Abstract
We have studied the in vivo tropism of human herpesvirus 6 (HHV-6) for hemopoietic cells in patients with latent HHV-6 infection. Having used a variety of cell purification, molecular, cytogenetic, and immunocytochemical procedures, we report the first evidence that HHV-6 latently infects early bone marrow progenitor cells and that HHV-6 may be transmitted longitudinally to cells which differentiate along the committed pathways.
We began our study to explore which peripheral blood or bone marrow cell types human herpesvirus 6 (HHV-6) infects, preferentially, in vivo.
We previously described six patients (two diagnosed with Hodgkin’s disease, three with non-Hodgkin’s lymphoma, and one with multiple sclerosis) who showed latent HHV-6 infection in blood cells and viral copy numbers so high that virus was detectable by Southern blot analysis (20). Although their conditions constitute a rare phenomenon, these patients provide a unique opportunity for the identification of the hematopoietic cell types latently infected by HHV-6 in vivo and for determining possible targets of initial latent infection. Our earlier molecular studies have shown that the HHV-6 variant B genome is integrated into the genome of the circulating peripheral blood mononuclear cells (PBMCs) of three of these patients (patients 1 through 3: one each with Hodgkin’s disease, non-Hodgkin’s lymphoma, and multiple sclerosis). Integration is apparently nonrandom, in that fluorescent in situ hybridization (FISH) studies showed a predominance of signals on the short arm of chromosome 17, specifically in band 17p13.3, in phytohemagglutinin-stimulated PBMCs of all three patients examined (20).
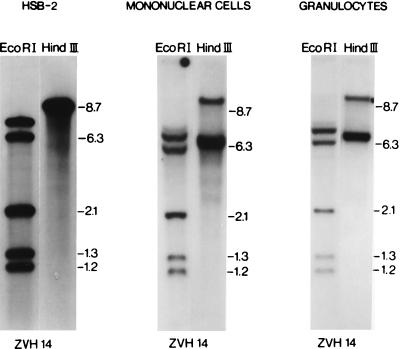
The first unexpected observation of the present study, given that HHV-6 is mainly a lymphotropic virus in vitro (1, 4, 9, 10), was the detection of HHV-6 DNA sequences in circulating granulocytes, which were separated on a Ficoll-Hypaque (Seromed, Berlin, Germany) gradient and further purified by sedimentation for 30 min at 37°C in Haemagel (Stholl Farmaceuticals, Modena, Italy), obtaining a purity of 95%. Granulocytes from these three subjects were positive for the presence of the HHV-6 genome by Southern blot analysis, after hybridization with the ZVH14 (6) (Fig. 1), ZVB70 (7), and HD12 (11) probe sequences (not shown), representative of both the left and the right ends of the viral genome. Furthermore, restriction fragment length polymorphism patterns were identical for both granulocytic and PBMC DNA with different enzymes (Fig. 1). HindIII restriction fragment sizes detected after hybridization with the ZVH14 probe were consistent with infection by HHV-6 variant B in all three patients (1 through 3) and of HHV-6 variant A in the infected HSB-2 cell line (Fig. 1). PCR with HHV-6 primers specific for the ZVH14 region (19) was also used to establish the relative distribution of viral sequences in cell fractions sorted from the total PBMC population of patients 1 through 3 (21). Blood samples (20 ml) were separated with a Ficoll-Hypaque (Pharmacia, Uppsala, Sweden) density gradient into PBMC and polymorphonuclear cell (PMNC) fractions. PBMCs were stained with anti-CD4 and anti-CD8 (Leu3aFITC + Leu2aPE [Becton Dickinson, San Jose, Calif.]) or anti-CD19 and anti-CD14 (OKB19aFITC [Ortho Diagnostic Systems, Raritan, N.J.] and IOM2PE [Immunotech, Marseille, France]) monoclonal antibodies. Cells in the Ficoll gradient pellet (enriched PMNCs and erythrocytes) were resuspended in erythrocyte-lysing buffer (150 mM NH4Cl, 10 mM KHCO3, 0.1 mM EDTA [pH 7.4]) and washed three times with phosphate-buffered saline. The remaining enriched PMNCs were stained with anti-CD16 and anti-CD14 (ION16FITC and IOM2PE; Immunotech) monoclonal antibodies. Cells were sorted on a flow cytometer (FACStarPLUS; Becton Dickinson) equipped for four-parameter analysis. A 2-W argon laser was operated at 205 mW for the 488-nm line. Total lymphocytes were sorted from Ficoll-purified mononuclear cells according to their low forward and side light-scattering characteristics and negative staining for CD14. Monocyte and lymphocyte populations were >98% pure. The sorted CD16+ granulocytes were >99% pure. For all three patients, HHV-6 DNA was detected in monocytes, B lymphocytes, and T lymphocytes—both in CD4+ and in CD8+ subsets—and also in granulocytes (Table 1). To estimate the viral load in the different cell populations, 10-fold dilutions of 105 highly purified cells (105, 104, 103, 102, and 10 cells) were analyzed for HHV-6 DNA. The lowest numbers of cells in which HHV-6 DNA was detected by PCR are shown in Table 1. For all three patients, this number in the granulocyte fraction was the same as the number in the monocyte fraction and was invariably lower than the numbers in the B- and T-lymphocyte fractions. This clearly shows that the HHV-6 DNA detected in purified CD16-positive granulocytes was present in this cell population and was not contributed by contaminating lymphocytes or monocytes. Long-range restriction map features of the HHV-6 integration sites in high-copy-number latently infected granulocyte DNAs from patients 1 through 3, and also in DNA of the HHV-6-infected cell line HSB-2, were examined by pulsed-field gel electrophoresis (PFGE), as previously reported (8). Granulocyte DNAs from patients 1 through 3, which were digested with the rare-cutting restriction enzymes NotI and MluI, showed ZVH14-positive fragment sizes longer than the length of the free viral genome (Fig. 2), consistent with integration and linkage of viral sequences with higher-molecular-weight cellular DNA (8, 20). The numbers and sizes of the long-range restriction fragments we have detected in granulocyte DNAs of these three subjects are similar to those described previously in PBMCs from the same subjects (Fig. 2a). In cytospins and/or smears of peripheral blood, weakly positive nuclear and cytoplasmic staining was detected only in rare granulocytes (less than 0.5%) with monoclonal antibody against the structural protein p101 of variant B (3, 23) (Fig. 3). The authenticity and specificity of the immunocytochemical results with this antibody have been previously reported (3). The detection of HHV-6 infection of granulocytes in vivo is of note. Of the herpesviruses, only human cytomegalovirus (HCMV), which shows high genetic homology to HHV-6, has been shown to infect granulocytes and to express immediate early and late mRNA in vivo (22). Epstein-Barr virus can infect granulocytes and disturb their functions in vitro (2), but Epstein-Barr virus infection and gene expression in human granulocytes in vivo have yet to be demonstrated.
FIG. 1.
Southern blot analysis of DNA extracted from PBMCs and granulocytes of patient 1, digested with the specified restriction enzymes, and hybridized with the ZVH14 probe. DNA extracted from the HHV-6-infected HSB-2 cell line, infected with the original strain of HHV-6, was used as a positive control. The estimated lengths of the bands obtained are reported in kilobases. The same restriction pattern was obtained for patients 2 and 3 (not shown). No hybridization signal was detected either in the uninfected HSB-2 cell line or in the PBMC and granulocyte samples derived from two HHV-6-negative blood donors (not shown).
TABLE 1.
Distribution of HHV-6 sequences in cell fractions sorted from the peripheral blood of subjects with high-copy-number latent viral infections
| Patient | HHV-6 DNA in sorted cell fractionsa
|
||||
|---|---|---|---|---|---|
| CD16+ granulocytes | CD14+ monocytes | CD19+ B lymphocytes | CD4+ T lymphocytes | CD8+ T lymphocytes | |
| 1 | 10 | 10 | 102 | 102 | 103 |
| 2 | 102 | 102 | 103 | 103 | 104 |
| 3 | 10 | 10 | 102 | 102 | 103 |
Monocyte and lymphocyte populations were >98% pure, while sorted CD16+ granulocytes were >99% pure. Tenfold dilutions of 105 FACS-purified cells (105, 104, 103, 102, and 10 cells) were tested for HHV-6 DNA by PCR, and the minimal numbers of positive cells are given.
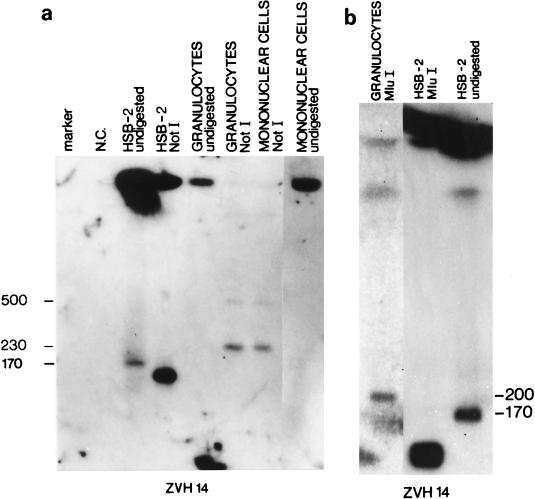
FIG. 2.
(a) Analysis by PFGE of DNA of PBMCs and granulocytes of patient 1 as well as of the infected HSB-2 cell line, before and after digestion with the NotI rare-cutting restriction enzyme. The negative control (N.C.) is undigested DNA from an uninfected HSB-2 cell line and undigested DNA from two blood donors (not shown). Saccharomyces cerevisiae chromosomal DNA serves as the marker. The estimated lengths of the bands obtained are reported in kilobases. The ZVH14 probe sequence was used. Unrestricted HSB-2 DNA showed only one band, of approximately 170 kb, after hybridization of the ZVH14 sequence, a result consistent with free-migrating viral genomes. In contrast, granulocyte and mononuclear cell DNA from patient 1 showed no free-migrating bands and hybridized only in the starting well with the ZVH14 probe, suggesting that free viral molecules were either absent from these cells or present in numbers too small to be detected by this method. When digested with the rare-cutting enzyme NotI, infected HSB-2 DNA showed fragments shorter than the full-length 170-kb viral genome. After digestion with the rare-cutting restriction enzyme NotI, granulocyte and mononuclear cell DNA of patient 1 showed ZVH14-positive fragment sizes longer than the length of the free viral genome. The same long-range restriction pattern was obtained in patients 2 and 3 (not shown). No hybridization signal was detected either in the uninfected HSB-2 cell line or in the PBMC samples derived from two HHV-6-negative blood donors (not shown). (b) Analysis by PFGE of DNA of peripheral granulocytes of patient 1 digested with the MluI rare-cutting restriction enzyme as well as of DNA of the infected HSB-2 cell line, before and after digestion with the same enzyme. The estimated lengths of the bands obtained are reported in kilobases. The ZVH14 probe sequence was used. Unrestricted HSB-2 DNA showed only one band, of approximately 170 kb, after hybridization of the ZVH14 sequence, a result consistent with free-migrating viral genomes. When digested with the rare-cutting enzyme MluI, infected HSB-2 DNA showed fragments shorter than the full-length 170-kb viral genome. After digestion with the rare-cutting restriction enzyme MluI, granulocyte DNA of patient 1 showed a ZVH14-positive fragment longer than the 170-kb viral genome. The same long-range restriction pattern was obtained in patients 2 and 3 (not shown).
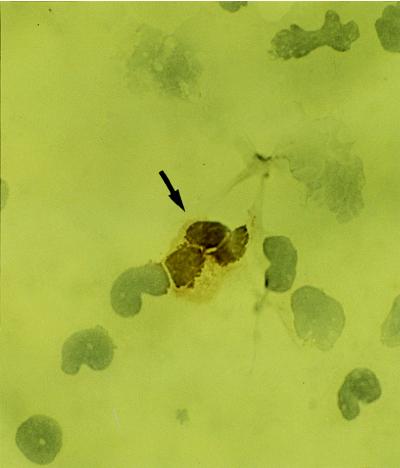
FIG. 3.
Positive staining with antibody against p101K in a peripheral blood granulocyte from patient 1 (arrow) (immunoperoxidase method). Original magnification, ×400.
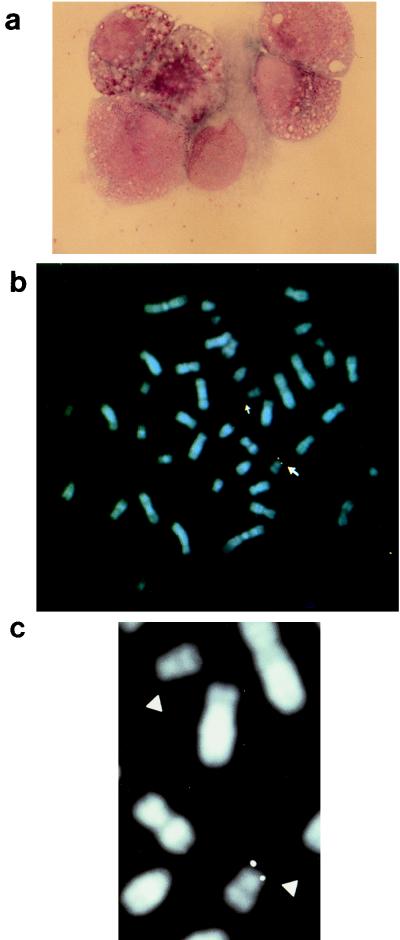
Granulocytes circulate in the blood for only a few hours, which is a much shorter period than the estimated few days required for progression from virus infection to expression of the late proteins, such as p101 (23). We therefore speculated that bone marrow myeloid progenitor cells might be infected with HHV-6 earlier in their development. To verify this hypothesis, individually plucked CFU granulocyte-macrophage (CFU-GM) and burst-forming unit erythroid (BFU-E) colonies were isolated from the bone marrow of the subjects (17) and examined for the presence of viral sequences by PCR (19). For all three patients, most of the CFU-GM and BFU-E colonies were positive for HHV-6 (Fig. 4; Table 2), while no HHV-6 DNA was detected in a comparable number of single colonies derived from the PCR-negative bone marrow specimens of 10 control subjects. We did not detect HHV-6 DNA in methylcellulose that contained no colonies, ruling out the possibility that diffusion of HHV-6, or the presence of rare HHV-6-infected mature cells interspersed among the colonies, could be responsible for the positivity in the CFU-GM and BFU-E colonies. The numbers of CFU evaluated after standard CFU outgrowth assays of the 3 patients are within the ranges obtained for the 10 control bone marrow patients with the same assay, showing that the growth rate of marrow progenitors was not influenced by the latent HHV-6 infection. In addition, bone marrow cells of patient 2, restored from liquid nitrogen, were cultured in the presence of GM colony-stimulating factor, interleukin-3, and stem cell factor for 6 days and analyzed by FISH, with the ZVB70 sequence as a probe (7). FISH, including immunofluorescence procedures, was essentially as described previously (14). Standard cytologic and cytochemical examinations showed 90% of immature myeloid progenitors with proliferative capacity, at various stages of differentiation, and only 2 to 3% of lymphocytes and 7 to 8% of monocytes/macrophages (Fig. 5a). A specific hybridization signal was detected on one or both chromatids of the terminal short arm of one chromosome 17, specifically in band 17p13.3, in 18 of 23 (78%) metaphase cells analyzed (Fig. 5b and c). Our previous FISH studies have shown that PBMCs which are HHV-6 negative by PCR show no detectable signals after hybridization with the pZVB70 probe (20). In bone marrow smears from patients 1 through 3, undifferentiated myeloid precursor cells and cells of the erythroid lineage showed no reactivity with anti-p101 (not shown).
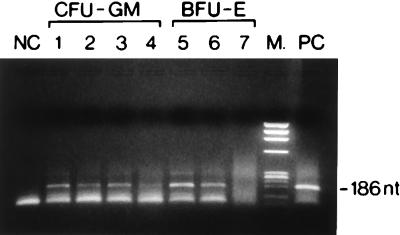
FIG. 4.
Ethidium bromide-stained agarose gel of representative HHV-6-positive CFU-GM (lanes 1 to 4) and BFU-E (lanes 5 and 6) colonies, as well as of one HHV-6-negative BFU-E colony (lane 7), from patients 1 through 3, as determined by PCR assays with primers and probe derived from the ZVH14 sequence. NC, HSB-2 uninfected DNA as a negative control; PC, HHV-6-infected HSB-2 cell line DNA as a positive control; M, Marker IX (Boehringer Mannheim). The length of the amplified segment is indicated.
TABLE 2.
Detection of HHV-6 DNA in single CFU-GM and BFU-E colonies from bone marrow of subjects with high-copy-number latent viral infections
| Patient | No. of HHV-6 DNA-positive colonies/total
|
|
|---|---|---|
| CFU-GM | BFU-E | |
| 1 | 18/20 | 15/21 |
| 2 | 21/23 | 20/26 |
| 3 | 20/24 | 19/23 |
FIG. 5.
(a) Bone marrow culture from patient 2 showing immature progenitors of myeloid lineage (May-Grünwald-Giemsa). Original magnification, ×600. (b) Chromosome fluorescence in situ hybridization experiments performed on metaphase chromosomes of cultured bone marrow cells from patient 2, with pZVB70 used as the viral probe. A specific hybridization signal was detected on the terminal short arm of one chromosome 17 (17p13.3). Chromosomes 17 are indicated by arrows. (c) Partial and enlarged representation of the two chromosomes 17, indicated by arrowheads.
In vitro studies have recently shown that HHV-6 can infect two hematopoietic progenitor cell lines, TF-1 (erythromyeloid) and KG-1 (lymphomyeloid), and, less efficiently, human CD34+ hematopoietic progenitors isolated from the bone marrow of healthy donors (5). We have demonstrated, for the first time, that bone marrow GM and erythroid progenitors may be infected with HHV-6 in vivo. Notably, FISH analyses also showed integration of the viral genome into chromosome 17, band 17p13.3, in a significant proportion of immature myeloid bone marrow progenitor metaphase cells of patient 2. This chromosomal integration site is identical to that previously detected in phytohemagglutinin-stimulated peripheral blood T cells of this and the two remaining patients with high-viral-copy-number latency (20). PFGE analyses of PBMC and granulocyte fractions were consistent with the FISH results in that similar HHV-6-specific restriction fragment patterns were detected in the different cell populations, indicative of HHV-6 integration at a similar genomic location in those cells. On the basis of these findings and collective assessment of PCR studies of sorted hematopoietic cell types, we consider it likely that HHV-6 has infected and integrated its genome into a very early self-renewing bone marrow precursor in the patients with high-copy-number latency. The integrated viral genome on chromosome 17p may then be longitudinally transmitted to cells which differentiate along the committed myeloid and lymphoid pathways without late gene expression (p101 antigen), which would allow it to escape cell-mediated immune response. As the virus can infect early clonogenic progenitors without eliminating their abilities to proliferate and differentiate, HHV-6 can create a pool of infected bone marrow progenitors that can serve as a reservoir of latent virus. However, upon differentiation of these cells to mature granulocytes, at least the expression of one late antigen is switched on in a small percentage of cells. It is conceivable that, in vivo, when the pressure exerted by the immune system declines (i.e., after immunosuppressive therapy), virus released from latently infected granulocytes may be a source of HHV-6 dissemination and possibly of clinical disease. Of note, the pattern of HHV-6 antigen expression is similar to that of HCMV, which replicates and expresses specific mRNA upon differentiation of monocytes to macrophages or in granulocytes (18, 22). It should be noted that HCMV, which causes myelosuppression in immunodeficient patients, is the only other human herpesvirus capable of infecting myeloid progenitors in vitro (24). Moreover, the presence of HCMV DNA has been recently documented either in CFU-GM colonies or in purified CD34+ cells from peripheral blood and bone marrow in vivo (13, 15, 21).
In order to provide further evidence of HHV-6 tropism for bone marrow progenitors, CD34-positive cells were separated by magnetic cell sorting (12), with a purity of 85 to 98%, from leukapheresis products of 10 HHV-6-seropositive autologous peripheral blood progenitor cell (PBPC) transplant patients (patients 4 through 13). DNA was extracted from the CD34-positive and -negative cell fractions and analyzed for the presence of HHV-6 viral DNA by PCR for the ZVH14 region (19). Results are summarized in Table 3. In four patients (patients 6, 7, 12, and 13), HHV-6 DNA was detected in the purified CD34-positive cells. Tenfold dilutions of 105 cells (105, 104, 103, 102, and 10 cells) from the CD34-positive and the CD34-negative cell fractions of these four patients were PCR assayed. The lowest numbers of cells in which HHV-6 DNA was detected by PCR are shown in Table 3. This number in the CD34-positive cell fraction was the same as (for patients 7 and 12) or invariably lower than (for patients 6 and 13) the number in the CD34-negative cell fraction. These titration experiments clearly show that the HHV-6 DNA detected in purified CD34-positive cells was present in this cell population and was not contributed by the contaminating mature leukocytes of the CD34-negative cell fraction. Of note, plasma viremia was not detected in serial serum-plasma samples collected from the 10 patients at the time of leukapheresis and up to 5 months later. Leukapheresis products obtained from two HHV-6-seronegative autologous PBPC transplant patients (patients 14 and 15 [Table 3]) were negative for the presence of HHV-6 DNA. Leukapheresis products were obtained also from four healthy, HHV-6-seropositive allogeneic PBPC donors, and purified CD34-positive and CD34-negative cell fractions (purity, >90%) were analyzed for the presence of HHV-6 DNA by PCR. For two healthy donors (patients 18 and 19 [Table 3]), HHV-6 DNA was detected in both fractions. Tenfold dilutions of 105 cells (105, 104, 103, 102, and 10 cells) from the CD34-positive and the CD34-negative cell fractions were PCR assayed. The lowest number of cells in which HHV-6 DNA was detected in the CD34-positive cell fraction was invariably lower than the number in the CD34-negative cell fraction (Table 3). This finding clearly shows that HHV-6 DNA detected in purified CD34-positive cells was present in this cell population and was not contributed by contaminating mature leukocytes of the CD34-negative cell fraction, even in healthy individuals. In order to verify HHV-6 infection of CD34-positive cells by an alternative approach, individually plucked CFU-GM and BFU-E colonies were isolated from the highly purified CD34-positive cells from two transplant patients (patients 6 and 7) and from one healthy donor (patient 19). Given the low rate of infection in these patients, colonies were pooled and, for each patient, 10 colony samples, each consisting of an average of 20 to 30 pooled colonies, were examined by PCR. For all patients, HHV-6 DNA was detected in four to six samples of pooled colonies. Altogether, these results suggest that HHV-6 infection of mobilized hematopoietic progenitors may occur in vivo, in the absence of concomitant viremia, and also in the more prevalent cases of latent infection characterized by a low copy number of viral DNA, detectable only by PCR, not only in immunosuppressed transplant patients but also in healthy subjects. This finding is significant because HHV-6 may cause myelosuppression and other clinical manifestations in patients who have received transplants (16) and, although the source of the virus is unknown, it has been proposed that most infections are caused by reactivation of a virus already latent in the recipient (16).
TABLE 3.
Detection of HHV-6 DNA in purified CD34-positive and CD34-negative cell fractions from leukapheresis products of HHV-6-seropositive and -seronegative autotransplanted patients, as well as of healthy HHV-6-seropositive allogeneic PBPC donorsa
| Patient | CD34-positive cell fraction | CD34-negative cell fraction |
|---|---|---|
| 4 | Neg | Neg |
| 5 | Neg | Neg |
| 6 | Pos (103) | Pos (104) |
| 7 | Pos (104) | Pos (104) |
| 8 | Neg | Pos (105) |
| 9 | Neg | Pos (105) |
| 10 | Neg | Neg |
| 11 | Neg | Neg |
| 12 | Pos (105) | Pos (105) |
| 13 | Pos (104) | Pos (105) |
| 14 | Neg | Neg |
| 15 | Neg | Neg |
| 16 | Neg | Pos (105) |
| 17 | Neg | Neg |
| 18 | Pos (103) | Pos (105) |
| 19 | Pos (104) | Pos (105) |
Tenfold dilutions of 105 FACS-purified cells (105, 104, 103, 102, and 10 cells) were tested for HHV-6 DNA by PCR, and the minimal numbers of positive cells are given. Neg, negative; Pos, positive.
Acknowledgments
This work was supported by the Associazione Italiana per la Ricerca sul Cancro (AIRC), Milan, Italy, and the Cancer Society of New Zealand. M.L. is a recipient of a fellowship for AIDS research from the Istituto Superiore di Sanità, Rome, Italy.
REFERENCES
- 1.Ablashi D V, Lusso P, Lung C L, Salahuddin S Z, Josephs S F, Llana T, Kramarsky B, Biberfeld P, Markham P D, Gallo R C. Utilization of human hematopoietic cell lines for the propagation and characterization of HBLV (human herpesvirus 6) Int J Cancer. 1988;42:787–791. doi: 10.1002/ijc.2910420526. [DOI] [PubMed] [Google Scholar]
- 2.Beaulieu A D, Paquin R, Gosselin J. Epstein-Barr virus modulates de novo protein synthesis in human neutrophils. Blood. 1995;86:2789–2798. [PubMed] [Google Scholar]
- 3.Challoner P B, Smith K T, Parker J D, MacLeod D L, Coulter S N, Rose T M, Schultz E, Bennett J L, Garber R L, Chang M, Schad P A, Stewart P M, Nowinski R C, Brown J P, Burmer G C. Plaque-associated expression of human herpesvirus 6 in multiple sclerosis. Proc Natl Acad Sci USA. 1995;92:7440–7444. doi: 10.1073/pnas.92.16.7440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Flamand L, Gosselin J, Stefanescu I, Ablashi D, Menezes J. Immunosuppressive effect of human herpesvirus 6 on T-cell functions: suppression of interleukin-2 synthesis and cell proliferation. Blood. 1995;85:1263–1271. [PubMed] [Google Scholar]
- 5.Furlini G, Vignoli M, Ramazzotti E, Re M C, Visani G, La Placa M. A concurrent human herpesvirus-6 infection renders two human hematopoietic progenitor (TF-1 and KG-1) cell lines susceptible to human immunodeficiency virus type-1. Blood. 1996;87:4737–4745. [PubMed] [Google Scholar]
- 6.Josephs S F, Salahuddin S Z, Ablashi D V, Schachter F, Wong-Staal F, Gallo R C. Genomic analysis of the human B-lymphotropic virus (HBLV) Science. 1986;234:601–603. doi: 10.1126/science.3020691. [DOI] [PubMed] [Google Scholar]
- 7.Kishi M, Harada H, Takahashi M, Tanaka A, Hayashi M, Nonoyama M, Josephs S F, Buchbinder A, Schachter F, Ablashi D V, Wong-Staal F, Salahuddin S Z, Gallo R C. A repeat sequence, GGGTTA, is shared by DNA of human herpesvirus 6 and Marek’s disease virus. J Virol. 1988;62:4824–4827. doi: 10.1128/jvi.62.12.4824-4827.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Luppi M, Marasca R, Barozzi P, Ferrari S, Ceccherini-Nelli L, Batoni G, Merelli E, Torelli G. Three cases of human herpesvirus-6 latent infection: integration of viral genome in peripheral blood mononuclear cell DNA. J Med Virol. 1993;40:44–52. doi: 10.1002/jmv.1890400110. [DOI] [PubMed] [Google Scholar]
- 9.Lusso P, Markham P D, Tschachler E T, di Marzo Veronese F, Salahuddin S Z, Ablashi D V, Pahwa S, Korhn K, Gallo R C. In vitro cellular tropism of human B-lymphotropic virus (human herpesvirus 6) J Exp Med. 1988;167:1659–1670. doi: 10.1084/jem.167.5.1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lusso P, Malnati M S, Garzino-Demo A, Crowley R W, Long E O, Gallo R C. Infection of natural killer cells by human herpesvirus 6. Nature. 1993;362:458–462. doi: 10.1038/362458a0. [DOI] [PubMed] [Google Scholar]
- 11.Martin M E D, Thomson B J, Honess R W, Craxton M A, Gompels U A, Liu M-Y, Littler E, Arrand J R, Teo I, Jones M D. The genome of human herpesvirus 6: maps of unit-length and concatemeric genomes for nine restriction endonucleases. J Gen Virol. 1991;72:157–168. doi: 10.1099/0022-1317-72-1-157. [DOI] [PubMed] [Google Scholar]
- 12.Miltenyl S, Muller W, Weichel W, Radbruch A. High gradient magnetic cell separation with MACS. Cytometry. 1990;11:231–238. doi: 10.1002/cyto.990110203. [DOI] [PubMed] [Google Scholar]
- 13.Minton E J, Tysoe C, Sinclair J H, Sissons J G P. Human cytomegalovirus infection of the monocyte/macrophage lineage in bone marrow. J Virol. 1994;68:4017–4021. doi: 10.1128/jvi.68.6.4017-4021.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Morris C, Courtay C, Geurts van Kessel A, ten Hoeve J, Heisterkamp N, Groffen J. Localization of a gamma-glutamyl-transferase-related gene family on chromosome 22. Hum Genet. 1993;91:31–36. doi: 10.1007/BF00230218. [DOI] [PubMed] [Google Scholar]
- 15.Sindre H, Tjonnfjord G E, Rollag H, Rannenberg-Nilsen T, Veiby O P, Beck S, Degrè M, Hestdal K. Human cytomegalovirus suppression of and latency in early hematopoietic progenitor cells. Blood. 1996;88:4526–4533. [PubMed] [Google Scholar]
- 16.Singh N, Carrigan D R. Human herpesvirus-6 in transplantation: an emerging pathogen. Ann Intern Med. 1996;124:1065–1071. doi: 10.7326/0003-4819-124-12-199606150-00007. [DOI] [PubMed] [Google Scholar]
- 17.Talpaz M, Estrov Z, Kantarjian H, Ku S, Foteh A, Kurzrock R. Persistence of dormant leukemic progenitors during interferon-induced remission in chronic myelogenous leukemia. Analysis by polymerase chain reaction of individual colonies. J Clin Investig. 1994;94:1383–1389. doi: 10.1172/JCI117473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Taylor-Wiedeman J, Sissons P, Sinclair J. Induction of endogenous human cytomegalovirus gene expression after differentiation of monocytes from healthy carriers. J Virol. 1994;68:1597–1604. doi: 10.1128/jvi.68.3.1597-1604.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Torelli G, Marasca R, Luppi M, Selleri L, Ferrari S, Narni F, Mariano M T, Federico M, Ceccherini-Nelli L, Bendinelli M, Montagnani G, Montorsi M, Artusi A. Human herpesvirus-6 in human lymphomas: identification of specific sequences in Hodgkin’s lymphomas by polymerase chain reaction. Blood. 1991;77:2251–2258. [PubMed] [Google Scholar]
- 20.Torelli G, Barozzi P, Marasca R, Cocconcelli P, Merelli E, Ceccherini-Nelli L, Ferrari S, Luppi M. Targeted integration of human herpesvirus 6 in the p arm of chromosome 17 of human peripheral blood mononuclear cells in vivo. J Med Virol. 1995;46:178–188. doi: 10.1002/jmv.1890460303. [DOI] [PubMed] [Google Scholar]
- 21.von Laer D, Meyer-Koenig U, Serr A, Finke J, Kanz L, Fauser A A, Neumann-Haefelin D, Brugger W, Hufert F T. Detection of cytomegalovirus DNA in CD34+ cells from blood and bone marrow. Blood. 1995;86:4086–4090. [PubMed] [Google Scholar]
- 22.von Laer D, Serr A, Meyer-König U, Kirste G, Hufert F T, Haller O. Human cytomegalovirus immediate early and late transcripts are expressed in all major leukocyte populations in vivo. J Infect Dis. 1995;172:365–370. doi: 10.1093/infdis/172.2.365. [DOI] [PubMed] [Google Scholar]
- 23.Yamamoto M, Black J B, Stewart J A, Lopez C, Pellett P E. Identification of a nucleocapsid protein as a specific serological marker of human herpesvirus 6 infection. J Clin Microbiol. 1990;28:1957–1962. doi: 10.1128/jcm.28.9.1957-1962.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhuravskaya T, Maciejewski J P, Netski D M, Bruening E, Mackintosh F R, St. Jeor S. Spread of human cytomegalovirus (HCMV) after infection of human hematopoietic progenitor cells: model of HCMV latency. Blood. 1997;90:2482–2491. [PubMed] [Google Scholar]