Abstract
Circular RNA (circRNA) is a type of non-coding RNA that forms a covalently closed, uninterrupted loop. The expression of circRNA differs among cell types and tissues, and various circRNAs are aberrantly expressed in a variety of diseases, including cancer. Aberrantly expressed circRNAs contribute to disease progression by acting as microRNA sponges, functional protein sponges, or novel templates for protein translation. Recent studies have shown that circRNAs are enriched in exosomes. Exosomes are spherical bilayer vesicles released by cells into extracellular spaces that mediate intercellular communication by delivering cargoes. These cargoes include metabolites, proteins, lipids, and RNA molecules. Exosome-mediated cell-cell or cell-microenvironment communications influence the progression of carcinogenesis by regulating cell proliferation, angiogenesis, metastasis as well as immune escape. In this review, we summarize the current knowledge about exosomal circRNAs in cancers and discuss their specific functions in tumorigenesis. Additionally, we discuss the potential value of exosomal circRNAs as diagnostic biomarkers and the potential applications of exosomal circRNA-based cancer therapy.
Keywords: Exosome, circRNAs, Tumorigenesis, Disgnostic biomarker, Cancer treatment
Introduction
Cancer is a leading cause of death worldwide, particularly because of its high morbidity and mortality, and it has caused enormous pain to individuals, imposed a tremendous burden on families and health systems [1]. Conventional blood biomarkers are widely used for cancer diagnosis, but their low sensitivity and specificity limit their application. The early symptoms of many malignant tumors are not obvious, and most patients are diagnosed at an advanced stage of the disease [2, 3]. Therefore, it is crucial to explore new non-invasive biomarkers for the early diagnosis of malignant cancers. Currently, liquid biopsy was developed for detecting novel, highly accurate biomarkers in human body fluids [4, 5]. It is non-invasive, simpler, faster, and more accurate compared to traditional histological biopsy [6]. In addition, more dynamic monitoring of disease progression and recurrence is allowed through repeated sampling via liquid biopsy [7, 8].
Exosomes are spherical bilayer vesicles released by a variety of cells into extracellular spaces. They mediate the cell-cell or cell-environments’ communications by delivering cargoes, such as circular RNAs (circRNAs), microRNAs, mRNAs, DNAs, long non-coding RNAs (lncRNAs), proteins, and lipids [9, 10]. Exosomes are one of the main detection materials for liquid biopsy because they are present in almost all body fluids, including blood, saliva, urine, and cerebrospinal fluid [11]. CircRNA is a type of non-coding RNA with a covalently closed, uninterrupted loop [12]. Due to their special loop structure, circRNAs are relatively stable and not easily degraded when compared to linear RNAs [13]. Moreover, circRNAs are enriched in exosomes, and their expression remarkably changes under physiological or pathological conditions [14, 15]. These studies suggest that circRNAs in the exosomes of body fluids potentially represent novel biomarkers for monitoring cancer progression and predicting prognosis [16].
In this review, we summarize the biological functions of exosomal circRNAs and their significance in cancer progression. We also review the potential clinical applications of exosomal circRNAs as biomarkers in cancer diagnosis, disease judgement, and prognosis observation. In addition, we discuss the potential value of exosome-based circRNA delivery for targeted cancer treatment.
Exosomes
Exosome is one kind of extracellular vehicles with a spherical bilayer membrane structure and a diameter of approximately 50–150 nm [17] (Fig. 1). Traditionally, exosomes are formed from endosomal compartment invaginations and are secreted from the plasma membrane [18]. It was found that almost all types of cells can normally secrete exosomes, which play a crucial role in regulating communication among cells, organs, tissues, and cellular microenvironments. Exosomes contain various molecular constituents, such as circRNAs, microRNAs, DNAs, long non-coding RNAs (lncRNA), proteins, lipids, and so on [19]. The special lipid bilayer structure of exosomes ensures that these contents cannot be degraded and can be easily absorbed by recipient cells [20]. Several studies have reported that the contents of exosomes change remarkably under pathological conditions and that cells can regulate each other’s biological processes via exosomes [21, 22]. For example, tumor-derived exosomes can contribute to angiogenesis and tumor metastasis by delivering these contents to human vascular endothelial cells [23]. Cancer-associated fibroblasts (CAFs) promote chemotherapy resistance of tumor cells via delivering microRNAs through exosomes [24]. CAFs-derived exosomal lncRNA H19 promotes the stemness and chemoresistance of colorectal cancer (CRC) [25]. Moreover, exosomes are widely present in body fluids including blood, saliva, urine, cerebrospinal fluid, and synovial fluid, implying that they could serve as primary detection materials for liquid biopsy [26, 27]. For example, Lydia et al. reported the role of exosomes and circulating miRNAs as a source of liquid biopsy biomarkers in ovarian cancer diagnosis [28]. Xiao et al. showed that circulating plasma exosomal lncRNAs could serve as prospective biomarkers in acute myeloid leukemia [29]. Exosomal circ-SCL38A1 can distinguish bladder cancer patients from healthy individuals, with a diagnostic accuracy of 0.878 [30]. These studies indicate that exosomes, especially exosomal RNA molecules, play an important role in cancer diagnosis and treatment.
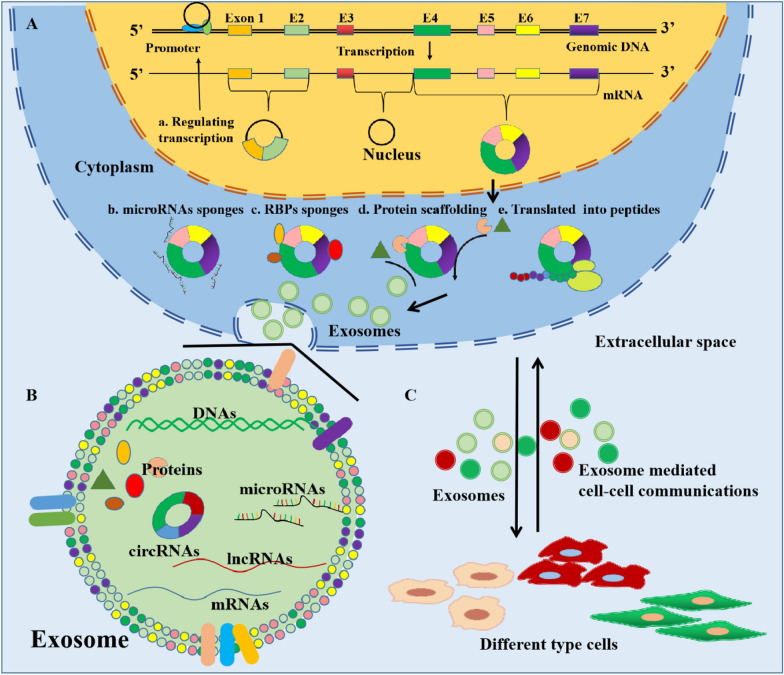
Fig. 1.
Structural schematic diagram of circRNA and exosomes. A The formation process of covalently closed, uninterrupted loop circRNAs and their biological functions; B the lipid bilayer structure of exosomes and its molecules contents; C Exosomes mediate the communications among different type cells
Biological functions of exosomal circRNAs in cancer
General characteristics of circRNA
CircRNA is a type of non-coding RNA formed by back-splicing in which a downstream splice donor site is joined with an upstream splice acceptor site to form a covalently closed, uninterrupted loop [31, 32] (Fig. 1). It was first reported by Dr. Hsu, and it was thought to have no valuable biological functions [33]. However, some recent studies revealed that more than 180,000 circRNAs are present in human transcriptomes and that their expression is associated with both normal cellular biological processes and disease progression [34, 35]. Based on their origin, circRNAs are classified into three major types: circular intronic RNAs, exon-intron circRNAs and exonic circRNAs [12, 36]. CircRNA were confirmed to play multiple roles in the biological processes through acting microRNAs or RNA binding proteins sponges to regulate target gene expression, regulating gene transcription or splicing and acting as templates for protein translation [37–39]. Research has shown that dysregulated circRNAs are associated with the pathogenesis of many human diseases, particularly cancer. Such as, circRNAs has been reported contribute to cancer metastasis and immune escape [40, 41].
Recently, circRNAs were found to be localized to exosomes and capable of being transferred between cells via exosomes, thereby affecting tumor progression. For example, exosome-derived circ-TFDP2 promotes the proliferation of prostate cancer (PC) cells by inhibiting caspase-3-dependent cleavage of PARP1 and DNA damage [42]. Furthermore, Zhao et al. reported that exosome-mediated transfer of circ_0000338 enhances 5-fluorouracil resistance in CRC by regulating microRNA-217/485-3p [43]. Exosomal circ-GSE1 promoteS immune escape of hepatocellular carcinoma (HCC) by inducing the expansion of regulatory T cells via the regulation of miR-324-5p/TGFBR1/Smad3/Tregs axis [44]. Importantly, circRNAs have the potential to serve as biomarkers for cancer diagnosis due to their exosome localization and enrichment. Such as, exosomal circ_0004771 has been reported to be overexpressed in CRC, with area under the curve (AUC) values of 0.86 and 0.88 used to differentiate stage I/II CRC patients and CRC patients from healthy controls, respectively [45].
Exosomal circRNAs and proliferation of cancer cells
Various exosomal circRNAs have been reported to regulate the proliferation of cancer cells. For example, exosomal circ-PDK1 promotes pancreatic cancer (PCa) cell proliferation by sponging miR-628-3p to activate the BPTF/c-Myc axis during hypoxia [46]. Furthermore, exosomal circ-PRRX1 promotes cell proliferation in vitro and tumor growth in vivo by sponging miR-596 and activating the NF-κB signaling pathway in gastric cancer (GC) [47]. According to a previous study, cancer-derived exosomal circ-SERPINE2 is shuttled to tumor-associated macrophages (TAMs), and it enhances IL-6 secretion, leading to increased proliferation of breast cancer cells [48]. TAM-secreted exosomal circ_0020256 promotes the proliferation and progression of cholangiocarcinoma by modulating the miR-432-5p/E2F3 axis [49]. In renal cell carcinoma (RCC), tumor-derived exosomal circ-PPKCI increases tumor cell proliferation via the miR-545-3p/CCND1 signaling pathway [50]. In HCC, adipocyte-derived exosomal circ-DB promotes tumor growth by suppressing miR-34a and activating the USP7/Cyclin A2 signaling pathway [51]. Furthermore, hepatic stellate cell-derived exosomal circ-WDR25 facilitates HCC cell proliferation by regulating the miR-4474-3p/ALOX15 axis [52]. Exosomal circ-RACGAP1 recruiteS PTBP1 to induce RIF1 deacetylation, which then activates the Wnt/β-catenin pathway and prmotes the proliferation of non-small cell lung cancer (NSCLC) cells [53]. Interesting, multiple myeloma (MM)-derived exosomal circ-HNRNPU encodes a novel 603-aa peptide, which regulates the bone marrow microenvironment and promotes cell proliferation [54].
However, Circ-LPAR1 expression in plasma exosomes was decreased in CRC and it suppressed the tumor cell proliferation by suppressing the translation of oncogene BRD4 [55]. Exosomal circ-PTPRA induced CRC cell cycle arrest and inhibited cell proliferation by enriching the level of SMAD4 via competitively binding to miR-671-5p [56]. Chen et al. reported that circ_0051443 was transmitted from normal cells to HCC cells via exosomes and suppressed the cell proliferation and malignant biological progression [57]. In oral squamous cell carcinoma (OSCC), exosomal circ-GDI2 was downregulated and its upregulation weakened the cell proliferation by regulating miR-424-5p/SCAI axis [58]. In addition, Chen et al. reported that tumor-suppressive circ-RHOBTB3 could be excreted out of CRC cells via exosomes and circ-RHOBTB3 suppressed cell growth and metastasis [59]. Besides, exosomal circ-BTG2 or circ_0004658 secreted from RBP-J overexpressed-macrophages inhibited glioma or HCC progression by regulating miR-25-3p/PTEN or miR-499b-5p/JAM3 pathway, respectively [60, 61].
Exosomal circRNAs in metastasis
Exosomal circRNAs also have crucial function in regulating tumor metastasis. Circ-PACRGL is secreted by CRC cells, and acts as a miR-142-3p/ miR-506-3p sponge to activate the TGF-β-related signaling and promote metastasis [62]. In HCC, exosome-transmitted circMMP2 induced metastasis by sponging miR-136-5p and increasing MMP2 expression [63]. Moreover, exosomal circRAPGEF5 promoted the metastasis of lung adenocarcinoma through the miR-1236-3p/ZEB1 axis [64]. Tumor-derived exosomal circPSMA1 facilitated the metastasis in triple-negative breast cancer through the regulation of miR-637/Akt1/β-catenin regulatory axis [65]. Furthermore, exosomal circ_0081234 promoted the epithelial-mesenchymal transition (EMT) of PC cells [66]. Circ_0003028 induced EMT of HCC cells by exosome pathway via microRNA-498/ODC1 signaling [67]. And exosomal circ_007293 promoted EMT of papillary thyroid carcinoma cells via the regulation of the miR-653-5p/PAX6 axis [68]. In addition, the metastatic ability of HCC cells could be enhanced by transferring exosomal circRNA-100,338 to human umbilical vein endothelial cells (HUVECs), and promoting angiogenesis [69]. In GC, tumor-derived exosomal circ_0044366 promoted tube formation of HUVECs and enhanced cancer migration [70]. In ovarian cancer, exosomal circ-NFIX increased angiogenesis via miR-518a-3p/TRIM44/JAK/STAT1 pathway [71]. In esophageal squamous carcinoma, exosomal circ_0026611 contributed to LNM by interacting with N-α-acetyltransferase 10 (NAA10) to inhibit NAA10-mediated PROX1 acetylation [72].
However, Chen et al. reported that CAFs directly transferred circ-IFNGR2 into ovarian cancer cells and suppressed metastasis by activating miR-378/ST5 [73]. Moreover, bone marrow mesenchymal stem cell-derived exosomal circ_0006790 suppressed metastasis of pancreatic ductal adenocarcinoma by binding to CBX7 and regulating S100A11 DNA methylation [74]. Lin et al. found that exosomal circ_0072088 suppressed migration and invasion of hepatic carcinoma cells by regulating miR-375/MMP-16 [75]. In GC, the expression of exosomal circ-ITCH and circ-STAU2 were significantly downregulated, they suppressed the metastasis of GC by regulating miR-199a-5p/Klotho axis or miR-589/ CAPZA1 respectively [76, 77].
Exosomal circRNAs in drug resistance
Exosomal circRNAs were associated with the drug resistance of cancers. Exosomal circ_0076305 promoted cisplatin (DDP) resistance of non-small cell lung cancer cell (NSCLC) by enhancing ABCC1 expression [78]. Circ-VMP1 and circ_0014235 were elevated in DDP-resistant NSCLC exosomes, they facilitated DPP resistance by regulating miR-524-5p/METTL3/SOX2 or miR-520a-5p/CDK4 axis, respectively [79]. In osteosarcoma, exosomal circ_103801 conferred DDP resistance by increasing the expression of MRP1 and p-glycoprotein [80]. Warburg effect promoted temozolomide (TMZ) resistant glioma cells releasing exosomal circ_0072083, which induced TMZ resistance of sensitive cells by regulating miR-1252-5p/NANOG [81]. Circ-ZNF91 was remarkably increased in exosomes of PCa under hypoxia condition and promoted gemcitabine resistance of normoxic PCa cells via regulating miR-23b-3p/SIRT1 and enhancing glycolysis [82]. In neuroblastoma, exosomal circ-DLGAP4 enhanced glycolysis and doxorubicin resistance via miR-143-HK2 axis [83]. Oxaliplatin-resistant CRC cells delivered exosomal circ_0005963 to sensitive cells, promoted drug resistance by miR-122 sponging and PKM2 upregulation [84]. Furthermore, exosomal circ_0091741 promoted oxaliplatin resistance of GC cells via the miR-330-3p/ TRIM14/Dvl2/Wnt/β-catenin pathway [85]. Exosomal circ-SFMBT2 and circ-XIAP were upregulated in docetaxel-resistant PC cells, their knockdown enhanced docetaxel sensitivity by regulating miR-136-5p/TRIB1 or miR-1182/TDP52 axis [86, 87]. Pan et al. reveled that exosomal circATG4B induced oxaliplatin resistance in CRC by encoding a novel protein to increase autophagy [88].
However, Xu et al. found that exosomal circ-FBXW7 led resistant cells sensitive to oxaliplatin and suppressed oxaliplatin efflux via sponging miR-18b-5p in CRC [89]. Moreover, circRNA-CREIT could be packaged into exosomes and disseminate doxorubicin sensitivity among TNBC cells by destabilizing PKR [90]. In liver cancer, transarterial chemoembolization increased the expression of exosomal circ-G004213, which promoted DDP sensitivity by regulating miR-513b-5p/PRPF39 axis [91].
We summarized exosomal circRNAs and their function in tumorigenesis in Table 1.
Table 1.
Exosomal circRNAs and their function in tumorigenesis
| Tumor type | circRNA | Target molecules | Function | References |
|---|---|---|---|---|
| NSCLC | Circ-RACGAP1 | Wnt/β-catenin | Proliferation | [53] |
| NSCLC | Circ_0076305 | miR-186-5p/ABCC1 | DDP resistance | [78] |
| NSCLC |
Circ-VMP1 Circ_0014235 |
miR-524-5p/SOX2 miR-520a-5p/CDK4 |
DDP resistance | [79] |
| NSCLC | Circ-STAB2 | miR-330-5p/PEAK1 | Progression | [92, 93] |
| NSCLC | Circ_0007385 | miR-1253/FAM83A | Proliferation, stemness | [94] |
| NSCLC | Circ_0008717 | miR-1287-5p/PAK2 | Tumorigenicity | [95] |
| NSCLC | Circ-ARHGAP10 | miR-638/FAM83F | Progression | [96] |
| NSCLC | Circ_102481 | miR-30a-5p/ROR1 | EGFR-TKIs resistance | [97] |
| NSCLC | Circ-PLK1 | miR-1294/HMGA1 | Progression | [98] |
| NSCLC | Circ_0014235 | miR-520a-5p/CDK4 | DDP resistance | [99] |
| NSCLC | Circ_0002130 | miR-498 | Osimertinib resistance | [100] |
| NSCLC | Circ-CCDC134 | miR-625-5p/NFAT5 | Progression | [101] |
| Lung cancer | Circ-DNER | miR-139-5p/ITGB8 | Paclitaxel resistance | [102] |
| LUAD | CircRAPGEF5 | miR-1236-3p/ZEB1 | Metastasis | [64] |
| CRC | Circ-PACRGL | miR-142-3p/miR-506-3p | Metastasis | [62] |
| CRC | Circ_0005963 | miR-122 | Oxaliplatin resistance | [84] |
| CRC | CircATG4B | Autophagy | Oxaliplatin resistance | [88] |
| CRC | Circ_0007334 | miR/KLF12 | Progression | [103] |
| CRC | Circ-COG2 | miR-1305/TGF-β2/smad3 | Progression | [104] |
| CRC | Circ-FMN2 | miR-338-3p/MSI1 | Progression | [105] |
| CRC | CircCOL1A2 | miR-665/LASP1 | Progression | [106] |
| CRC | Circ_0005615 | miR-873-5p/FOSL2 | Progression | [107] |
| CRC | Circ_0000395 | miR-432-5p/MYH9 | Progression | [108] |
| CRC | Circ-TUBGCP4 | miR-146b-3p/PDK/Akt | Metastasis | [109] |
| CRC | Circ-PABPC1 | miR-874/microRNA-1929 | Metastasis | [110] |
| CRC | Circ-133a | miR-133a/GEF-H1/RhoA | Metastasis | [111] |
| HCC | Circ-DB | miR-34a/USP7/Cyclin A2 | Proliferation | [51] |
| HCC | Circ-WDR25 | miR-4474-3p/ALOX15 | Proliferation | [52] |
| HCC | CircMMP2 | miR-136-5p/MMP2 | Metastasis | [63] |
| HCC | Circ_0003028 | miR-498/ODC1 | EMT process | [67] |
| HCC | Circ_100338 | Angiogenesis | Metastasis | [69] |
| HCC | Circ-Cdr1as | miR-1270 | Progression | [112] |
| HCC | Circ-TTLL5 | miR-136-5p/KIAA1522 | Metastasis | [113] |
| HCC | Circ-SORE | YBX1 | Sorafenib resistance | [114] |
| HCC | Circ-PAK1 | YAP | Lenvatinib resistance | [115] |
| HCC | Circ-ZFR | STAT3/NF-κB pathway | DDP resistance | [116] |
| Breast cancer | Circ-SERPINE2 | / | Proliferation | [48] |
| Breast cancer | CircPSMA1 | miR-637/Akt1/β-catenin | Metastasis | [65] |
| Breast cancer | Circ-MMP11 | miR-153-3P/ANLN | Lapatinib resistance | [117] |
| Breast cancer | CCirc-UBE2D2 | miR-200a-3p | Tamoxifen resistance | [118] |
| Breast cancer | Circ-CARM1 | miR-1252-5p/PFKFB2 | Glycolysis, progression | [119] |
| Breast cancer | Circ-EGFR | miR-1299/EGFR | Pirarubicin resistance | [120] |
| Gastric cancer | Circ-PRRX1 | miR-596 | Proliferation | [47] |
| Gastric cancer | Circ_0044366 | / | Metastasis | [70] |
| Gastric cancer | Circ_0091741 | miR-330-3p/ TRIM14 | Oxaliplatin resistance | [85] |
| Gastric cancer | Circ-NRIP1 | miR-145-5p/AKT1/mTOR | Metastasis | [121] |
| Gastric cancer | Circ_0001789 | miR-140-3p/PAK2 | Progression | [122] |
| Gastric cancer | Circ_0063562 | miR-449a/SHMT2 | DDP resistance | [123] |
| Gastric cancer | Circ-PVT1 | miR-301-5p/YAP1 | DDP resistance | [124] |
| Gastric cancer | Circ-LDLRAD3 | miR-588/SOX5 | DDP resistance | [125] |
| Gastric cancer | Circ_0032821 | miR-515-5p/SOX9 | Oxaliplatin resistance | [126] |
| Glioma | Circ_0072083 | miR-1252-5p/NANOG | TMZ resistance | [81] |
| Glioma | Circ-WDR62 | miR-370-3p/MGMT | TMZ resistance | [127] |
| Glioma | Circ-GLS3 | miR − 548 m/MED31 | TMZ resistance | [128] |
| Glioma | Circ_0043949 | miR-876-3p/ITGA1 | TMZ resistance | [129] |
| Glioblastoma | Circ-AHCY | miR-1294/ Wnt/β-catenin | Proliferation | [130] |
| Glioblastoma | Circ_0012381 | miR-340-5p/CCL2/CCR2 | Proliferation | [131] |
| Glioblastoma | Circ-KIF18A | FOXC2/PI3K/AKT | Angiogenesis | [132] |
| Prostate cancer | Circ_0081234 | / | EMT process | [66] |
| Prostate cancer | Circ-SFMBT2 | miR-136-5p/TRIB1 | Docetaxel resistance | [86] |
| Prostate cancer | Circ-XIAP | miR-1182/TDP52 | Docetaxel resistance | [87] |
| Prostate cancer | Circ-KDM4A | miR-338-3p/CUL4B | Malignancy | [133] |
| Ovarian cancer | Circ-NFIX | miR-518a-3p/TRIM44 | Angiogenesis | [71] |
| Ovarian cancer | Circ-PIP5K1A | miR-942/NFIB | DDP resistance | [134] |
| Ovarian cancer | Circ-Foxp1 | miR-22/miR-150-3p | DDP resistance | [135] |
| Ovarian cancer | Circ_0007841 | miR-532-5p/NFIB | DDP resistance | [136] |
| PCa | Circ-PDK1 | miR-628-3p/BPTF/c-Myc | Proliferation | [46] |
| PCa | Circ-ZNF91 | miR-23b-3p/SIRT1 | Gemcitabine resistance | [82] |
| PCa | Circ-IARS | miR-122 | Metastasis | [137] |
| EC | Circ_0000337 | miR-337-3p | DDP resistance | [138] |
| CCA | Circ_0020256 | miR-432-5p/E2F3 | Proliferation | [49] |
| RCC | Circ-PRKCI | miR-545-3p/CCND1 | Proliferation | [50] |
| MM | Circ-HNRNPU | / | Proliferation | [54] |
| PTC | Circ_007293 | miR-653-5p/PAX6 | EMT process | [68] |
| ESCC | Circ_0026611 | / | LNM | [72] |
| Osteosarcoma | Circ_103801 | / | DDP resistance | [80] |
| Neuroblastoma | Circ-DLGAP4 | miR-143-HK2 | Doxorubicin resistance | [83] |
| Cervical cancer | Circ_0074269 | miR-485-5p/TUFT1 | DDP resistance | [139] |
| Melanoma | Circ_0001005 | miRs sponges | Vemurafenib resistance | [140] |
| NPC | Circ-PARD3 | miR-579-3p/SIRT1 | Cisplatin resistance | [141] |
| CCA | Circ-CCAC1 | EZH2 | Angiogenesis | [142] |
Exosomal circRNAs in tumor immunity
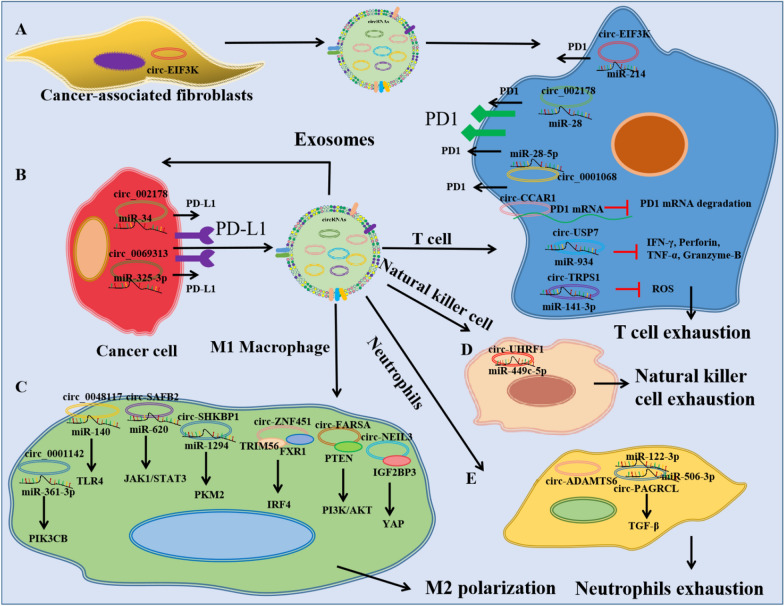
Exosomal circRNAs mediate the communication between tumor cells and immune cells (Fig. 2). In bladder cancer, exosome-derived circ-TRPS1 promotes CD8 + T cell exhaustion and the malignant phenotype by sponging miR-141-3p [143]. In NSCLC, upregulated plasma exosomal circ-USP7 inhibites CD8 + T cell function by sponging miR-934 and increasing SHP2 expression [144]. In LUAD, exosomal circ_002178 can be delivered to CD8 + T cells to induce PD1 expression and T cell exhaustion [145]. In ovarian cancer, exosomal circ-0001068 can be delivered to T cells and induced PD1 expression by sponging miR-28-5p [146]. In HCC, exosomal circ-CCAR1 promotes CD8 + T cell dysfunction by stabilizing the PD1 protein [147]. In OSCC, the transfer of circ_0069313 to Treg cells promotes immune escape by inhibiting miR-325-3p-induced Foxp3 degradation [148]. Moreover, CAF-derived exosomal circ-EIF3K increases the PD-L1 expression in CRC [149].
Fig. 2.
Exosomal circRNAs mediate the communication between tumor cells and immune cells. A The effect of cancer-associated fibroblasts-derived exosomal circRNAs on T cells; B The effect of cancer cell-derived exosomal circRNAs on tumor cells; C The effect of cancer cell-derived exosomal circRNAs on Macrophage cells; D The effect of cancer cell-derived exosomal circRNAs on Natural killer cells; E: The effect of cancer cell-derived exosomal circRNAs on Neutrophils
In NSCLC, exosomal circ-SHKBP1 or circ-FARSA promotes M2 polarization and cancer progression via the miR-1294/PKM2 or PTEN/PI3K/AKT pathway [150, 151]. In glioma, exosomal circ-NEL3 induces macrophage immunosuppressive polarization by stabilizing the oncogenic protein IGF2BP3 [152]. In LUAD, exosomal circ-ZNF451 restrains anti-PD1 treatment by polarizing macrophages and complexing with TRIM56 and FXR1 [153]. In breast cancer, exosomal circ_0001142 is released by cancer cells under endoplasmic reticulum stress, and it induces M2 polarization of macrophages [154]. In RCC, exosomal circ-SAFB2 reshapes the tumor environment, mediates M2 macrophage polarization, and promotes tumor progression [155]. In esophageal squamous cell carcinoma, tumor-derived exosomal circ_0048117 facilitates M2 macrophage polarization by regulating microRNA-140/TLR4 axis [156].
In HCC, cancer cells secrete exosomal circ-UHRF1, which induces natural killer cell exhaustion and promotes immune therapy resistance by regulating the miR-449c-5p/TIM3 axis [157]. CRC-derived exosomal circ-PACRGL regulates the differentiation of N1/N2 neutrophils [62]. Wang et al. reported that upregulated expression of plasma exosomal circ-ADAMTS6 is positively related to neutrophil extracellular traps in cholangiocarcinoma [158].
Potential clinical applications of exosomal circRNAs
Exosomal circRNAs in cancer diagnosis
CircRNAs have a special stable tertiary structure, and it has been reported that their expression is not significantly altered after 24 h of incubation at room temperature [14]. Furthermore, circRNAs were found to be dysregulated under pathological conditions and enriched in exosomes, which could be detected in body fluids such as blood, serum, urine, saliva, and cerebrospinal fluid [14, 15]. These features indicate that exosomal circRNAs can serve as biomarkers for cancer diagnosis. Xu et al. found that the expression of circ_0109046 and circ_0002577 were higher in exosomes isolated from serum samples of patients with stage III endometrial adenocarcinoma compared to healthy controls [159]. Xu et al. reported that circ-SHKBP1 is a promising circulating biomarker for GC diagnosis and prognosis due to its upregulation in serum and positive relationship with advanced TNM stage and poor survival [160]. Deng et al. reported that oral squamous cell carcinoma patients with higher expression of exosomal circ_047733 showed a lower risk of LNM [161]. Plasma exosome-derived circ_0055202, circ_0074920, and circ_0043722 are upregulated in glioblastoma multiforme and associated with tumor progression [162]. Furthermore, Hong et al. revealed that circ_0006220 and circ_0001666 are highly expressed in exosomes in the plasma of PCa patients compared to healthy controls and that they are associated LNM and tumor size. The AUC values were 0.7817 for circ_0006220, 0.8062 for circ_0001666, and 0.884 for the combined diagnosis [163]. The expressions of circ_0001492, circ_0001439, and circ_0000896 were significantly higher in the serum exosomes of LUAD patients, and the combination of these exosomal circRNAs had diagnostic sensitivity and specificity with an AUC value of 0.805 [164]. Furthermore, circ_0028861 was identified as a novel biomarker for HCC diagnosis, with an AUC of 0.79, and was capable of detecting small (AUC = 0.81), early-stage (AUC = 0.82), and AFP-negative (AUC = 0.78) tumors [165]. What’s more, exosomal circ_0015286 has an oncogenic function in GC, and its expression is closely associated with tumor size, TNM stage, LNM, and overall survival of GC patients [166]. Besides, clinical data have shown that exosomal circ_0000437 is enriched in the serum of GC patients and associated with LNM [167]. In addition, Wang et al. identified circ-SLC38A1 in the serum exosomes of bladder cancer patients, which could distinguish bladder cancer patients from healthy individuals with a diagnostic accuracy of 0.878 [30].
Other exosomal circRNAs that could serve as potential biomarkers for cancer diagnosis are summarized in Table 2.
Table 2.
Exosomal circRNAs in body fluids for cancer diagnosis
| Cancer | circRNAs | Level | Function | References |
|---|---|---|---|---|
| CRC | Circ-LPAR1 | Down | Diagnostic biomarker (AUC 0.875) | [55] |
| CRC | Circ-GAPVD1 | Up | Diagnostic biomarker (AUC 0.7662) | [168] |
| CRC | Circ-HIPK3 | Up | Diagnostic biomarker (AUC 0.771) | [169] |
| CRC | Circ-PNN | Up | Early-stage diagnosis (AUC 0.854) | [170] |
| GC | Circ_0015286 | Up | Diagnostic biomarker | [166] |
| GC | Circ_0000437 | Up | Associated with LNM | [167] |
| GC | Circ-CDR1as | Up | Diagnostic biomarker (AUC 0.536) | [171] |
| GC | Circ_0065149 | Down | Early diagnosis and prognosis prediction (AUC 0.64) | [172] |
| GC | Circ-KIA1244 | Down | TNM stage and lymphatic metastasis (AUC 0.7481) | [173] |
| GC | Circ_0000419 | Down | Diagnostic biomarker (AUC 0.84) | [174] |
| BC | Circ-MMP11 | Up | Diagnostic biomarker (AUC 0.9444) | [117] |
| BC | Circ-HIF1A | Up | Diagnostic biomarker (AUC 0.897) | [175] |
| BC | Circ_0000615 | Up | Diagnostic biomarker (AUC 0.904) | [176] |
| NSCLC | Circ_0047921, Circ_0056285, Circ_0007761 | - | Diagnostic biomarker in the Chinese population (AUC 0.89, 0.820) | [177] |
| NSCLC | Circ_0048856 | Up | Diagnostic biomarker (AUC 0.943) | [178] |
| NSCLC | Circ_0069313 | Up | Diagnostic biomarker (AUC 0.749) | [179] |
| NSCLC | Circ-ERBB2IP | Up | Positively correlated with malignant (AUC 0.9168) | [180] |
| LUAD | Circ_0001492, Circ_0001439, Circ_0000896 | Up | Diagnostic biomarker (AUC 0.805) | [164] |
| LUAD | Circ_0056616 | Up | Biomarker for lymph node metastasis (AUC 0.812) | [181] |
| LUAD | Circ_0013958 | Up | TNM stage and lymphatic metastasis (AUC 0.815) | [182] |
| LUSC | Circ_0014235, Circ_0025580 | Up | Diagnostic biomarker (AUC 0.8) | [183] |
| Lung cancer | Circ_0002490, Circ_0087357, Circ_0004891, Circ_0074368 | Down | Diagnostic biomarker (AUC 0.833, 0.793, 0.773, 0.730) | [184] |
| HCC | Circ_0051443 | Down | Diagnostic biomarker (AUC 0.8089) | [57] |
| HCC | Circ_0028861 | Down | Diagnostic biomarker | [165] |
| HCC | Circ-SMARCA5 | Down | Diagnostic biomarker (AUC 0.862) | [185] |
| HCC | Circ_0006602 | Up | Diagnostic biomarker (AUC 0.907) | [186] |
| HCC | Circ_0004001, Circ_0004123, Circ_0075792 | – | Positively correlated with the TNM stage and tumor size | [187] |
| ESCC | Circ_0026611 | Up | Lymph node-metastatic biomarker (AUC 0.724) | [188] |
| ESCC | Circ_0001946 | Up | Predict the recurrence and prognosis (AUC 0.894) | [189] |
| MM | Circ-MYC | Up | Recurrence and Bortezomib resistance (AUC 0.924) | [190] |
| Ovarian | Circ_0001068 | Up | Diagnostic biomarker (AUC 0.9697) | [146] |
| AC |
Circ_0109049 Circ_0002577 |
Up | Diagnostic stage III biomarker | [159] |
| OSCC | Circ_047733 | Down | Negatively with LNM | [161] |
| GBM | Circ_0055202, Circ_0074920, Circ_0043722 | Up | Predict the tumor progression | [162] |
| PCa | Circ_0006220 Circ_0001666 | Up | Diagnostic biomarker (AUC 0.884) | [163] |
Exosome-based circRNA delivery for cancer therapy
Exosomes can transport RNA molecules and deliver therapeutic drugs to cancer cells with good histocompatibility, high efficiency, and low cytotoxicity. Researchers have reported that some circRNAs have tumor suppressor functions, and the therapeutic delivery of exosomal circRNAs could suppress the proliferation, metastasis, drug resistance and progression of malignant tumors. Circ-EPB41L2 is downregulated in the exosomes of CRC patients, and exosome-mediated circ-EPB41L2 suppresses tumor progression by regulating the PTEN/AKT signaling pathway [191]. Zhang et al. reported that exosome-delivered circ-STAU2 inhibites the progression of GC by targeting the miR-589/CAPZA1 axis [77]. Moreover, Sang et al. reported that the exosomal transmission of circ-RELL1 suppresses the proliferation, invasion, and migration of GC cells [192]. Circ-DIDO1 is downregulated in GC, and circ-loaded, RGD-modified engineering exosomes significantly inhibit the proliferation, migration, and invasion of GC cells both in vivo and in vitro [193]. Furthermore, Circ-CREIT is aberrantly downregulated in doxorubicin-resistant TNBC cells and is associated with a poor prognosis. The exosomal transmission of circ-CREIT could disseminate doxorubicin sensitivity among these cells by destabilizing PKR [90]. Circ_0094343 is significantly downregulated in CRC, and exosome-carried circ_0094343 playes a tumor suppressor role and improves the chemosensitivity of tumor cells to 5-fluorouracil, oxaliplatin and doxorubicin [194].
Tumor microenvironment-associated cells also play tumor suppressor roles by delivering exosomal circRNAs to cancer cells. For example, CAF-derived exosomes deliver circ-IFNGR2 to ovarian cancer cells and inhibit malignant tumor progression by regulating the microRNA-378/ST5 axis [73]. Moreover, RBP-J-overexpressed- macrophage-derived exosomal circ-BTG2 or circ_0004658 inhibit glioma or HCC progression [60, 61]. Furthermore, Yao et al. reported that exosomal circ_0030167 derived from bone marrow-derived mesenchymal stem cells (BM-MSCs) exhibit significant tumor suppressor function in PCa by sponging microRNA-338-3p and targeting the Wif1/Wnt8/β-catenin axis [195]. BM-MSC-derived exosomal circ_0006790 inhibits growth, metastasis, and immune escape in pancreatic ductal adenocarcinoma [74].
Besides, Nanoparticles or exosomes mediated circRNAs silencing also a potential strategy for cancer treatment. For example, nanoparticles delivery si-circ-ROBO1 to hepatocellular carcinoma cells circ-ROBO1 inhibited tumor progression by modulating circ-ROBO1/miR-130a-5p/CCNT2 Axis[196]. And natural compound matrine blocked circ-SLC7A6 exosome secretion from CAFs, and then inhibited CRC cell proliferation and invasion[197]. These studies indicate that exosomal delivery of tumor-suppressing circRNAs or exosomal circRNAs-based engineering of exosomes or exosome circRNAs release inhibition may be novel cancer therapies.
The recent data reporter about “exosome-based circRNA delivery for cancer therapy” were summarized in Table 3.
Table 3.
Exosome-based circRNA delivery for cancer therapy
| Cancer | circRNAs | Source | Function | References |
|---|---|---|---|---|
| SCLC | Circ-SH3PXD2A | Circ-SH3PXD2A-overexpressing cells | Decreased chemoresistance and cell proliferation | [198] |
| Lung | Circ-RABL2B | Circ-RABL2B-overexpressing cells | Impoverished stemness, and promoted erlotinib sensitivity | [199] |
| CRC | Circ-PTPRA | Circ-PTPRA transfected cells | Inhibited tumorigenesis and promoted radiosensitivity | [56] |
| CRC | Circ-RHOBTB3 | ASOs treated CRC | Inhibited CRC growth and metastasis | [59] |
| CRC | Circ-FBXW7 | circ-FBXW7-transfected FHC cells | Ameliorated chemoresistance to oxaliplatin | [89] |
| CRC | Circ-EPB41L2 | Circ-EPB41L2 transfected cells | Inhibited proliferation and metastasis | [191] |
| CRC | Circ_0094343 | NCM460 | Improved chemosensitivity | [194] |
| HCC | Circ_0051443 | HL-7702 cell | Suppressed tumor progression | [57] |
| HCC | Circ_0004658 | RBP-J-overexpressed- macrophage | Inhibited the progression | [61] |
| HCC | Circ_0072088 | HCC cells | Suppressed the metastasis | [75] |
| HCC | Circ-G004213 | / | Promoted cisplatin sensitivity | [91] |
| PDAC | Circ_0006790 | BMSC | Inhibited growth, metastasis, and immune escape | [74] |
| PDAC | Circ_0012634 | Pancreatic ductal epithelial cel1l | Restrained PDAC progression | [200] |
| Gastric | Circ-ITCH | Circ-ITCH-transfected cells | Suppressed the metastasis | [76] |
| Gastric | CircSTAU2 | GES-1 cells | Inhibited the progression | [77] |
| Gastric | Circ_0017252 | GC cells | Inhibited macrophage M2 polarization | [201] |
| Gastric | Circ-RELL1 | / | Suppressed the malignant behavior | [192] |
| Gastric | Circ-DIDO1 | Circ-DIDO1 transfected 293T | Suppressed tumor progression | [193] |
| Glioma | Circ-BTG2 | RBP-J-overexpressed- macrophage | Inhibited the progression | [60] |
| Ovarian | CircIFNGR2 | CAF | Inhibited the malignant progression | [73] |
| PCa | Circ_0030167 | BMSCs | Inhibited the stemness | [195] |
| TNBC | Circ-CREIT | / | Overcome doxorubicin resistance | [90] |
| OSCC | Cicr-GDI2 | Circ-GDI2-transfected CAL27 cells | Suppressed tumor progression | [58] |
| RCC | Circ-SPIRE1 | Circ-SPIRE1 over-expressed cells | Suppressed angiogenesis and metastasis | [202] |
| NPC | Circ-FIP1L1 | Guggulsterone treated HNE1 cells | Repressed HUVECs angiogenesis | [203] |
Discussion and conclusion
In this review, we comprehensively summarized current knowledge about the crucial function of exosomal circRNAs in tumor cell proliferation, metastasis, drug resistance, and progression. Several studies have mainly focused their research on tumor-derived exosomal circRNAs, but cancer cells exist in a complex and comprehensive microenvironment, and tumor progression involves the participation of various types of cells. Further research needs to focus on the role of exosomal circRNAs that derived from CAF, TAM, and other immune cells in tumor initiation, development, and progression.
Although numerous studies have revealed the abundance and diverse contributions of exosomal circRNAs to tumorigenesis, many questions remain unanswered. CircRNAs are mainly synthesized and retained in the nucleus, and the regulatory mechanisms of exosomes localization of circRNAs are not fully understood. A recent study reported that N6-methyladenosine modification facilitates the cytoplasmic export of circRNAs [204], indicating that m6A modification may regulate the exosome sorting of circRNAs. Moreover, it has been reported that some RNA-binding proteins, such as Argonaute and mannose-binding lectin can bind to circRNAs [205], and exosome sorting of microRNAs is dependent on the ESCRT complex, with Ago2 being the critical protein [206], indicating that exosome-associated RBPs may regulate the exosome sorting of circRNAs. In addition, hnRNPA2B1 mediates the exosome sorting of circ-NEIL3 and circ-CCAR1 [147, 152]. Additional studies are needed to illustrate the regulatory mechanisms of exosomes localization of circRNAs.
Currently, a large number of studies have proved that exosomal circRNAs have a potential value in cancer diagnosis and prognosis observation due to their highly conserved structure and tissue-specific expression patterns. More experimental verification, larger cohorts, and sufficient theoretical results are warranted to prove the clinical applicable of exosomal circRNAs as biomarkers. Besides, research into engineered exosomes as an approach for targeted cancer treatment is still in its infancy, future efforts should focus on identifying specific exosomal circRNAs and developing efficient and safe engineered exosomes for clinical application.
In conclusion, we comprehensively reviewed current knowledge about the crucial function of exosomal circRNAs in cancer progression, discussed their potential value in cancer diagnosis and prognosis observation, and described the potential utility of engineered exosomes for targeted cancer treatment.
Acknowledgements
We thank Bullet Edits Limited for the linguistic editing and proofreading of the manuscript.
Abbreviations
- circRNA
Circular RNA
- lncRNAs
Long non-coding RNAs
- AUC
Area under the curve
- CAFs
Cancer-associated fibroblasts
- CRC
Colorectal cancer
- EC
Esophageal cancer
- BC
Breast cancer
- PC
Prostate cancer
- HCC
Hepatocellular carcinoma
- PCa
Pancreatic cancer
- GC
Gastric cancer
- TAMs
Tumor-associated macrophages
- RCC
Renal cell carcinoma
- NSCLC
Non-small cell lung cancer
- SCLC
Small cell lung cancer
- MM
Multiple myeloma
- EMT
Epithelial-mesenchymal transition
- HUVECs
Human umbilical vein endothelial cells
- NAA10
N-α-acetyltransferase 10
- TMZ
Temozolomide
- BM-MSCs
Bone marrow-derived mesenchymal stem cells
- PDAC
Pancreatic ductal adenocarcinoma
- NPC
Nasopharyngeal carcinoma
- CCA
Cholangiocarcinoma
- AC
Endometrial adenocarcinoma
- OSCC
Oral squamous cell carcinoma
- GBM
Glioblastoma multiforme
- PTC
Papillary thyroid carcinoma
- ESCC
Esophageal squamous carcinoma
- CCA
Cholangiocarcinoma
Author contributions
Jiaji Yue, Houyin Shi and Yang Liu performed the literature search, Qian Yi and Weichao Sun prepared the first draft of the manuscript; Qian Yi and Weichao Sun wrote and edited the manuscript; Jianguo Feng supervised and Wei Sun polished the manuscript. All of the authors have read and agreed to published version of the manuscript.
Funding
This work was supported by funds from the National Natural Sciences Foundation of China (No.82003126), Shenzhen Science and Technology Projects (No. JCYJ20210324103604013), Scientific Research Foundation of Southwest Medical University (No. 2021ZKMS009; No. 2021ZKZD011), Luzhou Science and Technology Program (No. 2021-JYJ-71), Sichuan Science and Technology Program (No. 2022NSFSC1368; No. 2022NSFSC1594; No.2022YFS0609).
Availability of data and materials
Not applicable.
Declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Jianguo Feng, Email: fengjianguo@swmu.edu.cn.
Weichao Sun, Email: weichaosunshine@163.com.
References
- 1.Collaborators GBDRF Global, regional, and national comparative risk assessment of 79 behavioural, environmental and occupational, and metabolic risks or clusters of risks, 1990–2015: a systematic analysis for the global burden of disease study 2015. Lancet. 2016;388(10053):1659–724. doi: 10.1016/S0140-6736(16)31679-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schiffman JD, Fisher PG, Gibbs P. Early detection of cancer: past, present, and future. Am Soc Clin Oncol Educ Book. 2015;57–65. 10.14694/EdBook_AM.2015.35.57. [DOI] [PubMed]
- 3.Torre LA, et al. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65(2):87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 4.Reimers N, Pantel K. Liquid biopsy: novel technologies and clinical applications. Clin Chem Lab Med. 2019;57(3):312–316. doi: 10.1515/cclm-2018-0610. [DOI] [PubMed] [Google Scholar]
- 5.Alix-Panabieres C, Pantel K. Liquid biopsy: from discovery to clinical application. Cancer Discov. 2021;11(4):858–873. doi: 10.1158/2159-8290.CD-20-1311. [DOI] [PubMed] [Google Scholar]
- 6.Mathai RA, et al. Potential utility of liquid biopsy as a diagnostic and prognostic tool for the assessment of solid tumors: implications in the precision oncology. J Clin Med. 2019;8(3):373. doi: 10.3390/jcm8030373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Heitzer E, et al. Current and future perspectives of liquid biopsies in genomics-driven oncology. Nat Rev Genet. 2019;20(2):71–88. doi: 10.1038/s41576-018-0071-5. [DOI] [PubMed] [Google Scholar]
- 8.Pardini B, et al. Noncoding RNAs in extracellular fluids as cancer biomarkers: the new frontier of liquid biopsies. Cancers (Basel). 2019;11(8):1170. doi: 10.3390/cancers11081170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kok VC, Yu CC. Cancer-derived exosomes: their role in cancer biology and biomarker development. Int J Nanomed. 2020;15:8019–36. doi: 10.2147/IJN.S272378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kalluri R, LeBleu VS. The biology, function, and biomedical applications of exosomes. Science. 2020;367(6478):eaau6977. doi: 10.1126/science.aau6977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yu W, et al. Exosome-based liquid biopsies in cancer: opportunities and challenges. Ann Oncol. 2021;32(4):466–477. doi: 10.1016/j.annonc.2021.01.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yi Q, et al. Circular RNAs in chemotherapy resistance of lung cancer and their potential therapeutic application. IUBMB Life. 2023;75(3):225–237. doi: 10.1002/iub.2624. [DOI] [PubMed] [Google Scholar]
- 13.Xia X, Tang X, Wang S. Roles of CircRNAs in Autoimmune Diseases. Front Immunol. 2019;10:639. doi: 10.3389/fimmu.2019.00639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li Y, et al. Circular RNA is enriched and stable in exosomes: a promising biomarker for cancer diagnosis. Cell Res. 2015;25(8):981–984. doi: 10.1038/cr.2015.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Qu S, et al. Circular RNA: a new star of noncoding RNAs. Cancer Lett. 2015;365(2):141–148. doi: 10.1016/j.canlet.2015.06.003. [DOI] [PubMed] [Google Scholar]
- 16.Yu D, et al. Exosomes as a new frontier of cancer liquid biopsy. Mol Cancer. 2022;21(1):56. doi: 10.1186/s12943-022-01509-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huang T, et al. The roles of extracellular vesicles in gastric cancer development, microenvironment, anti-cancer drug resistance, and therapy. Mol Cancer. 2019;18(1):62. doi: 10.1186/s12943-019-0967-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Baietti MF, et al. Syndecan-syntenin-ALIX regulates the biogenesis of exosomes. Nat Cell Biol. 2012;14(7):677–685. doi: 10.1038/ncb2502. [DOI] [PubMed] [Google Scholar]
- 19.Zhang H, et al. Exosome-induced regulation in inflammatory bowel disease. Front Immunol. 2019;10:1464. doi: 10.3389/fimmu.2019.01464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.van Niel G, D’Angelo G, Raposo G. Shedding light on the cell biology of extracellular vesicles. Nat Rev Mol Cell Biol. 2018;19(4):213–28. doi: 10.1038/nrm.2017.125. [DOI] [PubMed] [Google Scholar]
- 21.Wu P, et al. Exosomes derived from hypoxic glioma cells reduce the sensitivity of glioma cells to temozolomide through carrying miR-106a-5p. Drug Des Devel Ther. 2022;16:3589–3598. doi: 10.2147/DDDT.S382690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Elewaily MI, Elsergany AR. Emerging role of exosomes and exosomal microRNA in cancer: pathophysiology and clinical potential. J Cancer Res Clin Oncol. 2021;147(3):637–648. doi: 10.1007/s00432-021-03534-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.He L, et al. Ovarian cancer cell-secreted exosomal miR-205 promotes metastasis by inducing angiogenesis. Theranostics. 2019;9(26):8206–8220. doi: 10.7150/thno.37455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hu JL, et al. CAFs secreted exosomes promote metastasis and chemotherapy resistance by enhancing cell stemness and epithelial-mesenchymal transition in colorectal cancer. Mol Cancer. 2019;18(1):91. doi: 10.1186/s12943-019-1019-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ren J, et al. Carcinoma-associated fibroblasts promote the stemness and chemoresistance of colorectal cancer by transferring exosomal lncRNA H19. Theranostics. 2018;8(14):3932–3948. doi: 10.7150/thno.25541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cao Y, et al. Exosomes and cancer immunotherapy: a review of recent cancer research. Front Oncol. 2022;12:1118101. doi: 10.3389/fonc.2022.1118101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhou Y, et al. The role of exosomes and their applications in Cancer. Int J Mol Sci. 2022;22(22):12204. doi: 10.3390/ijms222212204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Giannopoulou L, et al. Liquid biopsy in ovarian cancer: the potential of circulating miRNAs and exosomes. Transl Res. 2019;205:77–91. doi: 10.1016/j.trsl.2018.10.003. [DOI] [PubMed] [Google Scholar]
- 29.Xiao Q, et al. Circulating plasma exosomal long non-coding RNAs LINC00265, LINC00467, UCA1, and SNHG1 as biomarkers for diagnosis and treatment monitoring of acute myeloid leukemia. Front Oncol. 2022;12:1033143. doi: 10.3389/fonc.2022.1033143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li P, et al. Characterization of circSCL38A1 as a novel oncogene in bladder cancer via targeting ILF3/TGF-beta2 signaling axis. Cell Death Dis. 2023;14(1):59. doi: 10.1038/s41419-023-05598-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Meng X, et al. Circular RNA: an emerging key player in RNA world. Brief Bioinform. 2017;18(4):547–57. doi: 10.1093/bib/bbw045. [DOI] [PubMed] [Google Scholar]
- 32.Dragomir M, Calin GA. Circular RNAs in cancer - lessons learned from microRNAs. Front Oncol. 2018;8:179. doi: 10.3389/fonc.2018.00179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hsu MT, Coca-Prados M. Electron microscopic evidence for the circular form of RNA in the cytoplasm of eukaryotic cells. Nature. 1979;280(5720):339–340. doi: 10.1038/280339a0. [DOI] [PubMed] [Google Scholar]
- 34.Dong R, et al. CIRCpedia v2: an updated database for comprehensive circular RNA annotation and expression comparison. Genomics Proteom Bioinf. 2018;16(4):226–33. doi: 10.1016/j.gpb.2018.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Arnaiz E, et al. CircRNAs and cancer: biomarkers and master regulators. Semin Cancer Biol. 2019;58:90–99. doi: 10.1016/j.semcancer.2018.12.002. [DOI] [PubMed] [Google Scholar]
- 36.Chen LL. The biogenesis and emerging roles of circular RNAs. Nat Rev Mol Cell Biol. 2016;17(4):205–211. doi: 10.1038/nrm.2015.32. [DOI] [PubMed] [Google Scholar]
- 37.Pamudurti NR, et al. Translation of CircRNAs. Mol Cell. 2017;66(1):9–21e7. doi: 10.1016/j.molcel.2017.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kristensen LS, et al. The biogenesis, biology and characterization of circular RNAs. Nat Rev Genet. 2019;20(11):675–691. doi: 10.1038/s41576-019-0158-7. [DOI] [PubMed] [Google Scholar]
- 39.Wang C, et al. CircRNAs in lung cancer - biogenesis, function and clinical implication. Cancer Lett. 2020;492:106–115. doi: 10.1016/j.canlet.2020.08.013. [DOI] [PubMed] [Google Scholar]
- 40.Wang X, et al. Emerging roles of circular RNAs in gastric cancer metastasis and drug resistance. J Exp Clin Cancer Res. 2022;41(1):218. doi: 10.1186/s13046-022-02432-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Carlos-Reyes A, et al. Role of circular RNAs in the regulation of Immune cells in response to Cancer Therapies. Front Genet. 2022;13:823238. doi: 10.3389/fgene.2022.823238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ding L, et al. Exosome-derived circTFDP2 promotes prostate cancer progression by preventing PARP1 from caspase-3-dependent cleavage. Clin Transl Med. 2023;13(1):e1156. doi: 10.1002/ctm2.1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhao K, et al. Exosome-mediated transfer of circ_0000338 enhances 5-fluorouracil resistance in colorectal cancer through regulating microRNA 217 (miR-217) and miR-485-3p. Mol Cell Biol. 2021;41(5):e00517–20. doi: 10.1128/MCB.00517-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Huang M, Huang X, Huang N. Exosomal circGSE1 promotes immune escape of hepatocellular carcinoma by inducing the expansion of regulatory T cells. Cancer Sci. 2022;113(6):1968–1983. doi: 10.1111/cas.15365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pan B, et al. Identification of serum exosomal hsa-circ-0004771 as a novel diagnostic biomarker of colorectal cancer. Front Genet. 2019;10:1096. doi: 10.3389/fgene.2019.01096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lin J, et al. Hypoxia-induced exosomal circPDK1 promotes pancreatic cancer glycolysis via c-myc activation by modulating miR-628-3p/BPTF axis and degrading BIN1. J Hematol Oncol. 2022;15(1):128. doi: 10.1186/s13045-022-01348-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.He Y, et al. Exosomal circPRRX1 functions as a ceRNA for miR-596 to promote the proliferation, migration, invasion, and reduce radiation sensitivity of gastric cancer cells via the upregulation of NF-kappaB activating protein. Anticancer Drugs. 2022;33(10):1114–25. doi: 10.1097/CAD.0000000000001358. [DOI] [PubMed] [Google Scholar]
- 48.Zhou B, et al. Targeting tumor exosomal circular RNA cSERPINE2 suppresses breast cancer progression by modulating MALT1-NF. J Exp Clin Cancer Res. 2023;42(1):48. doi: 10.1186/s13046-023-02620-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chen S, et al. Tumor-associated macrophages promote cholangiocarcinoma progression via exosomal Circ_0020256. Cell Death Dis. 2022;13(1):94. doi: 10.1038/s41419-022-04534-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Qian Y et al. Tumor cell-derived exosomal circ-PRKCI promotes proliferation of renal cell carcinoma via regulating miR-545-3p/CCND1. Axis Cancers (Basel). 2022; 15(1). [DOI] [PMC free article] [PubMed]
- 51.Zhang H, et al. Exosome circRNA secreted from adipocytes promotes the growth of hepatocellular carcinoma by targeting deubiquitination-related USP7. Oncogene. 2019;38(15):2844–2859. doi: 10.1038/s41388-018-0619-z. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 52.Liu L, et al. Hepatic stellate cell exosome-derived circWDR25 promotes the progression of hepatocellular carcinoma via the miRNA-4474-3P-ALOX-15 and EMT axes. Biosci Trends. 2022;16(4):267–281. doi: 10.5582/bst.2022.01281. [DOI] [PubMed] [Google Scholar]
- 53.Xiong H, et al. circ_rac GTPase-activating protein 1 facilitates stemness and metastasis of non-small cell lung cancer via polypyrimidine tract-binding protein 1 recruitment to promote Sirtuin-3-mediated replication timing regulatory factor 1 deacetylation. Lab Invest. 2023;103(1):100010. doi: 10.1016/j.labinv.2022.100010. [DOI] [PubMed] [Google Scholar]
- 54.Tang X, et al. A novel protein encoded by circHNRNPU promotes multiple myeloma progression by regulating the bone marrow microenvironment and alternative splicing. J Exp Clin Cancer Res. 2022;41(1):85. doi: 10.1186/s13046-022-02276-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zheng R, et al. Exosomal circLPAR1 functions in colorectal cancer diagnosis and tumorigenesis through suppressing BRD4 via METTL3-eIF3h interaction. Mol Cancer. 2022;21(1):49. doi: 10.1186/s12943-021-01471-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yang Y, Yang N, Jiang J. Exosomal circ_PTPRA inhibits tumorigenesis and promotes radiosensitivity in colorectal cancer by enriching the level of SMAD4 via competitively binding to miR-671-5. Cytotechnology. 2022;74(1):51–64. doi: 10.1007/s10616-021-00506-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chen W, et al. Exosome-transmitted circular RNA hsa_circ_0051443 suppresses hepatocellular carcinoma progression. Cancer Lett. 2020;475:119–128. doi: 10.1016/j.canlet.2020.01.022. [DOI] [PubMed] [Google Scholar]
- 58.Zhang Y, et al. Exosomal CircGDI2 suppresses oral squamous cell carcinoma progression through the regulation of MiR-424-5p/SCAI Axis. Cancer Manag Res. 2020;12:7501–14. doi: 10.2147/CMAR.S255687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chen C, et al. Tumor-suppressive circRHOBTB3 is excreted out of cells via exosome to sustain colorectal cancer cell fitness. Mol Cancer. 2022;21(1):46. doi: 10.1186/s12943-022-01511-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Shi L, et al. Exosomal circRNA BTG2 derived from RBP-J overexpressed-macrophages inhibits glioma progression via miR-25-3p/PTEN. Cell Death Dis. 2022;13(5):506. doi: 10.1038/s41419-022-04908-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhang L, et al. Exosomal hsa_circ_0004658 derived from RBPJ overexpressed-macrophages inhibits hepatocellular carcinoma progression via miR-499b-5p/JAM3. Cell Death Dis. 2022;13(1):32. doi: 10.1038/s41419-021-04345-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Shang A, et al. Exosomal circPACRGL promotes progression of colorectal cancer via the miR-142-3p/miRi>-506-3p- TGF-beta1 axis. Mol Cancer. 2020;19(1):117. doi: 10.1186/s12943-020-01235-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Liu D, et al. Exosome-transmitted circ_MMP2 promotes hepatocellular carcinoma metastasis by upregulating MMP2. Mol Oncol. 2020;14(6):1365–80. doi: 10.1002/1878-0261.12637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhou H, et al. CircRAPGEF5 promotes the proliferation and metastasis of lung adenocarcinoma through the miR-1236-3p/ZEB1 Axis and serves as a potential biomarker. Int J Biol Sci. 2022;18(5):2116–2131. doi: 10.7150/ijbs.66770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yang SJ, et al. Tumor-derived exosomal circPSMA1 facilitates the tumorigenesis, metastasis, and migration in triple-negative breast cancer (TNBC) through miR-637/Akt1/beta-catenin (cyclin D1) axis. Cell Death Dis. 2021;12(5):420. doi: 10.1038/s41419-021-03680-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhang G, et al. Inhibition of circ_0081234 reduces prostate cancer tumor growth and metastasis via the miR-1/MAP 3 K1 axis. J Gene Med. 2022;24(8):e3376. doi: 10.1002/jgm.3376. [DOI] [PubMed] [Google Scholar]
- 67.Zhang T, et al. Circular RNA circ_0003028 regulates cell development through modulating miR-498/ornithine decarboxylase 1 axis in hepatocellular carcinoma. Anticancer Drugs. 2023;34(4):507–518. doi: 10.1097/CAD.0000000000001457. [DOI] [PubMed] [Google Scholar]
- 68.Lin Q, et al. Exosomal circular RNA hsa_circ_007293 promotes proliferation, migration, invasion, and epithelial-mesenchymal transition of papillary thyroid carcinoma cells through regulation of the microRNA-653-5p/paired box 6 axis. Bioengineered. 2021;12(2):10136–10149. doi: 10.1080/21655979.2021.2000745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Huang XY, et al. Exosomal circRNA-100338 promotes hepatocellular carcinoma metastasis via enhancing invasiveness and angiogenesis. J Exp Clin Cancer Res. 2020;39(1):20. doi: 10.1186/s13046-020-1529-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Li S, et al. Gastric cancer derived exosomes mediate the delivery of circRNA to promote angiogenesis by targeting miR-29a/VEGF axis in endothelial cells. Biochem Biophys Res Commun. 2021;560:37–44. doi: 10.1016/j.bbrc.2021.04.099. [DOI] [PubMed] [Google Scholar]
- 71.Ye H, et al. Exosomal circNFIX promotes angiogenesis in ovarian cancer via miR-518a-3p/TRIM44 axis. Kaohsiung J Med Sci. 2023;39(1):26–39. doi: 10.1002/kjm2.12615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yao W, et al. Exosomal circ_0026611 contributes to lymphangiogenesis by reducing PROX1 acetylation and ubiquitination in human lymphatic endothelial cells (HLECs) Cell Mol Biol Lett. 2023;28(1):13. doi: 10.1186/s11658-022-00410-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Chen X, et al. Exosome-transmitted circIFNGR2 modulates ovarian cancer metastasis via miR-378/ST5 Axis. Mol Cell Biol. 2023;43(1):22–42. doi: 10.1080/10985549.2022.2160605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Gao G, Wang L, Li C. Circ_0006790 carried by bone marrow mesenchymal stem cell-derived exosomes regulates S100A11 DNA methylation through binding to CBX7 in pancreatic ductal adenocarcinoma. Am J Cancer Res. 2022;12(5):1934–59. [PMC free article] [PubMed] [Google Scholar]
- 75.Lin Y, et al. Tumor cell-derived exosomal Circ-0072088 suppresses migration and invasion of hepatic carcinoma cells through regulating MMP-16. Front Cell Dev Biol. 2021;9:726323. doi: 10.3389/fcell.2021.726323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wang Y, et al. Circular RNA ITCH suppresses metastasis of gastric cancer via regulating miR-199a-5p/Klotho axis. Cell Cycle. 2021;20(5–6):522–536. doi: 10.1080/15384101.2021.1878327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zhang C, et al. Exosome-delivered circSTAU2 inhibits the progression of gastric cancer by targeting the miR-589/CAPZA1 Axis. Int J Nanomed. 2023;18:127–42. doi: 10.2147/IJN.S391872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wang X, et al. Circular RNAcirc_0076305 promotes cisplatin (DDP) resistance of non-small cell lung cancer cells by regulating ABCC1 through miR-186-5p. Cancer Biother Radiopharm. 2021;38(5):293–304. doi: 10.1089/cbr.2020.4153. [DOI] [PubMed] [Google Scholar]
- 79.Xie H, et al. Exosome-transmitted circVMP1 facilitates the progression and cisplatin resistance of non-small cell lung cancer by targeting miR-524-5p-METTL3/SOX2 axis. Drug Deliv. 2022;29(1):1257–1271. doi: 10.1080/10717544.2022.2057617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Pan Y, Lin Y, Mi C. Cisplatin-resistant osteosarcoma cell-derived exosomes confer cisplatin resistance to recipient cells in an exosomal circ_103801-dependent manner. Cell Biol Int. 2021;45(4):858–68. doi: 10.1002/cbin.11532. [DOI] [PubMed] [Google Scholar]
- 81.Ding C, et al. Warburg effect-promoted exosomal circ_0072083 releasing up-regulates NANGO expression through multiple pathways and enhances temozolomide resistance in glioma. J Exp Clin Cancer Res. 2021;40(1):164. doi: 10.1186/s13046-021-01942-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zeng Z, et al. Hypoxic exosomal HIF-1alpha-stabilizing circZNF91 promotes chemoresistance of normoxic pancreatic cancer cells via enhancing glycolysis. Oncogene. 2021;40(36):5505–17. doi: 10.1038/s41388-021-01960-w. [DOI] [PubMed] [Google Scholar]
- 83.Tan WQ, et al. Exosome-delivered circular RNA DLGAP4 induces chemoresistance via mir-143-HK2 axis in neuroblastoma. Cancer Biomark. 2022;34(3):375–384. doi: 10.3233/CBM-210272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Wang X, et al. Exosome-delivered circRNA promotes glycolysis to induce chemoresistance through the mir-122-PKM2 axis in colorectal cancer. Mol Oncol. 2020;14(3):539–55. doi: 10.1002/1878-0261.12629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Chen Y, et al. Exosomal circ_0091741 promotes gastric cancer cell autophagy and chemoresistance via the miR-330-3pi>/TRIM14/Dvl2/Wnt/beta-catenin axis. Hum Cell. 2023;36(1):258–275. doi: 10.1007/s13577-022-00790-6. [DOI] [PubMed] [Google Scholar]
- 86.Tan X, et al. Exosomal circRNA Scm-like with four malignant brain tumor domains 2 (circ-SFMBT2) enhances the docetaxel resistance of prostate cancer via the microRNA-136-5p/tribbles homolog 1 pathway. Anticancer Drugs. 2022;33(9):871–882. doi: 10.1097/CAD.0000000000001365. [DOI] [PubMed] [Google Scholar]
- 87.Zhang H, et al. Exosomal Circ-XIAP promotes docetaxel resistance in prostate cancer by regulating miR-1182/TPD52 Axis. Drug Des Dev Ther. 2021;15:1835–49. doi: 10.2147/DDDT.S300376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Pan Z, et al. A novel protein encoded by exosomal CircATG4B induces oxaliplatin resistance in colorectal cancer by promoting autophagy. Adv Sci (Weinh) 2022;9(35):e2204513. doi: 10.1002/advs.202204513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Xu Y, et al. Exosomal transfer of circular RNA FBXW7 ameliorates the chemoresistance to oxaliplatin in colorectal cancer by sponging miR-18b-5p. Neoplasma. 2021;68(1):108–118. doi: 10.4149/neo_2020_200417N414. [DOI] [PubMed] [Google Scholar]
- 90.Wang X, et al. CircRNA-CREIT inhibits stress granule assembly and overcomes doxorubicin resistance in TNBC by destabilizing PKR. J Hematol Oncol. 2022;15(1):122. doi: 10.1186/s13045-022-01345-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Qin L, et al. Hsa–circRNA–G004213 promotes cisplatin sensitivity by regulating miR–513b–5p/PRPF39 in liver cancer. Mol Med Rep. 2021;23(6):421–421. doi: 10.3892/mmr.2021.12060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Zhang N, et al. Circular RNA circSATB2 promotes progression of non-small cell lung cancer cells. Mol Cancer. 2020;19(1):101. doi: 10.1186/s12943-020-01221-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Zhu J, et al. Exosome-delivered circSATB2 targets the miR-330-5p/PEAK1 axis to regulate proliferation, migration and invasion of lung cancer cells. Thorac Cancer. 2022;13(21):3007–17. doi: 10.1111/1759-7714.14652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Ning Z, et al. Exosomal circ_0007385 enhances non-small cell lung cancer cell proliferation and stemness via regulating miR-1253/FAM83A axis. Anticancer Drugs. 2022;33(1):61–74. doi: 10.1097/CAD.0000000000001103. [DOI] [PubMed] [Google Scholar]
- 95.Wang H, et al. Cancer-released exosomal circular RNA circ_0008717 promotes cell tumorigenicity through microRNA-1287-5p/P21-activated kinase 2 (PAK2) axis in non-small cell lung cancer. Bioengineered. 2022;13(4):8937–8949. doi: 10.1080/21655979.2022.2056822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Fang K, et al. Serum-derived exosomes-mediated circular RNA ARHGAP10 modulates the progression of Non-Small Cell Lung Cancer through the miR-638/FAM83F Axis. Cancer Biother Radiopharm. 2022;37(2):96–110. doi: 10.1089/cbr.2019.3534. [DOI] [PubMed] [Google Scholar]
- 97.Yang B, et al. Tumor-derived exosomal circRNA_102481 contributes to EGFR-TKIs resistance via the miR-30a-5p/ROR1 axis in non-small cell lung cancer. Aging. 2021;13(9):13264–13286. doi: 10.18632/aging.203011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Li C, et al. Upregulation of exosomal circPLK1 promotes the development of non-small cell lung cancer through the miR-1294/ high mobility group protein A1 axis. Bioengineered. 2022;13(2):4185–4200. doi: 10.1080/21655979.2022.2026727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Xu X, et al. Exosome-transferred hsa_circ_0014235 promotes DDP chemoresistance and deteriorates the development of non-small cell lung cancer by mediating the miR-520a-5p/CDK4 pathway. Cancer Cell Int. 2020;20(1):552. doi: 10.1186/s12935-020-01642-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Ma J, Qi G, Li L. A novel serum exosomes-based biomarker hsa_circ_0002130 facilitates osimertinib-resistance in non-small cell lung cancer by sponging miR-498. Onco Targets Ther. 2020;13:5293–307. doi: 10.2147/OTT.S243214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Tong Z, et al. A novel molecular mechanism mediated by circCCDC134 regulates non-small cell lung cancer progression. Thorac Cancer. 2023;14(20):1958–1968. doi: 10.1111/1759-7714.14942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Li J, et al. Exosomal circDNER enhances paclitaxel resistance and tumorigenicity of lung cancer via targeting miR-139-5p/ITGB8 Thorac. Cancer. 2022;13(9):1381–1390. doi: 10.1111/1759-7714.14402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Bai L, et al. Circular noncoding RNA circ_0007334 sequestrates miR-577 to derepress KLF12 and accelerate colorectal cancer progression. Anticancer Drugs. 2022;33(1):e409–22. doi: 10.1097/CAD.0000000000001221. [DOI] [PubMed] [Google Scholar]
- 104.Gao L, et al. Exosome-transmitted circCOG2 promotes colorectal cancer progression via miR-1305/TGF-beta2/SMAD3 pathway. Cell Death Discov. 2021;7(1):281. doi: 10.1038/s41420-021-00680-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Yu Q, et al. Exosomal Circ_FMN2 derived from the serum of colorectal cancer patients promotes cancer progression by miR-338-3p/MSI1 axis. Appl Biochem Biotechnol. 2023 doi: 10.1007/s12010-023-04456-3. [DOI] [PubMed] [Google Scholar]
- 106.Miao Z, Zhao X, Liu X. Exosomal circCOL1A2 from cancer cells accelerates colorectal cancer progression via regulating miR-665/LASP1 signal axis. Eur J Pharmacol. 2023;950:175722. doi: 10.1016/j.ejphar.2023.175722. [DOI] [PubMed] [Google Scholar]
- 107.Yu L, Zhang F, Wang Y. Circ_0005615 regulates the progression of colorectal cancer through the miR-873-5p/FOSL2 signaling pathway. Biochem Genet. 2023 doi: 10.1007/s10528-023-10355-3. [DOI] [PubMed] [Google Scholar]
- 108.Fan L, Li W, Jiang H. Circ_0000395 promoted CRC progression via elevating MYH9 expression by sequestering miR-432-5p. Biochem Genet. 2023;61(1):116–137. doi: 10.1007/s10528-022-10245-0. [DOI] [PubMed] [Google Scholar]
- 109.Chen C, et al. Exosomal circTUBGCP4 promotes vascular endothelial cell tipping and colorectal cancer metastasis by activating akt signaling pathway. J Exp Clin Cancer Res. 2023;42(1):46. doi: 10.1186/s13046-023-02619-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Li Y, et al. Exosomal circPABPC1 promotes colorectal cancer liver metastases by regulating HMGA2 in the nucleus and BMP4/ADAM19 in the cytoplasm. Cell Death Discov. 2022;8(1):335. doi: 10.1038/s41420-022-01124-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Yang H, et al. Hypoxia induced exosomal circRNA promotes metastasis of colorectal cancer via targeting GEF-H1/RhoA axis. Theranostics. 2020;10(18):8211–8226. doi: 10.7150/thno.44419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Su Y, et al. CircRNA Cdr1as functions as a competitive endogenous RNA to promote hepatocellular carcinoma progression. Aging. 2019;11(19):8183–203. doi: 10.18632/aging.102312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Liu C, et al. Exosome-mediated circTTLL5 transfer promotes hepatocellular carcinoma malignant progression through miR-136-5p/KIAA1522 axis. Pathol Res Pract. 2023;241:154276. doi: 10.1016/j.prp.2022.154276. [DOI] [PubMed] [Google Scholar]
- 114.Xu J, et al. CircRNA-SORE mediates sorafenib resistance in hepatocellular carcinoma by stabilizing YBX1. Signal Transduct Target Ther. 2020;5(1):298. doi: 10.1038/s41392-020-00375-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Hao X, et al. CircPAK1 promotes the progression of hepatocellular carcinoma via modulation of YAP nucleus localization by interacting with 14-3-3zeta. J Exp Clin Cancer Res. 2022;41(1):281. doi: 10.1186/s13046-022-02494-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Zhou Y, et al. Cancer-associated fibroblast exosomes promote chemoresistance to cisplatin in hepatocellular carcinoma through circZFR targeting signal transducers and activators of transcription (STAT3)/ nuclear factor -kappa B (NF-kappaB) pathway. Bioengineered. 2022;13(3):4786–97. doi: 10.1080/21655979.2022.2032972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Wu X, et al. Circular RNA circ-MMP11 contributes to lapatinib resistance of breast cancer cells by regulating the miR-153-3p/ANLN Axis. Front Oncol. 2021;11:639961. doi: 10.3389/fonc.2021.639961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Hu K, et al. Exosomes mediated transfer of Circ_UBE2D2 enhances the resistance of breast cancer to tamoxifen by binding to MiR-200a-3p. Med Sci Monit. 2020;26:e922253. doi: 10.12659/MSM.922253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Liu Y, et al. Correction: exosomal circCARM1 from spheroids reprograms cell metabolism by regulating PFKFB2 in breast cancer. Oncogene. 2022;41(14):2137. doi: 10.1038/s41388-022-02217-w. [DOI] [PubMed] [Google Scholar]
- 120.Ma J, et al. CircEGFR reduces the sensitivity of pirarubicin and regulates the malignant progression of triple-negative breast cancer via the miR-1299/EGFR axis. Int J Biol Macromol. 2023;244:125295. doi: 10.1016/j.ijbiomac.2023.125295. [DOI] [PubMed] [Google Scholar]
- 121.Zhang X, et al. Circular RNA circNRIP1 acts as a microRNA-149-5p sponge to promote gastric cancer progression via the AKT1/mTOR pathway. Mol Cancer. 2019;18(1):20. doi: 10.1186/s12943-018-0935-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.You J, et al. Circular RNA 0001789 sponges mir-140-3p and regulates PAK2 to promote the progression of gastric cancer. J Transl Med. 2023;21(1):83. doi: 10.1186/s12967-022-03853-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Yang G, et al. Exosome-mediated transfer of circ_0063526 enhances cisplatin resistance in gastric cancer cells via regulating miR-449a/SHMT2 axis. Anticancer Drugs. 2022;33(10):1047–1057. doi: 10.1097/CAD.0000000000001386. [DOI] [PubMed] [Google Scholar]
- 124.Yao W, et al. Exosome-derived Circ-PVT1 contributes to cisplatin resistance by regulating autophagy, invasion, and apoptosis via miR-30a-5p/YAP1 axis in gastric cancer cells. Cancer Biother Radiopharm. 2021;36(4):347–59. doi: 10.1089/cbr.2020.3578. [DOI] [PubMed] [Google Scholar]
- 125.Liang Q, et al. circ-LDLRAD3 knockdown reduces cisplatin chemoresistance and inhibits the development of gastric cancer with cisplatin resistance through miR-588 enrichment-mediated SOX5 inhibition. Gut Liver. 2022;17:389–403. doi: 10.5009/gnl210195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Zhong Y, et al. Circular RNA circ_0032821 contributes to oxaliplatin (OXA) resistance of gastric cancer cells by regulating SOX9 via miR-515-5. Biotechnol Lett. 2021;43(2):339–351. doi: 10.1007/s10529-020-03036-3. [DOI] [PubMed] [Google Scholar]
- 127.Geng X, et al. Exosomal circWDR62 promotes temozolomide resistance and malignant progression through regulation of the miR-370-3p/MGMT axis in glioma. Cell Death Dis. 2022;13(7):596. doi: 10.1038/s41419-022-05056-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Li G, Lan Q. Exosome-mediated transfer of circ-GLIS3 enhances Temozolomide resistance in glioma cells through the miR-548m/MED31 Axis. Cancer Biother Radiopharm. 2023;38(1):62–73. doi: 10.1089/cbr.2021.0299. [DOI] [PubMed] [Google Scholar]
- 129.Li X, et al. Hsa_circ_0043949 reinforces temozolomide resistance via upregulating oncogene ITGA1 axis in glioblastoma. Metab Brain Dis. 2022;37(8):2979–2993. doi: 10.1007/s11011-022-01069-3. [DOI] [PubMed] [Google Scholar]
- 130.Li Y, et al. Exosomal circ-AHCY promotes glioblastoma cell growth via Wnt/beta-catenin signaling pathway. Ann Clin Transl Neurol. 2023;10(6):865–78. doi: 10.1002/acn3.51743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Zhang C, et al. Radiated glioblastoma cell-derived exosomal circ_0012381 induce M2 polarization of microglia to promote the growth of glioblastoma by CCL2/CCR2 axis. J Transl Med. 2022;20(1):388. doi: 10.1186/s12967-022-03607-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Jiang Y, et al. Glioblastoma-associated microglia-derived exosomal circKIF18A promotes angiogenesis by targeting FOXC2. Oncogene. 2022;41(26):3461–3473. doi: 10.1038/s41388-022-02360-4. [DOI] [PubMed] [Google Scholar]
- 133.Huang G, et al. Exosomal circKDM4A induces CUL4B to promote prostate cancer cell malignancy in a mir-338-3p-Dependent manner. Biochem Genet. 2023;61(1):390–409. doi: 10.1007/s10528-022-10251-2. [DOI] [PubMed] [Google Scholar]
- 134.Sheng H, Wang X. Knockdown of circ-PIP5K1A overcomes resistance to cisplatin in ovarian cancer by miR-942-5p/NFIB axis. Anticancer Drugs. 2023;34(2):214–26. doi: 10.1097/CAD.0000000000001406. [DOI] [PubMed] [Google Scholar]
- 135.Luo Y, Gui R. Circulating exosomal circFoxp1 confers cisplatin resistance in epithelial ovarian cancer cells. J Gynecol Oncol. 2020;31(5):e75. doi: 10.3802/jgo.2020.31.e75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Gao Y, Huang Y. Circ_0007841 knockdown confers cisplatin sensitivity to ovarian cancer cells by down-regulation of NFIB expression in a mir-532-5p-dependent manner. J Chemother. 2023;35(2):117–130. doi: 10.1080/1120009X.2022.2056995. [DOI] [PubMed] [Google Scholar]
- 137.Li J, et al. Circular RNA IARS (circ-IARS) secreted by pancreatic cancer cells and located within exosomes regulates endothelial monolayer permeability to promote tumor metastasis. J Exp Clin Cancer Res. 2018;37(1):177. doi: 10.1186/s13046-018-0822-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Zang R, et al. Exosomes mediated transfer of Circ_0000337 contributes to cisplatin (CDDP) resistance of esophageal cancer by regulating JAK2 via miR-377-3p front cell. Dev Biol. 2021;9:673237. doi: 10.3389/fcell.2021.673237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Chen J, et al. Hsa_circ_0074269-mediated upregulation of TUFT1 through mir-485-5p increases cisplatin resistance in cervical cancer. Reprod Sci. 2022;29(8):2236–2250. doi: 10.1007/s43032-022-00855-9. [DOI] [PubMed] [Google Scholar]
- 140.Wang X, Cheng Q. Suppression of exosomal hsa_circ_0001005 eliminates the Vemurafenib resistance of melanoma. J Cancer Res Clin Oncol. 2023;149(9):5921–5936. doi: 10.1007/s00432-022-04434-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Ai J, et al. Exosomes loaded with circPARD3 promotes EBV-miR-BART4-induced stemness and cisplatin resistance in nasopharyngeal carcinoma side population cells through the miR-579-3p/SIRT1/SSRP1 axis. Cell Biol Toxicol. 2023;39(2):537–556. doi: 10.1007/s10565-022-09738-w. [DOI] [PubMed] [Google Scholar]
- 142.Xu Y, et al. A circular RNA, cholangiocarcinoma-associated circular RNA 1, contributes to cholangiocarcinoma progression, induces angiogenesis, and disrupts vascular endothelial barriers. Hepatology. 2021;73(4):1419–35. doi: 10.1002/hep.31493. [DOI] [PubMed] [Google Scholar]
- 143.Yang C, et al. Exosome-derived circTRPS1 promotes malignant phenotype and CD8 + T cell exhaustion in bladder cancer microenvironments. Mol Ther. 2022;30(3):1054–1070. doi: 10.1016/j.ymthe.2022.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Chen SW, et al. Cancer cell-derived exosomal circUSP7 induces CD8(+) T cell dysfunction and anti-PD1 resistance by regulating the miR-934/SHP2 axis in NSCLC. Mol Cancer. 2021;20(1):144. doi: 10.1186/s12943-021-01448-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Wang J, et al. circRNA-002178 act as a ceRNA to promote PDL1/PD1 expression in lung adenocarcinoma. Cell Death Dis. 2020;11(1):32. doi: 10.1038/s41419-020-2230-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Wang X, Yao Y, Jin M. Circ-0001068 is a novel biomarker for ovarian cancer and inducer of PD1 expression in T cells. Aging. 2020;12(19):19095–19106. doi: 10.18632/aging.103706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Hu Z, et al. Exosome-derived circCCAR1 promotes CD8 + T-cell dysfunction and anti-PD1 resistance in hepatocellular carcinoma. Mol Cancer. 2023;22(1):55. doi: 10.1186/s12943-023-01759-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Chen Y, et al. CircRNA has_circ_0069313 induced OSCC immunity escape by mir-325-3p-Foxp3 axes in both OSCC cells and Treg cells. Aging. 2022;14(10):4376–4389. doi: 10.18632/aging.204068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Yang K, Zhang J, Bao C. Exosomal circEIF3K from cancer-associated fibroblast promotes colorectal cancer (CRC) progression via miR-214/PD-L1 axis. BMC Cancer. 2021;21(1):933. doi: 10.1186/s12885-021-08669-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Chen W, et al. Exosomal circSHKBP1 participates in non-small cell lung cancer progression through PKM2-mediated glycolysis. Mol Ther Oncolytics. 2022;24:470–85. doi: 10.1016/j.omto.2022.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Chen T, et al. Tumor-derived exosomal circFARSA mediates M2 macrophage polarization via the PTEN/PI3K/AKT pathway to promote non-small cell lung cancer metastasis. Cancer Treat Res Commun. 2021;28:100412. doi: 10.1016/j.ctarc.2021.100412. [DOI] [PubMed] [Google Scholar]
- 152.Pan Z, et al. EWSR1-induced circNEIL3 promotes glioma progression and exosome-mediated macrophage immunosuppressive polarization via stabilizing IGF2BP3. Mol Cancer. 2022;21(1):16. doi: 10.1186/s12943-021-01485-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Gao J, et al. Exosomal circZNF451 restrains anti-PD1 treatment in lung adenocarcinoma via polarizing macrophages by complexing with TRIM56 and FXR1. J Exp Clin Cancer Res. 2022;41(1):295. doi: 10.1186/s13046-022-02505-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Lu C, et al. Endoplasmic reticulum stress promotes breast cancer cells to release exosomes circ_0001142 and induces M2 polarization of macrophages to regulate tumor progression. Pharmacol Res. 2022;177:106098. doi: 10.1016/j.phrs.2022.106098. [DOI] [PubMed] [Google Scholar]
- 155.Huang X, et al. Exosomal Circsafb2 reshaping tumor environment to promote renal cell carcinoma progression by mediating M2 macrophage polarization. Front Oncol. 2022;12:808888. doi: 10.3389/fonc.2022.808888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Lu Q, et al. Hypoxic tumor-derived exosomal Circ0048117 facilitates M2 macrophage polarization acting as miR-140 sponge in esophageal squamous cell carcinoma. Onco Targets Ther. 2020;13:11883–97. doi: 10.2147/OTT.S284192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Zhang PF, et al. Cancer cell-derived exosomal circUHRF1 induces natural killer cell exhaustion and may cause resistance to anti-PD1 therapy in hepatocellular carcinoma. Mol Cancer. 2020;19(1):110. doi: 10.1186/s12943-020-01222-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Wang X, et al. Exosomal circ-PTPN22 and circ-ADAMTS6 mark T cell exhaustion and neutrophil extracellular traps in asian intrahepatic cholangiocarcinoma. Mol Ther Nucleic Acids. 2023;31:151–63. doi: 10.1016/j.omtn.2022.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Xu H, et al. Circular RNA expression in extracellular vesicles isolated from serum of patients with endometrial cancer. Epigenomics. 2018;10(2):187–197. doi: 10.2217/epi-2017-0109. [DOI] [PubMed] [Google Scholar]
- 160.Xie M, et al. Exosomal circSHKBP1 promotes gastric cancer progression via regulating the miR-582-3p/HUR/VEGF axis and suppressing HSP90 degradation. Mol Cancer. 2020;19(1):112. doi: 10.1186/s12943-020-01208-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Deng Q, et al. Exosomal hsa_circRNA_047733 integrated with clinical features for preoperative prediction of lymph node metastasis risk in oral squamous cell carcinoma. J Oral Pathol Med. 2023;52(1):37–46. doi: 10.1111/jop.13379. [DOI] [PubMed] [Google Scholar]
- 162.Xia D, Gu X. Plasmatic exosome-derived circRNAs panel act as fingerprint for glioblastoma. Aging. 2021;13(15):19575–19586. doi: 10.18632/aging.203368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Hong L, et al. Exosomal circular RNA hsa_circ_0006220, and hsa_circ_0001666 as biomarkers in the diagnosis of pancreatic cancer. J Clin Lab Anal. 2022;36(6):e24447. doi: 10.1002/jcla.24447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Kang Y, et al. Serum and serum exosomal CircRNAs hsa_circ_0001492, hsa_circ_0001439, and hsa_circ_0000896 as diagnostic biomarkers for lung adenocarcinoma. Front Oncol. 2022;12:912246. doi: 10.3389/fonc.2022.912246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Wang Y, et al. The potential of serum exosomal hsa_circ_0028861 as the Novel Diagnostic Biomarker of HBV-Derived Hepatocellular Cancer. Front Genet. 2021;12:703205. doi: 10.3389/fgene.2021.703205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Zheng P, et al. Plasma exosomal hsa_circ_0015286 as a potential diagnostic and prognostic biomarker for gastric Cancer. Pathol Oncol Res. 2022;28:1610446. doi: 10.3389/pore.2022.1610446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Shen X, et al. Hsa_circ_0000437 promotes pathogenesis of gastric cancer and lymph node metastasis. Oncogene. 2022;41(42):4724–4735. doi: 10.1038/s41388-022-02449-w. [DOI] [PubMed] [Google Scholar]
- 168.Li T, et al. Plasma exosome-derived circGAPVD1 as a potential diagnostic marker for colorectal cancer. Transl Oncol. 2023;31:101652. doi: 10.1016/j.tranon.2023.101652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Barbagallo C, et al. LncRNA UCA1, upregulated in CRC Biopsies and downregulated in serum exosomes, controls mRNA expression by RNA-RNA interactions. Mol Ther Nucleic Acids. 2018;12:229–41. doi: 10.1016/j.omtn.2018.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Xie Y, et al. RNA-Seq profiling of serum exosomal circular RNAs reveals Circ-PNN as a potential biomarker for human colorectal Cancer. Front Oncol. 2020;10:982. doi: 10.3389/fonc.2020.00982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Li R, et al. CircRNA CDR1as: a novel diagnostic and prognostic biomarker for gastric cancer. Biomarkers. 2023;28(5):448–457. doi: 10.1080/1354750X.2023.2206984. [DOI] [PubMed] [Google Scholar]
- 172.Shao Y, et al. Hsa_circ_0065149 is an Indicator for early gastric cancer screening and prognosis prediction. Pathol Oncol Res. 2020;26(3):1475–1482. doi: 10.1007/s12253-019-00716-y. [DOI] [PubMed] [Google Scholar]
- 173.Tang W, et al. CircRNA microarray profiling identifies a novel circulating biomarker for detection of gastric cancer. Mol Cancer. 2018;17(1):137. doi: 10.1186/s12943-018-0888-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Tao X, et al. Clinical significance of hsa_circ_0000419 in gastric cancer screening and prognosis estimation. Pathol Res Pract. 2020;216(1):152763. doi: 10.1016/j.prp.2019.152763. [DOI] [PubMed] [Google Scholar]
- 175.Chen T, et al. CircHIF1A regulated by FUS accelerates triple-negative breast cancer progression by modulating NFIB expression and translocation. Oncogene. 2021;40(15):2756–2771. doi: 10.1038/s41388-021-01739-z. [DOI] [PubMed] [Google Scholar]
- 176.Liu J, et al. The diagnostic value of serum exosomal Has_circ_0000615 for breast cancer patients. Int J Gen Med. 2021;14:4545–54. doi: 10.2147/IJGM.S319801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Xian J, et al. Identification of three circular RNA cargoes in serum exosomes as diagnostic biomarkers of non-small-cell lung cancer in the Chinese population. J Mol Diagn. 2020;22(8):1096–1108. doi: 10.1016/j.jmoldx.2020.05.011. [DOI] [PubMed] [Google Scholar]
- 178.He Y, et al. Exosomal circ_0048856 derived from non-small cell lung cancer contributes to aggressive cancer progression through downregulation of miR-1287-5p. Pathol Res Pract. 2022;232:153659. doi: 10.1016/j.prp.2021.153659. [DOI] [PubMed] [Google Scholar]
- 179.Chen Y, et al. Serum exosomal hsa_circ_0069313 has a potential to diagnose more aggressive non-small cell lung cancer. Clin Biochem. 2022;102:56–64. doi: 10.1016/j.clinbiochem.2022.01.005. [DOI] [PubMed] [Google Scholar]
- 180.Peng X, et al. Exosomal ERBB2IP contributes to tumor growth via elevating PSAT1 expression in non-small cell lung carcinoma. Thorac Cancer. 2023;14(19):1812–1823. doi: 10.1111/1759-7714.14926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.He F, et al. Plasma exo-hsa_circRNA_0056616: a potential biomarker for lymph node metastasis in lung adenocarcinoma. J Cancer. 2020;11(14):4037–4046. doi: 10.7150/jca.30360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Zhu X, et al. hsa_circ_0013958: a circular RNA and potential novel biomarker for lung adenocarcinoma. FEBS J. 2017;284(14):2170–2182. doi: 10.1111/febs.14132. [DOI] [PubMed] [Google Scholar]
- 183.Wang Y, et al. Circular RNA expression profile of lung squamous cell carcinoma: identification of potential biomarkers and therapeutic targets. Biosci Rep. 2020;40(4):BSR20194512. doi: 10.1042/BSR20194512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Yang G, et al. Has_Circ_0002490 circular RNA: a potential novel biomarker for lung cancer. Genet Test Mol Biomark. 2022;26(1):1–7. doi: 10.1089/gtmb.2021.0173. [DOI] [PubMed] [Google Scholar]
- 185.Zhang X, et al. The circular RNA hsa_circ_0001445 regulates the proliferation and migration of hepatocellular carcinoma and may serve as a diagnostic biomarker. Dis Markers. 2018;2018:3073467. doi: 10.1155/2018/3073467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Guo S, et al. Circular RNA 0006602 in plasma exosomes: a new potential diagnostic biomarker for hepatocellular carcinoma. Am J Transl Res. 2021;13(6):6001–15. [PMC free article] [PubMed] [Google Scholar]
- 187.Sun XH, et al. Serum-derived three-circRNA signature as a diagnostic biomarker for hepatocellular carcinoma. Cancer Cell Int. 2020;20:226. doi: 10.1186/s12935-020-01302-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Liu S, et al. Upregulated expression of serum exosomal hsa_circ_0026611 is associated with lymph node metastasis and poor prognosis of esophageal squamous cell carcinoma. J Cancer. 2021;12(3):918–926. doi: 10.7150/jca.50548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Fan L, et al. Circular RNA profiling and its potential for esophageal squamous cell cancer diagnosis and prognosis. Mol Cancer. 2019;18(1):16. doi: 10.1186/s12943-018-0936-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Luo Y, Gui R. Circulating exosomal CircMYC is associated with recurrence and bortezomib resistance in patients with multiple myeloma. Turk J Haematol. 2020;37(4):248–62. doi: 10.4274/tjh.galenos.2020.2020.0243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Jiang Z, et al. Exosomal circEPB41L2 serves as a sponge for mir-21-5p and mir-942-5p to suppress colorectal cancer progression by regulating the PTEN/AKT signalling pathway. Eur J Clin Invest. 2021;51(9):e13581. doi: 10.1111/eci.13581. [DOI] [PubMed] [Google Scholar]
- 192.Sang H, et al. Exosomal circRELL1 serves as a miR-637 sponge to modulate gastric cancer progression via regulating autophagy activation. Cell Death Dis. 2022;13(1):56. doi: 10.1038/s41419-021-04364-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Guo Z, et al. Engineered exosome-mediated delivery of circDIDO1 inhibits gastric cancer progression via regulation of MiR-1307-3p/SOCS2 Axis. J Transl Med. 2022;20(1):326. doi: 10.1186/s12967-022-03527-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Li C, Li X. Exosome-derived Circ_0094343 promotes chemosensitivity of colorectal cancer cells by regulating glycolysis via the miR-766-5p/TRIM67 Axis. Contrast Media Mol Imaging. 2022;2022:2878557. doi: 10.1155/2022/2878557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Yao X, et al. Exosomal circ_0030167 derived from BM-MSCs inhibits the invasion, migration, proliferation and stemness of pancreatic cancer cells by sponging miR-338-5p and targeting the Wif1/Wnt8/betai>-catenin axis. Cancer Lett. 2021;512:38–50. doi: 10.1016/j.canlet.2021.04.030. [DOI] [PubMed] [Google Scholar]
- 196.Meng H, et al. Nanoparticles mediated circROBO1 silencing to inhibit Hepatocellular Carcinoma Progression by modulating miR-130a-5p/CCNT2 Axis. Int J Nanomed. 2023;18:1677–93. doi: 10.2147/IJN.S399318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Gu C, Lu H, Qian Z. Matrine reduces the secretion of exosomal circSLC7A6 from cancer-associated fibroblast to inhibit tumorigenesis of colorectal cancer by regulating CXCR5. Biochem Biophys Res Commun. 2020;527(3):638–645. doi: 10.1016/j.bbrc.2020.04.142. [DOI] [PubMed] [Google Scholar]
- 198.Chao F, et al. Extracellular vesicles derived circSH3PXD2A inhibits chemoresistance of small cell lung cancer by miR-375-3p/YAP1. Int J Nanomed. 2023;18:2989–3006. doi: 10.2147/IJN.S407116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Lu L, et al. EIF4a3-regulated circRABL2B regulates cell stemness and drug sensitivity of lung cancer via YBX1-dependent downregulation of MUC5AC expression. Int J Biol Sci. 2023;19(9):2725–2739. doi: 10.7150/ijbs.78588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Wang L, et al. Exosome-transmitted hsa_circ_0012634 suppresses pancreatic ductal adenocarcinoma progression through regulating miR-147b/HIPK2 axis. Cancer Biol Ther. 2023;24(1):2218514. doi: 10.1080/15384047.2023.2218514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Song J, et al. Exosomal hsa_circ_0017252 attenuates the development of gastric cancer via inhibiting macrophage M2 polarization. Hum Cell. 2022;35(5):1499–1511. doi: 10.1007/s13577-022-00739-9. [DOI] [PubMed] [Google Scholar]
- 202.Shu G, et al. Exosomal circSPIRE1 mediates glycosylation of E-cadherin to suppress metastasis of renal cell carcinoma. Oncogene. 2023;42(22):1802–1820. doi: 10.1038/s41388-023-02678-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Zhou X, et al. EIF4A3-induced circFIP1L1 represses miR-1253 and promotes radiosensitivity of nasopharyngeal carcinoma. Cell Mol Life Sci. 2022;79(7):357. doi: 10.1007/s00018-022-04350-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Chen RX, et al. N(6)-methyladenosine modification of circNSUN2 facilitates cytoplasmic export and stabilizes HMGA2 to promote colorectal liver metastasis. Nat Commun. 2019;10(1):4695. doi: 10.1038/s41467-019-12651-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Du WW, et al. Identifying and characterizing circRNA-protein interaction. Theranostics. 2017;7(17):4183–4191. doi: 10.7150/thno.21299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.McKenzie AJ, et al. KRAS-MEK signaling controls Ago2 sorting into exosomes. Cell Rep. 2016;15(5):978–987. doi: 10.1016/j.celrep.2016.03.085. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.