Abstract
Nicotinamide adenine dinucleotide (NAD) levels are essential for the normal physiology of the cell and are strictly regulated to prevent pathological conditions. NAD functions as a coenzyme in redox reactions, as a substrate of regulatory proteins, and as a mediator of protein-protein interactions. The main objectives of this study were to identify the NAD-binding and NAD-interacting proteins, and to uncover novel proteins and functions that could be regulated by this metabolite. It was considered if cancer-associated proteins were potential therapeutic targets. Using multiple experimental databases, we defined datasets of proteins that directly interact with NAD – the NAD-binding proteins (NADBPs) dataset – and of proteins that interact with NADBPs – the NAD-protein–protein interactions (NAD-PPIs) dataset. Pathway enrichment analysis revealed that NADBPs participate in several metabolic pathways, while NAD-PPIs are mostly involved in signalling pathways. These include disease-related pathways, namely, three major neurodegenerative disorders: Alzheimer’s disease, Huntington’s disease, and Parkinson’s disease. Then, the complete human proteome was further analysed to select potential NADBPs. TRPC3 and isoforms of diacylglycerol (DAG) kinases, which are involved in calcium signalling, were identified as new NADBPs. Potential therapeutic targets that interact with NAD were identified, that have regulatory and signalling functions in cancer and neurodegenerative diseases.
Keywords: cancer, interactome, NAD metabolism, neurodegenerative disorders, signalling
1. Introduction
Nicotinamide adenine dinucleotide (NAD) is a crucial metabolite in the cell, generally known for its function as cofactor in oxidation-reduction reactions responsible for energy production in the form of ATP, where it alternates between the oxidized (NAD+) and the reduced (NADH) forms. By transferring electrons between reactions, NAD participates in a multitude of metabolic processes that are key to the normal physiology of the cell including glycolysis, the citric acid cycle, fatty acids beta-oxidation and mitochondrial electron transport. Additionally, NAD is a substrate for proteins involved in cell survival, DNA damage repair, calcium signalling, or transcription regulation. NAD-dependent enzymes include sirtuins (SIRTs) [1], poly- and mono-(ADP-ribose) polymerases (PARPs and MARTs) [2, 3], and cyclic ADP-ribose hydrolases, such as CD38 [4]. Maintenance of NAD cellular levels depends on a balance between its production and its depletion, for which the interconversion between NAD+/NADH and NADP/NADPH is not accounted.
Another role for NAD was acknowledged more recently, where NAD would function as a direct modulator of protein–protein interactions (PPIs), through its binding to the NUDIX domain [5]. The NUDIX domain is a 23 amino acid long general structure of a Nucleoside Diphosphate linked to a variable moiety X, with catalytic activity on nucleotides. Through their activity, many NUDIX proteins contribute to cellular homeostasis by cleaning the cell from deleterious compounds. Others regulate the concentrations of several metabolites, such as NAD, NADP and ADP-ribose. Others remove 5′-cap from RNA and control the stability of mRNA, as well as gene expression. Nevertheless, several NUDIX enzymes remain uncharacterized [6–8].
NAD binding to the NUDIX homology domain (NHD) of the Deleted in Breast Cancer 1 (DBC1) protein prevented its interaction with PARP1 [5], and the DBC1-PARP1 interaction inhibits PARP1 normal function in the DNA damage repair process. Conversely, DBC1 regulates the activity of several proteins such as the transcription factor p53; the androgen and estrogen receptors (AR and ER), that are involved in hormone signalling; the BRCA1, which is also a DNA damage repair protein; and other NAD-dependent proteins that are epigenetic regulators, such as SIRT1 and HDAC3 [9].
The PARP catalytic domain is an example of a conserved protein domain that is common to all proteins within the PARP family, in which resides their main function of transferring the ADP-ribose moiety from its substrate (NAD) to carboxylate groups of aspartic and glutamic residues [10].
In this study, we aimed to characterize the NAD interactome, due to multitude of NAD cellular functions and relevance of NAD metabolism in normal and pathological conditions. Considering the NAD role in regulating PPIs, we focused on NAD-binding proteins and their interactions. Multiple experimental databases were surveyed to define an NAD-binding dataset, that was characterized through pathway enrichment analysis and protein structural domains analysis. The full human proteome was then screened, and a selection of potential NAD-binding proteins were further analysed. As previously reported in [11], we identified new proteins that potentially interact with NAD. Here, we described in detail the NADBPs dataset, we predicted NAD interacting residues of known NADBPs to serve as a reference and we further analysed the NUDIX containing proteins. We also uncovered NADBPs that are cancer-associated and potential drug targets. In addition, we performed molecular docking to predict the NAD-binding to potential NADBPs.
2. Workflow
2.1. Data collection
2.1.1. NAD-binding proteins (NADBPs) dataset
The first dataset was composed by proteins that directly interact with NAD, obtained from several databases of experimentally validated data. Namely, the following databases were searched: Human Metabolome Database (https://hmdb.ca/), STITCH (https://stitch4.embl.de/), Protein Data Bank (https://www.rcsb.org/), ChEMBL (https://www.ebi.ac.uk/chembl/), PubChem (https://pubchem.ncbi.nlm.nih.gov/) and DrugBank (https://go.drugbank.com/). The NADBPs dataset was composed by the sum of the proteins identified in the interactions from the six chemical databases used, as described in [11].
2.1.2. NAD-related proteins dataset
To study the proteins potentially related to NAD, an NAD-related dataset was defined. This was made using “NAD” as a keyword search, which considered all proteins that have “NAD” in protein name or in any field of description, such as protein family names, gene description, function or ontology classification. All human reviewed proteins obtained through UniProt (https://www.uniprot.org/) [12] and from IMEX Consortium database (http://www.imexconsortium.org/) [13] were considered.
2.1.3. NAD-protein–protein interactions (NAD-PPIs) dataset
A dataset composed by the proteins that interact with the NADBPs was built using three sources: BIOGRID (https://thebiogrid.org/) [14], STRING (https://string-db.org/) v.10 [15] and IMEX Consortium, as previously described [11].
2.2. Gene ontology (GO) analysis of the protein datasets
GO analysis was performed on PANTHER (http://pantherdb.org/) [16], through an overrepresentation test (Fisher’s exact, False Discovery Rate correction), using the Pathways annotation dataset (version 13.0). The NAD-binding, the NAD-related and the NAD-PPIs datasets were analysed.
2.3. Identification of putative NAD-binding proteins
The most frequent protein domains and protein families within the NADBPs dataset were identified [11]. The total of 20,303 human reviewed proteins from the Uniprot database were considered as a reference dataset, and the 50,588 unreviewed proteins as a test dataset. Proteins that presented at least one of the most frequent NADBPs domains were retrieved from both reference and test datasets. The genes/proteins that were found exclusively within the test dataset of unreviewed proteins were identified and further analysed using the NADbinder (http://crdd.osdd.net/raghava/nadbinder/) [17] to predict the number of NAD interacting residues, and the STRING database (https://string-db.org/) v. 11 [15], to obtain the interactions of each of those proteins.
2.4. Molecular docking
To evaluate the potential binding of NAD to the top target, an automated in silico molecular docking analysis was performed using SwissDock web server (http://www.swissdock.ch), as described by Grosdidier and collaborators [18]. NAD ligand was used as provided by ZINC database (https://zinc.docking.org/), with the ID ZINC8214766, and the protein 3D structures of the top target were retrieved from AlphaFold database (https://alphafold.ebi.ac.uk/) [19].
2.5. Cancer associated proteins and potential drug targets
Proteins from the NADBPs dataset were compared with catalogues of protein-coding genes from the subproteomes of the Human Protein Atlas (https://www.proteinatlas.org/) [20]. Namely, the cancer proteome, that contains a list of 569 mutated proteins strongly implicated in cancer, as defined through the catalogue of somatic mutations in cancer (COSMIC), and the druggable proteome, that contains a list of 754 proteins targeted directly by an FDA approved drug, were considered. Currently, approximately four thousand protein-coding genes in the UniProt database have experimental evidence of involvement in several disease conditions, including cancer, neurologic, systemic and cardiovascular disease. From those, a list of 1326 proteins annotated in The Human Protein Atlas as potential drug targets, was also considered, as they belong to known drug target protein classes, such as enzymes, transporters, receptors and ion-channels, and are not yet targets for FDA approved or experimental drugs in the Drugbank database.
3. Results
3.1. NAD-binding and NAD-related proteins differ in their predominant cellular roles
After collecting data from six different databases, we obtained a NADBPs dataset composed by a total of 439 proteins (Figure 1 and Appendix A). The NAD metabolite was found under different forms and names, and both oxidized and reduced forms were included. The highest numbers of interactions with NAD were found on the databases STITCH, DrugBank and the Human Metabolome Database.
Figure 1:
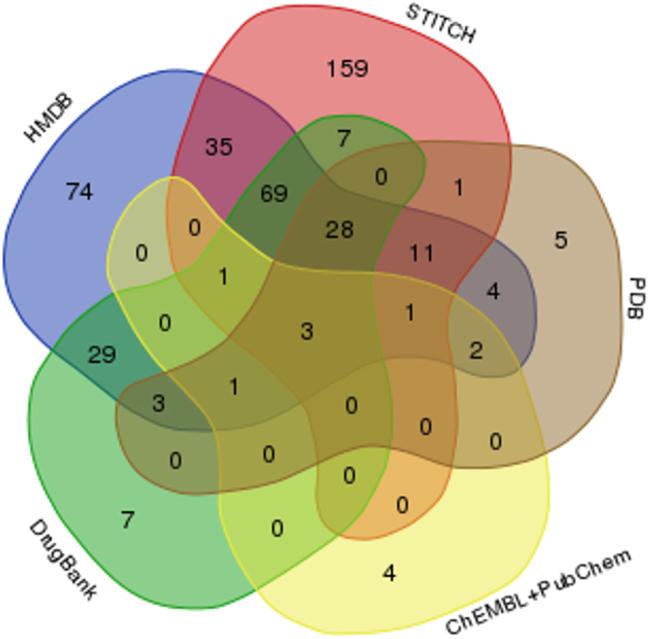
Venn-diagram showing the number of NAD-binding proteins obtained from each source.
The analysis of the 439 NADBPs showed that around 80% of these proteins were enzymes, most with catalytic activity, involved in metabolite interconversion. The major protein classes were dehydrogenases (92 proteins), from which over 30 were NADH dehydrogenase, and oxidoreductases (55 proteins), but several others were identified, as shown in Figure 2. More than one hundred proteins were mitochondrial isoforms of enzymes, which participate in the chain of reactions responsible for ATP production. Adding to enzymes that use NAD as cofactor in redox reactions, we also found all PARPs and all SIRTs, which are enzymes that use NAD as a substrate. Regarding their molecular function, a small number of proteins involved in regulation or transporter activities was also found.
Figure 2:
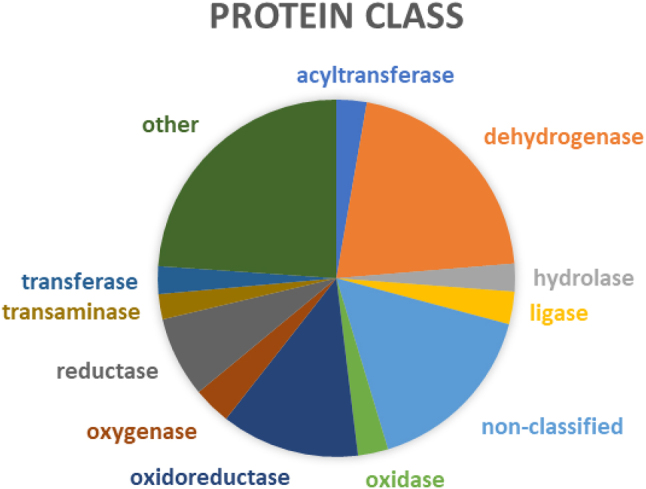
Classification of the NAD-binding proteins according to the protein class from PANTHER (www.pantherdb.org). The graphics represent the classes with more than 10 proteins. The remaining 43 classes comprising a total of 105 proteins are under the label “other” and 71 proteins remained non-classified.
The NAD-related datasets included all proteins potentially related to NAD, either by protein names or by any field of description. For the “NAD-related” dataset, we obtained 456 proteins from UniProt and 1907 from IMEX. In a total of 2125 proteins, only 238 were common to both sources. We then identified 279 proteins that were also present in the NADBP dataset, leaving a total of 1846 NAD-related proteins that do not bind NAD directly.
We performed a GO analysis on the 439 proteins of the NADBPs dataset (Table 1) and on the 1846 proteins of the NAD-related dataset, to compare the results of the enriched pathways obtained in each one. Only two pathways were common to the two datasets, Glycolysis and the FAS signalling pathway. We found 31 pathways specific of the NADBPs dataset, that were not enriched on the NAD-related dataset. Those included pathways related to biosynthesis or metabolism of nucleic acids, carbohydrates, and amino acids.
Table 1:
NADBPs dataset pathway enrichment.
| Pathway | Fold enrichment | |
|---|---|---|
| (min. 3.04–max. 47.93) | ||
| Biosynthesis | Adrenaline and noradrenaline biosynthesis | 6.18 |
| Alanine biosynthesis | 47.93 | |
| Androgen/estrogene/progesterone biosynthesis | 23.97 | |
| Asparagine and aspartate biosynthesis | 35.95 | |
| De novo purine biosynthesis | 6.39 | |
| Formyltetrahydroformate biosynthesis | 29.96 | |
| Gamma-aminobutyric acid synthesis | 23.97 | |
| Histidine biosynthesis | 47.93 | |
| Isoleucine biosynthesis | 23.97 | |
| Leucine biosynthesis | 47.93 | |
| Methionine biosynthesis | 47.93 | |
| O-antigen biosynthesis | 19.17 | |
| Proline biosynthesis | 28.76 | |
| Serine glycine biosynthesis | 38.35 | |
| Tetrahydrofolate biosynthesis | 19.17 | |
| Valine biosynthesis | 31.95 | |
| Basic metabolism | 5-Hydroxytryptamine degradation | 38.35 |
| Acetate utilization | 31.95 | |
| Aminobutyrate degradation | 47.93 | |
| Fructose galactose metabolism | 19.97 | |
| Glutamine glutamate conversion | 23.97 | |
| Glycolysis | 14.38 | |
| Methylmalonyl pathway | 19.17 | |
| Phenylethylamine degradation | 35.95 | |
| Purine metabolism | 20.54 | |
| Pyruvate metabolism | 30.5 | |
| TCA cycle | 38.35 | |
| Xanthine and guanine salvage pathway | 23.97 | |
| Signaling | Dopamine receptor mediated signaling pathway | 4.79 |
| Endothelin signaling pathway | 5.46 | |
| FAS signaling pathway | 7.05 | |
| GABA-B receptor II signaling | 11.28 | |
| Heterotrimeric G-protein signaling pathway-Gi alpha and Gs alpha mediated pathway | 3.04 |
In the NAD-related dataset, we found 36 pathways that did not appeared in the NADBPs dataset, that were mostly related to signalling. The highest fold enrichment values were found in the pentose phosphate pathway (the highest fold = 10.1), the JAK/STAT signalling, and four pathways related to p53 signalling. Of note, disease related pathways arose in the NAD related dataset, such as Alzheimer, Huntington, and Parkinson diseases. Also, signalling pathways related to angiogenesis, inflammation, and apoptosis, which are disease related mechanisms, were identified within the results.
3.2. Proteins that interact with NADBPs comprise about half of the human proteome
Then, the NAD-protein–protein interactions (NAD-PPIs), i.e., the proteins that interacted with the NADBPs were studied. Using the 439 proteins from the NADBPs dataset, 9823 pairs of proteins from STRING database were obtained, that corresponded to a total of 7815 unique gene name identifiers, 19,682 pairs from BIOGRID database, that corresponded to a total of 6479 unique gene IDs, and 5594 pairs from IMEX, that corresponded to a total of 3301 unique IDs. After mapping each type of ID retrieved from each database to the UniProtKB ID, with reviewed annotation (either using automatic tools or manually, in the case of automatically unmapped IDs), the duplicated entries we removed that were mainly due to gene or protein alternative names, or disease names associated to those genes. From STRING, a total of 7533 proteins were successfully mapped and 75 elements remained unmapped. From BIOGRID, a total of 5752 proteins were mapped and 54 remained unmapped. Most of these unmapped IDs were pseudogenes. From IMEX, 2500 proteins were mapped, and 90 elements remained unmapped. We found 40 CHEBI IDs, that were retrieved from CHEBI database for identification, but were not included for further analysis, since they corresponded to chemical compounds that interact with NADBPs and not protein-protein interactions, as it was intended. The proteins common to the three sources of PPIs were identified, and a final list of 10,020 proteins involved in PPIs with NADBPs remained.
As this represents about half of the human proteins annotated so far, according to the most recent version of UniProt Knowledge Database (UniProtKB 2020_06, [21]), with 20,379 reviewed proteins on the human proteome, the 1368 proteins common to all databases (Figure 3) were further analysed. With this, the selection of the most validated interactions was assured.
Figure 3:
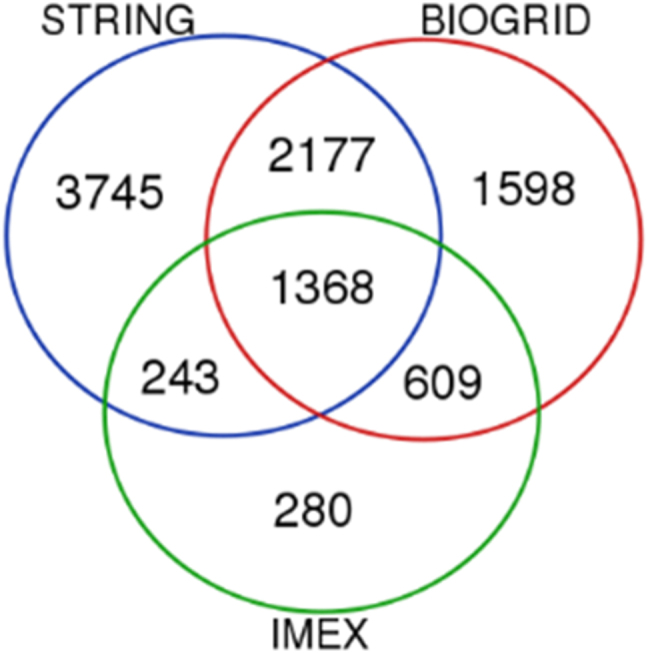
Venn-diagram representing the protein-protein interactions from STRING, BIOGRID and IMEX databases, where the 439 NADBPs were used as query.
A GO analysis was performed on the 1368 proteins from the NAD-PPIs dataset and compared with the results from the NADBPs dataset described previously (Table 1). Similarly to the NAD-related dataset, the NAD-PPIs dataset presented an enrichment in several signalling pathways, as compared to the NADBPs dataset. The pathways with the highest number of genes (over 50) were related to hormone receptors signalling, namely for gonadotropin and for the gastrointestinal peptide hormones cholecystokinin and gastrin, followed by the Wnt signalling and angiogenesis pathways. Several other pathways were related to hormone or growth factor signalling, and disease pathways also emerged, namely three major neurodegenerative diseases, Alzheimer’s, Huntington’s, and Parkinson’s.
3.3. Overview of NADBPs protein structural domains
Protein domains analysis was performed on the 439 NADBPs through PFAM database and all matches that achieved an expectation value (E-value) below 1 (max. 0.88) were selected. The results show the top hit domain for each protein and how many hits were found. Within the 439 proteins, 1101 identifications were made, which corresponded to a total of 412 different domains. Two proteins didn’t have an identified domain (NDUFA11 and GPAT2) and, in the remaining 437 proteins, 222 different domains were identified as top hit. More than half of the proteins (56% – 247 proteins) belonged to the FAD/NAD(P)-binding Rossmann fold superfamily, and 27% belonged to the Ankyrin repeat superfamily. In our approach, the top 15 more common domains, which appeared in more than 10 proteins (Table 2), were selected. Five different ankyrin repeats were among these top domains found. Others were the short chain dehydrogenase, the aldehyde dehydrogenase family, the cytochrome P450 and the poly(ADP-ribose) polymerase (PARP) catalytic domain.
Table 2:
The top 15 domains identified in the NADBPs dataset.
| Domain hmm ID | Domain name | Number of proteins | Description |
|---|---|---|---|
| PF00023.29 | Ank | 30 | Ankyrin repeat |
| PF13637.5 | Ank_4 | 25 | Ankyrin repeat |
| PF13857.5 | Ank_5 | 24 | Ankyrin repeat |
| PF00106.24 | adh_short | 23 | Short chain dehydrogenase |
| PF13561.5 | adh_short_C2 | 22 | Enoyl-(Acyl carrier protein) reductase |
| PF13606.5 | Ank_3 | 21 | Ankyrin repeat |
| PF08659.9 | KR | 20 | KR domain |
| PF12796.6 | Ank_2 | 20 | Ankyrin repeat |
| PF00211.19 | Guanylate_cyc | 18 | Adenylate and Guanylate cyclase catalytic domain |
| PF00171.21 | Aldedh | 16 | Aldehyde dehydrogenase family |
| PF00644.19 | PARP | 15 | Poly(ADP-ribose) polymerase catalytic domain |
| PF01370.20 | Epimerase | 14 | NAD dependent epimerase/dehydratase family |
| PF00153.26 | Mito_carr | 12 | Mitochondrial carrier protein |
| PF00107.25 | ADH_zinc_N | 10 | Zinc-binding dehydrogenase |
| PF00067.21 | p450 | 10 | Cytochrome P450 |
We also identified 65 proteins that had one of the 43 domains containing the term “NAD” in their names or descriptions. Eighteen proteins contain specifically one of the six different “NAD binding domain”, namely the D-isomer specific 2-hydroxyacid dehydrogenase, the 3-hydroxyacyl-CoA dehydrogenase, the lactate/malate dehydrogenase, the malic enzyme, the UDP-glucose/GDP-mannose dehydrogenase family, and the 6-phosphogluconate dehydrogenase NAD binding domains. The NUDIX domain was found only in two proteins from the NAD-binding dataset, namely NUDT12 and NUDT7.
3.4. Identification of 13 new NAD-binding proteins based on protein domains
We searched for the 15 domains that were identified in ten or more proteins from the NADBPs dataset (Table 2) within the dataset of the full human proteome unreviewed proteins (test dataset) and obtained 901 protein sequences. After removing all protein fragments and duplicates, 255 proteins were identified, which corresponded to 204 single genes. A similar approach was performed in the reference dataset yielding 474 genes. Given our aim to identify uncharacterized proteins, from the 204 genes, 195 that were also identified in the reference dataset were excluded and 8 genes remained, corresponding to 13 protein sequences, found uniquely in the test dataset (Table 3).
Table 3:
Proteins identified from the test dataset.
| UniProt ID | Status | Gene name | Protein name | Length | Number of NAD |
|---|---|---|---|---|---|
| interacting | |||||
| residues | |||||
| A0A087WV00 | Unreviewed | DGKI | Diacylglycerol kinase (DAG kinase) | 932 | 21 |
| E7EM72 | Unreviewed | DGKI | Diacylglycerol kinase (DAG kinase) | 1047 | 29 |
| E7EWQ4 | Unreviewed | DGKI | Diacylglycerol kinase (DAG kinase) | 1078 | 31 |
| E9PFX6 | Unreviewed | DGKI | Diacylglycerol kinase (DAG kinase) | 734 | 23 |
| E9PNL8 | Unreviewed | DGKZ | Diacylglycerol kinase (DAG kinase) | 707 | 26 |
| E9PK39 | Unreviewed | LRRK1 | Leucine-rich repeat serine/threonine-protein kinase 1 | 650 | 12 |
| E9PMK9 | Unreviewed | LRRK1 | Leucine-rich repeat serine/threonine-protein kinase 1 | 689 | 7 |
| Q495V5 | Unreviewed | POTEB | POTE ankyrin domain family member B (POTEB protein) | 301 | 16 |
| D6R9P2 | Unreviewed | SLC9B2 | Sodium/hydrogen exchanger 9B2 | 112 | 11 |
| D6RC49 | Unreviewed | TRPC3 | Short transient receptor potential channel 3 | 276 | 5 |
| J3QTB0 | Unreviewed | TRPC3 | Short transient receptor potential channel 3 | 793 | 33 |
| A0A087WV96 | Unreviewed | CYP3A7-CYP3A51P | CYP3A7-CYP3A51P readthrough | 506 | 27 |
| V9GXZ4 | Unreviewed | FPGT-TNNI3K | FPGT-TNNI3K readthrough | 949 | 24 |
Among the 13 proteins, there were five isoforms of the Diacylglycerol (DAG) kinase, four encoded by the DGKI gene, and one encoded by DGKZ gene. There were two other kinase isoforms, from the Leucine-rich repeat serine/threonine-protein kinase 1, encoded by the LRRK1 gene. There were also two proteins related to membrane transport, the Sodium/hydrogen exchanger 9B2 (SLC9B2) and two isoforms of a short transient receptor potential channel encoded by the TRPC3 gene. A smaller isoform of the POTEB member of the ankyrin family was also found. Of note, POTEB was the only protein that presented simultaneously two of the 15 domains (Ank_2 e Ank_5). Additionally, there were two proteins resultant from the readthrough of two genes, CYP3A7-CYP3A51P, which belong to a subfamily of the Cytochrome P450, and FPGT-TNNI3K, from the neighbouring fucose-1-phosphate guanylyltransferase (FPGT) and TNNI3 interacting kinase (TNNI3K) genes.
To evaluate the possibility that NAD has an impact on the interactions between these proteins, we further searched for the interactions of each of the proteins. DGKI and SLC9B2 had no reported interactions, as well as the proteins resultant from the two readthrough events. LRRK1 had the highest number of interactions, followed by TRPC3.
3.5. Number of NAD interacting residues in new and known NADBPs
The 13 identified proteins were further analysed using the NADbinder software (Table 3). Here, instead of the protein structure, the protein sequence is considered. The highest number of NAD-interacting residues was 33 and was identified in the longest isoform of TRPC3, with 793 amino acids, followed by the longest isoform of DGKI with 1078 amino acids, where 31 residues were identified. The five DAG kinase isoforms retrieved more than 20 NAD-interacting residues, as well as the two readthrough proteins. A positive correlation was observed between the amino acid length and the number of NAD-interacting residues identified.
To serve as a reference, six proteins known to be involved in NAD metabolism were additionally scanned for the number of NAD interacting residues. Namely, two enzymes that consume NAD intracellularly (PARP1 and SIRT1), two enzymes that consume NAD extracellularly (CD38 and CD73), and two enzymes that participate in NAD biosynthesis (NAMPT and NAPRT) were analysed. Among these sequences, there was no significant correlation between the number of NAD-binding residues and protein length. According to the NADbinder analysis results, CD73 had the highest number of NAD interacting residues (51), and the remaining NAD-consuming enzymes had between 37 and 43 residues, which were higher than the ones identified in the 13 previously studied proteins. NAMPT and NAPRT don’t interact directly with the NAD molecule, and presented 39 and 18 NAD interacting residues, respectively. However, they bind nicotinamide and nicotinic acid, similar molecules that are NAD precursors, and are responsible for the first steps of their conversion into NAD.
3.6. NUDIX containing proteins in NADBPs dataset
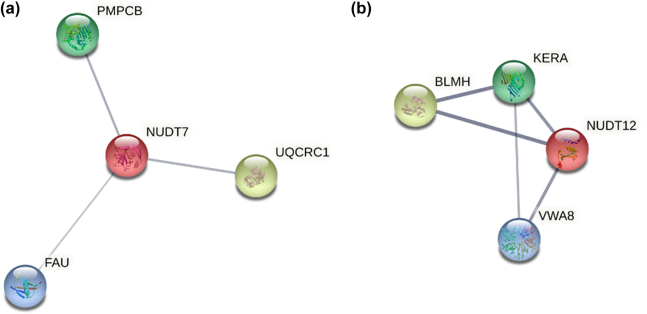
As it was previously described that NUDIX domain directly interacts with NAD, the proteins within NADBPs dataset that are NUDIX hydrolases, NUDT7 and NUDT12, were also studied. In NUDT7, 21 NAD interacting residues were identified and, in NUDT12, only six residues were detected. We also searched for their interactors, considering only experimentally validated physical interactions, and found three proteins that interact with NUDT7 and three proteins that interact with NUDT12 (Figure 4).
Figure 4:
Protein–protein interactions of NUDIX proteins from the NAD-binding proteins dataset. (a) NUDT7 and (b) NUDT12. Queried proteins are represented by red nodes and the line thickness indicates the confidence level of the interaction. Only physical interactions are represented. The network was obtained through STRING (string-db.org).
3.7. Identification of NADBPS as potential new drug targets mutated in cancer
We found 122 proteins that are NAD-binding and potential drug targets. Two of them also belong to the set of cancer mutated genes, fumarate hydratase (FH) and 5′-nucleotidase, cytosolic II (NT5C2). Additionally, three other NADBPs, which mutations are implicated in cancer, were found in the catalogue of FDA approved targets, namely, 5-aminoimidazole-4-carboxamide ribonucleotide formyltransferase/IMP cyclohydrolase (ATIC), androgen receptor (AR) and isocitrate dehydrogenase 2 (IDH2).
4. Discussion
NAD binds to a large number of different proteins in order to perform a diversity of functions within the cell. In those reactions, NAD can: (1) act as an enzymatic cofactor in redox reactions, (2) be degraded by NAD-dependent enzymes, and (3) mediate protein-protein interactions, therefore regulating several cellular processes. Our approach in this study to identify potential NAD-binding proteins, drove us to a global analysis of the NAD interactome. We integrated data from various sources to include a large dataset of proteins that were already known to interact with the NAD molecule, or that were in some way related to NAD functions. We functionally characterized the protein datasets through gene ontology and protein structural domains analysis.
Through the analyses of enriched pathways, based on gene ontology annotations, we found that NADBPs are involved in a diversity of cellular pathways. The comparison with the NAD-related or the NAD-PPIs datasets emphasised that NADBPs are central in basic metabolism and biosynthetic processes. Nonetheless, essential metabolic pathways, such as glycolysis and TCA cycle, and signalling pathways mediated by GABA or dopamine receptors, were found in all datasets. Conversely, the proteins that participate in NAD-PPIs are involved in signalling pathways, from development and apoptosis to general immune and hormone responses, and including many disease pathways, showing the extension of the action of this small molecule.
Analysis of the protein structural domains showed that the ankyrin repeats were the most frequent, with some proteins presenting more than one ankyrin repeat in their structures. The ankyrin domain is very frequent in all human proteome as it mediates protein-protein interactions [22] and regulates the function of other proteins [23]. Confirming their high frequency, in the unreviewed dataset here obtained from the full human proteome, based on the UniProt database, 448 proteins have at least one of the five ankyrin repeats.
Adding to protein structural domains, the number of NAD interacting residues was considered, given that the direct binding of NAD at specific sites of a protein ultimately determines its action [17]. NAD binding to the NUDIX homology domain of DBC1 regulates its action on PARP1, by preventing the interaction between the two proteins [5]. In this study, no more than 10 residues were identified within the NUDIX domain that are conserved across several species. Considering the presence of a specific domain with a folding favourable to an interaction with a small molecule, only a small number of residues might be responsible for the actual interaction. The identification of NAD interacting residues within the sequence of known NADBPs, revealed that, while some NAD consuming enzymes had around 40 residues, the two NUDIX-containing domain proteins had lower numbers (21 and 6).
The role of the NAD-capped RNA hydrolase NUDT12 is directly associated with NAD, also known as deNADding enzyme, and it interacts with Bleomycin Hydrolase (BLMH) through the ankyrin repeats of NUDT12 [24]. The known role of the peptidase BLMH is to cleave the anti-cancer peptide Bleomycin, reducing the intracellular levels of the drug, but its primary biological function remains unknown.
Among the new proteins that might potentially bind NAD identified in our study, TRPC3 (UniProt ID J3QTB0) had the ankyrin repeat domain and had the highest number of NAD-interacting residues. The molecular docking performed revealed a potential NAD-binding location on TRPC3. From a total of 31 clusters of docking positions obtained, 26 were placed within a same location, including the ones with the best scoring and lowest estimated energy (Figure 5).
Figure 5:
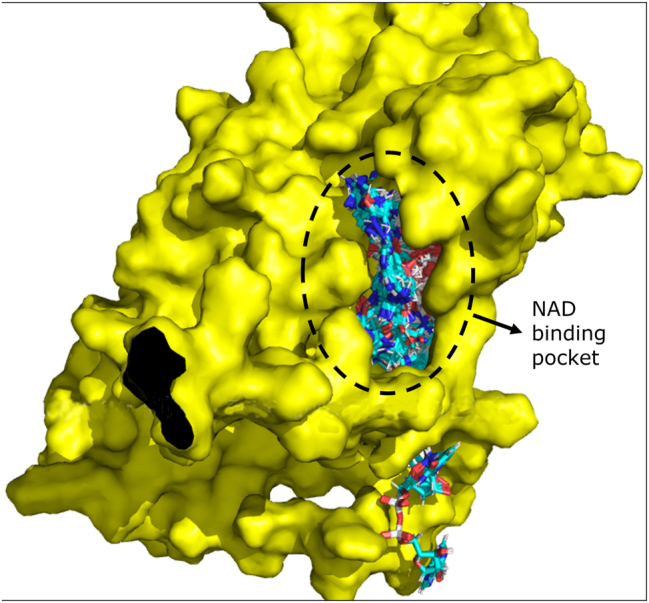
Docking results for NAD ligand on TRPC3 as a target (D6RC49 and J3QTB0). Protein 3D structures were obtained from AlphaFold (https://alphafold.ebi.ac.uk/) and visualized using Pymol software.
The corresponding reviewed protein (UniProt ID: Q13507) of TRPC3 is longer than the two isoforms detected here, with 836 amino acids. Its known interactions were found to be mostly involved in signal transduction, response to stress, anatomical structure development, and transport processes, many of them related to calcium transport and signalling, such as the inositol trisphosphate (IP3) receptors ITPR1 and ITPR3, and the Sodium/calcium exchanger SLC8A1.
TRPC3 is a member of the transient receptor potential (TRP) channels family, which regulates intracellular calcium concentration [25] and is directly activated by lipids, specifically diacylglycerol (DAG). Together with IP3, DAG is a product of the hydrolysis of a phospholipid catalysed by the phospholipase C (PLC) enzymes. PLC gamma enzymes are key components of intracellular signalling, and some PLCG1 functions have been associated to a specific protein domain that directly interacts with TRPC3 and PLCG1, regulating calcium entry [26]. Very recently, the role of PLC gamma enzymes in disease development has been explored [27]. Of note, PLCG1 was also found in our dataset of NAD-PPIs, showing that it already binds other NADBPs, and several unreviewed isoforms of DAG kinases were identified in this study as potential NADBPs.
Both NAD-dependent signalling and calcium-dependent signalling are essential in the cell and therefore their dysregulation is often associated with disease. In particular, the role of NAD as a regulator of calcium channels has been recently reviewed, due to its impact on cancer treatment research [28], where calcium channels emerge as potential targets for anticancer therapy. In addition to cancer, the TRP channels, namely the TRPC3 group, regulate functions in neurons and are involved in various neurological and psychiatric disorders [29].
Interestingly, only one ion channel was identified in the primary NADBPs dataset in our study, named Transient receptor potential cation channel subfamily M member 2 (TRPM2). Although it was not identified in the domain analysis through Pfam, the presence of NUDIX domains in the structure of TRPM2 has been described in the literature and associated to its conformational changes and gating functions [30]. The activation of TRPM2 by NAD has been documented for over two decades and is one example of the relation between NAD and calcium metabolism [31].
In a final step of this research, we decided to investigate whether some of the NADBPs were potential therapeutic drug targets. We found FH and NT5C2, which are directly involved in NAD related reactions: the former participates in the TCA cycle and the latter in the NAD synthesis, specifically by catalysing the hydrolysis of NMN into NR or NAMN into NAR. Both enzymes are altered in cancer and are also associated with neurological diseases [32–34]. In addition, from the NADBPs dataset ATIC, AR and IDH2 are already being used as therapeutic targets. ATIC participates in purine biosynthesis, where it catalyses the last two steps of the pathway [35]. IDH2 is the mitochondrial isoform of the isocitrate dehydrogenases family of enzymes, that depends on NADP and calcium binding to perform the oxidative decarboxylation of isocitrate, one of the steps of the TCA cycle. Therefore, alterations in these enzymes will have an important impact in metabolism. IDH1 and IDH2 mutations have been described in different types of cancer, including glioblastoma, and are being targeted for acute myeloid leukaemia [36, 37]. The androgen receptor act as a transcription factor and, when activated by the hormone androgen, binds to target genes, and directly regulates gene transcription of a high number of genes. SIRT1, an NAD-dependent deacetylase, regulates AR activity, linking NAD metabolism to ligand-induced hormone signalling [38]. Aberrant expression of AR contributes to the progression of prostate cancer, making this protein a recognized therapeutic target in this context [39]. Alterations in AR have also been associated to neurological diseases, from developmental deficiencies to neurodegenerative disorders [40].
5. Conclusions
Concluding, this global study of the NAD interactome resulted in the identification of new potentially NAD-binding proteins, including TRPC3 and a few isoforms of DGA kinases, which are involved in calcium signalling. NADBPs participate in several metabolic pathways and signalling processes in the cell, while proteins interacting with NADPBs (NAD-PPIs) are mostly involved in signalling pathways. Furthermore, we identified NADBPs that are known (ATIC, AR and IDH2),as well as potential new drug targets in cancer (FH and NT5C2).
Appendix A: List of the proteins that compose the NAD-binding proteins (NADBPs) dataset
| UniProt ID | Gene name | Length | UniProt ID | Gene name | Length |
|---|---|---|---|---|---|
| Q8NSZO | AADAT | 425 | P30838 | ALDH3A1 | 453 |
| Q4L235 | AASDH | 1098 | P51648 | ALDH3A2 | 485 |
| Q9NRN7 | AASDHPPT | 309 | P43353 | ALDH3B1 | 468 |
| Q9UDR5 | AASS | 926 | P48448 | ALDH3B2 | 385 |
| P80404 | ABAT | 500 | P30038 | ALDH4A1 | 563 |
| P33897 | ABCD1 | 745 | P51649 | ALDHSA1 | 535 |
| P09110 | ACAA1 | 424 | Q02252 | ALDH6A1 | 535 |
| P42765 | ACAA2 | 397 | P49419 | ALDH7A1 | 539 |
| Q9UKU7 | ACAD8 | 415 | Q9H2A2 | ALDH8A1 | 487 |
| P28330 | ACADL | 430 | P49189 | ALDH9A1 | 494 |
| P11310 | ACADM | 421 | P04075 | ALDOA | 364 |
| P16219 | ACADS | 412 | P05062 | ALDOB | 364 |
| P45954 | ACADSB | 432 | P09972 | ALDOC | 364 |
| P24752 | ACAT1 | 427 | Q96NU7 | AMDHD1 | 426 |
| Q9BWD1 | ACAT2 | 397 | P48728 | AMT | 403 |
| Q8TDX5 | ACMSD | 336 | P19801 | AOC1 | 751 |
| P21399 | ACO1 | 889 | O75106 | AOC2 | 756 |
| Q99798 | ACO2 | 780 | Q16853 | AOC3 | 763 |
| O14734 | ACOT8 | 319 | Q06278 | AOX1 | 1338 |
| P33121 | ACSL1 | 698 | P10275 | AR | 920 |
| Q9NUB1 | ACSS1 | 689 | P62330 | ARF6 | 175 |
| Q9NR19 | ACSS2 | 701 | P15848 | ARSB | 533 |
| Q08828 | ADCY1 | 1119 | P51689 | ARSD | 593 |
| Q08462 | ADCY2 | 1091 | P54793 | ARSF | 590 |
| O60266 | ADCY3 | 1144 | QSFYA8 | ARSH | 562 |
| Q8NFM4 | ADCY4 | 1077 | QSFYB1 | ARSI | 569 |
| O95622 | ADCY5 | 1261 | QSFYBO | ARS3 | 599 |
| O43306 | ADCY6 | 1168 | Q6UVVY0 | ARSK | 536 |
| P51828 | ADCY7 | 1080 | P51690 | ARSL | 589 |
| P40145 | ADCY8 | 1251 | P52961 | ART1 | 327 |
| O60503 | ADCY9 | 1353 | Q13508 | ARTS | 389 |
| P07327 | ADH1A | 375 | Q93070 | ART4 | 314 |
| P00325 | ADH1B | 375 | Q96L15 | ARTS | 291 |
| P00326 | ADH1C | 375 | A6ND91 | ASPDH | 283 |
| P08319 | ADH4 | 380 | P31939 | ATIC | 592 |
| P11766 | ADHS | 374 | O75936 | BBOX1 | 387 |
| P28332 | ADH6 | 368 | P54687 | BCAT1 | 386 |
| P40394 | ADH7 | 386 | O15382 | BCAT2 | 392 |
| Q63HM1 | AFMID | 303 | P12694 | BCKDHA | 445 |
| P23526 | AHCY | 432 | P21953 | BCKDHB | 392 |
| O43865 | AHCYL1 | 530 | Q02338 | BDH1 | 343 |
| Q96HN2 | AHCYL2 | 611 | Q9BUT1 | BDH2 | 245 |
| O95831 | AIFM1 | 613 | Q93088 | BHMT | 406 |
| P14550 | AKR1A1 | 325 | P53004 | BLVRA | 296 |
| P15121 | AKR1B1 | 316 | P30043 | BLVRB | 206 |
| O60218 | AKR1B10 | 316 | Q10588 | BST1 | 318 |
| Q04828 | AKR1C1 | 323 | Q13137 | CALC00O2 | 446 |
| P52895 | AKR1C2 | 323 | P04040 | CAT | 527 |
| P42330 | AKR1C3 | 323 | Q8N4T8 | CBR4 | 237 |
| P17516 | AKR1C4 | 323 | P28907 | CD38 | 300 |
| P51857 | AKR1D1 | 326 | Q16878 | CD01 | 200 |
| P00352 | ALDH1A1 | 501 | P28329 | CHAT | 748 |
| O94788 | ALDH1A2 | 518 | Q8NE62 | CHDH | 594 |
| P47895 | ALDH1A3 | 512 | P21964 | COMT | 271 |
| P30837 | ALDH1B1 | 517 | P43155 | CRAT | 626 |
| Q3SY69 | ALDH1L2 | 923 | Q9UKG9 | CROT | 612 |
| P05091 | ALDH2 | 517 | Q9Y2S2 | CRYL1 | 319 |
| O75390 | CS | 466 | PO4406 | GAPDH | 335 |
| Q9Y600 | CSAD | 493 | O14556 | GAPDHS | 408 |
| Q13363 | CTBP1 | 440 | O75600 | GCAT | 419 |
| P56545 | CTBP2 | 445 | Q92947 | GCDH | 438 |
| P32929 | CTH | 405 | P23434 | GCSH | 173 |
| P00167 | CYBSA | 134 | Q9Y2T3 | GDA | 454 |
| Q9UHQ9 | CYBSR1 | 305 | Q13630 | GFUS | 321 |
| Q6BCY4 | CYBSR2 | 276 | P23378 | GLDC | 1020 |
| P00387 | CYB5R3 | 301 | P00367 | GLUD1 | 558 |
| Q7L1T6 | CYBSR4 | 521 | P49448 | GLUD2 | 558 |
| Q6IPT4 | CYBSRL | 315 | O60547 | GMDS | 372 |
| Q53TN4 | CYBRD1 | 286 | P49915 | GMPS | 693 |
| P08574 | CYC1 | 325 | P17174 | GOT1 | 413 |
| P99999 | CYCS | 105 | Q8NHS2 | GOT1L1 | 421 |
| P05093 | CYP17A1 | 508 | P00505 | GOT2 | 430 |
| Q02318 | CYP27A1 | 531 | Q9HCL2 | GPAM | 828 |
| P05181 | CYP2E1 | 493 | Q6NUI2 | GPAT2 | 795 |
| Q6VVX0 | CYP2R1 | 501 | Q86UL3 | GPAT4 | 456 |
| Q9NYLS | CYP39A1 | 469 | P21695 | GPD1 | 349 |
| Q02928 | CYP4A11 | 519 | Q8N335 | GPD1L | 351 |
| QSTCH4 | CYP4A22 | 519 | P43304 | GPD2 | 727 |
| P22680 | CYP7A1 | 504 | Q9UBQ7 | GRHPR | 328 |
| O75881 | CYP7B1 | 506 | P00390 | GSR | 522 |
| Q9UNU6 | CYP8B1 | 501 | H3C1 | ||
| P11182 | DBT | 482 | H3C2 | ||
| Q16698 | DECR1 | 335 | H3C3 | ||
| O15121 | DEGS1 | 323 | H3C4 | ||
| Q9UBM7 | DHCR7 | 475 | H3C6 | ||
| P00374 | DHFR | 187 | P68431 | H3C7 | 136 |
| Q86XFO | DHFR2 | 187 | H3C8 | ||
| P49366 | DHPS | 369 | H3C10 | ||
| Q96HY7 | DHTKD1 | 919 | H3C11 | ||
| P10515 | DLAT | 647 | H3C12 | ||
| P09622 | DLD | 509 | O95479 | H6PD | 791 |
| P36957 | DLST | 453 | Q9U383 | HACL1 | 578 |
| Q9NRD9 | DUOX1 | 1551 | Q16836 | HADH | 314 |
| Q9NRD8 | DUOX2 | 1548 | P40939 | HADHA | 763 |
| P30084 | ECHS1 | 290 | P55084 | HADHB | 474 |
| P42126 | ECI1 | 302 | P42357 | HAL | 657 |
| Q08426 | EHHADH | 723 | P31937 | HIBADH | 336 |
| Q9GZV4 | EIFSA2 | 153 | Q6NVY1 | HIBCH | 386 |
| Q9NXB9 | ELOVL2 | 296 | P35914 | HMGCL | 325 |
| Q9NYP7 | ELOVLS | 299 | PO4035 | HMGCR | 888 |
| Q8TC92 | ENOX1 | 643 | P09601 | HMOX1 | 288 |
| Q16206 | ENOX2 | 610 | P30519 | HMOX2 | 316 |
| P22413 | ENPP1 | 925 | P15428 | HPGD | 266 |
| O14638 | ENPP3 | 875 | P28845 | HSD11B1 | 292 |
| O95864 | FADS2 | 444 | P80365 | HSD11B2 | 405 |
| Q8WVX9 | FAR1 | 515 | P14061 | HSD17B1 | 328 |
| Q96K12 | FAR2 | 515 | Q99714 | HSD17B10 | 261 |
| Q96IV6 | FAXDC2 | 333 | Q8NBQS | HSD17B11 | 300 |
| P37268 | FDFT1 | 417 | Q53GQ0 | HSD17B12 | 312 |
| P07954 | FH | 510 | Q9BPX1 | HSD17B14 | 270 |
| P98177 | FOX04 | 505 | P37059 | HSD17B2 | 387 |
| Q6ZNAS | FRRS1 | 592 | P37058 | HSD17B3 | 310 |
| Q14376 | GALE | 348 | P51659 | HSD17B4 | 736 |
| P07902 | GALT | 379 | O14756 | HSD17B6 | 317 |
| P56937 | HSD17B7 | 341 | O15239 | NDUFA1 | 70 |
| Q92506 | HSD17B8 | 261 | O95299 | NDUFA10 | 355 |
| P14060 | HSD3B1 | 373 | Q86Y39 | NDUFA11 | 141 |
| P26439 | HSD3B2 | 372 | Q9UI09 | NDUFAl2 | 145 |
| Q9H2F3 | HSD3B7 | 369 | Q9P0J0 | NDUFA13 | 144 |
| P48735 | IDH2 | 452 | O43678 | NDUFA2 | 99 |
| P50213 | IDH3A | 366 | O95167 | NDUFA3 | 84 |
| O43837 | IDH3B | 385 | O00483 | NDUFA4 | 81 |
| P51553 | IDH3G | 393 | Q9NRX3 | NDUFA4L2 | 87 |
| P20839 | IMPDH1 | 514 | Q16718 | NDUFAS | 116 |
| P12268 | IMPDH2 | 514 | P56556 | NDUFA6 | 128 |
| Q9NPH2 | ISYNA1 | 558 | O95182 | NDUFA7 | 113 |
| O15229 | KMO | 486 | P51970 | NDUFA8 | 172 |
| Q16719 | KYNU | 465 | Q16795 | NDUFA9 | 377 |
| Q9H9P8 | L2HGDH | 463 | O14561 | NDUFAB1 | 156 |
| P00338 | LDHA | 332 | O75438 | NDUFB1 | 58 |
| Q6ZMR3 | LDHAL6A | 332 | O96000 | NDUFB10 | 172 |
| Q9BYZ2 | LDHAL6B | 381 | Q9NX14 | NDUFB11 | 153 |
| P07195 | LDHB | 334 | O95178 | NDUFB2 | 105 |
| P07864 | LDHC | 332 | O43676 | NDUFB3 | 98 |
| Q86WU2 | LDHD | 507 | O95168 | NDUFB4 | 129 |
| O00214 | LGALS8 | 317 | O43674 | NDUFB5 | 189 |
| P01229 | LHB | 141 | O95139 | NDUFB6 | 128 |
| P18858 | LIG1 | 919 | P17568 | NDUFB7 | 137 |
| P49916 | LIG3 | 1009 | O95169 | NDUFB8 | 186 |
| P49917 | LIG4 | 911 | Q9Y6M9 | NDUFB9 | 179 |
| P21397 | MAOA | 527 | O43677 | NDUFC1 | 76 |
| P27338 | MAOB | 520 | O95298 | NDUFC2 | 119 |
| P40925 | MDH1 | 334 | P28331 | NDUFS1 | 727 |
| P40926 | MDH2 | 338 | O75306 | NDUFS2 | 463 |
| P48163 | ME1 | 572 | O75489 | NDUFS3 | 264 |
| P23368 | ME2 | 584 | O43181 | NDUFS4 | 175 |
| Q16798 | ME3 | 604 | O43920 | NDUFSS | 106 |
| Q15800 | MSMO1 | 293 | O75380 | NDUFS6 | 124 |
| P00156 | MT- CYB | 380 | O75251 | NDUFS7 | 213 |
| P11586 | MTHFD1 | 935 | O00217 | NDUFS8 | 210 |
| P13995 | MTHFD2 | 350 | P49821 | NDUFV1 | 464 |
| Q9H903 | MTHFD2L | 347 | P19404 | NDUFV2 | 249 |
| P42898 | MTHFR | 656 | P56181 | NDUFV3 | 108 |
| P49914 | MTHFS | 203 | Q9HAN9 | NMNAT1 | 279 |
| Q13613 | MTMR1 | 665 | Q9BZQ4 | NMNAT2 | 307 |
| Q13614 | MTMR2 | 643 | Q96T66 | NMNAT3 | 252 |
| Q9Y217 | MTMR6 | 621 | Q9NWW6 | NMRK1 | 199 |
| Q9Y216 | MTMR7 | 660 | Q9NPI5 | NMRK2 | 230 |
| P03886 | MT-ND1 | 318 | P40261 | NNMT | 264 |
| P03891 | MT-ND2 | 347 | Q13423 | NNT | 1086 |
| P03897 | MT-ND3 | 115 | P15559 | NQO1 | 274 |
| P03905 | MT-ND4 | 459 | P16083 | NQO2 | 231 |
| P03901 | MT-ND4L | 98 | Q15738 | NSDHL | 373 |
| P03915 | MT-NDS | 603 | P49902 | NTSC2 | 561 |
| P03923 | MT-ND6 | 174 | Q9BQG2 | NUDT12 | 462 |
| Q99707 | MTR | 1265 | P00O24 | NUDT7 | 238 |
| O95544 | NADK | 446 | Q6D104 | NXN | 435 |
| Q4GON4 | NADK2 | 442 | Q04671 | OCA2 | 838 |
| Q6IA69 | NADSYN1 | 706 | Q02218 | OGDH | 1023 |
| P43490 | NAMPT | 491 | Q9ULDO | OGDHL | 1010 |
| Q6XQN6 | NAPRT | 538 | P55809 | OXCT1 | 520 |
| Q53GL7 | PARP10 | 1025 | Q9NRC8 | SIRT7 | 400 |
| Q9NR21 | PARP11 | 338 | P53007 | SLC25A1 | 311 |
| Q9HOJ9 | PARP12 | 701 | Q9UBX3 | SLC25A10 | 287 |
| Q460N5 | PARP14 | 1801 | Q02978 | SLC25A11 | 314 |
| Q460N3 | PARP15 | 678 | Q9BQT8 | SLC25A21 | 299 |
| Q8N5Y8 | PARP16 | 322 | O14975 | SLC27A2 | 620 |
| Q9UGNS | PARP2 | 583 | Q9Y2P5 | SLC27A5 | 690 |
| Q9Y6F1 | PARP3 | 533 | Q9NTN3 | SLC35D1 | 355 |
| Q9U10C3 | PARP4 | 1724 | Q8N808 | SLC35G3 | 338 |
| Q2NL67 | PARP6 | 630 | Q9Y6L6 | SLCO1B1 | 691 |
| Q8N3A8 | PARP8 | 854 | Q9NPDS | SLCO1B3 | 702 |
| Q8IXQ6 | PARP9 | 854 | P84022 | SMAD3 | 425 |
| P11498 | PC | 1178 | Q13485 | SMAD4 | 552 |
| P61457 | PCBD1 | 104 | Q00796 | SORD | 357 |
| Q9HONS | PCBD2 | 130 | Q9UHE8 | STEAP1 | 339 |
| P05165 | PCCA | 728 | Q8NFT2 | STEAP2 | 490 |
| P05166 | PCCB | 539 | Q658P3 | STEAP3 | 488 |
| P35558 | Pau | 622 | Q687X5 | STEAP4 | 459 |
| P08559 | PDHA1 | 390 | P08842 | STS | 583 |
| P29803 | PDHA2 | 388 | Q9P2R7 | SUCLA2 | 463 |
| P11177 | PDHB | 359 | P53597 | SUCLG1 | 346 |
| O00330 | PDHX | 501 | Q96I99 | SUCLG2 | 432 |
| P00558 | PGK1 | 417 | Q8NBK3 | SUMF1 | 374 |
| P07205 | PGK2 | 417 | P17735 | TAT | 454 |
| O43175 | PHGDH | 533 | O15178 | TBXT | 435 |
| Q9NRX4 | PHPT1 | 125 | O95455 | TGDS | 350 |
| Q9POZ9 | PIPDX | 390 | Q7Z3E1 | TIPARP | 657 |
| P00491 | PNP | 289 | O95271 | TNKS | 1327 |
| P01189 | POMC | 267 | Q9H2K2 | TNKS2 | 1166 |
| O43272 | PRODH | 600 | P60174 | TPI1 | 249 |
| Q9Y617 | PSAT1 | 370 | P51580 | TPMT | 245 |
| Q14914 | PTGR1 | 329 | O94759 | TRPM2 | 1503 |
| Q8N8N7 | PTGR2 | 351 | Q86TN4 | TRPT1 | 253 |
| Q8TE99 | PXYLP1 | 480 | P14679 | TYR | 529 |
| P32322 | PYCR1 | 319 | P17643 | TYRP1 | 537 |
| Q96C36 | PYCR2 | 320 | O60701 | UGDH | 494 |
| Q53H96 | PYCR3 | 274 | Q16851 | UGP2 | 508 |
| P09417 | QDPR | 244 | Q9UDW1 | UQCR10 | 63 |
| Q15274 | QPRT | 297 | P14927 | UQCRB | 111 |
| P11233 | RALA | 206 | P31930 | UQCRC1 | 480 |
| Q8IZVS | RDH10 | 341 | P22695 | UQCRC2 | 453 |
| O75452 | RDH16 | 317 | P47985 | UQCRFS1 | 274 |
| Q92781 | RDHS | 318 | P07919 | UQCRFI | 91 |
| Q6NUM9 | RETSAT | 610 | O14949 | UQCRQ | 82 |
| P12271 | RLBP1 | 317 | Q96N76 | UROC1 | 676 |
| Q16518 | RPE65 | 533 | Q8NBZ7 | UXS1 | 420 |
| O75845 | SCSD | 299 | P47989 | XDH | 1333 |
| Q8NBX0 | SCCPDH | 429 | |||
| P22307 | SCP2 | 547 | |||
| Q8N3Y7 | SDR16C5 | 309 | |||
| P34896 | SHMT1 | 483 | |||
| P34897 | SHMT2 | 504 | |||
| Q96EB6 | SIRT1 | 747 | |||
| Q81)06 | SIRT2 | 389 | |||
| Q9NTG7 | SIRT3 | 399 | |||
| Q9Y6E7 | SIRT4 | 314 | |||
| Q9NXA8 | SIRTS | 310 |
Footnotes
Author contribution: All authors have accepted responsibility for the entire content of this manuscript and approved its submission.
Research funding: This work was supported by FEDER (Programa Operacional Factores de Competi-tividade – COMPETE) and by FCT (Fundação para a Ciência e Tecnologia), within the grant SFRH-BD-108890-2015 to SDP, and the projects UIDB/04501/2020 (PO-CI-01-0145-FEDER-007628) to iBiMED, UIDB/00127/2020 to IEETA and UIDB/04279/2020 to CIIS. FCT/MCTES and UCP support CEEC institutional funding of RMS (CEECINST/00137/2018/CP1520/CT0012).
Conflict of interest statement: Authors state no conflict of interest.
References
- 1.Haigis MC, Sinclair DA. Mammalian sirtuins: biological insights and disease relevance. Annu Rev Pathol. 2010;5:253–95. doi: 10.1146/annurev.pathol.4.110807.092250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gupte R, Liu Z, Kraus WL. PARPs and ADP-ribosylation: recent advances linking molecular functions to biological outcomes. Genes Dev. 2017;31:101–26. doi: 10.1101/gad.291518.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bütepage M, Eckei L, Verheugd P, Lüscher B. Intracellular mono-ADP-ribosylation in signaling and disease. Cells. 2015;4:569–95. doi: 10.3390/cells4040569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chini EN. CD38 as a regulator of cellular NAD: a novel potential pharmacological target for metabolic conditions. Curr Pharmaceut Des. 2009;15:57–63. doi: 10.2174/138161209787185788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li J, Bonkowski MS, Moniot S, Zhang D, Hubbard BP, Ling AJ, et al. A conserved NAD+ binding pocket that regulates protein-protein interactions during aging. Science. 2017;355:1312–7. doi: 10.1126/science.aad8242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carreras-Puigvert J, Zitnik M, Jemth AS, Carter M, Unterlass JE, Hallström B, et al. A comprehensive structural, biochemical and biological profiling of the human NUDIX hydrolase family. Nat Commun. 2017;8:1541. doi: 10.1038/s41467-017-01642-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kulikova VA, Nikiforov AA. Role of NUDIX hydrolases in NAD and ADP-ribose metabolism in mammals. Biochemistry (Mosc) 2020;85:883–94. doi: 10.1134/S0006297920080040. [DOI] [PubMed] [Google Scholar]
- 8.McLennan AG. The Nudix hydrolase superfamily. Cell Mol Life Sci. 2006;63:123–43. doi: 10.1007/s00018-005-5386-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Santos L, Colman L, Contreras P, Chini CC, Carlomagno A, Leyva A, et al. A novel form of Deleted in breast cancer 1 (DBC1) lacking the N-terminal domain does not bind SIRT1 and is dynamically regulated in vivo. Sci Rep. 2019;9:14381. doi: 10.1038/s41598-019-50789-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Amé JC, Spenlehauer C, de Murcia G. The PARP superfamily. Bioessays. 2004;26:882–93. doi: 10.1002/bies.20085. [DOI] [PubMed] [Google Scholar]
- 11.Duarte-Pereira S, Matos S, Oliveira JL, Silva RM. The NAD interactome, identification of putative new NAD-binding proteins. In: Fdez-Riverola F, Rocha M, Mohamad MS, Caraiman S, Gil-González AB, editors. Practical Applications of Computational Biology and Bioinformatics, 16th International Conference (PACBB 2022) Lecture Notes in Networks and Systems. 1st ed. Cham: Springer; 2022. [Google Scholar]
- 12.The UniProt Consortium UniProt: the universal protein knowledgebase. Nucleic Acids Res. 2017;45:D158–69. doi: 10.1093/nar/gkw1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Orchard S, Kerrien S, Abbani S, Aranda B, Bhate J, Bidwell S, et al. Protein interaction data curation: the International Molecular Exchange (IMEx) consortium. Nat Methods. 2012;9:345–50. doi: 10.1038/nmeth.1931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stark C, Breitkreutz BJ, Reguly T, Boucher L, Breitkreutz A, Tyers M. BioGRID: a general repository for interaction datasets. Nucleic Acids Res. 2006;34:D535–9. doi: 10.1093/nar/gkj109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Szklarczyk D, Gable AL, Lyon D, Junge A, Wyder S, Huerta-Cepas J, et al. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019;47:D607–13. doi: 10.1093/nar/gky1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mi H, Muruganujan A, Huang X, Ebert D, Mills C, Guo X, et al. Protocol Update for large-scale genome and gene function analysis with the PANTHER classification system (v.14.0) Nat Protoc. 2019;14:703–21. doi: 10.1038/s41596-019-0128-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ansari HR, Raghava GP. Identification of NAD interacting residues in proteins. BMC Bioinf. 2010;11:160. doi: 10.1186/1471-2105-11-160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Grosdidier A, Zoete V, Michielin O. SwissDock, a protein-small molecule docking web service based on EADock DSS. Nucleic Acids Res. 2011;39:W270–7. doi: 10.1093/nar/gkr366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jumper J, Evans R, Pritzel A, Green T, Figurnov M, Ronneberger O, et al. Highly accurate protein structure prediction with AlphaFold. Nature. 2021;596:583–9. doi: 10.1038/s41586-021-03819-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Uhlén M, Fagerberg L, Hallström BM, Lindskog C, Oksvold P, Mardinoglu A, et al. Proteomics. Tissue-based map of the human proteome. Science. 2015;347:1260419. doi: 10.1126/science.1260419. [DOI] [PubMed] [Google Scholar]
- 21.Consortium U. UniProt: a worldwide hub of protein knowledge. Nucleic Acids Res. 2019;47:D506–15. doi: 10.1093/nar/gky1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Barz MJ, Hof J, Groeneveld-Krentz S, Loh JW, Szymansky A, Astrahantseff K, et al. Subclonal NT5C2 mutations are associated with poor outcomes after relapse of pediatric acute lymphoblastic leukemia. Blood. 2020;135:921–33. doi: 10.1182/blood.2019002499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li J, Mahajan A, Tsai MD. Ankyrin repeat: a unique motif mediating protein-protein interactions. Biochemistry. 2006;45:15168–78. doi: 10.1021/bi062188q. [DOI] [PubMed] [Google Scholar]
- 24.Kumar A, Balbach J. Folding and stability of ankyrin repeats control biological protein function. Biomolecules. 2021;11:840. doi: 10.3390/biom11060840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wu H, Li L, Chen KM, Homolka D, Gos P, Fleury-Olela F, et al. Decapping enzyme NUDT12 partners with BLMH for cytoplasmic surveillance of NAD-capped RNAs. Cell Rep. 2019;29:4422–34. doi: 10.1016/j.celrep.2019.11.108. [DOI] [PubMed] [Google Scholar]
- 26.Samanta A, Hughes TET, Moiseenkova-Bell VY. Transient receptor potential (TRP) channels. Subcell Biochem. 2018;87:141–65. doi: 10.1007/978-981-10-7757-9_6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wen W, Yan J, Zhang M. Structural characterization of the split pleckstrin homology domain in phospholipase C-gamma1 and its interaction with TRPC3. J Biol Chem. 2006;281:12060–8. doi: 10.1074/jbc.M600336200. [DOI] [PubMed] [Google Scholar]
- 28.Liu Y, Bunney TD, Khosa S, Macé K, Beckenbauer K, Askwith T, et al. Structural insights and activating mutations in diverse pathologies define mechanisms of deregulation for phospholipase C gamma enzymes. EBioMedicine. 2020;51:102607. doi: 10.1016/j.ebiom.2019.102607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yu P, Cai X, Liang Y, Wang M, Yang W. Roles of NAD+ and its metabolites regulated calcium channels in cancer. Molecules. 2020;25:4826. doi: 10.3390/molecules25204826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huang Q, Wang X, Lin X, Zhang J, You X, Shao A. The role of transient receptor potential channels in blood-brain barrier dysfunction after ischemic stroke. Biomed Pharmacother. 2020;131:110647. doi: 10.1016/j.biopha.2020.110647. [DOI] [PubMed] [Google Scholar]
- 31.Wang L, Fu TM, Zhou Y, Xia S, Greka A, Wu H. Structures and gating mechanism of human TRPM2. Science. 2018;362:eaav4809. doi: 10.1126/science.aav4809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sano Y, Inamura K, Miyake A, Mochizuki S, Yokoi H, Matsushime H, et al. Immunocyte Ca2+ influx system mediated by LTRPC2. Science. 2001;293:1327–30. doi: 10.1126/science.1062473. [DOI] [PubMed] [Google Scholar]
- 33.Schmidt C, Sciacovelli M, Frezza C. Fumarate hydratase in cancer: a multifaceted tumour suppressor. Semin Cell Dev Biol. 2020;98:15–25. doi: 10.1016/j.semcdb.2019.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Duarte RRR, Bachtel ND, Côtel MC, Lee SH, Selvackadunco S, Watson IA, et al. The psychiatric risk gene NT5C2 regulates adenosine monophosphate-activated protein kinase signaling and protein translation in human neural progenitor cells. Biol Psychiatr. 2019;86:120–30. doi: 10.1016/j.biopsych.2019.03.977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vergis JM, Beardsley GP. Catalytic mechanism of the cyclohydrolase activity of human aminoimidazole carboxamide ribonucleotide formyltransferase/inosine monophosphate cyclohydrolase. Biochemistry. 2004;43:1184–92. doi: 10.1021/bi035139b. [DOI] [PubMed] [Google Scholar]
- 36.Biedermann J, Preussler M, Conde M, Peitzsch M, Richter S, Wiedemuth R, et al. Mutant IDH1 differently affects redox state and metabolism in glial cells of normal and tumor origin. Cancers. 2019;11:2028. doi: 10.3390/cancers11122028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cerchione C, Romano A, Daver N, DiNardo C, Jabbour EJ, Konopleva M, et al. IDH1/IDH2 inhibition in acute myeloid leukemia. Front Oncol. 2021;11:639387. doi: 10.3389/fonc.2021.639387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Houtkooper RH, Cantó C, Wanders RJ, Auwerx J. The secret life of NAD+: an old metabolite controlling new metabolic signaling pathways. Endocr Rev. 2010;31:194–223. doi: 10.1210/er.2009-0026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tan MH, Li J, Xu HE, Melcher K, Yong EL. Androgen receptor: structure, role in prostate cancer and drug discovery. Acta Pharmacol Sin. 2015;36:3–23. doi: 10.1038/aps.2014.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bianchi VE, Rizzi L, Bresciani E, Omeljaniuk RJ, Torsello A. Androgen therapy in neurodegenerative diseases. J Endocr Soc. 2020;4:bvaa120. doi: 10.1210/jendso/bvaa120. [DOI] [PMC free article] [PubMed] [Google Scholar]