Abstract
Infectious hematopoietic necrosis virus (IHNV) is a rhabdovirus that produces an acute, lethal infection in rainbow trout (Oncorhynchus mykiss). Fish that survive infection cease to produce detectable infectious virus at approximately 46 days after infection, yet there is evidence that survivor fish continue to harbor virus particles (B. S. Drolet, P. P. Chiou, J. Heidel, and J. C. Leong, J. Virol. 69:2140–2147, 1995). In an effort to determine the biological function of these particles, the kidneys and livers from IHNV survivors were harvested and divided into samples for nested reverse transcriptase PCR analysis and explant culture. Sequences for the IHNV nucleoprotein and polymerase genes were detected in 50 and 89%, respectively, of the organs from survivor fish. When explant tissue cultures were infected with purified standard IHNV, the liver tissues from survivor fish produced up to 10-fold less virus than naive control fish liver tissues. In addition, immunosorbent electron microscopy analysis of the supernatant media from the cultured explants of survivor fish revealed truncated particles, whereas the control tissue supernatants contained only standard viral particles. These results suggest that the truncated IHNV particles observed in persistently infected fish are defective interfering particles that may mediate virus persistence.
Infectious hematopoietic necrosis virus (IHNV) is the prototype rhabdovirus for a newly named genus of Rhabdoviridae, Novirhabdovirus. The IHNV genome is comprised of six viral genes in the order 3′-N-P-M-G-NV-L-5′, which code for the nucleocapsid (N), phosphoprotein (P, formerly M1), matrix (M, formerly M2), glycoprotein (G), nonvirion protein (NV), and polymerase (L) (20, 21). The NV gene, which encodes a nonvirion protein, distinguishes this genus from the other Rhabdoviridae (20, 30). The virus normally produces an acute infection in rainbow trout (Oncorhynchus mykiss) fry that eventually results in death, and mortality rates in hatcheries can reach as high as 80 to 90%. What has intrigued biologists about this disease is that IHNV cannot be isolated from survivors within 2 months after the onset of the epizootic. When the fish migrate to the ocean, there is still no detectable infectious virus in any of the tissues. As these anadromous fish return to spawn as adults, the animals are initially negative for infectious virus, and as they mature to spawning age, the prevalence of virus-positive animals increases (25). There are two models that have been proposed to account for the reappearance of infectious virus in these animals. One model proposes that the survivors clear all virus after the acute infection and that when these fish return from the sea to their spawning grounds, they are severely immunosuppressed and susceptible to reinfection from virus present in resident fish or in contaminated sediment in the river. There has been, however, no convincing evidence of any other virus carriers in the river system, and virus has not been isolated from river sediment near the spawning grounds. The second model proposes that survivors carry the virus in a latent state until they are immunosuppressed by the stress of the spawning migration. Infectious virus is then produced and released in the spawning fluids. Neither model had much experimental support until it was found that survivors of an IHNV epizootic contained viral proteins and viral nucleic acid (10).
Truncated viral particles resembling rhabdovirus defective interfering particles (DIPs) were observed in fish that had survived an IHNV epizootic 1 and 2 years after infectious virus was no longer detectable in the survivors (10). The evidence included immunogold staining of these particles with monoclonal antibody specific to the viral proteins, detection of viral nucleic acid by PCR amplification, and detection of viral protein by immunohistochemistry. Because these fish had been reared for 2 years in virus-free well water under quarantine conditions, the evidence of virus persistence in these animals without detectable infectious virus prompted an investigation into the nature and biological significance of these truncated particles.
In the present study, we examined the tissues of IHNV survivor fish for truncated particles and attempted to determine the biological function of these particles. Through techniques of explant culture, plaque assay, immunosorbent electron microscopy (ISEM), and nested reverse transcriptase PCR (RT-PCR), we have confirmed our earlier observations that virus does persist in survivors of IHNV infection and that superinfection of the tissues of survivor fish results in the production of truncated particles, a finding consistent with a model that postulates that DIPs are mediators of virus persistence.
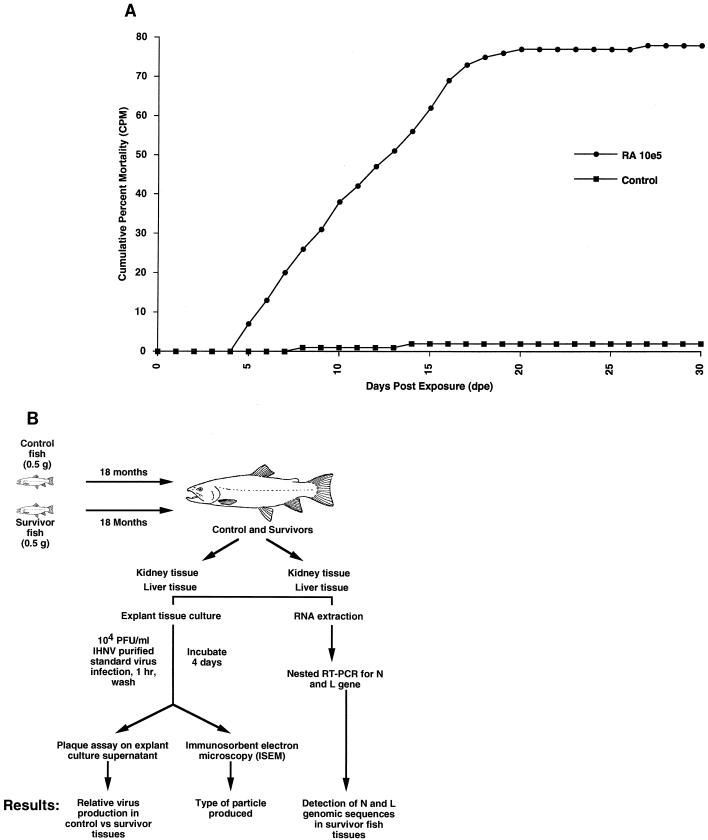
We obtained fish that were persistently infected with IHNV by exposing specific-pathogen-free rainbow trout fry, obtained from the Fish Toxicology Laboratory at Oregon State University, to IHNV. Each treatment group consisted of three tanks of 50 fish each. Fish in one group of 15-liter tanks were treated with 105 PFU of IHNV-RA (Rangen isolate) per ml, and those in another group were treated with phosphate-buffered saline. The fish were exposed to each treatment in a volume of 2 liters for a period of 6 h, after which the volume of water in the tank was raised to 20 liters. At the end of 30 days, the fish remaining in each group of three tanks were combined and moved to 100-liter tanks, so that there were two 100-liter tanks (virus survivors and control fish) where the fish were maintained initially. As the fish grew over an 18-month period, the group was once again split into four 100-liter tanks where the survivors were kept separate from the control fish. The fish were reared at the Salmon Disease Laboratory at Oregon State University. This facility is supplied with specific-pathogen-free well water. Thus, laboratory conditions and the water source precluded environmental viral contamination. In 6 years of continuous operation, there has never been a virus outbreak in control fish at the facility. After 6 days, a majority of the infected fish began to show clinical signs of disease, with extended abdomens, petechial hemorrhages, and a whirling form of swimming. Dead fish were removed daily for the duration of the experiment, although fish mortality reached its maximum by 21 days postexposure, and no further mortalities were observed in the virus-infected groups (Fig. 1A). The cumulative percent mortality for the virus-infected group was 78%. In the control group, 2% (3 fish) died during the experiment.
FIG. 1.
Mortality of rainbow trout challenged with the Rangen isolate of IHNV. (A) Triplicate tanks of 100 fish each were infected with 105 PFU of IHNV per ml. The average mortality for each virus isolate is shown on the ordinate and is plotted against the number of days postexposure. The range in the mortalities did not vary significantly between tanks and thus is not shown. (B) Flowchart of experimental procedures for the processing of tissues from survivor and control fish for ISEM, plaque assay, and nested RT-PCR.
After 18 months, the survivor and control fish were sampled for viral RNA by nested RT-PCR. RNA was extracted from the tissues by the guanidinium thiocyanate method carried out with Stat 60 (Tel-Test, Inc., Friendswood, Tex.) as previously described (10). Briefly, the entire liver and kidneys from each fish were removed and minced, and the minced tissue was divided into six equal portions. One tissue sample was used for RNA extraction, and the other five samples were used for explant tissue culture. For RNA extraction, tissues were homogenized with 1 ml of Stat 60 directly in 2-ml storage tubes with disposable plastic pestles to prevent contamination of the samples. The samples were incubated at room temperature for 5 min before 200 μl of chloroform was added to each sample and each sample was processed as recommended by the manufacturer.
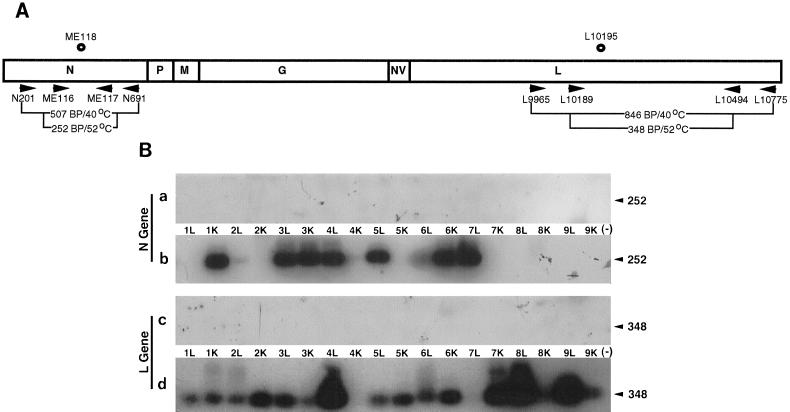
The primers that were used for these nested RT-PCRs were based on the IHNV complete genome sequence submitted by Morzunov et al. (26). First-strand synthesis for the RT-PCR was primed by either the primer N691 (5′-CGATCTTGGCCAGGACTCC-3′), located at positions 709 to 691 for the N gene product, or the primer L10775 (5′-AGCGTCGGTGTCCCACACT-3′), located at positions 10793 to 10775 for the L gene product (Fig. 2A). These primers were specific for only the negative-sense IHNV genome and not the positive-sense IHNV mRNA. The reaction mixture included 5 μM primer, 1 mM deoxynucleoside triphosphates, 1 U of RNasin (Promega, Inc., Madison, Wis.), 10 mM dithiothreitol, 2 U of avian myeloblastosis virus (AMV) RT, and 1× AMV buffer in a 25-μl volume. The samples were incubated at 42°C for 60 min, heated to 95°C for 5 min, and then placed on ice. One unit of RNase H was added to each sample, and each sample was incubated for 20 min at 37°C, heated to 95°C for 5 min, and then diluted to a final volume of 500 μl.
FIG. 2.
(A) The schematic diagram of the IHNV genome indicates the location of the oligonucleotide primers and probes used for the RT-PCR experiments and Southern blotting. The IHNV genome is shown with its six viral genes in the order 3′-N-P-M-G-NV-L-5′. Arrowheads show the polarity of the primers, and circles represent the oligonucleotides used as end-labeled probes for hybridization analysis. (B) Southern blot analysis of nested RT-PCR products of the IHNV nucleoprotein and polymerase genes. Panels a and c (control) and b and d (survivor) show the nested RT-PCR products for the N gene and L gene. Samples are identified by fish number and tissue type (kidney [K] or liver [L]). (−), RNA sample from uninfected CHSE-214 tissue culture cells. The products were analyzed by Southern blotting with internal oligonucleotides specific for either the N or L gene. Numbers on the right are molecular sizes, in base pairs.
The cDNAs were subsequently used as templates for nested PCR within either the N gene or the L gene (Fig. 2A). For the primary PCR the mixture contained the following: 10 μl of cDNA, 0.9 mM MgCl2, 200 μM deoxynucleoside triphosphates, 1 μM primer N691 or L10775, 1 μM primer N201 (5′-CGCACTCAGAGAGACGTTC-3′) (positions 183 to 201) or primer L9965 (5′-AACCCTATTCGACGAACGG-3′) (positions 9947 to 9965), and 1 U of Taq polymerase (Promega, Inc.). The DNAs were denatured at 95°C for 1 min, annealed at 40°C for 1 min, and extended at 74°C for 2 min for 35 cycles in an automatic thermal cycler (Coy Laboratory Products, Grass Lake, Mich.).
For the nested PCR the mixture was the same as that described above, with 10 μl of the primary PCR mixture serving as the template for the nested reaction and with primers ME116 (5′-TTCGCAGATCCCAACAACAA-3′) (positions 493 to 512) and ME117 (5′-CTTGGTGAGCTTCTGTCCA-3′) (positions 744 to 727) for the N gene and primers L10189 (5′-AATCGAGGGGCCCCAGAATCCCTTCT-3′) (positions 10164 to 10189) and L10494 (5′-GAAGCGCGGGGAGGGACCA-3′) (positions 10512 to 10494) for the L gene. The DNAs were denatured at 95°C for 1 min, annealed at 52°C for 1 min, and extended at 74°C for 2 min for 35 cycles in an automatic thermal cycler. ME118 (5′-TTTTGGCAGTATGTGGCCATCTTGTC-3′) (positions 626 to 601) and L10195 (5′-GCTCGCTCCAAGACCTCTT-3′) (positions 10195 to 10213), internal primers for the N and L genes, respectively, were 5′ end labeled with [γ-32P]ATP and used as probes in Southern blot analysis (18).
The results of the nested RT-PCR are shown in Fig. 2B. Of the nine fish examined, all harbored IHNV nucleic acid from either the N or L gene in the kidney or liver tissues. The N gene was detected in the liver tissues of fish assigned numbers 2 to 7 and in the kidney tissues of fish 1, 3, and 6. The L gene was detected in the liver tissues of fish 1 to 6, 8, and 9 and in the kidney tissues of fish 1 to 3 and 5 to 9. Only the kidney tissue of fish 4 was negative for both the N and L genes, but the liver of fish 4 was positive for both. Tissues of all control fish were negative for both the N and L genes. In the previous study by Drolet et al. (10), the IHNV N gene was detected in only one in six survivor fish tested (17%). In this study we were able to detect the IHNV N gene in 9 of 18 tissues from survivor fish (50%). The IHNV L gene was detectable in 16 of 18 tissues from survivor fish (89%).
Tissue culture-propagated truncated and standard virus particles were purified for these experiments. Chinook salmon embryo (CHSE-214) cell monolayers were infected with IHNV-RA at a multiplicity of infection of 0.01 and grown as previously described (10). The medium was harvested when viral cytopathic effects were apparent (usually within 48 to 72 h). Cellular debris was removed by low-speed centrifugation. This clarified medium was subsequently used to purify truncated and standard IHNV. Virus purification involved ultracentrifugation to concentrate virus particles, followed by velocity sedimentation through a 5 to 50% sucrose gradient. The truncated virus particles banded at a lower density than the standard IHNV. Two sequential purification procedures were carried out in order to remove contaminating truncated or standard virus from each of the respective bands. To ensure that the purified particles were indeed truncated or standard virus, electron microscopic (EM) examination of the separated preparations was performed. Short, round to ovoid viral structures with diameters of approximately 50 to 60 nm were observed in preparations of the lower-density band. In the standard virus preparation, bullet-shaped virus particles measuring 50 by 130 nm were observed.
To determine if the truncated particles that were in the tissues of the survivor fish could interfere with viral replication, an explant culture assay for virus interference was developed. The entire kidneys and livers from survivor fish were harvested aseptically and placed in medium with 2× antibiotics and amphotericin B (Fungizone) modified Eagle medium with penicillin, streptomycin, and gentamicin). The tissues were minced into 1-mm2 pieces and then divided equally into six 6-ml tubes (Fisher, Pittsburgh, Pa.). One tube of tissue was used for RNA extraction as indicated above. To four of these tubes, IHNV at 4 × 105 PFU/mg of tissue was added and allowed to adsorb for 1 h, at which time tissues were washed with medium twice and resuspended in 2 ml of medium. One tube from each sample set received no virus to ensure that no resident infectious virus was harbored in the tissues of the survivor fish. The five tubes were gently rocked for 4 days, and the supernatant media were collected.
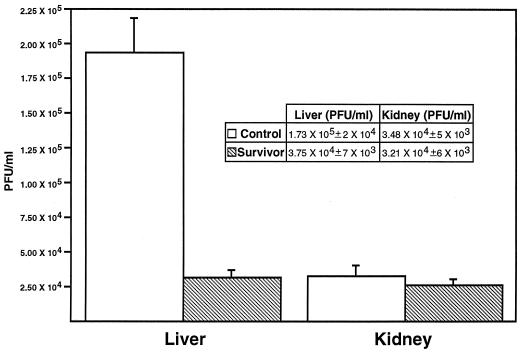
The supernatants were stored at −80°C until the virus titer could be measured in plaque assays as previously described (10). Liver tissues from the survivor fish produced an average of 3.75 × 104 PFU/ml, whereas those from the control fish produced 1.73 × 105 PFU/ml, an approximately 10-fold difference (Fig. 3). The statistical software program Statview (Abacus Concepts, Berkeley, Calif.) was used to determine the difference in mean values of the control and survivor groups by analysis of variance. The mean difference in virus production between the control and survivor liver tissue was statistically significant (P < 0.0001). In contrast, the virus titers from the kidney tissue of survivor and control fish were essentially the same (Fig. 3). The presence of antibody in the survivor fish was assessed by enzyme-linked immunosorbent assay. We saw no difference between sera from survivor and control fish even at a 1:20 dilution in an assay system with a detection limit of 1:81,920 for positive fish antisera. In addition, the sera from survivor and control fish were indistinguishable from a known negative-control serum in this assay. A nonspecific immune response, such as interferon production, could explain this plaque reduction. However, interferon would be elicited in both survivor and control fish tissues; it seemed reasonable to assume that interferon inhibition of virus production in both survivor and control would be observed. Fresh liver tissue and explant liver tissue from both survivor and control fish were examined histologically. No evidence of liver damage was observed in fish from either group. There was also no lymphocytic infiltration in the tissues. In addition, the explant tissues after 4 days in culture were examined by EM, and again, tissues from the two groups were indistinguishable and in good condition.
FIG. 3.
Virus titers produced from liver and kidney tissues of IHNV survivor and control fish. Supernatants from explant cultures of liver and kidney tissues from survivor or control fish were used in plaque assays to assess the ability to replicate IHNV. The mean difference in virus production between the control and survivor livers was statistically significant (P < 0.0001). The data, with bars indicating standard errors, is shown for 27 different samples in the survivor and control groups.
To detect IHNV in the explant tissue supernatants, EM grids were incubated with monoclonal antibody 136J, which is specific for the IHNV glycoprotein, for 1 h and washed once with distilled water to remove the non-virus glycoprotein cell debris. The supernatants were filtered through a 0.22-μm-pore-size Acrodisc filter (Gelman, Ann Arbor, Mich.) and then ultracentrifuged for 2 h in a SW 50.1 rotor (Beckman, Fullerton, Calif.). The virus pellets were resuspended in 40 μl of distilled water and subsequently incubated with the antibody-coated grid for 24 h at 4°C. After being washed once with distilled water, the grids were stained with phosphotungstic acid for 1 min and examined with a transmission electron microscope (Philips).
Purified standard and truncated virus served as controls for the ISEM micrographs. Standard virus possessed the shape (bullet) and size prototypical for IHNV (Fig. 4A). The purified truncated virus was a short, round to ovoid structure with a diameter of approximately 50 to 60 nm, and it showed the envelope glycoprotein studding the surface of the particle (Fig. 4C). When the explant culture supernatants were concentrated by centrifugation and analyzed by ISEM, truncated-virus-like structures were observed in the samples taken from the survivor tissues (Fig. 4D). These particles were the same size and shape as those in the purified truncated-virus preparations. In addition, the particles often contained the hallmark rhabdoviral indentation on one end, as in the top particle shown in Fig. 4D, as well as the envelope glycoproteins. As few as one truncated virus particle/grid and as many as four truncated virus particles/grid were observed among the standard virus particles in the preparations. In contrast, only standard virus particles were observed in the preparations from the infected control tissues, with up to 10 particles/square on the nickel grids (Fig. 4B). The fact that truncated virus was observed only in the survivor fish and not in the control fish is consistent with the theory that defective interfering virus is responsible for the plaque reduction in the survivor fish.
FIG. 4.
Immunosorbent electron micrographs of IHNV standard and truncated particles. Particles were selected by ISEM from a virus preparation grown in cell culture (A) and in control, uninfected fish liver explant culture (B). Truncated particles were selected from a virus preparation grown in cell culture (C) and in survivor fish liver explant culture (D). The EM grids were coated with a monoclonal antibody (136J) specific for the IHNV glycoprotein, incubated with the culture supernatant samples, washed with distilled water, and then examined by transmission EM. The white bar in each panel represents 50 nm.
The number of particles in each explant culture sample was approximated. If we assume that 1 PFU is equal to 200 particles in an IHNV plaque assay (23), then in the plaque assays conducted for the liver tissues of the control fish (1.7 × 105 PFU/ml), there are approximately 7 × 107 particles of IHNV in the 2 ml of explant culture medium. Since as many as 10 standard virus particles were observed on each square of the grid and there were 100 squares/grid, then 1,000 particles/grid represented 7 × 107 IHNV standard particles in the control sample. In the survivor samples, there were approximately 10 to 100 standard particles/grid and 1 to 4 truncated virus particles/grid, or 7 × 104 to 28 × 104 truncated particles per survivor sample. The amount of truncated virus particles ranged from 4 to 10% of the total particles in the survivor samples.
An attempt to detect DIPs directly from survivor fish tissues without the addition of purified standard virus was made. The culture media from the explant cultures were concentrated by centrifugation and analyzed for the presence of truncated virus. No truncated virus was observed in either the control or survivor fish liver and kidney explants. More than likely, there were too few particles in the samples for direct detection by ISEM.
The report of truncated particles in fish surviving an IHNV epizootic provided the first evidence that interference by viral DIPs might be the answer to a long-standing paradox in the life cycle of this virus (10). In the present study, the persistence of viral RNA in survivor fish was confirmed by nested RT-PCR. The difference in the detection frequencies for the primers for the N (50%) and the L (89%) RNA was striking. Since the primers selected for first-strand synthesis were specific for the IHNV genome, the molar ratios of the N and L cDNAs should have been equal. There are several possible explanations for this difference, including (i) differences in primer binding affinity, (ii) the loss of or less efficient production of cDNA during first-strand synthesis, and (iii) the presence of DIPs in the survivor tissue. This last explanation is attractive since panhandle-type structures (29) missing the 3′ end of the genome make up the majority of the DIPs for vesicular stomatitis virus (35). In this case, there would be more L gene sequences than N gene sequences in a DIP preparation. The prevalence of the L gene in the samples might indicate the presence of more DIP genomes in the tissues of survivor fish.
The 10-fold decrease in virus titer observed only in the liver tissue of survivor fish might be accounted for by an immune response in the survivor fish (11, 34). However, the same observation was not made in the kidney tissues, which contain high concentrations of lymphoid cells in fish. Liver, in contrast, has relatively few immune cells, and any background nonspecific immune response would be minimal in this tissue. There was no detectable anti-IHNV antibody in either control or survivor fish, and we were unable to obtain reliable measurements of the cellular immune response to IHNV in these fish. Any in vitro induction of the cellular immune response would not have been possible in the short time period (4 days) used for the explant culture. Interferon production could also account for the plaque reductions; however, interferon should be elicited in both the control as well as the survivor tissues. An explanation consistent with these observations is that the reduction in virus production seen specifically in the liver tissue of survivor fish was mediated by virus interference through DIPs.
Although these results support the hypothesis that truncated IHNV particles are biologically active in survivor fish, the observation of these particles by ISEM is particularly compelling. The ISEM technique affords sensitivity as well as specificity, because cellular debris is washed away and only specific viral particles remain on the grid for negative staining. ISEM has been used to detect virus from samples that were contaminated with other viruses and extraneous cellular and bacterial debris that would impede detection of the virus (7, 8, 19). As few as one to four truncated particles/grid were observed in each of the survivor fish samples. Even with so few particles in each sample, ISEM made it possible to detect these truncated particles.
DIPs have been postulated to be key elements in disease modulation and persistence (15). Since most viruses have been shown to produce DIPs upon passage in tissue culture resulting in a reduction in virus yield and amelioration of cell cytopathicity, DIP production has been an attractive explanation for the modulation of virus pathogenesis. Over the years, numerous attempts to understand how DIPs moderate virus infections in vivo have been made (2, 13, 16, 22, 27, 28, 31, 32). These studies have shown that simultaneous infection of experimental animals with DIPs and homologous standard virus alters the pathology and time course of the ensuing disease. However, these studies have never provided any indication of how DIPs change the course of virus pathogenesis in natural infections. To date there has been no unequivocal report of the detection of DIPs in any natural infection in animals or humans. It has long been debated whether DIPs are actually present and involved in disease modulation in natural infections or merely represent laboratory culture phenomena. The failure to observe DIPs is due largely to the fact that detection of them relies primarily on their capacity to interfere with standard virus replication, which is a time-consuming assay (17), or on the detection of deleted genomic RNA, which requires 5 to 50 pg (5).
Although most viruses are thought to produce DIPs in tissue culture cells and some of these DIPs have been shown to modulate infections in animals, much of the proposed impact of these particles in natural infections has been speculative. There have been no unequivocal reports of the detection of DIPs from any natural virus infection in mammals. Nevertheless, the possibility that DIPs exist in nature and play a role in modulating infection has been strengthened by studies with hepatitis virus, rotavirus, and influenza virus. Robinson suggested that the establishment of persistent infections caused by hepatitis B virus might involve DIPs (31), and Ruiz-Opazo et al. (33) reported evidence that supercoiled DNA was present in infectious hepatitis B virus virions and that relaxed circular DNA may be the form present in DIPs. For hepatitis A virus, defective genomes have been found in fecal specimens and viremic blood collected in the course of hepatitis A virus infection in humans (27). The deletions in the viral genome coincided with two of the deletions detected in particles grown in vitro in cell cultures and have been shown to interfere with the replication of standard hepatitis A virus virions. Pedley et al. isolated rotavirus from two chronically infected immunodeficient children (28). The genomes of these two isolates had an unusual structure analogous to that of DIP genomes. Serial undiluted passage of bovine rotavirus in tissue culture produced virus particles with extra RNA segments similar to the virus isolated from the immunodeficient children. These particles could be DIPs; however, the human rotaviruses could not be adapted to grow in tissue culture, and it was not possible to demonstrate their interfering activity.
Bean et al. (3) found that an avirulent type A influenza virus isolated from chickens contained small DIP-type RNAs which were not present in a lethal form of the virus. There were no differences in the antigenic or genomic structure of the isolates to account for the difference in virulence. Interfering activity was shown by the reduction in the numbers of deaths in chickens infected with mixtures of the virulent strain and the avirulent, small-RNA-containing strain. The data suggested that the presence of the small RNAs resulted in the avirulence. However, the virus strains were passaged twice in embryonated eggs before analysis; virus isolated directly from the infected chickens was not analyzed. It is possible that the small RNAs were synthesized by the avirulent strain during the isolation procedure. Evidence has also been sought to implicate DIPs in the persistence of arenaviruses in small rodents (4), infectious pancreatic necrosis virus in salmon (9, 24), measles virus with subacute sclerosing panencephalitis (6, 12), and vesicular stomatitis virus in convalescent hamsters (1).
The data presented in this paper suggests that the truncated particles observed by EM in fish persistently infected with IHNV (10) have biological activity as DIPs. With IHNV infections in trout, we can examine virus infection during the acute phase, recovery, and persistence. Indeed, it appears that this system is suited for determining the events that might trigger the path to persistence rather than virus clearance in natural (noninjected) virus infections. We propose that IHNV persistence in trout is an exceptional model system for studying RNA virus persistence and DIP effects on virus pathogenesis.
Acknowledgments
We thank Sandra Ristow (Washington State University, Pullman) for monoclonal antibodies used for this study and Pat Buckley for discussion and technical assistance with EM and photography. In addition, we gratefully acknowledge Linda Bootland, Grant Trobridge, and Dan Mourich for their helpful discussions.
Portions of this work were supported by the USDA National Research Initiative Competitive Grants Program (grant 9603020), the United States Department of Agriculture to the Western Regional Aquaculture Consortium (grant 683021), and the National Oceanic and Atmospheric Administration, Office of Sea (grants BA36RGO451 and J0098653).
Footnotes
Oregon Agriculture Experiment Station technical paper 11278.
REFERENCES
- 1.Barrera J C, Letchworth G J. Persistence of vesicular stomatitis virus New Jersey RNA in convalescent hamsters. Virology. 1996;219:453–464. doi: 10.1006/viro.1996.0271. [DOI] [PubMed] [Google Scholar]
- 2.Barrett A D, Dimmock N J. Defective interfering viruses and infections of animals. Curr Top Microbiol Immunol. 1986;128:55–84. doi: 10.1007/978-3-642-71272-2_2. [DOI] [PubMed] [Google Scholar]
- 3.Bean W J, Kawaoka Y, Wood J M, Pearson J E, Webster R G. Characterization of virulent and avirulent A/chicken/Pennsylvania/83 influenza A viruses: potential role of defective interfering RNAs in nature. J Virol. 1985;54:151–160. doi: 10.1128/jvi.54.1.151-160.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Buckmeier M J, Welsh R M, Dutko F J, Oldstone M B A. The virology and immunobiology of lymphocytic choriomeningitis virus infection. Adv Immunol. 1980;30:275–331. doi: 10.1016/s0065-2776(08)60197-2. [DOI] [PubMed] [Google Scholar]
- 5.Cave D R, Hagen F S, Palma E L, Huang A S. Detection of vesicular stomatitis virus RNA and its defective-interfering particles in individual mouse brains. J Virol. 1984;50:86–91. doi: 10.1128/jvi.50.1.86-91.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cernescu C, Sorodoc Y. Subacute sclerosing panencephalitis and defective interfering measles virus particles. Virology. 1980;31:3–8. [PubMed] [Google Scholar]
- 7.Derrick K S. Quantitative assay for plant viruses using serological specific electron microscopy. Virology. 1973;23:119–124. doi: 10.1016/0042-6822(73)90068-8. [DOI] [PubMed] [Google Scholar]
- 8.Doane F W. Immunoelectron microscopy in diagnostic virology. Ultrastruct Pathol. 1987;11:681–686. doi: 10.3109/01913128709048454. [DOI] [PubMed] [Google Scholar]
- 9.Dobos P, Roberts T E. The molecular biology of infectious pancreatic necrosis virus: a review. Can J Microbiol. 1983;29:377–384. doi: 10.1139/m83-062. [DOI] [PubMed] [Google Scholar]
- 10.Drolet B S, Chiou P P, Heidel J, Leong J C. Detection of truncated virus particles in a persistent RNA virus infection in vivo. J Virol. 1995;69:2140–2147. doi: 10.1128/jvi.69.4.2140-2147.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Grace M F, Manning J J. Histogenesis of the lymphoid organs in rainbow trout, Salmo gairdneri Rich. 1836. Dev Comp Immunol. 1980;4:255–264. doi: 10.1016/s0145-305x(80)80029-2. [DOI] [PubMed] [Google Scholar]
- 12.Hall W W, Choppin P W. Evidence for lack of synthesis of the M polypeptide of measles virus in brain cells in subacute sclerosing panencephalitis. Virology. 1979;99:443–447. doi: 10.1016/0042-6822(79)90026-6. [DOI] [PubMed] [Google Scholar]
- 13.Holland J J. Defective interfering rhabdoviruses. In: Wagner R R, editor. The rhabdoviruses. New York, N.Y: Plenum Press; 1987. pp. 297–360. [Google Scholar]
- 14.Hsiung G D. Diagnostic virology illustrated by light and electron microscopy. New Haven, Conn: Yale University Press; 1982. [Google Scholar]
- 15.Huang A S, Baltimore D. Defective viral particles and viral disease processes. Nature (London) 1970;226:325–327. doi: 10.1038/226325a0. [DOI] [PubMed] [Google Scholar]
- 16.Huang A S, Baltimore D. Defective interfering animal viruses. In: Fraenkel-Conrat H, Wagner R R, editors. Comprehensive virology. Vol. 10. New York, N.Y: Plenum Press; 1977. pp. 73–116. [Google Scholar]
- 17.Kawai A, Matsumoto S. A sensitive bioassay for detecting defective interfering particles of rabies virus. Virology. 1982;122:98–108. doi: 10.1016/0042-6822(82)90380-4. [DOI] [PubMed] [Google Scholar]
- 18.Kim C H, Casey J W. Genomic variation and segregation of equine infectious anemia virus during acute infection. J Virol. 1992;66:3879–3882. doi: 10.1128/jvi.66.6.3879-3882.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kjeldsberg E. Immunonegative stain techniques for electron microscopic detection of viruses in human faeces. Ultrastruct Pathol. 1986;10:553–560. doi: 10.3109/01913128609007212. [DOI] [PubMed] [Google Scholar]
- 20.Kurath G, Leong J C. Characterization of infectious hematopoietic necrosis virus mRNA species reveals a nonvirion rhabdovirus protein. J Virol. 1985;53:462–468. doi: 10.1128/jvi.53.2.462-468.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kurath G, Ahern K G, Pearson G D, Leong J C. Molecular cloning of the six mRNA species of infectious hematopoietic necrosis virus, a fish rhabdovirus, and gene order determination by R-loop mapping. J Virol. 1985;53:469–476. doi: 10.1128/jvi.53.2.469-476.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lazzarini R A, Keene J D, Schubert M. The origins of defective interfering particles of the negative-strand RNA viruses. Cell. 1981;26:145–154. doi: 10.1016/0092-8674(81)90298-1. [DOI] [PubMed] [Google Scholar]
- 23.Leong J C. Molecular basis of immunity and disease. J Fish Biol. 1995;47:61–75. [Google Scholar]
- 24.MacDonald R D, Yamamoto T. Quantitative analysis of defective interfering particles in infectious pancreatic necrosis virus preparations. Arch Virol. 1978;57:77–89. doi: 10.1007/BF01315639. [DOI] [PubMed] [Google Scholar]
- 25.Meyers T R, Thomas J B, Follett J E, Saft R R. Infectious hematopoietic necrosis virus: trends in prevalence and the fish management approach in Alaskan sockeye salmon culture. J Aquat Anim Health. 1990;2:85–98. [Google Scholar]
- 26.Morzunov S P, Winton J R, Nichol S T. The complete genome structure and phylogenetic relationship of infectious hematopoietic necrosis virus. Virus Res. 1995;38:175–192. doi: 10.1016/0168-1702(95)00056-v. [DOI] [PubMed] [Google Scholar]
- 27.Nuesch J P, de Chastonay J, Siegl G. Detection of defective genomes in hepatitis A virus particles present in clinical specimens. J Gen Virol. 1989;70:3475–3480. doi: 10.1099/0022-1317-70-12-3475. [DOI] [PubMed] [Google Scholar]
- 28.Pedley S, Hundley F, Chrystie I, McCrae M A, Desselberger U. The genomes of rotaviruses isolated from chronically infected immunodeficient children. J Gen Virol. 1984;65:1141–1150. doi: 10.1099/0022-1317-65-7-1141. [DOI] [PubMed] [Google Scholar]
- 29.Perrault J. Origin and replication of defective interfering particles. Curr Top Microbiol Immunol. 1981;93:151–207. doi: 10.1007/978-3-642-68123-3_7. [DOI] [PubMed] [Google Scholar]
- 30.Peters D. Divergent evolution of Rhabdoviridae and Bunyaviridae in plants and animals. Semin Virol. 1991;2:27–38. [Google Scholar]
- 31.Robinson W S. Persistent infection with hepatitis B virus. In: Stevens J G, Todaro G J, Fox C F, editors. Persistent viruses. New York, N.Y: Academic Press; 1978. pp. 485–497. [Google Scholar]
- 32.Roux L, Simon A E, Holland J J. Effects of defective interfering viruses on virus replication and pathogenesis in vitro and in vivo. Adv Virus Res. 1991;40:181–211. doi: 10.1016/S0065-3527(08)60279-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ruiz-Opazo N, Chakraborty P R, Shafritz D A. Evidence for supercoiled hepatitis B virus DNA in chimpanzee liver and serum dane particles: possible implications in persistent HBV infection. Cell. 1982;29:129–138. doi: 10.1016/0092-8674(82)90097-6. [DOI] [PubMed] [Google Scholar]
- 34.Scapigliati G, Romano N, Abelli L. Cellular techniques in aquaculture: antibodies against Ig and T- and B-lymphocytes of the sea bass Dicentrarchus labrax define the basic immunological profile, abstr. G11. Dev Comp Immunol. 1997;1997:140. [Google Scholar]
- 35.Villarreal L P, Breindl M, Holland J J. Determination of molar ratios of vesicular stomatitis virus induced RNA species in BHK21 cells. Biochemistry. 1976;15:1663–1667. doi: 10.1021/bi00653a012. [DOI] [PubMed] [Google Scholar]