Abstract
The human immunodeficiency virus type 1 (HIV-1) Vpu and Env proteins are expressed from a bicistronic mRNA. To address the biological significance of the coordinated expression of vpu and env, we compared the relative effects on particle release of HIV-1 isolates containing an intact vpu gene or carrying point mutations in its initiation codon or internal deletions, respectively. We found that the primary AD8 isolate, which is unable to express vpu due to a mutation in its translation initiation codon, was able to replicate in primary macrophages and peripheral blood mononuclear cells with efficiency similar to that of an isogenic variant expressing Vpu. Interestingly, AD8 lacking a vpu initiation codon produced higher levels of Env protein than its Vpu-expressing isogenic variant. In contrast, disabling Vpu without removing the vpu initiation codon did not alter Env expression but significantly reduced virus production. AD8 Env when provided in trans was capable of enhancing release not only of AD8 particles but also of viruses of the T-cell-tropic NL4-3 isolate. We conclude that AD8 Env encodes a Vpu-like activity similar to that previously reported for HIV-2 Env proteins and is thus able to augment virus secretion. When expressed at elevated levels, i.e., following mutation of the vpu initiation codon, AD8 Env was able to compensate for the lack of Vpu and thereby ensure efficient virus release. Thus, the ability to regulate virus release is redundant in AD8 and can be controlled by either Vpu or Env. Since Vpu controls several independent functions, including CD4 degradation, our results suggest that some HIV-1 isolates may have evolved a mechanism to regulate Vpu activity without compromising their ability to efficiently replicate in the host cells.
Human immunodeficiency virus type 1 (HIV-1) is a complex retrovirus that encodes at least nine structural and nonstructural genes. All genes are expressed from a primary transcript that is initiated from a single promoter located in the 5′ long terminal repeat. Except for the gag gene, which is located near the 5′ end of the primary transcript, translation of all downstream genes requires posttranscriptional mechanisms such as partial or full splicing of the primary transcript (38), ribosomal frameshifting (22, 58), or, in the case of vpu and env, leaky scanning of a bicistronic mRNA (46). Coregulation of gag and pol gene expression by means of ribosomal frameshifting is functionally significant inasmuch as it ensures the balanced expression of HIV structural proteins, which may be critical for proper virus assembly (23); for a review, see reference 27). In contrast, the significance of a coordinated expression of vpu and env from a bicistronic mRNA has thus far been obscure.
Vpu and Env are both integral membrane proteins. However, the principal functions of these two proteins are quite distinct. The Env protein is one of the main virion components, and its primary function is to act as a ligand for binding of virus particles to CD4 and coreceptor molecules on target cells. In addition, the Env protein of certain HIV-2 isolates has the capacity to regulate virus release (7, 8, 39) in a Vpu-like manner (8). Unlike Env, Vpu appears to be largely restricted to intracellular membranes (25, 43) and has so far not been found in association with virions. Vpu has several independent functions, the best characterized of which is its ability to induce CD4 degradation (55, 56). This function requires phosphorylation of two conserved serine residues in the cytoplasmic domain of Vpu (18, 32, 35, 40); it further involves the formation of multiprotein complexes containing CD4, Vpu, h-βTrCP, and Skp1 (32) and leads to the ubiquitin-dependent proteolysis of CD4 by proteasomes (19, 44). Another well-characterized function of Vpu is its role in regulating virus release from infected cells (for reviews, see references 21, 33, 48, and 51). This function of Vpu is correlated with its ability to form cation-conductive membrane pores (16, 42); for a review, see reference 28) and is regulated by Vpu from a post-endoplasmic reticulum (ER) compartment (40). Aside from that, Vpu was found to interfere with an early step in the biosynthesis of major histocompatibility complex (MHC) class I molecules (24), to affect cell surface delivery of certain glycoproteins (52), and to influence the syncytium-inducing ability of HIV-1 (59).
We have recently analyzed the importance of Vpu for replication of various macrophage-tropic HIV-1 isolates in primary human cells. We observed that Vpu augmented virus release from infected macrophages about four- to sixfold, while virus production from infected peripheral blood mononuclear cells (PBMC) was increased two- to threefold in the presence of Vpu (41). All of the Vpu-deficient variants used in that study carried internal mutations in vpu which retained the bicistronic nature of the vpu/env mRNAs. It is interesting, however, that several Vpu-defective natural HIV-1 isolates, including the macrophage-tropic AD8 isolate (50), do not carry internal mutations in their vpu genes but contain a point mutation in the vpu translation initiation codon (ATGvpu) (34). Based on the ribosome-scanning model (for a review, see reference 26), expression of Env is thought to occur by leaky scanning of the vpu/env mRNA (46). Consequently, mutation of the ATGvpu is expected not only to abolish vpu expression but to have a positive effect on env expression.
We used AD8 as a model system to study the potential significance of the coordinated expression of env and vpu from a common bicistronic mRNA. We found that AD8, despite the lack of an ATGvpu [as in the construct pAD8(−)], efficiently released virus particles and exhibited replication kinetics both in primary macrophage cultures and in PBMC that were comparable to those of an AD8 variant expressing functional Vpu. In contrast, virus release from cells infected with a vpu-deficient variant of AD8 containing an intact ATGvpu but carrying an internal deletion in Vpu was significantly reduced. pAD8(−) was found to produce higher levels of Env protein than AD8 variants expressing functional Vpu or carrying an internal deletion in the vpu gene. We found that expression of elevated levels of AD8 Env protein could compensate for the lack of Vpu and thus ensure efficient virus release. Our results suggest that AD8 encodes a redundant Vpu-like activity that is revealed upon mutation of the Vpu initiation codon and the concomitant increase in Env synthesis. The fact that the AD8 Vpu and Env proteins are both able to regulate virus release suggests that mutating the ATGvpu may function as a molecular switch that enables the virus to turn off Vpu expression without compromising its ability to efficiently replicate in host cells.
MATERIALS AND METHODS
Molecular HIV-1 clones and plasmid constructions.
Plasmid pNL4-3 is an infectious molecular clone of the T-cell-tropic isolate NL4-3 (1). pNL-ATG is a vpu-deficient derivative of pNL4-3 and was constructed by introducing an ATG-to-GTG mutation into the translation initiation codon of vpu. For this purpose, pNL-A1 (47) template DNA was amplified by PCR using the 5′ primer ATCAAGCTTCTCTATCAAAGCAGTAAGTAGTACATGTAGTGCAACC carrying the desired mutation and the 3′ primer CTTTGTCTTAATATTTTCC. The resulting 141-bp PCR product was digested with HindIII and SspI, cloned first into the HindIII/SspI sites of pSP-8 (47), leading to pSP-8/U-ATG, and then subcloned as an EcoRI/KpnI fragment into pNL4-3, leading to pNL-ATG. Another derivative of pNL4-3, vpuDEL-1, carrying a 41-bp out-of-frame deletion in vpu, has been described elsewhere (25). pNLenv1-UDEL1 is a derivative of vpuDEL-1 and carries, in addition to the deletion in vpu, a 1,264-bp out-of-frame deletion in env (KpnI-BglII). pNLenv1-UDEL1 expresses all HIV-1 proteins except Vpu and Env. A 5,032-bp BssHII-EcoRI fragment of plasmid pAD8(+) (positions 940 to 5972 [50]) was cloned into the corresponding sites of pNLenv1-UDEL1, resulting in plasmid pAD8env1-UDEL1. pAD8env1-UDEL1 encodes AD8-specific gag, pol, and vif genes, while tat, rev, and nef genes are derived from NL4-3. Construction of the vpu-positive macrophage-tropic molecular clone pAD8 [referred to here as pAD8(+)] and of its vpu-deficient variant pAD8-2 [referred to here as pAD8(−)] has been described elsewhere (50). pAD8(−) is an infectious molecular clone of the macrophage-tropic HIV-1AD87 isolate which carries a point mutation (ATG to ATA) in the vpu translation initiation codon and, as a result, does not express Vpu. pAD8(+) is an isogenic variant of pAD8(−) in which the vpu ATG was restored by oligonucleotide-directed mutagenesis (50). Construction of pAD8-UDEL2, carrying a 81-bp deletion plus an 8-bp linker insertion in vpu (Fig. 1), has been described previously (41). pCM10-Env is a Rev-dependent vector for expression of the HIV-2ROD10 envelope protein under the transcriptional control of the cytomegalovirus (CMV) early promoter. This construct is identical to the previously described pCM5-Env (7) except for the absence of an NcoI site at position 467 in the CMV promoter. Plasmid pCM10.env1 is an env-defective control vector derived from pCM10-Env. A frameshift mutation was created by inserting a 10-bp XhoI linker (New England BioLabs, Beverly, Mass.) into the BsaBI site of pCM10-Env at position 944. The resulting construct produces a truncated ROD10 Env protein consisting of only the first 60 N-terminal amino acids plus 15 missense residues. pCM2-AD2Env is a Rev-dependent construct that expresses the full-length HIV-1 AD8 envelope protein under the transcriptional control of the CMV promoter. To generate pCM2-AD2Env, a 2,821-bp fragment consisting of the full-length vpu and env genes flanked by XbaI and BamHI sites was PCR amplified from pAD8(−). The PCR product was digested with XbaI-BamHI and cloned into the XbaI and BglII sites of the previously described plasmid pCMV-CD8/cyto4 (57). This strategy replaced all CD8 and CD4 sequences from the original clone with the HIV-1 AD8 vpu (ATG deficient) and env genes.
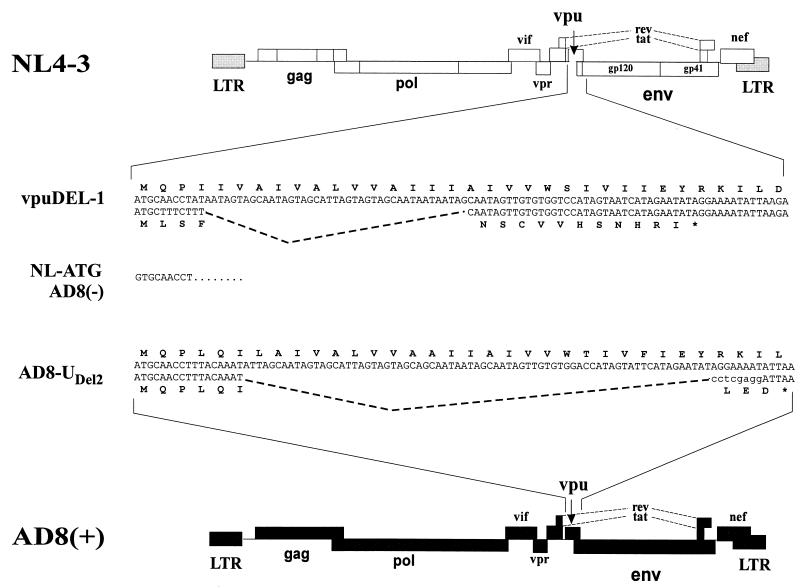
FIG. 1.
Schematic representation of viruses used in this study. Schematic outlines of the parental isolates NL4-3 and AD8(+) are shown at the top and bottom, respectively. The nucleotide sequences and deduced amino acid sequences for wild-type (top row of each sequence pair) and mutant (bottom row) vpu genes are shown for comparison. The deletion in the vpu gene of vpuDEL-1 corresponds to a natural deletion found in the NY-5 isolate of HIV-1 (25). The mutant was constructed by oligonucleotide-directed mutagenesis as described elsewhere (25). Note that vpuDEL-1 does not express any vpu-specific sequences (25, 43). AD8-UDEL2 was constructed as described in Materials and Methods. This mutant has the potential to express six vpu-specific and three missense amino acids. The sequences of NL-ATG and AD8(−) are identical to the corresponding wild-type sequences except for the presence of a ATG-to-GTG mutation which eliminates the vpu translation initiation codon. LTR, long terminal repeat.
Cell culture, transfection, and infection.
HeLa cells (ATCC CCL2) were propagated in Dulbecco’s modified Eagle’s medium (DMEM) containing 10% fetal bovine serum (FBS). A3.01 cells, a derivative of the CEM human T-lymphocytic cell line which was selected for high-level expression of CD4 (17), were cultivated in complete RPMI 1640 supplemented with 10% FBS (RPMI 1640-FBS). Stocks of PBMC were prepared from gradient-isolated lymphocytes of a healthy, HIV-seronegative individual by using lymphocyte separation medium (Organon Teknika-Cappel, Durham, N.C.) and stored in liquid nitrogen. For each infection, 5 × 106 cells were stimulated for 2 days with phytohemagglutinin (PHA; 1 mg/ml; Sigma Chemical Co., St. Louis, Mo.) in the presence of purified human interleukin-2 (hIL-2; 20 U/ml; Boehringer, Mannheim, Germany). PBMC cultures were maintained in RPMI 1640-FBS supplemented with hIL-2.
For transfection, HeLa cells were grown to near confluence in 25-cm2 flasks (5 × 106 cells per flask). Two hours prior to transfection, the medium was replaced with 5 ml of fresh DMEM-FBS. Calcium phosphate-precipitated plasmid DNA (25 to 30 μg) was added to the cells. After 6 h, the medium was removed, and the cells were subjected to a glycerol shock for 2.5 min as described elsewhere (41). The cultures were then washed once with phosphate-buffered saline (PBS; 10 mM phosphate buffer [pH 7.4], 100 mM NaCl) (53) and maintained in 5 ml of DMEM-FBS.
Virus stocks were prepared in HeLa cells transfected with plasmid DNAs of individual molecular clones. Virus-containing supernatants were clarified by centrifugation (1,000 × g, 5 min) and filtered through a 0.45-μm-pore-size filter to remove residual cells and debris. Virions were pelleted by ultracentrifugation in a Beckman SW55 rotor (1 h, 35,000 rpm; Beckman Instruments, Fullerton, Calif.), resuspended in 1 ml of RPMI 1640-FBS, and filtered. The virus stocks were assayed for reverse transcriptase (RT) activity by an assay using [32P]TTP incorporation with an oligo(dT)-poly(A) template as described previously (54).
Routinely, 106 RT units were used to infect 106 PBMC or monocyte-derived macrophages (MDM). Following 15 h of adsorption, the medium was changed completely to remove residual input virus. PBMC were maintained in RPMI 1640-FBS at a density of ca. 106 cells/ml and were fed by replacing 80% of the medium every other day. Infection of the cultures was monitored by determining the RT activity released into the supernatant fluid as described above. The cultures were also examined by light microscopy for syncytium formation and scored by counting the number of syncytia per field.
Monocyte isolation and culture.
PBMC were isolated from the leukopheresed blood of HIV-seronegative donors after gradient separation using lymphocyte separation medium (Organon Teknika-Cappel). Monocytes were isolated by countercurrent centrifugal elutriation using a Beckman system. Elutriated monocytes were >99% viable as determined by trypan blue exclusion and were >95% pure as determined by morphological analysis using Giemsa staining of representative cytocentrifuge preparations. Monocytes were precultured as adherent cell monolayers by using a modification of the procedure of Lazdins et al. (29). Briefly, 3 × 106 cells were suspended in 2 ml of high-glucose (4.5 g/liter) DMEM (Gibco, Grand Island, N.Y.) supplemented with 50 U of penicillin and 50 mg of streptomycin (Gibco) per ml, 2 mM l-glutamine (Gibco), 1 mM sodium pyruvate (Gibco), and 10% pooled human serum. Cultures were then incubated at 37°C in a 5% CO2 humidified incubator for 7 days, supplemented with an additional 2 ml of DMEM, and incubated further. After 10 to 14 days, MDM were obtained by harvesting culture supernatants, then adding 2 ml of cold, Ca2+/Mg2+-free PBS to adherent cell monolayers, and incubating the cells for 45 min at 4°C prior to removal with a plastic cell scraper. All cells were pooled, pelleted by centrifugation (10 min, 1,000 × g), and then resuspended in fresh DMEM at a concentration of 0.5 × 106 cells/ml. Harvested cells (1.5 ml) were finally replated into 24-well tissue culture plates (Nunc), allowed to adhere, and infected within 3 days.
Antisera and antibodies.
Serum from an asymptomatic HIV-1-seropositive patient was used to detect HIV-1-specific proteins by immunoprecipitation and Western blotting. In addition, a Vpu-specific polyclonal antiserum (sheep) raised against a synthetic peptide comprising residues 41 to 58 of Vpu (40) (gift of T. Porstmann) as well as a polyclonal Vpu antiserum (rabbit) directed against the hydrophilic C-terminal domain of Vpu expressed in Escherichia coli (31) were used for detection of Vpu. A polyclonal rabbit antiserum recognizing gp160 and gp120 was described recently (55).
Metabolical labeling and immunoprecipitation.
For pulse-chase experiments using transfected HeLa cells, adherent cultures were washed once with PBS and starved for 10 min in methionine-free RPMI 1640 (Specialty Media, Inc., Lavalette, N.J.). Cells were pulse-labeled with [35S]methionine (2 mCi/ml; Du Pont Inc., Boston, Mass.) for various times as indicated in the text. The medium was then removed, the cells were washed once in PBS, and equal aliquots were added to 500 μl of prewarmed RPMI 1640-FBS for each time point of the chase period and incubated at 37°C while gently shaking. At the indicated time points, cells were collected and stored on dry ice. For studying virus particle release, the supernatants were centrifuged (2 min, 16,000 × g) and filtered through a 0.45-μm-pore-size Spin-X filter tube (Costar, Cambridge, Mass.). Cell-free virus particles were pelleted in a refrigerated Eppendorf Microfuge (4°C, 100 min, 16,000 × g) as described elsewhere (41). The pelleted virions were lysed in a buffer containing 300 mM NaCl, 50 mM Tris-hydrochloride (pH 7.4), and 0.1% (vol/vol) Triton X-100 by shaking at room temperature for 20 min. Cells were lysed in a buffer containing 50 mM Tris-hydrochloride (pH 8.0), 5 mM EDTA, 100 mM NaCl, 0.5% (wt/vol) CHAPS {3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate}, and 0.2% (wt/vol) deoxycholate (DOC) (3). Cell lysates were precleared by incubation at 4°C for 1 h with GammaBind-G–Sepharose beads (Pharmacia LKB Biotechnology, Piscataway, N.J.) preadsorbed with 20 μg of immunoglobulin G from rabbit preimmune serum.
For steady-state metabolic labeling, transfected HeLa cells (5 × 106) were washed with PBS and preincubated for 10 min in methionine-free RPMI to deplete the intracellular pool of methionine. Cells were then washed and incubated in 1 ml of methionine-free RPMI containing 450 μCi of [35S]methionine. After 8 h, 450 μCi of [35S]methionine in 0.3 ml of methionine-free RPMI 1640 was added, and the labeling was continued for an additional 16 h. Cells were lysed in CHAPS-DOC buffer.
For pulse-chase metabolic labeling of MDM, a procedure similar to a previously described protocol (41) was followed. In general, cells remained attached to the tissue culture flasks during the pulse-chase period. Different wells of infected MDM cultures were used for individual time points. Parallel cultures of 7.5 × 105 MDM were seeded in each well of a 24-well tissue culture plate (Costar), and one well was used for each time point during the chase period. Cells were infected with 7.5 × 105 RT units of purified virus stocks, and spread of infection was measured as RT activity released into the culture supernatants. Medium change was performed in 3-day intervals. At the time of maximal virus production, adherent cells were metabolically labeled as follows. Cells were preincubated for 25 min in methionine-free DMEM (Specialty Media) supplemented with 5% heat-inactivated, pooled human serum to deplete the intracellular pool of methionine. Cells were then washed with 1 ml of methionine-free DMEM and pulse-labeled for 30 min in 0.2 ml of methionine-free DMEM containing 350 μCi of [35S]methionine. Cells were washed and chased in the absence of radioactive amino acids for various time periods. The supernatants (1.3 ml) were harvested and then passed through 0.45-μm-pore-size Spin-X filter tubes (Costar). Cell debris was removed by centrifugation for 2 min at 16,000 × g, and virions were pelleted by centrifugation (100 min, 16,000 × g, 4°C) in a refrigerated Eppendorf Microfuge. Immunoglobulins in the clarified supernatants were removed by two rounds of preabsorption using a 1:1 mixture of protein A-Sepharose and GammaBind-G–Sepharose beads (Pharmacia). Pelleted virions were lysed in a buffer containing 300 mM NaCl, 50 mM Tris-hydrochloride (pH 7.4), and 0.1% (vol/vol) Triton X-100 by shaking at room temperature for 20 min. Cells attached to the tissue culture plates were lysed in 210 μl of a buffer containing 50 mM Tris-hydrochloride (pH 8.0), 5 mM EDTA, 100 mM NaCl, 0.5% (wt/vol) CHAPS, and 0.2% (wt/vol) DOC, incubated for 5 min at 37°C, and transferred into 1.5-ml Eppendorf tubes. Cell debris was removed by centrifugation (15 min, 16,000 × g, 4°C) followed by filtration through 0.45-μm-pore-size Spin-X filter tubes (Costar). Lysates of cells and virions were further clarified by two rounds of incubation with protein A- and protein G-Sepharose, preadsorbed with normal rabbit serum. Viral proteins from 50% of the clarified lysates of cells and virions, and from the clarified supernatants, were immunoprecipitated with a 1:1 mixture of HIV-1-positive human serum and a polyclonal rabbit serum specific for HIV-1 proteins gp160 and gp120 preadsorbed to protein G-Sepharose. Viral proteins immunoprecipitated from the cell, virion, or supernatant fractions were solubilized by boiling in sample buffer (2% sodium dodecyl sulfate [SDS], 1% β-mercaptoethanol, 1% glycerol, 65 mM Tris-hydrochloride [pH 6.8]) and separated in 10% polyacrylamide or 8% acrylamide–Acryl-Aide (FMC Corp., Rockland, Maine) SDS-gels. Gels were fixed for 30 min by incubation in 40% methanol–10% acetic acid, rinsed with water, soaked in 1 M sodium salicylic acid for 30 min, and dried. Radioactive bands were visualized by fluorography. Quantitation of the radioactivity of each band was performed with a Fuji BAS 2000 Bio-Image Analyzer.
Immunoblotting.
Infected MDM (7.5 × 105) or PBMC (106) were lysed in 210 μl of lysis buffer (50 mM Tris-HCl [pH 7.5], 5 mM EDTA, 150 mM NaCl, 0.5% [wt/vol] CHAPS, 0.2% [wt/vol] DOC), adjusted with 200 μl of sample buffer, boiled for 5 min, separated in an SDS–10% polyacrylamide gel, and transferred to Immobilon polyvinylidene difluoride membranes (Millipore Corp., Bedford, Mass.). Membranes were incubated with the appropriate combination of antibodies, and binding of the antibodies was identified by using 125I-protein A (0.1 μCi/ml; New England Nuclear, Du Pont, Wilmington, Del.) followed by autoradiography. For analysis of virus-associated proteins, viral particles from 1 ml of culture supernatants were pelleted (100 min, 16,000 × g, 4°C), washed with 0.5 ml of PBS, and lysed in 20 μl of sample buffer.
RESULTS
A naturally occurring Vpu-defective AD8 isolate efficiently replicates in primary macrophages and PBMC.
We previously reported that efficient virus production from HIV-1-infected primary macrophage cultures requires the activity of Vpu (41). This conclusion is based on the analysis of several virus isolates, including the HIV-1 AD8 primary monocyte-tropic isolate, expressing functional Vpu, and its isogenic variant, AD8-UDEL2, carrying an internal deletion in its vpu gene (41). However, more recent characterization of the original AD8 isolate [referred to here as AD8(−)], which carries a point mutation in the ATGvpu instead of an internal deletion, revealed only minor differences in the replication kinetics of Vpu-deficient and Vpu-expressing AD8 (50). A schematic representation of the various vpu mutants used in this study is shown in Fig. 1. To verify these observations, we compared the replication profiles of HIV-1AD8 [hereafter referred to as AD8(+)] and AD8(−) in primary human macrophages (Fig. 2A). Parallel cultures of MDM were each infected at 1 RT unit per cell with purified virus stocks generated in HeLa cells transfected with pAD8(+) or pAD8(−) plasmid DNA. As a control for potential contamination of MDM cultures with CD4+ T lymphocytes, the T-cell-tropic isolate NL4-3, which cannot efficiently replicate in this macrophage system (41, 50), was included in this and all subsequent experiments involving MDM. Aliquots of culture supernatants were collected at 3-day intervals over a period of 1 month, and secretion of virus particles into culture supernatants was subsequently determined by measuring the virus-associated RT activity. The resulting replication profiles (Fig. 2A) are consistent with the results reported by Theodore et al. (50), confirming that the lack of Vpu expression in AD8(−) has no significant effect on the ability of the virus to replicate in MDM. These results were confirmed by Western blot analysis of virions released into the culture supernatants (not shown). In all experiments performed with MDM cultures, replication of NL4-3 was undetectable, attesting to the absence of CD4+ T lymphocytes in the MDM preparation.
FIG. 2.
Effect of ATGvpu on virus replication of HIV-1AD8 in macrophage cultures. (A) Parallel cultures of 7.5 × 105 MDM were infected with equal RT units of AD8(+) or AD8(−) virus stocks obtained from transfected HeLa cells. As a negative control, cells were infected with virus stocks of the T-cell-tropic isolate NL4-3 or were mock infected. Supernatants were collected at 3-day intervals, and release was measured by determining the virus-associated RT activity in an in vitro assay (54). (B and C) Parallel cultures of 7.5 × 105 MDM (B) or 5 × 106 PHA–hIL-2-stimulated PBMC (C) were infected with equal RT units of purified virus stocks of AD8(+), AD8(−), or AD8-UDEL2. As a negative control, a parallel MDM culture was either infected with NL4-3 virus or mock infected. Virus production was monitored beyond the peaks of virus production in each infection experiment, and the amount of progeny viruses released into the supernatant medium was detected by RT assay.
To directly compare the effect of the lack of a Vpu initiation codon in AD8(−) with that of an internal deletion in Vpu (AD8-UDEL2) which retains the ATGvpu, we assessed the replication profiles of AD8(+), AD8(−), and AD8-UDEL2 viruses in primary macrophage cultures derived from different donors (Fig. 2B). Parallel cultures of MDM were infected with equal RT units of purified virus stocks prepared from transfected HeLa cells. Culture supernatants were harvested at 3-day intervals for a period of 2 months, and virus production was assessed by determining the virus-associated RT activity (Fig. 2B). As before, AD8(+) and AD8(−) viruses replicated with comparable kinetics. In contrast, virus production from AD8-UDEL2-infected cultures was significantly reduced and consistent with the three- to fourfold reduction reported previously (41). Similar results were observed with MDM derived from several independent donors, thus eliminating the possibility of experimental artifacts due to donor variation (not shown).
We next analyzed virus replication of the AD8-type viruses in PBMC, another major target for HIV-1 infection in vivo which are susceptible to infection by monocyte-tropic HIV-1 isolates. Parallel cultures of PHA- and hIL-2-stimulated PBMC were infected with the AD8-type viruses with the same multiplicity of infection as used for infection of macrophages. Aliquots of culture supernatants were collected at 2-day intervals, and the replication profiles were determined as before. The results are consistent with those obtained in macrophages inasmuch as they revealed virtually indistinguishable replication profiles for the AD8(+) and AD8(−) viruses, compared to an approximately twofold-reduced virus production from AD8-UDEL2-infected cultures (Fig. 2C). Thus, inactivation of Vpu in AD8 does not automatically result in impaired virus secretion; rather, the resulting phenotype is dependent on the nature of the vpu mutation.
Differential effects of ATGvpu and UDEL2 mutations on short-term virus release kinetics from infected macrophages and transfected HeLa cells.
To confirm that the differential effects associated with the mutation of the AD8 ATGvpu or the introduction of an internal deletion in Vpu (UDEL2) on virus replication are due to differences in the particle release kinetics, we performed pulse-chase analyses of transfected HeLa cells (Fig. 3A and B). For comparison, the effects of similar mutations in vpu on the release kinetics of the T-cell-tropic isolate NL4-3 were also analyzed in transfected HeLa cells (Fig. 3C). Since in earlier studies the NL4-3 Env protein had no effect on virus release, even when overexpressed in trans (8), we did not expect to observe differences in the virus release kinetics of an NL4-3 variant lacking the ATGvpu versus a variant carrying an internal deletion in vpu (UDEL1).
FIG. 3.
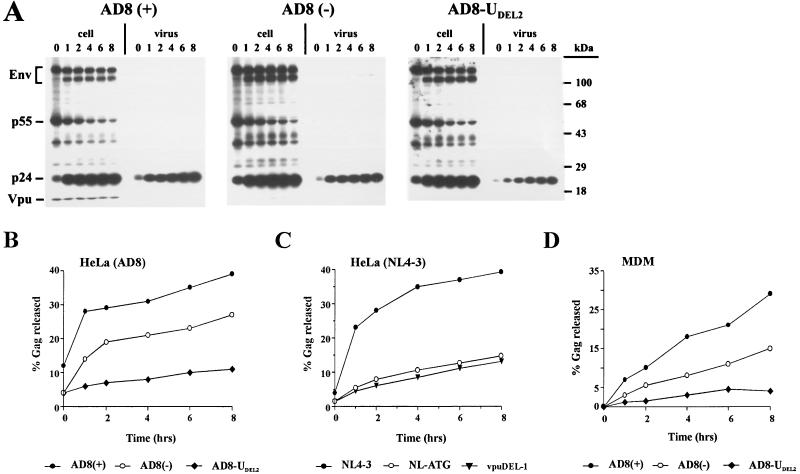
Effect of ATGvpu mutations on short-term virus release kinetics of AD8-type viruses in transfected HeLa cells or infected macrophages. (A to C) Parallel cultures of HeLa cells (1.5 × 107) were transfected with 75 μg of pAD8(+), pAD8(−), or pAD8-UDEL2 (A and B) or of pNL4-3, pNL-ATG, or vpuDEL-1 (C) plasmid DNA. Twenty-four hours posttransfection, cells were labeled for 30 min with [35S]methionine and chased for up to 8 h. Viral proteins from the cell lysates, pelleted virions, or the clarified supernatants were immunoprecipitated with an HIV-1-reactive human serum complemented with a polyclonal antiserum specific for HIV-1 Env glycoproteins. Immunoprecipitates were separated by SDS-PAGE (8% Acryl-Aide) and analyzed by fluorography. (A) Fluorographs for HeLa cells transfected with pAD8(+), pAD8(−), or pAD8-UDEL2. Positions of Vpu, Env, p55gag, p24gag are indicated on the left; positions of molecular size standards are indicated on the right. (B and C) Gag-specific proteins (p24gag and Pr55gag) detected in the cell lysates, virus pellets, and clarified supernatants were quantitated by image analysis. Relative amounts of Gag proteins in the virus pellets were calculated as the percentage of total p24gag and Pr55gag for each time point and were plotted as a function of time. (D) For pulse-chase analysis in macrophages, parallel cultures of MDM were plated in 24-well tissue culture plates (7.5 × 105 cells per well) and infected with equal RT units of AD8(+), AD8(−), or AD8-UDEL2 virus stock. At peak virus production, cultures were pulse-labeled for 30 min with [35S]methionine (2 mCi/ml) and chased for up to 8 h while cells remained attached to the tissue culture plates. Cells were harvested, and viral proteins from detergent lysates of cells, pelleted virions, or clarified supernatants were immunoprecipitated, separated, and analyzed as for panel B.
HeLa cells were transfected with plasmid DNAs encoding AD8- or NL4-3-type viruses or their Vpu variants. Cells were pulse-labeled for 30 min and chased for up to 8 h. At each time point, aliquots of cells were harvested and virions released into the supernatants were collected by centrifugation and processed for immunoprecipitation with an HIV-1-reactive human serum as described in Materials and Methods. Immunoprecipitates were separated by SDS-polyacrylamide gel electrophoresis (PAGE) and visualized by fluorography. Figure 3A shows a representative set of fluorographs depicting the pulse-chase analysis of the AD8-type viruses. The relative amounts of p24gag and Pr55gag released from cells producing AD8-type (Fig. 3B) or NL4-3-type (Fig. 3C) viruses were quantified by image analysis, and secretion of viral particles was calculated as the percentage of Gag proteins present in the viral pellet relative to the sum of proteins detected intra- and extracellularly (Fig. 3B and C). Consistent with our previous report (41), particle release from pAD8-UDEL2-transfected cells was three- to fourfold lower than in cultures transfected with the Vpu-expressing pAD8(+) molecular clone. Virus release from pAD8(−)-transfected cells was significantly (two- to threefold) higher than that from pAD8-UDEL2-transfected cells but lower than that from pAD8(+)-transfected cells (Fig. 3B). As expected, mutation of the ATGvpu in NL4-3 did not result in increased virus release (Fig. 3C). In fact, the virus release kinetics of the NL-ATG variant, lacking the ATGvpu, were indistinguishable from those of the vpuDEL-1 variant, carrying an internal deletion in vpu similar to that of AD8-UDEL2 (Fig. 3C). Thus, the partial rescue of the virus release defect in the AD8 ATGvpu mutant is a specific feature of the AD8 isolate.
Similar results were obtained when short-term virus release kinetics of AD8-type isolates were determined in infected macrophages (Fig. 3D). MDM were infected with equal RT units of purified AD8-type viruses as described for Fig. 2, and pulse-chase experiments were conducted as detailed in Materials and Methods. Briefly, replicate wells of MDM cultures were used for each time point of the pulse-chase experiment. Cells were left attached during the 30-min pulse and subsequent chase periods. Viral proteins from detergent lysates of cells, pelleted virions, and clarified supernatants were immunoprecipitated, separated by SDS-PAGE, and quantified by image analysis (not shown). The relative amounts of Pr55gag and p24gag present in the viral pellet were calculated as for Fig. 3B and C and plotted as a function of time (Fig. 3D). Consistent with the results for transfected HeLa cells, virus release from AD8(−)-infected MDM was at an intermediate level between release from AD8(+)- and AD8-UDEL2-producing cells. Thus, the relative short-term release kinetics of AD8-type viruses are cell type independent and consistent with the virus replication profiles observed for MDM or PBMC (Fig. 2B and C). We conclude that inactivation of the AD8 vpu gene through mutation of the ATGvpu has significantly less severe consequences for virus production than an internal deletion in vpu.
Mutation of ATGvpu affects AD8 Env expression.
One of the well-characterized functions of Vpu is its ability to enhance HIV-1 virus release. Consistent with that notion is the above finding that introduction of an internal deletion in the vpu gene significantly reduced virus secretion. On the other hand, inactivation of vpu by mutating its initiation codon, while clearly abolishing Vpu synthesis (see Fig. 3A), did not result in a significant drop in virus particle release. One of the differences observed for the two vpu mutants was an apparent increase in the level of Env protein produced from the AD8(−)-expressing cultures relative to the cultures expressing AD8(+) or AD8-UDEL2 (Fig. 3A).
To directly verify the effect of the ATGvpu mutation on expression of AD8 Env and to demonstrate that expression of Vpu itself did not contribute to the differential expression of Env, relative levels of AD8 Env proteins were analyzed by steady-state labeling of HeLa cells transfected with pAD8(+), pAD8(−), and pAD8-UDEL2 plasmid DNAs. Transfected HeLa cells were labeled for 12 h with [35S]methionine. Radiolabeled viral proteins from cell lysates were immunoprecipitated with a 1:1 mixture of an HIV-1-reactive human serum and a polyclonal rabbit antiserum against HIV-1 Env, separated by SDS-PAGE, and analyzed by fluorography (Fig. 4). The relative amounts of Env glycoproteins gp120 and gp160 were determined by image analysis and normalized to the level of Gag precursor protein Pr55gag present in the cells. We found that inactivation of the ATGvpu in AD8(−) resulted in an approximately threefold increase of cell-associated Env. In contrast, inactivation of Vpu by internal deletion of the vpu gene did not affect Env expression. This result demonstrates that the level of Env in AD8-expressing cells is not regulated by a biological activity of Vpu but is dependent on the presence or absence of the ATGvpu.
FIG. 4.
Mutation of the ATGvpu affects AD8 Env expression. Parallel cultures of HeLa cells (5 × 106) were transfected with 25 μg of pAD8(+), pAD8(−), or pAD8-UDEL2 plasmid DNA. Cells were harvested 24 h after transfection and then metabolically labeled with [35S]methionine for 12 h. Viral proteins were immunoprecipitated with a 1:1 mixture of an HIV-reactive human serum and a polyclonal rabbit serum directed against HIV-1 Env proteins gp160 and gp120, separated by SDS-PAGE (8% Acryl-Aide), and analyzed by fluorography. Positions of molecular weight marker proteins are indicated in kilodaltons on the left. The relative amounts of Pr55gag and the Env gp120 and gp160 were quantitated with an image analyzer, and the ratios of Gag/Env proteins relative to the AD8(+) sample are indicated at the bottom.
Overexpression of AD8 Env enhances virus secretion and can compensate for the lack of Vpu.
We and others recently reported that the Env proteins of certain HIV-2 isolates have a Vpu-like activity in that they are capable of increasing virus secretion (7, 8, 39). At least in the case of the Env protein of HIV-2ROD10, this activity is not limited to HIV-2 virus production but, like Vpu, can affect secretion of HIV-1 or SIV particles as well (8). While no such activity has thus far been ascribed to Env proteins of HIV-1 isolates, the observed correlation between increased expression of AD8 Env and enhanced virus secretion could be indicative of such a Vpu-like activity of the AD8 Env protein.
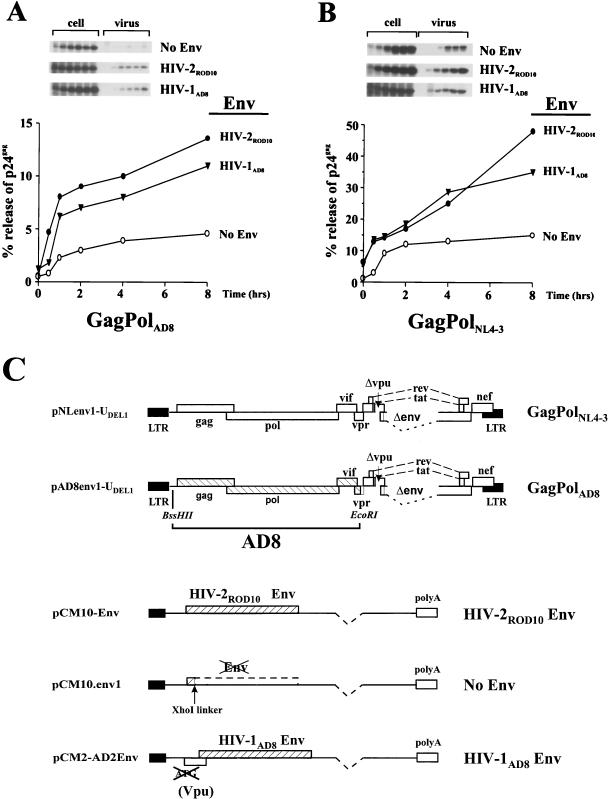
To test this hypothesis, we performed a series of experiments involving Vpu- and Env-deficient subgenomic constructs encoding gag-pol from either HIV-1AD8 (Fig. 5A) or HIV-1NL4-3 (Fig. 5B). Env proteins from AD8 or, for comparison, HIV-2ROD10 were provided in trans from separate expression vectors. Plasmid pCM10-Env was used for the expression of HIV-2ROD10 Env, while plasmid pCM2-AD2Env was used for the expression of HIV-1AD8 Env. To preserve the original structure of the vpu/env mRNA, pCM2-AD2Env also contains the vpu gene but lacks an ATGvpu. The absence of Vpu expression in pCM2-AD2Env-transfected cells was established in independent experiments using a mixture of different Vpu antibodies (not shown). Plasmid pCM10.env1, an Env-deficient variant of pCM10-Env, was included as a negative control. In all cases, expression of Env was under the control of the CMV early promoter. A schematic representation of the plasmids involved in this experiment is shown in Fig. 5C. HeLa cells transfected with pNLenv1-UDEL1 expressing NL4-3 Gag and Pol polyproteins (Fig. 5A) or pAD8env1-UDEL2 expressing AD8 Gag and Pol polyproteins (Fig. 5B) in the presence of either pCM7-Env (Fig. 5, HIV-2ROD10), pCM2-AD2Env (Fig. 5, HIV-1AD8), or pCM10.env1 (Fig. 5, No Env) plasmid DNA were pulse-labeled for 15 min and subjected to a chase for up to 8 h. At each time point, aliquots of cells were harvested and virions released into the supernatants were collected by centrifugation as described in Materials and Methods. Cell lysates and pelleted virions were subjected to immunoprecipitation with an HIV-1-reactive human serum. Immunoprecipitated proteins were separated by SDS-PAGE and visualized by fluorography. To estimate the efficiency of particle release, bands corresponding to p24gag (Fig. 5A and B, insets) were quantified, and the percentage of virion-associated Gag proteins was calculated and plotted as a function of time. In the absence of Vpu and Env proteins, release of virus particles containing Gag and Pol proteins derived from AD8 or NL4-3 was significantly impaired (Fig. 5A and B, No Env). However, expression of either HIV-1AD8 Env or HIV-2ROD10 Env protein in trans led to a distinct and similar increase of virus particle release. AD8 Env was only slightly less efficient in supporting virus secretion than ROD10 Env. In agreement with our previous finding for HIV-2ROD10 Env (8), this effect was independent of the origin of the HIV Gag and Pol proteins, as both Env proteins enhanced secretion of NL4-3- and AD8-type virus particles alike. We therefore conclude that the Env protein of HIV-1AD8, like that of HIV-2ROD10, can augment virus secretion and thus compensate for the lack of Vpu.
FIG. 5.
AD8 Env expresses a Vpu-like virus release activity. The release kinetics for HIV-1 AD8-type (A) or NL4-3-type (B) particles were analyzed either in the presence of Env glycoproteins derived from HIV-1AD8 or HIV-2ROD10 or in the absence of Env. HeLa cells (1.5 × 107) were transfected with 50 μg of pAD8env1-UDEL1 (A) or pNLenv1-UDEL1 (B) in combination with either 15 μg of pCM10-Env (HIV-2ROD10), pCM2-AD2Env (HIV-1AD8), or pCM10.env1 (No Env). Cells were pulse-labeled for 15 min with [35S]methionine (2 mCi/ml) and chased for up to 8 h. At each time point, aliquots of the cells and the virus-containing supernatants were harvested, and virus particles present in the culture supernatants were pelleted by centrifugation. Detergent lysates were prepared from each sample, and viral proteins were recovered by immunoprecipitation from cells, virus pellets, and clarified supernatant fractions. Immunoprecipitates were separated on SDS-PAGE (8% Acryl-Aide) and visualized by fluorography. Radioactive bands corresponding to p24gag present in each sample (insets) were quantified with an image analyzer. The relative amount of Gag protein present in the viral pellet at each time point was calculated as described for Fig. 2 and plotted as a function of time. (C) Schematic representation of molecular clones used for the expression of HIV-1AD8 and HIV-1NL4-3 particles. Both clones carry deletions in the vpu and env genes that abolish expression of these proteins. Env proteins were expressed in trans under the control of the CMV promoter. LTR, long terminal repeat; polyA, simian virus 40 polyadenylation signal.
DISCUSSION
One of the consequences of Vpu and Env expression from a bicistronic mRNA is the coordinate expression of the two proteins in close physical proximity. Because of the known function of Vpu on destabilizing Env-CD4 complexes in the ER (56), the formation of which not only traps CD4 in the ER (5, 14, 20) but also blocks gp160 cleavage and maturation (5), we have previously assumed that the biological significance of a coordinated expression of Vpu and Env is to ascertain that newly synthesized Env proteins are not trapped in the ER due to their interaction with CD4 but are efficiently processed and transported to the cell surface. Our finding that the macrophage-tropic AD8 isolate of HIV-1 encodes two genes, vpu and env, that can regulate virus release has important implications for the possible biological significance of a coordinated expression of Vpu and Env. Indeed, the fact that the original AD8 isolate, AD8(−), which is unable to express Vpu due to a mutation in its initiation codon, exhibits replication kinetics similar to those of its Vpu-expressing isogenic variant suggests that this virus has evolved a mechanism to inactivate Vpu without compromising its ability to efficiently replicate in host cells.
Our results show that the lack of a Vpu initiation codon in AD8(−) is correlated with increased Env synthesis. The fact that the vpu initiation codon is located upstream of the env initiation codon on the bicistronic vpu/env mRNA is likely to contribute to this phenomenon by increasing translation efficiency of the distal env gene. However, it is also possible that, due to the close proximity of AUGvpu to the tat/rev 5′ splice site, mutation of AUGvpu could inadvertently affect splicing and lead to a relative increase in the amount of env mRNA in AD8(−) cultures. Indeed, competitive RT-PCR analyses suggest that splicing at the tat/rev 5′ splice site is reduced in AD8(−) cells and thus could at least in part account for the observed increase in Env synthesis (data not shown).
The observation that AD8 has evolved a mechanism that preserves efficient virus secretion even when Vpu function is turned off could be an indication that under certain circumstances, elimination of Vpu may constitute a selective advantage. One possible advantage of abolishing Vpu expression could be the reduction or elimination of potential cytotoxic effects that may be associated with Vpu expression. Since Vpu has the ability to form ion-conducting membrane pores (16, 42), it is not difficult to envision how expression of a viral ion channel could disturb the ionic milieu of the host cell and, as a result, induce cytotoxic effects. While such a phenomenon may have only minor consequences for infection of short-lived peripheral blood lymphocytes, it is conceivable that the ability of a virus to limit cytotoxic effects could constitute a selective advantage for targeting of long-lived cells such as tissue macrophages. Thus, it is possible that the reversible inactivation of Vpu through simple point mutation of its initiation codon has evolved as a mechanism to support the establishment of a chronic HIV infection.
Alternatively, it is possible that the ability of AD8 isolates to inactivate Vpu without losing the ability to efficiently replicate in human cells has evolved as a mechanism to prevent degradation of CD4. However, this possibility seems unlikely in light of the fact that HIV makes a significant effort to down-modulate CD4 from the surface of infected cells and involves the activities of no fewer than three HIV-1 gene products, i.e., Nef, Env, and Vpu (11; for reviews, see references 6, 33, and 51). Thus, inactivating Vpu for the purpose of inhibiting cell surface down-modulation of CD4 would be effective only if the detrimental effects of Env and Nef on cell surface CD4 were simultaneously lost as well. Finally, it is conceivable that inactivating Vpu in certain cell types or at specific times during HIV-infection could be important to eliminate possible effects of Vpu on other cellular factors such as MHC class 1 molecules which represent an additional target for Vpu (24).
The fact that Vpu exerts several independent functions has raised the question of which of the activities of Vpu is more important, regulation of virus release or degradation of CD4. Based on the data presented in this study, it can be argued that the ability to increase virus release is a crucial function which some viruses maintain even when Vpu itself is inactivated. This may be particularly true for macrophages, where we previously found optimal virus release to be more dependent on Vpu than in PBMC (41). The comparatively lower dependence of virus release on Vpu in PBMC could then explain why Env proteins of T-cell-tropic HIV-1 isolates may not have evolved the ability to functionally substitute for Vpu. At any rate, the fact that in principle, regulation of virus release can be accomplished by proteins other than Vpu, i.e., AD8 Env or HIV-2 Env, suggests that Vpu may have evolved not primarily as a regulator of virus release but rather to perform additional tasks, such as induce CD4 degradation or down-regulate MHC class I complexes.
It remains to be resolved why the ability of the AD8 Env to enhance virus secretion is apparent only after overexpression of the protein. However, if one assumes that Vpu and AD8 Env function through similar biochemical mechanisms, it can be speculated that such activity of the AD8 Env is correlated with an ability to form ion-conductive pores on cellular membranes as has been demonstrated for Vpu (16, 42, 43). It is clear that the ability of Vpu to form an ion channel is linked to its ability to form homo-oligomers. Similarly, the ability of HIV Env molecules to form oligomeric structures is a well-known phenomenon (15, 36). Although, there is currently no direct experimental support for the notion that HIV-1 Env can form ion-conductive pores, it was previously reported that the gp41 transmembrane subunit of Env could affect the permeability of E. coli membranes (2). In addition, a peptide corresponding to a carboxy-terminal fragment of the HIV-1 envelope protein was reported to interact with the cell membrane of infected T cells to create lipid pores which increase membrane permeability (13). Furthermore, both HIV-1 gp120 and HIV-2 gp120 were reported to affect membrane transport processes in human astrocytes (4). Finally, with respect to the HIV-2 Env proteins, we have evidence that the ability to regulate virus release is correlated with the formation of distinct oligomeric structures (9). It is possible that the potential formation of membrane pores by AD8 Env molecules requires local concentrations of Env protein that may not be achieved with normal expression levels. Critical threshold levels of Env may be obtained only after mutation of the ATGvpu, which, as demonstrated in this study, can result in an increase in Env production. Indeed, trans-complementation studies using the HIV-2 model suggest that threshold levels of HIV-2 Env protein are required for their ability to regulation of virus release (10). A similar principle applies for activation of the HIV protease, which requires dimerization of its Gag/Pol precursor molecules. Dimerization of Gag and Pol is concentration dependent and thus depends on the accumulation of threshold levels of Gag and Pol products which is favored at the site of viral budding. This ensures that processing of the Gag/Pol precursor proteins does not occur until sufficient protein has been synthesized to allow for the production of infectious virions (for a review, see reference 27). Alternatively, it is possible that Vpu and AD8 Env are both capable of modulating an as yet unknown cellular factor or complex and thus indirectly affect secretion of virus particles from the cell surface.
The results presented in Fig. 5 clearly demonstrate that the AD8 Env protein is capable of enhancing virus secretion in trans with efficiency similar to that of the HIV-2ROD10 Env protein. This activity is not restricted to the release of AD8 particles but was observed for the release of particles of the NL4-3 T-cell-tropic isolate as well. It remains to be resolved, however, whether the ability of HIV-1 Env proteins to substitute for Vpu function is a phenomenon that is restricted to monocyte/macrophage-tropic virus isolates or, for that matter, is a peculiarity of the AD8 isolate itself. While there are T-cell-tropic isolates of HIV-1 that carry mutations in the ATGvpu (34), it is unclear whether the Env products of such isolates exhibit a Vpu-like activity. In fact, reconstitution of Vpu expression in the originally Vpu-deficient HXB2 isolate (carrying a mutation in the ATGvpu) significantly increased virus production (49), suggesting that the HXB2 Env protein does not have a Vpu-like activity. Also, our results (Fig. 3C and reference 8) are consistent with those observations inasmuch as they reveal no Vpu-like activity for the NL4-3 Env protein. Since monocytes/macrophages generally express low to undetectable levels of CD4 (37), down-regulation of CD4 as a way to avoid potential interference with processes occurring at the cell surface may not be as important for virus replication in macrophages as in T lymphocytes, which express relatively high levels of CD4. Consequently, the inability of a virus isolate to induce CD4 degradation is likely to have less impact on virus replication in monocytes/macrophages than in PBMC. Thus, it is possible that inactivation of Vpu occurs primarily during replication in monocytes/macrophages. However, the effects of Vpu on HIV-1 replication in vivo are difficult to assess, due to the lack of an appropriate animal model. Hence, there is no information available as to whether there might be a selective pressure in vivo to favor reversion of mutations in the ATGvpu and whether such reversion would be correlated with a switch in the host range from macrophage tropism to T-cell or dual tropism, which has frequently been associated with disease progression (12, 45). Nevertheless, it is interesting that in studies involving simian immunodeficiency virus-HIV (30) chimeras, the level of viral RNA in plasma during acute infection and the frequency of virus isolation were higher for Vpu-positive viruses, suggesting that Vpu acts to increase virus replication in vivo (30).
ACKNOWLEDGMENTS
We thank Theodore S. Theodore for plasmids pAD8(+) and pAD8(−), Kathleen Clouse and Karis Faust for PBMC and MDM preparations, and Rao Bachoti and Alicia Buckler-White for oligonucleotide synthesis and sequence analysis.
Part of this work was supported by a grant to K.S. from the NIH Intramural AIDS Targeted Antiviral Program.
REFERENCES
- 1.Adachi A, Gendelman H E, Koenig S, Folks T, Willey R, Rabson A, Martin M A. Production of acquired immunodeficiency syndrome-associated retrovirus in human and nonhuman cells transfected with an infectious molecular clone. J Virol. 1986;59:284–291. doi: 10.1128/jvi.59.2.284-291.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arroyo J, Boceta M, Gonzales M E, Michel M, Carrasco L. Membrane permeabilization by different regions of the human immunodeficiency virus type 1 transmembrane glycoprotein gp41. J Virol. 1995;69:4095–4102. doi: 10.1128/jvi.69.7.4095-4102.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bennett M K, Calakos N, Scheller R H. Syntaxin: a synaptic protein implicated in docking of synaptic vesicles at presynaptic active zones. Science. 1992;257:255–259. doi: 10.1126/science.1321498. [DOI] [PubMed] [Google Scholar]
- 4.Benos D J, Hahn B H, Bubien J K, Ghosh S K, Mashburn N A, Chaikin M A, Shaw G M, Benveniste E N. Envelope glycoprotein gp120 of human immunodeficiency virus type 1 alters ion transport in astrocytes: implications for AIDS dementia complex. Proc Natl Acad Sci USA. 1994;91:494–498. doi: 10.1073/pnas.91.2.494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bour S, Boulerice F, Wainberg M A. Inhibition of gp160 and CD4 maturation in U937 cells after both defective and productive infections by human immunodeficiency virus type 1. J Virol. 1991;65:6387–6396. doi: 10.1128/jvi.65.12.6387-6396.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bour S, Geleziunas R, Wainberg M. The human immunodeficiency virus type 1 (HIV-1) CD4 receptor and its central role in promotion of HIV-1 infection. Microbiol Rev. 1995;59:63–93. doi: 10.1128/mr.59.1.63-93.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bour S, Schubert U, Peden K, Strebel K. The envelope glycoprotein of human immunodeficiency virus type 2 enhances particle release: a Vpu-like factor? J Virol. 1996;70:820–829. doi: 10.1128/jvi.70.2.820-829.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bour S, Strebel K. The human immunodeficiency virus (HIV) type 2 envelope protein is a functional complement to HIV type 1 Vpu that enhances particle release of heterologous retroviruses. J Virol. 1996;70:8285–8300. doi: 10.1128/jvi.70.12.8285-8300.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bour S, Aberham C, Perrin C, Strebel K. Lack of effect of cytoplasmic tail truncations on human immunodeficiency virus type 2 ROD Env particle release activity. J Virol. 1999;73:778–782. doi: 10.1128/jvi.73.1.778-782.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bour, S., and K. Strebel. 1998. Unpublished data.
- 11.Chen B K, Gandhi R T, Baltimore D. CD4 down-modulation during infection of human T cells with human immunodeficiency virus type 1 involves independent activities of Vpu, Env, and Nef. J Virol. 1996;70:6044–6053. doi: 10.1128/jvi.70.9.6044-6053.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen M, Singh M K, Balachandran R, Gupta P. Isolation and characterization of two divergent infectious molecular clones of HIV type 1 longitudinally obtained from a seropositive patient by a progressive amplification procedure. AIDS Res Hum Retroviruses. 1997;13:743–750. doi: 10.1089/aid.1997.13.743. [DOI] [PubMed] [Google Scholar]
- 13.Chernomordik L, Chanturiya A N, Suss-Toby E, Nora E, Zimmerberg J. An amphipathic peptide from the C-terminal region of the human immunodeficiency virus envelope glycoprotein causes pore formation in membranes. J Virol. 1994;68:7115–7123. doi: 10.1128/jvi.68.11.7115-7123.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Crise B, Buonocore L, Rose J K. CD4 is retained in the endoplasmic reticulum by the human immunodeficiency virus type 1 glycoprotein precursor. J Virol. 1990;64:5585–5593. doi: 10.1128/jvi.64.11.5585-5593.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Earl L P, Doms R W, Moss B. Oligomeric structure of the human immunodeficiency virus type 1 envelope glycoprotein. Proc Natl Acad Sci USA. 1990;87:648–652. doi: 10.1073/pnas.87.2.648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ewart G D, Sutherland T, Gage P W, Cox G B. The Vpu protein of human immunodeficiency virus type 1 forms cation-selective ion channels. J Virol. 1996;70:7108–7115. doi: 10.1128/jvi.70.10.7108-7115.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Folks T, Benn S, Rabson A, Theodore T, Hoggan D, Martin M, Lightfoote M, Sell K. Characterization of a continuous T-cell line susceptible to the cytopathic effects of the acquired immunodeficiency syndrome (AIDS)-associated retrovirus. Proc Natl Acad Sci USA. 1985;82:4539–4543. doi: 10.1073/pnas.82.13.4539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Friborg J, Ladha A, Goettlinger H, Haseltine W A, Cohen E A. Functional analysis of the phosphorylation sites on the human immunodeficiency virus type 1 Vpu protein. J Acquired Immune Defic Syndr Hum Retroviruses. 1995;8:10–22. [PubMed] [Google Scholar]
- 19.Fujita K, Omura S, Silver J. Rapid degradation of CD4 in cells expressing HIV-1 Env and Vpu is blocked by proteasome inhibitors. J Gen Virol. 1997;78:619–625. doi: 10.1099/0022-1317-78-3-619. [DOI] [PubMed] [Google Scholar]
- 20.Jabbar M A, Nayak D P. Intracellular interaction of human immunodeficiency virus type 1 (ARV-2) envelope glycoprotein gp160 with CD4 blocks the movement and maturation of CD4 to the plasma membrane. J Virol. 1990;64:6297–6304. doi: 10.1128/jvi.64.12.6297-6304.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jabbar M A. The human immunodeficiency virus type 1 Vpu protein: roles in virus release and CD4 downregulation. Curr Top Microbiol Immunol. 1995;193:107–120. doi: 10.1007/978-3-642-78929-8_6. [DOI] [PubMed] [Google Scholar]
- 22.Jacks T, Power M, Masiarz F R, Luciw P A, Barr P J, Varmus H E. Characterization of ribosomal frameshifting in HIV-1 gag-pol expression. Nature. 1988;331:280–283. doi: 10.1038/331280a0. [DOI] [PubMed] [Google Scholar]
- 23.Karacostas V, Wolffe E J, Nagashima K, Gonda M A, Moss B. Overexpression of the HIV-1 Gag-Pol polyprotein results in intracellular activation of HIV-1 protease and inhibition of assembly and budding of virus-like particles. Virology. 1993;193:661–671. doi: 10.1006/viro.1993.1174. [DOI] [PubMed] [Google Scholar]
- 24.Kerkau T, Bacik I, Bennink J R, Yewdell J W, Huenig T, Schimpl A, Schubert U. The human immunodeficiency virus type 1 (HIV-1) Vpu protein interferes with an early step in the biosynthesis of major histocompatibility complex (MHC) class I molecules. J Exp Med. 1997;185:1295–1305. doi: 10.1084/jem.185.7.1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Klimkait T, Strebel K, Hoggan M D, Martin M A, Orenstein J M. The human immunodeficiency virus type 1-specific protein Vpu is required for efficient virus maturation and release. J Virol. 1990;64:621–629. doi: 10.1128/jvi.64.2.621-629.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kozak M. The scanning model for translation: an update. J Cell Biol. 1989;108:229–241. doi: 10.1083/jcb.108.2.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Krausslich H G, Welker R. Intracellular transport of retroviral capsid components. Curr Top Microbiol Immunol. 1996;214:25–63. doi: 10.1007/978-3-642-80145-7_2. [DOI] [PubMed] [Google Scholar]
- 28.Lamb R A, Pinto L H. Do Vpu and Vpr of human immunodeficiency virus type 1 and NB of influenza B virus have ion channel activities in the viral life cycles? Virology. 1997;229:1–11. doi: 10.1006/viro.1997.8451. [DOI] [PubMed] [Google Scholar]
- 29.Lazdins J K, Woods-Cook K, Walker M, Alteri E. The lipophilic muramyl peptide MTP-PE is a potent inhibitor of HIV replication in macrophages. AIDS Res Hum Retroviruses. 1990;6:1157–1161. doi: 10.1089/aid.1990.6.1157. [DOI] [PubMed] [Google Scholar]
- 30.Li J T, Halloran M, Lord C I, Watson A, Ranchalis J, Fung M, Letvin N L, Sodroski J G. Persistent infection of macaques with simian-human immunodeficiency viruses. J Virol. 1995;69:7061–7071. doi: 10.1128/jvi.69.11.7061-7067.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Maldarelli F, Chen M Y, Willey R L, Strebel K. Human immunodeficiency virus type 1 Vpu protein is an oligomeric type 1 integral membrane protein. J Virol. 1993;67:5056–5061. doi: 10.1128/jvi.67.8.5056-5061.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Margottin F, Bour S P, Durand H, Selig L, Benichou S, Richard V, Thomas D, Strebel K, Benarous R. A novel human WD protein, h-βTrCP, that interacts with HIV-1 Vpu, connects CD4 to the ER degradation pathway through an F-box motif. Mol Cell. 1998;1:565–574. doi: 10.1016/s1097-2765(00)80056-8. [DOI] [PubMed] [Google Scholar]
- 33.Miller R, Sarver N. HIV accessory proteins as therapeutic targets. Nat Med. 1997;3:389–394. doi: 10.1038/nm0497-389. [DOI] [PubMed] [Google Scholar]
- 34.Myers G, Foley B, Mellors J W, Korber B, Jeang K T, Wain-Hobson S, editors. Human retroviruses and AIDS. Los Alamos, N.Mex: Los Alamos National Laboratory; 1996. [Google Scholar]
- 35.Paul M, Jabbar M A. Phosphorylation of both phosphoacceptor sites in the HIV-1 Vpu cytoplasmic domain is essential for Vpu-mediated ER degradation of CD4. Virology. 1997;232:207–216. doi: 10.1006/viro.1997.8541. [DOI] [PubMed] [Google Scholar]
- 36.Pinter A, Honnen W J, Tilley S A, Bona C, Zaghouani H, Gorny M K, Zolla-Pazner S. Oligomeric structure of gp41, the transmembrane protein of human immunodeficiency virus type 1. J Virol. 1989;496:2674–2679. doi: 10.1128/jvi.63.6.2674-2679.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Potts B J, Maury W, Martin M A. Replication of HIV-1 in primary monocyte cultures. Virology. 1990;175:465–476. doi: 10.1016/0042-6822(90)90431-p. [DOI] [PubMed] [Google Scholar]
- 38.Purcell D F J, Martin M A. Alternative splicing of human immunodeficiency virus type 1 mRNA modulates viral protein expression, replication, and infectivity. J Virol. 1993;67:6365–6378. doi: 10.1128/jvi.67.11.6365-6378.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ritter G D, Yamshchikov G, Cohen S J, Mulligan M J. Human immunodeficiency virus type 2 glycoprotein enhancement of particle budding: role of the cytoplasmic domain. J Virol. 1996;70:2669–2673. doi: 10.1128/jvi.70.4.2669-2673.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schubert U, Strebel K. Differential activities of the human immunodeficiency virus type 1-encoded Vpu protein are regulated by phosphorylation and occur in different cellular compartments. J Virol. 1994;68:2260–2271. doi: 10.1128/jvi.68.4.2260-2271.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schubert U, Clouse K A, Strebel K. Augmentation of virus secretion by the human immunodeficiency virus type 1 Vpu protein is cell type independent and occurs in cultured human primary macrophages and lymphocytes. J Virol. 1995;69:7699–7711. doi: 10.1128/jvi.69.12.7699-7711.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schubert U, Ferrer-Montiel A V, Oblatt-Montal M, Henklein P, Strebel K, Montal M. Identification of an ion channel activity of the Vpu transmembrane domain and its involvement in the regulation of virus release from HIV-1-infected cells. FEBS Lett. 1996;398:12–18. doi: 10.1016/s0014-5793(96)01146-5. [DOI] [PubMed] [Google Scholar]
- 43.Schubert U, Bour S, Ferrer-Montiel A V, Montal M, Maldarelli F, Strebel K. The two biological activities of human immunodeficiency virus type 1 Vpu protein involve two separable structural domains. J Virol. 1996;70:809–819. doi: 10.1128/jvi.70.2.809-819.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schubert U, Anton L C, Bacik I, Cox J H, Bour S, Bennink J R, Orlowski M, Strebel K, Yewdell J W. CD4 glycoprotein degradation induced by human immunodeficiency virus type 1 Vpu protein requires the function of proteasomes and the ubiquitin conjugating pathway. J Virol. 1998;72:2280–2288. doi: 10.1128/jvi.72.3.2280-2288.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schuitemaker H, Koot M, Kootstra N A, Dercksen M W, de Goede R E, van Steenwijk R P, Lange J M, Schattenkerk J K, Miedema F, Tersmette M. Biological phenotype of human immunodeficiency virus type 1 clones at different stages of infection: progression of disease is associated with a shift from monocytotropic to T-cell-tropic virus population. J Virol. 1992;66:1354–1360. doi: 10.1128/jvi.66.3.1354-1360.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schwartz S, Felber B K, Fenyö E M, Pavlakis G N. Env and Vpu proteins of human immunodeficiency virus type 1 are produced from multiple bicistronic mRNAs. J Virol. 1990;64:5448–5456. doi: 10.1128/jvi.64.11.5448-5456.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Strebel K, Klimkait T, Martin M A. A novel gene of HIV-1, vpu, and its 16-kilodalton product. Science. 1988;241:1221–1223. doi: 10.1126/science.3261888. [DOI] [PubMed] [Google Scholar]
- 48.Strebel K. Structure and function of HIV-1 Vpu, p. III-19–III-27. In: Myers G, Foley B, Mellors J W, Korber B, Jeang K T, Wain-Hobson S, editors. Human retroviruses and AIDS. Los Alamos, N.Mex: Los Alamos National Laboratory; 1996. [Google Scholar]
- 49.Terwilliger E F, Cohen E A, Lu Y, Sodroski J G, Haseltine W A. Functional role of human immunodeficiency virus type 1 vpu. Proc Natl Acad Sci USA. 1989;86:5163–5167. doi: 10.1073/pnas.86.13.5163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Theodore T S, Englund G, Buckler-Whitte A, Buckler C E, Martin M A, Peden K W C. Construction and characterization of a stable full-length macrophage-tropic HIV type 1 molecular clone that directs the production of high titers of progeny virions. AIDS Res Hum Retroviruses. 1996;12:191–194. doi: 10.1089/aid.1996.12.191. [DOI] [PubMed] [Google Scholar]
- 51.Trono D. HIV accessory proteins: leading roles for the supporting cast. Cell. 1995;82:189–192. doi: 10.1016/0092-8674(95)90306-2. [DOI] [PubMed] [Google Scholar]
- 52.Vincent M J, Jabbar M A. The human immunodeficiency virus type 1 Vpu protein: a potential regulator of proteolysis and protein transport in the mammalian secretory pathway. Virology. 1995;213:639–649. doi: 10.1006/viro.1995.0035. [DOI] [PubMed] [Google Scholar]
- 53.Wakefield J K, Kang S M, Morrow C D. Construction of a type 1 human immunodeficiency virus that maintains a primer binding site complementary to tRNAHis. J Virol. 1996;70:966–975. doi: 10.1128/jvi.70.2.966-975.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Willey R L, Smith D H, Lasky L A, Theodore T S, Earl P L, Moss B, Capon D J, Martin M A. In vitro mutagenesis identifies a region within the envelope gene of the human immunodeficiency virus that is critical for infectivity. J Virol. 1988;62:139–147. doi: 10.1128/jvi.62.1.139-147.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Willey R L, Maldarelli F, Martin M A, Strebel K. Human immunodeficiency virus type 1 Vpu protein induces rapid degradation of CD4. J Virol. 1992;66:7193–7200. doi: 10.1128/jvi.66.12.7193-7200.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Willey R L, Maldarelli F, Martin M A, Strebel K. Human immunodeficiency virus type 1 Vpu protein regulates the formation of intracellular gp160-CD4 complexes. J Virol. 1992;66:226–234. doi: 10.1128/jvi.66.1.226-234.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Willey R L, Buckler-White A, Strebel K. Sequences present in the cytoplasmic domain of CD4 are necessary and sufficient to confer sensitivity to the human immunodeficiency virus type 1 Vpu protein. J Virol. 1994;68:1207–1212. doi: 10.1128/jvi.68.2.1207-1212.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wilson W, Braddock M, Adams S E, Rathjen P D, Kingsman S M, Kingsman A J. HIV expression strategies: ribosomal frameshifting is directed by a short sequence in both mammalian and yeast systems. Cell. 1988;55:1159–1169. doi: 10.1016/0092-8674(88)90260-7. [DOI] [PubMed] [Google Scholar]
- 59.Yao X J, Garzon S, Boisvert F, Haseltine W A, Cohen E A. The effect of vpu on HIV-1-induced syncytia formation. J Acquired Immune Defic Syndr. 1993;6:135–141. [PubMed] [Google Scholar]