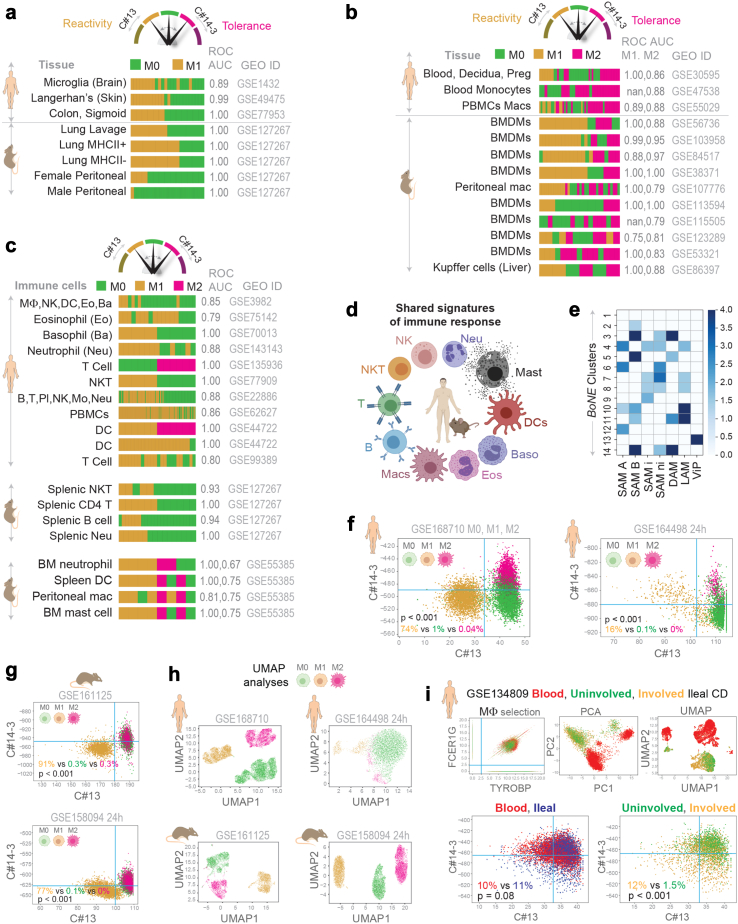
Fig. 2.
Definitions of “reactivity” and “tolerance” are conserved across tissues, organs, species, and diverse immune cell types. a and b) Validation studies assessing the ability of SMaRT genes to classify diverse tissue-resident macrophage datasets from both humans and mice. Performance is measured by computing ROC-AUC. Barplots show the ranking order of different sample types based on the composite scores of C#13 and path #14-3. c and d) Validation studies (c) assessing the ability of SMaRT genes to classify active vs inactive states of diverse immune cell types in both humans and mice. The schematic (d) summarizes findings in c. e) Published disease-associated macrophage gene signatures (see Supplemental Information 2) are analysed for significant overlaps with various gene clusters in the Boolean map of macrophage processes. Results are displayed as heatmaps of -Log10(p) values as determined by a hypergeometric test. f and g) Scatterplots of the composite score of C#13 and path #14-3 in human (f, GSE168710, GSE164498 24 h) and mouse (g, GSE161125, GSE158094 24 h) single cell RNASeq datasets with well defined macrophage polarization states (M0, M1, M2). Blue lines correspond to the StepMiner thresholds. Percentages of different cell types are reported in the bottom-left quadrant. Pvalue is computed by two tailed two proportions z-test for M1 vs M0. h) Traditional UMAP analysis of the single cell RNASeq datasets. i) PCA, UMAP and BoNE analysis of single cell RNASeq dataset GSE134809 that includes blood and ileal biopsy (uninvolved and involved) samples from Crohn’s disease (CD) patients. Macrophages were selected as the top right corner by using thresholds (2.5, blue lines) on TYROBP and FCER1G. Blue lines correspond to the StepMiner thresholds in the scatterplot between C#13 and C#14-3 (bottom plots). Bottom-left quadrant is evaluated for enrichment of cell types across tissue (blood vs ileal) and disease states (uninvolved vs involved CD). Percentages of different cell types are reported in the bottom-left quadrant. P value is computed by two tailed two proportions z-test.