Abstract
How cells sense and respond to environmental changes is still a key question. It has been identified that cellular metabolism is an important modifier of various epigenetic modifications, such as DNA methylation, histone methylation and acetylation and RNA N6-methyladenosine (m6A) methylation. This closely links the environmental nutrient availability to the maintenance of chromatin structure and gene expression, and is crucial to regulate cellular homeostasis, cell growth and differentiation. Cancer metabolic reprogramming and epigenetic alterations are widely observed, and facilitate cancer development and progression. In cancer cells, oncogenic signaling-driven metabolic reprogramming modifies the epigenetic landscape via changes in the key metabolite levels. In this review, we briefly summarized the current evidence that the abundance of key metabolites, such as S-adenosyl methionine (SAM), acetyl-CoA, α-ketoglutarate (α-KG), 2-hydroxyglutarate (2-HG), uridine diphospho-N-acetylglucosamine (UDP-GlcNAc) and lactate, affected by metabolic reprogramming plays an important role in dynamically regulating epigenetic modifications in cancer. An improved understanding of the roles of metabolic reprogramming in epigenetic regulation can contribute to uncover the underlying mechanisms of metabolic reprogramming in cancer development and identify the potential targets for cancer therapies.
Keywords: cancer metabolic reprogramming, DNA methylation, epigenetic modifications, histone acetylation, histone methylation, RNA m6A
Introduction
Cellular metabolism is a process that exists exclusively to meet energetic and biosynthetic needs. By intricately connecting to multiple cellular processes, metabolism plays an important role in cell growth, division and differentiation. Abnormal metabolism is connected to a variety of diseases including cancer [1], [2], [3]. A metabolic alteration that cancer cells show a notably increased consumption of glucose even in the presence of oxygen, was firstly observed by Otto Warburg. In the past decade, metabolic reprogramming becomes an emerging hallmark of cancer. Altered metabolism of lipids, amino acids and nucleotides in cancer cells has also been described. The progresses of studies on cancer metabolism have expanded our understanding of the functions and mechanisms of metabolic reprogramming at different stages of cancer development [3], [4], [5], [6]. Mutations in the genes encoding isocitrate dehydrogenase 1 and 2 (IDH1/2) are frequently occurred in gliomas and acute myeloid leukemias (AMLs). Mutated IDH1/2 lose the ability to catalyze the conversion of isocitrate to α-ketoglutarate (α-KG). Instead, IDH1/2 mutations lead to a novel ability to catalyze the conversion of α-KG to 2-hydroxyglutarate (2-HG). Due to its structural similarity to α-KG, 2-HG can act as a competitive inhibitor of α-KG-dependent dioxygenases including ten-eleven translocation (TET) family of 5-methylcytosine (5mC) hydroxylases and histone demethylases. Therefore, the accumulation of oncometabolite 2-HG caused by IDH1/2 mutations contributes to the development of gliomas and AMLs [7], [8], [9], [10], [11]. In addition, glutamine metabolism in cancer cells, a crucial component of cancer metabolism, has also been reported to exceed the use of other amino acids. In cancer cells, tricarboxylic acid (TCA) cycle is characterized by an efflux of substrates for use in biosynthetic processes. This synthetic activity depends on anaplerosis to continue TCA cycle. Interestingly, a high rate of glutamine metabolism can meet these needs in cancer [12, 13]. Acetyl-CoA, one of central metabolic intermediates, is widely used in energy production and biosynthesis. In normal cells, glycolysis, glutaminolysis and β-oxidation of fatty acids are conventional sources of acetyl-CoA. However, cancer cells can also use acetate as an alternative source of acetyl-CoA via acyl-CoA synthetase short chain family member 2 (ACSS2) to support cell survival and proliferation under stressed conditions, such as hypoxia [14, 15].
Epigenome is defined as the heritable and dynamic chemical changes to the genome that occurs independently of DNA sequence. The patterns of various epigenetic modifications on chromatins, such as DNA methylation, and histone acetylation, methylation, phosphorylation, ubiquitylation and glycation, are crucial to modulate chromatin structure and regulate chromatin accessibility and gene expression [16]. Recently, RNA methylation, mainly the methylation of adenosine residue at N-6 position (N6-methyladenosine, m6A), is emerging as a new hotspot in epigenetics. RNA m6A modification has been identified as one of post-transcriptional regulatory mechanism in different types of RNA. Furthermore, m6A modification has been documented to involve in regulating RNA stability, splicing and translation [17]. Given that epigenetic modifications are crucial to a lot of biological processes, dysregulation of epigenetic modifications has been closely linked to a variety of developmental diseases as well as the development of cancers [16], [17], [18]. DNA methylation can act as a switch controlling gene expression. Global DNA hypomethylation and CpG site-specific promoter hypermethylation are commonly observed in cancer [18, 19]. The mutations of DNA methyltransferase 3 alpha (DNMT3A), one of DNA methyltransferases, have been reported in hematopoietic malignancies. DNMT3A mutations can impair the enzymatic activity, resulting in aberrant DNA methylation [19, 20]. The co-occurrence of two histone methylation marks, activating H3K4me3 and repressive H3K27me3, constitutes the “bivalent domains”, which are originally thought to be important for the differentiation of embryonic stem cells [21]. The altered balance of these histone modifications may contribute to the pathogenesis of cancer via deregulating stem cell marker genes and epithelial-mesenchymal transition (EMT) genes [18, 22], [23], [24]. Epigenetic modification related genes are not only affected by mutations but also by abnormal expression. The expression of histone deacetylases (HDACs) is upregulated in multiple cancer types. The underlying mechanisms by which HDACs contribute to tumorigenesis are diverse due to a range of histone and nonhistone substrates. HDACs can either repress the expression of tumor suppressor genes or regulate oncogenic pathways by modifications of key molecules [19, 25]. Owing to the important role of m6A modification in gene expression regulation, aberrant m6A modification has been reported to be associated with various cancers. Alpha-ketoglutarate dependent dioxygenase FTO (FTO), an m6A demethylase, is overexpressed in AMLs and promotes leukemogenesis through regulating target gene expression by reducing m6A levels. In bladder cancer, the oncogenic role of another m6A modifier methyltransferase like 3 (METTL3), which is one of m6A methylases, is to reduce phosphatase and tensin homolog (PTEN) by accelerating the maturation of pri-miR221/222 [17, 26, 27].
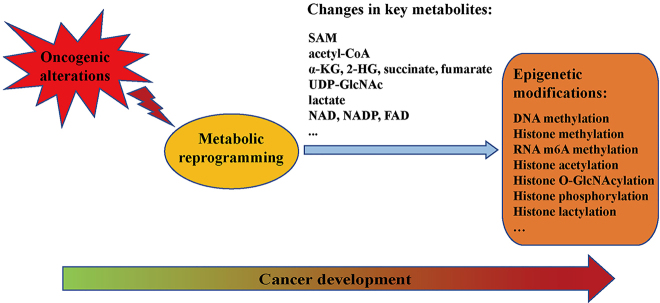
Metabolic reprogramming driven by the oncogenic alterations can contribute to altered epigenetic modifications in cancer. The metabolites can serve as substrates, cofactors or regulators for epigenetic enzymes, thereby leading to changes in DNA, RNA and histone methylation and histone acetylation, which then promote the development of cancer [28, 29]. For instance, 2-HG produced by IDH1/2 mutants affects the activities of α-KG-dependent 2-oxoglutarate-dependent dioxygenases (2-OGDDs), including histone N-methyl-lysine demethylases (KDMs), TETs and RNA m6A modification erasers, consequently causing histone, DNA and RNA modification changes to promote tumorigenesis [30], [31], [32]. Moreover, hypoxia is an important stimulus for various diseases such as cancer [33]. Hypoxia regulates the activities of levels of oxygen-dependent 2-OGDDs, leading to altered epigenetic modifications in cancer [30, 34]. In this review, we will give a brief summary for the effects of metabolic reprogramming on epigenetic modifications in cancer. We will discuss how the availability of specific metabolites, such as S-adenosyl methionine (SAM), acetyl-CoA, α-KG, uridine diphospho-N-acetylglucosamine (UDP-GlcNAc) and lactate, contributes to epigenetic modification changes and consequently promote the development of cancer.
Effect of metabolism on epigenetic modifications
Metabolism reprogramming, which may be both direct and indirect consequence of oncogenic alterations, contributes to tumorigenesis. Through metabolism reprogramming, cancer cells acquire the ability to uptake and utilize the necessary nutrients from a nutrient-deficient condition to both maintain viability and build new biomass [3, 5]. Cancer-associated metabolic alterations can affect the metabolite influx through shaping the way the nutrients are preferentially assigned to metabolic pathways that promote oncogenic properties. The emerging hallmarks of cancer-associated metabolic changes have been summarized: (1) abnormal uptake of glucose and amino acids; (2) opportunistic modes of nutrient acquisition; (3) biosynthesis and nicotinamide adenine dinucleotide phosphate (NADPH) production with glycolysis/TCA cycle intermediates; (4) high demand for nitrogen; (5) altered regulation of metabolite-driven gene expression; (6) metabolic interactions with tumor microenvironment [5, 35]. Since metabolites such as SAM, acetyl-CoA and α-KG are substrates, cofactors, donors and antagonists for the activities of epigenetic modifying enzymes, metabolites play crucial roles in modulating epigenetic modifications [36, 37]. Therefore, the metabolite changes caused by cancer-associated metabolic reprogramming have profound effects on gene expression (Figure 1) [5, 28]. In this section, we will summarize the effect of oncogenic metabolic reprogramming on epigenetic modifications in cancer.
Figure 1:
Connections between metabolic reprogramming and epigenetic modifications in cancer development. SAM, S-adenosyl methionine; α-KG, α-ketoglutarate; 2-HG, 2-hydroxyglutarate; UDP-GlcNAc, uridine diphospho-N-acetylglucosamine.
SAM level regulates cellular methylation
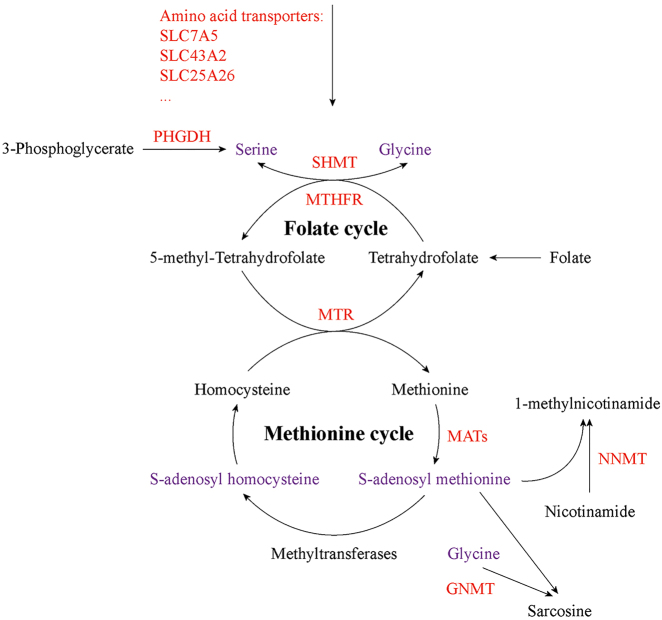
Regulation of cellular methylation by nutritional factors mainly involved in two mechanisms: changes of the available methyl donors and alterations in the enzymatic activity of corresponding enzymes [38]. SAM, synthesized by methionine adenosyltransferase (MAT) with methionine and adenosine tri-phosphate (ATP) (Figure 2), is a universal methyl donor for cellular methylation processes, including DNA and histone methylation as well as RNA m6A methylation. After transferring the methyl group from SAM to the substrate, S-adenosyl homocysteine (SAH) is produced along with the methylated substrate, and is then converted to homocysteine. Finally, homocysteine obtains a methyl group from 5-methyl tetrahydrofolate (mTHF) to regenerate methionine (Figure 2) [36, 39], [40], [41]. Besides, the methyltransferase domains of RNA m6A core writers METTL3 and methyltransferase like 14 (METTL14) can bind both SAM and SAH in the catalytic site [36, 40, 42]. Hence, the SAM/SAH ratio has an effect on the activities of methyltransferases in vivo. Alterations in SAM/SAH ratio is closely linked to aberrant histone methylation in cancers [29, 38, 43, 44]. Two genes methionine adenosyltransferase 1A (MAT1A) and methionine adenosyltransferase 2A (MAT2A) that encode the catalytic subunit of MAT, are dysregulated in several cancer types, including hepatocellular and colon cancer [43], [44], [45]. Recently, Villa et al. [46] reported that mechanistic target of rapamycin complex 1 (mTORC1) can stimulate the synthesis of SAM through the regulation of MAT2A expression. Furthermore, mTORC1 also increases the level of Wilms’ tumor 1-associating protein (WTAP), the positive regulatory subunit of RNA m6A methyltransferase complex. Consequently, mTORC1 stimulates RNA m6A modification to promote protein synthesis and cell proliferation. Moreover, MAT2A inhibition suppresses RNA m6A modification and tumor cell growth.
Figure 2:
Altered S-adenosyl methionine (SAM) metabolism in cancer. SAM, a universal methyl donor for cellular methylation reactions, is produced from methionine. Folate cycle can support SAM production with serine. In addition to providing methyl group for DNA, RNA and histone methylation, SAM is also consumed by NNMT and GNMT to generate 1-methylnicotinamide and sarcosine, respectively. Red indicates altered metabolic proteins, and purple indicates the key metabolites that affect methylation modifications in cancer. SLC7A5, solute carrier family 7 member 5; SLC43A2, solute carrier family 43 member 2; SLC25A26, solute carrier family 25 member 26; PHGDH, phosphoglycerate dehydrogenase; SHMT, serine hydroxymethyltransferase; MTHFR, methylene tetrahydrofolate reductase; MTR, methionine synthase; MATs, methionine adenosyltransferases; NNMT, nicotinamide N-methyltransferase; GNMT, glycine N-methyltransferase.
Additionally, alterations in one-carbon metabolism can affect SAM levels and hyperactivation of one-carbon metabolism is a driver of tumorigenesis and establishes a link to epigenetic status [47], [48], [49]. One-carbon metabolism, a network of pathways that donate and regenerate carbon units, involves methionine and folate cycles and integrates nutrients such as amino acids and vitamins to generate diverse outputs for methylation reactions, maintenance of redox status and biosynthesis of nucleotides, lipids and proteins. Folate metabolism, which is a universal metabolic process to support one-carbon metabolism, is important to activate and transfer one-carbon units for biosynthetic processes including nucleic acid synthesis and homocysteine methylation. Folate is a B vitamin and the biologically active form of folate is tetrahydrofolate (THF). In the body, folate is mainly present in the reduced form, typically mTHF (Figure 2) [38, 39, 48]. Folate metabolism enzymes are strongly upregulated in cancer [50, 51]. Serine hydroxymethyltransferase (SHMT) catalyzes the conversion of glycine to serine with 5,10-methylene-THF (Figure 2) [48]. Loss of the catalytic activity of mitochondrial SHMT2 leads to defective oxidative phosphorylation in cancer cells due to impaired mitochondrial translation. Mitochondrial SHMT2 has been found to provide methyl donors to produce the taurinomethyluridine base at the wobble position of mitochondrial tRNAs. In SHMT2-knockout cells, the lack of this mitochondrial tRNA modification causes defective translation, thereby affecting the expression of respiratory chain enzymes [52]. mTHF is produced by the cytosolic NADPH-dependent methylene tetrahydrofolate reductase (MTHFR) (Figure 2) [48]. In AML, the reduced-function polymorphisms of MTHFR reduce histone H3K27/K9 methylation [53]. mTHF can be consumed by vitamin B12-dependent methionine synthase (MTR) [48]. In colorectal cancer, the MTR 2756AG genotype is correlated with promoter hypermethylation of Ras association domain family member 1 A (RASSF1A) [54]. Nicotinamide N-methyltransferase (NNMT) can catalyze the production of 1-methylnicotinamide (1-MNA) from nicotinamide by using SAM as methyl donor, hence directly linking one-carbon metabolism to cellular methylation status and nicotinamide adenine dinucleotide (NAD+) level (Figure 2). NNMT upregulation is observed in a variety of cancers, including liver, kidney and colon cancers, and is associated with tumor progression. NNMT can act as a sink for SAM and NNMT upregulation strongly depletes cellular SAM pool, resulting in reduction in SAM/SAH ratio and making SAM unavailable for histone methylation. Accordingly, cells with NNMT overexpression show a notable decrease in histone methylation marks at H3K4, H3K9, H3K27 and H4K20 [29, 55, 56].
The carbon units provided by serine and glycine metabolism satisfy many of diverse biological functions. As an important one-carbon unit donor to the folate cycle, serine supports the metabolic processes which are crucial for nucleotide synthesis, methylation reaction and NADPH production. Aberrant serine and glycine metabolism links metabolic pathways to cancer biology, and serine and glycine can act as the drivers of tumorigenesis. Increasing evidence suggests that unbalanced availability of nutrients caused by altered one-carbon metabolism has an effect on histone and DNA methylation [39, 47, 48, 57]. Some cancer cells can utilize glucose-derived carbon through de novo biosynthesis of serine and glycine via phosphoglycerate dehydrogenase (PHGDH) (Figure 2). Consistently, PHGDH has been reported to be frequently amplified and overexpressed in various cancers. PHGDH inhibition decreases DNA and histone methylation in burkitt lymphoma [58, 59]. Moreover, serine metabolism can contribute to methionine cycle and DNA and RNA methylation through de novo ATP synthesis in cancer cells [60]. Since glycine N-methyltransferase (GNMT) can catalyze SAM-mediated methylation of glycine to produce sarcosine (Figure 2), it acts as a cellular buffer for maintaining the SAM level [48]. GNMT deletion causes increased hepatic SAM and methionine and induces abnormal methylation of DNA and histone, resulting in hepatocellular carcinoma (HCC) in mice [61]. In addition, amino acid transporters such as solute carrier family 7 member 5 (SLC7A5) and solute carrier family 43 member 2 (SLC43A2) are upregulated to increase the uptake of methionine in cancer cells [29, 62, 63]. Cells sorted for surface SLC7A5 exhibit activated enhancer of zeste 2 polycomb repressive complex 2 subunit (EZH2) and enrichment of methionine cycle intermediates in mice. Co-expression of SLC7A5 and EZH2 has been observed in lung cancer. SLC7A5 knockdown results in SAM reduction and decreases EZH2 activity [62]. Interestingly, tumor cells avidly consume methionine and outcompete T cells for methionine by the upregulation of SLC43A2. Inhibition of tumor SLC43A2 restores H3K79me2 in T cells and promotes spontaneous and checkpoint-induced tumor immunity [63]. Solute carrier family 25 member 26 (SLC25A26), which can catalyze the import of SAM into mitochondrial matrix, is downregulated in cervical cancer cells. Overexpression of SLC25A26 in cervical cancer cells increases mitochondrial SAM availability and promotes mitochondrial DNA hypermethylation [64].
Acetyl-CoA influences histone acetylation
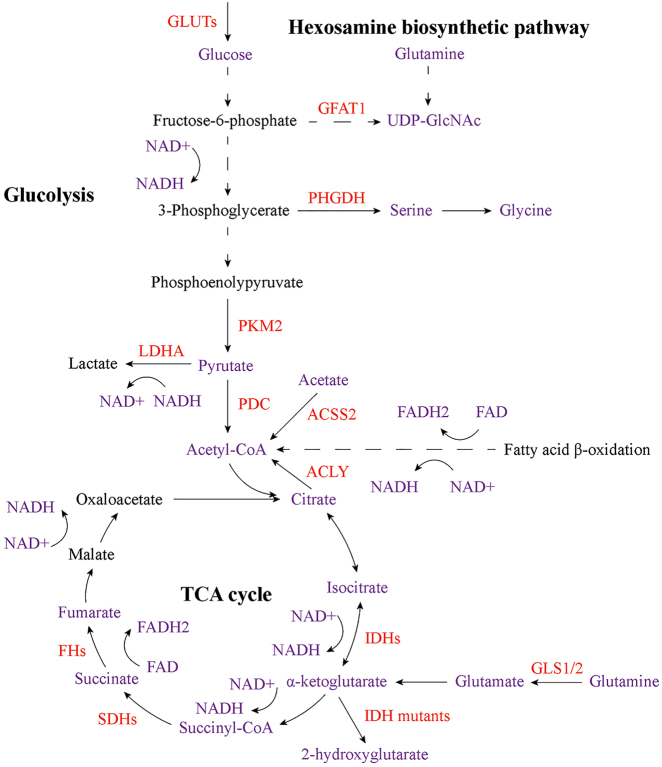
Histone acetylation, referring to add an acetyl group from acetyl-CoA to lysine residues in histones, is one of the widely studied histone modifications. Histone acetylation is dynamic and reversible process regulated by histone acetyltransferases (HATs) and HDACs, which involve in the addition and removal of the acetyl group from lysine residues, respectively [65]. Acetyl-CoA is both a central metabolic intermediate and the acetyl donor for acetylation modifications, and plays important roles in energy production, lipid metabolism and epigenetic modifications, consequently positioning it at the crossroads of metabolism and epigenetic regulation [28, 66]. Acetyl-CoA can be produced from pyruvate, citrate and acetate by pyruvate dehydrogenase complex (PDC), ATP-citrate lyase (ACLY) and ACSS2, respectively. It can also be generated from fatty acid β-oxidation, amino acid metabolism and ketone bodies (Figure 3) [65, 67]. Acetyl-CoA is used for biosynthesis in the cytosol, including de novo fatty acid and cholesterol synthesis, and is also needed for post-translational lysine acetylation throughout the cell [28]. Acetyl-CoA level changes with nutrient abundance, and thus it may serve as a signal to the cell of nutrient status [28, 67]. Intracellular acetyl-CoA concentrations are approximately 3–20 mM under normal physiological conditions, which may potentially limit the activity of some lysine acetyltransferases (KATs) such as p300. Moreover, many KATs are inhibited by CoA. Thus, the acetyl-CoA/CoA ratio is crucial to regulate acetylation in response to metabolic changes [28, 29, 68]. Nutrient limitation or glycolytic inhibition results in decreased acetyl-CoA levels and then reduces histone acetylation [28]. Both acetyl-CoA and acetyl-CoA/CoA ratio have been shown to regulate histone acetylation in cancer [29, 69, 70]. Cancer cells utilize glucose to produce lactate even in oxygenated conditions (Figure 3) [71]. Subsequent studies show that changes in glucose metabolism can affect histone acetylation [72, 73]. Cellular glucose availability is linked to acetyl-CoA abundance and histone acetylation. To facilitate the glucose uptake, glucose transporters such as glucose transporter type 1 and 4 (GLUT1/4) are dysregulated in cancer cells (Figure 3) [67, 74]. In colon cancer cells, the rate of glycolysis with corresponding changes in glycolytic intermediates is linked to abnormal histone acetylation. Dynamic histone acetylation has been found to be positively correlated with acetyl-CoA levels and be inversely associated with the acetyl-CoA/CoA ratio [73]. PDC decarboxylates pyruvate to produce acetyl-CoA, thereby linking glycolysis to TCA cycle and contributing to cellular biosynthesis and energy metabolism (Figure 3). Inhibition of PDC activity by pyruvate dehydrogenase kinase (PDK) via mediating the phosphorylation of PDC, has been reported to be associated with cancer. Perturbations of PDC/PDK axis can induce a “glycolytic shift,” resulting in ATP production via glycolysis over oxidative phosphorylation (OXPHOS) in cancer cells [75]. In prostate cancer, compartmentalized activities of PDC support lipid synthesis. Pyruvate dehydrogenase A1 (PDHA1), a subunit of PDC, and PDC activator pyruvate dehydrogenase phosphatase 1 (PDP1) are frequently amplified and overexpressed in prostate cancer. PDC localizes in both mitochondria and nucleus. Nuclear PDC regulates the expression of sterol regulatory element-binding transcription factor (SREBF)-target genes by affecting histone acetylation, and mitochondrial PDC contributes to the production of cytosolic citrate for lipogenesis [76]. The biosynthesis of citrate, which is one of the acetyl-CoA donors, is decreased due to aerobic glycolysis in cancer [77]. Availability of acetyl-CoA for HATs is also modulated by ACLY expression and activity, which probably contributes to nuclear acetyl-CoA pool [29]. ACLY is overexpressed in lung adenocarcinoma, and active phosphatidylinositol 3-kinase (PI3K)-protein kinase B (AKT) pathway enhances the phosphorylation of ACLY [78]. Activating Kirsten rat sarcoma viral oncogene homolog (KRAS) mutations have been observed in >90% cases of pancreatic ductal adenocarcinoma (PDAC). Acetyl-CoA abundance and histone H4 acetylation are increased in pancreatic acinar cells with KRAS mutations prior to the appearance of premalignant lesions. In PDAC cells, growth factors enhance AKT-ACLY signaling and histone acetylation. Thus, KRAS mutation-driven metabolic alterations promote the development of PDAC through acetyl-CoA-dependent processes [79]. These results suggest that oncogenic signaling can contribute to increased histone acetylation. Solid cancer cells frequently experience reduced oxygen availability [80]. Glucose is the main carbon source of acetyl-CoA in oxygenated cells. However, cancer cells shunt glucose-derived carbon to lactate during hypoxia [81, 82]. Subsequent studies reveal that cancer cells can utilize acetate as alternative acetyl-CoA source to support cell survival and proliferation under hypoxia [14, 15, 83, 84]. In hypoxic cancer cells, acetate promotes lipogenic genes acetyl-CoA carboxylase alpha (ACACA) and fatty acid synthase (FASN) expression by increasing histone H3 acetylation at their promoter regions, thereby enhancing lipogenesis. Acetyl-CoA synthetases ACSS1/2, which can catalyze the production of acetyl-CoA from acetate, contribute to the above acetate-mediated epigenetic regulation [15]. Besides, ACSS2 can fulfill different functions depending on the cellular location, and oxygen limitation increases the nuclear ACSS2 levels [84]. Moreover, the newly identified ACSS member ACSS3 is upregulated in bladder cancer, and involves in acetate utilization and histone acetylation in bladder cancer cells under metabolic stress [85]. Acyl-CoA thioesterase 12 (ACOT12), which is significantly downregulated in HCC, is vital to support tumor growth and EMT by regulating cellular acetyl-CoA levels and histone acetylation [86].
Figure 3:
Core cellular metabolic reprogramming that contributes to altered epigenetic modifications in cancer. Changes in metabolic proteins, which play important roles in glycolysis, TCA cycle and hexosamine biosynthetic pathway, affect the abundance of key metabolites, including acetyl-CoA, α-KG, succinate, fumarate, 2-hydroxyglutarate, UDP-GlcNAc, lactate and NAD+/NADH. These alterations have significant influences on DNA, RNA and histone methylation as well as histone acetylation, O-GlcNAcylation, phosphorylation and lactylation in cancer cells. Red indicates altered metabolic proteins, and purple indicates the key metabolites that affect epigenetic modifications in cancer. GLUTs, glucose transporters; GFAT1, glutamine fructose-6-phosphate amidotransferase 1; UDP-GlcNAc, uridine diphospho-N-acetylglucosamine; PHGDH, phosphoglycerate dehydrogenase; PKM2, pyruvate kinase 2; LDHA, lactate dehydrogenase A; PDC, pyruvate dehydrogenase complex; ACSS2, acyl-CoA synthetase short chain family member 2; ACLY, ATP-citrate lyase; IDHs, isocitrate dehydrogenases; SDHs, succinate dehydrogenases; FHs, fumarate hydratases; GLS1/2, glutaminase 1 and 2.
Additionally, other dysregulated metabolites that involve in the regulation of acetyl-CoA abundance also play important roles in cancer. Arginine and its metabolic products are crucial to cellular physiology, and their dysregulation is associated with cancer [87]. In prostate cancer cells, arginine also activates the expression of KATs and increases the acetyl-CoA level and histone acetylation, facilitating TEA domain transcription factor 4 (TEAD4) recruitment, which leads to upregulation of OXPHOS genes [88]. In normal colonocytes, butyrate is the important energy source, and is used to produce acetyl-CoA. However, glucose becomes the primary energy source in colon cancer cells due to Warburg effect. Therefore, butyrate accumulates and acts as an HDAC inhibitor. Although both glucose and butyrate can increase histone acetylation, different target genes are upregulated. Consequently, butyrate facilitates colon cancer cell proliferation when Warburg effect is prevented, whereas it inhibits the proliferation of colon cancer cells undergoing Warburg effect [89]. Moreover, primary mammary gland-derived adipocytes secret β-hydroxybutyrate that is required to contribute to the malignancy of breast cancer cells with monocarboxylate transporter 2 (MCT2) expression via upregulating global histone H3K9 acetylation [90].
Acidosis is considered as a hallmark of cancer. Some tumor metabolic peculiarities, including the release of lactate that is a byproduct from elevated aerobic glycolysis, account for low pH in tumor microenvironment [12, 91]. Fatty acid oxidation becomes the main source of acetyl-CoA in acidic pH-adapted cancer cells. Fatty acid-derived acetyl-CoA not only supports TCA cycle and cell respiration, but also contributes to mitochondrial protein hyperacetylation in tumor cells. Fatty acid oxidation and synthesis are regulated through the downregulation of acetyl-CoA carboxylase 2 (ACC2) via sirtuin-mediated histone deacetylation [92].
TCA cycle intermediates: α-KG, succinate and fumarate are crucial to cellular methylation
2-OGDDs are a superfamily of enzymes which require oxygen, reduced iron and 2-oxoglutarate (also known as α-KG) to function. 2-OGDDs play crucial roles in multiple biological processes, including regulation of hypoxia-inducible factor (HIF)-mediated adaptation to hypoxia and epigenetic modifications [30]. Several 2-OGDD members are important to reverse the methylation modifications in DNA, RNA and histone. Three isozymes of TET hydroxylase reverse DNA 5mC methylation. Similarly, alkB homolog 5 (ALKBH5) and FTO mediate the common m6A demethylation in RNA. KDMs, the 2-OGDDs that contains a Jumonji C (JmjC) catalytic domain, can target mono-, di- and tri-methylated lysine residues in histone to contribute to histone demethylation [30, 93]. Since these demethylation-related enzymes are α-KG-dependent, the cellular α-KG levels are crucial for the regulation of methylation. Moreover, two other TCA cycle intermediates succinate and fumarate are analogs of α-KG (Figure 3), and thereby they can act as the competitive inhibitors of multiple α-KG-dependent dioxygenases, including above-mentioned DNA and histone demethylation-related enzymes, such as TET1, TET2, KDM2B, KDM4A and KDM4B [30, 94].
The IDH family enzymes IDH1, IDH2 and IDH3 are three isozymes that catalyze the conversion of isocitrate to α-KG (Figure 3). IDH1 locates in cytosol, whereas IDH2 and IDH3 are distributed in mitochondria [95]. In TCA cycle, succinate is converted to fumarate catalyzed by succinate dehydrogenases (SDHs). Subsequently, fumarate hydratases (FHs) catalyze the hydration of fumarate into malate (Figure 3) [96]. The relative concentrations of α-KG, succinate and fumarate are crucial to cellular methylation. However, genes encoding IDHs, SDHs and FHs are frequently mutated in a variety of cancer types [7], [8], [9], [10, 97, 98]. Loss-of-function mutations lead to SDHs and FHs deficiency, thereby resulting in the intracellular accumulation of succinate and fumarate. Excess succinate and fumarate contribute to the inhibition of multiple α-KG-dependent dioxygenases, including TETs and KDMs, and consequent changes in genome-wide DNA and histone methylation in cancer [30, 94, 98, 99]. Interestingly, cancer-associated IDH1/2 mutations are heterozygous, with tumors retaining one wild type copy of IDH1 or IDH2 allele, suggesting that these mutations lead to an enzymatic gain of function. Therefore, IDH1/2 mutations alter the original catalytic activity of IDHs, and mutant enzymes can catalyze an irreversible reaction in which α-KG is reduced rather than carboxylated, resulting in the production of another oncometabolite 2-HG (Figure 3) [7, 8, 10, 30]. 2-HG is necessary and sufficient to mediate the oncogenic effects of mutant IDHs [30, 95]. Similar to succinate and fumarate, 2-HG is also an analog of α-KG, and thereby the accumulation of 2-HG can competitively inhibit the activities of α-KG-dependent dioxygenases. The catalytic activities of TET2 and KDM enzymes, such as KDM2A and KDM4C, are inhibited by 2-HG in IDH-mutant cancer cells, resulting in global DNA and histone methylation alterations [10, 11, 30, 100], [101], [102]. In addition to being an oncometabolite, 2-HG has been reported to exhibit a broad anti-tumor activity in AML cells by inhibiting cell proliferation and survival and by promoting cell cycle arrest and apoptosis. Mechanistically, 2-HG inhibits the activity of FTO, thereby increasing global RNA m6A modification, which consequently decreases the stability of MYC proto-oncogene, basic helix–loop–helix (bHLH) transcription factor/CCAAT enhancer binding protein alpha (MYC/CEBPA) transcripts [31]. Moreover, 2-HG also abrogates FTO/YTH N6-methyladenosine RNA binding protein 2 (YTHDF2)-mediated the RNA m6A modification of two critical glycolytic genes: phosphofructokinase platelet (PFKP) and lactate dehydrogenase B (LDHB) in AML cells, in turn leading to the upregulation of these two genes and thereby suppressing aerobic glycolysis [32].
Hexosamine biosynthetic pathway (HBP) contributes to histone O-GlcNAcylation
The addition of O-linked β-N-Acetylglucosamine (O-GlcNAc), a monosaccharide, onto serine and threonine residues, is one of the key post-translational modifications (PTMs) of nuclear and cytosolic proteins, termed O-GlcNAcylation. O-GlcNAcylation is involved in the nutrient flux through HBP, which integrates glucose, glutamine, fatty acid and nucleotide metabolism to produce UDP-GlcNAc, the donor substrate for O-GlcNAcylation [103, 104]. In this pathway, a portion of fructose-6-phosphate (fructose-6-P), the isomer of glucose-6-phosphate (glucose-6-P) generated from glucose, is utilized by glutamine: fructose-6-phosphate amidotransferase 1 (GFAT1), which is the first and key rate limiting enzyme of HBP. GFAT1 catalyzes the production of glucosamine-6-phosphate from fructose-6-P with an amine group from glutamine. In subsequent reactions, an acetyl group from acetyl-CoA and an UDP from UTP are added to generate UDP-GlcNAc (Figure 3). Hence, supply of glucose and glutamine is crucial to UDP-GlcNAc synthesis [29]. The addition and removal of O-GlcNAc in the substrate proteins are dynamic processes mediated by two enzymes which make the O-GlcNAc modification reversible and are crucial to maintain cellular O-GlcNAc balance. O-GlcNAc transferase (OGT) catalyzes the incorporation of O-GlcNAc from UDP-GlcNAc to target proteins, whereas O-GlcNAcase (OGA) involves in the removal of O-GlcNAc from the glycosylated substrate [105].
In addition to being dependent on nutrient availability, O-GlcNAcylation is also sensitive to various cellular stress such as heat shock and hypoxia. Therefore, O-GlcNAcylation has been proposed to function as a nutrient and stress sensor that regulates fundamental biological processes involved in transcription, translation, signal transduction and metabolism [103, 106]. From an epigenetic perspective, all four histones (H2A, H2B, H3, and H4) can be modified by O-GlcNAcylation to constitute part of the so-called “histone code” [29, 106, 107]. H2A O-GlcNAcylation at threonine 101 (T101) site can modulate chromatin structure through direct destabilization of H2A/H2B dimer in the nucleosome [108]. Fujiki et al. [109] reported that histone H2B O-GlcNAcylation at serine 112 (S112) residue by OGT promotes K120 monoubiquitination, in which the O-GlcNAc moiety serves as an anchor for histone H2B ubiquitin ligase. OGT can affect other histone modifications by interacting with chromatin remodelers, as exemplified by the study that overexpression of OGT alters histone H3 modifications at K9, K27, S10, and R17 residues [110]. Furthermore, despite OGT has no effects on 5-hydroxymethylcytosine activity, TET2 and TET3 promote OGT activity. The complex of TET2/3-OGT co-localizes at active promoters enriched for H3K4me3 and facilitates gene transcription. Mechanistically, host cell factor 1 (HCF1), a shared subunit of the H3K4 methyltransferase SET1/COMPASS, can be O-GlcNAcylated by TET2/3-OGT, and consequently impacts the integrity of SET1/COMPASS and the H3K4me3 level [107, 111]. In embryonic stem cells, TET1 is required for the binding of OGT to chromatin, which in turn affects TET1 activity to regulate CpG island methylation [112].
The dysregulation of O-GlcNAcylation caused by metabolic disorders contributes to multiple diseases including cancer. Upregulation of HBP including the GFAT1 level, which is associated with increased protein O-GlcNAcylation, is generally observed in cancer cells and is primarily driven by elevated glucose uptake and metabolism [29, 113], [114], [115], [116], [117], [118], [119]. In KRAS G12D-driven PDAC model, KRAS mutant promotes glucose utilization through upregulating the expression of GLUT1, hexokinase 1 and 2 (HK1/2), which then stimulates glycolytic flux into HBP and activates protein O-GlcNAcylation [29, 115]. In another study, Guillaumond et al. [116] reported that hypoxia increases the glycolysis and glutamine metabolism which is pivotal for PDAC cell survival. The authors further found that the transcription of HBP genes as well as the protein O-GlcNAcylation are upregulated under hypoxic condition. These results indicate that hypoxia-driven metabolic rewiring including high glycolytic rate and HBP activation, favors PDAC progression. Upregulated O-GlcNAcylation and OGT expression are essential for tumor growth and metastasis in papillary thyroid cancer (PTC). In details, Yes1 associated transcriptional regulator (YAP) serine 109 (S109) O-GlcNAcylation promotes the malignant phenotypes of PTC cells via promoting YAP serine 127 (S127) dephosphorylation and activation [119]. In breast cancer and prostate cancer, elevated OGT and O-GlcNAcylation contribute to tumor progression through indirect regulation of the oncogenic transcription factor forkhead box M1 (FOXM1) [117, 118]. In addition, O-GlcNAcylation has also been reported to be a key regulator of glucose metabolism in cancer cells. O-GlcNAcylation at serine 529 (S529) of phosphofructokinase 1 (PFK1) is induced in response to hypoxia, and inhibits PFK1 activity to redirect glycose flux through pentose phosphate pathway, thereby leading to a selective growth advantage on cancer cells [120]. Under glucose deprivation condition, Adenosine 5′-monophosphate (AMP)-activated protein kinase (AMPK) phosphorylates FH at serine 75 (S75), which in turn forms a complex with activating transcription factor 2 (ATF2) to stimulate promoter activation. FH-catalyzed fumarate in promoter regions inhibits KDM2A activity, thereby maintaining the H3K36me2 profile to facilitate gene expression for cell growth arrest. On the other hand, FH is observed to be O-GlcNAcylated at S75 residue, which impedes FH-ATF2-mediated downstream events in cancer cells that show high OGT activity [121]. The crosslink between O-GlcNAcylation and epigenetic modifications also play an important role in cancer. The O-GlcNAcylation of HDAC1 is upregulated in HCC, which in turn increases the activated phosphorylation of HDAC1 and simultaneously enhances its enzyme activity. The O-GlcNAc modified HDAC1 affects gene transcription such as p21 transcription via regulating histone acetylation level, and consequently facilitates the proliferation, invasion, and migration of HepG2 cells [122]. In breast cancer MCF7 cells, OGT mediates EZH2 O-GlcNAcylation at S75 site, which is required for the protein stability of EZH2, and in turn contributes to the formation of H3K27me3, by which several potential tumor suppressor genes are repressed [123].
Metabolism related enzymes are involved in regulating histone phosphorylation
In addition to O-GlcNAcylation, histones can also be modified by phosphorylation at serine and threonine residues, which are dynamically phosphorylated by a number of protein kinases and dephosphorylated by phosphatases [105, 124]. Histone H2AX phosphorylation at serine 139 (S139), which is generally referred to as γH2AX, is an important histone modification that is involved in DNA damage response. H2AX has also been reported to be phosphorylated at tyrosine 142 (Y142), which is regulated in a DNA damage-dependent manner and is critical to γH2AX maintenance. Phosphorylation of H2B at serine 14 (S14) site has also been detected upon DNA damage induced by ionizing radiation and shown to co-localize with γH2AX foci [124], [125], [126]. Moreover, a great number of phosphorylated histone residues are closely linked to the regulation of proliferative gene expression. For instance, H3 phosphorylation at serine 10 (S10) and serine 28 (S28) and H2B phosphorylation at serine 32 (S32) are associated with the regulation of epidermal growth factor (EGF)-responsive gene expression, while phosphorylation of S10 of H3 and S32 of H2B are linked to the expression of oncogenes [124]. Phosphorylation of threonine residues in H3 such as threonine 11 (T11) have also been reported to be implicated in transcription regulation [127]. Phosphorylation of H3 at S10, T11 and S28 can play a role in transcription activation via influencing H3 acetylation [124]. In addition, phosphorylation of histone H3T6 is a key event that prevents lysine specific demethylase 1 (LSD1, also known as KDM1A) from demethylating H3K4 during androgen receptor (AR)-dependent gene activation [128]. H3 phosphorylation exemplifies a histone phosphorylation event that is associated with chromosome compaction during mitosis, meiosis and apoptosis [124].
Upon appropriate signaling, chromatin remodelers facilitate chromatin relaxation, thereby allowing an access for the coregulatory complexes to the target genes. Signaling serine/threonine and tyrosine kinases in cancer cells, such as mitogen and stress activated kinases, p21-activated kinase 1, EGF receptor kinases and janus kinase 2, contribute to signaling-mediated phosphorylation of histones, which serves as a regulatory switch for histone modifications and connects to chromatin remodeling complexes into gene transcription, indicating the pivotal role of histone phosphorylation in the development of cancer [129]. Histone H2A threonine 120 (T120) phosphorylation stimulates abnormal gene expression, including upregulated cyclin D1, which promotes oncogenic transformation [130]. H3S28 phosphorylation regulated by p300-CBP-associated factor (PCAF), promotes autophagy by targeting autophagy related 5 and 7 (ATG5/7) in osteosarcoma cancer cells [131]. A role of tryptophan-aspartic (WD) repeat domain 5 (WDR5) in prostate cancer exemplifies that the interplay between histone phosphorylation and other modifications plays a major role in cancer cell proliferation. WDR5 facilitates the recruitment of the mixed lineage leukemia 1 (MLL1) complex and subsequent H3K4me3 via interacting with H3T11 phosphorylation upon androgen stimulation, indicating the critical role of WDR5 as an epigenomic integrator of histone phosphorylation and methylation [132].
AMPK, a serine/threonine kinase which serves as a sensor of cellular energy status consistent with the ratios of adenosine monophosphate (AMP)/ATP and adenosine diphosphate (ADP)/ATP, is activated by an increase in AMP or ADP but inactivated by ATP [105, 133]. As a nutrient sensor, AMPK plays an important role in regulating energy homeostasis and mediating cell adaptation and survival under a variety of stress conditions that produce energy deprivation such as hypoxia, exercise, nutrient starvation, and infection [134]. AMPK has been reported to activate transcription through phosphorylation of histone H2B at serine 36 (S36) residue to facilitate cellular adaptation to stress [135]. In addition, several metabolic enzymes can moonlight as protein kinases responsible for the phosphorylation of multiple protein substrates, which are crucial for key cellular functions, including the Warburg effect in cancer cells [136]. One of the key rate-limiting glycolytic enzymes pyruvate kinase 2 (PKM2) (Figure 3), which can act as a protein kinase mediating histone phosphorylation, is commonly upregulated in multiple cancers [137], [138], [139]. Upon activation of EGF receptor and platelet-derived growth factor receptor (PDGFR) in cancer cells, PKM2 is phosphorylated at serine 37 (S37) residue via activated extracellular signal-regulated kinase (ERK), resulting in conversion of PKM2 from a tetramer to a monomer and exposure of its nuclear localization signal [137, 138, 140]. Nuclear PKM2 binds to phosphorylated β-catenin, which guides PKM2 to the promoter regions of downstream genes, such as cyclin D1 (CCND1) and MYC, where PKM2 utilizes phosphoenolpyruvate (PEP) as a phosphate group donor and serves as a protein kinase phosphorylating histone H3 at T11 residue [137, 139, 141]. This phosphorylation is required for the removal of HDAC3 from the CCND1 and MYC promoter regions and subsequent histone H3K9 acetylation. Moreover, the PKM2-dependent H3 phosphorylation promotes EGF-induced expression of CCND1 and MYC, cell proliferation and tumorigenesis [139]. Furthermore, EGF treatment enhances PKM2-dependent H3T11 phosphorylation at the programmed death-ligand 1 (PD-L1) promoter region, consequently leading to the elevated expression of PD-L1 mRNA and protein in HCC cells [142]. In breast cancer cells, nuclear PKM2 interacts with and directly phosphorylates histone H2AX at serine 139 (S139) under DNA damage conditions, and consequently promoting genomic instability [136, 143].
Lactate is responsible for histone lactylation
Evidence indicates lactate, a main by-product of aerobic glycolysis in cancer cells (Figure 3) [71], plays an important role in tumor progression, as exemplified by the study conducted by Brand et al. [144] that lactate accumulation caused by elevated lactate dehydrogenase A (LDHA) expression in melanomas modulates immune cell function and promotes tumor invasion and metastasis. Moreover, lactate has also been reported to be a primary carbon source for TCA cycle in cancer [145, 146]. In HCT116 cells, lactate can inhibit the activity of HDACs and promote the expression of corresponding genes [147]. In addition to these oncogenic roles of lactate, Zhang et al. [148] reported a novel lactate-derived histone lysine modification that directly stimulates gene transcription from chromatin in T cells. Subsequent studies have shown that histone lactylation is widely observed in macrophages [149, 150]. Macrophages can use lactate to promote the lactylation of high mobility group box 1 (HMGB1), while lactate also stimulates HMGB1 acetylation through β-arrestin2-mediated recruitment of p300/CBP and Hippo/YAP-mediated suppression of sirtuin 1 (SIRT1). Then, the modified HMGB1 is released from macrophages through exosome secretion, and subsequently results in increased endothelium permeability [150]. During early stages of senescent cell reprogramming, the transcription factor GLIS family zinc finger 1 (GLIS1) directly binds to and opens chromatin at glycolytic genes to upregulate glycolysis, which subsequently increases acetyl-CoA and lactate levels, thereby enhancing H3K27 acetylation and H3K18 lactylation at pluripotency gene loci to promote cellular reprogramming [151]. In addition, Chai and colleagues [152] recently identified an oncogenic role of histone lactylation in ocular melanoma. The authors showed that the histone lactylation level is elevated in ocular melanoma and high histone lactylation is associated with shorter recurrence free survival of ocular melanoma patients. Mechanistically, histone lactylation contributes to the development of ocular melanoma by facilitating the expression of YTHDF2, which then recognizes the m6A modified mRNAs of period circadian regulator 1 (PER1) and tumor protein p53 (TP53) and promotes their degradation.
Reduction/oxidation (redox) status regulates epigenetic modifications
Cellular redox state plays important roles in regulating diverse physiological processes, including homeostasis, proliferation and apoptosis [153]. In cancer cells, dysregulated metabolism frequently involves in the reducing equivalent, generally including the reduced forms of coenzymes such as NADH, NADPH, and flavin adenine dinucleotide (FADH2) [99]. NAD+, an abundant metabolite that is vital for maintenance of cellular homeostasis, acts as a cofactor in multiple redox reactions related to glycolysis, TCA cycle, OXPHOS, fatty acid oxidation, and amino acid biosynthesis (Figure 3). Moreover, NAD+ is a substrate for multiple important enzymes such as sirtuins and poly ADP-ribose polymerases (PARPs) [154, 155]. The NAD+/NADH ratio can form a linkage between metabolism and epigenetic regulation of gene expression by histone acetylation [156]. Sirtuins, the class III HDACs, use NAD+ as a cofactor to catalyze the removal acetyl modifications of lysine residues in histones and other proteins. Thus, the activity of sirtuins depends on the availability of NAD+ or the NAD+/NADH ratio [154, 157]. Higher NAD+/NADH ratio is found in cancer cells compared to normal cells. Instead of transferring electrons from NADH to mitochondrial OXPHOS, a large proportion of NAD+ is regenerated by reducing pyruvate to lactate due to highly increased glycolysis in cancer cells [154, 157, 158]. In addition, a critical enzyme of the NAD+ salvage pathway, nicotinamide phosphoribosyltransferase (NAMPT) catalyzes the biosynthesis of NAD. NAMPT has been reported to be overexpressed in several cancer types and be able to function as an oncogene to promote tumor progression by increasing the NAD+ levels [154, 159].
NADPH, another essential electron donor, is a dispensable cofactor used to reserve and transfer the reduction potential for anabolic reactions, such as the NADPH-dependent reductive carboxylation of α-KG into isocitrate [156, 160]. An increasing body of evidence has shown that regeneration and maintenance of the cellular NADPH is strongly implicated in the development of cancer. Compared with non-tumor cells, tumor cells usually maintain elevated levels of NADPH, not only to power redox defense but also to use for biosynthetic reactions, such as the synthesis of nucleotide and fatty acids, to support rapid cell growth and survival [160], [161], [162]. NAD+ kinase (NADK), the rate limiting enzyme in NADPH production, catalyzes the conversion of NAD+ to NADP+ by using ATP as the phosphate donor. Subsequently, NADP+ is reduced to NADPH by glucose-6-phosphate dehydrogenase (G6PD) and the malic enzymes. Decrease in NADPH level leads to repression of HDAC1 activity in human hepatic adenocarcinoma SK-Hep1 cells [163]. NADK mutant, whose expression leads to gain-of-function enzymatic activity resulting in low cellular reactive oxygen species (ROS) and elevated NADPH levels, is found in PDAC [160, 162, 164]. As mentioned in previously, IDH1/2 mutations are commonly observed in multiple cancers, and mutant IDH1/2 enzymes catalyze the production of 2-HG from α-KG. NADPH is an essential cofactor for this reaction, and lowering of NADPH levels inhibits the growth of cancer cells with IDH1/2 mutations [160, 161].
FAD is derived from the vitamin riboflavin and acts as a cofactor for histone lysine-specific demethylases KDM1A and KDM1B (also known as LSD1 and LSD2), which are members of amine oxidase superfamily [165], [166], [167], [168]. The histone demethylation reaction involves the reduction of FAD to FADH2. FADH2 can be re-oxidized by oxygen with hydrogen peroxide as a byproduct. Cellular redox status might affect the availability of FAD and thereby the activity of KDM1A. FAD also functions as a coenzyme for other oxidative reactions such as fatty acid β-oxidation and electron transport chain. Therefore, KDM1A activity might be influenced by the above-mentioned FAD oxidation processes [165, 166, 168, 169].
Oxygen availability has an effect on epigenetic modifications
In addition to the metabolites mentioned above, oxygen availability also plays a critical role in epigenetic modifications. Oxygen is essential to support cellular activities by serving as a key substrate for numerous biochemical reactions. Examples of oxygen consuming reactions include mitochondrial ATP production. Moreover, the above-mentioned 2-OGDDs also need oxygen as a cofactor to function, which are involved in various metabolic reactions, including RNA, DNA and histone demethylation reactions. Therefore, the maintenance of oxygen homeostasis is crucial to most species. Decreased oxygen concentration (hypoxia) triggers complex adaptive responses at cellular, tissue and organismal levels to match oxygen availability with metabolic demand. A critical question is how cells sense changes in oxygen levels to coordinate diverse biological responses during hypoxia. One of the important and best-studied mechanisms for cell response to hypoxia involves HIFs, which are stabilized by low oxygen availability and regulate the expression of a number of genes involved in cell viability, glycolysis, angiogenesis and metastasis [33, 170].
Hypoxic regions arise in tumors due to the rapid cell proliferation and the exhaustion of available nutrients and oxygen supplies. Accordingly, hypoxia induces multiple pathways, genomic and epigenetic changes which make tumor cells to adapt to poor nutrition and stressed conditions for the development of tumors [33, 170], [171], [172]. Hypoxia-associated histone hypermethylation is a common phenomenon. Hypoxia can contribute to histone hypermethylation through both a direct inhibition of the activities of oxygen-sensitive histone KDMs and an indirect effect on the expression of KDMs [30, 34, 173, 174]. The histone H3K27 demethylase KDM6A is oxygen sensitive. KDM6A loss, like hypoxia, inhibits H3K27 demethylation and blocks cellular differentiation in a HIF- and 2-HG-independent manner. Inhibition of KDM5A induces histone H3K4 methylation that is similar with the effects of hypoxia [175, 176]. KDM5A has been reported to be downregulated in metastatic glioma and inhibited the invasive ability of glioma cells. Thus, hypoxia-associated inhibition of KDM5A activity might be an alternative mechanism for suppressing KDM5A in cancer. Similarly, KDM6A is mutated and downregulated in multiple cancers and has been shown a tumor suppressive role. Therefore, in tumors with wild-type KDM6A, hypoxia might inhibit the activity of KDM6A and thereby promotes tumor progression [30, 177], [178], [179], [180]. Accordingly, theoretically, tumor hypoxia should promote the inhibition of KDMs. Interestingly, the roles of KDMs in cancer are divergent and multiple KDMs act as oncogenes to contribute to the development of cancer [181]. Tumor cells employ specific mechanisms to maintain the activity of tumor-promoting KDMs under hypoxia, such as by increasing α-KG availability or by upregulating the expression of KDMs [30]. Posterior fossa A ependymomas are maintained under hypoxia, and are associated with increased histone H3K27 demethylation catalyzed by KDM6A and KDM6B. No differences in the mRNA expression levels of KDM6A and KDM6B were detected under hypoxia vs. normal O2 concentration. However, the α-KG/succinate ratio is highly elevated under hypoxia, leading to enhance the activities of KDM6A and KDM6B [172]. Various KDMs, such as KDM1A, KDM2B and KDM5B, are overexpressed in cancer and show oncogenic effects [181], [182], [183], [184]. In addition, numerous KDMs are transcriptional targets of HIFs. KDM3A, KDM4B, KDM4C, KDM5B are direct HIF-1 target genes with HIF-1 binding to their promoters and thereby are upregulated under hypoxic conditions in cancer cells. In contrast, KDM6B is a hypoxia response gene regulated by HIF-2α [185], [186], [187], [188], [189].
TET1 and TET2 have been reported to have the low O2 Km values, indicating that these two enzymes might not act as oxygen sensors. However, the activity of TET enzymes is reduced by tumor hypoxia in human and mouse cells, and thereby leads to DNA hypermethylation. This reduction in TET activity is independent of hypoxia-associated alterations in TET expression or HIF activity, and depends directly on O2 shortage. In patients with non-small cell lung cancer (NSCLC), tumor suppressor gene promoters are notably hypermethylated in hypoxic tumor tissue [30, 190]. Conversely, hypoxia contributes to the upregulation of TET enzymes in other cancer types. Hypoxia-mediated induction of TET1 and TET3 is observed in glioma cells, leading to the overexpression of octamer-binding protein 4 (OCT4, also known as POU5F1) and nanog homeobox (NANOG), thereby contributing to the formation of cancer stem cells. Moreover, hypoxia involves in HIF-1-dependent transcriptional activation of TET1, and thereby results in the induction of hypoxia-responsive genes and global DNA demethylation in neuroblastoma [191], [192], [193].
RNA m6A modifications also play an important role in hypoxic response in cancer [194]. The reduction in total m6A level of mRNAs has been observed in cancer cells during hypoxia, which might be mediated by the induction of ALKBH5. Hypoxia affects the level/activity of m6A modification writers, erasers and readers, such as upregulates METTL14 and ALKBH5 and downregulates YTH N6-methyladenosine RNA binding protein 3 (YTHDF3), thereby leading to the reduced m6A level and consequently upregulation of target transcripts in cancer cells [195, 196]. In hypoxic breast cancer cells, HIFs activate ALKBH5 expression that then induces m6A demethylation and stabilization of NANOG mRNA, resulting in promoting the cancer stem cell phenotype [197]. Similarly, HIF-dependent ALKBH5 level is significantly increased under hypoxia in endometrial cancer stem cells, and contributes to the upregulation of SRY-box transcription factor 2 (SOX2) via mRNA demethylation, resulting in restoring the stem-like state of differentiated endometrial cancer stem cells [198]. Moreover, hypoxia induced overexpression of m6A modification readers is common in cancer and plays an important role in cancer progression. HIF-1α-dependent overexpression of m6A modification reader YTH N6-methyladenosine RNA binding protein 1 (YTHDF1) facilitates hypoxia-induced autophagy and malignancy of HCC. Mechanistically, YTHDF1 promotes the translation of autophagy-related genes autophagy related 2A (ATG2A) and autophagy related 14 (ATG14) via binding to m6A modified ATG2A and ATG14 mRNA [199]. HIF1α binds to the hypoxia-response elements of YTHDF2 gene and contributes to the transactivity of the YTHDF2 promoter, and then upregulates YTHDF2 in t(8;21) AML cells, thereby promoting cell proliferation [200].
HDACs have also been observed to be altered under hypoxic conditions. Hypoxia involves in regulating the activity and expression of several HDACs [186]. HIF-1α-induced HDAC3 is crucial to hypoxia-induced EMT and metastatic phenotypes in cancer cells. Under hypoxic condition, HDAC3 interacts with hypoxia-induced WDR5, recruits the histone methyltransferases (HMTs) complex to upregulate H3K4-specific HMT activity, and thereby activates mesenchymal gene expression. Moreover, HDAC3 also acts as an corepressor to repress epithelial gene expression [201].
Interestingly, the accumulation of two above-mentioned oncometabolites succinate and fumarate, caused by loss-of-function mutations of SDHs and FHs, has an effect on HIFs. Aberrantly activated HIF-1α is one mechanism contributing to tumorigenesis due to succinate or fumarate accumulation. Excess succinate or fumarate inhibits HIF prolylhydroxylases, and consequently promotes the stabilization of HIF-1α in normoxic conditions, thereby activating the expression of HIF target genes that can involve in glucose metabolism and angiogenesis [28, 202, 203]. These results suggest that except for affecting the activities of 2-OGDDs, succinate and fumarate also conduct their oncogenic effects via regulating HIFs.
Target altered cellular metabolism for anticancer therapy
Given that cancer cells meet increased bioenergetic and biosynthetic needs, maintain redox balance and enhance epigenetic modification-dependent regulation of gene expression, through cellular metabolism rewiring, to facilitate cancer progression, targeting altered cancer metabolism represents an emerging therapeutic strategy [204]. One therapeutic opportunity that has been explored is the small molecule inhibition of key enzymes involved in aberrant metabolic processes. Several bioenergetic metabolic pathways altered in cancer, including glycolysis, TCA cycle, glutaminolysis and fatty acid oxidation, have been investigated as potential drug targets [205].
Glucose is one of the most abundant and essential nutrients commonly utilized by cancer cells [5]. Thus, a number of drugs that interfere with glucose metabolism such as glycolysis are being investigated as anticancer agents [206]. A large body of preclinical studies indicate that glucose transporters and glycolytic enzymes, such as GLUT1, PKM2 and LDHA, may be the targets for anticancer therapy. Moreover, some of these drugs are under clinical investigation [206]. Liu et al. [207] reported a novel representative compound WZB117 that decreases the level of GLUT1 protein in cancer cells. Treatment with WZB117 inhibits cancer growth in both cell lines and a mouse model. Furthermore, daily intraperitoneal injection of WZB117 at 10 mg/kg leads to an obvious reduction in the size of human lung cancer of A549 cell origin. Mechanistically, WZB117 inhibits glucose transport in human red blood cells in which GLUT1 acts as the sole glucose transporter. Therefore, WZB117 is a prototype for the development of anticancer strategy via targeting GLUT1-mediated glucose transport and metabolism [207]. HA344, which covalently binds to PKM2 and inhibits the final and rate-limiting step of glycolysis, efficiently targets vemurafenib-resistant cutaneous metastatic melanoma (CMM) cells and impairs CMM xenograft tumor growth in mice. Further, HA344 functions synergistically with B-Raf proto-oncogene, serine/threonine kinase (BRAF) inhibitors on CMM cells in vitro. Hence, HA344 provides attractive therapeutic avenues for CMM patients [208]. In triple-negative breast cancer (TNBC), phosphorylation of PKM2 at S37 is connected to a cyclin-dependent kinase (CDK) pathway. In parallel, pyruvate kinase activator TEPP-46 binds phosphorylated PKM2 and reduces its nuclear localization. In a TNBC mouse xenograft model, treatment with either TEPP-46 or the potent CDK inhibitor dinaciclib reduces tumor growth and diminishes PKM2 phosphorylation. Combination of dinaciclib with TEPP-46 inhibits cell invasion, impairs redox balance, and induces cancer cell death. Consequently, targeting the PKM2 regulatory axis is a potential treatment for TNBC with PKM2 S37 phosphorylation [209]. LDHA is remarkably upregulated in NSCLC, and associated with radioresistance in NSCLC patients. LDHA inhibition with oxamate significantly increases the radiosensitivity of cancer cells, and enhances ionizing radiation-induced apoptosis [210]. Moreover, oxamate can also promote the efficacy of anti-PD-1 treatment in NSCLC mouse model [211]. Interestingly, dual treatment of the PKM2 activator TEPP-46 and the LDHA inhibitor FX-11 synergistically inhibits pancreatic cancer cell proliferation and remarkably suppresses tumor growth in vivo without apparent toxicity [212]. Woodford et al. [213] have recently identified that the human tumor suppressor folliculin (FLCN), a dedicated intracellular protein, can act as a binding partner and uncompetitive inhibitor of LDHA, which regulates LDHA activity and metabolic homeostasis in normal cells. However, in cancer cells that experience the Warburg effect, FLCN dissociates from LDHA. Treatment of cancer cells with a decapeptide derive from FLCN loop region induces cell death. In addition, Targeting GFAT1, the key rate-limiting enzyme of HBP, with a small molecule glutamine analog DON decreases the self-renewal potential and metastatic ability of cancer cells. Further, treatment with DON sensitizes PDAC to anti-PD1 therapy, resulting in tumor regression and prolonged survival [214].
Glutamine metabolism has been confirmed as an attractive anticancer therapeutic target due to the dependence of cancer cells on this process. Many classes of compounds that target glutamine transport and conversion have been examined [215]. Among mechanisms underlying the elevated glutamine metabolism in cancer is enhanced glutamine uptake mediated by the glutamine transporters, with solute carrier family 1 member 5 (SLC1A5) shown to play a pivotal role. Consistently, high SLC1A5 expression is associated with poorer survival of melanoma patients. IMD-0354, which prevents the localization of SLC1A5, has been selected by an image-based screen. Further study showed that IMD-0354 can act as a potent inhibitor of glutamine uptake that sustains low intracellular glutamine level. Combination of IMD-0354 with glutaminase 1 (GLS1) or LDHA inhibitors enhances melanoma cell death, and IMD-0354 inhibits melanoma growth in a xenograft model [216]. GLS1, a key enzyme involved in glutaminolysis, is commonly overexpressed in cancer cells to fulfill high glutamine demand. Allosteric inhibitors of GLS1 such as CB-839 have shown promise in preclinical cancer models. CB-839 has shown an effective antiproliferative activity in TNBC and AML cells, and is currently the subject of several clinical trials [215, 217], [218], [219].
Additionally, drugs that target cancer IDH mutants also provide an effective approach for anticancer strategy. AG-120 (ivosidenib), an inhibitor of IDH1 mutant enzyme, exhibits a profound effect on lowering 2-HG in tumor models and influences differentiation of primary patient AML samples. Preliminary data from phase 1 clinical trials suggests that AG-120 has an acceptable safety profile and clinical activity [220]. Chaturvedi and colleagues have reported a novel pan-mutant IDH1 inhibitor BAY1436032, which specifically inhibits 2-HG production and colony growth, and induces myeloid differentiation of AML cells carrying a variety of IDH1 mutations. Oral administration of BAY1436032 leads to prolonged survival in two independent patient-derived xenograft IDH1 mutant AML mouse models. Together, BAY1436032 is highly effective against AML with major types of IDH1 mutant [221]. AG-221 is an orally available, selective, potent inhibitor of the mutant IDH2 enzyme. Treatment with AG-221 inhibits 2-HG production and induces differentiation in primary human IDH2 mutation-positive AML cells ex vivo and in xenograft mouse models. AG-221 also provides a significant survival benefit in an aggressive IDH2-mutant AML xenograft mouse model [222]. Another drug vorasidenib (AG-881), which is currently in clinical development for treatment of low-grade glioma with mutant IDHs, is a potent oral and brain-penetrant dual inhibitor of both mutant IDH1 and IDH2 and can significantly inhibit 2-HG production in glioma tissue in an orthotopic glioma mouse model [223].
Serine synthesis pathway, which contributes to multiple key cellular processes, is commonly dysregulated in cancer and plays an important role in changes in DNA, RNA and histone methylation of cancer cells. Enzymes involved in serine synthesis have become potential therapeutic targets of great interest for cancer treatment [224, 225]. Inhibition of PHGDH, the enzyme involved in the first step of serine synthesis pathway, cooperates with serine and glycine depletion to suppresses cancer growth. In vivo, combination of PHGDH inhibition with serine starvation shows therapeutic efficacy against tumors that are resistant to diet or drug alone, indicating that inhibition of PHGDH will enhance the therapeutic effect of a serine depleted diet [224]. SHIN2, an inhibitor of SHMT which is another important enzyme involved in serine synthesis, increases survival in notch receptor 1 (NOTCH1)-driven mouse primary T-cell ALL in vivo. As methotrexate is standard treatment for T-cell ALL, the utility of combination of SHIN2 and methotrexate in this disease was explored. Low dose methotrexate sensitizes human T-cell ALL cells to SHIN2, and cells rendering methotrexate resistant in vitro show increased sensitivity to SHIN2. Further, SHIN2 and methotrexate synergistically increase survival in mouse primary T-cell ALL and in a human patient-derived xenograft. Collectively, SHMT inhibition provides a complementary strategy in the treatment of T-cell ALL [225]. Besides, since modulation of amino acid availability plays an important role in cancer treatment, amino acid transporters are attractive potential targets for anticancer therapy [35]. The SLC7A5 (also known as LAT1) is widely overexpressed in numerous human neoplasms. SLC7A5 block by JPH203 suppresses cell proliferation and mTORC1 signaling in human thyroid cancer cells. In vivo, JPH203 treatment induces tumor growth arrest in mouse model of thyroid cancer [226].
Additionally, Miller et al. [227] have synthesized a small molecule inhibitor that can act as an analog to block ACSS2 activity in vitro and in vivo. Pharmacologic inhibition of ACSS2 impairs TNBC growth, suggesting that ACSS2 may be an effective therapeutic target for the treatment of patients with TNBC.
A large number of studies have shown that the dietary components demonstrate anti-proliferative effect against cancer, which have been proposed as an adjuvant for cancer [105]. In melanoma, tumor cells often experience glutamine starvation, which promote cell dedifferentiation, whereas dietary glutamine supplementation significantly suppresses tumor growth, prolongs survival in melanoma mouse model, and increases the sensitivity to a BRAF inhibitor. Thus, these results offer evidence that glutamine supplementation can serve as a potential dietary intervention to block melanoma tumor growth and sensitize tumor to targeted therapy [228]. Jeon et al. [229] reported that methionine deprivation shows remarkable effects on the invasion and migration of cancer cells. Moreover, Bian and colleagues [63] showed that tumor cells disrupt methionine metabolism in CD8+ T cells, thereby leading to lower intracellular levels of methionine and SAM and, consequently, loss of histone H3K79me2. Loss of H3K79me2 results in low expression of signal transducer and activator of transcription 5 (STAT5) and impairs T cell immunity. Methionine supplementation improves the expression of H3K79me2 and STAT5 in T cells, which is accompanied by increased T cell immunity in tumor-bearing mice and patients with colon cancer. Taken together, cancer methionine consumption is an immune evasion mechanism, and dietary methionine supplementation may provide a potential immunotherapeutic approach [63]. However, dietary restriction of serine and glycine suppresses the growth of cancer cells, due to their dependence on exogenous serine [105, 224, 230, 231]. In addition, dietary isothiocyanates, such as sulforaphane, phenethyl isothiocyanate, benzyl isothiocyanate and allyl isothiocyanate, can inhibit cancer progression via modulating epigenome [232].
Conclusions and future perspectives
Connections between metabolism and epigenetic regulation are crucial to cellular adaptation to nutritional and physiological status. Cancer metabolic reprogramming responds to cellular metabolic needs by direct regulations of metabolic reactions and pathways. Moreover, metabolic reprogramming contributes to cancer cell growth, proliferation and other activities by regulating gene expression [3, 5, 29, 65]. A growing body of evidence demonstrates the crucial roles of cell metabolism in regulating epigenetic modifications, such as DNA, RNA and histone methylation as well as histone acetylation and O-GlcNAcylation, which then affect target gene expression. Here, we discussed the effects of several altered metabolites caused by cancer metabolic reprogramming on the changes of epigenetic modifications in cancer and their ties to cancer development and progression. In addition, the links between other common histone modifications and various metabolites can be seen in other reviews [36, 65]. However, metabolism and epigenetic modifications are dynamic and complex processes that involves in multiple pathways and metabolites. Furthermore, the effects of metabolites on epigenetic modifications may be complex and cell-specific, as exemplified by the study that α-KG affect the level of 5mC by regulating the expression DNMT3A and DNMT3B rather than TETs via the transcription factor orthodenticle homeobox 2 (OTX2) in embryonic stem cells [233]. Collectively, more studies should be conducted to clearly uncover the exact roles of various metabolites in epigenetic regulation.
The research strategies in systems biology will play important roles in completely uncovering the complexity of the connections between metabolic variability and flexibility and epigenetic modifications in cancer. Advances in the next-generation sequencing and computational biology have facilitated the development of novel experimental and computational methods, such as Perturb-seq [234], which can be utilized in the exploration of the connections between metabolism and epigenetic modifications. In addition, whether a metabolic change leads to a biological outcome due to a specific effect on epigenetic modifications, is still largely unknown due to the fact that metabolites involved in epigenetic regulation are also connected to larger metabolic networks that affect nearly all aspects of biological processes. Combining defined metabolic perturbations with CRISPR-Cas9-based technologies may be a good way to uncover underlying mechanisms [235].
Recently, acetylation of cytidine in mRNA has been identified and involves in promoting gastric cancer metastasis [236, 237]. Ye et al. [238] showed that the methylation of phosphatidylethanolamine (PE) is a major consumer of SAM. The induction of phospholipid biosynthetic genes is accompanied by induction of the enzyme that hydrolyzes SAH. Besides, the methylation of PE promotes the usage of SAM for the synthesis of cysteine and glutathione. Conversely, cells that lack of PE methylation accumulate SAM, which leads to histone hypermethylation [238]. In addition, NADP has been reported to directly bind to FTO, and then independently increase FTO activity, consequently leading to RNA m6A demethylation and adipogenesis [239]. However, whether these regulations can contribute to cancer needs further exploration.
Mechanistically understanding how changes in the availability of metabolites regulate specific target genes, is crucial to further elucidate the underlying mechanisms by which metabolite-driven epigenetic regulations response to altered metabolism and contribute to the phenotypes and activities of cancer cells. Cancer driver events, such as KRAS mutation, active mTORC and PI3K-AKT pathways, drive metabolic alterations and induce relevant oncogene expression via various metabolism-driven epigenetic modifications, consequently promoting cancer development [46, 78, 79]. In addition to the metabolic roles, metabolic enzymes can perform nonmetabolic functions and drive the pathologic progression of cancer. For instance, nuclear PKM2 acts as a protein kinase to phosphorylate histone H3 at T11 with PEP as a phosphate group donor, resulting in promoting histone H3K9 acetylation and the expression of CCND1 and MYC [139]. Besides, metabolic enzymes can form complexes with histone modification enzymes and transcription factors. Nucleus-localized MAT2A functions as a transcriptional modulator by interacting with chromatin regulators and supplying SAM to methyltransferases for histone methylation, subsequently regulating gene expression [65, 137]. Thus, identifying the compartment-specific interaction molecules, the mechanisms of nuclear entry and location, and the dynamic posttranslational modifications, is also vital to better reveal the roles of metabolic enzymes in gene regulation [28].
Additionally, deeper identifying the landscape of the connections between metabolism and epigenetic regulations will shed light on our understanding of the underlying biological mechanisms for the development of numerous diseases including cancer. Moreover, metabolic processes become the promising targets for novel anticancer strategy. Targeting altered metabolism in cancer cells might affect the effect of epigenetic therapies. Given that epigenetic modifications, such as methylation and acetylation, are closely linked to nutrient availability, daily nutrition intake might influence cancer progression and therapies [28, 105, 181]. In addition, disrupting the nuclear transport of metabolic enzymes is also expected to be effective approaches to counteract cancer development. Structurally elucidating the nuclear target protein and DNA of nuclear metabolic enzymes will facilitate the identification of precise interventions that interfere with their nuclear activities [65]. Drugs that target a variety of metabolic processes have been tested in preclinical cancer models. Several drugs can directly target metabolic enzymes, and others may have a role in inhibiting pathways that are crucial to supply nutrients to cancer cells [206]. An emerging issue is that cancer cells show metabolic plasticity to alter their metabolic profile during cancer progression, by which cancer cells could develop resistance to inhibition of a particular metabolic enzyme or pathway by activating compensatory pathways. Consequently, an efficient therapeutic strategy should involve simultaneously targeting multiple metabolic pathways or targeting a particular metabolic pathway in combination with therapies against other oncogenic pathways [240].
In summary, it is now clear that metabolic reprogramming plays an important role in epigenetic modifications, thereby significantly contributing to cancer development. In order to comprehensively understand the underlying mechanisms of tumorigenesis, more studies should continue to elucidate the complex connections between metabolic reprogramming and epigenetic regulations in cancer. Additionally, understanding these connections will greatly promote the development of novel effective cancer therapies and prevention strategies.
Footnotes
Research funding: This work was supported by grants from National Natural Science Foundation of China (91749205, 92049302, 32088101), China Ministry of Science and Technology (2020YFA0804000, 2016YFE0108700) and Shanghai Municipal Science and Technology Major Project (2017SHZDZX01) to J.D.J.H. The funding organizations played no role in the study design; in the collection, analysis, and interpretation of data; in the writing of the report; or in the decision to submit the report for publication.
Author contributions: G.W. analyzed the literatures and wrote the first draft. J.-D.J.H. conceived the study and revised the manuscript.
Competing interests: The authors declare no conflicts of interest.
Ethical approval: Not applicable.
Informed consent: Not applicable.
References
- 1.Saggese P, Sellitto A, Martinez CA, Giurato G, Nassa G, Rizzo F, et al. Metabolic regulation of epigenetic modifications and cell differentiation in cancer. Cancers (Basel) 2020;12:3788. doi: 10.3390/cancers12123788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Powell EE, Wong VW, Rinella M. Non-alcoholic fatty liver disease. Lancet. 2021;397:2212–24. doi: 10.1016/s0140-6736(20)32511-3. [DOI] [PubMed] [Google Scholar]
- 3.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–74. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 4.Warburg O. On respiratory impairment in cancer cells. Science. 1956;124:269–70. doi: 10.1126/science.124.3215.269. [DOI] [PubMed] [Google Scholar]
- 5.Pavlova NN, Thompson CB. The emerging hallmarks of cancer metabolism. Cell Metabol. 2016;23:27–47. doi: 10.1016/j.cmet.2015.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bian X, Liu R, Meng Y, Xing D, Xu D, Lu Z. Lipid metabolism and cancer. J Exp Med. 2021;218 doi: 10.1084/jem.20201606. e20201606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dang L, White DW, Gross S, Bennett BD, Bittinger MA, Driggers EM, et al. Cancer-associated IDH1 mutations produce 2-hydroxyglutarate. Nature. 2009;462:739–44. doi: 10.1038/nature08617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ward PS, Patel J, Wise DR, Abdel-Wahab O, Bennett BD, Coller HA, et al. The common feature of leukemia-associated IDH1 and IDH2 mutations is a neomorphic enzyme activity converting alpha-ketoglutarate to 2-hydroxyglutarate. Cancer Cell. 2010;17:225–34. doi: 10.1016/j.ccr.2010.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yan H, Parsons DW, Jin G, McLendon R, Rasheed BA, Yuan W, et al. IDH1 and IDH2 mutations in gliomas. N Engl J Med. 2009;360:765–73. doi: 10.1056/nejmoa0808710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Figueroa ME, Abdel-Wahab O, Lu C, Ward PS, Patel J, Shih A, et al. Leukemic IDH1 and IDH2 mutations result in a hypermethylation phenotype, disrupt TET2 function, and impair hematopoietic differentiation. Cancer Cell. 2010;18:553–67. doi: 10.1016/j.ccr.2010.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xu W, Yang H, Liu Y, Yang Y, Wang P, Kim SH, et al. Oncometabolite 2-hydroxyglutarate is a competitive inhibitor of alpha-ketoglutarate-dependent dioxygenases. Cancer Cell. 2011;19:17–30. doi: 10.1016/j.ccr.2010.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xia L, Oyang L, Lin J, Tan S, Han Y, Wu N, et al. The cancer metabolic reprogramming and immune response. Mol Cancer. 2021;20:28. doi: 10.1186/s12943-021-01316-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.DeBerardinis RJ, Mancuso A, Daikhin E, Nissim I, Yudkoff M, Wehrli S, et al. Beyond aerobic glycolysis: transformed cells can engage in glutamine metabolism that exceeds the requirement for protein and nucleotide synthesis. Proc Natl Acad Sci USA. 2007;104:19345–50. doi: 10.1073/pnas.0709747104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schug ZT, Peck B, Jones DT, Zhang Q, Grosskurth S, Alam IS, et al. Acetyl-CoA synthetase 2 promotes acetate utilization and maintains cancer cell growth under metabolic stress. Cancer Cell. 2015;27:57–71. doi: 10.1016/j.ccell.2014.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gao X, Lin SH, Ren F, Li JT, Chen JJ, Yao CB, et al. Acetate functions as an epigenetic metabolite to promote lipid synthesis under hypoxia. Nat Commun. 2016;7:11960. doi: 10.1038/ncomms11960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lu C, Thompson CB. Metabolic regulation of epigenetics. Cell Metabol. 2012;16:9–17. doi: 10.1016/j.cmet.2012.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jiang X, Liu B, Nie Z, Duan L, Xiong Q, Jin Z, et al. The role of m6A modification in the biological functions and diseases. Signal Transduct Target Ther. 2021;6:74. doi: 10.1038/s41392-020-00450-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lu Y, Chan YT, Tan HY, Li S, Wang N, Feng Y. Epigenetic regulation in human cancer: the potential role of epi-drug in cancer therapy. Mol Cancer. 2020;19:79. doi: 10.1186/s12943-020-01197-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mancarella D, Plass C. Epigenetic signatures in cancer: proper controls, current challenges and the potential for clinical translation. Genome Med. 2021;13:23. doi: 10.1186/s13073-021-00837-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Russler-Germain DA, Spencer DH, Young MA, Lamprecht TL, Miller CA, Fulton R, et al. The R882H DNMT3A mutation associated with AML dominantly inhibits wild-type DNMT3A by blocking its ability to form active tetramers. Cancer Cell. 2014;25:442–54. doi: 10.1016/j.ccr.2014.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Blanco E, Gonzalez-Ramirez M, Alcaine-Colet A, Aranda S, Di Croce L. The bivalent genome: characterization, structure, and regulation. Trends Genet. 2020;36:118–31. doi: 10.1016/j.tig.2019.11.004. [DOI] [PubMed] [Google Scholar]
- 22.Zhao Z, Shilatifard A. Epigenetic modifications of histones in cancer. Genome Biol. 2019;20:245. doi: 10.1186/s13059-019-1870-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hahn MA, Li AX, Wu X, Yang R, Drew DA, Rosenberg DW, et al. Loss of the polycomb mark from bivalent promoters leads to activation of cancer-promoting genes in colorectal tumors. Cancer Res. 2014;74:3617–29. doi: 10.1158/0008-5472.can-13-3147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Malouf GG, Taube JH, Lu Y, Roysarkar T, Panjarian S, Estecio MR, et al. Architecture of epigenetic reprogramming following Twist1-mediated epithelial-mesenchymal transition. Genome Biol. 2013;14:R144. doi: 10.1186/gb-2013-14-12-r144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li Y, Seto E. HDACs and HDAC inhibitors in cancer development and therapy. Cold Spring Harb Perspect Med. 2016;6:a026831. doi: 10.1101/cshperspect.a026831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li Z, Weng H, Su R, Weng X, Zuo Z, Li C, et al. FTO plays an oncogenic role in acute myeloid leukemia as a N(6)-methyladenosine RNA demethylase. Cancer Cell. 2017;31:127–41. doi: 10.1016/j.ccell.2016.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Han J, Wang JZ, Yang X, Yu H, Zhou R, Lu HC, et al. METTL3 promote tumor proliferation of bladder cancer by accelerating pri-miR221/222 maturation in m6A-dependent manner. Mol Cancer. 2019;18:110. doi: 10.1186/s12943-019-1036-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Campbell SL, Wellen KE. Metabolic signaling to the nucleus in cancer. Mol Cell. 2018;71:398–408. doi: 10.1016/j.molcel.2018.07.015. [DOI] [PubMed] [Google Scholar]
- 29.Wong CC, Qian Y, Yu J. Interplay between epigenetics and metabolism in oncogenesis: mechanisms and therapeutic approaches. Oncogene. 2017;36:3359–74. doi: 10.1038/onc.2016.485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Losman JA, Koivunen P, Kaelin WG. 2-Oxoglutarate-dependent dioxygenases in cancer. Nat Rev Cancer. 2020;20:710–26. doi: 10.1038/s41568-020-00303-3. [DOI] [PubMed] [Google Scholar]
- 31.Su R, Dong L, Li C, Nachtergaele S, Wunderlich M, Qing Y, et al. R-2HG exhibits anti-tumor activity by targeting FTO/m(6)A/MYC/CEBPA signaling. Cell. 2018;172:90–105. doi: 10.1016/j.cell.2017.11.031. e23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Qing Y, Dong L, Gao L, Li C, Li Y, Han L, et al. R-2-hydroxyglutarate attenuates aerobic glycolysis in leukemia by targeting the FTO/m(6)A/PFKP/LDHB axis. Mol Cell. 2021;81:922–39. doi: 10.1016/j.molcel.2020.12.026. e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee P, Chandel NS, Simon MC. Cellular adaptation to hypoxia through hypoxia inducible factors and beyond. Nat Rev Mol Cell Biol. 2020;21:268–83. doi: 10.1038/s41580-020-0227-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Beyer S, Kristensen MM, Jensen KS, Johansen JV, Staller P. The histone demethylases JMJD1A and JMJD2B are transcriptional targets of hypoxia-inducible factor HIF. J Biol Chem. 2008;283:36542–52. doi: 10.1074/jbc.m804578200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pranzini E, Pardella E, Paoli P, Fendt SM, Taddei ML. Metabolic reprogramming in anticancer drug resistance: a focus on amino acids. Trends Cancer. 2021;7:682–99. doi: 10.1016/j.trecan.2021.02.004. [DOI] [PubMed] [Google Scholar]
- 36.Dai Z, Ramesh V, Locasale JW. The evolving metabolic landscape of chromatin biology and epigenetics. Nat Rev Genet. 2020;21:737–53. doi: 10.1038/s41576-020-0270-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhu J, Thompson CB. Metabolic regulation of cell growth and proliferation. Nat Rev Mol Cell Biol. 2019;20:436–50. doi: 10.1038/s41580-019-0123-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ferrari A, Longo R, Silva R, Mitro N, Caruso D, De Fabiani E, et al. Epigenome modifiers and metabolic rewiring: new frontiers in therapeutics. Pharmacol Ther. 2019;193:178–93. doi: 10.1016/j.pharmthera.2018.08.008. [DOI] [PubMed] [Google Scholar]
- 39.Locasale JW. Serine, glycine and one-carbon units: cancer metabolism in full circle. Nat Rev Cancer. 2013;13:572–83. doi: 10.1038/nrc3557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mentch SJ, Mehrmohamadi M, Huang L, Liu X, Gupta D, Mattocks D, et al. Histone methylation dynamics and gene regulation occur through the sensing of one-carbon metabolism. Cell Metabol. 2015;22:861–73. doi: 10.1016/j.cmet.2015.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jia G, Fu Y, He C. Reversible RNA adenosine methylation in biological regulation. Trends Genet. 2013;29:108–15. doi: 10.1016/j.tig.2012.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang P, Doxtader KA, Nam Y. Structural basis for cooperative function of Mettl3 and Mettl14 methyltransferases. Mol Cell. 2016;63:306–17. doi: 10.1016/j.molcel.2016.05.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Calvisi DF, Simile MM, Ladu S, Pellegrino R, De Murtas V, Pinna F, et al. Altered methionine metabolism and global DNA methylation in liver cancer: relationship with genomic instability and prognosis. Int J Cancer. 2007;121:2410–20. doi: 10.1002/ijc.22940. [DOI] [PubMed] [Google Scholar]
- 44.Cai J, Mao Z, Hwang JJ, Lu SC. Differential expression of methionine adenosyltransferase genes influences the rate of growth of human hepatocellular carcinoma cells. Cancer Res. 1998;58:1444–50. [PubMed] [Google Scholar]
- 45.Chen H, Xia M, Lin M, Yang H, Kuhlenkamp J, Li T, et al. Role of methionine adenosyltransferase 2A and S-adenosylmethionine in mitogen-induced growth of human colon cancer cells. Gastroenterology. 2007;133:207–18. doi: 10.1053/j.gastro.2007.03.114. [DOI] [PubMed] [Google Scholar]
- 46.Villa E, Sahu U, O’Hara BP, Ali ES, Helmin KA, Asara JM, et al. mTORC1 stimulates cell growth through SAM synthesis and m(6)A mRNA-dependent control of protein synthesis. Mol Cell. 2021;81:2076–93. doi: 10.1016/j.molcel.2021.03.009. e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yang M, Vousden KH. Serine and one-carbon metabolism in cancer. Nat Rev Cancer. 2016;16:650–62. doi: 10.1038/nrc.2016.81. [DOI] [PubMed] [Google Scholar]
- 48.Ducker GS, Rabinowitz JD. One-carbon metabolism in health and disease. Cell Metabol. 2017;25:27–42. doi: 10.1016/j.cmet.2016.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Reina-Campos M, Linares JF, Duran A, Cordes T, L’Hermitte A, Badur MG, et al. Increased serine and one-carbon pathway metabolism by PKClambda/iota deficiency promotes neuroendocrine prostate cancer. Cancer Cell. 2019;35:385–400. doi: 10.1016/j.ccell.2019.01.018. e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kim D, Fiske BP, Birsoy K, Freinkman E, Kami K, Possemato RL, et al. SHMT2 drives glioma cell survival in ischaemia but imposes a dependence on glycine clearance. Nature. 2015;520:363–7. doi: 10.1038/nature14363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nilsson R, Jain M, Madhusudhan N, Sheppard NG, Strittmatter L, Kampf C, et al. Metabolic enzyme expression highlights a key role for MTHFD2 and the mitochondrial folate pathway in cancer. Nat Commun. 2014;5:3128. doi: 10.1038/ncomms4128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Morscher RJ, Ducker GS, Li SH, Mayer JA, Gitai Z, Sperl W, et al. Mitochondrial translation requires folate-dependent tRNA methylation. Nature. 2018;554:128–32. doi: 10.1038/nature25460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Su A, Ling F, Vaganay C, Sodaro G, Benaksas C, Dal Bello R, et al. The folate cycle enzyme MTHFR is a critical regulator of cell response to MYC-targeting therapies. Cancer Discov. 2020;10:1894–911. doi: 10.1158/2159-8290.cd-19-0970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Coppede F, Migheli F, Lopomo A, Failli A, Legitimo A, Consolini R, et al. Gene promoter methylation in colorectal cancer and healthy adjacent mucosa specimens: correlation with physiological and pathological characteristics, and with biomarkers of one-carbon metabolism. Epigenetics. 2014;9:621–33. doi: 10.4161/epi.27956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Roberti A, Fernandez AF, Fraga MF. Nicotinamide N-methyltransferase: at the crossroads between cellular metabolism and epigenetic regulation. Mol Metabol. 2021;45:101165. doi: 10.1016/j.molmet.2021.101165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ulanovskaya OA, Zuhl AM, Cravatt BF. NNMT promotes epigenetic remodeling in cancer by creating a metabolic methylation sink. Nat Chem Biol. 2013;9:300–6. doi: 10.1038/nchembio.1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gao X, Locasale JW. Serine metabolism links tumor suppression to the epigenetic landscape. Cell Metabol. 2016;24:777–9. doi: 10.1016/j.cmet.2016.11.012. [DOI] [PubMed] [Google Scholar]
- 58.Locasale JW, Grassian AR, Melman T, Lyssiotis CA, Mattaini KR, Bass AJ, et al. Phosphoglycerate dehydrogenase diverts glycolytic flux and contributes to oncogenesis. Nat Genet. 2011;43:869–74. doi: 10.1038/ng.890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bialopiotrowicz E, Noyszewska-Kania M, Kachamakova-Trojanowska N, Loboda A, Cybulska M, Grochowska A, et al. Serine biosynthesis pathway supports MYC-miR-494-EZH2 feed-forward circuit necessary to maintain metabolic and epigenetic reprogramming of burkitt lymphoma cells. Cancers (Basel) 2020;12:580. doi: 10.3390/cancers12030580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Maddocks OD, Labuschagne CF, Adams PD, Vousden KH. Serine metabolism supports the methionine cycle and DNA/RNA methylation through De Novo ATP synthesis in cancer cells. Mol Cell. 2016;61:210–21. doi: 10.1016/j.molcel.2015.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Martinez-Chantar ML, Vazquez-Chantada M, Ariz U, Martinez N, Varela M, Luka Z, et al. Loss of the glycine N-methyltransferase gene leads to steatosis and hepatocellular carcinoma in mice. Hepatology. 2008;47:1191–9. doi: 10.1002/hep.22159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Dann SG, Ryskin M, Barsotti AM, Golas J, Shi C, Miranda M, et al. Reciprocal regulation of amino acid import and epigenetic state through Lat1 and EZH2. EMBO J. 2015;34:1773–85. doi: 10.15252/embj.201488166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bian Y, Li W, Kremer DM, Sajjakulnukit P, Li S, Crespo J, et al. Cancer SLC43A2 alters T cell methionine metabolism and histone methylation. Nature. 2020;585:277–82. doi: 10.1038/s41586-020-2682-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Menga A, Palmieri EM, Cianciulli A, Infantino V, Mazzone M, Scilimati A, et al. SLC25A26 overexpression impairs cell function via mtDNA hypermethylation and rewiring of methyl metabolism. FEBS J. 2017;284:967–84. doi: 10.1111/febs.14028. [DOI] [PubMed] [Google Scholar]
- 65.Li X, Egervari G, Wang Y, Berger SL, Lu Z. Regulation of chromatin and gene expression by metabolic enzymes and metabolites. Nat Rev Mol Cell Biol. 2018;19:563–78. doi: 10.1038/s41580-018-0029-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wang Z, Long H, Chang C, Zhao M, Lu Q. Crosstalk between metabolism and epigenetic modifications in autoimmune diseases: a comprehensive overview. Cell Mol Life Sci. 2018;75:3353–69. doi: 10.1007/s00018-018-2864-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sivanand S, Viney I, Wellen KE. Spatiotemporal control of acetyl-CoA metabolism in chromatin regulation. Trends Biochem Sci. 2018;43:61–74. doi: 10.1016/j.tibs.2017.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Fan J, Krautkramer KA, Feldman JL, Denu JM. Metabolic regulation of histone post-translational modifications. ACS Chem Biol. 2015;10:95–108. doi: 10.1021/cb500846u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kinnaird A, Zhao S, Wellen KE, Michelakis ED. Metabolic control of epigenetics in cancer. Nat Rev Cancer. 2016;16:694–707. doi: 10.1038/nrc.2016.82. [DOI] [PubMed] [Google Scholar]
- 70.Lee JV, Carrer A, Shah S, Snyder NW, Wei S, Venneti S, et al. Akt-dependent metabolic reprogramming regulates tumor cell histone acetylation. Cell Metabol. 2014;20:306–19. doi: 10.1016/j.cmet.2014.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Koppenol WH, Bounds PL, Dang CV. Otto Warburg’s contributions to current concepts of cancer metabolism. Nat Rev Cancer. 2011;11:325–37. doi: 10.1038/nrc3038. [DOI] [PubMed] [Google Scholar]
- 72.Sutendra G, Kinnaird A, Dromparis P, Paulin R, Stenson TH, Haromy A, et al. A nuclear pyruvate dehydrogenase complex is important for the generation of acetyl-CoA and histone acetylation. Cell. 2014;158:84–97. doi: 10.1016/j.cell.2014.04.046. [DOI] [PubMed] [Google Scholar]
- 73.Cluntun AA, Huang H, Dai L, Liu X, Zhao Y, Locasale JW. The rate of glycolysis quantitatively mediates specific histone acetylation sites. Cancer Metabol. 2015;3:10. doi: 10.1186/s40170-015-0135-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Echeverria C, Nualart F, Ferrada L, Smith GJ, Godoy AS. Hexose transporters in cancer: from multifunctionality to diagnosis and therapy. Trends Endocrinol Metabol. 2021;32:198–211. doi: 10.1016/j.tem.2020.12.006. [DOI] [PubMed] [Google Scholar]
- 75.Stacpoole PW. Therapeutic targeting of the pyruvate dehydrogenase complex/pyruvate dehydrogenase kinase (PDC/PDK) axis in cancer. J Natl Cancer Inst. 2017;109:djx071. doi: 10.1093/jnci/djx071. [DOI] [PubMed] [Google Scholar]
- 76.Chen J, Guccini I, Di Mitri D, Brina D, Revandkar A, Sarti M, et al. Compartmentalized activities of the pyruvate dehydrogenase complex sustain lipogenesis in prostate cancer. Nat Genet. 2018;50:219–28. doi: 10.1038/s41588-017-0026-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Icard P, Lincet H. The reduced concentration of citrate in cancer cells: an indicator of cancer aggressiveness and a possible therapeutic target. Drug Resist Updates. 2016;29:47–53. doi: 10.1016/j.drup.2016.09.003. [DOI] [PubMed] [Google Scholar]
- 78.Migita T, Narita T, Nomura K, Miyagi E, Inazuka F, Matsuura M, et al. ATP citrate lyase: activation and therapeutic implications in non-small cell lung cancer. Cancer Res. 2008;68:8547–54. doi: 10.1158/0008-5472.can-08-1235. [DOI] [PubMed] [Google Scholar]
- 79.Carrer A, Trefely S, Zhao S, Campbell SL, Norgard RJ, Schultz KC, et al. Acetyl-CoA metabolism supports multistep pancreatic tumorigenesis. Cancer Discov. 2019;9:416–35. doi: 10.1158/2159-8290.cd-18-0567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Brown JM, Wilson WR. Exploiting tumour hypoxia in cancer treatment. Nat Rev Cancer. 2004;4:437–47. doi: 10.1038/nrc1367. [DOI] [PubMed] [Google Scholar]
- 81.Papandreou I, Cairns RA, Fontana L, Lim AL, Denko NC. HIF-1 mediates adaptation to hypoxia by actively downregulating mitochondrial oxygen consumption. Cell Metabol. 2006;3:187–97. doi: 10.1016/j.cmet.2006.01.012. [DOI] [PubMed] [Google Scholar]
- 82.Kim JW, Tchernyshyov I, Semenza GL, Dang CV. HIF-1-mediated expression of pyruvate dehydrogenase kinase: a metabolic switch required for cellular adaptation to hypoxia. Cell Metabol. 2006;3:177–85. doi: 10.1016/j.cmet.2006.02.002. [DOI] [PubMed] [Google Scholar]
- 83.Kamphorst JJ, Chung MK, Fan J, Rabinowitz JD. Quantitative analysis of acetyl-CoA production in hypoxic cancer cells reveals substantial contribution from acetate. Cancer Metabol. 2014;2:23. doi: 10.1186/2049-3002-2-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Bulusu V, Tumanov S, Michalopoulou E, van den Broek NJ, MacKay G, Nixon C, et al. Acetate recapturing by nuclear acetyl-CoA synthetase 2 prevents loss of histone acetylation during oxygen and serum limitation. Cell Rep. 2017;18:647–58. doi: 10.1016/j.celrep.2016.12.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Zhang J, Duan H, Feng Z, Han X, Gu C. Acetyl-CoA synthetase 3 promotes bladder cancer cell growth under metabolic stress. Oncogenesis. 2020;9:46. doi: 10.1038/s41389-020-0230-3. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 86.Lu M, Zhu WW, Wang X, Tang JJ, Zhang KL, Yu GY, et al. ACOT12-dependent alteration of acetyl-CoA drives hepatocellular carcinoma metastasis by epigenetic induction of epithelial-mesenchymal transition. Cell Metabol. 2019;29:886–900. doi: 10.1016/j.cmet.2018.12.019. e5. [DOI] [PubMed] [Google Scholar]
- 87.Patil MD, Bhaumik J, Babykutty S, Banerjee UC, Fukumura D. Arginine dependence of tumor cells: targeting a chink in cancer’s armor. Oncogene. 2016;35:4957–72. doi: 10.1038/onc.2016.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Chen CL, Hsu SC, Chung TY, Chu CY, Wang HJ, Hsiao PW, et al. Arginine is an epigenetic regulator targeting TEAD4 to modulate OXPHOS in prostate cancer cells. Nat Commun. 2021;12:2398. doi: 10.1038/s41467-021-22652-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Donohoe DR, Collins LB, Wali A, Bigler R, Sun W, Bultman SJ. The Warburg effect dictates the mechanism of butyrate-mediated histone acetylation and cell proliferation. Mol Cell. 2012;48:612–26. doi: 10.1016/j.molcel.2012.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Huang CK, Chang PH, Kuo WH, Chen CL, Jeng YM, Chang KJ, et al. Adipocytes promote malignant growth of breast tumours with monocarboxylate transporter 2 expression via beta-hydroxybutyrate. Nat Commun. 2017;8:14706. doi: 10.1038/ncomms14706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Gatenby RA, Gillies RJ. A microenvironmental model of carcinogenesis. Nat Rev Cancer. 2008;8:56–61. doi: 10.1038/nrc2255. [DOI] [PubMed] [Google Scholar]
- 92.Corbet C, Pinto A, Martherus R, Santiago de Jesus JP, Polet F, Feron O. Acidosis drives the reprogramming of fatty acid metabolism in cancer cells through changes in mitochondrial and histone acetylation. Cell Metabol. 2016;24:311–23. doi: 10.1016/j.cmet.2016.07.003. [DOI] [PubMed] [Google Scholar]
- 93.Herr CQ, Hausinger RP. Amazing diversity in biochemical roles of Fe(II)/2-oxoglutarate oxygenases. Trends Biochem Sci. 2018;43:517–32. doi: 10.1016/j.tibs.2018.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Xiao M, Yang H, Xu W, Ma S, Lin H, Zhu H, et al. Inhibition of alpha-KG-dependent histone and DNA demethylases by fumarate and succinate that are accumulated in mutations of FH and SDH tumor suppressors. Genes Dev. 2012;26:1326–38. doi: 10.1101/gad.191056.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Dang L, Yen K, Attar EC. IDH mutations in cancer and progress toward development of targeted therapeutics. Ann Oncol. 2016;27:599–608. doi: 10.1093/annonc/mdw013. [DOI] [PubMed] [Google Scholar]
- 96.Eniafe J, Jiang S. The functional roles of TCA cycle metabolites in cancer. Oncogene. 2021;40:3351–63. doi: 10.1038/s41388-020-01639-8. [DOI] [PubMed] [Google Scholar]
- 97.Gill AJ. Succinate dehydrogenase (SDH)-deficient neoplasia. Histopathology. 2018;72:106–16. doi: 10.1111/his.13277. [DOI] [PubMed] [Google Scholar]
- 98.Schmidt C, Sciacovelli M, Frezza C. Fumarate hydratase in cancer: a multifaceted tumour suppressor. Semin Cell Dev Biol. 2020;98:15–25. doi: 10.1016/j.semcdb.2019.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Liu Y, Yang C. Oncometabolite in cancer: current understanding and challenges. Cancer Res. 2021;81:2820–3. doi: 10.1158/0008-5472.can-20-3730. [DOI] [PubMed] [Google Scholar]
- 100.Lu C, Ward PS, Kapoor GS, Rohle D, Turcan S, Abdel-Wahab O, et al. IDH mutation impairs histone demethylation and results in a block to cell differentiation. Nature. 2012;483:474–8. doi: 10.1038/nature10860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Turcan S, Rohle D, Goenka A, Walsh LA, Fang F, Yilmaz E, et al. IDH1 mutation is sufficient to establish the glioma hypermethylator phenotype. Nature. 2012;483:479–83. doi: 10.1038/nature10866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Kernytsky A, Wang F, Hansen E, Schalm S, Straley K, Gliser C, et al. IDH2 mutation-induced histone and DNA hypermethylation is progressively reversed by small-molecule inhibition. Blood. 2015;125:296–303. doi: 10.1182/blood-2013-10-533604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Yang X, Qian K. Protein O-GlcNAcylation: emerging mechanisms and functions. Nat Rev Mol Cell Biol. 2017;18:452–65. doi: 10.1038/nrm.2017.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Wellen KE, Lu C, Mancuso A, Lemons JM, Ryczko M, Dennis JW, et al. The hexosamine biosynthetic pathway couples growth factor-induced glutamine uptake to glucose metabolism. Genes Dev. 2010;24:2784–99. doi: 10.1101/gad.1985910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Sun L, Zhang H, Gao P. Metabolic reprogramming and epigenetic modifications on the path to cancer. Protein Cell. 2021 doi: 10.1007/s13238-021-00846-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Sakabe K, Wang Z, Hart GW. Beta-N-acetylglucosamine (O-GlcNAc) is part of the histone code. Proc Natl Acad Sci USA. 2010;107:19915–20. doi: 10.1073/pnas.1009023107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Wu D, Cai Y, Jin J. Potential coordination role between O-GlcNAcylation and epigenetics. Protein Cell. 2017;8:713–23. doi: 10.1007/s13238-017-0416-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Lercher L, Raj R, Patel NA, Price J, Mohammed S, Robinson CV, et al. Generation of a synthetic GlcNAcylated nucleosome reveals regulation of stability by H2A-Thr101 GlcNAcylation. Nat Commun. 2015;6:7978. doi: 10.1038/ncomms8978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Fujiki R, Hashiba W, Sekine H, Yokoyama A, Chikanishi T, Ito S, et al. GlcNAcylation of histone H2B facilitates its monoubiquitination. Nature. 2011;480:557–60. doi: 10.1038/nature10656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Sakabe K, Hart GW. O-GlcNAc transferase regulates mitotic chromatin dynamics. J Biol Chem. 2010;285:34460–8. doi: 10.1074/jbc.m110.158170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Deplus R, Delatte B, Schwinn MK, Defrance M, Mendez J, Murphy N, et al. TET2 and TET3 regulate GlcNAcylation and H3K4 methylation through OGT and SET1/COMPASS. EMBO J. 2013;32:645–55. doi: 10.1038/emboj.2012.357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Vella P, Scelfo A, Jammula S, Chiacchiera F, Williams K, Cuomo A, et al. Tet proteins connect the O-linked N-acetylglucosamine transferase Ogt to chromatin in embryonic stem cells. Mol Cell. 2013;49:645–56. doi: 10.1016/j.molcel.2012.12.019. [DOI] [PubMed] [Google Scholar]
- 113.Deen AJ, Arasu UT, Pasonen-Seppanen S, Hassinen A, Takabe P, Wojciechowski S, et al. UDP-sugar substrates of HAS3 regulate its O-GlcNAcylation, intracellular traffic, extracellular shedding and correlate with melanoma progression. Cell Mol Life Sci. 2016;73:3183–204. doi: 10.1007/s00018-016-2158-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Yang C, Peng P, Li L, Shao M, Zhao J, Wang L, et al. High expression of GFAT1 predicts poor prognosis in patients with pancreatic cancer. Sci Rep. 2016;6:39044. doi: 10.1038/srep39044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Ying H, Kimmelman AC, Lyssiotis CA, Hua S, Chu GC, Fletcher-Sananikone E, et al. Oncogenic Kras maintains pancreatic tumors through regulation of anabolic glucose metabolism. Cell. 2012;149:656–70. doi: 10.1016/j.cell.2012.01.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Guillaumond F, Leca J, Olivares O, Lavaut MN, Vidal N, Berthezene P, et al. Strengthened glycolysis under hypoxia supports tumor symbiosis and hexosamine biosynthesis in pancreatic adenocarcinoma. Proc Natl Acad Sci USA. 2013;110:3919–24. doi: 10.1073/pnas.1219555110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Caldwell SA, Jackson SR, Shahriari KS, Lynch TP, Sethi G, Walker S, et al. Nutrient sensor O-GlcNAc transferase regulates breast cancer tumorigenesis through targeting of the oncogenic transcription factor FoxM1. Oncogene. 2010;29:2831–42. doi: 10.1038/onc.2010.41. [DOI] [PubMed] [Google Scholar]
- 118.Lynch TP, Ferrer CM, Jackson SR, Shahriari KS, Vosseller K, Reginato MJ. Critical role of O-linked beta-N-acetylglucosamine transferase in prostate cancer invasion, angiogenesis, and metastasis. J Biol Chem. 2012;287:11070–81. doi: 10.1074/jbc.m111.302547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Li X, Wu Z, He J, Jin Y, Chu C, Cao Y, et al. OGT regulated O-GlcNAcylation promotes papillary thyroid cancer malignancy via activating YAP. Oncogene. 2021;40:4859–71. doi: 10.1038/s41388-021-01901-7. [DOI] [PubMed] [Google Scholar]
- 120.Yi W, Clark PM, Mason DE, Keenan MC, Hill C, Goddard WA, et al. Phosphofructokinase 1 glycosylation regulates cell growth and metabolism. Science. 2012;337:975–80. doi: 10.1126/science.1222278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Wang T, Yu Q, Li J, Hu B, Zhao Q, Ma C, et al. O-GlcNAcylation of fumarase maintains tumour growth under glucose deficiency. Nat Cell Biol. 2017;19:833–43. doi: 10.1038/ncb3562. [DOI] [PubMed] [Google Scholar]
- 122.Zhu G, Tao T, Zhang D, Liu X, Qiu H, Han L, et al. O-GlcNAcylation of histone deacetylases 1 in hepatocellular carcinoma promotes cancer progression. Glycobiology. 2016;26:820–33. doi: 10.1093/glycob/cww025. [DOI] [PubMed] [Google Scholar]
- 123.Chu CS, Lo PW, Yeh YH, Hsu PH, Peng SH, Teng YC, et al. O-GlcNAcylation regulates EZH2 protein stability and function. Proc Natl Acad Sci USA. 2014;111:1355–60. doi: 10.1073/pnas.1323226111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Rossetto D, Avvakumov N, Cote J. Histone phosphorylation: a chromatin modification involved in diverse nuclear events. Epigenetics. 2012;7:1098–108. doi: 10.4161/epi.21975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Xiao A, Li H, Shechter D, Ahn SH, Fabrizio LA, Erdjument-Bromage H, et al. WSTF regulates the H2A.X DNA damage response via a novel tyrosine kinase activity. Nature. 2009;457:57–62. doi: 10.1038/nature07668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Fernandez-Capetillo O, Allis CD, Nussenzweig A. Phosphorylation of histone H2B at DNA double-strand breaks. J Exp Med. 2004;199:1671–7. doi: 10.1084/jem.20032247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Metzger E, Yin N, Wissmann M, Kunowska N, Fischer K, Friedrichs N, et al. Phosphorylation of histone H3 at threonine 11 establishes a novel chromatin mark for transcriptional regulation. Nat Cell Biol. 2008;10:53–60. doi: 10.1038/ncb1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Metzger E, Imhof A, Patel D, Kahl P, Hoffmeyer K, Friedrichs N, et al. Phosphorylation of histone H3T6 by PKCbeta(I) controls demethylation at histone H3K4. Nature. 2010;464:792–6. doi: 10.1038/nature08839. [DOI] [PubMed] [Google Scholar]
- 129.Kumar R, Deivendran S, Santhoshkumar TR, Pillai MR. Signaling coupled epigenomic regulation of gene expression. Oncogene. 2017;36:5917–26. doi: 10.1038/onc.2017.201. [DOI] [PubMed] [Google Scholar]
- 130.Aihara H, Nakagawa T, Mizusaki H, Yoneda M, Kato M, Doiguchi M, et al. Histone H2A T120 phosphorylation promotes oncogenic transformation via upregulation of cyclin D1. Mol Cell. 2016;64:176–88. doi: 10.1016/j.molcel.2016.09.012. [DOI] [PubMed] [Google Scholar]
- 131.Kong D, Ying B, Zhang J, Ying H. PCAF regulates H3 phosphorylation and promotes autophagy in osteosarcoma cells. Biomed Pharmacother. 2019;118:109395. doi: 10.1016/j.biopha.2019.109395. [DOI] [PubMed] [Google Scholar]
- 132.Kim JY, Banerjee T, Vinckevicius A, Luo Q, Parker JB, Baker MR, et al. A role for WDR5 in integrating threonine 11 phosphorylation to lysine 4 methylation on histone H3 during androgen signaling and in prostate cancer. Mol Cell. 2014;54:613–25. doi: 10.1016/j.molcel.2014.03.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Hardie DG, Schaffer BE, Brunet A. AMPK: an energy-sensing pathway with multiple inputs and outputs. Trends Cell Biol. 2016;26:190–201. doi: 10.1016/j.tcb.2015.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Gongol B, Sari I, Bryant T, Rosete G, Marin T. AMPK: an epigenetic landscape modulator. Int J Mol Sci. 2018;19:3238. doi: 10.3390/ijms19103238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Bungard D, Fuerth BJ, Zeng PY, Faubert B, Maas NL, Viollet B, et al. Signaling kinase AMPK activates stress-promoted transcription via histone H2B phosphorylation. Science. 2010;329:1201–5. doi: 10.1126/science.1191241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Lu Z, Hunter T. Metabolic kinases moonlighting as protein kinases. Trends Biochem Sci. 2018;43:301–10. doi: 10.1016/j.tibs.2018.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Xu D, Shao F, Bian X, Meng Y, Liang T, Lu Z. The evolving landscape of noncanonical functions of metabolic enzymes in cancer and other pathologies. Cell Metabol. 2021;33:33–50. doi: 10.1016/j.cmet.2020.12.015. [DOI] [PubMed] [Google Scholar]
- 138.Yang W, Xia Y, Cao Y, Zheng Y, Bu W, Zhang L, et al. EGFR-induced and PKCε monoubiquitylation-dependent NF-κB activation upregulates PKM2 expression and promotes tumorigenesis. Mol Cell. 2012;48:771–84. doi: 10.1016/j.molcel.2012.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Yang W, Xia Y, Hawke D, Li X, Liang J, Xing D, et al. PKM2 phosphorylates histone H3 and promotes gene transcription and tumorigenesis. Cell. 2012;150:685–96. doi: 10.1016/j.cell.2012.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Yang W, Zheng Y, Xia Y, Ji H, Chen X, Guo F, et al. ERK1/2-dependent phosphorylation and nuclear translocation of PKM2 promotes the Warburg effect. Nat Cell Biol. 2012;14:1295–304. doi: 10.1038/ncb2629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Yang W, Xia Y, Ji H, Zheng Y, Liang J, Huang W, et al. Nuclear PKM2 regulates beta-catenin transactivation upon EGFR activation. Nature. 2011;480:118–22. doi: 10.1038/nature10598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Wang X, Liang C, Yao X, Yang RH, Zhang ZS, Liu FY, et al. PKM2-induced the phosphorylation of histone H3 contributes to EGF-Mediated PD-L1 transcription in HCC. Front Pharmacol. 2020;11:577108. doi: 10.3389/fphar.2020.577108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Xia L, Qin K, Wang XR, Wang XL, Zhou AW, Chen GQ, et al. Pyruvate kinase M2 phosphorylates H2AX and promotes genomic instability in human tumor cells. Oncotarget. 2017;8:109120–34. doi: 10.18632/oncotarget.22621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Brand A, Singer K, Koehl GE, Kolitzus M, Schoenhammer G, Thiel A, et al. LDHA-associated lactic acid production blunts tumor immunosurveillance by T and NK cells. Cell Metabol. 2016;24:657–71. doi: 10.1016/j.cmet.2016.08.011. [DOI] [PubMed] [Google Scholar]
- 145.Faubert B, Li KY, Cai L, Hensley CT, Kim J, Zacharias LG, et al. Lactate metabolism in human lung tumors. Cell. 2017;171:358–71. doi: 10.1016/j.cell.2017.09.019. e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Hui S, Ghergurovich JM, Morscher RJ, Jang C, Teng X, Lu W, et al. Glucose feeds the TCA cycle via circulating lactate. Nature. 2017;551:115–8. doi: 10.1038/nature24057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Latham T, Mackay L, Sproul D, Karim M, Culley J, Harrison DJ, et al. Lactate, a product of glycolytic metabolism, inhibits histone deacetylase activity and promotes changes in gene expression. Nucleic Acids Res. 2012;40:4794–803. doi: 10.1093/nar/gks066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Zhang D, Tang Z, Huang H, Zhou G, Cui C, Weng Y, et al. Metabolic regulation of gene expression by histone lactylation. Nature. 2019;574:575–80. doi: 10.1038/s41586-019-1678-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Dichtl S, Lindenthal L, Zeitler L, Behnke K, Schlosser D, Strobl B, et al. Lactate and IL6 define separable paths of inflammatory metabolic adaptation. Sci Adv. 2021;7 doi: 10.1126/sciadv.abg3505. eabg3505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Yang K, Fan M, Wang X, Xu J, Wang Y, Tu F, et al. Lactate promotes macrophage HMGB1 lactylation, acetylation, and exosomal release in polymicrobial sepsis. Cell Death Differ. 2021 doi: 10.1038/s41418-021-00841-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Li L, Chen K, Wang T, Wu Y, Xing G, Chen M, et al. Glis1 facilitates induction of pluripotency via an epigenome-metabolome-epigenome signalling cascade. Nat Metabol. 2020;2:882–92. doi: 10.1038/s42255-020-0267-9. [DOI] [PubMed] [Google Scholar]
- 152.Yu J, Chai P, Xie M, Ge S, Ruan J, Fan X, et al. Histone lactylation drives oncogenesis by facilitating m(6)A reader protein YTHDF2 expression in ocular melanoma. Genome Biol. 2021;22:85. doi: 10.1186/s13059-021-02308-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Wu R, Li S, Hudlikar R, Wang L, Shannar A, Peter R, et al. Redox signaling, mitochondrial metabolism, epigenetics and redox active phytochemicals. Free Radic Biol Med. 2020;S0891–5849:31670–1. doi: 10.1016/j.freeradbiomed.2020.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Navas LE, Carnero A. NAD(+) metabolism, stemness, the immune response, and cancer. Signal Transduct Target Ther. 2021;6:2. doi: 10.1038/s41392-020-00354-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Chini CCS, Zeidler JD, Kashyap S, Warner G, Chini EN. Evolving concepts in NAD(+) metabolism. Cell Metabol. 2021;33:1076–87. doi: 10.1016/j.cmet.2021.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Hitchler MJ, Domann FE. Redox regulation of the epigenetic landscape in cancer: a role for metabolic reprogramming in remodeling the epigenome. Free Radic Biol Med. 2012;53:2178–87. doi: 10.1016/j.freeradbiomed.2012.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Chiarugi A, Dolle C, Felici R, Ziegler M. The NAD metabolome--a key determinant of cancer cell biology. Nat Rev Cancer. 2012;12:741–52. doi: 10.1038/nrc3340. [DOI] [PubMed] [Google Scholar]
- 158.Liberti MV, Locasale JW. The Warburg effect: how does it benefit cancer cells? Trends Biochem Sci. 2016;41:211–8. doi: 10.1016/j.tibs.2015.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Chen H, Wang S, Zhang H, Nice EC, Huang C. Nicotinamide phosphoribosyltransferase (Nampt) in carcinogenesis: new clinical opportunities. Expert Rev Anticancer Ther. 2016;16:827–38. doi: 10.1080/14737140.2016.1190649. [DOI] [PubMed] [Google Scholar]
- 160.Ju HQ, Lin JF, Tian T, Xie D, Xu RH. NADPH homeostasis in cancer: functions, mechanisms and therapeutic implications. Signal Transduct Target Ther. 2020;5:231. doi: 10.1038/s41392-020-00326-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Rather GM, Pramono AA, Szekely Z, Bertino JR, Tedeschi PM. In cancer, all roads lead to NADPH. Pharmacol Ther. 2021;226:107864. doi: 10.1016/j.pharmthera.2021.107864. [DOI] [PubMed] [Google Scholar]
- 162.Tedeschi PM, Bansal N, Kerrigan JE, Abali EE, Scotto KW, Bertino JR. NAD+ kinase as a therapeutic target in cancer. Clin Cancer Res. 2016;22:5189–95. doi: 10.1158/1078-0432.ccr-16-1129. [DOI] [PubMed] [Google Scholar]
- 163.Ho HY, Lin YT, Lin G, Wu PR, Cheng ML. Nicotinamide nucleotide transhydrogenase (NNT) deficiency dysregulates mitochondrial retrograde signaling and impedes proliferation. Redox Biol. 2017;12:916–28. doi: 10.1016/j.redox.2017.04.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Tsang YH, Dogruluk T, Tedeschi PM, Wardwell-Ozgo J, Lu H, Espitia M, et al. Functional annotation of rare gene aberration drivers of pancreatic cancer. Nat Commun. 2016;7:10500. doi: 10.1038/ncomms10500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Li L, Li W. Epithelial-mesenchymal transition in human cancer: comprehensive reprogramming of metabolism, epigenetics, and differentiation. Pharmacol Ther. 2015;150:33–46. doi: 10.1016/j.pharmthera.2015.01.004. [DOI] [PubMed] [Google Scholar]
- 166.Shi Y, Lan F, Matson C, Mulligan P, Whetstine JR, Cole PA, et al. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell. 2004;119:941–53. doi: 10.1016/j.cell.2004.12.012. [DOI] [PubMed] [Google Scholar]
- 167.Ciccone DN, Su H, Hevi S, Gay F, Lei H, Bajko J, et al. KDM1B is a histone H3K4 demethylase required to establish maternal genomic imprints. Nature. 2009;461:415–8. doi: 10.1038/nature08315. [DOI] [PubMed] [Google Scholar]
- 168.Forneris F, Battaglioli E, Mattevi A, Binda C. New roles of flavoproteins in molecular cell biology: histone demethylase LSD1 and chromatin. FEBS J. 2009;276:4304–12. doi: 10.1111/j.1742-4658.2009.07142.x. [DOI] [PubMed] [Google Scholar]
- 169.Hino S, Sakamoto A, Nagaoka K, Anan K, Wang Y, Mimasu S, et al. FAD-dependent lysine-specific demethylase-1 regulates cellular energy expenditure. Nat Commun. 2012;3:758. doi: 10.1038/ncomms1755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Nakazawa MS, Keith B, Simon MC. Oxygen availability and metabolic adaptations. Nat Rev Cancer. 2016;16:663–73. doi: 10.1038/nrc.2016.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Xie H, Simon MC. Oxygen availability and metabolic reprogramming in cancer. J Biol Chem. 2017;292:16825–32. doi: 10.1074/jbc.r117.799973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Michealraj KA, Kumar SA, Kim LJY, Cavalli FMG, Przelicki D, Wojcik JB, et al. Metabolic regulation of the epigenome drives lethal infantile ependymoma. Cell. 2020;181:1329–45. doi: 10.1016/j.cell.2020.04.047. e24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Lu Y, Chu A, Turker MS, Glazer PM. Hypoxia-induced epigenetic regulation and silencing of the BRCA1 promoter. Mol Cell Biol. 2011;31:3339–50. doi: 10.1128/mcb.01121-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Chervona Y, Costa M. The control of histone methylation and gene expression by oxidative stress, hypoxia, and metals. Free Radic Biol Med. 2012;53:1041–7. doi: 10.1016/j.freeradbiomed.2012.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Chakraborty AA, Laukka T, Myllykoski M, Ringel AE, Booker MA, Tolstorukov MY, et al. Histone demethylase KDM6A directly senses oxygen to control chromatin and cell fate. Science. 2019;363:1217–22. doi: 10.1126/science.aaw1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Batie M, Frost J, Frost M, Wilson JW, Schofield P, Rocha S. Hypoxia induces rapid changes to histone methylation and reprograms chromatin. Science. 2019;363:1222–6. doi: 10.1126/science.aau5870. [DOI] [PubMed] [Google Scholar]
- 177.Dai B, Huang H, Guan F, Zhu G, Xiao Z, Mao B, et al. Histone demethylase KDM5A inhibits glioma cells migration and invasion by down regulating ZEB1. Biomed Pharmacother. 2018;99:72–80. doi: 10.1016/j.biopha.2018.01.020. [DOI] [PubMed] [Google Scholar]
- 178.Wang L, Shilatifard A. UTX mutations in human cancer. Cancer Cell. 2019;35:168–76. doi: 10.1016/j.ccell.2019.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Andricovich J, Perkail S, Kai Y, Casasanta N, Peng W, Tzatsos A. Loss of KDM6A activates super-enhancers to induce gender-specific squamous-like pancreatic cancer and confers sensitivity to BET inhibitors. Cancer Cell. 2018;33:512–26. doi: 10.1016/j.ccell.2018.02.003. e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Ntziachristos P, Tsirigos A, Welstead GG, Trimarchi T, Bakogianni S, Xu L, et al. Contrasting roles of histone 3 lysine 27 demethylases in acute lymphoblastic leukaemia. Nature. 2014;514:513–7. doi: 10.1038/nature13605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Hojfeldt JW, Agger K, Helin K. Histone lysine demethylases as targets for anticancer therapy. Nat Rev Drug Discov. 2013;12:917–30. doi: 10.1038/nrd4154. [DOI] [PubMed] [Google Scholar]
- 182.Hayami S, Kelly JD, Cho HS, Yoshimatsu M, Unoki M, Tsunoda T, et al. Overexpression of LSD1 contributes to human carcinogenesis through chromatin regulation in various cancers. Int J Cancer. 2011;128:574–86. doi: 10.1002/ijc.25349. [DOI] [PubMed] [Google Scholar]
- 183.Tzatsos A, Paskaleva P, Ferrari F, Deshpande V, Stoykova S, Contino G, et al. KDM2B promotes pancreatic cancer via polycomb-dependent and -independent transcriptional programs. J Clin Invest. 2013;123:727–39. doi: 10.1172/JCI64535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Hayami S, Yoshimatsu M, Veerakumarasivam A, Unoki M, Iwai Y, Tsunoda T, et al. Overexpression of the JmjC histone demethylase KDM5B in human carcinogenesis: involvement in the proliferation of cancer cells through the E2F/RB pathway. Mol Cancer. 2010;9:59. doi: 10.1186/1476-4598-9-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Kim I, Park JW. Hypoxia-driven epigenetic regulation in cancer progression: a focus on histone methylation and its modifying enzymes. Cancer Lett. 2020;489:41–9. doi: 10.1016/j.canlet.2020.05.025. [DOI] [PubMed] [Google Scholar]
- 186.Choudhry H, Harris AL. Advances in hypoxia-inducible factor biology. Cell Metabol. 2018;27:281–98. doi: 10.1016/j.cmet.2017.10.005. [DOI] [PubMed] [Google Scholar]
- 187.Xia X, Lemieux ME, Li W, Carroll JS, Brown M, Liu XS, et al. Integrative analysis of HIF binding and transactivation reveals its role in maintaining histone methylation homeostasis. Proc Natl Acad Sci USA. 2009;106:4260–5. doi: 10.1073/pnas.0810067106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Pollard PJ, Loenarz C, Mole DR, McDonough MA, Gleadle JM, Schofield CJ, et al. Regulation of Jumonji-domain-containing histone demethylases by hypoxia-inducible factor (HIF)-1α. Biochem J. 2008;416:387–94. doi: 10.1042/bj20081238. [DOI] [PubMed] [Google Scholar]
- 189.Guo X, Tian Z, Wang X, Pan S, Huang W, Shen Y, et al. Regulation of histone demethylase KDM6B by hypoxia-inducible factor-2α. Acta Biochim Biophys Sin (Shanghai) 2015;47:106–13. doi: 10.1093/abbs/gmu122. [DOI] [PubMed] [Google Scholar]
- 190.Thienpont B, Steinbacher J, Zhao H, D’Anna F, Kuchnio A, Ploumakis A, et al. Tumour hypoxia causes DNA hypermethylation by reducing TET activity. Nature. 2016;537:63–8. doi: 10.1038/nature19081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Matuleviciute R, Cunha PP, Johnson RS, Foskolou IP. Oxygen regulation of TET enzymes. FEBS J. 2021 doi: 10.1111/febs.15695. [DOI] [PubMed] [Google Scholar]
- 192.Prasad P, Mittal SA, Chongtham J, Mohanty S, Srivastava T. Hypoxia-mediated epigenetic regulation of stemness in brain tumor cells. Stem Cell. 2017;35:1468–78. doi: 10.1002/stem.2621. [DOI] [PubMed] [Google Scholar]
- 193.Mariani CJ, Vasanthakumar A, Madzo J, Yesilkanal A, Bhagat T, Yu Y, et al. TET1-mediated hydroxymethylation facilitates hypoxic gene induction in neuroblastoma. Cell Rep. 2014;7:1343–52. doi: 10.1016/j.celrep.2014.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Gu Y, Wu X, Zhang J, Fang Y, Pan Y, Shu Y, et al. The evolving landscape of N(6)-methyladenosine modification in the tumor microenvironment. Mol Ther. 2021;29:1703–15. doi: 10.1016/j.ymthe.2021.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Wang YJ, Yang B, Lai Q, Shi JF, Peng JY, Zhang Y, et al. Reprogramming of m(6)A epitranscriptome is crucial for shaping of transcriptome and proteome in response to hypoxia. RNA Biol. 2021;18:131–43. doi: 10.1080/15476286.2020.1804697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Panneerdoss S, Eedunuri VK, Yadav P, Timilsina S, Rajamanickam S, Viswanadhapalli S, et al. Cross-talk among writers, readers, and erasers of m(6)A regulates cancer growth and progression. Sci Adv. 2018;4 doi: 10.1126/sciadv.aar8263. eaar8263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Zhang C, Samanta D, Lu H, Bullen JW, Zhang H, Chen I, et al. Hypoxia induces the breast cancer stem cell phenotype by HIF-dependent and ALKBH5-mediated m(6)A-demethylation of NANOG mRNA. Proc Natl Acad Sci USA. 2016;113:E2047–56. doi: 10.1073/pnas.1602883113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Chen G, Liu B, Yin S, Li S, Guo Y, Wang M, et al. Hypoxia induces an endometrial cancer stem-like cell phenotype via HIF-dependent demethylation of SOX2 mRNA. Oncogenesis. 2020;9:81. doi: 10.1038/s41389-020-00265-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Li Q, Ni Y, Zhang L, Jiang R, Xu J, Yang H, et al. HIF-1α-induced expression of m6A reader YTHDF1 drives hypoxia-induced autophagy and malignancy of hepatocellular carcinoma by promoting ATG2A and ATG14 translation. Signal Transduct Target Ther. 2021;6:76. doi: 10.1038/s41392-020-00453-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Chen Z, Shao YL, Wang LL, Lin J, Zhang JB, Ding Y, et al. YTHDF2 is a potential target of AML1/ETO-HIF1α loop-mediated cell proliferation in t(8;21) AML. Oncogene. 2021;40:3786–98. doi: 10.1038/s41388-021-01818-1. [DOI] [PubMed] [Google Scholar]
- 201.Wu MZ, Tsai YP, Yang MH, Huang CH, Chang SY, Chang CC, et al. Interplay between HDAC3 and WDR5 is essential for hypoxia-induced epithelial-mesenchymal transition. Mol Cell. 2011;43:811–22. doi: 10.1016/j.molcel.2011.07.012. [DOI] [PubMed] [Google Scholar]
- 202.Selak MA, Armour SM, MacKenzie ED, Boulahbel H, Watson DG, Mansfield KD, et al. Succinate links TCA cycle dysfunction to oncogenesis by inhibiting HIF-α prolyl hydroxylase. Cancer Cell. 2005;7:77–85. doi: 10.1016/j.ccr.2004.11.022. [DOI] [PubMed] [Google Scholar]
- 203.Isaacs JS, Jung YJ, Mole DR, Lee S, Torres-Cabala C, Chung YL, et al. HIF overexpression correlates with biallelic loss of fumarate hydratase in renal cancer: novel role of fumarate in regulation of HIF stability. Cancer Cell. 2005;8:143–53. doi: 10.1016/j.ccr.2005.06.017. [DOI] [PubMed] [Google Scholar]
- 204.Han A, Schug ZT, Aplin AE. Metabolic alterations and therapeutic opportunities in rare forms of melanoma. Trends Cancer. 2021;7:671–81. doi: 10.1016/j.trecan.2021.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Galluzzi L, Kepp O, Vander Heiden MG, Kroemer G. Metabolic targets for cancer therapy. Nat Rev Drug Discov. 2013;12:829–46. doi: 10.1038/nrd4145. [DOI] [PubMed] [Google Scholar]
- 206.Martinez-Outschoorn UE, Peiris-Pages M, Pestell RG, Sotgia F, Lisanti MP. Cancer metabolism: a therapeutic perspective. Nat Rev Clin Oncol. 2017;14:11–31. doi: 10.1038/nrclinonc.2016.60. [DOI] [PubMed] [Google Scholar]
- 207.Liu Y, Cao Y, Zhang W, Bergmeier S, Qian Y, Akbar H, et al. A small-molecule inhibitor of glucose transporter 1 downregulates glycolysis, induces cell-cycle arrest, and inhibits cancer cell growth in vitro and in vivo . Mol Cancer Therapeut. 2012;11:1672–82. doi: 10.1158/1535-7163.mct-12-0131. [DOI] [PubMed] [Google Scholar]
- 208.Zerhouni M, Martin AR, Furstoss N, Gutierrez VS, Jaune E, Tekaya N, et al. Dual covalent inhibition of PKM and IMPDH targets metabolism in cutaneous metastatic melanoma. Cancer Res. 2021;81:3806–21. doi: 10.1158/0008-5472.can-20-2114. [DOI] [PubMed] [Google Scholar]
- 209.Apostolidi M, Vathiotis IA, Muthusamy V, Gaule P, Gassaway BM, Rimm DL, et al. Targeting pyruvate kinase M2 phosphorylation reverses aggressive cancer phenotypes. Cancer Res. 2021;81:4346–59. doi: 10.1158/0008-5472.can-20-4190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Yang Y, Chong Y, Chen M, Dai W, Zhou X, Ji Y, et al. Targeting lactate dehydrogenase a improves radiotherapy efficacy in non-small cell lung cancer: from bedside to bench. J Transl Med. 2021;19:170. doi: 10.1186/s12967-021-02825-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Qiao T, Xiong Y, Feng Y, Guo W, Zhou Y, Zhao J, et al. Inhibition of LDH-A by oxamate enhances the efficacy of Anti-PD-1 treatment in an NSCLC humanized mouse model. Front Oncol. 2021;11:632364. doi: 10.3389/fonc.2021.632364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Mohammad GH, Vassileva V, Acedo P, Olde Damink SWM, Malago M, Dhar DK, et al. Targeting pyruvate kinase M2 and lactate dehydrogenase a is an effective combination strategy for the treatment of pancreatic cancer. Cancers (Basel) 2019;11:1372. doi: 10.3390/cancers11091372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Woodford MR, Baker-Williams AJ, Sager RA, Backe SJ, Blanden AR, Hashmi F, et al. The tumor suppressor folliculin inhibits lactate dehydrogenase A and regulates the Warburg effect. Nat Struct Mol Biol. 2021;28:662–70. doi: 10.1038/s41594-021-00633-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Sharma NS, Gupta VK, Garrido VT, Hadad R, Durden BC, Kesh K, et al. Targeting tumor-intrinsic hexosamine biosynthesis sensitizes pancreatic cancer to anti-PD1 therapy. J Clin Invest. 2020;130:451–65. doi: 10.1172/JCI127515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Altman BJ, Stine ZE, Dang CV. From Krebs to clinic: glutamine metabolism to cancer therapy. Nat Rev Cancer. 2016;16:749. doi: 10.1038/nrc.2016.114. [DOI] [PubMed] [Google Scholar]
- 216.Feng Y, Pathria G, Heynen-Genel S, Jackson M, James B, Yin J, et al. Identification and characterization of IMD-0354 as a glutamine carrier protein inhibitor in melanoma. Mol Cancer Therapeut. 2021;20:816–32. doi: 10.1158/1535-7163.mct-20-0354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Gross MI, Demo SD, Dennison JB, Chen L, Chernov-Rogan T, Goyal B, et al. Antitumor activity of the glutaminase inhibitor CB-839 in triple-negative breast cancer. Mol Cancer Therapeut. 2014;13:890–901. doi: 10.1158/1535-7163.mct-13-0870. [DOI] [PubMed] [Google Scholar]
- 218.Jacque N, Ronchetti AM, Larrue C, Meunier G, Birsen R, Willems L, et al. Targeting glutaminolysis has antileukemic activity in acute myeloid leukemia and synergizes with BCL-2 inhibition. Blood. 2015;126:1346–56. doi: 10.1182/blood-2015-01-621870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Yu W, Yang X, Zhang Q, Sun L, Yuan S, Xin Y. Targeting GLS1 to cancer therapy through glutamine metabolism. Clin Transl Oncol. 2021;23:2253–68. doi: 10.1007/s12094-021-02645-2. [DOI] [PubMed] [Google Scholar]
- 220.Popovici-Muller J, Lemieux RM, Artin E, Saunders JO, Salituro FG, Travins J, et al. Discovery of AG-120 (Ivosidenib): a first-in-class mutant IDH1 inhibitor for the treatment of IDH1 mutant cancers. ACS Med Chem Lett. 2018;9:300–5. doi: 10.1021/acsmedchemlett.7b00421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Chaturvedi A, Herbst L, Pusch S, Klett L, Goparaju R, Stichel D, et al. Pan-mutant-IDH1 inhibitor BAY1436032 is highly effective against human IDH1 mutant acute myeloid leukemia in vivo . Leukemia. 2017;31:2020–8. doi: 10.1038/leu.2017.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Yen K, Travins J, Wang F, David MD, Artin E, Straley K, et al. AG-221, a first-in-class therapy targeting acute myeloid leukemia harboring oncogenic IDH2 mutations. Cancer Discov. 2017;7:478–93. doi: 10.1158/2159-8290.cd-16-1034. [DOI] [PubMed] [Google Scholar]
- 223.Konteatis Z, Artin E, Nicolay B, Straley K, Padyana AK, Jin L, et al. Vorasidenib (AG-881): a first-in-class, brain-penetrant dual inhibitor of mutant IDH1 and 2 for treatment of glioma. ACS Med Chem Lett. 2020;11:101–7. doi: 10.1021/acsmedchemlett.9b00509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Tajan M, Hennequart M, Cheung EC, Zani F, Hock AK, Legrave N, et al. Serine synthesis pathway inhibition cooperates with dietary serine and glycine limitation for cancer therapy. Nat Commun. 2021;12:366. doi: 10.1038/s41467-020-20223-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Garcia-Canaveras JC, Lancho O, Ducker GS, Ghergurovich JM, Xu X, da Silva-Diz V, et al. SHMT inhibition is effective and synergizes with methotrexate in T-cell acute lymphoblastic leukemia. Leukemia. 2021;35:377–88. doi: 10.1038/s41375-020-0845-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Hafliger P, Graff J, Rubin M, Stooss A, Dettmer MS, Altmann KH, et al. The LAT1 inhibitor JPH203 reduces growth of thyroid carcinoma in a fully immunocompetent mouse model. J Exp Clin Cancer Res. 2018;37:234. doi: 10.1186/s13046-018-0907-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Miller KD, Pniewski K, Perry CE, Papp SB, Shaffer JD, Velasco-Silva JN, et al. Targeting ACSS2 with a transition-state mimetic inhibits triple-negative breast cancer growth. Cancer Res. 2021;81:1252–64. doi: 10.1158/0008-5472.can-20-1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Ishak Gabra MB, Yang Y, Li H, Senapati P, Hanse EA, Lowman XH, et al. Dietary glutamine supplementation suppresses epigenetically-activated oncogenic pathways to inhibit melanoma tumour growth. Nat Commun. 2020;11:3326. doi: 10.1038/s41467-020-17181-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Jeon H, Kim JH, Lee E, Jang YJ, Son JE, Kwon JY, et al. Methionine deprivation suppresses triple-negative breast cancer metastasis in vitro and in vivo . Oncotarget. 2016;7:67223–34. doi: 10.18632/oncotarget.11615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Muthusamy T, Cordes T, Handzlik MK, You L, Lim EW, Gengatharan J, et al. Serine restriction alters sphingolipid diversity to constrain tumour growth. Nature. 2020;586:790–5. doi: 10.1038/s41586-020-2609-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Maddocks ODK, Athineos D, Cheung EC, Lee P, Zhang T, van den Broek NJF, et al. Modulating the therapeutic response of tumours to dietary serine and glycine starvation. Nature. 2017;544:372–6. doi: 10.1038/nature22056. [DOI] [PubMed] [Google Scholar]
- 232.Sundaram MK, Preetha R, Haque S, Akhter N, Khan S, Ahmed S, et al. Dietary isothiocyanates inhibit cancer progression by modulation of epigenome. Semin Cancer Biol. 2021 doi: 10.1016/j.semcancer.2020.12.021. [DOI] [PubMed] [Google Scholar]
- 233.Betto RM, Diamante L, Perrera V, Audano M, Rapelli S, Lauria A, et al. Metabolic control of DNA methylation in naive pluripotent cells. Nat Genet. 2021;53:215–29. doi: 10.1038/s41588-020-00770-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Dixit A, Parnas O, Li B, Chen J, Fulco CP, Jerby-Arnon L, et al. Perturb-Seq: dissecting molecular circuits with scalable single-cell RNA profiling of pooled genetic screens. Cell. 2016;167:1853–66. doi: 10.1016/j.cell.2016.11.038. e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Reid MA, Dai Z, Locasale JW. The impact of cellular metabolism on chromatin dynamics and epigenetics. Nat Cell Biol. 2017;19:1298–306. doi: 10.1038/ncb3629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Arango D, Sturgill D, Alhusaini N, Dillman AA, Sweet TJ, Hanson G, et al. Acetylation of cytidine in mRNA promotes translation efficiency. Cell. 2018;175:1872–86. doi: 10.1016/j.cell.2018.10.030. e24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Zhang Y, Jing Y, Wang Y, Tang J, Zhu X, Jin WL, et al. NAT10 promotes gastric cancer metastasis via N4-acetylated COL5A1. Signal Transduct Target Ther. 2021;6:173. doi: 10.1038/s41392-021-00489-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Ye C, Sutter BM, Wang Y, Kuang Z, Tu BP. A metabolic function for phospholipid and histone methylation. Mol Cell. 2017;66:180–93. doi: 10.1016/j.molcel.2017.02.026. e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Wang L, Song C, Wang N, Li S, Liu Q, Sun Z, et al. NADP modulates RNA m(6)A methylation and adipogenesis via enhancing FTO activity. Nat Chem Biol. 2020;16:1394–402. doi: 10.1038/s41589-020-0601-2. [DOI] [PubMed] [Google Scholar]
- 240.DeBerardinis RJ, Chandel NS. Fundamentals of cancer metabolism. Sci Adv. 2016;2:e1600200. doi: 10.1126/sciadv.1600200. [DOI] [PMC free article] [PubMed] [Google Scholar]