Abstract
The Epstein-Barr virus (EBV) EBNA2 protein is a transcriptional activator that controls viral latent gene expression and is essential for EBV-driven B-cell immortalization. EBNA2 is expressed from the viral C promoter (Cp) and regulates its own expression by activating Cp through interaction with the cellular DNA binding protein CBF1. Through regulation of Cp and EBNA2 expression, EBV controls the pattern of latent protein expression and the type of latency established. To gain further insight into the important regulatory elements that modulate Cp usage, we isolated and sequenced the Cp regions corresponding to nucleotides 10251 to 11479 of the EBV genome (−1079 to +144 relative to the transcription initiation site) from the EBV-like lymphocryptoviruses found in baboons (herpesvirus papio; HVP) and Rhesus macaques (RhEBV). Sequence comparison of the approximately 1,230-bp Cp regions from these primate viruses revealed that EBV and HVP Cp sequences are 64% conserved, EBV and RhEBV Cp sequences are 66% conserved, and HVP and RhEBV Cp sequences are 65% conserved relative to each other. Approximately 50% of the residues are conserved among all three sequences, yet all three viruses have retained response elements for glucocorticoids, two positionally conserved CCAAT boxes, and positionally conserved TATA boxes. The putative EBNA2 100-bp enhancers within these promoters contain 54 conserved residues, and the binding sites for CBF1 and CBF2 are well conserved. Cp usage in the HVP- and RhEBV-transformed cell lines was detected by S1 nuclease protection analysis. Transient-transfection analysis showed that promoters of both HVP and RhEBV are responsive to EBNA2 and that they bind CBF1 and CBF2 in gel mobility shift assays. These results suggest that similar mechanisms for regulation of latent gene expression are conserved among the EBV-related lymphocryptoviruses found in nonhuman primates.
Epstein-Barr virus (EBV) is a human lymphocryptovirus associated with various malignancies, including Burkitt’s lymphoma (BL), Hodgkin’s disease, nasopharyngeal carcinoma, and lymphomas in immunosuppressed individuals (34). EBV establishes a lifelong infection in the human host, where B lymphocytes are the primary latent reservoir from which occasional virus reactivation occurs (34).
EBV also establishes latent infection in B lymphocytes in vitro and transforms them into continuously proliferating lymphoblastoid cell lines (LCLs). Molecular genetic techniques have demonstrated that several EBV nuclear antigens (EBNAs) and an integral membrane protein (LMP1) are required for lymphocyte transformation in vitro (19, 36). EBNA2 plays a central role in the process of transformation, as it activates the expression of the other EBNAs, LMP1 and LMP2 proteins, and various cellular proteins associated with the transformed phenotype (1, 5, 7, 20, 25, 35, 41, 52–56, 60, 61).
The regulation of EBNA expression in EBV-infected cells is likely to be important in viral persistence and the development of EBV-associated malignancies. With the exception of EBNA1, all EBNAs possess epitopes which induce a strong cytotoxic T-cell response (22, 34). Downregulation of EBNA expression may allow a latently infected B cell to escape immune surveillance and also permit proliferation of malignant EBV-infected cells. In healthy individuals, latent EBV infection appears to be primarily confined to resting B cells. The only EBV gene expressed in these cells is LMP2a, a pattern of gene expression termed latency 0 (29, 30, 51). In BL cells, only EBNA1 is expressed (latency I) (34, 51). In Hodgkin’s disease, nasopharyngeal carcinoma, and T-cell lymphomas, EBNA1 and one or both LMPs are expressed (latency II) (34, 51). All EBNAs and LMPs are expressed during acute infectious mononucleosis, in lymphoproliferative syndromes in immunocompromised individuals, and in LCLs (latency III) (34, 51). The association of these specific patterns of EBNA expression with various physiologic and pathologic states underscores the importance of EBNA gene regulation in EBV persistence and EBV-associated oncogenesis.
Upon in vitro infection of B lymphocytes, the first EBV genes expressed are EBNA-LP and EBNA2, transcribed from the W promoter (Wp) as bicistronic mRNAs (2, 3, 40, 56). Thirty-six hours postinfection, transcription of EBNA2 and the other EBNAs switches to the upstream C promoter (Cp) (2, 3, 40, 57). EBNA2 protein is first detectable at 12 h postinfection and reaches maximal levels between 32 and 46 h (2). These findings are consistent with a role for EBNA2 in activating Cp and mediating the switch from Wp transcription. Elucidating the mechanisms that regulate transcription of EBNA2 and the other EBNAs is therefore important for understanding the process of EBV-mediated B-lymphocyte transformation.
Transcription from Cp leads to concomitant expression of EBNA2 and the other EBNAs. Thus, Cp activity is the major difference between latency III and the other latency programs. Several mechanisms that may regulate Cp activity have been described. First, EBNA1 binds to an enhancer 3 kb upstream of Cp and activates Cp in transfection assays (32, 33, 47). Second, glucocorticoid response elements (GREs) about 900 bp upstream of Cp respond to glucocorticoids and activate Cp in vitro but are not required for transformation in vitro (21, 45, 46, 49). Transient transfections have also demonstrated that the Cp GREs are capable of responding to glucocorticoids (21, 45, 49). Third, the EBNA2-responsive enhancer (E2RE) in Cp is critical for Cp function in vitro and in vivo (17, 23, 48, 59). Deletion of E2RE from the viral genome results in loss of Cp activity in infected cells (59). Finally, methylation of CpG sequences in the promoter region downregulates Cp (28, 38, 44). Whereas LCLs (latency III) show no evidence of Cp methylation, B cells in latencies I and II are hypermethylated in different regions of Cp (28, 38, 44). Consistent with a role for methylation in the silencing of Cp, in vitro treatment of type I BL cells with 5-azacytidine, an inhibitor of DNA methylation, led to activation of Cp (27, 38).
In order to gain further insight into the significance of Cp and the transcriptional elements that regulate its expression, a segment of the genome from the BCRF1 poly(A) site to the end of the first Cp exon was sequenced from the EBV-like lymphocryptoviruses herpesvirus papio (HVP) and rhesus macaque EBV (RhEBV). These sequences were compared to EBV sequences, and the abilities of these promoters to respond to EBNA2 and to bind CBF1 and CBF2 cellular proteins were evaluated.
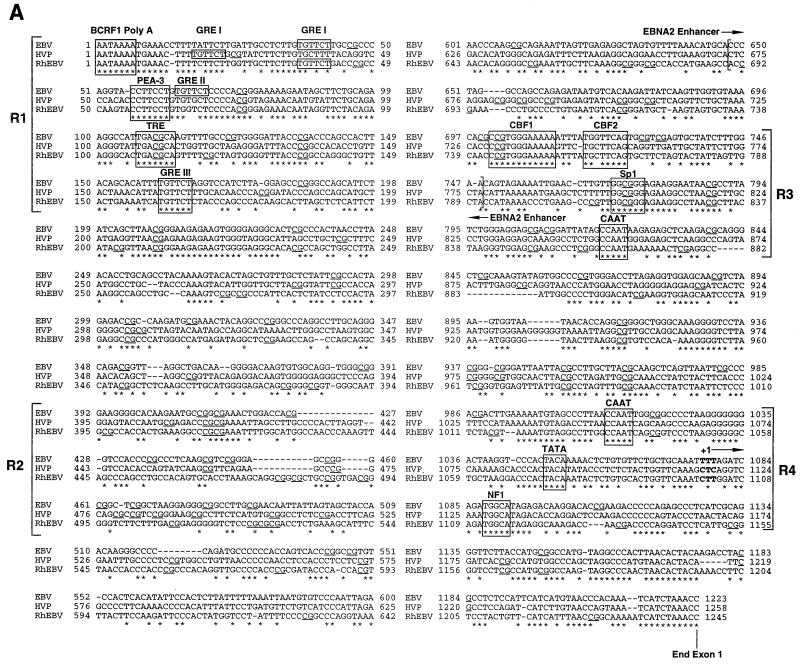
DNA sequences of the Cp from HVP and RhEBV and their alignment with the EBV Cp.
Several cell lines were used for our studies. DG75 and CA46 are EBV-negative BL cell lines. B95-8 is a type 1-EBV-transformed marmoset cell line, and P3HR1 is a type 2-EBV-transformed cell line containing an immortalization-defective EBV strain. 26CB-1 (obtained from the American Type Culture Collection as CRL-1495) and Ba65 are baboon cell lines transformed with HVP (12). H254 (gift from Micheal Murphy-Corb) and LCL 8664 (CRL-1805) are rhesus macaque cell lines transformed with RhEBV. Previous sequence analysis of a cosmid clone derived from the Ba65 cell line revealed sequences identical to B95-8 EBV DNA in the C1 exon of the HVP Cp (unpublished data and reference 25). Based on this and preliminary analysis by rapid amplification of cDNA 5′ ends of the BCRF2 open reading frame of HVP (unpublished observations), PCR primers that would amplify the putative BCRF2 and Cp sequences from HVP were designed. Briefly, 26CB-1 or BA65 cells were lysed in PBS–1% Triton X-100, boiled, and treated with proteinase K for 1 h. Samples were boiled again for 10 min, and the cleared lysates were used for PCR analysis. From each of the cell lines a 2.0-kb DNA molecule was amplified and cloned directly into pT7Blue-2 (Novagen). The resulting plasmids, pCRL-3 and pBA65 Cp, were derived from 26CB-1 and BA65 cells, respectively. Plasmid inserts were sequenced on both strands with an automated sequencing system (Applied Biosystems). With the sequence information from the HVP clones, two primers were designed for common sequences also present in EBV that amplify RhEBV Cp sequences from the end of the BCRF2 open reading frame to the end of the putative C1 exon. LCL 8664 and H254 were lysed, and DNAs were prepared for PCR amplification as described above. Two clones, pPDL 380A and pPDL 380B, from PCR products of independent cell preparations from H254 were obtained. Two clones, pPDL 394A and pPDL 394B, were also obtained from LCL 8664 in a similar manner. During all PCR amplifications, control samples containing all essential PCR components except cell lysates were consistently negative. Both strands of the RhEBV plasmid inserts were also sequenced with an Applied Biosystems automated sequencing system.
For HVP, clones pCRL-3 and pBA65 Cp were sequenced. The sequence for CRL-3 is shown in Fig. 1A. Six nucleotide differences were observed between these particular isolates and are listed in Fig. 1B. Since only one clone was analyzed from each cell line, it is unclear whether these changes are strain specific or the result of PCR error. For RhEBV, both clones derived from the H254 cell line in independent PCRs were identical. Surprisingly, clones isolated from RhEBV LCL 8664 were also identical in sequence to those derived from H254. Both cell lines were established at the Delta Regional Primate Center and may contain identical or similar viral strains. While the passage history of H254 is somewhat unclear, it may have been established from a virus obtained from LCL 8664 or an animal in the same colony. A sequence alignment of the Cp sequences from the three related viruses is shown in Fig. 1A.
FIG. 1.
(A) Nucleotide sequences of aligned EBV, HVP, and RhEBV Cp sequences from the poly(A)+ site of BCR2 to the end of the C1 exon. Asterisks indicate conserved nucleotides. Binding sites for transcription factors are indicated by boxed sequences. The boundaries of the EBNA2 enhancer are indicated with brackets inside the sequence diagram, and the different functional regions of the Cp are denoted with brackets outside the sequence diagram and are labeled as R1 to R4. The horizontal arrow above the bases shown in bold indicates the transcriptional initiation site. The CpG dinucleotides are underlined. (B) Nucleotide differences between HVP Cp sequences obtained from the 26CB-1 and Ba65 cell lines. Shown are the nucleotide positions from panel A where base differences were detected and the nucleotides present in each strain.
Sequence comparison of the Cps derived from these primate viruses reveals that EBV and HVP Cp sequences are 64% conserved, EBV and RhEBV Cp sequences are 66% conserved, and HVP and RhEBV Cp sequences are 65% conserved relative to each other. The overall GC content of each sequence is 52, 52, and 54% for the EBV, HVP, and RhEBV Cps, respectively. Several sequence elements in the EBV Cp that have been proposed to be important for promoter function have been described. Among them are three GREs (GRE I, II, and III), E2RE, distal and proximal CCAAT boxes, and the TATA box. The sequence alignment in Fig. 1 shows that there are four regions that contain clusters of either functional or putative transcription factor binding sites (regions 1, 3, and 4 [R1, R3, and R4, respectively) and a clustered region containing a high concentration of CpG dinucleotides (R2).
R1 contains putative binding sites for PEA-3 as well as a TPA response element (TRE) (8, 13, 26). Three GREs have been described in the EBV Cp, which is localized about 900 bp upstream of the transcription initiation site (21, 45). The in vivo importance of these elements remains unclear. Only one of these GREs is conserved in both HVP and RhEBV (GRE III). However, HVP Cp has one other potential GRE localized just upstream of GRE I while RhEBV appears to have positionally retained GRE I. Neither primate sequence appears to have conserved GRE II.
R2 contains an island of CpGs in all three sequences even though the levels of sequence homology are significantly diverged between EBV and the primate viruses. The overall GC content of R2 is slightly higher than for the Cp overall and is 65, 63, and 58% for EBV, HVP, and RhEBV, respectively. However, while the average CpG frequencies for the Cp sequences in Fig. 1 are 4.3, 3.3, and 4.0% for EBV, HVP, and RhEBV, respectively, the CpG frequencies nearly double or triple in R2 (11.0, 9.0, and 8.0% for EBV, HVP, and RhEBV, respectively). In Cp-negative cell lines such as Rael, nearly all CpG within the first upstream 400 bp proximal to the transcription initiation site are methylated (37, 38, 44). Extensive analysis of CpG methylation within R2 has not been performed with these cells. Addition of 5-azacytidine, which inhibits methyltransferases, results in demethylation and activation of Cp (27, 38). In addition, previous studies have shown that methylation primarily targets two regions, called methylation-hypersensitive region I (MHRI; which encompasses the EBNA2 enhancer) and MHRII (from −220 to −25 or positions 826 to 1023 in Fig. 1) (37). MHRI seems to control Cp transcriptional activation by EBNA2, while MHRII controls the basal activity of the promoter. The reported important CpG elements in MHRI are localized downstream of the CBF2 binding site, and these sites do not appear to be conserved. However, some of the potential methylation sites present in MHRII are conserved in both HVP and RhEBV.
R3 is part of the EBNA2 enhancer and contains the binding sites for the cellular DNA binding proteins CBF1 and CBF2 (11, 24). The CBF1 recognition sequence is fully conserved in the three promoters, supporting the important role of this protein in Cp activation by EBNA2. We have recently published that the most critical region contributing to CBF2 binding is the GGTTCA sequence found downstream of the CBF1 site (11). In addition to this sequence, the surrounding nucleotides also contribute to the affinity of binding. With the exception of a G→C change found in the primate viral sequences, our mutagenesis results are consistent with the fact that critical sequences required for CBF2 binding are conserved. A distal CCAAT box identified as functionally significant is also conserved (32). Also within this region is a putative Sp1 binding site.
R4 contains the proximal CCAAT box, TATA box, and the transcription initiation site. Just downstream of the TATA box a conserved NF1 site is present (15). The conservation of sequences surrounding these important elements suggests that they are also important for Cp expression.
Analysis of Cp expression from HVP- and RhEBV-transformed B lymphocytes.
Analysis of Cp usage and mapping of the site for initiation of transcription in HVP and RhEBV B cell lines were performed by S1 nuclease protection analysis. Based on the sequence analysis in Fig. 1, oligonucleotide probes that could detect Cp usage in the primate and EBV-transformed cell lines were synthesized. Poly(A)+ RNA was prepared with a fastTrack kit (version 2.0; Invitrogen). Fifty to 100 fmol of kinased oligonucleotide was hybridized to 2 μg of poly(A)+ RNA in 50 μl of hybridization buffer {40 mM PIPES [piperazine-N,N′-bis(2-ethanesulfonic acid), pH 6.4], 1 mM EDTA, 0.4 M NaCl, 0.1% sodium dodecyl sulfate, 50% (vol/vol) formamide} at 37°C for 12 h. The hybridization reaction mixture was diluted to 400 μl with buffer with final concentrations of 0.28 M NaCl, 50 mM NaOAc (pH 4.6), and 4.5 mM ZnSO4 and digested with 20 U of S1 nuclease (Gibco BRL) for 30 min at 37°C. The reaction mixtures were phenol-chloroform extracted and ethanol precipitated. The resulting precipitate was resuspended in loading buffer and fractionated on a 10% denaturing polyacrylamide gel.
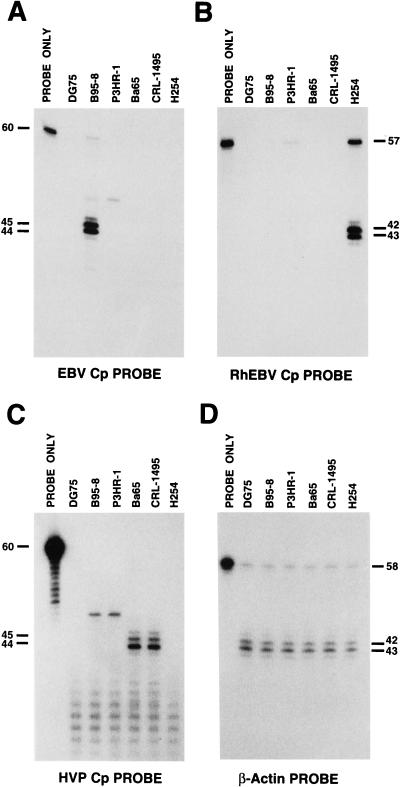
As shown in Fig. 2A, an EBV-specific probe detected transcripts from B95-8 (type 1 EBV) of the predicted sizes that were not detected in the EBV-negative B-cell line DG75 or the other baboon or macaque cell lines. This result is consistent with the results of other studies, and the start site is indicated in Fig. 1 (4, 57). In contrast to previously published work, we detected a minor RNA species in both B95-8 and P3HR1 that was approximately 3 bp larger than the major 44- and 45-bp protected fragments. More stringent hybridization conditions failed to abolish these protected species. It is unclear at this time whether these RNAs were derived from the Cp or were some other cross-reactive RNA derived from another part of the viral genome. Wp-specific probes detected W-specific transcripts from P3HR1 but not B95-8 (data not shown). As shown in panels B and C of Fig. 2, similar patterns of Cp transcripts were detected in RhEBV- and HVP-transformed cells with the Cp probe specific for those viruses. Again the minor RNA species detected in both B95-8 and P3HR1 were also detected with the HVP Cp probe but not the RhEBV Cp probe. The HVP probe also had some nonspecific hybridization with apparent cellular RNA that appears as a ladder of bands in both virus-positive and -negative cells. As a control for RNA abundance and quality, a cellular β-actin probe was also used to detect β-actin RNA in the cell lines (Fig. 2D). The transcription start sites for HVP and RhEBV are indicated in Fig. 1. Even though there is some sequence divergence around the EBV transcription initiation site and among the TATA boxes, it appears that HVP- and RhEBV-transformed cells also initiate Cp transcripts at an analogous location.
FIG. 2.
S1 nuclease protection experiments with specific oligonucleotides. Identical amounts of poly(A)+-selected RNAs derived from the indicated cell lines were subjected to analysis with a C1 oligonucleotide as a hybridization probe. Specific protection fragment sizes are indicated in bases at the side of each panel. (A) EBV Cp probe, 5′-CTCTGGGGGTCTTCGGTGTCCTTGTCTCTATGCCATCTGATCTAAAATTTGCAGCAGAAC-3′; (B) RhEBV Cp probe, 5′-TC CTGGGGTCGTTGGTCTTTGCCTCTATGCCATCTGATCCAAGATTTGA ACCAGTGC-3′; (C) HVP Cp probe, 5′-TACTGGGGGGTCTTGGAGTCCTGGTGTCTATGCCATTTGACCTGAGCTTTGAACCAGTAG-3′; (D) β-actin probe, 5′-ACATAGGAATCCTTCTGACCCATGCCCACCATCACGCCCTGGGAAGGAAAGGACAAGA-3′.
Analysis of CBF1 and CBF2 binding to the HVP and RhEBV Cp sequences.
The sequence alignment of Fig. 1A shows that the EBNA2 enhancer lies in R3. The CBF1 binding site is fully conserved in HVP and RhEBV. The CBF2 binding site has a single change in a critical region for binding. Electrophoretic mobility shift assays (EMSA) were performed as described previously to test the ability of these enhancers to bind CBF1 and CBF2.
By using the full-length HVP and RhEBV EBNA2 enhancers as probes, we found that CBF1 binds with high affinity and that EBNA2 interacts with CBF1 as detected in gel mobility shift assays (data not shown). The CBF2 activity associated with the EBV Cp enhancer was stronger than that associated with the HVP and RhEBV enhancers, indicating that these promoters possess a reduced affinity for this protein.
To quantify the affinity of CBF2 for the response elements present in HVP and RhEBV, competition assays were performed. The affinities of the HVP and RhEBV CBF2 sites were measured by quantifying their ability to compete for CBF2 binding on the EBV sequence. An example of the competitions is shown in Fig. 3A, and a summary of the data is presented in Fig. 3B. This analysis showed that while only 3.2 nM EBV CBF2 oligonucleotide is required to compete 50% of the binding, 15.7 and 12.5 nM HVP and RhEBV oligonucleotides, respectively, are required for the same level of competition. This represents 4.7-fold (HVP) and 3.9-fold (RhEBV) reduced affinities for binding compared with that for the EBV binding site.
FIG. 3.
Affinities of HVP and RhEBV EBNA2 enhancers for CBF2. Nuclear extracts from CA46 cells were mixed with 0-, 2.5-, 5-, 25-, and 125-fold molar excesses of the different unlabeled mutant oligonucleotides. A fixed amount of 32P-EBV CBF2 binding oligonucleotide was then added to the shift reaction mixtures. The reaction mixtures were separated on 4.5% nondenaturing polyacrylamide gels, dried, and autoradiographed. Amounts of CBF2 complex were quantified with a PhophorImager (Molecular Dynamics). (A) The EMSA gel shows binding of nuclear extract containing CBF2 to an EBV 36-mer oligonucleotide probe from positions −339 to −368 alone or with increasing (triangle) amounts of the indicated equivalent EBV or HVP cold competitor oligonucleotide. (B) Summary of competition results. The sequences shown represent the central 30 bp of the 36-mer oligonucleotide. The crucial sequences for CBF2 binding are in bold, with the mutational changes underlined. The numbers in column a are concentrations of unlabeled oligonucleotide required for 50% competition. The numbers in column b are percentages representing the ability of each oligonucleotide to compete for CBF2 relative to that of the wild-type element, which was set at 100%. (C) Direct binding of HVP CBF2 wild-type and mutant oligonucleotides. Nuclear extracts from CA46 cells were incubated with radiolabeled HVP wild-type and mut.1 and mut.2 CBF2 oligonucleotides in the presence or absence of a 100 M excess of competitor oligonucleotide and analyzed by EMSA. For lanes 1 to 5, the HVP CBF2 probe was used; for lanes 6 to 8, the HVP mut.1 CBF2 probe was used; and for lanes 9 to 11, the HVP mut.2 probe was used. Lanes 1, 6, and 9, probe only; lanes 2, 7, and 10, CA46 extract; lane 3, CA46 extract and cold HVP CBF2 oligonucleotide; lanes 5 and 8, CA46 and cold HVP mut.1 CBF2 oligonucleotide; lanes 4 and 11, CA46 and cold HVP mut.2 CBF2 oligonucleotide. Results are averages of values from three independent experiments.
We have previously published that a double transversion mutation in the EBV CBF2 binding site from TT to GG (GGTTCA to GGGGCA) abolishes CBF2 binding (11). When this mutation is introduced into the Cp, the ability to respond to EBNA2 transactivation in transient-transfection assays is strongly reduced. Oligonucleotides representing the CBF2 binding site sequences from HVP and RhEBV were synthesized, and a TT→GG change was introduced into both the HVP and RhEBV CBF2 sequences (Fig. 3B). These mutants were called HVP mut.1 CBF2 and RhEBV mut.1 CBF2 (Fig. 3B). Interestingly, the HVP sequence has two putative CBF2 binding sites. One site lies in the same position as in EBV, and the other site is 3 nucleotides downstream of the first site (site 1, GCTTCA, and site 2, GGTTGA) (Fig. 3B). An oligonucleotide carrying the TT→GG change in the putative second site was also synthesized, and the resulting mutant was called HVP mut.2 CBF2. This mutant was also analyzed in the competition experiments, as shown in Fig. 3B. Competition experiments with HVP mut.1 and mut.2 showed that, as in EBV, changes in the TTGGCA sequence result in decreased affinity for CBF2. RhEBV mut.1 had a 25-fold-reduced binding affinity. In HVP, both sites were found to contribute to CBF2 binding, with the upstream site being the most important. Mutation of the first site completely abolished the binding, while mutation of the downstream site did not abolish binding although it led to a 2.7-fold-reduced activity compared with that of the HVP wild-type sequence.
Figure 3C shows the direct binding of HVP CBF2 wild-type and mutant oligonucleotides in the gel retardation assay. As expected, HVP mut.1 CBF2 was not able to bind CBF2 (lane 7) while HVP mut.2 was still able to bind but with reduced affinity (lane 10). These results confirm that, although the upstream site is the most important for binding, the second site contributes to the binding of the wild-type sequences.
Activation of HVP and RhEBV Cp by EBNA2.
The 1-kb regions from the HVP and RhEBV sequences that correspond to the EBV Cp construction (Cp −1021 to +3) were PCR amplified, and these fragments were cloned upstream of the luciferase reporter gene in the pGL3-Basic vector (Promega). DNA transfections were carried out by using the DEAE dextran method. DG75 cells (5 × 106) were transfected with 2 μg of target plasmid and the amounts of effector plasmid indicated in the legend to Fig. 3. The final DEAE dextran concentration in the transfection reaction mixture was 1 mg/ml. Transfected cells were harvested after 2 days of incubation and lysed, and the luciferase activity was measured according to the instructions of the manufacturer. A constitutively expressing chloramphenicol acetyltransferase reporter vector (pCAT control; Promega) was used as the internal control for transfections, and the values from luciferase assays were normalized to the chloramphenicol acetyltransferase activity.
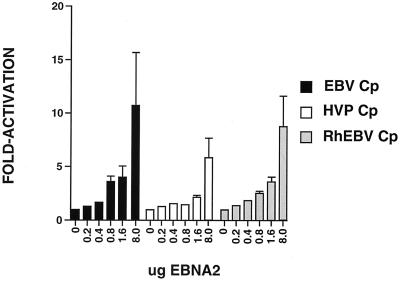
DG75 cells were transfected with the Cp constructs, and their ability to respond to an EBNA2-expressing plasmid was evaluated. EBNA2 activated the EBV Cp to a maximum of 11-fold, while it activated the HVP and RhEBV promoters 6-fold and 9-fold, respectively (Fig. 4). At the highest doses of EBNA2 these differences between the promoters are not statistically significant. At lower doses of EBNA2 (0.8 and 1.6 μg of EBNA2), there was a statistically significant difference in the ability of EBNA2 to transactivate the EBV Cp about 5-fold and it was able to transactivate the HVP and RhEBV Cp promoters only 1.5- to 3-fold.
FIG. 4.
Analysis of the EBNA2 responsiveness of EBV, HVP, and RhEBV Cps. DG75 cells were transfected with 2 μg of target plasmid and 0, 0.2, 0.4, 0.8, 1.6, and 8.0 μg of effector EBV EBNA2-expressing plasmid (24). Results are averages of values from five independent experiments. Standard errors of the means are indicated with the T bars.
In summary, to gain further insight into the regulatory elements that modulate the expression of the viral latency Cp, the Cp regions corresponding to nucleotides 10251 to 11479 of the EBV genome (+144 to −1079 relative to the transcription initiation site) from the EBV-like lymphocryptoviruses HVP and RhEBV were cloned and sequenced. The Cp regions from these viruses are about 65% identical. Earlier studies have shown that HVP and EBV have approximately 40% overall DNA homology and indicate that Cp is more highly conserved than the rest of the genomes (9, 16). No experiments to determine general hybridization kinetics between EBV and RhEBV have been carried out. Among some of the HVP gene sequences, the DNA identities between LMP1 (43%), Fp (also called Qp) (86%), EBNA1 (63%), and ori-Lyt (89%) range from 43 to 89% compared to the identities between similar regions in EBV (10, 42, 43, 58). The RhEBV sequences for LMP1 (44%), Fp/Qp (80%), EBNA2 (41%), and GP350 (57%) range from 44 to 80% compared to the identities between similar regions of EBV (10, 14, 31, 43). While these genes represent only a small portion of the genome, it appears that Cps are somewhat less homologous than other regulatory elements but more homologous than latent protein coding regions. The significance of this is unclear, since the evolutionary pressures that are present for regulatory regions may be different than those for coding regions or genes involved in the lytic cycle. The fact that Cp is generally more conserved than the rest of the genome suggests that there are universal regulatory elements within Cp which are required for the virus life cycle.
An alignment of these sequences suggests that the function and regulation of these promoters have been conserved. Initial surveys of the individual Cp sequences by computer analysis reveals that there are many potential transcription factor binding sites in each promoter. Given the relative degrees of homology between EBV and the other lymphocryptoviruses, we reasoned that crucial cis-acting elements required for Cp function are conserved between these viral promoters. Using these criteria, we have identified three regions within the Cp that contain clusters of conserved transcription factor binding sites and a fourth region that contains a high frequency of CpG residues. For ease of discussion we have called these regions R1 to R4.
Previous studies identified three GREs in the EBV Cp located between nucleotides 10240 and 10440 (−1077 to −894 relative to the transcription start site) of the EBV genome and demonstrated that this region could confer responsiveness to glucocorticoids both in the context of Cp and when fused to a heterologous promoter (21, 45). In contrast, using a large plasmid construction containing both Cp and OriP, Woisetschlaeger et al. found that deletions of sequences containing the GRE had little overall effect on basal or activated promoter activity in transient-transfection experiments (56). Finally, recombinant EBV containing a deletion encompassing the three GREs display on average fivefold-higher expression from Cp than wild-type recombinant viruses (6). In light of these data, the biological role of the GRE remains unclear. R1 contains two conserved glucocorticoid elements. One of these, GRE III, appears to be positionally conserved, while GRE I in the HVP Cp is not. No obvious counterpart to EBV GRE II was identified in our sequence comparisons. While the role of glucocorticoids in modulating Cp activity during the immortalization process remains unclear, the conservation of two GRE sites suggests an important role for these elements in vivo. Two other conserved sequence elements that resemble ETS/PEA3 and TRE were also identified in R1 (8, 13, 26). The roles these factors have for Cp function remain unknown, but previous results obtained with mutants with R1 deleted may also be due to deletion of ETS or TRE. Introduction of more subtle point mutations in the GRE and putative ETS and TRE sites will clarify the role of these control elements in Cp function.
Several studies have described a role for CpG methylation in silencing of Cp activity (28, 37–39, 44, 50). CpG methylation of the EBNA2 enhancer region as well as downstream sequences from the enhancer to the TATA box approaches 100% of available CpG in Rael cells, a type I-phenotype cell line (38, 44). Using a methylated cassette assay, Robertson and Ambinder have demonstrated that CpG methylation of Cp sequences in the EBNA2 enhancer silence EBNA2-mediated Cp activity (37). In addition, CpG methylation from the EBNA2 enhancer downstream to the TATA box abolished Cp activity completely in transient-transfection assays (37). Site-specific methylation has been proposed to inhibit binding of cellular factors required for Cp activity (37, 38). R2 contains a high proportion of CpG sites. However, these previous studies have not shown a role for this region in regulation of Cp activity. A recent analysis of the methylation sites in two Mutu clones (BL cell lines) that differ from each other in the EBV latent gene expression pattern, have shown that all the CpG sites present in the region homologous to R2 are methylated in the latency type I clone (50). No differential methylation was found in any other region of the promoter, including the CBF1 and CBF2 binding sites. Site-specific methylation has been proposed to inhibit binding of cellular factors required for CBF2 activity (37, 38). However, an alternative mechanism for how methylation may work to silence promoter activity is through recruitment of DNA binding proteins with affinities for methylated CpG sites. These proteins may function as transcriptional repressors by (i) altering the structure of neighboring chromatin, (ii) excluding positive transcription factors from binding their cognate recognition sites, or (iii) directly exposing a repressor domain (18). The nature of chromatin assembly on plasmid DNA during transient-transfection analysis may not reflect the status of EBV chromosomes in infected cells, and methylation effects on chromatin structure may be manifest only in the context of the viral genome in immortalized cells. The contribution of R2 to Cp silencing may be revealed only under these circumstances.
R3 retains elements for the binding of the cellular factors CBF1 and CBF2, which along with EBNA2 are the primary mediators of Cp EBNA2 enhancer function. We have recently reported that the requirement for a functional CBF2 binding site is more apparent at low concentrations of EBNA2 (11). Quantification of CBF2 binding to HVP and RhEBV promoters revealed 4.7- and 3.9-fold-reduced affinities for CBF2, respectively. However, this analysis was done with extracts derived from a human B-cell line and it is possible that the affinities of the HVP and RhEBV Cps for human CBF2 do not accurately reflect their affinities for the CBF2 homologue present in their natural host cells. When testing these promoters in transient-transfection assays with increasing amounts of EBNA2, we observed a reduction of the ability of the HVP and RhEBV promoters to respond to low concentrations (0.8 μg of effector) (Fig. 4) of EBNA2 compared with the responsiveness of the EBV Cp. At higher concentrations of EBNA2, the three promoters had similar levels of induction. A similar phenomenon was also observed when the wild-type EBV Cp was compared to the EBV Cp containing mutations in the CBF2 binding site (11). The physiological significance of the reduced ability of the EBV-related viruses to respond to low concentrations of EBNA2 is uncertain. The conservation of the CBF2 binding site in the Cp EBNA2 enhancer of HVP- and RhEBV-related viruses argues in favor of CBF2 serving an important role in promoter function.
R4 contains a second conserved CCAAT box, also important for Cp function, as well as the TATA box (32). S1 nuclease protection assays revealed that all of the nonhuman primate cell lines immortalized by their species-specific lymphocryptovirus use Cp and that the transcription start sites are located in analogous positions relative to the TATA box, even though there is some heterogeneity in the nucleotides surrounding the +1 initiation site.
The presence of transcriptional control elements conserved in both HVP and RhEBV Cp sequences that were previously identified as important mediators of Cp activity, combined with data that Cp is used in cells immortalized by these viruses and that HVP and RhEBV Cp reporter constructions are transactivated by EBNA2, suggests that these nonhuman lymphocryptoviruses have retained similar mechanisms for regulation of latency gene expression. The presence of conserved transcriptional control elements that have not been examined in the context of Cp will provide opportunities to identify additional cellular factors that regulate Cp activity. The Cp sequence comparison also reveals several additional conserved stretches of nucleotides that will aid in identifying new or novel transcription factors that regulate Cp activity.
Acknowledgments
This work was supported by NIH grants R29 CA69437 (P.D.L.) and R29 CA60067 (S.S.) and an award from the William Stamps Farish Foundation (P.D.L.).
We thank Elliott Kieff for the Ba65 cell line and Fred Wang for RhEBV LCL 8664.
REFERENCES
- 1.Abbot S D, Rowe M, Cadwallader K, Ricksten A, Gordon J, Wang F, Rymo L, Rickinson A B. Epstein-Barr virus nuclear antigen 2 induces expression of the virus-encoded latent membrane protein. J Virol. 1990;64:2126–2134. doi: 10.1128/jvi.64.5.2126-2134.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alfieri C, Birkenbach M, Kieff E. Early events in Epstein-Barr virus infection of human B lymphocytes. Virology. 1991;181:595–608. doi: 10.1016/0042-6822(91)90893-g. [DOI] [PubMed] [Google Scholar]
- 3.Allday M J, Crawford D H, Griffin B E. Epstein-Barr virus latent gene expression during the initiation of B cell immortalization. J Gen Virol. 1989;70:1755–1764. doi: 10.1099/0022-1317-70-7-1755. [DOI] [PubMed] [Google Scholar]
- 4.Bodescot M, Perricaudet M, Farrell P J. A promoter for the highly spliced EBNA family of RNAs of Epstein-Barr virus. J Virol. 1987;61:3424–3430. doi: 10.1128/jvi.61.11.3424-3430.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cordier M, et al. Stable transfection of Epstein-Barr virus (EBV) nuclear antigen 2 in lymphoma cells containing the EBV P3HR1 genome induces expression of B-cell activation molecules CD21 and CD23. J Virol. 1990;64:1002–1013. doi: 10.1128/jvi.64.3.1002-1013.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Evans T J, Farrell P J, Swaminathan S. Molecular genetic analysis of Epstein-Barr virus Cp promoter function. J Virol. 1996;70:1695–1705. doi: 10.1128/jvi.70.3.1695-1705.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fahraeus R, Jansson A, Ricksten A, Sjoblom A, Rymo L. Epstein-Barr virus-encoded nuclear antigen 2 activates the viral latent membrane protein promoter by modulating the activity of a negative regulatory element. Proc Natl Acad Sci USA. 1990;87:7390–7394. doi: 10.1073/pnas.87.19.7390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Faisst S, Meyer S. Compilation of vertebrate-encoded transcription factors. Nucleic Acids Res. 1992;20:3–26. doi: 10.1093/nar/20.1.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Falk L, Deinhardt F, Nonoyama M, Wolfe L G, Bergholz C. Properties of a baboon lymphotropic herpesvirus related to Epstein-Barr virus. Int J Cancer. 1976;18:798–807. doi: 10.1002/ijc.2910180611. [DOI] [PubMed] [Google Scholar]
- 10.Franken M, Devergne O, Rosenzweig M, Annis B, Kieff E, Wang F. Comparative analysis identifies conserved tumor necrosis factor receptor-associated factor 3 binding sites in the human and simian Epstein-Barr virus oncogene LMP1. J Virol. 1996;70:7819–7826. doi: 10.1128/jvi.70.11.7819-7826.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fuentes-Panana E M, Ling P D. Characterization of the CBF2 binding site within the Epstein-Barr virus latency C promoter and its role in modulating EBNA2-mediated transactivation. J Virol. 1998;72:693–700. doi: 10.1128/jvi.72.1.693-700.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gerber P, Kslter S S, Schidllovsky G, Peterson W D, Daniel M D. Biologic and antigenic characteristics of Epstein-Barr virus-related herpesviruses of chimpanzees and baboons. Int J Cancer. 1977;20:448–459. doi: 10.1002/ijc.2910200318. [DOI] [PubMed] [Google Scholar]
- 13.Goding C R, Temperley S M, Fischer F. Multiple transcription factors interact with the adenovirus-2 EII-late promoter: evidence for a novel CCAAT recognition factor. Nucleic Acids Res. 1987;15:7761–7780. doi: 10.1093/nar/15.19.7761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gordadze, A., and P. D. Ling. Unpublished data.
- 15.Gounari F, De Francesco R, Schmitt J, van der Vliet P C, Cortese R, Stunnenberg H. Amino-terminal domain of NF1 binds to DNA as a dimer and activates adenovirus DNA replication. EMBO J. 1990;9:559–566. doi: 10.1002/j.1460-2075.1990.tb08143.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Heller M, Kieff E. Colinearity between the DNAs of Epstein-Barr virus and herpesvirus papio. J Virol. 1981;37:821–826. doi: 10.1128/jvi.37.2.821-826.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jin X W, Speck S H. Identification of critical cis elements involved in mediating Epstein-Barr virus nuclear antigen 2-dependent activity of an enhancer located upstream of the viral BamHI C promoter. J Virol. 1992;66:2846–2852. doi: 10.1128/jvi.66.5.2846-2852.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kass S U, Pruss D, Wolffe A P. How does DNA methylation repress transcription? Trends Genet. 1997;13:444–449. doi: 10.1016/s0168-9525(97)01268-7. [DOI] [PubMed] [Google Scholar]
- 19.Kieff E. Epstein-Barr virus and its replication. In: Fields B N, Knipe D M, Howley P M, editors. Fields Virology. 3rd ed. Philadelphia, Pa: Lippincott-Raven Publishers; 1996. pp. 107–172. [Google Scholar]
- 20.Knutson J C. The level of c-fgr RNA is increased by EBNA-2, an Epstein-Barr virus gene required for B-cell immortalization. J Virol. 1990;64:2530–2536. doi: 10.1128/jvi.64.6.2530-2536.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kupfer S R, Summers W C. Identification of a glucocorticoid-responsive element in Epstein-Barr virus. J Virol. 1990;64:1984–1990. doi: 10.1128/jvi.64.5.1984-1990.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Levitskaya J, Coram M, Levitsky V, Imreh S, Steigerwald-Mullen P M, Klein G, Kurilla M G, Masucci M G. Inhibition of antigen processing by the internal repeat region of the Epstein-Barr virus nuclear antigen-1. Nature. 1995;375:685–688. doi: 10.1038/375685a0. [DOI] [PubMed] [Google Scholar]
- 23.Ling P D, Hsieh J J, Ruf I K, Rawlins D R, Hayward S D. EBNA-2 upregulation of Epstein-Barr virus latency promoters and the cellular CD23 promoter utilizes a common targeting intermediate, CBF1. J Virol. 1994;68:5375–5383. doi: 10.1128/jvi.68.9.5375-5383.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ling P D, Rawlins D R, Hayward S D. The Epstein-Barr virus immortalizing protein EBNA-2 is targeted to DNA by a cellular enhancer-binding protein. Proc Natl Acad Sci USA. 1993;90:9237–9241. doi: 10.1073/pnas.90.20.9237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ling P D, Ryon J J, Hayward S D. EBNA-2 of herpesvirus papio diverges significantly from the type A and type B EBNA-2 proteins of Epstein-Barr virus but retains an efficient transactivation domain with a conserved hydrophobic motif. J Virol. 1993;67:2990–3003. doi: 10.1128/jvi.67.6.2990-3003.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Martin M E, Piette J, Yaniv M, Tang W J, Folk W R. Activation of the polyomavirus enhancer by a murine activator protein 1 (AP1) homolog and two contiguous proteins. Proc Natl Acad Sci USA. 1988;85:5839–5843. doi: 10.1073/pnas.85.16.5839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Masucci M G, Contreras-Salazar B, Ragnar E, Falk K, Minarovits J, Ernberg I, Klein G. 5-Azacytidine up regulates the expression of Epstein-Barr virus nuclear antigen 2 (EBNA-2) through EBNA-6 and latent membrane protein in the Burkitt’s lymphoma line Rael. J Virol. 1989;63:3135–3141. doi: 10.1128/jvi.63.7.3135-3141.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Minarovits J, Hu L F, Minarovits K S, Klein G, Ernberg I. Sequence-specific methylation inhibits the activity of the Epstein-Barr virus LMP1 and BCR2 enhancer-promoter regions. Virology. 1994;200:661–667. doi: 10.1006/viro.1994.1229. [DOI] [PubMed] [Google Scholar]
- 29.Miyashita E M, Yang B, Babcock G J, Thorley-Lawson D A. Identification of the site of Epstein-Barr virus persistence in vivo as a resting B cell. J Virol. 1997;71:4882–4891. doi: 10.1128/jvi.71.7.4882-4891.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Miyashita E M, Yang B, Lam K M, Crawford D H, Thorley-Lawson D A. A novel form of Epstein-Barr virus latency in normal B cells in vivo. Cell. 1995;80:593–601. doi: 10.1016/0092-8674(95)90513-8. [DOI] [PubMed] [Google Scholar]
- 31.Moghaddam A, Koch J, Annis B, Wang F. Infection of human B lymphocytes with lymphocryptoviruses related to Epstein-Barr virus. J Virol. 1998;72:3205–3212. doi: 10.1128/jvi.72.4.3205-3212.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Puglielli M T, Woisetschlaeger M, Speck S H. oriP is essential for EBNA gene promoter activity in Epstein-Barr virus-immortalized lymphoblastoid cell lines. J Virol. 1996;70:5758–5768. doi: 10.1128/jvi.70.9.5758-5768.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Reisman D, Sugden B. trans activation of an Epstein-Barr viral transcriptional enhancer by the Epstein-Barr viral nuclear antigen 1. J Virol. 1986;6:3838–3846. doi: 10.1128/mcb.6.11.3838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rickinson A B, Kieff E. Epstein-Barr virus. In: Fields B N, Knipe D M, Howley P M, editors. Fields Virology. 3rd ed. Philadelphia, Pa: Lippincott-Raven Publishers; 1996. pp. 2397–2446. [Google Scholar]
- 35.Rickinson A B, Young L S, Rowe M. Influence of the Epstein-Barr virus nuclear antigen EBNA 2 on the growth phenotype of virus-transformed B cells. J Virol. 1987;61:1310–1317. doi: 10.1128/jvi.61.5.1310-1317.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Robertson E S, Kieff E D. Genetic analysis of Epstein-Barr virus in B lymphocytes. Epstein-Barr Virus Rep. 1995;2:73–80. [Google Scholar]
- 37.Robertson K D, Ambinder R F. Mapping promoter regions that are hypersensitive to methylation-mediated inhibition of transcription: application of the methylation cassette assay to the Epstein-Barr virus major latency promoter. J Virol. 1997;71:6445–6454. doi: 10.1128/jvi.71.9.6445-6454.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Robertson K D, Hayward S D, Ling P D, Samid D, Ambinder R F. Transcriptional activation of the Epstein-Barr virus latency C promoter after 5-azacytidine treatment: evidence that demethylation at a single CpG site is crucial. Mol Cell Biol. 1995;15:6150–6159. doi: 10.1128/mcb.15.11.6150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Robertson K D, Swinnen L J, Zong J C, Gulley M L, Ambinder R F. CpG methylation of the major Epstein-Barr virus latency promoter in Burkitt’s lymphoma and Hodgkin’s disease. Blood. 1996;88:3129–3136. [PubMed] [Google Scholar]
- 40.Rooney C, Howe J G, Speck S H, Miller G. Influence of Burkitt’s lymphoma and primary B cells on latent gene expression by the nonimmortalizing P3J-HR-1 strain of Epstein-Barr virus. J Virol. 1989;63:1531–1539. doi: 10.1128/jvi.63.4.1531-1539.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rooney C M, Brimmell M, Buschle M, Allan G, Farrell P J, Kolman J L. Host cell and EBNA-2 regulation of Epstein-Barr virus latent-cycle promoter activity in B lymphocytes. J Virol. 1992;66:496–504. doi: 10.1128/jvi.66.1.496-504.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ryon J J, Fizman E D, Houchens C, Zong J, Lieberman P M, Chang Y-N, Hayward G S, Hayward S D. The lytic origin of herpesvirus papio is highly homologous to Epstein-Barr virus ori-Lyt: evolutionary conservation of transcriptional activation and replication signals. J Virol. 1993;67:4006–4016. doi: 10.1128/jvi.67.7.4006-4016.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sample, J. Personal communication.
- 44.Schaefer B C, Strominger J L, Speck S H. Host-cell-determined methylation of specific Epstein-Barr virus promoters regulates the choice between distinct viral latency programs. J Virol. 1997;17:364–377. doi: 10.1128/mcb.17.1.364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schuster C, Chasserot-Golaz S, Urier G, Beck G, Sergeant A. Evidence for a functional glucocorticoid responsive element in the Epstein-Barr virus genome. Mol Endocrinol. 1991;5:267–272. doi: 10.1210/mend-5-2-267. [DOI] [PubMed] [Google Scholar]
- 46.Sinclair A J, Brimmell M, Farrell P J. Reciprocal antagonism of steroid hormones and BZLF1 in switch between Epstein-Barr virus latent and productive cycle gene expression. J Virol. 1992;66:70–77. doi: 10.1128/jvi.66.1.70-77.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sugden B, Warren N. A promoter of Epstein-Barr virus that can function during latent infection can be transactivated by EBNA-1, a viral protein required for viral DNA replication during latent infection. J Virol. 1989;63:2644–2649. doi: 10.1128/jvi.63.6.2644-2649.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sung N S, Kenney S, Gutsch D, Pagano J S. EBNA-2 transactivates a lymphoid-specific enhancer in the BamHI C promoter of Epstein-Barr virus. J Virol. 1991;65:2164–2169. doi: 10.1128/jvi.65.5.2164-2169.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Swaminathan S. Characterization of Epstein-Barr virus recombinants with deletions of the BamHI C promoter. Virology. 1996;217:532–541. doi: 10.1006/viro.1996.0148. [DOI] [PubMed] [Google Scholar]
- 50.Takacs M, Myohanen S, Altoik E, Minarovits J. Analysis of methylation patterns in the regulatory region of the latent Epstein-Barr virus promoter BCR2 by automated fluorescent sequencing. Biol Chem. 1998;379:417–422. doi: 10.1515/bchm.1998.379.4-5.417. [DOI] [PubMed] [Google Scholar]
- 51.Thorley-Lawson D A, Miyashita E M, Khan G. Epstein-Barr virus and the B cell: that’s all it takes. Trends Microbiol. 1996;4:204–208. doi: 10.1016/s0966-842x(96)90020-7. [DOI] [PubMed] [Google Scholar]
- 52.Tsang S F, Wang F, Izumi K M, Kieff E. Delineation of the cis-acting element mediating EBNA-2 transactivation of latent infection membrane protein expression. J Virol. 1991;65:6765–6771. doi: 10.1128/jvi.65.12.6765-6771.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang F, Gregory C, Sample C, Rowe M, Liebowitz D, Murray R, Rickinson A, Kieff E. Epstein-Barr virus latent membrane protein (LMP1) and nuclear proteins 2 and 3C are effectors of phenotypic changes in B lymphocytes: EBNA-2 and LMP1 cooperatively induce CD23. J Virol. 1990;64:2309–2318. doi: 10.1128/jvi.64.5.2309-2318.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang F, Gregory C D, Rowe M, Rickinson A B, Wang D, Birkenbach M, Kikutani H, Kishimoto T, Kieff E. Epstein-Barr virus nuclear antigen 2 specifically induces expression of the B-cell activation antigen CD23. Proc Natl Acad Sci USA. 1987;84:3452–3456. doi: 10.1073/pnas.84.10.3452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang F, Kikutani H, Tsang S F, Kishimoto T, Kieff E. Epstein-Barr virus nuclear protein 2 transactivates a cis-acting CD23 DNA element. J Virol. 1991;65:4101–4106. doi: 10.1128/jvi.65.8.4101-4106.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Woisetschlaeger M, Jin X W, Yandava C N, Furmanski L A, Strominger J L, Speck S H. Role for the Epstein-Barr virus nuclear antigen 2 in viral promoter switching during initial stages of infection. Proc Natl Acad Sci USA. 1991;88:3942–3946. doi: 10.1073/pnas.88.9.3942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Woisetschlaeger M, Yandava C N, Furmanski L A, Strominger J L, Speck S H. Promoter switching in Epstein-Barr virus during the initial stages of infection of B lymphocytes. Proc Natl Acad Sci USA. 1990;87:1725–1729. doi: 10.1073/pnas.87.5.1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yates J L, Camiolo S M, Ali S, Ying A. Comparison of EBNA1 proteins of Epstein-Barr virus and herpesvirus papio in sequence and function. Virology. 1996;222:1–13. doi: 10.1006/viro.1996.0392. [DOI] [PubMed] [Google Scholar]
- 59.Yoo L I, Mooney M, Puglielli M T, Speck S H. B-cell lines immortalized with an Epstein-Barr virus mutant lacking the Cp EBNA2 enhancer are biased toward utilization of the oriP-proximal EBNA gene promoter Wp1. J Virol. 1997;71:9134–9142. doi: 10.1128/jvi.71.12.9134-9142.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zimber-Strobl U, Kremmer E, Grasser F, Marschall G, Laux G, Bornkamm G W. The Epstein-Barr virus nuclear antigen 2 interacts with an EBNA2 responsive cis-element of the terminal protein 1 gene promoter. EMBO J. 1993;12:167–175. doi: 10.1002/j.1460-2075.1993.tb05642.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zimber-Strobl U, Suentzenich K O, Laux G, Eick D, Cordier M, Calender A, Billaud M, Lenoir G M, Bornkamm G W. Epstein-Barr virus nuclear antigen 2 activates transcription of the terminal protein gene. J Virol. 1991;65:415–423. doi: 10.1128/jvi.65.1.415-423.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]