Abstract
Respiratory syncytial virus (RSV) cpts530/1030 is an attenuated, temperature-sensitive subgroup A vaccine candidate derived previously from cold-passaged RSV (cpRSV) by two sequential rounds of chemical mutagenesis and biological selection. Here, cpts530/1030 was shown to be highly attenuated in the upper and lower respiratory tracts of seronegative chimpanzees. However, evaluation in seropositive children showed that it retains sufficient replicative capacity and virulence to preclude its direct use as a live attenuated vaccine. Nucleotide sequence analysis of the genome of cpts530/1030 showed that it had acquired two nucleotide substitutions (compared to its cpts530 parent), both of which were in the L gene: a silent mutation at nucleotide position 8821 (amino acid 108) and a missense mutation at nucleotide position 12458 resulting in a tyrosine-to-asparagine change at amino acid 1321, herein referred to as the 1030 mutation. It also contained the previously identified 530 missense mutation at nucleotide 10060 in the L gene. The genetic basis of attenuation of cpts530/1030 was defined by the introduction of the 530 and 1030 mutations into a cDNA clone of cpRSV, from which recombinant RSV was derived and analyzed to determine the contribution of each mutation to the temperature sensitivity (ts) and attenuation (att) phenotypes of cpts530/1030. The 530 mutation, derived from cpts530, was previously shown to be responsible for the ts and att phenotypes of that virus. In the present study, the 1030 mutation was shown to be responsible for the increased temperature sensitivity of cpts530/1030. In addition, the 1030 mutation was shown to be responsible for the increased level of attenuation of cpts530/1030 in the upper and lower respiratory tracts of mice. The 530 and 1030 mutations were additive in their effects on the ts and att phenotypes. It was possible to introduce the 1030 mutation, but not the 530 mutation, into an attenuated vaccine candidate with residual reactogenicity in very young infants, namely, cpts248/404, by use of reverse genetics. The inability to introduce the 530 mutation into the cpts248/404 virus was shown to be due to its incompatibility with the 248 missense mutation at the level of L protein function. The resulting rA2cp248/404/1030 mutant virus was more temperature sensitive and more attenuated than the cpts248/404 parent virus, making it a promising new RSV vaccine candidate created by use of reverse genetics to improve upon an existing vaccine virus.
Respiratory syncytial virus (RSV), a single-stranded, negative-sense RNA virus of the family Paramyxoviridae, remains the most important viral cause of bronchiolitis and pneumonia in young children and infants (5). Currently, attenuated mutants of RSV are being developed for use as live-virus vaccines to be administered during the first month of life to prevent serious lower respiratory tract disease. To be effective, such vaccine candidates must achieve a delicate balance between attenuation and immunogenicity. This balance has been difficult to achieve, and a variety of live attenuated RSV vaccine candidates which have been evaluated in human infants and children were found to be either underattenuated (16, 18) or overattenuated and thus insufficiently immunogenic (15, 27). Nonetheless, live attenuated RSV vaccines still represent promising vaccine candidates because of their ability to (i) immunize in the presence of passively derived RSV antibodies (9), (ii) induce both local immunity and systemic immunity, and (iii) elicit a protective immune response without the disease enhancement which was observed following immunization with formalin-inactivated RSV (2) and which also appears to be associated with immunization of experimental animals with purified RSV antigen (13, 21).
To achieve the goal of developing a satisfactorily attenuated RSV vaccine strain, the genetic basis for the attenuation and temperature sensitivity has been determined for several vaccine candidates, namely, cold-passaged RSV (cpRSV), cpts248/404, and cpts530 (14, 25, 26). As previously reported, the nucleotide sequence of the genome for each of these viruses was determined and compared to that of its A2 parent strain. The individual contribution of each of the identified mutations to the overall temperature sensitivity and attenuation of the virus was defined. From this research effort and other efforts to create new attenuating mutations, such as deletion of the SH gene (1), a menu of attenuating mutations is being assembled. The intent is to use the technique of reverse genetics with combinations of the well-characterized mutations from this menu to create novel vaccine candidates which are satisfactorily attenuated and immunogenic and retain these properties following replication in susceptible human infants and children.
In the present study, the genetic basis for the attenuation and temperature sensitivity of RSV vaccine candidate cpts530/1030 was investigated. As previously described, this virus was derived from cpRSV by two sequential treatments with the mutagen 5-fluorouracil. The first round of mutagenesis and biological selection produced the cpts530 virus, which is temperature sensitive and attenuated in mice (8), and the second round produced the cpts530/1030 virus, which exhibits increased temperature sensitivity and attenuation (9). In this paper, we report that nucleotide sequence analysis of cpts530/1030 showed that it had acquired, in addition to the mutations of its cpts530 parent (14), a unique missense mutation in the L gene, referred to as the 1030 mutation. This mutation was shown to contribute to the temperature sensitivity (ts) and attenuation (att) phenotypes of cpts530/1030, with the 530 and 1030 mutations being additive in their effects. Importantly, the addition of the 1030 mutation to a recombinant version of vaccine candidate cpts248/404, the most promising candidate identified to date, produced a virus that was more temperature sensitive and more attenuated. This finding illustrates the feasibility of adding mutations to an underattenuated vaccine candidate to generate a vaccine with the desired level of attenuation.
MATERIALS AND METHODS
Cells and viruses.
Wild-type (wt) RSV strain A2 HEK-7 and its biologically derived mutants, cpts530, cpts530/1030, and cpts248/404, were grown in HEp-2 or Vero cells as previously described (6–8). Cell monolayer cultures were maintained in Opti-MEM I (Life Technologies, Inc., Gaithersburg, Md.) supplemented with 4% fetal bovine serum (Summit Biotechnology, Fort Collins, Colo.) and 0.05 mg of gentamicin (Quality Biological, Inc., Gaithersburg, Md.) per ml. Virus suspensions for clinical trials were produced in Vero cells and found to be free of adventitious agents by Louis Potash (Dyncorp/PRI, Rockville, Md.). When necessary, the viruses were diluted in L-15 medium (BioWhittaker, Walkersville, Md.) immediately prior to use. The modified vaccinia virus Ankara recombinant expressing the bacteriophage T7 RNA polymerase (MVA/T7 pol) was provided by L. Wyatt and B. Moss and grown in primary chicken embryo cells (28).
Studies with chimpanzees.
Evaluation of the replication of cpts530/1030 in the upper and lower respiratory tracts of chimpanzees was performed as previously described (9). Briefly, a pair of 2-year-old RSV-seronegative chimpanzees was inoculated by both the intranasal and the intratracheal routes with 104 PFU of cpts530/1030 in a 1.0-ml dose at each site. For virus quantitation following inoculation, nasopharyngeal swab samples were collected daily for 10 days, and tracheal lavage samples were collected on days 2, 4, 6, 8, and 10. Virus titers were determined by a plaque assay on HEp-2 cell monolayers. The extent of rhinorrhea, a marker of upper respiratory tract disease, was estimated daily and assigned a score of 0 to 4 (0, none; 1, trace; 2, mild; 3, moderate; and 4, severe). Twenty-eight days following inoculation, chimpanzees were challenged intranasally and intratracheally with 104 PFU of wt RSV strain A2 in a 1.0-ml dose. Samples were collected as described above, and virus titers were determined.
Clinical studies with humans.
The guidelines for human experimentation set by the Joint Committee for Clinical Investigation of The Johns Hopkins University School of Medicine were followed for conducting clinical studies with infants and children. The cpts530/1030 vaccine candidate was evaluated in randomized, double-blind, placebo-controlled phase I trials with RSV-seropositive infants and children 15 to 59 months of age at The Johns Hopkins University Center for Immunization Research. Pretrial serum antibody screening, intranasal vaccination, and nasal wash sample collection and processing were performed as previously described (16). Study participants were evaluated at The Johns Hopkins University Center for Immunization Research for respiratory and febrile illnesses as previously described (16, 17).
Sequence analysis.
Three overlapping reverse transcription (RT)-PCR fragments representing the complete genome of cpts530/1030 were generated from virion-derived RNA by use of the SuperScript preamplification system (Life Technologies) and the Advantage cDNA polymerase mix (Clontech Laboratories, Palo Alto, Calif.). Fragments 1, 2, and 3 were amplified from nucleotides (nt) 28 to 5131, 5067 to 10751, and 10413 to 15179, respectively. Fragments 2 and 3 were cloned into pUC119 prior to sequence analysis. The complete nucleotide sequence of each clone was determined for both strands by automated Taq dideoxy terminator cycle sequencing (ABI, Foster City, Calif.) from an M13 random library constructed for each cDNA plasmid insert from sonicated DNA. Uncloned PCR fragment 1 was used directly for construction of an M13 random library. Nucleotides that were found to differ from the published sequences of parent viruses cpRSV (6) and cp530 (14) were confirmed with a second, independent RT-PCR fragment, thereby eliminating clone-specific differences arising from errors introduced during RT and PCR amplification. Clones containing the 3′ leader region were prepared by polyadenylation of purified viral RNA followed by RT-PCR, and clones containing the 5′ trailer region were prepared from viral RNA by reverse transcription, terminal transferase tailing, and PCR (11). The 3′- and 5′-end clones were sequenced with Sequenase 2.0 (U.S. Biochemical Corp., Cleveland, Ohio).
Site-directed mutagenesis and assembly of cDNA clones.
The two nucleotide substitutions specific to cpts530 and cpts530/1030, referred to as the 530 and 1030 mutations, were introduced singly or together into a modified cDNA representing the cpRSV parent genome as previously described (26). The sequence changes shown in Table 1 were made with subclones of the L gene by site-directed mutagenesis (19) and then introduced into the full-length cpRSV cDNA (D53cp) to create constructs suitable for generating recombinant RSV. The 530 and 1030 mutations were each tagged by the presence or absence of an adjacent restriction enzyme cleavage site which was designed to be translationally silent. All sequence changes were confirmed by nucleotide sequencing. In the final D53cp-based cDNA constructs, the presence of the 530 or 1030 mutation, as well as the 13 other mutations found in D53cp (26), was confirmed by restriction enzyme analysis.
TABLE 1.
Introduction of the 530 and 1030 mutations into full-length antigenome cDNA clones to create recombinant RSV
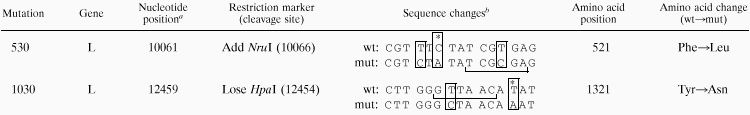 |
The nucleotide position shown is for the nucleotide change (see footnote b) leading to an amino acid substitution in the biologically derived mutant of wt RSV. The numbering is relative to that for the recombinant virus and is 1 nt greater than that for the biologically derived virus because the recombinant virus contains a 1-nt insertion in the NS2-N intergenic region (3).
Nucleotide changes (positive sense) are boxed. Restriction enzyme recognition sites which were modified for use as genetic markers are bracketed. The sequence is presented in coding triplets, and each single nucleotide change in the biologically derived virus resulting in a missense mutation is indicated with an asterisk. The first boxed nucleotide change in the 530 mutant, which was silent, was introduced to decrease the likelihood of same-site reversion to the wt codon. mut, mutant.
To construct full-length cDNA clones of rA2cp248/404 (25) bearing the 530 and/or 1030 mutations, subclones of the L gene derived from D53cp248/404 were mutagenized as described previously (25) and cloned back into D53cp248/404 to create three novel constructs: D53cp248/404/530, D53cp248/404/1030, and D53cp248/404/530/1030. The presence of the mutations in cDNA constructs was confirmed by nucleotide sequencing and restriction enzyme analysis.
Production of recombinant RSV.
Transfection and recovery of recombinant RSV were performed as previously described (3). Briefly, HEp-2 cell monolayers infected with MVA/T7 pol were transfected with a full-length cDNA clone along with the pTM1-based N, P, L, and M2-1 expression plasmids by use of LipofectACE reagent (Life Technologies). Following 3 days of incubation at 32°C, clarified cell culture supernatants were passaged in fresh HEp-2 cell monolayers for the purpose of virus amplification. Virus present in HEp-2 cell culture supernatants 5 days later was quantified by plaque titration with monoclonal antibody-horseradish peroxidase staining as described previously (21). To ensure a homogeneous virus population, recombinant viruses were biologically cloned by three successive plaque purifications and then amplified two or three times in HEp-2 cell monolayers to produce virus suspensions suitable for characterization.
L gene functional assay.
Four L gene expression plasmids based on pTM1-L were constructed to test the effect of mutations on L gene function in a minigenome system. These pTM1-L-based plasmids were constructed to contain L genes from the following sources: (i) wt RSV, as present in cDNA clone D53 (26); (ii) cDNA D53 sites, which contains six translationally silent restriction site markers in the L gene (26); (iii) cDNA D53cp248/404, which contains the cp, 248, and 404 mutations (25); and (iv) D53cp248/404/530, as constructed in the present study. The ability of the L proteins expressed from these plasmids to support the replication of an RSV minigenome carrying a luciferase reporter gene (4) was assayed. Briefly, MVA-T7 pol-infected HEp-2 cell monolayers were transfected with minigenome C2L (4), the N, P, and M2-1 expression plasmids, and either wt or mutant plasmid pTM1-L. Following 3 days of incubation at 32°C, cells were harvested, lysed, and assayed for luciferase activity (20).
Virus characterization.
The presence of the introduced mutations in the genome of each recombinant virus was confirmed by RT-PCR and restriction enzyme analysis as previously described (25).
The ts phenotype of each recombinant virus was evaluated by determining the efficiency of plaque formation at various temperatures as previously described (10). Plaque titration was performed with HEp-2 cell monolayers incubated for 5 days at 32, 35, 36, 37, 38, or 39°C in temperature-controlled water baths. Plaques were enumerated by immunostaining as indicated above.
The replication of each recombinant virus in the upper and lower respiratory tracts of mice was evaluated as described previously (8, 25). Briefly, respiratory pathogen-free, 12-week-old BALB/c mice in groups of five were inoculated intranasally under methoxyflurane anesthesia on day 0 with 106 PFU of RSV delivered in a 0.1-ml dose. On day 4, mice were sacrificed by carbon dioxide inhalation and nasal turbinates and lung tissues were harvested. Clarified tissue homogenates were assayed by virus plaque titration; titers are expressed as mean log10 PFU per gram of tissue ± standard error.
RESULTS
Replication of cpts530/1030 in chimpanzees.
The cpts530/1030 virus was previously shown to be attenuated in mice (9). Prior to initiating clinical studies with humans, the level of replication of cpts530/1030 in two young, seronegative chimpanzees was evaluated. The cpts530/1030 virus was administered simultaneously by intranasal and intratracheal instillation. Nasal swab and tracheal lavage samples were collected over the next 10 days, RSV titers were determined, and the results were compared to those in previous studies with parent virus cpts530 and wt strain A2 (9) (Table 2). The mean peak RSV titer of cpts530/1030 was reduced 40-fold in the upper respiratory tract compared to that of cpts530. In the lower respiratory tract, replication of cpts530 was already reduced by more than 104-fold, and further restriction of replication was not observed with cpts530/1030. Significantly, cpts530/1030, unlike cpts530 or wt RSV, did not induce rhinorrhea or cough in either of the animals tested. Chimpanzees receiving cpts530/1030 were completely protected against subsequent challenge with wt RSV, as indicated by the failure to recover challenge virus from the upper and lower respiratory tracts (data not shown). Thus, the 1030 mutation acquired by cpts530/1030 rendered it more attenuated in the upper respiratory tract and decreased its ability to cause disease in this pair of chimpanzees.
TABLE 2.
Mutant virus cpts530/1030 is highly attenuated in the upper and lower respiratory tracts of chimpanzees
| Inoculuma | No. of chimpanzees | Reference | Mean peak virus titer (log10 PFU/ml) in:
|
Rhinorrhea score
|
No. of days with cough | ||
|---|---|---|---|---|---|---|---|
| Nasal wash | Tracheal lavage | Peak | Meanb | ||||
| A2 (wt) | 2 | 7, 8 | 5.0 ± 0.35 | 5.5 ± 0.40 | 3.0 | 1.6 | 1.0 |
| cpRSV | 2 | 7, 8 | 4.7 ± 0.40 | 2.9 ± 0.10 | 1.0 | 0.6 | 0.0 |
| cpts530 | 4 | 9 | 4.1 ± 0.44 | 1.1 ± 0.50 | 2.0 | 0.5 | 1.0 |
| cpts530/1030 | 2 | This study | 2.5 ± 0.01 | 0.8 ± 0.14 | 0.0 | 0.0 | 0.0 |
Chimpanzees were inoculated simultaneously by the intranasal and intratracheal routes with 104 PFU of the indicated virus at each site. Nasal wash samples were collected daily for 10 days, and tracheal lavage samples were collected on days 2, 5, 6, 8, and 10.
Mean of the scores obtained during the 8 days of peak virus shedding.
Response of seropositive infants and children to cpts530/1030.
Encouraged by the level of attenuation of cpts530/1030 in mice and chimpanzees, we initiated clinical trials with seropositive humans. For the purpose of comparison, the clinical evaluation of cpts530/1030 is presented in Table 3 in the context of the evaluation of other attenuated viruses in seropositive children (16). The cpts530/1030 virus infected 50% of the subjects tested. Upper respiratory tract illness was observed in 25% of the vaccinees but not in placebo recipients who were studied concomitantly. In addition, a single child experienced lower respiratory tract illness which likely was caused by an adenovirus isolated from the child. The frequency and magnitude of cpts530/1030 virus shedding were similar to those observed in studies of the cpts248/955 virus, which was previously found to be insufficiently attenuated for seronegative vaccinees (16); therefore, the cpts530/1030 virus was not further evaluated in seronegative humans.
TABLE 3.
Responses of seropositive infants and children to cpts248/955, cpts530/1030, or cpts530/1009 mutant virus or placeboa
| RSV or placebo administered | Study | Dose (log10 PFU) | No. of subjects (% infected) | Virus isolation (nasal wash)
|
% with indicated illness
|
|||
|---|---|---|---|---|---|---|---|---|
| % Shedding virus | Mean ± SE peak titer shedb (log10 PFU/ml) | Fever | URI | LRI | ||||
| cpts248/955 | 1c | 5.0 | 13 (62) | 38 | 2.7 ± 0.64 | 15 | 7 | 0 |
| cpts530/1030 | 2 | 5.0 | 12 (50) | 42 | 1.6 ± 0.44 | 25 | 25 | 8d |
| cpts530/1009 | 3c | 5.0 | 13 (31) | 0 | ≤0.6 | 15 | 0 | 0 |
| Placebo | 1 | 0.0 | 9 (0) | 0 | ≤0.6 | 44 | 0 | 0 |
| Placebo | 2 | 0.0 | 6 (0) | 0 | ≤0.6 | 0 | 0 | 0 |
| Placebo | 3 | 0.0 | 7 (0) | 0 | ≤0.6 | 14 | 0 | 0 |
RSV-seropositive children and infants, 15 to 59 months old, were enrolled in these studies. Seropositive subjects had an RSV serum plaque reduction neutralizing antibody titer of >1:40. URI, upper respiratory tract illness; LRI, lower respiratory tract illness.
Calculated for infected subjects only.
See reference 16.
Associated with the isolation of adenovirus and thus not likely due to the RSV vaccine.
Sequence analysis of cpts530/1030.
Although the results described above indicated that the cpts530/1030 virus itself would not be a satisfactory vaccine, it was possible that mutations contained in this virus might be useful additions to recombinant versions of other vaccine candidates to achieve a further increment in attenuation. Therefore, the genome of cpts530/1030 was sequenced in its entirety. This analysis confirmed the presence of each of the five cp mutations and the 530 missense mutation which were present in the cpts530 parent. Two additional mutations, both in the L gene coding sequence, were identified: a silent mutation at nucleotide position 8821 (amino acid 108) and a missense mutation at nucleotide position 12458. The T-to-A (positive-sense) missense mutation, herein designated the 1030 mutation, resulted in a tyrosine-to-asparagine change at amino acid 1321 in the L protein. Because this single mutation is the only significant genetic difference between cpts530 and cpts530/1030, it seemed reasonable to assume that the ts and att phenotypic differences between these viruses could be attributed to the presence of this mutation.
Recombinant RSV.
The role of the 530 and 1030 missense mutations in specifying the ts and att phenotypes of cpts530/1030 was evaluated by their introduction into recombinant cpRSV, which contains the genetic background from which the cpts530 and cpts530/1030 viruses were originally derived. As previously described, this recombinant cpRSV, designated rA2cp, contains the five identified cp mutations, the six L gene restriction site markers (designated “sites” mutations), and the two F gene mutations (designated “HEK” mutations) required to bring the coding region of the recombinant wt clone into agreement with that of the human embryonic kidney (HEK) cell-passaged progenitor of cpRSV (26). Three recombinant viruses containing the 530 and 1030 mutations, singly or in combination, were generated for analysis. The mutations were introduced singly into rA2cp to produce rA2cp530 and rA2cp1030 and were introduced together to produce rA2cp530/1030. By use of RT-PCR and restriction fragment analysis, each of the recombinant viruses was confirmed to contain the restriction site markers which were introduced adjacent to the 530 and 1030 mutations, as well as each of the cp, sites, and HEK mutations present in the rA2cp parent virus (data not shown).
Temperature sensitivity of recombinant viruses.
The level of temperature sensitivity of the recombinant viruses, as determined by their ability to form plaques at a range of temperatures, is presented in Table 4. The temperature sensitivity of rA2cp530 (39°C shutoff temperature) was the same as that of its biologically derived counterpart cpts530, in agreement with previously published results (14). The rA2cp1030 mutant virus was more temperature sensitive (38°C shutoff temperature) than rA2cp530, and the mutant virus rA2cp530/1030 was more temperature sensitive (36°C shutoff temperature) than virus containing either of the mutations alone, indicating the additive effect of the 530 and 1030 mutations on the ts phenotype. In addition, the temperature sensitivity of rA2cp530/1030 was equivalent to that of its biologically derived counterpart, confirming that the 530 and 1030 mutations are the determinants of the ts phenotype of cpts530/1030.
TABLE 4.
The 530 and 1030 mutations independently confer the ts and att phenotypes
| Virusa | Presence (+) or absence (−) of the following mutation:
|
Mean virus titer (log10 PFU/ml) at temp (°C) ofb:
|
Shutoff temp (°C)c | Mean ± SE titer (log10 PFU/g) ind:
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| cp | 530 | 1030 | 32 | 35 | 36 | 37 | 38 | 39 | Nasal turbinates | Lungs | ||
| A2 (wt) (HEK-7) | − | − | − | 4.7 | 4.6 | 4.4 | 4.4 | 4.5 | 4.3 | >39 | 4.4 ± 0.07 | 5.0 ± 0.06 |
| rA2cp | + | − | − | 6.0 | 5.9 | 6.0 | 5.7 | 5.6 | 4.8e | >39 | 3.8 ± 0.11 | 4.1 ± 0.10 |
| rA2cp530 | + | + | − | 5.5 | 5.4 | 5.2 | 4.9 | 4.8e | <0.7 | 39 | 2.4 ± 0.12 | 3.9 ± 0.05 |
| cpts530 | + | + | − | 5.5 | 5.4 | 5.4 | 4.9 | 5.2e | <0.7 | 39 | 3.3 ± 0.04 | 4.2 ± 0.04 |
| rA2cp1030 | + | − | + | 5.5 | 5.6 | 5.3 | 4.6e | 2.2e | <0.7 | 38 | 2.4 ± 0.19 | 2.9 ± 0.07 |
| rA2cp530/1030 | + | + | + | 5.6 | 4.7e | <0.7 | <0.7 | <0.7 | <0.7 | 36 | <2.0 ± 0.00 | <1.7 ± 0.00 |
| cpts530/1030 | + | + | + | 4.5f | 3.5e | <0.7 | <0.7 | <0.7 | <0.7 | 36 | <2.0 ± 0.00 | <1.7 ± 0.00 |
Recombinant (r) viruses each contained the “HEK” F gene mutations and the six silent L gene “sites” mutations (26). A2 (wt) (HEK-7), cpts530, and cpts530/1030 were biologically derived viruses.
Boldfacing indicates the virus titer at the shutoff temperature.
Defined as the lowest restrictive temperature at which a 100-fold or greater reduction in titer was observed.
Mice in groups of five were administered 106 PFU of the indicated virus intranasally under light anesthesia on day 0 and sacrificed on day 4.
Pinpoint plaque size.
Small plaque size.
Levels of replication of recombinant viruses in BALB/c mice.
The levels of replication of wt RSV, the biologically derived mutant viruses, and the recombinant viruses in the nasal turbinates and lungs of BALB/c mice following intranasal inoculation with 106 PFU of virus were compared (Table 4). Recombinant viruses rA2cp530 and rA2cp1030 grew to comparable levels in the upper respiratory tract and exhibited a 25-fold decrease in replication compared to that of rA2cp. In the lower respiratory tract, the replication of rA2cp530 was not significantly different from that of rA2cp; however, rA2cp1030 showed a 10-fold decrease in replication compared to that of rA2cp. Consistent with its effect on the ts phenotype, the combination of the 530 and 1030 mutations rA2cp530/1030 created a virus more restricted in replication than either of the viruses bearing single mutations. Like that of its biologically derived counterpart, rA2cp530/1030 replication was not detectable in the upper or lower respiratory tract of mice.
The 248 and 530 mutations are incompatible.
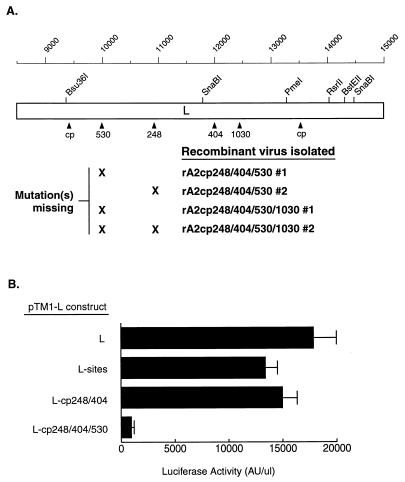
Ongoing evaluation of the vaccine candidate cpts248/404 in very young infants indicated that it has characteristics of attenuation, stability, and immunogenicity that make it the most promising vaccine candidate identified to date; however, it retained the ability to cause nasal stuffiness that interfered with feeding for a majority of vaccinees less than 3 months old, indicating that a more-attenuated derivative is needed for this target age group (26a). To test the ability of the 530 and 1030 mutations to further attenuate a recombinant version of cpts248/404, these mutations were introduced singly and in combination into plasmid D53cp248/404; in each instance, putative rA2cp248/404 derivatives were obtained and characterized for the presence of the introduced mutations. RT-PCR and restriction site marker analysis indicated that each virus which was designed to contain both the 530 and the 248 mutations, namely, the rA2cp248/404/530 and rA2cp248/404/530/1030 viruses, in fact was missing one or both mutations (Fig. 1A). The other combination of mutations, in virus rA2cp248/404/1030, was successfully recovered from the infectious recombinant virus.
FIG. 1.
Incompatibility of the 248 and 530 mutations. (A) To achieve an increase in the level of attenuation of vaccine candidate cpts248/404, the 530 and 1030 mutations were introduced into rA2cp248/404 to generate putative recombinant viruses rA2cp248/404/530, rA2cp248/404/1030, and rA2cp248/404/530/1030. Following biological cloning of these viruses, RT-PCR and restriction enzyme analysis were used to verify the presence of each introduced mutation. The L gene is illustrated as an open rectangle. The positions of the six translationally silent “sites” mutations are indicated by restriction enzyme names. The positions of the mutations derived from cpts248/404 (cp, 248, and 404) as well as the 530 and 1030 mutations are shown. Mutations confirmed to be missing from the recombinant viruses are marked with an X. Nucleotide positions relative to the full-length genome are indicated on the scale at the top. (B) To test the effects of these mutations on the function of the L protein, expression plasmid pTM1-L was modified to contain the sites mutations as well as mutations from rA2cp248/404 and/or the 530 mutation. The transcription and replication levels of the minigenome are indicated as luciferase activity measured as transfection supported by the various pTM1-L constructs. AU, arbitrary units. Error bars indicate standard errors.
Sequence analysis of the antigenome cDNA constructs bearing the 248 and 530 mutations confirmed that each DNA contained the correct sequence. We therefore presume that the substitution of the wild-type assignment(s) into the recombinant viruses was due to vaccinia virus-mediated recombination between the mutant antigenome cDNA and the wild-type pTM1-L support plasmid, as has been described for antigenome and support plasmids for Sendai virus (12) and RSV (25). These findings may indicate that the 248 and 530 mutations are incompatible. If so, the most likely scenario is that a combination of the two mutations renders the L protein unstable, nonfunctional, or both.
The functionality of the L protein bearing the 248 and 530 mutations was evaluated by use of a reconstituted replication and transcription system with a minigenome bearing the luciferase marker gene. The following pTM1 plasmids were evaluated: L-wt, L-sites, L-cp248/404, and L-cp248/404/530. The transcription and replication levels of the minigenome were indicated by luciferase activity in transfection reactions supported by the various pTM1-L constructs (Fig. 1B). L protein expressed from pTM1-L-cp248/404/530 was essentially nonfunctional; we interpret this finding as indicating structural and/or functional incompatibility between the 248 and 530 mutations. Alternative explanations, such as the possibility that these mutations somehow altered the expression, stability, or translation of the encoded mRNA, seem unlikely.
Characterization of rA2cp248/404/1030.
The rA2cp248/404/1030 virus was readily recovered and found to contain each of the introduced mutations (data not shown). The ts and att phenotypes of this virus in vitro and in mice were characterized. The addition of the 1030 mutation to rA2cp248/404 increased its temperature sensitivity, as indicated by a shift in the shutoff temperature from 37 to 36°C (Table 5). Mutant virus rA2cp248/404/1030 also formed slightly smaller plaques than rA2cp248/404 on HEp-2 cell monolayers at the 32°C permissive temperature (data not shown). The level of replication of rA2cp248/404/1030 in the nasal turbinates and lungs of BALB/c mice was below the level of detection, reflecting a decrease compared to the already low level of replication of rA2cp248/404 (Table 5). Virus was not recovered from any of the mice inoculated with the rA2cp248/404/1030 virus; this result represents a significant decrease compared to the frequency of recovery of rA2cp248/404 (Fisher’s exact test, P = 0.001). Clearly, the introduction of the 1030 mutation into the cpts248/404 background rendered the virus more attenuated, as indicated by its decrease in plaque size at the permissive temperature, its lower shutoff temperature, and its lower frequency of recovery from animals administered the virus.
TABLE 5.
The 1030 mutation increases the ts and att phenotypes of rA2cp248/404
| Virusa | Virus titer (log10 PFU/ml) at temp (°C) ofb:
|
Shutoff temp (°C)c | No. of mice with virus ind:
|
Mean ± SE titer (log10 PFU/g) ind:
|
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 32 | 35 | 36 | 37 | 38 | 39 | Nasal turbinates | Lungs | Nasal turbinates | Lungs | ||
| A2 (wt) (HEK-7) | 5.8 | 5.6 | 5.7 | 5.6 | 5.6 | 5.4 | >40 | 6 | 6 | 4.8 ± 0.07 | 4.7 ± 0.08 |
| cpts248/404 | 5.4 | 5.1 | 4.6e | <0.7 | <0.7 | <0.7 | 37 | 4 | 6 | 2.2 ± 0.13 | 2.2 ± 0.09 |
| rA2cp248/404 | 5.2 | 4.8 | 4.1e | <0.7 | <0.7 | <0.7 | 37 | 4 | 4 | 2.0 ± 0.02 | 1.8 ± 0.12 |
| rA2cp248/404/1030 | 5.5f | 4.0e | <0.7 | <0.7 | <0.7 | <0.7 | 36 | 0 | 0 | <2.0 ± 0.00 | <1.7 ± 0.00 |
Recombinant (r) viruses each contained the “HEK” F gene mutations and the L gene “sites” mutations (26). A2 (wt) (HEK-7) and cpts248/404 were biologically derived viruses.
Boldfacing indicates virus titer at the shutoff temperature.
Defined as the lowest restrictive temperature at which a 100-fold or greater reduction of titer was observed.
Mice in groups of six were administered 106 PFU of virus intranasally under light anesthesia on day 0 and sacrificed on day 4.
Pinpoint plaque size.
Small plaque size.
DISCUSSION
The goals of the present study were to examine the suitability of the biologically derived cpts530/1030 virus for use as a live attenuated RSV vaccine in humans as well as to determine the genetic basis of its ts and att phenotypes. Since cpts530/1030 was highly attenuated in mice and chimpanzees and manifested a high level of temperature sensitivity in vitro, it was thought that it would be an appropriate vaccine virus. However, the cpts530/1030 virus readily infected seropositive children, 42% of whom shed virus and 25% of whom experienced upper respiratory tract illness. The level of infectivity and virus shedding of this vaccine candidate in seropositive children were similar to those of another candidate, the cpts248/955 virus, which was found to retain the capacity to cause mild bronchiolitis when evaluated in seronegative children (16). Based on these observations, we concluded that cpts530/1030 was unacceptable as a vaccine candidate, and further clinical studies with this virus were not pursued. However, several conclusions can be drawn from this clinical study. First, the cpts530/1030 virus is unusual because it is the first vaccine candidate for which a high degree of attenuation in chimpanzees was not reproduced in young children and infants. A likely explanation is that humans are more permissive for RSV replication than nonhuman animals, even chimpanzees. Thus, accurate evaluation of an RSV vaccine candidate clearly requires clinical studies. Second, this study illustrates that clinical studies with seropositive subjects remain a very important step for the safe evaluation of novel RSV vaccine candidates.
The genetic basis of attenuation of the cpts530/1030 virus was identified by determining the nucleotide sequence of its genome followed by the staged introduction of the identified mutations into recombinant RSV. This analysis indicated that the 530 and 1030 mutations each contributed to the ts and att phenotypes of cpts530/1030, with the 1030 mutation conferring higher levels of attenuation and temperature sensitivity than the 530 mutation. The levels of temperature sensitivity and attenuation of rA2cp530/1030 were higher than those of rA2cp530 or rA2cp1030, indicating that the effects of the 530 and 1030 mutations on these phenotypes were additive. This situation is not always the case, as was seen recently for the ts and att mutation in the M2 gene start cis-acting sequence of cpts248/404, which were shown to be the dominant ts and att mutation that masked the effects of the ts and att mutations in the L protein of the cpts248/404 virus (25). The levels of temperature sensitivity and attenuation of rA2cp530/1030 were equivalent to those of its biologically derived counterpart, confirming that the 530 and 1030 mutations are the primary determinants of the temperature sensitivity and attenuation of cpts530/1030.
As indicated above, we have been compiling a menu of attenuating mutations from which specific mutations can be selected for placement, via reverse genetics, into incompletely attenuated RSV vaccine candidates to generate more-attenuated derivatives. This concept was evaluated in the present study with the 530 and 1030 mutations as specific examples. Since it will be necessary to further attenuate cpts248/404 for use in very young infants, the 530 and 1030 mutations were introduced singly and together into rA2cp248/404. Unexpectedly, the 248 and 530 mutations were found to be incompatible, and recombinants possessing this combination of mutations could not be isolated. This incompatibility was shown to be at the functional level of the L protein. However, recombinant virus rA2cp248/404/1030 was viable and was shown to be more temperature sensitive and more attenuated in mice than rA2cp248/404. At least four possible outcomes have been observed in the process of combining two or more ts mutations into a single virus: (i) they can be additive, as shown here for the 530 and 1030 mutations and as previously found for ts mutations in parainfluenza virus and influenza virus (23, 24); (ii) they can be nonadditive, as with rA2cp248/404, with the temperature sensitivity of the double mutant reflecting that of its more-temperature-sensitive member (25); (iii) the combination of two ts mutations can result in a virus that is less temperature sensitive than either parent (22, 23); and (iv) the combination of ts mutations derived from different viruses can be lethal, as shown here for a combination of the 248 and 530 mutations in the L protein. Thus, each new combination of mutations will require careful individual study.
Even though cpts530/1030 was unacceptable as a vaccine candidate for young children, study of this virus identified a mutation that was able to further attenuate the vaccine candidate cpts248/404. Mutations with this capability are important to ongoing RSV vaccine research, since conventional methods, such as the chemical mutagenesis used to derive the cpts vaccine candidates tested to date, have failed to produce more-attenuated viruses that are phenotypically stable (7; unpublished data). Reverse-genetics techniques, such as those used here to generate rA2cp248/404/1030, and information derived from concurrent clinical studies are allowing us to systematically create new RSV vaccine candidates. Thus, the rA2cp248/404/1030 virus fulfills the need to further attenuate the vaccine candidate cpts248/404 and represents the most attenuated member of the current lineage. Future clinical evaluations will determine whether this new vaccine candidate has lost the residual virulence of cpts248/404 without compromising immunogenicity.
ACKNOWLEDGMENTS
We thank Robert Chanock for careful review of the manuscript and Roberta Casey, Barbara Burns, Pamela Nehring, Victoria Hodgins, and Jean Froehlich for assistance with the clinical trials and related laboratory work.
This research is part of a continuing program of research and development with Wyeth-Lederle Vaccines and Pediatrics through CRADA grants AI-000030 and AI-000087.
REFERENCES
- 1.Bukreyev A, Whitehead S S, Murphy B R, Collins P L. Recombinant respiratory syncytial virus from which the entire SH gene has been deleted grows efficiently in cell culture and exhibits site-specific attenuation in the respiratory tract of the mouse. J Virol. 1997;71:8973–8982. doi: 10.1128/jvi.71.12.8973-8982.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chanock R M, Murphy B R. Past efforts to develop safe and effective RSV vaccines. In: Meigner B, Murphy B, Ogra P, editors. Animal models of respiratory syncytial virus infections. Lyon, France: Merieux Foundation; 1991. pp. 35–42. [Google Scholar]
- 3.Collins P L, Hill M G, Camargo E, Grosfeld H, Chanock R M, Murphy B R. Production of infectious human respiratory syncytial virus from cloned cDNA confirms an essential role for the transcription elongation factor from the 5′ proximal open reading frame of the M2 mRNA in gene expression and provides a capability for vaccine development. Proc Natl Acad Sci USA. 1995;92:11563–11567. doi: 10.1073/pnas.92.25.11563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Collins P L, Hill M G, Cristina J, Grosfeld H. Transcription elongation factor of respiratory syncytial virus, a nonsegmented negative-strand RNA virus. Proc Natl Acad Sci USA. 1996;93:81–85. doi: 10.1073/pnas.93.1.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Collins P L, McIntosh K, Chanock R M. Respiratory syncytial virus. In: Fields B N, Knipe D M, Howley P M, Chanock R M, Melnick J L, Monath T P, Roizman B, Straus S E, editors. Fields virology. 3rd ed. Vol. 2. Philadelphia, Pa: Lippincott-Raven; 1996. pp. 1313–1352. [Google Scholar]
- 6.Connors M, Crowe J E, Jr, Firestone C-Y, Murphy B R, Collins P L. A cold-passaged, attenuated strain of human respiratory syncytial virus contains mutations in the F and L genes. Virology. 1995;208:478–484. doi: 10.1006/viro.1995.1178. [DOI] [PubMed] [Google Scholar]
- 7.Crowe J E, Jr, Bui P T, Davis A R, Chanock R M, Murphy B R. A further attenuated derivative of a cold-passaged temperature-sensitive mutant of human respiratory syncytial virus retains immunogenicity and protective efficacy against wild-type challenge in seronegative chimpanzees. Vaccine. 1994;12:783–790. doi: 10.1016/0264-410x(94)90286-0. [DOI] [PubMed] [Google Scholar]
- 8.Crowe J E, Jr, Bui P T, London W T, Davis A R, Hung P P, Chanock R M, Murphy B R. Satisfactorily attenuated and protective mutants derived from a partially attenuated cold-passaged respiratory syncytial virus mutant by introduction of additional attenuating mutations during chemical mutagenesis. Vaccine. 1994;12:691–699. doi: 10.1016/0264-410x(94)90218-6. [DOI] [PubMed] [Google Scholar]
- 9.Crowe J E, Jr, Bui P T, Siber G R, Elkins W R, Chanock R M, Murphy B R. Cold-passaged, temperature-sensitive mutants of human respiratory syncytial virus (RSV) are highly attenuated, immunogenic, and protective in seronegative chimpanzees, even when RSV antibodies are infused shortly before immunization. Vaccine. 1995;13:847–855. doi: 10.1016/0264-410x(94)00074-w. [DOI] [PubMed] [Google Scholar]
- 10.Crowe J E, Jr, Collins P L, London W T, Chanock R M, Murphy B R. A comparison in chimpanzees of the immunogenicity and efficacy of live attenuated respiratory syncytial virus (RSV) temperature-sensitive mutant vaccines and vaccinia virus recombinants that express the surface glycoproteins of RSV. Vaccine. 1993;11:1395–1404. doi: 10.1016/0264-410x(93)90168-w. [DOI] [PubMed] [Google Scholar]
- 11.Crowe J E, Jr, Firestone C Y, Whitehead S S, Collins P L, Murphy B R. Acquisition of the ts phenotype by a chemically mutagenized cold-passaged human respiratory syncytial virus vaccine candidate results from the acquisition of a single mutation in the polymerase (L) gene. Virus Genes. 1996;13:269–273. doi: 10.1007/BF00366988. [DOI] [PubMed] [Google Scholar]
- 12.Garcin D, Pelet T, Calain P, Roux L, Curran J, Kolakofsky D. A highly recombinogenic system for the recovery of infectious Sendai paramyxovirus from cDNA: generation of a novel copy-back nondefective interfering virus. EMBO J. 1995;14:6087–6094. doi: 10.1002/j.1460-2075.1995.tb00299.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Graham B S, Henderson G S, Tang Y W, Lu X, Neuzil K M, Colley D G. Priming immunization determines T helper cytokine mRNA expression patterns in lungs of mice challenged with respiratory syncytial virus. J Immunol. 1993;151:2032–2040. [PubMed] [Google Scholar]
- 14.Juhasz K, Whitehead S S, Bui P T, Biggs J M, Boulanger C A, Collins P L, Murphy B R. The temperature-sensitive (ts) phenotype of a cold-passaged (cp) live attenuated respiratory syncytial virus vaccine candidate, designated cpts530, results from a single amino acid substitution in the L protein. J Virol. 1997;71:5814–5819. doi: 10.1128/jvi.71.8.5814-5819.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Karron R A, Buonagurio D A, Georgiu A F, Whitehead S S, Adamus J E, Clements-Mann M L, Harris D O, Randolph V B, Udem S A, Murphy B R, Sidhu M S. Respiratory syncytial virus (RSV) SH and G proteins are not essential for viral replication in vitro: clinical evaluation and molecular characterization of a cold-passaged, attenuated RSV subgroup B mutant. Proc Natl Acad Sci USA. 1997;94:13961–13966. doi: 10.1073/pnas.94.25.13961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Karron R A, Wright P F, Crowe J E, Jr, Clements M L, Thompson J, Makhene M, Casey R, Murphy B R. Evaluation of two live, cold-passaged, temperature-sensitive respiratory syncytial virus (RSV) vaccines in chimpanzees, adults, infants and children. J Infect Dis. 1997;176:1428–1436. doi: 10.1086/514138. [DOI] [PubMed] [Google Scholar]
- 17.Karron R A, Wright P F, Hall S L, Makhene M, Thompson J, Burns B A, Tollefson S, Steinhoff M C, Wilson M H, Harris D O, Clements M L, Murphy B R. A live attenuated bovine parainfluenza virus type 3 vaccine is safe, infectious, immunogenic, and phenotypically stable in infants and children. J Infect Dis. 1995;171:1107–1114. doi: 10.1093/infdis/171.5.1107. [DOI] [PubMed] [Google Scholar]
- 18.Kim H W, Arrobio J O, Pyles G, Brandt C D, Camargo E, Chanock R M, Parrott R H. Clinical and immunological response of infants and children to administration of low-temperature adapted respiratory syncytial virus. Pediatrics. 1971;48:745–755. [PubMed] [Google Scholar]
- 19.Kunkel T A, Roberts J D, Zakour R A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Methods Enzymol. 1987;154:367–382. doi: 10.1016/0076-6879(87)54085-x. [DOI] [PubMed] [Google Scholar]
- 20.Kuo L, Fearns R, Collins P L. The structurally diverse intergenic regions of respiratory syncytial virus do not modulate sequential transcription by a dicistronic minigenome. J Virol. 1996;70:6143–6150. doi: 10.1128/jvi.70.9.6143-6150.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Murphy B R, Sotnikov A V, Lawrence L A, Banks S M, Prince G A. Enhanced pulmonary histopathology is observed in cotton rats immunized with formalin-inactivated respiratory syncytial virus (RSV) or purified F glycoprotein and challenged with RSV 3-6 months after immunization. Vaccine. 1990;8:497–502. doi: 10.1016/0264-410x(90)90253-i. [DOI] [PubMed] [Google Scholar]
- 22.Ramig R F, Fields B N. Revertants of temperature-sensitive mutants of reovirus: evidence for frequent extragenic suppression. Virology. 1979;92:155–167. doi: 10.1016/0042-6822(79)90221-6. [DOI] [PubMed] [Google Scholar]
- 23.Skiadopoulos M H, Durbin A P, Tatem J M, Wu S L, Paschalis M, Tao T, Collins P L, Murphy B R. Three amino acid substitutions in the L protein of the human parainfluenza virus type 3 cp45 live attenuated vaccine candidate contribute to its temperature-sensitive and attenuation phenotypes. J Virol. 1998;72:1762–1768. doi: 10.1128/jvi.72.3.1762-1768.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Subbarao E K, Park E J, Lawson C M, Chen A Y, Murphy B R. Sequential addition of temperature-sensitive missense mutations into the PB2 gene of influenza A transfectant viruses can effect an increase in temperature sensitivity and attenuation and permits the rational design of a genetically engineered live influenza A virus vaccine. J Virol. 1995;69:5969–5977. doi: 10.1128/jvi.69.10.5969-5977.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Whitehead S S, Firestone C Y, Collins P L, Murphy B R. A single nucleotide substitution in the transcription start signal of the M2 gene of respiratory syncytial virus vaccine candidate cpts248/404 is the major determinant of the temperature-sensitive and attenuation phenotype. Virology. 1998;247:232–239. doi: 10.1006/viro.1998.9248. [DOI] [PubMed] [Google Scholar]
- 26.Whitehead S S, Juhasz K, Firestone C Y, Collins P L, Murphy B R. Recombinant respiratory syncytial virus (RSV) bearing a set of mutations from cold-passaged RSV is attenuated in chimpanzees. J Virol. 1998;72:4467–4471. doi: 10.1128/jvi.72.5.4467-4471.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26a.Wright, P. F. Personal communication.
- 27.Wright P F, Belshe R B, Kim H W, Van Voris L P, Chanock R M. Administration of a highly attenuated, live respiratory syncytial virus vaccine to adults and children. Infect Immun. 1982;37:397–400. doi: 10.1128/iai.37.1.397-400.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wyatt L S, Moss B, Rozenblatt S. Replication-deficient vaccinia virus encoding bacteriophage T7 RNA polymerase for transient gene expression in mammalian cells. Virology. 1995;210:202–205. doi: 10.1006/viro.1995.1332. [DOI] [PubMed] [Google Scholar]