The massive surge of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has had significant implications for the population of the entire world during the ongoing coronavirus disease 2019 (COVID-19) pandemic. The SARS-CoV-2 virus, a species of the family Coronaviridae, is a single-stranded RNA virus. Unlike DNA viruses, RNA viruses are prone to mutations. These mutations are very rapid and are required to be a part of natural selection1. Importantly, evidence suggests that in the past decades, the rapid spread of a pandemic was usually altered by such mutations through natural selection. This natural selection, followed by the mutations, reduces the virulence of the virus, which is, in turn, beneficial for society. The same scenario was noticeable in the case of the SARS-CoV-2 virus2. According to the significant classifications made by the WHO and Centers for Disease Control and Prevention, there are several emerging variants; some are called variants of concern, while the rest are called variants of interest. These emerging variants, in turn, alter several types of viral characteristics, like transmissibility, vaccine activity, rate of infection, etc.3,4. In all these emerging SARS-CoV-2 variants, the D614G mutation is the most common mutation found in the viral S-protein3. According to certain data, the scientists implied that this mutation had altered several virus properties. The increased fitness and transmissibility of the virus are solely responsible for this mutation. Due to this mutation, the viral replication also increases, making the virus more transmissible5. This change in characteristics raises the question of when the ongoing COVID-19 pandemic will end. Besides, the increased transmissibility is directly proportional to the rate of infection. Thus, the researchers aim to comprehensively analyze the entire genome of the virus to develop efficient therapeutics to eradicate the pandemic. Most importantly, natural selection has always been a critical factor in regulating the evolution of various emerging mutants’.
Evolution is a classical factor that acts as a driving force to select the organisms that will be the best choice for nature. It has become prevalent in the population in line with the theory of Charles Darwin’s natural selection2. Scientists have cited that the D614G mutation is a part of the positive selection6. It is not an outcome of any ancillary factors like genetic drift. The mutation has been beneficial for nature. The transmissibility has increased significantly, which may be considered a limitation, but the drop in mortality rate is the scientific community’s primary concern. Nevertheless, the molecular basis of evolution should follow the same line where the transmission rate will increase, but the virulence will drop significantly. A more detailed understanding of the D614G mutation will help scientists to predict diseases’ progression by the upcoming variants.
The naturally selected mutations are observed in Alpha, Beta, Delta, Gamma, and the most recent emerging variant, Omicron; the D614G mutation is also one of the significant mutations in S-protein. The D614G mutation is found in all the variants of SARS-CoV-2 and is also present in very high frequency (Fig. 1). Thus, this mutation is so widespread throughout the world. Importantly, all these naturally selected mutations not only give extra fitness to the virus to enhance its capacity to transmit but also mitigate many of the mutations that could have been more deadly. Natural selection is the key to deciding these factors depending on the well-being of the population. For instance, reducing the virulence of the SARS-CoV-2 was possible by this D614G driven mutation. The disease progression is somewhat inversely proportional to the viral virulence. The naturally selected D614G mutation is an ideal example of it. The presence of this mutation in the viral genome has increased the transmission rate of the SARS-CoV-2 virus to a greater extent. However, the mortality rate has dropped significantly, suggesting a potential decrease in viral virulence. The Omicron variant is the perfect example of it. This variant is the most transmissible and least severe7–9. Simultaneously, the effective reproductive number(R0) of the Omicron variant is noted as 8.2, which is about 3.8 times superior in terms of transmissibility compared to the Delta variant10,11. However, scientists concluded that the D614G mutation helps the variant become a more transmissible variant and might reduce the virulence along with the other mutations.
Figure 1.
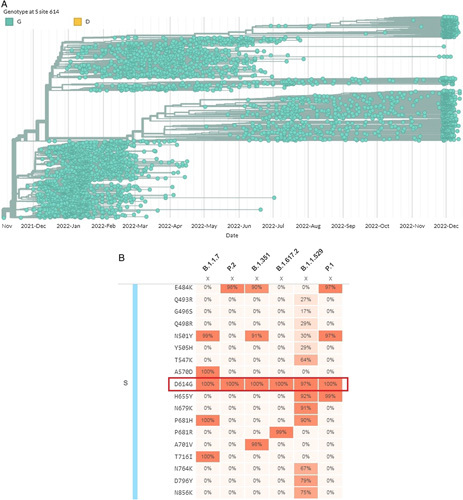
The phylogenic tree of the D614G mutation containing genome and mutation frequencies of different mutations of S-protein of significant variants of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). (a) The phylogenic tree of D614G mutation of SARS-CoV-2 genome shows the mutation present in most of the variants genome of SARS-CoV-2. (b) Mutation frequencies of the S-protein of significant variants of SARS-CoV-2 show D614G mutation to be present in all the variants in very high frequency. The figure was developed through COVIDCG and GISAID databases.
The structural alterations have always been a disadvantage for the interaction of any specific protein with the receptor. The notable features observed in the D614G mutation that alters the nature of transmission and virulence of the viruses are generally implied by the change in the structural conformation. Due to evolution, there has been an alteration in the furin cleavage site of the ACE2 receptor, which is increasing the surface area of the domain surrounding it. The variation in the conformational plasticity enlarges the volume of the cleavage site, which increases the interaction of the virus with the host receptor12. The D614G spike protein’s structure is altered globally as a result of the introduction of the mutation, which organizes the 630-loop structure.
In contrast to the disordered structures in the wild-type protein, the organized 630-loop reduces the strength of the local interactions between the 614 residue and the adjacent residues. The mutation creates a mobile, asymmetric down conformation and speeds up transitions to an up conformation by allosterically changing the connections between receptor-binding regions. The mutation often stabilizes the proximal region of the fusion peptide, including the spike protein protomer. The fusion peptide enables membrane fusion by causing the deletion of the salt bridge between the K854 and D614 residues13. Moreover, Gellenoncourt et al.14 elucidated that SARS-CoV-2 variants generating highly fusogenic spikes emerged due to the D614G mutation, which stabilized the S1/S2 interaction and allowed for the selection of those mutations that are capable of boosting the S1/S2 cleavage. This structural difference is a critical factor for the SARS-CoV-2 virus to interact with the ACE2 receptor of the host. This suggests that evolution gives additional fitness to the virus in infecting the host.
Out of the several mutations residing in the receptor-binding domain, the D614G is widespread for all the SARS-CoV-2 variants of interest and variants of concern. The main question reviving among the researchers’ minds is the alteration of the specific properties of the virus. This mutation became very prevalent in the entire world within a short period of time. By taking a closer look at the mechanism of entry of the SARS-CoV-2 inside the host cell, it is evident that spike glycoprotein plays a vital role in enhancing the virus’ interaction with the ACE2 receptors of the host cell. In order to develop efficient therapeutics, understanding the structural and functional landscape of the spike protein complex is essential. Several studies highlight that this D614G mutation has given evidence of excessive viral loads that are predominantly present in the upper parts of the respiratory system15. The presence of this mutation has been a boon for the virus, enhancing replication and providing fitness. Due to these acquired properties, the transmission rates of the SARS-CoV-2 virus have increased to a greater extent. As discussed, this rapid progression of the virus is accelerated by the naturally selected mutations. These paradigm shifts suggest natural selection. This, in turn, depends on several ancillary factors like the bottleneck effect, population density, and genetic drift. The naturally selected mutations lasted in the environment for a longer period of time. These enhanced qualities mitigate one mutation over the other, suggesting its superiority will prevail in the environment16. This evolutionary evidence will also help scientists develop more efficient therapeutics. The mutations, though they can be a part of natural selection, might develop some properties that no longer neutralize the antigens. Thus, a detailed study of the mutation will be helpful for the community in many ways.
Ethical approval
This article does not require any human/animal subjects to acquire such approval.
Sources of funding
This study received no specific grant from any funding agency in the public, commercial, or not-for-profit sectors.
Author contribution
C.C.: conceived and designed analysis, original draft writing. S.C., M.B., H.C., P.B.: original draft writing. M.A.I., K.D.: review and editing final version.
Conflicts of interest disclosure
All authors report no conflicts of interest relevant to this article.
Research registration unique identifying number (UIN)
Name of the registry: not applicable.
Unique identifying number or registration ID: not applicable.
Hyperlink to your specific registration (must be publicly accessible and will be checked): not applicable.
Guarantor
Md. Aminul Islam, COVID-19 Diagnostic Lab, Department of Microbiology, Noakhali Science and Technology University, Noakhali 3814, Bangladesh. E-mail: aminulmbg@gmail.com
Provenance and peer review
Not commissioned, internally peer-reviewed.
Data statement
The data in this correspondence article is not sensitive in nature and is accessible in the public domain. The data is therefore available and not of a confidential nature. No specific data was collected for the above manuscript.
Acknowledgments
All authors are thankful to their respective institutes and universities.
Footnotes
C.C., S.C., M.B. contributed equally for this study.
Sponsorships or competing interests that may be relevant to content are disclosed at the end of this article.
Published online 16 February 2023
Contributor Information
Chiranjib Chakraborty, Email: drchiranjib@yahoo.com.
Srijan Chatterjee, Email: srijan221199@gmail.com.
Manojit Bhattacharya, Email: mbhattacharya09@gmail.com.
Hitesh Chopra, Email: chopraontheride@gmail.com.
Prosun Bhattacharya, Email: prosun@kth.se.
Md. Aminul Islam, Email: aminulmbg@gmail.com.
Kuldeep Dhama, Email: kdhama@rediffmail.com.
References
- 1. Kadam SB, Sukhramani GS, Bishnoi P, et al. SARS-CoV-2, the pandemic coronavirus: molecular and structural insights. J Basic Microbiol 2021;61:180–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Chakraborty C, Saha A, Sharma AR, et al. D614G mutation eventuates in all VOI and VOC in SARS-CoV-2: Is it part of the positive selection pioneered by Darwin? Mol Ther Nucleic Acids 2021;26:237–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Chakraborty C, Sharma AR, Bhattacharya M, et al. Evolution, mode of transmission, and mutational landscape of newly emerging SARS-CoV-2 variants. mBio 2021;12:e0114021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Chakraborty C, Bhattacharya M, Sharma AR. Present variants of concern and variants of interest of severe acute respiratory syndrome coronavirus 2: their significant mutations in S-glycoprotein, infectivity, re-infectivity, immune escape and vaccines activity. Rev Med Virol 2022;32:e2270. [Google Scholar]
- 5. Bhattacharya M, Chatterjee S, Sharma AR, et al. D614G mutation and SARS-CoV-2: impact on S-protein structure, function, infectivity, and immunity. Appl Microbiol Biotechnol 2021;105:9035–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hou Y, Zhao S, Liu Q, et al. Ongoing positive selection drives the evolution of SARS-CoV-2 genomes. Genom Proteom Bioinform 2022. doi: 10.1016/j.gpb.2022.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Christensen PA, Olsen RJ, Long SW, et al. Signals of significantly increased vaccine breakthrough, decreased hospitalization rates, and less severe disease in patients with coronavirus disease 2019 caused by the Omicron variant of severe acute respiratory syndrome coronavirus 2 in Houston, Texas. Am J Pathol 2022;192:642–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Islam F, Dhawan M, Nafady MH, et al. Understanding the omicron variant (B.1.1.529) of SARS-CoV-2: mutational impacts, concerns, and the possible solutions. Ann Med Surg (Lond) 2022;78:103737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Torjesen I. Covid-19: Omicron may be more transmissible than other variants and partly resistant to existing vaccines, scientists fear. BMJ 2021;375:n2943. [DOI] [PubMed] [Google Scholar]
- 10. Liu Y, Rocklöv J. The effective reproductive number of the Omicron variant of SARS-CoV-2 is several times relative to Delta. J Travel Med 2022;29:taac037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bhattacharya M, Chatterjee S, Sharma AR, et al. Delta variant (B.1.617.2) of SARS-CoV-2: current understanding of infection, transmission, immune escape, and mutational landscape. Folia Microbiol (Praha) 2022:1–12. doi: 10.1007/s12223-022-01001-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Trucchi E, Gratton P, Mafessoni F, et al. Population dynamics and structural effects at short and long range support the hypothesis of the selective advantage of the G614 SARS-CoV-2 spike variant. Mol Biol Evol 2021;38:1966–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Dokainish HM, Sugita Y. Structural effects of spike protein D614G mutation in SARS-CoV-2. Biophys J 2022. doi: 10.1016/j.bpj.2022.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Gellenoncourt S, Saunders N, Robinot R, et al. The spike-stabilizing D614G mutation interacts with S1/S2 cleavage site mutations to promote the infectious potential of SARS-CoV-2 variants. J Virol 2022;96:e0130122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ozono S, Zhang Y, Ode H, et al. SARS-CoV-2 D614G spike mutation increases entry efficiency with enhanced ACE2-binding affinity. Nat Commun 2021;12:848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Volz E, Hill V, McCrone JT, et al. Evaluating the effects of SARS-CoV-2 spike mutation D614G on transmissibility and pathogenicity. Cell 2021;184:64–75e11. [DOI] [PMC free article] [PubMed] [Google Scholar]


