Abstract
Vaccinia virus genes are expressed in a sequential fashion, suggesting a role for negative as well as positive regulatory mechanisms. A potential down regulator of gene expression was mapped by transfection assays to vaccinia virus open reading frame D10, which encodes a protein with no previously known function. Inhibition was independent of the promoter type used for the reporter gene, indicating that the mechanism did not involve promoter sequence recognition. The inhibition was overcome, however, when the open reading frame of the reporter gene was preceded by the encephalomyocarditis virus internal ribosome entry site, which excludes the possibility of nonspecific metabolic or other antiviral effects and suggests that capped mRNAs or cap-dependent translation might be the target of the D10 product. The inducible overexpression of the D10 gene by a recombinant vaccinia virus severely inhibited viral protein synthesis, decreased the steady-state level of viral late mRNA, and blocked the formation of infectious virus.
Vaccinia virus contains nearly 200 genes (20) that can be divided into three classes—early, intermediate, and late—based on their times of expression (29, 33), promoter consensus sequences (2, 11, 12), and specific transcription factor requirements (9, 22, 41). Early gene expression is unique in that it does not require the de novo synthesis of DNA or proteins since the template, viral RNA polymerase, and early transcription factors are present within the infectious virus particle (21, 30). The early transcription factors, like virion structural proteins, are synthesized at late times (1, 8, 17). The viral proteins needed for intermediate transcription are products of early genes (34, 35, 41, 42), whereas at least some of those required for late transcription are products of intermediate genes (19, 22, 32, 47), providing the basis for sequential regulation.
At present, only positive regulators of poxvirus gene expression have been described. However, the rapidity of the shifts in cellular and viral protein synthesis raises the possibility of a role for negative regulators. The sharp increase in the rates of decay of host and viral mRNAs during vaccinia virus infection may accelerate progression through the stages of host and early, intermediate, and late viral gene expression (3, 31, 37). Stage-specific repressors of transcription and translation also have been suggested but without supporting evidence.
Inhibition of reporter gene expression by the D10 ORF.
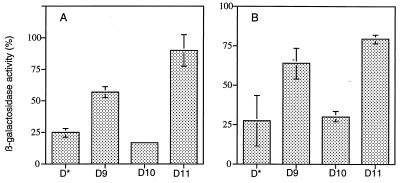
The present study originated from an unbiased screen of the vaccinia virus genome for transactivators of viral late gene expression (22). In cells infected with vaccinia virus in the presence of cytosine arabinoside, an inhibitor of DNA replication, reporter gene expression was activated upon transfection of specific combinations of six cosmids containing cloned fragments that spanned the entire vaccinia virus genome. The activators were mapped to the G8, A1, and A2 open reading frames (ORFs) (22), and the proteins were later shown to be transcription factors. We had noted in some experiments that the omission of cosmid 10 increased reporter gene activity severalfold in this assay (data not shown). Since the HindIII D DNA segment of the vaccinia virus genome was unique to cosmid 10, multiple plasmids that contained DNA from that region were tested for inhibitory activity in transfection assays using reporter plasmids with Escherichia coli lacZ under control of the vaccinia virus late P11 promoter (4). These experiments were performed with cells infected with vaccinia virus in the absence of cytosine arabinoside. In contrast to the vector alone, one plasmid containing a 2.9-kbp segment that included the D9, D10, and D11 ORFs inhibited lacZ expression by the reporter plasmid (Fig. 1A). The 2.9-kbp DNA was subcloned into segments that contained single ORFs. When these plasmids were transfected, D10 was as inhibitory as the fragment containing all three ORFs whereas D9 and D11 had lesser effects (Fig. 1A). Similar results were obtained when the reporter gene was regulated by the intermediate P30/300 (2) promoter (Fig. 1B). As a control, frameshift insertion mutations were made by cleaving the D10 and D9 genes at unique HpaI and EcoRV sites, respectively, and ligating an 8-mer EcoRI oligonucleotide linker. Plasmids containing frameshifted D10 or D9 ORFs did not inhibit late or intermediate promoter-regulated gene expression (data not shown). The effects of D10 on early promoters was not evaluated because genes regulated by early promoters are poorly expressed in transfection assays (10).
FIG. 1.
Repression of reporter gene expression by cotransfected plasmids. Human 293 cells (106) were infected with 10 PFU of vaccinia virus per cell and transfected 1 h later with calcium phosphate precipitates containing 2 μg of p11Xβ (A) or p30/300 (B) and 5 μg of pUC19 or derivatives containing vaccinia virus DNA. After 18 h, lysates were prepared from duplicate wells and assayed for β-galactosidase activity (22). The β-galactosidase activities obtained with the pUC19 vector and p11Xβ or p30/300 were assigned the value of 100%. The percentage of that activity obtained by transfections with the other plasmids were determined, and the mean values from two separate experiments are shown. p11Xβ, lacZ gene under control of late P11 promoter; p30/300, lacZ gene under control of G8R intermediate promoter; D*, 2.9-kbp BamHI fragment containing the D9, D10, and D11 ORFs; D9, BamHI/BglII fragment containing the D9 ORF; D10, SpeI/RsaI fragment containing the D10 ORF; D11, BglII/BamHI fragment containing the D11 ORF.
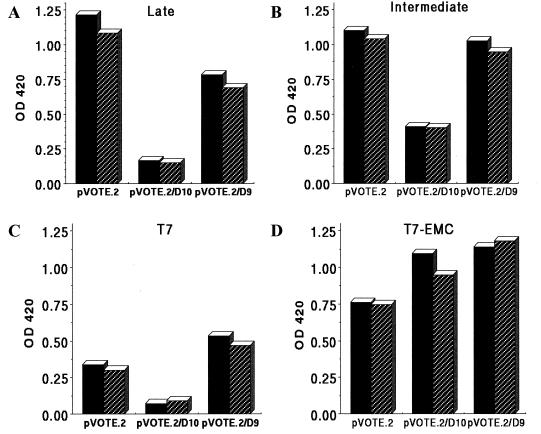
In the experiments described above, the transfected D10 and D9 genes were regulated by their natural promoters. Niles and coworkers (24–26) had reported that the D9 and D10 genes have early and late promoters, respectively. The difference in promoter type, and presumably level of expression, may have affected the relative activities of these genes in transfection assays. We therefore cloned the D9 and D10 ORFs into the plasmid vector, pVOTE.2, which contains the bacteriophage T7 promoter and an encephalomyocarditis (EMC) virus leader that provides enhanced cap-independent translation (13, 38). The resulting plasmids were called pVOTE.2/D9 and pVOTE.2/D10. To provide T7 RNA polymerase, we used vaccinia virus vTF7-3, which has the bacteriophage T7 RNA polymerase gene regulated by the vaccinia virus early-late P7.5 promoter (15). Cells were infected with vTF7-3 and then transfected with the late reporter plasmid and either the control pVOTE.2 vector, pVOTE.2/D10, or pVOTE.2/D9 plasmid. We found that D10 severely inhibited expression of the reporter gene regulated by either the late (Fig. 2A) or intermediate (Fig. 2B) promoter.
FIG. 2.
Effects of promoters and EMC leader sequences on repression of reporter gene expression. The D9 and D10 ORFs, with NdeI and XhoI restriction endonuclease sites, were directionally cloned into the pVOTE.2 expression vector (43) to form pVOTE.2/D9 and pVOTE.2/D10, respectively. BS-C-1 cells in six-well dishes were infected with vTF7-3 (15) at a multiplicity of infection of 10 PFU/cell and transfected 1 h later with DOTAP (Boehringer Mannheim) containing 2.5 μg of reporter plasmid and 2.5 μg of pVOTE.2, pVOTE.2/D9, or pVOTE.2/D10 per well. Extracts, prepared 24 h after transfection, were analyzed by using a β-galactosidase enzyme assay (Promega) and a SpectraMax 250 microplate spectrophotometer (Molecular Devices Corporation) set at A420. A minimum of two concentrations of extract were used to ensure linearity. Transfections were performed in duplicate and repeated a minimum of three times. Duplicates from a typical experiment are shown. The promoter type (late, intermediate, T7, or T7-EMC) used for regulation of the β-galactosidase expression is indicated above each panel. OD420, optical density at 420 nm.
Specificity of inhibition.
To evaluate the specificity of the inhibitory effect for vaccinia virus promoters, two lacZ reporter plasmids with bacteriophage T7 promoters were employed: pT7lacZ and pT7EMClacZ. As implied by its name, the latter contains a cDNA copy of the EMC leader to provide cap-independent translation (14). D10 inhibited expression by pT7lacZ, indicating that the effect was not specific for vaccinia virus promoters (Fig. 2C). Remarkably, reporter gene expression mediated by the T7 promoter and EMC leader was not inhibited (Fig. 2D), suggesting that capped mRNAs or cap-dependent translation is the target of the D10 product and excluding the possibility of nonspecific metabolic or other antiviral effects.
Construction of recombinant vaccinia viruses that inducibly overexpress the D10 or D9 genes.
To compare the effects of the D10 and D9 proteins on viral gene expression, we constructed vaccinia viruses with an inducible T7 RNA polymerase gene and an extra copy of the D9 or D10 ORF under an inducible T7 promoter and EMC virus leader. By employing the EMC virus leader, we sought to prevent negative autoregulation of D10 or D9 expression. This VOTE system was originally developed to inducibly express heterologous recombinant genes (43) but has been successfully employed to regulate vaccinia virus genes as well (18, 46). The pVOTE.2/D10 and pVOTE.2/D9 plasmids were transfected into cells that had been infected with vT7lacOI, a vaccinia virus that expresses the E. coli lac repressor and an inducible copy of the T7 RNA polymerase (43), and recombinant viruses vT7lacOI/D10 and vT7lacOI/D9 were isolated by antibiotic selection and plaque purified several times in succession. The presence of the original D10 or D9 gene, as well as the inducible copy, was verified by PCR and Southern blotting (data not shown).
Inducible overexpression of the D10 and D9 genes at 20 h after infection was demonstrated by Western blotting with rabbit polyclonal antiserum made to the C-terminal peptide (CYYESLKKLTEDD) of D10 and to the N-terminal peptide (MGITMDEEVIFETPRC) of D9. The amounts of D10 and D9 made by the parental vT7lacOI or by the recombinant viruses in the absence of inducer were below the level of detection with these antibodies. However, prominent D10 (Fig. 3A) and D9 (Fig. 3B) bands were visualized from extracts of infected cells that had been induced with IPTG (isopropyl-β-d-thiogalactopyranoside). Preimmune serum, from one of the two rabbits used for peptide immunization, reacted with an unidentified protein with a higher molecular weight than that of D10 in the lysates of all infected cells (Fig. 3A).
FIG. 3.
Inducible expression of D9 or D10 by recombinant vaccinia viruses. BS-C-1 cells in six-well dishes were mock infected or infected with 10 PFU of vT7lacOI per cell and 10 PFU of vT7lacOI/D10 (A) or vT7lacOI/D9 (B) per cell in the presence or absence of 100 μM IPTG. The infected cells were harvested at 22 h and lysed in 62.5 mM Tris-HCl (pH 6.8)–12.5% glycerol–1.25% sodium dodecyl sulfate–0.001% bromophenol blue–1.5% β-mercaptoethanol. The viscosity of the extracts was reduced by passage through a 25-gauge needle, and the material was applied to a 12.5% polyacrylamide gel. After electrophoresis, the proteins were transferred to a nitrocellulose membrane (Micron, Inc.) and incubated with preimmune or anti-D10 or anti-D9 rabbit serum as indicated. Western blots were developed with a BCIP-NBT (5-bromo-4-chloro-3-indolylphosphate–nitroblue tetrazolium) phosphatase substrate (Kirkegaard & Perry Laboratories Inc.). Standard markers are shown on the left of each blot, with masses in kilodaltons. The arrows point to the D10 and D9 bands.
Effects of D10 overexpression on virus replication.
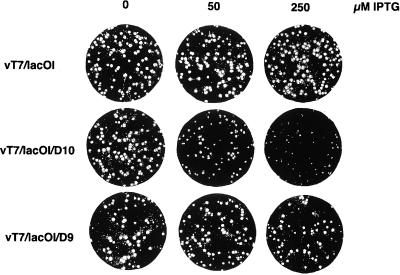
The sizes of plaques formed by vT7lacOI/D10 were moderately reduced with 50 μg of IPTG per ml and severely reduced at 250 μg/ml (Fig. 4), suggesting that large amounts of the D10 gene product had a negative effect on virus replication. In contrast, the sizes of vT7lacOI and vT7lacOI/D9 plaques were unaffected by inducer (Fig. 4). The selective effect of IPTG on replication of vT7lacOI/D10 was confirmed under single-step growth conditions (Fig. 5). In this experiment, we included recombinant virus vGW3 which contains the lacZ gene under control of the inducible T7 promoter and EMC virus leader (43).
FIG. 4.
Effect of D10 overexpression on plaque formation. BS-C-1 cells were inoculated with vT7/lacOI, vT7/lacOI/D10, or vT7/lacOI/D9 in the presence of 0, 50, or 250 μM IPTG. The cells were stained with crystal violet after 2 days (13).
FIG. 5.
Effect of D10 overexpression on virus yield. BS-C-1 cells were inoculated with 5 PFU of vT7lacoI/D10 (I/D10), vT7lacOI/D9 (I/D9), or vGW3 per cell in the presence (solid lines) or absence (broken lines) of IPTG. At the indicated times, the infected cells were harvested and the plaque titers were determined in duplicate on BS-C-1 cells in the absence of IPTG. Mean values were plotted.
Effects of D10 and D9 overexpression on viral protein synthesis.
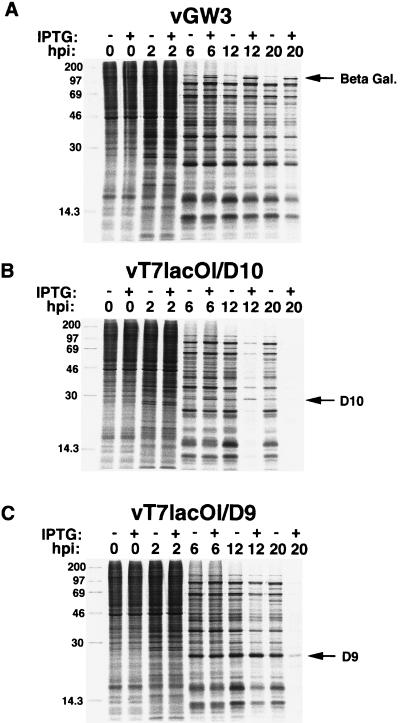
Cells were infected in the presence or absence of IPTG and pulse-labeled with [35S]methionine at various times to determine the effects of D10 and D9 expression on viral protein synthesis. Overexpression of D10 severely inhibited viral protein synthesis at 12 and 20 h after infection, whereas D9 inhibited protein synthesis only at the later time (Fig. 6), which probably accounts for the lack of effect of D9 overexpression on virus replication (Fig. 4 and 5). The D10 and D9 bands were difficult to resolve by pulse-labeling because of comigration of other viral proteins. However, bands of the expected sizes appeared to have a greater intensity, relative to that of proteins of other sizes, in the presence of IPTG than in its absence. In contrast, the β-galactosidase band induced by vGW3 (43) was readily discerned and there was little effect on the synthesis of viral proteins, excluding the possibility of nonspecific effects due to overexpression of any protein in this system (Fig. 6).
FIG. 6.
Effects of D10 or D9 overexpression on viral protein synthesis. BS-C-1 cells in six-well dishes were infected with 5 PFU of vGW3, vT7lacOI/D10, or vT7lacOI/D9 per cell in the presence (+) or absence (−) of 250 μM IPTG. At the indicated hours postinfection (hpi), the cells were incubated for 30 min in methionine-free medium and then incubated for 30 min with 50 μCi of [35S]methionine and harvested. The lysates were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and autoradiography. β-Galactosidase (Beta Gal.) could be seen as a distinct band in the IPTG-containing lanes and is indicated by an arrow. The D10 and D9 bands could not be resolved, but their approximate positions based on Western blots (Fig. 3) are indicated by arrows. The positions of standard markers with masses in kilodaltons are shown on the left.
Effects of D10 and D9 on steady-state levels of late mRNA.
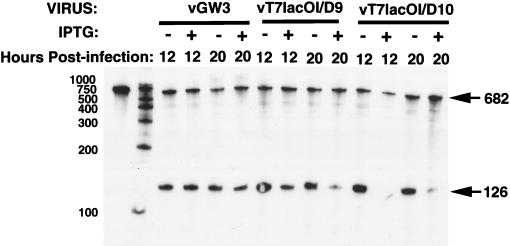
RNase protection assays were performed to determine the effects of the D10 and D9 proteins on the steady-state levels of a late mRNA. The transcript of the F17 gene, encoding the 11,000-molecular-weight structural protein, has been well characterized and used in previous studies as a representative late mRNA (3, 5, 36, 45). BS-C-1 cells infected with vGW3 served as a control. The 682-nucleotide biotinylated RNA probe extended from −556 to +126, relative to the RNA start site at +1. A properly initiated 126-nucleotide RNase-protected band as well as some undigested full-length probe was detected at 12 and 20 h after infection with vGW3; the addition of IPTG had little or no effect (Fig. 7). In cells infected with vT7lacOI/D9, IPTG induced a moderate reduction in the amount of the RNase-protected band at 12 h and a severe reduction at 20 h (Fig. 7). In cells infected with vT7lacOI/D10, IPTG caused a severe reduction at both 12 and 20 h (Fig. 7). The reductions in the amounts of the late mRNA corresponded to the effects on viral protein synthesis and replication shown in the previous sections.
FIG. 7.
Effect of D10 and D9 overexpression on viral RNA. BS-C-1 cells were infected as described in the legend to Fig. 6. Infected cells were harvested after 12 or 20 h by using a Direct Protect Lysate RPA kit (Ambion Inc.). A 682-nucleotide biotinylated RNA probe complementary to sequences upstream and downstream of the start site of the F17R transcript encoding the 11,000-molecular-weight protein was made in vitro by using the BrightStar Biotinscript kit (Ambion Inc.). The lysate (49 μl) was hybridized with an excess of the probe (0.2 to 1 ng) overnight at 37°C. RNase T1 and A were used to digest unhybridized RNA and then were inactivated by treatment with 10% Sarkosyl and proteinase K. The remaining RNA was precipitated with isopropanol and separated on an 8 M urea–6% polyacrylamide denaturing gel. The RNA was transferred to a BrightStar-Plus positively charged nylon membrane (Ambion Inc.) and immobilized by UV cross-linking. The RNA was detected by using the Ambion BrightStar BioDetect nonisotopic detection kit. The 682- and 126-nucleotide biotinylated RNA species are indicated by arrows. The numbers on the left refer to molecular size standards in nucleotides.
Conclusions.
A transfection approach, successfully used to identify the ORFs encoding activators of vaccinia virus late gene expression (22), has led us to an ORF that decreases gene expression. The inhibitory effect of D10 was abrogated by interrupting its reading frame, indicating that the protein product was required for activity. The inhibitory effect of D10 was manifested regardless of whether an intermediate or late vaccinia virus promoter or a bacteriophage T7 promoter was used for the reporter gene, indicating that the mechanism did not involve promoter sequence recognition. The specificity of the mechanism was demonstrated by using a reporter gene with an EMC virus leader sequence which allows cap-independent translation. In this case, expression was resistant to D10, thereby excluding indirect metabolic effects. Further studies demonstrated that the inducible overexpression of D10, and to a lesser extent D9, by recombinant vaccinia viruses inhibited viral gene expression. No such effect occurred when cells were infected with a similarly constructed recombinant virus expressing β-galactosidase, which excludes nonspecific effects of protein overexpression. The inhibition mediated by D10 correlated with decreases in the steady-state concentrations of a representative late mRNA.
Global alignments indicated that the vaccinia virus D10 and D9 proteins are 25% identical to each other and highly conserved among other poxviruses. Homologues are present in variola virus (27, 39), fowlpox virus (7), Shope fibroma virus (40), and molluscum contagiosum virus (38). The inability to isolate infectious D10 deletion mutants of fowlpox virus (7) or vaccinia virus (39a) is consistent with an essential function. By contrast, the neighboring D9 gene was deleted from both fowlpox virus (7) and vaccinia virus (39a) without abrogating infectivity. If D9 and D10 have overlapping functions, as suggested by their sequence similarity, D10 may be able to compensate for the absence of D9 but not vice versa. Although we succeeded in deleting the original D10 gene from vT7lacOI/D10, replication still occurred in the absence of IPTG, suggesting insufficiently stringent repression of the T7 promoter-controlled D10 gene and a requirement for only very small amounts of the D10 product (39a).
The finding of MutF motifs within both the D9 and D10 genes (23) provides a possible clue to their function and a direction for further research. This amino acid signature was originally identified as an important functional region of the E. coli MutT and the Streptococcus pneumoniae MutX antimutator proteins (28). Subsequent computer searches revealed a 20- to 30-amino-acid sequence with 6 to 10 positions that have highly conserved amino acids in a variety of proteins encoded by eubacteria, eukaryotes, and members of two virus families, poxviruses and African swine fever virus (6, 23). The characterized nonviral proteins in this family have widely differing functions due to their ability to hydrolyze a nucleoside diphosphate linked to some moiety, such as nucleoside triphosphates, coenzymes, nucleotide sugars, and dinucleoside polyphosphates. Since the cap structure of mRNA is a dinucleoside triphosphate (16, 44), our finding that cap-independent reporter gene expression was not inhibited by the D10 product is intriguing. The D10 protein might bind to or hydrolyze cap structures and thereby affect the stability or translatability of mRNAs. The decreased amounts of late vaccinia virus mRNA detected when D10 was overexpressed could be a direct consequence of D10 activity or secondary to a translational effect on the synthesis of other viral or cellular proteins.
Acknowledgments
We thank Joe Baldick for carrying out preliminary transfection and RNA analysis experiments.
REFERENCES
- 1.Ahn B-Y, Moss B. RNA polymerase-associated transcription specificity factor encoded by vaccinia virus. Proc Natl Acad Sci USA. 1992;89:3536–3540. doi: 10.1073/pnas.89.8.3536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baldick C J, Keck J G, Moss B. Mutational analysis of the core, spacer and initiator regions of vaccinia virus intermediate class promoters. J Virol. 1992;66:4710–4719. doi: 10.1128/jvi.66.8.4710-4719.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baldick C J, Jr, Moss B. Characterization and temporal regulation of mRNAs encoded by vaccinia virus intermediate-stage genes. J Virol. 1993;67:3515–3527. doi: 10.1128/jvi.67.6.3515-3527.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bertholet C, Drillien R, Wittek R. One hundred base pairs of 5′ flanking sequence of a vaccinia virus late gene are sufficient to temporally regulate late transcription. Proc Natl Acad Sci USA. 1985;82:2096–2100. doi: 10.1073/pnas.82.7.2096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bertholet C, Van Meir E, ten Heggeler-Bordier B, Wittek R. Vaccinia virus produces late mRNAs by discontinuous synthesis. Cell. 1987;50:153–162. doi: 10.1016/0092-8674(87)90211-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bessman M J, Frick D N, O’Handley S F. The MutT proteins or “Nudix” hydrolases, a family of versatile, widely distributed, “housecleaning” enzymes. J Biol Chem. 1996;271:25059–25062. doi: 10.1074/jbc.271.41.25059. [DOI] [PubMed] [Google Scholar]
- 7.Binns M M, Britton B S, Mason C, Boursnell M E G. Analysis of the fowlpox virus genome region corresponding to the vaccinia virus D6 to A1 region—location of, and variation in, non-essential genes in poxviruses. J Gen Virol. 1990;71:2873–2881. doi: 10.1099/0022-1317-71-12-2873. [DOI] [PubMed] [Google Scholar]
- 8.Broyles S S, Fesler B S. Vaccinia virus gene encoding a component of the viral early transcription factor. J Virol. 1990;64:1523–1529. doi: 10.1128/jvi.64.4.1523-1529.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Broyles S S, Yuen L, Shuman S, Moss B. Purification of a factor required for transcription of vaccinia virus early genes. J Biol Chem. 1988;263:10754–10760. [PubMed] [Google Scholar]
- 10.Cochran M A, Mackett M, Moss B. Eukaryotic transient expression system dependent on transcription factors and regulatory DNA sequences of vaccinia virus. Proc Natl Acad Sci USA. 1985;82:19–23. doi: 10.1073/pnas.82.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Davison A J, Moss B. The structure of vaccinia virus early promoters. J Mol Biol. 1989;210:749–769. doi: 10.1016/0022-2836(89)90107-1. [DOI] [PubMed] [Google Scholar]
- 12.Davison A J, Moss B. The structure of vaccinia virus late promoters. J Mol Biol. 1989;210:771–784. doi: 10.1016/0022-2836(89)90108-3. [DOI] [PubMed] [Google Scholar]
- 13.Earl P L, Cooper N, Moss B. Preparation of cell cultures and vaccinia virus stocks. In: Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. Vol. 2. New York, N.Y: Greene Publishing Associates & Wiley Interscience; 1991. pp. 16.16.1–16.16.7. [Google Scholar]
- 14.Elroy-Stein O, Fuerst T R, Moss B. Cap-independent translation of mRNA conferred by encephalomyocarditis virus 5′ sequence improves the performance of the vaccinia virus/bacteriophage T7 hybrid expression system. Proc Natl Acad Sci USA. 1989;86:6126–6130. doi: 10.1073/pnas.86.16.6126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fuerst T R, Niles E G, Studier F W, Moss B. Eukaryotic transient-expression system based on recombinant vaccinia virus that synthesizes bacteriophage T7 RNA polymerase. Proc Natl Acad Sci USA. 1986;83:8122–8126. doi: 10.1073/pnas.83.21.8122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Furuichi Y, Muthukrishnan S, Shatkin A J. 5′-Terminal m-7G(5′)ppp(5′)G-m-p in vivo: identification in reovirus genome RNA. Proc Natl Acad Sci USA. 1975;72:742–745. doi: 10.1073/pnas.72.2.742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gershon P D, Moss B. Early transcription factor subunits are encoded by vaccinia virus late genes. Proc Natl Acad Sci USA. 1990;87:4401–4405. doi: 10.1073/pnas.87.11.4401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hu X, Carroll L J, Wolffe E J, Moss B. De novo synthesis of the early transcription factor 70-kDa subunit is required for morphogenesis of vaccinia virions. J Virol. 1996;70:7669–7677. doi: 10.1128/jvi.70.11.7669-7677.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hubbs A E, Wright C F. The A2L intermediate gene product is required for in vitro transcription from a vaccinia virus late promoter. J Virol. 1996;70:327–331. doi: 10.1128/jvi.70.1.327-331.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Johnson G P, Goebel S J, Paoletti E. An update on the vaccinia virus genome. Virology. 1993;196:381–401. doi: 10.1006/viro.1993.1494. [DOI] [PubMed] [Google Scholar]
- 21.Kates J R, McAuslan B R. Poxvirus DNA-dependent RNA polymerase. Proc Natl Acad Sci USA. 1967;58:134–141. doi: 10.1073/pnas.58.1.134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Keck J G, Baldick C J, Moss B. Role of DNA replication in vaccinia virus gene expression: a naked template is required for transcription of three late transactivator genes. Cell. 1990;61:801–809. doi: 10.1016/0092-8674(90)90190-p. [DOI] [PubMed] [Google Scholar]
- 23.Koonin E V. A highly conserved sequence motif defining the family of MutT-related proteins from eubacteria, eukaryotes and viruses. Nucleic Acids Res. 1993;21:4847. doi: 10.1093/nar/21.20.4847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee-Chen G J, Bourgeois N, Davidson K, Condit R C, Niles E G. Structure of the transcription initiation and termination sequences of seven early genes in the vaccinia virus HindIII D fragment. Virology. 1988;163:64–79. doi: 10.1016/0042-6822(88)90234-6. [DOI] [PubMed] [Google Scholar]
- 25.Lee-Chen G J, Niles E G. Map positions of the 5′ ends of eight mRNAs synthesized from the late genes in the vaccinia virus HindIII D fragment. Virology. 1988;163:80–92. doi: 10.1016/0042-6822(88)90235-8. [DOI] [PubMed] [Google Scholar]
- 26.Lee-Chen G J, Niles E G. Transcription and translation mapping of the 13 genes in the vaccinia virus HindIII D fragment. Virology. 1988;163:52–63. doi: 10.1016/0042-6822(88)90233-4. [DOI] [PubMed] [Google Scholar]
- 27.Massung R F, Liu L-I, Qi J, Knight J C, Yuran T E, Kerlavage A R, Parsons J M, Venter J C, Esposito J. Analysis of the complete genome of smallpox Variola major virus strain Bangladesh-1975. Virology. 1994;201:215–240. doi: 10.1006/viro.1994.1288. [DOI] [PubMed] [Google Scholar]
- 28.Méjean V, Salles C, Bullions L C, Bessman M J, Claverys J-P. Characterization of the mutX gene of Streptococcus pneumoniae as a homologue of Escherichia coli mutT, and tentative definition of a catalytic domain of the dGTP pyrophosphohydrolases. Mol Microbiol. 1994;11:323–330. doi: 10.1111/j.1365-2958.1994.tb00312.x. [DOI] [PubMed] [Google Scholar]
- 29.Moss B, Salzman N P. Sequential protein synthesis following vaccinia virus infection. J Virol. 1968;2:1016–1027. doi: 10.1128/jvi.2.10.1016-1027.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Munyon W E, Paoletti E, Grace J T., Jr RNA polymerase activity in purified infectious vaccinia virus. Proc Natl Acad Sci USA. 1967;58:2280–2288. doi: 10.1073/pnas.58.6.2280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Oda K, Joklik W K. Hybridization and sedimentation studies on “early” and “late” vaccinia messenger RNA. J Mol Biol. 1967;27:395–419. doi: 10.1016/0022-2836(67)90047-2. [DOI] [PubMed] [Google Scholar]
- 32.Passarelli A L, Kovacs G R, Moss B. Transcription of a vaccinia virus late promoter template: requirement for the product of the A2L intermediate-stage gene. J Virol. 1996;70:4444–4450. doi: 10.1128/jvi.70.7.4444-4450.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pennington T H. Vaccinia virus polypeptide synthesis: sequential appearance and stability of pre- and post-replicative polypeptides. J Gen Virol. 1974;25:433–444. doi: 10.1099/0022-1317-25-3-433. [DOI] [PubMed] [Google Scholar]
- 34.Rosales R, Harris N, Ahn B-Y, Moss B. Purification and identification of a vaccinia virus-encoded intermediate stage promoter-specific transcription factor that has homology to eukaryotic transcription factor SII (TFIIS) and an additional role as a viral RNA polymerase subunit. J Biol Chem. 1994;269:14260–14267. [PubMed] [Google Scholar]
- 35.Sanz P, Moss B. A new vaccinia virus intermediate transcription factor. J Virol. 1998;72:6880–6883. doi: 10.1128/jvi.72.8.6880-6883.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schwer B, Visca P, Vos J C, Stunnenberg H G. Discontinuous transcription or RNA processing of vaccinia virus late messengers results in a 5′ poly(A) leader. Cell. 1987;50:163–169. doi: 10.1016/0092-8674(87)90212-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sebring E D, Salzman N P. Metabolic properties of early and late vaccinia messenger ribonucleic acid. J Virol. 1967;1:550–575. doi: 10.1128/jvi.1.3.550-558.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Senkevich T G, Koonin E V, Bugert J J, Darai G, Moss B. The genome of molluscum contagiosum virus: analysis and comparison with other poxviruses. Virology. 1997;233:19–42. doi: 10.1006/viro.1997.8607. [DOI] [PubMed] [Google Scholar]
- 39.Shchelkunov S N, Blinov V M, Totmenin A V, Marennikova S S, Kolykhalov A A, Frolov I V, Chizhikov V E, Gytorov V V, Gashikov P V, Belanov E F, Belavin P A, Resenchuk S M, Andzhaparidze O G, Sandakhchiev L S. Nucleotide sequence analysis of variola virus HindIII M,L, I genome fragments. Virus Res. 1993;27:25–35. doi: 10.1016/0168-1702(93)90110-9. [DOI] [PubMed] [Google Scholar]
- 39a.Shors, T., and B. Moss. Unpublished data.
- 40.Strayer D S, Jerng H H, Oconnor K. Sequence and analysis of a portion of the genomes of Shope fibroma virus and malignant rabbit fibroma virus that is important for viral replication in lymphocytes. Virology. 1991;185:585–595. doi: 10.1016/0042-6822(91)90529-k. [DOI] [PubMed] [Google Scholar]
- 41.Vos J C, Sasker M, Stunnenberg H G. Promoter melting by a stage-specific vaccinia virus transcription factor is independent of the presence of RNA polymerase. Cell. 1991;65:105–114. doi: 10.1016/0092-8674(91)90412-r. [DOI] [PubMed] [Google Scholar]
- 42.Vos J C, Sasker M, Stunnenberg H G. Vaccinia virus capping enzyme is a transcription initiation factor. EMBO J. 1991;10:2553–2558. doi: 10.1002/j.1460-2075.1991.tb07795.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ward G A, Stover C K, Moss B, Fuerst T R. Stringent chemical and thermal regulation of recombinant gene expression by vaccinia virus vectors in mammalian cells. Proc Natl Acad Sci USA. 1995;92:6773–6777. doi: 10.1073/pnas.92.15.6773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wei C M, Moss B. Methylated nucleotides block 5′-terminus of vaccinia virus mRNA. Proc Natl Acad Sci USA. 1975;72:318–322. doi: 10.1073/pnas.72.1.318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wittek R, Hanggi M, Hiller G. Mapping of a gene coding for a major late structural polypeptide on the vaccinia virus genome. J Virol. 1984;49:371–378. doi: 10.1128/jvi.49.2.371-378.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wolffe E J, Moore D M, Peters P J, Moss B. Vaccinia virus A17L open reading frame encodes an essential component of nascent viral membranes that is required to initiate morphogenesis. J Virol. 1996;70:2797–2808. doi: 10.1128/jvi.70.5.2797-2808.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wright C F, Keck J G, Moss B. A transcription factor for expression of vaccinia virus late genes is encoded by an intermediate gene. J Virol. 1991;65:3715–3720. doi: 10.1128/jvi.65.7.3715-3720.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]