Abstract
Objective:
This study aimed to explore the incidence and antimicrobial resistance (AMR) of Escherichia coli, Staphylococcus aureus, and Bacillus cereus in raw milk and some Egyptian dairy products, namely Kariesh cheese and rice with milk.
Material and Methods:
112 samples (70 raw milk, 30 Kariesh cheese, and 12 rice with milk) were randomly collected from different districts in Cairo and Giza, Egypt. The samples were examined for E. coli, S. aureus, and B. cereus presence. The susceptibility of the obtained isolates was tested against 11 antimicrobials using the disk diffusion method, and further, the presence of AMR genes was examined.
Results:
The incidences of E. coli, S. aureus, and B. cereus were 69.64%, 12.5%, and 16.7% in the examined samples, respectively. The antibiogram indicated that E. coli isolates (n = 60) were resistant to gentamycin (73.33%), ampicillin (AM, 53.3%), and cefotaxime (CTX, 16.66%). Multidrug-resistant (MDR) E. coli strains (n = 5) were tested for β-lactams resistance genes. blaTEM was detected in all isolates, and two of them additionally carried blaCTX-M. Staphylococcus aureus isolates (n = 10) were resistant to AM (100%), followed by tetracycline (TE), CTX, and gentamycin (60% each). All MDR S. aureus strains (n = 4) carried blaZ and tetK, and three of them additionally carried aac(6’)-aph (2’’). Bacillus cereus isolates (n = 30) showed resistance to AM (100%), amoxicillin (20%), and TE (6.66%). bla and tetA genes were detected in all MDR B. cereus isolates (n = 6).
Conclusion:
Our findings denote the high incidence of potential health hazards in raw milk and some of its products and the existence of AMR bacteria, including MDR strains, which can cause human illnesses that are difficult to treat.
Keywords: Antimicrobial resistance, dairy products, potential pathogens, public health
Introduction
Milk is the most complete food, as it is a good source of many essential nutrients that make it one of the most fundamental foods for all age categories and plays a key role in the diet of over 6 billion people in the world [1]. On the other side, it provides favorable environmental conditions for microbial growth, especially for pathogenic bacterial species [2].
Several bacterial pathogens, such as Staphylococcus aureus, Bacillus cereus, and Escherichia coli, were isolated from raw milk and dairy products [3]. The occurrence of such pathogens in milk constitutes a public health threat, specifically among individuals who have weakened immune systems or those who consume unpasteurized raw milk or its products [4].
Bacterial contamination of milk with pathogenic microorganisms may result from many factors, including the milking process, utensils, environment, and personnel. In addition, it may be contaminated during unhygienic storage, and transportation [5].
The presence of enteric bacteria, including E. coli, in food is a reliable indicator of fecal contamination [6]. Although most E. coli present as commensals, many are opportunistic pathogens that can cause gastrointestinal illness and can be used as a bio-indicator of antimicrobial resistance (AMR) [7].
Staphylococcus aureus is categorized as the third causal agent of foodborne diseases worldwide. It is considered one of the most important foodborne pathogens isolated from milk and dairy products. Its isolation is an indicator of neglected hygienic measures employed during the production, handling, and distribution of milk and dairy products or contamination due to mastitis or contaminated food handlers [8].
Bacillus cereus is a major foodborne pathogen that has a bad impact on heat-treated milk as its thermophilic endospores can withstand the pasteurization process and can germinate and produce spoilage enzymes, leading to off flavors in the pasteurized milk. Ingestion of food contaminated with B. cereus or its toxins can cause severe gastrointestinal illness with diarrhea and without significant upper intestinal symptoms, which is the commonly known manifestation of the disease [9].
Multidrug-resistant (MDR) microorganisms have become a great threat to public health worldwide [3]. The uncontrolled use of antibiotics, in therapeutic and sub-therapeutic points, in dairy cows increases the incidence of MDR pathogens in raw milk and the subsequent incidence in its products [10].
The antibiotic-resistant pathogens could transmit from animals to humans in different ways, including through the ingestion of contaminated milk and dairy products directly or through cross-contamination [11]. The AMR genes spread among microbes in the dairy environment pose a risk that may come into contact with humans through the processing steps or consumption of contaminated dairy products [12].
Raw milk and products made from it are considered one of the focal sources for outbreaks with antibiotic-resistant pathogens in developing countries, because of the presence of several contamination sources due to poor hygienic practices, inadequate regulations concerning food safety, insufficient resources, neglected food management systems, and bad personal hygiene by handlers [3]. Therefore, together with investigating the incidence rates, investigating the AMR resistance phenotypes of the pathogens obtained from food sources is crucial. Furthermore, the available data about AMR prevalence and its molecular basis in bacteria from Egyptian food is still scarce. Therefore, the purpose of the current study was to investigate the incidence of some pathogens such as E. coli, S. aureus, and B. cereus in raw milk and some Egyptian dairy products, namely Kariesh cheese and rice with milk, and examine the AMR of the isolated bacteria.
Material and Methods
Study samples
A total of 112 samples, including 70 samples of raw milk, 30 Kariesh cheese (Egyptian soft cheese) samples, and 12 rice with milk (a traditional Egyptian dessert). Samples were randomly collected in their retail containers during the period from December 2020 to April 2021 at the consumer level from street vendors, grocery stores, and dairy shops at different markets and from dairy farms in Cairo and Giza governorates, Egypt. The samples were collected and transported to the laboratory in an insulated icebox with the minimum delay.
Sample preparation, isolation, and identification of pathogenic bacteria (E. coli, S. aureus, and B. cereus)
Twenty-five milliliters of raw milk or 25 gm of the other dairy products were prepared according to International Organization for Standardization (ISO) 6887-5 [13]. All samples were examined for the incidence of E. coli, S. aureus, and B. cereus as described previously [14–16]. Biochemical identification of the obtained isolates was done according to the methods recommended by APHA [17,18] and ISO 7932 [16].
Antimicrobial susceptibility testing
Antimicrobial susceptibility patterns of the isolates were determined using the Kirby–Bauer disk diffusion method, and the results were interpreted according to CLSI guidelines [19].
Escherichia coli isolates were tested against eight commercially available antimicrobial disks: ampicillin (AM, 10 μg), amoxicillin-clavulanic acid (AMC, 30 μg), cefotaxime (CTX, 30 μg), ceftazidime (CAZ, 30 μg), chloramphenicol (C, 30 μg), ciprofloxacin (CIP, 5 μg), tetracycline (TE, 30 μg), and trimethoprim–sulfamethoxazole (SXT-25 μg).
For testing S. aureus isolates, eight antibiotics that are frequently used in veterinary and human illnesses were selected, such as AM (10 μg), AMC (30 μg), CTX (30 μg), CIP (5 μg), Gentamicin (GM, 10 μg), TE (30 μg), SXT (25 μg), and Vancomycin (VA, 30 μg). Bacillus cereus isolates were tested against amoxicillin (AX, 25 μg), AM (10 μg), TE (30 μg), and VA (30 μg) (Oxoid, UK).
Detection of AMR genes
Escherichia coli, S. aureus, and B. cereus isolates that exhibited MDR phenotypes were examined for the presence of AMR genes relevant to each main phenotype. The presence of genes linked with β-lactam resistance [blaTEM, blaCTX-M for E. coli; blaZ for S. aureus and bla for B. cereus], TE resistance [tetK in S. aureus and tetA in B. cereus], and aminoglycosides [aac(6’)aph (2’’) in S. aureus] were detected using polymerase chain reaction (PCR) as previously described [20].
The predicted sizes of PCR products for different AMR genes and primer sequences used for their detection are mentioned in Table 1. The PCR products were visualized under UV light (Alpha Innotech, San Leandro, USA) after electrophoresis using 1.5% agarose gels (ABgene, Surrey, UK).
Table 1. Primers for the detection of antimicrobial-resistant genes and for the identification of gene cassettes.
| Reference | Amplicon size | Primer sequence (5’–3’) | Gene | Target |
|---|---|---|---|---|
| [21] | 593 bp | ATGTGCAGYACCAGTAARGTKATGGC | bla CTX-M | E. coli |
| TGGGTRAARTARGTSACCAGAAYCAGCGG | ||||
| [22] | 516 bp | ATCAGCAATAAACCAGC | blaTEM | |
| CCCCGAAGAACGTTTTC | ||||
| [23] | 833 bp | TACAACTGTAATATCGGAGGG | blaZ | S. aureus |
| CATTACACTCTTGGCGGTTTC | ||||
| [24] | 360 bp | GTAGCGACAATAGGTAATAGT | tetK | |
| GTAGTGACAATAAACCTCCTA | ||||
| 491 bp | GAAGTACGCAGAAGAGA | aac(6’)aph (2’’) | ||
| ACATGGCAAGCTCTAGGA | ||||
| [25] | 502 bp | GGCGGTCTTCTTCATCATGC | tetA | B. cereus |
| CGGCAGGCAGAGCAAGTAGA | ||||
| [26] | 680 bp | CATTGCAAGTTGAAGCGAAA | bla | |
| TGTCCCGTAACTTCCAGCTC |
Results
Incidence of pathogenic bacteria (E. coli, S. aureus, and B. cereus) in the examined samples
The results of bacterial isolation from 112 samples (70 raw milk, 30 Kariesh cheese, and 12 rice with milk) are represented in Table 2. Out of the 112 tested samples, E. coli was isolated from 58 (82.85%), 16 (53.33%), and 4 (33.33%) raw milk, Kariesh cheese, and rice with milk samples, respectively, with an overall isolation percentage of 78 (69.64%).
Table 2. Incidence of pathogenic bacteria in examined raw milk and dairy products samples.
| Sample type | No. of samples | E. coli | S. aureus | B. cereus | |||
|---|---|---|---|---|---|---|---|
| No. of positive samples (%) | No. of isolates | No. of positive samples (%) | No. of isolates | No. of positive samples (%) | No. of isolates | ||
| Raw milk | 70 | 58 (82.85%) | 60 | 12(17.14%) | 16 | 8 (11.42%) | 14 |
| Kariesh cheese | 30 | 16 (53.33%) | 16 | 2 (6.66%) | 6 | 2 (6.66%) | 6 |
| Rice with milk | 12 | 4 (33.33%) | 6 | 0 (0%) | 0 | 8 (66.66%) | 24 |
| Total | 112 | 78 (69.64%) | 82 | 14 (12.5%) | 22 | 18 (16.07%) | 44 |
Staphylococcus aureus incidence in the three types of tested samples is shown in Table 2. Twelve raw milk samples (17.14%) and two (6.66%) of the cheese samples were contaminated with S. aureus, while all rice with milk samples (n = 12) were not contaminated. The overall occurrence of S. aureus in the different types of samples was 12.5%.
Concerning the B. cereus isolation rate, Table 2 shows that the highest incidence was in rice with milk samples (66.66%), while for raw milk, eight samples were positive with an incidence of 11.42%), and only two samples (6.66%) from Kariesh cheese were positive for B. cereus.
AMR phenotypes and genotypes of E. coli, S. aureus, and B. cereus isolates
The antibiogram of the isolates is displayed in Table 3. The results indicated a higher resistance rate of tested E. coli isolates (60 isolates) against gentamycin, where 44 (73.33%) isolates were resistant, followed by AM, for which 32 (53.3%) isolates were resistant, and 10 (16.66) were resistant to CTX. However, all isolates showed sensitivity to TE, CIP, and trimethoprim–sulfamethoxazole. Concerning S. aureus, all examined isolates (10 isolates) were resistant to AM, and 6 (60%) isolates were resistant to TE, CTX, and gentamycin. On the other hand, all isolates were sensitive to CIP, AMC acid, and trimethoprim–sulfamethoxazole. Of the 30 B. cereus tested isolates, all were resistant to AM, followed by AX 6 (20%) and TE 2 (6.66). All isolates were sensitive to VA.
Table 3. Antibiotics sensitivity of isolated pathogens (E. coli, S. aureus, and B. cereus) from examined raw milk and dairy product samples.
| Antimicrobial agenta |
E. coli No. (%) of isolates (N = 60) |
S. aureus No. (%) of isolates (N = 10) |
B. cereus No. (%) of isolates (N = 30) |
||||||
|---|---|---|---|---|---|---|---|---|---|
| R (%) | I (%) | S (%) | R (%) | I (%) | S (%) | R (%) | I (%) | S (%) | |
| AM | 32 (53.3) | 24 (40) | 4 (6.66) | 10 (100) | 0 (0) | 0 (0) | 30 (100) | 0 (0) | 0 (0) |
| AMC | 2 (3.33) | 16 (26.66) | 42 (70) | 0 (0) | 0 (0) | 10 (100) | NA | NA | NA |
| GM | 44 (73.33) | 14 (23.33) | 2 (3.33) | 6 (60) | 4 (40) | 0 (0) | NA | NA | NA |
| TE | 0 (0) | 0 (0) | 60 (100) | 6 (60) | 0 (0) | 4 (40) | 2 (6.66) | 0 (0) | 28 (93.33) |
| CTX | 10 (16.66) | 0 (0) | 50 (83.33) | 6 (60) | 0 (0) | 4 (40) | NA | NA | NA |
| CAZ | 8 (13.33) | 0 (0) | 52 (86.66) | NA | NA | NA | NA | NA | NA |
| C | 0 (0) | 10 (16.66) | 50 (83.33) | 0 (0) | 2 (20) | 8 (80) | NA | NA | NA |
| CIP | 0 (0) | 0 (0) | 60 (100) | 0 (0) | 0 (0) | 10 (100) | NAb | NA | NA |
| SXT | 0 (0) | 0 (0) | 60 (100) | 0 (0) | 0 (0) | 10 (100) | NA | NA | NA |
| AX | NA | NA | NA | NA | NA | NA | 6 (20) | 18 (60) | 6 (20) |
| VA | NA | NA | NA | NA | NA | NA | 0 (0) | 0 (0) | 30 (100) |
AM, ampicillin (10 μg); AMC, amoxicillin-Clavulanic A, (30 μg); AX, amoxicillin (25 μg); C, chloramphenicol (30 μg); CAZ, ceftazidime (30 μg); CIP, ciprofloxacin (5 μg); CTX, cefotaxime (30 μg); GM, gentamycin (10 μg); SXT, trimethoprim–sulfamethoxazole (25 μg); TE, tetracycline (30 μg); VA, vancomycin (30 μg).
NA, Not applicable.
Results in Table 4 illustrate the incidence of MDR strains among the recovered isolates. The highest MDR rate was met with S. aureus, of which 4 out of 10 S. aureus isolates (40%) conferred resistance to 3 antimicrobials belonging to different categories in 2 isolates, and the other 2 were resistant to 4 agents. After which, 6 isolates out of 30 (20%) of B. cereus showed an MDR pattern. On the other hand, 5 out of 60 E. coli isolates (8.33%), all of which were isolated from raw milk, showed an MDR profile.
Table 4. MDR isolates of E. coli, S. aureus, and B. cereus isolated from examined raw milk and dairy product samples.
| Isolated pathogen | No. of isolates with MDR pattern | MDR patternsa |
|---|---|---|
| E. coli (n = 5) | 3 2 |
AM, CTX, CAZ CTX, GM, CAZ |
| S. aureus (n = 4) | 2 | AM, GM, TE |
| 2 | AM, CTX, GM, TE | |
| B. cereus (n = 6) | 6 | AM, AX, TE |
AM, ampicillin (10 μg); AX, amoxicillin (25 μg); CAZ, ceftazidime (30 μg); CTX, cefotaxime (30 μg); GM, gentamycin (10 μg); TE, tetracycline (30 μg).
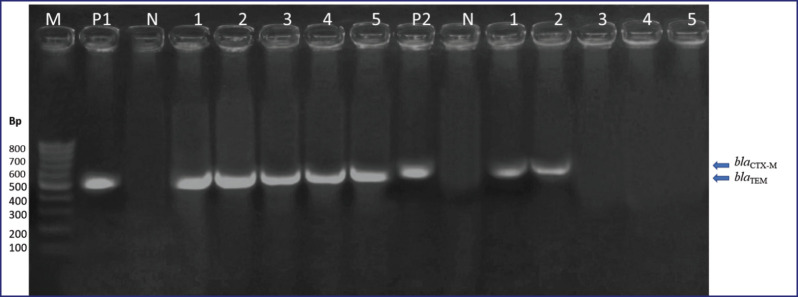
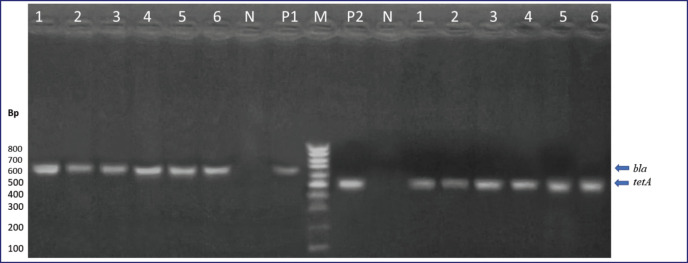
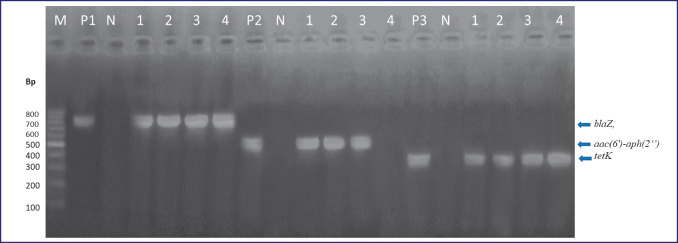
As represented in Table 5, five E. coli isolates with MDR patterns were subjected to genotyping; all five isolates had blaTEM genes, and 40% of them were positive for blaCTX-M. The presence of the blaZ gene and tetK were confirmed in all MDR isolates of S. aureus (four isolates) with 100% incidence, and the aac(6’)aph (2’’) gene was positive in three (75%) isolates. These three genes are the most involved in the antibiotic resistance of S. aureus strains. For B. cereus, all six isolates possessed the bla and tetA genes (100%). The resistance gene PCR product patterns are shown in Figures 1–3.
Table 5. Distribution of resistance genes in the selected MDR isolates of E. coli, S. aureus, and B. cereus isolated from examined raw milk and dairy products samples.
| Target genes | E. coli (na = 5) | S. aureus (n = 4) | B. cereus (n = 6) |
|---|---|---|---|
| bla TEM | 5 (100%) | - | - |
| bla CTX-M | 2 (40%) | - | - |
| blaZ | - | 4 (100%) | - |
| aac(6’)aph (2’’) | - | 3 (75%) | - |
| tetK | - | 4 (100%) | - |
| bla | - | - | 6 (100%) |
| tetA | - | - | 6 (100%) |
n = total number of isolates.
Figure 1. β-lactams resistance genes (blaTEM, blaCTX-M) in MDR E. coli isolates. Lanes: M, marker, 100-bp ladder; P1, positive control for blaTEM; P2, positive control for blaCTX-M; N, negative control DW; 1–5, examined E. coli isolates.
Figure 3. bla and tetA genes in MDR B. cereus isolates. Lanes: M, marker, 100-bp ladder; P1, positive control for bla; P2, positive control for tetA; N, negative control DW; 1–6, examined B. cereus isolates.
Discussion
Milk and its products are vital sources of food for the human population all over the world. Despite their numerous health benefits, milk is an optimum medium for numerous bacteria that could represent human health hazards, such as E. coli, S. aureus, and B. cereus, which are involved in most foodborne illnesses. In addition to the danger of food poisoning, milk and its products may be potential sources of MDR bacterial strains due to the misuse of antibacterial therapeutics in dairy farm management. These MDR strains constitute a huge complication for consumers at the level of antibacterial therapeutic efficiency. The MDR determinants of transmission to other bacterial pathogens have clinical significance [20].
Figure 2. blaZ, aac(6’)-aph (2’’) and tetK genes in MDR S. aureus isolates. Lanes: M, marker, 100-bp ladder; P1, positive control for blaZ; P2, positive control for aac(6’)-aph(2’’); P3, positive control for tetK; N, negative control DW; 1–4, examined S. aureus isolates.
The Egyptian standard (ES) ES:154-1 [27] asserts that raw milk must be free from pathogenic bacteria and their toxins. The results of the current study highlighted that E. coli was the most prevalent among the investigated pathogens in the examined milk and cheese samples, which could be due to the wide spreading of E. coli on the animal body and in the environment as the organism is a common fecal resident. Out of 112 tested samples, E. coli was isolated from 58 (82.85%), 16 (53.33%), and 4 (33.33%) from raw milk, Kariesh cheese, and rice with milk samples, respectively, with an overall isolation percentage of 78 (69.64%) (Table 2). The occurrence of E. coli in raw milk and its products is an indicator of poor hygienic measures during the milking process, insufficient cleaning and sanitation of dairy utensils, worker hands, animal udders, or post-process contamination. The findings of Sultana et al. [28], who isolated E. coli from yogurt samples, support the isolation of E. coli from rice with milk despite being heat treated, which may result in post-process contamination.
The high incidence of E. coli in milk and cheese samples was previously reported by Selvamalar et al. [29] and by Ibrahim et al. [30], who stated 52% and 48% prevalence rates of E. coli, respectively, in raw milk in Sudan and Pakistan. On the other hand, the prevalence of E. coli was high in other reports, but with lesser rates than ours (26% by Sultana et al. [28], 24% by Selvamalar et al. [29]), while other studies failed to isolate E. coli from raw milk and cheese samples as mentioned by Awaad et al. [31]. This variation in the incidence rates of E. coli could be due to differences in the origin of samples, the techniques used for their collection and transportation, or environmental conditions.
Considering the incidence of S. aureus in the examined samples as tabulated in Table 2, S. aureus was isolated from raw milk at a higher rate (17.14%) than from Kariesh cheese (6.66%), while it failed to be detected in rice with milk samples. The overall incidence of S. aureus in the different types of samples was 12.5%. These results may be attributed to the contamination of raw milk during production, which could be easily controlled during dairy product processing either via a high acidity percentage as in the case of Kariesh cheese or via heat treatment as in the case of desserts. Zeinhom et al. [32] indicated that the high incidence of S. aureus in raw milk samples from Beni Suef, Egypt, was due to contamination from the environment, cross-contamination, and poor handling in milk collection centers or during transportation. In addition, the udders of infected animals were also blamed. Also, Awaad et al. [31] isolated S. aureus from raw milk produced in Fayoum farms, Egypt, before cheese production, but with a higher incidence level (40%), this incidence was decreased to 8% after ripening of cheese produced from the same milk, indicating the effect of ripening changes on S. aureus survival.
According to the ES:154-1 [27], for raw milk, the S. aureus count must not be more than 1 × 102 colony forming unit/ml, while for other dairy products such as Kariesh cheese, it must be free from it or its toxins. However, in the current study, 6.7% of Kariesh cheese samples were contaminated with the organism; Zeinhom et al. [32] isolated S. aureus from Kariesh cheese in Egypt with a higher incidence rate (18%). This variation in the results could be attributed to the fact that this product is usually produced on a small scale or homemade, in addition to the absence of a heating process or pasteurization of milk during its manufacturing and its low salt content.
Looking at the incidence of B. cereus, it was clear that the organism was more prevalent in the rice with milk than in other samples. Data presented in Table 2 showed that eight samples were positive from the raw milk samples, with an incidence of 11.42%. Only two samples from Kariesh cheese were positive for B. cereus, with an incidence percentage of 6.66%, while the highest incidence for B. cereus was in rice with milk samples (66.66%). Bacillus cereus contamination is related to the efficacy of the hygienic measures applied during the processing and distribution of milk products.
The low prevalence of B. cereus in Kariesh cheese samples examined in the current study comes in agreement with previous reports that indicated that the acidity of such kinds of cheese acts as a control measure [33]. On the contrary, the high prevalence of B. cereus in rice and milk samples could be due to the slight boiling during cooking, which leads to the destruction of most of the vegetative bacterial species, leaving the heat-resistant B. cereus spores to flourish after cooling.
Rice pudding milk is a popular dairy dessert in Egypt among people of different ages due to its pleasant and satiating power. However, its contamination with B. cereus is high due to its unhygienic processing, storage, and distribution. Bacillus cereus is one of the most isolated pathogens from this product [34].
Together with investigating the incidence rates, investigating the antibiotic resistance phenotypes of the pathogens isolated from food sources is crucial; therefore, it was determined for isolated pathogens in our study as shown in Table 3.
Escherichia coli isolates showed high resistance rates against GM (73.33%), AM (53.3%), and CTX (16.66%) and were moderately sensitive to chloramphenicol, CAZ, CTX, and AX with clavulanic acid, while all the isolates showed sensitivity to TE, trimethoprim–sulfamethoxazole, and CIP.
All isolates of S. aureus (100%) were resistant to AM, with TE, CTX, and gentamycin (60%) coming in second and third place, respectively. TEs are broad-spectrum antimicrobial agents often used in the treatment of infections in dairy animals. The high prevalence of resistance against TE has been reported previously among E. coli and S. aureus isolated from raw milk and dairy products from different countries [35]. On the other hand, all the isolates (100%) were sensitive to CIP, AX, clavulanic acid, and trimethoprim–sulfamethoxazole.
Regarding B. cereus, all isolates (100%) showed resistance to AM, followed by AX (20%) and TE (6.66%). All isolates (100%) were sensitive to VA.
Our findings coincide with some previous studies, such as Zeinhom et al.’s [32] study, in which S. aureus isolates from raw milk and dairy products such as cheese were resistant to AM (72%), and TE (60%).
MDR is defined as acquiring non-susceptibility to at least one agent in three or more antimicrobial classes [36]. Results in Table 4 illustrate the incidence of MDR strains among the recovered isolates. The highest MDR rate was met with S. aureus, of which 4 isolates out of 10 (40%) conferred resistance to 3 antimicrobials belonging to different categories, with 1 of them even resistant to 4 agents. On the other hand, only 5 out of 60 E. coli isolates (8.33%) and 6 out of 30 B. cereus isolates (20%) showed a MDR profile.
Concerning E. coli, the prevalence of MDR isolates detected in the present study is not as frightening as what was reported by Ombarak et al. [20], who reported that 50% of E. coli isolates recovered from raw milk and cheese samples in Egypt was MDR. The absolute resistance of B. cereus to AM agrees with the report of Osama et al. [33]. Detection of even one MDR isolate of B. cereus, as in the current study, is alarming as the bacterium is a spore-former and able to reside in the environment for long periods while carrying this transmissible criterion.
The high resistance levels, with a high proportion for beta-lactams, among the examined isolates obtained in this study may be due to the increased usage of beta-lactams in the veterinary field.
In this study, all the tested resistant isolates of E. coli were found to carry the blaTEM gene (n = 5) at 100% (Table 5), which agrees with the previous findings [33] from food and food-producing animals. In the same context, Yu et al. [37] reported that 83.1% of E. coli isolates from raw milk samples had the blaTEM gene, while 40% of these isolates carried the blaCTX-M gene. Hassani et al. [3] found that 50% of E. coli isolates had blaTEM. The same genes were detected in resistant E. coli from dairy animals [38]. It was noted that all isolates possessing both genes showed resistance to AM, AX, and CAZ in phenotypic experiments.
All tested resistant S. aureus isolates (n = 4) possessed the blaZ and TetK genes, whereas the aac6’-aph 2’’ gene was found in only three isolates, as presented in Table 5. Ronco et al. [39] researched the prevalence of resistance genes in S. aureus isolates from dairy cows with clinical mastitis and bulk tank milk and detected blaZ in 17.2% of isolates. Liu et al. [40] stated in their study that 100% of S. aureus isolates carried the aac6’-aph 2’’ gene, and the tet gene was detected in 14.3%.
The detection of MDR B. cereus isolates possessing the bla and tetA genes is worrisome, as the emergence of MDR pathogenic bacteria can be a serious hazard [41]. Ranjbar and Sami [42] highlighted the risk of different extended spectrum β-lactamases genes (antibiotic resistance genes) being transferred among bacteria in the environment, with the resultant threat to the efficacy of available antibiotics currently used in medical applications.
Further studies with a high number of samples and different dairy products are needed to elucidate the incidence of MDR pathogens in different dairy products produced from different locations in Egypt. Also, investigating the routes of raw milk contamination with AMR bacteria is important to uncover whether these bacteria access the milk from the animal or contaminate the milk during, after, or during processing. Moreover, an investigation of the antibiotic resistance of the MDR pathogens is required.
Conclusion
The safety of raw milk and milk products investigated in the current study is not satisfactory as MDR-potential pathogens were detected. Sequentially, such products constitute a potential public health hazard. Moreover, the existence of MDR strains exaggerates the problem and calls for attention to the urgent need for decisive rules and regulations to face the increasing misuse of antibacterial therapeutics in dairy herd management and emphasize the need for new natural antimicrobial therapeutic agents.
Acknowledgment
This study did not receive any specific grant from any funding agency.
List of abbreviations
AMR: Antimicrobial resistance; APHA, American Public Health Association; CLSI, Clinical and Laboratory Standards Institute; ES, Egyptian Standards; ISO, International Organization for Standardization; MDR: Multi-drug resistant; PCR: Polymerase chain reaction.
Conflict of interest
The authors declare that there is no conflict of interest.
Authors’ contributions
DA and RAO designed the study, interpreted the data, and drafted the manuscript. ABA was involved in collection of data and contributed to manuscript preparation. AS, ABA, and RAO took part in critical checking of this manuscript.
References
- [1].Górska-Warsewicz H, Rejman K, Laskowski W, Czeczotko M. Milk and dairy products and their nutritional contribution to the average Polish diet. Nutrients. 2019;11(8):1771. doi: 10.3390/nu11081771. https://doi.org/10.3390%2Fnu11081771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Sobeih A, AlHawary I, Khalifa ES, Ebied N. Prevalence of Enterobacteriaceae in raw milk and some dairy products. Kafrelshheikh Vet Med J. 2020;18(2):9–13. http://dx.doi.org/10.21608/kvmj.2020.39992.1009. [Google Scholar]
- [3].Hassani S, Moosavy MH, Gharajalar SN, Khatibi SA, Hajibemani A, Barabadi Z. High prevalence of antibiotic resistance in pathogenic foodborne bacteria isolated from bovine milk. Sci Rep. 2022;12(1):3878. doi: 10.1038/s41598-022-07845-6. https://doi.org/10.1038/s41598-022-07845-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].FDA. The dangers of raw milk—unpasteurized milk can pose a serious health risk. 2018. [January 2023]. Available via https://www.fda.gov/food/buy-store-serve-safe-food/dangers-raw-milk-unpasteurized-milk-can-pose-serious-health-risk .
- [5].Hohmann MF, Wente N, Zhang Y, Krömker V. Bacterial load of the teat apex skin and associated factors at herd level. Animals (Basel) 2020;10(9):1647. doi: 10.3390/ani10091647. https://doi.org/10.3390%2Fani10091647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Li E, Saleem F, Edge TA, Schellhorn HE. Biological indicators for fecal pollution detection and source tracking: a review. Processes. 2021;9(11):2058. https://doi.org/10.3390/pr9112058. [Google Scholar]
- [7].Ramos S, Silva V, Dapkevicius MLE, Caniça M, Tejedor-Junco MT, Igrejas G, et al. Escherichia coli as commensal and pathogenic bacteria among food-producing animals: health implications of extended spectrum β-lactamase (ESBL) production. Animals (Basel) 2020;10(12):2239. doi: 10.3390/ani10122239. https://doi.org/10.3390%2Fani10122239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Eid HM, El-Mahallawy HS, Mohammed SR, Mohammed NEY, Eidaroos NH. Multidrug-resistant and enterotoxigenic methicillin-resistant Staphylococcus aureus isolated from raw milk of cows at small-scale production units. J Adv Vet Anim Res. 2022;9(1):113–21. doi: 10.5455/javar.2022.i575. https://doi.org/10.5455%2Fjavar.2022.i575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Vidic J, Chaix C, Manzano M, Heyndrickx M. Food sensing: detection of Bacillus cereus spores in dairy products. Biosensors. 2020;10(3):15. doi: 10.3390/bios10030015. https://doi.org/10.3390%2Fbios10030015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Kamaruzzaman EA, Abdul Aziz S, Bitrus AA, Zakaria Z, Hassan L. Occurrence and characteristics of extended-spectrum β-Lactamase-producing Escherichia coli from dairy cattle, milk, and farm environments in Peninsular Malaysia. Pathogens. 2020;9(12):1007. doi: 10.3390/pathogens9121007. https://doi.org/10.3390/pathogens9121007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Brown K, Mugoh M, Call DR, Omulo S. Antibiotic residues and antibiotic-resistant bacteria detected in milk marketed for human consumption in Kibera, Nairobi. PLoS One. 2020;15(5):e0233413. doi: 10.1371/journal.pone.0233413. https://doi.org/10.1371%2Fjournal.pone.0233413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Tóth AG, Csabai I, Krikó E, Tőzsér D, Maróti G, Patai ÁV, et al. Antimicrobial resistance genes in raw milk for human consumption. Sci Rep. 2020;10(1):7464. doi: 10.1038/s41598-020-63675-4. https://doi.org/10.1038%2Fs41598-020-63675-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].ISO6887-5. Geneva, Switzerland: ISO; 2020. Microbiology of food chain preparation of test samples, initial suspension, and decimal dilutions for microbiological examination—part 5: specific rules for the preparation of milk and milk products. [Google Scholar]
- [14].ISO16649-1. Geneva, Switzerland: ISO; 2018. Microbiology of the food chain—horizontal method for the enumeration of beta-glucuronidase-positive Escherichia coli—part 1: colony-count technique at 44 degrees C using membranes and 5-bromo-4-chloro-3-indolyl beta-D-glucuronide. [Google Scholar]
- [15].ISO6888-1. Geneva, Switzerland: ISO; 2021. Microbiology of the food chain—horizontal method for the enumeration of coagulase-positive staphylococci (Staphylococcus aureus and other species)—part 1: method using Baird-Parker agar medium. [Google Scholar]
- [16].ISO7932. Geneva, Switzerland: ISO; 2004. Microbiology of food and animal feeding stuff—horizontal method for the enumeration of presumptive Bacillus cereus—colony-count technique at 30 °C. [Google Scholar]
- [17].APHA. 16th. Washington, DC: American Public Health Association; 1992. Standard methods for the examination of dairy products. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].APHA. 5th. Washington, DC: APHA; 2015. Compendium of methods for the microbiological examination of foods; pp. 509–26. Chapter 39. [Google Scholar]
- [19].CLSI. CLSI document M100. 30th. Wayne, PA: Clinical and laboratory standards Institute; 2020. Performance standards for antimicrobial disk susceptibility tests-approved standard. [Google Scholar]
- [20].Ombarak RA, Hinenoya A, Elbagory AM, Yamasaki S. Prevalence and molecular characterization of antimicrobial resistance in Escherichia coli isolated from raw milk and raw milk cheese in Egypt. J Food Prot. 2018;81(2):226–32. doi: 10.4315/0362-028X.JFP-17-277. https://doi.org/10.4315/0362-028x.jfp-17-277. [DOI] [PubMed] [Google Scholar]
- [21].Archambault M, Petrov P, Hendriksen RS, Asseva G, Bangtrakulnonth A, Hasman H, et al. Molecular characterization and occurrence of extended-spectrum beta-lactamase resistance genes among Salmonella enterica serovar Corvallis from Thailand, Bulgaria, and Denmark. Microb Drug Resist. 2006;12(3):192–8. doi: 10.1089/mdr.2006.12.192. https://doi.org/10.1089/mdr.2006.12.192. [DOI] [PubMed] [Google Scholar]
- [22].Colom K, Pèrez J, Alonso R, Fernández-Aranguiz A, LariňoE Cisterna R. Simple and reliable multiplex PCR assay for detection of blaTEM, blaSHV and blaOXA–1 genes in Enterobacteriaceae. FEMS Microbiol Lett. 2003;223:147–51. doi: 10.1016/S0378-1097(03)00306-9. https://doi.org/10.1016/S0378-1097(03)00306-9. [DOI] [PubMed] [Google Scholar]
- [23].Bagcigil AF, Taponen S, Koort J, Bengtsson B, Myllyniemi A, Pyörälä S. Genetic basis of penicillin resistance of S. aureus isolated in bovine mastitis. Acta Vet Scand. 2012;54:69. doi: 10.1186/1751-0147-54-69. https://doi.org/10.1186/1751-0147-54-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Duran N, Ozer B, Duran GG, Onlen Y, Demir C. Antibiotic resistance genes & susceptibility patterns in staphylococci. Indian J Med Res. 2012;135(3):389–96. [PMC free article] [PubMed] [Google Scholar]
- [25].Rather MA, Aulakh RS, Gill JP, Mir AQ, Hassan MN. Detection and sequencing of plasmid encoded tetracycline resistance determinants (tetA and tetB) from food-borne Bacillus cereus isolates. Asian Pac J Trop Med. 2012;5(9):709–12. doi: 10.1016/S1995-7645(12)60111-4. https://doi.org/10.1016/s1995-7645(12)60111-4. [DOI] [PubMed] [Google Scholar]
- [26].Chen Y, Tenover FC, Koehler TM. Beta-lactamase gene expression in a penicillin-resistant Bacillus anthracis strain. Antimicrob Agents Chemother. 2004;48(12):4873–7. doi: 10.1128/AAC.48.12.4873-4877.2004. https://doi.org/10.1128%2FAAC.48.12.4873-4877.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].ES. Egyptian Organization for Standardization and Quality Control. Cairo, Egypt: Ministry of Industry; 2005. Egyptian specifications 154-1. Milk and milk products, part 1: raw milk. [Google Scholar]
- [28].Sultana T, Rabbi BR, Sarker BR, Islam MS, Begum MIA, Hossain KMM. Prevalence and antibiotic resistance patterns of Escherichia coli isolated from milk and milk products. J Bio Sci. 2021;29(2):81–91. http://dx.doi.org/10.3329/jbs.v29i2.54957. [Google Scholar]
- [29].Selvamalar AP, Sekar M, Porter K, Narayan R, Elango A, Kummar M, et al. Screening of E. coli for its antimicrobial susceptibility pattern in milk and dairy products in Chennai, India. Int J Chem Stud. 2018;6:35–7. [Google Scholar]
- [30].Ibrahim AH, Ali ME, Ahmed MF, Abdelkhalek A. Prevalence and characterization of Escherichia coli in raw milk and some dairy products at Mansoura City. J Adv Vet Res. 2022;12(4):363–70. [Google Scholar]
- [31].Awaad SS, Moawad AA, Abdelsalam AB, Sallam SS. Impact of raw materials and processing techniques on the microbiological quality of Egyptian Domiati cheese. Int J Vet Sci Med. 2020;9(4):505–10. http://dx.doi.org/10.37422/IJVS/20.060. [Google Scholar]
- [32].Zeinhom M, Abed A. Prevalence, characterization, and control of Staphylococcus aureus isolated from raw milk and Egyptian soft cheese. J Vet Med Res. 2020;27(2):152–60. https://doi.org/10.21608/jvmr.2021.146885. [Google Scholar]
- [33].Osama R, Ahmed M, Abdulmawjood A, Al-Ashmawy M. Prevalence and antimicrobial resistance of Bacillus cereus in milk and dairy products. Mansoura Vet Med J. 2020;21(2):11–8. https://doi.org/10.21608/mvmj.2020.2.202. [Google Scholar]
- [34].Ahmed A, El-Gamal A, Ibrahim A. Prevalence of Bacillus cereus in some dairy desserts in Egypt. Egypt J Food Safety. 2018;5(1):1–11. https://doi.org/10.21608/ejfsj.2018.138034. [Google Scholar]
- [35].Haulisah NA, Hassan L, Bejo SK, Jajere SM, Ahmad NI. High levels of antibiotic resistance in isolates from diseased livestock. Front Vet Sci. 2021;8:652351. doi: 10.3389/fvets.2021.652351. https://doi.org/10.3389/fvets.2021.652351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Alagna L, Palomba E, Mangioni D, Bozzi G, Lombardi A, Ungaro R, et al. Multidrug-resistant Gram-negative bacteria decolonization in immunocompromised patients: a focus on fecal microbiota transplantation. Int J Mol Sci. 2020;21(16):5619. doi: 10.3390/ijms21165619. https://doi.org/10.3390/ijms21165619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Yu ZN, Wang J, Ho H, Wang YT, Huang SN, Han RW. Prevalence and antimicrobial-resistance phenotypes and genotypes of Escherichia coli isolated from raw milk samples from mastitis cases in four regions of China. J Global Antimicrob Resist. 2020;22:94–101. doi: 10.1016/j.jgar.2019.12.016. https://doi.org/10.1016/j.jgar.2019.12.016. [DOI] [PubMed] [Google Scholar]
- [38].Rubab M, Oh DH. Molecular detection of antibiotic resistance genes in Shiga toxin-producing E. coli isolated from different sources. Antibiotics. 2021;10(4):344. doi: 10.3390/antibiotics10040344. https://doi.org/10.3390/antibiotics10040344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Ronco T, Klaas IC, Stegger M, Svennesen L, Astrup LB, Farre M, et al. Genomic investigation of Staphylococcus aureus isolates from bulk tank milk and dairy cows with clinical mastitis. Vet Microbiol. 2018;215:35–42. doi: 10.1016/j.vetmic.2018.01.003. https://doi.org/10.1016/j.vetmic.2018.01.003. [DOI] [PubMed] [Google Scholar]
- [40].Liu H, Li S, Meng L, Dong L, Zhao S, Lan X, et al. Prevalence, antimicrobial susceptibility, and molecular characterization of Staphylococcus aureus isolated from dairy herds in northern China. J Dairy Sci. 2017;100(11):8796–803. doi: 10.3168/jds.2017-13370. https://doi.org/10.3168/jds.2017-13370. [DOI] [PubMed] [Google Scholar]
- [41].Ghazaei C. Phenotypic and molecular detection of beta-lactamase enzyme produced by Bacillus cereus isolated from pasteurized raw milk. J Med Bacteriol. 2019;8(3–4):1–7. [Google Scholar]
- [42].Ranjbar R, Sami M. Genetic investigation of beta-lactam associated antibiotic resistance among Escherichia coli strains isolated from water sources. Open Microbiol J. 2017;11:203–10. doi: 10.2174/1874285801711010203. https://doi.org/10.2174/1874285801711010203. [DOI] [PMC free article] [PubMed] [Google Scholar]