Abstract
In the past several years, nanopore sequencing technology from Oxford Nanopore Technologies (ONT) and single-molecule real-time (SMRT) sequencing technology from Pacific BioSciences (PacBio) have become available to researchers and are currently being tested for cancer research. These methods offer many advantages over most widely used high-throughput short-read sequencing approaches and allow the comprehensive analysis of transcriptomes by identifying full-length splice isoforms and several other posttranscriptional events. In addition, these platforms enable structural variation characterization at a previously unparalleled resolution and direct detection of epigenetic marks in native DNA and RNA. Here, we present a comprehensive summary of important applications of these technologies in cancer research, including the identification of complex structure variants, alternatively spliced isoforms, fusion transcript events, and exogenous RNA. Furthermore, we discuss the impact of the newly developed nanopore direct RNA sequencing (RNA-Seq) approach in advancing epitranscriptome research in cancer. Although the unique challenges still present for these new single-molecule long-read methods, they will unravel many aspects of cancer genome complexity in unprecedented ways and present an encouraging outlook for continued application in an increasing number of different cancer research settings.
Keywords: alternative splicing, application, cancer genome, epigenome, third-generation sequencing
Introduction
Genome sequencing has become increasingly available over the past few decades. The second generation of sequencing technologies, known as next-generation sequencing (NGS), “massive-parallel,” or “high-throughput” sequencing, made DNA sequencing dramatically simpler and faster by employing microscopic, spatially separated DNA templates to massively parallelize the capture of data [1]. The sequencing process uses various platforms, such as DNA sequencing, RNA sequencing (RNA-Seq), single-cell RNA and DNA sequencing. DNA sequencing, such as whole-exome sequencing initially allows the detection of single nucleotide variants (SNVs) and copy number variations (CNVs) [2]. Similarly, RNA-Seq data analysis can produce information about gene expression levels, alternative splicing (AS), allelic silencing or differential allelic expression, gene fusions, and RNA editing [3, 4]. Single cell sequencing is emerging as a powerful tool for profiling cell-to-cell variability on a genomic scale [5]. Intratumor heterogeneity is a confirmed major cause of treatment failure and drug resistance in cancer. Single-cell sequencing of tumor cells addresses this issue by identifying subpopulations of cancer cells and immune cells within a single patient. Furthermore, spatial transcriptomics research is expected to generate highly detailed maps of single-cell gene expression at any tissue coordinate in cancer [6]. To date, NGS has become a standard tool for many applications in basic biology as well as for clinical and agronomical research. However, the read length from NGS data remains a bottleneck for biological studies.
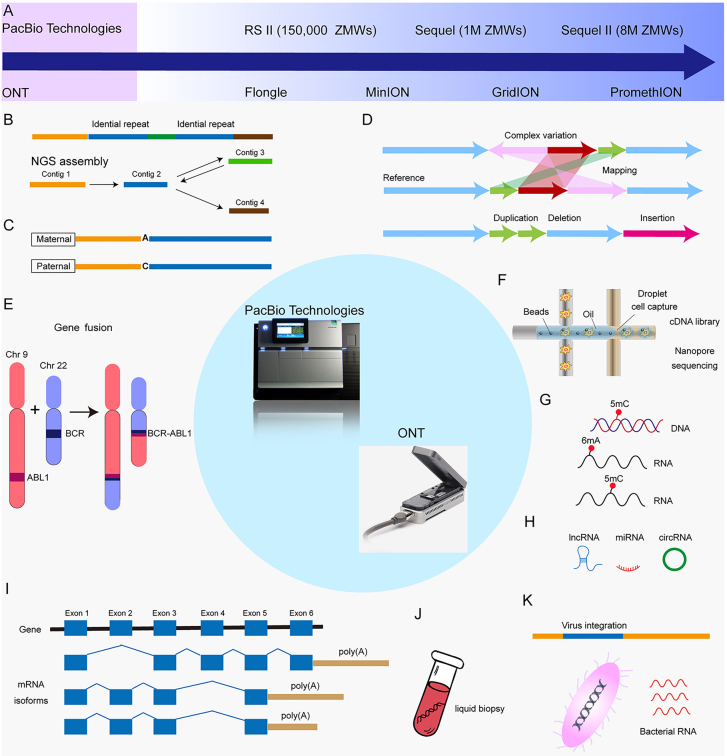
With the development of sequencing technology, DNA sequencing has inched the era of single-molecule sequencing (SMS) or third-generation sequencing (TGS) [7]. When DNA sequencing, PCR amplification is not needed to obtain the individual sequence of each DNA molecule and produces long-read in a real-time sequencing process [8]. The two major available TGS technologies are Pacific Biosciences (PacBio) single molecule real-time (SMRT) sequencing and the Oxford Nanopore Technologies (ONT) sequencing platform [9], [10], [11]. These long-read technologies permit sequencing/assembly through repetitive and complex elements, direct variant phasing, and even direct detection of epigenetic modifications that provide preference for certain applications [12], [13], [14]. The methodologies for these technologies (Figure 1) and comparison of the performance of NGS short-read and TGS long-read methods in terms of read accuracy, throughput and cost were comprehensively described in recent reviews [8, 15]; herein, we will focus on their potential use in advancing our understanding of cancer genomics, and applications in cancer research. In this review, we first provide a brief description of NGS in cancer research and outline the limitations of NGS and then describe the developments in TGS technologies. Next, we look at the bioinformatics for TGS data and describe the challenges in the development of bioinformatics tools. We also provide examples of how TGS has been adapted to investigate key aspects of cancer genomics for applications including the analysis of complicated cancer genomes, AS, fusion genes, exogenous RNA, and epigenetic marks. We highlight the unique challenges still present for TGS technologies in cancer research and provide some suggestions to overcome these challenges. We finish by briefly discussing the comparison of advantages between TGS and NGS and provide certain applications suited for each platform.
Figure 1:
Overview of NGS short-read and TGS long-read methods.
(A). In NGS by Illumina technology, DNA is fragmented into manageable sizes, and these fragments are ligated to adapters. After library preparation, individual DNA molecules are sequencing for short reads. Following a sequencing run, raw sequence reads were aligned to a reference genome. (B). In PacBio SMRT sequencing, DNA fragment is ligated to hairpin adapters to form a topologically circular molecule, known as SMRTbell. It is loaded onto a SMRT Cell and bound by a DNA polymerase for sequencing. In ONT sequencing, DNA is tagged with sequencing adapters preloaded with a motor protein on one or both ends. The DNA is combined with tethering proteins and loaded onto the flow cell for sequencing. Following a sequencing run, raw sequence reads were aligned to a reference genome. NGS, next generation sequencing; TGS, third generation sequencing.
A brief description of NGS in oncology
NGS has been widely used in new mutation screening, gene expression profiling, molecular classification, neoantigen prediction, and liquid biopsy in cancers [16], [17], [18]. These capabilities could impact therapy selection by offering insights regarding therapeutic sensitivity or resistance, or factors affecting diagnosis or prognosis [19]. For example, SNVs, insertion/deletion (indels) mutations, structural alterations (CNVs, translocations, inversions), loss of heterozygosity and aneuploidy can all be detected in a whole-genome data set comparing tumor DNA and normal DNA [20, 21]. In addition to genome characterization, NGS has also been deployed to characterize the cancer transcriptome using RNA-seq. RNA-seq can be able to provide information on RNA expression, as well as to detect alternative splicing and fusions [22]. Furthermore, single-cell sequencing through NGS has the advantages of assessing of tumor heterogeneity and separate cell types, such as immune cells [23]. The Cancer Genome Atlas (TCGA), funded jointly by the National Cancer Institute and National Human Genome Research Institute of the National Institutes of Health and the International Cancer Genome Consortium (ICGC), contains data for a series of large-scale studies in different countries across the world that were funded by the governments of each country and have helped establish the importance of cancer genomics and transformed our understanding of cancer [24]. Comprehensive analyses of these cancer genomic data provide unique opportunities for understanding cell-of-origin patterns, oncogenic processes, and signaling pathways [25, 26].
Although the availability of whole-genome (WGS), -exome (WES), or -transcriptome sequencing (RNA-seq) has been increasing, targeted gene sequencing is the method of choice for cancer diagnosis in clinical laboratories [27, 28]. With increasing numbers of cancer driver mutations genes now known, NGS panels are commonly used in hospitals for a wide range of cancers to ensure optimal sequencing quality (read depth and coverage, variant characterization, reporting), cost-effectiveness, and turnaround time [29]. In addition to NGS panels, identified biomarkers are also being used for cancer diagnosis, prognosis, and therapeutic applications in clinical laboratories [30], [31], [32]. In our Precision Cancer Medical Center in Fudan University Shanghai Cancer Center (FUSCC), large custom-designed NGS panels have been developed for breast cancer and other solid tumors for the detection of SNVs, indels, and CNVs [33]. The FUSCC-BC panel is used to detect somatic and germline mutations in breast cancer-specific genes in clinical settings, and identified the mutation characteristics, and potential molecular targets for breast cancer in China.
Another application of NGS in oncology is the identification and enrollment of patients for appropriate clinical treatments, such as liquid biopsy [34]. Blood contains many types of biological materials like circulating cells, extracellular vesicles (EVs), non-coding RNAs (ncRNAs), and cell-free DNA (cfDNA). The minimally invasive procedure for sample acquisition for this type of DNA or RNA assessment has been coined a “liquid biopsy”. Tumor cells in the body can release cfDNA through apoptosis, necrosis, or activate release [35]. Furthermore, cancers infrequently shed cells into the circulation, known as circulating tumor cells (CTCs) [36]. Therefore, DNA can be obtained from cancer patient blood samples or other bodily fluids (cerebral spinal fluid, cervical mucus, and urine). In addition, EVs are found in various body fluids and serve for intercellular communication by delivering their cargo molecules to other cells [37]. The emergence of EVs analytics in combination with liquid biopsy sampling opened a plethora of new possibilities for the detection of tumors [38]. RNA-seq, which can provide accurate and comprehensive gene expression profiling of liquid biopsies including genomic components of extracellular vesicles (EVs) and ncRNAs from blood, dramatically enhancer the probability of ncRNAs as biomarkers [39]. Therefore, the detection of ncRNAs using RNA-seq is a noninvasive, innovative approach for diagnosis [40]. The tumor-specific signatures in these samples can act as a new type of cancer biomarker and help to identify cancer patients from a group of healthy individuals [18]. Facilitated by the rapid development of NGS technologies, liquid biopsy can achieve much higher sensitivity than tissue biopsy and can be designed for different purposes [41]. However, there are several challenges due to its low concentration of DNA and high fragmentation (ranging from 100 to 10,000 bp fragments) in these samples [42], as well as the hard to build accurate classification model, using liquid biopsy for cancer screening and early detection remained to be solved.
Drawbacks of NGS methods
NGS is advantageous in many aspects, such as low cost, high speed and high yield. However, NGS methods also have some limitations. One of the most obvious limitations of NGS is the short-read length. This limits the precision of many biological studies, especially large genome assembly studies and precise specific isoforms analysis [43], [44], [45]. The read length of 100–200 bases is too short in the context of a vast genome, which makes it extremely difficult to accurately assemble the genome sequences from billions of short-reads [1, 46]. In addition, larger structural variations (SVs) are more challenging to detect and characterize using short-reads. Despite advances in sequencing technologies and bioinformatics, de novo assembly of large genomes remains challenging [3].
NGS platforms rely on clonal amplification to create multiple molecules, and do not have the sensitivity to detect nucleotides at a single molecule level. Transcriptomes based on the NGS platforms and RNA samples are typically subject to RNA fragmentation to a certain size range. Then, the cDNA fragments are sequenced in a high-throughput manner to obtain millions of short sequences. During sample preparation section, reverse transcription, PCR and size selection add base incorporation errors in individual molecules within a cluster [47]. The amplification process also creates an underrepresentation of bases in areas of high or low GC contents [48]. Therefore, during both the sequence processes and computational analysis phases, imperfections and biases may be introduced. This process greatly reduces the accuracy in the analysis of AS, gene fusion, and paralogous regions. Although generating high-read output and developing new computational approaches would be particularly beneficial for the quantitative analysis of transcript isoforms, other biases and limitations will result from the myriad of computational methods [3, 15].
NGS is also limited by its inability to directly sequence RNA. Short-read platforms rely on converting mRNA to cDNA before sequencing; as such, they are typically blind to nucleotide modifications. Consequently, by using NGS, indirect methods are required to identify transcriptome-wide RNA modifications. However, these methods cannot provide quantitative estimates of the modification at a given site and are often unable to identify the underlying RNA molecule that is modified [49].
TGS/long-read sequencing
PacBio and ONT technologies provide alternative long-read technologies that enable SMS of complete individual RNA molecules after conversion to cDNA. These two platforms both permit sequencing of non-amplified DNA of exceptionally long linear read lengths, high throughput and fast sequencing times (Table 1). PacBio utilizes a sequencing-by-synthesis approach by which polymerases incorporate distinguishable fluorescently labeled nucleotides to single DNA molecules and the fluorescent signals are recorded in real time in the zero-mode waveguides (ZMWs) [50]. Nanopore sequencing uses the minor changes in ionic current when nucleotide bases of single-stranded DNA/RNA molecules pass through protein nanopores to identify different nucleotides [51, 52]. The development and general features of PacBio and ONT sequencers are listed in Table 1 and Figure 2A.
Table 1:
Throughput of different third-generation sequencing platforms.
| Platform | Sequel | Sequel II | Flongle | MinION | GridION | PromethION |
|---|---|---|---|---|---|---|
| Company | PacBio | PacBio | ONT | ONT | ONT | ONT |
| Throughput | 15 Gba | 96 Gba | 1.8 Gb | 42 Gb | 210 Gbb | 5.88–11.76 Tbc |
| Run time | Up to 20 h | Up to 30 h | Up to 16 h | Up to 72 h | Up to 72 h | Up to 72 h |
Numbers are based on company website documentation (https://nanoporetech.com and https://www.pacb.com, both accessed April 29, 2021). aOne SMRT cell. bFive flow cells. cTwenty-four or forty-eight flow cells.
Figure 2:
Overview of cancer applications by PacBio and ONT.
(A) The development platforms of third generation sequencing (TGS); (B) Sequencing a region with two nearly identical repeats (blue) separated by a unique sequence (green) will generate reads corresponding to the upstream region (yellow) in short-read sequencing. Assembly programs for NGS will assemble these reads into a single contig; (C) Haplotype phasing. SNPs (single nucleotide: A or C) between maternal and paternal alleles; (D) Illustration of the use of direct mapping of long-read to a reference genome to resolve complex structural variations, including insertions, deletions, duplications, and complex variation; (E) Detecting of fusion genes; (F) Long-read single cell sequencing; (G) Base modification in a DNA and RNA molecule (represented by a red cycle) give rise to specific signals in TGS data that can be identified using computational methods; (H) Detecting of ncRNAs, including lncRNA, miRNA, and circRNA; (I) A multitude of mRNA transcript isoforms can be generated from a single gene through alternative intron splicing and alternative polyadenylation. Long-read sequencing covers the entire transcript and detects the whole alternative splicing events; (J) TGS-based DNA/RNA analysis for liquid biopsy; (K) Detecting of exogenous RNAs, including viral RNA and bacterial RNA. NGS: next generation sequencing; PacBio: Pacific Bioscience; Oxford Nanopore Technologies (ONT).
Long-read are an advantage of TGS technology. This technology overcomes some of the issues associated with short-read approaches and greatly improves the quality of genome assembly [8]. The high rate of false-positive splice junction detection by NGS is reduced with TGS, and the computational methods for de novo transcriptome analysis are also simplified, which leads to a more complete capture of isoform diversity, alternative polyadenylation (APA), fusion transcripts, structural variations, and paralogous regions [53]. Furthermore, another advantage presented by these platforms is that they have the ability to detect base modifications in native DNA. For instance, PacBio SMRT sequencing can directly detect and differentiate between base modifications such as 5-methylcytosine (5mC) and 5-hydroxymethylcytosine (5hmC) [54]. The ONT method determines the sequence of nucleic acids in a molecule directly without the need for amplification, sequencing by synthesis, or modification. This approach, termed dRNA-Seq, removes the biases generated by these processes, and permits the identification of RNA modifications and determination of poly-A tail length [55, 56]. Consequently, ONT technology can be used not only to sequence DNA but also to sequence RNA, including miRNA directly [50, 57].
However, PacBio and ONT share a common disadvantage of a high error rate of ∼5–20% randomly distributed errors before correction [12, 58]. Thus, it is necessary to reduce the error rate before subsequent utilization. The technology behind these systems is considerably different. For higher accuracy, these two platforms developed their own methods to achieve the accurate sequence. ONT developed a method to sequence both strands of a double-stranded DNA molecule, which was called “two-directional” (2D) sequencing. For PacBio SMRT sequencing, the library preparation on this platform creates a circular input molecule. The molecular can be sequenced many times, and the random errors can be mitigated by increasing the read depth using circular consensus sequence (CCS) for high-fidelity (HiFi) sequence reads. Furthermore, self-correction software, such as long-read multiple aligner (LoRMA), which needs high coverage to obtain accurate correction, can be used for error correction [59]. Alternatively, the hybrid error correction strategy uses short-reads from NGS to correct long-read [60]. As such, a hybrid sequencing strategy combining second- and third-generation sequencing technologies could reduce the error rate and quantify transcript isoforms or fusion genes [61, 62].
Bioinformatics for TGS data analysis
As mentioned above, TGS offers a number of advantages over short-read sequencing. However, the data from these platforms is qualitatively different from NGS, thus necessitating tailored analysis tools. In addition, raw reads from PacBio SMRT sequencing and Oxford nanopore sequencing have high error rates with most errors occurring due to false insertions or deletions [50,63], [64], [65]. The error mode in SMRT sequencing is remarkably stochastic in nature [50]. For nanopore sequencing on the other hand, the error profile has been reported to be biased. For example, A-to-T and T-to-A errors were estimated to be very low [66, 67]. Therefore, they require new bioinformatics approaches to overcome their complex errors and modalities. The development of bioinformatics approaches that could take full advantage of long-read sequencing has become one of the most important issues in bioinformatics. We now have various tools for base calling, error correction and polishing, de novo genome assembly, mapping, and phasing using long-read data. Here we present an overview of the analysis pipelines for PacBio and ONT data and highlight popular tools in cancer research (Table 2). There are also many existing tools that detect variants, gene isoform resolution and epigenetic modifications from long-read. Recent reviews focusing on long-read bioinformatics tools can be found in the literature [68], [69], [70]. For example, Amarasinghe et al. [70] presented an overview of the analysis pipelines and popular tools for TGS data. They also introduced a complementary open-source catalogue of long-read analysis tools: long-read-tools.org, which allows users to search and filter tools based on various parameters.
Table 2:
Typical long-read analysis pipelines for PacBio and ONT data.
| Application | PacBio | ONT |
|---|---|---|
| Basecalling | Generate CCS [71] | Guppy [92] |
| Quality control | Isoseq3 [71] | NanoQCa, NanoStata |
| Read error correction | ||
| Non-hybrid methods | LoRMA [59] | Canu [198], LoRMA |
| Hybrid methods | LORDEC [199], proovread [200], LSC [201], FMLRC [202] | Nanocorr [203], FMLRC |
| Polishing | Arrow [71], Raconb | Nanopolish [110], Racon |
| Alignment | Pbmm2 [71], BLASR [71], minimap2c | Pbmm2 |
| Structural variation analysis | CORGi [81], PBHoney [82], pbsv [71], Sniffles [83], SMRT- SV [84], SVIM-asm [85] |
NanoSV [86], Picky [87], Sniffles, SVIM-asm |
| Isoform detection | IsoSeq3, Cupcaked, IsoCon [88] | FLAIR [90], Pinfish [92] |
| Isoform quantification | Salmon [91], FLAIR, featureCounts [93], Wub [92] | |
| Differential analysis of isoform | DEseq2 [204], limma [205], edgeR [206] | |
| APA events detection | TAPIS [97], PRIPI [98] | |
| Fusion transcript detection | IDP-fusion [151] | |
| Base modifications detection | SMRT LINK, Tombo [112] | NanoMod [111], Tombo, Nanopolish, signalAlign [113], D-Nascent [114], DeepSignal [115], mCaller [116], DeepMod [117] |
| circRNA transcripts detection | PRAPI | isoCirc [192], CIRI-long [193] |
aNanopack, https://github.com/wdecoster/nanopack, accessed June 9, 2021. b https://github.com/isovic/racon, accessed June 9, 2021. c https://github.com/lh3/minimap2, accessed June 9, 2021. d https://github.com/Magdoll/cDNA_Cupcake, accessed June 9, 2021.
The challenges in the development of bioinformatics tools
Basecalling
The first step in any long-read analysis is basecalling, the computational process of translating raw data to nucleotide sequences. This step is more standardized and usually performed using proprietary software. SMRT sequences detect fluorescence events that correspond to the addition of one specific nucleotide by a polymerase. The template of SMRT sequencing is circular, and the polymerase go over the DNA fragment multiple times. During basecalling process, a continuous long-read is converted from the pulses, then split into subreads. These subreads are aligned together, and derive a circular consensus sequence using workflow CCS [71]. ONT sequencers measure the ionic current fluctuations when single-stranded nucleotide passes through the nanopores. Basecalling for ONT is more complicated than PacBio basecalling, and more options are available. Wick et al. [72] examined the performance of different basecalling tools, and Guppy basecaller performs well overall, with good accuracy and fast performance.
Error correction
Despite increasing accuracy of both TGS platforms, error correction remains an important step in long-read analysis pipelines. In the case of SMRT sequencing, the CCS quality is heavily dependent on the number of times the fragment is read. As mentioned above, the random errors can be mitigated by increasing the read depth using CCS. Long HiFi reads with an average length of 13.5 kb generated using the CCS mode on the PacBio Sequel Systems, can provide base-level resolution with >99.8% single-molecule read accuracy [73, 74]. CCS reads enable structural variant detection and de novo assembly at similar contiguity. However, CCS reads retain a major residual error and exhibit a bias for indels in homopolymers [74]. Furthermore, the current tools, such GATK, which was designed for short-reads, does not properly model the CCS error profile. For ONT on the other hand, indels and substitutions are frequent in nanopore data. The error profile has been reported to be biased, which tend to occur in homopolymer regions [66]. These error characteristics pose a challenge for long-read data analyses, and error correction algorithms are needed to be developed for fixing sequencing error.
Two groups of methods to error correct long-reads can be employed: methods that only use long-reads (non-hybrid methods) and methods that leverage the accuracy of additional short-read data (hybrid methods). A few studies have showcased comparisons among rapidly evolving error correction algorithms to evaluate the quality and computational resource requirements of these tools [75], [76], [77]. Non-hybrid methods perform self-correction with long-reads alone. All reads are first aligned to each other and generate consensus sequences using overlap information. This consensus sequences are used to correct individual reads, which can be taken forward for assembly or other applications. However, the performance of non-hybrid methods deteriorated significantly when sequencing depth was decreased. Effective error correction requires high sequencing coverage, which needs both cost and time consumed during sequencing and analysis. Hybrid error correction strategy, which uses short-reads is still outperforming long-read-only correction. The same biological sample must be sequenced using both technologies in certain applications require high base-level accuracy. Hybrid error correction methods can be further divided into two categories according to how short-reads are used, short-read-alignment-based methods and short-read-assembly-based methods, respectively. In short-read-alignment-based methods, the short-reads are directly aligned to the long-read using a variety of aligners, to generate corrected long-read. In short-read-assembly-based methods, the short-reads are first used to generate contigs using an existent assembler, or only build the de Bruijn graph (DBG). Then the long-read are corrected by aligning to the assembly or by traversing the DBG.
Compared with non-hybrid methods, hybrid methods aided by short accurate reads can achieve better correction quality, especially when handling low coverage-depth long-read. The relative cost per base pair using TGS is still several folds higher when compared to the NGS. Users are recommended to choose hybrid methods in certain applications. Despite continuous improvements in the accuracy of long-read, error correction remains indispensable in many applications. Complexities resulting from GC-rich regions, tandem repeats and highly variable gene complexes cannot be accurately sequenced using short-reads sequencing [78]. Therefore, repeats, or complex regions may not be correctly handled using hybrid methods. For future work, removing the need for short-reads, better and more efficient self-correction algorithms are expected to reduce the cost and complexity of genomic projects.
Structural variation analysis
Long-read help to increase the detection of SVs as they considerably ease de novo genome assembly and mapping. Two recent reviews described the algorithms of structural variant calling from long-read data in detail [79, 80]. For instance, algorithms detect SVs from SMRT data by leveraging intra-read and inter-read signatures. CORGi [81], PBHoney [82], PBSV, Sniffles [83], SMRT-SV [84] and SVIM-asm [85] detect SVs through combinations of these two signatures. Due to higher operational costs and a large input DNA requirement, long-read have been mostly applied to single-genome assemblies. In the case of ONT, the signatures to detect SVs are similar to those used in PacBio data. Callers that detect SVs from nanopore data include NanoSV [86], Picky [87], Sniffles and SVIM-asm. Compared with PacBio sequencing, ONT provides improved read lengths, lower adaptation costs and higher throughput. It is more effective to detect many SVs. However, indels are frequent in nanopore data, which make it less suitable for smaller SVs. Furthermore, evaluating the performance of long-read SV callers is complicated by the fact that benchmark data sets may be missing SVs in their annotation.
Long-read transcriptome analysis
The goal of long-read transcriptome analysis mainly consists of the following three parts, which are isoform detection, quantification, and differential analysis. PacBio supplies the IsoSeq3 analysis pipeline for the analysis of their cDNA CCS reads, allowing the assembly of full-length transcripts. Cupcake provides scripts for collapsing redundant isoforms and merging Iso-Seq runs from different batches. Iso-Con [88] and SQANTI [89] pipelines attempt to mitigate the erroneous merging of similar transcripts of the Iso-Seq pipeline. For assaying the sequences of highly-similar gene families, IsoCon detects isoforms from genes with significant alternate splicing and minor shifts in the splice junctions. In the case of ONT, both Pinfish and FLAIR are intended for nanopore data. However, full-length alternative isoform analysis of RNA (FLAIR) needs to use short-read reads to improve junction annotation [90]. In addition, their accuracy has not yet been extensively verified. Several methods, such as Salmon [91], Wub [92], featureCounts [93], FLAIR [90], etc., can be used to quantify the abundance of transcripts. However, these methods rely on a complete and accurate isoform annotation. Therefore, quantifying and performing differential expression analysis of transcript levels on the isoform instead of the gene level remains significant challenges. It is hard to decide at which point a known and a newly identified isoform, and systematically differentiate allele-specific isoform expression.
AS events, APA and alternative transcription initiation (ATI) are major processes that contribute to transcriptome diversity. ONT directed RNA sequencing was used to measure the length of poly(A) and identify a range of novel transcript isoforms including those with AS, ATI, and APA [94], [95], [96]. For the analysis of post-transcriptional regulation based on long-read sequencing, transcriptome analysis pipeline for isoform sequencing (TAPIS) pipeline [97] and Post-transcriptional Regulation Analysis Pipeline for Iso-Seq (PRAPI) [98] are two main bioinformatics tools that use PacBio reads to identify AS and APA. For instance, the survey of APA events using TAPIS is becoming a landmark for annotation in sorghum (Sorghum bicolor L. Moench) [97], moso bamboo (Phyllostachys edulis) [99], wild apple (Malus sieversii) [100], Chinese cabbage (Brassica rapa L. ssp. pekinensis) [101], cotton (Gossypium spp.) [102], Ricinus communis [103], silkworm (Bombyx mori) [104], Lateolabrax maculatus [105], red clover (Trifolium pratense L.) [106], Gnetum luofuense [107], and perennial ryegrass (Lolium perenne) [108]. In addition, PRAPI can also identify several other events, such as production of circular RNAs (circRNAs). However, at present, quantification analysis of AS or APA still depends on NGS due to the low coverage of PacBio and ONT sequencing. In the future, it is expected that both TGS can be used for quantification analysis once the throughput increases.
Base modifications detection
In SMRT sequencing, base modifications in DNA are inferred from the interpulse duration (IPD) between fluorescence pulses [54]. mA, mC and hmC in a DNA template alter the kinetic characteristics, such as the IPD between two successive base incorporations [54]. These changes of kinetic signatures can be analyzed directly via the SMRT Portal for base modification detection. However, reliable calling of these base modifications requires high sequence coverage per strand. For instance, reliable calling of 4 mA and 6 mC requires 25× coverage per strand, whereas 250× coverage is required for 5 mC and 5 mhC [109]. Such high coverage is not realistic for large genomes and does not allow single molecule epigenetic analysis. ONT sequencing detect base modifications owing to the signal shifts caused by the modified RNA or DNA bases as they pass through the nanopore [49, 110]. Several computational tools have been developed to detect base modifications on the basis of these characteristic disruptions: NanoMod [111], Tombo [112], Nanopolish [110], signalAlign [113], D-NAscent [114], DeepSignal [115], mCaller [116], and DeepMod [117]. However, these methods may suffer from a high false discovery rate, since no experimental exist systematically validate the detection of the large variety of modifications present in RNA. In addition, many modifications do not influence the TGS’s dynamics sufficiently to be detected at a useful sensitivity. Therefore, continued efforts in developing and benchmarking tools are required to obtain accurate, and complete (including base modifications) genomes and transcriptomes.
Applications of TGS technologies in cancer
Detecting complicated cancer genomes
Human genome is considered to be one of the most complete mammalian reference assemblies. However, it contains many regions of high and low complexity that have relevance to human’s disease, such as low-complexity tandem repeats, pseudogenes, high GC content, and extremely copy number variable regions [118]. Sequencing those DNA elements is difficult with short-read sequencing, and TGS is a great tool to resolve these gaps. Cancer studies using long-read information to decipher allele-resolution mutation statuses and the complete structures of complicated cancer genomes have been rapidly increasing and continuously progressing. With the application of long-read sequencing alone or in combination with more accurate conventional short-read sequencing, it is possible to detect cancer mutations occurring at a low frequency. For example, PacBio has been applied to screen for the emergence of actionable mutations in the tumor suppressor TP53 [119]. In acute myeloid leukemia (AML) and myelodysplastic syndrome (MDS), many patients harbor multiple TP53 mutations in their tumors, and these TP53 variants are located in different alleles. Further, long-read are also utilized in the detection of genomic mutations at single-allele resolution. For instance, Suzuki et al. [120] applied long-read MinION and found that 72% of the reads harbored two epidermal growth factor receptor (EGFR) mutations (T790M and L858R), and 22% of the reads harbored neither mutation in H1975 cells (lung adenocarcinoma cell line). Then, they also detected aberrantly spliced RNAs in neurofibromatosis type 1 (NF1) and gene fusion transcripts, such as coiled-coil domain containing 6-rearranged during transfection (CCDC6-RET) and echinoderm microtubule-associated protein-like 4-anaplastic lymphoma kinase (EML4-ALK). Finally, they successfully applied this developed approach to characterize the mutation genotypes of eight clinical samples.
Particularly for cancer applications, cancer-associated SVs, such as large insertions, deletions, inversions, duplications, and translocations of variable genomic sequences, could be detected by long-read sequencing approaches [121], [122], [123]. Considering the limitations of NGS, long-read sequencing can improve the validation, resolution, and classification of germline SVs [124]. For example, in chromophobe renal cell carcinoma, structural alterations in the telomerase reverse transcriptase (TERT) promoter region identified by WGS analysis can be validated by PacBio sequencing [125]. Although short-reads can also be assessed SVs, large and complex SVs and repetitive regions cannot be detected this way [43, 46]. Norris et al. [122] applied nanopore sequencing to detect a series of well-characterized SVs and successfully identified cancer-related SVs in the cyclin dependent kinase inhibitor 2A (CDKN2A) and Mothers against decapentaplegic homolog 4 (SMAD4) genes at a low level in pancreatic cancer. Interestingly, they demonstrated that nanopore sequencing can detect these SVs at dilutions as low as 1:100, with as few as 500 reads per sample, which indicates that this technology could become an ideal tool for the low-level detection of cancer-associated SVs. In addition, Williams et al. [126] applied targeted nanopore sequencing and identified ABCB1 structural variants in THP-1 AML cells and high-grade serous ovarian cancer cells. In the context of non-small-cell lung cancer, Sakamoto et al. [127] found that long-read sequencing (PromethION) was particularly useful for precisely identifying and characterizing structural aberrations and identified several medium-sized structural aberrations, consisting of complex combinations of local duplications, inversions, and microdeletions, in lung cancer cell lines and clinical samples. In the context of breast cancers, a pioneering study by Schatz’s group involved the sequencing of the SK-BR-3 breast cancer cell line genome using PacBio SMRT long-read sequencing and demonstrated that amplification of the Erb-B2 receptor tyrosine kinase 2 (ERBB2) oncogene (also known as HER2) appeared within complex rearrangements, which can only be precisely identified by long-read sequencing [123]. Further, they sequenced SK-BR-3 cells and patient-derived organoids representing tumor and matched normal cells from two breast cancer patients via ONT, PacBio, and Illumina/10× Genomics for the comprehensive analysis of SVs. Interestingly, they found that long-read sequencing allowed for substantially more accurate and sensitive SV detection and that hundreds of variants within known cancer-related genes were detectable only through long-read sequencing [128]. Recently, Lin et al. [129] identified five genomic regions on 17q as potential hotspots of chromothripsis in breast cancer. Nanopore sequencing further detected translocations between chromosomes 17q23 and 20q13 and confirmed complex rearrangements between them that harbor a dense estrogen receptor α (Erα) hub and their corresponding target loci, respectively. These findings highlight the need for long-read sequencing to enable an in-depth analysis of how SVs disrupt the genome, and they also shed new light on the complex mechanisms involved in cancer genome evolution.
Characterization of AS
AS in particular is known to affect more than half of all human genes [130]. However, assessment of the differences in mRNA isoform expression between tissues and determination of which mRNA splice isoforms are potentially deleterious can be challenging [131]. TGS of long-read has the potential to identify and quantify isoforms simply by sequencing cDNA or mRNA molecules end-to-end from 3′ polyA tail to 5′ cap. For example, de Jong et al. [132] performed MinION nanopore sequencing of long-range PCR amplicons to identify 20 novel breast cancer gene 1 (BRCA1) isoforms, 18 of which contained multiple individual splicing events, and found that these events can co-occur within single transcripts. In addition, nanopore sequencing can be used in single-cell analysis [133], [134], [135], [136], [137]. Singh et al. [133] described a rapid high-throughput method to sequence full-length transcripts using targeted capture and Oxford nanopore sequencing of T-cell receptor and B-cell receptor mRNA transcripts and linked this with short-read transcriptome profiling. They revealed the clonal and transcriptional landscape of lymphocytes at single-cell resolution. This novel method, termed Repertoire and Gene Expression by Sequencing (RAGE-Seq), offers a new genomic toolkit for advanced single-cell analysis.
Particular splicing events are associated with many cellular processes, such as cellular growth, differentiation, tissue development, and oncogenesis [131]. Aberrant splicing events frequently occur in cancer and are associated with the hallmarks of cancer [138]. The importance of studying connections between AS and cancer is underscored by the possibility that some specific splice isoforms drive the oncogenic process and could represent attractive therapeutic targets. Nanopore sequencing has been used to characterize the global transcriptome signatures of mitochondrial and ribosomal gene expression in human cancer stem-like cell populations, which might provide a basis for the application of additional pathway-directed therapies such as those targeting mitochondria and ribosomes [139]. Moreover, unique splice variants or sets of splice variants are strongly associated with particular types of cancers and have diagnostic and prognostic value [138]. For example, RNA immunoprecipitation with a LINE 1-specific antibody followed by nanopore sequencing detected LINE1 transcripts of 90 individual elements in VM-Cub-1 UC cells. Further study showed that the expression of the individual variant long interspersed element 1 (LINE1s) is highly heterogeneous among cancer types [140].
Mutations in the splicing factor spliceosome factor 3b (SF3B1) have been associated with characteristic alterations in splicing. In a recent study, nanopore technology was applied to resequence a subset of tumor samples and normal samples from chronic lymphocytic leukemia (CLL) patient with wild-type SF3B1 or the K700E mutation and to sequence normal B-cell samples [90]. They demonstrated differential 3′ splice site changes associated with SF3B1 mutation, which is consistent with the known effects of SF3B1 mutation. They also observed a strong downregulation of intron retention events associated with SF3B1 mutation. This study of primary CLL samples by nanopore sequencing demonstrates the ability of the nanopore approach to identify and quantify cancer-specific transcript variants.
Long-read can completely cover full-length transcript sequences, so long-read sequencing is a superior approach over short-read RNA-Seq in detecting AS transcripts or transcript isoforms. Kohli et al. [141] applied modified RNA-Seq and found frequent coexpression of androgen receptor variant (AR-V) 9 and AR-V7 in prostate cancer. They further performed SMRT isoform sequencing (Iso-Seq) with a PacBio RSII to determine the full sequence of each AR mRNA transcript. In this study, they identified a common shared 3′ terminal exon as the molecular basis for frequent AR-V7 and AR-V9 coexpression in castration-resistant prostate cancer (CRPC). AR-V7 has been studied as a potential biomarker for drug resistance in prostate cancer. Thus, AR-V9 may also be a predictive biomarker for resistance. They further performed long-read RNA-Seq to analyze the effects on the expression of AR and truncated AR variants and found that AR gene rearrangements correlated with AR overexpression. Tumor-specific overexpression of AR-Vs indicated resistance to endocrine therapies, indicating that AR gene rearrangements are an important mechanism of resistance to endocrine therapies in CRPC [142]. Long-read SMRT sequencing also revealed that alternative isoforms and tumor-specific isoforms that arise from aberrant splicing are common during liver tumorigenesis. More excitingly, unannotated variants of ARHGEF2 (v1 and v3) were found to have biological significance in underscoring two major cancer hallmarks [143].
The splicing pattern of specific isoforms of numerous genes is altered as cells move through the oncogenic process. For instance, BRCA1 associated RING domain 1 (BARD1) interacts with BRCA1 and may act as a potent tumor suppressor. However, BARD1 splice isoforms show effects that are antagonistic to those of BARD1-full-length and have been associated with disease progression and a poor prognosis in multiple cancer types. Walker et al. [144] presented a comprehensive BARD1 mRNA splicing landscape by performing nanopore sequencing and splicing assays for 12 tissue types (normal and cancer tissue). Similarly, a recent study used a combination of nanopore sequencing, RNA-Seq, and RT-qPCR analyses to identify the details of BARD1 AS in melanoma [145]. These studies include the most comprehensive assessments of BARD1 mRNA splicing to date and provide information for the convenient assessment of the roles of these isoforms in human biology.
Indeed, in a pioneering study by Oka et al. [146], they performed MinION full-length DNA sequencing to characterize the alternatively spliced isoforms of lung cancer cell lines and then biologically validated the alternatively spliced isoforms. These aberrant transcripts were then found in non-small-cell lung cancer specimens. Interestingly, the authors applied liquid chromatography with tandem mass spectrometry (LC/MS/MS) and demonstrated that at least some alternatively spliced isoforms were truly translated into peptides and could play a role in producing neoantigens in cancer. Most importantly, these peptides derived from splicing isoforms and frameshift mutations could activate the T cell response through interaction with human leukocyte antigens (HLAs). These results clearly show that long-read sequencing can be used to identify novel isoforms and neoantigens that may be overlooked with the current short-read sequencing approaches. To improve the diversity of captured full-length isoforms, Hu et al. [147] developed a normalized single-molecule RNA-Seq method, and identified new cancer-specific transcriptome signatures in human gastric signet-ring cell carcinoma. This method can capture 3.2–6.0-fold more full-length high-quality isoform species for different human samples than the non-normalized capture procedure and provides a new option for specific projects. Taken together, these papers clearly highlight the potential of TGS to identify new isoforms and isoform features, which is essential for the precise identification of aberrant transcript structures in cancer cells.
Identification of fusion genes
Fusion transcripts are the result of a trans-splicing event that joins two separately encoded pre-RNAs into one transcript [148] and are known to be major driver events for carcinogenesis in several types of cancers. Many fusion genes are strong driver mutations in neoplasia and have provided fundamental insights into the disease mechanisms that are involved in tumorigenesis [149]. Many fusion genes are extremely likely to produce unique tumor neoantigens that are recognizable by immune cells; thus, they are an ideal marker for the selection of immune checkpoint inhibitors [150]. Compared with the existing tools, the integration of TGS long-read and NGS short-reads (named Isoform Detection and Prediction [IDP] fusion) to detect fusion genes provides a higher precision and a very low false positive rate [151]. These approaches can also be readily applied to the analysis of cancer-associated fusion transcripts. For instance, using long-read MinION, Suzuki et al. identified cancerous mutations in lung cancer cells and clinical samples and detected the major driver genes, which have diverse patterns, including point mutations and fusions [120]. A recent study identified TTYH1-C19MC fusions leading to the overexpression of microRNAs in embryonal tumor with multilayered rosettes. This effect in turn drives the expression of a brain-specific DNMT3B isoform and promotes tumourigenesis [152]. In another report, the authors applied MinION RNA-Seq to sequence full-length transcripts in lung cancer cell lines and detected a cancer driver fusion transcript of the CCDC6-RET gene from the LC2/ad cell line [153]. In prostate cancer, one study identified a novel fusion transcript comprising the RLN1 and RLN2 genes. The fusion transcript encodes a putative Relaxin 2 (RLN2) with a deleted secretory signal peptide, indicating a potentially biologically important alteration [154].
Transcripts with aberrant structures are extremely likely to produce chimeric proteins and serve as specific targets for treatment [149]. For example, chronic myelogenous leukemia (CML) is a blood cancer that is caused by a translocation between chromosomes 9 and 22, giving rise to BCR-ABL1. Imatinib, a potent inhibitor of the oncogenic tyrosine kinase BCR-ABL, has shown remarkable clinical activity in patients with CML [155]. Several studies have reported that PacBio sequencing and ONT sequencing can be applied to detect BCR-ABL1 fusion events and related tyrosine kinase inhibitor (TKI) resistance mutations in samples from CML patients both at diagnosis and during follow-up [156], [157], [158]. These technologies are also important for identifying other components involved in CML pathogenesis to can be exploited to overcome tyrosine kinase inhibitors (TKI) resistance. In addition, Jeck et al. [159] developed a nanopore-based sequencing assay that can decrease the turnaround time for the detection of BCR-ABL1 fusion transcripts, which may be a valid approach for laboratories with low specimen volumes and for cases in which rapid results are needed.
In the context of AML, FMS-like tyrosine kinase 3 (FLT3) mutation is the most common genetic alteration in patients, and most of these mutations are constitutively activating internal tandem duplication (ITD) mutations. Shah and colleagues applied SMRT sequencing and first confirmed the presence of activating mutations in FLT3 (FLT3-ITD), which are associated with a poor prognosis in AML. Secondary kinase domain (KD) mutations in FLT3-ITD can cause preclinical and acquired clinical resistance to the highly potent type II FLT3 inhibitor quizartinib [160]. Further Shah et al. [161] described the cocrystal structure of FLT3 with the TKI quizartinib and identified a novel FLT3 inhibitor, PLX3397, that retains activity against the F691L mutant. Further, FUsion Detection from Gene Enrichment (FUDGE) was developed to accurately identify fusion genes from low-coverage nanopore sequencing within 2 days. In this assay, Cas9-targeted enrichment of fusion genes is performed, and then the unknown fusion partner and precise breakpoint are identified by nanopore sequencing. FUDGE enables multiplexed enrichment for the simultaneous analysis of several genes in multiple samples in one sequencing run. The application of this assay in the clinic could allow for rapid gene fusion detection [162]. Hence, TGS can now be applied for gene fusion detection as a diagnostic and prognostic tool for therapy initiation and minimal residual disease monitoring following treatment.
Characterization of exogenous RNA
Although cancer is generally considered to be a disease of host genetics and environmental factors, microorganisms are implicated in ∼20% of human malignancies [163]. Microbes and microbiota can contribute to cancer development and progression and the responsiveness to cancer therapeutics [164]. It has been demonstrated that TGS has tremendous potential utility for WGS. The MinION platform was used with a 6-h sequencing run time, and sufficient data were generated to identify bacterial and viral samples down to the species level, which suggests that TGS can accurately identify and differentiate both viral and bacterial species present within biological samples via amplicon sequencing [165]. Hepatitis B virus (HBV) infection is the main cause of hepatocellular carcinoma (HCC) worldwide [166]. HBV can integrate into human DNA and promote carcinogenesis by insertional mutagenesis or by promoting genomic instability [167]. Recently, integrative analysis of HBV genomes based on NGS and TGS of tumor and nontumor liver tissues from HCC patients provided a comprehensive view of the integration process in liver tissues. Interestingly, replicating HBV DNA was more frequently detected in nontumor tissues than in tumor tissues and was associated with a higher number of nonclonal integrations. More importantly, integration of viral enhancers near a cancer driver gene may lead to strong overexpression of oncogenes. HBV integration can drive carcinogenesis by altering cancer driver genes (TERT, TP53, MYC) at a distance, and the number of HBV integration events is an independent prognostic factor in HBV-related HCC [168]. Similarly, Tatkiewicz et al. [169] assessed relative provirus expression in HERV-K (HML-2) via both short- and long-read sequencing in three mantle cell lymphoma cell lines (JVM2, Granta519 and REC1) and observed a strong tissue-specific pattern of provirus expression. Therefore, the development of TGS has enabled more precise characterization of the role of viral and bacterial genetic material in cancer diagnosis and prognosis.
Increasing numbers of studies have highlighted the key role of gut microbiota in mediating tumor responses to chemotherapeutic agents and immunotherapies targeting programmed death-ligand 1 (PD-L1) or cytotoxic T lymphocyte-associated protein 4 (CTLA-4) [170], [171], [172]. Nasal microbiota results at the genus level were compared using Illumina vs. nanopore 16S rRNA gene sequencing, and long-read sequencing had a higher efficiency than short-read sequencing in terms of the taxonomic classification of gut microbiota at the species level [173]. Therefore, this technology can be further adopted for gut microbial community research. The gastric pathogen Helicobacter pylori is the main causative agent for gastric cancer and gastric and duodenal ulcers [174]. In a study by Devi et al. [175], the authors demonstrated that low Bifidobacterium abundance among the lower gut microbiota is associated with H. pylori-related gastric ulcers and gastric cancer, indicating that long-read sequencing may serve as a noninvasive assessment method. In another study, Tetz et al. [176] used a combination of short-read and MinION long-read sequencing technologies to draft a complete genome sequence of Kluyvera intestine sp. nov. isolated from the stomach of a patient with gastric cancer. The identification of antibiotic resistance genes would enable us to understand the possible pathogenicity of these bacteria and their role in cancer. Interestingly, Gaiser et al. [177] performed real-time full-length 16S rRNA gene sequencing (PacBio single-molecule sequencing) on paired cyst fluid and plasma from patients with suspected pancreatic cystic neoplasms to assess the microbial composition and diversity in the cyst fluid. They showed that the intracystic bacterial DNA and interleukin-1β concentrations were significantly elevated and positively correlated with intraductal papillary mucinous neoplasm incidence and neoplastic grade. Recent work has also documented that the gut microbiota is related to the occurrence and development of colorectal cancer (CRC) [178]. Thus, classification of the gut microbial community in CRC should be applied in clinical settings to predict CRC development. MinION platforms for 16S rRNA sequencing have been applied to detect and classify microbial communities, and the MinION sequencing platform coupled with the corresponding algorithm could function as a practicable strategy for classifying the bacterial community down to the species level [179]. The authors further assessed the gut microbiota in clinical subjects, including healthy participants and CRC patients, and found significant differences in gut microbial communities between patients with adenomas and healthy subjects [180]. Taken together, these findings indicate that these technologies can be applied for classifying differential gut microbial communities in distinct clinical specimens and will be useful tools for rapid screening.
Direct identification of epigenetic marks in DNA and RNA
DNA modifications play an essential role in the regulation of a variety of biological processes, and deregulation of the epigenetic machinery has been directly implicated in tumorigenesis [181]. These epigenetic modifications can be interrogated directly by instruments from both PacBio [54, 182] and ONT [113]. For instance, long-read sequencing, including ONT and PacBio sequencing, can be used to concurrently assess the CpG methylation of novel and extant transposable element (TE) insertions in the hippocampus and heart, as well as in paired tumor and nontumor liver samples [183]. One study developed transposons from long DNA reads (TLDR) software to interrogate the methylation patterns of both nonreference and reference TE insertions and found pronounced demethylation of young long interspersed element 1 (LINE-1) retrotransposons in cancer. Finally, the authors recovered the complete sequences of tumor-specific LINE-1 insertion and demonstrated their retrotransposition functions. This approach demonstrated that long-read sequencing can simultaneously survey the epigenome and detect somatic TE mobilization.
In addition, McKelvey et al. [184] applied nanopore Cas9-targeted sequencing (nCATS), to characterize allele-specific methylation in thyroid cancer cell lines heterozygous for the TERT promoter mutation. They found that the mutant TERT promoter allele was significantly less methylated than the wild type allele and that the transcriptional activators GABPA and MYC bind only to the mutant TERT allele. Importantly, epigenetic information may have direct clinical value, as demonstrated in the study by Euskirchen and colleagues [121]. A study was designed to achieve same-day detection of IDH1, IDH2, H3F3A, TP53 and TERT promoters CNVs and methylation profiles using the nanopore MinION approach. A significant correlation was observed in the outcomes of nanopore sequencing and data generated from short-read exome sequencing, Sanger sequencing, SNP array, and/or genome-wide methylation microarray. Overall, the ONT method can be applied for precision medicine development for cancer patients in setting with limited resources within a short period of time and in a cost-effective manner. Moreover, Wongsurawat et al. [185] demonstrated that nCATS can be used to identify IDH1 and IDH2 mutations and simultaneously evaluate MGMT methylation levels not only at the promoter region but also at CpGs across the proximal promoter region within 2 days of surgical resection in fresh biopsies of diffuse glioma at high resolution. The nCATS technique provides a promising tool for enhancing precision cancer medicine with the potential for simultaneously assessing multiple molecular targets.
Furthermore, epigenetic modification affects both DNA and RNA, and these base modifications can have a functional effect on transcription and translation [186]. A study showed that direct RNA sequencing could be applied to detect 6 mA RNA modifications with high accuracy in terms of systematic errors and decreased base-calling qualities [49]. Therefore, direct RNA modification analysis by nanopore sequencing is rapidly developing and improving in reliability and has the potential to provide a complete view of RNA modifications such as N6-methyladenosine (6mA), 7-methylguanosine (7mG), and 5mC. However, extracting RNA modification information from ONT reads is still an unsolved challenge, and this technology has still not reached maturity for routine application in RNA epitranscriptomics.
Identification of non-coding RNAs
Non-coding RNAs (ncRNAs), which include long non-coding RNAs, microRNAs, and circular RNAs, represent functional regulatory molecules that control the development, promotion, and metastasis of cancers. These types of ncRNAs can be captured by long-read sequencing. For instance, nanopore-induced phase-shift sequencing (NIPSS) was developed to directly sequence microRNA, which can demonstrate single molecule sequencing of miRNA, such as the discriminations between different sequences, isoforms, and epigenetic modifications among synthetic miRNA sequences [57]. Since miRNAs are potential therapeutic targets or biomarkers in many human disease, such as Parkinson’s disease [187] and cancer [188]. Direct miRNA sequencing by NIPSS may be directly implemented in clinical applications. Further, Troskie et al. [189] sequenced RNA from normal mixed adult and foetal human tissues on a PacBio platform, and defined a complex tissue-specific pseudogene transcriptome, which can be utilized as a resource for transcriptomic analyses and to design functional screens. This study sets up a foundation for the use of long-read sequencing to comprehensively identify full-length pseudogene transcripts.
Although, circRNAs can be discovered and quantified using short-read RNA-seq data via identifying the occurrence of back-splice junctions (BSJs), the ability to reconstruct circRNAs is still limited to determine the full-length sequences and internal AS events with circRNAs [190, 191]. Long-read RNA-seq is a powerful tool for resolving full-length transcript isoforms. By combining circular reverse transcription and size selection strategy along with nanopore sequencing, isoCirc [192] and circRNA identifier using long-read sequencing data (CIRI-long) [193] are two major methods to effectively characterize the full-length circRNA isoforms. However, the application of these approach is limited at the current sequencing depth, due to the low throughput and high per-sequence cost for long-read sequencing.
Liquid biopsy
Liquid biopsy is a powerful technique that can be used to identify cancer patients to improve early diagnosis and improve intervention. In the area of NGS, it has been applied to sequence cfDNA. Unfortunately, TGS platforms are not designed for cfDNA analysis. Nevertheless, Martignano and colleagues [194] modified nanopore standard protocols to make them compatible with small cfDNA fragments. They sequenced cfDNA from cancer patients and healthy subjects, and successfully obtain a CNV profile from plasma cfDNA of cancer patients. By comparing the performance of nanopore sequencing with a standard NGS approach, nanopore sequencing has the same performance of NGS approaches. In addition, it would be possible to inspect the CNV profile in less than a working day while the run is still ongoing, which is unique to nanopore sequencing. The applications of this approach exploit the full potential of liquid biopsy for both research and clinical purposes. In the context of cervical cancer, human papilloma virus (HPV) is the major cause of the disease, and HPV16 and HPV18 are the two most prevalent high-risk HPV types worldwide [195]. Nanopore sequencing was used to detect HPV integration in cervical cancer samples and cervical liquid-based cytology samples [196, 197]. This approach could potentially be utilized as a diagnostic tool for cervical cancer.
The challenges of long-read sequencing in cancer research
As demonstrated by the multitude of applications mentioned above, TGS has several important advantages to NGS. However, there still remains significant challenges to be overcome.
As mentioned above, the long-read produced suffer a high error rate, which might hamper the accuracy of genome sequencing projects. The expression of short and long transcripts varies for each sample and each sample will only include a fraction of all transcripts in the reference annotation. This length bias is rooted in the way samples are prepared for sequencing. Thus, median-read length approaches are further constrained by challenges in sample preparation and biases in library preparation. The high accuracy rate for short-reads and the longer length of long-read can be combined to achieve better accuracy. Second, in contrast with short-read sequencing which have spent the last decade creating a large number of tools for data analysis, the tools for TGS are still in development. These challenges have necessitated new algorithms for the efficient analysis of longer reads, including isoform identification, quantification, and modification detection, etc.
Third, all current TGS approaches suffer from experimental artefacts caused by degraded DNA/RNA molecules. The integrity of DNA/RNA going into TGS experiments is most important. However, it is not always possible to obtain sufficiently large intact samples of DNA and full-length RNA from clinical samples. Surgical specimens and biopsies are commonly preserved as formalin-fixed paraffin-embedded (FFPE) tissues for histopathological staining and long-term storage. DNA/RNAs from FFPE samples are highly fragmented and damaged. The rapid growth of biobanks has enabled the collection of thousands of fresh frozen tissues. However, it is not yet clear what represents the best extraction and processing method for DNA/RNA. In our studies, we extract RNA from biobanking fresh frozen samples rely on physical disruption and trizol based protocols, followed by precipitations or column-based clean-up. The integrity of RNA was damaged, and the long RNA transcripts were degraded in some samples (the size of cDNA library was less than 1 kb). Long-read RNA-seq depend on long RNA molecules being present as full-length transcripts, so these samples were excluded for further long-read sequencing. As such, we need to carefully control the quality of the samples used for RNA extraction. Forth, TGS is ultimately limited by the number of reads available for analysis. For example, to truly explore the complexity of mammalian transcriptomes, hundreds of millions of reads covering full-length transcripts will be required per tissue or organ. Up to now, PacBio and ONT sequencers routinely generate 30 million of reads per $1,000 of human RNA sequencing. The relatively high costs associated with current single-molecule sequencing will be driven down as throughput increases.
Finally, despite all applications held by long-read sequencing technologies, they still have several barriers withholding their application in clinical sequencing settings. Most of the demonstrated applications mentioned above involved research studies. It takes a very long way to go for a transition into clinical routine. For instance, the evolving TGS platforms and associated primary analysis tools must be validated and confirmed for International Organization for Standardization (ISO) and associated administrations. In addition, the clinical adaptation is also hindered by a knowledge gap between genetic counselors and bioinformatics experts.
Conclusion and future perspectives
TGS platforms, such as those provided by PacBio and ONT, are rapidly advancing the field with improved reference genomes, more comprehensive variant identification and more complete views of transcriptomes and epigenomes (Figure 2). The use of TGS to address fundamental problems in cancer research presents an encouraging outlook for its continued application in an increasing number of different clinical and research settings (Table 3). Despite all advantages held by long-read sequencing technologies, they still have some limitations, as well. In particular, they suffer from much lower throughput and much high error rates than NGS platforms. The limitation of lower throughput makes them difficult to perform differential gene expression analysis. The key advantages of the TGS platforms are the long-read length and the ability to detect base modifications in native DNA and RNA. However, their ability to capture more of full-length transcripts are additionally dependent on having high-quality RNA libraries as input. The key advantages of the NGS platforms are their high accuracy, relative low cost, and high throughput, which make them the most popular platforms in both clinical and research settings currently. Therefore, each platform offers its own advantages that provide preference for certain applications (Table 4). In the near future, with developments in sample preparation protocols, sequencing accuracy, and computational tools, TGS long-read approaches alone or integrated with NGS short-read approaches will expand low-cost diagnostic sequencing to more loci at higher accuracies and pave the way for novel clinical applications.
Table 3:
Applications of TGS in human cancers.
| Application | Sequencing technology | Targets | Cancer |
|---|---|---|---|
| Detect mutations at a low frequency | PacBio | TP53 | AML and MDS [119] |
| Detect mutations at single-allele resolution | ONT | EGFR | Lung cancer [120] |
| Detect Low-level of cancer-associated SVs | ONT | CDKN2A and SMAD4 | Pancreatic cancer [122] |
| Detect complex rearrangements | ONT | Chr 17q23 and 20q13 | Breast cancer [129] |
| Detect full-length transcript sequences | PacBio | AR-V9 and AR-V7 | Prostate cancer [141] |
| Comprehensive assessment of isoforms | ONT | BARD1 | Melanoma and others [144, 145] |
| Detect neoantigens | ONT | NDST1, SENP2 et al. | Lung cancer [146] |
| Detect fusion genes | ONT | CCDC6-RET | Lung cancer [153] |
| Detect fusion genes | PacBio | RLN1 and RLN2 | Prostate cancer [154] |
| Detect fusion genes | PacBio/ONT | BCR-ABL1 | CML [156], [157], [158] |
| Virus integrative analysis | PacBio | Cancer driver genes | HCC [168] |
| Axonomic classification of gut microbiota | PacBio/ONT | NA | Gastric cancer, CRC [174], [175], [176, 178] |
| Transposable element epigenomic profiling | ONT | TE | HCC [183] |
| Same-day detection of CNVs and methylation | ONT | Cancer driver genes | Brain cancer [121] |
| Identification of miRNA and circRNA | ONT | NA | NA |
| Liquid biopsy | ONT | NA | Lung cancer [194] |
NA, not available.
Table 4:
Applications most suited for NGS, PacBio, and/or ONT in cancer research.
| Application | Optimal technology | Reason |
|---|---|---|
| WES, WGS, and GWAS | NGS | High throughput at low cost |
| Gene panels in cancer | NGS | High throughput at low cost |
| Cancer-specific biomarkers, such as MSI, TMB | NGS | High throughput at low cost |
| Complicated cancer genomes | PacBio or ONT | Read lengths can traverse most repeat structures of the genome |
| De novo genome assembly | PacBio or ONT | Long-read enable much higher N50s |
| Haplotype phasing | PacBio or ONT | Long-read permit direct phasing |
| Gene expression | NGS | RNA-seq with low cost |
| Identification of ncRNAs | NGS | RNA-seq with low cost |
| Alternative splicing/transcripts | PacBio or ONT | Full-length RNA transcripts sequencing |
| Identification of fusion genes | PacBio or ONT | Long-read permit characterizing SVs and full-length RNA transcripts |
| Epigenetics | PacBio or ONT | Direct detection of DNA modifications |
| RNA modification detection | ONT | Direct detection of RNA modifications |
| Microbial genome sequencing | PacBio or ONT | Read lengths can span the majority of the bacterial 16S rRNA gene |
| Single-cell RNA/DNA sequencing | NGS | High throughput at low cost |
Footnotes
Author contributions: Zhiao Chen: Writing original draft. Xianghuo He: Writing, reviewing and editing, Supervision. All authors have accepted responsibility for the entire content of this manuscript and approved its submission.
Research funding: The National Natural Science Foundation of China played a role in the collection and interpretation of data, as well as in the writing of the manuscript. Our work was supported by grants from the National Natural Science Foundation of China (82172937, 81972247, 81930123 and 81790252).
Competing interests: Authors state no conflict of interest.
Informed consent: Not applicable.
Ethical approval: Not applicable.
Contributor Information
Zhiao Chen, Email: zachen@fudan.edu.cn.
Xianghuo He, Email: xhhe@fudan.edu.cn.
References
- 1.Goodwin S, McPherson JD, McCombie WR. Coming of age: ten years of next-generation sequencing technologies. Nat Rev Genet. 2016;17:333–51. doi: 10.1038/nrg.2016.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mardis ER. The impact of next-generation sequencing technology on genetics. Trends Genet. 2008;24:133–41. doi: 10.1016/j.tig.2007.12.007. [DOI] [PubMed] [Google Scholar]
- 3.Stark R, Grzelak M, Hadfield J. RNA sequencing: the teenage years. Nat Rev Genet. 2019;20:631–56. doi: 10.1038/s41576-019-0150-2. [DOI] [PubMed] [Google Scholar]
- 4.Hrdlickova R, Toloue M, Tian B. RNA-Seq methods for transcriptome analysis. Wiley Interdiscip Rev RNA. 2017;8:e1364. doi: 10.1002/wrna.1364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Saliba AE, Westermann AJ, Gorski SA, Vogel J. Single-cell RNA-seq: advances and future challenges. Nucleic Acids Res. 2014;42:8845–60. doi: 10.1093/nar/gku555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Avila M, Meric-Bernstam F. Next-generation sequencing for the general cancer patient. Clin Adv Hematol Oncol. 2019;17:447–54. [PMC free article] [PubMed] [Google Scholar]
- 7.Schadt EE, Turner S, Kasarskis A. A window into third-generation sequencing. Hum Mol Genet. 2010;19:R227–40. doi: 10.1093/hmg/ddq416. [DOI] [PubMed] [Google Scholar]
- 8.van Dijk EL, Jaszczyszyn Y, Naquin D, Thermes C. The third revolution in sequencing technology. Trends Genet. 2018;34:666–81. doi: 10.1016/j.tig.2018.05.008. [DOI] [PubMed] [Google Scholar]
- 9.Roberts RJ, Carneiro MO, Schatz MC. The advantages of SMRT sequencing. Genome Biol. 2013;14:405. doi: 10.1186/gb-2013-14-7-405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Berlin K, Koren S, Chin CS, Drake JP, Landolin JM, Phillippy AM. Assembling large genomes with single-molecule sequencing and locality-sensitive hashing. Nat Biotechnol. 2015;33:623–30. doi: 10.1038/nbt.3238. [DOI] [PubMed] [Google Scholar]
- 11.Lu H, Giordano F, Ning Z. Oxford nanopore MinION sequencing and genome assembly. Dev Reprod Biol. 2016;14:265–79. doi: 10.1016/j.gpb.2016.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rhoads A, Au KF. PacBio sequencing and its applications. Dev Reprod Biol. 2015;13:278–89. doi: 10.1016/j.gpb.2015.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ardui S, Ameur A, Vermeesch JR, Hestand MS. Single molecule real-time (SMRT) sequencing comes of age: applications and utilities for medical diagnostics. Nucleic Acids Res. 2018;46:2159–68. doi: 10.1093/nar/gky066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Logsdon GA, Vollger MR, Eichler EE. Long-read human genome sequencing and its applications. Nat Rev Genet. 2020;21:597–614. doi: 10.1038/s41576-020-0236-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Midha MK, Wu M, Chiu KP. Long-read sequencing in deciphering human genetics to a greater depth. Hum Genet. 2019;138:1201–15. doi: 10.1007/s00439-019-02064-y. [DOI] [PubMed] [Google Scholar]
- 16.Mardis ER. The impact of next-generation sequencing on cancer genomics: from discovery to clinic. Cold Spring Harb Perspect Med. 2019;9 doi: 10.1101/cshperspect.a036269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hundal J, Carreno BM, Petti AA, Linette GP, Griffith OL, Mardis ER, et al. pVAC-Seq: a genome-guided in silico approach to identifying tumor neoantigens. Genome Med. 2016;8:11. doi: 10.1186/s13073-016-0264-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen M, Zhao H. Next-generation sequencing in liquid biopsy: cancer screening and early detection. Hum Genom. 2019;13:34. doi: 10.1186/s40246-019-0220-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Marco-Puche G, Lois S, Benitez J, Trivino JC. RNA-seq perspectives to improve clinical diagnosis. Front Genet. 2019;10:1152. doi: 10.3389/fgene.2019.01152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yu Z, Li A, Wang M. CLImAT-HET: detecting subclonal copy number alterations and loss of heterozygosity in heterogeneous tumor samples from whole-genome sequencing data. BMC Med Genom. 2017;10:15. doi: 10.1186/s12920-017-0255-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kurnit KC, Bailey AM, Zeng J, Johnson AM, Shufean MA, Brusco L, et al. “Personalized cancer therapy”: a publicly available precision oncology resource. Canc Res. 2017;77:e123–e6. doi: 10.1158/0008-5472.can-17-0341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kukurba KR, Montgomery SB. RNA sequencing and analysis. Cold Spring Harb Protoc. 2015;2015:951–69. doi: 10.1101/pdb.top084970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Macaulay IC, Ponting CP, Voet T. Single-cell multiomics: multiple measurements from single cells. Trends Genet. 2017;33:155–68. doi: 10.1016/j.tig.2016.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Consortium ITP-CAoWG Pan-cancer analysis of whole genomes. Nature. 2020;578:82–93. doi: 10.1038/s41586-020-1969-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vogelstein B, Papadopoulos N, Velculescu VE, Zhou S, Diaz LA, Kinzler KW. Cancer genome landscapes. Science. 2013;339:1546–58. doi: 10.1126/science.1235122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Alexandrov LB, Kim J, Haradhvala NJ, Huang MN, Tian Ng AW, Wu Y, et al. The repertoire of mutational signatures in human cancer. Nature. 2020;578:94–101. doi: 10.1038/s41586-020-1943-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Miller TE, Yang M, Bajor D, Friedman JD, Chang RYC, Dowlati A, et al. Clinical utility of reflex testing using focused next-generation sequencing for management of patients with advanced lung adenocarcinoma. J Clin Pathol. 2018;71:1108–15. doi: 10.1136/jclinpath-2018-205396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim H, Yun JW, Lee ST, Kim HJ, Kim SH, Kim JW, et al. Korean society for genetic diagnostics guidelines for validation of next-generation sequencing-based somatic variant detection in hematologic malignancies. Ann Lab Med. 2019;39:515–23. doi: 10.3343/alm.2019.39.6.515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Surrey LF, MacFarland SP, Chang F, Cao K, Rathi KS, Akgumus GT, et al. Clinical utility of custom-designed NGS panel testing in pediatric tumors. Genome Med. 2019;11:32. doi: 10.1186/s13073-019-0644-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Coccaro N, Anelli L, Zagaria A, Specchia G, Albano F. Next-generation sequencing in acute lymphoblastic leukemia. Int J Mol Sci. 2019;20 doi: 10.3390/ijms20122929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sood R, Kamikubo Y, Liu P. Role of RUNX1 in hematological malignancies. Blood. 2017;129:2070–82. doi: 10.1182/blood-2016-10-687830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Daver N, Schlenk RF, Russell NH, Levis MJ. Targeting FLT3 mutations in AML: review of current knowledge and evidence. Leukemia. 2019;33:299–312. doi: 10.1038/s41375-018-0357-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lang GT, Jiang YZ, Shi JX, Yang F, Li XG, Pei YC, et al. Characterization of the genomic landscape and actionable mutations in Chinese breast cancers by clinical sequencing. Nat Commun. 2020;11:5679. doi: 10.1038/s41467-020-19342-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhong Y, Xu F, Wu J, Schubert J, Li MM. Application of next generation sequencing in laboratory medicine. Ann Lab Med. 2021;41:25–43. doi: 10.3343/alm.2021.41.1.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stroun M, Lyautey J, Lederrey C, Olson-Sand A, Anker P. About the possible origin and mechanism of circulating DNA apoptosis and active DNA release. Clin Chim Acta. 2001;313:139–42. doi: 10.1016/s0009-8981(01)00665-9. [DOI] [PubMed] [Google Scholar]
- 36.Chemi F, Mohan S, Guevara T, Clipson A, Rothwell DG, Dive C. Early dissemination of circulating tumor cells: biological and clinical insights. Front Oncol. 2021;11:672195. doi: 10.3389/fonc.2021.672195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fonseka P, Marzan AL, Mathivanan S. Introduction to the community of extracellular vesicles. Subcell Biochem. 2021;97:3–18. doi: 10.1007/978-3-030-67171-6_1. [DOI] [PubMed] [Google Scholar]
- 38.Spilak A, Brachner A, Kegler U, Neuhaus W, Noehammer C. Implications and pitfalls for cancer diagnostics exploiting extracellular vesicles. Adv Drug Deliv Rev. 2021;175:113819. doi: 10.1016/j.addr.2021.05.029. [DOI] [PubMed] [Google Scholar]
- 39.Zaporozhchenko IA, Ponomaryova AA, Rykova EY, Laktionov PP. The potential of circulating cell-free RNA as a cancer biomarker: challenges and opportunities. Expert Rev Mol Diagn. 2018;18:133–45. doi: 10.1080/14737159.2018.1425143. [DOI] [PubMed] [Google Scholar]
- 40.De Rubis G, Rajeev Krishnan S, Bebawy M. Liquid biopsies in cancer diagnosis, monitoring, and prognosis. Trends Pharmacol Sci. 2019;40:172–86. doi: 10.1016/j.tips.2019.01.006. [DOI] [PubMed] [Google Scholar]
- 41.Wan JCM, Massie C, Garcia-Corbacho J, Mouliere F, Brenton JD, Caldas C, et al. Liquid biopsies come of age: towards implementation of circulating tumour DNA. Nat Rev Canc. 2017;17:223–38. doi: 10.1038/nrc.2017.7. [DOI] [PubMed] [Google Scholar]
- 42.Martignano F. Cell-free DNA: an overview of sample types and isolation procedures. Methods Mol Biol. 2019;1909:13–27. doi: 10.1007/978-1-4939-8973-7_2. [DOI] [PubMed] [Google Scholar]
- 43.Petersen BS, Fredrich B, Hoeppner MP, Ellinghaus D, Franke A. Opportunities and challenges of whole-genome and -exome sequencing. BMC Genet. 2017;18:14. doi: 10.1186/s12863-017-0479-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Martin JA, Wang Z. Next-generation transcriptome assembly. Nat Rev Genet. 2011;12:671–82. doi: 10.1038/nrg3068. [DOI] [PubMed] [Google Scholar]
- 45.Bayega A, Wang YC, Oikonomopoulos S, Djambazian H, Fahiminiya S, Ragoussis J. Transcript profiling using long-read sequencing technologies. Methods Mol Biol. 2018;1783:121–47. doi: 10.1007/978-1-4939-7834-2_6. [DOI] [PubMed] [Google Scholar]
- 46.Salzberg SL, Yorke JA. Beware of mis-assembled genomes. Bioinformatics. 2005;21:4320–1. doi: 10.1093/bioinformatics/bti769. [DOI] [PubMed] [Google Scholar]
- 47.Buermans HP, den Dunnen JT. Next generation sequencing technology: advances and applications. Biochim Biophys Acta. 2014;1842:1932–41. doi: 10.1016/j.bbadis.2014.06.015. [DOI] [PubMed] [Google Scholar]
- 48.Risso D, Schwartz K, Sherlock G, Dudoit S. GC-content normalization for RNA-Seq data. BMC Bioinf. 2011;12:480. doi: 10.1186/1471-2105-12-480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liu H, Begik O, Lucas MC, Ramirez JM, Mason CE, Wiener D, et al. Accurate detection of m(6)A RNA modifications in native RNA sequences. Nat Commun. 2019;10:4079. doi: 10.1038/s41467-019-11713-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Eid J, Fehr A, Gray J, Luong K, Lyle J, Otto G, et al. Real-time DNA sequencing from single polymerase molecules. Science. 2009;323:133–8. doi: 10.1126/science.1162986. [DOI] [PubMed] [Google Scholar]
- 51.Clarke J, Wu HC, Jayasinghe L, Patel A, Reid S, Bayley H. Continuous base identification for single-molecule nanopore DNA sequencing. Nat Nanotechnol. 2009;4:265–70. doi: 10.1038/nnano.2009.12. [DOI] [PubMed] [Google Scholar]
- 52.Jain M, Olsen HE, Paten B, Akeson M. The Oxford nanopore MinION: delivery of nanopore sequencing to the genomics community. Genome Biol. 2016;17:239. doi: 10.1186/s13059-016-1103-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Patel A, Schwab R, Liu YT, Bafna V. Amplification and thrifty single-molecule sequencing of recurrent somatic structural variations. Genome Res. 2014;24:318–28. doi: 10.1101/gr.161497.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Flusberg BA, Webster DR, Lee JH, Travers KJ, Olivares EC, Clark TA, et al. Direct detection of DNA methylation during single-molecule, real-time sequencing. Nat Methods. 2010;7:461–5. doi: 10.1038/nmeth.1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Garalde DR, Snell EA, Jachimowicz D, Sipos B, Lloyd JH, Bruce M, et al. Highly parallel direct RNA sequencing on an array of nanopores. Nat Methods. 2018;15:201–6. doi: 10.1038/nmeth.4577. [DOI] [PubMed] [Google Scholar]
- 56.Workman RE, Tang AD, Tang PS, Jain M, Tyson JR, Razaghi R, et al. Nanopore native RNA sequencing of a human poly(A) transcriptome. Nat Methods. 2019;16:1297–305. doi: 10.1038/s41592-019-0617-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang J, Yan S, Chang L, Guo W, Wang Y, Wang Y, et al. Direct microRNA sequencing using nanopore-induced phase-shift sequencing. iScience. 2020;23:100916. doi: 10.1016/j.isci.2020.100916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Weirather JL, de Cesare M, Wang Y, Piazza P, Sebastiano V, Wang XJ, et al. Comprehensive comparison of pacific biosciences and Oxford nanopore technologies and their applications to transcriptome analysis. F1000Research. 2017;6:100. doi: 10.12688/f1000research.10571.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Salmela L, Walve R, Rivals E, Ukkonen E. Accurate self-correction of errors in long reads using de Bruijn graphs. Bioinformatics. 2017;33:799–806. doi: 10.1093/bioinformatics/btw321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Koren S, Schatz MC, Walenz BP, Martin J, Howard JT, Ganapathy G, et al. Hybrid error correction and de novo assembly of single-molecule sequencing reads. Nat Biotechnol. 2012;30:693–700. doi: 10.1038/nbt.2280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ritz A, Bashir A, Sindi S, Hsu D, Hajirasouliha I, Raphael BJ. Characterization of structural variants with single molecule and hybrid sequencing approaches. Bioinformatics. 2014;30:3458–66. doi: 10.1093/bioinformatics/btu714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Au KF, Sebastiano V, Afshar PT, Durruthy JD, Lee L, Williams BA, et al. Characterization of the human ESC transcriptome by hybrid sequencing. Proc Natl Acad Sci USA. 2013;110:E4821–30. doi: 10.1073/pnas.1320101110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Quail MA, Smith M, Coupland P, Otto TD, Harris SR, Connor TR, et al. A tale of three next generation sequencing platforms: comparison of Ion Torrent, Pacific Biosciences and Illumina MiSeq sequencers. BMC Genom. 2012;13:341. doi: 10.1186/1471-2164-13-341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Jain M, Koren S, Miga KH, Quick J, Rand AC, Sasani TA, et al. Nanopore sequencing and assembly of a human genome with ultra-long reads. Nat Biotechnol. 2018;36:338–45. doi: 10.1038/nbt.4060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Carneiro MO, Russ C, Ross MG, Gabriel SB, Nusbaum C, DePristo MA. Pacific biosciences sequencing technology for genotyping and variation discovery in human data. BMC Genom. 2012;13:375. doi: 10.1186/1471-2164-13-375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Jain M, Fiddes IT, Miga KH, Olsen HE, Paten B, Akeson M. Improved data analysis for the MinION nanopore sequencer. Nat Methods. 2015;12:351–6. doi: 10.1038/nmeth.3290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ashton PM, Nair S, Dallman T, Rubino S, Rabsch W, Mwaigwisya S, et al. MinION nanopore sequencing identifies the position and structure of a bacterial antibiotic resistance island. Nat Biotechnol. 2015;33:296–300. doi: 10.1038/nbt.3103. [DOI] [PubMed] [Google Scholar]
- 68.Sedlazeck FJ, Lee H, Darby CA, Schatz MC. Piercing the dark matter: bioinformatics of long-range sequencing and mapping. Nat Rev Genet. 2018;19:329–46. doi: 10.1038/s41576-018-0003-4. [DOI] [PubMed] [Google Scholar]
- 69.Chu J, Mohamadi H, Warren RL, Yang C, Birol I. Innovations and challenges in detecting long read overlaps: an evaluation of the state-of-the-art. Bioinformatics. 2017;33:1261–70. doi: 10.1093/bioinformatics/btw811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Amarasinghe SL, Su S, Dong X, Zappia L, Ritchie ME, Gouil Q. Opportunities and challenges in long-read sequencing data analysis. Genome Biol. 2020;21:30. doi: 10.1186/s13059-020-1935-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Pacific Biosciences . [1 June 2021]. https://github.com/PacificBiosciences Accessed.
- 72.Wick RR, Judd LM, Holt KE. Performance of neural network basecalling tools for Oxford Nanopore sequencing. Genome Biol. 2019;20:129. doi: 10.1186/s13059-019-1727-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Pacific Biosciences . [1 June 2021]. https://www.pacb.com/ Accessed.
- 74.Wenger AM, Peluso P, Rowell WJ, Chang PC, Hall RJ, Concepcion GT, et al. Accurate circular consensus long-read sequencing improves variant detection and assembly of a human genome. Nat Biotechnol. 2019;37:1155–62. doi: 10.1038/s41587-019-0217-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zhang H, Jain C, Aluru S. A comprehensive evaluation of long read error correction methods. BMC Genom. 2020;21:889. doi: 10.1186/s12864-020-07227-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Fu S, Wang A, Au KF. A comparative evaluation of hybrid error correction methods for error-prone long reads. Genome Biol. 2019;20:26. doi: 10.1186/s13059-018-1605-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lima L, Marchet C, Caboche S, Da Silva C, Istace B, Aury JM, et al. Comparative assessment of long-read error correction software applied to nanopore RNA-sequencing data. Brief Bioinf. 2020;21:1164–81. doi: 10.1093/bib/bbz058. [DOI] [PubMed] [Google Scholar]
- 78.Alkan C, Sajjadian S, Eichler EE. Limitations of next-generation genome sequence assembly. Nat Methods. 2011;8:61–5. doi: 10.1038/nmeth.1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Mahmoud M, Gobet N, Cruz-Davalos DI, Mounier N, Dessimoz C, Sedlazeck FJ. Structural variant calling: the long and the short of it. Genome Biol. 2019;20:246. doi: 10.1186/s13059-019-1828-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ho SS, Urban AE, Mills RE. Structural variation in the sequencing era. Nat Rev Genet. 2020;21:171–89. doi: 10.1038/s41576-019-0180-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Stephens Z, Wang C, Iyer RK, Kocher JP. Detection and visualization of complex structural variants from long reads. BMC Bioinf. 2018;19:508. doi: 10.1186/s12859-018-2539-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.English AC, Salerno WJ, Reid JG. PBHoney: identifying genomic variants via long-read discordance and interrupted mapping. BMC Bioinf. 2014;15:180. doi: 10.1186/1471-2105-15-180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Sedlazeck FJ, Rescheneder P, Smolka M, Fang H, Nattestad M, von Haeseler A, et al. Accurate detection of complex structural variations using single-molecule sequencing. Nat Methods. 2018;15:461–8. doi: 10.1038/s41592-018-0001-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Huddleston J, Chaisson MJP, Steinberg KM, Warren W, Hoekzema K, Gordon D, et al. Discovery and genotyping of structural variation from long-read haploid genome sequence data. Genome Res. 2017;27:677–85. doi: 10.1101/gr.214007.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Heller D, Vingron M. SVIM-asm: structural variant detection from haploid and diploid genome assemblies. Bioinformatics. 2020 doi: 10.1093/bioinformatics/btaa1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Cretu Stancu M, van Roosmalen MJ, Renkens I, Nieboer MM, Middelkamp S, de Ligt J, et al. Mapping and phasing of structural variation in patient genomes using nanopore sequencing. Nat Commun. 2017;8:1326. doi: 10.1038/s41467-017-01343-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Gong L, Wong CH, Cheng WC, Tjong H, Menghi F, Ngan CY, et al. Picky comprehensively detects high-resolution structural variants in nanopore long reads. Nat Methods. 2018;15:455–60. doi: 10.1038/s41592-018-0002-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Sahlin K, Tomaszkiewicz M, Makova KD, Medvedev P. Deciphering highly similar multigene family transcripts from Iso-Seq data with IsoCon. Nat Commun. 2018;9:4601. doi: 10.1038/s41467-018-06910-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Tardaguila M, de la Fuente L, Marti C, Pereira C, Pardo-Palacios FJ, Del Risco H, et al. SQANTI: extensive characterization of long-read transcript sequences for quality control in full-length transcriptome identification and quantification. Genome Res. 2018;28:396–411. doi: 10.1101/gr.222976.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Tang AD, Soulette CM, van Baren MJ, Hart K, Hrabeta-Robinson E, Wu CJ, et al. Full-length transcript characterization of SF3B1 mutation in chronic lymphocytic leukemia reveals downregulation of retained introns. Nat Commun. 2020;11:1438. doi: 10.1038/s41467-020-15171-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Patro R, Duggal G, Love MI, Irizarry RA, Kingsford C. Salmon provides fast and bias-aware quantification of transcript expression. Nat Methods. 2017;14:417–9. doi: 10.1038/nmeth.4197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Oxford Nanopore Technologies Oxford nanopore technologies GitHub. . [1 June 2021]. https://github.com/nanoporetech Accessed.
- 93.Liao Y, Smyth GK, Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30:923–30. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- 94.Parker MT, Knop K, Sherwood AV, Schurch NJ, Mackinnon K, Gould PD, et al. Nanopore direct RNA sequencing maps the complexity of Arabidopsis mRNA processing and m(6)A modification. Elife. 2020;9:e49658. doi: 10.7554/eLife.49658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Soneson C, Yao Y, Bratus-Neuenschwander A, Patrignani A, Robinson MD, Hussain S. A comprehensive examination of nanopore native RNA sequencing for characterization of complex transcriptomes. Nat Commun. 2019;10:3359. doi: 10.1038/s41467-019-11272-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Zhang S, Li R, Zhang L, Chen S, Xie M, Yang L, et al. New insights into Arabidopsis transcriptome complexity revealed by direct sequencing of native RNAs. Nucleic Acids Res. 2020;48:7700–11. doi: 10.1093/nar/gkaa588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Abdel-Ghany SE, Hamilton M, Jacobi JL, Ngam P, Devitt N, Schilkey F, et al. A survey of the sorghum transcriptome using single-molecule long reads. Nat Commun. 2016;7:11706. doi: 10.1038/ncomms11706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Gao Y, Wang H, Zhang H, Wang Y, Chen J, Gu L. PRAPI: post-transcriptional regulation analysis pipeline for Iso-Seq. Bioinformatics. 2018;34:1580–2. doi: 10.1093/bioinformatics/btx830. [DOI] [PubMed] [Google Scholar]
- 99.Wang T, Wang H, Cai D, Gao Y, Zhang H, Wang Y, et al. Comprehensive profiling of rhizome-associated alternative splicing and alternative polyadenylation in moso bamboo (Phyllostachys edulis) Plant J. 2017;91:684–99. doi: 10.1111/tpj.13597. [DOI] [PubMed] [Google Scholar]
- 100.Liu X, Li X, Wen X, Zhang Y, Ding Y, Zhang Y, et al. PacBio full-length transcriptome of wild apple (Malus sieversii) provides insights into canker disease dynamic response. BMC Genom. 2021;22:52. doi: 10.1186/s12864-021-07366-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Tan C, Liu H, Ren J, Ye X, Feng H, Liu Z. Single-molecule real-time sequencing facilitates the analysis of transcripts and splice isoforms of anthers in Chinese cabbage (Brassica rapa L. ssp. pekinensis) BMC Plant Biol. 2019;19:517. doi: 10.1186/s12870-019-2133-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Feng S, Xu M, Liu F, Cui C, Zhou B. Reconstruction of the full-length transcriptome atlas using PacBio Iso-Seq provides insight into the alternative splicing in Gossypium australe. BMC Plant Biol. 2019;19:365. doi: 10.1186/s12870-019-1968-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Wang L, Jiang X, Wang L, Wang W, Fu C, Yan X, et al. A survey of transcriptome complexity using PacBio single-molecule real-time analysis combined with Illumina RNA sequencing for a better understanding of ricinoleic acid biosynthesis in Ricinus communis. BMC Genom. 2019;20:456. doi: 10.1186/s12864-019-5832-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Chen T, Sun Q, Ma Y, Zeng W, Liu R, Qu D, et al. A transcriptome atlas of silkworm silk glands revealed by PacBio single-molecule long-read sequencing. Mol Genet Genom. 2020;295:1227–37. doi: 10.1007/s00438-020-01691-9. [DOI] [PubMed] [Google Scholar]
- 105.Tian Y, Wen H, Qi X, Zhang X, Liu S, Li B, et al. Characterization of full-length transcriptome sequences and splice variants of Lateolabrax maculatus by single-molecule long-read sequencing and their involvement in salinity regulation. Front Genet. 2019;10:1126. doi: 10.3389/fgene.2019.01126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Chao Y, Yuan J, Li S, Jia S, Han L, Xu L. Analysis of transcripts and splice isoforms in red clover (Trifolium pratense L.) by single-molecule long-read sequencing. BMC Plant Biol. 2018;18:300. doi: 10.1186/s12870-018-1534-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Hou C, Lian H, Cai Y, Wang Y, Liang D, He B. Comparative analyses of full-length transcriptomes reveal Gnetum luofuense stem developmental dynamics. Front Genet. 2021;12:615284. doi: 10.3389/fgene.2021.615284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Xie L, Teng K, Tan P, Chao Y, Li Y, Guo W, et al. PacBio single-molecule long-read sequencing shed new light on the transcripts and splice isoforms of the perennial ryegrass. Mol Genet Genom. 2020;295:475–89. doi: 10.1007/s00438-019-01635-y. [DOI] [PubMed] [Google Scholar]
- 109.Pacific Biosciences Detecting DNA base modifications using single molecule, real-time sequencing. . [1 June 2021]. https://www.pacb.com/wp-content/uploads/2015/09/WP_Detecting_DNA_Base_Modifications_Using_SMRT_Sequencing.pdf Accessed.
- 110.Simpson JT, Workman RE, Zuzarte PC, David M, Dursi LJ, Timp W. Detecting DNA cytosine methylation using nanopore sequencing. Nat Methods. 2017;14:407–10. doi: 10.1038/nmeth.4184. [DOI] [PubMed] [Google Scholar]
- 111.Liu Q, Georgieva DC, Egli D, Wang K. NanoMod: a computational tool to detect DNA modifications using nanopore long-read sequencing data. BMC Genom. 2019;20:78. doi: 10.1186/s12864-018-5372-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Marcus Stoiber JQ, Egan R, Lee JE, Celniker S, Robert K, Neely NL, et al. De novo identification of DNA modifications enabled by genome-guided nanopore signal processing. bioRxiv. 2017 [Google Scholar]
- 113.Rand AC, Jain M, Eizenga JM, Musselman-Brown A, Olsen HE, Akeson M, et al. Mapping DNA methylation with high-throughput nanopore sequencing. Nat Methods. 2017;14:411–3. doi: 10.1038/nmeth.4189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Muller CA, Boemo MA, Spingardi P, Kessler BM, Kriaucionis S, Simpson JT, et al. Capturing the dynamics of genome replication on individual ultra-long nanopore sequence reads. Nat Methods. 2019;16:429–36. doi: 10.1038/s41592-019-0394-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Ni P, Huang N, Zhang Z, Wang DP, Liang F, Miao Y, et al. DeepSignal: detecting DNA methylation state from nanopore sequencing reads using deep-learning. Bioinformatics. 2019;35:4586–95. doi: 10.1093/bioinformatics/btz276. [DOI] [PubMed] [Google Scholar]
- 116.McIntyre ABR, Alexander N, Grigorev K, Bezdan D, Sichtig H, Chiu CY, et al. Single-molecule sequencing detection of N6-methyladenine in microbial reference materials. Nat Commun. 2019;10:579. doi: 10.1038/s41467-019-08289-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Liu Q, Fang L, Yu G, Wang D, Xiao CL, Wang K. Detection of DNA base modifications by deep recurrent neural network on Oxford nanopore sequencing data. Nat Commun. 2019;10:2449. doi: 10.1038/s41467-019-10168-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Schmidt MHM, Pearson CE. Disease-associated repeat instability and mismatch repair. DNA Repair (Amst) 2016;38:117–26. doi: 10.1016/j.dnarep.2015.11.008. [DOI] [PubMed] [Google Scholar]
- 119.Lode L, Ameur A, Coste T, Menard A, Richebourg S, Gaillard JB, et al. Single-molecule DNA sequencing of acute myeloid leukemia and myelodysplastic syndromes with multiple TP53 alterations. Haematologica. 2018;103:e13–6. doi: 10.3324/haematol.2017.176719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Suzuki A, Suzuki M, Mizushima-Sugano J, Frith MC, Makalowski W, Kohno T, et al. Sequencing and phasing cancer mutations in lung cancers using a long-read portable sequencer. DNA Res. 2017;24:585–96. doi: 10.1093/dnares/dsx027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Euskirchen P, Bielle F, Labreche K, Kloosterman WP, Rosenberg S, Daniau M, et al. Same-day genomic and epigenomic diagnosis of brain tumors using real-time nanopore sequencing. Acta Neuropathol. 2017;134:691–703. doi: 10.1007/s00401-017-1743-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Norris AL, Workman RE, Fan Y, Eshleman JR, Timp W. Nanopore sequencing detects structural variants in cancer. Canc Biol Ther. 2016;17:246–53. doi: 10.1080/15384047.2016.1139236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Nattestad M, Goodwin S, Ng K, Baslan T, Sedlazeck FJ, Rescheneder P, et al. Complex rearrangements and oncogene amplifications revealed by long-read DNA and RNA sequencing of a breast cancer cell line. Genome Res. 2018;28:1126–35. doi: 10.1101/gr.231100.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Thibodeau ML, O’Neill K, Dixon K, Reisle C, Mungall KL, Krzywinski M, et al. Improved structural variant interpretation for hereditary cancer susceptibility using long-read sequencing. Genet Med. 2020;22:1892–7. doi: 10.1038/s41436-020-0880-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Davis CF, Ricketts CJ, Wang M, Yang L, Cherniack AD, Shen H, et al. The somatic genomic landscape of chromophobe renal cell carcinoma. Canc Cell. 2014;26:319–30. doi: 10.1016/j.ccr.2014.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Williams MS, Basma NJ, Amaral FMR, Williams G, Weightman JP, Breitwieser W, et al. Targeted nanopore sequencing for the identification of ABCB1 promoter translocations in cancer. BMC Canc. 2020;20:1075. doi: 10.1186/s12885-020-07571-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Sakamoto Y, Xu L, Seki M, Yokoyama TT, Kasahara M, Kashima Y, et al. Long-read sequencing for non-small-cell lung cancer genomes. Genome Res. 2020;30:1243–57. doi: 10.1101/gr.261941.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Aganezov S, Goodwin S, Sherman RM, Sedlazeck FJ, Arun G, Bhatia S, et al. Comprehensive analysis of structural variants in breast cancer genomes using single-molecule sequencing. Genome Res. 2020;30:1258–73. doi: 10.1101/gr.260497.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Lin CL, Tan X, Chen M, Kusi M, Hung CN, Chou CW, et al. ERalpha-related chromothripsis enhances concordant gene transcription on chromosome 17q11.1–q24.1 in luminal breast cancer. BMC Med Genom. 2020;13:69. doi: 10.1186/s12920-020-0729-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 131.Wang ET, Sandberg R, Luo S, Khrebtukova I, Zhang L, Mayr C, et al. Alternative isoform regulation in human tissue transcriptomes. Nature. 2008;456:470–6. doi: 10.1038/nature07509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.de Jong LC, Cree S, Lattimore V, Wiggins GAR, Spurdle AB, kConFab I, et al. Nanopore sequencing of full-length BRCA1 mRNA transcripts reveals co-occurrence of known exon skipping events. Breast Canc Res. 2017;19:127. doi: 10.1186/s13058-017-0919-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Singh M, Al-Eryani G, Carswell S, Ferguson JM, Blackburn J, Barton K, et al. High-throughput targeted long-read single cell sequencing reveals the clonal and transcriptional landscape of lymphocytes. Nat Commun. 2019;10:3120. doi: 10.1038/s41467-019-11049-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Oguchi Y, Ozaki Y, Abdelmoez MN, Shintaku H. NanoSINC-seq dissects the isoform diversity in subcellular compartments of single cells. Sci Adv. 2021;7:eabe0317. doi: 10.1126/sciadv.abe0317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Fan X, Tang D, Liao Y, Li P, Zhang Y, Wang M, et al. Single-cell RNA-seq analysis of mouse preimplantation embryos by third-generation sequencing. PLoS Biol. 2020;18:e3001017. doi: 10.1371/journal.pbio.3001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Volden R, Palmer T, Byrne A, Cole C, Schmitz RJ, Green RE, et al. Improving nanopore read accuracy with the R2C2 method enables the sequencing of highly multiplexed full-length single-cell cDNA. Proc Natl Acad Sci USA. 2018;115:9726–31. doi: 10.1073/pnas.1806447115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Byrne A, Beaudin AE, Olsen HE, Jain M, Cole C, Palmer T, et al. Nanopore long-read RNAseq reveals widespread transcriptional variation among the surface receptors of individual B cells. Nat Commun. 2017;8:16027. doi: 10.1038/ncomms16027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Oltean S, Bates DO. Hallmarks of alternative splicing in cancer. Oncogene. 2014;33:5311–8. doi: 10.1038/onc.2013.533. [DOI] [PubMed] [Google Scholar]
- 139.Witte KE, Hertel O, Windmoller BA, Helweg LP, Hoving AL, Knabbe C, et al. Nanopore sequencing reveals global transcriptome signatures of mitochondrial and ribosomal gene expressions in various human cancer stem-like cell populations. Cancers (Basel) 2021;13:1136. doi: 10.3390/cancers13051136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Whongsiri P, Goering W, Lautwein T, Hader C, Niegisch G, Kohrer K, et al. Many different LINE-1 retroelements are activated in bladder cancer. Int J Mol Sci. 2020;21 doi: 10.3390/ijms21249433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Kohli M, Ho Y, Hillman DW, Van Etten JL, Henzler C, Yang R, et al. Androgen receptor variant AR-V9 is coexpressed with AR-V7 in prostate cancer metastases and predicts abiraterone resistance. Clin Canc Res. 2017;23:4704–15. doi: 10.1158/1078-0432.ccr-17-0017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Li Y, Yang R, Henzler CM, Ho Y, Passow C, Auch B, et al. Diverse AR gene rearrangements mediate resistance to androgen receptor inhibitors in metastatic prostate cancer. Clin Canc Res. 2020;26:1965–76. doi: 10.1158/1078-0432.ccr-19-3023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Chen H, Gao F, He M, Ding XF, Wong AM, Sze SC, et al. Long-read RNA sequencing identifies alternative splice variants in hepatocellular carcinoma and tumor-specific isoforms. Hepatology. 2019;70:1011–25. doi: 10.1002/hep.30500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Walker LC, Lattimore VL, Kvist A, Kleiblova P, Zemankova P, de Jong L, et al. Comprehensive assessment of BARD1 messenger ribonucleic acid splicing with implications for variant classification. Front Genet. 2019;10:1139. doi: 10.3389/fgene.2019.01139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.McDougall LI, Powell RM, Ratajska M, Lynch-Sutherland CF, Hossain SM, Wiggins GAR, et al. Differential expression of BARD1 isoforms in melanoma. Genes (Basel) 2021;12 doi: 10.3390/genes12020320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Oka M, Xu L, Suzuki T, Yoshikawa T, Sakamoto H, Uemura H, et al. Aberrant splicing isoforms detected by full-length transcriptome sequencing as transcripts of potential neoantigens in non-small cell lung cancer. Genome Biol. 2021;22:9. doi: 10.1186/s13059-020-02240-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Hu Y, Shu XS, Yu J, Sun MA, Chen Z, Liu X, et al. Improving the diversity of captured full-length isoforms using a normalized single-molecule RNA-sequencing method. Commun Biol. 2020;3:403. doi: 10.1038/s42003-020-01125-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Li H, Wang J, Mor G, Sklar J. A neoplastic gene fusion mimics trans-splicing of RNAs in normal human cells. Science. 2008;321:1357–61. doi: 10.1126/science.1156725. [DOI] [PubMed] [Google Scholar]
- 149.Mertens F, Johansson B, Fioretos T, Mitelman F. The emerging complexity of gene fusions in cancer. Nat Rev Canc. 2015;15:371–81. doi: 10.1038/nrc3947. [DOI] [PubMed] [Google Scholar]
- 150.Yang W, Lee KW, Srivastava RM, Kuo F, Krishna C, Chowell D, et al. Immunogenic neoantigens derived from gene fusions stimulate T cell responses. Nat Med. 2019;25:767–75. doi: 10.1038/s41591-019-0434-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Weirather JL, Afshar PT, Clark TA, Tseng E, Powers LS, Underwood JG, et al. Characterization of fusion genes and the significantly expressed fusion isoforms in breast cancer by hybrid sequencing. Nucleic Acids Res. 2015;43:e116. doi: 10.1093/nar/gkv562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Kleinman CL, Gerges N, Papillon-Cavanagh S, Sin-Chan P, Pramatarova A, Quang DA, et al. Fusion of TTYH1 with the C19MC microRNA cluster drives expression of a brain-specific DNMT3B isoform in the embryonal brain tumor ETMR. Nat Genet. 2014;46:39–44. doi: 10.1038/ng.2849. [DOI] [PubMed] [Google Scholar]
- 153.Seki M, Katsumata E, Suzuki A, Sereewattanawoot S, Sakamoto Y, Mizushima-Sugano J, et al. Evaluation and application of RNA-seq by MinION. DNA Res. 2019;26:55–65. doi: 10.1093/dnares/dsy038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Tevz G, McGrath S, Demeter R, Magrini V, Jeet V, Rockstroh A, et al. Identification of a novel fusion transcript between human relaxin-1 (RLN1) and human relaxin-2 (RLN2) in prostate cancer. Mol Cell Endocrinol. 2016;420:159–68. doi: 10.1016/j.mce.2015.10.011. [DOI] [PubMed] [Google Scholar]
- 155.Ren R. Mechanisms of BCR-ABL in the pathogenesis of chronic myelogenous leukaemia. Nat Rev Canc. 2005;5:172–83. doi: 10.1038/nrc1567. [DOI] [PubMed] [Google Scholar]
- 156.Cavelier L, Ameur A, Haggqvist S, Hoijer I, Cahill N, Olsson-Stromberg U, et al. Clonal distribution of BCR-ABL1 mutations and splice isoforms by single-molecule long-read RNA sequencing. BMC Canc. 2015;15:45. doi: 10.1186/s12885-015-1046-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Zhao H, Chen Y, Shen C, Li L, Li Q, Tan K, et al. Breakpoint mapping of a t(9;22;12) chronic myeloid leukaemia patient with e14a3 BCR-ABL1 transcript using nanopore sequencing. J Gene Med. 2021;23:e3276. doi: 10.1002/jgm.3276. [DOI] [PubMed] [Google Scholar]
- 158.Minervini CF, Cumbo C, Orsini P, Anelli L, Zagaria A, Impera L, et al. Mutational analysis in BCR-ABL1 positive leukemia by deep sequencing based on nanopore MinION technology. Exp Mol Pathol. 2017;103:33–7. doi: 10.1016/j.yexmp.2017.06.007. [DOI] [PubMed] [Google Scholar]
- 159.Jeck WR, Lee J, Robinson H, Le LP, Iafrate AJ, Nardi V. A nanopore sequencing-based assay for rapid detection of gene fusions. J Mol Diagn. 2019;21:58–69. doi: 10.1016/j.jmoldx.2018.08.003. [DOI] [PubMed] [Google Scholar]
- 160.Smith CC, Wang Q, Chin CS, Salerno S, Damon LE, Levis MJ, et al. Validation of ITD mutations in FLT3 as a therapeutic target in human acute myeloid leukaemia. Nature. 2012;485:260–3. doi: 10.1038/nature11016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Smith CC, Zhang C, Lin KC, Lasater EA, Zhang Y, Massi E, et al. Characterizing and overriding the structural mechanism of the quizartinib-resistant FLT3 “gatekeeper” F691L mutation with PLX3397. Canc Discov. 2015;5:668–79. doi: 10.1158/2159-8290.cd-15-0060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Stangl C, de Blank S, Renkens I, Westera L, Verbeek T, Valle-Inclan JE, et al. Partner independent fusion gene detection by multiplexed CRISPR-Cas9 enrichment and long read nanopore sequencing. Nat Commun. 2020;11:2861. doi: 10.1038/s41467-020-16641-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.de Martel C, Ferlay J, Franceschi S, Vignat J, Bray F, Forman D, et al. Global burden of cancers attributable to infections in 2008: a review and synthetic analysis. Lancet Oncol. 2012;13:607–15. doi: 10.1016/s1470-2045(12)70137-7. [DOI] [PubMed] [Google Scholar]
- 164.Garrett WS. Cancer and the microbiota. Science. 2015;348:80–6. doi: 10.1126/science.aaa4972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Kilianski A, Haas JL, Corriveau EJ, Liem AT, Willis KL, Kadavy DR, et al. Bacterial and viral identification and differentiation by amplicon sequencing on the MinION nanopore sequencer. GigaScience. 2015;4:12. doi: 10.1186/s13742-015-0051-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Villanueva A. Hepatocellular carcinoma. N Engl J Med. 2019;380:1450–62. doi: 10.1056/nejmra1713263. [DOI] [PubMed] [Google Scholar]
- 167.Levrero M, Zucman-Rossi J. Mechanisms of HBV-induced hepatocellular carcinoma. J Hepatol. 2016;64:S84–101. doi: 10.1016/j.jhep.2016.02.021. [DOI] [PubMed] [Google Scholar]
- 168.Peneau C, Imbeaud S, La Bella T, Hirsch TZ, Caruso S, Calderaro J, et al. Hepatitis B virus integrations promote local and distant oncogenic driver alterations in hepatocellular carcinoma. Gut. 2021;Online ahead of print doi: 10.1136/gutjnl-2020-323153. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Tatkiewicz W, Dickie J, Bedford F, Jones A, Atkin M, Kiernan M, et al. Characterising a human endogenous retrovirus(HERV)-derived tumour-associated antigen: enriched RNA-Seq analysis of HERV-K(HML-2) in mantle cell lymphoma cell lines. Mobile DNA. 2020;11:9. doi: 10.1186/s13100-020-0204-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Peled JU, Devlin SM, Staffas A, Lumish M, Khanin R, Littmann ER, et al. Intestinal microbiota and relapse after hematopoietic-cell transplantation. J Clin Oncol. 2017;35:1650–9. doi: 10.1200/JCO.2016.70.3348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Routy B, Le Chatelier E, Derosa L, Duong CPM, Alou MT, Daillere R, et al. Gut microbiome influences efficacy of PD-1-based immunotherapy against epithelial tumors. Science. 2018;359:91–7. doi: 10.1126/science.aan3706. [DOI] [PubMed] [Google Scholar]
- 172.Gopalakrishnan V, Spencer CN, Nezi L, Reuben A, Andrews MC, Karpinets TV, et al. Gut microbiome modulates response to anti-PD-1 immunotherapy in melanoma patients. Science. 2018;359:97–103. doi: 10.1126/science.aan4236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Heikema AP, Horst-Kreft D, Boers SA, Jansen R, Hiltemann SD, de Koning W, et al. Comparison of Illumina versus nanopore 16S rRNA gene sequencing of the human nasal microbiota. Genes (Basel) 2020;11 doi: 10.3390/genes11091105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Bik EM, Eckburg PB, Gill SR, Nelson KE, Purdom EA, Francois F, et al. Molecular analysis of the bacterial microbiota in the human stomach. Proc Natl Acad Sci USA. 2006;103:732–7. doi: 10.1073/pnas.0506655103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Devi TB, Devadas K, George M, Gandhimathi A, Chouhan D, Retnakumar RJ, et al. Low Bifidobacterium abundance in the lower gut microbiota is associated with Helicobacter pylori-related gastric ulcer and gastric cancer. Front Microbiol. 2021;12:631140. doi: 10.3389/fmicb.2021.631140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Tetz G, Vecherkovskaya M, Zappile P, Dolgalev I, Tsirigos A, Heguy A, et al. Complete genome sequence of Kluyvera intestini sp. nov., isolated from the stomach of a patient with gastric cancer. Genome Announc. 2017;5:e01184–17. doi: 10.1128/genomeA.01184-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Gaiser RA, Halimi A, Alkharaan H, Lu L, Davanian H, Healy K, et al. Enrichment of oral microbiota in early cystic precursors to invasive pancreatic cancer. Gut. 2019;68:2186–94. doi: 10.1136/gutjnl-2018-317458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Sobhani I, Bergsten E, Couffin S, Amiot A, Nebbad B, Barau C, et al. Colorectal cancer-associated microbiota contributes to oncogenic epigenetic signatures. Proc Natl Acad Sci USA. 2019;116:24285–95. doi: 10.1073/pnas.1912129116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Wei PL, Hung CS, Kao YW, Lin YC, Lee CY, Chang TH, et al. Characterization of fecal microbiota with clinical specimen using long-read and short-read sequencing platform. Int J Mol Sci. 2020:21. doi: 10.3390/ijms21197110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Wei PL, Hung CS, Kao YW, Lin YC, Lee CY, Chang TH, et al. Classification of changes in the fecal microbiota associated with colonic adenomatous polyps using a long-read sequencing platform. Genes (Basel) 2020;11:1374. doi: 10.3390/genes11111374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Baylin SB, Herman JG. DNA hypermethylation in tumorigenesis: epigenetics joins genetics. Trends Genet. 2000;16:168–74. doi: 10.1016/s0168-9525(99)01971-x. [DOI] [PubMed] [Google Scholar]
- 182.Yang Y, Sebra R, Pullman BS, Qiao W, Peter I, Desnick RJ, et al. Quantitative and multiplexed DNA methylation analysis using long-read single-molecule real-time bisulfite sequencing (SMRT-BS) BMC Genom. 2015;16:350. doi: 10.1186/s12864-015-1572-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Ewing AD, Smits N, Sanchez-Luque FJ, Faivre J, Brennan PM, Richardson SR, et al. Nanopore sequencing enables comprehensive transposable element epigenomic profiling. Mol Cell. 2020;80:915–28 e5. doi: 10.1016/j.molcel.2020.10.024. [DOI] [PubMed] [Google Scholar]
- 184.McKelvey BA, Gilpatrick T, Wang Y, Timp W, Umbricht CB, Zeiger MA. Characterization of allele-specific regulation of telomerase reverse transcriptase in promoter mutant thyroid cancer cell lines. Thyroid. 2020;30:1470–81. doi: 10.1089/thy.2020.0055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Wongsurawat T, Jenjaroenpun P, De Loose A, Alkam D, Ussery DW, Nookaew I, et al. A novel Cas9-targeted long-read assay for simultaneous detection of IDH1/2 mutations and clinically relevant MGMT methylation in fresh biopsies of diffuse glioma. Acta Neuropathol Commun. 2020;8:87. doi: 10.1186/s40478-020-00963-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Gilbert WV, Bell TA, Schaening C. Messenger RNA modifications: form, distribution, and function. Science. 2016;352:1408–12. doi: 10.1126/science.aad8711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Wang H. MicroRNAs, Parkinson’s disease, and diabetes mellitus. Int J Mol Sci. 2021;22:2953. doi: 10.3390/ijms22062953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Sempere LF, Azmi AS, Moore A. microRNA-based diagnostic and therapeutic applications in cancer medicine. Wiley Interdiscip Rev RNA. 2021;Online:e1662. doi: 10.1002/wrna.1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Troskie RL, Jafrani Y, Mercer TR, Ewing AD, Faulkner GJ, Cheetham SW. Long-read cDNA sequencing identifies functional pseudogenes in the human transcriptome. Genome Biol. 2021;22:146. doi: 10.1186/s13059-021-02369-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Szabo L, Salzman J. Detecting circular RNAs: bioinformatic and experimental challenges. Nat Rev Genet. 2016;17:679–92. doi: 10.1038/nrg.2016.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Gao Y, Zhao F. Computational strategies for exploring circular RNAs. Trends Genet. 2018;34:389–400. doi: 10.1016/j.tig.2017.12.016. [DOI] [PubMed] [Google Scholar]
- 192.Xin R, Gao Y, Gao Y, Wang R, Kadash-Edmondson KE, Liu B, et al. isoCirc catalogs full-length circular RNA isoforms in human transcriptomes. Nat Commun. 2021;12:266. doi: 10.1038/s41467-020-20459-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Zhang J, Hou L, Zuo Z, Ji P, Zhang X, Xue Y, et al. Comprehensive profiling of circular RNAs with nanopore sequencing and CIRI-long. Nat Biotechnol. 2021;39:836–45. doi: 10.1038/s41587-021-00842-6. [DOI] [PubMed] [Google Scholar]
- 194.Martignano F, Munagala U, Crucitta S, Mingrino A, Semeraro R, Del Re M, et al. Nanopore sequencing from liquid biopsy: analysis of copy number variations from cell-free DNA of lung cancer patients. Mol Canc. 2021;20:32. doi: 10.1186/s12943-021-01327-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Wardak S. Human Papillomavirus (HPV) and cervical cancer. Med Dosw Mikrobiol. 2016;68:73–84. [PubMed] [Google Scholar]
- 196.Quan L, Dong R, Yang W, Chen L, Lang J, Liu J, et al. Simultaneous detection and comprehensive analysis of HPV and microbiome status of a cervical liquid-based cytology sample using Nanopore MinION sequencing. Sci Rep. 2019;9:19337. doi: 10.1038/s41598-019-55843-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Yang W, Liu Y, Dong R, Liu J, Lang J, Yang J, et al. Accurate detection of HPV integration sites in cervical cancer samples using the nanopore MinION sequencer without error correction. Front Genet. 2020;11:660. doi: 10.3389/fgene.2020.00660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Koren S, Walenz BP, Berlin K, Miller JR, Bergman NH, Phillippy AM. Canu: scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome Res. 2017;27:722–36. doi: 10.1101/gr.215087.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Salmela L, Rivals E. LoRDEC: accurate and efficient long read error correction. Bioinformatics. 2014;30:3506–14. doi: 10.1093/bioinformatics/btu538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Hackl T, Hedrich R, Schultz J, Forster F. Proovread: large-scale high-accuracy PacBio correction through iterative short read consensus. Bioinformatics. 2014;30:3004–11. doi: 10.1093/bioinformatics/btu392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Au KF, Underwood JG, Lee L, Wong WH. Improving PacBio long read accuracy by short read alignment. PLoS One. 2012;7:e46679. doi: 10.1371/journal.pone.0046679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Wang JR, Holt J, McMillan L, Jones CD. FMLRC: hybrid long read error correction using an FM-index. BMC Bioinf. 2018;19:50. doi: 10.1186/s12859-018-2051-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Goodwin S, Gurtowski J, Ethe-Sayers S, Deshpande P, Schatz MC, McCombie WR. Oxford nanopore sequencing, hybrid error correction, and de novo assembly of a eukaryotic genome. Genome Res. 2015;25:1750–6. doi: 10.1101/gr.191395.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, et al. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Robinson MD, McCarthy DJ, Smyth GK. EdgeR: a bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–40. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]