Abstract
The capsid protein of feline calicivirus (FCV) was expressed by using plasmids containing cytomegalovirus, simian virus 40, or T7 promoters. The strongest expression was achieved with the T7 promoter and coinfection with vaccinia virus expressing the T7 RNA polymerase (MVA/T7pol). The FCV precursor capsid protein was processed to the mature-size protein, and these proteins were assembled in to virus-like particles.
Calicivirus capsids are structurally simple, with 180 copies of a single structural protein (capsid protein) assembled as 90 homodimers in a T=3 icosahedral symmetry (12, 13). The expression of the capsid protein gene differs among the members of this family. The leporid caliciviruses rabbit hemorrhagic disease virus and European brown hare syndrome virus assemble from capsid proteins expressed from a subgenomic RNA and are not further processed posttranslationally (6, 16). In other caliciviruses, such as San Miguel sea lion virus and feline calicivirus (FCV), the mature capsid protein is generated by posttranslational processing from a capsid protein precursor (8, 10, 11, 14). In FCV the 75-kDa precursor protein is composed of 668 amino acids and is processed by removal of the amino-terminal 124 amino acids to generate the mature 62-kDa capsid protein of 544 amino acids (11, 14, 15). We sought to answer the questions whether FCV capsid proteins can self-assemble into empty virus capsids and whether the processing of the capsid protein precursor is a host range determinant in vitro.
Cells, virus, and plasmid constructs.
Crandell feline kidney cells (CRFK), murine L929 cells, and Vero cells were grown in Dulbecco’s modified essential medium (DMEM) with 5% fetal calf serum (FCS). The FCV isolate FCV-KS20, initially isolated from a cat with chronic stomatitis in Germany (5), was used throughout this study and was passaged about five times in CRFK cells. Recombinant vaccinia virus MVA/T7pol (18) was routinely propagated and titered in primary chicken embryo fibroblasts grown in DMEM supplemented with 10% FCS. L929 cells and Vero cells were inoculated with FCV-KS20 at a multiplicity of infection (MOI) of 100, and no virus could be recovered even after three blind passages (not shown). These cell lines were therefore considered to be nonpermissive for FCV.
CRFK cells were infected with FCV-KS20 at an MOI of 10, and viral RNA was isolated 12 h postinfection by using the RNEasy-Mini-System following the instructions of the supplier (Qiagen, Hilden, Germany). Reverse transcription PCR was performed essentially as described previously (5) by using Taq and Pwo polymerases (Expand System; Boehringer, Mannheim, Germany) and the primer pair FCV-4 (5′-ATGTGCCAACCTGCGCTAA-3′) and FCV-2 (5′-TCTAATTGCATTTA ATTGATCGTCA-3′) for the amplification of the capsid precursor gene (nucleotides [nt] 5314 to 7616) and the primer pair FCV-26 (5′-CTTGGGTACCATTATGGCTGACGGAGATGGTTCCATC-3′) and FCV-2 for the amplification of the truncated “mature” capsid protein (nt 5686 to 7616). The viral sequence of primer FCV-26 is GCTGACGGAGATGGTTCCATC; the 5′-terminal sequences represent an introduced KpnI site and an ATG start codon for the initiation of translation. The amplicons were either cloned into pCR2.1 and subcloned into pcDNA3.1 by using the introduced KpnI and the pCR2.1 XbaI sites (pCAPI) or directly cloned into pCRIII.1 uni (pCAPIII) (all vectors from Invitrogen, San Diego, Calif.). The construct with the simian virus 40 (SV40) promoter (pCAPIV) was generated by cloning the mature capsid protein gene via blunted KpnI and XbaI sites into the blunted EcoRI-restricted pSG5 vector (Stratagene, La Jolla, Calif.) (Fig. 1). A 720-bp DNA fragment containing the complete coding sequence of green fluorescent protein (GFP) was prepared by PCR from pEGFP-1 (Clontech Laboratories, Heidelberg, Germany) and inserted into the NcoI and NotI sites of plasmid pIII T7emc to generate the expression plasmid pT7-GFP (Fig. 1). This construct allows T7 RNA polymerase-dependent expression of the GFP gene regulated by the T7 promoter and terminator sequences together with the encephalomyocarditis virus untranslated region (9).
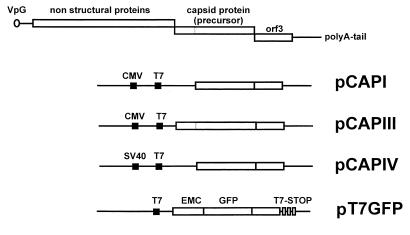
FIG. 1.
Summary of the capsid protein constructs that were used in this study. The truncated mature capsid protein (pCAPI) was cloned into pcDNA3.1, the entire precursor capsid protein gene (pCAPIII) was cloned into pCRII.uni, and the mature truncated capsid protein gene was cloned into pSG5 (pCAPIV). All FCV capsid protein gene constructs contain about 90% of the ORF3 gene. The pT7GFP vector is also depicted. Cloning details are given in the text. EMC, encephalomyocarditis virus leader sequence; T7 Stop, T7 polymerase stop motifs.
FCV capsid protein expression.
For capsid protein expression, 5 × 105 CRFK cells in 9-cm2 dishes were transfected with 1 to 2 μg of DNA by using Lipofectamine (Gibco-BRL, Eggenstein, Germany). DNA was diluted in 100 μl of OPTI-MEM, and 8 μl of Lipofectamine was added. The mixture was incubated at room temperature for 45 min and then added to the cells; after 5 h the medium was replaced by DMEM with 5% FCS. The efficiency of recombinant gene expression was monitored by using pT7-GFP and the recombinant modified vaccinia virus Ankara expressing the T7 bacteriophage RNA polymerase (MVA/T7pol). Accordingly, for FCV capsid protein expression cells were infected with MVA/T7pol (MOI between 10 and 100) 30 min prior to plasmid DNA transfection. Cells were lysed 48 h after transfection and analyzed by sodium dodecyl sulfate–10% polyacrylamide gel electrophoresis (SDS–10% PAGE) and Western blotting with the FCV capsid protein-specific monoclonal antibody (MAb) MAb K, raised against FCV-KS20, as described previously (5). For comparative Western blot analysis the cell lysates from transfections with the different constructs were standardized by cell number.
Transfection of both the precursor protein gene and the mature capsid protein gene revealed expression of proteins of the expected sizes of 75 and 62 kDa, respectively, that reacted with FCV-specific MAbs. The level of expression, however, as judged by the intensity of specific immunostaining achieved in the Western blot analysis of protein lysates, differed markedly between the constructs. Expression of cytomegalovirus (CMV) promoter-regulated genes in CRFK cells was weak, and examination by immunofluorescence revealed only few cells producing the FCV protein (data not shown). Plasmid vectors containing the mature capsid protein construct under the control of the SV40 promoter gave an improved but still moderate level of recombinant gene expression (Fig. 2 and 3).
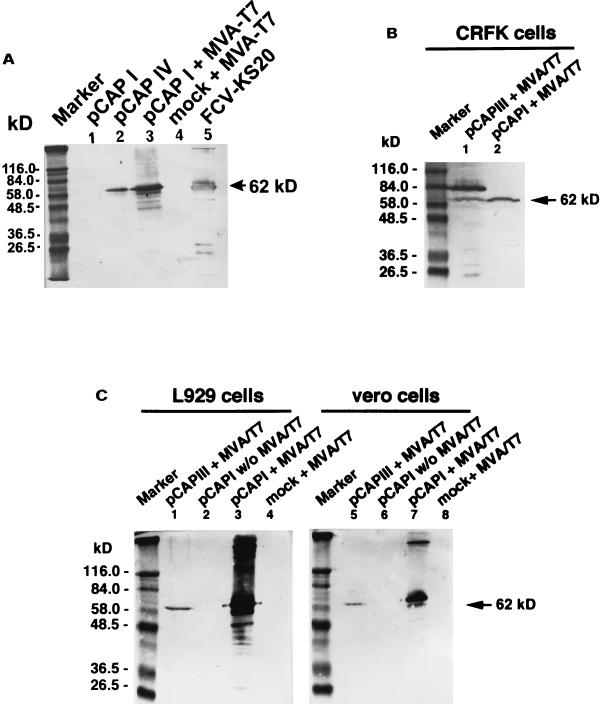
FIG. 2.
Western blots of cell lysates of CRFK cells transfected with various constructs and stained with FCV-specific Mab K. (A) Comparison of the expression efficiencies of the CMV, SV40, and T7 promoter constructs. Lane 1, pCAPI (CMV promoter) lysate of about 5 × 106 cells; lane 2, pCAPIV (SV40 promoter) lysate of about 5 × 106 cells; lane 3, pCAPI plus MVA/T7 (T7 promoter) lysate of about 5 × 105 cells; lane 4, MVA/T7-infected control cells (5 × 106 cells); lane 5, CsCl gradient-purified virus FCV-KS20. (B) Processing of the capsid protein precursor and the mature capsid protein in CRFK cells. Lane 1, precursor capsid protein gene expressed with MVA/T7. Note the processing of the capsid protein precursor to the 62-kDa mature-size protein; lane 2, expression of the mature truncated capsid protein gene expressed with MVA/T7. (C) Processing of the capsid protein precursor in nonpermissive cell lines: murine L929 cells (lanes 1 to 4) and Vero cells (lanes 5 to 8). Lanes 1 and 5, precursor gene expressed with MVA/T7; Lanes 2 and 6, mature truncated capsid protein gene expressed without MVA/T7; lanes 3 and 7, mature truncated capsid protein expressed with MVA/T7, lanes 4 and 8, MVA/T7-infected control cells.
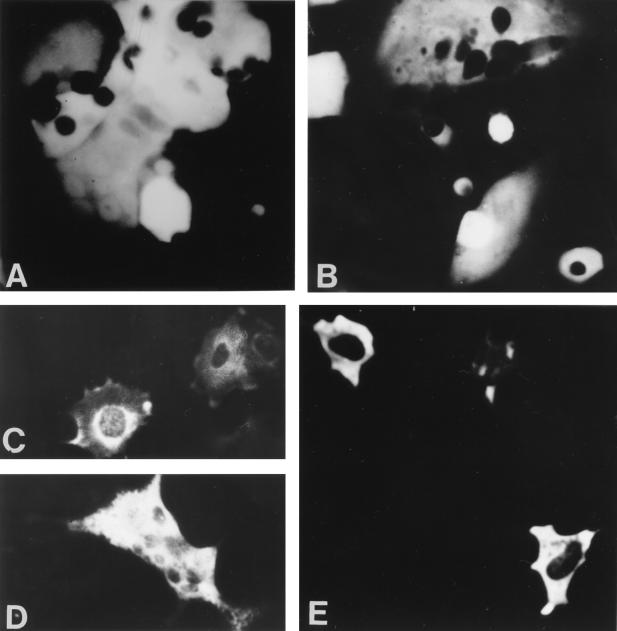
FIG. 3.
Immunofluorescence of CRFK cells transfected with pT7GFP and coinfection with MVA/T7pol (A and B) or transfected with the FCV capsid protein construct pCAPI (C and D), or pCAPIV (E) and coinfection with MVA/T7 pol. All cells were stained with MAb K. Note the syncytium formation due to MVA virus infection.
A strong expression was only observed with the MVA/T7pol system (Fig. 2 and 3). Transfecting MVA-T7pol-infected CRFK with pT7-GFP revealed high levels of GFP fluorescence in up to 80% of the cells (Fig. 3). The most efficient expression was achieved in experiments using 1.5 μg of plasmid DNA and an MOI of MVA/T7pol of 10 to 100 PFU. Recombinant FCV capsid protein gene expression in this system relies on the synthesis of the T7 RNA polymerase in the cytoplasm of MVA/T7pol-infected eukaryotic cells. Besides the high transcriptional activity of this enzyme, additional benefits of the MVA/T7pol-based expression system include its independence of a plasmid transport to the cellular nucleus and the avoidance of undesirable cell-specific splicing of recombinant mRNA. With regard to the cytoplasmic life cycle of caliciviruses the latter advantage seems most likely responsible for the efficient production of capsid protein, as several putative splicing donor and acceptor sites are located throughout the FCV precursor capsid protein gene. As has been described for MVA infection of most other mammalian cells (1, 4, 18) MVA/T7pol appeared to fail to replicate in CRFK as electronmicroscopic examination of inoculated tissues revealed no assembly of mature progeny virions (data not shown).
FCV capsid protein processing.
Transfection of the FCV capsid precursor gene construct (pCAPIII) in MVA/T7-infected CRFK cells and Western blot analysis of cell lysates prepared at 36 h after transfection revealed production of both precursor and mature capsid proteins (Fig. 2). The processing of the precursor capsid protein was also observed in other, nonpermissive mammalian cells, i.e., murine L929 cells and Vero cells (Fig. 2). FCV capsid protein expression is known to be initiated from a subgenomic RNA. Initially, a capsid precursor protein is synthesized which is processed posttranslationally to the mature capsid protein. This involves the cleavage of the amino-terminal 124 amino acids. Experiments with FCV and other caliciviruses strongly suggest that the cleavage is behind the glutamic acid residue at amino acid position 124 in the recognition sequence --FRLE-AD-- (2, 14). The proteases involved in the processing have not yet been identified but the viral 3C-like proteases that process the polyprotein precursor of the nonstructural proteins encoded by ORF1 have been shown to be able to cleave the precursor protein at position 124 (17). Our experiments show that the processing of the precursor capsid protein in FCV can also be mediated by a noncaliciviral protease. An autocatalytic activity of the precursor protein is unlikely as in vitro translation of the protein did not result in the generation of the mature 62-kDa protein (not shown). A cellular protease of CRFK cells is also not likely to be involved in the processing of the precursor protein as no cleavage was observed in the absence of MVA vaccinia virus (not shown). However, gene expression without MVA/T7 was weak and protein processing may have occurred at low levels that could have been missed in this study. Most likely, however, a protease provided by MVA vaccinia virus was responsible for the cleavage. The question whether the ability to cleave the capsid protein precursor is a host range determinant in vitro cannot be answered, as cleavage was observed in both permissive and nonpermissive cells after vaccinia virus coinfection.
FCV capsid assembly.
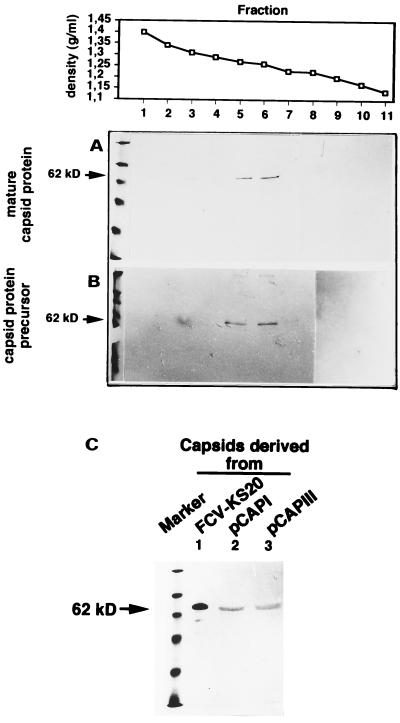
Transfection of both the precursor capsid protein gene and a truncated (mature) capsid protein gene resulted in the expression of the respective capsid proteins, the processing to the mature-size protein, and the assembly into virus-like particles (VLPs). FCV virions and VLPs were purified by CsCl gradient density centrifugation essentially as described by Zhou et al. (22). In brief, cells were inoculated with FCV, incubated for 2 days, and frozen. The supernatant was clarified by low-speed centrifugation, and the virions were pelleted after polyethylene glycol 6000 precipitation by centrifugation (10,000 × g for 30 min). The pellet was dissolved in borate buffer (pH 7.4) and loaded on a CsCl gradient (1.33 g/ml). The empty capsids were purified essentially by the same method, but instead of polyethylene glycol precipitation the capsids were pelleted by centrifugation at 150,000 × g for 12 h. The gradients were collected in 0.5-ml fractions, and the fractions were diluted 1:10 in borate buffer and centrifuged for 1 h at 450,000 × g. The pellet was dissolved in 50 μl of borate buffer and analyzed by immunoblotting and electron microscopy. When purified by CsCl gradient centrifugation the capsids derived from both the mature and the precursor protein gene constructs banded at a density of 1.26 g/ml (Fig. 4), consistent with the described density of empty calicivirus capsids (VLPs) reported elsewhere (1.22 to 1.31 g/ml) (6–8, 19, 22). In some gradients traces of capsid protein were also very rarely observed in fractions with higher densities. The nature of these particles is not known but we believe that their occurrence might be the result of a carry-over during the collection of the gradient fractions. Contamination with infectious virus can be excluded as all fractions were tested for the presence of infectious virus in three consecutive CRFK cell passages. All capsids appeared uniform in morphology and size (35 nm) and were indistinguishable from intact calicivirus virions (Fig. 5).
FIG. 4.
Immunoblots of CsCl density gradient fractions for purification of empty virus capsids after transfection of the mature capsid protein gene (A) or the capsid protein precursor gene (B). The densities of the respective fractions are indicated at the top. (C) Immunoblot of fraction 5 from both gradients along with purified virions separated in an SDS–10% PAGE gel and detected with FCV MAb K to indicate the size homologies of the respective capsid proteins.
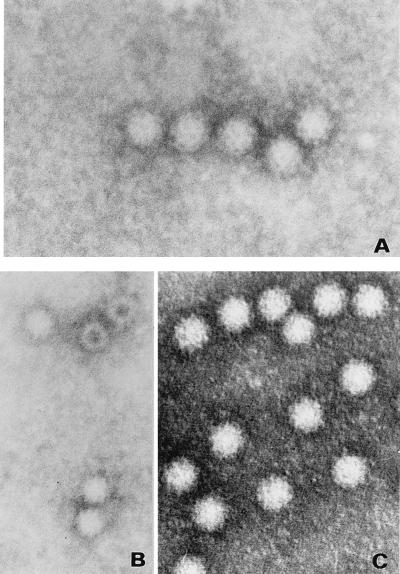
FIG. 5.
Negative-staining electron microscopy of empty virus capsids produced after transfection of the precursor capsid protein gene (A), after transfection of the mature capsid protein gene (B), and after transfection of authentic full virus particles (C).
The VLPs could only be purified from cell lysates. They were not detected in the supernatant of transfected cells by Western blot analysis or electron microscopy after 100-fold concentration. The capsids were not infectious and did not contain detectable levels of FCV RNA, as demonstrated by inoculation on CRFK cells and reverse transcription PCR with various primer pairs covering the capsid protein gene. Preliminary antigenic examination of the expressed capsids with two panels of MAbs (5) revealed no substantial differences from authentic virus particles when tested in Western blots (data not shown).
Capsid assembly has been recently described for FCV (3). The precursor capsid protein gene was expressed in a baculovirus system, and Western blots of the capsids revealed both 75-kDa species and 62-kDa species of the capsid protein. Although not supported by the figures presented in the paper, the authors stated that in their system the capsids were formed exclusively by the 75-kDa protein precursor in the absence of any processing to the mature 62-kDa protein. The reason for this discrepancy with our results presented here are unknown.
Beside the capsid protein gene sequences both constructs also encode about 90% of the ORF3 gene product. The function of this protein is still unknown although in rabbit hemorrhagic disease virus it is believed to represent a second structural protein (21). A role for the ORF3 moiety in capsid assembly in our FCV system therefore cannot be excluded. Studies of other caliciviruses with the baculovirus system, however, showed that the ORF3 protein is dispensable for capsid assembly (20).
In summary, our experiments showed that the precursor capsid protein is processed to a mature-size capsid protein after transfection into various permissive and nonpermissive mammalian cells and coinfection with MVA/T7 and that these proteins assemble into empty capsids.
Acknowledgments
The skillful technical assistance of Andrea Jahnke and Gaby Platzer is gratefully acknowledged. We thank Lutz Guertler for his interest in this work.
This study was supported by a grant from the Fondation Pierre Richard Dick-Virbac and a NATO collaborative research grant (CRG 960212). K.G. was the recipient of a fellowship from the German Academic Exchange Service (DAAD).
REFERENCES
- 1.Carroll M W, Moss B. Host range and cytopathogenicity of the highly attenuated MVA strain of vaccinia virus: propagation and generation of recombinant viruses in a non-human mammalian cell line. Virology. 1997;238:198–211. doi: 10.1006/viro.1997.8845. [DOI] [PubMed] [Google Scholar]
- 2.Carter M J, Milton I D, Turner P C, Meanger J, Bennett M, Gaskell R M. Identification and sequence determination of the capsid protein gene of feline calicivirus. Arch Virol. 1992;122:223–235. doi: 10.1007/BF01317185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.DeSilver D A, Guimond P M, Gibson J K, Thomsen D R, Wardley R C, Lowery D E. Expression of the complete capsid and the hypervariable region of feline calicivirus in the baculovirus expression system. In: Chasey D, Gaskell R M, Clarke I N, editors. Proceedings of the First International Symposium on Caliciviruses. Reading, United Kingdom: European Society for Veterinary Virology; 1997. pp. 131–143. [Google Scholar]
- 4.Drexler I, Heller K, Wahren B, Erfle V, Sutter G. Highly attenuated modified vaccinia virus Ankara replicates in baby hamster kidney cells, a potential host for virus propagation, but not in various human transformed and primary cells. J Gen Virol. 1998;79:347–352. doi: 10.1099/0022-1317-79-2-347. [DOI] [PubMed] [Google Scholar]
- 5.Geissler K, Schneider K, Platzer G, Truyen B, Kaaden O-R, Truyen U. Genetic and antigenic heterogeneity among feline calicivirus isolates from distinct disease manifestations. Virus Res. 1997;48:193–206. doi: 10.1016/s0168-1702(97)01440-8. [DOI] [PubMed] [Google Scholar]
- 6.Laurent S, Vautherot J-F, Madelaine M-F, Le Gall G, Rasschaert D. Recombinant rabbit hemorrhagic disease virus capsid protein expressed in baculovirus self-assembles into viruslike particles and induces protection. J Virol. 1994;68:6794–6798. doi: 10.1128/jvi.68.10.6794-6798.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Leite J P G, Ando T, Noel J S, Jiang B, Humphrey C D, Lew J F, Green K Y, Glass R I, Monroe S S. Characterization of Toronto virus capsid protein expressed in baculovirus. Arch Virol. 1996;141:865–875. doi: 10.1007/BF01718161. [DOI] [PubMed] [Google Scholar]
- 8.Li T-C, Yamakawa Y, Suzuki K, Tatsumi M, Razak M A A, Uchida T, Takeda N, Miyamura T. Expression and self-assembly of empty virus-like particles of hepatitis E virus. J Virol. 1997;71:7207–7213. doi: 10.1128/jvi.71.10.7207-7213.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Moss B, Elroy-Stein O, Mizukami T, Alexander W, Fuerst T. New mammalian expression vectors. Nature. 1990;348:91–92. doi: 10.1038/348091a0. [DOI] [PubMed] [Google Scholar]
- 10.Neill J D. Nucleotide sequence of the capsid protein gene of two serotypes of San Miguel sea lion virus: identification of conserved and non-conserved amino acid sequences among calicivirus capsid proteins. Virus Res. 1992;24:211–222. doi: 10.1016/0168-1702(92)90008-w. [DOI] [PubMed] [Google Scholar]
- 11.Neill J D, Reardon I M, Heinrikson R L. Nucleotide sequence and expression of the capsid protein gene of feline calicivirus. J Virol. 1991;65:5440–5447. doi: 10.1128/jvi.65.10.5440-5447.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Prasad B V, Matson D O, Smith A W. Three-dimensional structure of calicivirus. J Mol Biol. 1994;240:256–264. doi: 10.1006/jmbi.1994.1439. [DOI] [PubMed] [Google Scholar]
- 13.Prasad B V, Rothnagel R, Jiang X, Estes M K. Three-dimensional structure of baculovirus-expressed Norwalk virus capsids. J Virol. 1994;68:5117–5125. doi: 10.1128/jvi.68.8.5117-5125.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Seal B S, Ridpath J F, Mengeling W L. Analysis of feline calicivirus capsid protein genes: identification of variable antigenic determinant regions of the protein. J Gen Virol. 1993;74:2519–2524. doi: 10.1099/0022-1317-74-11-2519. [DOI] [PubMed] [Google Scholar]
- 15.Shin Y S, Tohya Y, Oshikamo R, Kawaguchi Y, Tomonaga K, Miyazawa T, Kai C, Mikami T. Antigenic analysis of feline calicivirus capsid precursor protein and its deleted polypeptides produced in a mammalian cDNA expression system. Virus Res. 1993;30:17–26. doi: 10.1016/0168-1702(93)90012-c. [DOI] [PubMed] [Google Scholar]
- 16.Sibilia M, Boniotti M B, Angoscini P, Capucci L, Rossi C. Two independent pathways of expression lead to self-assembly of the rabbit hemorrhagic disease virus capsid protein. J Virol. 1995;69:5812–5815. doi: 10.1128/jvi.69.9.5812-5815.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sosnovtsev S V, Sosnovtseva S A, Green K Y. Cleavage of the feline calicivirus capsid precursor is mediated by a virus-encoded proteinase. J Virol. 1998;72:3051–3059. doi: 10.1128/jvi.72.4.3051-3059.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sutter G, Ohlmann M, Erfle V. Non-replicating vaccinia vector efficiently expresses bacteriophages T7 RNA polymerase. FEBS Lett. 1995;28:9–12. doi: 10.1016/0014-5793(95)00843-x. [DOI] [PubMed] [Google Scholar]
- 19.White L J, Hardy M E, Estes M K. Biochemical characterization of a smaller form of recombinant Norwalk virus capsids assembled in insect cells. J Virol. 1997;71:8066–8072. doi: 10.1128/jvi.71.10.8066-8072.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Williams J C, Liu B L, Lambden P R, Clarke I N. Expression of SRSV ORFs 2 and 3: assembly of virus-like particles is independent of ORF3 activity. In: Chasey D, Gaskell R M, Clarke I N, editors. Proceedings of the First International Symposium on Caliciviruses. Reading, United Kingdom: European Society for Veterinary Virology; 1997. pp. 51–58. [Google Scholar]
- 21.Wirblich C, Thiel H-J, Meyers G. Genetic map of the calicivirus rabbit hemorrhagic disease virus as deduced from in vitro translation studies. J Virol. 1996;70:7974–7983. doi: 10.1128/jvi.70.11.7974-7983.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhou L, Yu Q, Luo M. Characterization of two density populations of feline calicivirus particles. Virology. 1994;205:530–533. doi: 10.1006/viro.1994.1674. [DOI] [PubMed] [Google Scholar]