Hemoglobinopathies, including sickle cell disease (SCD) and thalassemia syndromes, represent the commonest monogenic diseases in the world. Although their pathogenesis is established, the diverse clinical manifestations and the varying degree of severity are less understood and are thought to be governed, in part, by genetic modifiers. Previous studies have demonstrated the role of genetic modifiers in different hemoglobinopathy phenotypes, with co-inheritance of α-thalassemia and higher levels of fetal hemoglobin (HbF) being the best characterized disease modifiers. Several genome-wide analyses have identified three major quantitative trait loci modulating HbF levels: a promoter variant on HBG2 (XmnI-rs7482144), the HBS1L-MYB intergenic region and BCL11A, which together explain up to 50% of the genetic variation affecting HbF1,2. More recently, studies have identified genetic modifiers associated with laboratory and clinical markers of disease complications3,4. However, few of these modifiers have reached a level of clinical utility.
Importantly, most association studies in SCD have been restricted to patients that have not received disease-modifying therapies. Given that the influence of genetic disease modifiers may change with treatment, identification of genetic modifiers requires high-quality clinical, laboratory and treatment data to allow accurate genotype/phenotype correlation. Furthermore, with the emergence of novel targeted therapies for hemoglobinopathies, such as gene therapy, genetic modifiers can facilitate patient stratification and, also, influence the response to these treatments.
Currently, the ITHANET portal5 manually curates around 800 disease-modifying variants in over 420 genomic locations. However, most of these variants have not been validated with confirmatory or large-scale studies, and across diverse ethnic populations. In addition, data from different studies are not frequently reproducible and their possible effect size remains unknown6. Most importantly, with most studies having a sample size of less than 2000 patients, it is not possible to identify genetic modifiers with high confidence. As a result, the translation of these results into clinical practice has been limited. There are currently very few established polygenic risk scores related to disease complications, severity, or response to treatment, that can be used as an evidence base to disease stratify and offer patient specific treatment regimens in hemoglobinopathy patients7.
A validated standard for data collection and phenotypic definitions is crucial for the accurate comparison and pooling of data. The recently developed Sickle Cell Disease Ontology8 represents a positive step towards disease-specific standardization that can facilitate integration of datasets in the field. In parallel, other ongoing initiatives, such as patient registries by RADeep and SPARCO, are working on standardization of clinical data collection for hemoglobinopathies using well-established international standards, such as the Human Phenotype Ontology9. Despite these efforts, a common understanding and discussion among different initiatives is necessary to allow integration of data for large-scale clinical and genomic studies. Furthermore, there is a limited amount of high-throughput or genome-wide data available for further research, despite several genetic studies in the field. A large, international disease-specific data repository, compliant with the FAIR data principles10, would revolutionize research in the field of hemoglobinopathies towards evidence-based approaches that utilize data science and artificial intelligence.
In 2020, nine existing international or regional consortia recognized the need for global synergies to address the above challenges and they agreed to create the International Hemoglobinopathy Research Network (INHERENT) as an umbrella network focused on the study of genetic modifiers of hemoglobinopathies. Specifically, INHERENT brings together the following consortia:
ITHANET (https://ithanet.eu/),
Rare Anaemia Disorders European Epidemiological Platform (RADeep; https://www.radeepnetwork.eu/),
African Research and Innovative Initiative for Sickle Cell Education (ARISE; https://www.ariseinitiative.org/),
Sickle Pan-African Research Consortium (SPARCO),
Sickle Africa Data Coordinating Center (SADaCC; https://sickleinafrica.org/),
Réseau d’Etude de la Drépanocytose en Afrique Centrale (REDAC; https://redacnetwork.org/),
Human Variome Project Global Globin Network (HVP GGN; https://www.humanvariomeproject.org/gg2020),
International Health Repository (IHR),
ClinGen Hemoglobinopathy Variant Curation Expert Panel (Hemoglobinopathy VCEP; https://clinicalgenome.org/affiliation/50052/)
INHERENT is also endorsed by the European Reference Network on rare hematological diseases, ERN-EuroBloodNet.
The primary aim of INHERENT is to study the role of genetic modifiers in hemoglobinopathies through a large-scale, multi-ethnic genome-wide association study (GWAS). Therefore, INHERENT will address challenges of previous studies related to small sample sizes and low statistical power, while promoting participation of diverse populations worldwide. Specifically, INHERENT aims to: (a) discover new genetic modifiers of hemoglobinopathies, (b) validate previously reported genetic modifiers, (c) pool and analyze existing genomic data, (d) standardize phenotypic descriptions using established standards, aligned with international recommendations, (e) develop a case report form (CRF) to efficiently gather sufficient high-quality data across countries accounting for different resource capabilities, and (f) develop a research resource of disease-specific data generated in INHERENT, including genomic, phenotypic, and functional data.
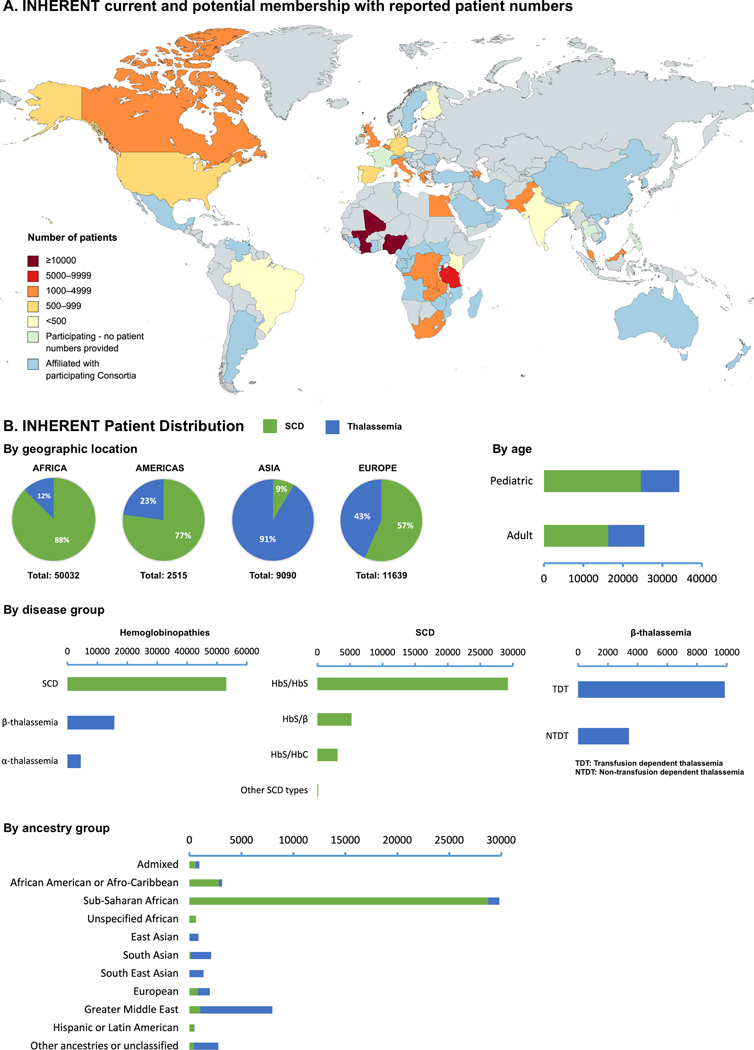
All INHERENT members agreed on the participation criteria, which are aimed at being inclusive and straightforward. Hence, participation in INHERENT is open for any group that can submit a minimum of thirty DNA samples with their core phenotypic description. Additional members that can significantly contribute to specific network activities, such as bioinformatics and biostatistical analyses, data management, genotyping, and regulatory/ethical issues, have also been invited to join the network. As a result, the current INHERENT membership is both international and interdisciplinary and includes over 160 experts from 89 organizations, spanning 36 countries worldwide (see Supplementary Table). Notably, based on the current membership of participating consortia, the projected membership of INHERENT can reach 73 countries, as shown in Figure 1A.
Figure 1:
An overview of the INHERENT and the reported data by its current membership. Panel A: geographic distribution of the current and and potential membership of INHERENT, illustrating the reported number of patients per country. Panel B: Number of patients reported by INHERENT members, distributed by geographic location, age, disease group, and self-reported ancestry group.
The target sample size of INHERENT is at least 30,000 individuals with hemoglobinopathies from diverse ancestries and geographic locations. Both pediatric and adult patients will be enrolled to study disease complications across all ages. To investigate the feasibility of the network’s primary goal to perform a large-scale, multi-ethnic GWAS, we performed a survey among all centers that participate in INHERENT. The survey requested the number of patients per center and their distribution per disease group, age, and ancestry group. A total of 81 participating centers (91% of all members) responded to the survey, with the results illustrated in Figure 1. Through its current membership, INHERENT has the potential to enrol over 73,200 individuals with hemoglobinopathies. Specifically, 53,100 people with SCD have been reported by the participating centers, with 54.8% being homozygous S/S, while other major disease groups (S/β and S/C) are being well-represented. In addition, around 15,600 people with β-thalassemia have been reported, with over 62% of them being transfusion-dependent, and 4,400 people with α-thalassemia. Basic genetic data are available for fewer than 45% of the patients, highlighting the need for genomic research in the field. Importantly, the diverse spectrum of ancestry groups for different disease groups clearly reflects the existing knowledge about the global hemoglobinopathy epidemiology, thus further supporting the multi-ethnic nature of the project. Some of the requested information was not readily available in the centers, and for large centers without an existing electronic patient registry the responses were approximations. Nevertheless, the results of the survey clearly support that the minimum target sample size set by INHERENT is realistic, highlighting the network’s potential to be the largest international network on hemoglobinopathies.
INHERENT is in a unique position to address limitations of previous GWAS studies in hemoglobinopathies by increasing the sample size and achieving sufficient power to detect associations at GWAS significant levels. The Supplementary Figure illustrates different hypothetical, yet realistic, scenarios for the analyses to be performed to estimate the power under different assumptions. Panels A, B, and C refer to the evaluation of binary phenotypes using logistic regression with a p-value of 5×10-8. Based on the survey results, the assumed total sample size of 10,000 is realistic for both SCD and β-thalassemia, while the case rate of 0.3 is applicable for several hemoglobinopathy complications, such as the prevalence of stroke11 in SCD, and osteoporosis in β-thalassemia12. We show that we have the statistical power to detect associations down to a minor allele frequency (MAF) of 0.05, with an Odds Ratio of 1.5, while detection of associations for less common disease complications is also possible. Similarly for the analyses using survival endpoints (Supplementary Figure, panels D, E, and F), we have sufficient power to detect associations even with 5000 samples, assuming an event rate of 30%, Hazard Ratio of 1.5, and MAF greater than 0.05.
INHERENT is coordinated by a Steering and Data Access Committee, which provides direction to the network and includes the working group (WG) chairs and one representative from each of the participating consortia. To achieve the main objectives, the activities of the network have been divided into five WGs: clinical, genotyping, data management and analysis, ethics, and knowledge translation.
In conclusion, by bringing together existing consortia and additional partners throughout the world, INHERENT avoids duplication of efforts and is focused on integration and consolidation of evidence as well as the generation and analysis of novel and large datasets. The increased sample size and the diversity in the studied populations can lead to novel discoveries and translational impact in the field of hemoglobinopathies. Moreover, the interdisciplinary and international nature of the network can result in synergistic research studies that promote innovative use of the collected and generated data as well as novel methodological approaches.
Supplementary Material
Supplementary Figure: Statistical power calculations under different scenarios. Panels A, B, C refer to the evaluation of binary phenotypes using logistic regression with a p-value of 5×10-8. Panels D, E, and F refer to power clalculations using survival endpoints with a p-value of 5×10-8.
Funding information:
The coordination of INHERENT, the ITHANET portal and the ClinGen Hemoglobinopathy VCEP are co-funded by the European Regional Development Fund and the Republic of Cyprus through the Research and Innovation Foundation (Project: EXCELLENCE/1216/256).
The ARISE project has received funding from the European Union’s Horizon 2020 research and innovation programme under the Marie Skłodowska-Curie grant agreement No. 824021.
INHERENT is endorsed by the European Reference Network on Rare Hematological Diseases (ERN-EuroBloodNet) “ERN-2016 – Framework Partnership Agreement 2017–2021.” FPA 739541.
Conflict of Interest Statement:
Kevin H.M. Kuo – Grants/Research Support: NHLBI/NIH: 1R33HL147845, Thalassemia Foundation Canada, Peter Munk Cardiac Centre, University of Toronto, Cincinnati Children’s Hospital Medical Center, Canadian Hematology Society, Pfizer; Consultancy: Agios, Alexion, Apellis, Aruvant, Bluebirdbio, Celgene/BMS, Novartis, Pfizer; Chair of Data Safety Monitoring Board: Bioverativ/Sanofi (ZFN gene editing trial for sickle cell disease NCT03653247); Research collaboration: Phoenicia Biosciences; Lead investigator: BENEFiTS (Phoenicia), AG348-C-017 (Agios); Site-PI (in alphabetical order): ACE-536-B-THAL-001, ACE-536-LTFU (Celgene/BMS); AG348-C-010, AG348-C-018, AG-348-C-020, (Agios); GBT440–031, GBT440–034 (Global Blood Therapeutics); ARU-1801 (CCMHC, Aruvant); APL2–302 (Apellis); CSEG101A2301 (Novartis); CTX001–131 (Vertex); Co-investigator for: APL2–307 (Apellis), CTX001–111, CTX001–121 (Vertex).
Footnotes
The full membership of INHERENT is provided in the Supplementary Table
Data Availability Statement:
Data sharing not applicable to this article as no datasets were generated or analysed during the current study.
References
- 1.Thein SL, Menzel S, Lathrop M. & Garner C. Control of fetal hemoglobin: new insights emerging from genomics and clinical implications. Hum. Mol. Genet 18, R216–R223 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Galarneau G. et al. Fine-mapping at three loci known to affect fetal hemoglobin levels explains additional genetic variation. Nat. Genet 42, 1049–1051 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Flanagan JM et al. Genetic predictors for stroke in children with sickle cell anemia. Blood 117, 6681–6684 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jacob SA et al. Thrombospondin-1 Gene Polymorphism is Associated with Estimated Pulmonary Artery Pressure in Patients with Sickle Cell Anemia. Am. J. Hematol 92, E31–E34 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kountouris P. et al. IthaGenes: An Interactive Database for Haemoglobin Variations and Epidemiology. PLoS ONE 9, (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stephanou C. et al. Genetic Modifiers at the Crossroads of Personalised Medicine for Haemoglobinopathies. J. Clin. Med 8, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rampersaud E. et al. A polygenic score for acute vaso-occlusive pain in pediatric sickle cell disease. Blood Adv. 5, 2839–2851 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.The Sickle Cell Disease Ontology: enabling universal sickle cell-based knowledge representation. Database 2019, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Köhler S. et al. The Human Phenotype Ontology in 2021. Nucleic Acids Res. 49, D1207–D1217 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wilkinson MD et al. The FAIR Guiding Principles for scientific data management and stewardship. Sci. Data 3, 160018 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ohene-Frempong K. et al. Cerebrovascular Accidents in Sickle Cell Disease: Rates and Risk Factors. Blood 91, 288–294 (1998). [PubMed] [Google Scholar]
- 12.Taher AT et al. Overview on practices in thalassemia intermedia management aiming for lowering complication rates across a region of endemicity: the OPTIMAL CARE study. Blood 115, 1886–1892 (2010). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure: Statistical power calculations under different scenarios. Panels A, B, C refer to the evaluation of binary phenotypes using logistic regression with a p-value of 5×10-8. Panels D, E, and F refer to power clalculations using survival endpoints with a p-value of 5×10-8.
Data Availability Statement
Data sharing not applicable to this article as no datasets were generated or analysed during the current study.