Abstract
Introduction
Sleep is vital to human health, and sleep staging is an essential process in sleep assessment. However, manual classification is an inefficient task. Along with the increased demand for portable sleep quality detection devices, lightweight automatic sleep staging needs to be developed.
Methods
This study proposes a novel attention-based lightweight deep learning model called LWSleepNet. A depthwise separable multi-resolution convolutional neural network is introduced to analyze the input feature map and captures features at multiple frequencies using two different sized convolutional kernels. The temporal feature extraction module divides the input into patches and feeds them into a multi-head attention block to extract time-dependent information from sleep recordings. The model's convolution operations are replaced with depthwise separable convolutions to minimize its number of parameters and computational cost. The model's performance on two public datasets (Sleep-EDF-20 and Sleep-EDF-78) was evaluated and compared with those of previous studies. Then, an ablation study and sensitivity analysis were performed to evaluate further each module.
Results
LWSleepNet achieves an accuracy of 86.6% and Macro-F1 score of 79.2% for the Sleep-EDF-20 dataset and an accuracy of 81.5% and Macro-F1 score of 74.3% for the Sleep-EDF-78 dataset with only 55.3 million floating-point operations per second and 180 K parameters.
Conclusion
On two public datasets, LWSleepNet maintains excellent prediction performance while substantially reducing the number of parameters, demonstrating that our proposed Light multiresolution convolutional neural network and temporal feature extraction modules can provide excellent portability and accuracy and can be easily integrated into portable sleep monitoring devices.
Keywords: Sleep staging, artificial intelligence, lightweight module, attention, EEG
Introduction
Human health depends on sleep. The ability to monitor sleep quality significantly impacts medical research 1 . The professional diagnosis of sleep disorders and diseases typically relies on polysomnography (PSG), which contains multiple signal channels during sleep, such as electroencephalography (EEG), electrocardiography (ECG), electrooculography (EOG), and electromyography (EMG). These signals typically represent sleep-related events and indicators.2,3 According to the American Academy of Sleep Medicine (AASM) manual, complete sleep recording is divided into 30 s epochs and classified into five stages: Wake (W), rapid eye movement (REM) and three non-REM stages (N1, N2, and N3). 4 Because this is a laborious and inefficient task, automatic sleep staging technology is required to assist physicians. Additionally, it is challenging to implement portable multi-channel sleep monitoring using PSG. Therefore, several studies have used single-channel signals (for example, EEG and EOG) for at-home monitoring to evaluate the quality of sleep. Additionally, the development of portable personal sleep monitoring devices has increased the demand for long-lasting, highly accurate, and cost-effective sleep-staging algorithms.
Many previous studies have used traditional machine learning methods to classify PSG recordings based on time, frequency, and time-frequency-domain features. These methods typically include two steps. First, the discriminative features are extracted and connected into a vector. Next, the vectors are fed into machine learning models, such as support vector machines5,6, decision trees, 7 and random forests, 8 to classify the recordings. These methods have low hardware requirements and high computational costs. However, manually capturing features relies on specialist knowledge, and discriminative features may vary across different devices and patients. Therefore, traditional machine learning models require expertise and perform inconsistently during sleep-stage classification.
The original time-series EEG recordings contain multiple discriminative features. Long-term dependencies also exist in the recording, such as the transition rules that sleep experts use to identify the next possible sleep stage from a PSG epochs' series. 4 Recently, numerous studies have analyzed sleep structures using the powerful feature-learning capabilities of convolutional neural networks (CNNs). CNNs were used to extract information from single-channel EEG spectrograms. 9 One-dimensional CNNs served as feature capture modules with single-channel EEG signals as inputs.10,11 However, these studies failed to consider temporal dependencies in sleep recordings. A few studies have used recurrent neural networks (RNNs) in sleep staging to capture time-dependent features.12,13 For example, 55 time- and frequency-domain features manually extracted from single-channel EEG signals have been used as inputs in the model. 13 Long short-term memory (LSTM) was cascaded using an RNN to construct a model to extract temporal features and classify them. Despite these promising results, these methods still rely on manually extracted features.
Several studies have combined CNN, RNN, and attention modules to extract temporal features to capture time-dependent information in sleep recordings and to improve the automatic sleep stage classification accuracy.14–18 AttnSleep 14 used a multi-resolution CNN (MRCNN) to extract multi-frequency features, which were then fed directly into a temporal feature encoder with an attention mechanism. DeepSleepNet 15 employed MRCNN as a feature extraction block and then fed the features into a bidirectional LSTM to extract temporal information. Bidirectional RNN and attention mechanisms were used as feature extractors in SeqSleepNet 16 . Then, the sequences of the epoch-wise feature vectors were modeled using a bidirectional LSTM block to encode long-term sequence information between epochs. 16 Such a method can automatically complete the feature capture and extraction of time dependencies without manual operation, resulting in high accuracy in end-to-end sleep staging. However, such models with many parameters and excessive computational costs are difficult to integrate into portable sleep monitoring devices.
Recently, several lightweight architectures have been proposed because of the growing demand for portable sleep monitoring devices.19,20 A CNN and channel shuffle model were incorporated into a lightweight model (LightSleepNet) 19 for sleep staging to improve accuracy. TinySleepNet 20 is composed of multiple CNN layers serving as a feature-grasping module and an LSTM serving as a sequence feature extraction module. These studies significantly reduced the model parameters and the computational cost. However, this comes at the expense of the algorithm's ability to classify different sleep stages.
Therefore, the development of portable sleep monitoring devices still faces three obstacles. First, how to capture discriminative features from sleep recordings. Second, the features of sleep recordings have time dependencies, for which the existing models have insufficient learning ability. Third, most studies achieved high accuracy with numerous model parameters and high computational costs, which cannot be applied to portable and long-term sleep monitoring devices.
To address these issues, we propose a novel lightweight sleep staging model, LWSleepNet, and conduct various experiments on two publicly available datasets to evaluate the performance of LWSleepNet. The main contributions of the proposed novel lightweight model are as follows.
It improves its accuracy while reducing the computational cost and number of parameters, enabling it to be integrated into portable sleep-monitoring devices.
It uses multi-headed attention (MHA) to capture temporal dependencies within the feature map.
It uses depthwise separable multi-resolution convolutions to extract features of multiple frequencies in the sleep recording at a low computational cost.
Methods
This section introduces LWSleepNet for automatic sleep staging using a single-channel EEG.
Overview of the model
Figure 1 illustrates the components of LWSleepNet. It consists of three components: Representation extraction, temporal feature extraction (TFE), and an output block.
Figure 1.
Overall structure of the proposed model for automatic sleep staging.
First, the signal is fed into the depthwise separable multiresolution convolution module (Light-MRCNN). It uses two different sizes of convolutional kernels to extract features from the signal. Large and small convolutional kernels extract the low- and high-frequency features, respectively. The output is concatenated and then recalibrated using a bottleneck block. Then, the feature map is fed into the TFE block after representation extraction. It contains a multi-head attention (MHA) module, from which the feature map's temporal features are extracted. Finally, the output module, which contains a pooling layer, a fully connected layer, and a Softmax layer, completes the classification decision. In the following subsections, the principles and functions of each component are described in detail.
Representation learning
Depthwise separable convolutions and bottleneck block
Pointwise and depthwise convolutions (dw Convs) are combined to form a depthwise separable convolution 21 . A pointwise convolution pw Conv, a convolution with a kernel size of one, generates a new feature map by weighting and summing the input feature maps. The output and the input feature maps have the same size. A pw Conv can perform both descending and ascending dimension operations (changing the dimensionality of the output) with less computational effort than a standard convolution and can mix information between channels. dw Conv is different from the standard convolution. The number of convolution kernels in each layer is equal to the number of channels in the input feature map. After the dw Conv operation, the number of channels in both the output and input feature maps is equal. However, this operation performs a convolution operation independently for each channel of the input layer, which ineffectively uses the feature information from various channels at the same spatial location. Therefore, pw Conv must generate new feature maps.
Depthwise separable convolution can reduce the number of parameters and computation required for the model compared with standard convolution. Suppose the size of the input feature map is and produce a feature map, where denotes the length of the input one-dimensional (1D) feature map, is the number of input channels, and denotes the number of output channels.
A convolution kernel of size , where denotes the size of the kernel, denotes the number of input channels, and denotes the number of output channels, parameterize the standard. Therefore, the computational cost of the standard convolution is computed as follows:
| (1) |
A combination of dw Conv and pw Conv is operated on the above inputs to output a feature map of the same size using depthwise separable convolution. A convolution kernel of size , where denotes the size of the kernel and denotes the number of input channels, is used to parameterize the dw Conv. Therefore, the computational cost of depthwise separable convolution is determined as follows:
| (2) |
Therefore, computation reduction is computed as follows:
| (3) |
Figure 2 illustrates the structure of the bottleneck block, which consists of pw Conv, dw Conv, Gaussian error linear unit (GELU) 22 and 1D batch normalization. Particularly, pw Conv(inp, n inp) in Figure 2 refers to a pw Conv with inp input channels and n inp output channels, and dw Conv(ks) refers to a dw Conv with a kernel size of ks.
Figure 2.
Structure of bottleneck block in LWSleepNet.
The bottleneck block was introduced in MobileNetV2 23 and has been adopted in MobileNetV3 24 and many other studies25,26. The bottleneck block uses an inverted residual structure, which resembles the structure of common residuals. However, the residual operation switches the descending and ascending dimensions in the bottleneck block. Pw Conv is used to first increase the number of channels in the input feature; then, information is extracted using dw Conv. Finally, the dimensions are descended using pw Conv. Simultaneously, a shortcut is used to connect the inputs and outputs. In LWSleepNet, the inverted residual structure is used for feature extraction and recalibration of the input 1D feature map, with a GELU 22 serving as the activation function.
Depthwise separable multi-resolution convolution
To extract features from multiple frequencies, a Light-MRCNN was developed, as shown in Figure 3. In the proposed feature-grabbing module, the features of the signals are extracted using a 1D dw Conv layer and a 1D pw Conv layer, each of which is followed by 1D batch normalization. The GeLU is used as the activation function, which has been demonstrated to be more capable of passing negative values 14 and is suitable for EEG signal processing. After concatenating the outputs, a bottleneck block was used as a recalibration to improve the module's performance, and dropout layers were used to reduce overfitting.
Figure 3.
Structure of Light-multi-resolution convolutional neural network (MRCNN).
Inspired by Eldele et al. 14 and Supratak et al. 15 a two-branch convolution module, which contains convolution kernels of different sizes, is implemented to extract the features of single-channel EEG signals at different frequencies. For example, we assumed that the sampling frequency of the input signal is 100 Hz. First, the convolution with a small kernel (receptive field size of five) has a window of 0.05 s to obtain the feature information of the signal at 20 Hz, which roughly corresponds to the frequency of the gamma wave in the EEG. Second, a large kernel (receptive field size of 50) was used to obtain waveform information at approximately 2 Hz, which corresponds to the frequency bands of theta and delta waves in the EEG. Additionally, both large and small convolutional kernels can capture time- and frequency-domains information. 15
Temporal feature extraction
Figure 4 illustrates the structure and working principle of the TFE block. The TFE block first divides (reshapes) the input into several patches and then learns the temporal dependence between each patch using MHA. This was inspired by several previous studies in the computer vision field that divided feature maps into patches and then fed them into multiple heads of attention for feature extraction27–29. Additionally, before and after the MHA block, deep and point-by-point convolutions are combined to capture features and project them into a higher dimensional space.
Figure 4.
The structure of proposed temporal ature extraction (TFE) block.
The MHA block matches the feature information of each patch in the EEG signal epoch with that of other patches. First, MHA uses multiple heads, each of which creates a weight between the different positions in the epoch, allowing for the analysis of the dependencies between the different positions. Then, the MHA adds up these weights to obtain the final weight for each position. These weights were used to encode the patches to obtain information about the temporal features in the EEG signal.
Particularly, we consider the input to be . After extracting the features using convolution, the output is and the feature map is reshaped into patches , where . Each patch is fed into the linear network to generate query ( ), key ( ), and value ( ) which are required by the attention block, that is, . The output value of attention can be calculated as follows:
| (4) |
where represents the ordinal number of the head in an MHA, represents the dimensions of , , , and the operator ( ) represents the dot product. This is referred to as the scaled dot-product attention, which was proposed in 30 . To improve the generalization of the model and extract useful information from the signal, the MHA based on the method described above was used. The first patches were divided into subspaces, where denotes the number of heads, that is, , . The values in each subspace were then separately subjected to the attention operation described above. Finally, the results are concatenated. The output of the MHA block is calculated as follows:
| (5) |
where , and denotes the number of heads.
This module extracts temporal information from the intra-EEG epoch for two distinct reasons. Firstly, extracting information from the 30-second epoch enables the capture of more nuanced features, which enhances sleep staging reliability. Second, the division of the input features into different patches increases the representation subspace compared to mapping the feature map sequences to vector inputs into the temporal feature capture module, for the MHA used by the TFE blocks. This division increases the representation subspace, resulting in attention weights that more accurately reflect the importance of each partition. Cascading these representations leads to a more comprehensive representation, which improves classification accuracy 14 . Additionally, in the experiments presented herein, we endeavored to capture inter-EEG epoch information. However, our results indicate a decrease in performance and an increase in the number of parameters of the model.
In LWSleepNet, the TFE module cuts the input into patches 25 in length and feeds them to the MHA. This module is set to repeat 3 times to extract enough temporal dependencies.
Training method
LWSleepNet was built and trained using Pytorch 1.12 31 . The hyperparameters of the model were derived based on the training results. The batch size was set to 120, and the unit length sequence was 30 s. We used AdamW 32 as the optimizer and set the beta 1, beta 2, and weight decay values to 0.9, 0.999, and 1 , respectively. Cross-entropy loss was used as the loss function, and label smoothing was set to 0.05 to reduce the effect of sample imbalance. We observed that the performance of the model stabilized as it approached 100 epochs. Therefore, the number of epochs was set at 100. A multi-step learning rate scheduler was used to divide the entire training process into three stages: Learning rates at 1–10, 11–90, and 91–100 and epoch were 1 , 1 , and 1 , respectively. The dw Conv kernel size in all bottleneck blocks was set as nine. For the TFE module, we set the number of MHA module heads to eight and the input channels to 64.
Statistical analysis
The model was evaluated epoch-by-epoch by calculating performance metrics, such as staging accuracy. This study assessed the agreement between PSG-based manual scoring and LWSleepNet automated scoring results using Cohen's kappa coefficient ( ) 33 . Confusion matrices were presented to account for scoring accuracy at each sleep stage, and precision, recall rate, and macro-F1 scores were calculated.
To further explore the performance and necessity of modules of the model, ablation experiments were conducted by replacing or removing certain modules. The accuracy and macro-F1 scores of the individual models were cross-validated on publicly available datasets, and a t-test was used to explore their variability compared to the original models. Therefore, as shown in Table 5, the statistical significance of differences between the performance of the replaced or disassembled models and the original model was examined.
Table 5.
Accuracy, MF1 (average standard deviation), and number of parameters of the algorithms in ablation study.
| Blocks | ACC | MF1 | Param |
|---|---|---|---|
| LWSleepNet | 86.6 2.7 | 79.2 2.6 | 1.8 |
| w/o depthwise separable convolutions | 86.5 2.9 | 79.4 2.8 | 6.0 |
| w/o Light-MRCNN | 81.7 2.5* | 69.9 2.2* | 1.6 |
| w/o TFE block | 76.6 3.5* | 62.4 3.1* | 7.9 |
| MRCNN with LSTM block | 81.3 1.8* | 75.9 2.3* | 6.6 |
MRCNN: multi-resolution convolutional neural network; TFE: temporal feature extraction; LSTM: long short-term memory; MF1: macro-averaged F1 score; Param: number of parameters.
*
Results
Datasets
In our experiments, we used the Sleep-EDF-20 and Sleep-EDF-78 public datasets 34 obtained from the PhysioBank 35 . Table 1 demonstrate the detail information including the distribution of epoch labels. Sleep-EDF-20 contained sleep PSG data files containing sleep data from 20 subjects, whereas Sleep-EDF-78 was an extended version containing sleep PSG data from 78 individuals over several nights. The data collected from the subjects included two channels of EEG signals (from the Fpz-Cz and Pz-Oz electrode locations), an EOG signal (horizontal), an EMG signal (chin), and an event marker. Additionally, both the EOG and EEG signals were sampled at 100 Hz. The data for the whole night were recorded in two files: a SC*PSG.edf file contained the data for each channel and a *Hypnogram.edf file contained the stage classification markers for each 30 s signal as described in the Rechtschaffen and Kales manual 4 . In the experiments, marker files and EEG signals from the Fpz-Cz channel were used as training and validation data, respectively.
Table 1.
Details of the datasets.
| Datasets | W | N1 | N2 | N3 | REM | #Total samples |
|---|---|---|---|---|---|---|
| Sleep-EDF-20 | 8285 | 2804 | 17,799 | 5703 | 7717 | 42,308 |
| 19.6% | 6.6% | 42.1% | 13.5% | 18.2% | ||
| Sleep-EDF-78 | 65,951 | 21,522 | 69,132 | 13,039 | 25,835 | 195,479 |
| 33.7% | 11.0% | 35.4% | 6.7% | 13.2% |
Performances
The model's performance was accurately evaluated using subject-wise 20-fold cross-validation. The Sleep-EDF-20 dataset was divided into 20 groups based on the subjects. For 20 rounds, 19 data points were used as the training set and the remaining one as the test set. Finally, we combined the predicted sleep stages from all 20 rounds of the test sample to calculate the various performance metrics.
We evaluated the model using the following metrics: Accuracy (ACC), precision (PR), recall (RE), macro-averaged F1 score (MF1), Cohen Kappa ( ), number of parameters (Param), and floating point of operations (FLOPs). Given the true positives ( ), false positives ( ), true negatives ( ), and false negatives ( ) of class , the following metrics: ACC, PR, RE, and MF1, can be calculated as follows:
| (6) |
| (7) |
where and .
The LWSleepNet was cross-validated on Sleep-EDF-20 and Sleep-EDF-78 datasets. The confusion matrix was counted and calculated for both datasets along with the corresponding PR, RE and F1 scores (F1) for each classification and the results are shown in Tables 2 and 3. Finally, the overall performances were calculated as follows. LWSleepNet had an ACC of 86.6% and MF1 of 79.2% for the Sleep-EDF-20 dataset and an ACC of 81.5% and MF1 of 74.3% for the Sleep-EDF-78 dataset.
Table 2.
Confusion matrix on Sleep-EDF-20.
| Predict | Performance | |||||||
|---|---|---|---|---|---|---|---|---|
| W | N1 | N2 | N3 | R | PR | RE | MF1 | |
| W | 7745 | 258 | 85 | 20 | 177 | 91.3 | 93.5 | 92.4 |
| N1 | 472 | 923 | 648 | 4 | 757 | 55.5 | 32.9 | 41.3 |
| N2 | 111 | 258 | 16389 | 469 | 572 | 88.3 | 92.1 | 90.2 |
| N3 | 17 | 0 | 769 | 4913 | 4 | 90.9 | 86.2 | 88.4 |
| R | 139 | 225 | 667 | 2 | 6684 | 81.6 | 86.7 | 84.0 |
PR: precision; RE: recall; MF1: macro-averaged F1 score.
Table 3.
Confusion matrix on Sleep-EDF-78.
| Predict | Performance | |||||||
|---|---|---|---|---|---|---|---|---|
| W | N1 | N2 | N3 | R | PR | RE | MF1 | |
| W | 58312 | 2945 | 771 | 83 | 3840 | 96.4 | 88.4 | 92.2 |
| N1 | 1604 | 7775 | 6177 | 1060 | 4906 | 45.4 | 36.1 | 40.2 |
| N2 | 278 | 4190 | 62541 | 470 | 1653 | 83.6 | 90.5 | 86.9 |
| N3 | 37 | 800 | 2518 | 9468 | 216 | 85.2 | 72.6 | 78.4 |
| R | 261 | 1422 | 2812 | 29 | 21311 | 66.8 | 82.5 | 73.8 |
PR: precision; RE: recall; MF1: macro-averaged F1 score.
To validate the proposed model in assisting medical diagnosis, we selected the first fold of the model in a cross-validation of two datasets as well as the test dataset to output sleep staging results and predict sleep latency (SL), REM sleep latency (RSL), and total sleep time (TST). Subsequently, the predicted and true parameters were used to calculate the root mean squared error (RMSE), as shown in Table 4. Based on the data presented in the table, it is evident that the model's predictions for SL and TST are relatively accurate, with a negligible error of 0 for SL in Sleep-EDF-20. However, in Sleep-EDF-78, the model's prediction error on RSL is considerably higher. This disparity may be attributed to the shorter duration the first REM period and the model's less capability of predicting the REM stages, which is indicated by Tables 2 and 3. Nevertheless, the model's capability to compute clinical relevant parameters has the potential to assist in clinical diagnosis. Therefore, it can be inferred that the model provides a valuable tool in the assessment of sleep-related disorders.
Table 4.
The measurement RMSE for several clinically relevant parameters.
| Datasets | SL (min) | RSL (min) | TST (min) |
|---|---|---|---|
| Sleep-EDF-20 | 0.0 | 0.5 | 24.2 |
| Sleep-EDF-78 | 9.8 | 80.1 | 9.7 |
RMSE: root mean squared error; SL: sleep latency; RSL: rapid eye movement sleep latency; TST: total sleep time.
To further demonstrate the above findings, a visualization of the output for a subject from the Sleep-EDF-20 dataset is shown in Figure 5. Figure 5(a) shows the probability curves for each category, Figure 5(b) shows the output Hypnogram predicted by the model while Figure 5(c) shows the classification results of the data by the sleep experts in the dataset. The in Figure 5(b) indicate the prediction results that differ from the experts. In Figure 5(a), the threshold value of confidence is 0.5. The majority of errors are observed during the transition period and at low confidence levels, while they occur less frequently throughout the duration of a stage.
Figure 5.
Visualization of the output of one subject of Sleep-EDF-20.
Ablation study
LWSleepNet integrates depthwise separable convolution, Light-MRCNN, and TFE blocks. Ablation experiments were conducted on the Sleep-EDF-20 dataset to validate the need for each module. We evaluated the LWSleepNet model, the model with all depthwise separable convolutions replaced with normal convolutions, the model without the TFE block, and the model without the Light-MRCNN. To further explore the superiority of intra-EEG epoch information in LWSleepNet, we added one model to the ablation experiment – replacing the TFE block with a LSTM module to capture the inter-EEG epoch features. Additionally, unlike other models' training methods, the input sequences of the MRCNN with LSTM model will not be shuffled and the cell states of the LSTM are reset for each subject. The models' performance was evaluated using the following metrics: ACC, MF1, and number of parameters (Figure 6).
Figure 6.
The result visualization of sensitivity analysis for number of temporal feature extraction (TFE) blocks.
According to the ablation study shown in Table 5, significant statistical differences can be found in the performance of these models ( ), and the following conclusions can be drawn. First, the depth-separable convolution drastically reduces the number of parameters in the model, which improves the portability of the model while having almost no impact on its accuracy. Second, the TFE block is adept at extracting temporally dependent information, which increases stage classification accuracy. Third, the automatic feature capture capability of the light-MRCNN improves the accuracy of the model. It can extract recording information from two frequencies to capture the most discriminative features. Moreover, we find that combining the MRCNN module with the LSTM model results in lower performance and portability compared to the original model, which demonstrates the superiority of our TFE block and the distinguishability of the intra-epoch information. In conclusion, the Light-MRCNN, TFE block, and depthwise separable convolution are crucial components of the model.
To further validate the impact of each newly proposed component, we replaced the representation extraction module and temporal information extraction module in AttnSleep 14 with Light-MRCNN and TFE block, respectively. Then, we trained and validated the combined model on Sleep-EDF-20, and the results were presented in Table 6. According to the table, it was evident that the proposed modules were not only applicable to LWSleepNet, but they could also improve the classification accuracy of models of the same type. Meanwhile, although these combined models have slightly higher accuracy than LWSleepNet, the proposed model still leads substantially in terms of number of parameters and MF1. Additionally, when Light-MRCNN was employed for AttnSleep, the number of parameters of the combined model substantially decreased due to the inclusion of depthwise separable convolutional layers. Notably, replacing the components did not result in a obvious change in the MF1 score of the model. Therefore, to address the problem of data imbalance in the future, it will be necessary to consider additional learning methods and loss functions.
Table 6.
Analysis of proposed components with AttnSleep.
| Blocks | ACC | MF1 | Param |
|---|---|---|---|
| AttnSleep | 84.4 | 78.1 | 5.2 |
| AttnSleep (with TFE block) | 87.2 | 78.5 | 5.6 |
| AttnSleep (with Light-MRCNN) | 86.8 | 77.8 | 1.8 |
MF1: macro-averaged F1 score; ACC: accuracy; MRCNN: multi-resolution convolutional neural network; TFE: temporal feature extraction; Param: number of parameters.
Sensitivity analysis for number of TFE blocks
As TFE block is a key component of our model, we repeated it to capture more temporal dependencies, and thus it is essential to study how the number of TFE blocks ( ) affects the model's accuracy and portability. In particular, we fix the other parameters and test different . With the other parameters set as described above, the model was trained in Sleep-EDF-20with from 1 to 7. The figure shows the variation of thousands of parameters (KParam), million FLOPs, and accuracy with the number of TFE blocks. As increases in the range of [1, 7], we can observe that the number of parameters and computational cost of the model increase substantially, while the accuracy only increases very slightly after is greater than 3. Therefore, to balance the portability and performance of the model, we set the number of TFE blocks to 3.
Discussion
Due to the development of portable sleep monitoring devices, the demand for lightweight sleep algorithms increase. In this study, we propose a novel lightweight sleep staging algorithm with single channel EEG, LWSleepNet. In this model, we use Light-MRCNN to capture discriminative features and use the TFE module to capture association between features. Therefore, the model is able to effectively extract features from EEG signals and temporal dependencies to accomplish 86.6% and 81.5% accuracy on Sleep-EDF-20 and Sleep-EDF-78 datasets, respectively. Finally, the necessity and performance of each module in the model are verified by ablation study and the optimal number of TFE modules from 1 to 7 is explored by sensitivity study.
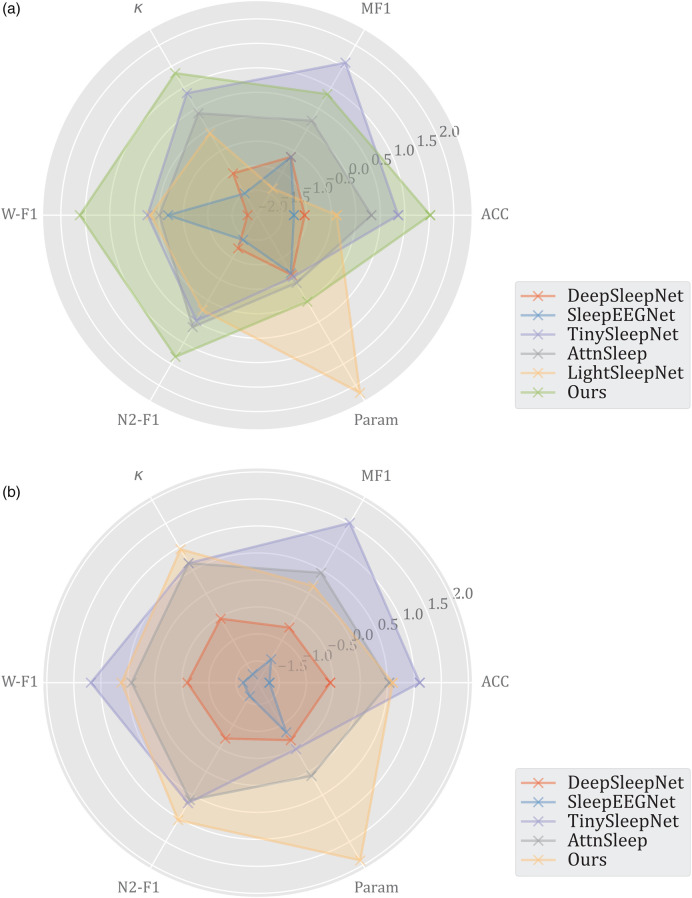
Table 7 compares and analyzes LWSleepNet and previous work on raw single-channel EEG or PSG-based automated sleep staging. Additionally, we normalized the data in Table 7 to obtain the radar map, as shown in Figure 7. All of these studies were based on deep learning networks for five classifications. Most of the researchers used CNN for feature extraction followed by extraction of temporal dependencies using RNN or MHA. A small number of studies used only CNNs for classification. Also, only a small part of these studies are exploring lightweight models. DeepSleepNet 15 implemented a multi-branch CNN, LSTM, and a residual connection for sleep staging. Also, SleepEEGNet 36 used the same CNN feature extraction structure as DeepSleepNet, followed by a coder-decoder structure with attention for sleep stage classification. Additionally, TinySleepNet 20 used the structure of DeepSleepNet and removed the CNN bypass to reduce the computation and number of parameters, which enabled lightweight sleep staging. AttnSleep 14 used a CNN with two resolutions as the automatic feature extraction module, followed by a temporal information encoder containing an attention mechanism to classify sleep stages. LightSleepNet 19 used CNN with a residual connection to construct a lightweight sleep stage classification model.
Table 7.
Performance comparison among LWSleepNet and previous work.
| Model | Dataset | Overall Metrics | Per-class F1-Score | Param | FLOPs | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| ACC | MF1 | W | N1 | N2 | N3 | REM | |||||
| DeepSleepNet | Sleep-EDF-20 | 81.9 | 76.6 | 0.76 | 86.7 | 45.5 | 85.1 | 83.3 | 82.6 | 2.6 | - |
| Sleep-EDF-78 | 77.8 | 71.8 | 0.70 | 90.9 | 45.0 | 79.2 | 72.7 | 71.1 | |||
| SleepEEGNet | Sleep-EDF-20 | 81.5 | 76.6 | 0.75 | 89.4 | 44.4 | 84.7 | 84.6 | 79.6 | 2.1 | - |
| Sleep-EDF-78 | 74.2 | 69.9 | 0.66 | 89.8 | 42.1 | 75.2 | 70.4 | 70.6 | |||
| TinySleepNet | Sleep-EDF-20 | 85.4 | 80.5 | 0.80 | 90.1 | 51.4 | 88.5 | 88.3 | 84.3 | 1.3 | - |
| Sleep-EDF-78 | 83.1 | 78.1 | 0.77 | 92.8 | 51.0 | 85.3 | 81.1 | 80.3 | |||
| AttnSleep | Sleep-EDF-20 | 84.4 | 78.1 | 0.79 | 89.7 | 42.6 | 88.8 | 90.2 | 79.0 | 5.2 | 60.9 M |
| Sleep-EDF-78 | 81.3 | 75.1 | 0.74 | 92.0 | 42.0 | 85.0 | 82.1 | 74.2 | |||
| LightSleepNet | Sleep-EDF-20 | 83.8 | 75.3 | 0.78 | 90.0 | 31.0 | 88.0 | 89.0 | 78.0 | 4.3 | 45.8 M |
| Ours | Sleep-EDF-20 | 86.6 | 79.2 | 0.81 | 92.4 | 41.3 | 90.2 | 88.4 | 84.0 | 1.8 | 55.3 M |
| Sleep-EDF-78 | 81.5 | 74.3 | 0.75 | 92.2 | 40.2 | 86.9 | 78.4 | 73.8 | |||
REM: rapid eye movement; Param: number of parameters; ACC: accuracy; MF1: macro-averaged F1 score; FLOPs: floating-point operations.
Figure 7.
Radar map for comparison.
According to the result of comparison, the proposed model employs fewer parameters and computations while achieves higher accuracy. It is noteworthy that the model exhibits superior accuracy compared to the majority of existing models, owing to the utilization of Light MRCNN and TFE block, and it has better performance in discriminating the W and N2 sleep stages. Additionally, because of the depth-separable convolution, LWSleepNet allows both accuracy and portability (number of parameters and FLOPs), indicating that the model achieves a better balance between the number of parameters and performance. However, the model tended to misclassify the N1 periods as W and N2, as indicated by the performance comparisons in Table 7 and the confusion matrices in Tables 2 and 3.
LWSleepNet has a similar structure to AttnSleep 14 , but has several differences in performance. First, the main reason is that all CNN layers are replaced with depthwise separable convolution layers in this study, which ensures that the model extracts features accurately while reducing the computational cost and the number of parameters. Second, the proposed model cuts the feature maps into several equal length patches and inputs them into the MHA before the temporal correlation extraction. Meanwhile, the training of AttnSleep 14 utilizes a class-aware cost-sensitive loss function to deal with the data imbalance problem. This illustrates the balanced performance of its model on each class. Meanwhile, Table 7 shows that LightSleepNet 19 has the best portability. The model was constructed using only several layers of CNNs, but the use of unsupervised learning for training allowed the model to improve its classification performance. However, the model could not achieve a higher accuracy because it did not consider the temporal dependence information present in the EEG signal. Furthermore, although the majority of the LWSleepNet's performance metrics are inferior to those of TinySleepNet 20 , which also aimed to enhance the model's portability. LWSleepNet substantially decreases the number of model parameters, while preserving fine performance metrics.
Despite the promising results, our study also has some limitations. First, the databases used in our experiments are from retrospective studies with biased sets of recordings collected by specific hardware. Therefore, we should use new recorded data in a prospective way in further studies. Secondly, the model is not sufficiently capable of handling imbalanced data where N1 stage is underrepresented. Data balancing and improved loss functions should be used to solve the problem. Finally, the portability of the model should be further improved to better suit the needs of mobile sleep monitoring devices. This can be achieved by reducing the number of modules and using methods such as semi-supervised and self-supervised learning.
Conclusion
We propose an automatic sleep staging model, LWSleepNet, which is based on a Light-MRCNN and a TFE block. The Light-MRCNN was used to automatically extract features, and the TFE block was used to extract temporal information. Depthwise separable convolution and bottleneck blocks were used to perform recalibration, dimensionality operations, and feature extraction operations. Cross-validation experiments were used to evaluate the model's performance and compare it with previous studies. The results demonstrate that our model substantially improve mobility while maintains the performance of the model and improves certain metrics to a certain extent. Finally, an ablation experiment was performed to verify the necessity and functionality of each module in the model.
In the future, we intend to explore ways to achieve higher accuracy while minimizing the number of model parameters and FLOPs to achieve a highly accurate sleep staging algorithm suitable for portable devices. Furthermore, we shall endeavor to find and employ larger datasets which is correspond to criterion adopted in sleep centers for the purpose of training and validating our models, thereby enabling our model to be applied in medical scenarios.
Acknowledgements
The authors would like to thank Editage (www.editage.cn) for English language editing.
Footnotes
Author Contributions: Chenguang Yang handled the methodology and software of the study. Baozhu Li handled the methodology and writing—review and editing of the study. Yamei Li handled the writing—review and editing of the study. Yixuan He handled the writing—review and editing of the study. Yuan Zhang handled the project administration and supervision of the study.
The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Ethical approval: This study was conducted using the Sleep-EDF dataset publicly available at Physionet and did not involve further ethical approval or patient consent.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was supported in part by the National Natural Science Foundation of China under Grant 62172340 and in part by the Chongqing Student Innovation and Entrepreneurship Training Program under Grant S202210635179.
Guarantor: Yuan Zhang.
ORCID iD: Chenguang Yang https://orcid.org/0000-0002-5002-7987
References
- 1.Luyster FS, Strollo PJ, Zee PC. et al. Sleep: a health imperative. Sleep 2012; 35: 727–734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Butkov N, Keenan SA. An overview of polysomnographic technique. Springer, 2017. pp. 267–294.
- 3.Keenan SA. An overview of polysomnography. Elsevier, 2005. pp. 33–50.
- 4.Moser D, Anderer P, Gruber G. et al. Sleep classification according to AASM and rechtschaffen & kales: effects on sleep scoring parameters. Sleep 2009; 32: 139–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhu G, Li Y, Wen P. Analysis and classification of sleep stages based on difference visibility graphs from a single-channel EEG signal. IEEE J Biomed Health Inform 2014; 18: 1813–1821. [DOI] [PubMed] [Google Scholar]
- 6.Sharma M, Dhiman HS, Acharya UR. Automatic identification of insomnia using optimal antisymmetric biorthogonal wavelet filter bank with ECG signals. Comput Biol Med 2021; 131: 104246. [DOI] [PubMed] [Google Scholar]
- 7.Lajnef T, Chaibi S, Ruby P. et al. Learning machines and sleeping brains: automatic sleep stage classification using decision-tree multi-class support vector machines. J Neurosci Methods 2015; 250: 94–105. [DOI] [PubMed] [Google Scholar]
- 8.Seifpour S, Niknazar H, Mikaeili M. et al. A new automatic sleep staging system based on statistical behavior of local extrema using single channel EEG signal. Expert Syst Appl 2018; 104: 277–293. [Google Scholar]
- 9.Kuo CE, Chen GT, Liao PY. An EEG spectrogram-based automatic sleep stage scoring method via data augmentation, ensemble convolution neural network, and expert knowledge. Biomed Signal Process Control 2021; 70: 102981. [Google Scholar]
- 10.Tsinalis O, Matthews PM, Guo Y. et al. Automatic sleep stage scoring with single-channel EEG using convolutional neural networks. arXiv preprint arXiv:161001683 2016;.
- 11.Li F, Yan R, Mahini R. et al. End-to-end sleep staging using convolutional neural network in raw single-channel EEG. Biomed Signal Process Control 2021; 63: 102203. [Google Scholar]
- 12.Wei Y, Qi X, Wang H. et al. A multi-class automatic sleep staging method based on long short-term memory network using single-lead electrocardiogram signals. IEEE Access 2019; 7: 85959–85970. [Google Scholar]
- 13.Michielli N, Acharya UR, Molinari F. Cascaded LSTM recurrent neural network for automated sleep stage classification using single-channel EEG signals. Comput Biol Med 2019; 106: 71–81. [DOI] [PubMed] [Google Scholar]
- 14.Eldele E, Chen Z, Liu C. et al. An attention-based deep learning approach for sleep stage classification with single-channel eeg. IEEE Trans Neural Syst Rehabil Eng 2021; 29: 809–818. [DOI] [PubMed] [Google Scholar]
- 15.Supratak A, Dong H, Wu C. et al. DeepSleepNet: A model for automatic sleep stage scoring based on raw single-channel EEG. IEEE Trans Neural Syst Rehabil Eng 2017; 25: 1998–2008. [DOI] [PubMed] [Google Scholar]
- 16.Phan H, Andreotti F, Cooray N. et al. Seqsleepnet: End-to-end hierarchical recurrent neural network for sequence-to-sequence automatic sleep staging. IEEE Trans Neural Syst Rehabil Eng 2019; 27: 400–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang H, Guo H, Zhang K. et al. Automatic sleep staging method of EEG signal based on transfer learning and fusion network. Neurocomputing 2022; 488: 183–193. [Google Scholar]
- 18.Urtnasan E, Park JU, Joo EY. et al. Deep convolutional recurrent model for automatic scoring sleep stages based on single-lead ECG signal. Diagnostics 2022; 12: 1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liao Y, Zhang C, Zhang M. et al. LightSleepNet: Design of a personalized portable sleep staging system based on single-channel EEG. IEEE Trans Circuits Syst II: Exp Briefs 2021; 69: 224–228. [Google Scholar]
- 20.Supratak A, Guo Y. Tinysleepnet: An efficient deep learning model for sleep stage scoring based on raw single-channel EEG. In 2020 42nd Annual International Conference of the IEEE Engineering in Medicine & Biology Society (EMBC). IEEE, pp. 641–644. [DOI] [PubMed]
- 21.Howard AG, Zhu M, Chen B. et al. Mobilenets: Efficient convolutional neural networks for mobile vision applications. arXiv preprint arXiv:170404861 2017.
- 22.Hendrycks D, Gimpel K. Gaussian error linear units (GELUs). arXiv preprint arXiv:160608415 2016.
- 23.Sandler M, Howard A, Zhu M. et al. Mobilenetv2: Inverted residuals and linear bottlenecks. In Proceedings of the IEEE conference on computer vision and pattern recognition. pp. 4510–4520.
- 24.Howard A, Sandler M, Chu G. et al. Searching for mobilenetv3. In Proceedings of the IEEE/CVF international conference on computer vision. pp. 1314–1324.
- 25.Saxen F, Werner P, Handrich S. et al. Face attribute detection with mobilenetv2 and nasnet-mobile. In 2019 11th International Symposium on Image and Signal Processing and Analysis (ISPA). IEEE, pp. 176–180.
- 26.Qian S, Ning C, Hu Y. MobileNetV3 for image classification. In 2021 IEEE second International Conference on Big Data, Artificial Intelligence and Internet of Things Engineering (ICBAIE). IEEE, pp. 490–497.
- 27.Dosovitskiy A, Beyer L, Kolesnikov A. et al. An image is worth 16x16 words: Transformers for image recognition at scale. arXiv preprint arXiv:201011929 2020.
- 28.Mehta S, Rastegari M. Separable self-attention for mobile vision transformers. arXiv preprint arXiv:220602680 2022.
- 29.Mehta S, Rastegari M. Mobilevit: light-weight, general-purpose, and mobile-friendly vision transformer. arXiv preprint arXiv:211002178 2021.
- 30.Vaswani A, Shazeer N, Parmar N. et al. Attention is all you need. In: Guyon I, Von Luxburg U, Bengio S, et al. (eds) Advances in Neural Information Processing Systems. Curran Associates, Inc., 2017; 30. https://proceedings.neurips.cc/paper_files/paper/2017/file/3f5ee243547dee91fbd053c1c4a845aa-Paper.pdf [Google Scholar]
- 31.Paszke A, Gross S, Massa F. et al. Pytorch: An imperative style, high-performance deep learning library. In: Wallach H, Larochelle H, Beygelzimer A, et al. (eds) Advances in Neural Information Processing Systems. Curran Associates, Inc., 2019; 32. https://proceedings.neurips.cc/paper_files/paper/2019/file/bdbca288fee7f92f2bfa9f7012727740-Paper.pdf [Google Scholar]
- 32.Loshchilov I, Hutter F. Decoupled weight decay regularization. arXiv preprint arXiv:171105101 2017.
- 33.Cohen J. A coefficient of agreement for nominal scales. Educ Psychol Meas 1960; 20: 37–46. [Google Scholar]
- 34.Kemp B, Zwinderman A, Tuk B. et al. Analysis of a sleep-dependent neuronal feedback loop: the slow-wave microcontinuity of the EEG. IEEE Trans Biomed Eng 2000; 47: 1185–1194. DOI: 10.1109/10.867928. [DOI] [PubMed] [Google Scholar]
- 35.Goldberger AL, Amaral LA, Glass L. et al. Physiobank, physiotoolkit, and physionet: components of a new research resource for complex physiologic signals. circulation 2000; 101: e215–e220. [DOI] [PubMed] [Google Scholar]
- 36.Mousavi S, Afghah F, Acharya UR. SleepEEGNet: Automated sleep stage scoring with sequence to sequence deep learning approach. PLoS ONE 2019; 14: e0216456. [DOI] [PMC free article] [PubMed] [Google Scholar]