Abstract
Human pluripotent stem cells (PSCs) have become popular tools within the research community to study developmental and model diseases. While many induced-PSCs (iPSCs) from various genetic background sources are currently available, scientific advancement has been hampered by the considerable phenotypic variations observed between different iPSC lines. A recent collaborative effort selected a novel iPSC line to address this and encourage the adoption of a standardized iPSC line termed KOLF2.1J. Here, leveraging the multiplexing power of isobaric labeling, we systematically investigate, at the 10k proteome level, the relative protein abundance profiles of the KOLF2.1J reference iPSC line upon two distinct cell state differentiation trajectories. In addition, we side-by-side systematically compare this line with the H9 line, an established embryonically derived PSC line that we previously characterized. We noticed differences in the basal proteome of the two cell lines and highlighted the differentially expressed proteins. While the difference between the cell line’s proteome subsisted upon differentiation, the global proteome remodeling trajectory was highly similar during the tested differentiation routes. We thus conclude that the KOLF2.1J line performs well at the proteome level upon the neuro and cardiomyogenesis differentiation protocol used. We believe this dataset will serve as a resource of value for the research community.
Keywords: FAIMS, H9 cells, induced-pluripotent stem cells (iPSCs), KOLF2.1J cells, TMTpro
1 │. INTRODUCTION
The mammalian biological system is a complex network consisting of a diverse array of cellular subtypes. However, human disorders and biological signaling processes are often studied using established immortalized cell lines which evade normal cellular senescence to pro-liferate indefinitely in vitro. Understanding the molecular mechanisms that direct cell type and disease-specific phenotypes is substantially limited when the immortalized cell lines are used. Therefore, the introduction of stem cell engineering has increased our understanding of biological processes by allowing researchers to generate diverse cell types relevant to model diseases. Specifically, advancements in directed differentiation of pluripotent stem cells (PSCs) into cell type(s) of interest, such as sub-types of neurons, muscle cells, etc., encourages researchers to develop a workflow to study human development and cell-type specific diseases.
Researchers have been exploiting mainly two sources of human stem cells: embryonic stem cell (ESC) lines and induced pluripotent stem cell (iPSC) lines. Human ESC (hESC) lines are derived from the inner cell mass of a blastocyst. They have an unlimited capacity for self-renewal and are pluripotent; thus, they can differentiate into any somatic cell type. In this case, additional genetic engineering is required to reflect the disease-causing mutations for specific diseases. On the other hand, human iPSC (hiPSC) lines are derived from skin or blood cells reprogrammed back into an embryonic-like pluripotent state. Commonly, distinct iPSC lines are derived from patients with either sporadic or familial diseases, and the control iPSC lines are derived from age/sex-matched control individuals. The research community quickly embraced working with iPSCs because they created the ability to study a patient’s disorder in disease-relevant cell types. While appealing at first, the usage of hiPSCs has been hampered by the fact that the inter-individual variability at the proteome level is considerably more pronounced than the mutation-induced variability when “control” and “disease” iPSC cells are compared. This has made the interpretation of a disease-causing mutation’s specific role(s) in an observed biological effect extremely challenging.
Advances in Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR)-mediated gene editing now enable precise mutation, mutation correction, or gene deletion in any line of interest, allowing isogenic controls (of the same or of closely similar genotypes) to be generated, instead of age/sex-matched controls. Recently, a new stem cell initiative from the NIH, the iPSC Neurodegenerative Disease Initiative (iNDI), was created to help provide high-quality and standardized iPSC models to the research community [1]. This thorough effort resulted in the selection of a novel reference iPSC line called KOLF2.1J, which has served as a foundation for ongoing and further large scale systematic gene editing to study disease-causing mutations [2]. Here, we aim to characterize the relative protein abundance profiles of this new reference hiPSC line side by side with another widely established reference line derived from hESC, named H9 (also referred to as WA09 or WAe009-A) [3]. Characterization and quality control of PSC lines and their respective differentiation potentials are most often focused on transcript level measurement combined with a handful of pre-defined cell state markers reflecting protein expression. While transcript expression profiling is an important part of the process, systematic proteome characterization, the end product of transcription and translation, is often overlooked. Notably, accumulating studies indicate that protein and transcript abundances have poor correlation (around ∼45% across many studies) [4–10] highlighting the need to also include a systematic proteome expression profiling layer, to current characterization workflows.
Using multiplexed proteomics, we performed in-depth quantitative proteome profiling of two genetically unmatched reference human iPSC and ESC and assessed (1) their steady state proteome differences, (2) their neural progenitor differentiation potential, and (3) their temporal cardiomyocyte differentiation potential. While strong differences between the two-reference cell lines in the pluripotent state are highlighted, this study also demonstrates a very strong correlation in their differentiation capacity based on their respective proteome remodeling trajectories upon differentiation into the two cell lineages studied. This dataset will serve as a resource for understanding possible differences/similarities at the molecular level upon directed differentiation of these two-reference pluripotent stem cell lines and will provide a framework to assess cell type specific protein expression throughout two unique differentiation routes.
2 │. EXPERIMENTAL SECTION
2.1 │. Materials
Tandem mass tag (TMT)-pro isobaric reagents were from Thermo Fisher Scientific (Waltham, MA). Trypsin was purchased from Thermo Fisher Scientific (Waltham, MA). Dulbecco’s modified Eagle’s medium (DMEM)/F12 and Roswell Park Memorial Institute (RPMI) media were obtained from Life Technologies (Waltham, MA).
2.2 │. Cell differentiation and harvesting
H9 (WiCell, Madison, WI) and KOLF2.1J (The Jackson Laboratory, Bar Harbor, ME) cells were maintained in a chemically defined in-house Essential 8 (E8) medium on Matrigel-coated (BD Bioscience) plates at 37 °C with 5% (v/v) CO2 and O2.
For differentiation of H9 and KOLF2.1J cells to cardiomyocytes, a chemically defined monolayer method described in a previous study was used [11]. Briefly, ∼90% confluent stem cells were incubated in RPMI-1640 with B-27 supplement (without insulin, Life Technologies) supplemented with CHIR99021 for 72 h. After that, cells were treated in insulin-minus RPMI-B27 with IWR-1, for 48 h. At day 7, the beating cardiomyocytes were observed, and media was replaced to RPMI-1640 with B27 supplement (With insulin, Life Technologies). At day 9, PSC-CM monolayers were purified for 2 days using RPMI-1640 without glucose and with B-27 supplement (Life Technologies). After purification, cells were maintained in RPMI-1640 with B-27 supplement.
For neuronal differentiation of H9 and KOLF2.1J cells, we followed another previously established protocol [12]. Briefly, the day before the start of the differentiation, stem cells were detached with 0.5 mM EDTA and seeded at high density. The neuronal induction with neural maintenance medium, 1 μM dorsomorphin (Peprotech), and 10 μM SB431542 (Peprotech) was started when the cells reached 100% confluence (day 0). The neural induction medium was changed daily for 8 days. The neural maintenance medium (1 L) consisted of 500 mL DMEM/F12 (Thermo Fisher Scientific), 0.25 mL Insulin (10 mg/mL, Life Technologies), 3.5 μL β-mercaptoethanol (14.26 M, Alfa Aesar), 5 mL non-essential amino acids (100×, Thermo Fischer Scientific), 5 mL sodium pyruvate (100 mM, Life Technologies), 2.5 mL Pens/Strep (10,000 U/μL, Life Technologies), 5 mL N2 (Life Technologies), 10 mL B27 (Life Technologies), 11.25 mL glutamax (100×, Life Technologies), and 500 mL Neurobasal Plus (Life Technologies) medium.
2.3 │. Cell lysis and protein digestion
Cells were washed with ice cold PBS twice and lysed in RIPA lysis buffer (50 mM Tris/HCl pH 7.5, 150 mM NaCl, 1% sodium deoxycholate, 0.1% SDS, 10 mM sodium pyrophosphate, 10 mM β-glycerol phosphate, 1% (v/v) NP-40, 0.5 mM TCEP, proteases inhibitor cocktail, phosphatases inhibitor cocktail) to produce whole cell extracts. Whole cell extracts were sonicated and clarified by centrifugation (16,000 × g for 10 min at 4°C) and protein concentrations were determined by Bradford assay.
Protein digestion for proteomics was performed as described in our previous study [13]. Protein extracts (100 μg) were subjected to disulfide bond reduction with 5 mM TCEP (25°C, 10 min) and alkylated with 20 mM 2-chloroacetamide (25°C, 20 min). Next, we performed methanol–chloroform precipitation to extract protein before trypsin digestion. To each sample four parts of neat methanol were added and vortexed, one part chloroform was then added and vortexed, and finally, three parts water was added and sample vortexed. Samples were centrifuged at 10,000 rpm for 5 min (room temperature) and washed with 100% methanol twice. Samples were resuspended in 100 mM EPPS pH 8.5 containing 0.1% RapiGest and digested overnight at 37°C with 1 μg of MS grade trypsin, after which digestion efficiency of a small aliquot was tested prior to labeling.
2.4 │. Tandem mass tag labeling
Trypsin digested samples (50 μg peptide input) were Tandem Mass Tag (TMT)pro labeled by adding 10 μL of 10 ng/μL of TMTpro reagent with acetonitrile to make a final acetonitrile concentration of about 30% (v/v) at room temperature (1 h). Labeling efficiency of a small aliquot was tested after incubation at room temperature for 1 h, and the reaction was then quenched with hydroxylamine to a final concentration of 0.5% (v/v) for 15 min. The TMTpro-labeled samples were combined together at a 1:1 ratio. The sample was acidified with formic acid, centrifuged at 10,000 × g for 5 min at room temperature and subjected to C18 solid-phase extraction (SPE) (50 mg, Sep-Pak, Waters).
2.5 │. Fe3+-NTA phospho-peptide enrichment
For phospho-peptide enrichment, a Fe3+-NTA phosphopeptide enrichment kit (Thermo, A32992) was used according to the manufacturer’s recommendations. In brief, dried peptides were enriched for phosphopeptides and eluted into a tube containing 25 μL of 10% formic acid (FA) to neutralize the pH of the elution buffer, then dried down. The unbound peptides (flow through) and washes were combined and saved for total proteome analysis.
2.6 │. Off-line basic Ph reversed-phase fractionation
Dried TMTpro-labeled sample was resuspended in 100 μl of 10 mM NH4HCO3 (pH 8.0) and fractionated using basic pH reversed-phase HPLC [14]. Briefly, samples were offline fractionated over a 90 min run, into 96 fractions by basic pH reverse-phase HPLC (Agilent LC1260) through an Aeris peptide xb-c18 column (Phenomenex; 250 mm × 3.6 mm), with mobile phase A containing 5% acetonitrile and 10 mM NH4HCO3 in LC-MS grade H2O, and mobile phase B containing 90% acetonitrile and 10 mM NH4HCO3 in LC-MS grade H2O (both pH 8.0). Next, the collected fractions were combined in a non-continuous manner into 24 fractions (as outlined in fig. S5 of [15]) and used for subsequent mass spectrometry analysis after desalting via C18 StageTip.
For Phospho-peptides, dried peptides were fractionated according to manufacturer’s instructions using a Basic pH reversed-phase peptide fractionation kit (Thermo Fisher Scientific, San Jose, CA) for a final six fractions and subjected to C18 StageTip desalting prior to MS analysis.
2.7 │. Liquid chromatography and tandem mass spectrometry – Proteome analysis
Mass spectrometry data were acquired using an Orbitrap Eclipse Tribrid mass spectrometer (Thermo Fisher Scientific, San Jose, CA) connected to an UltiMate 3000 RSLCnano system liquid chromatography (LC) pump (Thermo Fisher Scientific, San Jose, CA). Peptides were separated on a 100 μm inner diameter microcapillary column packed in-house with ∼30 cm of HALO Peptide ES-C18 resin (2.7 μm, 160 Å, Advanced Materials Technology, Wilmington, DE) with a gradient consisting of 5%–23% (0–75 min), 23–40% (75–110 min) (ACN, 0.1% FA) over a 120 min run at ∼500 nL/min. 3/10 of each fraction was loaded onto the column for analysis. Proteome analysis used Multi-Notch MS3-based TMT quantification [16], combined with Real Time Search analysis software [17, 18], and the FAIMS Pro Interface (using previously optimized 3 CV parameters [19]), to reduce ion interference. The scan sequence began with an MS1 spectrum (Orbitrap analysis; resolution 120,000 at 200 Th; mass range 400–1500 m/z; maximum injection time 50 ms; automatic gain control (AGC) target 4×105). For MS2 analysis precursors were selected based on a cycle time of 1.25 s/CV method (FAIMS CV = −40/−60/−80). MS2 analysis consisted of collision-induced dissociation (quadrupole ion trap analysis; rapid scan rate; AGC 1.0 × 104; isolation window 0.5 Th; normalized collision energy (NCE) 35; maximum injection time 35 ms). Monoisotopic peak assignment was used, and previously interrogated precursors were excluded using a dynamic window (180 s ± 10 ppm). Following acquisition of each MS2 spectrum, a synchronous-precursor-selection (SPS) API-MS3 scan was collected on the top 10 most intense ions b or y-ions matched by the online search algorithm in the associated MS2 spectrum [17, 18]. MS3 precursors were fragmented by high energy collision-induced dissociation (HCD) and analyzed using the Orbitrap (NCE 45; AGC 2.5×105; maximum injection time 200 ms, resolution was 50,000 at 200 Th). Closeout was set at two peptides per protein per fraction, so that MS3s were no longer collected for proteins having two peptide-spectrum matches (PSMs) that passed quality filters [18].
2.8 │. Liquid chromatography and tandem mass spectrometry – Phospho-peptide analysis
Mass spectrometry data were acquired using an Orbitrap Eclipse Tribrid mass spectrometer (Thermo Fisher Scientific, San Jose, CA) connected to an UltiMate 3000 RSLCnano system liquid chromatography (LC) pump (Thermo Fisher Scientific). Peptides were separated on a 100 μm inner diameter microcapillary column packed in-house with ∼30 cm of HALO Peptide ES-C18 resin (2.7 μm, 160 Å, Advanced Materials Technology, Wilmington, DE) with a gradient consisting of 3%–20% (0–90 min), 20%–35% (90–160 min) (ACN, 0.1% FA) over a 170 min run at ∼500 nL/min. For analysis, we loaded half of each fraction onto the column. Each analysis used the FAIMS Pro Interface, using previously optimized 3 CV parameters for TMT-labeled phosphopeptides [20] to reduce ion interference. The scan sequence began with an MS1 spectrum (Orbitrap analysis; resolution 120,000 at 200 Th; maximum injection time 50 ms; mass range 350–1500 m/z; automatic gain control (AGC) target 4 × 105). For MS2 analysis, precursors were selected using a cycle time of 1.25 s/CV method (FAIMS CV = −40/−60/−80). MS2 analysis consisted of high energy collision-induced dissociation (HCD) (Orbitrap analysis; resolution 50,000 at 200 Th; isolation window 0.5 Th; normalized collision energy (NCE) 38; AGC 2 × 105; maximum injection time 172 ms). Monoisotopic peak assignment was used, and previously interrogated precursors were excluded using a dynamic window (120 s ± 10 ppm).
2.9 │. Data analysis
Mass spectra were processed using a Comet-based (2019.01 rev. 5) software pipeline [21, 22]. Spectra were first converted to mzXML and monoisotopic peaks were re-assigned using Monocle software [23]. MS2 spectra were matched with peptide sequences with a composite sequence database including the Human Reference Proteome (2020–01 - SwissProt entries only) UniProt database, as well as sequences of common contaminants. This database was concatenated with one composed of all protein sequences in the reversed order. Analysis was performed using a 50 ppm precursor ion tolerance. Static modifications included, TMTpro tags on lysine residues and peptide N termini (+304.207 Da) and carbamidomethylation of cysteine residues (+57.021 Da). Oxidation of methionine residues (+15.995 Da) was set as a variable modification. For phosphorylation dataset search, phosphorylation (+79.966 Da) on serine, threonine or tyrosine and deamidation (+0.984 Da) on Asparagine or Glutamine were set as additional variable modifications. Peptide-spectrum matches (PSMs) were adjusted to a 1% false discovery rate (FDR) [24]. PSM filtering was performed using a linear discriminant analysis, [25], while considering the following parameters: XCorr, ΔCn, charge state, missed cleavages, precursor mass accuracy and peptide length. For protein-level comparisons, PSMs were identified, quantified, and collapsed to a 1% peptide false discovery rate (FDR) and then collapsed further to a final protein-level FDR of 1% [26]. TO generate the smallest set of proteins required to account for all observed peptides, the principles of parsimony were applied. For TMT-based reporter ion quantitation, summed signal-to-noise (S:N) ratio for each TMT channel was first extracted based on the closest matching centroid to the expected mass of the TMT reporter ion (integration tolerance of 0.003 Da). Isotopic impurities of the different TMT reagents provided by the manufacturer specifications, were used to adjust reporter ion intensities. Proteins were quantified by summing reporter ion signal-to-noise measurements across all matching PSMs, resulting in a “summed signal-to-noise” measurement. For total proteome, PSMs with poor quality, MS3 spectra with 7 (Figure 1), 14 (Figure 3) or more TMT reporter ion channels missing, or isolation specificity less than 0.75, or with TMT reporter summed signal-to-noise ratio that were less than 160 or had no MS3 spectra were excluded from quantification. For phospho proteome, PSMs with poor quality, MS2 spectra with 13 (Figure 2) or more TMT reporter ion channels missing, or isolation specificity less than 0.8, or with TMT reporter summed signal-to-noise ratio that were less than 160 were excluded from quantification. The AScore algorithm was used to determine the localization of phosphorylation sites [27]. AScore is a probability-based approach for high-throughput protein phosphorylation site localization. Precisely, a threshold of 13 relates to a 95% confidence in site localization.
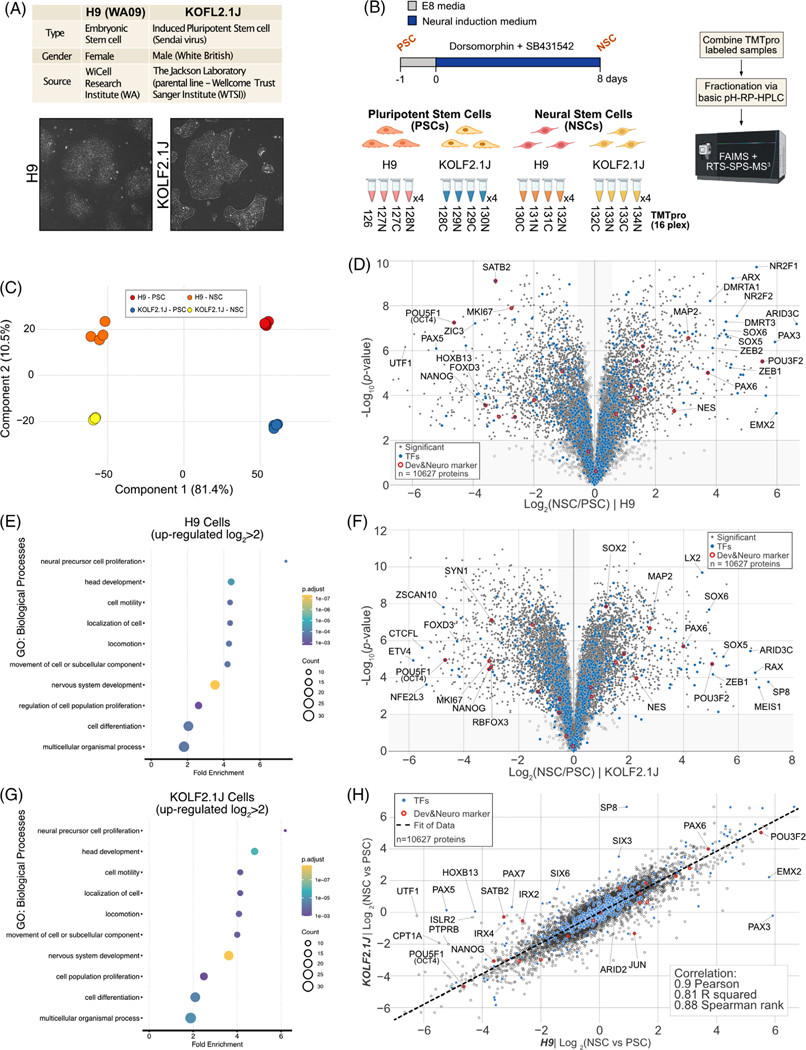
FIGURE 1.
Quantitative comparison of H9 and KOLF2.1 proteome remodeling upon Neural Stem Cell differentiation. (A) The information of H9 and KOLF2.1J reference cell lines (upper). Bright-field microscope images of H9 and KOLF2.1J cell lines (lower). (B) Workflow for neural stem cell differentiation from pluripotent stem cell and TMTpro based proteomics. 16plex proteomics was performed with biological quadruplicates per condition. (C) Principal component analysis (PCA) of proteomics data obtained from 16plex TMTpro based analysis. The inset indicates color coding for individual cells (Pluripotent Stem cell (PSC), Neural Stem Cell (NSC). (D) Volcano plot shows the H9 cell line -log10-transformed p-value versus the log2-transformed ratio of Neural Stem cells/ Pluripotent Stem cells for quantification. p-values were calculated by two-sided Welch’s t-test (adjusted with 1% false discovery rate (FDR) for multiple comparisons); for parameters, individual p-values and q-values, see Table S1. Individual proteins are shown in gray open circles, significantly changed protein: filled gray circle, transcription factors (TFs): filled blue circles, and developmental & neuronal markers: red open circles; 10627 proteins were quantified). (E) The enriched gene ontology (GO) terms for proteins whose abundance is significantly increased in H9 NSC compared to H9 PSC. (F) Volcano plot shows the KOLF2.1J cell line -log10-transformed p-value versus the log2-transformed ratio of Neural Stem cells/ Pluripotent Stem cells for quantification. p-values were calculated by two-sided Welch’s t-test (adjusted with 1% FDR for multiple comparisons); for parameters, individual p-values and q-values, see Table S1. Individual proteins are shown in gray open circles, significantly changed protein: filled gray circle, transcription factors (TFs): filled blue circles, and developmental and neuronal markers: red open circles, 10627 proteins were quantified). (G) The enriched gene ontology (GO) terms for proteins whose abundance is significantly increased in KOLF2.1J NSC compared to KOLF2.1J PSC. (H) Proteome level correlation plot of log2(NSC/PSC) from KOLF2.1J cell line (y axis) or H9 cell line (x axis) upon differentiation. The calculated correlation factors (Pearson, R squared, and Spearman rank) are indicated
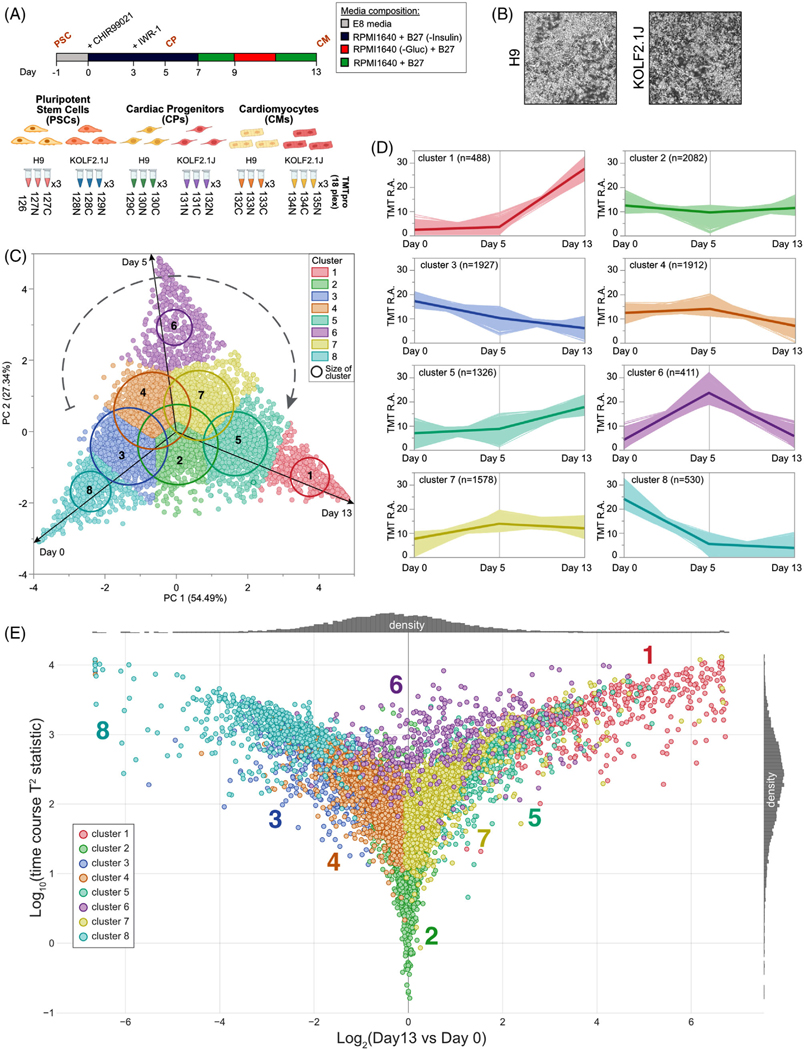
FIGURE 3.
Quantitative proteome remodeling analysis upon cardiomyogenesis. (A) Workflow for cardiac differentiation from pluripotent stem cell and TMTpro-based proteomics. 18plex proteomics was performed with biological triplicated per conditions. (B) Bright-field microscope images of H9 and KOLF2.1J cardiomyocytes (day 13). (C) H9 protein level principal component analysis of proteomics data obtained from 18plex TMTpro based analysis. The inset specifies the color coding used for each cluster. (D) A total of eight cluster profiles were isolated by k-means clustering analysis of the H9 cell data and the number of proteins per cluster is specified. For KOLF2.1J similar analysis was performed and available in Table S3. (E) Multivariate empirical Bayes analysis of the total proteome data of H9-derived PSC, CPs and CMs over the 13-day timespan. Plot shows log10(time course T2 statistic) versus log2(ratio day 13/day 0). Each dot illustrates the trajectory of changes in abundance for each individual quantified proteins during cardiac differentiation of H9 cells on the basis of nine independent abundance determinations. The eight individual clusters from (D) are also represented. Similar analysis was performed for KOLF2.1J and data are available in Table S3
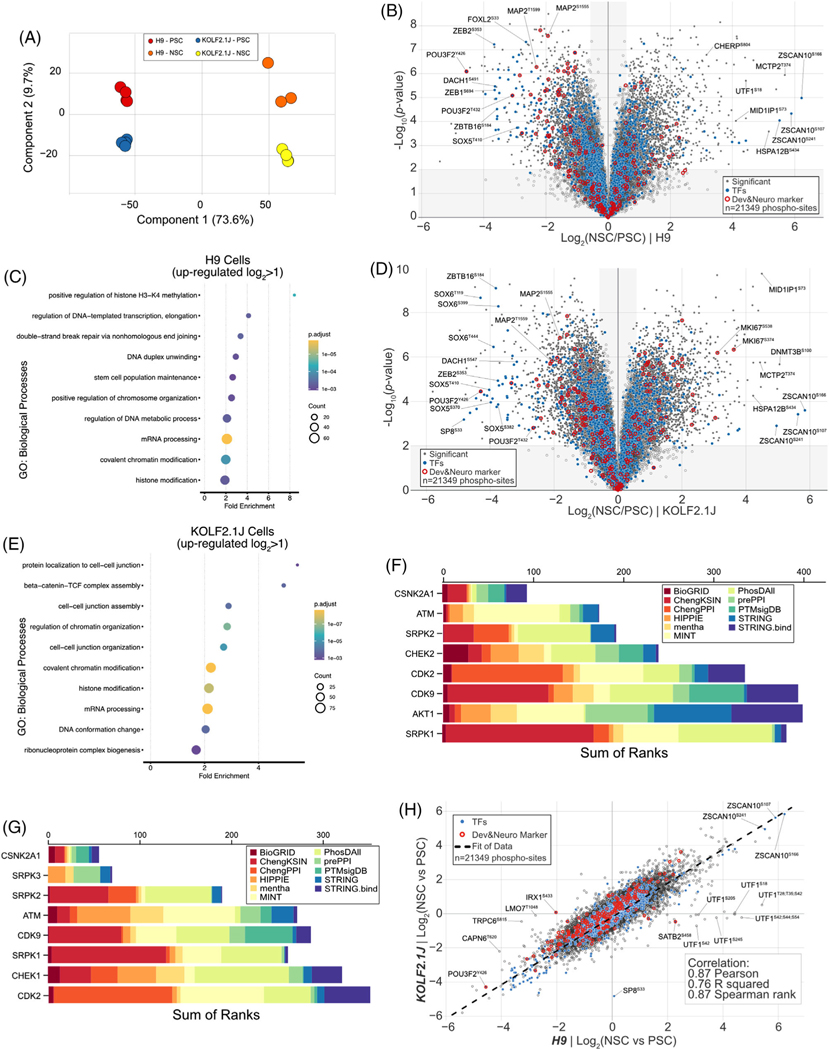
FIGURE 2.
Quantitative phosphoproteome analysis of H9 and KOLF2.1 upon neural stem cell differentiation. (A) Principal component analysis (PCA) of phospho-proteomics data obtained from 16plex TMTpro based analysis. The inset indicates color coding for individual cells (Pluripotent Stem cell (PSC), Neural Stem Cell (NSC). (B) Volcano plot shows the H9 cell line -log10-transformed p-value versus the log2-transformed ratio of Neural Stem cells/Pluripotent Stem cells for quantification. p-values were calculated by two-sided Welch’s t-test (adjusted with 1% FDR for multiple comparisons); for parameters, individual p-values and q-values, see Table S2. Individual proteins are shown in gray open circles, significantly changed protein: filled gray circle, transcription factors: filled blue circles, and developmental & neuronal markers: red open circles; 21,349 phospho-sites were quantified). (C) The enriched gene ontology (GO) terms for proteins whose abundance is significantly increased in H9 NSC compared to H9 PSC. (D) Volcano plot shows the KOLF2.1J cell line -log10-transformed p-value versus the log2-transformed ratio of neural stem cells/ pluripotent stem cells for quantification. p-values were calculated by two-sided Welch’s t-test (adjusted for multiple comparisons); for parameters, individual p-values and q-values, see Table S2. Individual proteins are shown in gray open circles, significantly changed protein: filled gray circle, transcription factors: filled blue circles, and developmental and neuronal markers: red open circles; 21349 phospho-sites were quantified). (E) The enriched gene ontology (GO) terms for proteins whose abundance is significantly increased in KOLF2.1J NSC compared to KOLF2.1J PSC. (F) MeanRank score from kinase enrichment analysis for H9 up-phosphorylated proteins in NSC to PSC comparison (Log2 > 1, p-value < 0.01). (G) MeanRank score from kinase enrichment analysis for KOLF2.1J up-phosphorylated proteins in NSC to PSC comparison (Log2 > 1, p-value < 0.01). (H) Phospho-proteome level correlation plot of log2(NSC/PSC) from KOLF2.1J cell line (y axis) or H9 cell line (x axis) upon differentiation. The calculated correlation factors (Pearson, R squared, and Spearman rank) are indicated
Protein or phospho-peptide quantification values were exported for further analysis in Microsoft Excel, R package (clusterProfiler 4.0 [28], pheatmap), Kinase Enrichment Analysis 3 (KEA3) [29] and Perseus [30]. Each reporter ion channel was summed across all quantified proteins and normalized assuming equal protein loading of all samples. Phospho-peptides were normalized to the corresponding protein abundance value (when available and indicated in supplementary tables). Maximum and minimum TMT ratio quantifiable were capped to 100-fold. Organellar protein marker annotations were compiled using the proteins which had scored with confidence “very high” or “high” from a previously published HeLa dataset [31] and additional entries from manually curated literature. Transcription factor annotation list was assembled from previously published database [32] and only transcription factors marked as “curated” were used.
Supplemental data tables list all quantified proteins and phospho-peptides as well as associated TMT reporter ratio to control channels used for quantitative analysis.
3 │. RESULTS AND DISCUSSION
Recent efforts from the iNDI [1] have led to the generation and selection of the KOLF2.1J cell line to be adopted by the research community as a novel common reference iPSC line [2]. This line underwent extensive quality control focused on morphology, proliferation, pluripotency, gene editing efficiency, in-depth RNA sequencing, as well as whole-genome sequencing. In addition, differentiation potential into different cell types was examined using single-cell RNA-sequencing. We have previously reported a proteomics workflow to characterize the temporal proteome remodeling of hESCs into NGN2-induced neurons (iNeurons) [13], using the established H9 (or WA09) reference hESC line. This resource detailed changes in protein abundance throughout differentiation from ESCs to neurons and provided critical insight into some of the molecular rewiring happening upon cell state differentiation. Here, we adapted a similar quantitative workflow to directly compare side by side the proteome’s developmental differentiation trajectories of the established H9 reference line with the novel KOLF2.1J reference line and thus provide additional resources to be used by the research community.
3.1 │. Global proteome variation between H9 and KOLF2.1J at PSC level
In an effort to further characterize constitutive expression differences at the protein level and to better understand how the proteome is remodeled upon neuronal state transition, we performed quantitative proteomics on both the H9 and the KOLF2.1J lines (Figure 1A). KOLF2.1J lines show a distinct morphology of undifferentiated status growing in the form of colony with large nucleus and scant cytoplasm similar to H9 (Figure 1A). Cells at PSC state were differentiated into Neural Stem Cell (NSC) using a previously established dual inhibition protocol [12] (Figure 1B). Total-cell extracts of cells at PSC (day 0) or NSC state (day 8) were collected in biological quadruplicates and subjected to 16plex TMTpro analysis [33] (Figure 1B). Principal component analysis revealed highly reproducible replicate data, with 81.4% of the variance being driven by differentiation and 10.5% driven by cell line background (Figure 1C), while the coefficient of variation across biological replicate was ∼5% for both cell line (Figure S1G). In total, we quantified 10,627 unique proteins across both cell lines and cell states (Table S1). As anticipated, we found profound inter-cell line variability at steady state (PSC – day 0) with ∼11.4% of the quantified proteome significantly different, of which 168 and 229 proteins were, respectively, upregulated or downregulated by more than two-fold (Figure S1A). Post-NSC differentiation (day 8) showed lower inter-cell line variability with ∼6.7% of the quantified proteome significantly different, of which 96 and 172 proteins were, respectively, upregulated or downregulated by more than two-fold (Figure S1B). Overall, about a dozen transcription factors were strongly differentially expressed between both cell lines. Of which several are linked to various aspect of embryonic development, such as members of the Paired box (PAX) [34], Iroquois homeobox factors (IRX) [35], and sine oculis (SIX) [36] family of transcription factors. Of note, the KOLF2.1J line is of male genotype, while the H9 line is of female genotype which could account for portion of the variability observed. We could quantify six Y-chromosome linked proteins (highlighted in green in Figure S1A–C) all virtually only expressed in KOLF2.1J and their level of expression was not affected by cell state transition based on a correlation plot (Figure S1C). A correlation plot of the two reference cell lines at both cell states displayed little correlation for both Pearson (0.4) and R squared correlation (0.16), indicating that the portion of the proteome that is differentially expressed between both lines is largely dissimilar (Figure S1C).
3.2 │. Quantifying proteome remodeling during conversion of hPSCs to neural stem cells
We next focused on quantifying how the proteome of each cell line remodeled upon differentiation into NSCs. From the more than 10,000 proteins quantified, we observed the predictable loss of pluripotency factors and increase in neurogenesis proteins, indicating that both H9 and KOLF2.1J cells underwent the expected differentiation program (Figure S1D). Three factors out of the 19 highlighted here showed a pattern affected by the cell line, namely IRX2, SATB2 and JUN (Figure S1D). Globally, for the H9 cell line ∼50.4% of the proteome was statistically different post-NSC differentiation (of which 1084 and 1240 proteins were, respectively, upregulated or downregulated by more than two-fold) (Figure 1D). While for the KOLF2.1J cell line, post-NSC differentiation, ∼70.8% of proteome was significantly different (of which 1356 and 1505 proteins were, respectively, upregulated or downregulated by more than two-fold) (Figure 1F). We next examined the behavior of ∼785 highly curated transcription factors upon cell state change. As previously reported for iNeuron differentiation [13], while the abundance of most of the transcriptional regulators quantified was largely unchanged in both cell lines, a cohort of factors linked with maintenance of pluripotency (e.g., OCT4, NANOG) was dramatically downregulated (log2(NSC/PSC) <−2.0) to similar levels in both cell lines (Figures 1D,F). On the other hand, the hPSC-to-hNSC transition in both cell lines is associated with a strong increase (log2(NSC/PSC) > 2.0) in the abundance of numerous proteins linked to nervous system development (e.g., MAP2, ZEB1/2, POU3F2) (Figures 1D,F). Next, to address what proteins might have their expression solely affected by the cell line backgrounds upon NSC differentiation, we plotted the remodeling fold change ratio (NSC/PSC) of the KOLF2.1J line versus the H9 line on a volcano plot (Figures S1E). This indicated that ∼11.8% of the remodeled proteome trajectory was significantly affected by the cell line backgrounds (of which 35 and 40 proteins were, respectively, upregulated or downregulated by more than four-fold (log2 ± 2.0)). These included transcript factors like UTF1, SP8, SATB2, SIX6/3 and PAX5 and PAX3 (Figures S1E). PAX5, associated with brain development and autism spectrum disorder (ASD) [37], and UTF1, which controls pluripotency and self-renewal in ESCs [38], are significantly decreased in H9 (NSC/PSC) but not in KOLF2.1J. SP8, which promotes the premature differentiation of NSC is increased in KOLF2.1J (NSC/PSC) [39]. KOLF2.1J was generated by correcting a mutation present in one copy of ARID2 in the parental KOLF2_C1 line [2]. Our quantitative analysis showed no significant differnces between the level of expression of ARID2 between H9 and KOLF2.1J at either cell state (Figure S1A,B,F). To get a broader view of the two reference cell lines’ proteome trajectories upon NSC transitioning from their respective PSC states, we used a correlation plot. Correlation plot metrics showed a very strong correlation for Pearson (0.9), R squared (0.81) and Spearman rank (0.88) correlations, indicating that, excepting minor outliers, the proteome remodeling trajectories of both reference lines are highly similar (Figure 1G).
3.3 │. Phospho-proteome remodeling analysis upon neural stem cell differentiation
In parallel, while quantifying the total proteome (Figure 1A), we also examined global phosphorylation using the previously established streamlined tandem mass tag workflow [20, 40], prior to proteome fractionation. In line with the total proteome, principal-component analysis revealed reproducible replicate data, with 73.6% of the variance being driven by differentiation and 9.7% driven by cell line background (Figure 2A). In total, we quantified 21,349 unique phospho-sites, in 5,181 proteins across both cell lines and cell states (Table S2). Of these, 17,440 sites were phosphorylated at a single site, 20,795 sites were normalized to their protein abundance and 18,524 sites were considered localized by having an AScore ≥ 13 (95% confidence in site localization) [27]. Globally, for the H9 cell line, ∼22% of the phospho-proteome was statistically different post-NSC differentiation (of which 288 and 214 sites were upregulated or downregulated, respectively, by more than four-fold (log2 ± 2.0) (Figure 2B). For the KOLF2.1J cell line, ∼44.3% of the phospho-proteome was statistically different post-NSC differentiation (of which 360 and 299 sites were upregulated or downregulated, respectively, by more than four-fold (log2 ± 2.0) (Figure 2D). A gene ontology analysis of the upregulated sites (log2 ratio > 1.0 and p < 0.01) for either cell line indicated enrichment in mRNA processing, chromosome remodeling and histone modification terms (Figure 2C,E). To confirm the upstream kinases of changed phospho-peptide (log2 ratio > 1.0 and p < 0.01) upon NSC differentiation, we used the Kinase Enrichment Analysis 3 (KEA3). This suggested that phosphorylation of ATM, CDK2, CDK9, and SRPK1/2 target proteins is elevated in NSC of either cell lines (Figure 2F,G). The activation of SRPK1/2, known to control of pre-mRNA splicing [41], and ATM & CDK2, regulator of DNA damage [42], is consistent with GO analysis. Including Akt substrates, which are inactivated by dorsomorphin treatment, phosphorylation of PLK1, CDK1/2 and AURKB substrates is commonly decreased during NSC differentiation (Figure S2A,B). Similar to our total proteome analysis, we used a correlation plot to gain a broader view of the two reference cell lines’ phospho-proteome trajectories upon NSC transition from their respective PSC states. Correlation plot metrics showed a very strong correlation for Pearson (0.87), R squared (0.76) and Spearman rank (0.87) correlations, indicating that, excepting a few outliers, the phospho-proteome remodeling trajectory of both reference line is highly similar (Figure 2H). Our data suggest that similar kinase pathway is regulated upon NSC differentiation of H9 and KOLF2.1J.
3.4 │. Global proteome remodeling analysis upon cardiomyogenesis differentiation
Having defined the overall broad similarities between the two reference PSC lines upon NPC differentiation, we next sought to further characterize them by subjecting them to a second, unrelated differentiation scheme. To our knowledge, an in-depth kinetic and quantitative analysis of changes in the proteome during conversion of hPSCs to cardiomyocytes has not been performed. We chose to benchmark the two reference lines upon cardiomyogenesis differentiation and include an additional temporal component (total of three time points) into our workflow to gain further statistical insight. We performed quantitative proteomics on whole-cell extracts of both H9 and KOLF2.1J lines subjected to a chemically defined monolayer differentiation method [11] (Figure 3A). Upon final differentiation (day 13), both cell lines differentiated into cardiomyocytes displaying pacemaker activity (Figure 3B, Video S1, S2). Total-cell extracts at PSC state (day 0), cardiac progenitor (CPs) state (day 5), and after final differentiation into cardiomyocytes (CMs) (day 13), all in biological quadruplicate where subjected to 18plex TMTpro analysis [43]. In total, we quantified 10,254 unique proteins across both cell lines and all three cell states (Table S3). Global principal-component analysis revealed highly reproducible replicate data, with 65.2% of the variance being driven by differentiation and 25% driven by cell line background (Figure S3A). In addition, for the H9 cell line we overlaid the loading plot to the score plot, resulting in a protein-level PCA map, displaying a clear clockwise temporal separation driven by differentiation (Figure 3C). Additionally, k-means clustering analysis performed at the cell line level, revealed eight shared discrete trajectory patterns (Figures 3C,D and Table S3). Then, we represented the time course “trajectories” of all the quantified proteins in Figure 3E. These trajectories are based on the Hotelling T-squared distribution (T2), a generalization of Student’s t distribution for multivariate hypothesis testing [44], in which each dot represents the statistical distribution for the changes in abundance of an individual protein over the 3-day time course [45]. Finally, we overlaid the k-means clustering analysis onto the Hotelling T2 scatter plot for easier global representation (Figure 3E). For example, we can clearly visualize that clusters 8 and 3 represent proteins for which the abundance decreased rapidly upon differentiation (Figure 3D,E). In contrast, clusters 1 and 5 correspond to numerous proteins upregulated 10- to 20-fold throughout cardiac differentiation (Figure 3D,E). Clusters centered within log2 (day 13/day 0) from −1 to +1 and within the bottom half of the y-axis involved proteins whose abundance is mostly unaffected (e.g., clusters 2, 4 and 7), while finally, those within the top half of the y-axis included proteins whose profile transiently increased (cluster 6) mid-differentiation (day 5) (Figure 3D,E). Thus, our deep profiling dataset during cardiomyogenesis provides a resource for uncovering factors important for this process.
3.5 │. Quantifying H9 and KOLF2.1J cardiomyogenesis differentiation potential
We next focused on quantifying how the proteome of each cell line remodeled upon differentiation into cardiomyocytes. From the more than 10,000 proteins quantified, we observed the expected temporal loss in pluripotency factors followed by the accumulation of cardiomyocyte proteins, indicating that both H9 and KOLF2.1J cells underwent the anticipated directed-differentiation program (Figure S3C). Of the 15 factors highlighted here, all of them display similar pattern across both cell lines. Next, based on the T2 hotelling scatter plot, we visualized the whole proteome of both reference lines as well as the mentioned developmental and cardiomyocyte markers and transcription factors (Figure 4A,B). We next examined the behavior of ∼550 quantified transcription factors upon cell state change. While the abundance of most of the transcriptional regulators quantified was largely downregulated by ∼50% in both cell lines (see violin plot insert - Figure 4A,B), a cohort of factors linked with maintenance of pluripotency (e.g., OCT4, NANOG) was more dramatically downregulated (log2(day 13/day 0) < −2.0) to similar levels in both cell lines (Figure 4A,B). On the other hand, the hPSC-to-CM transition in both cell lines is associated with a strong increase (log2(day 13/day 0) > 2.0) in the abundance of numerous proteins linked to cardiac muscle development (e.g., TBX5, GATA6, MEF2C) (Figures 4A,B).
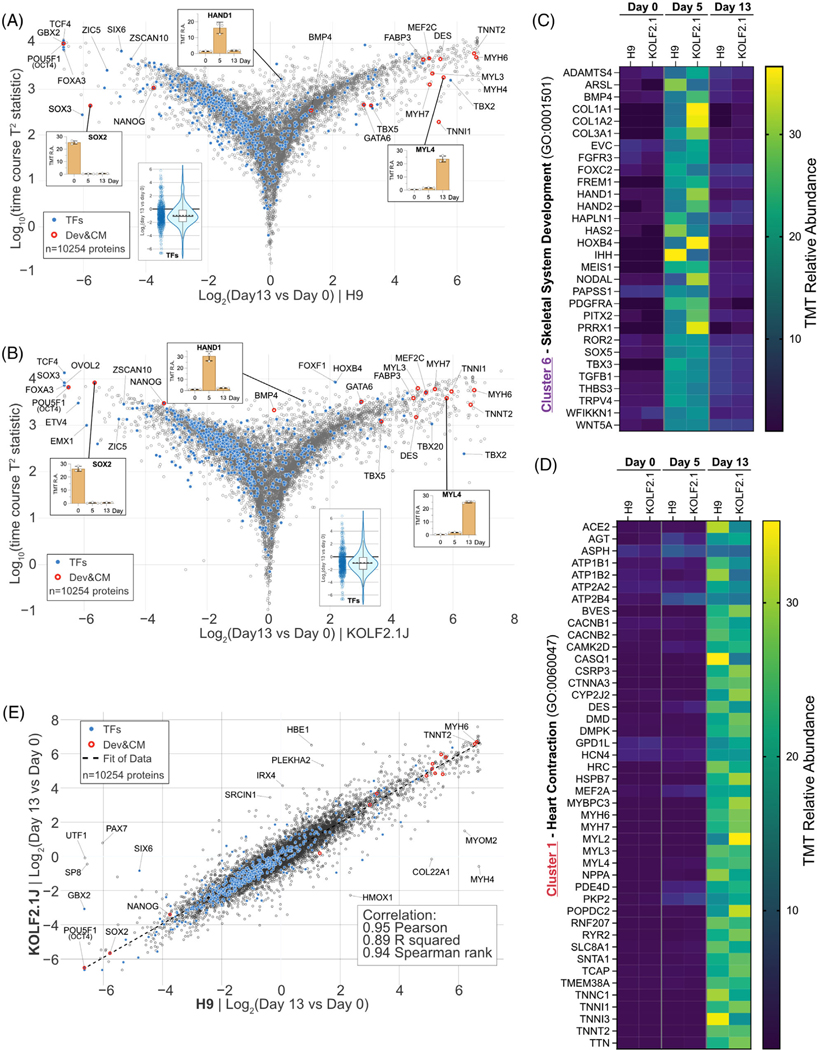
FIGURE 4.
Quantitative comparison of H9 and KOLF2.1J proteome remodeling upon cardiomyogenesis. (A) Multivariate empirical Bayes analysis of the total proteome data of H9 differentiation across the 13-day timespan. The plot displays log10((time course T2 statistics) versus log2(day 13/day 0). Individual proteins are displayed in gray open circles. Significantly changed protein: filled gray circle. Transcription factors: filled blue circles. Developmental & cardiac markers: red open circles. 10254 proteins were quantified. (B) Similar to (A) for KOLF2.1J dataset. (C) Heatmap of relative abundance with proteins in cluster 6 (CPs specific (day 5)) associated with the gene ontology term skeletal system development (GO: 0001501). (D) Heatmap of relative abundance with proteins in cluster 1 (CMs specific (day 13)) associated with the gene ontology term heart contraction (GO: 0060047). (E) Proteome level correlation plot for H9 (x axis) and KOLF2.1J (y axis) cells upon differentiation as determined by log2[ratio] in abundance comparing PSC versus CMs. The calculated correlation factors (Pearson, R squared, and Spearman rank) are indicated
To gain further insight into this unique programmed remodeling, we performed gene ontology analysis on the three clusters capturing the main temporal steps of the proteome remodeling (clusters 1, 6 and 8). Analysis revealed near perfect overlapping terms across both cell lines, with cluster 8 (enriched in PSC state (day 0)) showing enrichment for terms linked to cell division, mitosis and chromosomal regulation (Figure S4A,B). Cluster 2 (increasing temporarily at CPs stage only (day 5)) analysis revealed terms linked to skeletal system development and other extracellular organization processes (Figure S4C,D). Finally, cluster 1 (increasing strongly in CMs (day 13)) was enriched for terms related to muscle architecture, development, and contraction (Figure S4E,F). Next, we isolated the proteins linked to two GO terms of interest in clusters 6 and 1 in both H9 and KOLF2.1J cells. The skeletal system development term (GO:0001501), enriched in cluster 6 contained 30 unique proteins temporarily highly enriched at the cardiac progenitor state, in both cell lines (Figure 4C). These included, for example, the transcription factor HAND1/2, which is known to mark cardiac progenitor cells and to regulate their proliferation [46], and the tyrosine kinase receptor PDGFRα (Platelet-derived growth factor receptor) which is essential for the migration of myocardial precursors [47, 48]. The heart contraction term (GO:0060047), enriched in cluster 1, was composed of 44 unique proteins (Figure 4D). These included various myosin heavy and light chains (MYH6, MYH7, MYL2, MYL3 and MYL4), and RING finger protein RNF207, which is a regulator of cardiac excitation [49].
Having explored the nature of the proteome remodeling with this additional differentiation scheme, we turned to a correlation plot to measure how the two reference cell lines performed upon CMs transitioning from their respective PSC states. Correlation plot metrics showed an excellent correlation for Pearson (0.95), R squared (0.89) and Spearman rank (0.94) correlations, indicating that, excepting minor outliers, the proteome remodeling trajectory of both reference lines is similar (Figure 4E).
3.6 │. Concluding remarks
The present study surveyed the relative 10k proteome abundance profiles of two reference human pluripotent stem cell lines upon cell state remodeling. We employed a TMTpro-based multiplexing strategy along with FAIMS and real-time database search for quantitative accuracy and data completeness. Initially, we observed profound differences in the steady state proteome of each cell line at both pluripotent and differentiated cell states. These differences can be derived from the source of the reference lines and the inter-individual variability. Notably, however, the differentiation potential of both cell lines showed a remarkably high similarity based on their proteome remodeling trajectory during NSC or CM transition. These datasets can serve as a resource for further signaling analysis and assist molecular mechanistic studies involving directed differentiation of these two pluripotent stem cell lines.
DATA AVAILABILITY STATEMENT
RAW files are available upon request. In addition, the data have been deposited in the MassIVE public repository with the dataset identifier MSV000088833. In the supplementary information, we have included tables listing protein names, gene symbols, and TMT quantitation values for the datasets (Tables S1 and S3). We have also included lists of phospho-peptides, associated protein names, gene symbols, TMT quantification values, localization score, and motif for the datasets (Table S2).
Supplementary Material
Significance Statement.
The absence of a broadly adopted reference induced-pluripotent stem cell (iPSC) has complicated various efforts in the research community to interpret or reproduce specific observations made across various cell line settings. One primary explanation is thought to be the inherent inter-variability coming from different cell line background. To address this, following an extensive characterization KOLF2.1J, a human iPSC line, was recently offered to the research community as a reference iPSC line for future studies. Here we complement this characterization process by providing in depth unbiased quantitative protein abundance measurement in undifferentiated and differentiated cell-states. In parallel, we performed similar measures on a second line, termed H9 (WA09). We profiled the relative protein abundance of these two cell lines using a set of isobaric Tandem Mass Tag reagent (TMT) pro-based multiplexing approach combined with real-time database searching technologies for enhanced quantitative accuracy and data completeness. We highlight differences in the steady state proteome of each cell line and classify the proteins which were markedly regulated in each cell line. By providing comprehensive protein abundance measurements, our dataset can serve as a starting point to understand how signaling mechanisms or specific protein interactions are influenced in these two distinct cell lines.
ACKNOWLEDGEMENTS
This work was supported by Sloan Kettering Institute startup funds (A.O.) and Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education (2021R1A6A3A14038416 to K.H.N.). We thank William C. Skarnes (The Jackson Laboratory) and Michael E. Ward (National Institutes of Health) for discussion and providing access to the KOLF2.1J iPSC line and Keith Bartlett for editing assistance.
Funding information
National Research Foundation of Korea; Ministry of Education, Grant/Award Number: 2021R1A6A3A14038416
Abbreviations:
- ESC
embryonic stem cell
- PSC
pluripotent stem cell
- NSC
neural stem cell
- TMTpro
proline-based tandem mass tag
- FAIMS
field asymmetric ion mobility spectrometry
Footnotes
SUPPORTING INFORMATION
Additional supporting information may be found online https://doi.org/10.1002/pmic.202100246 in the Supporting Information section at the end of the article.
CONFLICT OF INTEREST
The authors declare no conflict of interest.
REFERENCES
- 1.Ramos DM, Skarnes WC, Singleton AB, Cookson MR, & Ward ME (2021). Tackling neurodegenerative diseases with genomic engineering: A new stem cell initiative from the NIH. Neuron, 109(7), 1080–1083. 10.1016/j.neuron.2021.03.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pantazis CB, Yang A, Lara E, McDonough JA, Blauwendraat C, Peng L, Oguro H, Zou J, Sebesta D, Pratt G, Cross E, Blockwick J, Buxton P, Kinner-Bibeau L, Medura C, Tompkins C, Hughes S, Santiana M, Faghri F, ... Merkle FT (2021). A reference induced pluripotent stem cell line for large-scale collaborative studies. bioRxiv. 10.1101/2021.12.15.472643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Thomson JA, Itskovitz-Eldor J, Shapiro SS, Waknitz MA, Swiergiel JJ, Marshall VS, & Jones JM (1998). Embryonic stem cell lines derived from human blastocysts. Science, 282(5391), 1145–1147. 10.1126/science.282.5391.1145 [DOI] [PubMed] [Google Scholar]
- 4.Gry M, Rimini R, Strömberg S, Asplund A, Pontén F, Uhlén M, & Nilsson P. (2009). Correlations between RNA and protein expression profiles in 23 human cell lines. Bmc Genomics [Electronic Resource], 10, 365. 10.1186/1471-2164-10-365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu Y, Beyer A, & Aebersold R. (2016). On the dependency of cellular protein levels on mRNA abundance. Cell, 165(3), 535–550. 10.1016/j.cell.2016.03.014 [DOI] [PubMed] [Google Scholar]
- 6.Buccitelli C, & Selbach M. (2020). mRNAs, proteins and the emerging principles of gene expression control. Nature Reviews Genetics, 21(10), 630–644. 10.1038/s41576-020-0258-4 [DOI] [PubMed] [Google Scholar]
- 7.Nusinow DP, Szpyt J, Ghandi M, Rose CM, Mcdonald ER, Kalocsay M, Jané-Valbuena J, Gelfand E, Schweppe DK, Jedrychowski M, Golji J, Porter DA, Rejtar T, Wang YK, Kryukov GV, Stegmeier F, Erickson BK, Garraway LA, Sellers WR, & Gygi SP (2020). Quantitative proteomics of the cancer cell line encyclopedia. Cell, 180(2), 387–402. e316. 10.1016/j.cell.2019.12.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Edfors F, Danielsson F, Hallström BM, Käll L, Lundberg E, Pontén F, Forsström B, & Uhlén M. (2016). Gene-specific correlation of RNA and protein levels in human cells and tissues. Molecular Systems Biology, 12(10), 883. 10.15252/msb.20167144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang B, Wang J, Wang X, Zhu J, Liu Q, Shi Z, Chambers MC, Zimmerman LJ, Shaddox KF, Kim S, Davies SR, Wang S, Wang P, Kinsinger CR, Rivers RC, Rodriguez H, Townsend RR, Ellis MJC, Carr SA, ... Liebler DC (2014). Proteogenomic characterization of human colon and rectal cancer. Nature, 513(7518), 382–387. 10.1038/nature13438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Roumeliotis TI, Williams SP, Gonã§Alves E, Alsinet C, Del Castillo Velasco-Herrera M, Aben N, Ghavidel FZ, Michaut M, Schubert M, Price S, Wright JC, Yu L, Yang M, Dienstmann R, Guinney J, Beltrao P, Brazma A, Pardo M, Stegle O, ... Choudhary JS (2017). Genomic determinants of protein abundance variation in colorectal cancer cells. Cell Reports, 20(9), 2201–2214. 10.1016/j.celrep.2017.08.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee J, Termglinchan V, Diecke S, Itzhaki I, Lam CK, Garg P, Lau E, Greenhaw M, Seeger T, Wu H, Zhang JZ, Chen X, Gil IP, Ameen M, Sallam K, Rhee J-W, Churko JM, Chaudhary R, Chour T, ... Wu JC (2019). Activation of PDGF pathway links LMNA mutation to dilated cardiomyopathy. Nature, 572(7769), 335–340. 10.1038/s41586-019-1406-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yi SA, Nam KH, Yun J, Gim D, Joe D, Kim YH, Kim H-J, Han J-W, & Lee J. (2020). Infection of brain organoids and 2D cortical neurons with SARS-CoV-2 pseudovirus. Viruses., 12(9), 1004. 10.3390/v12091004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ordureau A, Kraus F, Zhang J, An H, Park S, Ahfeldt T, Paulo JA, & Harper JW (2021). Temporal proteomics during neurogenesis reveals large-scale proteome and organelle remodeling via selective autophagy. Molecular Cell, 81(24), 5082–5098. e5011. 10.1016/j.molcel.2021.10.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang Y, Yang F, Gritsenko MA, Wang Y, Clauss T, Liu T, Shen Y, Monroe ME, Lopez-Ferrer D, Reno T, Moore RJ, Klemke RL, Camp DG, & Smith RD (2011). Reversed-phase chromatography with multiple fraction concatenation strategy for proteome profiling of human MCF10A cells. Proteomics, 11(10), 2019–2026. 10.1002/pmic.201000722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Paulo JA, O’connell JD, Everley RA, O’brien J, Gygi MA, & Gygi SP (2016). Quantitative mass spectrometry-based multiplexing compares the abundance of 5000 S. cerevisiae proteins across 10 carbon sources. Journal of Proteomics, 148, 85–93. 10.1016/j.jprot.2016.07.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mcalister GC, Nusinow DP, Jedrychowski MP, Wühr M, Huttlin EL, Erickson BK, Rad R, Haas W, & Gygi SP (2014). MultiNotch MS3 enables accurate, sensitive, and multiplexed detection of differential expression across cancer cell line proteomes. Analytical Chemistry, 86(14), 7150–7158. 10.1021/ac502040v [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Erickson BK, Mintseris J, Schweppe DK, Navarrete-Perea J, Erickson AR, Nusinow DP, Paulo JA, & Gygi SP (2019). Active instrument engagement combined with a real-time database search for improved performance of sample multiplexing workflows. Journal of Proteome Research, 18(3), 1299–1306. 10.1021/acs.jproteome.8b00899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schweppe DK, Eng JK, Yu Q, Bailey D, Rad R, Navarrete-Perea J, Huttlin EL, Erickson BK, Paulo JA, & Gygi SP (2020). Full-featured, real-time database searching platform enables fast and accurate multiplexed quantitative proteomics. Journal of Proteome Research, 19(5), 2026–2034. 10.1021/acs.jproteome.9b00860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schweppe DK, Prasad S, Belford MW, Navarrete-Perea J, Bailey DJ, Huguet R, Jedrychowski MP, Rad R, Mcalister G, Abbatiello SE, Woulters ER, Zabrouskov V, Dunyach J-J, Paulo JA, & Gygi SP (2019). Characterization and optimization of multiplexed quantitative analyses using high-field asymmetric-waveform ion mobility mass spectrometry. Analytical Chemistry, 91(6), 4010–4016. 10.1021/acs.analchem.8b05399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schweppe DK, Rusin SF, Gygi SP, & Paulo JA (2020). Optimized workflow for multiplexed phosphorylation analysis of TMT-labeled peptides using high-field asymmetric waveform ion mobility spectrometry. Journal of Proteome Research, 19(1), 554–560. 10.1021/acs.jproteome.9b00759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Eng JK, Hoopmann MR, Jahan TA, Egertson JD, Noble WS, & Maccoss MJ (2015). A deeper look into Comet–implementation and features. Journal of the American Society for Mass Spectrometry, 26(11), 1865–1874. 10.1007/s13361-015-1179-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Eng JK, Jahan TA, & Hoopmann MR (2013). Comet: An open-source MS/MS sequence database search tool. Proteomics, 13(1), 22–24. 10.1002/pmic.201200439 [DOI] [PubMed] [Google Scholar]
- 23.Rad R, Li J, Mintseris J, O’Connell J, Gygi SP, & Schweppe DK (2021). Improved monoisotopic mass estimation for deeper proteome coverage. Journal of Proteome Research, 20(1), 591–598. 10.1021/acs.jproteome.0c00563 [DOI] [PubMed] [Google Scholar]
- 24.Elias JE, & Gygi SP (2007). Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nature Methods, 4(3), 207–214. 10.1038/nmeth1019 [DOI] [PubMed] [Google Scholar]
- 25.Huttlin EL, Jedrychowski MP, Elias JE, Goswami T, Rad R, Beausoleil SA, Villén J, Haas W, Sowa ME, & Gygi SP (2010). A tissue-specific atlas of mouse protein phosphorylation and expression. Cell, 143(7), 1174–1189. 10.1016/j.cell.2010.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Savitski MM, Wilhelm M, Hahne H, Kuster B, & Bantscheff M. (2015). A scalable approach for protein false discovery rate estimation in large proteomic data sets. Molecular & Cellular Proteomics, 14(9), 2394–2404. 10.1074/mcp.M114.046995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Beausoleil SA, Villén J, Gerber SA, Rush J, & Gygi SP (2006). A probability-based approach for high-throughput protein phosphorylation analysis and site localization. Nature Biotechnology, 24(10), 1285–1292. 10.1038/nbt1240 [DOI] [PubMed] [Google Scholar]
- 28.Wu T, Hu E, Xu S, Chen M, Guo P, Dai Z, Feng T, Zhou L, Tang W, Zhan L, Fu X, Liu S, Bo X, & Yu G. (2021). clusterProfiler 4.0: A universal enrichment tool for interpreting omics data. Innovation (N Y), 2(3), 100141. 10.1016/j.xinn.2021.100141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kuleshov MV, Xie Z, London ABK, Yang J, Evangelista JE, Lachmann A, Shu I, Torre D, & Ma’ayan A. (2021). KEA3: Improved kinase enrichment analysis via data integration. Nucleic acids research, 49(W1), W304–W316. 10.1093/nar/gkab359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tyanova S, Temu T, Sinitcyn P, Carlson A, Hein MY, Geiger T, Mann M, & Cox J. (2016). The Perseus computational platform for comprehensive analysis of (prote)omics data. Nature Methods, 13(9), 731–740. 10.1038/nmeth.3901 [DOI] [PubMed] [Google Scholar]
- 31.Itzhak DN, Tyanova S, Cox J, & Borner GH (2016). Global, quantitative and dynamic mapping of protein subcellular localization. Elife, 5, e16950. 10.7554/eLife.16950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lambert SA, Jolma A, Campitelli LF, Das PK, Yin Y, Albu M, Chen X, Taipale J, Hughes TR, & Weirauch MT (2018). The human transcription factors. Cell, 172(4), 650–665. 10.1016/j.cell.2018.01.029 [DOI] [PubMed] [Google Scholar]
- 33.Li J, van Vranken JG, Pontano Vaites L, Schweppe DK, Huttlin EL, Etienne C, Nandhikonda P, Viner R, Robitaille AM, Thompson AH, Kuhn K, Pike I, Bomgarden RD, Rogers JC, Gygi SP, & Paulo JA (2020). TMTpro reagents: A set of isobaric labeling mass tags enables simultaneous proteome-wide measurements across 16 samples. Nature Methods, 17(4), 399–404. 10.1038/s41592-020-0781-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chi N, & Epstein JA (2002). Getting your Pax straight: Pax proteins in development and disease. Trends in Genetics, 18(1), 41–47. 10.1016/s0168-9525(01)02594-x [DOI] [PubMed] [Google Scholar]
- 35.Cavodeassi F, Modolell J, & Gómez-Skarmeta JL (2001). The Iroquois family of genes: From body building to neural patterning. Development (Cambridge, England), 128(15), 2847–2855. [DOI] [PubMed] [Google Scholar]
- 36.Kumar JP (2009). The sine oculis homeobox (SIX) family of transcription factors as regulators of development and disease. Cellular and Molecular Life Sciences, 66(4), 565–583. 10.1007/s00018-008-8335-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gofin Y, Wang T, Gillentine MA, Scott TM, Berry AM, Azamian MS, Genetti C, Agrawal PB, Picker J, Wojcik MH, Delgado MR, Lynch SA, Scherer SW, Howe JL, Bacino CA, Ditroia S, Vannoy GE, O’donnell-Luria A, Lalani SR, ... Scott DA (2022). Delineation of a novel neurodevelopmental syndrome associated with PAX5 haploinsufficiency. Human Mutation, 43(4), 461–470. 10.1002/humu.24332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jia J, Zheng X, Hu G, Cui K, Zhang J, Zhang A, Jiang H, Lu B, Yates J, Liu C, Zhao K, & Zheng Y. (2012). Regulation of pluripotency and self- renewal of ESCs through epigenetic-threshold modulation and mRNA pruning. Cell, 151(3), 576–589. 10.1016/j.cell.2012.09.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gaborieau E, Hurtado-Chong A, FernãąNdez M, Azim K, & Raineteau O. (2018). A dual role for the transcription factor Sp8 in postnatal neurogenesis. Science Reports, 8(1), 14560. 10.1038/s41598-018-32134-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Navarrete-Perea J, Yu Q, Gygi SP, & Paulo JA (2018). Streamlined tandem mass Tag (SL-TMT) protocol: An efficient strategy for quantitative (phospho)proteome profiling using tandem mass Tag-synchronous precursor selection-MS3. Journal of Proteome Research, 17(6), 2226–2236. 10.1021/acs.jproteome.8b00217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Aubol BE, Wu G, Keshwani MM, Movassat M, Fattet L, Hertel KJ, Fu X-D, & Adams JA (2016). Release of SR proteins from CLK1 by SRPK1: A symbiotic kinase system for phosphorylation control of Pre-mRNA splicing. Molecular Cell, 63(2), 218–228. 10.1016/j.molcel.2016.05.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Neganova I, Vilella F, Atkinson SP, Lloret M, Passos JF, von Zglinicki T, O’connor JE, Burks D, Jones R, Armstrong L, & Lako M. (2011). An important role for CDK2 in G1 to S checkpoint activation and DNA damage response in human embryonic stem cells. Stem Cells, 29(4), 651–659. 10.1002/stem.620 [DOI] [PubMed] [Google Scholar]
- 43.Li J, Cai Z, Bomgarden RD, Pike I, Kuhn K, Rogers JC, Roberts TM, Gygi SP, & Paulo JA (2021). TMTpro-18plex: The expanded and complete set of TMTpro reagents for sample multiplexing. Journal of Proteome Research, 20(5), 2964–2972. 10.1021/acs.jproteome.1c00168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hotelling H. (1931). The generalization of Student’s ratio. Annals of Mathematical Statistics, 2, 360–378. [Google Scholar]
- 45.Tai Y. (2007). Timecourse: statistical analysis for developmental microarray time course data. R package version 1.58(0). 10.18129/B9.bioc.timecourse [DOI] [Google Scholar]
- 46.Okubo C, Narita M, Inagaki A, Nishikawa M, Hotta A, Yamanaka S, & Yoshida Y. (2021). Expression dynamics of HAND1/2 in in vitro human cardiomyocyte differentiation. Stem Cell Reports, 16(8), 1906–1922. 10.1016/j.stemcr.2021.06.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.El-Rass S, Eisa-Beygi S, Khong E, Brand-Arzamendi K, Mauro A, Zhang H, Clark KJ, Ekker SC, & Wen X-Y (2017). Disruption of pdgfra alters endocardial and myocardial fusion during zebrafish cardiac assembly. Biology Open, 6(3), 348–357. 10.1242/bio.021212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Noseda M, Harada M, Mcsweeney S, Leja T, Belian E, Stuckey DJ, Abreu Paiva MS, Habib J, Macaulay I, de Smith AJ, Al-Beidh F, Sampson R, Lumbers RT, Rao P, Harding SE, Blakemore AIF, Eirik Jacobsen S, Barahona M, & Schneider MD (2015). PDGFRα demarcates the cardiogenic clonogenic Sca1+ stem/progenitor cell in adult murine myocardium. Nature Communication, 6, 6930. 10.1038/ncomms7930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Roder K, Werdich AA, Li W, Liu M, Kim TY, Organ-Darling LE, Moshal KS, Hwang JM, Lu Y, Choi B-R, Macrae CA, & Koren G. (2014). RING finger protein RNF207, a novel regulator of cardiac excitation. Journal of Biological Chemistry, 289(49), 33730–33740. 10.1074/jbc.M114.592295 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RAW files are available upon request. In addition, the data have been deposited in the MassIVE public repository with the dataset identifier MSV000088833. In the supplementary information, we have included tables listing protein names, gene symbols, and TMT quantitation values for the datasets (Tables S1 and S3). We have also included lists of phospho-peptides, associated protein names, gene symbols, TMT quantification values, localization score, and motif for the datasets (Table S2).