Abstract

In cystic fibrosis (CF), deletion of phenylalanine 508 (F508del) in the CF transmembrane conductance regulator (CFTR) is associated to misfolding and defective gating of the mutant channel. One of the most promising CF drug targets is the ubiquitin ligase RNF5, which promotes F508del-CFTR degradation. Recently, the first ever reported inhibitor of RNF5 was discovered, i.e., the 1,2,4-thiadiazol-5-ylidene inh-2. Here, we designed and synthesized a series of new analogues to explore the structure–activity relationships (SAR) of this class of compounds. SAR efforts ultimately led to compound 16, which showed a greater F508del-CFTR corrector activity than inh-2, good tolerability, and no toxic side effects. Analogue 16 increased the basal level of autophagy similar to what has been described with RNF5 silencing. Furthermore, co-treatment with 16 significantly improved the F508del-CFTR rescue induced by the triple combination elexacaftor/tezacaftor/ivacaftor in CFBE41o– cells. These findings validate the 1,2,4-thiadiazolylidene scaffold for the discovery of novel RNF5 inhibitors and provide evidence to pursue this unprecedented strategy for the treatment of CF.
1. Introduction
Cystic fibrosis (CF) is the most common genetic disorder in Caucasian populations1 caused by loss of function mutations in the CFTR gene encoding for the cystic fibrosis transmembrane conductance regulator (CFTR) protein.2 CFTR is a cAMP-regulated anion channel of primary importance for transepithelial chloride and bicarbonate ion transport in various organs, where it contributes to regulate salt and fluid homeostasis.3 While CF is a multisystem disease, the main clinical features are exocrine pancreatic insufficiency and bronchiectasis with chronic airway infection leading to respiratory failure and premature death.4 Until 10 years ago, the conventional therapy in use for CF was primarily based on controlling disease symptoms. Nowadays, the improved understanding of CFTR protein structure and of the consequence of gene mutations has opened the way to personalized treatments targeting specific defects.5
Currently, over 2000 different mutations in the CFTR have been described, although a pathogenetic role has been demonstrated only for approx. 400 of them, as reported in the Clinical and Functional Translation of CFTR2 database (https://cftr2.org, accessed on 23/03/2023). However, the most prevalent mutation is the deletion of a phenylalanine at position 508 (F508del), which affects ∼80% of CF patients worldwide, although with marked differences in frequency based on the ethnic origin. F508del-CFTR is responsible for three distinct defects of the mutant protein: (i) a trafficking defect due to misfolding of the F508del-CFTR, resulting in a reduced amount of channel present at the plasma membrane (PM);6−8 (ii) a decreased stability when the mutated channel is expressed on the plasma membrane;9 and (iii) a channel gating defect due to the reduced open-channel probability.10−12 Noteworthily, both F508del defects can be rescued, at least partially, using two classes of small-molecule CFTR modulators: correctors can help the transport of the misfolded CFTR to the cell surface,13 and potentiators can ameliorate the gating defect, helping to keep this ion channel open at the cell surface.14 Hence, combination therapies involving small molecules that synergistically aim at distinct structural defects are likely required to promote a marked F508del rescue.15
Intense research efforts in the CFTR modulators field resulted in the registration in 2012 of the potentiator ivacaftor (VX-770, Figure 1)16−18 under the trade name Kalydeco for patients with at least one copy of the G551D mutation, subsequently expanded to a selection of class III and IV mutations. It followed the 2015 marketing approval of the fixed dose combination Orkambi composed of ivacaftor and the corrector lumacaftor (VX-809, Figure 1)19 for CF patients carrying F508del mutation.19,20 Tezacaftor, also known as VX-661 (Figure 1), is an analogue of lumacaftor with improved pharmacokinetics and less side effects. The tezacaftor/ivacaftor co-therapy (trade name Symdeko) received marketing authorization in 2018 for both F508del homozygous patients and heterozygous F508del with G551D or with residual function mutations.21−24 More recently, Vertex Pharmaceuticals developed the next generation corrector elaxacaftor (VX-445, Figure 1), which showed additive or synergistic effects in combination with a first generation corrector (lumacaftor and tezacaftor) and with the potentiator ivacaftor.25,26 Strikingly, ivacaftor, tezacaftor, and elexacaftor are now included in the triple drug combination Trikafta for the treatment of CF patients aged 12 years and older carrying at least one F508del mutation or another mutation included in the list of 178 variants considered to be eligible to drug treatment (for the complete list of mutations, see Trikafta.com).27−29
Figure 1.
Structures of potentiators and correctors clinically approved.
Although both Orkambi and Symdeko have limited effects in clinical use,30 they still represent the standard care for many CF patients. Trikafta, despite undoubtedly representing a breakthrough in CF treatment by significantly slowing down CF progress with substantiated clinical benefits,31 fails to fully restore mutant CFTR function.26,32 As an example, treatment with Trikafta reduces only partially the ubiquitylation status of F508del-CFTR.32 Therefore, both academies and pharmaceutical companies have been involved in searching for small-molecule correctors33 and potentiators34 with different mechanisms or with ameliorated characteristics. Encouragingly, a number of emerging CFTR modulators are currently in the pipeline for preclinical models and early phase clinical trials, strengthening the restoration of CFTR function as a new therapeutic solution for CF.35 Moreover, a great part of CF research is now focusing on the discovery of active compounds affecting different CFTR-related targets (namely, proteostasis regulators), which can modify the CFTR proteostasis environment leading to beneficial effects on CFTR maturation and trafficking to the PM.36,37 This innovative strategy holds great promise as it can specifically target the steps in CFTR processing that create the main bottlenecks in its rescue. Furthermore, proteostasis regulator effects were seen to be additive with those of other types of correctors and therefore they may be useful to optimize combination therapies, especially for patients with mutations that still lack effective treatments.37
Several proteins have been identified that could represent useful drug targets for a CF therapy based on proteostasis modulation.36 Among them, the ubiquitin ligase RNF5/RMA1 is particularly interesting as it acts at early stages of CFTR biosynthesis and its loss by gene silencing synergizes with pharmacological correctors to correct folding defects in F508del-CFTR.38 Our group previously demonstrated that genetic suppression of RNF5 in vivo leads to an attenuation of intestinal pathological phenotypes due to malabsorption in F508del-CFTR mice and concomitantly increases CFTR activity in intestinal epithelial cells. This work validates the relevance of RNF5 as a novel drug target for CF, providing a strong basis for developing small molecules to inhibit RNF5 activity.39
As a further development of this project, using a computational approach based on ligand docking and virtual screening (VS), we recently identified the 1,2,4-thiadiazole derivative inh-2 (Figure 2A), a drug-like small molecule able to act as an RNF5 inhibitor. In in vitro experiments, inh-2 rescued F508del-CFTR activity in both CFBE41o– cells and human primary bronchial epithelia. Analysis of the inh-2 mechanism of action confirmed that it decreases ubiquitination and increases half-life of F508del-CFTR, further validating RNF5 as a drug target for CF and providing evidence to support its druggability.40
Figure 2.
Discovery of the first RNF5 inhibitor, the 1,2,4-thiadiazolylidene inh-2 by Sondo et al.(40) and chemical modification campaign around the 1,2,4-thiadiazol-5-ylidene scaffold. (A) Chemical structures and F508del-CFTR corrector activity of the RNF5 inhibitor inh-2 and the RNF5 activator analog-1. NA = not active. (B) Proposed binding mode of inh-2 into the RNF5 pocket. Blue dashed lines indicate the π–π stacking interactions, while yellow dashed lines indicate the hydrogen bonds. (C) Overview of the optimization strategy of inh-2 for SAR exploration.
Besides CF, given its important regulatory role in controlling cell differentiation, growth, and transformation, and its aberrant expression, RNF5 can be considered an interesting drug target also in pathological conditions, such as tumorigenesis.41 Previous studies identified an upregulation of RNF5 in breast cancer41 and tumor cell proliferation were inhibited after silencing of RNF5. Recently, RNF5 was correlated with glioma.42 In addition, modulation of RNF5 was demonstrated to be an effective treatment in neuroectodermal tumors.43 Taken together, these studies identify RNF5 as a valid candidate for the development of anti-cancer therapies.
Recently, a small-molecule inhibitor and degrader of RNF5 was discovered based on its ability to inhibit misfolded proteins from the ER lumen to the cytosol and to negatively regulate the RNF5 function.44 This finding further supports RNF5 druggability.
To discover more effective compounds, we here design and synthesize a library of new analogues (1–46, Table 1) of the 1,2,4-thiadiazole inh-2. In particular, we attempt to depict general structure–activity relationships (SAR) of 1–46 in inhibiting RNF5 and outline the biological profile of the most promising derivatives 6, 9–11, 14, 16, 17, 19, 21–25, 27–29, and 34.
Table 1. Structures of inh-2, Analog-1, and Compounds 1–46.
2. Results and Discussion
2.1. Design Approach
From a computational point of view, human RNF5 is a very challenging target; as to date, there are no structures available in the PDB of this E3 ligase. Moreover, there is very low identity with similar protein in the PDB for homology modeling endeavors. Therefore, to identify potential RNF5 inhibitors in a previous paper, we used two complementary approaches.40 First, we generated a homology model of RNF5 RING domain to perform VS based on ligand docking. In parallel, we used molecular fingerprinting to select a diversity set of compounds. With this strategy, we discovered the first ever reported RNF5 inhibitor inh-2 based on a 1,2,4-thiadiazole moiety, which displayed an EC50 of 2.6 μM in CFBE41o– cells from the HS-YFP assay.40 Notably, the same study showed that a close analog of inh-2 (analog-1, Figure 2A) had no activity as a CFTR corrector, whereas it elicited the opposite effects on RNF5 downstream targets as compared with inh-2, suggesting that small differences in the chemical structure may shift the effect of inh-2 analogues from RNF5 inhibition to activation. Furthermore, our group recently demonstrated that the RNF5 activator analog-1 can reduce neuroblastoma and melanoma tumor growth, both in vitro and in vivo models, suggesting that the activation of RNF5 may represent a potential anti-tumor treatment strategy.43 On the other hand, the biological effects of inh-2 are consistent with what has been described for RNF5 inhibition,39,40 although we cannot exclude that inh-2 may also affect other cellular targets. Therefore, inh-2 structural tuning is mandatory to gain a deeper knowledge on the handling of misfolded CFTR mutants by the quality control system of the cell.
The proposed binding mode based on docking simulations of inh-2 to the homology model of RNF5 shows (i) a H-bond between the amidine portion of the compound and ARG73, (ii) two π–π interactions among the thiadiazolidine, phenyl ring B, TRP48, and HIS52, and (iii) some hydrophobic interactions between the benzyl ring A and the hydrophobic pocket outlined by LEU51, VAL38, and VAL76 (Figure 2B). For a comparison with a hypothetical binding mode of analog-1, see Figure S1. However, despite the substantial margins of uncertainty of the docking pose of inh-2 due to the flexibility of RNF5, this binding mode offers the possibility to rationally modify it. Herein, to improve the inhibitory activity of inh-2, we conducted a chemical modification campaign around the 1,2,4-thiadiazol-5-ylidene scaffold. Figure 2C provides an overview of the structural variations introduced on the thiadiazole scaffold.
As the 3-methyl group of the thiadiazolidine central ring (pink region, Figure 2C) is shown to lie in a small hydrophobic pocket of the target, we first defined the optimal steric hindrance of this position by replacing the methyl group with ethyl, fluoromethyl, and isopropyl moieties (1–3, Table 1). Unfortunately, the removal of the methyl group was not possible due to poor chemical tractability of the 5-amino-1,2,4-thiadiazole and to the low reactivity of the functionalizable nitrogen atom of the ring. In the attempt to find the proper length of the alkyl chain connecting the thiadiazolidine and the phenyl ring A (blue region, Figure 2C), we replaced the methylene of inh-2 with ethylene or propylene tethers (4 and 5, Table 1). Ring A (red region, Figure 2C) is shown to interact with a large hydrophobic pocket of the target from the docking simulation (Figure 2B). Therefore, we investigated the role of this portion in possible hydrophobic interactions by introducing different EDGs and EWGs, such as methyl, methoxyl, hydroxyl, carboxyl, ethoxyl, isopropoxyl, trifluoromethyl, amino, methylamino, dimethylamino groups, and fluorine, chlorine, and bromine atoms in ortho, meta, or para positions (6–29, Table 1). The phenyl ring B was modified by introducing methoxyl or hydroxyl groups at different positions (30–33, Table 1) or replacing by 4-pyridyl, 3-pyridyl, or 2-furanyl moieties (34–36, Table 1). Indeed, it was reasoned that proton acceptor or donor groups on ring B could engage favorable interactions with HIS52 of the site, while different heterocycles could stabilize the T-shape-type π–π stacking interaction with TRP48. To assess the importance of ring C, the phenyl was replaced by m-tolyl, benzyl, cyclohexyl, and pyridyl moieties (37–41). To further explore the role of the N-phenylbenzamidine portion in possible π–π stacking interactions (green region, Figure 2C), both rings B and C were removed and replaced by pyridin-2-yl and 4-(2-methoxyethyl)pyridin-2-yl moieties (42 and 43, Table 1). Indeed, as suggested by docking simulation, the pyridin-2-yl group should maintain the H-bond stacking interaction with ARG73, while the methoxyethyl moiety could engage favorable H-bonds with HIS52. Last, modifications of the central 3-methyl-1,2,4-thiadiazolidine core (orange region, Figure 2C) were envisioned to investigate if different five-membered heterocycles could affect the π–π stacking interaction with TRP48 and HIS52 and therefore the inhibitory activity. Herein, 5-methyl-1,3,4-thiadiazolidine, 1,3,4-thiadiazolidine, and 1,3-thiazolidine moieties were explored at this position (44–46, Table 1).
2.2. Chemistry
Scheme 1 illustrates the common synthetic strategy for achieving the final desired compounds 1–41. The key intermediate imidoylthioureas 47a–78a underwent intramolecular cyclization by bromine oxidation, yielding thiadiazolium salts 47b–78b. Following, the hydrobromide salts were treated with the appropriate nitriles 79–82 under basic conditions (trimethylamine) to afford the 1,2,4-thiadiazolylidene final compounds 1–7, 9–11, 13–16, 19, 20, 23–31, and 33–41. Compounds 8, 17, and 32 were obtained by demethylation of the corresponding ether derivatives 7, 16, and 31 with BBr3. The carboxylic acid 12 was smoothly obtained by treatment of the bromo derivative 11 with n-buthyllithyum followed by the reaction with carbon dioxide. The isopropoxy derivative 18 was afforded by alkylation of the hydroxy derivative 17 with isopropyl bromide in the presence of K2CO3. Finally, alkylation of the primary amino group of derivative 20 with iodomethane under basic conditions gave the monomethylated and dimethylated derivatives 21 and 22.
Scheme 1. Synthesis of Final Compounds 1–41.

Reagents and conditions: (i) Br2, DCM/EtOAc (1:2 v/v), 5 °C to RT; then RT, 12 h, yield 33% - quantitative; (ii) TEA, reflux 30 min, yield 13–95%; (iii) BBr3, DCM, 0 °C, 30 min; then RT, 12 h, yield 23–59%; (iv) n-BuLi, THF, −78 °C, 30 min; then CO2, −78 °C to RT, yield 26%; (v) K2CO3, DMF, RT 1 h; then isopropyl bromide, 70 C°, 3 h, yield 18%; (vi) MeI, K2CO3, DMF, 50 °C, 24 h, yield 22–59%.
The E- and Z-configurations of the two imines of the newly synthetized compounds 1–41 were established through selective 1D Nuclear Overhauser Effect (NOE) experiments with compound 10 as a representative example (Figure S2). Irradiation of H-22 (δH 5.54 ppm) resulted in obvious enhancement of H-24,29 (δH 7.35 ppm) of the ring A and H-21 (δH 2.41 ppm) of the methyl group. Instead, no enhancement of protons of rings B or C were detectable, ascertaining that the benzyl group A and the N-phenylbenzamidine portion are opposite oriented and indicating that the geometry of the double bond N6=C5 is Z. Meanwhile, irradiation of H-14,10 (δH 6.75 ppm) of phenyl C resulted in predictable enhancement of H-13,11 (δH 7.23 ppm) and H-12 (δH 7.02 ppm) of the same ring. Notably, selective irradiation of H-14,10 caused strong enhancement of H-16,20 (δH 7.41 ppm) of phenyl B (Figure 3), indicating that the two rings are cis oriented and confirming the E geometry of the double bond N8=C7.
Figure 3.
Key NOE effect of compound 10.
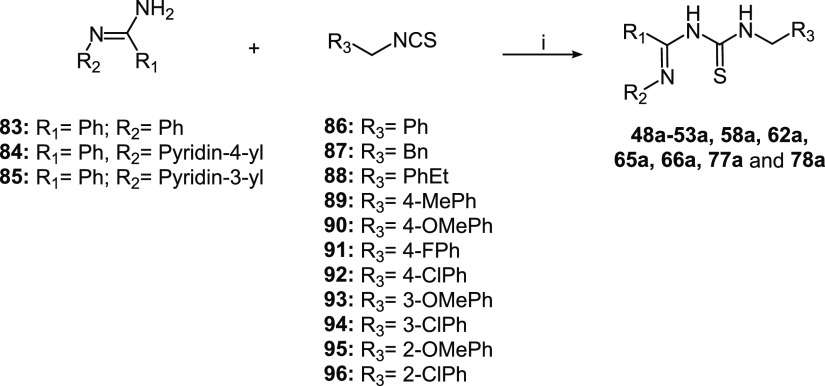
The imidoylthioureas 47a–78a necessary for final compound synthesis were prepared following two different strategies: 48a–53a, 58a, 62a, 65a, 66a, 77a, and 78a were obtained starting from the N-arylbenzamidines 83–85 that were reacted with substituted isothiocyanates 86–96 to form the desired imidoylthioureas (Scheme 2). The not commercially available N-arylbenzamidines 84 and 85 and isocyanates 89–96 were obtained following standard procedures as reported in the Supporting Information (Schemes S1 and S2).
Scheme 2. Synthesis of the Intermediate Imidoylthioureas 48a-53a, 58a, 62a, 65a, 66a, 77a, and 78a.
Reagents and conditions: (i) dry DCE, 55 °C, 22 h, yield 14–51%.
Unluckily in some cases, this strategy afforded inseparable byproducts that affected imidoylthioureas’ purification processes and reactions’ yields. Therefore, the imidoylthioureas 47a, 54a–57a, 59a–61a, 63a, 64a, and 67a–76a were synthesized by an alternative procedure as described in Scheme 3. The aromatic or heteroaromatic acyl chlorides 97–103 were reacted with the appropriate amines 104–107 to obtain the corresponding amides 108a–117a. The latter were converted into imidoyl chlorides 108b–117b through treatment with thionyl chloride or phosphorus pentachloride. Substitution of the chlorine atom by sodium thiocyanate followed by addition of the appropriate amines 118–129 afforded the desired thioureas.
Scheme 3. Synthesis of the Intermediate Imidoylthioureas 47a, 54a–57a, 59a–61a, 63a, 64a, and 67a–76a.
Reagents and conditions: (i) TEA, THF, 0 °C, 2-5 h, yield 85% - quantitative; (ii) treatment of compounds 108a–110a, 112a–117a: SOCl2, 70 °C, 2–2.5 h, yield 76–98%; treatment of compound 111a: PCl5, dry toluene, reflux, 4 h, yield 81%; (iii) NaSCN, dry acetone, −15 °C to 0 °C; then benzylamines 118–129, dry acetone, 0 °C to RT, yield 22–99%.
The 1,2,4-thiadiazolidines 42 and 43 bearing pyridylimino substituents were synthesized taking advantage of a synthetic strategy previously reported by Martinez et al. (Scheme 4).45 Reaction of appropriate pyridin-2-amines 130 and 131 with benzyl isothiocyanate 86 afforded the pyridinylthioureas 132a and 133a. Oxidation of 132a and 133a with bromine gave a regioselective ring-closure reaction, yielding the corresponding thiadiazolopyridinium bromides 132b and 133b in good yields. Finally, reaction of salts 132b and 133b in basic medium (diisopropylethylamine) with acetonitrile 79 at reflux temperature afforded the desired 5-pyridylimino 1,2,4-thiadiazolidines. The Z-configuration of compounds 42 and 43 was confirmed according to their 1H NMR spectroscopic data complemented with NOE experiments and in agreement with what was previously reported by Martinez et al.45
Scheme 4. Synthesis of Final N-(Pyridin-2-yl)-1,2,4-thiadiazolic Compounds 42 and 43.

Reagents and conditions: (i) benzyl isothiocyanate (86), dry DCE, 55 °C, 22 h, yield 68% - quantitative; (ii) Br2, DCM/EtOAc (1:2 v/v), 5 °C to RT; then RT, 12 h, yield 71–93%; (iii) ACN (79), DiPEA, reflux 2 h, yield 23–24%.
The 1,3,4-thiadiazolidines 44 and 45 and 1,3-thiazolidine 46 were readily synthesized through the synthetic procedure illustrated in Scheme 5, which was adapted from the synthesis reported by Nagao et al.46 Treatment of 2-amino-1,3,4-thiadiazoles 134a and 135a or 2-amino-1,3-thiazole 136a with trifluoroacetic anhydride afforded the corresponding 2-trifluoroacetylamino derivatives 134b–136b. Regioselective alkylation of heterocycles 134b–136b with benzyl bromide 137 in the presence of K2CO3 gave the corresponding 3-benzylthiadiazoline derivatives 134c and 135c or 3-benzylthiazoline 136c. After hydrolysis of the trifluoroacetyl-protecting group with 5% aqueous NaOH, the resulting 2-imino derivatives 134d–136d were reacted with N-phenylbenzimidoyl chloride 108b in the presence of pyridine to obtain the desired final compounds 44–46.
Scheme 5. Synthesis of Final 1,3,4-Thiadiazolylidenes 44 and 45 and 1,3-Thiazolylidene 46.
Reagents and conditions: (i) trifluoroacetic anhydride, toluene, 0 °C to RT; then 12 h, RT, yield 87% - quantitative; (ii) benzyl bromide (137), K2CO3, dry DMF, RT, 24 h, quantitative yield; (iii) NaOH 5%, THF, RT, yield 92% - quantitative; (vi) pyridine, DCM, 0 °C, 1 h; then RT, 12 h, yield 16–27%.
The proposed structures and isomerism of analogues 44–46 were confirmed by mono and two-dimensional NMR spectroscopy studies, including selective NOE (Figure S3) and Heteronuclear Multiple Bond Correlation (HMBC) experiments (Figure S4) with compound 44, taken as a representative example. A strong correlation in the NOE spectrum was observed between the H-14,10 (δH 6.65 ppm) of phenyl C and H-16,20 (δH 7.37 ppm) of phenyl B, indicating that these protons were proximal in the E-configuration, as illustrated in Figure 4. Conversely, no enhancement of protons of rings B or C was detectable after irradiation of H-22 (δH 5.49 ppm), indicating a Z geometry of the double bond N6=C2. Furthermore, observation of the HMBC cross peak for benzylic protons H-22 with C-2 (δc 159.75 ppm), but not for H-22 with C-5 (δc 153.71 ppm), confirmed that substitution with the benzyl moiety occurred exclusively at the nitrogen N-3 of the thiadiazole (Figure 4).
Figure 4.
Key NOE and HMBC effects of compound 44.
2.3. Biological Evaluation
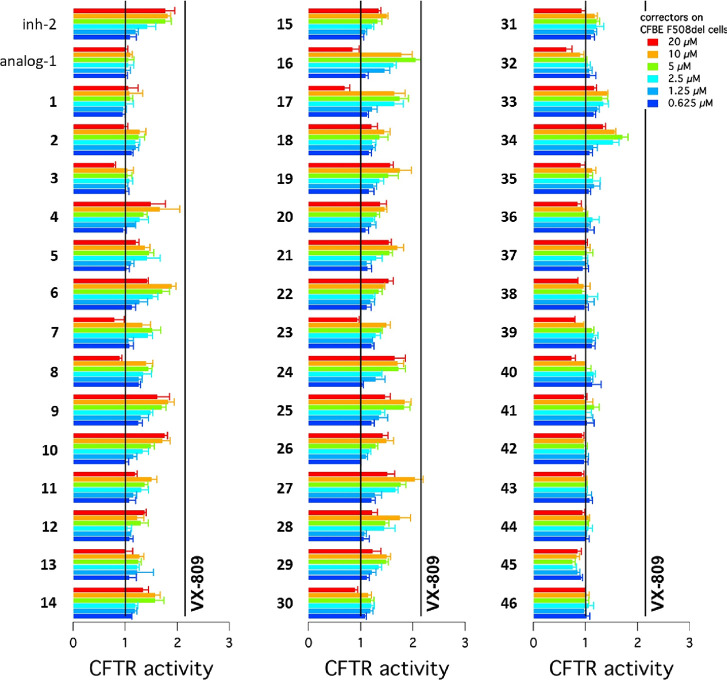
As a primary screen, the new thiadiazole derivatives 1–46 (Table 1) were tested for their ability to rescue the F508del-CFTR trafficking defect in CFBE41o– cells, stably co-expressing F508del-CFTR and the HS-YFP (Figure 5). This cell line has been extensively used by our group, in combination with the microfluorimetric assay based on the HS-YFP, to identify and characterize many CFTR correctors.39,47 This assay relies on the hypothesis that RNF5 inhibition results in increased mutant CFTR processing and activity. Therefore, it can easily identify putative RNF5 inhibitors, but not other types of RNF5-binding compounds that could have no effect or have a negative effect on CFTR function, such as RNF5 activators (see analog-1). Thus, we opted for this assay as our primary aim was the identification of RNF5 inhibitors as a possible therapeutic strategy in CF.
Figure 5.
Bar graph showing CFTR activity in CFBE41o– cells following 24 h treatment with vehicle alone or with analogs of inh-2 at the indicated concentrations. The vehicle alone (DMSO) and corrector VX-809 (1 μM) were used as negative and positive controls, respectively.
The substitution of the methyl group of the thiadiazoline core with larger lipophilic moieties as in compounds 1–3 (i.e., ethyl, fluoromethyl, and isopropyl, respectively) was not tolerated, suggesting that a methyl group best fits into the small lipophilic niche in the target pocket. Concerning the alkyl chain connecting phenyl ring A to the central core, the proper length of the tether emerged to be a methylene, as compounds 4 and 5, bearing, respectively, ethylene and propylene linkers, were devoid of significant F508del-CFTR corrector activity. This speculation was further corroborated by the lack of CFTR corrector activity previously shown by analog-1, in which the phenyl ring A is directly connected to the thiadiazoline core.40
Next, we examined the effect of different substituents on the benzyl group A, which showed to be allocated in a large lipophilic pocket of RNF5 protein. In the para-position, different lipophilic substituents such as a methyl group (6), fluoride (9), chloride (10), and bromide (11) atoms maintained the activity unaltered. Meanwhile, polar, EWG, and EDG substituents such as hydroxyl (8), amino (13), carboxyl (12), and methoxyl (7) groups resulted in a reduced potency. Regarding the meta- substituted series, derivatives 16, 17, and 24 carrying a methoxyl group, hydroxyl group, and a chloride atom, respectively, proved to have increased activity, while 15, 18, and 20 bearing trifluoromethyl, isopropoxyl, and amino groups, respectively, presented a decreased potency. Furthermore, the meta-methyl (14), ethoxyl (19), methylamino (21), and dimethylamino (22) groups and meta-fluoride atom (23) did not affect the activity. Among the explored ortho-substituents, the methyl (25) and methoxyl (27) groups caused a slight increase in activity while the trifluoromethyl (26) had a detrimental effect. Finally, 28 and 29 bearing a chloride atom and an amino moiety, respectively, showed a comparable activity to the unsubstituted original hit inh-2. Overall, the SARs of the new series of 1,2,4-thiadiazolidines with variations on ring A were rather complex to rationalize, and no clear pattern could be identified. However, compounds 16 and 17 showed the best activities of the series, with EC50 equal to 1.2 and 1.6 μM, respectively (inh-2, EC50 = 2.6 μM).40
Moving to the N-phenylbenzamidine portion, addition of methoxyl and hydroxyl groups in the ortho-, meta-, and para-positions of phenyl ring B (30–33) or replacement of ring B with pyridine-3-yl and furan-2-yl moieties (35 and 36, respectively) resulted in a drop of activity. Instead, the substitution of phenyl B with a pyridine-4-yl moiety, as in compound 34, was tolerated, maintaining the activity equal to inh-2. Hence, it appears that only a para-pyridine at this position could engage favorable hydrogen bond interactions with TRP48 of the RNF5 pocket, which may be responsible for the maintenance of the activity. The replacement of the phenyl ring C with meta-tolyl (37), benzyl (38), and cyclohexyl (39) rings or pyridines (40 and 41) led to inactive analogs. From the results obtained, it appears that both phenyl B and C of the N-phenylbenzamidine portion are necessary for π–π stacking interactions with the RNF5 pocket. This speculation was further corroborated by analogues 42 and 43, in which the removal of both rings and their replacement with pyridin-2-yl moieties led to inactive compounds, contrary to what was expected from the docking model.
Finally, the replacement of the central 1,2,4-thiadiazolidine core of the scaffold with different five-membered heterocycles, such as 5-methyl-1,3,4-thiadiazolidine (44), 1,3,4-thiadiazolidine (45), and 1,3-thiazolidine (46), abolished any corrector activity of the compounds. Therefore, we concluded that the 1,2,4-thiadiazolidine ring is mandatory for optimal π–π stacking interactions with TRP48 and HIS52 of the target pocket and thus for the activity of compounds.
In parallel to the SAR campaign, binding assays were also attempted to assess the putative physical interaction of the novel inhibitors with the RNF5 target. First, the hit inh-2, analog-1, and compounds 16 and 17, chosen as the most active of the series, were tested for their solubility and aggregation state by SPAM filter48 assays in PBS buffer (PBS pH 7.5, 1 μM ZnCl2, 1 mM DTT, 10% D2O, and 1% DMSO-d6) at different theoretical concentrations (20, 50, and 100 μM), using 4-(trifluoromethyl)benzenesulfonamide (200 μM) as an internal reference.
Unfortunately, the compounds showed a surprisingly poor solubility (Table 2) in spite of their four nitrogen atoms and hydroxyl group, which can be attributed to their expected near planarity.49 Under these circumstances, it was decided to explore the solubility of all the synthetized compounds, regardless of their biological activities, to find a suitable candidate for the validation of our working hypothesis, i.e., that the compounds exert their activities by directly binding to RNF5. For that, we first calculated the predicted octanol/water partition coefficient (log Po/w) and the predicted aqueous solubility (log S) of all the analogues by QikProp (Schrödinger Release 2022-1: QikProp, Schrödinger, LLC, New York, NY, 2021, Table S1). As expected, the obtained values of log Po/w > 1 suggest a hydrophobic nature of the studied compounds associated with a predicted poor aqueous solubility (Table S1).50 To shrink down the number of compounds for solubility experiments, just the ones that had −2.0 < log Po/w < 6 and −6.5 < log S <0.5 values were selected. Among them, the PBS solubility was determined for five representative analogues: (i) 12 and 29 bearing different substituents on the benzyl group A; (ii) 34 and 40 showing the replacement of rings B and C with pyridine-4-yl moieties; (iii) 42 in which the N-phenylbenzamidine portion was replaced with a pyridine-2-yl moiety (Table 1). The results reported in Table 2 highlighted that only compounds 12, 40, and 42 showed no aggregation and solubility suitable for in vitro/biophysical binding experiments.
Table 2. Computed Octanol/Water Log P Values, Log S Values (S in mol·dm–3), and Experimental Solubility (in μM) of Selected Compounds in PBS pH 7.5, 1 μM ZnCl2, 1 mM DTT, 10% D2O, and 1% DMSO-d6.
| solubility
and aggregation in PBS buffer by NMR |
||||||
|---|---|---|---|---|---|---|
| compound | Plog Po/w | Plog S | 20 μM | 50 μM | 100 μM | aggregation |
| inh-2 | 6.38 | –6.56 | <5 | <5 | <5 | no |
| analog-1 | 6.096 | –6.695 | <5 | <5 | <5 | no |
| 16 | 6.349 | –6.196 | <5 | <5 | <5 | no |
| 17 | 5.688 | -6.447 | <5 | <5 | <5 | no |
| 12 | 5.677 | -6.432 | 20 | 50 | 100 | no |
| 29 | 5.569 | -6.371 | <5 | <5 | <5 | no |
| 34 | 5.345 | –5.741 | 10 | 10 | 10 | no |
| 40 | 5.342 | –5.7 | 20 | 30 | 30 | no |
| 42 | 4.043 | –4.279 | 20 | 40 | 5 | no |
The bindings of compounds 12, 40, and 42 were initially evaluated by MicroScale Thermophoresis (MST) using a purified recombinant truncated form of RNF5 (aa 1–117), lacking the C-terminal transmembrane domains. The protein was covalently labeled with a red dye (NHS) on the primary amines (lysine residues). Although a complete affinity curve could not be built, evidence of direct binding (binding check tests) to the protein was observed only for compound 12 (see Figure S5). 1H Water-Ligand Observed via Gradient SpectroscopY (WaterLOGSY)51 and Saturation-Transfer Difference (STD)52 NMR experiments on the recombinant RNF5 form were further performed to test the direct binding of the three compounds to the protein with a more sensitive, label-free, independent approach. Analogs 12, 40, and 42 were tested at 50 μM in the absence and in the presence of 3 μM RNF5 (1–117) protein. As reported in Figure 6, the three compounds bind the recombinant RNF5 protein. Indeed, for all three compounds, the NMR signals of their methyls are present in the STD spectra, and in the WaterLOGSY spectra, their signals change from negative to positive in the presence of the protein, indisputably highlighting the formation of compound–protein binding events. Despite the lower F508del-CFTR activity compared to inh-2, compound 12 clearly binds RNF5 protein pointing at this protein as the target responsible for the observed in cell activity. Also compounds 40 and 42, although almost inactive as CFTR correctors, showed to interact with RNF5. It has to be noted that NMR is a very sensitive technique that allows detecting also compounds weakly binding to their target protein independent of the possible downstream biological effects. Indeed, we can speculate that compounds 40 and 42 may bind to RNF5 without inhibiting its ubiquitin ligase activity, thus not leading to mutant CFTR rescue. These compounds could even act as RNF5 activators, similar to analog-1. In conclusion, our data clearly confirm the ability of the synthesized compounds to directly bind RNF5, their putative target.
Figure 6.
1H NMR methyl signal of compounds 12 (left), 40 (middle), and 42 (right):1D spectrum (black), WaterLOGSY spectrum (red); WaterLOGSY spectrum in the presence of RNF5 (blue) and STD spectrum (green). The change, upon protein addition, of compounds NMR signals from negative to positive in WaterLOGSY spectra and the presence of compounds NMR signals in STD spectra indicate compounds binding to RNF5.
Even though the poor solubility demonstrated by most of the 1,2,4-thiadiazolylidene derivatives prevented us from performing NMR binding experiments on all the synthetized analogues, the SAR campaign allowed us to identify several compounds with promising corrector activity profiles, such as compounds 6, 9–11, 14, 16, 17, 19, 21–25, 27–29, and 34 that were selected for further cell-based studies.
We performed a preliminary evaluation of the effect of chronic treatment with selected analogues on the proliferation and apoptosis of CFBE41o– cells (Figure 7). Indeed, RNF5 can modulate cell motility and proliferation by regulating paxillin ubiquitylation and thus its degradation.53 For the proliferation assay, CFBE41o– cells stably co-expressing F508del-CFTR and the HS-YFP were treated with test compounds (5 μM), VX-809 (1 μM), or vehicle alone (DMSO) for 48 h. Cell proliferation was then monitored by measuring the area covered by the cells based on the YFP signal. For the apoptosis assay, 24 h after plating, CFBE41o– cells stably expressing F508del-CFTR were treated with test compounds (5, 20, and 100 μM), VX-809 (1 μM), MG132 (50 μM), or vehicle alone (DMSO) for 24 h. Cell nuclei were then counterstained with Hoechst 33342 and propidium iodide to visualize the total and apoptotic cell count, respectively, and imaged by using an Opera Phenix high-content screening system. None of the test compounds significantly altered cell proliferation (Figure 7A). However, compounds 17 and 27–29 markedly increased cell apoptosis, although only at a very high concentration (100 μM; Figure 7B). Notably, similar effects were observed upon treatment with VX-809 and analog-1 at the same concentration.
Figure 7.
Evaluation of the effect of putative RNF5 inhibitors on cell proliferation and apoptosis. (A) The bar graph shows the number of viable CFBE41o– cells following 48 h treatment with test compounds at 5 μM. (B) Bar graph shows the number of apoptotic CFBE41o– cells following 24 h treatment with test compounds at the indicated concentrations. Data are expressed as means ± SEM, n = 4–6. Asterisks indicate statistical significance: ***p < 0.001, **p < 0.01.
To further support our hypothesis that CFTR rescue following treatment with test compounds was indeed due to RNF5 inhibition, we considered additional biological processes known to be regulated by RNF5 ligase activity. To this aim, we focused our attention on ATG4B, a known regulator of basal autophagy, whose degradation is mediated by RNF5 activity.54In vivo ubiquitination experiments previously confirmed that inh-2 inhibits ATG4B degradation, thus increasing the basal level of autophagy.40 Therefore, induction of autophagy can be considered as additional evidence demonstrating the ability of test compounds to inhibit RNF5. We thus evaluated the effect of inh-2 analogues on induction of the autophagy pathway. To this aim, we monitored the formation of autophagic vacuoles in F508del-CFTR-expressing CFBE41o– cells (Figure 8) by using the autolysosome marker monodansylcadaverine (MDC). MDC accumulates inside autophagosomes. After fusion of autophagosomes with lysosomes, MDC fluorescence increases due to the acidic environment.55 Therefore, F508del-CFTR-expressing CFBE41o– cells were treated for 24 h with test compounds (5 μM), VX-809 (1 μM), or DMSO alone (vehicle). In the last 3 h of incubation, we added torin-1 (20 nM), a known autophagy inducer, and SAR-405 (2 μM), a potent inhibitor of the autophagic pathway. The cells were then loaded with MDC and evaluated by high-content confocal imaging to detect signal spots (corresponding to autophagic vacuoles) in each cell. Determination of signal spots clearly demonstrated that the number of autophagic vacuoles was significantly increased after treatment with torin-1, inh-2, or compounds 11, 16, and 21 (Figure 8). In contrast, incubation with SAR-405, analog-1, 9, and 34 significantly decreased the number of autophagic vacuoles (Figure 8).
Figure 8.
Evaluation of effects of putative RNF5 inhibitors on ATG4B-mediated basal autophagy. (A) Representative confocal microscopy images of F508del-CFTR expressing CFBE41o– cells treated with the indicated compounds and loaded with MDC. Scale bar = 50 μm. (B) The bar graph shows the quantification of the number of spots (resembling autophagic vesicles) in cells treated with the indicated compounds, normalized for the control condition. Data are expressed as means ± SEM, n = 4–6. Asterisks indicate statistical significance: ***p < 0.001, **p < 0.01, *p < 0.05.
From the abovementioned investigations, we could discard compounds exhibiting (i) lower or no CFTR corrector activity compared to inh-2 (1–5, 7, 8, 12, 13, 15, 18, 20, 26, 30–33, and 35–46), (ii) cytotoxic effects (17 and 27–29), and (iii) discrepancy between data obtained from MDC signal spot evaluation and the HS-YFP assay (6, 9, 10, 14, 17, 19, 22–25, 27–29, and 34). Therefore, 11, 16, and 21 resulted the most promising compounds of the library, and, among them, analogue 16 caused both a strong activation of basal autophagy and a greater F508del-CFTR rescue than inh-2.
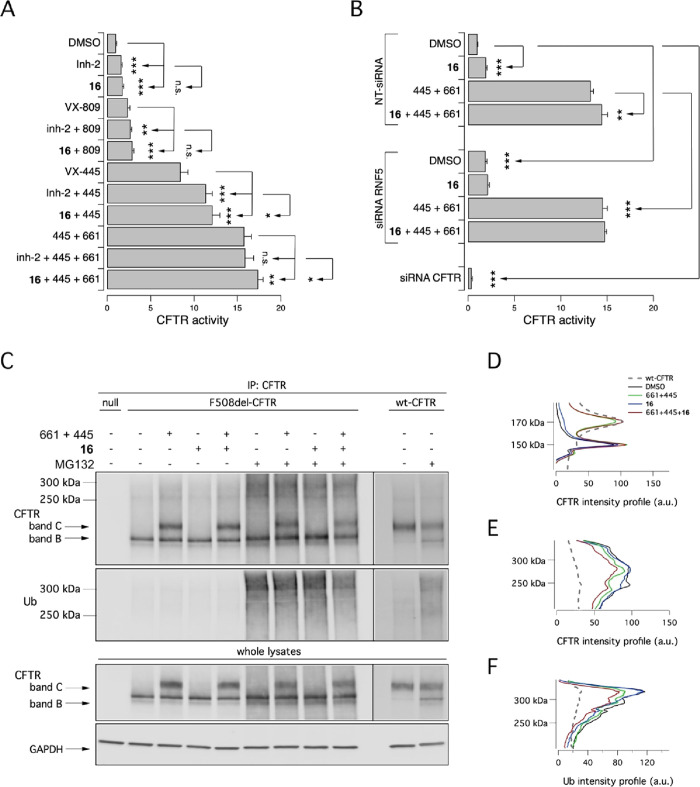
To further characterize the ability of RNF5 inhibitors to improve F508del rescue, we compared the efficacy of inh-2 and analogue 16 in combination with approved correctors. Both RNF5 inhibitors significantly increased mutant CFTR activity upon co-treatment with VX-809 or VX-445 (Figure 9A). However, only analogue 16 was able to further improve the rescue elicited by the double combination VX-661 + VX-445 (Figure 9A).
Figure 9.
Putative RNF5 inhibitors improve mutant CFTR rescue by approved correctors by decreasing CFTR ubiquitylation. (A) Bar graph showing CFTR activity in CFBE41o– cells following 24 h treatment with vehicle alone or with inh-2 (5 μM) and its analogue 16 (5 μM) as single agents or combined with correctors VX-809 (3 μM), VX-445 (3 μM), or VX-661 + VX-445 (10 μM + 3 μM). (B) Bar graph showing CFTR activity in CFBE41o– cells transfected with NT or RNF5 siRNA and treated for 24 h with DMSO, or analogue 16 (5 μM), or VX-661 + VX-445 (10 μM + 3 μM), or their combination. The effect of CFTR siRNA is shown as additional control of transfection efficiency. Asterisks indicate statistical significance: ***p < 0.001, **p < 0.01, *p < 0.05, while n.s. indicates “not significant”. (C) Biochemical analysis of CFTR ubiquitylation and expression pattern in CFTR immunoprecipitates from CFBE41o– cells after 24 h treatment with analogue 16 (5 μM), VX-661 + VX-445 (10 μM + 3 μM), and their combination in the absence or in the presence of MG-132 (10 μM; last 4 h) to block proteasomal degradation. Images for CFTR and Ub blots of F508del- and wt-CFTR samples are different exposures of the same membranes. (D–F) Analysis of intensity profiles of CFTR and ubiquitin (D) in the absence or (E and F) in the presence of MG-132.
We then aimed to indirectly confirm that the rescuing activity of analogue 16 was indeed due to RNF5 inhibition. We therefore tested the compound in F508del-CFTR-expressing CFBE41o– cells transfected with a non-targeting (NT) siRNA or an siRNA molecule targeting RNF5. We reasoned that the presence of an additive effect between treatment with analogue 16 and RNF5 silencing would have disproved the mechanism of action of analogue 16 (i.e., RNF5 inhibition). Interestingly, the extent of F508del-CFTR rescue was similar in 16-treated cells transfected with NT siRNA and in DMSO-treated cells transfected with RNF5 siRNA (Figure 9B). In addition, treatment with analogue 16 alone or combined with VX-661 + VX-445 increased the F508del-CFTR activity only in cells transfected with NT siRNA, but not in those transfected with RNF5 siRNA (Figure 9B).
Finally, we evaluated ubiquitylation of mutant CFTR in CFBE41o– cells following 24 h treatment with DMSO (vehicle), analogue 16, VX-661 + VX-445, and their combination. Subsequently, cells were treated for 4 h with DMSO alone or with MG-132 (10 μM; to block proteasomal degradation) and then lysed. Cell lysates were immunoprecipitated using an anti-CFTR antibody and then subjected to SDS-PAGE followed by Western blotting to evaluate CFTR expression and ubiquitylation status (Figure 9C–F). As previously reported, VX-661 + VX-445 caused a marked rescue of mutant CFTR, as shown by the appearance of the mature form of CFTR (band C; Figure 9C, CFTR blot) and evidenced also by the analysis of intensity profiles (Figure 9D), while the effect of analogue 16 was very modest (Figure 9C,D). Treatment with MG-132 caused the appearance of CFTR forms at high molecular weight (at 200–350 kDa, resembling ubiquitylated CFTR proteins) that decreased in the presence of test compounds, as evidenced by the analysis of intensity profiles (Figure 9E). MG-132 caused the accumulation of ubiquitylated CFTR, in particular under control (DMSO) conditions (Figure 9F, Ub blot), and markedly decreased upon treatment with VX-661 + VX-445, and, to a further extent, upon treatment with VX-661 + VX-445 plus the RNF5 inhibitor (Figure 9C,F). These data clearly demonstrate that the combination of approved drugs and an optimized RNF5 inhibitor can additively decrease ubiquitylation of mutant CFTR in immortalized bronchial cells.
Taken together, these findings suggest that 16 may represent the strongest RNF5 inhibitor of the series of tested compounds. The proposed binding mode based on docking simulations of compound 16 into the RNF5 homology model (Figure 10) suggests that 16 engages the same π–π and H-bond interactions as inh-2, but it might be hypothesized that the addition of a meta-methoxide group on the benzyl ring of the 1,2,4-thiadiazolylidene scaffold induces further favorable hydrophobic interactions with VAL38 and VAL76 (not shown in the figure) of the target pocket. Hence, these additional interactions may be responsible for the greater F508del-CFTR rescue activity elicited by compound 16 with respect to the parent compound inh-2. A comparison of the 16 and inh-2 binding modes to RNF5 is shown in Figure S6.
Figure 10.
Proposed binding mode of analogue 16 into the RNF5 pocket. Yellow dashed lines indicate the hydrogen bonds, while blue dashed lines indicate the π–π stacking interactions.
3. Conclusions
In CF, the most frequent autosomal recessive disease, the deletion of F508 in the CFTR anion channel is associated to misfolding and defective gating of the mutant protein. Among the known proteins involved in CFTR processing, one of the most promising drug targets is the ubiquitin ligase RNF5, which normally promotes F508del-CFTR degradation. In this context, a small molecule RNF5 inhibitor is expected to chemically mimic a condition of RNF5 silencing, thus preventing mutant CFTR degradation and causing its stabilization and plasma membrane trafficking. Hence, by exploiting a virtual screening (VS) campaign, the hit compound inh-2 was discovered as the first-in-class inhibitor of RNF5. Evaluation of inh-2 efficacy on CFTR rescue showed that it efficiently decreases ubiquitination of mutant CFTR and increases chloride current in human primary bronchial epithelia. More recently, another study aimed to identify compounds able to inhibit dislocation of misfolded proteins from the endoplasmic reticulum (ER) lumen to the cytosol in ER-associated degradation. This study led to the discovery of FX12 as an RNF5 E3 inhibitor and degrader that binds directly to RNF5 and inhibit its ligase activity in vitro.44 Consistent with this activity, and as reported for inh-2, FX12 decreases mutant CFTR ubiquitylation therefore rescuing CFTR channel activity, also enhancing the effect of the FDA-approved drugs VX809 and VX661. Finally, similar to inh-2, FX12 also modulates paxillin expression. However, FX12 does not improve mutant CFTR channel activity in human primary bronchial epithelia.44 A possible reason for this is that, while inh-2 is a RNF5 inhibitor, FX12 is not only an inhibitor but also a degrader of RNF5. This difference might impact mutant CFTR biogenesis, stability, and/or activity by affecting multiple pathways.
With the aim of gaining a better insight into the SAR of inh-2, a large library of analogs of the original hit compound was designed and synthetized. The optimization of general and versatile synthetic routes gave access to a series of novel 1,2,4-thiadiazolylidene-based compounds, which were subjected to biological activity evaluations as F508del-CFTR correctors. SAR efforts ultimately led to compound 16 that elicited a greater F508del-CFTR rescue than inh-2 in the HS-YFP functional assay. Evaluation of the effect of 16 on cell proliferation and apoptosis showed good tolerability and no toxic side effects of the putative ubiquitin ligase inhibitor. Interestingly, analogue 16 showed also an additive effect upon co-treatment with the highly effective triple combination elexacaftor/tezacaftor/ivacaftor, resulting in a decreased mutant CFTR ubiquitylation paralleled by an increased CFTR function. These results are particularly encouraging as small molecule ligase inhibitors could also act on ligases other than RNF5, leading to cytotoxic effects. On the contrary, RNF5 genetic suppression has no apparent negative effects in vitro and in vivo,39 and therefore putative RNF5 inhibitors are expected to lead to few side effects. The mechanism of action of analogue 16 was further investigated by exploiting known cellular targets of RNF5, such as the regulator of basal autophagy ATG4B. Functional evidence demonstrated that 16 strongly increases the basal level of autophagy of F508del-CFTR-expressing CFBE41o– cells, similar to the parent compound inh-2. Notably, there is an emerging interest for autophagy modulating compounds in controlling the pathogenesis of CF disease,56 and the restoration of autophagy has been proposed per se as a strategy to allow the rescue of F508del-CFTR trafficking.57−61 Furthermore, RNF5 knockout has shown to enhance autophagy-mediated clearance of bacterial infection,54 which is highly recommendable in CF patients having chronic lung infections. Although the induction of basal autophagy provided by 16 has to be considered as a secondary effect of RNF5 inhibition, it may have an additional positive effect on mutant CFTR rescue and innate host defense. These findings suggest that compound 16 may act as a stronger inhibitor of RNF5 ligase activity than inh-2 by directly binding to the RNF5 RING domain, as suggested by in silico prediction studies. Although the poor solubility of compound 16 hampered the experimental evidence of its direct binding to RNF5, biophysical evidence of direct protein interaction has been obtained for compound 16 analogs that are more soluble. However, the extent of rescue remains lower than that obtained with VX-809. This may be due to cellular QC mechanisms, which are functionally redundant. Indeed, the modulation of one of the cellular QC elements (such as the inhibition of the ubiquitin ligase RNF5) may have a lower effect than classic correctors on the global biological outcome due to adaptive responses. Nevertheless, it has been widely described that proteostasis regulators’ effects are additive with other correctors,37 and therefore, their combination with current therapies is expected to have higher therapeutic ceiling and expand pharmacological treatment applicability to CF patients bearing mutations poorly responsive to already developed modulators.
Taken together, our findings suggest that the 1,2,4-thiadiazolylidene scaffold could be further exploited for the discovery of novel RNF5 inhibitors able to rescue mutant CFTRs. Therefore, structural tuning will be further implemented to increase the promising corrector activity of 16 and its pharmacokinetic profile, ultimately providing a more drug-like 1,2,4-thiadiazolylidene derivative.
4. Experimental Section
4.1. Chemistry
4.1.1. General Chemical Methods
Solvents and reagents were purchased from commercial sources and used without further purification. If required, solvents were distilled prior to use. For simplicity, solvents and commonly used reagents are indicated as follows: acetonitrile (ACN), dichloromethane (DCM), 1,2-dichloroethane (DCE), diethyl ether (Et2O), petroleum ether (PE), dimethyl sulfoxide (DMSO), ethanol (EtOH), ethyl acetate (EtOAc), methanol (MeOH), N,N-dimethylformamide (DMF), tetrahydrofuran (THF), triethylamine (TEA), N,N-diisopropylethylamine (DIPEA), and 1,1′-thiocarbonyldiimidazole (TCDI). When stated, reactions were performed in an inert atmosphere. Reaction progress was monitored by thin layer chromatography (TLC) analyses on pre-coated silica gel plates (Kieselgel 60F254, Merck) and detected with UV light (254 nm) and/or KMnO4 stain. Flash column chromatography was carried out using a silica gel (particle size 40–63 μM, Merck) with the indicated solvent system as an eluent. NMR experiments were run on a Varian Gemini 401 MHz spectrometer (401.13 MHz for 1H and 100.62 MHz for 13C), equipped with a BBI probe and Z-gradients. Spectra were acquired at 300 K, using deuterated dimethyl sulfoxide (DMSO-d6) or deuterated chloroform (chloroform-d) as solvents. Chemical shifts (δ) for 1H and 13C spectra were recorded in parts per million (ppm) using the residual non-deuterated solvent as the internal standard (for chloroform-d, 1H 7.26 ppm; for DMSO-d6, 1H 2.50 ppm, 13C 39.52 ppm). Multiplicities are indicated using the following abbreviations: bs, broad signal; s, singlet; d, doublet; t, triplet; q, quartet; m, multiplet. The 2D experiments were acquired as follows: 1H-1H COSY (2 transients, 256 increments), 1H-13C HSQC (4 transients, 256 increments), 1H-13C HMBC (8 transients, 512 increments), and 2D NOESY (16 transients, 256 increments). The 1D NOESY experiment was performed with NOE DPFGSE pulse sequence at a mixing time of 2.5 s. UPLC/MS analyses of all the final compounds were run on a Waters ACQUITY UPLC-MS system consisting of an SQD (Single Quadrupole Detector) mass spectrometer equipped with an electrospray ionization (ESI) interface and a photodiode array detector (PDA). The PDA range was 210–401 nm. ESI in positive and negative mode was applied in the mass scan range 100–500 Da. Analyses were performed on an ACQUITY UPLC BEH C18 column (50 mm × 2.1 mm i.d., particle size 1.7 μm) with a VanGuard BEH C18 pre-column (5 mm × 2.1 mm i.d., particle size 1.7 μm) (log D > 1). The mobile phase was 10 mM NH4OAc in H2O at pH 5 adjusted with AcOH (A) and 10 mM NH4OAc in ACN/H2O (95:5) at pH 5 (B). Methods and gradients used were the following: Generic method. Linear gradient: 0–0.2 min, 5% B; 0.2–2.7 min, 5–95% B; 2.7–2.8 min, 95–100% B; 2.8–3.0 min, 100% B. Flow rate: 0.5 mL/min. Apolar method. Gradient: 0–0.2 min, 50% B; 0.2–2.7 min, 50–100% B; 2.7–3.0 min, 100% B. Flow rate: 0.5 mL/min. Compounds were named using the naming algorithm developed by CambridgeSoft Corporation and used in ChemBioDraw Ultra 19.0. All final compounds displayed ≥96% purity as determined by UPLC-MS analysis.
4.1.2. General Procedure A for the Synthesis of Final 1,2,4-Thiadiazole Derivatives 1–7, 9–11, 13–16, 19, 20, 23–31, and 33–43
To a suspension of thiadiazolium bromide salt 47b–78b, 132b, and 133b (1.0 equiv) in the appropriate nitrile 79, 80, and 82 (100 equiv) solvent, TEA (for 47b–78b, 1.5–2.5 equiv) or DiPEA (for 132b and 133b, 1 equiv) were added. For the 1,2,4-thiadiazolylidenes 2, fluoroacetonitrile 81 (3 equiv) was added to the suspension of 47b in THF (0.2 M) followed by TEA (1.5 equiv). The mixture became a clear solution, which was refluxed for 0.5–2 h and then quenched with ice. The crude was extracted with DCM (3 × 10 mL), washed with water (2 × 10 mL), and dried over Na2SO4, and the solvent was evaporated in vacuum. The crude material was purified via flash silica gel column chromatography or recrystallized from the appropriate solvent, unless otherwise noted.
4.1.3. General Procedure B for the Synthesis of 1,2,4-Thiadiazolylidenes 8, 17, and 32
To a stirring solution of the appropriate aryl methyl ether derivative 7, 16, and 31 (1 equiv) in dry DCM (0.2 M) and under an inert atmosphere, a solution 1 M BBr3 in DCM (2.0–2.5 equiv) was slowly added through the septum with a syringe at 0 °C. The reaction mixture was left to stir at the same temperature for 30 min and then overnight at room temperature. Afterward, the mixture was quenched with water, the crude was extracted with EtOAc (3 × 10 mL), washed with NaOH 1 M (2 × 10mL), and dried over Na2SO4, and the solvent was evaporated in vacuum. The crude material was purified via flash silica gel column chromatography, then washed with n-hexane, and filtered to give the desired compound, unless otherwise noted.
4.1.4. General Procedure C for the Synthesis of Final 1,3,4-Thiadiazolylidenes 44 and 45 and 1,3-Thiazolylidene 46
The appropriate 3-benzyl intermediate 134d-136b (1 equiv) was dissolved in dry DCM (0.2 M), and pyridine (1.2 equiv) was added to the solution under inert conditions. The mixture was cooled to 0 °C, and a solution of N-phenylbenzimidoyl chloride 108b (1.1 equiv) in dry DCM (0.3 M) was slowly added. When the addition was completed, the mixture was left stirring at 0 °C for 1 h and then at room temperature for additional 12 h. Afterward, the reaction mixture was quenched with water and extracted with DCM (3 × 20mL). The organic layers were combined, washed with water (2 × 20mL), dried over Na2SO4, and concentrated in vacuum. The remaining crude material was purified with flash silica gel column chromatography.
4.1.5. General Procedure D1 for the Synthesis of Imidoylthioureas 48a–53a, 58a, 62a, 65a, 66a, 77a, and 78a
A mixture of N-arylbenzamidines 83–85 (1.00 equiv) and the appropriate isothiocyanates 86–96 (1.00 equiv) in dry 1,2-dichloroethane (0.6 M) was heated at 55 °C for 22 h. Afterward, the reaction mixture was cooled to room temperature and the solvent was evaporated. The resulting crude material was purified via flash silica gel column chromatography to give the desired compound, unless otherwise noted.
4.1.6. General Procedure D2 for the Synthesis of Imidoylthioureas 47a, 54a–57a, 59a–61a, 63a, 64a, and 67a–76a
Over a solution of the appropriate imidoyl chloride 108b–117b (1.0 equiv) in anhydrous acetone (0.3 M) at −15 °C and under inert conditions, a solution of sodium thyocianate (1.1 equiv) in acetone (0.5 M) was slowly added. For the imidoylthioureas 71a and 72a, the corresponding imidoyl chloride was obtained as hydrochloride salt, and therefore, it was previously stirred with TEA (1 equiv) for 15 min at −15 °C. After the addition was completed, the mixture was allowed to reach 0 °C, and then, the appropriate primary amine 118–129 (1.1 equiv) was added. The resulting reaction mixture was stirred at room temperature for 12–24 h and then filtered through a plug of Celite. The solvent was removed in vacuum from the filtrate, and the remaining crude material was purified with flash silica gel column chromatography unless otherwise noted.
4.1.7. General Procedure E for the Synthesis of Hydrobromide Iminothiadiazoles 47b–78b, 132b, and 133b
To a solution of the appropriate thiourea 47a–78a, 132a, and 133a (1.0 equiv) in a mixture of DCM/EtOAc (1/2 v/v, 0.2 M), a 0.5 M solution of bromine (1.5–2.0 equiv) in EtOAc was added dropwise at 0 °C. Upon completion of the addition, PE was added (∼1 mL) and the resulting mixture was left stirring at 5 °C for 1 h and then RT for 12 h. The precipitate formed was isolated, washed with a mixture of PE/EtOAc (2/1 v/v), and dried in vacuum to afford the desired compound.
4.1.8. General Procedure F for the Synthesis of Amides 108a–117a
A solution of aromatic or heteroaromatic acyl chloride 97–103 (1.0 equiv) in dry THF (5 M) was slowly added at 0 °C to a solution of the appropriate amine 104–107 (1.05 equiv) and TEA (1.1–2 equiv) in dry THF (2 M). The reaction mixture was stirred for 2–6 h at room temperature. The formed triethylammonium chloride was removed by filtration and washed with THF. The solvent was removed in vacuum from the filtrate to give a solid residue that was washed with pentane and filtered to afford the desired compound.
4.1.9. General Procedure G for the Synthesis of Imidoyl Chlorides 108b–110b and 112b–117b
The appropriate amide 108a–110a and 112a–117a (1.0 equiv) was dissolved in thionyl chloride (4.3 equiv), and the resulting mixture was heated at 70 °C for 2–2.5 h. Then, the reaction mixture was cooled to room temperature, and the remaining thionyl chloride was removed in vacuum to afford the desired compound that was used in the next step without further purification.
Below, we report the characterization of compounds 108b and 112b. (See also the Supporting Information).
4.1.10. General Procedure H for the Synthesis of 2-Trifluoroacetylamino-1,3,4-thiadiazoles 134b and 135b and 2-Trifluoroacetylaminothiazole 136b
Commercially available 1,3,4-thiadiazoles 134a and 135a and thiazole 136a (1 equiv) were dissolved in dry toluene (0.3 M), and the mixture was cooled to 0 °C. Then, trifluoroacetic anhydride (1.2 equiv) was added dropwise under nitrogen. When the addition was completed, the mixture was warmed to room temperature and left stirring for additional 12 h. Afterward, the reaction mixture was quenched with water and the aqueous layer extracted with EtOAc (3 × 20mL). The organic phases were combined, washed with water (2 × 20mL), dried over Na2SO4, and concentrated in vacuum. The afforded trifluoroacetyl protected derivatives were used in the next step without further purification.
4.1.11. General Procedure I for the Synthesis of 2-Trifluoroacetylamino-3-benzyl-1,3,4-thiadiazoles 134c and 135c and 2-Trifluoroacetylamino-3-benzyl-thiazole 136c
The appropriate 2-trifluoroacetylamino-1,3,4-thiadiazoles 134b and 135b or 2-trifluoroacetylamino thiazole 136b (1 equiv) was dissolved in dry DMF (0.3 M), and K2CO3 (1.2 equiv) was added to the solution under an inert atmosphere. Then, (bromomethyl)benzene 137 (1.2 equiv) was added dropwise and the mixture was stirred at room temperature for 24 h. Afterward, the reaction mixture was quenched with water and extracted with EtOAc (3 × 20mL). The organic layers were combined, washed with water (2 × 20mL), dried over Na2SO4, and concentrated in vacuum. The afforded 2-trifluoroacetylamino-3-benzyl intermediates were used in the next step without further purification.
4.1.12. General Procedure J for the Synthesis of 3-Benzyl-1,3,4-thiadiazoles 134d and 135d and 3-Benzylthiazole 136d
To a stirring solution of the appropriate trifluoroacetyl-protected compounds 134c–136c (1 equiv) in THF (0.25 M), a 5% aqueous solution of NaOH (2 equiv) was added and the reaction mixture was stirred at room temperature until TLC indicated the total consumption of the starting material. Afterward, acetic acid was added to adjust to pH = 7, and the product was subsequently extracted with EtOAc (3 × 10mL). The organic layers were combined, dried over Na2SO4, and concentrated in vacuum to afford the corresponding 3-benzyl intermediates that were used in the next step without further purification.
4.1.12.1. (E)-N-((Z)-4-Benzyl-3-ethyl-1,2,4-thiadiazol-5(4H)-ylidene)-N′-phenylbenzimidamide (1)
N-Benzyl-2,3-diphenyl-1,2,4-thiadiazol-5(2H)-imine hydrobromide 47b (403 mg, 1.05 mmol), propionitrile 80 (7.5 mL, 105 mmol), and TEA (220 μL, 1.6 mmol) were reacted according to general procedure A. The crude product was purified by flash silica gel chromatography (PE/EtOAc in 95/5 ratio) to achieve the final compound 1 as a light yellow solid (205 mg, yield 49%). 1H NMR (401 MHz, DMSO-d6) δ 7.43–7.36 (m, 4H), 7.35–7.27 (m, 6H), 7.26–7.20 (m, 2H), 7.05–6.98 (m, 1H), 6.78–6.74 (m, 2H), 5.57 (s, 2H), 2.72 (q, J = 7.3 Hz, 2H), 1.18 (t, J = 7.3 Hz, 3H) ppm. 13C NMR (101 MHz, DMSO-d6) δ 171.02, 158.54, 157.56, 146.85, 135.98, 134.72, 129.48, 129.24 (2C), 128.94 (2C), 128.90 (2C), 127.98 (2C), 127.75, 126.85 (2C), 123.00, 122.04 (2C), 48.18, 22.43, 9.93 ppm. Rt = 4.65 min (apolar method); ESI-MS for C24H22N4S: calculated 398.16, found m/z 399.18 [M + H]+; UPLC-MS purity (UV 215 nm) >99.5%.
4.1.12.2. (E)-N-((Z)-4-Benzyl-3-(fluoromethyl)-1,2,4-thiadiazol-5(4H)-ylidene)-N′-phenylbenzimidamide (2)
N-Benzyl-2,3-diphenyl-1,2,4-thiadiazol-5(2H)-imine hydrobromide 47b (395 mg, 0.93 mmol), fluoroacetonitrile 81 (155 μL, 2.79 mmol), and TEA (194 μL, 1.4 mmol) were reacted according to general procedure A. The crude product was purified by flash silica gel chromatography (PE/EtOAc in 97/3 ratio) and washed with n-hexane to achieve the final compound 2 as a yellow solid (248 mg, yield 66%). 1H NMR (401 MHz, DMSO-d6) δ 7.43–7.38 (m, 3H), 7.37–7.32 (m, 4H), 7.30 (d, J = 7.6 Hz, 2H), 7.28–7.23 (m, 3H), 7.08–7.01 (m, 1H), 6.81–6.76 (m, 2H), 5.59 (d, J = 46.7 Hz, 2H), 5.59 (s, 2H) ppm. 13C NMR (101 MHz, DMSO-d6) δ 170.04, 158.32, 151.19 (d, JCF = 18.6 Hz), 146.18, 135.67, 134.20, 129.74, 129.27 (2C), 129.00 (2C), 128.66 (2C), 128.03 (2C), 127.77, 127.21 (2C), 123.37, 122.04 (2C), 77.38 (d, JCF = 168.1 Hz), 48.87 ppm. Rt = 4.22 min (apolar method); ESI-MS for C23H19FN4S: calculated 402.13, found m/z 403.25 [M + H]+; UPLC-MS purity (UV 215 nm) >99.5%.
4.1.12.3. (E)-N-((Z)-4-Benzyl-3-isopropyl-1,2,4-thiadiazol-5(4H)-ylidene)-N′-phenylbenzimidamide (3)
N-Benzyl-2,3-diphenyl-1,2,4-thiadiazol-5(2H)-imine hydrobromide 47b (1.2 g, 2.8 mmol), isobutyronitrile 82 (25.46 mL, 280 mmol), and TEA (600 μL, 4.2 mmol) were reacted according to general procedure A. The crude product was purified by flash silica gel chromatography (PE/EtOAc in 95/5 ratio) to achieve the final compound 3 as a yellow solid (401 mg, yield 35%). 1H NMR (401 MHz, DMSO-d6) δ 7.43–7.36 (m, 4H), 7.34–7.21 (m, 8H), 7.01 (tt, J = 7.3, 1.1 Hz, 1H), 6.75 (dd, J = 7.3, 1.2 Hz, 2H), 5.63 (s, 2H), 3.18 (hept, J = 6.7 Hz, 1H), 1.13 (d, J = 6.7 Hz, 6H) ppm. 13C NMR (101 MHz, DMSO-d6) δ 170.93, 161.45, 158.57, 146.83, 136.28, 134.71, 129.45, 129.20 (2C), 128.92 (2C), 128.85 (2C), 127.95 (2C), 127.68, 126.60 (2C), 123.00, 122.01 (2C), 48.25, 27.87, 20.94 ppm. Rt = 5.04 min (apolar method); ESI-MS for C25H24N4S: calculated 412.17, found m/z 413.38 [M + H]+; UPLC-MS purity (UV 215 nm) >99.5%.
4.1.12.4. (E)-N-((Z)-3-Methyl-4-phenethyl-1,2,4-thiadiazol-5(4H)-ylidene)-N′-phenylbenzimidamide (4)
N-Phenethyl-2,3-diphenyl-1,2,4-thiadiazol-5(2H)-imine hydrobromide 48b (200 mg, 0.46 mmol), ACN 79 (2.402 mL, 46 mmol), and TEA (96 μL, 0.69 mmol) were reacted according to general procedure A. The crude product was purified by flash silica gel chromatography (PE/EtOAc in 9/1 ratio) and washed with n-hexane to achieve the final compound 4 as a shiny yellow solid (115 mg, yield 63%). 1H NMR (401 MHz, DMSO-d6) δ 7.51–7.46 (m, 2H), 7.40–7.29 (m, 5H), 7.28–7.20 (m, 5H), 7.01 (t, J = 7.0 Hz, 1H), 6.77–6.73 (m, 2H), 4.42 (t, J = 7.2 Hz, 2H), 3.19–3.12 (m, 2H), 2.21 (s, 3H) ppm. 13C NMR (101 MHz, DMSO-d6) δ 169.97, 158.53, 153.61, 146.98, 138.14, 134.93, 129.48, 129.33 (2C), 128.96 (2C), 128.93 (2C), 128.60 (2C), 128.02 (2C), 126.72, 122.92, 122.04 (2C), 47.56, 33.26, 15.72 ppm. Rt = 4.32 min (apolar method); ESI-MS for C24H22N4S: calculated 398.16, found m/z 399.25 [M + H]+; UPLC-MS purity (UV 215 nm) 99%.
4.1.12.5. (E)-N-((Z)-3-Methyl-4-(3-phenylpropyl)-1,2,4-thiadiazol-5(4H)-ylidene)-N′-phenylbenzimidamide (5)
2,3-Diphenyl-N-(3-phenylpropyl)-1,2,4-thiadiazol-5(2H)-imine hydrobromide 49b (300 mg, 0.66 mmol), ACN 79 (3.447 mL, 66 mmol), and TEA (139 μL, 0.99 mmol) were reacted according to general procedure A. The crude product was purified by flash silica gel chromatography (PE/EtOAc in 9/1 ratio) and washed with n-hexane to achieve the final compound 5 as a light yellow solid (204 mg, yield 75%). 1H NMR (401 MHz, DMSO-d6) δ 7.43–7.38 (m, 2H), 7.38–7.25 (m, 7H), 7.25–7.17 (m, 3H), 6.99 (ddt, J = 7.7, 7.0, 1.2 Hz, 1H), 6.76–6.70 (m, 2H), 4.23 (t, J = 7.7 Hz, 2H), 2.75 (t, J = 7.4 Hz, 2H), 2.46 (s, 3H), 2.14 (p, J = 7.5 Hz, 2H) ppm. 13C NMR (101 MHz, DMSO-d6) δ 170.14, 158.29, 153.53, 146.97, 140.75, 134.75, 129.41, 129.26 (2C), 128.88 (2C), 128.32 (2C), 128.26 (2C), 127.93 (2C), 125.94, 122.83, 121.93 (2C), 45.43, 32.11, 28.63, 15.94 ppm. Rt = 4.61 min (apolar method); ESI-MS for C25H24N4S: calculated 412.17, found m/z 413.21 [M + H]+; UPLC-MS purity (UV 215 nm) >99.5%.
4.1.12.6. (E)-N-((Z)-3-Methyl-4-(4-methylbenzyl)-1,2,4-thiadiazol-5(4H)-ylidene)-N′-phenylbenzimidamide (6)
N-(4-Methylbenzyl)-2,3-diphenyl-1,2,4-thiadiazol-5(2H)-imine hydrobromide 50b (216 mg, 0.49 mmol), ACN 79 (2.559 mL, 49 mmol), and TEA (103 μL, 0.74 mmol) were reacted according to general procedure A. The crude product was purified by flash silica gel chromatography (PE/EtOAc in 9/1 ratio) and washed with n-hexane to achieve the final compound 6 as a shiny yellow solid (161 mg, yield 82%). 1H NMR (401 MHz, DMSO-d6) δ 7.45–7.40 (m, 2H), 7.37–7.26 (m, 3H), 7.26–7.17 (m, 6H), 7.04–6.98 (m, 1H), 6.78–6.73 (m, 2H), 5.51 (s, 2H), 2.41 (s, 3H), 2.28 (s, 3H) ppm. 13C NMR (101 MHz, DMSO-d6) δ 170.66, 158.46, 153.65, 146.80, 137.01, 134.71, 132.83, 129.44, 129.38 (2C), 129.20 (2C), 128.87 (2C), 127.95 (2C), 126.97 (2C), 122.94, 121.98 (2C), 48.30, 20.64, 16.20 ppm. Rt = 4.57 min (apolar method); ESI-MS for C24H22N4S: calculated 398.16, found m/z 399.16 [M + H]+; UPLC-MS purity (UV 215 nm) >99.5%.
4.1.12.7. (E)-N-((Z)-4-(4-Methoxybenzyl)-3-methyl-1,2,4-thiadiazol-5(4H)-ylidene)-N′-phenylbenzimidamide (7)
N-(4-Methoxybenzyl)-2,3-diphenyl-1,2,4-thiadiazol-5(2H)-imine hydrobromide 51b (114 mg, 0.25 mmol), ACN 79 (1.306 mL, 25 mmol), and TEA (53 μL, 0.38 mmol) were reacted according to general procedure A. The crude product was purified by flash silica gel chromatography (PE/EtOAc in 9/1 ratio) and washed with n-hexane to achieve the final compound 7 as a shiny yellow solid (62 mg, yield 60%). 1H NMR (401 MHz, DMSO-d6) δ 7.47–7.42 (m, 2H), 7.38–7.26 (m, 5H), 7.23 (t, J = 7.8 Hz, 2H), 7.01 (t, J = 7.4 Hz, 1H), 6.95 (d, J = 8.6 Hz, 2H), 6.76 (d, J = 7.0 Hz, 2H), 5.48 (s, 2H), 3.73 (s, 3H), 2.43 (s, 3H) ppm. 13C NMR (101 MHz, DMSO-d6) δ 170.69, 158.85, 158.49, 153.68, 146.83, 134.75, 129.47, 129.23 (2C), 128.91 (2C), 128.65 (2C), 127.99 (2C), 127.80, 122.96, 122.00 (2C), 114.25 (2C), 55.09, 48.05, 16.29 ppm. Rt = 3.93 min (apolar method); ESI-MS for C24H22N4OS: calculated 414.15, found m/z 415.41 [M + H]+; UPLC-MS purity (UV 215 nm) 99%.
4.1.12.8. (E)-N-((Z)-4-(4-Hydroxybenzyl)-3-methyl-1,2,4-thiadiazol-5(4H)-ylidene)-N′-phenylbenzimidamide (8)
(E)-N-((Z)-4-(4-Methoxybenzyl)-3-methyl-1,2,4-thiadiazol-5(4H)-ylidene)-N′-phenylbenzimidamide 7 (266 mg, 0.64 mmol) and 2.25 equiv of BBr3 (1.44 mmol) were reacted according to the general procedure B. The crude material was purified via flash silica gel column chromatography (PE/EtOAc in 8/2 ratio), then washed with n-hexane, and filtered to give the final compound 8 as a yellow solid (87 mg, yield 34%). 1H NMR (401 MHz, DMSO-d6) δ 9.50 (s, 1H), 7.49–7.40 (m, 2H), 7.32 (ddd, J = 14.1, 7.7, 5.9 Hz, 3H), 7.23 (t, J = 7.6 Hz, 2H), 7.18 (d, J = 8.4 Hz, 2H), 7.01 (t, J = 7.4 Hz, 1H), 6.76 (dd, J = 8.7, 2.3 Hz, 4H), 5.43 (s, 2H), 2.42 (s, 3H) ppm. 13C NMR (101 MHz, DMSO-d6) δ 170.72, 158.54, 157.06, 153.78, 146.83, 134.77, 129.49, 129.26 (2C), 128.92 (2C), 128.71 (2C), 128.01 (2C), 126.03, 122.98, 122.03 (2C), 115.56 (2C), 48.22, 18.90, 16.33 ppm. Rt = 5.26 min (generic method); ESI-MS for C23H20N4OS: calculated 400.14, found m/z 401.15 [M + H]+; UPLC-MS purity (UV 215 nm) 99%.
4.1.12.9. (E)-N-((Z)-4-(4-Fluorobenzyl)-3-methyl-1,2,4-thiadiazol-5(4H)-ylidene)-N′-phenylbenzimidamide (9)
N-(4-Fluorobenzyl)-2,3-diphenyl-1,2,4-thiadiazol-5(2H)-imine hydrobromide 52b (90 mg, 0.2 mmol), ACN 79 (1.045 mL, 20 mmol), and TEA (41 μL, 0.3 mmol) were reacted according to general procedure A. The crude product was purified by flash silica gel chromatography (PE/EtOAc in 9/1 ratio) and washed with n-hexane to achieve the final compound 9 as a yellow solid (48 mg, yield 60%). 1H NMR (401 MHz, DMSO-d6) δ 7.45–7.37 (m, 4H), 7.37–7.27 (m, 3H), 7.27–7.19 (m, 4H), 7.01 (tt, J = 7.3, 1.2 Hz, 1H), 6.79–6.73 (m, 2H), 5.54 (s, 2H), 2.42 (s, 3H) ppm. 13C NMR (101 MHz, DMSO-d6) δ 170.62, 161.62 (d, JCF = 244.0 Hz), 158.46, 153.58, 146.76, 134.66, 132.11 (d, JCF = 3.1 Hz), 129.49, 129.31 (d, JCF = 8.4 Hz, 2C), 129.22 (2C), 128.91 (2C), 127.99 (2C), 123.00, 121.98 (2C), 115.70 (d, JCF = 21.5 Hz, 2C), 47.85, 16.23 ppm. Rt = 4.12 min (apolar method); ESI-MS for C23H19FN4S: calculated 402.13, found m/z 403.09 [M + H]+; UPLC-MS purity (UV 215 nm) >99.5%.
4.1.12.10. (E)-N-((Z)-4-(4-Chlorobenzyl)-3-methyl-1,2,4-thiadiazol-5(4H)-ylidene)-N′-phenylbenzimidamide (10)
N-(4-Chlorobenzyl)-2,3-diphenyl-1,2,4-thiadiazol-5(2H)-imine hydrobromide 53b (70 mg, 0.15 mmol), ACN 79 (783 μL, 15 mmol), and TEA (32 μL, 0.23 mmol) were reacted according to general procedure A. The crude product was purified by flash silica gel chromatography (PE/EtOAc in 9/1 ratio) and washed with n-hexane to achieve the final compound 10 as a shiny yellow solid (50 mg, yield 80%). 1H NMR (401 MHz, DMSO-d6) δ 7.48–7.44 (m, 2H), 7.43–7.39 (m, 2H), 7.37–7.26 (m, 5H), 7.26–7.21 (m, 2H), 7.04–6.98 (m, 1H), 6.78–6.73 (m, 2H), 5.54 (s, 2H), 2.41 (s, 3H) ppm. 13C NMR (101 MHz, DMSO-d6) δ 170.58, 158.45, 153.55, 146.75, 134.89, 134.62, 132.40, 129.50, 129.22 (2C), 128.97 (2C), 128.92 (2C), 128.86 (2C), 127.98 (2C), 123.02, 121.98 (2C), 47.90, 16.19 ppm. Rt = 4.55 min (apolar method); ESI-MS for C23H19ClN4S: calculated 418.10, found m/z 419.13/421.16 [M + H]+; UPLC-MS purity (UV 215 nm) 99.5%.
4.1.12.11. (E)-N-((Z)-4-(4-Bromobenzyl)-3-methyl-1,2,4-thiadiazol-5(4H)-ylidene)-N′-phenylbenzimidamide (11)
N-(4-Bromobenzyl)-2,3-diphenyl-1,2,4-thiadiazol-5(2H)-imine hydrobromide 54b (558 mg, 1.11 mmol), ACN 79 (5.80 mL, 111 mmol), and TEA (232 μL, 1.67 mmol) were reacted according to general procedure A. The crude product was purified by flash silica gel chromatography (PE/EtOAc in 88/12 ratio) to achieve the final compound 11 as a yellow solid (380 mg, yield 74%). 1H NMR (401 MHz, DMSO-d6) δ 7.65–7.55 (m, 2H), 7.41 (dd, J = 6.8, 1.6 Hz, 2H), 7.38–7.24 (m, 5H), 7.28–7.19 (m, 2H), 7.01 (tt, J = 7.4, 1.2 Hz, 1H), 6.75 (dd, J = 8.3, 1.3 Hz, 2H), 5.53 (s, 2H), 2.41 (s, 3H) ppm. 13C NMR (101 MHz, DMSO-d6) δ 170.58, 158.45, 153.55, 146.75, 135.31, 134.61, 131.78 (2C), 129.51, 129.28 (2C), 129.23 (2C), 128.92 (2C), 127.99 (2C), 123.03, 121.98 (2C), 120.92, 47.95, 16.19 ppm. Rt = 4.63 min (apolar method); ESI-MS for C23H19BrN4S: calculated 462.05, found m/z 462.89/464.83 [M + H]+; UPLC-MS purity (UV 215 nm) >99.5%.
4.1.12.12. 4-(((Z)-3-Methyl-5-(((E)-phenyl(phenylimino)methyl)imino)-1,2,4-thiadiazol-4(5H)-yl)methyl)benzoic Acid (12)
To a solution of (E)-N-((Z)-4-(4-bromobenzyl)-3-methyl-1,2,4-thiadiazol-5(4H)-ylidene)-N′-phenylbenzimidamide 11 (148 mg, 0.32 mmol) in dry THF (5 mL) at −78 °C, 324 μL (0.81 mmol) of 2.5 M n-butyllithium in n-hexane was added dropwise. After being stirred at −78 °C for 30 min, the mixture was treated with an excess of dry ice and stirred for an additional 30 min at −78 °C. Then, the reaction mixture was warmed to room temperature, diluted with water, and adjusted to pH 3 with aqueous 2 N HCl. The crude was extracted with DCM (×3), the organic phase was washed with water (×2), and dried over Na2SO4, and the solvent was removed under reduced pressure. Purification was performed by direct phase flash chromatography (DCM/MeOH in 99.5/0.5 ratio, acetic acid 0.1%). Then, the solid was washed with a mixture of n-hexane/EtOAc (9/1) and filtered to achieve the final compound 12 as a light yellow solid (35 mg, yield 26%). 1H NMR (401 MHz, DMSO-d6) δ 12.25 (s, 1H), 7.96 (dd, J = 8.3, 5.3 Hz, 2H), 7.43–7.37 (m, 4H), 7.36–7.30 (m, 1H), 7.29 (dd, J = 6.9, 1.3 Hz, 2H), 7.26–7.21 (m, 2H), 7.05–6.98 (m, 1H), 6.78–6.73 (m, 2H), 5.63 (s, 2H), 2.40 (s, 3H) ppm. 13C NMR (101 MHz, DMSO-d6) δ 170.58, 167.00, 158.48, 153.62, 146.77, 140.61, 134.61, 130.53, 129.91 (2C), 129.52, 129.24 (2C), 128.94 (2C), 127.99 (2C), 126.98 (2C), 123.05, 122.01 (2C), 48.37, 16.19 ppm. Rt = 4.30 min (apolar method); ESI-MS for C24H20N4O2S: calculated 428.13, found m/z 429.27 [M + H]+, 427.43 [M – H]−; UPLC-MS purity (UV 215 nm) 98.5%.
4.1.12.13. (E)-N-((Z)-4-(4-Aminobenzyl)-3-methyl-1,2,4-thiadiazol-5(4H)-ylidene)-N′-phenylbenzimidamide (13)
4-(((2,3-Diphenyl-1,2,4-thiadiazol-5(2H)-ylidene)amino)methyl) aniline dihydrobromide 55b (137 mg, 0.26 mmol), ACN 79 (1.358 mL, 26 mmol), and TEA (65 μL, 0.47 mmol) were reacted according to general procedure A. The crude product was purified by flash silica gel chromatography (PE/EtOAc in 7/3 ratio) and washed with n-hexane to achieve the final compound 13 as a shiny yellow solid (42 mg, yield 40%). 1H NMR (401 MHz, DMSO-d6) δ 7.48–7.42 (m, 2H), 7.38–7.27 (m, 3H), 7.23 (t, J = 7.8 Hz, 2H), 7.05 (d, J = 8.4 Hz, 2H), 7.00 (tt, J = 7.3, 1.3 Hz, 1H), 6.75 (dd, J = 8.2, 1.3 Hz, 2H), 6.54 (d, J = 8.4 Hz, 2H), 5.35 (s, 2H), 5.13 (s, 2H), 2.43 (s, 3H) ppm. 13C NMR (101 MHz, DMSO-d6) δ 170.77, 158.54, 153.85, 148.48, 146.92, 134.87, 129.46, 129.27 (2C), 128.92 (2C), 128.46 (2C), 128.01 (2C), 122.93, 122.46, 122.04 (2C), 113.88 (2C), 48.50, 16.41 ppm. Rt = 3.02 min (apolar method); ESI-MS for C23H21N5S: calculated 399.15, found m/z 400.12 [M + H]+; UPLC-MS purity (UV 215 nm) 96.5%.
4.1.12.14. (E)-N-((Z)-3-Methyl-4-(3-methylbenzyl)-1,2,4-thiadiazol-5(4H)-ylidene)-N′-phenylbenzimidamide (14)
N-(3-Methylbenzyl)-2,3-diphenyl-1,2,4-thiadiazol-5(2H)-imine hydrobromide 56b (342 mg, 0.78 mmol), ACN 79 (4.07 mL, 78 mmol), and TEA (162 μL, 1.16 mmol) were reacted according to general procedure A. The crude product was purified by flash silica gel chromatography (PE/EtOAc in 93/7 ratio) to achieve the final compound 14 as a yellow solid (100 mg, yield 74%). 1H NMR (401 MHz, DMSO-d6) δ 7.46–7.38 (m, 2H), 7.37–7.19 (m, 6H), 7.17 (s, 1H), 7.13 (d, J = 7.6 Hz, 1H), 7.07 (d, J = 7.7 Hz, 1H), 7.01 (t, J = 7.4 Hz, 1H), 6.80–6.72 (m, 2H), 5.51 (s, 2H), 2.42 (s, 3H), 2.30 (s, 3H) ppm. 13C NMR (101 MHz, DMSO-d6) δ 170.64, 158.50, 153.72, 146.81, 138.04, 135.79, 134.73, 129.48, 129.22 (2C), 128.91 (2C), 128.81, 128.44, 127.98 (2C), 127.67, 123.99, 122.99, 122.01 (2C), 48.55, 21.01, 16.28 ppm. Rt = 4.39 min (apolar method); ESI-MS for C24H22N4S: calculated 398.16, found m/z 399.09 [M + H]+; UPLC-MS purity (UV 215 nm) >99.5%.
4.1.12.15. (E)-N-((Z)-3-Methyl-4-(3-(trifluoromethyl)benzyl)-1,2,4-thiadiazol-5(4H)-ylidene)-N′-phenylbenzimidamide (15)
N-(3-(Trifluoromethyl)benzyl)-2,3-diphenyl-1,2,4-thiadiazol-5(2H)-imine hydrobromide 57b (328 mg, 0.67 mmol), ACN 79 (3.48 mL, 67 mmol), and TEA (140 μL, 1 mmol) were reacted according to general procedure A. The crude product was purified by flash silica gel chromatography (PE/EtOAc in 96/4 ratio) to achieve the final compound 15 as a yellow solid (198 mg, yield 65%). 1H NMR (401 MHz, DMSO-d6) δ 7.87 (s, 1H), 7.71 (d, J = 7.7 Hz, 1H), 7.63 (t, J = 7.7 Hz, 1H), 7.57 (d, J = 7.8 Hz, 1H), 7.40–7.30 (m, 3H), 7.30–7.25 (m, 2H), 7.23 (t, J = 8.0 Hz, 2H), 7.06–6.97 (m, 1H), 6.79–6.71 (m, 2H), 5.62 (s, 2H), 2.47 (s, 3H) ppm. 13C NMR (101 MHz, DMSO-d6) δ 170.49, 158.40, 153.50, 146.72, 137.34, 134.62, 131.09 (q, JCF = 1.6 Hz), 130.09, 129.49, 129.15 (2C), 128.92 (2C), 127.93 (2C), 124.57 (q, JCF = 3.7, Hz), 122.88, 121.97 (2C), 48.26, 16.29 ppm. Rt = 4.40 min (apolar method); ESI-MS for C24H19F3N4S: calculated 452.13, found m/z 453.18 [M + H]+, 451.36 [M – H]−, 511.35 [M + OAc]−; UPLC-MS purity (UV 215 nm) >99.5%.
4.1.12.16. (E)-N-((Z)-4-(3-Methoxybenzyl)-3-methyl-1,2,4-thiadiazol-5(4H)-ylidene)-N′-phenylbenzimidamide (16)
N-(3-Methoxybenzyl)-2,3-diphenyl-1,2,4-thiadiazol-5(2H)-imine hydrobromide 58b (191 mg, 0.42 mmol), ACN 79 (2.194 mL, 42 mmol), and TEA (88 μL, 0.63 mmol) were reacted according to the general procedure. The crude product was purified by flash silica gel chromatography (PE/EtOAc in 9/1 ratio) and washed with n-hexane to achieve the final compound 16 as a shiny yellow solid (134 mg, yield 77%). 1H NMR (401 MHz, DMSO-d6) δ 7.45–7.40 (m, 2H), 7.36–7.21 (m, 6H), 7.04–6.98 (m, 1H), 6.95–6.87 (m, 2H), 6.85–6.80 (m, 1H), 6.78–6.74 (m, 2H), 5.52 (s, 2H), 3.72 (s, 3H), 2.42 (s, 3H) ppm. 13C NMR (101 MHz, DMSO-d6) δ 170.63, 159.47, 158.45, 153.70, 146.79, 137.41, 134.69, 130.09, 129.49, 129.22 (2C), 128.91 (2C), 127.98 (2C), 123.00, 122.00 (2C), 118.86, 113.15, 112.94, 55.01, 48.43, 16.22 ppm. Rt = 4.00 min (apolar method); ESI-MS for C24H22N4OS: calculated 414.15, found m/z 415.16 [M + H]+; UPLC-MS purity (UV 215 nm) >99.5%.
4.1.12.17. (E)-N-((Z)-4-(3-Hydroxybenzyl)-3-methyl-1,2,4-thiadiazol-5(4H)-ylidene)-N′-phenylbenzimidamide (17)
(E)-N-((Z)-4-(3-Methoxybenzyl)-3-methyl-1,2,4-thiadiazol-5(4H)-ylidene)-N′-phenylbenzimidamide 16 (136 mg, 0.33 mmol) and 2.25 equiv of BBr3 (0.75 mmol) were reacted according to the general procedure B. The crude material was purified via flash silica gel column chromatography (PE/EtOAc in 8/2 ratio), then washed with n-hexane, and filtered to give the final compound 17 as a yellow solid (78 mg, yield 59%). 1H NMR (401 MHz, DMSO-d6) δ 9.52 (s, 1H), 7.46–7.40 (m, 2H), 7.34–7.15 (m, 6H), 7.01 (t, J = 7.4 Hz, 1H), 6.79–6.63 (m, 5H), 5.49 (s, 2H), 2.39 (s, 3H) ppm. 13C NMR (101 MHz, DMSO-d6) δ 170.71, 158.52, 157.74, 153.77, 146.81, 137.22, 134.68, 129.95, 129.49, 129.26 (2C), 128.92 (2C), 127.98 (2C), 123.00, 122.01 (2C), 117.42, 114.76, 113.32, 48.37, 16.16 ppm. Rt = 5.39 min (generic method); ESI-MS for C23H20N4OS: calculated 400.14, found m/z 401.12 [M + H]+, 399.24 [M – H]−; UPLC-MS purity (UV 215 nm) 98%.
4.1.12.18. (E)-N-((Z)-4-(3-Isopropoxybenzyl)-3-methyl-1,2,4-thiadiazol-5(4H)-ylidene)-N′-phenylbenzimidamide (18)
(E)-N-((Z)-4-(3-Hydroxybenzyl)-3-methyl-1,2,4-thiadiazol-5(4H)-ylidene)-N′-phenylbenzimidamide 17 (270 mg, 0.47 mmol) was dissolved in dry DMF (0.2 M) and treated with K2CO3 (194 mg, 1.4 mmol) at room temperature for 1 h. Then, 2-bromopropane (129 μL, 1.4 mmol) was added and the reaction mixture was heated at 70 °C for 3 h. After completion of the reaction, the mixture was cooled to room temperature, diluted with H2O, and extracted with EtOAc (3 × 10mL). The organic phases were combined, washed with water (2 × 10mL) and brine (2 × 10mL), and dried over anhydrous Na2SO4, and the solvent was evaporated under reduced pressure. The crude product was purified by flash silica gel chromatography (PE/EtOAc in 92/8 ratio) to achieve the final compound 18 as a yellow solid (37 mg, yield 18%). 1H NMR (401 MHz, DMSO-d6) δ 7.44 (t, J = 1.5 Hz, 1H), 7.42 (d, J = 1.7 Hz, 1H), 7.37–7.32 (m, 1H), 7.31–7.26 (m, 3H), 7.26–7.20 (m, 2H), 7.01 (tdd, J = 7.4, 2.2, 1.1 Hz, 1H), 6.90 (t, J = 2.1 Hz, 1H), 6.86 (dd, J = 8.3, 2.4 Hz, 1H), 6.81 (dd, J = 7.6, 1.7 Hz, 1H), 6.79–6.74 (m, 2H), 5.51 (s, 2H), 4.50 (hept, J = 6.2 Hz, 1H), 2.43 (s, 3H), 1.21 (dq, J = 6.0, 0.5 Hz, 6H) ppm. 13C NMR (101 MHz, DMSO-d6) δ 170.63, 158.43, 157.64, 153.71, 146.81, 137.45, 134.70, 130.09, 129.51, 129.24 (2C), 128.93 (2C), 127.98 (2C), 123.01, 122.00 (2C), 118.71, 114.65, 114.52, 69.10, 48.48, 21.69, 16.25 ppm. Rt = 4.68 min (apolar method); ESI-MS for C26H26N4OS: calculated 442.18, found m/z 443.31 [M + H]+, 441.41 [M – H]−; UPLC-MS purity (UV 215 nm) 96.5%.
4.1.12.19. (E)-N-((Z)-4-(3-Ethoxybenzyl)-3-methyl-1,2,4-thiadiazol-5(4H)-ylidene)-N′-phenylbenzimidamide (19)
N-(3-Ethoxybenzyl)-2,3-diphenyl-1,2,4-thiadiazol-5(2H)-imine hydrobromide 59b (600 mg, 1.28 mmol), ACN 79 (6.67 mL, 128 mmol), and TEA (260 μL, 1.92 mmol) were reacted according to the general procedure. The crude product was purified by flash silica gel chromatography (PE/EtOAc in 90/10 ratio) to achieve the final compound 19 as a yellow solid (486 mg, yield 89%). 1H NMR (401 MHz, DMSO-d6) δ 7.45–7.40 (m, 2H), 7.37–7.26 (m, 4H), 7.26–7.21 (m, 2H), 7.01 (ddt, J = 8.6, 7.1, 1.2 Hz, 1H), 6.91–6.85 (m, 2H), 6.83 (ddd, J = 7.6, 1.7, 0.9 Hz, 1H), 6.79–6.74 (m, 2H), 5.51 (s, 2H), 3.96 (q, J = 7.0 Hz, 2H), 2.42 (s, 3H), 1.28 (t, J = 6.9 Hz, 3H) ppm. 13C NMR (101 MHz, DMSO-d6) δ 170.63, 158.72, 158.45, 153.72, 146.81, 137.39, 134.70, 130.09, 129.52, 129.24 (2C), 128.93 (2C), 127.99 (2C), 123.02, 122.01 (2C), 118.83, 113.53, 113.43, 62.96, 48.46, 16.25, 14.56 ppm. Rt = 4.37 min (apolar method); ESI-MS for C25H24N4OS: calculated 428.17, found m/z 429.31 [M + H]+; UPLC-MS purity (UV 215 nm) 99%.
4.1.12.20. (E)-N-((Z)-4-(3-Aminobenzyl)-3-methyl-1,2,4-thiadiazol-5(4H)-ylidene)-N′-phenylbenzimidamide (20)
3-(((2,3-Diphenyl-1,2,4-thiadiazol-5(2H)-ylidene)amino)methyl) aniline dihydrobromide 60b (1.757 g, 3.4 mmol), ACN 79 (17.65 mL, 338 mmol), and TEA (1.186 mL, 8.5 mmol) were reacted according to general procedure A. The crude product was purified by flash silica gel chromatography (PE/EtOAc in 70/30 ratio) to achieve the final compound 20 as a yellow solid (230 mg, yield 17%). 1H NMR (401 MHz, DMSO-d6) δ 7.46–7.40 (m, 2H), 7.36–7.26 (m, 3H), 7.26–7.20 (m, 2H), 7.01 (tt, J = 7.6, 1.1 Hz, 2H), 6.79–6.73 (m, 2H), 6.48 (ddd, J = 8.0, 2.2, 1.0 Hz, 1H), 6.42 (ddd, J = 7.4, 1.7, 1.0 Hz, 1H), 6.39 (t, J = 1.9 Hz, 1H), 5.42 (s, 2H), 5.17 (s, 2H), 2.39 (s, 3H) ppm. 13C NMR (101 MHz, DMSO-d6) δ 170.74, 158.56, 153.89, 149.20, 146.87, 136.40, 134.73, 129.46, 129.35, 129.28 (2C), 128.93 (2C), 127.98 (2C), 122.96, 122.02 (2C), 113.99, 113.23, 111.36, 48.65, 16.21 ppm. Rt = 5.41 min (generic method); ESI-MS for C23H21N5S: calculated 399.15, found m/z 400.3 [M + H]+, 398.47 [M – H]−; UPLC-MS purity (UV 215 nm) >99.5%.
4.1.12.21. (E)-N-((Z)-3-Methyl-4-(3-(methylamino)benzyl)-1,2,4-thiadiazol-5(4H)-ylidene)-N′-phenylbenzimidamide (21) and (E)-N-((Z)-4-(3-(Dimethylamino)benzyl)-3-methyl-1,2,4-thiadiazol-5(4H)-ylidene)-N′-phenylbenzimidamide (22)
In a screw-capped pressure tube, (E)-N-((Z)-4-(3-aminobenzyl)-3-methyl-1,2,4-thiadiazol-5(4H)-ylidene)-N′-phenylbenzimidamide 20 (250 mg, 0.63 mmol) was dissolved in dry DMF (4 mL), and then K2CO3 (113 mg, 0.81 mmol) and methyl iodide (47 μL, 0.75 mmol) were added. The vessel was then sealed and heated to 55 °C for 24 h. After reaction cooling, 5 mL of water was added into the mixture. The organic phase was separated, and the aqueous layer was extracted with EtOAc (3 × 10mL). The combined organic phase was washed with brine (2 × 10mL) and dried over Na2SO4. The solvent was removed under reduced pressure, and the crude product was purified by flash silica gel chromatography (A = PE, B = EtOAc, gradient 0–20% B). Yields: (21) yellow solid 90 mg, 35%; (22) yellow solid, 60 mg, 22%.
21) 1H NMR (401 MHz, DMSO-d6) δ 7.46–7.41 (m, 2H), 7.36–7.26 (m, 3H), 7.26–7.20 (m, 2H), 7.08 (t, J = 7.7 Hz, 1H), 7.01 (tt, J = 7.4, 1.1 Hz, 1H), 6.78–6.73 (m, 2H), 6.49–6.39 (m, 3H), 5.75 (q, J = 5.0 Hz, 1H), 5.44 (s, 2H), 2.61 (d, J = 5.1 Hz, 3H), 2.41 (s, 3H) ppm. 13C NMR (101 MHz, DMSO-d6) δ 170.72, 158.51, 153.86, 150.23, 146.87, 136.43, 134.75, 129.44, 129.25 (2C), 128.92 (2C), 127.96 (2C), 122.95, 122.00 (2C), 113.69, 110.65, 110.03, 48.77, 29.53, 16.22 ppm. Rt = 5.92 min (generic method); ESI-MS for C24H23N5S: calculated 413.17, found m/z 414.13 [M + H]+; UPLC-MS purity (UV 215 nm) 99%.
22) 1H NMR (401 MHz, DMSO-d6) δ 7.46–7.42 (m, 2H), 7.37–7.26 (m, 3H), 7.26–7.20 (m, 2H), 7.15 (dd, J = 8.3, 7.5 Hz, 1H), 7.03–6.98 (m, 1H), 6.80 (t, J = 2.1 Hz, 1H), 6.78–6.72 (m, 2H), 6.65 (dd, J = 8.1, 2.6 Hz, 1H), 6.48 (d, J = 7.8 Hz, 1H), 5.48 (s, 2H), 2.84 (s, 6H), 2.44 (s, 3H) ppm. 13C NMR (101 MHz, DMSO-d6) δ 170.69, 158.44, 153.82, 150.56, 146.87, 136.51, 134.77, 129.46, 129.43, 129.22 (2C), 128.91 (2C), 127.96 (2C), 122.96, 121.99 (2C), 114.16, 111.75, 111.26, 49.00, 16.30, 16.29 (2C) ppm. Rt = 4.37 min (apolar method); ESI-MS for C25H25N5S: calculated 427.18, found m/z 428.13 [M + H]+; UPLC-MS purity (UV 215 nm) 98%.
4.1.12.22. (E)-N-((Z)-4-(3-Fluorobenzyl)-3-methyl-1,2,4-thiadiazol-5(4H)-ylidene)-N′-phenylbenzimidamide (23)
(Z)-N-(3-Fluorobenzyl)-2,3-diphenyl-1,2,4-thiadiazol-5(2H)-imine hydrobromide 61b (416 mg, 0.94 mmol), ACN 79 (4.89 mL, 94 mmol), and TEA (197 μL, 1.41 mmol) were reacted according to general procedure A. The crude product was purified by flash silica gel chromatography (PE/EtOAc in 9/1 ratio) to achieve the final compound 23 as a yellow solid (308 mg, yield 81%). 1H NMR (401 MHz, DMSO-d6) δ 7.48–7.38 (m, 3H), 7.37–7.09 (m, 8H), 7.01 (t, J = 7.4 Hz, 1H), 6.79–6.73 (m, 2H), 5.56 (s, 2H), 2.42 (s, 3H) ppm. 13C NMR (101 MHz, DMSO-d6) δ 170.59, 162.24 (d, JCF = 244.2 Hz), 158.44, 153.58, 146.76, 138.70 (d, JCF = 7.5 Hz) 134.64, 131.00 (d, JCF = 8.3 Hz), 129.52, 129.22 (2C), 128.92 (2C), 127.99 (2C), 123.04, 122.95 (d, JCF = 2.8 Hz), 122.00 (2C), 114.68 (d, JCF = 20.8 Hz), 114.10 (d, JCF = 22.1 Hz), 48.06, 16.21 ppm. Rt = 3.99 min (apolar method); ESI-MS for C23H19FN4S: calculated 402.13, found m/z 403.09 [M + H]+, 401.21 [M – H]−; UPLC-MS purity (UV 215 nm) >99.5%.
4.1.12.23. (E)-N-((Z)-4-(3-Chlorobenzyl)-3-methyl-1,2,4-thiadiazol-5(4H)-ylidene)-N′-phenylbenzimidamide (24)
N-(3-Chlorobenzyl)-2,3-diphenyl-1,2,4-thiadiazol-5(2H)-imine hydrobromide 62b (133 mg, 0.29 mmol), ACN 79 (1.515 mL, 29 mmol), and TEA (61 μL, 0.43 mmol) were reacted according to general procedure A. The crude product was purified by flash silica gel chromatography (PE/EtOAc in 9/1 ratio) and washed with n-hexane to achieve the final compound 24 as a shiny yellow solid (115 mg, yield 95%). 1H NMR (401 MHz, DMSO-d6) δ 7.48 (d, J = 2.2 Hz, 1H), 7.46–7.37 (m, 4H), 7.37–7.27 (m, 3H), 7.26–7.20 (m, 3H), 7.05–6.98 (m, 1H), 6.78–6.73 (m, 2H), 5.54 (s, 2H), 2.44 (s, 3H) ppm. 13C NMR (101 MHz, DMSO-d6) δ 170.52, 158.43, 153.53, 146.73, 138.33, 134.64, 133.33, 130.83, 129.52, 129.20 (2C), 128.91 (2C), 127.99 (2C), 127.82, 127.32, 125.65, 123.04, 121.99 (2C), 48.04, 16.24 ppm. Rt = 4.53 min (apolar method); ESI-MS for C23H19ClN4S: calculated 418.10, found m/z 419.14/421.13 [M + H]+; UPLC-MS purity (UV 215 nm) 99%.
4.1.12.24. (E)-N-((Z)-3-Methyl-4-(2-methylbenzyl)-1,2,4-thiadiazol-5(4H)-ylidene)-N′-phenylbenzimidamide (25)
N-(2-Methylbenzyl)-2,3-diphenyl-1,2,4-thiadiazol-5(2H)-imine hydrobromide 63b (239 mg, 0.55 mmol), ACN 79 (2.872 mL, 55 mmol), and TEA (114 μL, 0.82 mmol) were reacted according to general procedure A. The crude product was purified by flash silica gel chromatography (PE/EtOAc in 92/8 ratio) and washed with n-hexane to achieve the final compound 25 as a yellow solid (111 mg, yield 51%). 1H NMR (401 MHz, DMSO-d6) δ 7.37–7.32 (m, 2H), 7.32–7.21 (m, 6H), 7.21–7.12 (m, 2H), 7.05–6.98 (m, 1H), 6.78–6.72 (m, 2H), 6.60 (dd, J = 7.5, 1.5 Hz, 1H), 5.53 (s, 2H), 2.42 (s, 3H), 2.35 (s, 3H) ppm. 13C NMR (101 MHz, DMSO-d6) δ 170.55, 158.58, 153.88, 146.77, 135.17, 134.64, 133.71, 130.45, 129.48, 129.21 (2C), 128.93 (2C), 127.96 (2C), 127.33, 126.46, 124.29, 123.04, 122.06 (2C), 46.45, 18.85, 15.95 ppm. Rt = 4.27 min (apolar method); ESI-MS for C24H22N4S: calculated 398.16, found m/z 399.33 [M + H]+; UPLC-MS purity (UV 215 nm) 99.5%.
4.1.12.25. (E)-N-((Z)-3-Methyl-4-(2-(trifluoromethyl)benzyl)-1,2,4-thiadiazol-5(4H)-ylidene)-N′-phenylbenzimidamide (26)
N-(2-(Trifluoromethyl)benzyl)-2,3-diphenyl-1,2,4-thiadiazol-5(2H)-imine hydrobromide 64b (375 mg, 0.76 mmol), ACN 79 (3.98 mL, 76 mmol), and TEA (159 μL, 1.14 mmol) were reacted according to general procedure A. The crude product was purified by flash silica gel chromatography (PE/EtOAc in 98/2 ratio) and washed with n-hexane to achieve the final compound 26 as a yellow solid (251 mg, yield 73%). 1H NMR (401 MHz, DMSO-d6) δ 7.85 (d, J = 7.8 Hz, 1H), 7.66 (t, J = 7.7 Hz, 1H), 7.54 (t, J = 7.7 Hz, 1H), 7.34–7.27 (m, 3H), 7.27–7.20 (m, 4H), 7.02 (t, J = 7.4 Hz, 1H), 6.88 (d, J = 7.8 Hz, 1H), 6.76 (dt, J = 7.4, 1.1 Hz, 2H), 5.70 (s, 2H), 2.37 (s, 3H) ppm. 13C NMR (101 MHz, DMSO-d6) δ 170.35, 158.28, 153.57, 146.69, 134.34, 133.89 (q, JCF = 1.5 Hz), 133.52, 129.57, 129.16 (2C), 128.95 (2C), 128.12, 127.92 (2C), 126.43 (q, JCF = 5.9 Hz), 126.22, 126.13, 125.76, 123.04, 121.93 (2C), 45.54, 15.82 ppm. Rt = 4.54 min (apolar method); ESI-MS for C24H19F3N4S: calculated 452.13, found m/z 453.17 [M + H]+; UPLC-MS purity (UV 215 nm) >99.5%.
4.1.12.26. (E)-N-((Z)-4-(2-Methoxybenzyl)-3-methyl-1,2,4-thiadiazol-5(4H)-ylidene)-N′-phenylbenzimidamide (27)
N-(2-Methoxybenzyl)-2,3-diphenyl-1,2,4-thiadiazol-5(2H)-imine hydrobromide 65b (235 mg, 0.52 mmol), ACN 79 (2.716 mL, 52 mmol), and TEA (108 μL, 0.78 mmol) were reacted according to general procedure A. The crude product was purified by flash silica gel chromatography (PE/EtOAc in 9/1 ratio) and washed with n-hexane to achieve the final compound 27 as a shiny yellow solid (97 mg, yield 45%). 1H NMR (401 MHz, DMSO-d6) δ 7.38–7.26 (m, 6H), 7.25–7.20 (m, 2H), 7.08 (dd, J = 8.4, 1.0 Hz, 1H), 7.00 (tt, J = 7.3, 1.3 Hz, 1H), 6.93 (td, J = 7.4, 1.0 Hz, 1H), 6.87 (dd, J = 7.6, 1.8 Hz, 1H), 6.76–6.71 (m, 2H), 5.45 (s, 2H), 3.87 (s, 3H), 2.41 (s, 3H) ppm. 13C NMR (101 MHz, DMSO-d6) δ 170.47, 158.46, 156.48, 153.84, 146.79, 134.70, 129.38, 129.15 (2C), 128.97, 128.85 (2C), 127.90 (2C), 127.15, 123.10, 122.91, 121.96 (2C), 120.48, 111.05, 55.46, 44.43, 15.87 ppm. Rt = 4.26 min (apolar method); ESI-MS for C24H22N4OS: calculated 414.15, found m/z 415.11 [M + H]+; UPLC-MS purity (UV 215 nm) 99%.
4.1.12.27. (E)-N-((Z)-4-(2-Chlorobenzyl)-3-methyl-1,2,4-thiadiazol-5(4H)-ylidene)-N′-phenylbenzimidamide (28)
N-(2-Chlorobenzyl)-2,3-diphenyl-1,2,4-thiadiazol-5(2H)-imine hydrobromide 66b (323 mg, 0.7 mmol), ACN 79 (3.656 mL, 70 mmol), and TEA (147 μL, 1.1 mmol) were reacted according to general procedure A. The crude product was purified by flash silica gel chromatography (PE/EtOAc in 9/1 ratio) and washed with n-hexane to achieve the final compound 28 as a shiny yellow solid (102 mg, yield 35%). 1H NMR (401 MHz, DMSO-d6) δ 7.56 (dd, J = 7.2, 2.0 Hz, 1H), 7.39–7.28 (m, 5H), 7.24 (q, J = 7.4 Hz, 4H), 7.01 (t, J = 7.4 Hz, 1H), 6.86 (dd, J = 7.0, 2.3 Hz, 1H), 6.75 (d, J = 7.8 Hz, 2H), 5.60 (s, 2H), 2.39 (s, 3H) ppm. 13C NMR (101 MHz, DMSO-d6) δ 170.31, 158.37, 153.57, 146.69, 134.46, 132.93, 131.54, 129.70, 129.51, 129.33, 129.17 (2C), 128.91 (2C), 127.93 (2C), 127.90, 126.98, 123.06, 121.96 (2C), 46.47, 15.92 ppm. Rt = 4.74 min (apolar method); ESI-MS for C23H19ClN4S: calculated 418.10, found m/z 419.11/421.11 [M + H]+; UPLC-MS purity (UV 215 nm) 96%.
4.1.12.28. (E)-N-((Z)-4-(2-Aminobenzyl)-3-methyl-1,2,4-thiadiazol-5(4H)-ylidene)-N′-phenylbenzimidamide (29)
N-(2-Aminobenzyl)-2,3-diphenyl-1,2,4-thiadiazol-5(2H)-imine dihydrobromide 67b (1.222 g, 2.35 mmol), ACN 79 (12.273 mL, 235 mmol), and TEA (820 μL, 5.88 mmol) were reacted according to general procedure A. The crude product was purified by flash silica gel chromatography (PE/EtOAc in 83/17 ratio) and washed with n-hexane to achieve the final compound 29 as a shiny yellow solid (195 mg, yield 21%). 1H NMR (401 MHz, DMSO-d6) δ 7.44–7.39 (m, 2H), 7.37–7.26 (m, 3H), 7.24 (t, J = 7.6 Hz, 3H), 7.01 (t, J = 7.5 Hz, 2H), 6.76 (d, J = 7.6 Hz, 3H), 6.70 (d, J = 7.8 Hz, 2H), 6.54 (t, J = 7.4 Hz, 1H), 5.42 (s, 2H), 5.40 (s, 2H), 2.38 (s, 3H) ppm. 13C NMR (101 MHz, DMSO-d6) δ 170.97, 158.44, 154.11, 146.70, 146.25, 134.65, 129.50, 129.11 (2C), 128.92 (2C), 128.51, 128.04 (2C), 127.32, 123.04, 122.05 (2C9, 118.07, 116.23, 115.33, 45.91, 16.28 ppm. Rt = 5.70 min (generic method); ESI-MS for C23H21N5S: calculated 399.15, found m/z 400.14 [M + H]+; UPLC-MS purity (UV 215 nm) 98%.
4.1.12.29. (E)-N-((Z)-4-Benzyl-3-methyl-1,2,4-thiadiazol-5(4H)-ylidene)-4-methoxy-N′-phenylbenzimidamide (30)
N-Benzyl-3-(4-methoxyphenyl)-2-phenyl-1,2,4-thiadiazol-5(2H)-imine hydrobromide 68b (300 mg, 0.66 mmol), ACN 79 (3.4 mL, 66 mmol), and TEA (138 μL, 0.99 mmol) were reacted according to general procedure A. The crude product was purified by recrystallization from EtOAc to achieve the final compound 30 as a yellow solid (100 mg, yield 37%). 1H NMR (401 MHz, DMSO-d6) δ 7.46–7.40 (m, 2H), 7.40–7.36 (m, 2H), 7.32 (d, J = 7.2 Hz, 3H), 7.26 (t, J = 7.8 Hz, 2H), 7.02 (td, J = 7.4, 1.3 Hz, 1H), 6.86–6.80 (m, 2H), 6.80–6.75 (m, 2H), 5.56 (s, 2H), 3.72 (s, 3H), 2.40 (s, 3H) ppm. 13C NMR (101 MHz, DMSO-d6) δ 170.37, 160.17, 157.56, 153.49, 147.09, 135.98, 131.13 (2C), 129.01 (2C), 128.87 (2C), 127.75, 126.98 (2C), 126.56, 122.86, 121.83 (2C), 113.33 (2C), 55.16, 48.51, 16.22 ppm. Rt = 3.86 min (apolar method); ESI-MS for C24H22N4OS: calculated 414.15, found m/z 415.10 [M + H]+; UPLC-MS purity (UV 215 nm) >99.5%.
4.1.12.30. (E)-N-((Z)-4-Benzyl-3-methyl-1,2,4-thiadiazol-5(4H)-ylidene)-3-methoxy-N′-phenylbenzimidamide (31)
N-Benzyl-3-(3-methoxyphenyl)-2-phenyl-1,2,4-thiadiazol-5(2H)-imine hydrobromide 69b (417 mg, 0.84 mmol), ACN 79 (4.387 mL, 84 mmol), and TEA (176 μL, 1.26 mmol) were reacted according to general procedure A. The crude product was purified by flash silica gel chromatography (PE/EtOAc in 85/15 ratio) and further recrystallized from EtOAc to achieve the final compound 31 as a yellow solid (105 mg, yield 30%). 1H NMR (401 MHz, DMSO-d6) δ 7.42–7.36 (m, 2H), 7.35–7.29 (m, 3H), 7.28–7.21 (m, 2H), 7.19 (t, J = 7.9 Hz, 1H), 7.05–6.98 (m, 2H), 6.95 (dd, J = 2.6, 1.5 Hz, 1H), 6.90 (ddd, J = 8.2, 2.7, 1.0 Hz, 1H), 6.79–6.74 (m, 2H), 5.55 (s, 2H), 3.57 (s, 3H), 2.42 (s, 3H) ppm. 13C NMR (101 MHz, DMSO-d6) δ 170.63, 158.58, 158.14, 153.68, 146.89, 135.90, 135.88, 129.05, 128.88 (2C), 128.84 (2C), 127.75, 126.98 (2C), 122.97, 121.90 (2C), 121.53, 115.22, 114.71, 54.81, 48.56, 16.22 ppm. Rt = 3.91 min (apolar method); ESI-MS for C24H22N4OS: calculated 414.15, found m/z 415.25 [M + H]+; UPLC-MS purity (UV 215 nm) 99.5%.
4.1.12.31. (E)-N-((Z)-4-Benzyl-3-methyl-1,2,4-thiadiazol-5(4H)-ylidene)-3-hydroxy-N′-phenylbenzimidamide (32)
(E)-N-((Z)-4-Benzyl-3-methyl-1,2,4-thiadiazol-5(4H)-ylidene)-3-methoxy-N′-phenylbenzimidamide 31 (258 mg, 0.63 mmol) and 3 equiv of BBr3 (1.9 mmol) were reacted according to the general procedure B. The crude material was purified via flash silica gel column chromatography (PE/EtOAc in 8/2 ratio). The product was further recrystallized from a mixture of EtOAc/n-hexane (9/1), then washed with n-hexane, and filtered to give the final compound 32 as a light yellow solid (58 mg, yield 23%). 1H NMR (401 MHz, DMSO-d6) δ 9.46 (s, 1H), 7.40 (ddt, J = 7.8, 6.3, 1.0 Hz, 2H), 7.32 (td, J = 6.6, 1.5 Hz, 3H), 7.27–7.20 (m, 2H), 7.08–6.98 (m, 2H), 6.91 (dd, J = 2.6, 1.5 Hz, 1H), 6.80–6.69 (m, 4H), 5.55 (s, 2H), 2.39 (s, 3H) ppm. 13C NMR (101 MHz, DMSO-d6) δ 170.58, 158.48, 156.91, 153.59, 146.70, 135.98, 135.87, 128.90 (2C), 128.88, 128.86 (2C), 127.78, 126.94 (2C), 122.93, 121.92 (2C), 119.96, 116.46, 116.13, 48.45, 16.20 ppm. Rt = 5.17 min (generic method); ESI-MS for C23H20N4OS: calculated 400.14, found m/z 401.08 [M + H]+, 399.18 [M – H]−; UPLC-MS purity (UV 215 nm) >99.5%.
4.1.12.32. (E)-N-((Z)-4-Benzyl-3-methyl-1,2,4-thiadiazol-5(4H)-ylidene)-2-methoxy-N′-phenylbenzimidamide (33)
N-Benzyl-3-(2-methoxyphenyl)-2-phenyl-1,2,4-thiadiazol-5(2H)-imine hydrobromide 70b (290 mg, 0.59 mmol), ACN 79 (3 mL, 59 mmol), and TEA (123 μL, 0.88 mmol) were reacted according to general procedure A. The crude product was purified by flash silica gel chromatography (PE/EtOAc in 84/16 ratio) and further recrystallized from EtOAc to achieve the final compound 33 as a yellow solid (92 mg, yield 38%). 1H NMR (401 MHz, DMSO-d6) δ 7.42–7.36 (m, 2H), 7.35–7.25 (m, 4H), 7.23 (dd, J = 7.5, 1.8 Hz, 1H), 7.17–7.10 (m, 2H), 6.95–6.86 (m, 3H), 6.71–6.66 (m, 2H), 5.48 (s, 2H), 3.40 (s, 3H), 2.39 (s, 3H) ppm. 13C NMR (101 MHz, DMSO-d6) δ 170.07, 158.61, 155.63, 153.51, 146.95, 135.87, 130.30, 129.58, 128.84 (2C), 128.21 (2C), 127.78, 127.03 (2C), 125.68, 122.94, 121.66 (2C), 120.22, 111.50, 54.95, 48.31, 16.20 ppm. Rt = 5.84 min (generic method); ESI-MS for C24H22N4OS: calculated 414.15, found m/z 415.27 [M + H]+; UPLC-MS purity (UV 215 nm) >99.5%.
4.1.12.33. (E)-N-((Z)-4-Benzyl-3-methyl-1,2,4-thiadiazol-5(4H)-ylidene)-N′-phenylisonicotinimidamide (34)
N-Benzyl-2-phenyl-3-(pyridin-4-yl)-1,2,4-thiadiazol-5(2H)-imine hydrobromide 71b (362 mg, 0.85 mmol), ACN 79 (4.439 mL, 85 mmol), and TEA (297 μL, 2.13 mmol) were reacted according to general procedure A. The crude product was purified by flash silica gel chromatography (DCM/MeOH in 99/1 ratio) to achieve the final compound 34 as a yellow solid (230 mg, yield 60%). 1H NMR (401 MHz, DMSO-d6) δ 8.53 (dd, J = 6.0, 1.6 Hz, 2H), 7.43–7.36 (m, 2H), 7.35–7.28 (m, 5H), 7.25 (td, J = 8.1, 7.6, 2.0 Hz, 2H), 7.04 (tt, J = 7.4, 1.2 Hz, 1H), 6.77 (dd, J = 7.3, 1.1 Hz, 2H), 5.56 (s, 2H), 2.43 (s, 3H) ppm. 13C NMR (101 MHz, DMSO-d6) δ 171.24, 156.91, 154.09, 149.71 (2C), 146.33, 142.14, 135.68, 129.01 (2C), 128.90 (2C), 127.83, 127.04 (2C), 123.48, 123.28 (2C), 121.95 (2C), 48.62, 16.25 ppm. Rt = 5.14 min (generic method); ESI-MS for C22H19N5S: calculated 385.14, found m/z 386.16 [M + H]+; UPLC-MS purity (UV 215 nm) >99.5%.
4.1.12.34. (E)-N-((Z)-4-Benzyl-3-methyl-1,2,4-thiadiazol-5(4H)-ylidene)-N′-phenylnicotinimidamide (35)
N-Benzyl-2-phenyl-3-(pyridin-3-yl)-1,2,4-thiadiazol-5(2H)-imine hydrobromide 72b (238 mg, 0.56 mmol), ACN 79 (2.925 mL, 56 mmol), and TEA (195 μL, 1.4 mmol) were reacted according to general procedure A. The crude product was purified by flash silica gel chromatography (DCM/MeOH in 99/1 ratio) to achieve the final compound 35 as a yellow solid (114 mg, yield 53%). 1H NMR (401 MHz, DMSO-d6) δ 8.58 (d, J = 2.0 Hz, 1H), 8.50 (dd, J = 4.8, 1.7 Hz, 1H), 7.75 (dt, J = 8.0, 2.0 Hz, 1H), 7.43–7.36 (m, 2H), 7.36–7.29 (m, 4H), 7.29–7.22 (m, 2H), 7.04 (tt, J = 7.3, 1.1, 0.6 Hz, 1H), 6.82–6.77 (m, 2H), 5.57 (s, 2H), 2.43 (s, 3H) ppm. 13C NMR (101 MHz, DMSO-d6) δ 171.02, 156.63, 153.97, 150.00, 149.84, 146.62, 136.64, 135.78, 130.56, 129.07 (2C), 128.89 (2C), 127.81, 127.01 (2C), 123.32, 123.11, 122.07 (2C), 48.62, 16.24 ppm. Rt = 5.19 min (generic method); ESI-MS for C22H19N5S: calculated 385.14, found m/z 386.34 [M + H]+; UPLC-MS purity (UV 215 nm) >99.5%.
4.1.12.35. (E)-N-((Z)-4-Benzyl-3-methyl-1,2,4-thiadiazol-5(4H)-ylidene)-N′-phenylfuran-2-carboximidamide (36)
N-Benzyl-3-(furan-2-yl)-2-phenyl-1,2,4-thiadiazol-5(2H)-imine hydrobromide 73b (270 mg, 0.65 mmol), ACN 79 (3.4 mL, 65 mmol), and TEA (136 μL, 0.98 mmol) were reacted according to general procedure A. The crude product was purified by flash silica gel chromatography (PE/EtOAc in 92/8 ratio) to achieve the final compound 36 as a light brown solid (213 mg, yield 88%). 1H NMR (401 MHz, DMSO-d6) δ 7.68–7.63 (m, 1H), 7.44–7.28 (m, 7H), 7.10 (t, J = 7.4 Hz, 1H), 6.89–6.82 (m, 2H), 6.64–6.58 (m, 1H), 6.50 (dq, J = 3.4, 1.6 Hz, 1H), 5.56 (s, 2H), 2.41 (s, 3H) ppm. 13C NMR (101 MHz, DMSO-d6) δ 170.51, 153.75, 148.40, 148.11, 147.20, 144.98, 135.95, 128.88 (2C), 128.78 (2C), 127.80, 127.13 (2C), 123.19, 121.26 (2C), 115.66, 111.60, 48.47, 16.25 ppm. Rt = 3.42 min (apolar method); ESI-MS for C21H18N4OS: calculated 374.12, found m/z 375.04 [M + H]+, 373.01 [M – H]−; UPLC-MS purity (UV 215 nm) >99.5%.
4.1.12.36. (E)-N-((Z)-4-Benzyl-3-methyl-1,2,4-thiadiazol-5(4H)-ylidene)-N′-(m-tolyl)benzimidamide (37)
N-Benzyl-3-phenyl-2-(m-tolyl)-1,2,4-thiadiazol-5(2H)-imine hydrobromide 74b (118 mg, 0.3 mmol), ACN 79 (1.6 mL, 30 mmol), and TEA (63 μL, 0.45 mmol) were reacted according to general procedure A. The crude product was purified by flash silica gel chromatography (PE/EtOAc in 92/8 ratio) to achieve the final compound 37 as a shiny yellow solid (134 mg, yield 77%). 1H NMR (401 MHz, DMSO-d6) δ 7.47–7.36 (m, 4H), 7.36–7.25 (m, 6H), 7.09 (t, J = 7.7 Hz, 1H), 6.83 (d, J = 7.5 Hz, 1H), 6.65 (s, 1H), 6.49 (d, J = 7.8 Hz, 1H), 5.56 (s, 2H), 2.41 (s, 3H), 2.22 (s, 3H) ppm. 13C NMR (101 MHz, DMSO-d6) δ 170.56, 158.17, 153.62, 146.68, 138.15, 135.91, 134.69, 129.49, 129.21 (2C), 128.88, (2C) 128.69, 127.96 (2C), 127.77, 126.97 (2C), 123.75, 122.59, 118.96, 48.52, 20.98, 16.22 ppm. Rt = 4.48 min (apolar method); ESI-MS for C24H22N4S: calculated 398.16, found m/z 399.18 [M + H]+; UPLC-MS purity (UV 215 nm) >99.5%.
4.1.12.37. (E)-N′-Benzyl-N-((Z)-4-benzyl-3-methyl-1,2,4-thiadiazol-5(4H)-ylidene)benzimidamide (38)
N,2-Dibenzyl-3-phenyl-1,2,4-thiadiazol-5(2H)-imine hydrobromide 75b (580 mg, 1.3 mmol), ACN 79 (6.5 mL, 130 mmol), and TEA (272 μL, 1.95 mmol) were reacted according to general procedure A. The crude product was purified by flash silica gel chromatography (PE/EtOAc in 91/9 ratio) and further recrystallized from EtOAc to achieve the final compound 38 as a white solid (199 mg, yield 38%). 1H NMR (401 MHz, DMSO-d6) δ 7.50–7.42 (m, 5H), 7.40–7.28 (m, 7H), 7.29–7.21 (m, 3H), 5.44 (s, 2H), 4.80 (s, 2H), 2.33 (s, 3H) ppm. 13C NMR (101 MHz, DMSO-d6) δ 168.55, 161.48, 152.98, 140.87, 136.02, 135.52, 129.11, 128.81 (2C), 128.27 (4C), 127.95 (2C), 127.65, 127.45 (2C), 126.84 (2C), 126.48, 53.53, 48.09, 16.14 ppm. Rt = 3.47 min (apolar method); ESI-MS for C24H22N4S: calculated 398.16, found m/z 399.33 [M + H]+; UPLC-MS purity (UV 215 nm) >99.5%.
4.1.12.38. (E)-N-((Z)-4-Benzyl-3-methyl-1,2,4-thiadiazol-5(4H)-ylidene)-N′-cyclohexylbenzimidamide (39)
N-Benzyl-2-cyclohexyl-3-phenyl-1,2,4-thiadiazol-5(2H)-imine hydrobromide 76b (192 mg, 0.45 mmol), ACN 79 (2.35 mL, 45 mmol), and TEA (94 μL, 0.67 mmol) were reacted according to general procedure A. The crude product was purified by flash silica gel chromatography (PE/EtOAc in 92/8 ratio) and further recrystallized from EtOAc and washed with n-hexane to achieve the final compound 39 as a white crystalline solid (65 mg, yield 37%). 1H NMR (401 MHz, DMSO-d6) δ 7.49–7.39 (m, 3H), 7.38–7.32 (m, 4H), 7.32–7.20 (m, 3H), 5.40 (s, 2H), 3.52 (tt, J = 13.0, 9.0, 3.5 Hz, 1H), 2.32 (s, 3H), 1.80–1.67 (m, 4H), 1.62–1.50 (m, 3H), 1.37–1.13 (m, 3H) ppm. 13C NMR (101 MHz, DMSO-d6) δ 167.77, 158.98, 152.62, 136.22, 136.10, 128.78 (2C), 128.66, 128.26 (2C), 127.61, 127.55 (2C), 126.84 (2C), 57.70, 48.00, 35.43 (2C), 25.32, 24.22 (2C), 16.11 ppm. Impossible to assign a Rt due broadening of the main peak; ESI-MS for C23H26N4S: calculated 390.19, found m/z 391.33 [M + H]+.
4.1.12.39. (E)-N-((Z)-4-Benzyl-3-methyl-1,2,4-thiadiazol-5(4H)-ylidene)-N′-(pyridin-4-yl)benzimidamide (40)
N-Benzyl-3-phenyl-2-(pyridin-4-yl)-1,2,4-thiadiazol-5(2H)-imine hydrobromide 77b (336 mg, 0.79 mmol), ACN 79 (4.126 mL, 79 mmol), and TEA (276 μL, 1.98 mmol) were reacted according to general procedure A. The crude product was purified by flash silica gel chromatography (DCM/MeOH in 99.5/0.5 ratio) to achieve the final compound 40 as a yellow solid (53 mg, yield 17%). 1H NMR (401 MHz, DMSO-d6) δ 8.33 (dd, J = 6.1, 3.1 Hz, 2H), 7.46–7.41 (m, 2H), 7.41–7.36 (m, 3H), 7.36–7.29 (m, 5H), 6.75 (dd, J = 6.2, 3.0 Hz, 2H), 5.58 (s, 2H), 2.44 (s, 3H) ppm. 13C NMR (101 MHz, DMSO-d6) δ 172.03, 159.92, 154.35, 154.04, 150.15 (2C), 135.64, 134.10, 130.00, 129.26 (2C), 128.91 (2C), 128.18 (2C), 127.85, 127.01 (2C), 117.41, 48.71, 16.27 ppm. Rt = 4.74 min (generic method); ESI-MS for C22H19N5S: calculated 385.14, found m/z 386.18 [M + H]+; UPLC-MS purity (UV 215 nm) 97%.
4.1.12.40. (E)-N-((Z)-4-Benzyl-3-methyl-1,2,4-thiadiazol-5(4H)-ylidene)-N′-(pyridin-3-yl)benzimidamide (41)
N-Benzyl-3-phenyl-2-(pyridin-3-yl)-1,2,4-thiadiazol-5(2H)-imine hydrobromide 78b (698 mg, 1.64 mmol), ACN 79 (8.565 mL, 164 mmol), and TEA (572 μL, 4.1 mmol) were reacted according to the general procedure. The crude product was purified by flash silica gel chromatography (DCM/MeOH in 99/1 ratio) to achieve the final compound 41 as a yellow solid (85 mg, yield 13%). 1H NMR (401 MHz, DMSO-d6) δ 8.20 (dd, J = 4.7, 1.6 Hz, 1H), 7.99 (dd, J = 2.6, 0.8 Hz, 1H), 7.43–7.37 (m, 4H), 7.37–7.29 (m, 6H), 7.26 (ddd, J = 8.1, 4.7, 0.8 Hz, 1H), 7.18 (ddd, J = 8.1, 2.5, 1.6 Hz, 1H), 5.57 (s, 2H), 2.43 (s, 3H) ppm. 13C NMR (101 MHz, DMSO-d6) δ 171.48, 160.38, 154.04, 143.87, 143.24, 143.07, 135.69, 134.45, 129.64, 129.23 (2C), 128.97, 128.87 (2C), 128.13 (2C), 127.79, 126.96 (2C), 123.64, 48.61, 16.22 ppm. Rt = 5.01 min (generic method); ESI-MS for C22H19N5S: 386.18 [M + H]+; UPLC-MS purity (UV 215 nm) 98%.
4.1.12.41. (Z)-4-Benzyl-3-methyl-N-(pyridin-2-yl)-1,2,4-thiadiazol-5(4H)-imine (42)
2-(Benzylamino)-[1,2,4]thiadiazolo[2,3-a]pyridin-4-ium bromide 132b (380 mg, 1.2 mmol), ACN 79 (7.5 mL, 120 mmol), and DiPEA (209 μL, 1.2 mmol) were reacted according to general procedure A. The crude product was purified by flash silica gel chromatography (PE/EtOAc in 9/1 ratio) to achieve the final compound 42 as a white solid (79 mg, yield 23%). 1H NMR (401 MHz, DMSO-d6) δ 8.47 (d, J = 4.9 Hz, 1H), 7.75 (td, J = 7.6, 1.9 Hz, 1H), 7.40–7.33 (m, 2H), 7.32–7.23 (m, 3H), 7.19 (d, J = 8.1 Hz, 1H), 7.04–6.96 (m, 1H), 5.45 (s, 2H), 2.33 (s, 3H) ppm. 13C NMR (101 MHz, DMSO-d6) δ 166.42, 157.30, 153.79, 145.26, 138.01, 136.17, 128.82 (2C), 127.59, 126.67 (2C), 119.30, 116.83, 47.71, 16.36 ppm. In agreement with that previously reported by Martinez et al.45 Rt = 4.95 min (generic method); ESI-MS for C15H14N4S: calculated 282.09, found m/z 283.1 [M + H]+; UPLC-MS purity (UV 215 nm) >99.5%.
4.1.12.42. (Z)-4-Benzyl-N-(4-(2-methoxyethyl)pyridin-2-yl)-3-methyl-1,2,4-thiadiazol-5(4H)-imine (43)
2-(Benzylamino)-7-(2-methoxyethyl)-[1,2,4]thiadiazolo[2,3-a]pyridin-4-ium bromide 133b (655 mg, 1.7 mmol), ACN 79 (8.878 mL, 170 mmol), and DiPEA (300 μL, 1.7 mmol) were reacted according to general procedure A. The crude product was purified by flash silica gel chromatography (PE/EtOAc in 8/2 ratio) to achieve the final compound 43 as a white solid (140 mg, yield 24%). 1H NMR (401 MHz, DMSO-d6) δ 8.34 (d, J = 5.3 Hz, 1H), 7.40–7.33 (m, 2H), 7.33–7.28 (m, 1H), 7.28–7.22 (m, 2H), 7.07 (s, 1H), 6.90 (dd, J = 5.4, 1.5 Hz, 1H), 5.44 (s, 2H), 3.57 (t, J = 6.5 Hz, 2H), 3.22 (s, 3H), 2.82 (t, J = 6.6 Hz, 2H), 2.32 (s, 3H) ppm. 13C NMR (101 MHz, DMSO-d6) δ 166.32, 157.36, 153.68, 150.31, 144.83, 136.18, 128.79 (2C), 127.54, 126.58 (2C), 119.27, 117.82, 71.47, 57.78, 47.64, 34.51, 16.31 ppm. Rt = 4.94 min (generic method); ESI-MS for C18H20N4OS: calculated 340.14, found m/z 341.05 [M + H]+; UPLC-MS purity (UV 215 nm) 99%.
4.1.12.43. (E)-N-((Z)-3-Benzyl-5-methyl-1,3,4-thiadiazol-2(3H)-ylidene)-N′-phenylbenzimidamide (44)
3-Benzyl-5-methyl-1,3,4-thiadiazol-2(3H)-imine 134d (500 mg, 2.43 mmol), N-phenylbenzimidoyl chloride 108b (625 mg, 2.9 mmol), and pyridine (243 μL, 2.9 mmol) were reacted according to general procedure C. The crude product was purified by flash silica gel chromatography (PE/EtOAc in 98/2 ratio) to achieve the final compound 44 as a yellow solid (222 mg, yield 23%). 1H NMR (401 MHz, DMSO-d6) δ 7.43–7.34 (m, 6H), 7.29 (dq, J = 14.5, 7.7, 7.1 Hz, 4H), 7.18 (t, J = 7.7 Hz, 2H), 6.93 (t, J = 7.4 Hz, 1H), 6.68 (d, J = 7.7 Hz, 2H), 5.53 (s, 2H), 2.47 (s, 3H) ppm. 13C NMR (101 MHz, DMSO-d6) δ 159.75, 157.61, 153.72, 148.77, 136.38, 135.85, 129.25 (2C), 129.15, 128.76 (2C), 128.63 (2C), 128.00 (2C), 127.83 (2C), 127.75, 122.17, 121.65 (2C), 52.51, 15.75 ppm. Rt = 4.77 min (apolar method); ESI-MS for C23H20N4S: calculated 384.14, found m/z 385.21 [M + H]+; UPLC-MS purity (UV 215 nm) 96%.
4.1.12.44. (E)-N-((Z)-3-Benzyl-1,3,4-thiadiazol-2(3H)-ylidene)-N′-phenylbenzimidamide (45)
3-Benzyl-1,3,4-thiadiazol-2(3H)-imine 135d (363 mg, 1.9 mmol), N-phenylbenzimidoyl chloride 108b (500 mg, 2.3 mmol), and pyridine (185 μL, 2.3 mmol) were reacted according to general procedure C. The crude product was purified by flash silica gel chromatography (PE/EtOAc in 97/3 ratio) to achieve the final compound 45 as a yellow solid (190 mg, yield 27%). 1H NMR (401 MHz, DMSO-d6) δ 8.78 (s, 1H), 7.43–7.34 (m, 6H), 7.34–7.24 (m, 4H), 7.18 (t, J = 7.8 Hz, 2H), 6.93 (t, J = 7.4 Hz, 1H), 6.69 (d, J = 7.7 Hz, 2H), 5.59 (s, 2H) ppm. 13C NMR (101 MHz, DMSO-d6) δ 158.58, 157.69, 148.75, 144.24, 136.22, 135.87, 129.23 (2C), 129.14, 128.77 (2C), 128.63 (2C), 128.05 (2C), 127.84 (2C), 127.81, 122.24, 121.63 (2C), 52.84, 16.07 ppm. Rt = 4.35 min (apolar method); ESI-MS for C22H18N4S: calculated 370.13, found m/z 371.15 [M + H]+, 369.43 [M – H]−; UPLC-MS purity (UV 215 nm) 97%.
4.1.12.45. (E)-N-((Z)-3-Benzylthiazol-2(3H)-ylidene)-N′-phenylbenzimidamide (46)
3-Benzylthiazol-2(3H)-imine 136d (790 mg, 4.1 mmol), N-phenyl benzimidoyl chloride 108b (1.06 g, 4.9 mmol), and pyridine (395 μL, 4.9 mmol) were reacted according to general procedure C. The crude product was purified by flash silica gel chromatography (DCM/MeOH in 99.5/0.5 ratio) to achieve the final compound 46 as a light yellow solid (240 mg, yield 16%). 1H NMR (401 MHz, DMSO-d6) δ 7.60 (d, J = 3.3 Hz, 1H), 7.42–7.29 (m, 7H), 7.29–7.21 (m, 3H), 7.14 (t, J = 7.7 Hz, 2H), 6.87 (t, J = 7.5 Hz, 1H), 6.84 (d, J = 3.3 Hz, 1H), 6.64 (d, J = 7.7 Hz, 2H), 5.41 (s, 2H) ppm. 13C NMR (101 MHz, DMSO-d6) δ 149.51, 136.93 (2C), 136.65, 129.16 (2C), 128.60 (4C), 127.95 (2C), 127.68 (4C), 127.30, 121.72 (2C), 121.56, 107.91, 50.47 ppm. Rt = 5.08 min (generic method); ESI-MS for C23H19N3S: calculated 369.13, found m/z 370.11 [M + H]+; UPLC-MS purity (UV 215 nm) >99.5%.
4.2. Biology
4.2.1. Fluorescence Assay for CFTR
Twenty-four hours after seeding on 96-well plates, CFBE41o– cells stably expressing F508del-CFTR and the HS-YFP were treated with test compounds at the desired concentration, one compound per well. Cells treated with vehicle alone (DMSO) and with corrector VX-809 (1 μM), respectively, served as negative and positive controls. At the time of the assay, cells were washed with PBS containing (in μM) 137 NaCl, 2.7 KCl, 8.1 Na2HPO4, 1.5 KH2PO4, 1 CaCl2, and 0.5 MgCl2. Cells were then incubated for 25 min with 60 mL of PBS plus forskolin (20 μM) and VX-770 (1 μM) to maximally stimulate F508del-CFTR. Cells were then transferred to a microplate reader (FluoStar Galaxy; BMG Labtech, Offenburg, Germany) for CFTR activity determination. The plate reader was equipped with high-quality excitation (HQ500/20X: 500 ± 10 nm) and emission (HQ535/30M: 535 ± 15 nm) filters for YFP (Chroma Technology). Each assay comprised a continuous 14 s fluorescence reading and 2 s before and 12 s after injection of 165 mL of an iodide-containing solution (PBS with Cl– replaced by I–; final I– concentration 100 μM). Data were normalized to the initial background-subtracted fluorescence. To determine the I– influx rate, the final 11 s of the data for each well was fitted with an exponential function to extrapolate initial slope (dF/dt). Reproducibility of results was confirmed by performing three independent experiments.
4.2.2. Gene Silencing by siRNA Transfection
CFBE41o– cells co-expressing F508del-CFTR and the HS-YFP were reverse-transfected with 50 nM (final concentration) siRNAs (Stealth, Life Technologies) in the presence of lipofectamine 2000 as a transfection agent. The following day, the medium was changed, and the cells were incubated at 37 °C for additional 24 h treated with test compounds at the desired concentration, prior to processing the cells for the YFP-based functional assay.
4.2.3. CFTR Immunoprecipitation (IP) Assay
CFBE41o– cells stably expressing wt- or F508del-CFTR were grown to confluence on 60 mm diameter dishes and treated for 24 h with the indicated compounds in the absence or in the presence of MG-132 (10 μM) in the last 4 h. CFTR immunoprecipitation was performed as previously reported.40 In brief, cells were lysed and, after centrifugation, the protein concentration in the supernatant was calculated using the BCA assay (Euroclone, Pero (MI), Italy) following supplier’s instructions An aliquot of supernatant corresponding to 600 μg of protein was incubated for 1 h with 2 μg/sample of rabbit polyclonal anti-CFTR antibody (Alomone Labs, Jerusalem, Israel), with rocking at room temperature (RT). Subsequently, the antibody–antigen mixture was precipitated with 25 μL/sample of Pierce Protein A/G Magnetic Beads (Thermo Fisher Scientific, Waltham, MA, USA) for 1 h rocking at RT, following manufacturer’s instructions. Immunoprecipitated proteins were eluted from the resin under reducing conditions with 100 μL of Laemli Sample Buffer 1× at RT. Equal amounts of IP products were analyzed by Western blotting (20 μL).
4.2.4. Western Blot
CFBE41o– cells were grown to confluence on 60 mm diameter dishes, treated for 24 h with the indicated compounds, lysed, and processed as previously reported.40 In brief, equal amounts of lysates (25 μg) or IP products (20 μL) were separated onto gradient 4–15% Criterion TGX Precast gels (Bio-rad laboratories Inc.), transferred to a nitrocellulose membrane with a Trans-Blot Turbo system (Bio-rad Laboratories Inc.), and analyzed by Western blotting. Proteins were detected using the following antibodies: mouse monoclonal anti-CFTR (cl.596; Cystic Fibrosis Therapeutics); mouse monoclonal anti-GAPDH (cl.6C5; Santa Cruz Biotechnology); mouse monoclonal anti-Ub (P4D1, Santa Cruz Biotechnology); or horseradish peroxidase (HRP)-conjugated anti-mouse IgG (Abcam). The proteins were then visualized by chemiluminescence using a SuperSignal West Femto Substrate (Thermo Scientific) and a Molecular Imager ChemiDoc XRS System. The molecular weight of the proteins (based on the Precision Plus Protein WesternC Standards, Bio-rad Laboratories Inc.) and the lane density profiles of ubiquitylated CFTR were calculated using the software Quantity one 4.6 of the Molecular Imager ChemiDoc XRS System.
4.2.5. Proliferation Study
CFBE41o– cells stably expressing F508del-CFTR and the HS-YFP were plated at a low density (5000 cell/well) on 96-well plates suitable for high-content imaging. After 6 h, cells were treated with different concentrations of test compounds or vehicle alone (DMSO). Cell proliferation was monitored (by exploiting the YFP signal to determine the area covered by cells) for 48 h using the Opera Phenix (PerkinElmer) high-content screening system. Alternatively, to monitor the cytotoxic effect of high concentrations of test compounds, CFBE41o– cells were plated (10,000 cell/well) and treated (after 6 h) with test compounds or vehicle alone (DMSO). After 24 h, cells were counterstained with Hoechst 33342 and propidium iodide to visualize total cells and apoptotic cells, respectively, and imaged by using the Opera Phenix (PerkinElmer) high-content screening system. Data are expressed as means ± SEM, n = 6. Reproducibility of results was confirmed by performing three independent experiments. Statistical significance was tested by parametric ANOVA followed by the Dunnet multiple comparisons test.
4.2.6. Labeling of Autophagic Vacuoles with Monodansylcadaverine (MDC)
CFBE41o– cells stably expressing F508del-CFTR and the HS-YFP were plated (50,000 cells/well) on good-quality clear-bottom 96-well black microplates suitable for high-content imaging. After 24 h, cells were treated with test compounds or DMSO (as the negative control). In the last 6 h of incubation, SAR405 2 μM (autophagy inhibitor) and torin1 20 nM (autophagy activator) were added to the cells. After 24 h, cells were incubated with 50 μM MDC (Sigma-Aldrich) in PBS at 37 °C for 10 min.62 After incubation, cells were washed three times with PBS and immediately analyzed. High-content imaging and data analysis were performed using the Opera Phenix (PerkinElmer) high-content screening system. Wells were imaged in confocal mode using a 40× water-immersion objective. The MDC signal was laser-excited at 405 nm, and the emission wavelengths were collected between 435 and 550 nm. Data analysis of MDC spot number was performed using the Harmony software (ver 4.5) of the Opera Phenix high-content system. Data are expressed as means ± SEM, n = 3 independent experiments. Statistical significance was tested by parametric ANOVA followed by the Dunnet multiple comparisons test (all groups against the control group).
4.2.7. RNF5 Expression and Purification
A construct coding for human RNF5 protein (aa 1–117) truncated to exclude the transmembrane domains, with an N-terminal GST-tag followed by the recognition site for TEV protease (pET vector backbone), was purchased from CliniSciences. A similar construct was recently described in the work of Ruan et al.44 The plasmid was first verified by sequencing and then transformed in BL21(DE3) Competent Cells (EC0114, Thermo Fischer Scientific). Overexpression of the protein was achieved in Escherichia coli by growing cells in LB medium at 37 °C to an OD600 ∼ 0.6 followed by induction with 0.2 mM isopropyl β-d-thiogalactopyranoside (IPTG) for 5 h at 37 °C. Bacterial pellets, obtained by centrifugation, were then stored at −80 °C until further processing. Once thawed, pellets were resuspended in lysis buffer [50 mM Tris–HCl pH 8, 1 mM EDTA, 1% Triton X-100, 5 mM DTT, 20 μM ZnCl2] supplemented with 200 μg/mL lysozyme, 10 μg/mL DNAse, 10 mM MgCl2, and protease inhibitor cocktail 1× (S8830, Sigma-Aldrich) and incubated for 45 min at 37 °C. Cell lysis was completed by sonication. The lysate was centrifuged, and the clear supernatant was incubated with Glutathione Sepharose 4 Fast Flow (FF) resin (17513202, Cytiva) for 1 h at RT. Resin was washed sequentially with lysis buffer and 50 mM Tris–HCl pH 8 and then eluted with GST elution buffer [50 mM Tris–HCl pH 8, 10 mM reduced glutathione]. Buffer was exchanged with PBS using a PD-10 desalting column (17085101, Cytiva), and TEV protease (T4455, Sigma-Aldrich) was added O/N at 4 °C to cleave the GST-tag. Undigested protein and soluble GST were removed by incubation with Glutathione Sepharose 4 FF resin for 1 h at RT. The unbound flow-through was further incubated with Ni-NTA agarose resin (30210, Qiagen) to remove the TEV protease, which contains a 6xHis-Tag. Finally, the unbound fraction was collected and concentrated using Amicon Ultra centrifugal filter devices with a cutoff of 3 kDa (Merck). Protein purity was verified by SDS-PAGE and blue Coomassie staining. Protein was either immediately used or stored at −80 °C in aliquots containing 5% glycerol.
4.2.8. MicroScale Thermophoresis (MST) Experiments
Purified RNF5 was labeled using a Monolith protein labeling kit RED-NHS 2nd generation (NanoTemper Technologies, München, Germany) following manufacturer’s instructions. Labeled protein (5 nM) was first tested in a pre-test assay for optimal fluorescence, absence of aggregation, and sticking to capillaries using MST buffer [PBS, 0.05% Tween-20, 0.1% PEG-8000, 1 μM ZnCl2] with or without 1 mM DTT. Compounds were tested in binding check experiments at the maximum concentration allowed based on their solubility. 5% DMSO was tested as the maximum final concentration. MST experiments were performed at medium MST-power and 20% laser excitation on standard capillaries. Experiments were performed on a Monolith Pico-Red/Nano-Blue Instrument (NanoTemper Technologies, München, Germany).
4.2.9. NMR Experiments
All the NMR experiments were recorded at 298 K using a 5 mm CryoProbe QCI 1H/19F–13C/15N–D quadruple resonance, a shielded z-gradient coil, and an automatic sample changer SampleJet NMR system with temperature control. The solubility of the compounds were evaluated by 1H 1D experiments and aggregation by WaterLOGSY, testing the compounds in buffer PBS pH 7,5, 1 μM ZnCl2, 1 mM DTT, 10% D2O (for the lock signal), and 1% DMSO-d6 at the theoretical concentrations of 20, 50, and 100 μM in the presence of 200 μM 4-trifluoromethyl benzoic acid (internal reference). For all samples, a 1D 1H NMR experiment was recorded, the water suppression was obtained using the standard NOESY (nuclear Overhauser effect spectroscopy) preset Bruker pulse sequence, with 64k data points, a spectral width (sw) of 30 ppm, 64 scans, acquisition time (aq) of 1.835 s, a relaxation delay (d1) of 4 s, and a mixing time of 10 ms. The WaterLOGSY experiments were achieved with a 7.5 ms long 180° Gaussian-shaped pulse, aq 0.852 s, mixing time of 1.7 s, relaxation delay of 2 s, and 256 scans. The data were multiplied by an exponential window function with 1 Hz line broadening prior to Fourier transformation. For NMR binding experiments, the compounds were tested at 50 μM in buffer PBS pH 7.5, 1 μM ZnCl2, 1 mM DTT, 10% D2O (for the lock signal), and 1% DMSO-d6 in the absence and in the presence of 3 μM RNF5. WaterLOGSY experiments were recorded with the same parameters used in solubility assays, but with a higher number of scans (512); STD experiments were recorded with 128 scans, with two on (0.7 and 1 ppm) and one off (−50 ppm) resonance spectra, a Gaussian-shaped train pulse of 50 ms each was employed, with a total saturation time of the protein envelope of 5, 3, 2, 1.5, 1, or 0.5 s. A T1ρ filter of 2 ms was employed in STD experiments to eliminate the background signals from the protein. The STD spectrum was obtained by subtraction of the on-resonance spectrum from the off-resonance spectrum. Subtraction was performed by phase cycling to minimize artifacts arising from magnet and temperature instabilities.
Acknowledgments
This work was supported by a grant from the Fondazione per la Ricerca sulla Fibrosi Cistica (FFC#09/2019; with the contribution of “Delegazione FFC di Genova con Gruppo di sostegno FFC di Savona Spotorno”, “Delegazione FFC di Valle Scrivia Alessandria”, “Delegazione FFC di Montescaglioso”, and “Delegazione FFC di Ascoli Piceno”), by the Istituto Italiano di Tecnologia (IIT) and the University of Bologna - Alma Mater Studiorum. Work in NP lab was also supported by Cystic Fibrosis Foundation grant PEDEMO20G0 and by the Italian Ministry of Health through Cinque per mille and Ricerca Corrente.
Glossary
Abbreviations Used
- CF
cystic fibrosis
- CFBE41o–
F508del-CFTR human CF bronchial epithelial cell line
- CFTR
cystic fibrosis transmembrane conductance regulator
- F508del
deletion of a phenylalanine 508
- HMBC
Heteronuclear Multiple Bond Correlation
- HS-YFP
halide-sensitive yellow fluorescent protein
- MDC
monodansylcadaverine
- MST
MicroScale Thermophoresis
- NOE
Nuclear Overhauser Effect
- PM
plasma membrane
- RNF5
RING finger protein 5
- STD
Saturation-Transfer Difference
- WaterLOGSY
1H Water-Ligand Observed via Gradient Spectros
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acs.jmedchem.3c00608.
Computed octanol/water log P values and log S values; computational methods; experimental procedures of chemical intermediates 47a–78a, 47b–78b, 108a–117a, 108b–117b, 132a, 132b, 133a, 133b, 134b–136b, 134c–136c, and 134d–136d; experimental procedures of N′-(pyridinyl)benzimidamides 84 and 85; experimental procedures of isothiocyanates 89–96; 1H and 13C NMR spectra of final compounds 1–46; HPLC–MS analysis of selected final compounds 6, 9–11, 14, 16, 17, 19, 21–25, 27–29, and 34; 1D NOE spectrum of compounds 10 and 44; and HMBC spectrum of compound 44 (PDF)
Molecular formula strings (CSV)
16 complex (PDB)
inh-2 complex (PDB)
Analog-1 complex (PDB)
The authors declare no competing financial interest.
Supplementary Material
References
- Farrell P. M. The prevalence of cystic fibrosis in the European Union. J. Cystic Fibrosis 2008, 7, 450–453. 10.1016/j.jcf.2008.03.007. [DOI] [PubMed] [Google Scholar]
- Riordan J. R.; Rommens J. M.; Kerem B.; Alon N.; Rozmahel R.; Grzelczak Z.; Zielenski J.; Lok S.; Plavsic N.; Chou J. L.; Drumm M. L.; Iannuzzi M. C.; Collins F. S.; Tsui L. C. Identification of the cystic fibrosis gene: cloning and characterization of complementary DNA. Science 1989, 245, 1066–1073. 10.1126/science.2475911. [DOI] [PubMed] [Google Scholar]
- Linsdell P. Functional architecture of the CFTR chloride channel. Mol. Membr. Biol. 2014, 31, 1–16. 10.3109/09687688.2013.868055. [DOI] [PubMed] [Google Scholar]
- Pilewski J. M.; Frizzell R. A. Role of CFTR in airway disease. Physiol. Rev. 1999, 79, S215–S255. 10.1152/physrev.1999.79.1.S215. [DOI] [PubMed] [Google Scholar]
- Pranke I.; Golec A.; Hinzpeter A.; Edelman A.; Sermet-Gaudelus I. Emerging Therapeutic Approaches for Cystic Fibrosis. From Gene Editing to Personalized Medicine. Front. Pharmacol. 2019, 10, 121. 10.3389/fphar.2019.00121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denning G. M.; Anderson M. P.; Amara J. F.; Marshall J.; Smith A. E.; Welsh M. J. Processing of mutant cystic fibrosis transmembrane conductance regulator is temperature-sensitive. Nature 1992, 358, 761–764. 10.1038/358761a0. [DOI] [PubMed] [Google Scholar]
- Lukacs G. L.; Mohamed A.; Kartner N.; Chang X. B.; Riordan J. R.; Grinstein S. Conformational maturation of CFTR but not its mutant counterpart (delta F508) occurs in the endoplasmic reticulum and requires ATP. Embo J. 1994, 13, 6076–6086. 10.1002/j.1460-2075.1994.tb06954.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelman M. S.; Kannegaard E. S.; Kopito R. R. A principal role for the proteasome in endoplasmic reticulum-associated degradation of misfolded intracellular cystic fibrosis transmembrane conductance regulator. J. Biol. Chem. 2002, 277, 11709–11714. 10.1074/jbc.M111958200. [DOI] [PubMed] [Google Scholar]
- Lukacs G. L.; Chang X. B.; Bear C.; Kartner N.; Mohamed A.; Riordan J. R.; Grinstein S. The delta F508 mutation decreases the stability of cystic fibrosis transmembrane conductance regulator in the plasma membrane. Determination of functional half-lives on transfected cells. J. Biol. Chem. 1993, 268, 21592–21598. 10.1016/S0021-9258(20)80582-1. [DOI] [PubMed] [Google Scholar]
- Dalemans W.; Barbry P.; Champigny G.; Jallat S.; Dott K.; Dreyer D.; Crystal R. G.; Pavirani A.; Lecocq J.-P.; Lazdunski M. Altered chloride ion channel kinetics associated with the delta F508 cystic fibrosis mutation. Nature 1991, 354, 526–528. 10.1038/354526a0. [DOI] [PubMed] [Google Scholar]
- Welsh M. J.; Smith A. E. Molecular mechanisms of CFTR chloride channel dysfunction in cystic fibrosis. Cell 1993, 73, 1251–1254. 10.1016/0092-8674(93)90353-R. [DOI] [PubMed] [Google Scholar]
- Sharma M.; Benharouga M.; Hu W.; Lukacs G. L. Conformational and temperature-sensitive stability defects of the delta F508 cystic fibrosis transmembrane conductance regulator in post-endoplasmic reticulum compartments. J. Biol. Chem. 2001, 276, 8942–8950. 10.1074/jbc.M009172200. [DOI] [PubMed] [Google Scholar]
- Lukacs G. L.; Verkman A. S. CFTR: folding, misfolding and correcting the ΔF508 conformational defect. Trends Mol. Med. 2012, 18, 81–91. 10.1016/j.molmed.2011.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentzsch M.; Mall M. A. Ion Channel Modulators in Cystic Fibrosis. Chest 2018, 154, 383–393. 10.1016/j.chest.2018.04.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okiyoneda T.; Veit G.; Dekkers J. F.; Bagdany M.; Soya N.; Xu H.; Roldan A.; Verkman A. S.; Kurth M.; Simon A.; et al. Mechanism-based corrector combination restores ΔF508-CFTR folding and function. Nat. Chem. Biol. 2013, 9, 444–454. 10.1038/nchembio.1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadida S.; Van Goor F.; Zhou J.; Arumugam V.; McCartney J.; Hazlewood A.; Decker C.; Negulescu P.; Grootenhuis P. D. J. Discovery of N-(2,4-di-tert-butyl-5-hydroxyphenyl)-4-oxo-1,4-dihydroquinoline-3-carboxamide (VX-770, ivacaftor), a potent and orally bioavailable CFTR potentiator. J. Med. Chem. 2014, 57, 9776–9795. 10.1021/jm5012808. [DOI] [PubMed] [Google Scholar]
- Condren M. E.; Bradshaw M. D. Ivacaftor: a novel gene-based therapeutic approach for cystic fibrosis. J. Pediatr. Pharmacol. Ther. 2013, 18, 8–13. 10.5863/1551-6776-18.1.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Goor F.; Hadida S.; Grootenhuis P. D. J.; Burton B.; Cao D.; Neuberger T.; Turnbull A.; Singh A.; Joubran J.; Hazlewood A.; et al. Rescue of CF airway epithelial cell function in vitro by a CFTR potentiator, VX-770. Proc. Natl. Acad. Sci. U. S. A. 2009, 106, 18825–18830. 10.1073/pnas.0904709106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Goor F.; Hadida S.; Grootenhuis P. D. J.; Burton B.; Stack J. H.; Straley K. S.; Decker C. J.; Miller M.; McCartney J.; Olson E. R.; et al. Correction of the F508del-CFTR protein processing defect in vitro by the investigational drug VX-809. Proc. Natl. Acad. Sci. U. S. A. 2011, 108, 18843–18848. 10.1073/pnas.1105787108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verkman A. S.; Galietta L. J. V. Chloride channels as drug targets. Nat. Rev. Drug Discovery 2009, 8, 153–171. 10.1038/nrd2780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerra L.; Favia M.; Di Gioia S.; Laselva O.; Bisogno A.; Casavola V.; Colombo C.; Conese M. The preclinical discovery and development of the combination of ivacaftor + tezacaftor used to treat cystic fibrosis. Expert Opin. Drug Discovery 2020, 15, 873–891. 10.1080/17460441.2020.1750592. [DOI] [PubMed] [Google Scholar]
- Boyle M. P.; Bell S. C.; Konstan M. W.; McColley S. A.; Rowe S. M.; Rietschel E.; Huang X.; Waltz D.; Patel N. R.; Rodman D. A CFTR corrector (lumacaftor) and a CFTR potentiator (ivacaftor) for treatment of patients with cystic fibrosis who have a phe508del CFTR mutation: a phase 2 randomised controlled trial. Lancet Respir. Med. 2014, 2, 527–538. 10.1016/s2213-2600(14)70132-8. [DOI] [PubMed] [Google Scholar]
- Donaldson S. H.; Pilewski J. M.; Griese M.; Cooke J.; Viswanathan L.; Tullis E.; Davies J. C.; Lekstrom-Himes J. A.; Wang L. T.; Tezacaftor/Ivacaftor in Subjects with Cystic Fibrosis and F508del/F508del-CFTR or F508del/G551D-CFTR. Am. J. Respir. Crit. Care Med. 2018, 197, 214–224. 10.1164/rccm.201704-0717OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowe S. M.; Daines C.; Ringshausen F. C.; Kerem E.; Wilson J.; Tullis E.; Nair N.; Simard C.; Han L.; Ingenito E. P.; et al. Tezacaftor–Ivacaftor in Residual-Function Heterozygotes with Cystic Fibrosis. N. Engl. J. Med. 2017, 377, 2024–2035. 10.1056/NEJMoa1709847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keating D.; Marigowda G.; Burr L.; Daines C.; Mall M. A.; McKone E. F.; Ramsey B. W.; Rowe S. M.; Sass L. A.; Tullis E.; et al. VX-445-Tezacaftor-Ivacaftor in Patients with Cystic Fibrosis and One or Two Phe508del Alleles. N. Engl. J. Med. 2018, 379, 1612–1620. 10.1056/NEJMoa1807120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veit G.; Roldan A.; Hancock M. A.; Da Fonte D. F.; Xu H.; Hussein M.; Frenkiel S.; Matouk E.; Velkov T.; Lukacs G. L. Allosteric folding correction of F508del and rare CFTR mutants by elexacaftor-tezacaftor-ivacaftor (Trikafta) combination. JCI Insight 2020, 5, e139983 10.1172/jci.insight.139983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoy S. M. Elexacaftor/Ivacaftor/Tezacaftor: First Approval. Drugs 2019, 79, 2001–2007. 10.1007/s40265-019-01233-7. [DOI] [PubMed] [Google Scholar]
- https://investors.vrtx.com/node/27601/pdf
- Ridley K.; Condren M. Elexacaftor-Tezacaftor-Ivacaftor: The First Triple-Combination Cystic Fibrosis Transmembrane Conductance Regulator Modulating Therapy. J. Pediatr. Pharmacol. Ther. 2020, 25, 192–197. 10.5863/1551-6776-25.3.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wainwright C. E.; Elborn J. S.; Ramsey B. W.; Marigowda G.; Huang X.; Cipolli M.; Colombo C.; Davies J. C.; De Boeck K.; Flume P. A.; et al. Lumacaftor–Ivacaftor in Patients with Cystic Fibrosis Homozygous for Phe508del CFTR. N. Engl. J. Med. 2015, 373, 220–231. 10.1056/NEJMoa1409547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heijerman H. G. M.; McKone E. F.; Downey D. G.; Van Braeckel E.; Rowe S. M.; Tullis E.; Mall M. A.; Welter J. J.; Ramsey B. W.; McKee C. M.; et al. Efficacy and safety of the elexacaftor plus tezacaftor plus ivacaftor combination regimen in people with cystic fibrosis homozygous for the F508del mutation: a double-blind, randomised, phase 3 trial. Lancet 2019, 394, 1940–1948. 10.1016/s0140-6736(19)32597-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capurro V.; Tomati V.; Sondo E.; Renda M.; Borrelli A.; Pastorino C.; Guidone D.; Venturini A.; Giraudo A.; Mandrup Bertozzi S.; et al. Partial Rescue of F508del-CFTR Stability and Trafficking Defects by Double Corrector Treatment. Int. J. Mol. Sci. 2021, 22, 5262. 10.3390/ijms22105262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spanò V.; Montalbano A.; Carbone A.; Scudieri P.; Galietta L. J. V.; Barraja P. An overview on chemical structures as ΔF508-CFTR correctors. Eur. J. Med. Chem. 2019, 180, 430–448. 10.1016/j.ejmech.2019.07.037. [DOI] [PubMed] [Google Scholar]
- Spanò V.; Venturini A.; Genovese M.; Barreca M.; Raimondi M. V.; Montalbano A.; Galietta L. J. V.; Barraja P. Current development of CFTR potentiators in the last decade. Eur. J. Med. Chem. 2020, 204, 112631 10.1016/j.ejmech.2020.112631. [DOI] [PubMed] [Google Scholar]
- Ghelani D. P.; Schneider-Futschik E. K. Emerging Cystic Fibrosis Transmembrane Conductance Regulator Modulators as New Drugs for Cystic Fibrosis: A Portrait of in Vitro Pharmacology and Clinical Translation. ACS Pharmacol. Transl. Sci. 2020, 3, 4–10. 10.1021/acsptsci.9b00060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balch W. E.; Morimoto R. I.; Dillin A.; Kelly J. W. Adapting proteostasis for disease intervention. Science 2008, 319, 916–919. 10.1126/science.1141448. [DOI] [PubMed] [Google Scholar]
- Brusa I.; Sondo E.; Falchi F.; Pedemonte N.; Roberti M.; Cavalli A. Proteostasis Regulators in Cystic Fibrosis: Current Development and Future Perspectives. J. Med. Chem. 2022, 65, 5212–5243. 10.1021/acs.jmedchem.1c01897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grove D. E.; Rosser M. F. N.; Ren H. Y.; Naren A. P.; Cyr D. M. Mechanisms for rescue of correctable folding defects in CFTRΔF508. Mol. Biol. Cell 2009, 20, 4059–4069. 10.1091/mbc.e08-09-0929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomati V.; Sondo E.; Armirotti A.; Caci E.; Pesce E.; Marini M.; Gianotti A.; Jeon Y. J.; Cilli M.; Pistorio A.; et al. Genetic Inhibition Of The Ubiquitin Ligase Rnf5 Attenuates Phenotypes Associated To F508del Cystic Fibrosis Mutation. Sci. Rep. 2015, 5, 12138. 10.1038/srep12138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sondo E.; Falchi F.; Caci E.; Ferrera L.; Giacomini E.; Pesce E.; Tomati V.; Mandrup Bertozzi S.; Goldoni L.; Armirotti A.; et al. Pharmacological Inhibition of the Ubiquitin Ligase RNF5 Rescues F508del-CFTR in Cystic Fibrosis Airway Epithelia. Cell Chem. Biol. 2018, 25, 891–905.e8. 10.1016/j.chembiol.2018.04.010. [DOI] [PubMed] [Google Scholar]
- Bromberg K. D.; Kluger H. M.; Delaunay A.; Abbas S.; DiVito K. A.; Krajewski S.; Ronai Z. Increased expression of the E3 ubiquitin ligase RNF5 is associated with decreased survival in breast cancer. Cancer Res. 2007, 67, 8172–8179. 10.1158/0008-5472.CAN-07-0045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y.; Xuan C.; Jin M.; An Q.; Zhuo B.; Chen X.; Wang L.; Wang Y.; Sun Q.; Shi Y. Ubiquitin ligase RNF5 serves an important role in the development of human glioma. Oncol. Lett. 2019, 18, 4659–4666. 10.3892/ol.2019.10801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Principi E.; Sondo E.; Bianchi G.; Ravera S.; Morini M.; Tomati V.; Pastorino C.; Zara F.; Bruno C.; Eva A.; et al. Targeting of Ubiquitin E3 Ligase RNF5 as a Novel Therapeutic Strategy in Neuroectodermal Tumors. Cancers (Basel) 2022, 14, 1802. 10.3390/cancers14071802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruan J.; Liang D.; Yan W.; Zhong Y.; Talley D. C.; Rai G.; Tao D.; LeClair C. A.; Simeonov A.; Zhang Y.; et al. A small-molecule inhibitor and degrader of the RNF5 ubiquitin ligase. Mol. Biol. Cell 2022, 33, ar120. 10.1091/mbc.E22-06-0233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez A.; Castro A.; Fonseca I.; Martinez-Ripoll M.; Cano F. H.; Albert A. New Synthetic Route to of 1,2,4-Thiadiazolines and 1,3-Thiazolines via Thiadiazolopyridinium Salts. Heterocycles 1996, 43, 2657. 10.3987/com-96-7593. [DOI] [Google Scholar]
- Nagao Y.; Iimori H.; Nam K. H.; Sano S.; Shiro M. Synthesis and Antibacterial Activity of New 1.BETA.-Methylcarbapenems Having the Potential for Intramolecular Nonbonded S...O Interactions. Chem. Pharm. Bull. 2001, 49, 1660–1661. 10.1248/cpb.49.1660. [DOI] [PubMed] [Google Scholar]
- Sondo E.; Tomati V.; Caci E.; Esposito A. I.; Pfeffer U.; Pedemonte N.; Galietta L. J. V. Rescue of the mutant CFTR chloride channel by pharmacological correctors and low temperature analyzed by gene expression profiling. Am. J. Physiol. Cell Physiol. 2011, 301, C872–C885. 10.1152/ajpcell.00507.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalvit C.; Caronni D.; Mongelli N.; Veronesi M.; Vulpetti A. NMR-based quality control approach for the identification of false positives and false negatives in high throughput screening. Curr. Drug Discovery Technol. 2006, 3, 115–124. 10.2174/157016306778108875. [DOI] [PubMed] [Google Scholar]
- Dahlgren M. K.; Schyman P.; Tirado-Rives J.; Jorgensen W. L. Characterization of biaryl torsional energetics and its treatment in OPLS all-atom force fields. J. Chem. Inf. Model. 2013, 53, 1191–1199. 10.1021/ci4001597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipinski C. A. Lead- and drug-like compounds: the rule-of-five revolution. Drug Discovery Today Technol. 2004, 1, 337–341. 10.1016/j.ddtec.2004.11.007. [DOI] [PubMed] [Google Scholar]
- Dalvit C.; Pevarello P.; Tatò M.; Veronesi M.; Vulpetti A.; Sundström M. Identification of compounds with binding affinity to proteins via magnetization transfer from bulk water. J. Biomol. NMR 2000, 18, 65–68. 10.1023/a:1008354229396. [DOI] [PubMed] [Google Scholar]
- Mayer M.; Meyer B. Characterization of Ligand Binding by Saturation Transfer Difference NMR Spectroscopy. Angew. Chem., Int. Ed. Engl. 1999, 38, 1784–1788. . [DOI] [PubMed] [Google Scholar]
- Didier C.; Broday L.; Bhoumik A.; Israeli S.; Takahashi S.; Nakayama K.; Thomas S. M.; Turner C. E.; Henderson S.; Sabe H.; et al. RNF5, a RING finger protein that regulates cell motility by targeting paxillin ubiquitination and altered localization. Mol. Cell. Biol. 2003, 23, 5331–5345. 10.1128/MCB.23.15.5331-5345.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuang E.; Okumura C. Y. M.; Sheffy-Levin S.; Varsano T.; Shu V. C.-W.; Qi J.; Niesman I. R.; Yang H.-J.; López-Otín C.; et al. Regulation of ATG4B Stability by RNF5 Limits Basal Levels of Autophagy and Influences Susceptibility to Bacterial Infection. PLoS Genet. 2012, 8, e1003007 10.1371/journal.pgen.1003007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vázquez C. L.; Colombo M. I.. Assays to Assess Autophagy Induction and Fusion of Autophagic Vacuoles with a Degradative Compartment, Using Monodansylcadaverine (MDC) and DQ-BSA. In Autophagy in Mammalian Systems, Part B; Klionsky D. J., Ed.; Methods in Enzymology; Elsevier Inc., 2009; pp. 85–95. [DOI] [PubMed] [Google Scholar]
- Bodas M.; Vij N. Adapting Proteostasis and Autophagy for Controlling the Pathogenesis of Cystic Fibrosis Lung Disease. Front. Pharmacol. 2019, 10, 20. 10.3389/fphar.2019.00020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vu C. B.; Bridges R. J.; Pena-Rasgado C.; Lacerda A. E.; Bordwell C.; Sewell A.; Nichols A. J.; Chandran S.; Lonkar P.; Picarella D.; et al. Fatty Acid Cysteamine Conjugates as Novel and Potent Autophagy Activators That Enhance the Correction of Misfolded F508del-Cystic Fibrosis Transmembrane Conductance Regulator (CFTR). J. Med. Chem. 2017, 60, 458–473. 10.1021/acs.jmedchem.6b01539. [DOI] [PubMed] [Google Scholar]
- Romani L.; Oikonomou V.; Moretti S.; Iannitti R. G.; D’Adamo M. C.; Villella V. R.; Pariano M.; Sforna L.; Borghi M.; Bellet M. M.; et al. Thymosin α1 represents a potential potent single-molecule-based therapy for cystic fibrosis. Nat. Med. 2017, 23, 590–600. 10.1038/nm.4305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stincardini C.; Renga G.; Villella V.; Pariano M.; Oikonomou V.; Borghi M.; Bellet M. M.; Sforna L.; Costantini C.; Goldstein A. L.; et al. Cellular proteostasis: a new twist in the action of thymosin α1. Expert Opin. Biol. Ther. 2018, 18, 43–48. 10.1080/14712598.2018.1484103. [DOI] [PubMed] [Google Scholar]
- Luciani A.; Villella V. R.; Esposito S.; Brunetti-Pierri N.; Medina D.; Settembre C.; Gavina M.; Pulze L.; Giardino I.; Pettoello-Mantovani M.; et al. Defective CFTR induces aggresome formation and lung inflammation in cystic fibrosis through ROS-mediated autophagy inhibition. Nat. Cell Biol. 2010, 12, 863–875. 10.1038/ncb2090. [DOI] [PubMed] [Google Scholar]
- Stefano D. D.; Villella V. R.; Esposito S.; Tosco A.; Sepe A.; Gregorio F. D.; Salvadori L.; Grassia R.; Leone C. A.; Rosa G. D.; et al. Restoration of CFTR function in patients with cystic fibrosis carrying the F508del-CFTR mutation. Autophagy 2014, 10, 2053–2074. 10.4161/15548627.2014.973737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biederbick A.; Kern H. F.; Elsässer H. P. Monodansylcadaverine (MDC) is a specific in vivo marker for autophagic vacuoles. Eur. J. Cell Biol. 1995, 66, 3–14. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.