Abstract
Herpes simplex virus type 1 DNA isomerization was studied by using a viral mutant, 5B8, lacking the unique SpeI site of its parent, SC16. In coinfected cells, SC16 genomic long segments flanked 5B8 genomes in all possible orientations with similar frequencies. Thus, recombination between progeny of different replication templates is sufficient to explain genomic isomerization.
Herpes simplex virus (HSV) genomes are linear molecules (20) comprising covalently linked long (L) and short (S) segments, each containing unique sequences (UL and US) bracketed by inverted repeats (26). The genome has three origins of replication (23), one in UL (OriL) and a diploid origin in the short repeated region (OriS). Replication intermediates are concatemers (1, 14–16) suggesting that a rolling circle mechanism is responsible for HSV DNA synthesis (21).
L and S segments freely invert relative to each other (4, 11), generating four equimolar genomic isomers. Cleavage of concatemers at alternative L-S junctions can generate two isomers from a single genomic length replication template (15), but formation of the remaining isomers requires homologous recombination between sequences in the large repeated regions of DNA flanking L-S junctions (17, 19, 24, 27, 29–32). Inversion of adjacent L segments in HSV concatemers has been observed 3 to 4 h after infection, when replicating DNA is still devoid of free genomic termini (34). These data raised the possibility that recombination might be an integral component of early DNA replication, and this hypothesis is supported by the knowledge that recombination is mediated by the viral replication machinery (24) and replication and recombination are temporally linked (5, 34). It has been suggested that recombination between one repeat region and its as-yet-unreplicated counterpart, generating a double rolling circle, might be the primary mechanism responsible for generating the concatemeric precursors of all four isomers (8, 9, 34).
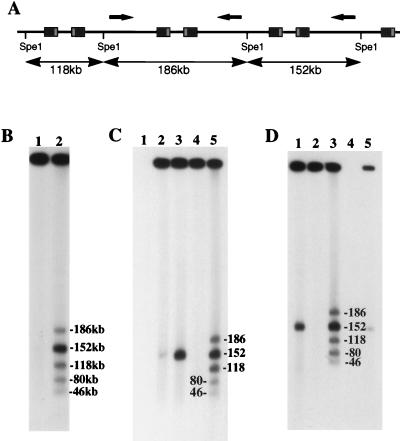
Double rolling circle replication is dependent on bidirectional initiation of DNA synthesis (8). While neither OriL nor OriS is exclusively required for HSV replication (13, 22), we reasoned that, if recombination is linked with the initial phase of HSV DNA replication, then the genomic location of one origin or the other might favor isomerization. To determine whether L-segment inversion in concatemeric DNA is dependent on OriL, we compared the structures of HSV concatemers produced by wild-type HSV-1 strain SC16 (12) and ts+7, a virus which lacks functional OriL (22). Agarose plugs, pre pared as described previously (34) from uninfected Vero cells (ATCC CCL81) or Vero cells infected for 6 h (20 PFU/cell), were electrophoresed with field inversion (3) to remove unit-length viral genomes, which otherwise complicate analysis of replication intermediates (34). After removal of genomic-length DNA, plugs containing replicating DNA were recovered from the gel and treated with SpeI, which cuts HSV DNA once only, in UL, approximately 46 kb from the left end of the prototype genomic configuration. Fragments were separated by field inversion gel electrophoresis (FIGE) and identified by Southern hybridization, as described (34). All three fragments associated with L-segment inversion (Fig. 1A) were detected in similar proportions in SC16 (Fig. 1B) and ts+7 (Fig. 1C) concatemers. It was concluded that L-segment inversion is not dependent on initiation of DNA synthesis by OriL. As previously reported (34), concatemers were flanked exclusively by genomic L segments (Fig. 1B and C). In a similar approach, strain R7713, lacking both copies of OriS (17), was used to show that frequent inversion of adjacent L segments in concatemeric DNA is not dependent on OriS (Fig. 1D).
FIG. 1.
Analysis of fragments generated by SpeI digestion of high-molecular-weight HSV DNA purified from infected cells by FIGE. (A) Schematic representation of SpeI fragments generated when adjacent L segments are in opposite orientations (arrows). (B) Southern blot of SpeI fragments from SC16-infected cells. Bands were sized, as marked, by comparison with concatemeric λ phage DNA and correspond to expected fragments (lane 2). Terminal fragments (80 and 46 kb) corresponded exclusively to L-segment termini, as previously described (34). Undigested material (lane 1) contained no residual genomic-length HSV DNA. (C) Southern blot showing fragments indicative of L-segment inversion (lane 5) in high-molecular-weight DNA purified from ts+7-infected cells. Terminal fragments were from the L segment only (80 and 46 kb). Removal of unit-length HSV DNA (152 kb) was shown to be complete by comparing undigested samples before and after FIGE purification of high-molecular-weight material (lanes 3 and 4, respectively). The migration of unit-length genomes was marked by using total DNA extracted 5.5 h after infection of cells with SC16 (lane 2). There was no background hybridization to uninfected-cell DNA (lane 1). (D) Southern blot showing L-segment inversion and L-segment termini in high-molecular-weight DNA from R7713-infected cells. Comparison between undigested total infected-cell DNA (lane 1) and high-molecular-weight DNA (lane 2) showed that unit-length genomes were removed from the analyzed sample. Lane 4, not loaded; lane 5, total undigested DNA 4 h after SC16 infection.
To determine whether adjacent, inverted L segments in HSV concatemers may be derived from different replication templates, a virus lacking an SpeI site was generated with a strategy similar to that used previously to eliminate XbaI sites (18). Briefly, BHK21 cells (ATCC CCL10) were transfected with SpeI-digested SC16 virion DNA by a standard calcium phosphate coprecipitation method and monitored until plaques were observed (2 days). To test transfection progeny for SpeI sites, SpeI-treated DNA from a proportion of the culture was analyzed by FIGE. Approximately 50% of the progeny DNA appeared to be resistant to digestion, and therefore, progeny viruses were plaque purified from the remainder of the infected cells. Viral DNA prepared from 5 of 11 plaque-purified viruses could not be cleaved with SpeI, and one virus, 5B8, was arbitrarily selected for further purification and study. In Vero cells, there was no discernible difference in the single-step replication kinetics (10 PFU/cell) of 5B8 and SC16 (Fig. 2A), indicating that replication in vitro was not impaired by deletion of the SpeI site.
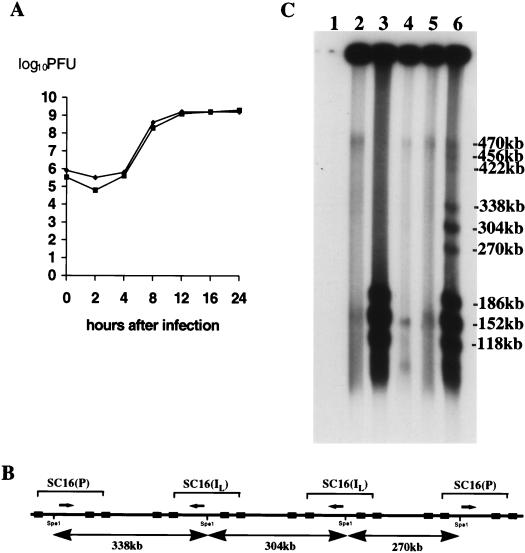
FIG. 2.
(A) Single-step growth curves (10 PFU/cell) of SC16 and 5B8 in Vero cells. (B) SC16-5B8 recombination; expected SpeI fragments from concatemers comprising L segments of 5B8 flanked by SC16. (C) FIGE and Southern blot analysis of fragments generated by SpeI digestion of high-molecular-weight infected-cell DNA (17 h after infection). Lane 1, uninfected Vero cells; lanes 2 and 3, high-molecular-weight SC16 DNA; lane 4, high-molecular-weight 5B8; lanes 5 and 6, mixed infection with SC16 and 5B8. Samples in lanes 3, 4, and 6 were treated with SpeI. Fragments, as marked, were sized by comparison with high-molecular-weight markers. A small amount of residual genomic-length DNA was detected in the sample loaded into lane 4. The filter was autoradiographed for 1.5 h.
Infected-cell DNA was analyzed by FIGE at various times after infection with SC16, 5B8, or both viruses (a total of 20 PFU/cell). In coinfected cells, the presence of SC16-5B8 recombinant concatemers was disclosed by the presence of SpeI fragments of 338, 304, and 270 kb (e.g., 17 h after infection) (Fig. 2B) and, according to phosphorimage analysis, the ratio between the intensities of fragments, in order of increasing size, was approximately 1:2:1. Fragments of 490, 456, and 422 kb were also detected, with a similar distribution of intensities. The intensities of each set of fragments decreased markedly with increasing size. It is thought that frequent branching of replicating DNA limits the amount that can be released in a noncomplex form by SpeI digestion (25), and hence, it is expected that the amount of digested DNA capable of entering a gel would decrease as the distance between SpeI sites increases. Consequently, it was not possible to determine directly the frequency with which SC16 and 5B8 genomes are adjacent by comparing the intensities of corresponding bands in each set. However, the relative ratios of the fragments in each set indicated that SC16 L segments are present in all possible orientations with equal frequency, irrespective of the number of intercalated 5B8 genomes. Similar results were obtained 6 and 13.5 h after infection (Fig. 3). Hence, the ratios between the amounts of adjacent SC16 L segments and adjacent SC16-5B8 L segments were the same at all times studied. While these data do not exclude the possibility that recombination might also occur between homologous sequences within the concatemeric product of a single template, they demonstrate that this is not essential for genome isomerization. These data strongly suggest that recombination between sequences derived from different replication templates is sufficiently frequent to account for the isomers found in infected cells. In prior experiments, cleavage of concatemers into unit-length progeny genomes was first detected ca. 6 h after infection. In the present work, the amount of high-molecular-weight infected-cell DNA recovered prior to 6 h was not sufficient to demonstrate the presence of recombinants by SpeI digestion. These data imply that complex branching of replicating DNA, which impairs electrophoretic migration of the majority of SpeI fragments, is a feature of not only late but also early replication.
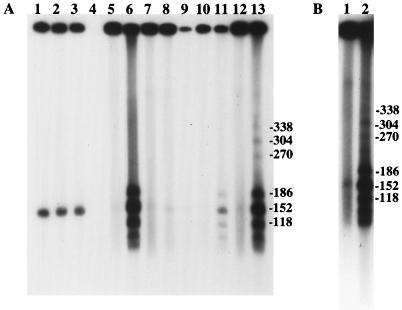
FIG. 3.
(A) Analysis of total and high-molecular-weight DNAs recovered from infected cells at various times after infection. Lanes 1 to 3, total infected-cell DNA recovered from cells infected with SC16, 5B8, or both viruses, respectively, demonstrating the appearance of unit-length progeny DNA (152 kb), 6 h after infection. Lane 4, uninfected-cell DNA; lanes 5 and 6, SC16; lanes 7 and 8, 5B8; lanes 9 to 13, mixed infection. Agarose plugs loaded into lanes 6, 8, 9, 11, and 13 were treated with SpeI. Lanes 5 to 8 and 12 and 13 show results 13.5 h after infection. Lanes 10 and 11 show results 6 h after infection. Lane 9, 4 h after infection, contained insufficient DNA for detection of putative recombinants. The filter was autoradiographed for 15 min. (B) Reanalysis of samples made 6 h after infection with SC16 and 5B8. Lane 1, uncut; lane 2, SpeI cut. The filter was autoradiographed for 5 h.
While these data are compatible with an uncomplicated rolling circle replication model, rolling circle DNA synthesis does not explain the structure of high-molecular-weight HSV DNA molecules found during the early stages of replication (34). Furthermore, the existence of three internal origins of DNA replication in the HSV genome and the dependence of early replication on topoisomerase II (7) suggest that DNA synthesis might be initiated via theta structures and generate catenated circles, but if this is the case, then none of the known origins is uniquely required. Rather, if replication of HSV DNA depends on OriL or OriS, then any one origin will alone suffice. In this work, the same result is shown for segment inversion, i.e., isomerization is not coupled to a replication strategy initiated exclusively by OriL or OriS.
A variation of the suggestion that plasmid amplification commences with theta structures is the idea that recombination creates a double rolling circle, akin to the replication of the 2μm plasmid of Saccharomyces cerevisiae (6, 10). In favor of a 2μm-like model is its ability to explain the apparent temporal (34) and functional (24) links between template amplification and segment inversion. However, if such a model is envisaged, it must incorporate the fact that recombination responsible for segment inversion in HSV is, unlike the 2μm plasmid, not site specific (19).
An intriguing possibility is that isomerization might be linked to the formation of circular replication templates from linear virion DNA. Linearized plasmids containing OriS and terminal a sequences can form circular replication templates in HSV-infected cells by recombination (28, 33), suggesting that recombination rather than fusion of free genomic termini is the primary mechanism responsible for circularization of viral DNA. Formation of circular molecules by recombination might be expected to generate multimeric replication templates, providing an alternative explanation for the early formation of hybrid SC16-5B8 concatemers. In such circumstances, isomerization could still be the sole result of homologous recombination between repeated sequences within the concatemers generated from a single, multimeric circle, fueling debate about a link between segment inversion and mechanisms responsible for template amplification (8, 9). While it has not been possible to visualize multimeric circular forms comprising intact HSV genomes, dimers have been observed following in vitro recombination between HSV amplicons and replication of amplicons in vivo (2, 5).
Acknowledgments
We thank B. Roizman for virus strain 7713 and P. Schaffer for ts+7.
This work was supported by the National Health and Medical Research Council of Australia.
REFERENCES
- 1.Ben-Porat T, Kaplan A S, Stehn B, Rubenstein A S. Replication of herpes DNA. II. Sedimentation characteristics of newly synthesized DNA. Virology. 1977;79:292–301. doi: 10.1016/0042-6822(77)90356-7. [DOI] [PubMed] [Google Scholar]
- 2.Bruckner R C, Dutch R E, Zemelman B V, Mocarski E S, Lehman J R. Recombination in vitro between herpes simplex virus type 1 a sequences. Proc Natl Acad Sci USA. 1992;89:10950–10954. doi: 10.1073/pnas.89.22.10950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Carle G F, Frank M, Olson M. Electrophoretic separation of large DNA molecules by periodic inversion of the electric field. Science. 1986;232:65–68. doi: 10.1126/science.3952500. [DOI] [PubMed] [Google Scholar]
- 4.Delius H, Clements J B. A partial denaturation map of herpes simplex virus type 1 DNA: evidence for inversions of the unique DNA regions. J Gen Virol. 1976;33:125–133. doi: 10.1099/0022-1317-33-1-125. [DOI] [PubMed] [Google Scholar]
- 5.Dutch R E, Bruckner R C, Mocarski E S, Lehman I R. Herpes simplex virus type 1 recombination: role of DNA replication and viral a sequences. J Virol. 1992;66:277–285. doi: 10.1128/jvi.66.1.277-285.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Futcher A B. Copy number amplification of the 2 μm circle plasmid of Saccharomyces cerevisiae. J Theor Biol. 1986;119:197–204. doi: 10.1016/s0022-5193(86)80074-1. [DOI] [PubMed] [Google Scholar]
- 7.Hammarsten O, Yao X, Elias P. Inhibition of topoisomerase II by ICRF-193 prevents efficient replication of herpes simplex virus type 1. J Virol. 1996;70:4523–4529. doi: 10.1128/jvi.70.7.4523-4529.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hammerschmidt W, Mankertz J. Herpes viral DNA replication: between the known and unknown. Semin Virol. 1991;2:257–269. [Google Scholar]
- 9.Hammerschmidt W, Sugden B. DNA replication of herpes viruses during the lytic phase of their life cycles. Mol Biol Med. 1990;7:45–57. [PubMed] [Google Scholar]
- 10.Hartley J L, Donelson J E. Nucleotide sequences of the yeast plasmid. Nature. 1980;286:860–865. doi: 10.1038/286860a0. [DOI] [PubMed] [Google Scholar]
- 11.Hayward G S, Jacob R J, Wadsworth S C, Roizman B. Anatomy of herpes simplex virus DNA: evidence for four populations of molecules that differ in the relative orientations of their long and short components. Proc Natl Acad Sci USA. 1975;72:4243–4247. doi: 10.1073/pnas.72.11.4243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hill T J, Field H J, Blyth W A. Acute and recurrent infection with herpes simplex virus in the mouse: a model for studying latency and recurrent disease. J Gen Virol. 1975;28:341–353. doi: 10.1099/0022-1317-28-3-341. [DOI] [PubMed] [Google Scholar]
- 13.Igarashi K, Fawl R, Roller R J, Roizman B. Construction and properties of a recombinant herpes simplex virus 1 lacking both S-component origins of DNA synthesis. J Virol. 1993;67:2123–2132. doi: 10.1128/jvi.67.4.2123-2132.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jacob R J, Roizman B. Anatomy of herpes simplex virus DNA. VIII. Properties of the replicating DNA. J Virol. 1977;23:394–411. doi: 10.1128/jvi.23.2.394-411.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jacob R J, Morse L S, Roizman B. Anatomy of herpes simplex virus DNA. XIII. Accumulation of head-to-tail concatemers in nuclei of infected cells and their role in the generation of the four isomeric arrangements of viral DNA. J Virol. 1979;29:448–457. doi: 10.1128/jvi.29.2.448-457.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jongeneel C V, Bachenheimer S L. Structure of replicating herpes simplex virus DNA. J Virol. 1981;39:656–660. doi: 10.1128/jvi.39.2.656-660.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Longnecker R, Roizman B. Generation of an inverting herpes simplex virus 1 mutant lacking the L-S junction a sequences, an origin of DNA synthesis, and several genes including those specifying glycoprotein E and the α47 gene. J Virol. 1986;58:583–591. doi: 10.1128/jvi.58.2.583-591.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.MacLean A R, Brown S M. Generation of a herpes simplex virus type 1 variant devoid of Xba1 sites. J Gen Virol. 1987;68:1165–1171. doi: 10.1099/0022-1317-68-4-1165. [DOI] [PubMed] [Google Scholar]
- 19.Martin D W, Weber P C. The a sequence is dispensable for isomerization of the herpes simplex virus type 1 genome. J Virol. 1996;70:8801–8812. doi: 10.1128/jvi.70.12.8801-8812.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McGeoch D J, Dalrymple M A, Davison A J, Dolan A, Frame M C, McNab D, Perry L J, Scott J E, Taylor P. The complete DNA sequence of the long unique region in the genome of herpes simplex virus type 1. J Gen Virol. 1988;69:1531–1574. doi: 10.1099/0022-1317-69-7-1531. [DOI] [PubMed] [Google Scholar]
- 21.Poffenberger K L, Roizman B. A noninverting genome of a viable herpes simplex virus 1: presence of head-to-tail linkages in packaged genomes and requirements for circularization after infection. J Virol. 1985;53:587–595. doi: 10.1128/jvi.53.2.587-595.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Polvino-Bodnar M, Orberg P K, Schaffer P A. Herpes simplex virus type 1 oriL is not required for virus replication or for the establishment and reactivation of latent infection in mice. J Virol. 1987;61:3528–3535. doi: 10.1128/jvi.61.11.3528-3535.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Roizman B, Sears A E. Herpes simplex viruses and their replication. In: Fields B, Knipe D M, editors. Virology. New York, N.Y: Raven Press Ltd.; 1990. pp. 1795–1841. [Google Scholar]
- 24.Sarisky R T, Weber P C. Requirement for double-strand breaks but not for specific DNA sequences in herpes simplex virus type 1 genome isomerization events. J Virol. 1994;68:34–47. doi: 10.1128/jvi.68.1.34-47.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Severini A, Morgan A R, Tovell D R, Tyrell D L. Study of the structure of HSV-1 replicative intermediates by pulsed-field gel electrophoresis. Virology. 1994;200:428–435. doi: 10.1006/viro.1994.1206. [DOI] [PubMed] [Google Scholar]
- 26.Sheldrick P, Berthelot N. Inverted repetitions in the chromosome of herpes simplex virus. Cold Spring Harbor Symp Quant Biol. 1975;39:667–678. doi: 10.1101/sqb.1974.039.01.080. [DOI] [PubMed] [Google Scholar]
- 27.Smiley J R, Duncan J, Howes M. Sequence requirements for DNA rearrangements induced by the terminal repeat of herpes simplex virus type 1 KOS DNA. J Virol. 1990;64:5036–5050. doi: 10.1128/jvi.64.10.5036-5050.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Smiley J R, Fong J R, Leung W C. Construction of a double-jointed herpes simplex viral DNA molecule: inverted repeats are required for segment inversion and direct repeats promote deletions. Virology. 1981;113:345–362. doi: 10.1016/0042-6822(81)90161-6. [DOI] [PubMed] [Google Scholar]
- 29.Smiley J R, Lavery C, Howes M. The herpes simplex virus type 1 (HSV-1) a sequence serves as a cleavage/packaging signal but does not drive recombinational genome isomerization when it is inserted into the HSV-2 genome. J Virol. 1992;66:7505–7510. doi: 10.1128/jvi.66.12.7505-7510.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Varmuza S L, Smiley J R. Signals for site-specific cleavage of HSV DNA: maturation involves two separate cleavage events at sites distal to the recognition sequences. Cell. 1985;41:793–802. doi: 10.1016/s0092-8674(85)80060-x. [DOI] [PubMed] [Google Scholar]
- 31.Weber P C, Challberg M D, Nelson N J, Levine M, Glorioso J C. Inversion events in the HSV-1 genome are directly mediated by the viral DNA replication machinery and lack sequence specificity. Cell. 1988;54:369–381. doi: 10.1016/0092-8674(88)90200-0. [DOI] [PubMed] [Google Scholar]
- 32.Weber P C, Levine M, Glorioso J C. Recombinogenic properties of herpes simplex virus type 1 DNA sequences resident in simian virus 40 minichromosomes. J Virol. 1990;64:300–306. doi: 10.1128/jvi.64.1.300-306.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yao X-D, Matecic M, Elias P. Direct repeats of the herpes simplex virus a sequence promote nonconservative homologous recombination that is not dependent on XPF/ERCC4. J Virol. 1997;71:6842–6849. doi: 10.1128/jvi.71.9.6842-6849.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang X, Efstathiou S, Simmons A. Identification of novel herpes simplex virus replicative intermediates by field inversion electrophoresis: implications for viral DNA amplification strategies. Virology. 1994;202:530–539. doi: 10.1006/viro.1994.1375. [DOI] [PubMed] [Google Scholar]