Abstract
Purpose:
To simultaneously register all the longitudinal images acquired in a radiotherapy course for analyzing patients’ anatomy changes for adaptive radiotherapy (ART).
Methods:
To address the unique needs of ART, we designed Seq2Morph, a novel deep learning-based deformable image registration (DIR) network. Seq2Morph was built upon VoxelMorph which is a general-purpose framework for learning-based image registration. The major upgrades are 1) expansion of inputs to all weekly CBCTs acquired for monitoring treatment responses throughout a radiotherapy course, for registration to their planning CT; 2) incorporation of 3D convolutional long short-term memory between the encoder and decoder of VoxelMorph, to parse the temporal patterns of anatomical changes; and 3) addition of bidirectional pathways to calculate and minimize inverse consistency errors (ICE). Longitudinal image sets from 50 patients, including a planning CT and six weekly CBCTs per patient were utilized for the network training and cross-validation. The outputs were deformation vector fields for all the registration pairs. The loss function was composed of a normalized cross-correlation for image intensity similarity, a DICE for contour similarity, an ICE, and a deformation regularization term. For performance evaluation, DICE and Hausdorff distance (HD) for the manual vs. predicted contours of tumor and esophagus on weekly basis were quantified and further compared with other state-of-the-art algorithms, including conventional VoxelMorph and Large Deformation Diffeomorphic Metric Mapping (LDDMM).
Results:
Visualization of the hidden states of Seq2Morph revealed distinct spatiotemporal anatomy change patterns. Quantitatively, Seq2Morph performed similarly to LDDMM, but significantly outperformed VoxelMorph as measured by GTV DICE: (0.799±0.078, 0.798±0.081, 0.773±0.078), and 50% HD (mm): (0.80±0.57, 0.88±0.66, 0.95±0.60). The per-patient inference of Seq2Morph took 22s, much less than LDDMM (~30 min).
Conclusion:
Seq2Morph can provide accurate and fast DIR for longitudinal image studies by exploiting spatial-temporal patterns. It closely matches the clinical workflow and has the potential to serve for both online and offline ART.
Keywords: Deep learning, Deformable image registration, Lung cancer, Adaptive Radiotherapy
I. INTRODUCTION
Deformable image registration (DIR) is a cornerstone of medical image analysis, and essential to many modern radiotherapy applications such as accumulating delivered radiation dose and analyzing anatomical and physiological changes in response to radiation.1–8 However, wide adoption of DIR in a routine clinical operation is still limited, due to concerns of inaccuracy and uncertainty in many DIR algorithms, and lack of efficient integration into a busy and demanding clinic workflow. For example, in an adaptive radiotherapy workflow, a series of multi-modal images including the planning CT (pCT) and weekly CBCTs acquired during treatment need to be simultaneously registered to the same frame of reference for subsequent analysis. Conventionally, a DIR algorithm involves two image sets, and maps a moving image to a fixed image to optimize the spatial correspondences. As a result, registrations of multiple image sets must be done sequentially. Time spent on registrations, especially with advanced algorithms such as Large Deformation Diffeomorphic Metric Mapping (LDDMM), 9 could mount to hours and becomes unacceptable for on-line applications. Furthermore, there exist considerable inconsistencies on the voxel level between mapping all the images backwards to the pCT and forwards to the latest weekly CBCT, which could cause significant uncertainties in accumulated radiation dose particularly for evaluating dose on a small volume such as D0.1cc. 10 Therefore there is a pressing need to develop an accurate, consistent, and fast DIR algorithm and facilitate the exact applications of adaptive radiotherapy and longitudinal image analysis on a large clinical scale.
While most DIR algorithms focus on spatial and image intensity features to determine deformations, there exists abundant and untapped information in a longitudinal imaging study that can be explored to improve the performance of a DIR. On a series of weekly images acquired during a radiotherapy course, systematic and graduate anatomical changes of structures of interest, such as gross tumor volume (GTV) and esophagus, have been identified. This information can be utilized to reduce the CBCT imaging dose, 11 or incorporated into either a regression model 12 or deep learning algorithms equipped with recurrent neural network 13–15 to predict how structures evolve spatially for the rest of a radiotherapy course. We hypothesize that these successes can also to be transferred to DIR: useful and robust features of patient’s anatomy can be reliably extracted in the temporal domain uniquely available in a longitudinal series, properly linked in a personalized and continuous fashion, and innovatively utilized to facilitate accurate and consistent mappings of multiple image samples in an image series in addition to the popular similarity measures of geometry and image intensity.
Deep learning based deformable image registration (DL-DIR) algorithms has been increasingly investigated and applied for radiotherapy. 16–20 Accuracies of DL-DIR methods have been substantially improved over the years and approaching to the benchmark of traditional methods such as LDDMM.21 A clear advantage of DL-DIR is its speed since it allocates time for data crunching in advance, and often completes one registration in seconds, two or three orders of magnitude faster than the conventional DIR algorithms.22 This makes DL-DIR well-suited for time-demanding tasks such as online adaptive radiotherapy. Furthermore, flexibility in the design of a neural network structure enables a DL-DIR to easily accommodate a time series with ever-increasing length,23 which is critical to analyzing all available images samples simultaneously and matching closely to the clinical workflow of adaptive radiotherapy. In this paper, we present a novel DL-DIR neural network to parse both spatial and temporal features in a longitudinal image series and perform accurate and timely DIR for patients receiving lung radiotherapy.
II. METHOD AND MATERIALS
We designed a new DL-DIR neural network, called Seq2Morph, to be integrated into clinical workflows as illustrated in Figure 1. The input of Seq2Morph includes a pCT, and all available weekly CBCTs with an increasing order of time acquired during a lung radiotherapy course for monitoring radiation responses. The output includes all the registration pairs’ deformation vector fields (DVF). Weekly CBCTs were warped to the pCT according to the corresponding DVFs and served subsequent clinical tasks such as response analysis. As radiotherapy progresses, new weekly CBCT appears one after another following a rigorous schedule. Seq2Morph was pre-trained to handle up to the latest 6 weekly CBCTs at once in a sliding window fashion and ensured a quick turnaround to facilitate a busy clinic.
Figure 1.
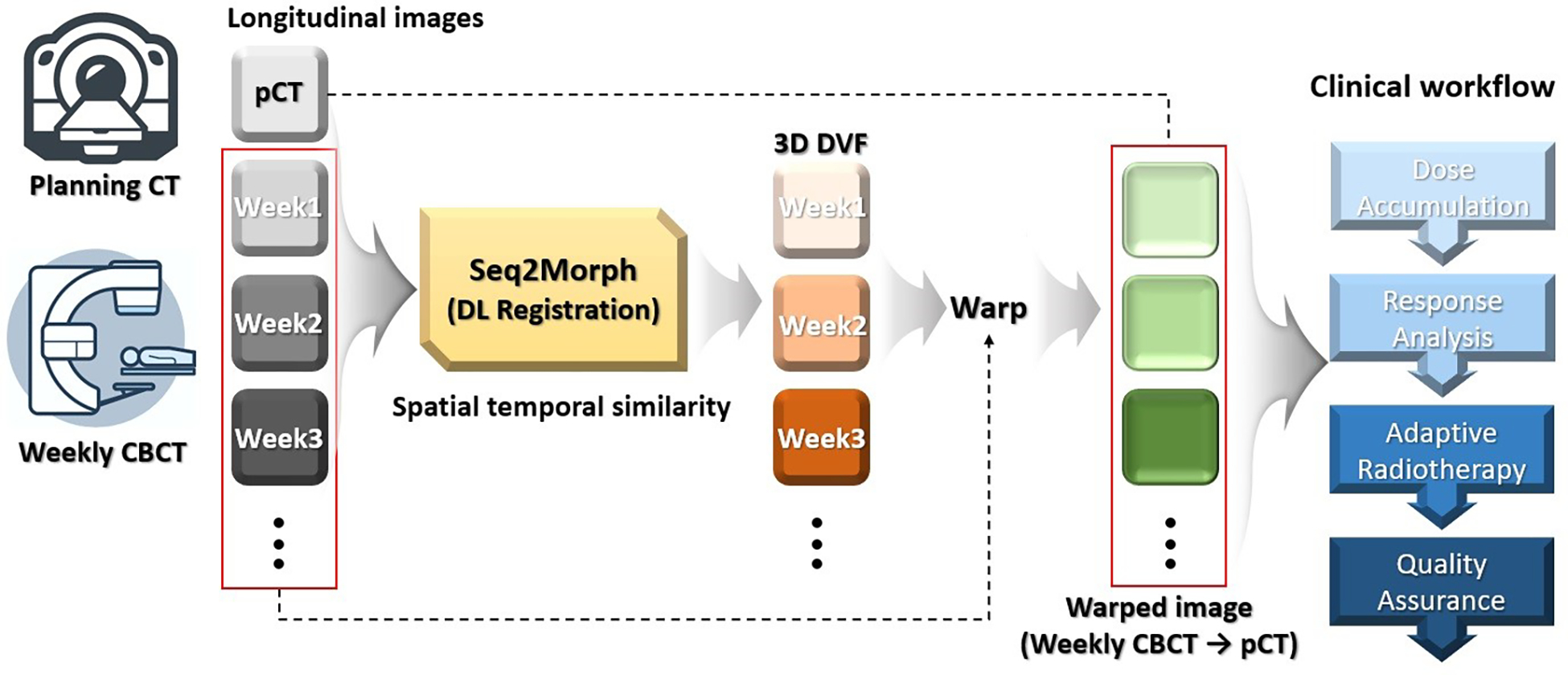
Seq2Morph is designed to register planning CT and weekly CBCTs acquired during radiotherapy for the longitudinal evaluation of anatomical changes of tumor and critical structures. This diagram shows its integration into the clinical workflow.
2.1. Overview of Seq2Morph
Seq2Morph as illustrated in Figure 2 was built upon a standard DL-DIR network, VoxelMorph, which is a general-purpose framework for learning-based image registration.24 The encoder of Seq2Morph (Figure 2.a) consisted of 4 sequential 3D convolutional (Conv3D) units with 2 strides to extract features in multiple resolutions. Each resolution layer in the encoder was activated by a ReLu activation function. The kernel size of the Conv3D layer was 3×3×3. The channel number was 16 at initialization and increased two times at each sequential 3D convolution layer. Batch normalization was also applied. As a result, the spatial and image intensity features relevant for registering a pCT to an individual weekly CBCT were parsed from fine to coarse resolutions levels in four steps, and the dimension of the feature map was reduced from 160×160×128 to 10×10×8 in the bottleneck.
Figure 2.
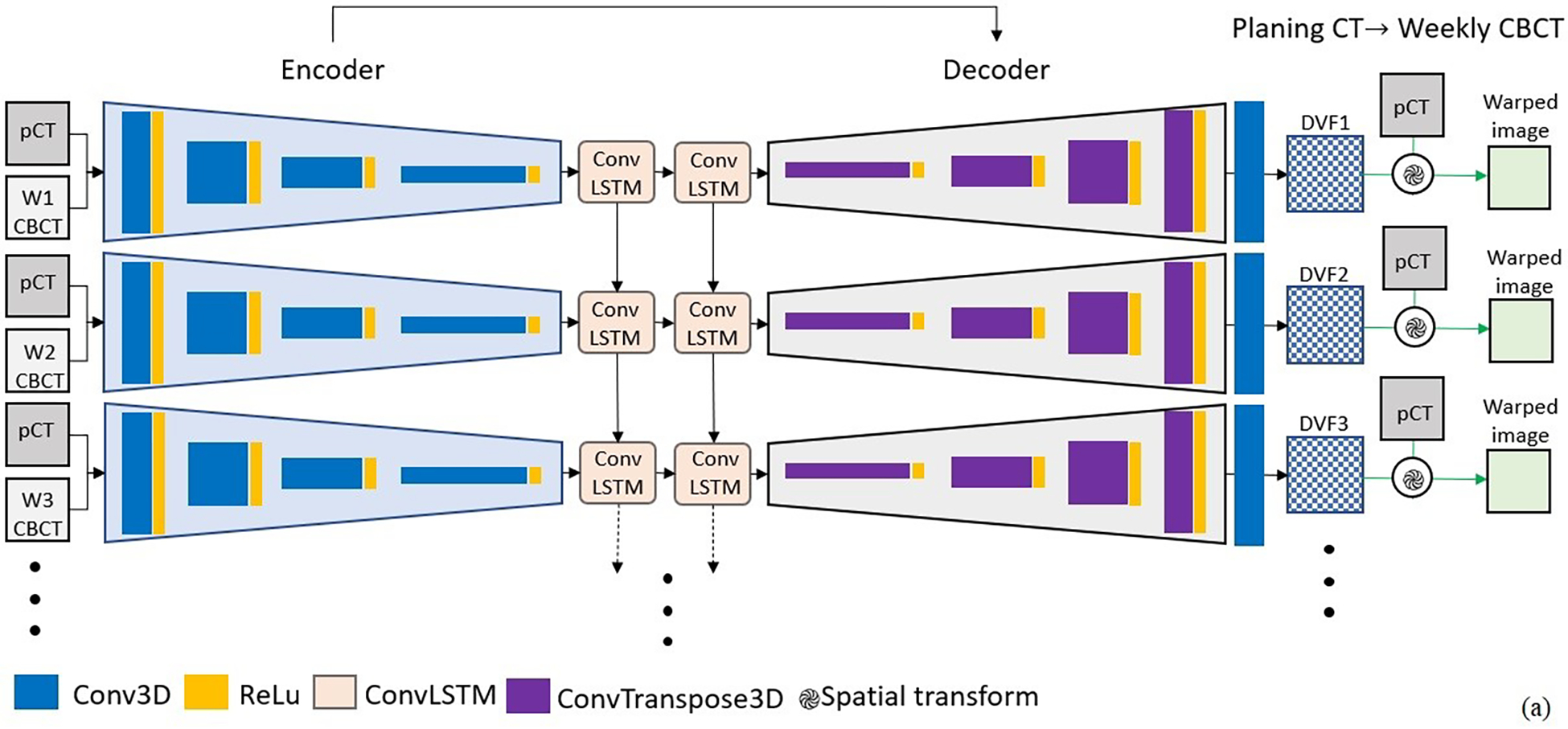

a) Seq2Morph is a hybrid network of VoxelMorph and recurrent neural network for the purpose of registering longitudinal image sets. b) convLSTM was used to parse the temporal features in addition to VoxelMorph which was used to parse the spatial features. c) Inverse consistency was implemented to minimize differences between forward and backward registrations.
We innovatively inserted a two-stacked 3D convolutional long short term memory (Conv-LSTM)15 between the encoder and decoder of VoxelMorph to parse the temporal patterns of anatomical changes among multiple image samples valuable to registration. Consequently, Seq2Morph expanded the search spaces to much bigger spatiotemporal domains and facilitated the challenging tasks of locating the best matches among image pairs. As illustrated in Figure 2.b, there are a cell state and 3 gates (forget, input, and output gates) in the ConvLSTM structure. Cell state is a key of ConvLSTM. With the assistance of the input, forget and output gate, the cell can decide what spatiotemporal feature should be retained or erased throughout the entire longitudinal dataset.
A decoder was used to calculate DVF. The decoder was constructed by 4 sequential 3D transposed convolution (ConvT3D) with 2 strides. It restored the dimension of the feature map back to 160×160×128. In addition, a shortcut connection was used to propagate features from encoder to decoder directly at the same resolution. The final Conv3D layer was used to calculate the DVF for each individual week. Subsequently we deformed the planning CT to generate a warped image and measure against each weekly CBCT.
To explicitly address the discrepancies between forward and backward registrations in a longitudinal imaging study, we implemented bidirectional pathways as illustrated in Figure 2.c to reduce inverse consistency errors.25 Registration of each image pair shared the same encoder and 3D Conv-LSTM to extract useful features but installed two decoders to generate DVFs for the forward and backward registration separately. Subsequently, two similarity measures, one between a deformed pCT and a weekly CBCT, and the other between a deformed weekly CBCT and a pCT, were calculated as metrics for registration.
2.2. Longitudinal Dataset
Seq2Morph was trained and cross-validated using longitudinal image set from 50 lung radiotherapy patients, 36 came from an internal MSKCC dataset, and 14 came from a publicly accessible lung tumor dataset. 14,26,27 Each patient dataset included a free-breathing 3D pCT and 6 consecutive weekly CBCTs acquired to monitor inter-fractional positional uncertainties and tumor changes during a conventional fractionated radiotherapy. GTV and esophagus were contoured on all images and reviewed by a clinical team. To pre-process these images, we first rigidly aligned the pCT and weekly CBCT according to spine and outer lung, and then cropped the images to form an isotropic 160×160×128 matrix with a voxel resolution of 2mm as region of interest (ROI). Boundary of GTV evolved during radiotherapy: averaged shrinkage measured by the effective diameter was 15.0±8.3mm, while the inter-fractional displacement of esophagus averaged 7.9±3.7 mm.
2.3. Training and cross-validation
For the pathway to register a series of weekly CBCTs as moving images to the pCT as a fixed image, the backward loss function was composed of a normalized cross-correlation (NCC) for image intensity similarity, a DICE for contour similarity, and a regularization term for smoothness of a DVF. It was calculated as:
| (1) |
where is the total number of weekly CBCTs available at the point of registration, is the individual week, is the deformable transformation for the image pair , smooth() is a diffusion regularizer on the spatial gradients of denotes the warping operation, and are contours from corresponding structures on and , and , and , are weighting factors, respectively. Similarly, for the pathway to register a as a moving image to a series of weekly as fixed images, the forward loss function was calculated as:
| (2) |
where is the deformable transformation for the image pair . Note that contours are only used in the unsupervised training, not for inference, because segmentations of GTV and organs at risk are not always available for the longitudinal image series in many clinical workflows. The weighting factor , and was set to 0.2 and 0.8, respectively, because this configuration led to superior prediction results in our previous study. 27 The weight for the smoothing term, , was set to 0.02 because it was selected in the original VoxelMorph study.22
We also calculated the inverse consistency error (ICE) on the reference coordinates as the absolute displacement after a round of forward and backward registrations. Given a voxel coordinate in the reference frame at week , its is calculated as:
| (3) |
Then we defined the ICE loss as:
| (4) |
where is the total number of voxels inside ROI. Finally, the total loss function was defined to simultaneously minimize the sum of the forward and backward loss as well as the inverse consistency loss weighted by :
| (5) |
We selected an equal weighting, , to train the deep learning model.
We divided the dataset into 5 subsets for 5-fold cross-validation to overcome overfitting issues. The PyTorch library was used for developing Seq2Morph. For the training Adam optimizer was used and the learning rate was 0.0001. For training and validation, we used a state-of-the-art high-performance cluster (HPC) equipped with an Intel(R) Xenon (R) Silver 4110 CPU @ 2.10GHz, 96 GB RAM, and NVIDIA A40 with 48 GB RAM. This computational infrastructure allowed us to implement a novel 3D Conv-LSTM, which required tremendous memory for the 3D CT and CBCT series.
2.4. Performance Evaluation
Using the approved GTV/esophagus segmentation on the pCT as the ground truth, we calculated the DICE, median Hausdorff distance (HD50), and 95% Hausdorff distance (HD95) as quantitative measures to evaluate performance. The set of results from the rigidly transferred manual weekly GTV/esophagus contours served as the baseline for comparison. Subsequently, we deformed and propagated the weekly GTV/esophagus to the pCT according to the weekly DVFs derived from Seq2Morph, VoxelMorph and LDDMM to assess the performance of the deformable registrations.
We compared Seq2Morph to the state-of-the-art DIR algorithm, LDDMM, to evaluate their potential applicability to adaptive radiotherapy. For LDDMM, a B-spline regularized diffeomorphic registration was performed between the two images using Advanced Normalization Tools (ANT).28 The transformation in LDDMM maps the corresponding points by finding a geodesic solution and the integrated regularization fits the DVF to a B-spline object to capture large deformations. This enforces free-form elasticity to the converging/diverging vectors that represents a morphological shrinkage/expansion. The hyperparameters were derived from a study optimizing the registration results for lung adaptive radiotherapy.29 Three levels of multi-resolution registration with B-spline mesh size of 64 mm at the coarsest level was used with Mutual Information as similarity metric. The mesh size was reduced by a factor of two at each sequential level. The optimization step size was set to 0.1 and the number of iterations (100, 70, 30) at each level. In this evaluation we generated the Jacobian maps for Seq2Morph and LDDMM and calculated the voxel-wise Pearson correlations to gain insights on how the two methods behaves on a voxel level, which is critical to analyze both global and local volume changes. We also trained and validated VoxelMorph with 5-fold cross validation from scratch using the same dataset on the same HPC to form benchmarks for evaluation.
To investigate the impact of insertions of Conv-LSTM, we visualized the hidden states of the network to check whether the spatiotemporal patterns of structural changes can be clearly captured through the stacked Conv-LSTM structure. Furthermore, we created an intermediate version of Seq2Morph that exclude inverse consistency errors inside the loss function (Seq2Morph\IC). Comparing the results of Seq2Morph\IC with VoxelMorph, we quantitively assessed the impact of expanding the search domain to cover both spatial and temporal features. Comparing the results of Seq2Morph\IC with Seq2Morph, we quantitively assessed the impact of confirming inverse consistency. 50 and 95 percentiles of ICE (ICE50 and ICE95) as well as the histogram of ICE were generated for the evaluation. Such a step-by-step investigation helped us understand how a deep learning network operates for the task of DIR.
III. RESULTS
It took ~48 hours to train and validate Seq2Morph on the HPC. The evolution of the feature maps for a particular patient is illustrated in Figure 3, which shows that the process converged well at around 150 epochs.
Figure 3.

Sequential evolutions of the feature maps in the training process indicates that the spatiotemporal patterns converged around 150 epochs for a particular patient.
A representative example of registration via Rigid, VoxelMorph, LDDMM, and Seq2Morph, is shown in Figure 4. At the early stage of radiotherapy all DIR algorithms performed equally well, as the weekly GTV changes were relatively small. However, gaps of performance between Seq2Morph and other algorithms became larger with respect to time as well as the magnitudes of GTV deformations, especially for week 5 and 6. Table 1 lists the performances of all algorithms measured by DICE, HD50, and HD95 between planning and deformed 6th weekly contours for GTV and Esophagus. Both Seq2Morph and Seq2Morph without inverse consistency (Seq2Morph\IC) outperformed VoxelMorph consistently across all GTV measures, with a significance of p<0.001 and p<0.05, calculated by a two-tailed t-test, respectively. This superiority demonstrated the advantage of adding temporal features to derive a deformation vector field. Although esophagus didn’t have as large deformations as observed in GTV, the performance gap between Seq2Morph and Voxelmorph was still statistically significant (p<0.05, two-tailed t-test).
Figure 4.

GTV/esophagus contours on the planning CT were propagated onto the weekly CBCTs according to various registration methods including rigid, VoxelMorph, LDDMM and Seq2Morph, and compared to the actual weekly contours for the evaluation of DIR accuracy.
Table 1.
Performance of investigated DIR algorithms measured by DICE, HD50, and HD95 between actual and deformed weekly contours for GTV and Esophagus.
| Rigid | VoxelMorph | LDDMM | Seq2Morph | Seq2Morph\IC | |
|---|---|---|---|---|---|
|
| |||||
| GTV DICE | 0.732±0.111 | 0.773±0.078 | 0.798±0.081 | 0.799±0.078 | 0.788±0.079 |
| GTV HD50 (mm) | 1.36±0.95 | 0.95±0.60 | 0.88±0.66 | 0.80±0.57 | 0.85±0.62 |
| GTV HD95 (mm) | 6.65±5.49 | 5.61±4.93 | 5.16±5.02 | 4.97±4.67 | 5.16±4.91 |
| Esophagus DICE | 0.674±0.113 | 0.688±0.113 | 0.695±0.108 | 0.695±0.110 | 0.685±0.111 |
| Esophagus HD50 (mm) | 3.12±3.89 | 3.09±3.88 | 2.96±3.79 | 2.63±3.61 | 2.93±3.89 |
| Esophagus HD95 (mm) | 15.07±13.63 | 14.91±13.63 | 14.56±13.46 | 12.88±12.89 | 14.15±13.50 |
We exported the extracted features at the hidden state of the last layer of the Seq2Morph decoders and illustrated the center cut of the regions of interest at each individual weekly decoder in the top row of Figure 5. The hidden state of VoxelMorph at the corresponding week were also shown in the bottom row for comparison. There are clear distinctions between Seq2Morph and VoxelMorph. VoxelMorph was able to detect obvious deformations at the GTV boundary, but the surrounding noises were intensive. In contrast, Seq2Morph with the assistance of 3D Conv-LSTM was able to amplify the systematic temporal patterns while surpassing the surrounding random deformations from noisy CBCTs and extract strong and relevant spatiotemporal features for calculating a deformation vector field.
Figure 5.
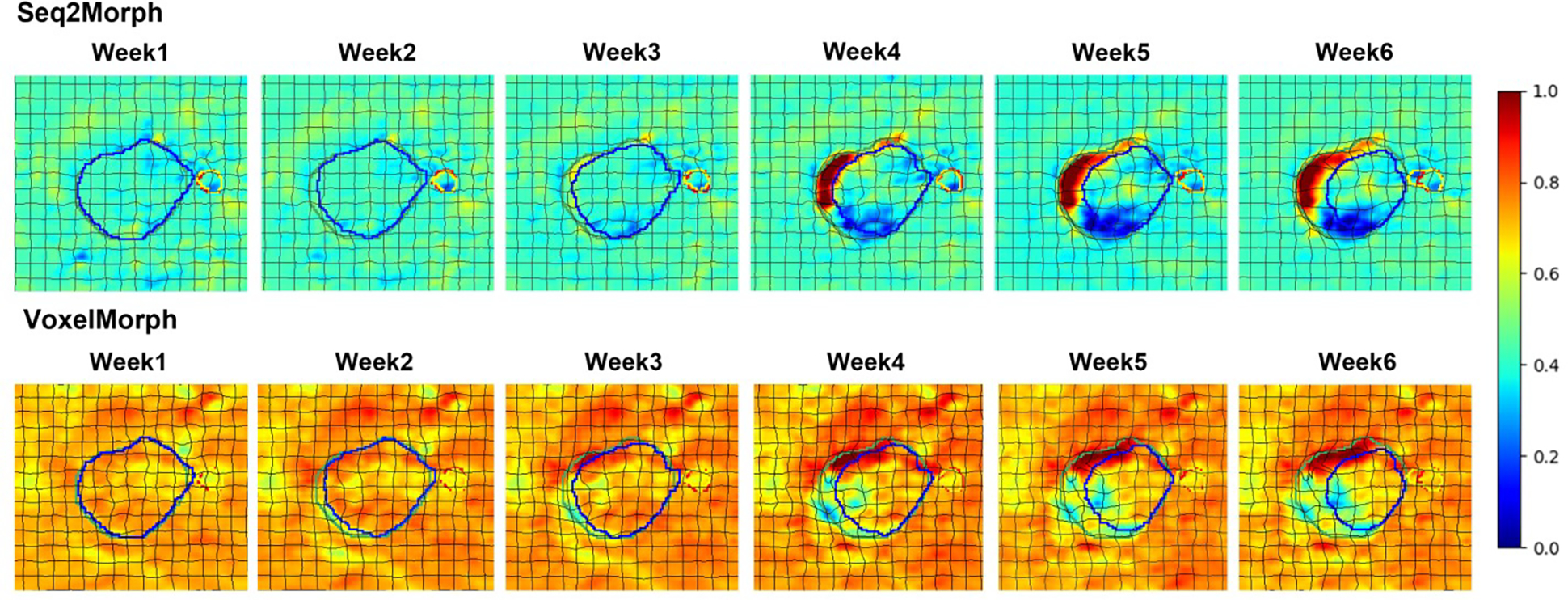
Seq2Morph shows clearer spatiotemporal patterns of deformations at the hidden state of decoder compared to VoxelMorph. Planning (green) and weekly (blue) GTV, planning (red) and weekly (yellow) esophagus, as well as deformation grid are superimposed for orientation.
An example of forward and backward registration via Seq2Morph as well as the histogram of ICE inside GTV is shown in Figure 6. The GTV ICE50 were 1.95mm, 4.33mm, and 6.60mm, and GTV ICE95 were 8.76mm, 11.85mm, and 16.17mm; the esophagus ICE50 were 1.29mm, 2.74mm, and 2.93mm, and the esophagus ICE95 were 4.13mm, 9.62mm, and 5.71mm, for Seq2Morph, Seq2Morph\IC and VoxelMorph, respectively. Seq2Morph had the smallest ICE due to the enforcement of inverse consistency (p<0.001, two-tailed t-test). ICE inside esophagus was smaller than GTV because esophagus is a tubular structure with smaller volume. The maximal ICE occurred at the region where image contrast is poor.
Figure 6.

An example of bi-direction DVFs. The green and red arrows in 3D rendering images are the forward (Weekly CBCT → planning CT) and the backward (planning CT → Weekly CBCT) DVFs, well reflecting the shrinkage of GTV. Histogram of ICE indicates Seq2Morph has the least inverse consistancy errors. Note that only DVFs with >2mm amplitude are displayed.
As shown in Table 1, the performances of Seq2Morph and LDDMM were comparable. The fraction of foldings for both methods, i.e., fraction of negative Jacobian, were 0, inside GTV and esophagus, as well as inside the 1cm ring around the two structures. The correlations between the Jacobian maps of Seq2Morph and LDDMM are 0.86±0.03, and 0.89±0.04, for GTV and esophagus, respectively. The strong correlations indicate the two algorithms reached to similar solutions even though the designs are totally different. The inference of Seq2Morph took 22 seconds for each patient with all the 6 weekly registrations, much faster than ~30 min with repeated LDDMM.
We investigated the influence of the training’s hyperparameter (Eq. 5), which is the relative weight between ICE and forward/backward registration loss, on the performance of Seq2Morph. We trained Seq2Morph with three different : 9, 1, and 0.11, calculated interferences respectively, and compared results. We found the best performance was reached with a weighting factor of 1.
To simulate the impact of intra- and inter-observer error/disagreement, we shifted the segmented contours of the training dataset by 2mm in the SI, LAT, and AP direction separately, retrained the registration network Seq2Morph, and recalculated the inference results with the validation set. We found that the average DICE resulted from the model trained with the shifted contours was slightly lower than that from the model trained with the original contour when registering the last weekly CBCT to pCT. The average difference was 0.004, not statistically significant (p=0.34, two-tailed t-test). This result indicates that the algorithm can tolerate small (~2mm) inter- or intra-observer variations.
IV. DISCUSSION
Cases of retreatment and adaptive radiotherapy increase significantly over the years since patients benefits tremendously from the advance of treatment paradigms and technologies. DIR is an essential tool to build a patient-specific storyline for the clinicians to review the longitudinal imaging data and select the best treatment option in a dynamic clinical process. Seq2Morph is a competent candidate since it (1) doesn’t require additional information such as structure contour or landmarks in interference; (2) handles multi-modal image data within variable time window as treatment proceeds without the need of retraining a network; and (3) ensuring inverse consistency thereafter causing less uncertainty in accumulating dose for different applications. We demonstrated that Seq2Morph can simultaneously register multiple CT/CBCT pairs acquired during a course of radiotherapy of lung cancers for the purpose of monitoring anatomical changes. Both accuracy and execution time of deformable image registrations performed by Seq2Morph is similar to the state-of-the-art studies summarized by Fu et al 21 in the review of deep learning-based lung registration methods. In addition, the inverse consistency error of Seq2Morph is comparable to what was reported in the literature of registering 4D-CT lung images via a DIR algorithm based on symmetric optical flow computation.30 We are in the process of integrating Seq2Morph into our automated watchdog for adaptive radiotherapy environment (AWARE), 31 specifically for the clinical site of lung cancer and head and neck cancer. Seq2Morph as an unsupervised and automated deep learning DIR algorithm will make a positive impact on our clinical operations.
We demonstrated that DIR can fundamentally benefit from expanding the search space to both spatial and temporal domain. Through the detailed illustration of the hidden state of the DIR network, we confirmed that Conv-LSTM was able to capture the graduate changes of GTV and esophagus with high fidelity. As a result, Seq2Morph is capable of closely modeling the weekly changes in small steps and generating accurate registrations even that the changes accumulated to a much larger amplitude at the end of radiotherapy. This behavior of Seq2Morph resembles LDDMM, where large deformations are achieved as a combination of small time and space steps. This similarity explains the high correlation between the DVFs obtained from the two independent algorithms. However, since the network model is pretrained, avoiding the time-consuming optimization process for the calculations of DVF on the fly as LDDMM, Seq2Morph completes registrations reasonably fast within half a minute and positions itself suitable to a much wider range of radiotherapy applications.
For future investigations, we will expand the scope of Seq2Morph to cover a larger timeline by registering images from previous treatments, diagnostic studies, and follow-up studies. We will also enhance the technical capability of Seq2Morph to handle other popular imaging modalities such as MRI and PET. Such development requires continuous accumulation and careful curation of large longitudinal image data sets. We are actively developing various software in our institution to create an infrastructure of a data lake and pool all the clinical data 32 to support both scientific research and clinical operations.
V. CONCLUSION
Seq2Morph can provide accurate and fast DIR for longitudinal image studies by exploiting spatial-temporal patterns. It closely matches the clinical workflow and has the potential to serve for both online and offline ART.
Acknowledgment:
This research was partially supported by the MSK Cancer Center Support Grant/Core Grant (P30 CA008748).
Reference:
- 1.Yang Deshan, Brame Scott, El Naqa Issam, Aditya Apte, Wu Yu, Goddu S. Murthy, Mutic Sasa, Deasy Joseph O., Daniel A. Low. Technical Note: DIRART-A software suite for deformable image registration and adaptive radiotherapy research. Medical Physics. 2010;38:67–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Veiga Catarina, McClelland Jamie, Moinuddin Syed, Lourenco Ana, Ricketts Kate, Annkah James, Modat March, Ourselin Sebastien, D’Souza Derek, Royle Gary. Toward adaptive radiotherapy for head and neck patients: Feasibility study on using CT-to-CBCT deformable registration for “dose of the day” calculations. Med Phys. 2014;41(3):031703. [DOI] [PubMed] [Google Scholar]
- 3.Choudhury A, Budgell G, MacKay R, Falk S, Faivre-Finn C, Dubec M, van Herk M, McWilliam A, The Future of image-guided Radiotherapy. Clinical Oncology. 2017; 29:662–666. [DOI] [PubMed] [Google Scholar]
- 4.Brock Kristy K., Mutic Sasa, McNutt Todd R., Li Hua, Kessler Marc L., Use of image registration and fusion algorithms and techniques in radiotherapy: Report of the AAPM Radiation Therapy Committee Task Group No. 132. Med Phys, 2018;44:e43–76. [DOI] [PubMed] [Google Scholar]
- 5.Cole AJ, Veiga C, Johnson U, D’Souza D, Lalli NK and McClelland JR. Toward adaptive radiotherapy for lung patients: feasibility study on deforming planning CT to CBCT to assess the impact of anatomical changes on dosimetry Phys. Med. Biol. 2018;63: 155014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chetty I, Rosu-Bubulac M. Deformable registration for dose accumulation. Semin Radiat Oncol. 2019;29(3):198–208. [DOI] [PubMed] [Google Scholar]
- 7.Rigaud B, Simon A, Castelli J, Lafond C, Acosta O, Haigron P, Cazoulat G and de Crevoisier R. Deformable image registration for radiation therapy: principle, methods, applications and evaluation. Acta Oncologica. 2019;58:1225–37. [DOI] [PubMed] [Google Scholar]
- 8.Yuen J, Barber J, Ralston A, Gray A, Walker A, Hardcastle N, Schmidt L, Harrison K, Poder J, Sykes J, Jameson M. An international survey on the clinical use of rigid and deformable image registration in radiotherapy. J Appl Clin Med Phys. 2020;21(10):10–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nadeem Saad, Zhang Pengpeng, Rimner Andreas, Sonke Jan-Jakob, Deasy Joseph O., Tannenbaum Allen, LDeform: Longitudinal deformation analysis for adaptive radiotherapy of lung cancer. Medical Physics. 2019; 47:132–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Karangelis Grigorios, Krishnan Karthik, Hui Susanta. A framework for deformable image registration validation in radiotherapy clinical applications. JACMP, 2013;14(1):192–213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yan Hao, Zhen Xin, Cerviño Laura, Jiang Steve B., Jia Xun. Progressive cone beam CT dosecontrol in image-guided radiation therapy. 2013; 40:060701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kavanaugh James, Roach Michael, Ji Zhen, Fontenot Jonas, Hugo Geoffrey D., A method for predictive modeling of tumor regression for lung adaptive radiotherapy. Medical Physics. 2020; 48: 2083–2094. [DOI] [PubMed] [Google Scholar]
- 13.Wang C, Rimner A, Hu Y, Tyagi N, Jiang J, Yorke E, Riyahi S, Mageras G, Deasy J, and Zhang P. Towards Predicting the Evolution of Lung Tumors During Radiotherapy Observed on a Longitudinal MR Imaging Study Via a Deep Learning Algorithm. Med Phys. 46(10):4699–4707, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang Chuang, Sadegh R Alam Siyuan Zhang, Hu Yu-Chi, Nadeem Saad, Tyagi Neelam, Rimmer Andreas, Lu Wei, Thor Maria, Zhang Pengpeng. Predicting spatial esophageal changes in a multimodal longitudinal imaging study via a convolutional recurrent neural network. Physics in Medicine and Biology. 2020;65:235027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee Donghoon, Alam Sadegh R., Jiang Jue, Zhang Pengpeng, Nadeem Saad, Hu Yu-chi, Deformation driven Seq2Seq longitudinal tumor and organ-at-risk prediction for radiotherapy. Medical Physics.2021;48:4784–4798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kearney Vasant, Haaf Samuel, Sudhyadhom Atchar, Valdes Gilmer and Timothy D Solberg. An unsupervised convolutional neural network based algorithm for deformable image registration. Physics in Medicine and Biology, 2018;63:185017. [DOI] [PubMed] [Google Scholar]
- 17.Jaing Zhuoran, Yin Fang-fang, Ge Yun and Ren Lei, A multi-scale framework with unsupervised joint training of convolutional neural networks for pulmonary deformable image registration, Physics in Medicine and Biology, 2020:65:015011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lei Yang, Fu Yabo, Wang Tonghe, Liu Yingzi, Patel Pretesh, Curran Walter J, Liu Tian and Yang Xiaofeng, 4D-CT deformable image registration using multiscale unsupervised deep learning. Physics in Medicine and biology. 2020;65:085003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fu Yabo, Lei Yang, Wang Tonghe, Higgins Kristin, Bradley Jeffrey D., Curran Walter J., Liu Tian, Yang Xiaofeng. LungRegNet: An unsupervised deformable image registration method for 4D-CT lung. Medical Physics; 2020; 47:1763–1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Han X, Hong J, Reyngold M, Crane C, Cuaron J, Hajj C, Mann J, Zinovoy M, Greer H, Yorke E, Mageras G, Niethammer M. Deep-learning-based image registration and automatic segmentation of organs-at-risk in cone-beam CT scans from high-dose radiation treatment of pancreatic cancer. Med Phys. 2021;48(6):3084–3095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fu Yabo, Lei Yang, Wang Tonghe, Walter J Curran Tian Liu, and Yang Xiaofeng. Deep learning in medical image registration: a review. Physics in Medicine and Biology. 2020;65:20TR01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Boveiri Hamid Reza, Khayami Raouf, Javidan Reza, Mehdizadeh Alireza. Medical Image registration using deep neural network: A comprehensive review. Computer & Electrical Engineering. 2020; 87:106767. [Google Scholar]
- 23.Lee D, Hu Y, Kuo L, Alam S, Yorke E, Li A, Rimner A, Zhang P. Deep learning driven predictive treatment planning for adaptive radiotherapy of lung cancer. Radiotherapy and Oncology. In press, 2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Balakrishnan Guha, Zhao Amy, Sabuncu Mert R., Guttag John, Dalca Adrian V. VoxelMorph: A Learning Framework for deformable medical image registration. IEEE Transactions on Medical Imaging; 2019;38:1788–1800. [DOI] [PubMed] [Google Scholar]
- 25.Kim Boah, Dong Hwan Kim Seong Ho Park, Kim Jieun, Lee June-Goo, Ye Jong Chul. CycleMorph: Cycle consistent unsupervised deformable image registration. Medical Image Analysis. 2021;71:102036. [DOI] [PubMed] [Google Scholar]
- 26.Hugo GD, Weiss E, Sleeman WC, Balik S, Keall PJ, Lu J, Williamson JF. A longitudinal four-dimensional computed tomography and cone beam computed tomography dataset for image-guided radiation therapy research in lung cancer, Medical Physics, 2017. 44(2):762–771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee D, Hu Y, Kuo L, Alam S, Yorke E, Li A, Rimner A, Zhang P. Deep learning driven predictive treatment planning for adaptive radiotherapy of lung cancer. Radiotherapy and Oncology. 169:57–63, 2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tustison NJ, Avants BB. Explicit b-spline regularization in diffeomorphic image registration. Front Neuroinform 2013;7:39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Alam S, Thor M, Rimner A, Tyagi N, Zhang S, Kuo L, Nadeem S, Lu W, Hu Y, Yorke E and Zhang P. Quantification of accumulated dose and associated anatomical changes of esophagus using weekly Magnetic Resonance Imaging acquired during radiotherapy of locally advanced lung cancer. Physics and Imaging in Radiation Oncology. 13: 36–43, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang Deshan, Li Hua, Low Daniel A, Deasy Joseph O, and El Naqa Issam. A fast inverse consistent deformable image registration method based on symmetric optical flow computation. Phys Med Biol. 2008. November 7; 53(21): 6143–6165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Aristophanous M, Aliotta E, Caringi A, Rochford B, Allgood N, Zhang P, Hu Y, Zhang P, Cervino L. A Clinically Implemented Watchdog Program to Streamline Adaptive LINAC-Based Head and Neck Radiotherapy. Medical Physics, 2021;48(6):e253. [Google Scholar]
- 32.Li A Workflow automation and radiotherapy platform (WARP) to integrate RT and non-RT EMR/HER data. Practical Big Data Workshop. Ann Arbor, 2021. [Google Scholar]


