Abstract
Background:
Recent progress in genomic analysis using next-generation sequencing (NGS) technology has enabled the comprehensive detection of mutations and tumor mutation burden (TMB) in patients. A high TMB (TMB-H) tumor is defined as one with high somatic mutational rates, which correlates with clinical responses to certain treatments such as immunotherapies. We determined TMB in lung adenocarcinoma and clarified the characteristics of patients with TMB-H in relation to common driver mutations and smoking history.
Materials and Methods:
Genomic aberrations and TMB were determined in Japanese patients with lung adenocarcinoma (N = 100) using NGS of 415 known cancer genes. TMB-H was defined as > 20 mutations per megabase (Mb) of sequenced DNA.
Results:
The median TMB was 13.5 (5–33) mutations/Mb. Ten out of 100 (10%) patients showed TMB-H, and the others showed low TMB (TMB-L). Only two out of 10 (20%) patients with TMB-H had one of the common driver mutations (ALK and ERBB2 mutation), while 57 out of 90 (63%) patients with TMB-L had one of the driver mutations, including ALK, EGFR, ERBB2, ROS, RET, and MET (P < 0.05). Notably, no EGFR mutation was observed in patients with TMB-H. Eight out of 10 (80%) patients with TMB-H had recent smoking history, whereas only 17 out of 90 (19%) patients with TMB-L had recent smoking history (P < 0.001).
Conclusion:
We found that TMB-H is associated with smoking history, whereas TMB-L is associated with the common driver mutations in lung adenocarcinoma, which may impact treatment strategies for these patients.
Keywords: next generation sequencing, tumor mutation burden, lung adenocarcinoma, immune checkpoint inhibitors, anti-PD-1
INTRODUCTION
Lung cancer is the leading cause of cancer death worldwide.1, 2 Adenocarcinoma is the most common histologic type, which accounts for approximately 40% of lung cancers.3 Lung adenocarcinoma usually arises in the distal airway and frequently occurs in current or former smokers.4 However, interestingly, adenocarcinoma is also the most frequent cancer among lung cancers in non-smokers.5 The proportion of lifelong non-smokers in lung cancer patients is reported to be high in Asia, and the population is thought to be genetically predisposed to lung cancer.6 More recently it has been found that lung cancer in never-smoker Asian females is driven by oncogenic mutations such as EGFR.7
Immune checkpoint inhibitors, such as the anti-PD-1 therapy developed by Dr. Honjo,8, 9 show dramatic effects on several types of solid tumors, including lung cancer.10, 11 Although the expectation of immune checkpoint inhibitors is tremendously high based on its dramatic efficacy, only a limited number of patients respond.12 Furthermore, there is no definite biomarker for identifying which tumors respond to this therapy.12
Recent progress in genomic analysis using the next-generation sequencing (NGS) technology has enabled the comprehensive detection of mutations and the definition of tumor mutation burden (TMB) in cancer patients.13-15 A high TMB (TMB-H) tumor is defined as one with high somatic mutational rates, which correlates with the generation of neo-antigens and also to potential clinical responses to immunotherapies.13 Interestingly, it has been reported that there is an association between smoking history and TMB-H in non-small cell lung cancer.12
Although sequencing studies of lung adenocarcinoma have been reported,12, 16 there is a paucity of data elucidating the relationship between comprehensive genomic data and clinical histories, such as smoking. We hypothesized that TMB-H is associated with smoking history in some patients with lung adenocarcinoma. The aim of this study was to identify TMB in patients with lung adenocarcinoma utilizing NGS and to seek association with smoking history and with common oncogenic driver mutations in lung adenocarcinoma.
MATERIALS AND METHODS
Patients
We analyzed a total of 100 patients diagnosed with stage I–IV lung adenocarcinoma according to American Joint Committee on Cancer (AJCC) Cancer Staging Manual 7th edition 17 who had received an operation at Niigata University Medical and Dental Hospital from January 2008 to December 2014 or at Kyushu University from October 2013 to August 2015. Patients who satisfied following criteria were selected to be analyzed: a tumor content of >20% based on pathological examination of primary tumor with hematoxylin and eosin (H&E) staining, radiological confirmation of lung adenocarcinoma with a consolidation/tumor ratio (C/T ratio) >0.5 using thin-section computed tomography (CT), and successful extraction of ≥150 ng DNA from the tumor sample. This study was approved by the Institutional Review Boards of Niigata University and Kyusyu University Hospital, and written informed consent was obtained from all patients.
Sequencing library preparation
We analyzed archival tissues of primary cancer from surgical specimens that were confirmed to be of high quality in the form of formalin-fixed, paraffin-embedded (FFPE) slides.18 An independent pathologist evaluated the tumor content on H&E stained slides for each study sample to ensure that > 20% tumor content was present. Where applicable, unstained slides were macro-dissected to enrich for tumor content, and genomic DNA was extracted using the BiOstic FFPE Tissue DNA Isolation Kit (Mo Bio Laboratories, Inc; Carlsbad, CA). DNA quality of the samples was confirmed by QC ratio as previously described.18 Briefly, the quality of an adequate quantity of DNA was assessed by determining the Q-ratio, in which 41 bp and 129 bp targets were amplified by qPCR and compared using a KAPA Human Genomic DNA Quantification and QC Kit (KAPA Biosystems). The 41 bp assay was used for absolute quantification of DNA samples. DNA with a Q-ratio (129 bp/41 bp) > 0.1 was designated as being of high enough quality for NGS analysis based on our previous results.18 All sample preparation, NGS, and analytics were performed in a CLIA/CAP-accredited laboratory (KEW Inc; Cambridge, MA).
NGS analysis and determination of tumor mutation burden
FFPE genomic DNA (150 ng) was converted into libraries and was enriched for the 415 genes with CANCERPLEX (KEW Inc; Cambridge, MA), as described previously.14, 19 Briefly, sequencing was performed on the Illumina MiSeq and NextSeq platforms with an average 500× sequencing depth. Genomic data were then processed through a proprietary bioinformatics platform and knowledgebase to identify multiple classes of genomic abnormalities, including single nucleotide substitutions (SNPs), small insertions/deletions (indels), copy number variations (CNV), and translocations in ALK, RET, and ROS1. A threshold of 10% allelic fraction was used for SNPs and indels. Thresholds of > 2.5-fold and < 0.5-fold were used for gains and loss. To assess the somatic status of mutations in a tumor-only setting, we employed a filtering strategy similar to one recently published 20 with minor differences. The TMB was determined by frequency of nonsynonymous SNPs per Mb. We considered nonsynonymous SNPs that were present in the tumor with a population frequency of < 1% in the NCBI dbSNP database and the 1000 genomes database for the TMB calculation. High TMB was defined as > 20 mutations/Mb of sequenced DNA. Common driver mutations, including ALK, EGFR, ERBB2, ROS, RET, and MET, and smoking status were compared with TMB to examine their association. For smoking status, a current smoker was defined as a patient who had not stopped smoking before outpatient referral visit. Former smoker was defined as a patient who had stopped smoking before outpatient referral visit. In our institutions, the period from referral visit to surgery is about one month. In this study, we considered any smoking history, regardless of smoking amount and exposure time.
Statistical analysis
Categorical variables were compared by Fisher’s exact test or Pearson’s X2 test, whereas continuous variables were compared by the Mann-Whitney U-test between two groups and by the Kruskal-Wallis test among three groups. All statistical analyses were performed using GraphPad Prism (GraphPad Software, Inc.). All tests were two-tailed, and P < 0.05 was considered significant.
RESULTS
One tenth of analyzed Japanese lung adenocarcinomas had a high mutation burden (TMB-H)
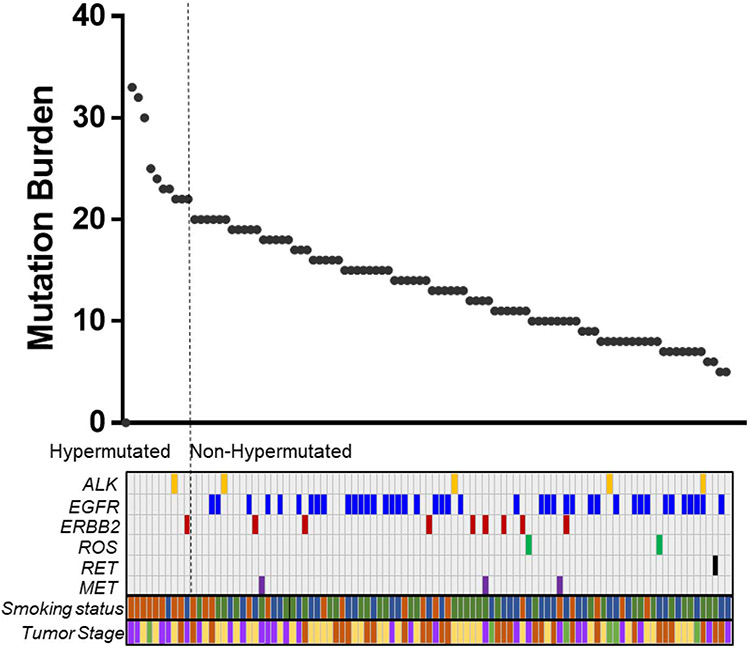
We determined TMB in Japanese patients with lung adenocarcinoma utilizing an NGS-based analysis (Fig. 1). The median TMB was 13.5 (5–33) mutations/Mb among the 100 patients with lung adenocarcinoma examined. Ten out of 100 (10%) patients showed TMB-H, and 90 (90%) patients showed low TMB (TMB-L; Fig. 1). This ratio was smaller than the ratio in a previous report on the US population.21
Figure 1. Inverse association of mutation burden with common oncodrivers in Japanese lung cancer.
The mutation burdens for 100 Japanese patients are shown in the graph above. The bars under the graph of mutation burden indicate the existence of common driver mutations, smoking status, and tumor stage for the 100 patients. For smoking status, the colors indicated the following: Brown, current smoker; Blue, former smoker; Green, never smoker. For tumor stage, the colors indicated the following: Brown, Stage I; Purple, Stage II; Yellow, Stage III; Green, Stage IV.
Current smokers demonstrated the highest TMB
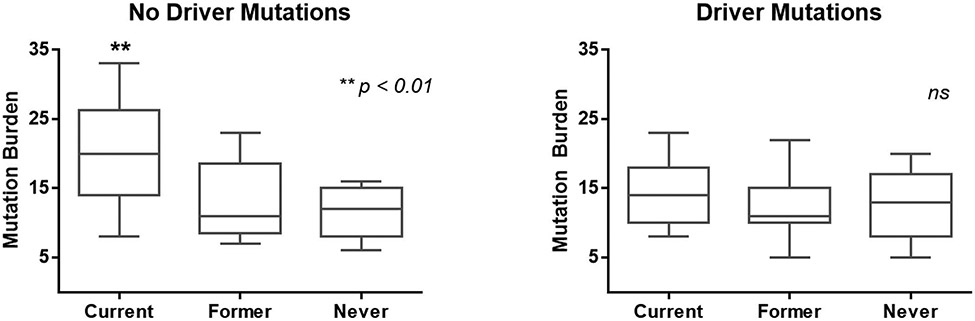
Next, we examined the relationship between TMB and smoking history. Eight out of 10 (80%) patients with TMB-H were current smokers, which was significantly more common than in patients with TMB-L (17 out of 90 (19%) patients were current smokers, P < 0.001; Table 1). Furthermore, we compared mutation burden among groups of current, former, and never smokers in patients with driver mutations or no driver mutations. Interestingly, we found that a group of current smokers without driver mutations had the highest TMB in our cohort (P < 0.01; Fig. 2). However, in the group of patients with driver mutations, there was no difference in TMB based on smoking history (Fig. 2). We further examined the association between smoking history and TMB using total tobacco exposure using pack years of exposure as a continuous variable; however, we were not able to find any association between total tobacco exposure and TMB, or a particular threshold of total tobacco exposure that determines the TMB-H group (data not shown).
Table 1.
Tumor mutation burden and smoking history in patients with lung adenocarcinoma
| Smoking history |
||||
|---|---|---|---|---|
| Positive | Negative | Total | P-value | |
| Hypermutated | 8 | 2 | 10 | < 0.001 |
| Non-hypermutated | 17 | 73 | 90 | |
Figure 2. Current smokers without driver mutations have the highest mutation burden in the Japanese cohort.
The mutation burden was determined in the patients who were current smokers, former smokers, and never-smokers in a group of patients with no driver mutations (left), and patients with driver mutations. Significance was determined by the Kruskal-Wallis test and a one-way ANOVA, **, P < 0.01.
Inverse correlation of mutation burden with common driver mutations of lung adenocarcinoma
Only two out of 10 (20%) patients with TMB-H had one of the common driver mutations (one patient had ALK, and the other had ERBB2 mutation), whereas 57 out of 90 (63%) patients with TMB-L had one of the driver mutations, including ALK, EGFR, ERBB2, ROS, RET, and MET (Fig. 1). There were significantly more patients with common driver mutations in the TMB-L group than in the TMB-H group (P < 0.05, Table 2). Notably, no EGFR mutations were observed in the TMB-H group (Fig. 1).
Table 2.
Tumor mutation burden and existence of oncogenic driver mutation in patients with lung adenocarcinoma
| Driver mutation |
||||
|---|---|---|---|---|
| Positive | Negative | Total | P-value | |
| Hypermutated | 2 | 8 | 10 | < 0.05 |
| Non-hypermutated | 57 | 33 | 90 | |
DISCUSSION
Given that only limited number of patients with lung cancer can benefit from the dramatic effect of immunotherapies, the role of TMB in relation to the efficacy of the immunotherapies has been attracting more attention.13, 14, 22 In the current study we found that there were significantly more current smokers in TMB-H group than in the TMB-L group. Furthermore, we found a significant inverse association between the existence of common driver mutations and TMB-H in patients with lung adenocarcinoma in Asia.
Our study showed that current smoker status is associated with TMB-H. In colorectal and gastric cancers, microsatellite instability (MSI) is closely associated with TMB-H. For instance, we have previously reported that 92% of colorectal cancer patients with MSI showed hypermutation.14 MSI and TMB are now considered as good predictive markers for immunotherapies, and FDA approved MSI and TMB as companion diagnostic indicators for pembrolizumab in CRC patients.13, 23, 24 However, it has been reported that there are few patients with MSI in lung adenocarcinoma.25 Indeed, we found no MSI in the 100 patients with lung adenocarcinoma. Due to the lack of good predictive biomarker for lung adenocarcinoma, information regarding smoking history is very informative for clinicians to speculate about TMB and potential response to immunotherapy without NGS analysis. Given that not all the lung adenocarcinoma patients can undergo NGS analysis due to the cost restrictions, this clinical information is critical for patient outcomes. Although further prospective trials are needed, it might be possible for clinicians to predict responses to immunotherapies more efficiently without NGS.
We also found that there were significantly more patients with common driver mutations in the TMB-L group than in the TMB-H group. It has been reported that lung cancer in never-smoker Asian females is driven by oncogenic mutations, such as in EGFR.7 Considering that never-smoker status is associated with TMB-L, it is possible that common driver mutations and TMB-L are clinical characteristics of never-smokers. In contrast, no common driver mutations and TMB-H are the characteristics of smokers, who may benefit from immunotherapies, such as anti-PD-1 therapy.
This study had some limitations. Firstly, this is a retrospective study with a small number of patients. Since it was difficult to calculate power in advance for hypermutation in Japanese lung adenocarcinoma, which has never been investigated previously, we did not conduct a prior power analysis. However, the power of Fisher’s exact test for the two sets (TMB vs. smoking history, and TMB vs. existence of oncogenic driver mutation) was calculated with 1000 randomization simulation after undertaking this study; The power was 0.98 and 0.76, respectively. The first set was considered to be an acceptable sample size. The second set was insufficient in terms of the common cutoff of power 0.8, but the resultant value of 0.76 was very close to the cutoff, and the statistical result for the second set was assumed to be relatively important.
Secondly, we have not investigated factors which potentially affect status of TMB, other than smoking history and common driver mutation. In addition, there is no standard cut-off for mutation burden. Based on previous findings for other solid tumors, such as colorectal cancer, we considered tumors with >20 mutations/Mb of sequenced DNA as a TMB-H. Our study included these limitations, however, we demonstrated that smoking history and no common driver mutation are significantly associated with TMB-H, which suggest useful clinical information for immunotherapy.
CONCLUSIONS
We found that patients with TMB-H are characterized by current smoking and no common driver mutations, which suggests that smoking history may be useful for the selection of treatment strategies for these patients.
ACKNOWLEGMENTS
This project was supported by funding from Denka Co., Ltd. The authors would like to acknowledge funding from the Japan Society for the Promotion of Science (JSPS) Grant-in-Aid for Scientific Research, Grant Numbers 15H05676 and 15K15471 for M.N., and 15H04927 and 16K15610 for T.W. M.N. is also supported by the Takeda Science Foundation. K.Takabe is supported by an NIH/NCI grant R01CA160688 and a Susan G. Komen Investigator Initiated Research Grant IIR12222224.
Footnotes
The author disclosure statement: S.L. is an employee of KEW Inc, who has been granted stock options by KEW Inc. The remaining authors declare that they have no competing interests.
REFERENCES
- 1.Ferlay J, Shin HR, Bray F, Forman D, Mathers C, et al. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. International Journal of Cancer. Journal International du Cancer 2010:127:2893–2917. [DOI] [PubMed] [Google Scholar]
- 2.Jemal A, Bray F, Center MM, Ferlay J, Ward E, et al. Global cancer statistics. CA: A Cancer Journal for Clinicians 2011:61:69–90. [DOI] [PubMed] [Google Scholar]
- 3.Denisenko TV, Budkevich IN, Zhivotovsky B. Cell death-based treatment of lung adenocarcinoma. Cell Death & Disease 2018:9:117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hanna JM, Onaitis MW. Cell of origin of lung cancer. Journal of carcinogenesis 2013:12:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Govindan R, Ding L, Griffith M, Subramanian J, Dees ND, et al. Genomic landscape of non-small cell lung cancer in smokers and never-smokers. Cell 2012:150:1121–1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Toh CK, Wong EH, Lim WT, Leong SS, Fong KW, et al. The impact of smoking status on the behavior and survival outcome of patients with advanced non-small cell lung cancer: a retrospective analysis. Chest 2004:126:1750–1756. [DOI] [PubMed] [Google Scholar]
- 7.Ha SY, Choi SJ, Cho JH, Choi HJ, Lee J, et al. Lung cancer in never-smoker Asian females is driven by oncogenic mutations, most often involving EGFR. Oncotarget 2015:6:5465–5474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ishida Y, Agata Y, Shibahara K, Honjo T. Induced expression of PD-1, a novel member of the immunoglobulin gene superfamily, upon programmed cell death. The EMBO Journal 1992:11:3887–3895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shinohara T, Taniwaki M, Ishida Y, Kawaichi M, Honjo T. Structure and chromosomal localization of the human PD-1 gene (PDCD1). Genomics 1994:23:704–706. [DOI] [PubMed] [Google Scholar]
- 10.Robert C, Schachter J, Long GV, Arance A, Grob JJ, et al. Pembrolizumab versus Ipilimumab in Advanced Melanoma. The New England Journal of Medicine 2015:372:2521–2532. [DOI] [PubMed] [Google Scholar]
- 11.Borghaei H, Paz-Ares L, Horn L, Spigel DR, Steins M, et al. Nivolumab versus Docetaxel in advanced nonsquamous non-small-cell lung cancer. The New England Journal of Medicine 2015:373:1627–1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rizvi NA, Hellmann MD, Snyder A, Kvistborg P, Makarov V, et al. Cancer Immunology. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science (New York, N.Y.) 2015:348:124–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yuza K, Nagahashi M, Watanabe S, Takabe K, Wakai T. Hypermutation and microsatellite instability in gastrointestinal cancers. Oncotarget 2017:8:112103–112115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nagahashi M, Wakai T, Shimada Y, Ichikawa H, Kameyama H, et al. Genomic landscape of colorectal cancer in Japan: clinical implications of comprehensive genomic sequencing for precision medicine. Genome Medicine 2016:8:136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ichikawa H, Nagahashi M, Shimada Y, Hanyu T, Ishikawa T, et al. Actionable gene-based classification toward precision medicine in gastric cancer. Genome Medicine 2017:9:93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.The Cancer Genome Atlas Research Network. Comprehensive molecular profiling of lung adenocarcinoma. Nature 2014:511:543–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Edge SB, Compton CC, Fritz AG, Greene FL, Trotti A (eds). AJCC Cancer Staging Manual. 7th ed. New York: Springer; 2010. [Google Scholar]
- 18.Nagahashi M, Shimada Y, Ichikawa H, Nakagawa S, Sato N, et al. Formalin-fixed paraffin-embedded sample conditions for deep next generation sequencing. The Journal of Surgical Research 2017:220:125–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sato S, Nagahashi M, Koike T, et al. Impact of Concurrent Genomic Alterations Detected by Comprehensive Genomic Sequencing on Clinical Outcomes in East-Asian Patients with EGFR-Mutated Lung Adenocarcinoma. Scientific Reports 2018:8:1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Garofalo A, Sholl L, Reardon B, Taylor-Weiner A, Amin-Mansour A, et al. The impact of tumor profiling approaches and genomic data strategies for cancer precision medicine. Genome Medicine 2016:8:79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Roszik J, Haydu LE, Hess KR, Oba J, Joon AY, et al. Novel algorithmic approach predicts tumor mutation load and correlates with immunotherapy clinical outcomes using a defined gene mutation set. BMC Medicine 2016:14:168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sinnadurai M, McDonald KL Immune checkpoint inhibition and its relationship with hypermutation phenoytype as a potential treatment for Glioblastoma. Journal of Neuro-oncology 2017:132:359–372. [DOI] [PubMed] [Google Scholar]
- 23.Le DT, Uram JN, Wang H, Bartlett BR, Kemberling H, et al. PD-1 Blockade in Tumors with Mismatch-Repair Deficiency. The New England Journal of Medicine 2015:372:2509–2520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kalyan A, Kircher S, Shah H, Mulcahy M, Benson A Updates on immunotherapy for colorectal cancer. Journal of gastrointestinal oncology 2018:9:160–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Takamochi K, Takahashi F, Suehara Y, Sato E, Kohsaka S, et al. DNA mismatch repair deficiency in surgically resected lung adenocarcinoma: Microsatellite instability analysis using the Promega panel. Lung Cancer (Amsterdam, Netherlands) 2017:110:26–31. [DOI] [PubMed] [Google Scholar]