Abstract
Sharing data has a crucial role in the advancement of biological science. The use of different types of data within and between scientific fields complicates establishing a single standard for making data publicly accessible. To address this difficulty, we make specific suggestions on how to share different kinds of biological data, including genomics data, proteomics data, microscopy and imaging data, and structural biology data. We provide suggestions for specialist and general repositories for depositing your data. We also provide a checklist to ensure that you share your data in standards consistent with the Findable, Accessable, Interpretable, Reuseable (FAIR) guiding principles for data management.
Introduction
Data sharing is an essential element of the scientific method, imperative to ensure transparency and reproducibility. Other researchers often reuse shared data for meta-analyses or to accompany new data. Different areas of research collect fundamentally different types of data, such as tabular data, sequence data, and image data. These types of data differ greatly in size and require different approaches for sharing. Here, we outline good practices to make your biological data publicly accessible and usable, generally and for several specific kinds of data.
FAIR principles.
Sharing data proves more useful when others can easily find and access, interpret, and reuse the data. To maximize the benefit of sharing your data, follow the Findable, Accessable, Interpretable, Reuseable (FAIR) guiding principles of data sharing1 (Box 1), which optimize reuse of generated data. The FAIR principles outline clear standards for ensuring that others can find and access your data, and that once accessed, users can easily understand and reuse the data. The FAIR principles provide a clear collection of important details to include within your data and metadata (see “Data, metadata, and documentation”).
Box 1: FAIR data sharing principles.
Findable
The first step in (re)using data is to find them. Metadata and data should be easy to find for both humans and computers. Machine-readable metadata are essential for automatic discovery of datasets and services, so this is an essential component of the FAIRification process.
-
F1
(Meta)data are assigned a globally unique and persistent identifier
-
F2
Data are described with rich metadata (defined by R1 below)
-
F3
(Meta)data clearly and explicitly include the identifier of the data they describe
-
F4
(Meta)data are registered or indexed in a searchable resource
Accessible
Once the user finds the required data, she/he needs to know how can they be accessed, possibly including authentication and authorisation.
-
A1(Meta)data are retrievable by their identifier using a standardised communications protocol
-
A1.1The protocol is open, free, and universally implementable
-
A1.2The protocol allows for an authentication and authorisation procedure, where necessary
-
A1.1
-
A2
Metadata are accessible, even when the data are no longer available
Interoperable
The data usually need to be integrated with other data. In addition, the data need to interoperate with applications or workflows for analysis, storage, and processing.
-
I1
(Meta)data use a formal, accessible, shared, and broadly applicable language for knowledge representation
-
I2
(Meta)data use vocabularies that follow FAIR principles
-
I3
(Meta)data include qualified references to other (meta)data
Reusable
The ultimate goal of FAIR is to optimise the reuse of data. To achieve this, metadata and data should be well-described so that they can be replicated and/or combined in different settings.
-
R1(Meta)data are richly described with a plurality of accurate and relevant attributes
-
R1.1(Meta)data are released with a clear and accessible data usage license
-
R1.2(Meta)data are associated with detailed provenance
-
R1.3(Meta)data meet domain-relevant community standards
-
R1.1
By GO FAIR1 (https://www.go-fair.org/fair-principles/), provided under the Creative Commons Attribution 4.0 International license.
The repositories and practices we recommend below fulfill some of these principles and make it easier for you to follow others. This will not only help others using your data, but can also save you time in the future (see “The benefits of sharing data to individual researchers”).
The National Institutes of Health (NIH), Canadian Institutes of Health Research (CIHR), Monarch Initiative,2,3 and the Research Data Alliance (https://www.rd-alliance.org/) all recommend FAIR principles for data sharing. Amendments to these recommendations that add measures for traceability (such as evidence and provenance), licensing, and connectedness (such as identifiers and versioning) further improve data reusability.4,5
Why share?
The benefits of sharing data to science and society
Sharing data allows for transparency in scientific studies, and allows one to fully understand what occurred in an analysis and reproduce the results. Without complete data, metadata (see “Data, metadata, and documentation”), and information about resources used to generate the data, reproducing a study proves impossible.6,7
Within the biological sciences we have a problem of data waste—ostensibly shared data that no one ever uses. Many otherwise useful datasets go underused because researchers can not effectively reuse the data. The inability to reuse arises from lack of discoverability, lack of important information provided, inconsistencies in data and metadata, and licensing issues.
When shared effectively, we can multiply the benefits of large datasets that cost large amounts of funds and research time. Combining previously shared biological data accelerates development of analytical methods used to analyze biological data. Reusing rare samples increases the sample impact. Combining data together in meta-analyses increases study power. Data sharing also leads to fewer duplicate studies. Researchers can build on previous studies to corroborate or falsify their findings rather than repeating the same experiment. Many research projects rely on data from resources such as the Encyclopedia of DNA Elements (ENCODE) Project.8,9 The existence of a large collection of accessible data also aids in the development of cross-cutting analyses such as recount2.10
Published manuscripts with reusable data will garner more citations and have more long-term impact on scientific knowledge.11 As such, many funders now require that grant proposals include a data management and sharing plan describing biological data and metadata.12,13 Many journals have also implemented policies making public data sharing a requirement upon publication.
The benefits of sharing data to individual researchers
Sharing data increases the impact of a researcher’s work and reputation for sound science.14 Awards for those with an excellent record of data sharing15 (https://researchsymbionts.org/) or data reuse16 (https://researchparasite.com/) can exemplify this reputation.
Demonstrating a track record of excellence in resource sharing benefits you when applying for funding. A commitment to and detailed plan for sharing data publicly increases the perception of a grant proposal’s impact.14 A detailed data sharing plan outlines the types of data you will share, available metadata available, and in which repositories you will deposit the data.
Preparing to share data publicly reduces unintentional errors within your own research group. When preparing the data for sharing, providing detailed metadata and documentation will eliminate guesswork, lost details, and maintain tacit knowledge that might otherwise remain unrecorded. Posting data on public repositories with links to the publication and links to data deposited within your publication ensure findability of your data.
Data citation standards now allow directly citing datasets in journal reference lists.17 Citable datasets provide an important incentive to data sharing since those using your shared data can now properly attribute citations to your dataset.
Addressing common concerns about data sharing
Despite the clear benefits of sharing data, some researchers still have concerns about doing so. Some worry that sharing data may decrease the novelty of their work and their chance to publish in prominent journals. You can address this concern by sharing your data only after publication. You can also choose to preprint your manuscript when you decide to share your data. Furthermore, you only need to share the data and metadata required to reproduce your published study.
Time spent on sharing data.
Some have concerns about the time it takes to organize and share data publicly. Many add “data available upon request” to manuscripts instead of depositing the data in a public repository in hopes of getting the work out sooner. It does take time to organize data in preparation for sharing, but sharing data publicly may save you time. Sharing data in a public repository that guarantees archival persistence means that you will not have to worry about storing and backing up the data yourself.
You can consider putting off data sharing tasks as incurring a form of “sharing debt”, by analogy with the concept of technical debt used in software engineering. Delaying these tasks may appear to save you time in the short run, but sharing the data later will take at least as much time as doing it now. You may also incur interest, as it can take longer in the long run to handle individual requests for data availability. Taking a few hours now to organize data and submit it to a repository will save you much of this time.
Human subject data.
Sharing of data on human subjects requires special ethical, legal, and privacy considerations. Existing recommendations18-24 largely aim to balance the privacy of human participants with the benefits of data sharing by de-identifying human participants and obtaining consent for sharing. Sharing human data poses a variety of challenges for analysis, transparency, reproducibility, interoperability, and access.18-24
Sometimes you cannot publicly post all human data, even after de-identification.25 We suggest three strategies for making these data maximally accessible. First, deposit raw data files in a controlled-access repository, such as the European Genome-phenome Archive (EGA).26 Controlled-access repositories allow only qualified researchers who apply to access the data. Second, even if you cannot make individual level raw data available, you can make as much processed data available as possible. This may take the form of summary statistics such as means and standard deviations, rather than individual-level data. Third, you may want to generate simulated data distinct from the original data but statistically similar to it. Simulated data would allow others to reproduce your analysis without disclosing the original data or requiring the security controls needed for controlled-access data.21
Data, metadata, and documentation
Data and metadata.
Data consist of recorded observations of the biological artifacts or models studied. Metadata describe the primary data and the resources used to generate it.
In a biological context, metadata often provide additional information on samples such as sex, disease, and tissue source site. Metadata often include information about resources such as cell lines and antibodies.
You should share metadata alongside every dataset. A lack of clear metadata for your specific dataset makes it more difficult to understand.27 This may make it more difficult to reproduce the research or reuse the data. For example, roughly half of >1700 evaluated research studies lacked sufficient specificity in describing resources such as cell lines, organisms, and antibodies to make the study reproducible.6
In addition to information about samples, metadata also describe experimental protocols and bioinformatic processes. These include tools used to generate the data, hardware and software versions, processing batch information, and details necessary for understanding data generation.
Most biological disciplines have specific metadata standards that describe the information expected to accompany datasets. For example, genomics researchers have benefited enormously from consistent minimum standard of metadata reporting. The Minimum Information About a Microarray Experiment (MIAME)28 and Minimum Information About a Next-generation Sequencing Experiment (MINSEQE)29 (http://fged.org/projects/minseqe/) guidelines have enabled large-scale efforts to combine and harmonize data, promoting reuse. These guidelines require descriptive standards, experimental design information, essential sample information such as tissue or sex, and bioinformatics processing protocols. Repositories of gene expression data, such as Gene Expression Omnibus (GEO),26 have mandated use of these guidelines. We discuss metadata standards for individual biological disciplines below.
Using controlled vocabularies or ontologies can improve the rigor of describing biological concepts in your metadata. Ontologies are controlled vocabularies that include both human- and machine-readable semantic relationships between concepts. Widely-used biological ontologies include the Gene Ontology30,31 (http://geneontology.org/) used to annotate gene function and the Uberon anatomy ontology32 (https://uberon.github.io/). Many repositories or consortium projects require the use of a controlled vocabulary in their metadata standard or data model. For example, the ENCODE Project suggests using Uberon to describe the source of biological tissues.
The formally-defined linkages between concepts in an ontology further support interoperability and reusability beyond a simple controlled vocabulary. For example, there exists a logical relationship defining the Gene Ontology term “dentate gyrus development” (GO:0021542) using a term from Uberon, “dentate gyrus of hippocampal formation” (UBERON:0001885).
Well-constructed controlled vocabularies and ontologies use globally unique persistent identifiers to refer to each concept. This eliminates ambiguity and makes it easier to link uses of the concept across the whole scientific endeavor. To refer to any controlled vocabulary or ontology term, use a persistent identifier, and version, if applicable.
Documentation.
Document your data in three ways: (1) with your manuscript, (2) with description fields in the metadata collected by repositories, and (3) with README files. README files provide abbreviated information about a collection of files. README files associated with biological data should explain organization, file locations, observations and variables present in each file, details on the experimental design, and details on bioinformatic processes.
We regard README files as essential for making your data easy to navigate. Below, we include specific README files for different types of biological data.
Source code.
Ideally, readers should have all materials needed to completely reproduce the study described in a publication, not just data. These materials includes source code, preprocessing, and analysis scripts. Guidelines for organization of computational biology projects33,34 can help you arrange your data and scripts in a way that will make it easier for you and other to access and re-use them.
Licensing.
Clear licensing information attached to your data avoids any questions of whether others may reuse it. While copyright law does not protect facts themselves, permission to reuse compilations of facts such as databases may seem less clear without an explicit license. Many data resources turn out not to be as reusable as the providers intended, due to lack of clarity in licensing or restrictive licensing choices.35
Accompany your data with a license that allows reuse and possibly redistribution. We recommend dedicating your data to the public domain with the CC0 Universal Public Domain Dedication (https://creativecommons.org/choose/zero/). Using CC0 maximizes the ability for others to reuse and remix the data. Other guidelines recommend CC04,36 and many journals and repositories require it.
For non-data artifacts associated with your manuscript you may wish to use a license with more restrictions than CC0. Relevant licenses include the GNU General Public License (https://www.gnu.org/licenses/gpl-3.0.html) for code and Creative Commons licenses (https://creativecommons.org/choose/) for documents.
When to share
We encourage you to share any data underlying a manuscript by the time of its publication. Many publishers and funding agencies such as NIH37,38 now make data sharing an explicit requirement. In addition to sharing all relevant data by publication time, some researchers will go further and make it available when posting a preprint.
Reviewers should have access to underlying data and code when assessing a manuscript.5 It may seem tempting to restrict data access so that only assigned reviewers can see it during manuscript peer review but this has hidden costs and uncertain benefits. Making the data and code public when submitting the manuscript can avoid this hassle, with few drawbacks. Posting a preprint of the associated manuscript at the same time provides a public record of priority.
How to share: tabular data
Researchers commonly store data in tabular format, an intuitive way to describe multiple similar observations. Tabular format stores information in a structure of rows and columns. Usually, rows contain observations and columns contain variables. In biological data, observations usually refer to samples, replicates, or genes. Variables consist of quantitative or qualitative properties assessed for each observation.
File format.
Researchers often save tabular data as spreadsheets. Especially when you have multiple supplementary tables to attach to a manuscript, save the data as a single XLSX workbook39 with a data dictionary sheet at the beginning of the document. Saving tabular data as XLSX allows for download of all supplementary tables at once. Most programming languages have libraries that make it easy to import and read XLSX workbooks.
Despite the advantages of XLSX workbooks, Microsoft Excel works poorly with certain types of data. Famously, Microsoft Excel changes some gene names to dates.40,41 This posed a sufficiently severe issue that geneticists changed the gene symbol nomenclature to prevent this mishap.42 Eluding Excel’s mangling of gene symbols can prove complicated. When your data has gene symbols and you have any uncertainty about avoiding corrupting these symbols when saving XLSX workbooks, use non-XLSX formats instead.
When depositing data in public repositories, rather than including it in a manuscript or on the journal’s supplementary data web site, save the data in tab-separated values (TSV) format. This format separates variables with a tab character and separates observations of multiple variables with a newline character. Many programs and programming environments can easily use TSV data.
Avoid comma-separated values (CSV) format, when possible. CSV format has the disadvantage of using commas to separate variables, when commas often occur within variables themselves. This leads to ambiguity and different, incompatible format variants that attempt to solve this problem.
Organization.
Certain organizational tactics make data much more interpretable and reduce errors. Broman & Woo43 and Ellis & Leek44 provide excellent suggestions on how to organize tabular data. First, ensure that you use the same labels in all areas of your data. For example, inconsistent sex labels, such as “female”, “Female”, “F”, “f”, and “0”, make the data hard to read and to reanalyze. Second, pick one representation of data nomenclature and remain consistent throughout your data and documentation. Third, ensure that you use consistent missing value notation, such as “NA”. Fourth, avoid using spaces in file and column names as this complicates use in many analyses.44 Incorporating these recommendations make your data easily interpretable and usable by yourself and others.
Data dictionary.
Data dictionaries have a crucial role in organizing your data, especially explaining the variables and their representation. When using XLSX workbooks, add a data dictionary as a separate sheet. When using TSV files, add an additional TSV file containing the data dictionary. Your data dictionaries should provide short names for each variable, a longer text label for the variable, a definition for each variable, data type (such as floating point number, integer, or string), measurement units, and expected minimum and maximum values. Data dictionaries can make explicit what future users would otherwise have to guess about the representation of data.
Where to share.
Share the tabular data most important for interpreting your manuscript as a table within the manuscript itself. You can supply more voluminous data or data less crucial for interpretation as supplementary data attached to the manuscript. Sharing data through the manuscript publisher this way can have three limitations. First, a publisher may limit the size of data you can include. Second, publishers may make the data difficult to download, especially to download many datasets at once. Third, sometimes publishers have misplaced supplementary data making it difficult to access later, or have placed it behind a paywall. To avoid these problems, share especially larger or more complex tabular data in generalist repositories such as Zenodo (https://zenodo.org/; see “How to share: everything else”).
How to share: genomics
File format.
Genomics data comes in many formats with many different associated biological and technical variables. Usually raw genomics data consist of sequences stored in FASTA45 (https://faculty.Virginia.edu/wrpearson/fasta/) or FASTQ format.46 When possible, deposit raw data in CRAM format47 (https://samtools.github.io/hts-specs/), with unaligned reads included. CRAM files contain sequence information, similar to binary alignment/map (BAM)48 files, but take up much less space47 than either a BAM file or FASTQ file. With unaligned reads included, you should have the ability to reproduce a FASTQ file from a CRAM file.
When possible, deposit your processed data as CRAM, browser extensible data (BED)49 (https://genome.ucsc.edu/FAQ/FAQformat.html), or tab-delimited files. Format data with genomic regions as BED files instead of tab-delimited files. BED files store genomic coordinates of genomic region of interest in the first three columns. The BED format allows additional annotations in subsequent columns, making BED files great for working with genes, binned windows, CpG sites, or transcript data, such as experimental results from genomics assays such as RNA-seq,50-53 chromatin immunoprecipitation-sequencing (ChIP-seq),54 and assay for transposase-accessible chromatin (ATAC-seq).55 Using BED formats makes it easy to perform quick analyses on your data with software such as BEDTools56 (https://bedtools.readthedocs.io/) or Bioawk (https://github.com/lh3/bioawk). Use the bedGraph57 variant of BED when saving continuous-value data in track format.
In microarray analyses, use CEL58 (Affymetrix; https://www.affymetrix.com/support/developer/powertools/changelog/gcos-agcc/cel.html) or IDAT59 (Illumina) file formats for raw data. For storing processed microarray data, store information about genomic regions such as transcripts or CpG sites in BED format.
Compression.
Compress large-scale genomic data to minimize the amount of computational storage used. Use gzip (https://www.gnu.org/software/gzip/) compression for single files, and ZIP archives for collections of files. Text-based file formats easily compress.
Reference assemblies.
Most genomic data has coordinates defined by alignment to a reference genome for a species. Note the reference genome assembly version you align your data to in your manuscript and README file (see “Data, metadata, and documentation”. With advancement of sequencing technologies, genomic coordinates will vary between reference assemblies. For example, a number of parts of the genome changed coordinates between the GRCh37/hg1960 and GRCh38/hg3861 genome assemblies. Thus, without knowing the reference assembly used for an aligned file, the genomic coordinates hold no value.
Unfortunately, some file formats, such as BED, do not require reference genome assembly metadata. In these cases, make sure to explicitly note which reference assembly you used to align your samples.
Where to share.
Public repositories make datasets easily findable by interested parties (Table 1). The GEO26 repository (https://www.ncbi.nlm.nih.gov/geo/) houses public gene expression and gene regulation data.67 This includes data on DNA methylation, histone modifications, chromatin organization, and interactions between the genome and proteins such as transcription factors. The submission form requires you to specify both data files and relevant metadata, such as experimental details. After successful deposition, GEO provides you with an accession number for your manuscript. You can place an embargo on your data to withhold public access until publication of your manuscript. GEO will allow an embargo of up to 3 yr, but you can change the release date at any time.
Table 1:
Genomics repositories.
| Repository | Purpose | Formats |
|---|---|---|
| GEO26 | Quantitative gene expression, gene regulation, and epigenomics data, including data from RNA-seq,50-53 ChIP-seq,54 Hi-C,62 bisulfite sequencing,63 and microarrays | CRAM,47 BAM,48 SFF, HDF5, FASTQ, bedGraph, bigBed, WIG, bigWig, GFF, GTF, GEOarchive |
| SRA64 | Unassembled, high-throughput sequencing reads | CRAM,47 BAM,48 SFF, HDF5, FASTQ |
| EGA65 | All kinds of genomics data that contain private genetic or phenotype information on human participants | CRAM,47 BAM,48 FASTQ, VCF, SFF, HDF5 |
| GenBank66 | Other DNA and RNA sequences | FASTA |
Deposit high-throughput sequencing reads that don’t fit into GEO in the Sequence Read Archive (SRA)64 (https://www.ncbi.nlm.nih.gov/sra/). GEO will actually submit raw data files to SRA on your behalf, so you need not submit to both.
Deposit data that contains purely DNA or RNA sequence, rather than quantitative data, in GenBank66 (https://www.ncbi.nlm.nih.gov/genbank/). These data include sequence of genomic DNA, mRNA, noncoding RNA (ncRNA), plasmids, and synthetic constructs.
GenBank and the SRA make up part of the International Nucleotide Sequence Database Collaboration (INSDC)68 (https://www.insdc.org/), which also includes DNA Data Bank of Japan (DDBJ)69 (https://www.ddbj.nig.ac.jp/) and European Nucleotide Archive (ENA)70 (https://www.ebi.ac.uk/ena/). The INSDC members take data submitted to any of these repositories and automatically make it available in the others.
For sensitive genetic and phenotypic information from human participants, EGA65 (https://ega-archive.org/), a controlled-access genomics archive only permits qualified researchers you approve to access the data. Each dataset must have an associated data access committee that approves access requests and ensures responsible use of the data.25
How to share: proteomics
File format.
Like genomics data, mass spectrometry proteomics experiments generate both raw data and processed data. You should share both. Raw data typically come in a proprietary vendor file format, such as .raw (Thermo Scientific), .wiff (SCIEX), or .d (Agilent). Besides the raw data in their original format, also share peak files in the standard mzML file format71 (https://www.psidev.info/mzML).
Processed data include (1) identification results consisting of peptide-spectrum matches and protein identifications, and (2) quantification results consisting of determined amounts for the identified proteins. Public repositories, such as the ProteomeXchange consortium,72 require raw data and identification data for “complete” submissions. Provide identification data and quantification data in the standard mzTab format73 (https://www.psidev.info/mztab). Storing proteomics data in this format, a TSV variant, allows for use of various programming languages without the use of specialized libraries.
Also share other essential files besides the data itself used during the analysis. These include FASTA files with protein sequences or spectral libraries used for spectrum identification.
Metadata and documentation.
Provide a README with comprehensive metadata about the experiment, including sample metadata (such as organism and tissues), technical metadata (such as instrument model), and experimental design (such as number of technical and biological replicates). Use the Sample and Data Relationship Format for Proteomics (SDRF-Proteomics; https://github.com/bigbio/proteomics-metadata-standard)78 to encode this information in a structured fashion.
Use free text metadata to describe the study, the sample processing protocol, and the data processing protocol. Comprehensively describe all sample processing steps, including full analytical details. Provide full information on the bioinformatics tools used to process the data, including tool names, version numbers, the organism name, and version information of the FASTA files used for spectrum identification. Also provide the details of any statistical tests and thresholds employed.
For a reanalysis, describe the tools used and how the results differ from the originally deposited data. Do this both in free text metadata and in a README document.
Where to share.
The ProteomeXchange consortium72 (https://www.proteomexchange.org/), which includes the main proteomics data repositories such as Proteomics Identifications Database (PRIDE)74 (https://www.ebi.ac.uk/pride/) and Mass Spectrometry Interactive Virtual Environment (MassIVE) (https://massive.ucsd.edu/), provide a centralized system for sharing mass spectrometry proteomics data (Table 2). To submit data to ProteomeXchange member repositories, you must specify the data type of all files and link raw files to their corresponding peak files and identification results. By default, ProteomeXchange makes submitted datasets private and you can wait until publication time to make the data public. You can include a username and password in scientific manuscripts so that manuscript reviewers can still access the data.
Table 2:
Mass spectrometry proteomics repositories.
Most ProteomeXchange member repositories take any kind of mass spectrometry proteomics data, whereas some focus on a specific type of data. For example, PeptideAtlas SRMexperiment library (PASSEL)75 (http://www.peptideatlas.org/passel/) and Panorama Public76 (https://panoramaweb.org/) only accept deposition of targeted proteomics data.
Some repositories, including MassIVE, store the results of reanalysis of publicly available datasets also. MassIVE makes deposition of data reanalyses simple, as it does not require re-uploading original raw data files already available in public repositories. MassIVE will automatically link the new results to the original data.
How to share: microscopy
Microscopy image data use large amounts of disk space. Microscopy images also have complex associated metadata with great heterogeneity across datasets. The extreme heterogeneity comes from many sources, both biological and technical. Biologists acquire images in two or three spatial dimensions, and sometimes across time via live cell imaging experiments. Biologists also acquire images at different magnifications and across multiple light wavelength channels. The biological substrate captured varies in size (x, y) and depth (z). Biological substrates range from single molecules to whole organisms. Sample preparation before image acquisition also varies widely. Different biologists acquire images using different microscopes with different settings, often using proprietary software and file formats to save output image data with different resolutions, bit depths, and colors. These complexities pose unique data sharing challenges.79,80
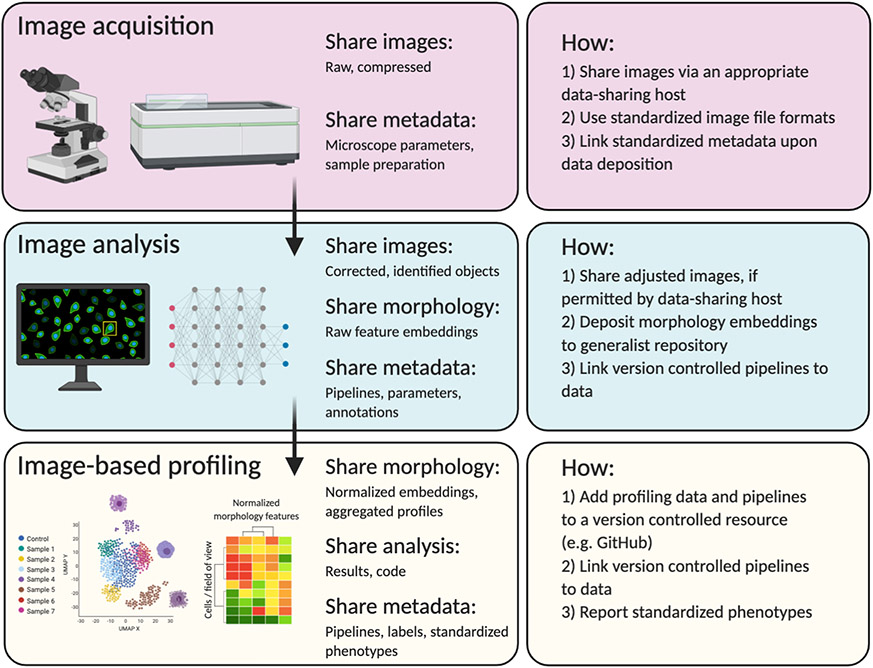
We provide guidelines on how to share microscopy images, intermediate data types, and metadata (Figure 1). These guidelines have three distinct themes:
Figure 1: You should share raw images, corrected images, image-based readouts, analysis pipelines, and comprehensive metadata.
While sometimes challenging, this process will ultimately increase the value of your data. Figure made with BioRender.
Use standardized file formats.
Select an appropriate repository.
Share high-value intermediate data and data processing pipelines.
Following these guidelines will enable the use of your microscopy data in secondary analyses, which will increase the impact of your data.
Compression.
Always share images with at least lossless compression. Lossless compression uses less disk space but loses no information as one can expand the compressed file into something identical to the original. Lossy compression, by contrast, loses information.
For very large microscopy datasets, using lossy compression may provide storage and access benefits without losing much vital biological information.87 Biologists often cringe at losing image resolution or information, but if the loss only marginally decreases analysis performance while increasing access speed and decreasing cost, only sharing the compressed formats may prove the best option. While microscopy data repositories currently offer high ceilings for dataset size (Table 3), this may change as microscopy images datasets grow in size and velocity.
Table 3:
Microscopy image repositories.
| Repository | Purpose | Substrate | Maximum size | Formats |
|---|---|---|---|---|
| IDR81 | Large and complete benchmark microscopy image datasets associated with a publication | Cells and tissues | 1000 GB, but you can ask to increase limit | Any Bio-Formats,82 OME-TIFF preferred |
| EMPIAR83 | Electron microscopy image data | High-resolution subcellular structures | Tens of TB | TIFF, HDF5, MRC, MRCS, DM4, IMAGIC, SPIDER, FEI |
| BioImage Archive84 | Link microscopy image data to associated publications | All non-medical images not suitable for IDR or EMPIAR | Tens of TB | Any Bio-Formats,82 OME-TIFF preferred |
| CellImageLibrary85 | Cell images and movies | Cells and intracellular structures | Tens of TB | Any Bio-Formats,82 OME-TIFF preferred |
| SSBD86 | Analysis of experimental and computationally-simulated biological image data | Any microscopic biological entity from single molecules to organelles and cells | Tens of TB | Any Bio-Formats,82 OME-TIFF preferred |
Intermediate data.
To maximize the value and impact of your microscopy studies, also share high-value intermediate processed data such as illumination-corrected images. You must correct for uneven illumination around the edges of each microscopy field of view, called shading or vignetting, using computational tools before measuring intensity-related continuous phenotypes.88,89 Typically, additional downstream analyses will use these adjusted images instead of the raw images. Ask the repository if it requires image adjustments before submission.
Image analysis.
Depending on experimental goals and strategies, you can also apply an image analysis pipeline. Image analysis produces summary data describing the images, such as morphology feature embeddings. These summary data take much less disk space than the original images. You can extract this summary data either manually or with specialized software.
Manual annotations provide a gold standard for benchmarking many computational approaches. Researchers generally create such annotations only for small image subsets, and these annotations include few phenotypic measurements.90 Nevertheless, if you create such annotations, you should make them public. When doing so, include important metadata such as the images used to derive the annotation, the annotator, time collected, and annotation batch (see “How to share: tabular data”).
To more rapidly and consistently measure a richer phenotypic landscape in larger datasets, avoid manual annotation and instead use a computational image analysis pipeline. Many free software packages perform image analysis and extract measurements, including CellProfiler,91 ImageJ,92 Icy,93 PhenoRipper,94 Wndcharm,95 and EBImage.96 These tools can perform many analyses, including segmenting and counting cells exhibiting a specific phenotype, identifying colocalization of molecules with fluorescent tags, and measuring cell morphology in an unbiased fashion.97
Image-based profiles.
Following image analysis, certain experiments result in high-dimensional readouts that require additional data processing. In these experiments, one extracts image-based profiles. Image-based profiles lack specificity for any target biology—instead, the profiles have no bias towards any biological hypothesis and represent the samples’ morphological states. This approach has evolved into a field known as image-based profiling, in which scientists discover biological insights through the aggregation and normalization of morphology features derived from image-analysis tools.97-99
Metadata.
To share microscopy data, first catalog experimental metadata in a standard format. Well-structured metadata provide a vital ingredient enabling others to find and use your data.82
Metadata standardization initiatives provide guidelines on what metadata to share. For example, the Open Microscopy Environment (OME) data model has proposed generic standards and developed software, such as Bio-Formats,82 for standardized metadata reporting and interoperable output file formats.100 The 4D Nucleome101 Imaging Standards Working group extended these guidelines to promote rigorous data sharing standards.102
Individual research communities have augmented these general standards. For example, communities have produced specialized guidelines for reporting cell migration data,103 time lapse data,104 3D microscopy images of whole brains (https://www.doryworkspace.org/), and fluorescence microscopy.105
Follow reporting standards for describing cell phenotypes106 and cell behavior.107 To increase the interoperability and value of your data, annotate your images using consistent ontologies.
Where to share.
Deposit your data in an appropriate repository108 (Table 3). Each microscopy data repository has a focused purpose, and accepts data that meet certain size, format, and biological sample conditions. For example, Image Data Resource (IDR)81 (https://idr.openmicroscopy.org/) accepts benchmark datasets with likely future secondary data analyses and in additional data integration efforts. Electron Microscopy Public Image Archive (EMPIAR)83 (https://www.ebi.ac.uk/pdbe/emdb/empiar/) accepts high-resolution images from subcellular compartments and biological structures. Bioimage Archive84 (https://www.ebi.ac.uk/bioimage-archive/) provides a home for all other microscopy image datasets, often those of a smaller size. CellImageLibrary85 (http://www.cellimagelibrary.org) hosts a wide variety of biological images and movies for research and education purposes. The Systems Science of Biological Dynamics (SSBD) database86 (http://ssbd.qbic.riken.jp) also hosts a variety of images, and even provides a home for computationally-simulated microscopy images.
To determine the appropriate repository, align your microscopy image dataset to the repository with the best-aligned purpose, biological substrate, file size, and output file format (Table 3). When in doubt, contact the repository to determine the suitability of your data. Together, the repositories described here provide a home for all microscopy image datasets.
Depositing your images only in a journal Portable Document Format (PDF) file or pasted in a Microsoft Word or PowerPoint document does not satisfy the FAIR principles (Box 1). This practice will result in low-quality images and compression artifacts and will make future analysis impossible. Do not share data by shipping physical storage devices to requesters or by using cloud provider links.109 Do not share data using a custom solution either (see “How not to share: do not use custom, in-house solutions”).
For sharing image data, we usually do not recommend using generalist repositories such as Figshare and Zenodo (see “How to share: everything else”). These repositories store data that do not have domain-specific resources. They therefore lack the special focus necessary to sufficiently catalog the complexities of microscopy images. Image-based profiles, which consist in small, intermediate data representing morphology feature embeddings, provide the only exception to this. For now, generalist repositories serve as the best place to deposit image-based profiles.
How to share: structural biology
Structural biology encompasses a range of different techniques, including X-ray crystallography, nuclear magnetic resonance (NMR) and multiple kinds of electron microscopy methods, such as single-particle cryogenic electron microscopy (EM) (cryo-EM), cryogenic electron tomography (cryo-ET),110,111 and microcrystal electron diffraction (microED).112 Each technique derives information from distinct initial raw data using unique processing approaches.
Where to share.
The various structural biology techniques exhibit vast differences in raw data types, files sizes, and paths toward final results. As such, each scientific community developed independent repositories for storing the input and output of these experiments (Table 4). In addition to sharing the final atomic coordinates, each structural biology field developed an individual path to sharing raw and processed data.
Table 4:
Structural biology repositories.
| Repository | Purpose | Substrate | Maximum size | Formats |
|---|---|---|---|---|
| PDB | Atomic coordinates and ensembles | Subcellular structures | Tens of MB | PDB, mmCIF |
| EMDB | 3D reconstructions from processed EM data | Subcellular structure | GB | MRC, CCP4 |
| EMPIAR | Electron microscopy raw and processed image data | Subcellular structures (single-particle cryo-EM, cryo-ET) | Tens of TB | TIFF, HDF5, MRC, MRCS, DM4, EER, IMAGIC, SPIDER, SCIPION, EMDB-SFF, AMIRA, STL, VTK, VTP, OBJ, AVI, JPEG, PNG, EMX, BLENDER, TXT |
| IRRMC | X-ray diffraction raw data | Subcellular structures | Hundreds of MB | Raw diffraction data formats |
| BMRB | NMR raw data | Subcellular structures | GB | CCPN, mmCIF, PDB, NMR-STAR, X-PLOR |
Protein Data Bank (PDB)113 (https://rcb.org/) serves as a repository for atomic coordinates of nucleic acids, proteins, and larger assemblies. Most journals require structural biology manuscripts to include unique PDB identifiers. Upon deposition of the finalized coordinate files and metadata, authors obtain an unique PDB identifier. Deposition involves creation of a unique identifier and a password. This keeps the files and metadata visible only to the authors and the database operators.
You can place an embargo on your PDB deposition to withhold public access until publication of your manuscript, or 1 yr, whichever comes first. We recommend immediate access at the time of publication. Moreover, some journals currently also require coordinate files at the time of submission, or by reviewer request. We encourage you to make all your data accessible upon acceptance of the manuscript.
Associate X-ray crystallography structure factors directly with your PDB entries. Store raw diffraction data in the Integrated Resource for Reproducibility in Macromolecular Crystallography (IRRMC)114,115 (https://www.proteindiffraction.org/).
Deposit NMR structural ensembles in the Biological Magnetic Resonance Bank (BMRB)116 (https://bmrb.io/). The BMRB usually represents multiple chains under a single identifier. For many experiments, you can deposit raw NMR data, in the form of restraints, in the NMR Restraints Grid116,117 (https://restraintsgrid.bmrb.wise.edu/).
The most important element of sharing EM data consists in the deposition of the raw, unprocessed data into EMPIAR83 (https://www.ebi.ac.uk/pdbe/emdb/empiar/). Modern direct detection cameras can generate thousands of images each day. Each image file can contain tens or hundreds of movie frames. Depending on the file format, raw data from a single session can range from 1 TB–10 TB. Deposit the raw, uncorrected movie stacks, as well as summed, motion-corrected micrographs, final particle coordinates, and final alignment files in EMPIAR. This greatly simplifies data validation and reproducibility. It also simplifies software development—having access to raw and processed training data improves heterogeneity classification and machine learning approaches.118-120
Cryo-EM microscopy uses Coulomb potential densities to build and refine coordinate files. Thus, the final coordinate model depends on the quality of the EM reconstructions, on your individual choices, and on the model-refinement approaches used. Thus, providing final filtered and unfiltered maps has great importance to the validation claims made in a manuscript. Deposit the coordinates and final calculated Coulomb potential maps in Electron Microscopy Data Bank (EMDB)121 (https://www.ebi.ac.uk/pdbe/emdb/), obtaining unique PDB andEMDB identifiers.
When a cryo-EM dataset reveals structural variability, provide a consensus output and deposit all the associated models and maps needed to support the claims of the manuscript in separate depositions. In addition, if multiple 3D variability clusters result in a large number of intermediate maps, add them to an EMPIAR deposition, along with the raw data. As new technologies enable collecting more data in shorter times, the focus on describing motion will increase. Accordingly, computational structural heterogeneity analysis approaches will become more sophisticated.
How to share: everything else
For some types of data not covered above, no specialized repositories exist. Deposit these kinds of data in generalist repositories that can manage many different types of data.
Organization.
Organize your data depositions with raw data separate from results. Use ZIP archives to collect your data so that viewers can preview individual files in repositories such as Zenodo. To make your data clear and interpretable, include a README with a detailed description of the project, and an explanation of what each of the files contain.
Where to share.
First, see whether an appropriate data repository exists in the re3data directory (https://www.re3data.org/). For cases where no such repository exists, we recommend Zenodo (https://zenodo.org/), a generalist repository that allows for deposition of data, code, analysis and manuscripts and has robust semantic versioning as well as a persistence guarantee.
Open Science Framework (OSF) (https://osf.io/) provides a system for organizing scientific projects, including data, code, and protocols. It also serves as a generalist repository, allowing you to share data and other materials simply by making you OSF project publicly available.
How not to share: do not use custom, in-house solutions
Hosting your data using a customized solution you create may seem attractive. For example, some share their data using public Amazon Web Services Simple Storage Service (S3) links or even building a new repository specifically for their project. Using a custom solution provides an illusion of complete control. In reality, custom efforts usually result in something fragile with uncertain permanence, and in difficulty for tracking attribution and citation.
Don’t reinvent the wheel. Third-party repositories have more permanence, exist outside the control of the original data generators, provide storage and infrastructure maintenance cost savings. Third-party repositories also enforce metadata standards that facilitate FAIR sharing principles (Box 1). These repositories have access to funding streams and institutional commitments that individual investigators lack. Users interact with third-party repositories frequently, and many have had negative experiences by custom hosting efforts, which generate more problems than solutions.
Discussion
We suggest a four-step checklist for biological researchers to complete when submitting a manuscript:
Deposit raw and processed data. Use a specialist repository if possible. Dedicate these datasets to the public domain with CC0.
Deposit code to a generalist repository.
Deposit all miscellaneous files to generalist repository.
Put all repository accession numbers and license information in your manuscript.
We encourage you to create a lab publication checklist that contains the necessary steps for a lab member to prepare a manuscript and associated artifacts for publication. Use the four-step checklist as a starting point, and add details specific to you and the kinds of data you work with.
We understand that some will find the above recommendations difficult or overwhelming at first. We encourage you to do what you can. Improving your data management and sharing practices gradually will still provide great value for you and other researchers. Finally, the intent to share matters.
Acknowledgments
We thank Erin Weisbart (0000-0002-6437-2458) and Anne E. Carpenter (0000-0003-1555-8261) (Broad Institute) for helpful discussions on how to share microscopy and image data. This work was supported by the Natural Sciences and Engineering Research Council of Canada (RGPIN-2015-03948 to M.M.H.), the CIHR Fellowship (MFE-171256 to S.L.W.), the Research Foundation—Flanders (12W0418N to W.B.) and the NIH (R24OD011883 to M.A.H.).
Footnotes
Competing interests
The authors declare no competing interests.
References
- 1.Wilkinson MD, Dumontier M, Aalbersberg IJ, Appleton G, Axton M, Baak A, Blomberg N, Boiten J-W, da Silva Santos LB, Bourne PE, et al. The FAIR Guiding Principles for scientific data management and stewardship. Scientific Data 3, 160018 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.McMurry JA, Köhler S, Washington NL, Balhoff JP, Borromeo C, Brush M, Carbon S, Conlin T, Dunn N, Engelstad M, et al. Navigating the phenotype frontier: The Monarch Initiative. Genetics 203, 1491–1495 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shefchek KA, Harris NL, Gargano M, Matentzoglu N, Unni D, Brush M, Keith D, Conlin T, Vasilevsky N, Zhang XA, et al. The Monarch Initiative in 2019: an integrative data and analytic platform connecting phenotypes to genotypes across species. Nucleic Acids Research 48, D704–D715 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Haendel M, Su A, McMurry J, Chute CG, C M, B G, C W, S M, H H, Peter R, M H, M B, D S, M M-T, G J, H L, P M & T C FAIR-TLC: Metrics to Assess Value of Biomedical Digital Repositories: Response to RFI NOT-OD-16-133 10.5281/zenodo.203295. [DOI] [Google Scholar]
- 5.McMurry JA, Juty N, Blomberg N, Burdett T, Conlin T, Conte N, Courtot M, Deck J, Dumontier M, Fellows DK, et al. Identifiers for the 21st century: How to design, provision, and reuse persistent identifiers to maximize utility and impact of life science data. PLOS Biology 15, e2001414 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vasilevsky NA, Brush MH, Paddock H, Ponting L, Tripathy SJ, LaRocca GM & Haendel MA On the reproducibility of science: unique identification of research resources in the biomedical literature. Peer J 1, e148 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ford J Unreliable research: trouble at the lab. Economist 409, 26–31 (2013). [Google Scholar]
- 8.ENCODE Project Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature 489, 57–74 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Moore JE, Purcaro MJ, Pratt HE, Epstein CB, Shoresh N, Adrian J, Kawli T, Davis CA, Dobin A, Kaul R, et al. Expanded encyclopaedias of DNA elements in the human and mouse genomes. Nature 583, 699–710 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Collado-Torres L, Nellore A, Kammers K, Ellis SE, Taub MA, Hansen KD, Jaffe AE, Langmead B & Leek JT Reproducible RNA-seq analysis using recount2. Nature Biotechnology 35, 319–321 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Piwowar HA, Day RS & Fridsma DB Sharing detailed research data is associated with increased citation rate. PLOS ONE 2, e308 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.National Institutes of Health. Final NIH Policy for Data Management and Sharing. NIH Guide to Grants and Contracts, NOT-OD-21–013 (2020). [Google Scholar]
- 13.National Institutes of Health. Supplemental information to the NIH Policy for Data Management and Sharing: selecting a repository for data resulting from NIH-supported research. NIH Guide to Grants and Contracts, NOT-OD-21–016 (2020). [Google Scholar]
- 14.Pierce HH, Dev A, Statham E & Bierer BE Credit data generators for data reuse. Nature 570, 30–32 (2019). [DOI] [PubMed] [Google Scholar]
- 15.Byrd JB & Greene CS Data-sharing models. New England Journal of Medicine 376,2305 (2017). [DOI] [PubMed] [Google Scholar]
- 16.Greene CS, Garmire LX, Gilbert JA, Ritcfile MD & Hunter LE Celebrating parasites. Nature Genetics 49, 483–484 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Starr J, Castro E, Crosas M, Dumontier M, Downs RR, Duerr R, Haak LL, Haendel M, Herman I, Hodson S, et al. Achieving human and machine accessibility of cited data in scholarly publications. PeerJ Computer Science 1, e1 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Knoppers BM & Thorogood AM Ethics and big data in health. Current Opinion in Systems Biology 4, 53–57 (2017). [Google Scholar]
- 19.World Health Organization & Council for International Organizations of Medical Sciences. International ethical guidelines for health-related research involving humans (2016).
- 20.Clayton EW, Evans BJ, Hazel JW & Rothstein MA The law of genetic privacy: applications, implications, and limitations. Journal of Law and the Biosciences 6, 1–36 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Haibe-Kains B, Adam GA, Hosny A, Khodakarami E, Waldron L, Wang B, McIntosh C, Goldenberg A, Kundaje A, Greene CS, et al. Transparency and reproducibility in artificial intelligence. Nature 586, E14–E16 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zook M, Barocas S, Boyd D, Crawford K, Keller E, Gangadharan SP, Goodman A, Hollander R, Koenig BA, Metcalf J, et al. Ten simple rules for responsible big data research. PLOS Computational Biology 3, e1005399 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Deverka PA, Majumder MA, Villanueva AG, Anderson M, Bakker AC, Bardill J, Boerwinkle E, Bubela T, Evans BJ, Garrison NA, et al. Creating a data resource: what will it take to build a medical information commons? Genome Medicine 9, 84 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Malin B, Goodman K, et al. Between access and privacy: challenges in sharing health data. Yearbook of Medical Informatics 27, 55–59 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Byrd JB, Greene AC, Prasad DV, Jiang X & Greene CS Responsible, practical genomic data sharing that accelerates research. Nature Reviews Genetics 21, 615–629 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Clough E & Barrett T The Gene Expression Omnibus database. Statistical Genomics, 93–110 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Leipzig J, Nüst D, Hoyt CT, Soiland-Reyes S, Ram K & Greenberg J The role of metadata in reproducible computational research. arXiv 2006, 08589 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brazma A, Hingamp P, Quackenbush J, Sherlock G, Spellman P, Stoeckert C, Aach J, Ansorge W, Ball CA, Causton HC, Gaasterland T, Glenisson P, Holstege FC, Kim IF, Markowitz V, Matese JC, Parkinson H, Robinson A, Sarkans U, Schulze-Kremer S, Stewart J, Taylor R, Vilo J &Vingron M Minimum information about a microarray experiment (MIAME)—toward standards for microarray data. Nature Genetics 29, 365–371 (2001). [DOI] [PubMed] [Google Scholar]
- 29.Brazma A Minimum Information About a Microarray Experiment (MIAME) – Successes, Failures, Challenges. The Scientific World 9, 420–423 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. Gene Ontology: tool for the unification of biology. Nature Genetics 25, 25–29 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gene Ontology Consortium. The Gene Ontology resource: enriching a GOld mine. Nucleic Acids Research 49, D325–D334 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Haendel MA, Balhoff JP, Bastian FB, Blackburn DC, Blake JA, Bradford Y, Comte A, Dahdul WM, Dececchi TA, Druzinsky RE, et al. Unification of multi-species vertebrate anatomy ontologies for comparative biology in Uberon. Journal of Biomedical Semantics 5, 21 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Noble WS A quick guide to organizing computational biology projects. PLOS Computational Biology 5, e1000424 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wilson G, Bryan J, Cranston K, Kitzes J, Nederbragt L & Teal TK Good enough practices in scientific computing. PLOS Computational Biology 13, e1005510 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Carbon S, Champieux R, McMurry JA, Winfree L, Wyatt LR & Haendel MA An analysis and metric of reusable data licensing practices for biomedical resources. PLOS ONE 14, e0213090 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schofield PN, Bubela T, Weaver T, Portilla L, Brown SD, Hancock JM, Einhorn D, Tocchini-Valentini G, de Angelis MH & Rosenthal N Post-publication sharing of data and tools. Nature 461, 171–173 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.National Institute of Allergy and Infectious Diseases. Data Sharing for Grants—Final Research Data SOP http://www.niaid.nih.gov/research/grants-data-sharing-final-research (2020).
- 38.National Institute of Allergy and Infectious Diseases. Genomic Data Sharing Plan Examples http://www.niaid.nih.gov/research/gds-plan-examples (2020).
- 39.Information technology — document description and processing languages — Office Open XML file formats — part 1: fundamentals and markup language reference en. Standard ISO/IEC 29500-1:2016 (International Organization for Standardization, Geneva, 2016). [Google Scholar]
- 40.Zeeberg BR, Riss J, Kane DW, Bussey KJ, Uchio E, Linehan WM, Barrett JC & Weinstein JN Mistaken identifiers: gene name errors can be introduced inadvertently when using Excel in bioinformatics. BMC Bioinformatics 5, 1–6 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ziemann M, Eren Y & El-Osta A Gene name errors are widespread in the scientific literature. Genome Biology 17, 177 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bruford EA, Braschi B, Denny R, Jones TE, Seal RL & Tweedie S Guidelines for human gene nomenclature. Nature Genetics 52, 754–758 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Broman KW & Woo KH Data organization in spreadsheets. The American Statistician 72, 2–10 (2018). [Google Scholar]
- 44.Ellis SE & Leek JT How to share data for collaboration. The American Statistician 72, 53–57 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lipman DJ & Pearson WR Rapid and sensitive protein similarity searches. Science 227, 1435–1441 (1985). [DOI] [PubMed] [Google Scholar]
- 46.Cock PJ, Fields CJ, Goto N, Heuer ML & Rice PM The Sanger FASTQ file format for sequences with quality scores, and the SolexaXS/Illumina FASTQ variants. Nucleic Acids Research 38, 1767–1771 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fritz MH-Y, Leinonen R, Cochrane G & Birney E Efficient storage of high throughput DNA sequencing data using reference-based compression. Genome Research 21, 734–740 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G & Durbin R The sequence alignment/map format and SAMtools. Bioinformatics 25, 2078–2079 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM & Haussler D The human genome browser at UCSC. Genome Research 12, 996–1006 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lister R, O’Malley RC, Tonti-Filippini J, Gregory BD, Berry CC, Millar AH & Ecker JR Highly integrated single-base resolution maps of the epigenome in Arabidopsis. Cell 133, 523–536 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wilhelm BT, Marguerat S, Watt S, Schubert F, Wood V, Goodhead I, Penkett CJ, Rogers J & Bähler J Dynamic repertoire of a eukaryotic transcriptome surveyed at single-nucleotide resolution. Nature 453, 1239–1243 (2008). [DOI] [PubMed] [Google Scholar]
- 52.Cloonan N, Forrest AR, Kolle G, Gardiner BB, Faulkner GJ, Brown MK, Taylor DF, Steptoe AL, Wani S, Bethel G, et al. Stem cell transcriptome profiling via massive-scale mRNA sequencing. Nature Methods 5, 613–619 (2008). [DOI] [PubMed] [Google Scholar]
- 53.Mortazavi A, Williams BA, McCue K, Schaeffer L & Wold B Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nature Methods 5, 621–628 (2008). [DOI] [PubMed] [Google Scholar]
- 54.Johnson DS, Mortazavi A, Myers RM & Wold B Genome-wide mapping of in vivo protein-DNA interactions. Science 316, 1497–1502 (2007). [DOI] [PubMed] [Google Scholar]
- 55.Buenrostro JD, Giresi PG, Zaba LC, Chang HY & Greenleaf WJ Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nature Methods 10, 1213–1218 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Quinlan AR & Hall IM BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–842 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Karolchik D, Hinrichs AS & Kent WJ The UCSC Genome Browser. Current Protocols in Bioinformatics 40, 1.4.1–1.4.33 (2012). [DOI] [PubMed] [Google Scholar]
- 58.Affymetrix Developer Network https://www.affymetrix.com/support/developer/powertools/changelog/gcos-agcc/cel.html (2021).
- 59.Smith ML, Baggerly KA, Bengtsson H, Ritchie ME & Hansen KD illuminaio: An open source IDAT parsing tool for Illumina microarrays. F1000Research 2, 264 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Church DM, Schneider VA, Graves T, Auger K, Cunningham F, Bouk N, Chen H-C, Agarwala R, McLaren WM, Ritchie GR, et al. Modernizing reference genome assemblies. PLOS Biology 9, e1001091 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Schneider VA, Graves-Lindsay T, Howe K, Bouk N, Chen H-C, Kitts PA, Murphy TD, Pruitt KD, Thibaud-Nissen F, Albracht D, et al. Evaluation of GRCh38 and de novo haploid genome assemblies demonstrates the enduring quality of the reference assembly. Genome Research 27, 849–864 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lieberman-Aiden E, Van Berkum NL, Williams L, Imakaev M, Ragoczy T, Telling A, Amit I, Lajoie BR, Sabo PJ, Dorschner MO, et al. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science 326, 289–293 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Frommer M, McDonald LE, Millar DS, Collis CM, Watt F, Grigg GW, Molloy PL & Paul CL A genomic sequencing protocol that yields a positive display of 5-methylcytosine residues in individual DNA strands. Proceedings of the National Academy of Sciences of the United States of America 89, 1827–1831 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kodama Y, Shumway M & Leinonen R The Sequence Read Archive: explosive growth of sequencing data. Nucleic Acids Research 40, D54–D56 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lappalainen I, Almeida-King J, Kumanduri V, Senf A, Spalding JD, Saunders G, Kandasamy J, Caccamo M, Leinonen R, Vaughan B, et al. The European Genome-phenome Archive of human data consented for biomedical research. Nature Genetics 47, 692–695 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sayers EW, Cavanaugh M, Clark K, Pruitt KD, Schoch CL, Sherry ST & Karsch-Mizrachi I GenBank. Nucleic Acids Research 49, D92–D96 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Barrett T, Wilhite SE, Ledoux P, Evangelista C, Kim IF, Tomashevsky M, Marshall KA, Phillippy KH, Sherman PM, Holko M, et al. NCBI GEO: archive for functional genomics data sets—update. Nucleic Acids Research 41, D991–D995 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Arita M, Karsch-Mizrachi I & Cochrane G The International Nucleotide Sequence Database Collaboration. Nucleic Acids Researched 49, D121–D124 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Fukuda A, Kodama Y, Mashima J, Fujisawa T & Ogasawara O DDBJ update: streamlining submission and access of human data. Nucleic Acids Research 49, D71–D75 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Harrison PW, Ahamed A, Aslam R, Alako BT, Burgin J, Buso N, Courtot M, Fan J, Gupta D, Haseeb M, et al. The European Nucleotide Archive in 2020. Nucleic Acids Research 49, D82–D85 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Martens L, Chambers M, Sturm M, Kessner D, Levander F, Shofstahl J, Tang WH, Römpp A, Neumann S, Pizarro AD, Montecchi-Palazzi L, Tasman N, Coleman M, Reisinger F, Souda P, Hermjakob H, Binz P-A & Deutsch EW mzML—a Community Standard for Mass Spectrometry Data. Molecular & Cellular Proteomics 10, R110.000133 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Deutsch EW, Bandeira N, Sharma V, Perez-Riverol Y, Carver JJ, Kundu DJ, García-Seisdedos D, Jarnuczak AE, Hewapathirana S, Pullman BS, Wertz J, Sun Z, Kawano S, Okuda S, Watanabe Y, Hermjakob H, MacLean B, MacCoss MJ, Zhu Y, Ishihama Y & Vizcaíno JA The ProteomeXchange Consortium in 2020: enabling ’big data’ approaches in proteomics. Nucleic Acids Research 48, D1145–D1152 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Griss J, Jones AR, Sachsenberg T, Walzer M, Gatto L, Hartler J, Thallinger GG, Salek RM, Steinbeck C, Neuhauser N, Cox J, Neumann S, Fan J, Reisinger F, Xu Q-W, del Toro N, Perez-Riverol Y, Ghali F, Bandeira N, Xenarios I, Kohlbacher O, Vizcaíno JA & Hermjakob H The mzTab data exchange format: communicating mass-spectrometry-based proteomics and metabolomics experimental results to a wider audience. Molecular & Cellular Proteomics 13, 2765–2775 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Perez-Riverol Y, Csordas A, Bai J, Bernal-Llinares M, Hewapathirana S, Kundu DJ, Inuganti A, Griss J, Mayer G, Eisenacher M, Pérez E, Uszkoreit J, Pfeuffer J, Sachsenberg T, Yilmaz ş., Tiwary S, Cox J, Audain E, Walzer M, Jarnuczak AF, Ternent T, Brazma A & Vizcaíno JA The PRIDE database and related tools and resources in 2019: improving support for quantification data. Nucleic Acids Research 47, D442–D450 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Farrah T, Deutsch EW, Kreisberg R, Sun Z, Campbell DS, Mendoza L, Kusebauch U, Brusniak M-Y, Hüttenhain R, Schiess R, Selevsek N, Aebersold R & Moritz RL PASSEL: The PeptideAtlas SRMexperiment Library. Proteomics 12, 1170–1175 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sharma V, Eckels J, Schilling B, Ludwig C, Jaffe JD, MacCoss MJ & MacLean B Panorama Public: a public repository for quantitative data sets processed in Skyline. Molecular & Cellular Proteomics 17, 1239–1244 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.MacLean B, Tomazela DM, Shulman N, Chambers M, Finney GL, Frewen B, Kern R, Tabb DL, Liebler DC & MacCoss MJ Skyline: an open source document editor for creating and analyzing targeted proteomics experiments. Bioinformatics 26, 966–968 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Perez-Riverol Y & European Bioinformatics Community for Mass Spectrometry. Toward a sample metadata standard in public proteomics repositories. Journal of Proteome Research 19, 3906–3909 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zaritsky A Sharing and reusing cell image data. Molecular Biology of the Cell 29, 1274–1280 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Marqués G, Pengo T & Sanders MA Imaging methods are vastly underreported in biomedical research. eLife 9, e55133 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Williams E, Moore J, Li SW, Rustici G, Tarkowska A, Chessel A, Leo S, Antal B, Ferguson RK, Sarkans U, Brazma A, Carazo Salas RE & Swedlow JR Image Data Resource: a bioimage data integration and publication platform. Nature Methods 14, 775–781 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Linkert M, Rueden CT, Allan C, Burel J-M, Moore W, Patterson A, Loranger B, Moore J, Neves C, MacDonald D, Tarkowska A, Sticco C, Hill E, Rossner M, Eliceiri KW & Swedlow JR Metadata matters: access to image data in the real world. Journal of Cell Biology 189, 777–782 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Iudin A, Korir PK, Salavert-Torres J, Kleywegt GJ & Patwardhan A EMPIAR: a public archive for raw electron microscopy image data. Nature Methods 13, 387–388 (2016). [DOI] [PubMed] [Google Scholar]
- 84.Ellenberg J, Swedlow JR, Barlow M, Cook CE, Sarkans U, Patwardhan A, Brazma A & Birney E A call for public archives for biological image data. Nature Methods 15, 849–854 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Orloff DN, Iwasa JH, Martone ME, Ellisman MH & Kane CM The cell: an image library-CCDB: a curated repository of microscopy data. Nucleic Acids Research 41, D1241–D1250 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Tohsato Y, Ho KHL, Kyoda K & Onami S SSBD: a database of quantitative data of spatiotemporal dynamics of biological phenomena. Bioinformatics 32, 3471–3479 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Balázs B, Deschamps J, Albert M, Ries J & Hufnagel L A real-time compression library for microscopy images. bioRxiv, 164624 (2017). [Google Scholar]
- 88.Singh S, Bray M-A, Jones T & Carpenter A Pipeline for illumination correction of images for high-throughput microscopy. Journal of Microscopy 256, 231–236 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Peng T, Thorn K, Schroeder T, Wang L, Theis FJ, Marr C & Navab N A BaSiC tool for background and shading correction of optical microscopy images. Nature Communications 8, 14836 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Ljosa V, Sokolnicki KL & Carpenter AE Annotated high-throughput microscopy image sets for validation. Nature Methods 9, 637–637 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.McQuin C, Goodman A, Chernyshev V, Kamentsky L, Cimini BA, Karhohs KW, Doan M, Ding L, Rafelski SM, Thirstrup D, Wiegraebe W, Singh S, Becker T, Caicedo JC & Carpenter AE CellProfiler 3.0: Next-generation image processing for biology. PLOS Biology 16, e2005970 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Rueden CT, Schindelin J, Hiner MC, DeZonia BE, Walter AE, Arena ET & Eliceiri KW ImageJ2: ImageJ for the next generation of scientific image data. BMC Bioinformatics 18, 529 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.De Chaumont F, Dallongeville S, Chenouard N, Hervé N, Pop S, Provoost T, Meas-Yedid V, Pankajakshan P, Lecomte T, Montagner YL, Lagache T, Dufour A & Olivo-Marin J-C Icy: an open bioimage informatics platform for extended reproducible research. Nature Methods 9, 690–696 (2012). [DOI] [PubMed] [Google Scholar]
- 94.Rajaram S, Pavie B, Wu LF & Altschuler SJ PhenoRipper: software for rapidly profiling microscopy images. Nature Methods 9, 635–637 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Shamir L, Orlov N, Eckley DM, Macura T, Johnston J & Goldberg IG Wndchrm — an open source utility for biological image analysis. Source Code for Biology and Medicine 3, 13 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Pau G, Fuchs F, Sklyar O, Boutros M & Huber W EBImage–an R package for image processing with applications to cellular phenotypes. Bioinformatics 26, 979–981 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Caicedo JC, Cooper S, Heigwer F, Warchal S, Qiu P, Molnar C, Vasilevich AS, Barry JD, Bansal HS, Kraus O, Wawer M, Paavolainen L, Herrmann MD, Rohban M, Hung J, Hennig H, Concannon J, Smith I, Clemons PA, Singh S, Rees P, Horvath P, Linington RG & Carpenter AE Data-analysis strategies for image-based cell profiling. Nature Methods 14, 849–863 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Scheeder C, Heigwer F & Boutros M Machine learning and image-based profiling in drug discovery. Current Opinion in Systems Biology 10, 43–52 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Chandrasekaran SN, Ceulemans H, Boyd JD & Carpenter AE Image-based profiling for drug discovery: due for a machine-learning upgrade? Nature Reviews Drug Discovery 20, 1–15 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Goldberg IG, Allan C, Burel J-M, Creager D, Falconi A, Hochheiser H, Johnston J, Mellen J, Sorger PK & Swedlow JR The Open Microscopy Environment (OME) Data Model and XML file: open tools for informatics and quantitative analysis in biological imaging. Genome Biology 6, R47 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Dekker J, Belmont AS, Guttman M, Leshyk VO, Lis JT, Lomvardas S, Mirny LA, O’shea CC, Park PJ, Ren B, et al. The 4D Nucleome project. Nature 549, 219–226 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Huisman M, Hammer M, Rigano A, Farzam F, Gopinathan R, Smith C, Grunwald D & Strambio-De-Castillia C Minimum Information guidelines for fluorescence microscopy: increasing the value, quality, and fidelity of image data. arXiv 1910, 11370 (2020). [Google Scholar]
- 103.Gonzalez-Beltran AN, Masuzzo P, Ampe C, Bakker G-J, Besson S, Eibl RH, Friedl P, Gunzer M, Kittisopikul M, Dévédec SEL, Leo S, Moore J, Paran Y, Prilusky J, Rocca-Serra P, Roudot P, Schuster M, Sergeant G, Strömblad S, Swedlow JR, van Erp M, Van Troys M, Zaritsky A, Sansone S-A & Martens L Community standards for open cell migration data. GigaScience 9, giaa041 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Burek P, Scherf N & Herre H Ontology patterns for the representation of quality changes of cells in time. Journal of Biomedical Semantics 10, 16 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Lee J-Y & Kitaoka M A beginner’s guide to rigor and reproducibility in fluorescence imaging experiments. Molecular Biology of the Cell 29, 1519–1525 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Jupp S, Malone J, Burdett T, Heriche J-K, Williams E, Ellenberg J, Parkinson H & Rustici G The Cellular Microscopy Phenotype Ontology. Journal of Biomedical Semantics 7, 28 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Sluka JP, Shirinifard A, Swat M, Cosmanescu A, Heiland RW & Glazier JA The Cell Behavior Ontology: describing the intrinsic biological behaviors of real and model cells seen as active agents. Bioinformatics 30, 2367–2374 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Dance A Find a home for every imaging data set. Nature 579, 162–163 (2020). [DOI] [PubMed] [Google Scholar]
- 109.Andreev A & Koo DE Practical guide to storage of large amounts of microscopy data. Microscopy Today 28, 42–45 (2020). [Google Scholar]
- 110.Grimm R, Bärmann M, Häckl W, Typke D, Sackmann E & Baumeister W Energy filtered electron tomography of ice-embedded actin and vesicles. Biophysical Journal 72, 482–489 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Dierksen K, Typke D, Hegerl R, Walz J, Sackmann E & Baumeister W Three-dimensional structure of lipid vesicles embedded in vitreous ice and investigated by automated electron tomography. Biophysical Journal 68, 1416–1422 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Shi D, Nannenga BL, Iadanza MG & Gonen T Three-dimensional electron crystallography of protein microcrystals. eLife 2, e01345 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN & Bourne PE The Protein Data Bank. Nucleic Acids Research 28, 235–242 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Grabowski M, Langner KM, Cymborowski M, Porebski PJ, Sroka P, Zheng H, Cooper DR, Zimmerman MD, Elsliger MA, Burley SK & Minor W A public database of macromolecular diffraction experiments. Acta Crystallographica Section D: Structural Biology 72, 1181–1193 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Grabowski M, Cymborowski M, Porebski PJ, Osinski T, Shabalin IG, Cooper DR & Minor W The Integrated Resource for Reproducibility in Macromolecular Crystallography: Experiences of the first four years. Structural Dynamics 6, 064301 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Ulrich EL, Akutsu H, Doreleijers JF, Harano Y, Ioannidis YE, Lin J, Livny M, Mading S, Maziuk D, Miller Z, Nakatani E, Schulte CF, Tolmie DE, Kent Wenger R, Yao H & Markley JL BioMagResBank. Nucleic Acids Research 36, 402–408 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Doreleijers JF, Nederveen AJ, Vranken W, Lin J, Bonvin AMJJ, Kaptein R, Markley JL & Ulrich EL BioMagResBank databases DOCR and FRED containing converted and filtered sets of experimental NMR restraints and coordinates from over 500 protein PDB structures. Journal of Biomolecular NMR 32, 1–12 (2005). [DOI] [PubMed] [Google Scholar]
- 118.Wagner T, Merino F, Stabrin M, Moriya T, Antoni C, Apelbaum A, Hagel P, Sitsel O, Raisch T, Prumbaum D, Quentin D, Roderer D, Tacke S, Siebolds B, Schubert E, Shaikh TR, Lill R, Gatsogiannis C & Raunser S SPHIRE-crYOLO is a fast and accurate fully automated particle picker for cryo-EM. Communications Biology 2, 218 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Bepler T, Kelley K, Noble AJ & Berger B Topaz-Denoise: general deep denoising models for cryoEM and cryoET. Nature Communications 11, 5208 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Tegunov D & Cramer P Real-time cryo-electron microscopy data preprocessing with Warp. Nature Methods 16, 1146–1152 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Lawson CL, Baker ML, Best C, Bi C, Dougherty M, Feng P, van Ginkel G, Devkota B, Lagerstedt I, Ludtke SJ, Newman RH, Oldfield TJ, Rees I, Sahni G, Sala R, Velankar S, Warren J, Westbrook JD, Henrick K, Kleywegt GJ, Berman HM & Chiu W EMDataBank.org: unified data resource for cryoEM. Nucleic Acids Research 39, D456–D464 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]