Abstract
Me31B/DDX6 is a DEAD-box family RNA helicase playing roles in post-transcriptional RNA regulation in different cell types and species. Despite the known motifs/domains of Me31B, the in vivo functions of the motifs remain unclear. Here, we used the Drosophila germline as a model and used CRISPR to mutate the key Me31B motifs/domains: helicase domain, N-terminal domain, C-terminal domain, and FDF-binding motif. Then, we performed screening characterization on the mutants and report the effects of the mutations on the Drosophila germline, on processes such as fertility, oogenesis, embryo patterning, germline mRNA regulation, and Me31B protein expression. The study indicates that the Me31B motifs contribute different functions to the protein and are needed for proper germline development, providing insights into the in vivo working mechanism of the helicase.
Graphical Abstract
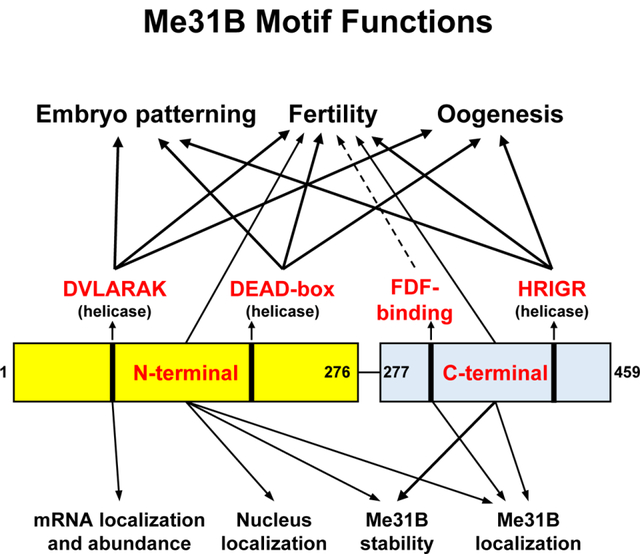
Me31B RNA helicase is essential for Drosophila germline development. This mutational analysis targets six key motifs/domains of Me31B: three motifs involved in the helicase activity (DVLARAK, DEAD-box, and HRIGR), N-terminal domain, C-terminal domain, and FDF-binding motif. These motifs/domains are found to play important functions in embryo patterning, fertility, oogenesis, germline mRNA localization and abundance, and Me31B protein stability and subcellular localization.
Introduction
Drosophila Me31B is an evolutionarily conserved, ATP-dependent RNA helicase that is essential for Drosophila germline development and fertility [1–5]. The helicase has orthologs like CGH-1 (worms) [6, 7], DjCBC-1 (planarians) [8, 9], Xp54 (frogs) [10, 11], p54 (mouse) [12, 13], and DDX6/Rck (humans) [14–18] playing similar roles in diverse cell types and animal species. In these animal cells, the main role of these helicases lies in their post-transcriptional RNA regulations including RNA storage, transportation, translational regulation, stabilization, and decay. This ensures the expression of the mRNAs at the correct time and location, which eventually leads to proper germline functioning and development [14, 19–23].
Drosophila germline has been often employed as a model to study the roles of Me31B/DDX6 helicase. Importantly, Me31B is essential for Drosophila germline growth: me31B loss-of-function mutation or strong knockdown (KD) causes severe egg chamber development defects or early oogenesis arrest [1, 24]. In normal egg chambers, Me31B proteins express and aggregate into ribonucleoprotein (RNP) complexes (granular, membrane-less assemblies of proteins and RNAs). In these granules, Me31B complexes with other partner proteins like Tral and Cup (two translational repressor proteins that usually complex with Me31B in various types of germline RNPs) to render post-transcriptional controls on the RNAs within the granules [3, 25–29]. So far, our understanding of Me31B’s role in the germline mostly came from the use of complete loss-of-function me31B alleles, strains with a significant loss of Me31B protein expression, or in vitro biochemical analysis. To gain insights into Me31B’s molecular-level working mechanism, this study investigates the in vivo roles of key Me31B motifs and aims to understand how the motifs contribute to Me31B’s whole-protein functions in the context of Drosophila germline development.
Me31B (459 AA) has been structurally identified as a DEAD-box-family RNA helicase (reviews on DEAD-box helicases in [19, 30]). Like other helicases in the family, Me31B’s helicase activity is enabled by the cooperative actions of its N-terminal (N-ter) RecA-like domain (AA 1 – 267) and C-terminal (C-ter) RecA-like domain (AA 268 – 459) [19, 31–33]. And, three well-conserved sequence motifs, DVLARAK motif (AA 97 – 103, within the N-ter), DEAD motif (also known as the “DEAD” box, AA 207 – 210, within the N-ter), and HRIGR (AA 381 – 385, within the C-ter) (see their location illustrations in Figure 1) are crucial for the helicase activity of the protein. Previous in vitro studies showed that mutations in any of the three motifs (DVLARAK→DVLAAAA, DEAD→DAAD, or HRIGR→HRIGQ) strongly disrupt the helicase activity [34–36]. Therefore, our study uses the above three substitution mutations individually to gauge the in vivo role of Me31B’s helicase activity. Besides the helicase activity, we analyze the in vivo roles of the N-ter RecA-like domain (independently), C-ter RecA-like domain (independently), and the FDF-binding motif (CLNTL, AA 285 – 289) because they are key components and contributors to Me31B’s whole-protein functions. The N-ter and C-ter domains, besides their involvement in the helicase activity, participate in other functions including partner-protein interactions and Me31B assembly into RNP granules [19, 34, 37]; The FDF-binding motif enables Me31B to physically bind to the FDF motifs on translational repressor partners like Tral and EDC3 [32, 33, 37, 38]. To mutate the above target motifs, we used the CRISPR technique and created Drosophila strains with point mutations or deletion mutations in them (Figure 1). Analysis of the resulting me31B mutants revealed that the helicase activity, the N-ter domain, the C-ter domain, and the FDF-binding motif are needed for proper Drosophila germline development and fertility, yet they play different molecular-level roles in localizing germline RNA, maintaining germline RNA level, stabilizing Me31B protein, localizing the protein, and interacting with partner proteins.
Figure 1. Diagram of the Me31B wildtype and mutant proteins in the Drosophila strains generated in this study.
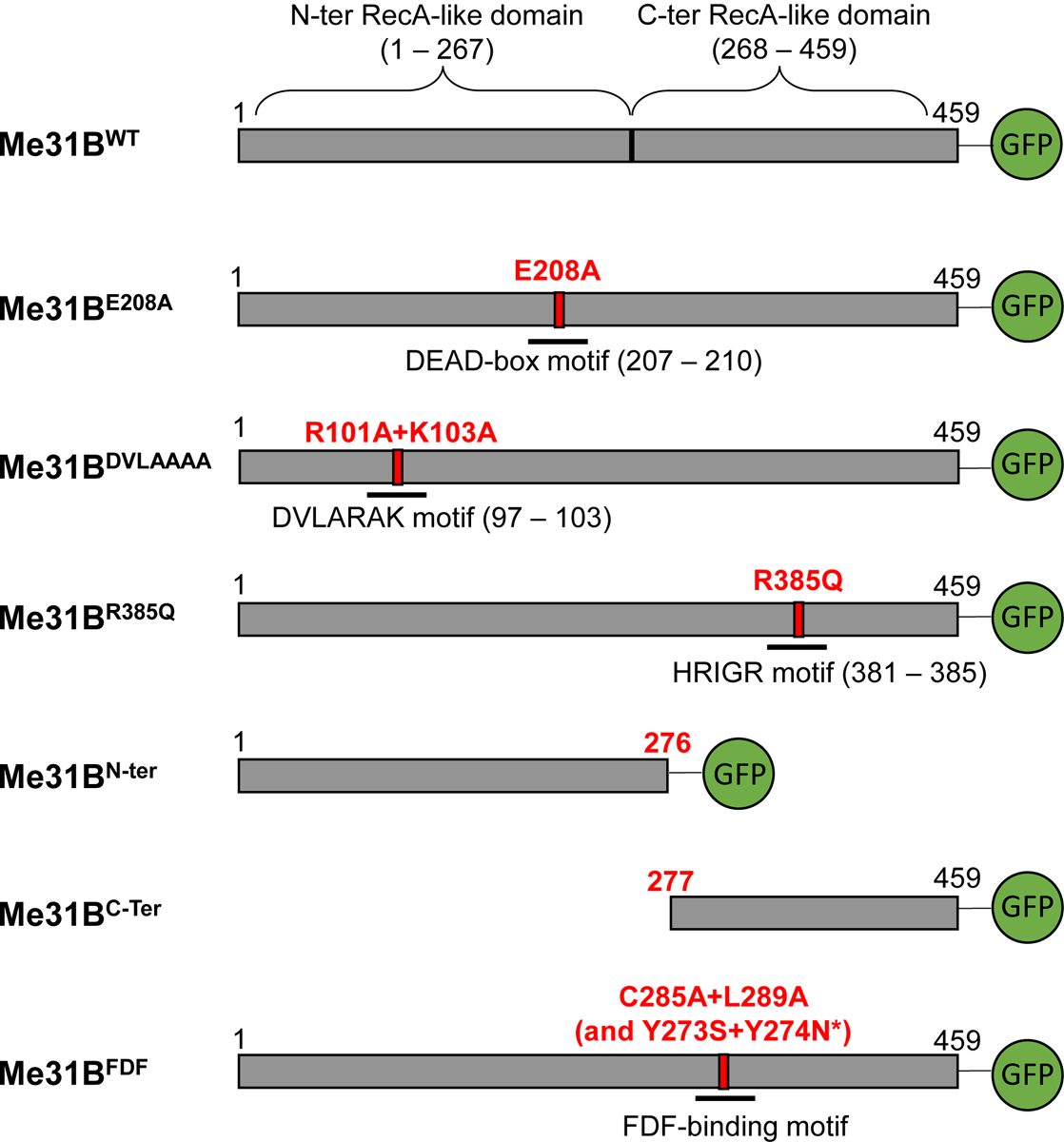
The bar after each Me31B protein name represents the primary structure of the protein. The numbers on top of the bars mark the number of amino acids and their positions in the protein. Green Fluorescence Protein (GFP) tags are expressed at the C-terminal end of the constructed Me31B proteins. The point mutations are as follows: E208A, glutamic acid 208 replaced by alanine; R101A, arginine 101 replaced by alanine; K103A, lysine 103 replaced by alanine; R385Q, arginine 385 replaced by glutamine; C285A, cysteine 285 replaced by alanine; L289A, leucine 289 replaced by alanine; Y273S, tyrosine 273 replaced by serine; Y274N, tyrosine 274 replaced by asparagine. Me31BN-ter protein (deletion of amino acids 277 to 459) contains the first 276 amino acids of the wild-type protein. Me31BC-ter protein (deletion of amino acids 1 to 276) contains the last 183 amino acids of the wild-type protein.
*Note that two unintended, missense mutations were detected in the me31BFDF strain when sequencing its me31B gene. The two mutations (Y273S and Y274N) are outside the FDF-binding motif.
Results
Me31B helicase mutations lead to dominant female sterility.
To elucidate the in vivo roles of Me31B’s helicase activity, we used CRISPR and generated mutations individually in the three motifs (DEAD, DVLARAK, and HRIGR motifs) that are crucial for Me31B’s helicase activities including ATP hydrolysis, RNA binding, and RNA unwinding [11, 19, 33, 34, 36, 39, 40]. Specifically, the substitution mutations were: E208A (DEAD→DAAD), R101A and K103A (DVLARAK→DVLAAAA), and R385Q (HRIGR→HRIGQ), and these substitutions were made according to previous studies showing that these mutations strongly disrupt the helicase activities of Me31B or structurally similar DEAD-box helicases [34–36, 40–43]. For simplicity, the three motifs hereafter will be referred to as helicase motifs. The resulting me31B alleles are named me31BE208A, me31BDVLAAAA, and me31BR385Q, respectively (illustrated in Figure 1). After obtaining the three mutant strains, we first noticed that all three alleles dominantly caused female sterility. In detail, me31BE208A/+, me31BDVLAAAA/+, and me31BR385Q/+ heterozygous females (in the presence of wildtype w1118 males) do not produce any viable progeny. In contrast, me31BE208A/+, me31BDVLAAAA/+, and me31BR385Q/+ heterozygous males can fertilize w1118 females and produce viable progenies bearing the mutant me31B alleles. At the same time, the me31BWT control allele (generated in this study by the same CRISPR procedure without altering the me31B gene sequence) showed wildtype-like fertility. Therefore, we conclude that the three helicase alleles cause dominant sterility in females and suggest that Me31B’s helicase activity is integral for female fertility.
Me31B helicase mutations lead to maternal-effect embryo patterning and oogenesis defects
To analyze what caused the sterility in the helicase mutants (me31BE208A/+, me31BDVLAAAA/+, and me31BR385Q/+), we screened the three strains for egg laying, morphology, and development. The me31BWT/+ control showed similar phenotypes in the above areas as the w1118 strain (Supplementary Table 1 and Figure 2). In contrast, although egg laying was observed from all three helicase mutants, none (0%) of their eggs (n = 541 for me31BE208A/+, n = 100 for me31BDVLAAAA/+, and n = 17 for me31BR385Q/+) were able to develop into larva or later stages (Supplementary Table 1; See the Materials and Methods for the hatchability assay conditions). Interestingly, different portions of the mutants’ un-hatchable embryos showed morphological defects: 78% of the me31BE208A/+ embryos, 33% of the me31BDVLAAAA/+ embryos, and 100% of the me31BR385Q/+ embryos had one or more types of patterning defects like fused dorsal appendages, very short, or no dorsal appendages (Supplementary Table 1 and Figure 2A). Although the me31BE208A and me31BDVLAAAA mutants were able to produce normal-appearance embryos, none of them could develop into larvae or hatch. Among the three helicase mutants, me31BR385Q/+ showed the most severe phenotype: the flies laid few eggs, and all the embryos (100%) had drastic morphological defects including the complete missing of dorsal appendages (Figure 2A).
Figure 2. Defective embryo patterning and oogenesis in heterozygous me31BE208A, me31BDVLAAAA, and me31BR385Q strains.
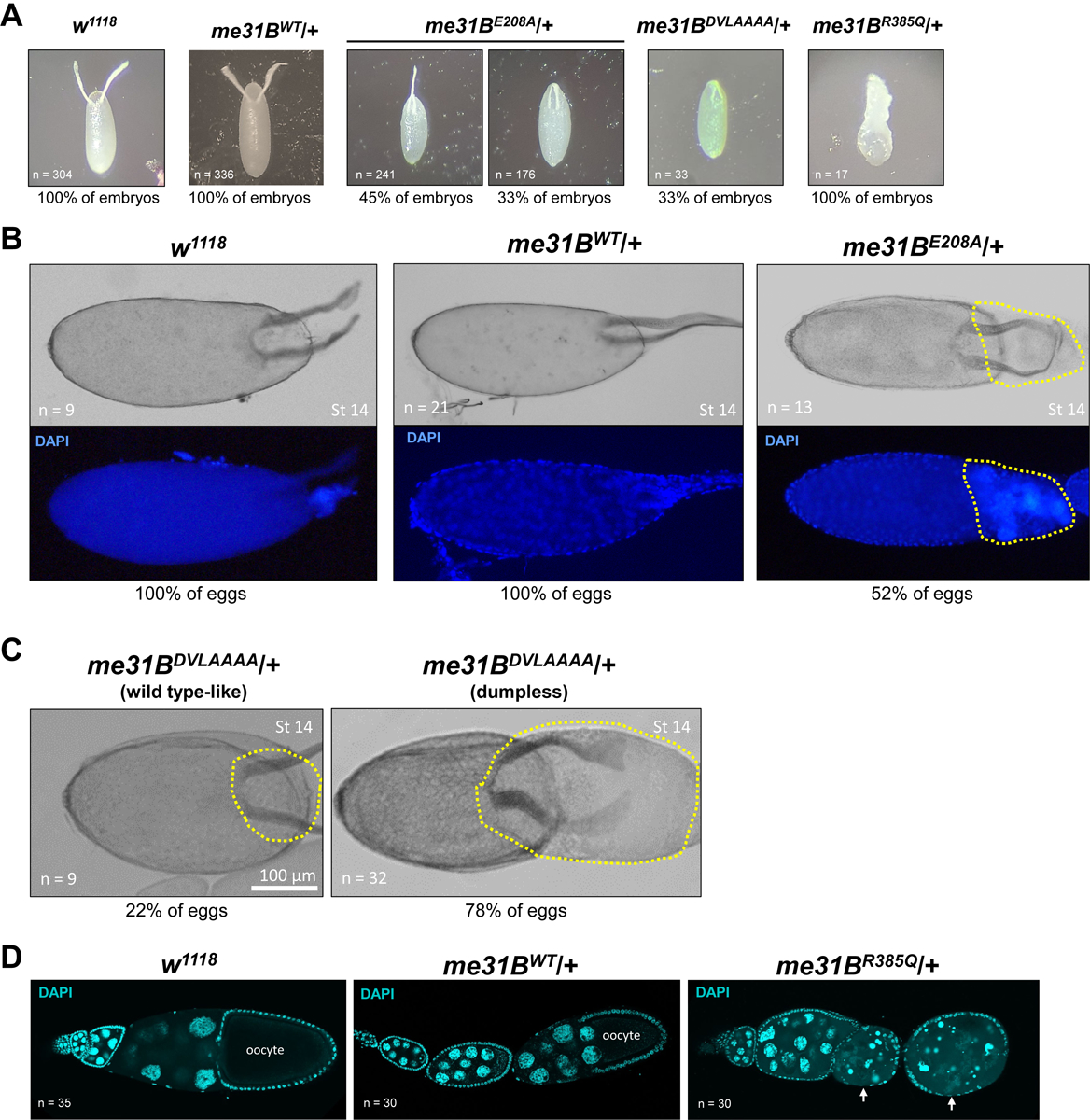
(A) Embryos laid by w1118 and me31BWT/+ control females all had normal (100%) dorsal appendages. In contrast, embryos laid by me31BE208A/+ females had fused (45%), shortened (33%), and normal (22%, Supplementary Table 1, picture not shown) dorsal appendages; embryos laid by me31BDVLAAAA/+ females had shortened (33%) or normal (67%, not shown) dorsal appendages; embryos laid by me31BR385Q/+ females all had severe morphological defects (100%). (B) In 52% of late-stage (stage 14) eggs of me31BE208A/+ mutant (right panel), dumping of the nurse cell content into the oocyte was incomplete (see the highlighted region with yellow dashed lines), in contrast to the successful dumping in the w1118 and me31BWT/+ control eggs. DAPI staining of the same eggs was shown at the bottom panels, and nurse cell nuclei-like materials were present in the “un-dumped” region of the me31BE208A/+ mutant egg. (C) Similar dumpless phenotypes were observed in 78% of me31BDVLAAAA/+ mutant’s late-stage (stage 14) eggs. The “un-dumped” regions (highlighted with yellow dashed lines) were broader than the me31BE208A/+ mutant in (B). Only 22% of the me31BDVLAAAA/+ mutant’s eggs appeared normal. (D) Ovarioles from w1118, me31BWT/+, and me31BR385Q/+ females were stained by DAPI stain. The ovarioles are oriented so that the early-stage egg chambers are on the left and the later-stage egg chambers are on the right. Early-to-mid-stage egg chamber degenerations in me31BR385Q/+ mutant (right panel) are indicated by arrows, in contrast to the successfully developed early- and mid-stage egg chambers in the w1118 and me31BWT/+ controls.
The embryo patterning defects in the helicase mutants indicate that their oogenesis could be abnormal, so we examined their ovarian tissues. Indeed, in me31BE208A and me31BDVLAAAA ovaries, we observed abnormal nurse cell dumping, a process in which the nurse cells completely expel all their cytoplasm into late-stage eggs (reviewed in [44]). In the me31BE208A mutant, 52% of the stage-14 eggs (n = 13) were still connected with egg chamber tissues that contain nurse cell materials (Figure 2B), indicating incomplete nurse cell dumping. This “dumpless” phenotype is more prominent in the me31BDVLAAAA mutant: approximately 78% of the me31BDVLAAAA stage-14 egg chambers (n = 32) did not complete dumping, and the “un-dumped” regions appeared larger than that in the me31BE208A mutant (Figure 2C). The most severe oogenesis defects were observed in the me31BR385Q mutant. Besides the dumpless phenotype (Supplementary Figure 2), this mutant showed frequent egg chamber degenerations even at early-to-mid stages (Figure 2D), consistent with this mutant’s few egg lay and sterility. These data indicate that Me31B helicase activity is needed for proper oogenesis.
Me31B helicase mutations decrease Me31B protein levels from the alleles
To determine whether the helicase mutants’ sterility was associated with the levels of mutant Me31B expression, we performed anti-GFP Western blots to quantify the proteins expressed from me31BE208A, me31BDVLAAAA, me31BR385Q, and me31BWT (control) alleles in heterozygous flies (homozygotes cannot be obtained for the helicase alleles because of their dominant sterility effect). The Me31BE208A-GFP, Me31BDVLAAAA-GFP, and Me31BR385Q-GFP protein levels were 68% (p > 0.05), 53% (p < 0.05), and 23% (p < 0.05) of the Me31BWT-GFP control, respectively (Figure 3 and Supplementary Figure 1). With these results, we conclude that Me31B helicase-motif mutations decrease the expression of the proteins, though to different extents (Me31BE208A < Me31BDVLAAAA < Me31BR385Q, decreasing effects from mild to strong). However, we reason that the protein level decreases cannot fully explain the mutant me31B alleles being dominant, especially when these mutant proteins were expressed in a heterozygous background. Therefore, we suggest that it is likely the altered Me31B protein functions rather than abundance that caused the dominant sterility. This notion is also indirectly supported by another me31B mutant strain which expresses a low amount of Me31B protein (40% of wildtype level) but remains fertile (see the me31BN-ter strain results below).
Figure 3. Western blot quantification of mutant Me31B protein levels in me31BE208A, me31BDVLAAAA, and me31BR385Q heterozygous strains.
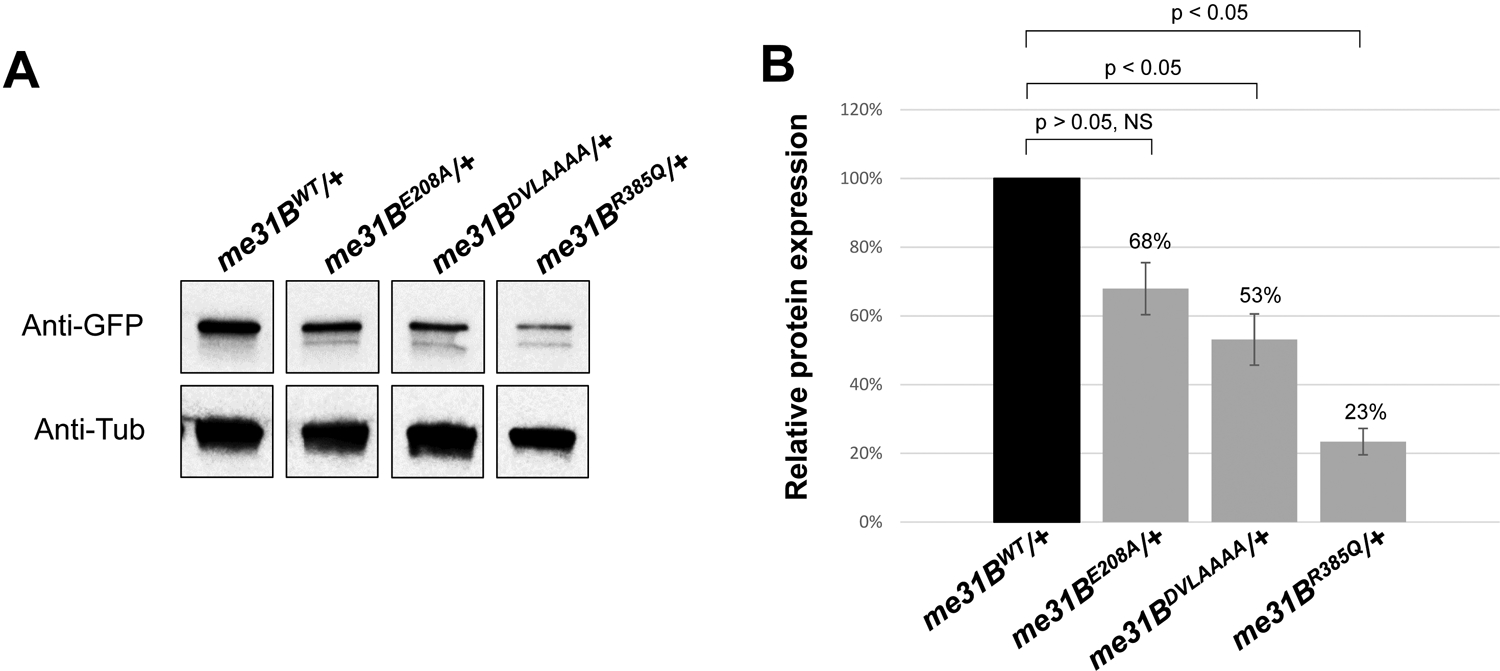
(A) Anti-GFP Western blots were used to quantify the Me31BWT-GFP, Me31BE208A-GFP, Me31BDVLAAAA-GFP, and Me31BR385Q-GFP proteins in the ovaries of the corresponding heterozygous strains. Anti-tubulin Western blots were used as loading controls. The expression levels of Me31BE208A-GFP, Me31BDVLAAAA-GFP, and Me31BR385Q-GFP proteins were lower than the Me31BWT control. The images shown are representative images of three biological replicates. The additional, uncropped biological replicate images are presented in Supplementary Figure 1. (B) The Me31BE208A-GFP, Me31BDVLAAAA-GFP, and Me31BR385Q-GFP protein levels are 68% (p > 0.05), 53% (p < 0.05), and 23% (p < 0.05) relative to the Me31BWT-GFP control, respectively. Western blot image analysis was performed with ImageJ and protein quantification was normalized by using the alpha-tubulin proteins. NS, not statistically significant. Error bar represents the standard error of the mean.
Me31B helicase activity functions in nos localization by modulating osk and nos transcript levels
Next, we probed further into the molecular underpinnings of the helicase mutants’ sterility. Considering Me31B’s known role in post-transcriptional RNA regulations, we hypothesized that mutations in Me31B helicase activity could affect the localization and/or stability of important germline mRNAs such as nanos (nos). Normal nos localization occurs through the Oskar-dependent formation of homotypic clusters within germ granules, aggregates that contain multiple copies of nos transcripts [45, 46]. Transcripts of nos begin to localize to their germ plasm destination during mid-oogenesis in a stochastic seeding and self-recruitment process that continues until the early embryo [46] (germ plasm is a special cytoplasm at the posterior pole of late-stage eggs and early embryos; germ plasm contains mRNAs needed for processes including embryo patterning and germ cell formation. For reviews, see [47–51]). To test whether nos localization is affected by helicase activity mutations, we performed single-molecule fluorescent in situ hybridization (smFISH) on stage-13 oocytes to identify both unlocalized single nos transcripts that reside outside the germ plasm and nos homotypic clusters found within the germ plasm. Using previously established imaging and image analysis techniques [45, 46] (See Material and Methods), we first determined that there was an average of 7.82 ± 0.56 nos transcripts per cluster in yellow-white (yw) flies, which was consistent with the previously published value [46]. The yw average nos cluster size was not significantly different from the control CRISPR strain (me31BWT-GFP) that generated an average nos cluster size of 6.42 ± 0.21 (p > 0.46), demonstrating that our CRISPR genome modifications do not affect nos localization. Using the me31BDVLAAAA/+ strain as a representative for disruption of Me31B helicase activity, we found that the average number of nos transcripts found in a homotypic cluster was 2.39 ± 0.21, which was a significant reduction when compared to the me31BWT-GFP control (p < 0.002). Together, these data suggest that the Me31B helicase activity influences nos localization to the germ plasm.
We next explored the mechanisms by which Me31B influences nos localization. Recently, it has been shown using computational modeling and experimental validation that the localization of nos is influenced by 1) the number of nos transcripts expressed, 2) the amount of osk in the system, which contributes to germ granules’ carrying capacity, and 3) nos’s clustering factor, a quantifiable effect that, in conjunction with nos transcript and osk levels, regulates the number of transcripts that can accumulate within a homotypic cluster [46, 52]. To identify whether me31BDVLAAAA affects one or more of these mechanisms, we first conducted qRT-PCR to measure the levels of nos and osk RNAs in the ovaries. We found that nos transcript levels were reduced to 61% ± 7% of wild-type levels and osk levels were reduced to 56% ± 4% when compared to wild-type (p < 0.05) (Figure 4C). To determine whether these changes can account for the observed reduction of nos homotypic cluster sizes in the me31BDVLAAAA mutant, we used computational modeling to recapitulate nos localization in silico by adjusting a previously published model’s parameters [52] from 1.00 to 0.61 for nos transcript levels and from 1.00 to 0.56 for germ granule carrying capacity (See Material and Methods). Modeled nos homotypic clusters contained an average of 2.58 ± 0.06 transcripts per cluster which was not significantly different from the average produced by the me31BDVLAAAA mutant (p > 0.99) (Figure 4B). Together, these data suggest that the mechanism creating the nos localization defect in the me31BDVLAAAA mutant is caused by a combined effect generated by the simultaneous reduction of nos and osk levels.
Figure 4. smFISH analysis of nos mRNA in me31BDVLAAAA heterozygous mutant and quantitative RT-PCR analysis of nos and osk mRNA levels in heterozygous me31BE208A, me31BDVLAAAA, and me31BR385Q mutants.
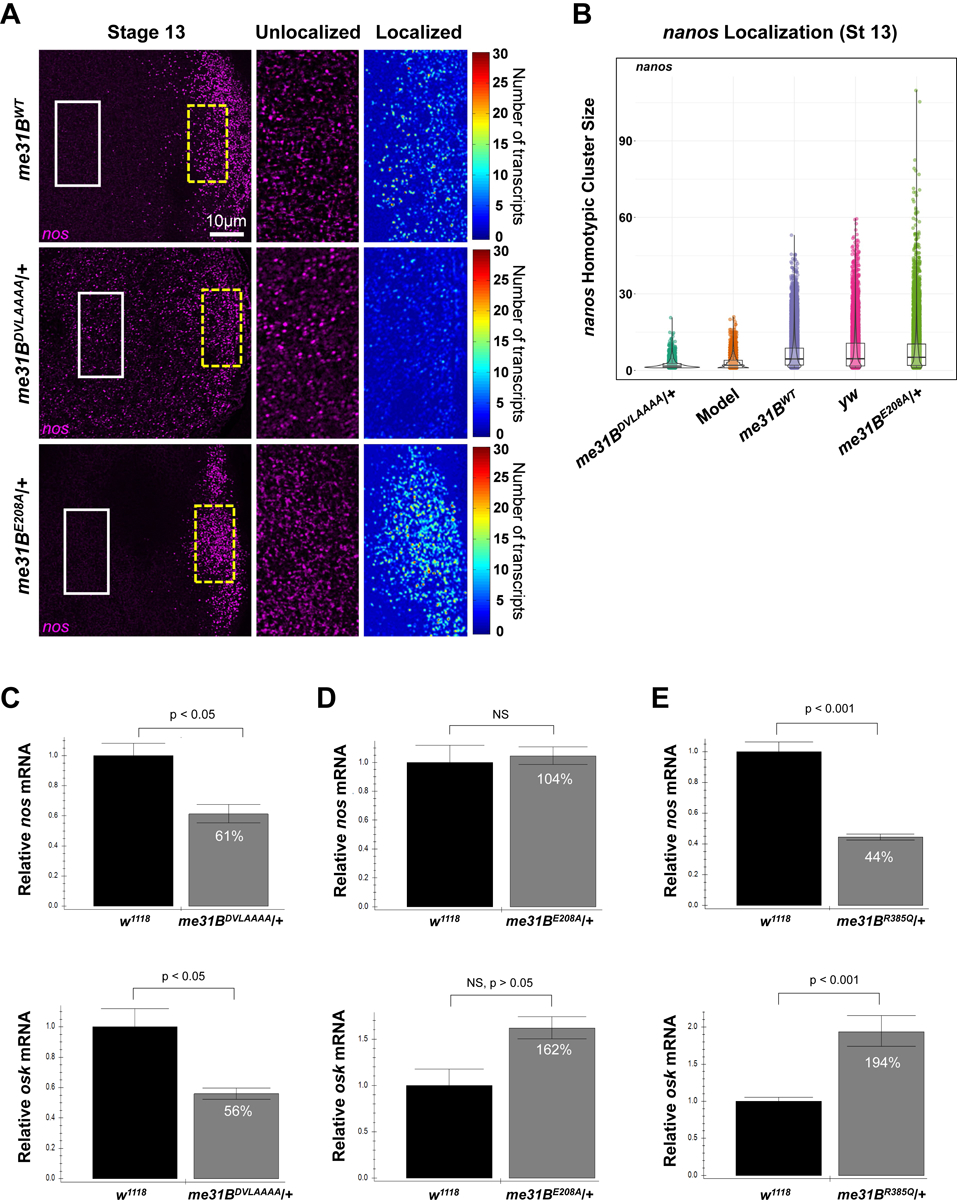
(A) Stage 13 oocytes from me31BWT, me31BDVLAAAA, and me31BE208A flies, with nos (magenta) mRNAs detected using smFISH. The white solid box indicates the unlocalized single transcripts of nos that are found in the bulk cytoplasm (enlarged in the second panel) while the broken yellow box highlights nos that have localized to the germ plasm by forming homotypic clusters (enlarged and shown as a heatmap in the third panel). (B) The distribution of nos cluster size (number of nos transcripts calculated within a granule) found in each genotype’s germ plasm and the computational model. Clusters were identified and analyzed from a minimum of four germ plasm replicates and included a total of 3,295 clusters from me31BDVLAAAA, 5,551 from the computational model, 19,195 from me31BWT, 10,383 from yellow-white (yw), and 13,131 from me31BE208A. Of note, only oocytes that did not display a dumpless phenotype were included in this analysis. (C) In me31BDVLAAAA/+ ovaries, nos transcript level is 61% ± 7% of that in the w1118 control (p < 0.05), and osk transcript level is 56% ± 4% of that in the control (p < 0.05). (D) In me31BE208A/+ ovaries, nos transcript level is 104% ± 7% of that in the w1118 control (NS) and osk transcript level is 162% ± 12% of that in the control (NS, p > 0.05). (E) In me31BR385Q/+ ovaries, nos transcript level is 44% ± 2% of that in the w1118 control (p < 0.001), and osk transcript level is 194% ± 12% of that in the control (p < 0.001). NS, not statistically significant. Error bar represents the standard error of the mean. Three biological replicates (each with three technical replicates) were performed for each strain’s RT-PCR experiments.
Next, we wondered whether these nos and osk transcript level changes also occur in the other two helicase mutants (me31BE208A/+ and me31BR385Q/+). To this end, we measured the transcript levels of osk and nos in the two mutants. In the me31BE208A/+ mutant, both nos and osk levels were not significantly different from the w1118 control (p > 0.05) (Figure 4D). Using smFISH, we determined that me31BE208A/+ flies had an average nos cluster size of 6.67 ± 0.88, which was not significantly different from the me31BWT-GFP control (p > 0.99). Interestingly, we identified a small number of nos clusters from me31BE208A/+ flies, 20 out of 13,131, that had nos cluster sizes that were larger than expected when compared to yw and/or the me31BWT-GFP control (Figure 4B), suggesting that a small subset of germ granules may have a larger carrying capacity in the me31BE208A/+ mutant (See Discussion). In the me31BR385Q/+ mutant, the nos mRNA showed a significant decrease (44% ± 2%), while the osk level showed a significant increase (194% ± 12%) when compared to the control (Figure 4E). Of note, smFISH experiments on me31BR385Q/+ oocytes did not produce quantifiable results because the strain produces severe oocyte defects (Supplementary Figure 2) and lays few eggs that have severe morphological defects (Figure 2A). Comparing the changes of nos and osk between the three helicase mutants (compare Figure 4C, D, and E), it appears that the three helicase mutations caused non-uniform effects on the levels of nos and osk. This suggests that the three mutations likely rendered different impacts on Me31B’s helicase activity and its substrate mRNAs. This notion is consistent with the three mutants’ different sterility severity described earlier. Moreover, we cautiously reason that the dysregulation of nos and osk levels was not likely to fully explain the sterility of the helicase mutants, therefore, further studies are needed to reveal and contrast how the helicase mutations globally affect germline RNAs.
Me31B N-ter motif, C-ter motif, and FDF-binding motif mutations decrease female fertility
Next, we went on to investigate the in vivo roles of Me31B N-ter domain, C-ter domain, and FDF-binding motif in the Drosophila germline. Earlier studies showed that the N-ter RecA-like domain contributes to the protein’s ATP binding, helicase activity, and P-body assembly [19, 34]; The C-ter RecA-like domain contributes to the protein’s ATP hydrolysis, helicase activity, RNA translational repression, partner-protein binding, and P-body assembly [32, 34, 37, 53]; The FDF-binding motif allows Me31B to physically interact with FDF motifs in partner RNA-repressor proteins like Tral and EDC3 [3, 4, 28, 33, 37, 38]. To study these motifs, we generated me31BN-ter, me31BC-ter, and me31BFDF alleles containing the N-ter coding sequence only (AA 1 – 276), the C-ter coding sequence only (AA 277 – 459), and substitution mutations in the FDF-binding motif (CLNTL→ALNTA, according to [32]), respectively (illustrated in Figure 1). We do note that our many attempts to generate the original N-ter (AA 1 – 267) and C-ter (AA 268 – 459) mutations were not successful, but we were able to generate alternative N-ter (AA 1 – 276) and C-ter (AA 277 – 459) mutations instead, and the latter CRISPR constructs were used in this study. We found that flies homozygous for me31BN-ter, me31BC-ter, and me31BFDF alleles were viable as adults, so we used homozygous mutants from those alleles in the rest of the study. Prompted by the helicase-motif mutations’ strong effects on fly fertility, we first screened the me31BN-ter, me31BC-ter, and me31BFDF female flies’ fertility by using a previously reported fertility assay [54]. In this assay, three to five biological replicates of single-fly crosses (see Materials and Methods for the assay procedure) were conducted to quantify the egg laying and progenies hatched (hatchability). The assays revealed that the me31BN-ter, me31BC-ter, and me31BFDF flies had a significant fertility decrease when compared to the me31BWT control (Figure 5A and Supplementary Table 2). Specifically, for the egg laying, a me31BWT control female fly laid an average of 117 eggs during the assay period, while a me31BN-ter, me31BC-ter, and me31BFDF female laid an average of 68, 29, and 77 eggs, respectively, a significant decrease of 42.9%, 75.2%, and 34.2%, respectively (Figure 5A). For the progenies hatched, a me31BWT control female produced an average of 106 progeny flies, while a me31BN-ter, me31BC-ter, and me31BFDF female produced 47, 0, and 75 progenies, a significant decrease of 55.7 %, 100%, and an insignificant decrease of 29.2%, respectively (Figure 5B). The surprising absence of progenies from the me31BC-ter mutant indicated that the me31BC-ter females were sterile. Indeed, we examined a separate batch of eggs (n = 97) from the me31BC-ter females (accompanied by w1118 males) and observed that none of the eggs developed into larvae or later stages. Furthermore, all the eggs from the me31BC-ter mothers showed morphological defects including fused dorsal appendages (n = 66, 68% of the eggs) or no dorsal appendages (n = 31, 32% of the eggs) (Figure 5D), consistent with me31BC-ter females being sterile. Then, using the above data, we calculated the egg hatchability rate by dividing the number of progenies hatched by the number of eggs laid. The egg hatchability rates from me31BN-ter and me31BC-ter mothers are 68.3% and 0%, respectively, a significant decrease when compared to the 90.8% hatchability from the me31BWT control (Figure 5C). For the me31BFDF females, their egg hatchability (97.1%) is not significantly different from the control (Figure 5C).
Figure 5. Fertility assays of me31BN-ter, me31BC-ter, me31BFDF mutants.
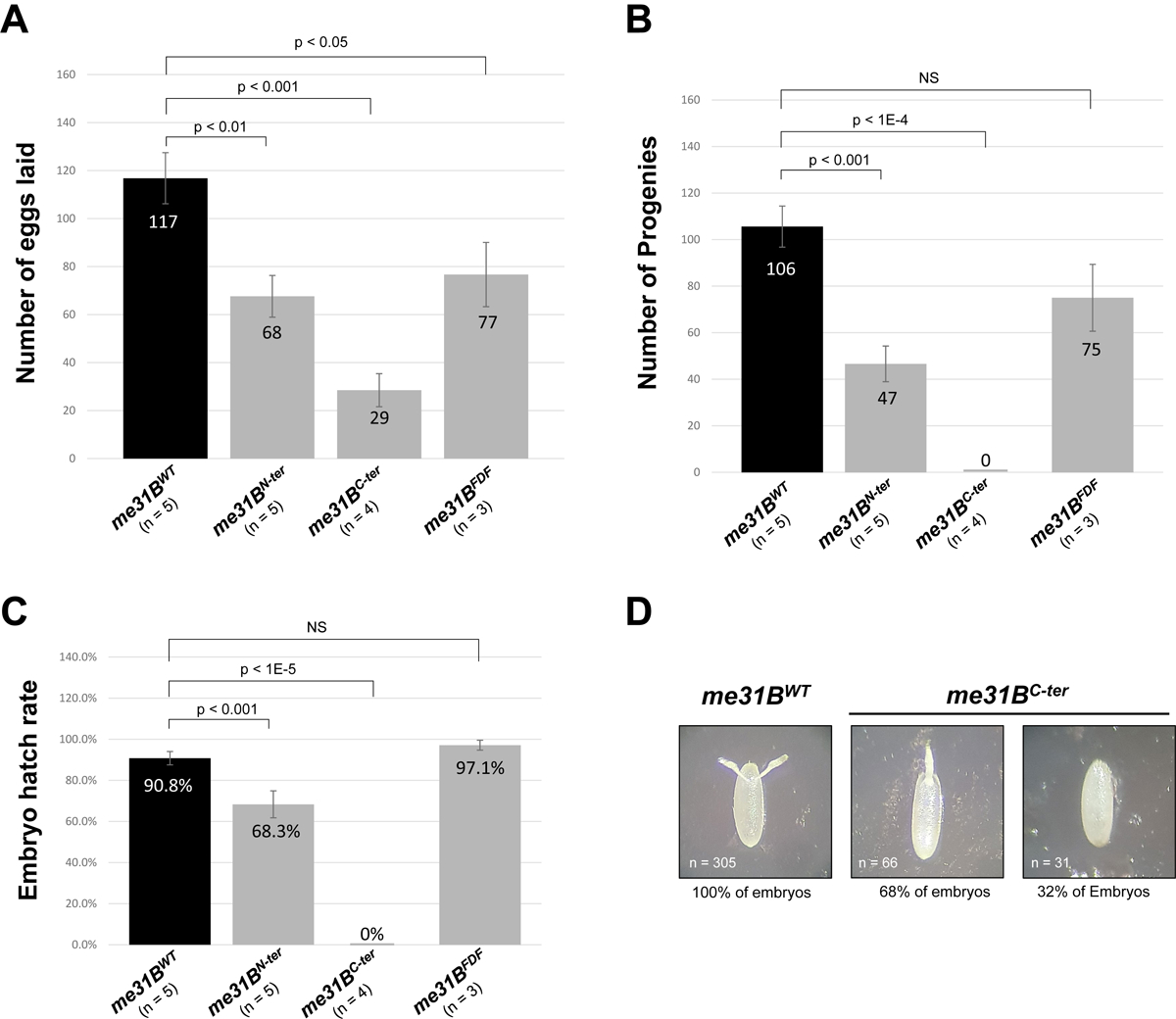
(A) A me31BWT (control), me31BN-ter, me31BC-ter, and me31BFDF female fly lay an average of 117, 68, 29, and 77 eggs, respectively. The egg number of me31BN-ter, me31BC-ter, and me31BFDF mutants is 58% (p < 0.01), 25% (p < 0.001), and 66% (p < 0.05) of the me31BWT (control), respectively. (B) A me31BWT (control), me31BN-ter, me31BC-ter, and me31BFDF female fly produced an average of 106, 47, 0, and 75 progenies, respectively. The progeny number of me31BN-ter, me31BC-ter, and me31BFDF mutants is 44% (p < 0.001), 0% (p < 0.0001), and 71% (NS) of the control, respectively. NS, not statistically significant. (C) The average egg hatch rates for me31BWT (control), me31BN-ter, me31BC-ter, and me31BFDF strains are 90.8%, 68.3%, 0%, and 97.1%, respectively. Compared to me31BWT, the hatch rate decreases in me31BN-ter and me31BC-ter mutants are statistically significant but not significant in me31BFDF. (D) Embryos laid by me31BC-ter flies had fused dorsal appendages (68%) or missing dorsal appendages (32%), in contrast to the normal dorsal appendages (100%) in me31BWT (control). Homozygous me31BWT (control), me31BN-ter, me31BC-ter, and me31BFDF flies were used in this and other experiments of this study unless otherwise specified. Error bar represents the standard error of the mean. The n number represents the biological replicates performed (full data in Supplementary Table 2)
We conclude that Me31B’s N-ter or C-ter motifs (when expressed alone) and mutation in the FDF-binding motif cause Drosophila female fertility to decrease. Interestingly, although both me31BN-ter and me31BC-ter mutants caused decreases in egg-lay and hatchability, only the me31BC-ter behaved as a recessive-sterility allele, suggesting that the two domains have different roles in fertility. Comparatively, the me31BFDF mutant showed a very mild fertility decrease with no measurable defects in egg hatchability. This could mean that the Me31B-partner-protein interactions mediated by the FDF-binding motif only have a small influence on fly fertility (more in Discussion).
Me31B N-ter motif and C-ter motif mutations decrease the Me31B protein level.
We hypothesized that the fertility decreases in the me31BN-ter, me31BC-ter, and me31BFDF mutants could be caused by Me31B protein abundance changes. To test this, we used anti-GFP Western blots to quantify the Me31B proteins expressed in their ovaries. We observed that the Me31BN-ter and Me31BC-ter protein levels were 40% (p < 0.01) and 11% (p < 0.001) of the Me31BWT control, respectively (Figure 6, with additional biological replicate images shown in Supplementary Figure 3). However, the Me31BFDF protein level (114%, p > 0.05) was comparable to the Me31BWT control. These protein level changes were not caused by me31B mRNA level changes as our RT-PCR data indicated that the me31B transcripts levels in the me31BN-ter, me31BC-ter, and me31BFDF mutants were not significantly different from the wildtype control (Supplementary Figure 4A). We conclude that the N-ter and C-ter motifs are important for maintaining the Me31B protein level and deleting either motif likely destabilizes the protein. The data also suggest that the Me31BC-ter protein is less stable than the Me31BN-ter protein. Together with the fertility experiments above (Figure 5), we noticed that the mutants’ fertility and their protein expression levels shared a similar trend, with the me31BC-ter mutant showing complete sterility and the least protein expression and the me31BFDF mutant showing a mild fertility decrease and near wildtype-level protein expression (compare Figure 5 and Figure 6B). This leads us to speculate that the fertility decreases in the me31BN-ter, me31BC-ter, and me31BFDF mutants could be the combined effects of Me31B function and abundance changes.
Figure 6. Western blot quantification of Me31B protein levels in me31BN-ter, me31BC-ter, me31BFDF mutants.
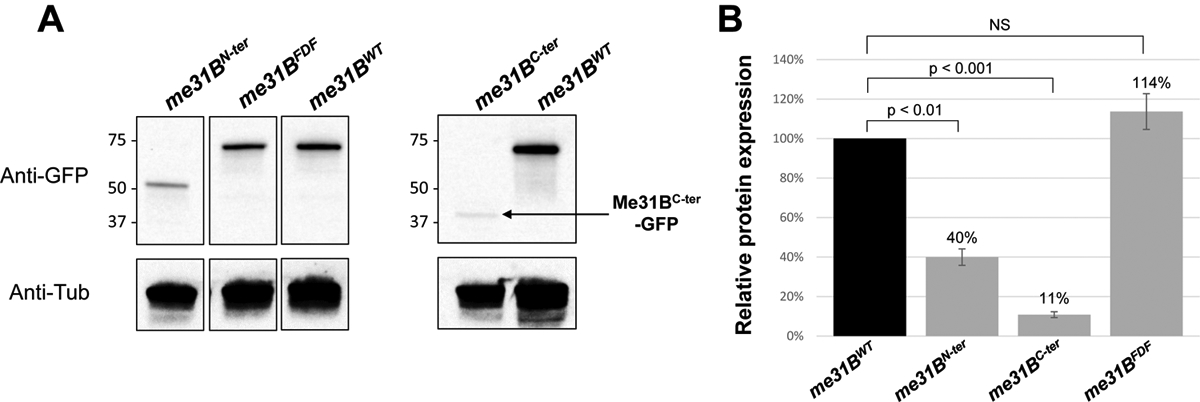
(A) Anti-GFP Western blots were used to quantify the Me31BWT-GFP, Me31BN-ter-GFP, Me31BC-ter-GFP, and Me31BFDF-GFP proteins in the ovaries of the corresponding fly strains. Anti-tubulin Western blots were used as loading controls. The expression levels of Me31BN-ter and Me31BC-ter proteins were conspicuously lower than the Me31BWT control protein, while the Me31BFDF protein level is comparable to the control. The images shown are representative images of multiple biological replicates. The additional, uncropped biological replicate images are presented in Supplementary Figure 3. (B) The Me31BN-ter-GFP and Me31BC-ter-GFP protein levels are 40% (p < 0.01) and 11% (p < 0.001) relative to the Me31BWT-GFP control protein, respectively. The Me31BFDF-GFP protein level is 114% relative to the control (NS). Western blot image analysis was performed with ImageJ and protein quantification was normalized by using the alpha-tubulin proteins. NS, not statistically significant. Error bar represents the standard error of the mean.
Me31B N-ter motif, C-ter motif, and FDF-binding motif mutations alter Me31B subcellular localization.
Me31B has particular localization patterns in Drosophila germline cells, and its localization is believed to reflect the working sites of the protein. Therefore, we asked whether the fertility decrease of the me31BN-ter, me31BC-ter, and me31BFDF mutants could be associated with altered Me31B subcellular localization. Previous research established that wildtype Me31B is expressed abundantly in the cytoplasm of nurse cells and developing oocytes in Drosophila ovaries. In these cells, Me31B complexes with partners like Tral and Cup and aggregate together into RNP granules such as perinuclear granules (nuage granules) in nurse cells, P-body/sponge body granules in nurse cells and oocytes, and germ plasm granules in oocytes [1, 24, 28]. To screen for potential localization abnormalities, we took advantage of the GFP tags on the mutant Me31B proteins to visualize them and used anti-Tral (and anti-Cup) immunostaining to mark the germline RNP granules in the egg chambers.
In the early-stage egg chambers (Figure 7, left panel, A through D”), wildtype Me31BWT-GFP and Tral both localize to the nuage granules and the P body/sponge body granules in the nurse cells and developing oocytes (Figure 7, A to A”). The two proteins showed extensive overlap at these granules, suggesting likely colocalization. This localization pattern of Me31BWT-GFP and Tral is indistinguishable from that in the Oregon-R (OR) strain or the GFP-Trap strain carrying wildtype me31B genes [1, 24], confirming that the CRISPR me31BWT control strain maintains wildtype Me31B activities. In contrast, the three mutant Me31B proteins, Me31BN-ter-GFP, Me31BC-ter-GFP, and Me31BFDF-GFP, were conspicuously more diffused in the nurse cells and oocytes, and no longer overlap with Tral whose localization remained similar to the control (Figure 7, B through (D”). Furthermore, the three mutant proteins each showed distinct, subcellular localization/distribution patterns. Me31BN-ter-GFP proteins were surprisingly found in the nurse cell nucleus (Figure 7B), which was further confirmed by co-staining the egg chambers with DNA stain DAPI (Supplementary Figure 5). The Me31BC-ter-GFP formed a small number of aggregates (Figure 7C), but the aggregates were smaller in size than the Me31BWT-GFP aggregates and did not overlap with the Tral aggregates. Interestingly, the Me31BC-ter-GFP proteins were found in small, ring-like structures that appear to be ring canals (yellow dashed squares in Figure 7C and C”), structures that connect the cytoplasm between nurse cells and oocytes and allow for intracellular transportations. The Me31BFDF-GFP completely failed to aggregate into any granules in the cytoplasm (Figure 7D) but, like the Me31BC-ter-GFP, accumulate at the ring canal-like structures (yellow dashed squares in Figure 7D and D”, also H and H”). These observations indicate that Me31B’s proper subcellular localizations, notably the localization to germline RNP granules, require the N-ter, C-ter, and FDF-binding motifs. We point out that the Me31BFDF-GFP’s failure to localize to RNP granules was independent of its protein expression level (Figure 6). This evidence, together with the mild fertility decrease of the me31BFDF strain (Figure 5), suggests that Me31B’s localization to germline RNPs only has a small influence on fertility.
Figure 7. Mutant Me31B proteins in me31BN-ter, me31BC-ter, and me31BFDF strain show altered localization patterns in developing egg chambers.
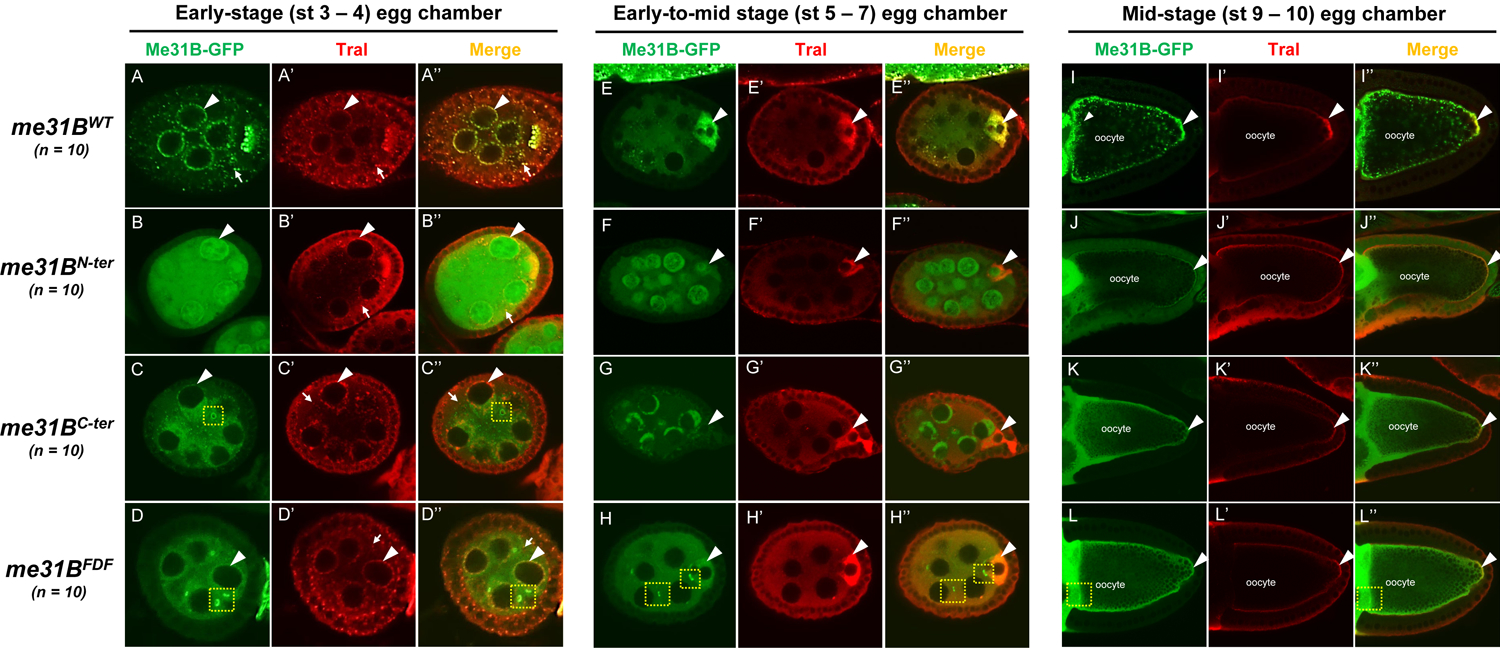
In early-stage egg chambers (left panel, A through D”), mutant Me31B-GFP proteins (green channel) in me31BN-ter, me31BC-ter, and me31BFDF strains are much more diffused in the nurse cell and oocytes, in contrast to the aggregated status of Me31BWT in RNP granules like nuage granules and P-bodies. And none of the three mutant proteins overlap with partner protein Tral. Unlike the Me31B mutant proteins, Tral (Red channel) localization is not affected in the three mutants. Note that Tral was slightly under-stained towards the center of the egg chamber in B’. Particularly, Me31BN-ter-GFP proteins are present in the nuclei of nurse cells. Me31BC-ter-GFP proteins form fewer numbers and smaller size granules than Me31BWT-GFP, and the Me31BC-ter granules do not associate with Tral-marked granules. Nurse cell perinuclear regions (nuage) are indicated by arrowheads. P-body granules marked by Tral are indicated by arrows. Note that Me31BC-ter-GFP and Me31BFDF-GFP proteins were found in ring-like structures that appear to be ring canals (yellow dashed squares in C, C”, D, D”, H, and H”). In early-to-mid stage egg chambers (middle panel, E through H”), mutant Me31B-GFP proteins (green channel) in me31BN-ter and me31BC-ter strains do not enrich in the developing oocytes like that in the me31BWT control. Me31BFDF-GFP protein’s enrichment in the developing oocytes is weaker than that in the control. Tral (red channel)’s enrichment in the oocytes is not affected in the three mutants. Developing oocytes are indicated by arrowheads. In mid-stage egg chambers (right panel, I through L”), mutant Me31B-GFP proteins of me31BN-ter, me31BC-ter, and me31BFDF strains localize to the cortex and the germplasm area at the posterior of oocytes, like the control. However, all three mutant Me31B proteins appear more diffused than the aggregated Me31BWT-GFP proteins in the above areas. The germplasm areas are indicated by arrowheads. Tral proteins (Red channel) remain localized to the cortex and germplasm area in the three mutants. Tral enrichment in the posterior, germ plasm area (over the cortex area) in the me31BN-ter, me31BC-ter, and me31BFDF oocytes appeared less than that in the me31BWT control, and follow-up quantifications showed that only the enrichment in me31BN-ter was significantly decreased (Supplementary Figure 6). For better visualization, the green fluorescence signals of the Me31BN-ter-GFP and Me31BC-ter-GFP proteins were tuned up relative to other images because of the lower expression levels of the proteins.
In the early-to-mid-stage egg chambers (Figure 7, middle panel, E through H”), we observed that Me31B enrichment in developing oocytes was abolished in the three mutants. In the me31BWT control egg chambers, Me31BWT-GFP and Tral both were highly enriched in the developing oocytes, with the two proteins extensively overlapping (Figure 7, E to E”). However, in the me31BN-ter and me31BC-ter egg chambers, the mutant Me31B proteins show no obvious enrichment in the future oocytes (Figure 7, F to F” and G to G”), while Tral’s enrichment pattern remained unaffected. In the me31BFDF mutants, the Me31BFDF-GFP proteins still showed enrichment in the oocytes, but it was much weaker than the Me31BWT-GFP control (Figure 7, H to H”). We conclude that the N-ter, C-ter, and FDF-binding motifs are needed for Me31B’s proper accumulation into future oocytes at early-to-mid oogenesis.
In the mid-stage egg chambers (Figure 7, right panel, I through L”), Me31BWT proteins were found along the cortex of the oocytes and localized to the germ plasm area (Figure 7, I to I”), similar to the wildtype strains previously reported [28]. In the me31BN-ter, me31BC-ter, and me31BFDF mutants, the Me31B proteins still localized to the above areas, but the localized proteins appeared diffused and had less granularity (Figure 7, J through L”). In the three mutants, Tral localized along the oocyte cortex and became enriched in the oocyte posterior (germ plasm area), but the enrichment appeared weaker than in the Me31BWT control. We quantified the enrichment level differences by measuring the ratio of the average Tral intensity in the oocyte posterior over that along the cortex (See Materials and Methods and [28]). Compared with Tral’s enrichment ratio of 2.56 in the me31BWT control, the ratios in me31BN-ter, me31BC-ter, and me31BFDF mutants were 1.26 (−50.8%, p < 0.01), 2.25 (−12.6%, p > 0.4), and 2.22 (−13.5%, p > 0.1), respectively, with only the decrease in me31BN-ter being significant (Supplementary Figure 6).
To further validate that the above localization phenotypes of the Me31BN-ter-GFP, Me31BC-ter-GFP, and Me31BFDF-GFP proteins were not a result of defective germline RNP formation, we performed immunostaining against another germline RNP marker, Cup. We found that Cup’s localization and aggregation into germline RNPs remained unchanged in the me31B mutants (Supplementary Figure 7).
Discussion
This study aims to investigate the in vivo roles of key functional motifs of DEAD-box RNA helicase Me31B in the context of Drosophila germline development and fertility. We generated Drosophila strains mutant for six key motifs and performed screening characterizations. The results are briefly summarized in Figure 8. The three helicase motifs (DEAD-box, DVLARAK, and HRIGR) essential for the helicase activity are conserved among the members of the Me31B protein family, and mutations in each of them resulted in alleles (me31BE208A, me31BDVLAAAA, and me31BR385Q) causing dominant female sterility. Their sterility is associated with oogenesis defects (like nurse cell dumping defects or egg chamber degeneration) and embryo patterning defects. Interestingly, the nurse cell dumping defects mimic previously reported “dumpless” phenotypes in the dcp-1 (encoding a caspase involved in apoptosis) mutant egg chambers, which also led to early embryo development arrest and female sterility ([55] and a later correction of the reference clarifying that the ovary phenotype was due to the combined effect of dcp-1 and CG3941). These dcp-1 phenotypes and sterility were associated with abnormal nucleus envelope breakdown and cytoskeleton organization during nurse cell apoptosis and dumping [55]. Whether the helicase mutations had the same underlying mechanisms remains to be analyzed.
Figure 8. Summary of the Me31B motif functions.
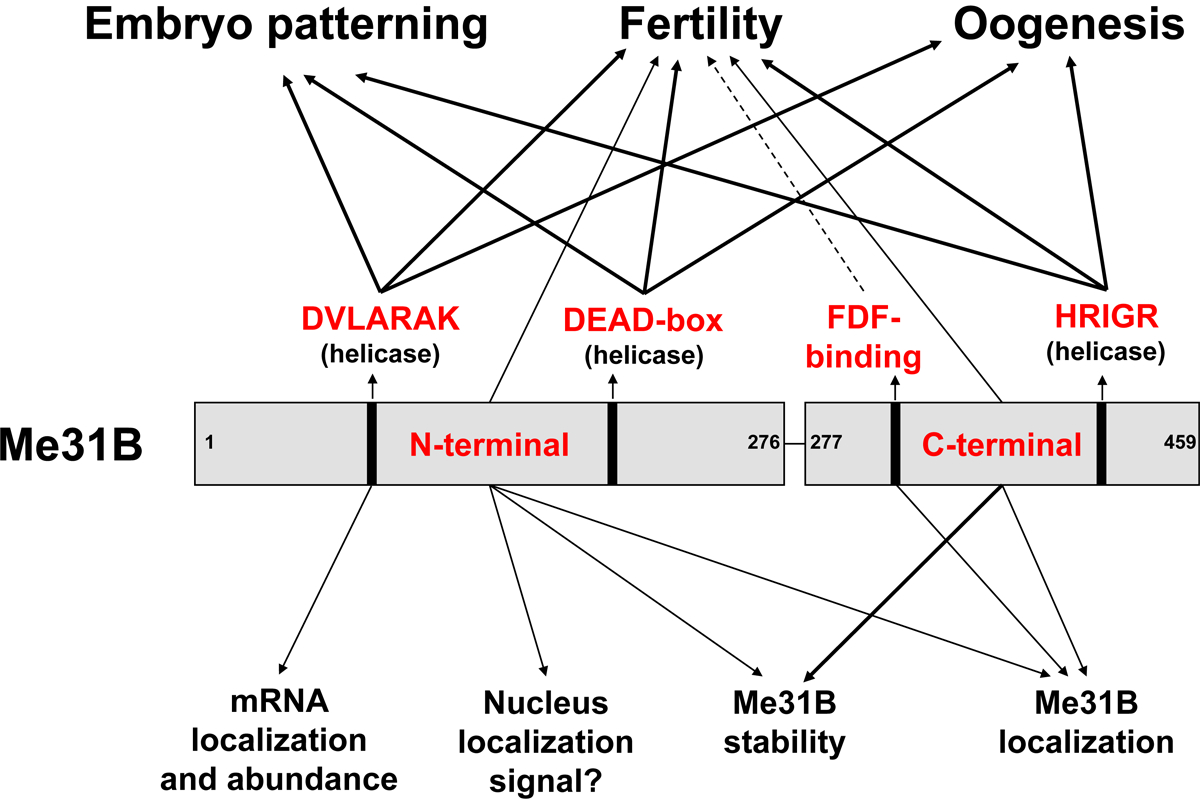
This study uses a target-motif-mutation approach to investigate the functionally important domains/motifs of Drosophila Me31B. Our characterization of the me31B mutants revealed Me31B helicase activity’s role in female fertility, oogenesis, and embryo patterning (DVLARARK, DEAD-box, and HRIGR motifs). An in-depth analysis of the DVLARAK motif mutation uncovered its function in maintaining nos mRNA localization and the transcript level of nos and osk mRNA levels. We further showed the Me31B N-terminal motif, C-terminal motif, and FDF-binding motif’s role in female fertility and their different functions in maintaining Me31B protein level and subcellular localization. Effect strengths are indicated with different arrow lines (mild effects by dashed lines, mid-level effects by thin lines, and strong effects by thick lines).
The sterility and germline defects in all three helicase mutants strongly suggest that each motif (DEAD-box, DVLARAK, and HRIGR) is integral for Me31B’s helicase activity, which, in turn, is needed for Drosophila germline development. An explanation for the helicase mutations being dominant could be that the mutations introduced detrimental functions to the Me31B protein. This speculation is supported by the effects of similar substitution mutations in other DEAD-box helicases. For example, in eIF4A (another DEAD-box RNA helicase important for germline mRNA translational control [56, 57]), DEAD→DQAD and HRIGR→HRIGQ mutations cause the protein to become dominant negative in translation in vitro [58–60]. As another example, in an assay of Xp54 (Me31B ortholog in Xenopus)’s effect on tethered RNAs, DEAD→DQAD mutation, DILARAK (homologous to Me31B’s DVLARAK) →DILAAAA mutation, and HRIGR→HRIGQ mutations changed the protein’s regulation on substrate RNAs from translational repression to activation [34]. This in vitro evidence, together with our in vivo data, makes us speculate that the me31BE208A, me31BDVLAAAA, and me31BR385Q mutations could have altered Me31B’s RNA-translation and RNA-stability control activities. Then, considering Me31B’s physical association with most maternal mRNAs [2], the mutated Me31B proteins may have dominantly caused a global dysregulation of the germline RNAs, eventually leading to hatching failures.
When comparing the germline phenotypes of the three helicase mutants, it was noticed that the HRIGR→HRIGQ mutation caused much more severe phenotypes such as mid-oogenesis apoptosis than the other two (DEAD→DAAD and DVLARAK→DVLAAAA) mutations. We suspect this may be because 1) substituting a motif’s key amino acid from arginine (R) to glutamine (Q) causes more detrimental effects than to alanine (A), or 2) the HRIGR motif contributes a distinct and more important role than the DEAD and DVLARAK motifs. Previous research in other systems provided conceptual support for both explanations. For the first, DQAD mutation in the DEAD-box motif of GLH-1 (a conserved germ granule RNA helicase in C. elegans) led to a more severe fertility decrease and embryo-arrest phenotypes than DAAD mutation [61, 62]. For the second, DEAD→DQAD mutation in the DEAD-box helicase eIF4A abolished the protein’s ATPase activity but retained a small amount of RNA-binding activity, while the HRIGR→HRIGQ mutation abolished the RNA-binding ability but retained some ATPase activity [58], suggesting that the HRIGR motif may play certain unique, non-redundant enzymatic steps for the helicase.
In our attempt to find a molecular-level explanation for the sterility of the helicase mutants (me31BE208A, me31BDVLAAAA, and me31BR385Q), we found that the me31BDVLAAAA mutation caused decreased levels of important germline mRNAs nos and osk, which were the underlying factors for the altered nos mRNA localization in the germ plasm. This indicates the role of Me31B’s helicase activity in maintaining germline mRNA transcript levels and therefore, their subcellular localization. Combining our new data with previously established findings that Me31B (whole protein) functions in post-transcriptional mRNA regulation like RNA stability control [1–3, 28, 38], we speculate that Me31B functions in vivo in stabilizing unlocalized germline RNPs containing nos and osk, allowing for stable mRNAs to reach and become incorporated into the germ plasm. We further speculate that disruption of the helicase motif DVLARAK results in a destabilization of mRNAs that have not localized to the germ plasm, resulting in a decrease in nos and osk that are available to form the germ plasm and ultimately reducing the nos localization. In the me31BE208A/+, ~ 0.15% of nos clusters had extra-large cluster phenotypes, suggesting that some germ granules have larger than normal carrying capacities (Figure 4B). However, altering carrying capacity through osk levels using computational modeling generates uniform effects on all germ granules [52]. Thus, we were unable to further investigate how a subset of granules obtain extra-large nos cluster phenotypes in me31BE208A/+. Future studies should explore how granules’ carrying capacity may be non-uniformly influenced to better understand how nos cluster size is regulated. Additional germ plasm mRNAs such as cycB, pgc, and gcl also diffuse to the posterior RNPs containing a single transcript and form homotypic clusters within germ granules [45]. Thus, future studies should explore whether Me31B’s helicase activity is specific to nos or has a more global role in stabilizing unlocalized germ plasm mRNAs.
Besides the helicase motifs, we screened Me31B’s N-ter motif, C-ter motif, and FDF-binding motif for their potential phenotypes in fertility, Me31B expression, and Me31B subcellular localization. We found that the me31BN-ter, me31BC-ter, and me31BFDF mutations caused fertility decreases to different extents, with the me31BC-ter allele showing the strongest effect for being a recessive sterility allele. The three mutations also caused different phenotypes in Me31B protein expression level and subcellular localizations (without causing significant changes to the transcript levels of me31B mRNA or germline mRNAs such as nos or osk. See Supplementary Figure 4). To avoid repeating the results already described, we only discuss a few striking observations next.
We were surprised to find that the two large deletion mutations, me31BN-ter and me31BC-ter, produced viable, homozygous flies. This surprise arises from previous knowledge that me31B is an essential gene and needed for the development of fly tissues beyond the germline: Complete loss-of-function mutation of me31B (me31BΔ) causes larval-stage lethal but me31BΔ germline clone (me31BΔ in the germline only) produces viable flies (although with germline tissue defects) [1], suggesting that Me31B also supports soma growth. Therefore, Me31B’s N-ter or C-ter motif, when expressed alone, must be able to provide certain essential, full-length Me31B functions needed for fly growth. How the two non-overlapping motifs provide such functions awaits future investigation.
The me31BN-ter and me31BC-ter homozygous mutations support fly viability but caused fertility to decrease, suggesting that they are likely hypomorphic alleles of me31B. Their fertility-decrease phenotypes may be explained by these alleles’ decreased protein expression levels (which somewhat parallel the extent of fertility decreases in the mutants) and/or the mutant Me31B proteins’ distinct subcellular localizations. Future studies should explore whether these hypomorphic alleles cause changes in other Me31B functions such as regulating the translational status of important germline genes like nos and, if yes, whether these changes also contribute to the decreased fertility.
Regarding Me31B’s localization to germline RNPs, the mutant proteins, Me31BN-ter-GFP, Me31BC-ter-GFP, and Me31BFDF-GFP, all showed defective localization and appeared rather diffused in the egg chambers. However, the diffused status of the Me31B proteins did not seem to affect the formation of germline RNPs (marked by Tral or Cup). This suggests that Me31B’s N-ter, C-ter, and FDF-binding motifs do not individually influence germline RNP formation, but rather play roles in localizing Me31B to the RNPs. Most interestingly, the me31BFDF mutation abolished Me31B’s localization to germline RNPs, but only mildly decreased fly fertility, suggesting that Me31B’s localization to the RNPs is not essential for fertility. This further suggests that there may be flexibility in the functioning of germline RNPs that does not completely rely on RNP-localized Me31B. Considering that Me31B is very abundantly expressed in the germline, dispersive Me31B proteins such as Me31BFDF can exist in large quantities in the proximity of germline RNPs, allowing the protein to interact with and thus provide Me31B-protein functions to the RNPs. This interaction may be more transient and not as extensive as wild-type Me31B proteins that aggregate on the RNPs, but it could be sufficient for the RNPs to perform essential functions such as post-transcriptional regulations on the constituent mRNAs, which, in turn, enables proper germline development and fertility. The above speculation is conceptually supported by that, although the RNAs within germline RNPs are found to be very stable [46, 52], the proteins in the RNPs such as Osk display liquid-like behavior as they have been shown to dynamically exchange with the environment [52, 61, 63–65]. This liquid characteristic may provide grounds for Me31BFDF to freely enter and interact with (although not retained in) the RNPs. Regarding Me31B protein’s aggregational status, another aggregation factor worth discussing is Me31B’s two predicted Intrinsically Disordered Regions (IDRs) at the N-ter end (AA 1–53) and the C-ter ends (AA 431 – 459), respectively. A previous in vitro study showed that deleting the IDRs caused rapid Me31B self-assembly into aggregate-like structures, and therefore the IDRs were suggested to attenuate the interactions between the folded, more rigid N-ter and C-ter motifs [66]. Together with the three motifs’ (N-ter, C-ter, and FDF-binding motifs) role in Me31B localization to RNP granules and their previously established functions in helicase activity, RNA interactions, and partner-protein binding, we speculate that Me31B’s localization to germline RNPs (and aggregation on them) could be a complex interplay between Me31B-RNA interacting, Me31B-partner protein binding, and IDR modulation.
The subcellular localization screening analysis revealed the confounding nuclear localization of Me31BN-ter proteins, in sharp contrast to Me31B’s established role as a cytoplasmic protein [1, 2, 24]. An explanation on trial is that Me31B may be a nucleocytoplasmic shuttling protein with a Nuclear Localization Signal (NLS) sequence in the N-terminal motif. This NLS sequence leads Me31B into the nucleus, and then the C-terminal motif mediates the protein’s export and retention in the cytoplasm. This speculation is conceptually supported by the nucleocytoplasmic shuttling activities of DDX6 (human homolog of Me31B) [53] and Xp54 (Xenopus homolog) [11] in cell culture models. However, our efforts to identify a Nuclear Localization Signal (NLS) sequence by using prediction tools (NLStradamus [67], cNLS Mapper [68], and SeqNLS Prediction Server [69]) were unsuccessful. Although we were able to locate amino acid sequences in Me31B (KSKLKLPPKDNRFK, AA 35 – 48, and CIPVLEQIDP, AA 113 – 122) that are homologous to the putative NLS and Nuclear Export Signals (NES) sequences in DDX6, respectively [53], experimental evidence is still needed to validate their in vivo functionality. We do not exclude the possibility that the N-terminal NLS is just a non-functional sequence that is normally masked in folded, full-length Me31B protein, and the sequence was accidentally exposed to nucleus transportation machinery upon the deletion of the C-terminal motif.
All in all, this study generated novel me31B Drosophila strains mutant for the protein’s key functional motifs, explored the fertility and germline phenotypes brought about by these mutations, and provided insights into the molecular-level mechanisms underlying the phenotypes. For the conserved nature of Me31B and its orthologs/homologs, our report paved the road to understanding the in vivo functions of Me31B, its key motifs, and other DEAD-box helicases in different cell types and species.
Materials and Methods
Fly strain generation by CRISPR gene editing
Mutant me31B Drosophila strains were generated by using the previously reported CRISPR procedure [70, 71]. Specifically, CRISPR Optimal Target Finder [70] and DRSC/TRiP Functional Genomics Resources (Harvard Medical School) were used to find gRNA cutting sites flanking the me31B gene in the Drosophila genome. Then, the found gRNA sequences (GAACGGGACTTAGGAACGACAGG and AAATCCTAGATTTACTTACATGG) were cloned into gRNA-expressing plasmid vector pCFD5 (Addgene) according to the suggested protocols. HDR donor plasmids carrying wild type me31B gene were constructed by cloning me31B gene DNA into the pHD-sfGFP-ScarlessDsRed cloning vector (Addgene) according to suggested protocols. In the constructed HDR plasmids, the super-fold GFP (sfGFP) gene is positioned in-frame and downstream of the me31B gene so that the sfGFP protein is tagged to the C-terminal end of the expressed Me31B proteins. The DsRed marker gene is positioned in the intergenic region downstream of the me31B gene. HDR plasmid with mutant me31B genes was generated by mutating the me31B wild-type gene in the wildtype HDR donor plasmid by using the Site-Directed Mutagenesis kit (New England Biolabs) according to the manufacturer’s recommended protocols. The resulting HDR plasmids containing different me31B alleles are named after the carried mutations (Figure 1). The gRNA-expressing plasmid and the HDR donor plasmids were co-injected into a Cas9-expressing strain (Genetivision) to generate desired wild type and mutant me31B strains, which were crossed with a 2nd chromosome balancer to establish balanced stocks when possible. The plasmid vectors constructed and the obtained me31B strains were validated by sequencing.
Genetic crosses
Balanced me31B wild type and mutant strains were self-crossed to obtain homozygous mutant strains. For the dominant female sterile me31B strains, males carrying the mutant allele were crossed with w1118 (Bloomington Drosophila Stock Center 3605) females to obtain heterozygous strains.
Immunohistochemistry
Drosophila ovary immunostaining was performed as previously described [24, 72, 73]. The following antibody dilutions were used: Rabbit-anti-Tral (1:1,000), Mouse-anti-Cup (1:1000), Donkey-anti-mouse-Cy3 and Donkey-anti-Rabbit-Cy3 secondary antibodies (Jackson ImmunoResearch) were used at 1:500. Images were captured by an Olympus FV3000 confocal laser scanning microscope.
Western blots
Western blot antibodies were used at the following dilutions: rabbit-anti-GFP (1:100,000), and mouse-anti-α-Tubulin (1:100,000). Secondary antibodies were used at the following dilutions: mouse-anti-rabbit HRP (Jackson ImmunoResearch) (1:10,000 for rabbit-anti-GFP), goat-anti-mouse HRP (Santa Cruz Biotechnology) (1: 50,000 for mouse-anti-α-Tubulin primary antibody). The protein band quantification analysis was performed by using ImageJ (https://imagej.nih.gov/ij/)
Fertility assay (for me31BWT, me31BN-ter, me31BC-ter, and me31BFDF strains)
Fertility assays were performed according to previously established protocols [54]. Briefly, virgin females from me31BWT, me31BN-ter, me31BC-ter, and me31BFDF strains were collected and allowed to age for 3 – 4 days separately in fly food vials. Each female was then put in a vial with a w1118 male. After 24 hours, the males were removed from the vials, and the fertilized females were transferred to a new vial every day for the next 10 days. The eggs laid and the progenies hatched from each vial were counted. The hatch rate of each vial is calculated by dividing the progenies hatched by the number of eggs laid. An equal amount (15 ml) of fly food medium (Molasses Formulation, Genesee Scientific) was used in each vial.
Embryo morphology and embryo hatchability (me31BE208A, me31BDVLAAAA, and me31BR385Q strains)
To observe the morphology and the hatchability of the embryos from me31BE208A/+, me31BDVLAAAA/+, me31BR385Q/+, and w1118 control strains, approximately 80 females from each strain were put into a small embryo collection cage in the presence of 20 w1118 males, and the embryos were collected on grape agar plates at 25°C after 24 hours. The embryos were counted and analyzed under a dissection microscope for their morphology. The embryos’ hatchability was calculated 72 hours after they were laid by counting the number of those that developed into larvae (or later stages) or by counting those that failed to develop and then subtracting the failed ones from the total number of eggs laid.
RNA extraction, cDNA synthesis, and quantitative RT-PCR
Three biological replicates (each with three technical replicates) were performed for each strain’s RT-PCR experiments. Ovarian total RNA was extracted from 5 μl freshly dissected fly ovaries by using Qiagen RNeasy Purification Kit (Qiagen) according to the manufacturer’s instructions. The obtained RNA samples’ concentrations were measured by using NanoDrop 2000c. The RNAs were reversely transcribed to cDNAs by using the High-Capacity cDNA Reverse Transcription Kit (ThermoFisher) according to the manufacturer’s instructions. The synthesized cDNAs were then used for quantitative PCR by using Luna Universal qPCR Master Mix (New England Biolabs). The following PCR Primers were used in this study: nos forward 5’ GTCACCAGCAAACGGACGAGATT −3’, nos reverse CGGAGCACTCCCGTAGGACAT, osk forward 5’- TTGCTGAGCCACGCCCAGAA −3’, osk reverse 5’- ACATTGGGAATGGTCAGCAGGAAATC −3’, rp49 forward 5’- GCTAAGCTGTCGCACAAA, rp49 reverse 5’- TCCGGTGGGCAGCATGTG −3’. rp49 RNA was used as the reference. Data analysis was conducted by using the CFX Manager Software (BioRad) and Microsoft Excel.
smFISH, image analysis, and computational modeling
smFISH was carried out as previously described using published nos probe sequences [46, 52, 74]. In summary, ovaries were dissected from yeast-fed females in cold PBS in under 10 minutes and tissues were fixed for 30 minutes in 4% paraformaldehyde and PBS solution. Tissue was incubated with smFISH probes overnight at 37 °C in the hybridization solution previously described [46]. For imaging, egg chambers were mounted in Prolong Glass (Life Technologies) and were allowed to cure for 72 hours at room temperature [46, 52]. A Leica STELLARIS 5 confocal microscope was used for imaging nos smFISH experiments that are described in detail [52]. The identification and quantification of unlocalized single nos transcripts and localized nos homotypic clusters were carried out using a custom MATLAB (Mathworks) program that has been previously described [46, 52]. Confocal images shown in Figure 4 are maximum projections that were filtered by a balanced circular difference-of-Gaussian with a center radius size of 1.2 pixels and surround size of 2.2 pixels as previously done for other germ plasm studies [45, 52]. The total number of homotypic clusters identified is reported in the figure legends. For modeling experiments, we employed a previously published model that simulates the formation of germ granules, including nos homotypic clusters [52]. The only modeling parameters that were adjusted were 1) carrying capacity, which is influenced by osk levels, was set from 1 (wild type) to 0.56, and 2) the pool of nos transcript expression was set from 1 (wild type) to 0.61 to mimic the RT-PCR levels reported in our results section [52].
Immunofluorescence image analysis
Fluorescence images were analyzed by using ImageJ (https://imagej.nih.gov/ij/) as previously described [28]. Briefly, the Tral posterior enrichment ratio was calculated by dividing the mean Tral signal intensity of the posterior portion by that of the cortex portion. The average intensity of the lateral and dorsal cortex was used for the ratio calculation. In the case one side of the cortex was apparently under-stained, only the other side of the cortex was used for the ratio calculation. We do note that these measurements and ratio calculations can only be used to estimate Tral enrichment and are subjected to technical limitations like the quality of antibody staining and the selection of the microscope focal planes.
Statistical Analysis
Reported p-values between average nos homotypic cluster sizes were performed using ANOVA test with a post-hoc Tukey test that was calculated using R statistical programming in R Studio using the function aov and TukeyHSD functions within the DescTools package [75]. Violin plots were created using the ggplot and ggstatsplot packages [76, 77]. Heatmaps of germ plasms were generated as previously described [46].
Supplementary Material
Acknowledgments
We thank the members of the Gao Lab for discussing and revising this manuscript. We thank Dr. Olivia Rissland and Dr. Akira Nakamura for their kind gifts of antibodies. We thank the Center for Biological Imaging at Kean University for assisting with image acquisition and the members and the Niepielko Lab for their helpful comments and fruitful discussions. Research reported in this publication was supported by the Eunice Kennedy Shriver National Institute of Child Health & Human Development of the National Institutes of Health under award no. R15HD102960 to MGN and 1R15HD092925-01A1 to MG.
List of Abbreviations
- Me31B
maternal expression at 31B
- CRISPR
clustered regularly interspaced short palindromic repeats
- KD
knockdown
- AA
amino acid
- RNP
ribonucleoprotein
- GFP
green fluorescent protein
- smFISH
single-molecule fluorescent in situ hybridization
- qRT-PCR
quantitative reverse-transcription polymerase chain reaction
- RT-PCR
reverse-transcription polymerase chain reaction
- P bodies
processing bodies
- OR
Oregon-R
- DAPI
4′,6-diamidino-2-phenylindole
- IDRs
intrinsically disordered regions
- NLS
nuclear localization signal
- NES
nuclear export signal
- gRNA
guide RNA
- HDR
homology-directed repair
- sfGFP
super-fold green fluorescent protein
- HRP
horseradish peroxidase
- cDNA
complementary DNA
- N-ter
N-terminal
- C-ter
C-terminal
- ANOVA
analysis of variance
- NS
not statistically significant
Footnotes
Competing Interests
The authors declare no competing interests.
Data Availability
Strains and plasmids are available upon request. The authors affirm that all data necessary for confirming the conclusions of the article are present within the article, figures, and tables.
References
- 1.Nakamura A, Amikura R, Hanyu K & Kobayashi S (2001) Me31B silences translation of oocyte-localizing RNAs through the formation of cytoplasmic RNP complex during Drosophila oogenesis, Development. 128, 3233–42. [DOI] [PubMed] [Google Scholar]
- 2.Wang M, Ly M, Lugowski A, Laver JD, Lipshitz HD, Smibert CA & Rissland OS (2017) ME31B globally represses maternal mRNAs by two distinct mechanisms during the Drosophila maternal-to-zygotic transition, Elife. 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gotze M, Dufourt J, Ihling C, Rammelt C, Pierson S, Sambrani N, Temme C, Sinz A, Simonelig M & Wahle E (2017) Translational repression of the Drosophila nanos mRNA involves the RNA helicase Belle and RNA coating by Me31B and Trailer hitch, RNA. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kugler JM, Chicoine J & Lasko P (2009) Bicaudal-C associates with a Trailer Hitch/Me31B complex and is required for efficient Gurken secretion, Dev Biol. 328, 160–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu L, Qi H, Wang J & Lin H (2011) PAPI, a novel TUDOR-domain protein, complexes with AGO3, ME31B and TRAL in the nuage to silence transposition, Development. 138, 1863–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Langerak S, Trombley A, Patterson JR, Leroux D, Couch A, Wood MP & Schisa JA (2019) Remodeling of the endoplasmic reticulum in Caenorhabditis elegans oocytes is regulated by CGH-1, Genesis. 57, e23267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ko S, Kawasaki I & Shim YH (2013) PAB-1, a Caenorhabditis elegans poly(A)-binding protein, regulates mRNA metabolism in germline by interacting with CGH-1 and CAR-1, PLoS One. 8, e84798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kashima M, Kumagai N, Agata K & Shibata N (2016) Heterogeneity of chromatoid bodies in adult pluripotent stem cells of planarian Dugesia japonica, Dev Growth Differ. 58, 225–37. [DOI] [PubMed] [Google Scholar]
- 9.Yoshida-Kashikawa M, Shibata N, Takechi K & Agata K (2007) DjCBC-1, a conserved DEAD box RNA helicase of the RCK/p54/Me31B family, is a component of RNA-protein complexes in planarian stem cells and neurons, Dev Dyn. 236, 3436–50. [DOI] [PubMed] [Google Scholar]
- 10.Alves-Rodrigues I, Mas A & Diez J (2007) Xenopus Xp54 and human RCK/p54 helicases functionally replace yeast Dhh1p in brome mosaic virus RNA replication, J Virol. 81, 4378–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Smillie DA & Sommerville J (2002) RNA helicase p54 (DDX6) is a shuttling protein involved in nuclear assembly of stored mRNP particles, J Cell Sci. 115, 395–407. [DOI] [PubMed] [Google Scholar]
- 12.Aravin AA, van der Heijden GW, Castaneda J, Vagin VV, Hannon GJ & Bortvin A (2009) Cytoplasmic compartmentalization of the fetal piRNA pathway in mice, PLoS Genet. 5, e1000764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Swetloff A, Conne B, Huarte J, Pitetti JL, Nef S & Vassalli JD (2009) Dcp1-bodies in mouse oocytes, Mol Biol Cell. 20, 4951–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lin F, Wang R, Shen JJ, Wang X, Gao P, Dong K & Zhang HZ (2008) Knockdown of RCK/p54 expression by RNAi inhibits proliferation of human colorectal cancer cells in vitro and in vivo, Cancer Biol Ther. 7, 1669–76. [DOI] [PubMed] [Google Scholar]
- 15.Marcon BH, Rebelatto CK, Cofre AR, Dallagiovanna B & Correa A (2020) DDX6 Helicase Behavior and Protein Partners in Human Adipose Tissue-Derived Stem Cells during Early Adipogenesis and Osteogenesis, Int J Mol Sci. 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang Y, Arribas-Layton M, Chen Y, Lykke-Andersen J & Sen GL (2015) DDX6 Orchestrates Mammalian Progenitor Function through the mRNA Degradation and Translation Pathways, Mol Cell. 60, 118–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hansen M, Zeddies S, Meinders M, di Summa F, van Alphen FPJ, Hoogendijk AJ, Moore KS, Halbach M, Gutierrez L, van den Biggelaar M, Thijssen-Timmer DC, Auburger GWJ, van den Akker E, von Lindern M & Rollmann E (2020) The RNA-Binding Protein ATXN2 is Expressed during Megakaryopoiesis and May Control Timing of Gene Expression, Int J Mol Sci. 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Di Stefano B, Luo EC, Haggerty C, Aigner S, Charlton J, Brumbaugh J, Ji F, Rabano Jimenez I, Clowers KJ, Huebner AJ, Clement K, Lipchina I, de Kort MAC, Anselmo A, Pulice J, Gerli MFM, Gu H, Gygi SP, Sadreyev RI, Meissner A, Yeo GW & Hochedlinger K (2019) The RNA Helicase DDX6 Controls Cellular Plasticity by Modulating P-Body Homeostasis, Cell Stem Cell. 25, 622–638 e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Weston A & Sommerville J (2006) Xp54 and related (DDX6-like) RNA helicases: roles in messenger RNP assembly, translation regulation and RNA degradation, Nucleic Acids Res. 34, 3082–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chu CY & Rana TM (2006) Translation repression in human cells by microRNA-induced gene silencing requires RCK/p54, PLoS Biol. 4, e210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Barckmann B, Pierson S, Dufourt J, Papin C, Armenise C, Port F, Grentzinger T, Chambeyron S, Baronian G, Desvignes JP, Curk T & Simonelig M (2015) Aubergine iCLIP Reveals piRNA-Dependent Decay of mRNAs Involved in Germ Cell Development in the Early Embryo, Cell Rep. 12, 1205–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jeske M, Moritz B, Anders A & Wahle E (2011) Smaug assembles an ATP-dependent stable complex repressing nanos mRNA translation at multiple levels, EMBO J. 30, 90–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sudhakaran IP, Hillebrand J, Dervan A, Das S, Holohan EE, Hulsmeier J, Sarov M, Parker R, VijayRaghavan K & Ramaswami M (2014) FMRP and Ataxin-2 function together in long-term olfactory habituation and neuronal translational control, Proc Natl Acad Sci U S A. 111, E99–E108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.DeHaan H, McCambridge A, Armstrong B, Cruse C, Solanki D, Trinidad JC, Arkov AL & Gao M (2017) An in vivo Proteomic Analysis of the Me31B Interactome in Drosophila Germ Granules, FEBS Lett. [DOI] [PubMed] [Google Scholar]
- 25.Arkov AL & Ramos A (2010) Building RNA-protein granules: insight from the germline, Trends Cell Biol. 20, 482–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Voronina E, Seydoux G, Sassone-Corsi P & Nagamori I (2011) RNA granules in germ cells, Cold Spring Harb Perspect Biol. 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rangan P, DeGennaro M, Jaime-Bustamante K, Coux RX, Martinho RG & Lehmann R (2009) Temporal and spatial control of germ-plasm RNAs, Curr Biol. 19, 72–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McCambridge A, Solanki D, Olchawa N, Govani N, Trinidad JC & Gao M (2020) Comparative Proteomics Reveal Me31B’s Interactome Dynamics, Expression Regulation, and Assembly Mechanism into Germ Granules during Drosophila Germline Development, Sci Rep. 10, 564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sankaranarayanan M, Emenecker RJ, Jahnel M, Trussina IREA, Wayland M, Alberti S, Holehouse AS & Weil TT (2021) The arrested state of processing bodies supports mRNA regulation in early development, bioRxiv, 2021.03.16.435709. [Google Scholar]
- 30.Linder P & Jankowsky E (2011) From unwinding to clamping - the DEAD box RNA helicase family, Nat Rev Mol Cell Biol. 12, 505–16. [DOI] [PubMed] [Google Scholar]
- 31.Lee J, Yoo E, Lee H, Park K, Hur JH & Lim C (2017) LSM12 and ME31B/DDX6 Define Distinct Modes of Posttranscriptional Regulation by ATAXIN-2 Protein Complex in Drosophila Circadian Pacemaker Neurons, Mol Cell. 66, 129–140 e7. [DOI] [PubMed] [Google Scholar]
- 32.Peter D, Ruscica V, Bawankar P, Weber R, Helms S, Valkov E, Igreja C & Izaurralde E (2019) Molecular basis for GIGYF-Me31B complex assembly in 4EHP-mediated translational repression, Genes Dev. 33, 1355–1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tritschler F, Braun JE, Eulalio A, Truffault V, Izaurralde E & Weichenrieder O (2009) Structural basis for the mutually exclusive anchoring of P body components EDC3 and Tral to the DEAD box protein DDX6/Me31B, Mol Cell. 33, 661–8. [DOI] [PubMed] [Google Scholar]
- 34.Minshall N, Kress M, Weil D & Standart N (2009) Role of p54 RNA helicase activity and its C-terminal domain in translational repression, P-body localization and assembly, Mol Biol Cell. 20, 2464–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Barbee SA, Estes PS, Cziko AM, Hillebrand J, Luedeman RA, Coller JM, Johnson N, Howlett IC, Geng C, Ueda R, Brand AH, Newbury SF, Wilhelm JE, Levine RB, Nakamura A, Parker R & Ramaswami M (2006) Staufen- and FMRP-containing neuronal RNPs are structurally and functionally related to somatic P bodies, Neuron. 52, 997–1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Minshall N, Thom G & Standart N (2001) A conserved role of a DEAD box helicase in mRNA masking, RNA. 7, 1728–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tritschler F, Eulalio A, Helms S, Schmidt S, Coles M, Weichenrieder O, Izaurralde E & Truffault V (2008) Similar modes of interaction enable Trailer Hitch and EDC3 to associate with DCP1 and Me31B in distinct protein complexes, Mol Cell Biol. 28, 6695–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hara M, Lourido S, Petrova B, Lou HJ, Von Stetina JR, Kashevsky H, Turk BE & Orr-Weaver TL (2018) Identification of PNG kinase substrates uncovers interactions with the translational repressor TRAL in the oocyte-to-embryo transition, Elife. 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cheng Z, Coller J, Parker R & Song H (2005) Crystal structure and functional analysis of DEAD-box protein Dhh1p, RNA. 11, 1258–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Intile PJ, Balzer GJ, Wolfgang MC & Yahr TL (2015) The RNA Helicase DeaD Stimulates ExsA Translation To Promote Expression of the Pseudomonas aeruginosa Type III Secretion System, J Bacteriol. 197, 2664–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liao SE, Kandasamy SK, Zhu L & Fukunaga R (2019) DEAD-box RNA helicase Belle posttranscriptionally promotes gene expression in an ATPase activity-dependent manner, RNA. 25, 825–839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mugler CF, Hondele M, Heinrich S, Sachdev R, Vallotton P, Koek AY, Chan LY & Weis K (2016) ATPase activity of the DEAD-box protein Dhh1 controls processing body formation, Elife. 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Story RM, Li H & Abelson JN (2001) Crystal structure of a DEAD box protein from the hyperthermophile Methanococcus jannaschii, Proc Natl Acad Sci U S A. 98, 1465–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Buszczak M & Cooley L (2000) Eggs to die for: cell death during Drosophila oogenesis, Cell Death Differ. 7, 1071–4. [DOI] [PubMed] [Google Scholar]
- 45.Little SC, Sinsimer KS, Lee JJ, Wieschaus EF & Gavis ER (2015) Independent and coordinate trafficking of single Drosophila germ plasm mRNAs, Nat Cell Biol. 17, 558–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Niepielko MG, Eagle WVI & Gavis ER (2018) Stochastic Seeding Coupled with mRNA Self-Recruitment Generates Heterogeneous Drosophila Germ Granules, Curr Biol. 28, 1872–1881 e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lehmann R (2016) Germ Plasm Biogenesis--An Oskar-Centric Perspective, Curr Top Dev Biol. 116, 679–707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gao M & Arkov AL (2013) Next generation organelles: structure and role of germ granules in the germline, Mol Reprod Dev. 80, 610–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jamieson-Lucy A & Mullins MC (2019) The vertebrate Balbiani body, germ plasm, and oocyte polarity, Curr Top Dev Biol. 135, 1–34. [DOI] [PubMed] [Google Scholar]
- 50.Bilinski SM, Jaglarz MK & Tworzydlo W (2017) The Pole (Germ) Plasm in Insect Oocytes, Results Probl Cell Differ. 63, 103–126. [DOI] [PubMed] [Google Scholar]
- 51.Wang JT & Seydoux G (2013) Germ cell specification, Adv Exp Med Biol. 757, 17–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Valentino M, Ortega BM, Ulrich B, Doyle DA, Farnum ED, Joiner DA, Gavis ER & Niepielko MG (2022) Computational modeling offers new insight into Drosophila germ granule development, Biophys J. 121, 1465–1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Huang JH, Ku WC, Chen YC, Chang YL & Chu CY (2017) Dual mechanisms regulate the nucleocytoplasmic localization of human DDX6, Sci Rep. 7, 42853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mossman JA, Mabeza RMS, Blake E, Mehta N & Rand DM (2019) Age of Both Parents Influences Reproduction and Egg Dumping Behavior in Drosophila melanogaster, J Hered. 110, 300–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.McCall K & Steller H (1998) Requirement for DCP-1 caspase during Drosophila oogenesis, Science. 279, 230–4. [DOI] [PubMed] [Google Scholar]
- 56.Shen R, Weng C, Yu J & Xie T (2009) eIF4A controls germline stem cell self-renewal by directly inhibiting BAM function in the Drosophila ovary, Proc Natl Acad Sci U S A. 106, 11623–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Oberer M, Marintchev A & Wagner G (2005) Structural basis for the enhancement of eIF4A helicase activity by eIF4G, Genes Dev. 19, 2212–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pause A, Methot N & Sonenberg N (1993) The HRIGRXXR region of the DEAD box RNA helicase eukaryotic translation initiation factor 4A is required for RNA binding and ATP hydrolysis, Mol Cell Biol. 13, 6789–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pause A & Sonenberg N (1992) Mutational analysis of a DEAD box RNA helicase: the mammalian translation initiation factor eIF-4A, EMBO J. 11, 2643–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pause A, Methot N, Svitkin Y, Merrick WC & Sonenberg N (1994) Dominant negative mutants of mammalian translation initiation factor eIF-4A define a critical role for eIF-4F in cap-dependent and cap-independent initiation of translation, EMBO J. 13, 1205–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chen W, Hu Y, Lang CF, Brown JS, Schwabach S, Song X, Zhang Y, Munro E, Bennett K, Zhang D & Lee HC (2020) The Dynamics of P Granule Liquid Droplets Are Regulated by the Caenorhabditis elegans Germline RNA Helicase GLH-1 via Its ATP Hydrolysis Cycle, Genetics. 215, 421–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Marnik EA, Fuqua JH, Sharp CS, Rochester JD, Xu EL, Holbrook SE & Updike DL (2019) Germline Maintenance Through the Multifaceted Activities of GLH/Vasa in Caenorhabditis elegans P Granules, Genetics. 213, 923–939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bose M, Lampe M, Mahamid J & Ephrussi A (2022) Liquid-to-solid phase transition of oskar ribonucleoprotein granules is essential for their function in Drosophila embryonic development, Cell. 185, 1308–1324 e23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Fare CM, Villani A, Drake LE & Shorter J (2021) Higher-order organization of biomolecular condensates, Open Biol. 11, 210137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Trcek T, Douglas TE, Grosch M, Yin Y, Eagle WVI, Gavis ER, Shroff H, Rothenberg E & Lehmann R (2020) Sequence-Independent Self-Assembly of Germ Granule mRNAs into Homotypic Clusters, Mol Cell. 78, 941–950 e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sankaranarayanan M, Emenecker RJ, Wilby EL, Jahnel M, Trussina I, Wayland M, Alberti S, Holehouse AS & Weil TT (2021) Adaptable P body physical states differentially regulate bicoid mRNA storage during early Drosophila development, Dev Cell. 56, 2886–2901 e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Nguyen Ba AN, Pogoutse A, Provart N & Moses AM (2009) NLStradamus: a simple Hidden Markov Model for nuclear localization signal prediction, BMC Bioinformatics. 10, 202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kosugi S, Hasebe M, Tomita M & Yanagawa H (2009) Systematic identification of cell cycle-dependent yeast nucleocytoplasmic shuttling proteins by prediction of composite motifs, Proc Natl Acad Sci U S A. 106, 10171–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lin JR & Hu J (2013) SeqNLS: nuclear localization signal prediction based on frequent pattern mining and linear motif scoring, PLoS One. 8, e76864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gratz SJ, Ukken FP, Rubinstein CD, Thiede G, Donohue LK, Cummings AM & O’Connor-Giles KM (2014) Highly specific and efficient CRISPR/Cas9-catalyzed homology-directed repair in Drosophila, Genetics. 196, 961–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Gratz SJ, Rubinstein CD, Harrison MM, Wildonger J & O’Connor-Giles KM (2015) CRISPR-Cas9 Genome Editing in Drosophila, Curr Protoc Mol Biol. 111, 31 2 1–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Creed TM, Loganathan SN, Varonin D, Jackson CA & Arkov AL (2010) Novel role of specific Tudor domains in Tudor-Aubergine protein complex assembly and distribution during Drosophila oogenesis, Biochem Biophys Res Commun. 402, 384–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Arkov AL, Wang JY, Ramos A & Lehmann R (2006) The role of Tudor domains in germline development and polar granule architecture, Development. 133, 4053–62. [DOI] [PubMed] [Google Scholar]
- 74.Eagle WVI, Yeboah-Kordieh DK, Niepielko MG & Gavis ER (2018) Distinct cis-acting elements mediate targeting and clustering of Drosophila polar granule mRNAs, Development. 145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Andri et mult. al S (2022) DescTools: Tools for Descriptive Statistics.
- 76.Wickham H (2016) ggplot2: Elegant Graphics for Data Analysis, pringer-Verlag; New York. [Google Scholar]
- 77.Patil I (2021) Visualizations with statistical details: The ggstatsplot approach, Journal of Open Source Software. 6, 3167. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Strains and plasmids are available upon request. The authors affirm that all data necessary for confirming the conclusions of the article are present within the article, figures, and tables.


