., solubility issues, a “dissociation value” at the concentration of 50 μM is presented for the sake of comparability to other inhibitors. The dissociation value represents the percentage of the PD-1/PD-L1 complex that is undissolved.
Promega PD-1/PD-L1 Blockade Assay
The aAPCs were seeded in white flat bottom 96-well plates at the density of 10 000 cells/well in the culture medium 24 h before the assay and grown overnight in a 5% CO2 37 °C incubator. On the day of the experiment, serial dilutions of tested compounds were prepared first in DMSO and then formulated in RPMI 1640-containing 1% FBS to keep the constant DMSO concentration of 0.1% (v/v). As a negative control, DMSO was used in assay buffer at the concentration of 0.1% (v/v). As a positive control, a serial dilution of durvalumab (an anti-PD-L1 antibody) in assay buffer was used. Meanwhile, the medium of the aAPCs cells was removed, and the compounds’ dilutions were added to the plate (40 μL/well). PD-1 effector Jurkat cells were pelleted and resuspended in the assay buffer. The cell suspension was distributed over the inner 60 wells of the assay plate (20 000 cells/well, 40 μL). The plates were incubated for 6 h in a 5% CO2 incubator at 37 °C. Once the assay time was over, the plate was taken out of the incubator to equilibrate to room temperature. Before measurements, a 75 μL per well of Bio-Glo luciferase substrate was added to the inner 60 wells of the plate. 75 μL was also added to selected wells for background measurement. The measurement was done on a Spark microplate reader (Tecan), using the standard settings for luminescence reading.
Cytotoxicity Assay
5000 ECs (Jurkat T cell line overexpressing PD-1 with a luciferase reporter under the control of NFAT promoter) were seeded on transparent 96-well plates and cultured for 48 h in the presence of increasing concentrations of the compounds or DMSO as a control (the concentration of DMSO was kept constant in all samples). Following the treatment, a metabolic activity test was performed with the use of Biolog Redox Dye Mix MB (BioLog), according to the manufacturer’s instructions.
Molecular Docking
The structure of 4a, 9a, and 9b compounds was prepared and minimized in UCSF Chimera software with AutoDock Vina.41,42 The structure of the compound and PD-L1 dimer (PDB: 6R3K) were prepared in PyMol and UCSF Chimera. All water molecules and original ligand (BMS1166) were removed and polar hydrogens atoms were added to the receptor. A grid box of dimensions 20 × 20 × 20 Å and following coordinates x: −8.746, y = 18.049, and z = −21.934 was placed at the interface of PD-L1 homodimer. Docking was carried out with exhaustiveness = 8.11 The obtained binding poses were carefully visually inspected in PyMol and UCSF Chimera.
Statistics
All values were expressed as mean ± SD unless stated otherwise. Each value is the mean of at least three independent experiments in each group. Statistical analyses were performed in GraphPad Prism version 7.0 (GraphPad Software) using the Mann–Whitney test (2 groups, nonparametric) or one-way analysis of variance (ANOVA) with Tukey Post-Hoc test (>2 groups, nonparametric). The asterisk (*) indicates the values that are significantly different from control (*p < 0.05, **p < 0.051 and ***p < 0.001).
Acknowledgments
This research was funded (to T.A.H.) by the project POIR.04.04.00-00-420F/17-00, which is carried out within the TEAM program of the Foundation for Polish Science co-financed by the European Union under the European Regional Development Fund, COFUNDs ALERT (grant agreement No 665250), and KWF Kankerbestrijding grant (grant agreement No 10504). Further, we acknowledge the MCB Structural Biology Core Facility (supported by the TEAM TECH CORE FACILITY/2017-4/6 grant from the Foundation for Polish Science) for valuable support. J.K.-K. acknowledges the fellowship with project no. POWR.03.02.00-00-I013/16 from the National Center for Research and Development. A.D. acknowledges the ERA Chair grant from the European Union.
Glossary
Abbreviations
- aAPCs
antigen-presenting cells
- CDR
complementarity-determining region
- CTLA-4
cytotoxic T cell antigen 4
- G-3CR
Gewald three component reaction
- HTRF
homogeneous time resolved fluorescence
- IL-2
interleukin-2
- MST
microscale thermophoresis
- NFAT
nuclear factor of activated T-cells
- PD-1
programmed cell death protein 1
- PD-L1
programmed death-ligand 1
- SMI
small molecular weight inhibitor
- SPR
surface plasmon resonance.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acs.jmedchem.3c00254.
1H, 13C NMR spectra, HRMS, and UPLC chromatograms for the synthesized compounds; 1H–15N HMQC NMR spectra of PD-L1 and PD-L1 with inhibitor, Jurkat cell line viability in PD-1/PD-L1 ICB assay, and MST affinity analysis for 4a, 9a, 9b; labeling optimization, HPLC chromatograms, and stability in solution and serum for [18F]4a and [18F]9a; and flow cytometry and Western blot for wild-type and PD-L1 knock-out of H358 and ES2 cancer cell lines, ex vivo autoradiography of cancer cell lines, and tonsils incubated with [18F]4a and [18F]9a (PDF)
Molecular formula strings for the synthesized compounds (CSV)
Author Contributions
○ M.A.W. and R.B. made equal contributions to this work.
The authors declare no competing financial interest.
Special Issue
Published as part of the Journal of Medicinal Chemistry virtual special issue “Diagnostic and Therapeutic Radiopharmaceuticals”.
Supplementary Material
References
- Pardoll D. M. The blockade of immune checkpoints in cancer immunotherapy. Nat. Rev. Cancer 2012, 12, 252–264. 10.1038/nrc3239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michot J. M.; Bigenwald C.; Champiat S.; Collins M.; Carbonnel F.; Postel-Vinay S.; Berdelou A.; Varga A.; Bahleda R.; Hollebecque A.; Massard C.; Fuerea A.; Ribrag V.; Gazzah A.; Armand J. P.; Amellal N.; Angevin E.; Noel N.; Boutros C.; Mateus C.; Lambotte O. Immune-related adverse events with immune checkpoint blockade: a comprehensive review. Eur. J. Cancer 2016, 54, 139–148. 10.1016/j.ejca.2015.11.016. [DOI] [PubMed] [Google Scholar]
- European Medicines Agency . EMEA/H/C/003985—EPAR Product Information 2022. https://www.ema.europa.eu/en/documents/product-information/opdivo-epar-product-information_en.pdf (accessed Sept 01, 2022).
- Opdivo (nivolumab) price, Farmacotherapeutisch Kompas. https://www.farmacotherapeutischkompas.nl/bladeren/preparaatteksten/n/nivolumab#kosten (accessed Sept 01, 2022).
- Queva C.; Morrow M.; Hammond S.; Alimzhanov M.; Babcock J.; Foltz I.; Kang J. S.; Sekirov L.; Boyle M.; Chodorge M.. Targeted binding agents against b7-h1. WO 2011066389 A1, 2011.
- Nastri H. G.; Iffland C.; Leger O.; An Q.; Cartwright M.; Mckenna S. D.; Sood V. D.; Hao G.. Anti-pd-l1 antibodies and uses thereof. WO 2013079174 A1, 2013.
- Irving B.; Chiu H.; Maecker H.; Mariathasan S.; Lehar S. M.; Wu Y.; Cheung J.. Anti-PD-L1 antibodies, compositions and articles of manufacture. U.S. Patent 8,217,149 B2, 2012.
- Zak K. M.; Grudnik P.; Guzik K.; Zieba B. J.; Musielak B.; Dömling A.; Dubin G.; Holak T. A. Structural basis for small molecule targeting of the programmed death ligand 1 (PD-L1). Oncotarget 2016, 7, 30323–30335. 10.18632/oncotarget.8730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F.; Wei H.; Wang X.; Bai Y.; Wang P.; Wu J.; Jiang X.; Wang Y.; Cai H.; Xu T.; Zhou A. Structural basis of a novel PD-L1 nanobody for immune checkpoint blockade. Cell Discovery 2017, 3, 17004. 10.1038/celldisc.2017.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magiera-Mularz K.; Skalniak L.; Zak K. M.; Musielak B.; Rudzinska-Szostak E.; Berlicki Ł.; Kocik J.; Grudnik P.; Sala D.; Zarganes-Tzitzikas T.; Shaabani S.; Dömling A.; Dubin G.; Holak T. A. Bioactive Macrocyclic Inhibitors of the PD-1/PD-L1 Immune Checkpoint. Angew. Chem., Int. Ed. 2017, 56, 13732–13735. 10.1002/anie.201707707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chupak L. S.; Ding M.; Martin S. W.; Zheng X.; Hewawasam P.; Connolly T. P.; Xu N.; Yeung K.-S.; Zhu J.; Langley D. R.; Scola P. M.. Compounds useful as immunomodulators. WO 2015034820 A1, 2015.
- Wang Y.; Gu T.; Tian X.; Li W.; Zhao R.; Yang W.; Gao Q.; Li T.; Shim J. H.; Zhang C.; Liu K.; Lee M. H. A small molecule antagonist of PD-1/PD-L1 interactions acts as an immune checkpoint inhibitor for NSCLC and melanoma immunotherapy. Front. Immunol. 2021, 12, 654463. 10.3389/fimmu.2021.654463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxinovel Pty. Ltd. . MAX-10181 Given Orally to Patients With Advanced Solid Tumor (NCT 04122339). https://clinicaltrials.gov/ct2/show/NCT04122339 (accessed Sept 01, 2022).
- Gilead Sciences . Study to Evaluate Safety, Tolerability, Pharmacokinetics, and Efficacy of GS-4224 in Participants With Advanced Solid Tumors (NCT04049617). https://www.clinicaltrials.gov/ct2/show/NCT04049617 (accessed Sept 01, 2022).
- Konstantinidou M.; Zarganes-Tzitzikas T.; Magiera-Mularz K.; Holak T. A.; Dömling A. Immune Checkpoint PD-1/PD-L1: Is There Life Beyond Antibodies. Angew. Chemie Int. Ed. 2018, 57, 4840–4848. 10.1002/anie.201710407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butera R.; Ważyńska M.; Magiera-Mularz K.; Plewka J.; Musielak B.; Surmiak E.; Sala D.; Kitel R.; de Bruyn M.; Nijman H. W.; Elsinga P. H.; Holak T. A.; Dömling A. Design, Synthesis, and Biological Evaluation of Imidazopyridines as PD-1/PD-L1 Antagonists. ACS Med. Chem. Lett. 2021, 12, 768–773. 10.1021/acsmedchemlett.1c00033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaabani S.; Huizinga H. P. S.; Butera R.; Kouchi A.; Guzik K.; Magiera-Mularz K.; Holak T. A.; Dömling A. A patent review on PD-1/PD-L1 antagonists: small molecules, peptides, and macrocycles (2015-2018). Expert Opin. Ther. Pat. 2018, 28, 665–678. 10.1080/13543776.2018.1512706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guzik K.; Tomala M.; Muszak D.; Konieczny M.; Hec A.; Błaszkiewicz U.; Pustuła M.; Butera R.; Dömling A.; Holak T. A. Development of the Inhibitors that Target the PD-1/PD-L1 Interaction-A Brief Look at Progress on Small Molecules, Peptides and Macrocycles. Molecules 2019, 24, 2071. 10.3390/molecules24112071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muszak D.; Surmiak E.; Plewka J.; Magiera-Mularz K.; Kocik-Krol J.; Musielak B.; Sala D.; Kitel R.; Stec M.; Weglarczyk K.; Siedlar M.; Dömling A.; Skalniak L.; Holak T. A. Terphenyl-Based Small-Molecule Inhibitors of Programmed Cell Death-1/Programmed Death-Ligand 1 Protein-Protein Interaction. J. Med. Chem. 2021, 64, 11614–11636. 10.1021/acs.jmedchem.1c00957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McRee D. E.Computational Techniques, Practical Protein Crystallography, 2nd ed.; Academic Press, 1999; p 91. [Google Scholar]
- Gewald K.; Jablokoff H.; Hentschel M. Synthese und Reaktionen von 2-Hydroxy-3-cyan-thiophenen. J. Prakt. Chem. 1975, 317, 861–866. 10.1002/prac.19753170521. [DOI] [Google Scholar]
- Huang Y.; Dömling A. The Gewald multicomponent reaction. Mol. Diversity 2011, 15, 3–33. 10.1007/s11030-010-9229-6. [DOI] [PubMed] [Google Scholar]
- Wang K.; Kim D.; Dömling A. Cyanoacetamide MCR (III): three-component Gewald reactions revisited. J. Comb. Chem. 2010, 12, 111–118. 10.1021/cc9001586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y.; Dömling A. 1,4-Thienodiazepine-2,5-diones via MCR (II): scaffold hopping by Gewald and Ugi-deprotection-cyclization strategy. Chem. Biol. Drug Des. 2010, 76, 130–141. 10.1111/j.1747-0285.2010.00990.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y.; Wolf S.; Bista M.; Meireles L.; Camacho C.; Holak T. A.; Dömling A. 1,4-Thienodiazepine-2,5-diones via MCR (I): synthesis, virtual space and p53-Mdm2 activity. Chem. Biol. Drug Des. 2010, 76, 116–129. 10.1111/j.1747-0285.2010.00989.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skalniak L.; Zak K. M.; Guzik K.; Magiera K.; Musielak B.; Pachota M.; Szelazek B.; Kocik J.; Grudnik P.; Tomala M.; Krzanik S.; Pyrc K.; Dömling A.; Dubin G.; Holak T. A. Small-molecule inhibitors of PD-1/PD-L1 immune checkpoint alleviate the PD-L1-induced exhaustion of T-cells. Oncotarget 2017, 8, 72167–72181. 10.18632/oncotarget.20050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surmiak E.; Magiera-Mularz K.; Musielak B.; Muszak D.; Kocik-Krol J.; Kitel R.; Plewka J.; Holak T. A.; Skalniak L. PD-L1 Inhibitors: Different Classes, Activities, and Mechanisms of Action. Int. J. Mol. Sci. 2021, 22, 11797. 10.3390/ijms222111797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mossine A. V.; Brooks A. F.; Makaravage K. J.; Miller J. M.; Ichiishi N.; Sanford M. S.; Scott P. J. Synthesis of [18F]Arenes via the Copper-Mediated [18F]Fluorination of Boronic Acids. Org. Lett. 2015, 17, 5780–5783. 10.1021/acs.orglett.5b02875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng T.; Zhao Y.; Li X.; Lin F.; Xu Y.; Zhang X.; Li Y.; Wang R.; Lai L. Computation of Octanol-Water Partition Coefficients by Guiding an Additive Model with Knowledge. J. Chem. Inf. Model. 2007, 47, 2140–2148. 10.1021/ci700257y. [DOI] [PubMed] [Google Scholar]
- Brown J. A.; Dorfman D. M.; Ma F. R.; Sullivan E. L.; Munoz O.; Wood C. R.; Greenfield E. A.; Freeman G. J. Blockade of programmed death-1 ligands on dendritic cells enhances T cell activation and cytokine production. J. Immunol. 2003, 170, 1257–1266. 10.4049/jimmunol.170.3.1257. [DOI] [PubMed] [Google Scholar]
- Ma J.; Li J.; Qian M.; Han W.; Tian M.; Li Z.; Wang Z.; He S.; Wu K. PD-L1 expression and the prognostic significance in gastric cancer: a retrospective comparison of three PD-L1 antibody clones (SP142, 28–8 and E1L3N). Diagn. Pathol. 2018, 13, 91. 10.1186/s13000-018-0766-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y. J.; Sun W. P.; Peng J. H.; Deng Y. X.; Fang Y. J.; Huang J.; Zhang H. Z.; Wan D. S.; Lin J. Z.; Pan Z. Z. Programmed death-ligand 1 expression correlates with diminished CD8+ T cell infiltration and predicts poor prognosis in anal squamous cell carcinoma patients. Cancer Manage. Res. 2017, 10, 1–11. 10.2147/CMAR.S153965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasikumar P. G.; Sudarshan N. S.; Adurthi S.; Ramachandra R. K.; Samiulla D. S.; Lakshminarasimhan A.; Ramanathan A.; Chandrasekhar T.; Dhudashiya A. A.; Talapati S. R.; Gowda N.; Palakolanu S.; Mani J.; Srinivasrao B.; Joseph D.; Kumar N.; Nair R.; Atreya H. S.; Gowda N.; Ramachandra M. PD-1 derived CA-170 is an oral immune checkpoint inhibitor that exhibits preclinical anti-tumor efficacy. Commun. Biol. 2021, 4, 699. 10.1038/s42003-021-02191-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musielak B.; Kocik J.; Skalniak L.; Magiera-Mularz K.; Sala D.; Czub M.; Stec M.; Siedlar M.; Holak T. A.; Plewka J. CA-170 - A Potent Small-Molecule PD-L1 Inhibitor or Not?. Molecules 2019, 24, 2804. 10.3390/molecules24152804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blevins D. J.; Hanley R.; Bolduc T.; Powell D. A.; Gignac M.; Walker K.; Carr M. D.; Hof F.; Wulff J. E. In Vitro Assessment of Putative PD-1/PD-L1 Inhibitors: Suggestions of an Alternative Mode of Action. ACS Med. Chem. Lett. 2019, 10, 1187–1192. 10.1021/acsmedchemlett.9b00221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganesan A.; Ahmed M.; Okoye I.; Arutyunova E.; Babu D.; Turnbull W. L.; Kundu J. K.; Shields J.; Agopsowicz K. C.; Xu L.; et al. Comprehensive in vitro characterization of PD-L1 small molecule inhibitors. Sci. Rep. 2019, 9, 12392. 10.1038/s41598-019-48826-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasikumar P. G.; Ramachandra M.; Prasad A.; Naremaddepalli S. S. S.. 3-substituted-1,2,4-oxadiazole and thiadiazole compounds as immunomodulators. WO 2016142886 A3, 2016.
- Chen F. F.; Li Z.; Ma D.; Yu Q. Small-molecule PD-L1 inhibitor BMS1166 abrogates the function of PD-L1 by blocking its ER export. Oncoimmunology 2020, 9, 1831153. 10.1080/2162402X.2020.1831153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Liao Y.; Chen L.; Feng Y.; Shen J.; Gao Y.; Cote G.; Choy E.; Harmon D.; Mankin H.; Hornicek F.; Duan Z. Targeting programmed cell death ligand 1 by CRISPR/Cas9 in osteosarcoma cells. Oncotarget 2017, 8, 30276–30287. 10.18632/oncotarget.16326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen E. F.; Goddard T. D.; Huang C. C.; Couch G. S.; Greenblatt D. M.; Meng E. C.; Ferrin T. E. UCSF Chimera--a visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- Trott O.; Olson A. J. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2009, 31, 455–461. 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.