Abstract
A cellular protein, previously described as p55, binds specifically to the plus strand of the mouse hepatitis virus (MHV) leader RNA. We have purified this protein and determined by partial peptide sequencing that it is polypyrimidine tract-binding protein (PTB) (also known as heterogeneous nuclear ribonucleoprotein [hnRNP] I), a nuclear protein which shuttles between the nucleus and cytoplasm. PTB plays a role in the regulation of alternative splicing of pre-mRNAs in normal cells and translation of several viruses. By UV cross-linking and immunoprecipitation studies using cellular extracts and a recombinant PTB, we have established that PTB binds to the MHV plus-strand leader RNA specifically. Deletion analyses of the leader RNA mapped the PTB-binding site to the UCUAA pentanucleotide repeats. Using a defective-interfering RNA reporter system, we have further shown that the PTB-binding site in the leader RNA is critical for MHV RNA synthesis. This and our previous study (H.-P. Li, X. Zhang, R. Duncan, L. Comai, and M. M. C. Lai, Proc. Natl. Acad. Sci. USA 94:9544–9549, 1997) combined thus show that two cellular hnRNPs, PTB and hnRNP A1, bind to the transcription-regulatory sequences of MHV RNA and may participate in its transcription.
Mouse hepatitis virus (MHV) is a single-stranded, positive-sense RNA virus and belongs to the Coronaviridae family. MHV contains an RNA genome of approximately 31 kb (22), which encodes seven to eight viral genes. MHV synthesizes six to seven subgenomic mRNAs, all of which share 5′ and 3′ ends identical to those of the genomic RNA (18). The common 5′ end, termed leader sequence (72 to 77 nucleotides [nt] long), is derived from the 5′ end of the genomic RNA (20, 38). The 3′ end of the leader RNA contains a consensus UCUAA repeat sequence (30), and the copy number of UCUAA repeats varies among different MHV strains. Another consensus sequence, UCUAAAC or a similar sequence, termed intergenic (IG) sequence, is present between each viral gene (5). The IG sequence, which is homologous to the UCUAA repeat of the leader RNA, serves as the initiation site for subgenomic mRNA synthesis (14, 29). Each of the seven subgenomic mRNAs starts from a different IG site and extends to the 3′ end of the genome; thus, the subgenomic mRNAs have a 3′-coterminal, nested-set structure (17, 23). Furthermore, the subgenomic mRNA synthesis involves fusion of two stretches of noncontiguous sequence by a discontinuous-transcription mechanism, such that the leader RNA is joined to the 5′ end of each mRNA (13, 25, 43). This fusion occurs between the UCUAA repeats of the leader and the consensus IG sequence. Based on this unique subgenomic mRNA structure, several models have been proposed to explain the discontinuous nature of coronavirus RNA transcription (16, 18, 35). All of these models require that the leader sequence interact with the IG sequence in the minus-strand template RNA or plus-strand genomic RNA. Such an interaction has been inferred from the findings that the efficiency of mRNA transcription is affected by different combinations of the leader and IG sequences, particularly in the region of UCUAA repeats (21, 36, 43). Interestingly, this interaction appears to occur between the leader and IG sequences in two different RNA molecules (in trans) as well as within the same RNA molecule (in cis) (43). Furthermore, the 3′-end sequence of MHV RNA is required for viral mRNA transcription (26), suggesting that the 3′ untranslated region (UTR) of viral RNA may also interact with the upstream transcription-regulatory sequences. Since sequence complementarity is not always present in these interacting RNA regions, these interactions may be mediated by protein-RNA and protein-protein interactions, involving either viral or cellular proteins (19, 42).
By UV cross-linking assays, several cellular proteins have been detected to bind specifically to the transcription-regulatory regions of MHV RNA (7, 27, 41, 44). Among these proteins, p35/38 binds to the minus-strand leader RNA and minus-strand IG sequences (7, 44), while another protein, p55, binds to the plus-strand leader RNA (7). The binding of p35/38 to the minus-strand IG sequence correlates with the efficiency of subgenomic mRNA transcription (44), suggesting that p35/38 may act as a transcription factor. Recently, this protein has been identified as heterogeneous nuclear ribonucleoprotein (hnRNP) A1 (24), a nuclear protein involved in the alternative splicing of cellular RNAs (6). It is hypothesized that the binding of hnRNP A1 to both the leader and IG sequences on the minus-strand template may bring these two distant RNA domains together and facilitate the formation of either an MHV-specific RNA transcription complex or an MHV RNA processing (splicing) complex.
Since plus-strand leader RNA interacts with the template RNA during MHV RNA transcription, p55 may serve as a mediator of this interaction. Therefore, we are interested in understanding the nature of this protein. In this report, we identified the p55 as polypyrimidine tract-binding protein (PTB), also known as hnRNP I. This protein has previously been shown to interact with hnRNP A1 in a pre-mRNA splicing complex in normal cells (3). Thus, the binding of hnRNP A1 and PTB to the minus-strand leader and IG site and the plus-strand leader RNA, respectively, may bring together these RNAs, fulfilling a requirement for MHV RNA transcription.
Purification of MHV plus-strand leader RNA-binding protein p55.
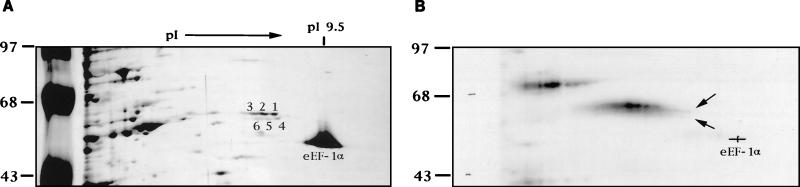
Although p55 was originally found in the cytoplasm of a murine cell line (DBT) (12), a similar protein was detected in both the nuclear and the cytoplasmic fractions of HeLa cells (data not shown). Therefore, HeLa cell lysates were used for protein purification according to the previously published procedures (24). After three steps of purification (ammonium sulfate precipitation and Q-Sepharose and heparin agarose chromatography), partially purified proteins were UV cross-linked to 32P-labeled plus-strand leader RNA, 182(+) RNA (7), representing the 5′-end 182 nt of viral genomic RNA, as previously described (24). The RNA-protein complex was digested with RNase A and analyzed by nonequilibrium two-dimensional (2-D) gel electrophoresis according to the procedure of O’Farrell et al. (31). The silver-stained protein profile of the 2-D gel (Fig. 1A) was then compared with the autoradiogram of the UV cross-linked 32P-labeled proteins (Fig. 1B). Two streaks of radiolabeled spots ranging from 55 to 57 kDa, each with a correspondingly decreasing isoelectric point (pI) that represents different numbers of nucleotides linking to the same protein (24), were detected on the autoradiogram (Fig. 1B). Since nucleotide attachment to a protein will increase the molecular weight and correspondingly lower the pI value of the protein, the radiolabeled spot with the lowest molecular weight and the highest pI (indicated by two arrows; Fig. 1B) represents the minimally cross-linked protein, which corresponded to spot no. 1 and spot no. 4 identified on the silver-stained gel (Fig. 1A). These two spots, thus, are the most likely candidates for p55. Four other protein spots, no. 2, 3, 5, and 6 (Fig. 1A), are in close proximity to spot no. 1 and no. 4 and have the same molecular weights; they may represent isoforms of p55. Because these six protein spots were not well separated, they were collected as two groups: p55a (spot no. 1 to 3) and p55b (spot no. 4 to 6). Each of these six protein spots has a pI value close to that of the translation factor eEF-1α (50 kDa, pI 9.5) (4) (Fig. 1A), and all were detected in both cytoplasmic and nuclear fractions (data not shown). Another streak of radiolabeled protein represents p70 (44). The identity of this protein was not further pursued in this study.
FIG. 1.
2-D PAGE analysis of the partially purified proteins from HeLa cytoplasmic extracts. HeLa cytoplasmic extract was separated by ammonium sulfate precipitation and Q-Sepharose and heparin agarose chromatography. The partially purified proteins were UV cross-linked to 32P-labeled plus-strand leader [5′-182(+)] RNA and separated by 2-D nonequilibrium pH gradient gel electrophoresis according to the procedures of O’Farrell et al. (31), by using Ampholine pH 3 to 10 (Bio-Rad) and pH 7 to 9 (Pharmacia) at a 1:1 ratio and separation for 5 h at 650 V for the first dimension. The second dimension was performed by SDS-PAGE on 10% polyacrylamide gels. (A) Silver-stained gel. p55 (spots 1 to 6) and molecular mass markers in kilodaltons are indicated, and eEF-1α is identified. (B) Autoradiogram of the same gel as in panel A. The two arrows indicate the radiolabeled spots that match spots no. 1 and no. 4 in the silver-stained gel.
Identification of p55 as PTB or hnRNP I.
The identity of p55a and p55b was determined by partial peptide sequencing and amino acid composition analysis (performed by W. M. Keck Foundation Biotechnology Resource Laboratory of Yale University). The high-pressure liquid chromatography patterns of the tryptic peptides of p55a and p55b were very similar (data not shown), suggesting that they were related proteins. One tryptic peptide from p55a, which was shared with p55b, was selected for peptide sequencing. The sequence of this peptide, DYGNSPLHR, matched the C-terminal sequence (amino acid 429 to 437) of a human PTB, also known as hnRNP I. Upon further analysis, the high-pressure liquid chromatography profiles of tryptic peptides of p55a and p55b were found to be similar to that of PTB (data not shown), further indicating the identity of p55 as PTB. PTB has been reported to consist of at least four isoforms (9, 10, 32), due to alternative splicing, and their molecular masses range from 57 to 62 kDa, and their pI range from 8.0 to 8.5 (6). The p55 (PTB) protein pattern in the silver-stained 2-D gel (Fig. 1A) was also similar to that of the immunopurified hnRNP I complex (9). The amino acid sequence of human PTB shows greater than 97% identity to its murine counterpart (3).
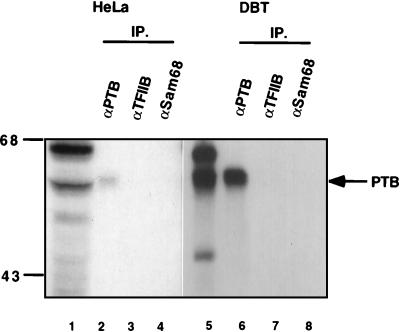
The identity of p55 as PTB was then tested by immunoprecipitation studies of UV cross-linked protein with a monoclonal antibody specific for human PTB (kindly provided by E. Wimmer, State University of New York, Stony Brook). The cytoplasmic extracts from HeLa and DBT cells were UV cross-linked with the MHV plus-strand leader RNA, and the RNA-protein complex was immunoprecipitated with antibodies against several different nuclear proteins (PTB, TFIIB, and Sam68). Only the PTB-specific monoclonal antibody (Fig. 2, lanes 2 and 6), and not the other two antibodies (Fig. 2, lanes 3, 4, 7, and 8), could precipitate the UV cross-linked p55 from both HeLa and DBT cells. These data established that the MHV plus-strand leader RNA-binding protein (p55) is PTB.
FIG. 2.
Immunoprecipitation of UV cross-linked proteins. Cytoplasmic extracts from HeLa (lanes 1 to 4) and DBT (lanes 5 to 8) cells were UV cross-linked with 32P-labeled plus-strand leader RNA; digested with RNase A; and precipitated with monoclonal antibody for PTB (lanes 2 and 6), monoclonal antibody for TFIIB (lanes 3 and 7), or polyclonal antibody for Sam68 (Santa Cruz Biotechnology) (lanes 4 and 8). The precipitated proteins were collected with protein A-Sepharose beads (Zymed) and analyzed by SDS-PAGE. Lanes 1 and 5 are UV cross-linked proteins without immunoprecipitation (IP). Numbers at left show molecular mass in kilodaltons.
RNA-binding property of the recombinant murine PTB (GST-PTB).
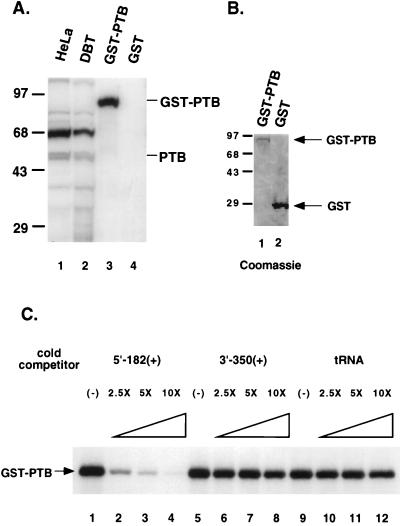
To further confirm that PTB is the protein that binds to the plus-strand MHV 5′ UTR, a recombinant glutathione S-transferase (GST)-fused murine PTB (GST-PTB) was prepared and used for UV cross-linking to the 32P-labeled 182-nt plus-strand 5′ UTR. The results showed that the purified GST-PTB fusion protein (Fig. 3B, lane 1) bound to this RNA (Fig. 3A, lane 3), whereas the purified GST protein (Fig. 3B, lane 2) did not bind the same RNA (Fig. 3A, lane 4). The binding of GST-PTB to the 32P-labeled 5′-end leader RNA [5′-182(+)] could be competed completely by a 10-fold excess of the cold homologous RNA but not by other RNAs, such as 3′ end of MHV RNA [3′-350(+)] or tRNA (Fig. 3C). These results indicate that PTB is indeed the protein that binds specifically to the 5′-end leader RNA.
FIG. 3.
UV cross-linking analysis of recombinant PTB. Plasmid pGEX-4T-1/PTB, which expresses a GST-murine PTB fusion protein, was constructed by subcloning a reverse-transcribed PCR-synthesized cDNA fragment (1,587 bp), containing the entire open reading frame of murine PTB (GenBank/EMBL data bank with accession no. X52101 [3]), made from the RNA of DBT cells. The expression of GST fusion protein in Escherichia coli was induced by addition of 0.2 mM isopropyl-1-thio-β-d-galactoside (IPTG) for 3 to 5 h at 37°C. GST fusion protein was purified from bacterial lysates with glutathione-Sepharose 4B beads (Pharmacia). (A) Purified recombinant PTB (GST-PTB) (lane 3) and GST (lane 4) were UV cross-linked with 32P-labeled plus-strand leader RNA and separated by SDS-PAGE. Cytoplasmic extracts from HeLa (lane 1) and DBT (lane 2) cells cross-linked to the same RNA were included as controls. (B) Coomassie blue-stained gel of the purified GST-PTB (lane 1) and GST (lane 2). (C) Competition analysis of GST-PTB binding to 5′-182(+) RNA. The purified GST-PTB was first incubated with increasing concentrations of unlabeled RNAs (cold competitors, 2.5×, 5×, or 10× molar excess over the labeled RNA) and then with 32P-labeled 5′-182(+) RNA. The UV cross-linked proteins were separated by SDS-PAGE. Numbers to the left of panels A and B show molecular mass in kilodaltons.
Mapping of the PTB-binding site on the plus-strand MHV 5′ UTR.
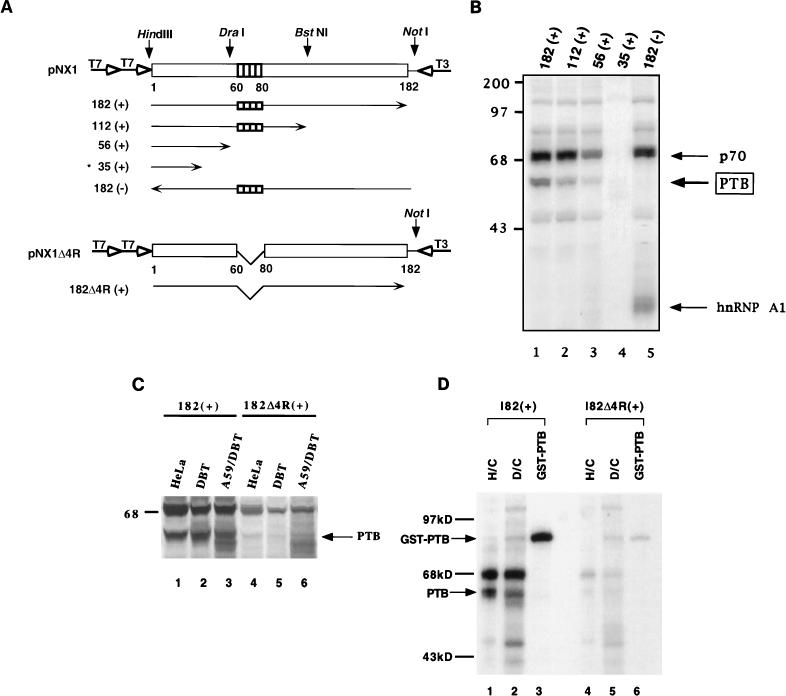
To locate the binding site of PTB on the MHV plus-strand 5′ UTR, various truncated, 32P-labeled RNAs of this region (Fig. 4A) were UV cross-linked to HeLa cell extract, followed by RNase A digestion and sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). Figure 4B shows that the amounts of PTB bound to 182(+) (lane 1, 100%) and 112(+) (lane 2, 70%) RNAs were comparable but that the amount of its binding to RNA 56(+) (lane 3) was significantly lower (19%). No PTB was found to bind RNA 35(+) (lane 4). As expected, PTB did not bind to the minus-strand, 182-nt RNA (lane 5), further confirming that PTB binds specifically to plus-strand 5′ UTR. These data indicated that nt 56 to 112 of the plus-strand leader RNA are the sequence most critical for the binding of PTB. Within this region, there is a stretch of four UCUAA repeats (nt 60 to 80 from the 5′ end), which has been demonstrated to be important for the MHV transcription (2, 21, 37, 43). The region between nt 35 and 56 from the 5′ end may also contribute to PTB binding to some extent since 56(+) RNA bound PTB weakly; this region includes part of the UCUAA repeat sequence and has two sequences similar to UCUU, which has been reported to be one of the PTB-binding consensus sequences (33). However, the extent of PTB binding to this region was variable. In some experiments, very little PTB binding was detected (see below) (Fig. 4C). Thus, PTB binding to nt 35 to 56 was very weak at best. Therefore, the major PTB-binding site is within nt 56 to 112 from the 5′ end. There was a slight difference in PTB binding between the 182(+) and 112(+) RNAs, suggesting that the extent of PTB binding to nt 56 to 112 may be affected by other RNA sequences or overall RNA conformation.
FIG. 4.
Mapping of PTB-binding sites on the MHV 5′-UTR RNA. (A) Schematic diagrams of the structure of plasmids and their transcribed RNAs used in this study. The unfilled box represents the MHV 5′ UTR, and shaded boxes indicate the four UCUAA repeats in the RNA. Vertical arrows represent the enzyme sites for linearizing the plasmids. The numbers under the boxes denote nucleotide numbers from the 5′ end of the MHV RNA. Horizontal arrows represent the in vitro transcribed RNAs. The open arrows represent T7 and T3 RNA polymerase promoters. The RNA *35 (+) was transcribed from synthetic DNA oligomers containing the T7 promoter. pNX1Δ4R is identical to pNX1 but lacks the four UCUAA repeats. (B) UV cross-linking of HeLa cytoplasmic extracts with various 5′-end RNAs. The amounts of PTB binding were quantitated by using the Ambis Radioanalytic Imaging System. (C) UV cross-linking of 5′-UTR RNA with or without the UCUAA repeat. Cytoplasmic extracts from HeLa (lanes 1 and 4), DBT (lanes 2 and 5), and A59-infected DBT (lanes 3 and 6) cells were used. (D) UV cross-linking of GST-PTB with 5′-182(+) and 5′-182Δ4R(+) RNAs. H/C and D/C represent cytoplasmic extracts of HeLa and DBT cells, respectively. Numbers at left of panels B and C show molecular mass in kilodaltons.
To investigate whether the UCUAA pentanucleotide repeat sequence in the leader RNA was responsible for the PTB binding, a mutant 5′-UTR RNA, 182Δ4R(+), in which the four UCUAA repeats have been deleted (Fig. 4A), was used to perform a UV cross-linking assay. The results showed that this mutant leader RNA bound very little PTB from the lysates of uninfected or MHV-A59-infected HeLa or DBT cells (Fig. 4C, lanes 4 to 6). This was significantly less than that bound to the wild-type leader RNA (lanes 1 to 3). Similarly, GST-PTB also bound strongly to 182(+) RNA but only weakly to 182Δ4R(+) RNA (Fig. 4D). These results combined thus indicate that the UCUAA repeat sequence is most crucial for PTB binding.
The PTB-binding site is crucial for the viral subgenomic mRNA transcription.
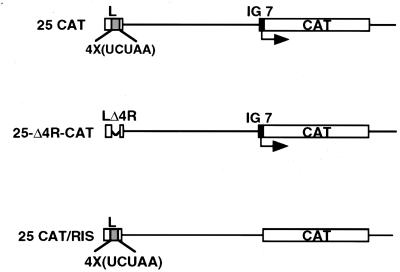
Previous studies have shown that the number of the UCUAA repeats at the 3′ end of the leader sequence determines the efficiency of transcription of viral subgenomic mRNAs in both a trans- and a cis-acting manner (21, 40, 43). Since PTB binds to this UCUAA region, we asked whether the elimination of this PTB-binding site would affect the transcription of MHV subgenomic mRNA. We constructed a variant of defective-interfering (DI) RNA reporter vector p25-Δ4R-CAT, in which the four UCUAA repeats in the leader sequence were deleted but which contains a chloramphenicol acetyltransferase (CAT) reporter gene placed behind an IG sequence (25) (Fig. 5). Previous studies have shown that the CAT activity detected from this DI RNA in the presence of a helper virus reflects mRNA transcription from the IG site (25, 44). Various DI RNAs, including p25CAT (with the wild-type leader), p25-Δ4R-CAT (with a mutant leader), and p25CAT/RIS (without the IG site) (Fig. 5), were transfected into A59-infected DBT cells at 1 h postinfection. Cell lysates were collected at 9 h postinfection for CAT assay. The results indicated that the CAT activity expressed by p25-Δ4R-CAT was approximately 15% of that of the wild-type DI vector, whereas the RNA without the IG sequence (p25CAT/RIS) was totally devoid of the CAT activity (data not shown). These results are consistent with the previous reports that the UCUAA repeat sequence in the leader RNA modulates mRNA transcription in cis and in trans (25, 43). Since PTB binds to this repeat sequence, PTB may play a role in this regulation.
FIG. 5.
Structure of various DI RNAs. 25CAT RNA (25) contains a wild-type leader RNA sequence; 25-Δ4R-CAT contains a mutant leader RNA without the four UCUAA repeats. IG 7 is the promoter sequence for gene 7; 25CAT/RIS is similar to 25 CAT but lacks the IG site (25). The plasmids for these RNAs were made by PCR amplification of p25CAT with appropriate primers.
The results presented here have demonstrated that PTB binds to the MHV leader RNA at the UCUAA repeat region. This UCUAA repeat is the site where the leader RNA interacts with the IG sequences on the template RNA in both a trans- and a cis-acting manner and where the leader joins subgenomic mRNAs (43). The deletion of the UCUAA repeat sequence resulted in the loss of PTB binding and the corresponding reduction of mRNA transcription from a downstream IG site. This finding further confirmed that the leader sequence has a cis-acting effect on subgenomic mRNA transcription (43) and that the UCUAA sequence plays an important role in mRNA initiation. Conceivably, the effect of the UCUAA repeats on transcription is mediated through PTB.
PTB is predominantly a nuclear protein, particularly localized in a discrete perinucleolar structure (9). It contains four repeated domains, each of which consists of 80 amino acids, constituting RNA-binding domains (9). Therefore, one molecule of PTB can potentially interact with several different RNA regions. PTB binds preferentially to the polypyrimidine tract of pre-mRNAs, near the 3′ splice site, and is involved in alternative RNA splicing and RNA transport in normal cells (8–10, 32, 33). In addition, PTB has been shown to regulate translation of some viral RNAs (1, 11, 15) and cap-independent cellular mRNAs (28). So far, we have identified two cellular proteins, hnRNP A1 (24) and PTB, which bind to several different regions of MHV RNA. hnRNP A1 binds to minus-strand leader sequence and the IG site, whereas PTB binds to plus-strand leader. Previous studies have suggested that these three regions may interact with each other to regulate mRNA transcription (19). It is conceivable that these interactions are mediated through hnRNP A1 and PTB by protein-RNA and protein-protein interactions. Our preliminary data have, indeed, shown that minus-strand leader sequence and the IG site can interact with each other through hnRNP A1 (45). Furthermore, PTB and hnRNP A1 have been shown to interact with each other in spliceosome complexes in normal cells (3). Therefore, once PTB and hnRNP A1 bind to plus-strand leader RNA and minus-strand IG and leader sequences, respectively, these two proteins may bring together these RNAs to form a ribonucleoprotein complex.
One intriguing feature about these proteins is that both hnRNP A1 and PTB are primarily nuclear proteins (6, 8, 9, 32); yet, coronavirus replicates in the cytoplasm. However, both hnRNP A1 and PTB are shuttled between the nucleus and cytoplasm in normal cells (11, 34), and all proteins are synthesized in the cytoplasm. These cytoplasmic proteins, hnRNP A1 and PTB, have been shown to promote translation in normal cells by facilitating ribosome binding via a 5′-end, cap-mediated mechanism (39). We have previously found that hnRNP A1 is translocated from the nucleus to the cytoplasm in MHV-infected cells (24). However, there is only a marginal increase in the amount of PTB in the cytoplasm of MHV-infected cells (unpublished observation). Nevertheless, since PTB participates in the regulation of mRNA translation in normal cells, sufficient PTB may be present in the cytoplasm of MHV-infected cells to allow its participation in MHV RNA transcription. The functional role of the cytoplasmic PTB in normal and MHV-infected cells should be of great interest.
Acknowledgments
We thank Daphne Shimoda for assistance in preparing the manuscript.
This study was supported by grant 19244 from the National Institutes of Health. M. M. C. Lai is an Investigator of the Howard Hughes Medical Institute.
REFERENCES
- 1.Ali N, Siddiqui A. Interaction of polypyrimidine tract-binding protein with the 5′ noncoding region of the hepatitis C virus RNA genome and its functional requirement in internal initiation of translation. J Virol. 1995;69:6367–6375. doi: 10.1128/jvi.69.10.6367-6375.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baker S C, Lai M M C. An in vitro system for the leader-primed transcription of coronavirus mRNAs. EMBO J. 1990;9:4173–4179. doi: 10.1002/j.1460-2075.1990.tb07641.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bothwell A L M, Ballard D W, Philbrick W M, Lindwall G, Mather S E, Bridgett M M, Jamison S F, Garcia-Blanco M A. Murine polypyrimidine tract binding protein. J Biol Chem. 1991;266:24657–24663. [PubMed] [Google Scholar]
- 4.Bravo R, Celis J E. Up-dated catalogue of HeLa cell proteins: percentages and characteristics of the major cell polypeptides labeled with a mixture of 16 14C-labeled amino acids. Clin Chem. 1982;28:766–781. [PubMed] [Google Scholar]
- 5.Budzilowicz C J, Wilczynski S P, Weiss S R. Three intergenic regions of coronavirus mouse hepatitis virus strain A59 genome RNA contain a common nucleotide sequence that is homologous to the 3′ end of the viral mRNA leader sequence. J Virol. 1985;53:834–840. doi: 10.1128/jvi.53.3.834-840.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dreyfuss G, Mantunis M J, Pinol-Roma S, Burd C G. hnRNP proteins and the biogenesis of mRNA. Annu Rev Biochem. 1993;62:289–321. doi: 10.1146/annurev.bi.62.070193.001445. [DOI] [PubMed] [Google Scholar]
- 7.Furuya T, Lai M M C. Three different cellular proteins bind to complementary sites on the 5′-end-positive and 3′-end-negative strands of mouse hepatitis virus RNA. J Virol. 1993;67:7215–7222. doi: 10.1128/jvi.67.12.7215-7222.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Garcia-Blanco M A, Jamison S F, Sharp P A. Identification and purification of a 62,000-dalton protein that binds specifically to the polypyrimidine tract of introns. Genes Dev. 1989;3:1874–1886. doi: 10.1101/gad.3.12a.1874. [DOI] [PubMed] [Google Scholar]
- 9.Ghetti A, Piñol-Roma S, Michael W M, Morandi C, Dreyfuss G. hnRNP I, the polypyrimidine tract-binding protein: distinct nuclear localization and association with hnRNAs. Nucleic Acids Res. 1992;20:3671–3678. doi: 10.1093/nar/20.14.3671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gil A, Sharp P A, Jamison S F, Garcia-Blanco M A. Characterization of cDNAs encoding the polypyrimidine tract-binding protein. Genes Dev. 1991;5:1224–1236. doi: 10.1101/gad.5.7.1224. [DOI] [PubMed] [Google Scholar]
- 11.Hellen C U T, Witherell G W, Schmid M, Shin S H, Pestova T V, Gil A, Wimmer E. A cytoplasmic 57-kDa protein that is required for translation of picornavirus RNA by internal ribosomal entry is identical to the nuclear pyrimidine tract-binding protein. Proc Natl Acad Sci USA. 1993;90:7642–7646. doi: 10.1073/pnas.90.16.7642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hirano N, Fujiwara K, Hino S, Matsumoto M. Replication and plaque formation of mouse hepatitis virus (MHV-2) in mouse cell line DBT culture. Arch Gesamte Virusforsch. 1974;44:298–302. doi: 10.1007/BF01240618. [DOI] [PubMed] [Google Scholar]
- 13.Jeong Y S, Makino S. Evidence for coronavirus discontinuous transcription. J Virol. 1994;68:2615–2623. doi: 10.1128/jvi.68.4.2615-2623.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Joo M, Makino S. Mutagenic analysis of the coronavirus intergenic consensus sequence. J Virol. 1992;66:6330–6337. doi: 10.1128/jvi.66.11.6330-6337.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kaminski A, Hunt S L, Patton J G, Jackson R J. Direct evidence that polypyrimidine tract binding protein (PTB) is essential for internal initiation of translation of encephalomyocarditis virus RNA. RNA. 1995;1:924–938. [PMC free article] [PubMed] [Google Scholar]
- 16.Lai M M C. Coronavirus leader RNA-primed transcription: an alternative mechanism to RNA splicing. BioEssays. 1986;5:257–260. doi: 10.1002/bies.950050606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lai M M C, Brayton P R, Armen R C, Patton C D, Pugh C, Stohlman S A. Mouse hepatitis virus A59: mRNA structure and genetic localization of the sequence divergence from the hepatotropic strain MHV-3. J Virol. 1981;39:823–834. doi: 10.1128/jvi.39.3.823-834.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lai M M C, Cavanagh D. The molecular biology of coronaviruses. Adv Virus Res. 1997;48:1–100. doi: 10.1016/S0065-3527(08)60286-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lai M M C, Liao C-L, Lin Y-J, Zhang X. Coronavirus: how a large RNA viral genome is replicated and transcribed. Infect Agents Dis. 1994;3:98–105. [PubMed] [Google Scholar]
- 20.Lai M M C, Patton C D, Baric R S, Stohlman S A. The presence of leader sequences in the mRNA of mouse hepatitis virus. J Virol. 1983;46:1027–1033. doi: 10.1128/jvi.46.3.1027-1033.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.La Monica N, Yokomori K, Lai M M C. Coronavirus mRNA synthesis: identification of novel transcription initiation signals which are differentially regulated by different leader sequences. Virology. 1992;188:402–407. doi: 10.1016/0042-6822(92)90774-J. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee H-J, Shieh C-K, Gorbalenya A E, Koonin E V, La Monica N, Tuler J, Bagdzhadzhyan A, Lai M M-C. The complete sequence (22 kilobases) of murine coronavirus gene 1 encoding the putative proteases and RNA polymerase. Virology. 1991;180:567–582. doi: 10.1016/0042-6822(91)90071-I. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Leibowitz J L, Wilhelmsen K C, Bond C W. The virus-specific intracellular RNA species of two murine coronaviruses: MHV-A59 and MHV-JHM. Virology. 1981;114:39–51. doi: 10.1016/0042-6822(81)90250-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li H-P, Zhang X, Duncan R, Comai L, Lai M M C. Heterogeneous nuclear ribonucleoprotein A1 binds to the transcription-regulatory region of mouse hepatitis virus RNA. Proc Natl Acad Sci USA. 1997;94:9544–9549. doi: 10.1073/pnas.94.18.9544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liao C-L, Lai M M C. Requirement of the 5′ end genomic sequence as an upstream cis-acting element for coronavirus subgenomic mRNA transcription. J Virol. 1994;68:4727–4737. doi: 10.1128/jvi.68.8.4727-4737.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lin Y-J, Zhang X, Wu R C, Lai M M C. The 3′ untranslated region of coronavirus RNA is required for subgenomic mRNA transcription from a defective interfering RNA. J Virol. 1996;70:7236–7240. doi: 10.1128/jvi.70.10.7236-7240.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu Q, Yu W, Leibowitz J L. A specific host cellular protein binding element near the 3′ end of mouse hepatitis virus genomic RNA. Virology. 1997;232:74–85. doi: 10.1006/viro.1997.8553. [DOI] [PubMed] [Google Scholar]
- 28.Macejak D G, Sarnow P. Internal initiation of translation mediated by the 5′ leader of a cellular mRNA. Nature. 1991;353:90–94. doi: 10.1038/353090a0. [DOI] [PubMed] [Google Scholar]
- 29.Makino S, Joo M, Makino J K. A system for study of coronavirus mRNA synthesis: a regulated, expressed subgenomic defective-interfering RNA results from intergenic site insertion. J Virol. 1991;65:6031–6041. doi: 10.1128/jvi.65.11.6031-6041.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Makino S, Lai M M-C. Evolution of the 5′-end of genomic RNA of murine coronaviruses during passages in vitro. Virology. 1989;169:227–232. doi: 10.1016/0042-6822(89)90060-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.O’Farrell P Z, Goodman H M, O’Farrell P H. High resolution two-dimensional gel electrophoresis of basic as well as acidic proteins. Cell. 1977;12:1133–1142. doi: 10.1016/0092-8674(77)90176-3. [DOI] [PubMed] [Google Scholar]
- 32.Patton J G, Mayer S A, Tempst P, Nadal-Ginard B. Characterization and molecular cloning of polypyrimidine tract-binding protein: a component of a complex necessary for pre-mRNA splicing. Genes Dev. 1991;5:1237–1251. doi: 10.1101/gad.5.7.1237. [DOI] [PubMed] [Google Scholar]
- 33.Perez I, Lin C H, McAfee J G, Patton J G. Mutation of PTB binding sites causes misregulation of alternative 3′ splice site selection in vivo. RNA. 1997;3:764–778. [PMC free article] [PubMed] [Google Scholar]
- 34.Piñol-Roma S, Dreyfuss G. Shuttling of pre-mRNA binding proteins between nucleus and cytoplasm. Nature. 1992;355:730–732. doi: 10.1038/355730a0. [DOI] [PubMed] [Google Scholar]
- 35.Sawicki S G, Sawicki D L. Coronavirus transcription: subgenomic mouse hepatitis virus replicative intermediates function in RNA synthesis. J Virol. 1990;64:1050–1056. doi: 10.1128/jvi.64.3.1050-1056.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shieh C-K, Lee H J, Yokomori K, La Monica N, Makino S, Lai M M C. Identification of a new transcriptional initiation site and the corresponding functional gene 2b in the murine coronavirus RNA genome. J Virol. 1989;63:3729–3736. doi: 10.1128/jvi.63.9.3729-3736.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shieh C-K, Soe L H, Makino S, Chang M-F, Stohlman S A, Lai M M C. The 5′-end sequence of the murine coronavirus genome: implications for multiple fusion sites in leader-primed transcription. Virology. 1987;156:321–330. doi: 10.1016/0042-6822(87)90412-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Spaan W, Delius H, Skinner M, Armstrong J, Rottier P, Smeekens S, van der Zeijst B A M, Siddell S G. Coronavirus mRNA synthesis involves fusion of non-contiguous sequences. EMBO J. 1983;2:1839–1844. doi: 10.1002/j.1460-2075.1983.tb01667.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Svitkin Y V, Ovchinnikov L P, Dreyfuss G, Sonenberg N. General RNA binding proteins render translation cap dependent. EMBO J. 1996;15:7147–7155. [PMC free article] [PubMed] [Google Scholar]
- 40.Yokomori K, Banner L R, Lai M M C. Heterogeneity of gene expression of hemagglutinin-esterase (HE) protein of murine coronaviruses. Virology. 1991;183:647–657. doi: 10.1016/0042-6822(91)90994-M. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yu W, Leibowitz J L. A conserved motif at the 3′ end of mouse hepatitis virus genomic RNA required for host protein binding and viral RNA replication. Virology. 1995;214:128–138. doi: 10.1006/viro.1995.9947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang X, Lai M M C. Unusual heterogeneity of leader-mRNA fusion in a murine coronavirus: implications for the mechanism of RNA transcription and recombination. J Virol. 1994;68:6626–6633. doi: 10.1128/jvi.68.10.6626-6633.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang X, Liao C-L, Lai M M C. Coronavirus leader RNA regulates and initiates subgenomic mRNA transcription, both in trans and in cis. J Virol. 1994;68:4738–4746. doi: 10.1128/jvi.68.8.4738-4746.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang X, Lai M M C. Interactions between the cytoplasmic proteins and the intergenic (promoter) sequence of mouse hepatitis virus RNA: correlation with the amounts of subgenomic mRNA transcribed. J Virol. 1995;69:1637–1644. doi: 10.1128/jvi.69.3.1637-1644.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang, X., H.-P. Li, W.-M. Xue, and M. M. C. Lai. Unpublished observation.