Background and aims:
Warm ischaemic injury (WII) stems from incorrect energy metabolism and is the main cause of graft dysfunction. Mitochondria, as the centre of cellular metabolic activities, may be the key in identifying accurate indicators for evaluating the quality of grafts. Our research focuses on the screening, clinical application, and mechanism of the optimal WII mitochondrion biomarker.
Approach and results:
Using a 100% hepatic warm ischaemia mouse model, without reperfusion, transmission electron microscopy demonstrated evident morphological changes of hepatic mitochondria at 15 min of ischaemia. However, all 13 mt-mRNAs could not display continuously upregulated consistency at 0–15–30–60 min during WII. High-throughput analysis of miRNA expression in both purified mitochondria and liver tissues suggested miR-23b-5p was a potential mitochondrial microRNA (mitomiR) biomarker with high sensitivity and 0-15-30-60 min change consistency. Fluorescence in-situ hybridization and reverse transcription quantitative polymerase chain reaction (RT-qPCR) further confirmed the results. Through overexpression and inhibition, the functionality of this mitomiR during WII was identified as a protective regulator in vitro and then verified in Dicer1fl/flAlbCre mice by downregulation of other miRNAs and supplementation of mature mitomiR-23b-5p. Dual-luciferase reporter assay and the Seahorse XF analyzer determined that mitomiR-23b-5p reduced mitochondrial respiratory function by silencing mt-RNR2 (16S). Clinically, mitomiR-23b-5p was positively correlated with serum alanine aminotransferase levels 3 days after the operation (P=0.032), and the C-statistic for 90-day graft survival rate was 0.698.
Conclusions:
MitomiR-23b-5p plays a protective regulatory role and implements a special mitochondrial regulation mechanism not yet reported in WII. These clinical results further support the experimental result that the expression of MitomiR-23b-5p is closely related to the prognosis of clinical liver transplantation patients. This is a promising new biomarker for WII evaluation of donor livers.
Keywords: donation after brain death, donation after cardiac death, donor liver, liver transplantation, mitomiR, warm ischaemic injury
Introduction
Highlights
Systematic searching for warm ischaemia injury (WII) biomarkers in liver mitochondria.
Precise display of miR-23b-5p functions in the liver of Dicer1fl/flAlbCre mice.
A special mitochondrial regulation mechanism for miR-23b-5p not yet reported in WII.
MiR-23b-5p promising in solving WII evaluation difficulties of donor liver.
Liver transplantation (LT) is the only treatment for end-stage liver disease. The incidence of primary graft nonfunction for donation after circulatory death has decreased from 15.5% in the 1990s to 2.1% in recent years1. In all donations, the primary graft nonfunction levels have declined from 3.8 to 2.2%, respectively. However, after LT, the incidence of early graft dysfunction is 7.2–27%2. If liver graft function fails without re-transplantation, the chance of death reaches 100%; the high re-transplantation cost should also be considered. Therefore, it is important to formulate high-precision evaluation criteria for donor liver quality3.
Presently, the selection criteria for liver grafts at most transplantation centres are based on clinical information such as age, transaminase, total bilirubin, ICU stay time, ischaemia time, pathology (such as steatosis), and imaging examination of potential donors. However, these indicators are inaccurate under the current organ acquisition mode4. Research has revealed various new biomarkers, including glutamate dehydrogenase, cytochrome c oxidase, caspase activity, achmg1, and caspase cleaved/full-length cytokeratin-18, which were shown to have lower comprehensive specificity and sensitivity than alanine aminotransferase (ALT) or aspartate aminotransferase5–7. Neutrophil gelatinase-associated lipocalin is a newly discovered blood marker of ischaemia injury, which can distinguish the reperfusion phase8. Molecular markers involved in liver injury include hepatocyte-derived microRNAs, such as miR-122, miR-148a, and miR-194, which are more specific than traditional molecular biological indicators. A previous study determined that miR-122 was an earlier and more sensitive indicator than ALT in a pig model of liver injury caused by cardiogenic shock9. However, the response time of miR-122 to hepatic warm ischaemic injury (WII) was slow, and 20 min after human donor hepatic warm ischaemia, the expression level dropped considerably, taking a further 10–20 min to drop to 50%10; most transplant centres believe that liver grafts with ischaemia for longer than 30 min are unsafe.
Ischaemia-reperfusion injury (I/RI) is the main reason for “marginal donor liver” and causes graft dysfunction after LT. The duration of WII in donor organs directly affects the severity of I/RI and thus considerably affects the prognosis of LT 11. The core of WII is incorrect energy metabolism, and mitochondria are the centre of cellular metabolic activities. Mitochondria may be the key to finding more accurate indicators of WII. During WII, the abnormal expression level of miRNAs in mitochondria, namely mitomiRs, may directly and rapidly lead to the abnormal function of mitochondria. In contrast, swollen and cracked mitochondria may release mitomiRs into the cytoplasm or the serum, which could be an early warning indicator of mitochondrial damage. However, most mitomiRs involved in mitochondrial energy metabolism were found by gene chip screening, and most of their functions are unclear. According to a systematic search, our research focused on the optimal WII mitochondrion biomarker as a potential solution for WII evaluation difficulty, while also clarifying its specific mechanism in WII.
Methods
Donors and recipients of LT
A total of 219 donor liver samples met the inclusion criteria. See supporting information (S1, Supplemental Digital Content 5, http://links.lww.com/JS9/A496) for inclusion criteria, exclusion criteria, detailed cohort data, and ethics. A total of 148 samples were included in the test set, and followed up until December 2020. The average follow-up date was 548.10±240.46 days postoperation. The other 71 samples that met the requirements comprised the validation set and were followed up until April 2022. The minimum follow-up time was no less than 90 days.
Animals and treatments
Our work has been reported in accordance with the ARRIVE guidelines (Animals in Research: Reporting In Vivo Experiments) 12. Details are described in S1, Supplemental Digital Content 5, http://links.lww.com/JS9/A496.
Statistical analyses
R (version 4.0.5) was used to analyze the relevant data. Categorical data were presented as numbers (percentage) and compared using Pearson’s χ2 and Fisher’s exact tests. Continuous variables were expressed as the mean value±SD and analyzed using a t-test and repeated measure analysis of variance. Overall graft survival was estimated via the Kaplan–Meier method. P less than 0.05 was considered to be statistically significant.
Other research methods and details
Further details are described in S1, Supplemental Digital Content 5, http://links.lww.com/JS9/A496.
Results
Early changes during the ischaemic stage in I/RI: Focus on the mitochondrion
We used immunohistochemistry and immunofluorescence to detect major apoptosis indicators in livers of wild-type C57BL/6 mice during WII. After reperfusion, the results of ischaemia for 60 min (I-60 min) and I-30 min displayed evident apoptotic damage in liver tissue (Figure 1 A; Supporting Fig. S1, Supplemental Digital Content 2, http://links.lww.com/JS9/A493). Western blot identified that after reperfusion the expression of Caspase-9 and Caspase-3 at I-30 min was significantly higher and Bcl-2 was significantly lower than that in normal liver tissue (P<0.01; Figure 1 D). However, at I-15 min after reperfusion, in accordance with our immunohistochemistry and immunofluorescence results (Figure 1C), classical apoptosis indices (Caspase-9, Caspase-3, and Bax) were unable to indicate any obvious injury; only the apoptosis mitochondrial pathway upstream factor Bcl-2 was markedly reduced. Therefore, the appearance of early evident injury after reperfusion may take about I-30 min; detection for the degree of WII within I-30 min is probably the key to evaluate graft quality. For I-60 min without reperfusion, no significant difference in apoptosis (Caspase-3 and TUNEL), oxidation (superoxide dismutase level, which is closely related to tissue oxidation level in WII14), or optical morphology was found (Figure 1A, C). Still, without reperfusion, there were no significant morphological, immunohistochemical, immunofluorescent under a light microscope, or western blot changes within I-30 min. However, without reperfusion, transmission electron microscopy demonstrated evident morphological changes in mitochondria at I-15 min (Figure 1C). Using high-throughput sequencing for mRNA expression at each ischaemia time point (three samples in the normal group and eight samples in either I-15, I-30, or I-60 min), we found a total of 316 upregulated genes at I-15 min (P<0.05 and fold change ≥2); signal path enrichment analysis suggested that the signal path that changed through each ischaemia time point was the mitochondrial apoptosis path (Figure 1B, Supporting Fig. 2, Supplemental Digital Content 3, http://links.lww.com/JS9/A494). Other detailed data of high-throughput sequencing results are shown in S2, Supplemental Digital Content 6, http://links.lww.com/JS9/A497.
Figure 1.
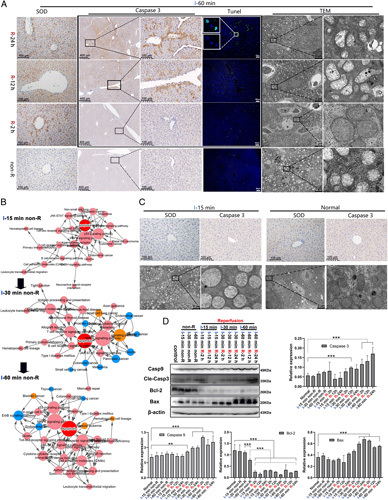
Mitochondrial changes in apoptosis possibly possess a more sensitive response to hepatic ischaemia. Warm ischaemia (I), Reperfusion (R). (A, C) The degree of tissue oxidation- [Immunohistochemistry (IHC)-SOD], apoptosis- (IHC-Caspase-3), and morphological change-related indicators in different ischaemic and reperfusion periods of mice livers [IHC, transmission electron microscope (TEM)]. The obvious injury occurred at I-30 and I-60 min (Supporting Fig. S1, Supplemental Digital Content 2, http://links.lww.com/JS9/A493) after reperfusion. There were no significant morphological, immunohistochemical, or immunofluorescent changes observed under a light microscope at I-15, I-30, or I-60 min without reperfusion. Via TEM, the mitochondria of the I-60 min without reperfusion conformed with Flameng 13 2–3 points, whereas those of the I-60 min with R-2 h conformed with Flameng 3–4 points. We were able to observe evident structural changes in the mitochondria at I-15 (Flameng 2 points). (B) High-throughput sequencing of the transcriptome at each ischaemia time point (three samples in the normal group and eight samples in either I-15, I-30, or I-60 min group). Signal pathway enrichment analysis suggested that the signal pathway that changed throughout each ischaemia time point was the mitochondrial apoptosis pathway. (D) Western blot analysis of the apoptosis-related molecules Caspase-9, Caspase-3, Bcl-2, and Bax.
Screening and identification of mitomiR-23b-5p as the target biomarker
The purity and integrity of mitochondrial extraction are particularly important for the relative quantitative analysis of the changes in biological macromolecules in mitochondria. Transmission electron microscopy showed that the mitochondria extracted during this study met the requirements of high purity and complete structure (Figure 2A). To explore biomarkers within I-30 min in mitochondria, we extracted mitochondria and performed (Reverse Transcription Quantitative Polymerase Chain Reaction (RTqPCR) analysis on all 13 mitochondrial mRNAs (mt-mRNAs; S3, Supplemental Digital Content 7, http://links.lww.com/JS9/A498; n=3 for each mRNA), which showed that the expression of 12 mt-mRNAs during WII at 0, 15, and 30 min lacked consistency; only mt-ND5 displayed a trend of continuous increase at I-15 min and I-30 min (Figure 2B). Generally, the half-life period of mRNA in vivo is short. For instance, the half-life of histone mRNA in the S phase is ~10–40 min15. This is consistent with our observed mt-RNA results, which showed a lack of sustained upregulated expression at I-60 min. Interestingly, although the stability of different miRNAs with corresponding half-lives ranging from 1.5 h to more than 13 h varies greatly among cells, it is potentially considerably higher than that of non-miRNAs in general16.
Figure 2.
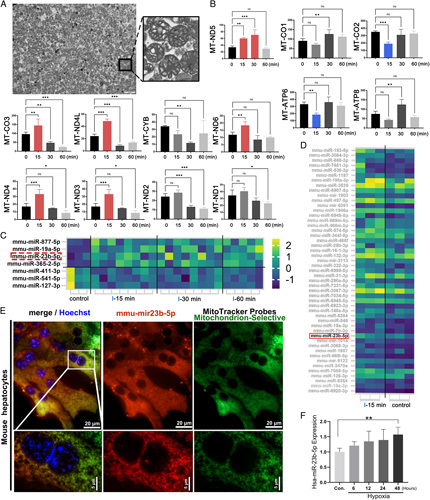
Targeting molecular biomarkers of warm ischaemia. (A) Mitochondria were extracted from mouse liver tissue, and the structure of mitochondria was complete and highly enriched. (B) Among the 13 mitochondrial genomic mRNAs, the expressions of mt-ND3, ND4L, and ND4 were significantly upregulated after 15 min of ischaemia. Only mt-ND5 had a consistent upregulation trend from 0 to 30 min. (C) Compared with that in the control group, the expression of 25 miRNAs at I-15 min, 44 miRNAs at I-30 min, and 40 miRNAs at I-60 was significantly upregulated, whereas four types of the same miRNAs existed among the I-15, I-30, and I-60 min groups. (D) To preliminarily screen the miRNA enriched in the mitochondria after extraction, miRNA chip showed the relative expression of miRNAs in normal and I-15 min groups. The miRNA shown in the red box has human-mouse homology and is consistent with the chip and sequencing results. (E) The primary liver cells were extracted from a mouse liver and cultured in a petri dish for FISH. (F) The expression of mitomiR-23b-5p increased gradually under 1% hypoxia induction; *P<0.1, **P<0.05, ***P<0.01.
Further, after mitochondrial extraction, miRNA chips (Affymetrix GeneChip miRNA 4.0) at I-15 min (n=3) demonstrated 49 upregulated miRNAs (P<0.05 and fold change ≥1.5); these miRNAs are probably mitochondria enriched miRNAs (mitomiRs) with high sensitivity to I-15 min. However, the method of mitochondrial extraction is slow and inefficient, resulting in inefficient rapid detection of donor liver. Thus, we tried to observe whether these upregulated miRNAs (possibly mitomiRs) could still show considerable upregulated expression without mitochondrial extraction. Through miRNA sequencing of mouse liver tissue (non-extracted mitochondria), compared with normal tissue (n=3), there were 25, 44, and 40 cases of upregulated miRNA expression at I-15 (n=8), I-30 (n=8), and I-60 min (n=5), respectively, of which 4 cases had the same 0–15–30–60 min increasing trend (Figure 2C). Only one miRNA expression change detected via an miRNA chip (fold change ≥1.5) in the extracted mitochondria was matched with miRNAs detected via high-throughput sequencing without extracted mitochondria (Figure 2D). This match suggests that this miRNA may be a mitomiR rather than common miRNA, possess a constant I-15–30–60 min increasing trend, and can be effectively extracted and detected in tissues without mitochondrial extraction. This miRNA (miR-23b-5p) also has human and mouse homology for the nucleotide sequence. In the primary culture of mice liver cells, miR-23b-5p was confirmed by FISH under a confocal microscope to be a mitomiR with a Pearson’s coefficient of 0.838 and an overlap coefficient of 0.915 (Figure 2E). Reverse transcription quantitative polymerase chain reaction (RT-qPCR) detection under 1% hypoxia culture also verified that expression of miR-23b-5p was positively correlated with hypoxia culture time (Figure 2F). Sample quality control, correlation detection, and other detailed data of high-throughput sequencing and chip results are presented in S4, Supplemental Digital Content 8, http://links.lww.com/JS9/A499.
Localization, inhibition, and overexpression of mitomiR-23b-5p in human cells
To verify the external consistency of the function of target miRNAs, we performed relevant tests in human cells. Through FISH, fluorescence microscopy showed that mitomiR-23b-5p was localized and enriched in the mitochondria of human cells (HeLa), having a Pearson’s coefficient of 0.958 and an overlap coefficient of 0.964 (Figure 3A). We used the lentivirus vector to inhibit the target miRNA in 293T cells; the specific viral design is shown in S1, Supplemental Digital Content 5, http://links.lww.com/JS9/A496. The lentivirus transfection efficiency of miR-23b-5p was greater than 80% (Figure 3B). Because the transfection occupied the fluorescent channel, we used Annexin V-APC single-staining flow cytometry for the detection of apoptosis (Figure 3C). On the third day after transfection (transfection efficiency ≥ 80%), the cells were cultured at 1% hypoxia for 24 h. Subsequently, flow cytometry demonstrated that inhibition of miR-23b-5p significantly increased the degree of apoptosis after hypoxic culture (8.64±0.06% vs. 1.09±0.16%; P<0.01). However, we did not observe any significant differences in the degree of apoptosis after miR-23b-5p expression was upregulated (2.22±0.63% vs. 1.55±0.19%; P=0.16). Upon further study via western blotting, the Bcl-2 expression level was significantly reduced under normoxic conditions after miR-23b-5p inhibition (P=0.009). Under 1% hypoxia induction, the difference in Bcl-2 expression was more significant (P<0.001). Additionally, we did not observe significant differences under normoxic or hypoxic conditions for the protein expression levels of Caspase-3, Caspase-9, and Bax (Figure 3D). Overall, 293T cells cultured under 1% hypoxia conditions after the inhibition of miR-23b-5p exhibited an increased apoptotic proportion, possibly due to the decreased expression of Bcl-2.
Figure 3.
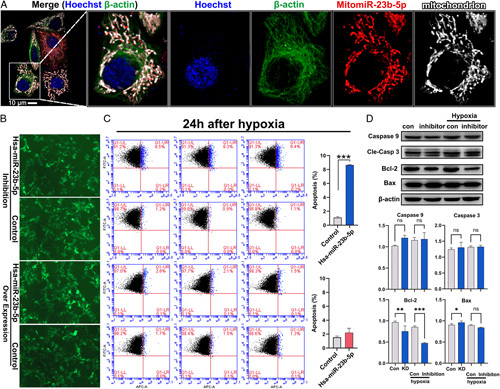
Overexpression and inhibition of mitomiR-23b-5p in 293T under hypoxia. (A) Human HeLa cells under confocal super-resolution optical microscope. (B) The transfection efficiency of miR-23b-5p overexpression (OE) and the inhibition by lentivirus in 293T cells were both higher than 80%. (C) Annexin V-APC single staining for apoptosis detection. After 24 h of 1% hypoxia culture, there was a significant increase in apoptosis in the inhibited group detected by flow cytometry. (D) Western blot analysis was used to detect the expression of the main functional molecules in the apoptosis pathway (Caspase-9, Caspase-3, Bcl-2, and Bax) in the inhibited group after being cultured for 24 h under 1% hypoxia and normoxia. The expression of Bcl-2 in the miR-23b-5p-inhibited group was significantly reduced under normoxia, and the downregulation of Bcl-2 expression was more significant under hypoxia.
In-vivo studies confirmed that mitomiR-23b-5p exerts a protective effect in I/RI
Because I/RI is the result of complex interactions between many factors, the results we observed needed to be verified in vivo (n=3 for each point in time). Wild-type C57 mice were transfected with adeno-associated virus (AAV) 9-mmu-miR-23b-5p inhibitors to achieve the target miRNA inhibition. Virus construction is demonstrated in S1, Supplemental Digital Content 5, http://links.lww.com/JS9/A496. Figure 4A shows the transfection efficiency of several frozen mouse liver tissue sections, all of which were greater than 80%.
Figure 4.
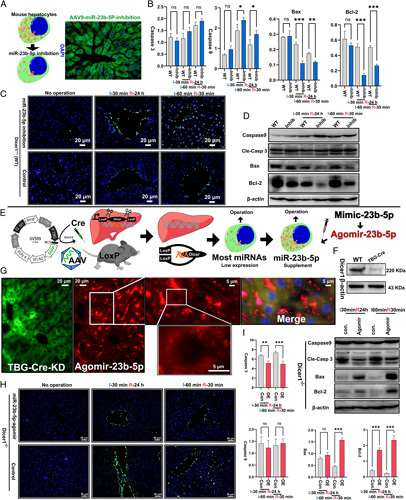
The function of mitomiR-23b-5p in mice during ischaemia-reperfusion injury (I/RI). (A) After 28 days of transfection of AAV-miR-23b-5p-inhibitor into wild-type C57 mice, the transfection efficiency was shown by frozen-section fluorescence microscopy. (C) In TUNEL, we observed that the apoptosis in miR-23b-5p-inhibited mice did not change significantly after WII. (B, D) Western blot detection of apoptosis-related molecules showed that miR-23b-5p could significantly downregulate Bax and Bcl-2 expression; however, the effect on Bcl-2 was greater. (E) By transfection with TBG-Cre-AAV9, knocking out Dicer1, most miRNA expression in hepatocytes was downregulated; then, by reinfusion of the mature miR-23b-5p (agomiR), the specific function of this miRNA in the liver was observed. (F) Western blot analysis showing Dicer1 knockout efficiency (***P<0.01). (G) The transfection efficiency of the virus and agomiR-miR-23b-5p was more than 80%. There was a high concentration of agomiR-miR-23b-5p in hepatic sinusoids, whereas agomiR-miR-23b-5p around the nucleus could be seen in hepatocytes. (H) The miR-23b-5p–upregulated group showed a lower apoptosis level in TUNEL. (I) Western blot analysis showed that the miR-23b-5p–upregulated group could upregulate Bax and Bcl-2 expression during the process of I/RI; however, the upregulation of Bcl-2 expression was more significant.
There was no significant difference in the TUNEL results in mitomiR-23b-5p-inhibited wild-type mice under ischaemic or normal conditions (Figure 4C). However, western blot results showed that the expression level of the Bcl-2 protein was notably downregulated after WII (Figure 4 D). Interestingly, Bax, which was expected to increase, decreased considerably (Figure 4D). Considering the possible functional overlap and interaction between miRNAs, the resulting in-vivo phenotypes of wild-type mice may be considerably affected. Dicer1 encodes a key processing protein converting pre-miRNA to mature miRNA. Knocking out Dicer1 reduces the expression of most miRNAs. By supplementing specific mature miRNAs after knocking out Dicer1, the effect of target miRNAs can be observed separately. Therefore, we selected Dicer1fl/flAlbCre inducible knockout transgenic mice. AAV9-TBG-Cre was used to induce the hepatic-specific expression of the Cre enzyme, and Dicer1 was knocked out in the liver cells. The specific viral design is shown in S1, Supplemental Digital Content 5, http://links.lww.com/JS9/A496 and Figure 4E. Western blot results reflected a Dicer1 knockout efficiency greater than 90% (Figure 4F). After Dicer1 knockout, we used more stable agomiR-miR-23b-5p for the supplementation of target miRNA. Fluorescence microscopy showed that agomiRs entered the cytoplasm of the liver around the nucleus (Figure 4G). As a result, after mouse liver cells were transfected with the mitomiR-23b-5p, a low apoptosis level was observed in the TUNEL (Figure 4H) and western blot results (Figure 4I). Overall, these results suggest that mitomiR-23b-5p has a protective effect in the process of hepatic I/RI in mice.
Expression of miR-23b-5p in human donor liver and prognosis of liver transplant recipients
Among the 238 donor liver samples we collected as test set, only 148 met the inclusion criteria (S1, Supplemental Digital Content 5, http://links.lww.com/JS9/A496). The median value of target miRNA expression was used for experimental grouping. The basic clinical information of the donors and recipients is shown in Figure 5A. As per the baseline comparison, no notable difference between the groups (high and low expression of mitomiR-23b-5p) was observed. Compared with that in the low-expression group, the ALT level at postoperative day 3 (ALT-POD3) in the high mitomiR-23b-5p-expression group was higher (P=0.032, Figure 5C), with a positive correlation trend (P=0.022, Figure 5D). The GSR of the high-expression group at 20 months was 89.2% (17.93±0.69), whereas that of the low-expression group was 98.6% (19.76±0.24); log-rank (Mantel–Cox) test, P=0.016 (Figure 5F). The receiver operating characteristic curve of the GSR of mitomiR-23b-5p at 90 d after the operation showed an area under the curve (AUC; C-statistic) of 0.698 (Figure 5E). In the validation set of 71 samples meeting the criteria, the AUC was 0.691 (Supporting Figure 3, Supplemental Digital Content 4, http://links.lww.com/JS9/A495). When the minimum P value was obtained from the GSR Kaplan–Meier survival curve (P<0.01), the optimal cut-off value of this biomarker (logarithm) for 90-day GSR was 1.95. The 20-month GSR in the high-expression group was 88.6%, whereas that in the low-expression group was 98.7% (P<0.01; Figure 5G). In the univariate analysis for GSR, mitomiR-23b-5p expression after logarithmic treatment was the unique statistically significant factor compared with the expression of conventional indicators, and only this factor reached P less than 0.05 via multivariate analysis. According to the results, it was suggested that mitomiR-23b-5p could be an independent risk factor for the survival of LT grafts (Figure 5B).
Figure 5.
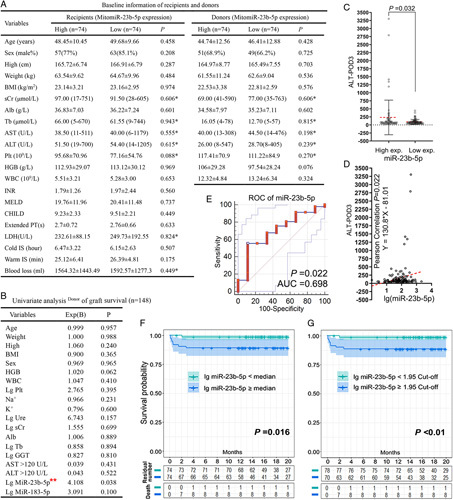
Prognostic markers of liver transplantation. (A) Relevant clinical baseline data of donors and recipients; According to the median value, miR-23b-5p was divided into high-expression and low-expression groups, respectively. Tb, sCr, AST, ALT [median (range)]. * Mann–Whitney U. (B) In univariate analysis, a significant statistical difference in the expression of mitomiR-23b-5p after logarithmic treatment was observed compared with other conventional indicators, and only this factor showed P<0.05 in multivariate analysis (**). (C, D) Pearson’s correlation test showed that the logarithmic expression levels of two biomarkers were closely related to the expression level of POD3-ALT (P=0.022). (E) Receiver operating characteristic curves for 90-day GSR with a significant statistical difference (AUC=0.698; P=0.022). (F, G) The 20-month GSR. The best cut-off value of mitomiR-23b-5p was 1.97, as calculated by X-tile software. ALB, albumin; AST, aspartate aminotransferase; HGB, haemoglobin, INR, international normalized ratio; IS, ischaemia; LDH, lactate dehydrogenase; MELD, model end-stage liver disease; Plt, platelet; sCr, creatinine; TB, total bilirubin; WBC, white blood cell.
Hsa-miR-23b-5p affects mitochondrial respiratory function primarily via mitochondrial RNR2 silencing
To further clarify the specific biological role of mitomiR-23b-5p in mitochondria, we performed transcriptome sequencing after using lentiviral vector-transfected 293T cells for the overexpression (OE) and inhibition of mitomiR-23b-5p. Detailed sequence data are presented in S5, Supplemental Digital Content 9, http://links.lww.com/JS9/A500. The results showed that mitomiR-23b-5p inhibition could increase the expression levels of mt-RNR1 and mt-RNR2 in mitochondria, whereas mitomiR-23b-5p-OE had the opposite effect (Figure 6C). The possible binding sites of RNR1 and RNR2 are demonstrated in Figure 6A. Detailed binding site analysis is demonstrated in S6, Supplemental Digital Content 10, http://links.lww.com/JS9/A501. In the follow-up dual-luciferase report, we found that human miR-23b-5p could silence RNR2 after mutating seven putative binding sites (Figre. 6 B; specific sites are shown in S6, Supplemental Digital Content 10, http://links.lww.com/JS9/A501). We also selected three sites for the mutation of RNR1; unfortunately, the results did not indicate that miR-23b-5p could silence RNR1 (S6, Supplemental Digital Content 10, http://links.lww.com/JS9/A501).
Figure 6.
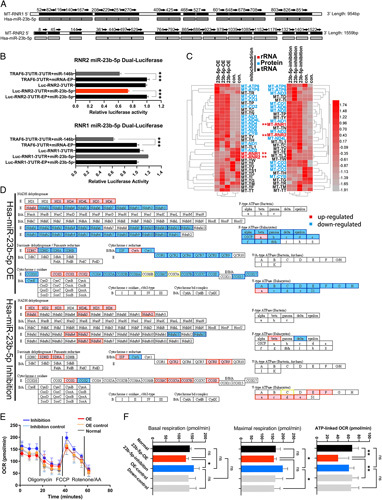
A potential mechanism of miR-23b-5p in mitochondria. (A) The top 20 potential binding sites of mt-RNR1 and mt-RNR2 to miR-23b-5p. (B) EP: Empty plasmid. Dual-luciferase reporting showed that miR-23b-5p could not bind to the target gene rRNA1 3’UTR and could not inhibit its expression; the experimental group (Luc-RNR2-3’UTR + miR-23b-5p), compared with the control group, had a significant decrease (P<0.05), indicating that the miRNA could bind to the target gene and inhibit its expression. (C) Transcriptome sequencing was performed in the inhibition and overexpression (OE) groups. Mitochondria encoded a total of 13 mitochondrial proteins and two mitochondrial ribosomal RNAs. Among them, the mitochondrial genomic transcriptional RNAs conforming to the classical miRNA silencing mode—after mitomiR-23b-5p expression was upregulated, target RNA expression was downregulated, and vice versa—were mt-RNR1 and mt-RNR2. (D) Mitochondrial respiratory functional gene expression. After the upregulation of miR-23b-5p expression, the expression of most mitochondria function–related transcriptome mRNAs was significantly downregulated, and vice versa. (E, F) ATP production decreased in the OE group, whereas basal respiration and maximal respiratory capacity increased in the inhibition group compared with those in the OE, normal, and control groups.
We analyzed the changes of miR-23b-5p in mitochondrial function via transcriptome sequencing. The specific mitochondrial genes encoding protein complexes I–V in the electron transport chain are presented in Figure 6D. For miR-23b-5p-OE, the encoded gene for the constituent proteins of each complex was generally downregulated; however, after miR-23b-5p expression was inhibited, gene transcription for each complex component protein was generally upregulated. To verify whether the sequencing results reflected an actual functional role in mitochondria, we tested the mitochondrial respiratory function of 293T cells. The Seahorse XF mitochondrial pressure test showed that ATP production was markedly reduced in miR-23b-5p-OE cells. The basal respiration rate and maximum respiratory capacity of the miR-23b-5p–inhibited cells were considerably upregulated (Figure 6E). The correspondence between the mitochondrial respiratory function and the quantitative analysis results are demonstrated in Figure 6F. Overall, the upregulation of mitomiR-23b-5p expression inhibited ATP produced by mitochondrial respiration; conversely, its inhibition increased the maximum respiratory capacity of mitochondria. These results were consistent with the mitochondrial function analysis via sequencing.
Discussion
WII is one of the most difficult injuries to measure or detect in donor livers. In the absence of high-precision quality index parameters, conservative evaluation results in a high abandonment rate of donor organs. Optimized evaluation of donor liver will help transplant physicians distinguish the high-risk “marginal donor liver” from the high-quality donor liver to improve the survival rate of transplanted organs and reduce the rate of organ abandonment. The main finding of our study was an optimal mitochondrial biomarker that can assist the current donor liver evaluation method, revealing promising results in solving WII evaluation difficulty; we also revealed a new mechanism where mitomiR silences mitochondrial ribosomal RNA (rRNA) to regulate mitochondrial function. Although there are many available prognostic models for LT, the basic elements adopted by most models are common as clinical detection indicators. To meet high accuracy, scoring models usually include more than 10 influencing factors, and most of these models include information after LT, such as recipient surgery and early prognosis information17. The current multi-factor prediction model is not ideal in practicality and multicenter verification, having a C-statistic within 0.54–0.7018–22; the main reason remains the lack of high-precision biomarkers. The newly discovered biomarker mitomiR-23b-5p is closely related to WII of the donor liver and is a preoperative predictor with independent prediction potential, with a potential AUC of 0.698 (90-day GSR).
MitomiRs and mt-rRNA silencing
Unlike other organelles, mitochondria are semi-autonomous and contain certain functional genes. mtDNA gene expression is regulated coordinately before and after transcription, and it has been found that interference with its expression leads to various human diseases. The ribosome is the core component of the protein synthesis apparatus and is one of the most conserved and complex molecular machines in cells. Each ribosomal subunit contains many ribosomal proteins; however, rRNA plays the most critical functional role. Interestingly, mitochondria are also organelles with ribosomes, and ribosomes in mitochondria are called mitochondrial ribosomes. The rRNA in mitochondria comes from the mitochondrial genome itself, namely mt-RNR1 (12S) and mt-RNR2 (16S). Changes in the mechanisms of these two rRNAs will significantly affect mitochondrial function. Currently, 14 mitochondrial small-subunit 12S rRNA methylation sites are functional, whereas 24 large subunit sites have been found23; any changes within these subunits significantly affect mitochondrial respiratory function and lead to various diseases24. These findings suggest that mitochondrial rRNA plays a vital role in mitochondria, suggesting that miRNAs may interfere with the transcription of genes related to mitochondrial respiratory function by regulating rRNA. However, whether mitomiRs play an mRNA silencing role in mitochondria, as they do in the cytoplasm, depends on whether mitochondria have RNA-induced silencing complex (RISC). Interestingly, RISC formation occurs in the cytoplasm; however, recent studies have shown that RISC is also found in mitochondria25–31. Theoretically, this implies that mitomiRs can silence mRNAs produced by mitochondrial DNA.
Phenotypic and functional studies of miR-23b-5p lacking reports on mitochondrial function
Many studies have reported various miRNAs participating and playing important roles in the regulation of I/RI. For instance, miR-21 has a protective effect on I/RI, and miR-122 is closely related to I/RI32. miR-23b is a member of the miR-196s family and is generated by an independent gene, miR-23b, located on chromosome 7.733. Studies have found that miR-23b is closely related to cell proliferation, tumour invasion, apoptosis, and WII34,35. miR-23b has two types of mature bodies and can exercise the function of miRNA, which are processed from the 5’ and the 3’ ends of miR-23b-5p and miR-23b-3p, respectively. At present, there are only a few reports on miR-23b-5p. Most studies are at the stage of phenotype research. The sequencing results suggest that the expression level of this miRNA is closely related to a variety of diseases, such as late-onset preeclampsia36, lung adenocarcinoma37, Graves’ disease38, and myeloid leukaemia39. The research on miR-23b-5p in terms of warm ischaemia remains scarce. Still, in recent years, some studies have observed that its precursor miR-23b is closely related to WII, and even in peripheral blood, it can be used as a marker of myocardial infarction40. These views are consistent with our findings.
In a functional study, Zhao et al. 41 found that miR-23b may increase I/RI, primarily via the p53 pathway . Boureima and colleagues knocked down and overexpressed miR-23b-5p in vivo and selected cardiomyocytes as research targets. The results showed a marked negative correlation between the expression level of miR-23b-5p in myocardial tissue and cardiac size and function42. In recent years, miR-23b-5p has been found to inhibit the occurrence and development of tumours in gastric cancer, oesophageal adenocarcinoma, prostate cancer, colorectal cancer, and lung cancer43–47. Hu et al. 37 found that miR-23b-5p can inhibit the proliferation and migration of h1975 cells and miR-23b-5p inhibition promotes the proliferation and migration of A549 cells . Additionally, miR-23b-5p was reportedly related to the Wnt/b-catenin48 and ERK signalling pathways in tumour cells46 and was suggested to be related to the HMGB2 pathway in cardiomyocytes42. Using dual-luciferase reporter analysis and RNA immunoprecipitation analysis, Dong and colleagues found direct evidence that miR-23b-5p can bind to CCAT2 37.
The similarity between these studies is that the downregulation of miR-23b-5p expression could promote tumour proliferation, while conversely, inhibition of proliferation can occur. These phenotypic and functional results are theoretically consistent with our findings. Additionally, these pathways exist outside the mitochondria, and there is no report on the function of miR-23b-5p in the mitochondria. Combined with our mechanism-research results, we speculate that miR-23b-5p may promote the expression of mitochondrial functional genes after downregulation, which is manifested as the improvement of mitochondrial respiratory capacity, increase of oxygen consumption, and increase of ATP production, thus leading to the rapid proliferation of tumour cells. We did not observe an increase in apoptosis under normoxia but speculate that the downregulation of miR-23b-5p expression in the warm ischaemia environment will increase apoptosis. Furthermore, we believe that miR-23b-5p may decrease oxygen consumption in its environment, downregulate the mitochondrial respiratory capacity, and further reduce the local ROS production of mitochondria. Because the changes in the mitochondria of subcellular organelles are difficult to explore by conventional research methods, many breakthroughs in technology are still required for accurate measurement. In future work, we aim to clarify the direct mechanism of miR-23b-5p.
Limitations and application scope
The indicator mitomiR-23b-5p is systematically detected in mitochondria and can be used for direct detection of hepatic WII without extracting mitochondria; there may be other highly effective markers, indicating a need for further relevant research. Compared with traditional indicators, our newly found indicator has application limitations that are not completely consistent with the traditional models. Importantly, because this new biomarker is a target marker of WII, it requires the transplant centre to have a mature transplant system and stable surgical technical support to reduce other interference factors, such as surgical complications. The shift of focus away from the traditional selection criteria, such as ignoring severe fatty liver, may notably affect the proportion of I/RI damage and thus reduce the predictive role of this biomarker. The practical significance of this indicator may compensate for the limitation that the warm ischaemia time cannot reflect the WII caused by hypoperfusion in the donor under the abovementioned conditions. Within certain applicability, this indicator may serve a better purpose than the warm ischaemia time, realizing the unification of the judgment of the WII results in each centre. However, this prospect needs further multicenter large sample clinical validation. Figure 7F.
Figure 7.
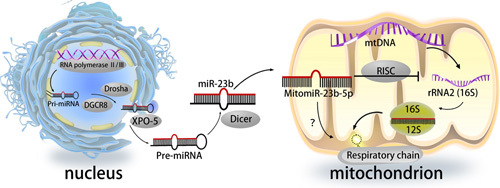
Mechanism of miR-23b-5p in mitochondrial respiration. miRNAs localized in the mitochondria are known as mitomiRs49. Their classical pattern is as follows: Pri-miRNAs are transcribed from the nucleus and then cleaved into pre-miRNAs by Drosha; subsequently, they exit the nucleus into the cytoplasm, where they are processed into mature miRNAs by Dicer and are finally transported into the mitochondria50. Mt-RNR1 is bound and silenced by the RNA-induced silencing complex (RISC), which interferes with the translation of relevant mitochondrial genomic functional genes.
Conclusions
As a mitomiR, miR-23b-5p plays a protective regulatory role and performs a unique mitochondrial regulatory mechanism in WII. It can reduce mitochondrial respiratory function by silencing mt-RNR2 (16S). Our clinical results further support the experimental result that the expression of MitomiR-23b-5p is closely related to the prognosis of clinical liver transplantation patients. This is a promising new biomarker for WII evaluation of donor livers.
Ethical approval
This study was approved by the ethics committee of West China Hospital. The use of clinical data was approved by the ethics committee of West China Hospital of Sichuan University, judgement's reference number No. 2020 review (No. 385). In our study, the grafts for LT were from donation after the death of citizens according to a new organ acquisition and distribution policy established in China after 2012. No prisoners were included as donors. All follow-up and clinical data were uploaded to the China Liver Transplant Registry (http://www.cltr.org/). Informed written consent was obtained from the family members of each liver donor for LT. Finally, the study protocol was in accordance with the Declaration of Helsinki of the World Medical Association. The animal care and experimental procedures were conducted in accordance with national and international laws and policies; they were also approved by the Animal Care and Use Committee of Sichuan University and complied with the criteria outlined in the National Institutes of Health Guide for the Care and Use of Laboratory Animals.
Sources of funding
This work was supported by Sichuan Natural Science Foundation (2022NSFSC0805); National Natural Science Foundation of China (81770653); The Sichuan University postdoctoral interdisciplinary Innovation Fund; China Postdoctoral Science Foundation (2022M720102);2022YFC2304700.
Author contribution
L.K., Y.S., and Jiayin Y designed the study; L.J., Jian Y, T.L., L.K., J.Y., and Y.Z. performed the research and collected the data; L.K. analyzed and interpreted the data; L.K. wrote the first draft of the manuscript; all authors edited the manuscript and approved the final draft; the acquisition of funding was completed by Jiayin Y and L.K.
Conflicts of interest disclosure
The authors declared that they have no conflicts of interest to this work.
Research registration unique identifying number (UIN)
1. Name of the registry: http://www.medresman.org.cn (Chinese Clinical Trial Registry chictr.org.cn) 2. Unique Identifying number or registration ID: ChiCTR2000032141 3. Hyperlink to your specific registration (must be publicly accessible and will be checked): http://www.chictr.org.cn/showproj.aspx?proj=52598
Guarantor
Dr. Jiayin Yang and Dr. Lingxiang Kong are the people who accept full responsibility for the work and/or the conduct of the study, had access to the data, and controlled the decision to publish.
Data statement
All human-related data of our centre are stored in the Chinese Liver Transplant Registry, a platform for unified management of liver transplantation centres in mainland China (CLTR: http://cltr.cotr.cn). The data that support the findings of this study are available from the Chinese Liver Transplant Registry (CLTR: http://cltr.cotr.cn), but restrictions apply to the availability of these data, which were used under license for the current study, and so are not publicly available. But all related data in this study are available from the corresponding author on reasonable request. Non-human research data are uploaded in the part of Attachments/ Supplementary Item.
Supplementary Material
Footnotes
Sponsorships or competing interests that may be relevant to content are disclosed at the end of this article.
Supplemental Digital Content is available for this article. Direct URL citations are provided in the HTML and PDF versions of this article on the journal's website, www.lww.com/international-journal-of-surgery.
Published online 13 May 2023
Contributor Information
Lingxiang Kong, Email: 870304402@qq.com.
Yongjie Zhou, Email: yongjiezhou@scu.edu.cn.
Jingsheng Yuan, Email: 811711491@qq.com.
Tao Lv, Email: 447756367@qq.com.
Jian Yang, Email: Yangjian19820810@hotmail.com.
Yujun Shi, Email: shiyujun@scu.edu.cn.
Jiayin Yang, Email: docjackyang@126.com.
References
- 1. Hartog H, Hann A, Perera M. Primary nonfunction of the liver allograft. Transplantation 2022;106:117–28. [DOI] [PubMed] [Google Scholar]
- 2. Salviano MEM, Lima AS, Tonelli IS, et al. Primary liver graft dysfunction and non-function: integrative literature review. Rev Col Bras Cir 2019;46:e2039. [DOI] [PubMed] [Google Scholar]
- 3. Bae C, Henry SD, Vedula G, et al. Wagener G. The Marginal Liver Donor and Organ Preservation Strategies. Liver Anesthesiology and Critical Care Medicine New York, NY. Springer; 2012:181–193. [Google Scholar]
- 4. EASL Clinical Practice Guidelines: Liver transplantation. J Hepatol 2016;64:433–85. [DOI] [PubMed] [Google Scholar]
- 5. Weemhoff JL, Woolbright BL, Jenkins RE, et al. Plasma biomarkers to study mechanisms of liver injury in patients with hypoxic hepatitis. Liver Int 2017;37:377–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Starkey Lewis PJ, Dear J, Platt V, et al. Circulating microRNAs as potential markers of human drug-induced liver injury. Hepatology 2011;54:1767–1776. [DOI] [PubMed] [Google Scholar]
- 7. Roderburg C, Benz F, Vargas Cardenas D, et al. Elevated miR-122 serum levels are an independent marker of liver injury in inflammatory diseases. Liver Int 2015;35:1172–1184. [DOI] [PubMed] [Google Scholar]
- 8. Yildirim D, Sarac F, Degerli MS, et al. Rat model investigation on the role of biomarkers in hepatic ischemia-reperfusion injury. Exp Clin Transplant 2021. (Epub ahead of print). [DOI] [PubMed] [Google Scholar]
- 9. Andersson P, Gidlof O, Braun OO, et al. Plasma levels of liver-specific miR-122 is massively increased in a porcine cardiogenic shock model and attenuated by hypothermia. Shock 2012;37:234–238. [DOI] [PubMed] [Google Scholar]
- 10. Farid WR, Pan Q, van der Meer AJ, et al. Hepatocyte-derived microRNAs as serum biomarkers of hepatic injury and rejection after liver transplantation. Liver Transpl 2012;18:290–297. [DOI] [PubMed] [Google Scholar]
- 11. Lowalekar SK, Cao H, Lu XG, et al. Sub-normothermic preservation of donor hearts for transplantation using a novel solution, Somah: a comparative pre-clinical study. J Heart Lung Transplant 2014;33:963–970. [DOI] [PubMed] [Google Scholar]
- 12. Kilkenny C, Browne WJ, Cuthill IC, et al. Improving bioscience research reporting: the ARRIVE guidelines for reporting animal research. PLoS Biol 2010;8:e1000412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Seguchi R, Watanabe G, Kato H, et al. Subzero 12-hour nonfreezing cryopreservation of porcine heart in a variable magnetic field. Transplant Direct 2015;1:e33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Feng C, Qian D, Chen C. A meta-analysis and systematic review of propofol on liver ischemia-reperfusion injury protection during hepatocellular carcinoma anesthesia surgery. Ann Palliat Med 2021;10:6726–35. [DOI] [PubMed] [Google Scholar]
- 15. Ross J. mRNA stability in mammalian cells. Microbiol Rev 1995;59:423–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Coenen-Stass AML, Pauwels MJ, Hanson B, et al. Extracellular microRNAs exhibit sequence-dependent stability and cellular release kinetics. RNA Biol 2019;16:696–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Rana A, Hardy MA, Halazun KJ, et al. Survival outcomes following liver transplantation (SOFT) score: a novel method to predict patient survival following liver transplantation. Am J Transplant 2008;8:2537–2546. [DOI] [PubMed] [Google Scholar]
- 18. Desai NM, Mange KC, Crawford MD, et al. Predicting outcome after liver transplantation: utility of the model for end-stage liver disease and a newly derived discrimination function. Transplantation 2004;77:99–106. [DOI] [PubMed] [Google Scholar]
- 19. Gyori GP, Silberhumer GR, Zehetmayer S, et al. Dynamic changes in MELD score not only predict survival on the waiting list but also overall survival after liver transplantation. Transpl Int 2012;25:935–940. [DOI] [PubMed] [Google Scholar]
- 20. Schrem H, Reichert B, Fruhauf N, et al. The Donor-Risk-Index, ECD-Score and D-MELD-Score all fail to predict short-term outcome after liver transplantation with acceptable sensitivity and specificity. Ann Transplant 2012;17:5–13. [DOI] [PubMed] [Google Scholar]
- 21. Pearson ACS, Subramanian A, Schroeder DR, et al. Adapting the Surgical Apgar Score for Perioperative Outcome Prediction in Liver Transplantation: a Retrospective Study. Transplant Direct 2017;3:e221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kong L, Lv T, Jiang L, et al. A simple four-factor preoperative recipient scoring model for prediction of 90-day mortality after adult liver Transplantation: a retrospective cohort study. Int J Surg 2020;81:26–31. [DOI] [PubMed] [Google Scholar]
- 23. Sergiev PV, Aleksashin NA, Chugunova AA, et al. Structural and evolutionary insights into ribosomal RNA methylation. Nat Chem Biol 2018;14:226–235. [DOI] [PubMed] [Google Scholar]
- 24. Van Haute L, Hendrick AG, D’Souza AR, et al. METTL15 introduces N4-methylcytidine into human mitochondrial 12S rRNA and is required for mitoribosome biogenesis. Nucleic Acids Res 2019;47:10267–10281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Barrey E, Saint-Auret G, Bonnamy B, et al. Pre-microRNA and mature microRNA in human mitochondria. PLoS One 2011;6:e20220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Das S, Ferlito M, Kent OA, et al. Nuclear miRNA regulates the mitochondrial genome in the heart. Circ Res 2012;110:1596–1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Das S, Bedja D, Campbell N, et al. miR-181c regulates the mitochondrial genome, bioenergetics, and propensity for heart failure in vivo. PLoS One 2014;9:e96820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Dasgupta N, Peng Y, Tan Z, et al. miRNAs in mtDNA-less cell mitochondria. Cell Death Discov 2015;1:15004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Jagannathan R, Thapa D, Nichols CE, et al. Translational regulation of the mitochondrial genome following redistribution of mitochondrial microRNA in the diabetic heart. Circ Cardiovasc Genet 2015;8:785–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Srinivasan H, Das S. Mitochondrial miRNA (MitomiR): a new player in cardiovascular health. Can J Physiol Pharmacol 2015;93:855–861. [DOI] [PubMed] [Google Scholar]
- 31. Zhang X, Zuo X, Yang B, et al. MicroRNA directly enhances mitochondrial translation during muscle differentiation. Cell 2014;158:607–619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Qin Y, Yu Y, Dong H, et al. MicroRNA 21 inhibits left ventricular remodeling in the early phase of rat model with ischemia-reperfusion injury by suppressing cell apoptosis. Int J Med Sci 2012;9:413–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Geng J, Luo H, Pu Y, et al. Methylation mediated silencing of miR-23b expression and its role in glioma stem cells. Neurosci Lett 2012;528:185–189. [DOI] [PubMed] [Google Scholar]
- 34. Majid S, Dar AA, Saini S, et al. miR-23b represses proto-oncogene Src kinase and functions as methylation-silenced tumor suppressor with diagnostic and prognostic significance in prostate cancer. Cancer Res 2012;72:6435–6446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kou Y, Zheng WT, Zhang YR. Inhibition of miR-23 protects myocardial function from ischemia-reperfusion injury through restoration of glutamine metabolism. Eur Rev Med Pharmacol Sci 2016;20:4286–4293. [PubMed] [Google Scholar]
- 36. Mavreli D, Lykoudi A, Lambrou G, et al. Deep sequencing identified dysregulated circulating microRNAs in late onset preeclampsia. In Vivo 2020;34:2317–2324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Hu GD, Wang CX, Wang HY, et al. Long noncoding RNA CCAT2 functions as a competitive endogenous RNA to regulate FOXC1 expression by sponging miR-23b-5p in lung adenocarcinoma. J Cell Biochem 2019;120:7998–8007. [DOI] [PubMed] [Google Scholar]
- 38. Hiratsuka I, Yamada H, Munetsuna E, et al. Circulating microRNAs in Graves’ disease in relation to clinical activity. Thyroid 2016;26:1431–1440. [DOI] [PubMed] [Google Scholar]
- 39. Barrera-Ramirez J, Lavoie JR, Maganti HB, et al. Micro-RNA profiling of exosomes from marrow-derived mesenchymal stromal cells in patients with acute myeloid leukemia: implications in leukemogenesis. Stem Cell Rev Rep 2017;13:817–825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Zhang J, Li Y, Zhao Q. Circulating miR-23b as a novel biomarker for early risk stratification after ST-elevation myocardial infarction. Med Sci Monit 2018;24:1517–1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Zhao Z, Guan JZ, Wu M, et al. Downregulation of microRNA-23b protects against ischemia-reperfusion injury via p53 signaling pathway by upregulating MDM4 in rats. J Cell Biochem 2019;120:4599–4612. [DOI] [PubMed] [Google Scholar]
- 42. Boureima Oumarou D, Ji H, Xu J, et al. Involvement of microRNA-23b-5p in the promotion of cardiac hypertrophy and dysfunction via the HMGB2 signaling pathway. Biomed Pharmacother 2019;116:108977. [DOI] [PubMed] [Google Scholar]
- 43. Qi P, Xu MD, Shen XH, et al. Reciprocal repression between TUSC7 and miR-23b in gastric cancer. Int J Cancer 2015;137:1269–1278. [DOI] [PubMed] [Google Scholar]
- 44. Janikova M, Zizkova V, Skarda J, et al. Prognostic significance of miR-23b in combination with P-gp, MRP and LRP/MVP expression in non-small cell lung cancer. Neoplasma 2016;63:576–587. [DOI] [PubMed] [Google Scholar]
- 45. Kou CH, Zhou T, Han XL, et al. Downregulation of mir-23b in plasma is associated with poor prognosis in patients with colorectal cancer. Oncol Lett 2016;12:4838–4844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Farina NH, Zingiryan A, Akech JA, et al. A microRNA/Runx1/Runx2 network regulates prostate tumor progression from onset to adenocarcinoma in TRAMP mice. Oncotarget 2016;7:70462–70474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Warnecke-Eberz U, Chon SH, Hölscher AH, et al. Exosomal onco-miRs from serum of patients with adenocarcinoma of the esophagus: comparison of miRNA profiles of exosomes and matching tumor. Tumour Biol 2015;36:4643–4653. [DOI] [PubMed] [Google Scholar]
- 48. Dong L, Deng J, Sun ZM, et al. Interference with the β-catenin gene in gastric cancer induces changes to the miRNA expression profile. Tumour Biol 2015;36:6973–6983. [DOI] [PubMed] [Google Scholar]
- 49. Bandiera S, Ruberg S, Girard M, et al. Nuclear outsourcing of RNA interference components to human mitochondria. PLoS One 2011;6:e20746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Ambros V. The functions of animal microRNAs. Nature 2004;431:350–355. [DOI] [PubMed] [Google Scholar]


