Abstract
The morpholino oligomer-based knockdown system has been used to identify the function of various gene products through loss or reduced expression. Morpholinos (MOs) have the advantage in biological stability over DNA oligos because they are not susceptible to enzymatic degradation. For optimal effectiveness, MOs are injected into 1–4 cell stage embryos. The temporal efficacy of knockdown is variable, but MOs are believed to lose their effects due to dilution eventually. Morpholino dilution and injection amount should be closely controlled to minimize the occurrence of off-target effects while maintaining on-target efficacy. Additional complementary tools, such as CRISPR/Cas9 should be performed against the target gene of interest to generate mutant lines and to confirm the morphant phenotype with these lines. This article will demonstrate how to design, prepare, and microinject a translation-blocking morpholino against hand2 into the yolk of 1–4 cell stage zebrafish embryos to knockdown hand2 function and rescue these “morphants” by co-injection of mRNA encoding the corresponding cDNA. Subsequently, the efficacy of the morpholino microinjections is assessed by first verifying the presence of morpholino in the yolk (co-injected with phenol red) and then by phenotypic analysis. Moreover, cardiac functional analysis to test for knockdown efficacy will be discussed. Finally, assessing the effect of morpholino-induced blockage of gene translation via western blotting will be explained.
Introduction
The utilization of zebrafish as a model for the study of cardiovascular development and disease offers a variety of advantages, including high conservation of gene function, optical transparency, rapid cardiovascular development, and cheaper cost when compared to traditional in vivo models1. Morpholino oligonucleotides (MOs) are the most commonly used antisense gene knockdown tools for the zebrafish model. MOs are frequently used to determine a phenotype or to probe gene function. Dr. James Summerton initially developed the morpholino delivery system for the in vivo inhibition of mRNA translation as an attempt to develop therapeutics for human developmental defects2,3. MOs have been used for in vitro and in vivo model organisms to knockdown genes and investigate the consequence of this knockdown on phenotype. This is done by observing alterations in the development of specific organs, for example, the heart. Knockdown of heart-specific genes in WT zebrafish embryos led to the failure of a proper heartbeat, attesting to the indispensable function of these genes for heart development4,5. These phenotypes were rescued by coinjection of mRNAs for the specific genes. A study involving cardiac troponin T (Tnnt2) showed that the expression of full-length tnnt2 mRNA could rescue sarcomeric phenotypes caused by morpholino knockdown6. Another study revealed that the integrity of A-bands and Z-discs could be restored by overexpression of the regulatory myosin light chain ortholog (cmlc2) mRNA in cmcl2 morphants7.
MOs are commonly used to knock down gene expression by targeting pre-mRNA splicing or by blocking translation. Splice blocking MOs bind and inhibit pre-mRNA by inhibiting the splicesome. Translational blocking occurs when the MO binds to the 5’-untranslated region of complementary mRNA to hinder the ribosome assembly. MOs are the most widely used gene-specific method to knockdown gene expression for in vivo models; they are also the most efficient mRNA blocking agents used in cell cultures. The morpholino itself typically consists of a short-chain (around 25) of morpholino subunit bases. Each MO subunit includes a nucleic acid base, a morpholine ring, and a non-ionic phosphorodiamidate. The different mechanisms of action for the two types of MOs necessitate different tests to verify the efficacy of the knockdown. For translation blocking MOs, a western blot analysis is the most reliable test of efficacy, as the protein of interest should not be produced due to blockage of the ATG translation start site. MOs do not directly degrade their target mRNA; instead, they bind to specific regions and inhibit the expression until naturally degraded. However, the splice blocking MOs modify the pre-mRNA by inducing splice modification, which can be assayed by reverse-transcriptase polymerase chain reaction (RT-PCR) and gel electrophoresis.
Three crucial parts of the MOs screening process must be standardized: (i) The MO dose curve must be tuned for phenotypic recognition. The dose curve also shows the lethal dose 50 (LD50: the dose at which 50% of injected embryos die) for each MO tested to improve the ability to optimize phenotypic ‘signal’ versus off-target ‘noise’3. (ii) The phenotyping nomenclature that was adapted should be well documented; a precise and easily understandable phenotypic description is critical to provide extensive explanations based on existing literature and investigator experience to facilitate information sharing among those who did not directly examine the embryos. (iii) Having well-defined language makes it easy to collect data centrally from Morpholino Database8.
In MO knockdown studies for cardiac genes, animals’ heart activity and blood flow dynamics must be monitored in order to determine the impact of MO knockdown experiments on cardiovascular system function. Such analyses require real-time visualizing of the cardiovascular system at high resolution. Zebrafish skin is transparent for the first week of development, enabling visualization of the heart and blood circulation via microscopy. For assessment of heart function, the most calculated physiological parameters are heart rate and cardiac output as well as fractional shortening, fractional area change, and ejection fraction. Blood flow velocities can be measured by tracking moving RBCs, and these measurements are used to determine shear stress levels, a crucial mechanobiological factor on endothelial cells. Such an assessment requires recording time-lapse movies for beating heart and flowing blood via an inverted or a stereomicroscope equipped with a high-speed camera.
This paper shows how to design, prepare, and microinject a translational-blocking morpholino against a gene of interest into the yolk of freshly fertilized zebrafish embryos to knock down gene function. It will also show rescuing these “morphants” by co-injection of mRNA encoding this gene. We will then analyze the efficacy of the morpholino microinjections through phenotypic characterizations as well as cardiac structural and functional analyses. This approach will be demonstrated on a widely studied cardiac gene, hand2.
Protocol
All experiments were carried out in accordance with the accepted standards of humane animal care under the regulation of the IACUC at QU; animals were held in the zebrafish facility under Qatar University Biomedical Research Center (QU-BRC). All animals used in these experimental studies were under 3 days post-fertilization (dpf).
NOTE: For each experimental group, it is advisable to use at least 30 embryos for statistical rigor. The experimental groups are as follows:
Control group:
This group includes embryos cultured in egg water without any injections. Results here will form the control baseline.
Negative Control group:
This group includes embryos cultured in egg water injected with scrambled MOs.
Injected group:
This group includes embryos injected with hand2 MO alone and hand2 MO with hand2 mRNA to rescue the phenotype. Results here will confirm that observed phenotypes appeared due to injected MOs. Comparison of experimental groups will enable assessment of the influence of inhibiting and rescue of hand2 on heart function precisely.
1. Morpholino designs for hand2.
NOTE: MO sequences can be adapted from the literature9,10,11. Alternatively, these oligos can be designed online by Gene-tools. Gene-tools offers a free and fast online design service, which can be accessed through their website12. A custom MO can readily be designed by providing information about the genes of interest, such as sequence information or accession numbers. The following specific steps summarize how to design MOs against hand2 in zebrafish:
First, search for details of the gene of interest from the NCBI database. For hand2, in zebrafish13.
Get GenBank mRNA transcript ID from NCBI14.
-
Obtain the mRNA sequence from NCBI15.
NOTE: Morpholino (MO)-modified antisense oligonucleotides for the zebrafish hand2 gene are available and were previously published. hand2 MO sequence9,10,11: 5’-CCTCCAACTAAACTCATGGCGACAG-3’, MO sequences used in the study are listed in the Table of Materials.
Get a negative control from either a mismatched or scrambled MO (similar sequences with random base pair changes) from gene tools16 to be injected. This would help attest to the specificity of the phenotype (s) observed in specific MO injections and minimize the risk that the observed effects are from an artifact of the injection procedure.
Materials.
| Name | Company | Catalog Number | Comments |
|---|---|---|---|
| Acrylamide 40% | Sigma | Sigma, cat. no. C977M88 | |
| Agarose | Sigma-Aldrich | Sigma-Aldrich cat. no A9539–250G | |
| All Prep DNA/RNA Mini Kit | Qiagen | Qiagen cat. no. 80204. | |
| alpha Tubulin | Abcam | Abcam- ab4074 | Rabbit polyclonal to alpha Tubulin lot GR3 180877–1 (50 kDa) |
| Ammonium persulfate molecular grade | Sigma | Sigma, cat. no C991U65 | |
| BV10 capillary beveller | Sutter Instruments Product | Sutter Instruments Product Catalog # BV10 | |
| Chemiluminescence Imaging Gene Gnome | SYNGENE | SYNGENE | |
| Cleaver Scientific Blotting | CVS10D_OmniPAGEMini | CVS10D_OmniPAGEMini | |
| Coomassie | Thermo Fisher | Thermo Fisher cat. no C861C44 | |
| Electrochemiluminescence (ECL) kit | Abcam Biochemicals | Abcam Biochemicals cat. no ab65623 | |
| Glycine | Sigma | Sigma, cat. no C988U91 | |
| Goat anti Rabbit | Abcam | Abcam- ab6721 | Goat Anti-Rabbit IgG H&L (HRP) 2nd antibodies lot GR3179871–1 |
| HAND2 | Gene tools | Custom made for HAND2 (NM_021973) | 5’-CCTCCAACTAAACTCATGGCGACAG-3’ |
| Hand2 | Abcam | Abcam- ab10131 | Rabbit polyclonal Anti-HAND2 antibody lot GR143200–9 (24– 26 kDa) |
| HAND2 (NM_021973) Human Tagged ORF Clone | OriGene Technologies, Inc | RC224436L3 | Vector: pLenti-C-Myc-DDK-P2A-Puro (PS100092) |
| IBI DNA/RNA/Protein Extraction Kit | IBI Scientific | IBI Scientific cat. no -r IB47702 | |
| Imaging System | iBright | iBright CL1000 Imaging System | |
| Isopropanol | Sigma-Aldrich | Sigma-Aldrich cat. no 278475–2L | |
| Laemmli sample loading buffer (4x) | Sigma-Aldrich | Sigma-Aldrich cat. no 70607 | |
| Mercaptoethanol | Sigma | Sigma, cat. no M6250–1L | |
| Microplate Spectrophotometer with the Gen5 Data Analysis software interface | Epoch | Epoch | |
| Microscope | Ziess SteREO Lumar V12 Flourescence Microscope | Ziess SteREO Lumar V12 Flourescence Microscope | |
| Mineral oil | Fisher Scientific | Fisher Scientific cat. no 0121–1 | |
| mMESSAGE mMACHINE T7/T3/SP6 Transcription Kit | Thermo Fisher | Thermo Fisher cat. no.AM1340 | for mRNA generation |
| Nuclease-free water | New England Biolabs | New England Biolabs cat. no B1500L | |
| PC-100 Micropipette Puller | NARISHIGE GROUP Product | NARISHIGE GROUP Product Catalog # PC-100 | |
| Phenol red | Sigma | Sigma, cat. no. P-0290 | |
| Picolitre Injector | Harvard Apparatus | Harvard Apparatus cataloge # PLI-90A | |
| Pierce Bicinchoninic acid assay (BCA) Protein Assay kit | Thermo Fisher | Thermo Fisher cat. no 23227 | |
| PMSF, Protease inhibitor as protease inhibitors | Thermo Fisher | Thermo Fisher cat. no 36978 | |
| Ponceau S | Sigma-Aldrich | Sigma-Aldrich cat. no 10165921001 | |
| Protease Inhibitor Cocktail | Thermo Fisher | Thermo Fisher cat. no 88668 | |
| Protein ladder | SMOBiO | SMOBiO cat. no PM2500 | |
| Radioimmunoprecipitation Assay (RIPA) | Thermo Fisher | Thermo Fisher cat. no 89900 | |
| Ringer’s solution | Thermofisher | Catalog No.S25513 | |
| SDS | Sigma | Sigma, cat. no 436143 | |
| Standard Control | Gene tools | SKU: PCO-StandardControl-100 | 5’-CCTCTTACCTCAGTTACAATTTATA-3’- that targets a human betaglobin intron mutation |
| Stripping buffer | Sigma-Aldrich | Sigma-Aldrich cat. 21059 | |
| Temed | IBI scientific | IBI scientific cat. no C000A52 | |
| Tris Base | Thermo Fisher | Thermo Fisher cat. no BP-152–500 | |
| Tween | sigma life science | sigma life science cat. no P2287 | |
| Zebra Box Revolution-Danio Track system chamber with the EthoVision XT 11.5 software | Noldus Information Technology, NL | Noldus Information Technology, NL | |
| Zeiss Axiocam ERc 5s | Zeiss | Stemi 508 Zeiss | |
| Zeiss Stemi 2000-C | Zeiss | Stemi 2000-C |
2. Preparation of morpholino injection
-
Stock preparation
-
Dissolve MO stocks with ddH2O; Diethyl pyrocarbonate (DEPC) can damage MOs and be toxic to embryos. Hence, use DEPC-free ddH2O. Upon ordering, MOs are delivered in a vial at a concentration of 300 nM.
NOTE: While Gene Tools recommend 1 mM stock solutions (~8 ng/nL), this can be too low, especially if the morpholino requires a high dose or is mixed with other MOs. Therefore, it is ideal for making various concentrations of MO stock solutions (2 or 3 mM).
Prepare a 0.2 mM working solution by sub-diluting the working solution (i.e., for 2 mM stock, 1:10 dilution). Prepare 10 μL of working solution by adding Danieaux’s solution (7 μL), stock morpholino (1 μL), and of phenol red (2 μL from 10x stock). Phenol red gives the solution a dark red color to help trace the injected material in the embryo.
-
Keep MOs at room temperature (RT) in airtight containers to prevent evaporation. Do not keep MOs on ice on the benchtop, as the solution may precipitate. RT handling is most appropriate.
NOTE: Gene Tools recommends either of two storage methods for MOs: The working solution can be kept at −20 °C (for many months to years) or RT in a sealed tube for long-term storage. If kept at −20 °C, heat the oligonucleotide solution at 65 °C for 10 min and vortex to completely dissolve the morpholino before use (no need to warm it up if kept at RT).
-
-
In vitro transcription of mRNA
NOTE: The in vitro transcription was used to generate mRNA of Hand2 from Plasmid HAND2 (NM_021973) Human Tagged ORF Clone to be microinjected into zebrafish embryos for rescuing the phenotype due to knocking down zebrafish hand2 using morpholinos, plasmid details used in the study are listed in the Table of Materials.
Linearize the plasmid DNA by an appropriate restriction enzyme and purify using a DNA purification kit (Table of Materials). Use the purified linearized DNA as a template for in vitro transcription to generate mRNA for injection as per the manufacturer’s instructions (Table of Materials).
Finally, inject 250 pg of human mRNA for the gene of interest to perform the rescue of phenotype to test whether the lack of a phenotypeis indeed from the loss of zebrafish gene by MO.
-
Preparing to inject
Pull a needle using a micropipettepuller (Table of Materials), following steps 2.3.2–2.3.6.
Use 1 mm capillary tubes with filaments and a micropipette Puller. Turn the machine on and open the lid
Turn the mode selection knob to NO.2 Heater. Using NO.2 heater, adjust the knob to bring the heat to 68 °C
Turn the mode selection knob back to Step 1 position. Use four weights (2 Type Light / 2 Type Heavy)
-
Place the capillary on the holder and close the lid. Press the Start button to get the pulled needle.
NOTE: This will generate capillary needles with a long taper
Cut the tip of the needles briefly, using forceps. Sharpen the tip of the pulled needle for easy penetration into zebrafish embryos using a capillary beveler for about 30 s, resulting in tips with diameters of about 15–25 μm.
Calibrate the needle to estimate the amount of injection following steps 2.3.8–2.3.13.
Briefly mix mRNA/morpholino solution before loading it into the needle to dissolve the particles that could clog the needle fully.
Add ~3–4 μL of MO solution to the back part of the injection needle using a micron-tipped pipet tip. The injection needles are equipped with a small trench to facilitate capillary backfilling.
Confirm that there are no bubbles in the injection needle. Mount the needle to the microinjector.
Check that the tip of the needle is not clogged and that MO solution can flow out of the needle by pressing the injection pedal.
Calibrate the volume of injection by measuring the droplet size. Put the tip of the needle in mineral oil on a micrometer slide. Press the injection pedal and adjust the injection (Eject) pressure (psi) and/or injection time (ms) until the ejected droplet diameter is 0.1 mm, corresponding to 0.5 nL.
Ensure that the injection volume is around 0.5–1 nL. Adjust the volume by changing the injection (Eject) pressure (psi) or time (ms).
For picoliter injector setup protocol, follow steps 2.3.15–2.3.16
Use picoliter injector for zebrafish injections. Turn on the injector’s feed pump.
-
Prepare the injector according to the requirements shown below:
P-balance: Ensure that P-balance is around 0. Ensure that it is slightly negative to prevent yolk flow back into the needle, diluting the MO solution in the needle. Conversely, if the backpressure is too high, the MO will constantly flow out of the needle even without the exertion of pressure on the pedal, resulting in variability and inconsistencies in the injected dose and the observed phenotypes.
P-inject: 20–25 psi is ideal; change this to adjust injection volume. This can range from 10–30 psi but start with ~20 psi to check whether it delivers the desired volume.
Injection time: Ensure to reduce the injection time to 300 ms. Prior to adjusting the injection volume, make adjustments to the time of injection.
For morpholino injection, follow steps 2.3.18–2.3.19.
-
Prepare the injection chamber as follows: Make 2% agarose from 0.6 g of agarose and 30 mL of E3 medium. Pour the agarose into a Petri dish and place an injection mold on the agarose using a TU-1 injection mold. Once cooled in a freezer, furrows will form for placing and stabilizing embryos (Figure 1A).
NOTE: The 60x E3M stock solution recipe consists of 5.0 mM NaCl, 0.17 mM KCl, 0.16 mM MgSO4·7H2O, 0.4 mM CaCl2·2H2O in 1 L ddH2O with a final pH of 7.6.
Transfer the collected embryos to the furrows using a transfer pipette, remove all excess E3 medium around embryos to prevent the floating of embryos (Figure 1B).
Figure 1: Steps for injection of MO solution into the yolk.
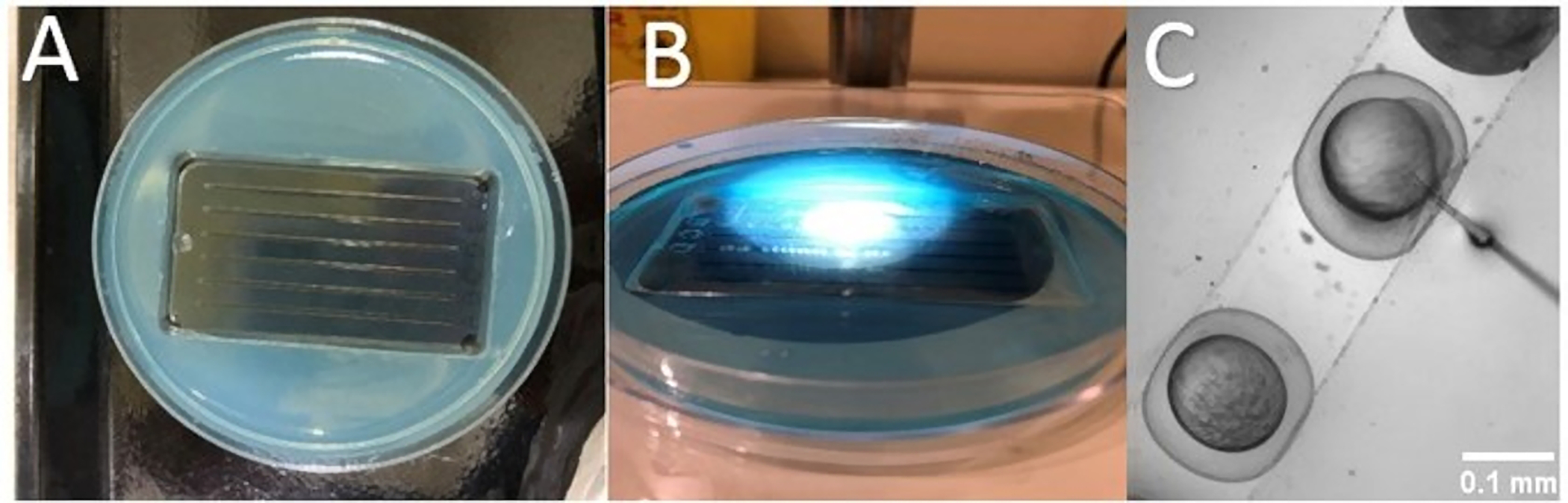
(A) Injection chamber. (B) Using a transfer pipette, transfer the collected embryos to the furrows. (C) Insert the needle through the chorion, inject near the boundary of cell/yolk to the embryo.
3. Injection of MO and mRNA solution into the yolk
-
Insert the needle through the chorion, inject near the boundary of cell/yolk to the embryo side and press the injection pedal (Figure 1C).
NOTE: The injected MO solution/phenol red will not diffuse immediately within the yolk. A red spot will be observed in the yolk that gradually dissipates. The injected material should not exceed more than 10% of the embryo size. MOs can be microinjected into the yolk of newly-laid zebrafish eggs at up to the 8-cell stage because the MOs can easily be taken up in the cytoplasmic stream of the yolk sac by the developing cells17,18. In plasmid or capped mRNA injections, the injection should be performed into a single blastomere of a one-cell stage embryo.
-
Transfer the embryos into a Petri dish with E3 medium and incubate them at 28 °C until 3 dpf, with daily replenishment of E3 medium. At that stage, perform the phenotypic assessment using a microscope.
NOTE: MO solutions must be carefully diluted for each investigation to establish the lowest concentration needed to induce a specific phenotype. Embryonic lethality for each MOs must also be determined. At higher concentrations (above 4–10 ng, depending on the MO), MOs tend to elicit non-specific effects, such as brain or general cell death.
Co-inject MO with 250 pg of mRNA of human hand2 in the same way to confirm the rescue of phenotype induced by loss of zebrafish hand2 gene by MO.
4. Western blot to verify the success of morpholino knockdown
Collect the embryos at specific time points (48 hpf, 72 hpf) and dechorionate if needed using the Pronase enzyme.
-
Deyolk the embryos to remove Vitellogenin using a previously known methodology19.
NOTE: Vitellogenin is a phospholipo-glycoprotein that serves as a source of nutrients for the growing embryo. High-resolution 2D gel electrophoresis and vastly improved western blotting are made possible by deyolking the embryos and removing Vitellogenin.
Perform western blotting as described previously20.
5. Cardiac structure and function assessment:
-
Live imaging of zebrafish: Imaging the ventricle
NOTE: Several heart function/hemodynamics parameters can be calculated by visualizing the beating heart of zebrafish embryo21,22. These parameters include cardiac output (CO), ejection fraction (EF), stroke volume (SV), fractional shortening (FS), and fractional area change (FAC) (please see previous papers involving detailed protocols for cardiac function assessment for zebrafish embryos22,23). The following steps briefly explain how to calculate those parameters for zebrafish embryos at ~3 dpf, a stage when the skin of these organisms is transparent, enabling visualization with bright-field microscopy.
Put a drop of 3% methylcellulose solution (RT) in the concave well imaging slide.
-
Position the Zebrafish embryo in the well using a suitable plastic dropper (Figure 2A, B)
NOTE: Overfilling the well may cause fish displacement out of the well.
-
Gently mix the 3% methylcellulose drop with the E3 medium to stabilize the hatched embryo.
NOTE: To prepare 3% methylcellulose solution, dissolve 3 g of methylcellulose powder in 100 mL of PBS, or other mounting media, in a flask. Place a stirring magnet in the mixture’s flask and place the flask on a magnetic stirring plate. Set the speed to “low” and keep it at 4 °C for ~1 day to dissolve all the clumps. Once the methylcellulose is completely dissolved, aliquot into small tubes and store at −20 °C.
Position the fish on its left, with its right side facing up and anterior point to the left to facilitate unambiguous imaging for the ventricle (Figure 2C).
Under the microscope, zoom in on the embryo heart with 100x magnification and start recording for ~5 s. Ensure that the ventricle borders are inside the imaging window (Figure 2D).
Record time-lapse movies using a high-speed camera and stereomicroscope at about 100 frames per second (fps) of the whole embryo (Figure 2C), beating ventricle (Figure 2D), and moving red blood cells (RBC) in major vessels, such as the dorsal aorta or the Posterior Cardinal Vein (Figure 2E), for cardiac function analysis.
Save the movie in either AVI movie format or TIFF (or JPEG) image sequences format.
-
Calculate the heart rate
Calculate the time needed to record two consecutive frames. For 100 fps the time interval is equal to 0.01 s.
Choose a known point from any recorded cardiac cycle (i.e., end-diastole or end-systole). Calculate the number of frames needed to repeat the cycle.
Multiply the time interval from step 5.2.1 with the number of frames from step 5.2.2. The result is the time duration (in seconds) for one heartbeat.
-
To calculate beats per minute, divide 60 by the number obtained in the previous step. For normal 3 dpf embryos, this should be around 150 bpm (2.5 Hz).
NOTE: Many software applications can calculate heart rate automatically from beating heart recordings such as ViewPoint24 and DanioVision25.
-
Analyze the cardiac structure using a record time-lapse movie at about 100 fps to check the whole heart to detect the presence of cardiac edema or any other structural defect.
NOTE: Figure 3A shows a normal 3 dpf embryo heart, and Figure 3B shows a 3 dpf heart with cardiac edema and heart looping defect (elongated ventricle).
-
Calculate the fractional area change (FAC):
NOTE: Fractional area change (FAC) is a parameter used to compare ventricular end-diastole and end-systole areas to assess contractility of the ventricle.
Use a record time-lapse movie at about 100 fps to determine frames that represent one cardiac cycle. Extract the frames that show the end of both end-systole and end-diastole.
Calculate both end-diastole area () and end-systole area () using ImageJ or a similar image analysis software.
-
Calculate the fractional shortening ():
NOTE: Fractional shortening () is another parameter used to evaluate the contractility of the ventricle; to determine FS, the end-diastolic and end-systolic diameters of the ventricles must be measured21.
Repeat step 5.3.1
Use ImageJ or any other equivalent image analysis program to measure the diameters of the ventricular walls at the end-diastole and the end-systole points. In most cases, short-axis diameters are employed to determine the fractional smoothness () (Figure 4).
-
Calculation of stroke volume (SV):
NOTE: For each heartbeat, the SV is the amount of blood pumped from the ventricle, which is easily computed from the end-diastole and end-systole volumes of the ventricles28.
-
Calculate the ejection fraction ():
NOTE: is defined as the fraction of blood ejected from the ventricle with each heartbeat 28.
From the above formulas, extract and for the fish.
-
Calculate the cardiac output :
NOTE: is the volume of blood being pumped by the heart28. has a value of 10–55 nL/min for 2–6 dpf embryos29.
Calculate SV and HR as mentioned in the previous subsections 5.5 and 5.2.
-
Measure the RBCs velocities:
NOTE: Cells speed determination is vital for evaluating flow rate, vessel diameter, and calculating the shear stress exerted on the vessel’s endothelial cells (i.e., Shear stress, on the other hand, is the frictional force imposed on the endothelial cells by moving blood). For 2–5 dpf embryos, the average red blood cells (RBC) velocity is about 300–750μm/s29. To measure the RBCs velocities:
Under the microscope, zoom in on the animal tail at a magnification of 100x. RBCs movement should be visible at this magnification (Figure 5A, B).
Start recording for about 8 s. Ensure that dorsal aorta (DA) and posterior cardinal vein (), the two most important axial arteries, are 100–120 fps inside the imaging window. Track individual cells from sequential frames (Figure 5A, B).
Extract several frames using ImageJ or other similar image analysis software.
Calculate the difference in distance that an individual RBC moves .
Determine the difference in time between the consecutive frames .
-
NOTE: Maximum and average velocity can be extracted from the repetition of consecutive frames for individual cells21. RBCs velocity also represents blood flow velocity. Alternatively, a variety of commercially available software applications can be used to measure RBCs’ velocity automatically. ViewPoint24, as well as DanioVision25, have such applications (Figure 5C). Furthermore, there are few available plugins with ImageJ, such as TrackMate30 and MTrackJJ31, compatible with movies recorded above 100 fps.
-
Calculation of shear stress from measured cell velocities Measured RBC velocities in blood vessels also represent blood flow velocities (Figure 5C). From these measurements, calculate the shear stress in a vessel of interest as follows with Poiseuille Flow assumption22,27 :
Where is the average blood velocity , is the blood viscosity (dynes/cm2 ), and is the vessel diameter . For 3 dpf embryos, shear stress in DA22,26,27 is about 4 dynes/cm2.
Figure 2: Mounting and imaging Zebrafish embryos.
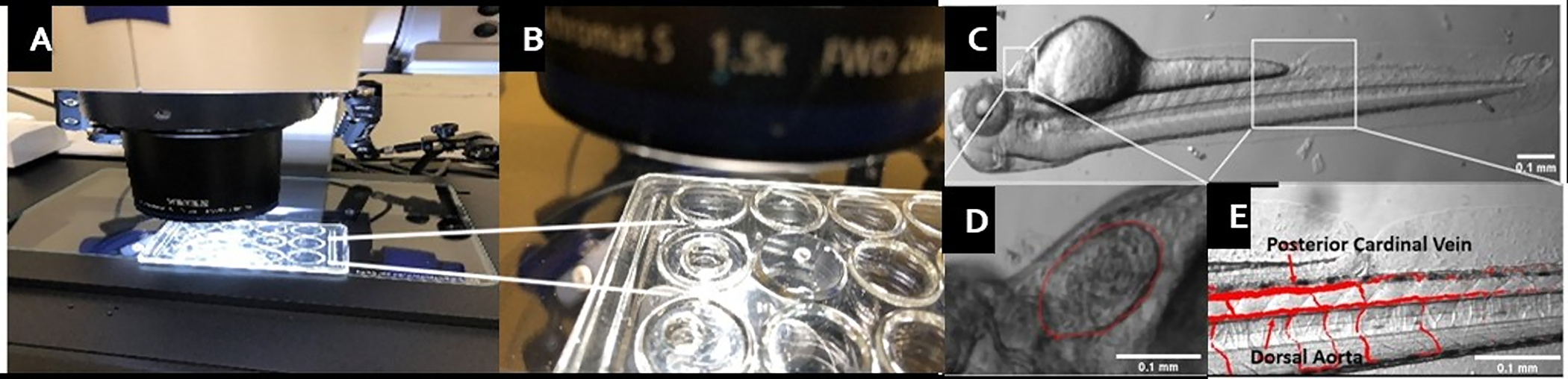
(A) The concave well imaging slide with zebrafish embryo under the microscope. (B) Magnified view of the well. Fill the wells ¾ of volume with E3 medium. (C) Positioning of the embryo should be on its left side. (D) Ventricle can be seen clearly in this configuration (zoomed image on left, ventricle borders are highlighted). Anterior is to the left. (E) Moving red blood cells (RBC) in major vessels, such as the dorsal aorta or the Posterior Cardinal Vein. Anterior is to the left.
Figure 3: Analysis of the whole heart to detect the presence of cardiac edema and/or any structural defects.
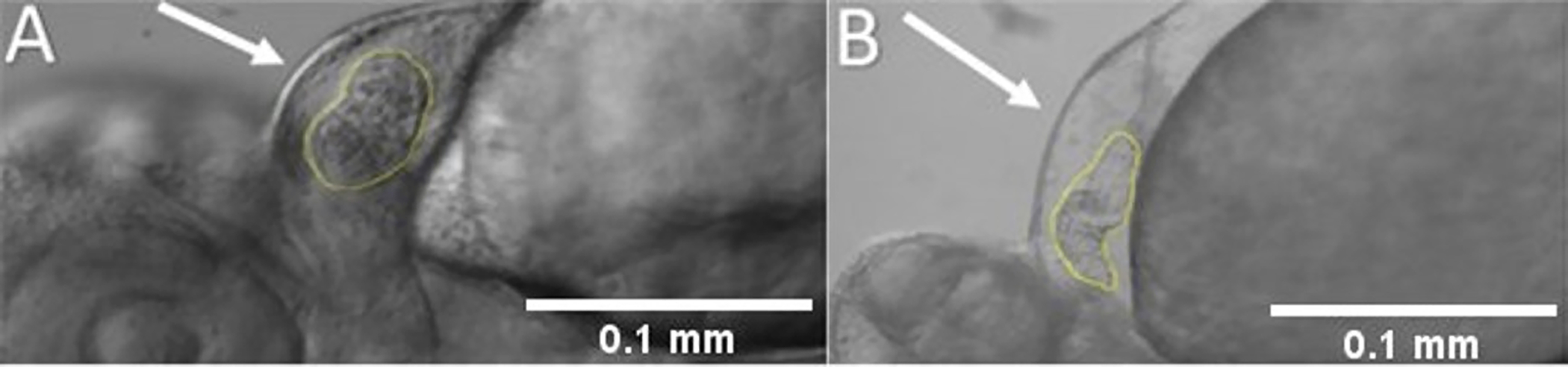
(A) A normal 3 dpf embryo heart. (B) Cardiac edema and looping defect in 3 dpf zebrafish embryo. Anterior is to the left.
Figure 4: Measurement of ventricle size.
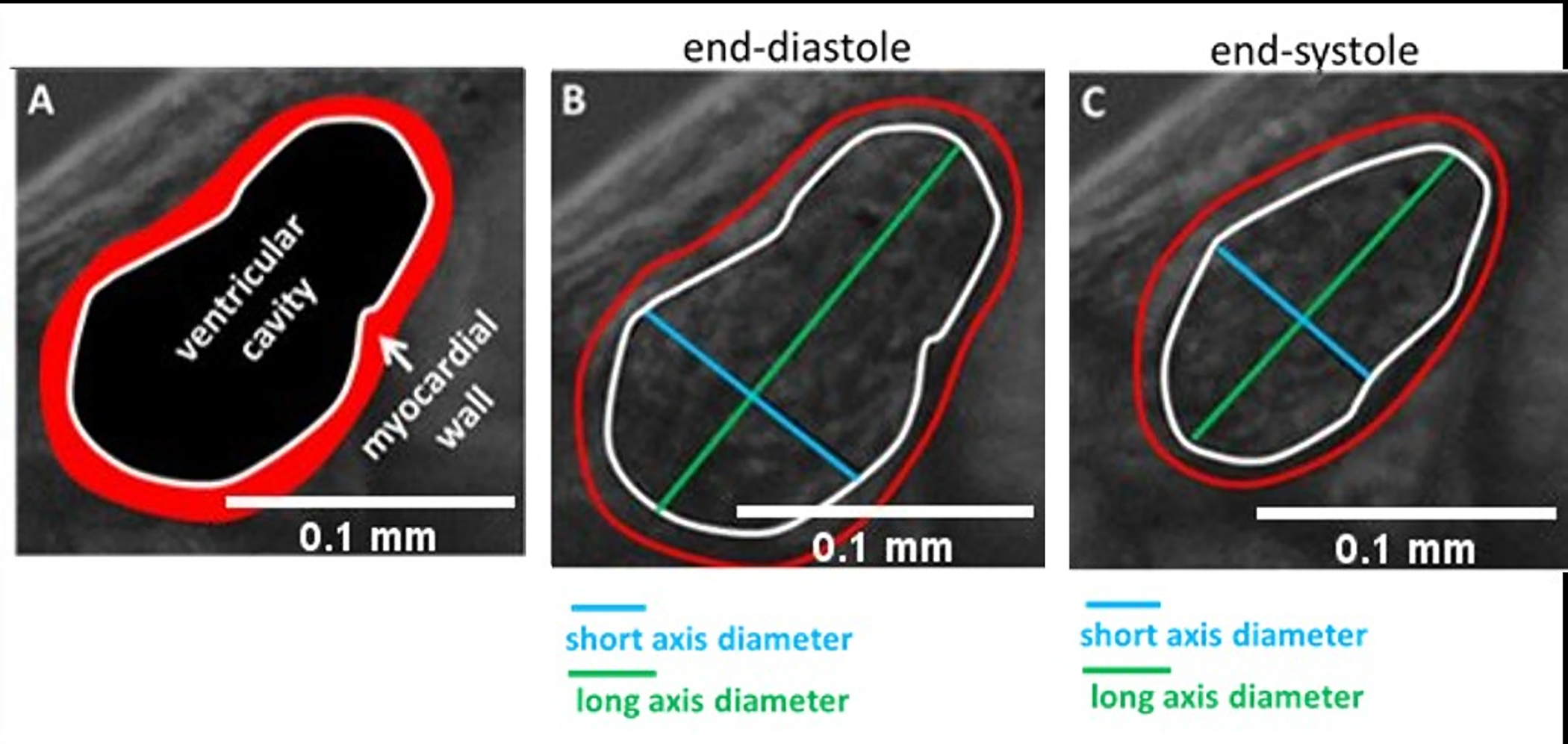
(A) Ventricular cavity and myocardial wall are highlighted. Long axis and short axis diameters are seen for (B) end-diastole and (C) end-systole.
Figure 5: Automatic detection of vessels using ZebraLab.
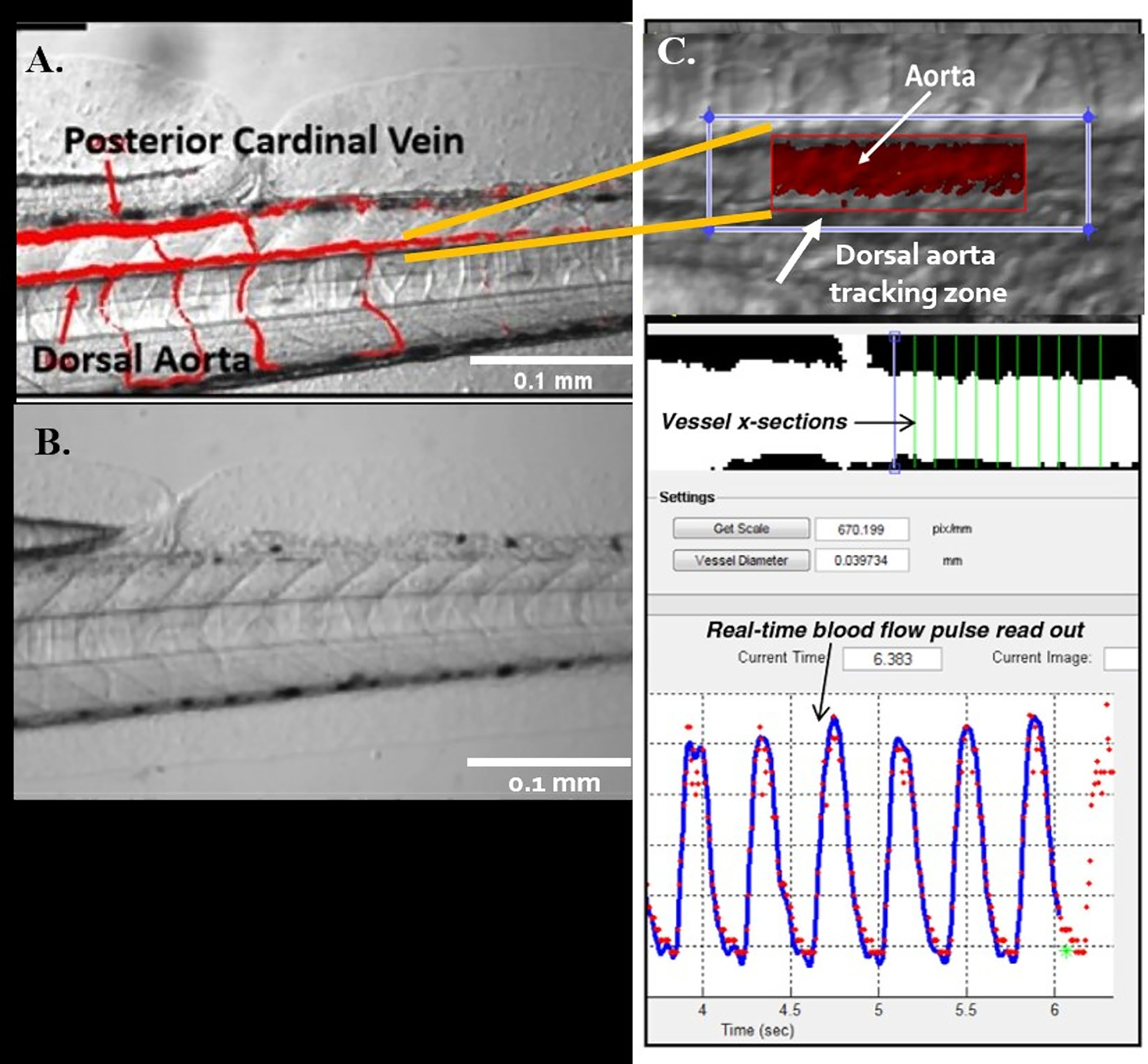
(A,B) The two major blood vessels in zebrafish: the dorsal aorta (DA) and the posterior cardinal vein (PCV). (C) Viewpoint program used to quantify blood flow and vessel diameter in DA.
Representative Results
The graph in Figure 6 illustrates the average percent of embryos surviving at 24, 48, and 72 hpf for both HAND2- specific MO and control scrambled MO-injected embryos. The 1 mM (8 ng/μL) and 8 mM (6.4 ng/μL) MO-injected embryos showed a significant reduction in survival percentage compared to control scrambled MO-injected embryos. This was observed across each measured time point where lethality or malformation was observed. The results indicated that a high concentration of HAND2MO had a significantly lower survival percentage due to off-target effects and toxicity of MO. While 0.4 mM (3.2 ng/nL) HAND2 MO-injected embryos did not show a significant reduction in survival percentage; however, some similar phenotypes in the experimental group were noted, such as delayed in heart development, elongated tub-like structure heart, and pericardial edema in comparison to control scrambled MO-injected embryos (Figure 6).
Figure 6: Hand2 MO titration.
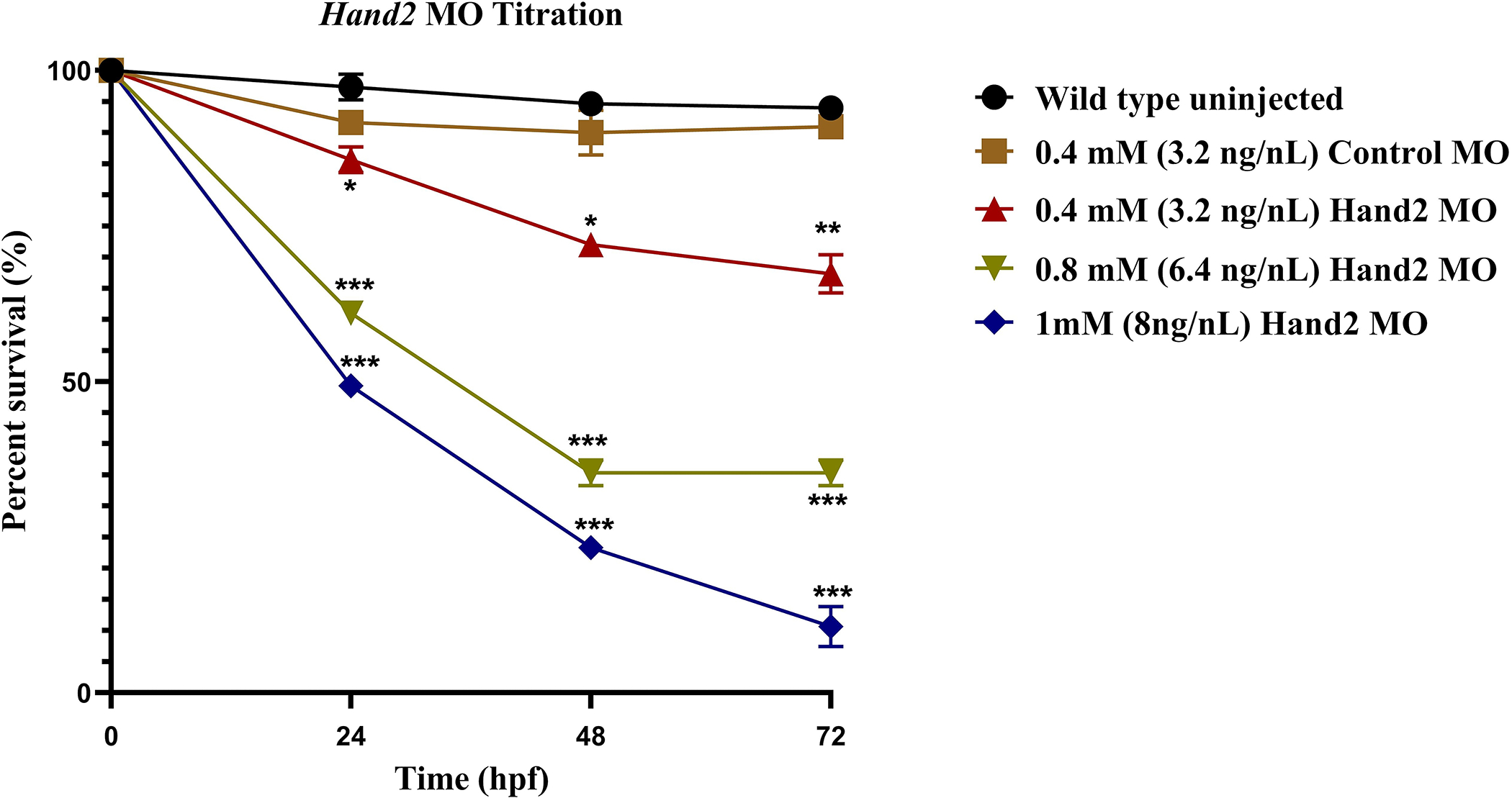
Embryonic survival for different Hand2 MO concentrations was converted to a percentage, averaged, and plotted. The percent survival of MO-injected embryos was calculated at 24, 48, and 72 hpf. All data points represent mean ± SEM (100 embryos were used in each group; experiment was performed in triplicate). The analysis was done by two-way-ANOVA with Dunnett test. *p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.0001.
HAND2 knockdown embryos exhibited distinct and specific phenotypes. Phenotypes of the morphants were examined between 24 and 72 hpf under a Light Microscope. HAND2 loss of function gave rise to embryos with a delay in heart development and an elongated heart, shaped like a tube with pericardial edema (Figure 7). As a result, embryos with this defect in heart formation behaved differently in terms of cardiac performance in comparison to scrambled MO (Figure 8).
Figure 7: Hand2 morphants exhibit distinct and specific phenotypes.
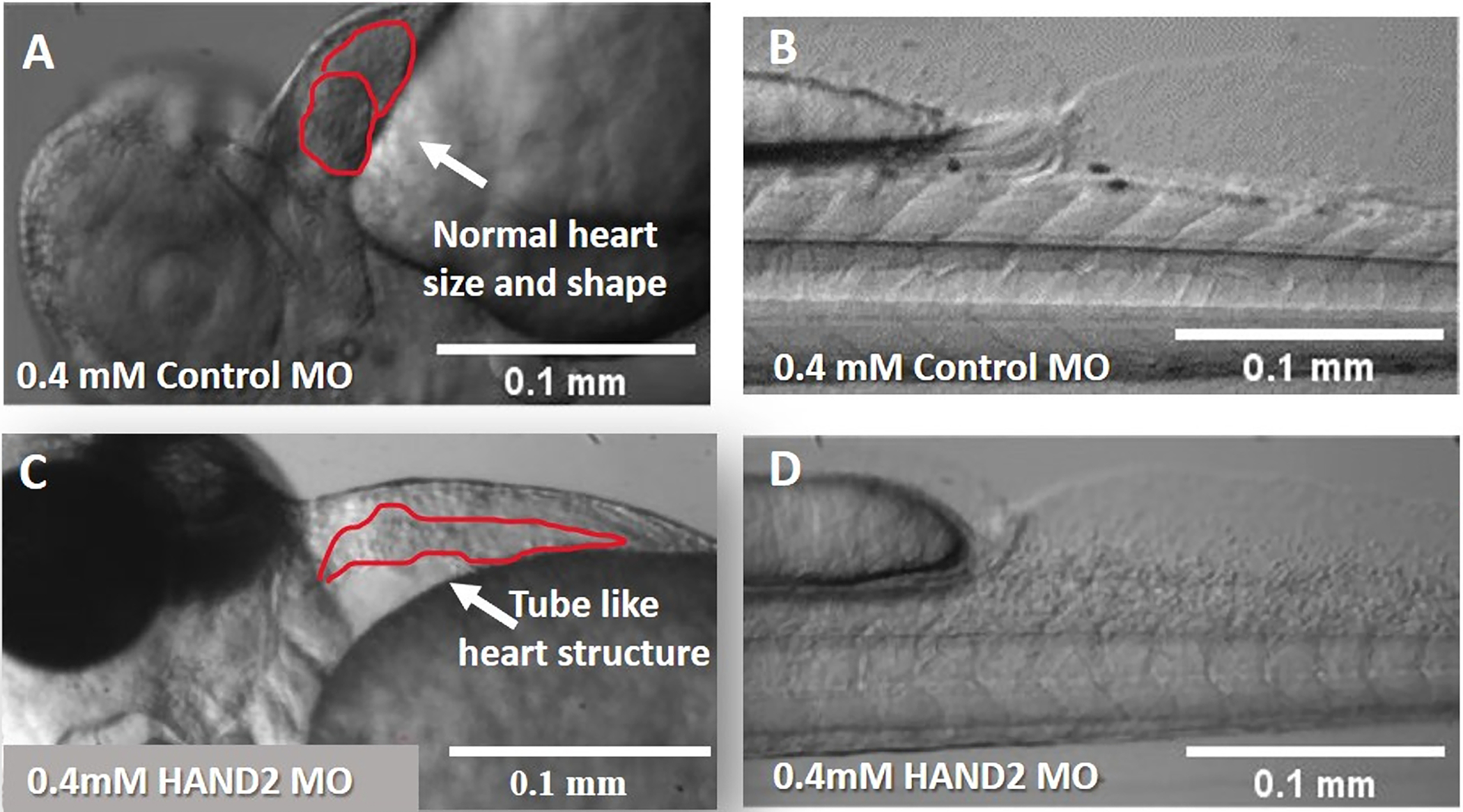
Hearts and tails of 72 hpf embryos were examined under a light microscope (100x magnification). (A,B) Control scrambled MO shows normal heart development without pericardial edema. (C,D) 0.4 mM HAND2 MO-injected embryos as positive control shows tube-like heart structure.
Figure 8: Assessment of cardiac function.
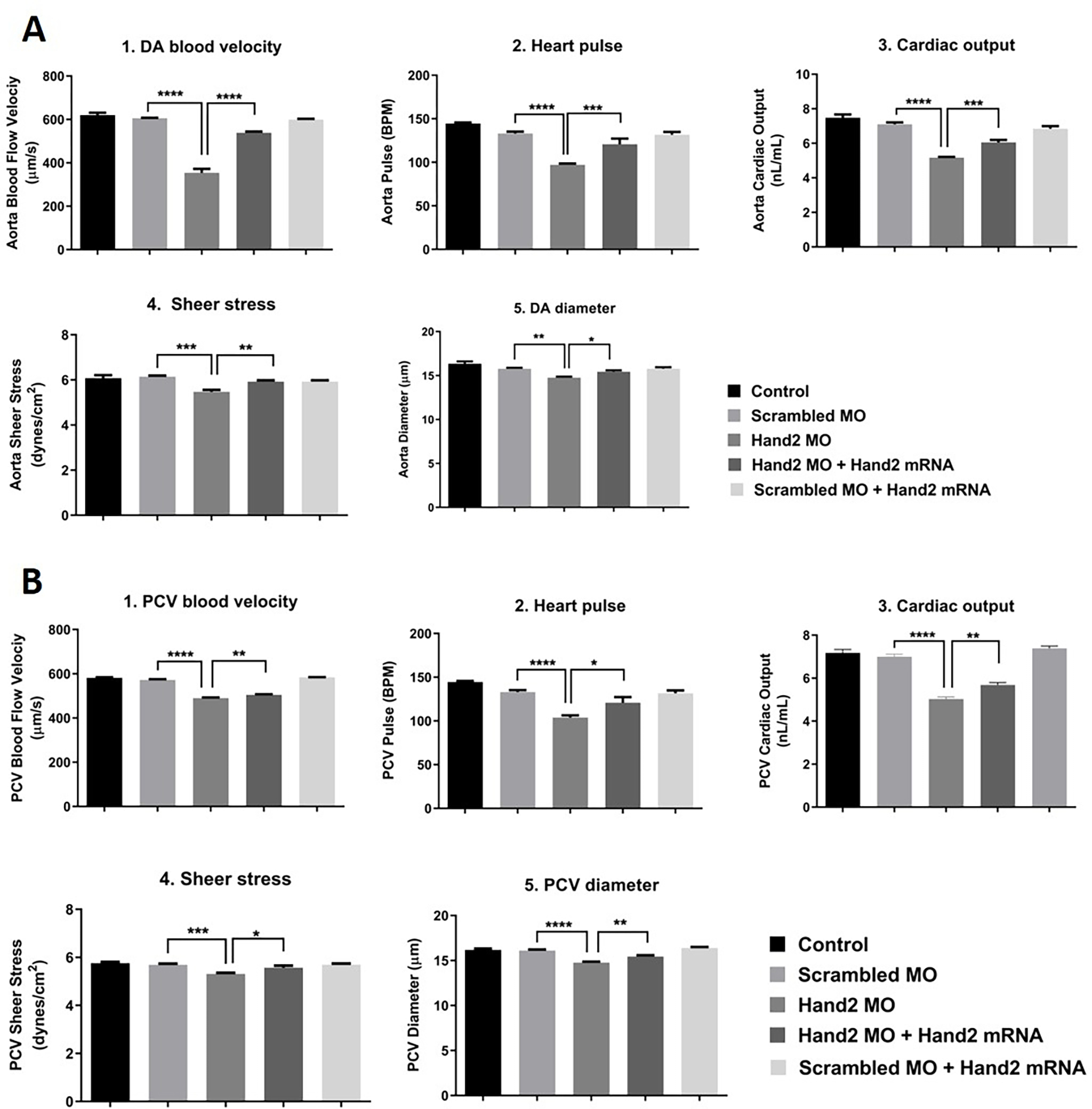
(A) Dorsal aorta blood flow analysis, (B) Posterior Cardinal Vein (PCV) blood flow analysis. 1–4 cell stage zebrafish embryos were injected as groups with scrambled MO, Hand2 MO, Hand2 MO + Hand2 mRNA rescue, and scrambled MO + Hand2 mRNA rescue. Un-injected embryos were used as control. All data are presented as mean ± SEM (6 embryos were used in each group; experiment was performed in triplicate). The analysis was done by one-way ANOVA with Sidak post hoc test. *p < 0.05, **p < 0.01, ***p < 0.001 and ****p < 0.0001.
In our investigation, we have characterized the changes in the zebrafish heart shape, cardiac output, and blood flow in control and HAND2 knockdown animals. Blood flow velocity was measured by tracking down moving RBCs, which were used to determine shear stress levels, an important mechanobiological factor on endothelial cells.
Cardiac output and blood flow analyses showed that HAND2 knockdown triggered a delay in heart development in comparison to wild-type embryos. This alteration resulted in a decrease in DA blood velocity, a drop in heart pulse, and a reduction in cardiac output (Figure 8A). In agreement with the negative impact of HAND2 knockdown on cardiac function, shear stress was also reduced significantly (Figure 8A) in comparison to the control scrambled MO-injected embryos. Most of the heart function failure was rescued when HAND2 MO was co-injected with human HAND2 mRNA, which resulted in an increase in DA blood velocity, increase in heart pulse, and restoration in cardiac output (Figure 8A). These results are in agreement with cardiac output and blood flow analysis results for PCV (Figure 8B). Collectively, the data suggested that HAND2 might play a critical role in cardiomyogenesis during zebrafish development.
Discussion
Morpholino (MO) technology has been extensively used in zebrafish, xenopus, sea urchins, and more recently in cell culture model systems. With most methods, along with the benefits, there are also pitfalls that the experimenter should be aware of. One of the major pitfalls with MO technology includes the concern that phenotypic effects observed by the MO-mediated gene knockdown approach are not due to the loss of function associated with the primary gene product but that some other genes along with the primary gene or independent of the primary gene that has been targeted, and thus this so-called “off-target” effect is the reason for the “observed” morphant phenotype. This point came to the forefront when poor correlation was bserved between morphants and mutants for the same gene in zebrafish32. Thus, several recommendations and guidelines for morpholino use in zebrafish field2,3 have been established, and it is recommended that experimenters follow these guidelines. In our view, when possible, a second independent complementary approach (CRISPR) confirming the morphant or mutant phenotype is desirable. A second limitation includes the ability to selectively knockdown a gene in space and time using MOs. Yes, photoactivatable MOs33 are available, but genes with complex expression patterns and those that have distinct functions at different times in development (early vs. late) will be difficult to target using this method. Further, the use of high light intensities to uncage/activate the MOs may damage the embryonic tissue, which precludes its widespread use. A third limitation of MO is a general one for knockdown technology wherein only transient blocking of gene transcription is possible, and from a model system perspective, MOs will only work effectively in organisms like zebrafish, Xenopus, sea urchins where organogenesis occurs within a short time (2–3 days) period. This time frame limits the use of the technology in higher order organisms, and in adults. Some reports have emerged for in vivo MO use in mice34 although, such approaches have not gained widespread adoption. Finally, non-coding RNAs that comprise the bulk of our genome is a challenge to target with MOs. MOs have successfully targeted short non-coding RNAs (miRNAs)35 but long (>200 bp) non-coding RNAs without translational start site or definitive exon-intron splicing sequence information remains a challenge.
In this study, we investigate HAND2, a transcription factor that regulates cardiac development. Its expression domain determines the heart-forming region in the anterior lateral plate mesoderm (ALPM)36,37,38. HAND2 has an early impact on cardiac progenitor cells formation; consequently, HAND2 knockdown embryos were used as a positive control to assess defective heart development. To examine the involvement of HAND2 in zebrafish embryonic heart development, its translation was interrupted using antisense MO that was designed to target the 5’ untranslated region (UTR). The lack of evidence describing the optimal concentration of MO required for HAND2 silencing in zebrafish necessitated the optimization of the MO dosage.
Zebrafish injected with scrambled MO had a similar survival percentage as compared to un-injected embryos. This finding rules out any embryo lethality attributed to damage from the morpholino injection solution. However, the low survival percentage before 12 hpf could be attributed to the injection of unfertilized embryos. Several concentrations were tested to ascertain the optimal dosage in which knockdown would be achieved while maintaining the normal development of the fish. Fish should appear healthy and not developmentally stunted, which would indicate a delicate balance between phenotypic ‘signal’ versus off-target ‘noise has been obtained3. For clarity, only the optimum amount used is shown in Figure 6.
After successfully optimizing the condition for MO injection, we further examined whether HAND2 knockdown affected heart function. Cardiac defects in HAND2 knockdown zebrafish embryos generally manifested as pericardial edema and faulty circulation at an early stage39. For functional analysis, we began with studying heart rate, as it is an important factor for the assessment of pathological heart function in zebrafish27,40. HAND2 MO-injected embryos exhibited a significant decrease in heart rate, confirming that HAND2 is important for heart function. Successful rescue of HAND2 expression was confirmed at both structural and functional levels by assessing the protein level as well as the embryo’s phenotype, as shown in cardiac function analyses. The results in Figure 8 show that HAND2 expression was significantly diminished and that expression was significantly rescued when the HAND2 MO was co-injected with human HAND2mRNA. This result indicated that HAND2 knockdown is the cause of the induced phenotype.
Our investigations confirmed that in comparison to wildtype, HAND2 knockdown embryos exhibited a delay in heart development accompanied by a reduction in the number of red blood cells and an overall decrease in blood flow and cardiac output.
The MO-based knockdown system has been used to identify the roles played by various gene products through a lack of functional testing. It has the advantage in biological stability over DNA oligos since MOs are not susceptible to enzymatic degradation. For optimal effectiveness, they are injected into 1–4 cell stage embryos. The temporal efficacy of knockdown is variable, and the oligo is believed to eventually lose its effect due to dilution. This protocol details the biological analysis and structural/functional assessment steps to determine if a specific phenotypic abnormality is produced when a cardiac-specific gene is targeted via MOs in zebrafish.
Acknowledgments
The publication of this article was covered with a generous support from BARZAN HOLDINGS. RR is partly supported by R61HL154254 and funds from Department of Pediatrics and Children’s Hospital.
Footnotes
A complete version of this article that includes the video component is available at http://dx.doi.org/10.3791/63324.
Disclosures
The authors declareno financial interests or other conflicts of interest.
References
- 1.Bakkers J Zebrafish as a model to study cardiac development and human cardiac disease. Cardiovascular Research. 91 (2), 279–288 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bill BR, Petzold AM, Clark KJ, Schimmenti LA, Ekker SC A primer for morpholino use in zebrafish. Zebrafish. 6 (1), 69–77 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stainier DY et al. Guidelines for morpholino use in zebrafish. PLoS Genetics. 13 (10), e1007000 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhou B et al. Epicardial progenitors contribute to the cardiomyocyte lineage in the developing heart. Nature. 454 (7200), 109–113 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Eve AM, Place ES, Smith JC Comparison of Zebrafish tmem88a mutant and morpholino knockdown phenotypes. PLoS One. 12 (2), e0172227 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huang W, Zhang R, Xu X Myofibrillogenesis in the developing zebrafish heart: A functional study of tnnt2. Developmental Biology. 331 (2), 237–249 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen Z et al. Depletion of zebrafish essential and regulatory myosin light chains reduces cardiac function through distinct mechanisms. Cardiovascular Research. 79 (1), 97–108 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bedell VM, Westcot SE, Ekker SC Lessons from morpholino-based screening in zebrafish. Briefings in Functional Genomics. 10 (4), 181–188 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Reichenbach B et al. Endoderm-derived Sonic hedgehog and mesoderm Hand2 expression are required for enteric nervous system development in zebrafish. Developmental Biology. 318 (1), 52–64 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Maves L, Tyler A, Moens CB, Tapscott S. J. J. D. b. Pbx acts with Hand2 in early myocardial differentiation. Developmental Biology. 333 (2), 409–418 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hinits Y et al. Zebrafish Mef2ca and Mef2cb are essential for both first and second heart field cardiomyocyte differentiation. Developmental Biology. 369 (2), 199–210 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gene Tools, custom-sequence Morpholinos. at <https://oligodesign.gene-tools.com/> (2011). [Google Scholar]
- 13.hand2 heart and neural crest derivatives expressed 2 [ Danio rerio (zebrafish). at <https://www.ncbi.nlm.nih.gov/gene/58150> (2021).
- 14.Danio rerio heart and neural crest derivatives expressed 2 (hand2), mRNA. <https://www.ncbi.nlm.nih.gov/nuccore/NM_131626.3> (2021).
- 15.NCBI reference sequence -Danio rerio heart and neural crest derivatives expressed 2 (hand2), mRNA FASTA. at <https://www.ncbi.nlm.nih.gov/nuccore/NM_131626.3?report=fast> (2021).
- 16.Prepared control oligos. at <https://store.gene-tools.com/prepared-control-oligos> (2021). [Google Scholar]
- 17.Schubert S, Keddig N, Hanel R, Kammann U Microinjection into zebrafish embryos (Danio rerio)-a useful tool in aquatic toxicity testing? Environmental Sciences Europe. 26 (1), 32 (2014). [Google Scholar]
- 18.Moulton JD Guide for morpholino users: toward therapeutics. Journal of Drug Discovery, Development, and Delivery. 3 (2), 1023 (2016). [Google Scholar]
- 19.Link V, Shevchenko A, Heisenberg C-P Proteomics of early zebrafish embryos. BMC Developmental Biology. 6 (1), 1 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Eslami A, Lujan J Western blotting: sample preparation to detection. Journal of Visualized Experiments: JoVE. 44, e2359 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shin JT, Pomerantsev EV, Mably JD, MacRae CA High-resolution cardiovascular function confirms functional orthology of myocardial contractility pathways in zebrafish. Physiological Genomics. 42 (2), 300–309 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yalcin HC, Amindari A, Butcher JT, Althani A, Yacoub M Heart function and hemodynamic analysis for zebrafish embryos. Developmental Dynamics: An Officical Publication of the American Association of Anatomists. 246 (11), 868–880 (2017). [DOI] [PubMed] [Google Scholar]
- 23.Zakaria ZZ et al. Using zebrafish for investigating the molecular mechanisms of drug-induced cardiotoxicity. Biomed Research International. 2018, 1642684 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Parker T et al. A multi-endpoint in vivo larval zebrafish (Danio rerio) model for the assessment of integrated cardiovascular function. Journal of Pharmacological and Toxicological Methods. 69 (1), 30–38 (2014). [DOI] [PubMed] [Google Scholar]
- 25.Grone BP et al. Epilepsy, behavioral abnormalities, and physiological comorbidities in syntaxin-binding protein 1 (STXBP1) mutant zebrafish. PLoS One. 11 (3), e0151148 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Salman HE, Yalcin HC Advanced blood flow assessment in Zebrafish via experimental digital particle image velocimetry and computational fluid dynamics modeling. Micron. 130, 102801 (2020). [DOI] [PubMed] [Google Scholar]
- 27.Benslimane FM et al. Cardiac function and blood flow hemodynamics assessment of zebrafish (Danio rerio) using high-speed video microscopy. Micron. 136, 102876 (2020). [DOI] [PubMed] [Google Scholar]
- 28.DeGroff CG Doppler echocardiography. Pediatric Cardiology. 23 (3), 307–333 (2002). [DOI] [PubMed] [Google Scholar]
- 29.Bagatto B, Burggren W A three-dimensional functional assessment of heart and vessel development in the larva of the zebrafish (Danio rerio). Physiological and Biochemical Zoology. 79 (1), 194–201 (2005). [DOI] [PubMed] [Google Scholar]
- 30.Tinevez J -Y. et al. TrackMate: An open and extensible platform for single-particle tracking. Methods (San Diego, Calif.). 115, 80–90 (2017). [DOI] [PubMed] [Google Scholar]
- 31.Meijering E, Dzyubachyk O, Smal I Methods for cell and particle tracking. Methods in Enzymology. 504, 183–200 (2012). [DOI] [PubMed] [Google Scholar]
- 32.Kok FO et al. Reverse genetic screening reveals poor correlation between morpholino-induced and mutant phenotypes in zebrafish. Developmental Cell. 32 (1), 97–108 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sumanas S Inducible inhibition of gene function with photomorpholinos. Methods in Molecular Biology (Clifton, N.J.). 1565, 51–57 (2017). [DOI] [PubMed] [Google Scholar]
- 34.Kyritsi K et al. Knockdown of hepatic gonadotropin-releasing hormone by vivo-morpholino decreases liver fibrosis in multidrug resistance gene 2 knockout mice by down-regulation of miR-200b. The American Journal of Pathology. 187 (7), 1551–1565 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Flynt AS, Rao M, Patton JG Blocking zebrafish microRNAs with morpholinos. Methods in Molecular Biology (Clifton, N.J.). 1565, 59–78 (2017). [DOI] [PubMed] [Google Scholar]
- 36.Schoenebeck JJ, Keegan BR, Yelon D Vessel and blood specification override cardiac potential in anterior mesoderm. Developmental Cell. 13 (2), 254–267 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lu F, Langenbacher A, Chen J-N. Transcriptional regulation of heart development in zebrafish. Journal of Cardiovascular Development and Disease. 3 (2), 14 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Laurent F et al. HAND2 target gene regulatory networks control atrioventricular canal and cardiac valve development. Cell Reports. 19 (8), 1602–1613 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Miura GI, Yelon D A guide to analysis of cardiac phenotypes in the zebrafish embryo. Methods in Cell Biology. 101, 161–180 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.De Luca E et al. ZebraBeat: a flexible platform for the analysis of the cardiac rate in zebrafish embryos. Scientific Reports. 4, 4898 (2014). [Google Scholar]


